Summary
The two‐component signal transduction system PhoBR regulates the adaptation to phosphate limitation and the virulence of many animal bacterial pathogens. However, PhoBR in phytopathogens has rarely been investigated. In this study, we found that PhoBR in Xanthomonas oryzae pv. oryzae (Xoo), the pathogen of rice bacterial leaf blight, also regulates the adaptation to phosphate starvation. Unexpectedly, rice leaves infected by the phoBR‐deleted mutant and wild‐type PXO99A showed similar lesions, indicating that PhoBR is unnecessary for the virulence of Xoo. phoBR was found to be silenced during host infection, whereas artificially constitutive PhoBR expression attenuated virulence on host rice and growth in phosphate‐rich media. RNA‐sequencing (RNA‐seq) was then performed to investigate the global effect caused by constitutive PhoBR activation. RNA‐seq and further experiments revealed that the PhoBR regulon in Xoo comprised a wide range of genes. Nutrient transport and metabolism readjustments that resulted from PhoBR regulon activation may be responsible for growth attenuation. Our findings suggest that growth reduction regulated by PhoBR is a fitness cost of adaptation to phosphate starvation. PhoBR in Xoo is activated under phosphate‐limited conditions, which could exist in epiphytic and saprophytic surviving phases, and is strictly repressed within phosphate‐rich host plants to minimize fitness costs.
Keywords: growth, phosphate limitation, plant‐pathogenic bacteria, two‐component signal transduction system, virulence, Xanthomonas
Introduction
Phosphate is an essential nutrient for all living organisms. Phosphate is found in several molecules, including membrane lipids, complex sugars and nucleic acids. Phosphate also plays important roles in phosphoryl group transfer‐dependent biochemical reactions, such as signal transduction and energetic metabolism. PhoBR, a two‐component signal transduction system consisting of histidine kinase (HK) PhoR and response regulator (RR) PhoB, is widely known for the regulation of phosphate uptake and assimilation in Escherichia coli and other bacteria (VanBogelen et al., 1996).
The inner membrane sensor protein PhoR responds to periplasmic phosphate concentration variations probably by interaction with a phosphate transport system. When environmental phosphate is limited (below 4 µm), PhoR is autophosphorylated on a histidine residue, and this phosphoryl group is transferred to an aspartate residue of its cognate RR protein PhoB. The phosphorylated PhoB then acts as a transcriptional regulator to activate the expression of phosphate regulon genes (Lamarche et al., 2008). The phosphate regulon in E. coli comprises at least 47 genes (Baek and Lee, 2006), and the phosphate‐specific transport (Pst) system is conserved in phosphate regulons. Pst comprises a periplasmic phosphate binding protein (PstS), two transmembrane proteins (PstA and PstC) and an ATP binding protein (PstB), forming an ABC transporter to transport phosphate with high affinity into the cytosol (Van Dien and Keasling, 1998). PhoBR regulates not only phosphate transport, but also the virulence of animal‐pathogenic bacteria. PhoB of Vibrio cholerae controls virulence genes by direct modulation of the expression of tcpPH, a key upstream transcriptional regulator (Pratt et al., 2010). The phoB mutant strain of V. cholerae is less able than wild‐type V. cholerae to colonize rabbit intestine (von Kruger et al., 1999). The production of pyocyanin toxin in Pseudomonas aeruginosa is positively activated by PhoB in phosphate‐limited environments (Jensen et al., 2006). The opportunistic pathogen P. aeruginosa can apply phosphate deficiency as the environmental cues of host to activate a lethal phenotype (Zaborin et al., 2009).
Although studies have focused on PhoBR in animal pathogens, functional studies on PhoBR in phytopathogenic bacteria are limited. In the plant pathogen Agrobacterium tumefaciens, the homologue of PhoB (ChvI) is essential for virulence (Mantis and Winans, 1993), but its regulatory mechanism remains unknown. To determine the characteristics of PhoBR in phytopathogens, we investigated the intrinsic regulatory mechanism of PhoBR in Xanthomonas oryzae pv. oryzae (Xoo). Rice bacterial leaf blight caused by Xoo has been reported to reduce yields from 10% to 50% in some areas (Verdier et al., 2012). We found that the PhoBR regulatory system is essential for the adaptation of Xoo to phosphate limitation stress, but not for its full virulence. The expression of PhoBR in different niches was investigated, and PhoBR regulon genes were identified and analysed. Our findings suggest that the adaptation of Xoo to phosphate limitation is costly, and the strict regulation of PhoBR regulon genes by environmental phosphate availability is a strategy to minimize fitness costs.
Results
PhoBR is essential for the adaptation to phosphate limitation
To study the PhoBR system in Xoo, we found PhoB and PhoR orthologues in Xoo PXO99A using blastp. PXO_RS19870, which was annotated as a DNA binding RR in the genome of Xoo PXO99A (NC_010717.2), is homologous to PhoB in E. coli str. K‐12 with 99% coverage and 59% identity. PXO_RS19875, annotated as an HK, was found to be the orthologue of PhoR. Similar to its orthologue PhoB in E. coli, PXO_RS19870 comprises the signal receiver domain (REC) and the DNA binding domain Trans_reg_C, analysed using SMART (Letunic et al., 2012) (Fig. 1A). The conserved domain architecture of PXO_RS19875 is also similar to that of orthologue PhoR in E. coli. For consistency, PXO_RS19870 and PXO_RS19875 are also named PhoB and PhoR, respectively, in this study. Gene members of two‐component signal transduction systems are always located in the same operon. Reverse transcription‐polymerase chain reaction (RT‐PCR) confirmed that phoB and phoR are co‐transcribed (Fig. 1B).
Figure 1.
