Summary
To deploy durable plant resistance, we must understand its underlying molecular mechanisms. Type III effectors (T3Es) and their recognition play a central role in the interaction between bacterial pathogens and crops. We demonstrate that the Ralstonia solanacearum species complex (RSSC) T3E ripAX2 triggers specific resistance in eggplant AG91‐25, which carries the major resistance locus EBWR9. The eggplant accession AG91‐25 is resistant to the wild‐type R. pseudosolanacearum strain GMI1000, whereas a ripAX2 defective mutant of this strain can cause wilt. Notably, the addition of ripAX2 from GMI1000 to PSS4 suppresses wilt development, demonstrating that RipAX2 is an elicitor of AG91‐25 resistance. RipAX2 has been shown previously to induce effector‐triggered immunity (ETI) in the wild relative eggplant Solanum torvum, and its putative zinc (Zn)‐binding motif (HELIH) is critical for ETI. We show that, in our model, the HELIH motif is not necessary for ETI on AG91‐25 eggplant. The ripAX2 gene was present in 68.1% of 91 screened RSSC strains, but in only 31.1% of a 74‐genome collection comprising R. solanacearum and R. syzygii strains. Overall, it is preferentially associated with R. pseudosolanacearum phylotype I. RipAX2GMI1000 appears to be the dominant allele, prevalent in both R. pseudosolanacearum and R. solanacearum, suggesting that the deployment of AG91‐25 resistance could control efficiently bacterial wilt in the Asian, African and American tropics. This study advances the understanding of the interaction between RipAX2 and the resistance genes at the EBWR9 locus, and paves the way for both functional genetics and evolutionary analyses.
Keywords: bacterial wilt, disease resistance, functional genetics, host–pathogen interactions, plant immunity, plant‐pathogenic bacteria, Solanum melongena
INTRODUCTION
In agrosystems, plants are constantly challenged by various pathogens, such as bacteria, viruses, fungi and nematodes. Monoculture as it is currently practised in modern agro‐ecosystems not only leads to a reduction in plant genetic variability, but also to the selection of highly specialized and aggressive pathogens (Stukenbrock and McDonald, 2008). The use of plant resistant germplasm appears to be the best strategy to control plant diseases, as it is cost‐effective and environmentally friendly (Boyd et al., 2013). However, an increasing number of studies have regularly reported cases of resistance breakdown (Daverdin et al., 2012; Kiyosawa, 1982; Montarry et al., 2006; Moury, 2010; Palloix et al., 2009; Zeigler et al., 1994), which have questioned the durability of plant resistance. The prediction of this durability can be achieved by deciphering the evolutionary potential of the pathogen (McDonald and Linde, 2002). However, first, we need to understand the molecular bases of plant–microbe interactions in order to assess the evolutionary constraints on their stability and dynamics. Bacterial type III effectors (T3Es) are key components in this interaction as they can both subvert and trigger the plant immune system (Buttner, 2016). As such, these molecules are players in the identification, description and therefore development of sustainable resistance strategies (Clarke et al., 2015; Vleeshouwers and Oliver, 2014).
Bacterial wilt affects more than 54 botanical families, including several crops of major interest (tomato, potato, banana, groundnut, eucalyptus) (Hayward, 1991, 1994). Its causative agent is the soil‐borne Betaproteobacterium Ralstonia solanacearum, a species complex composed of four phylotypes of specific geographical origins: Asia and Africa for phylotype I, Africa for phylotype III, America for phylotype II and Indonesia for phylotype IV (Fegan and Prior, 2005; Wicker et al., 2012). A recent taxonomic update (Safni et al., 2014) has proposed that R. solanacearum should be renamed the Ralstonia solanacearum species complex (RSSC), subdivided into three distinct species: R. pseudosolanacearum (phylotypes I and III), R. solanacearum (phylotypes IIA and IIB) and R. syzygii (phylotype IV and BDB).