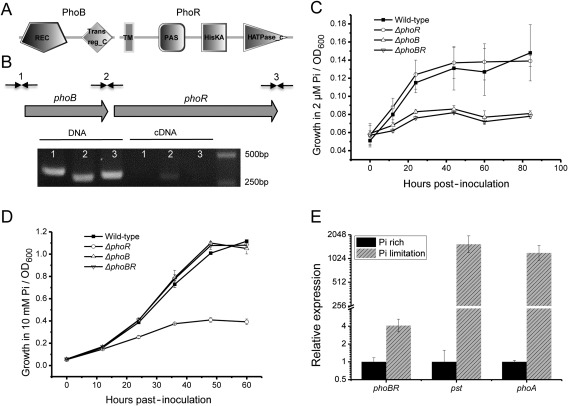
PhoBR is essential for the adaptation to phosphate (Pi) limitation. (A) The conserved domain architecture of PhoB and PhoR in Xanthomonas oryzae pv. orzyzae (Xoo). REC, signal receiver domain; Trans_reg_C, transcriptional regulatory protein, C‐terminal; TM, transmembrane region; PAS, Per‐ARNT‐Sim sensor domain; HisKA, histidine kinase A domain; HATPase_c, histidine kinase‐like ATPases domain. (B) phoB and phoR were co‐transcribed, confirmed by reverse transcription‐polymerase chain reaction (RT‐PCR) using primers across two adjacent genes. (C) Growth curves of phoR‐deleted mutant ΔphoR, phoB‐deleted mutant ΔphoB, phoBR operon‐deleted mutant ΔphoBR and wild‐type strain cultured in XOM2 adjusted with 2 µm phosphate. Bacterial density was measured by the absorbance at λ = 600 nm, and shown as the mean of three experimental repeats ± standard deviation. OD600, optical density at 600 nm. (D) Growth of ΔphoR, ΔphoB, ΔphoBR and the wild‐type strain cultured in phosphate‐rich (10 mm) XOM2. (E) Relative mRNA abundance of phoBR, pst and phoA in Xoo cultured in phosphate‐rich (10 mm) and phosphate‐limited (2 μm) XOM2 medium measured by quantitative RT‐PCR.
The PhoBR system is widely known for the regulation of adaptation to phosphate limitation. To verify whether PhoBR is essential for the phosphate starvation response in Xoo, we deleted the RR gene phoB, HK gene phoR and phoBR operon, and determined the abilities of the phoB‐deleted mutant ΔphoB, phoR‐deleted mutant ΔphoR and phoBR operon‐deleted mutant ΔphoBR to adapt to environmental phosphate limitation. Figure 1C shows that phoB absence and phoBR operon deficiency resulted in growth deficiency when environmental phosphate was limited (2 µm), whereas no evident growth difference was observed between ΔphoB, ΔphoBR and the wild‐type strain when environmental phosphate was rich (10 mm; Fig. 1D). In contrast, growth reduction occurred in the absence of phoR when environmental phosphate was not limited (10 mm; Fig. 1D). However, no evident growth difference between ΔphoR and wild‐type PXO99A was observed when environmental phosphate was limited (2 µm; Fig. 1C).
The Pst system and the alkaline phosphatase PhoA are well‐known members of the phosphate regulon and are very important for high‐affinity phosphate assimilation (Van Dien and Keasling, 1998). The relative transcription of phoBR, pst (indicated by pstB, PXO_RS09785) and phoA (PXO_RS23610) was quantified through quantitative RT‐PCR (qRT‐PCR). In Fig. 1E, phoBR, pst and phoA were highly induced by phosphate limitation stress. These results indicate that the PhoBR system in Xoo responds to environmental phosphate limitation and is essential for the adaptation to phosphate limitation, and PhoR is necessary to help Xoo survive in phosphate‐rich environments.
PhoR is essential for the full virulence of Xoo
The phosphate assimilation regulatory system PhoBR regulates the virulence of animal‐pathogenic bacteria (Mohammed Chekabab et al., 2014). To explore the regulatory function of PhoBR in Xoo during host plant infection, we assayed the virulence of the phoB‐deleted mutant ΔphoB, phoR‐deleted mutant ΔphoR and phoBR operon‐deleted mutant ΔphoBR on susceptible rice breed MH63. Unexpectedly, rice bacterial blight lesions caused by ΔphoB and ΔphoBR were similar to those caused by the wild‐type strain PXO99A over the whole infection stage (Fig. 2A,B), indicating that phoB or phoBR deletion did not affect the virulence of Xoo. In contrast, rice leaf lesions caused by ΔphoR were 50% shorter than those caused by the wild‐type strain PXO99A. The virulence attenuation of ΔphoR could be rescued by the ectopic expression of phoR controlled by its native promoter, indicating that the absence of phoR reduces the virulence of Xoo (Fig. 2C,D). All the above findings show that, although PhoBR is unnecessary for Xoo infection of the host plant, PhoR is essential for the full virulence of Xoo.
Figure 2.
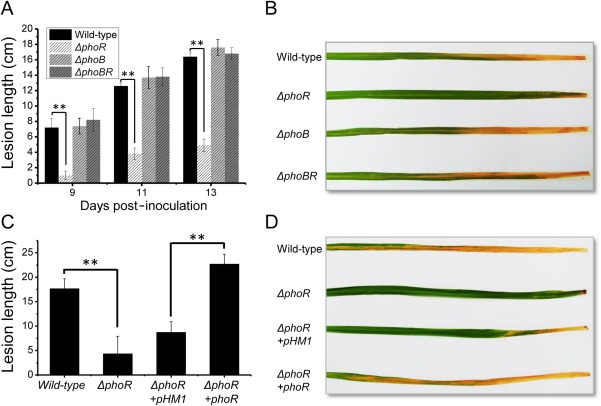
PhoR is essential for the full virulence of Xanthomonas oryzae pv. oryzae (Xoo). (A) The virulence of the phoR‐deleted mutant ΔphoR, phoB‐deleted mutant ΔphoB, phoBR operon‐deleted mutant ΔphoBR and wild‐type strain was assayed on susceptible rice. Leaf blight caused by the Xoo strains was quantified by the lesion length at 9, 11 and 13 days post‐inoculation. (B) Photographs of blight symptoms of representative infected rice leaves were taken at 13 days post‐inoculation. (C) Virulence of ΔphoR and phoR rescued strain was assayed on susceptible rice. Leaf blight caused by the Xoo strains was quantified by the lesion length at 14 days post‐inoculation. (D) Blight symptoms of representative infected rice leaves. All the virulence assays were performed at least three times, and one representative result is shown as the average lesion length ± standard deviation. Significant growth and virulence differences are marked with asterisks, indicating P < 0.01, statistically analysed by analysis of variance (ANOVA).
Absence of phoR results in the constitutive expression of PhoB
Growth in phosphate‐rich medium and virulence on host rice were reduced in the phoR‐deleted mutant ΔphoR, whereas these phenotypes were not affected by the absence of phoB or phoBR. To explain the different phenotypic changes caused by the absence of phoR and phoB, we measured the relative mRNA abundance of the phoBR operon and the phosphate assimilation genes pst and phoA in ΔphoR cultured in phosphate‐rich medium by qRT‐PCR. In Fig. 3A, the relative expression of phoBR was increased by 21‐fold in ΔphoR. The deficiency of phoR also sharply increased the transcription of pst and phoA, although Xoo strains were cultured in phosphate‐rich XOM2 medium (Fig. 3A). This finding suggests that phoR absence in Xoo results in the constitutive expression of phoB and other phosphate assimilation‐associated genes.
Figure 3.
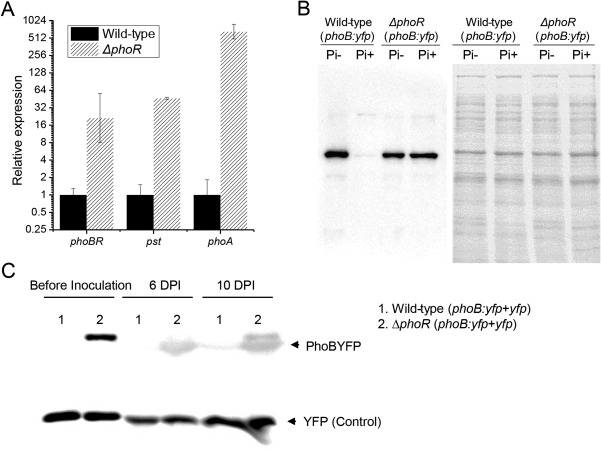
Absence of phoR results in the constitutive expression of PhoB. (A) Relative expression of phoBR, pst (pstB) and phoA in the phoR‐deleted mutant ΔphoR cultured in phosphate (Pi)‐rich XOM2 determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). (B) phoB in wild‐type strain and ΔphoR was tagged with the yellow fluorescent protein gene (yfp). Relative expression of PhoB in Xanthomonas oryzae pv. orzyzae (Xoo) strains cultured in phosphate‐rich and phosphate‐limited medium indicated by the intensity of the western blot band. The result of Coomassie brilliant blue staining on the right shows that a similar amount of bacterial total protein was loaded on each lane. (C) Expression of PhoB (tagged with YFP) in ΔphoR and the wild‐type strain determined by western blot. Additional YFP controlled by the lac promoter was used as internal control. Total proteins of leaves infected by Xoo strains at 6 and 10 days post‐inoculation (DPI) were extracted and assayed.
To confirm this conclusion, we determined the relative expression of PhoB in the phoR‐deleted mutant ΔphoR cultured in phosphate‐limited and phosphate‐rich media through western blot. PhoB in the ΔphoR mutant and wild‐type strain PXO99A was tagged with yellow fluorescent protein (YFP) through gene knock‐in. Western blot using anti‐green fluorescent protein (anti‐GFP) antibody showed that PhoB expression in wild‐type PXO99A was very low under phosphate‐rich conditions (10 mm), but strongly induced when environmental phosphate was limited (Fig. 3B). This observation is consistent with the qRT‐PCR result shown in Fig. 1E. However, PhoB was expressed strongly in ΔphoR under both phosphate‐rich and phosphate‐limited conditions (Fig. 3B).
Virulence on susceptible rice and growth in phosphate‐rich medium were reduced in the phoR‐deleted mutant, whereas neither of these phenotypes was changed in phoB‐ or phoBR‐deleted mutants. The coherence between the virulence on susceptible rice and the growth in phosphate‐rich medium implied that there may be a phosphate‐rich environment within rice leaf tissues. PhoB expression in Xoo infecting its host was determined through western blot. In Fig. 3C, PhoB in the wild‐type strain cultured in rich medium was silenced, whereas the absence of phoR de‐repressed PhoB. Similar to the expression pattern of PhoB in Xoo cultured in vitro, the expression of PhoB was detected only in the phoR‐deleted mutant, but not in the wild‐type strain, at 6 and 10 days post‐inoculation on rice leaves. These results suggest that PhoBR is expressed in phosphate‐limited environments, and the absence of phoR results in the constitutive expression of phoB even in phosphate‐rich environments, including within host niches.
Constitutive expression of PhoB is deleterious
The absence of phoR reduces the growth of Xoo when environmental phosphate is rich. Growth reduction in the phoR‐deleted mutant ΔphoR may be a result of the overexpression of PhoB, because PhoB is constitutively expressed in ΔphoR. To verify this hypothesis, we constitutively overexpressed PhoB in Xoo. PhoB controlled by the lac promoter was cloned to the cosmid pHM1. However, cosmid pHM1 possessing constitutively expressed phoB cannot be imported into the Xoo strain (Fig. 4A). The alignment between PhoB in Xoo and its orthologues in other bacteria revealed that the 55‐aspartate of PhoB is conserved in all sequences. A previous study on E. coli has demonstrated that this aspartate residue of PhoB is a key site for phosphorylation (Zundel et al., 1998). In our study, this key site was mutated to alanine, generating PhoBMT (Fig. 4B). In contrast with the case in PhoB, PhoBMT could be easily overexpressed in Xoo (Fig. 4A). These results indicate that constitutive PhoB is deleterious to Xoo in phosphate‐rich environments, and PhoB expression should be accurately controlled for normal physiological function.
Figure 4.
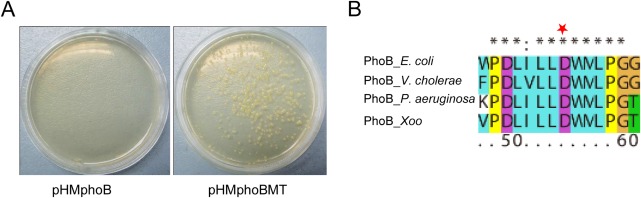
Constitutive expression of PhoB is deleterious. (A) Cosmids pHMphoB and pHMphoBMT, in which phoB and phoBMT (key aspartate residue mutant), respectively, are under the control of the lac promoter, were electroporated into Xanthomonas oryzae pv. orzyzae (Xoo) PXO99A. (B) Alignment between PhoB in Xoo PXO99A and its orthologues in other bacteria. Key aspartate residue mutated in phoBMT is marked with a red star. E. coli, Escherichia coli; P. aeruginosa, Pseudomonas aeruginosa; V. cholerae, Vibrio cholerae.