The search for and breeding for plant resistance against RSSC are critical for food security in numerous countries. However, the genetic bases of plant resistance to this major plant pathogen still remain unknown, except for the model species Arabidopsis thaliana (Bernoux et al., 2008; Deslandes and Genin, 2014; Deslandes et al., 2002; Le Roux et al., 2015; Tasset et al., 2010) and Medicago truncatula (Ben et al., 2013; Lohou et al., 2014; Turner et al., 2009; Vailleau et al., 2007). Interactions between RSSC strains and Solanaceae genotypes (tomato, eggplant and pepper) have already been investigated (Lebeau, 2010; N'Guessan, 2013) and formalized into six patterns, called pathoprofiles (Lebeau et al., 2011). This screening established that the highest resistance levels against RSSC were found in the eggplant germplasm. The resistance of the AG91‐25 accession (Ano et al., 1991, 2002) has been mapped and led to the identification of the major resistance locus ERs1 (Lebeau et al., 2013), later named EBWR9 (Salgon et al., 2017). In order to characterize the durability of AG91‐25 resistance, it is necessary to understand the molecular basis of the interaction between RSSC and the AG91‐25 eggplant immune system.
In this work, we validated the contribution of the GMI1000 allele of ripAX2 to the control of disease by AG91‐25. We showed that the immunity triggered by RipAX2 does not require a putative zinc (Zn)‐binding motif, previously shown to be critical to trigger immunity on Solanum torvum leaves (Nahar et al., 2014). Finally, we investigated the prevalence and allelic variability of ripAX2 alleles in pathogen populations in order to predict the durability of EBWR9 resistance if it were to be deployed in specific R. solanacearum‐infested areas.
RESULTS
RipAX2GMI1000 triggers AG91‐25 resistance
Disease control by a single resistance (R) gene is often caused by the specific recognition of a single pathogen effector, which elicits plant defences (Buttner, 2016). In order to investigate whether AG91‐25 resistance is associated with an effector from GMI1000, 41 single mutants of effectors described in Cunnac et al. (2004) were screened for disease appearance on a few GMI1000‐resistant AG91‐25 plants, after inoculation by soil drenching [108 colony‐forming units (CFU)/mL, 10 mL per plant]. Susceptible MM738 plants were also included in the tests. From this screen, only the ripAX2 mutant GMI1000 ripAX2::pCZ367 was found to cause disease on AG91‐25 plants. To confirm the role of RipAX2 in AG91‐25 disease resistance, a complemented strain was made