The identification of the PhoBR regulon
To explore the global effect of the constitutive expression of PhoB, we performed RNA‐sequencing (RNA‐seq) analysis between the phoR‐deleted mutant ΔphoR and the wild‐type strain PXO99A cultured in phosphate‐rich XOM2. RNA‐seq revealed that 372 genes were down‐regulated and 524 genes were up‐regulated in ΔphoR compared with those in PXO99A (Table S2, see Supporting Information). These observations imply that a wide range of genes are differentially expressed because of the up‐regulated PhoB.
The interaction between the PhoB protein and the putative promoter of differentially expressed genes (DEGs) was detected through electrophoretic mobility shift assay (EMSA) to clarify the directly regulated gene members of the PhoBR regulon. First, the direct interaction between PhoB and the phoBR operon promoter was determined. 6‐Carboxy‐fluorescein (FAM)‐labelled phoBR promoter DNA was used as a probe for EMSA, and the non‐labelled phoBR promoter DNA probe was used as a competitor. In Fig. 5A, the shifted bands were observed when phoBR promoter DNA was incubated with increasing amounts of PhoB, and these bands progressively disappeared as the amount of added non‐labelled probe to the competitive assay system increased. These results demonstrate that PhoB specifically binds to its promoter DNA and directly self‐feedback controls the transcription of phoBR.
Figure 5.
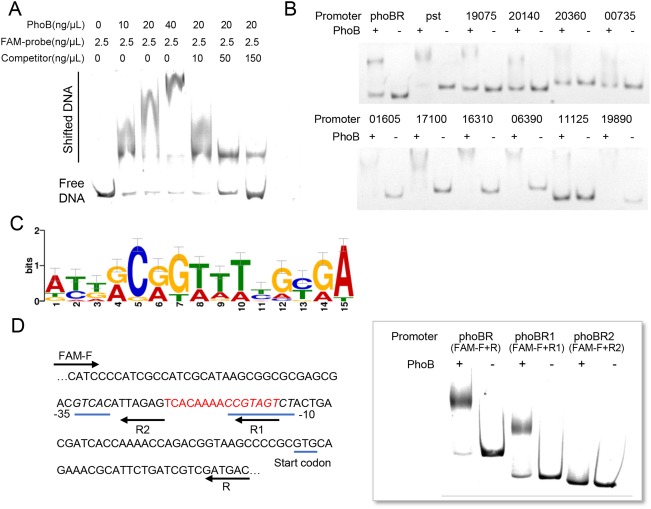
The identification of the PhoBR regulon. (A) The direct interaction between the PhoB protein and phoBR operon promoter DNA tagged with 6‐carboxy‐fluorescein (FAM), detected by electrophoretic mobility shift assay (EMSA). The final concentrations of PhoB and DNA in the 20‐µL reaction system are shown above each lane. Shifted bands indicate the probe DNA which interacts with the PhoB protein. Promoter DNA without tag was added to the incubation system to compete with the interaction between PhoB and the FAM‐tagged probe. (B) The direct interaction between PhoB protein and the promoter DNA of PhoBR regulon genes, detected by EMSA. The numbers above the electrophoretograms are the abbreviations of the gene locus, e.g. 19075 is the abbreviation for PXO_RS19075. (C) DNA binding motif of PhoB in Xanthomonas oryzae pv. orzyzae (Xoo). (D) The putative PhoB binding position in the phoBR promoter is highlighted in red, and the putative −10 and −35 regions of the phoBR promoter are shown in italic and underlined. The direct interaction between the PhoB protein and the truncated phoBR operon promoter DNA (with or without the putative PhoB binding region) was detected by EMSA.
Further EMSA results showed that PhoB could directly interact at least with the promoter of the pst operon, tonB‐dependent receptors (PXO_RS19075, PXO_RS20140, PXO_RS20360 and PXO_RS00735), glycosyl transferase gene PXO_RS19890, β‐1,4‐xylanase PXO_RS01605, endonuclease PXO_RS11125, Oar protein PXO_RS16310, hypothetical protein PXO_RS06390 and membrane protein PXO_RS17100, in addition to its own promoter of phoBR (Fig. 5B). The conserved motif of these binding DNAs was discovered by MEME software (Bailey et al., 2009) (Fig. 5C), and the PhoB DNA binding sequence was located between −35 and −10 regions of the phoBR promoter (Fig. 5D). To confirm the binding sequence of PhoB in phoBR promoter DNA, we truncated the probe DNA in EMSA. In Fig. 5D, the full‐length promoter DNA phoBR and the truncated phoBR1 could directly interact with the PhoB protein. However, the probe DNA phoBR2 without the putative binding sequence could not bind to the PhoB protein. This indicates that ‘TCACAAAACCGTAGT’ of the phoBR promoter is essential for the interaction with the PhoB protein. The identified consensus sequence was then used to scan the genome of Xoo PXO99A through FIMO software (Bailey et al., 2009). The PhoB DNA binding motif was mapped to the upstream regions (300 bp) of 157 genes. In addition to the genes confirmed by EMSA, 47 of these 157 genes were also in the DEGs list of the phoR‐deleted mutant ΔphoR (Table S2), suggesting that these genes could be regulated directly by PhoBR.
Nutrient transport and metabolism are readjusted in ΔphoR
RNA‐seq indicated that all of the Pst system‐encoding genes (PXO_RS09760 to PXO_RS09795) and alkaline phosphatase‐encoding genes (PXO_RS23610 and PXO_RS23615) were dramatically up‐regulated in the phoR‐deleted mutant ΔphoR, and this result was consistent with the qRT‐PCR result shown in Fig. 3A. In addition to the genes associated with phosphate uptake, tonB‐dependent receptor‐encoding genes were differentially expressed in ΔphoR. TonB‐dependent receptors are bacterial outer membrane proteins that are important transporters for ferric ions, vitamin B12, nickel complexes and carbohydrates (Noinaj et al., 2010). There are 37 genes annotated as tonB‐dependent receptors in the genome of Xoo PXO99A. Eight of these 37 genes were down‐regulated and 11 were up‐regulated because of the absence of phoR. This finding suggests that the transport of many nutrients in Xoo is under the control of the PhoBR regulatory system. This could also be deduced by gene ontology (GO) enrichment analysis. The GO term iron ion binding was remarkably over‐represented with a corrected P‐value of 0.028 (Table S3, see Supporting Information). Cellular transition metal ion homeostasis, carbohydrate transport and membrane protein are amongst the 20 most enriched GO terms shown in Fig. 6A.
Figure 6.
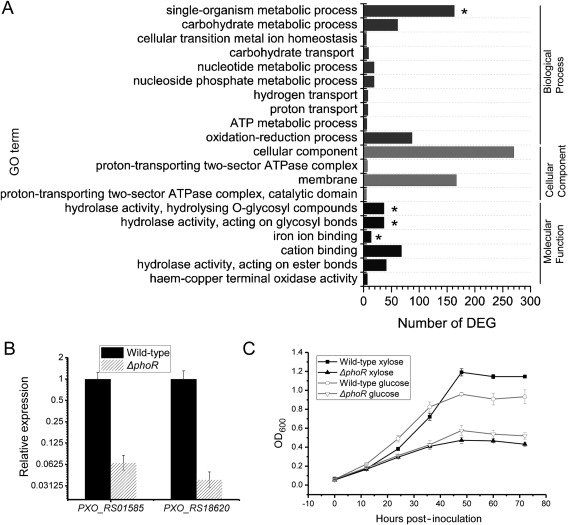
Nutrient transport and metabolism are readjusted in the phoR‐deleted mutant ΔphoR. (A) Differentially expressed genes (DEGs) in ΔphoR were annotated by gene ontology (GO). The 20 most enriched GO terms are shown. The significant over‐represented GO terms with a corrected P‐value of <0.05 are marked with an asterisk. (B) The relative expression of the tonB‐dependent receptor genes PXO_RS18620 and PXO_RS01585 in ΔphoR compared with the wild‐type strain cultured in XOM2 was confirmed by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). (C) Growth deficiency of ΔphoR was partly rescued by the replacement of the carbon source. Xylose in XOM2 medium was replaced by an equal amount of glucose. The growth of ΔphoR and the wild‐type strain in XOM2 and glucose‐replaced XOM2 was compared. OD600, optical density at 600 nm.
The tonB‐dependent receptors XCC2828 and XCC4120 in X. campestris pv. campestris are induced by xylose and xylan (Blanvillain et al., 2007). PXO_RS18620 and PXO_RS01585, which are the respective orthologues of XCC2828 and XCC4120, were down‐regulated by 22.63‐ and 19.56‐fold, respectively, in the phoR‐deleted mutant ΔphoR compared with the levels in the wild‐type strain PXO99A. This differential expression was further confirmed by qRT‐PCR (Fig. 6B). Xylose is the main carbon source for XOM2 medium used for the culture of Xoo. Furthermore, xylose isomerase PXO_RS01665, an important enzyme in xylose metabolism, was down‐regulated in ΔphoR (Table S2). These findings imply that the transport and catabolism of xylose are greatly restrained in ΔphoR. This could be one of the reasons for the growth weakness of ΔphoR. To confirm this hypothesis, we replaced the xylose in XOM2 medium with the same amount of glucose, and detected the growth of phoR mutant and the wild‐type strain. In Fig. 6C, the wild‐type strain PXO99A grew better in XOM2 than in the glucose‐replaced medium. However, the growth deficiency of ΔphoR was partly rescued by glucose. This finding indicates that the decline in xylose utilization is one of the factors reducing the growth of ΔphoR under phosphate‐rich conditions.
Carbohydrate metabolic process was one of the most enriched GO terms in the list of DEGs. In addition to xylose‐processing genes, glycometabolism‐related genes, such as α‐N‐arabinofuranosidase PXO_RS01575 and glucan 1,4‐α‐glucosidase PXO_RS01635, were differentially expressed in the phoR‐deleted mutant ΔphoR. GO:0004553 (hydrolase activity, hydrolysing O‐glycosyl compounds) and GO:0016798 (hydrolase activity, acting on glycosyl bonds) were also significantly over‐represented. This result implies that glycometabolism could be one of the physiological processes controlled by the PhoBR system. In addition to carbohydrate metabolic process, nucleotide metabolic process and lipid metabolic process were regulated by PhoBR. Nineteen genes involving nucleotide metabolic process and 45 genes involving lipid metabolic process were regulated by PhoBR (Table S3). Single‐organism metabolic process was the most over‐represented with a corrected P‐value of 0.0133. A total of 163 genes involving single‐organism metabolic processes were in the list of DEGs that resulted from the absence of phoR, thereby indicating that metabolic processes were remarkably changed in the phoR‐deleted mutant ΔphoR compared with that in the wild‐type strain.
In summary, PhoBR not only regulates the Pst system to transport inorganic phosphate with high affinity, but also controls the transport and metabolism of many other nutrients to adapt to phosphate limitation. Nutrient transport and metabolic process readjustments could be responsible for the growth reduction.
Discussion
Phosphate is an essential nutrient for bacteria, and adaptation to phosphate limitation is crucial to help bacteria survive in hostile environments. This study shows that the two‐component signal transduction system PhoBR in Xoo is responsible for the phosphate starvation response. PhoB is activated by phosphorylated PhoR and small molecules, such as acetyl phosphate, under phosphate‐limited conditions (Lamarche et al., 2008). Activated PhoB regulates the expression of phosphate regulon genes to adapt to phosphate starvation and initiates a feedback loop. During growth shift in a phosphate‐rich environment, PhoB is inactivated, probably by PhoR, acting as a phosphatase (Carmany et al., 2003). The deficiency of PhoR interrupts the deactivation of PhoB and increases the expression of phoB and other PhoBR regulon genes, regardless of environmental phosphate availability.