by introducing a wild‐type copy of ripAX2 in the mutant (GMI1000 ripAX2::pCZ367::ripAX2). The GMI1000 wild‐type strain and the GMI1000 ripAX2::pCZ367::ripAX2 strain were not able to trigger disease on AG91‐25, whereas the ripAX2 mutant caused disease 4 days after inoculation (Fig. 1). The strains GMI1000, GMI1000 ripAX2::pCZ367 and GMI1000 ripAX2::pCZ367::ripAX2 were still able to induce disease on the susceptible control MM738, with similar wilting rates (Fig. 1). Biological replicates performed on AG91‐25 and on the MM738 susceptible control gave similar results (Figs S1 and S2, see Supporting Information).
Figure 1.
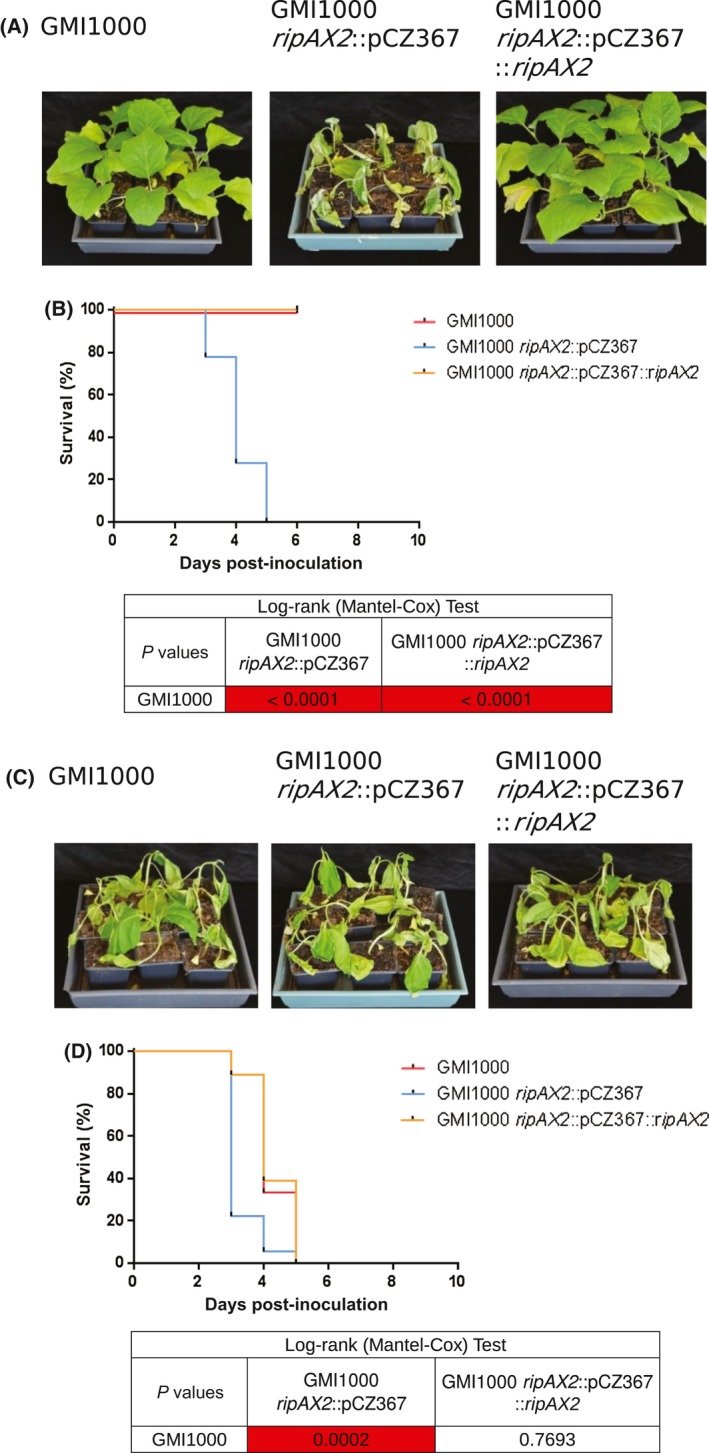
The RipAX2 type III effector of Ralstonia pseudosolanacearum is necessary for resistance in AG91‐25 eggplants. (A) Photographs taken at 6 days post‐inoculation. The R. pseudosolanacearum GMI1000 wild‐type strain and the GMI1000 ripAX2::pCZ367::ripAX2 complemented strain are not able to trigger disease on AG91‐25, whereas the GMI1000 ripAX2::pCZ367 mutant is able to do so. (B) Survival curves representing the wilting of AG91‐25 eggplants after root inoculation with the GMI1000 wild‐type strain, GMI1000 ripAX2::pCZ367 mutant and the complemented strain [108 colony‐forming units (CFU)/mL, 10 mL per plant, 18 plants per strain]. Log‐rank (Mantel–Cox) test P values compared with GMI1000 are shown below the graph. (C) Photographs taken at 6 days post‐inoculation. The strains GMI1000, GMI1000 ripAX2::pCZ367 and GMI1000 ripAX2::pCZ367::ripAX2 are all able to wilt MM738 susceptible eggplants. (D) Survival curves representing the wilting of MM738 eggplants after root inoculation. Log‐rank (Mantel–Cox) test P values compared with GMI1000 are shown below the graph. [Colour figure can be viewed at wileyonlinelibrary.com]
The eggplant pathogenic strain PSS4 expressing ripAX2GMI1000 becomes avirulent on AG91‐25
The PSS4 strain is pathogenic on AG91‐25 and its genome lacks any ripAX2 allele (Guinard et al., 2016). In order to investigate whether the presence of ripAX2 is sufficient for AG91‐25 to control disease, a PSS4 strain expressing ripAX2 GMI1000 was generated, and inoculated by soil drenching as mentioned above. This strain was not able to cause disease on AG91‐25, but still able to wilt the MM738 susceptible control (Fig. 2). Thus, we established that the expression of ripAX2 GMI1000 in an AG91‐25 pathogenic strain is sufficient to trigger AG91‐25 resistance. Biological replicates performed on AG91‐25 and on the MM738 susceptible control gave similar results (Figs S1 and S2).
Figure 2.