The pathogenic role of the PhoBR regulatory system has been explored in many animal pathogens, and PhoBR is considered to be part of a complex network important for bacterial virulence (Lamarche et al., 2008). In P. aeruginosa, PhoBR senses phosphate deficiency as an environmental signal of the host Caenorhabditis elegans to activate pyocyanin toxin production (Zaborin et al., 2009). In this study, PhoBR was repressed in the wild‐type Xoo within the host rice, and the absence of phoB or phoBR did not affect the virulence of Xoo. We conclude that PhoBR helps Xoo to adapt to phosphate starvation stress, which could exist in epiphytic and saprophytic survival phases, but PhoBR is unnecessary for Xoo infection of its host. Consistent with our findings that rice leaf tissues may contain rich phosphate, a previous study has shown that phosphate concentration in plant tissues is rich (5–20 mm) (Raghothama, 1999), indicating that the pathogenic function of PhoBR uncovered in Xoo could be extended to other phytopathogens.
The silence of PhoB during host plant infection confirms that rice leaf tissues represent a phosphate‐rich environment. Thus, we deduced that the attenuation of the virulence of the phoR‐deleted mutant ΔphoR is mainly the result of growth reduction in the phosphate‐rich environment. We also examined the virulence factors. Type III secretion system and effectors encoded by hrp genes are important virulence factors in the infection of host plants (Buttner and He, 2009). The expression of hrp genes is under the control of the key RR HrpG and the transcriptional activator HrpX (Wengelnik and Bonas, 1996; Wengelnik et al., 1996). Although no evident differences in hrpG or hrpX expression between the ΔphoR mutant and wild‐type PXO99A strain were observed, many hrp genes were down‐regulated in ΔphoR (Table S2). Down‐regulated genes in ΔphoR, ΔhrpG and ΔhrpX mutants were further compared (RNA‐seq data of ΔhrpG and ΔhrpX mutants are not shown). Of the 372 down‐regulated genes in ΔphoR, 75 overlapped with those in ΔhrpG and/or ΔhrpX (Fig. S1, see Supporting Information), suggesting that these hrp genes are fine‐tuned by PhoBR. The decrease in hrp gene expression may also contribute to the virulence attenuation of ΔphoR.
Several physiological and biochemical strategies are used by bacteria to cope with phosphate limitation (Bieleski, 1973). The absence of phoR misleads Xoo to undergo a phosphate starvation response state regardless of environmental phosphate concentrations. Physiological and biochemical changes in the phoR‐deleted mutant ΔphoR can be regarded as strategies employed by Xoo to survive phosphate limitation. The up‐regulation of alkaline phosphatase PhoA can be a strategy to increase environmental phosphate availability. The most important strategy is the efficient uptake of phosphate into cells, and the up‐regulation of the pst operon in ΔphoR implies that this strategy is also employed by Xoo. The absence of phoR also triggers the readjustment of other physiological processes, such as nutrient transport and metabolism. Under environmental stress, many bacteria switch to a slow‐growing persistence status, which tolerates environmental stress (Balaban, 2011). The growth reduction of the phoR mutant implies that Xoo specifically switches to a persistence‐like growth status under phosphate‐limited conditions. This was also verified by a previous proteomic study on X. citri, which showed that transport and metabolism‐associated proteins were over‐represented amongst the differentially expressed proteins identified from X. citri cultured in phosphate‐rich and phosphate‐limited media (Pegos et al., 2014). We propose that growth reduction, which resulted from nutrient transport and metabolic process readjustments, can be another strategy to adapt to phosphate limitation. Our findings imply that adaptation to phosphate limitation is costly, and PhoBR regulon expression is strictly controlled to minimize fitness costs.
Experimental Procedures
Strains, plasmids and culture conditions
The bacterial strains, plasmids and primers used in this study are listed in Table S1 (see Supporting Information). All Xoo strains and E. coli strains were stored at −80 °C with the protection of 20% glycerol for long‐term maintenance. The wild‐type Xoo strain PXO99A, which was genome sequenced in 2008 (Salzberg et al., 2008), and its derivative strains were cultured at 28 °C in NB medium (beef extract, 3 g/L; yeast extract, 1 g/L; polypeptone, 5 g/L; sucrose, 10 g/L) or NA medium (NB with 1.5% agar), except for special circumstances. Escherichia coli strains were grown in Luria–Bertani (LB) medium or on LB agar medium at 37 °C. The final concentrations of antibiotics used were as follows: kanamycin, 25 µg/mL; spectinomycin, 50 µg/mL.
Genetic manipulation
Gene knock‐out and knock‐in in Xoo were implemented by allelic homologous recombination as described previously (Zheng et al., 2016). Briefly, homologous DNA fragments between the target gene for deletion were cloned to the suicide plasmid pK18mobsacB. In the case of gene knock‐in, homologous DNA fragments between the insert site were ligated to flanks of the target gene by overlapping PCR and cloned to pK18mobsacB. The recombinant plasmid was subsequently imported and integrated to the Xoo genome to generate one cross‐over mutant, followed by the second cross‐over exchange when culturing on NAS medium (NA containing 10% sucrose).
For phoR complementation analysis, putative promoter and terminator DNA segments of the phoBR operon, together with the open reading frame (ORF) region of phoR, were amplified and ligated by overlapping PCR (Table S1). phoR controlled by its native promoter and terminator was then cloned to the broad‐host‐range cosmid pHM1 and electroporated into the phoR‐deleted mutant to analyse the complementation.
phoB, the 55‐aspartate mutant phoBMT generated by overlapping PCR and yfp were cloned to pBBR1MCS4_start to express PhoB, PhoBMT and YFP under the control of the lac promoter. The lac promoter, inserted ORF and the terminator region were subcloned from pBBR1MCS4_start to pHM1 by One Step Cloning using exnase (Vazyme Biotech, Nanjing, China) according to the manufacturer's instructions, and expressed in PXO99A. All the plasmid constructions, except for the special circumstances mentioned, were performed by typical gene cloning methods using TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China), endonuclease and T4 ligase commercially purchased from Takara Bio (Kusatsu, Japan). Sanger sequencing was performed after plasmid construction to confirm that there was no additional mutation in the recombined plasmids.
Xoo growth curve in phosphate‐limited and phosphate‐rich XOM2 medium
Phosphate in XOM2 medium (Tsuge et al., 2002) was adjusted to the required concentrations, generating phosphate‐limited (d‐xylose, 0.18%; d,l‐methionine, 670 µm; sodium l(+)‐glutamate, 10 mm; KH2PO4, 2 μm; MnSO4, 40 µm; Fe(iii)‐EDTA, 240 µm; MgCl2, 5 mm; pH 6.8 Tris buffer, 10 mm) and phosphate‐rich media. Xoo strains were cultured in NB for about 24 h to the middle logarithmic phase. Xoo cells were then harvested, washed by XOM2 medium or XOM2 derivative medium, and finally adjusted to an optical density at 600 nm (OD600) of 2.5. The resuspended Xoo culture was inoculated to XOM2 medium or XOM2 derivative medium by 2%. The absorbance at λ = 600 nm was measured at certain times, and the values are expressed as the mean of three biological repeats ± standard deviation.
RT‐PCR and qRT‐PCR
Xoo strains were cultured in XOM2 and phosphate concentration‐adjusted XOM2 medium to the middle logarithmic phase. RNA was extracted using an EasyPure RNA Kit (TransGen Biotech) according to the manufacturer's instructions, followed by digestion with DNases I (RNase‐free) and a PCR procedure to ensure that the RNA samples were DNA‐free; 2 μg of total RNA was then reverse transcribed to cDNA using a cDNA synthesis kit (Takara Bio) according to the manufacturer's instructions. cDNA synthesized from Xoo cultured in phosphate‐limited medium was used as PCR template (RT‐PCR) to confirm the co‐transcription of phoBR. qRT‐PCR was performed as described previously (Zheng et al., 2016).
Virulence assay
Virulence assay was performed as described previously (Zheng et al., 2016). Briefly, susceptible cultivar MH63 rice plants were grown in a glasshouse (28–32 ºC during the day and 25–28 ºC during the night, with 75% relative humidity) for 4–5 weeks. Xoo strains were cultured in NB medium for 24–48 h to the late logarithmic growth phase and adjusted to OD600 = 1.0. Rice leaves were then clipped about 2 cm from the tip with scissors dipped in bacterial culture. The lesion lengths of the infected rice leaves were measured at 9–14 days post‐inoculation. All of the virulence assays were performed in at least three biological repeats. In each experiment, 10 leaves in five plants were inoculated, and the values are expressed as the mean lesion length ± standard deviation.
Quantitative analysis of PhoB expression by Western blot
The YFP encoding sequence was knocked into the 3′ terminus of phoB, generating wild‐type (phoB:yfp) strain, and ΔphoR (phoB:yfp) was obtained by knocking out phoR in the wild‐type (phoB:yfp). Wild‐type (phoB:yfp) and ΔphoR (phoB:yfp) were then cultured in XOM2 or phosphate‐limited XOM2 medium to the middle logarithmic growth phase. Total soluble protein was extracted from ultrasonic processor‐disrupted Xoo cells and separated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). Western blot was performed using standard protocols with anti‐GFP monoclonal antibody and horseradish peroxidase (HRP)‐conjugated secondary antibody (Proteintech, Wuhan, China), and shown by a chemiluminescent detector system using Pierce ECL (Thermo Fisher Scientific, Waltham, MA, USA) as an HRP substrate.
The expression of PhoB in Xoo during rice leaf infection was determined by western blot. To acquire a reference expression signal, pHMYFP plasmid was electroporated into wild‐type (phoB:yfp) and ΔphoR (phoB:yfp), generating wild‐type (phoB:yfp+yfp) and ΔphoR (phoB:yfp+yfp), respectively. Rice leaves infected by wild‐type (phoB:yfp+yfp) and ΔphoR (phoB:yfp+yfp) were sampled at 6 and 10 days post‐inoculation of Xoo strains and homogenized in liquid nitrogen. Total protein of rice leaves and Xoo strains was extracted by acetone, followed by SDS‐PAGE and western blot, as described above.
RNA‐seq and data analysis
Xoo strains were grown in XOM2 to the middle logarithmic phase (OD600 = 0.4 for the phoR‐deleted mutant ΔphoR; OD600 = 0.8 for the wild‐type PXO99A; each Xoo strain was performed with two biological repeats). RNA extracted as described above was reverse transcribed to cDNA and used for Illumina sequencing after rRNA removal and fragmentation (sequenced at Novogene, Beijing, China). Clean read data were obtained after sequencing and data filtering. Gene expression indicated by FPKM (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) was counted by Htseq (Anders et al., 2015). Gene expression in ΔphoR and PXO99A was compared by Deseq (Anders and Huber, 2010). A negative binomial distribution test was employed, and genes with an adjusted P‐value of <0.05 were defined as DEGs exported as a tabular file (Table S2). DEGs were then annotated with GO terms, and further analysed by GO enrichment using the Goseq program based on the Wallenius non‐central hypergeometric distribution test (Young et al., 2010). GO terms with a corrected P‐value of <0.05 were significantly over‐represented statistically (Table S3).
EMSA
The phoB ORF was cloned to the pET28a vector to heterogeneously express hexa‐histidine tagged PhoB. PhoB expressed in BL21 (DE3) was separated and purified using an Ni‐NTA affinity column (Beijing CoWin Biotech, Beijing, China) according to the manufacturer's instructions. The purified protein was then dialysed in 10 mm, pH 7.4, Tris‐HCl buffer to remove the residual imidazole.
The phoBR promoter DNA probe was amplified with FAM‐labelled primers and purified with a Gel Extraction Kit (Omega Bio‐tek, Norcross, GA, USA). Labelled probe was incubated with PhoB proteins in EMSA buffer [50 mm Tris‐HCl, pH 7.4, 10 mm MgCl2, 100 mm NaCl, 1 mm dithiothreitol (DTT) and 1% glycerol] at 25 °C for 2 h (Tang et al., 2014). For competitive assay, different amounts of non‐labelled probe were added to the EMSA system. The incubation mixture was then subjected to 6% native polyacrylamide gel electrophoresis at 120 V for 45 min. The position of the labelled probe in the polyacrylamide gel was shown by a Typhoon Scanner (GE Healthcare, Chicago, IL, USA).