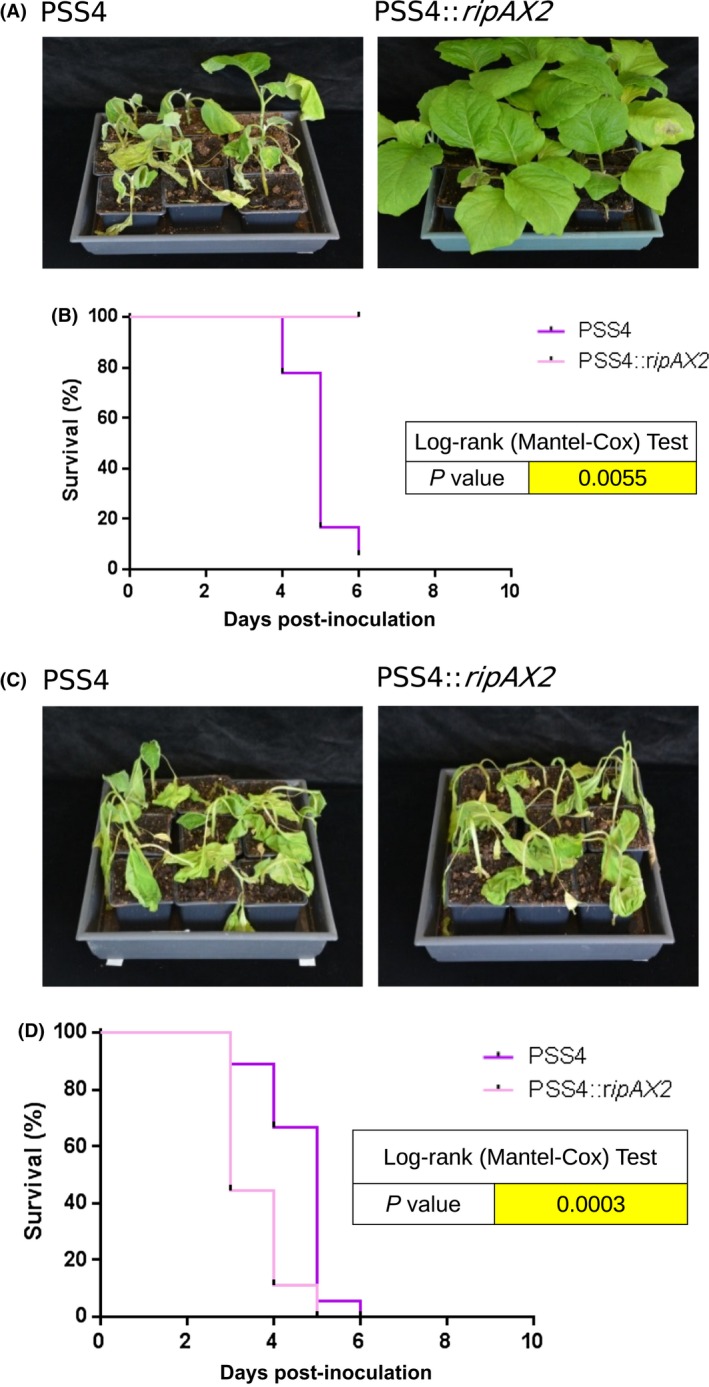
The trans‐complementation of the AG91‐25 pathogenic strain PSS4 by RipAX2 permits AG91‐25 resistance. (A) Photographs taken at 6 days post‐inoculation. (B) Survival curves representing the wilting of AG91‐25 eggplants after soil drenching inoculation [108 colony‐forming units (CFU)/mL, 10 mL per plant] with wild‐type PSS4 and PSS4::ripAX2. (C) Photographs taken at 6 days post‐inoculation. The strains PSS4 and PSS4::ripAX2 are able to wilt MM738 susceptible eggplants. (D) Survival curves representing the wilting of MM738 eggplants after root inoculation. Log‐rank (Mantel–Cox) test P values compared with GMI1000 are shown beside the graphs for (B) and (D). [Colour figure can be viewed at wileyonlinelibrary.com]
RipAX2‐triggered AG91‐25 resistance restricts plant colonization only after root infection
From the soil, R. solanacearum can enter the root system by wounds or lateral root emerging points. It then colonizes the cortex, invades the xylem and spreads to aerial parts, where it causes wilting (Denny, 2006). This multistep infection process makes it challenging to determine where resistance actually occurs.