Promoter DNAs of other DEGs were amplified and incubated with PhoB protein, as described above, to further assay the interaction between PhoB and promoter DNAs; 20 ng/µL PhoB and 2.5 ng/µL promoter DNAs were incubated at 25 °C for 2 h, subsequently separated by 6% native polyacrylamide gel, stained with ethidium bromide (EB), followed by image display under ultraviolet light.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of strains, plasmids and primers used in this study.
Table S2 List of differentially expressed genes in phoR‐deleted mutant ΔphoR compared with the wild‐type strain.
Table S3 Gene ontology enrichment analysis of differentially expressed genes in the phoR‐deleted mutant ΔphoR compared with the wild‐type strain.
Fig. S1 Down‐regulated genes in hrpG‐, hrpX‐ and phoR‐deleted mutants relative to the wild‐type strain are compared and shown in a Venn diagram.
Acknowledgements
Thanks are due to the National Natural Science Foundation of China (Project 2017YFD0201103) and Science and Technology Department of Hubei Province (Project 2017CFB438) for financial support.
References
- Anders, S. and Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq—a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek, J.H. and Lee, S.Y. (2006) Novel gene members in the Pho regulon of Escherichia coli . FEMS Microbiol. Lett. 264, 104–109. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L. , Boden, M. , Buske, F.A. , Frith, M. , Grant, C.E. , Clementi, L. , Ren, J. , Li, W.W. and Noble, W.S. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban, N.Q. (2011) Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21, 768–775. [DOI] [PubMed] [Google Scholar]
- Bieleski, R.L. (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu. Rev. Plant Physiol. 24, 225–252. [Google Scholar]
- Blanvillain, S. , Meyer, D. , Boulanger, A. , Lautier, M. , Guynet, C. , Denancé, N. , Vasse, J. , Lauber, E. and Arlat, M. (2007) Plant carbohydrate scavenging through TonB‐dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One, 2, e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, D. and He, S. (2009) Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmany, D.O. , Hollingsworth, K. and McCleary, W.R. (2003) Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185, 1112–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, V. , Lons, D. , Zaoui, C. , Bredenbruch, F. , Meissner, A. , Dieterich, G. , Munch, R. and Haussler, S. (2006) RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB‐dependent and ‐independent pathways. J. Bacteriol. 188, 8601–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kruger, W.M. , Humphreys, S. and Ketley, J.M. (1999) A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate‐limitation response and intestinal colonization. Microbiology, 145, 2463–2475. [DOI] [PubMed] [Google Scholar]
- Lamarche, M.G. , Wanner, B.L. , Crépin, S. and Harel, J. (2008) The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473. [DOI] [PubMed] [Google Scholar]
- Letunic, I. , Doerks, T. and Bork, P. (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantis, N.J. and Winans, S.C. (1993) The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J. Bacteriol. 175, 6626–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed Chekabab, S. , Harel, J. and Dozois, C.M. (2014) Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence, 5, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj, N. , Guillier, M. , Barnard, T.J. and Buchanan, S.K. (2010) TonB‐dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegos, V.R. , Nascimento, J.F. , Sobreira, T.J. , Pauletti, B.A. , Paes‐Leme, A. and Balan, A. (2014) Phosphate regulated proteins of Xanthomonas citri subsp. citri: a proteomic approach. J. Proteomics, 108, 78–88. [DOI] [PubMed] [Google Scholar]
- Pratt, J.T. , Ismail, A.M. and Camilli, A. (2010) PhoB regulates both environmental and virulence gene expression in Vibrio cholerae . Mol. Microbiol. 77, 1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama, K.G. (1999) Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.‐A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A . BMC Genomics, 9, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Q. , Li, X. , Zou, T. , Zhang, H. , Wang, Y. , Gao, R. , Li, Z. , He, J. and Feng, Y. (2014) Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol. Microbiol. 94, 1006–1023. [DOI] [PubMed] [Google Scholar]
- Tsuge, S. , Furutani, A. , Fukunaka, R. , Oku, T. , Tsuno, K. , Ochiai, H. , Inoue, Y. , Kaku, H. and Kubo, Y. (2002) Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68, 363–371. [Google Scholar]
- VanBogelen, R.A. , Olson, E.R. , Wanner, B.L. and Neidhardt, F.C. (1996) Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli . J. Bacteriol. 178, 4344–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dien, S.J. and Keasling, J.D. (1998) A dynamic model of the Escherichia coli phosphate‐starvation response. J. Theor. Biol. 190, 37–49. [DOI] [PubMed] [Google Scholar]
- Verdier, V. , Vera Cruz, C. and Leach, J.E. (2012) Controlling rice bacterial blight in Africa: needs and prospects. J. Biotechnol. 159, 320–328. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- Young, M.D. , Wakefield, M.J. , Smyth, G.K. and Oshlack, A. (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol. 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborin, A. , Romanowski, K. , Gerdes, S. , Holbrook, C. , Lepine, F. , Long, J. , Poroyko, V. , Diggle, S.P. , Wilke, A. , Righetti, K. , Morozova, I. , Babrowski, T. , Liu, D.C. , Zaborina, O. and Alverdy, J.C. (2009) Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. USA, 106, 6327–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D. , Yao, X. , Duan, M. , Luo, Y. , Liu, B. , Qi, P. , Sun, M. and Ruan, L. (2016) Two overlapping two‐component systems in Xanthomonas oryzae pv. oryzae contribute to full fitness in rice by regulating virulence factors expression. Sci. Rep. 6, 22 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zundel, C.J. , Capener, D.C. and McCleary, W.R. (1998) Analysis of the conserved acidic residues in the regulatory domain of PhoB. FEBS Lett. 441, 242–246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of strains, plasmids and primers used in this study.
Table S2 List of differentially expressed genes in phoR‐deleted mutant ΔphoR compared with the wild‐type strain.
Table S3 Gene ontology enrichment analysis of differentially expressed genes in the phoR‐deleted mutant ΔphoR compared with the wild‐type strain.
Fig. S1 Down‐regulated genes in hrpG‐, hrpX‐ and phoR‐deleted mutants relative to the wild‐type strain are compared and shown in a Venn diagram.