In order to better characterize AG91‐25 resistance, bacterial multiplication in the stem was quantified after root inoculation and stem injection. After root inoculation, the bacterial load (log10 CFU/g fresh weight) reached high levels in AG91‐25 infected with PSS4 and GMI1000 ripAX2::pCZ367, 5 days after inoculation (means of 10.3 and 9.7, respectively; Fig. 3). No bacteria were detected in the stems of plants inoculated with PSS4 expressing ripAX2 and with GMI1000 ripAX2::pCZ367::ripAX2 at 5 or 10 days post‐inoculation. A few plants inoculated with GMI1000 showed some bacterial multiplication in their stem at 5 or 10 days post‐inoculation (Fig. 3); one of these plants showed wilting symptoms, suggesting that resistance did not occur in some rare cases. After stem injection, a high bacterial load was detected after 3 days in AG91‐25 inoculated with GMI1000, GMI1000 ripAX2::pCZ367 and GMI1000 ripAX2::pCZ367::ripAX2 (Fig. 4). The levels of bacteria were not significantly different between plants injected with GMI1000 and GMI1000 ripAX2::pCZ367 and high in all plants sampled, contrasting strongly with the results after root inoculation shown in Fig. 3. Furthermore, we observed wilting symptoms at 3 days after stem injection by all strains on some of the plants before destructive sampling (33% of AG91‐25 inoculated with GMI1000, 53% of AG91‐25 inoculated with GMI1000 ripAX2::pCZ367 and 40% of AG91‐25 inoculated with GMI1000 ripAX2::pCZ367::ripAX2), indicating that the bacteria had reached, colonized and clogged the xylem vessels of the stem after injection.
Figure 3.
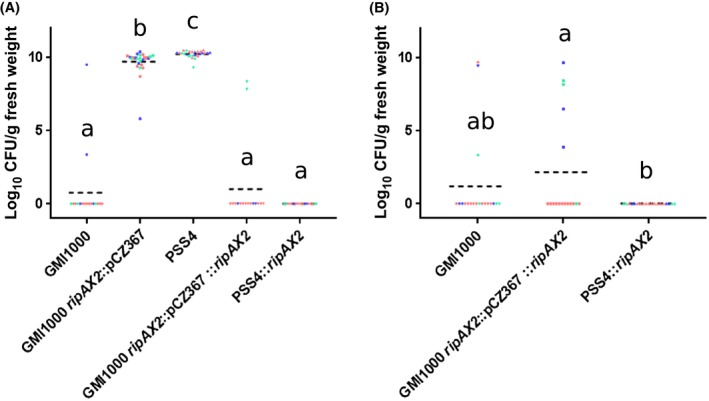
In planta growth of Ralstonia pseudosolanacearum strains GMI1000, GMI1000 ripAX2::pCZ367, PSS4, GMI1000 ripAX2::pCZ367::ripAX2 and PSS4::ripAX2 inoculated on AG91‐25 eggplants at 5 days (A) and 10 days (B) post‐inoculation. Each strain was inoculated by soil drenching [~108 colony‐forming units (CFU)/mL, 10 mL per plant]. Each point corresponds to the value obtained from a single plant. The three biological replicates are shown in different colours. If bacterial colonies were counted on plates, but their number was inferior to 10, the points were taken off as the actual bacterial quantity could not be assessed precisely. Statistical analyses for grouping were performed by Kruskal–Wallis test with α = 0.05. [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 4.
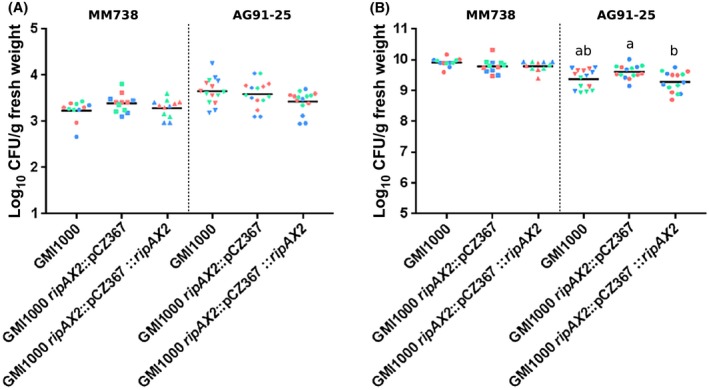
In planta growth of Ralstonia pseudosolanacearum after stem injection of strains GMI1000, GMI1000 ripAX2::pCZ367 and GMI1000 ripAX2::pCZ367::ripAX2 on MM738 and AG91‐25 eggplants, just after injection (A) and 3 days after injection (B). Each strain was inoculated by injection into the stem [106 colony‐forming units (CFU)/mL, 10 µL per plant]. Each point corresponds to the value obtained from a single plant. The three biological replicates are shown in different colours. Statistical analyses for grouping were performed by Kruskal–Wallis test with α = 0.05. No statistical differences were found just after injection by the different strains in MM738 and AG91‐25. No statistical difference was found 3 days after injection in MM738. [Colour figure can be viewed at wileyonlinelibrary.com]
Collectively, these results indicate that the key biological process inducing resistance to bacterial multiplication most probably occurs before the stem multiplication stage.
The conserved putative Zn‐finger domain is not critical for RipAX2‐triggered immunity
The putative Zn‐finger domain has been shown to be required for the immunity triggered by RipAX2RS1000 (initially named Rip36) on S. torvum (Nahar et al., 2014). RipAX2RS1000 and RipAX2GMI1000 have exactly the same amino acid sequence (Fig. S3, see Supporting Information). We generated the same point mutant (E149A) and showed that the integration of this mutated version (through the integrative plasmid pAM123) in both GMI1000 ripAX2::pCZ367 and PSS4 backgrounds (inoculated by soil drenching) restored the avirulence phenotype on AG91‐25 eggplant (Fig. 5A,B), unlike on S. torvum (Nahar et al., 2014).
Figure 5.
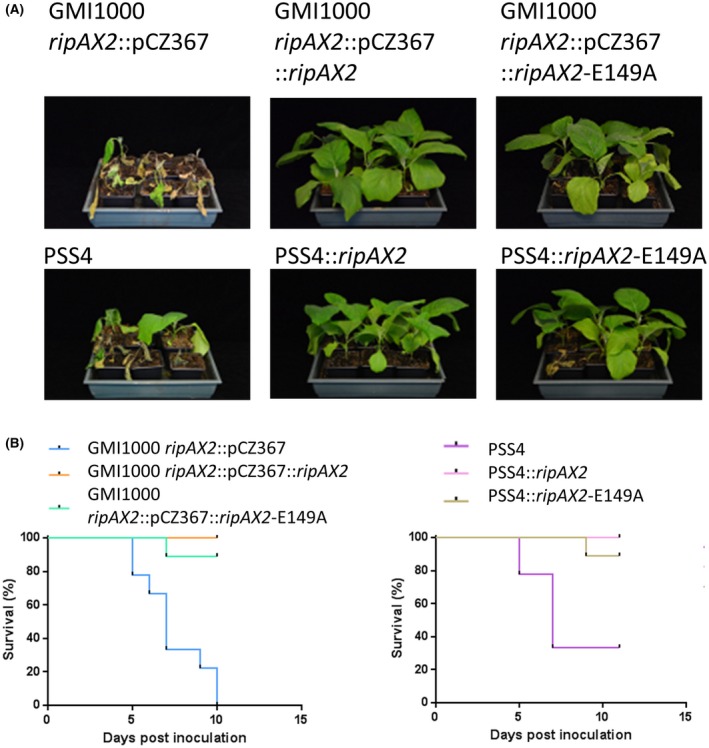
RipAX2‐triggered immunity does not require the conserved putative zinc‐finger domain. (A) Photographs taken at 14 days post‐inoculation for GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2 and GMI1000 ripAX2::pCZ367::ripAX2‐E149A, and at 15 days post‐inoculation for PSS4, PSS4::ripAX2 and PSS4::ripAX2‐E149A, inoculated by soil drenching [~108 colony‐forming units (CFU)/mL, 10 mL per plant]. Strains complemented with RipAX2 mutated in the putative zinc‐finger domain (point mutation E149A) were not able to trigger disease on AG91‐25 eggplants. (B) Survival curves representing the wilting of AG91‐25 eggplants after Ralstonia pseudosolanacearum inoculation. Log‐rank (Mantel–Cox) test shows that the wilting rates of plants inoculated with the GMI1000 ripAX2::pCZ367 mutant and the two complemented strains (with RipAX2 or with RipAX2‐E149A) are significantly different (P values of 0.004 and 0.009, respectively). The wilting rates of the plants inoculated with PSS4 and the two trans‐complemented strains (with RipAX2 or with RipAX2‐E149A) are also significantly different (P < 0.0001 and 0.0003, respectively). Each strain was inoculated by soil drenching on three replicates of nine plants. [Colour figure can be viewed at wileyonlinelibrary.com]
Biological replicates performed on AG91‐25 and on the MM738 susceptible control gave similar results, as detailed in Figs S4 and S5 (see Supporting Information), respectively.
ripAX2 is highly prevalent in RSSC and is preferentially associated with phylotype I
The ripAX2 gene was successfully amplified and sequenced in 68.1% of the world RSSC collection considered (n = 91). This proportion was lower (31.1%) in the RSSC genomes available in the RalstoT3E database (n = 74), most probably as a result of the difference in phylotype composition: our collection contains a majority of R. pseudosolanacearum phylotype I, whereas the RalstoT3E database contains a majority of R. solanacearum phylotype II and R. syzygii phylotype IV (Table 1). Based on the 15 strains common to both collections, polymerase chain reaction (PCR)‐based and genome sequencing‐based screenings gave highly concordant results (Table S3, see Supporting Information).
Table 1.
Distribution of ripAX2 within the Ralstonia solanacearum species complex (RSSC) world collection as estimated by polymerase chain reaction (PCR) sequencing (A), and within the RalstoT3E database of genomic sequences as estimated by automatic annotation (B). The gene was considered to be present if annotated as functional (PROT), frameshifted (FS) or pseudogene (PS). Results are expressed in strain numbers (percentage). Sample distributions per state (presence/absence) and phylotype were compared using a χ 2 test.
| Phylotype | Absence | Presence | Total (% of the collection) | P value (χ 2 test)† |
|---|---|---|---|---|
| ||||
| I | 12 (18.2) | 54 (81.8) | 66 (72.5) | 0.017* |
| IIA | 5 (55.6) | 4 (44.4) | 9 (9.9) | 0.127NS |
| IIB | 10 (100.0) | 0 (0) | 10 (11.0) | <0.0001*** |
| III | 2 (33.3) | 4 (66.7) | 6 (6.6) | 0.938NS |
| Total | 29 (31.9) | 62 (68.1) | 91 | |
| P value (χ 2 test)‡ | 0.0002*** | 0.025* | ||
| ||||
| I | 13 (48.1) | 14 (51.9) | 27 (36.5) | 0.019* |
| IIA | 5 (83.3) | 1 (16.7) | 6 (8.1) | 0.445NS |
| IIB | 16 (66.7) | 8 (33.3) | 24 (32.4) | 0.811NS |
| III | 2 (100) | 0 (0) | 2 (2.7) | 0.342NS |
| IV | 15 (100) | 0 (0) | 15 (20.3) | 0.009** |
| Total | 51 (68.9) | 23 (31.1) | 74 | |
| P value (χ 2 test)‡ | 0.37NS | 0.05NS | ||
†The H0 tested is: ‘Respective frequencies of ripAX2 presence and absence within each phylotype are equal to their expected values based on the total frequencies’.
‡The H0 tested is: ‘Respective distributions of phylotypes within each ripAX2 state (presence, absence) are equal to their expected values based on the total frequencies’.
NS, not significant; *significant (0.05 > P ≥ 0.01); **highly significant (0.01 > P ≥ 0.001); ***very highly significant (P < 0.001).
Because both collection compositions were unbalanced across phylotypes, we compared ripAX2‐per‐phylotype distributions using χ 2 tests. We observed that ripAX2 was significantly more present in R. pseudosolanacearum phylotype I than expected in both collections, whereas it was significantly more absent in R. syzygii phylotype IV (Table 1). ripAX2 was less commonly present in R. solanacearum phylotypes IIA and IIB, but this difference was not statistically supported. Hence, ripAX2 seemed to be preferentially associated with phylotype I.
Focusing on phylotype I, in which sample sizes were highest, we assessed whether the ripAX2 distribution could be associated with region of origin (defined as Africa, Asia or South America). Despite the low sample size, RipAX2 seemed to be significantly less prevalent in South America (six strains of 12; χ2 P = 0.004**).
Among 19 RipAX2 identified alleles, RipAX2GMI1000 is the most prevalent
Although most ripAX2 DNA sequences were 657 base pairs in length (219 amino acids), four were highly divergent (608 to 666 amino acids) and could not be aligned with the others. After the first 103 base pairs, the sequence amplified in RUN1546, RUN1740 and RUN1743 contained a very long insertion and several early stop codons, suggesting that these alleles code for non‐functional proteins (Fig. S6, see Supporting Information). Meanwhile, the RUN1994 sequence was incomplete (206 amino acids), with also several early stop codons. Fifteen other RipAX2 alleles were identified (Fig. 6), among which seven could be considered as non‐functional because of early stop codons generating a truncated protein by more than 20% of the reference effector protein (RipAX2GMI1000). Amongst these 19 alleles, the 11 non‐functional alleles were not considered in the analysis below.
Figure 6.
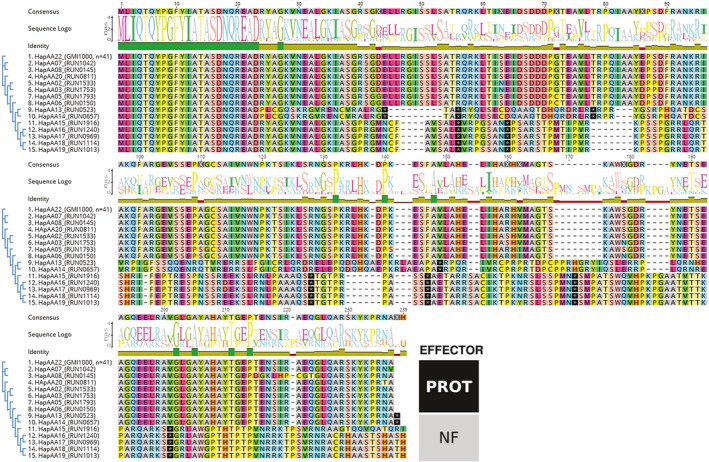
Proteic alignment of 15 RipAX2 alleles (of 19) identified within the world Ralstonia solanacearum species complex (RSSC) collection. Each allele number is linked to its representative strain and to the number of strains in which it was found. Proteic sequences were aligned (MUSCLE with default settings) and clustered using the FastTree method, with optimization of the Gamma20 likelihood at 20 rate categories of sites. The zinc (Zn)‐binding motif (HEXXH) described by Nahar et al. (2014) is marked by dark shading (positions 152–158). The effector sequence is considered to be non‐functional (NF) if its length is less than 80% of the GMI1000 reference sequence (219 amino acids). [Colour figure can be viewed at wileyonlinelibrary.com]
Amongst the eight probably functional alleles, RipAX2GMI1000 (coded HapAA22; Table S5, see Supporting Information) was the most prevalent, and found in both R. pseudosolanacearum phylotype I (n = 40) and R. solanacearum phylotype IIA (n = 1). Apart from RipAX2GMI1000, all seven alleles were phylotype specific: three were specifically found in phylotype I (HapAA02, HapAA07, HapAA20), whereas two alleles were phylotype III specific (HapAA05 and HapAA08) and two were phylotype IIA specific (HapAA03 and HapAA06).
The analysis of the geographical distribution of the different RipAX2 alleles was hampered by the large sample imbalances between countries. To take into account these sample unbalances, we looked not only at the number of alleles per region, but also at the genotype‐per‐strain rate (also named G/N). Across the three regions represented in the collection, Africa displayed the highest G/N (meaning that, with a set amount of strains, more different alleles were found), followed by Asia and then South America (Fig. S7, see Supporting Information). The RipAX2GMI1000 allele was prevalent within each region, and its distribution was not significantly associated with any country or region (Table S6, see Supporting Information). Apart from RipAX2GMI1000, no other allele was found to be present in more than one region.
DISCUSSION
We have demonstrated that ripAX2 GMI1000 is the determinant triggering resistance to RSSC in the AG91‐25 eggplant line. Moreover, the naturally virulent PSS4 strain no longer triggers disease once it expresses the RipAX2GMI1000 allele, without significantly affecting its aggressiveness on the susceptible control MM738. Our research was supported by previous results indicating that RipAX2 is associated with resistance in solanaceous plants (eggplant AG91‐25, tomato Hawaii 7996; Pensec et al., 2015).
Furthermore, we showed that plants recognizing RipAX2 largely excluded bacteria from entering the stems after root colonization, suggesting that the resistance occurs in the early stages of infection. This resistance could occur anywhere from the infected roots to further root and lower stem colonization.
Recently, an orthologue of ripAX2 GMI1000 in the RS1000 strain (ripAX2 RS1000) has been shown to induce a strong hypersensitive response (HR) in the leaves of a wild relative of eggplant (S. torvum cv. torubamubiga), but not on eggplant (Solanum melongena cv. senryo‐nigou) (Nahar et al., 2014). However, as no R gene has been characterized to date in these two cultivars, the R gene involved in the recognition of RipAX2RS1000 has not yet been identified. In this model, the recognition of the effector was dependent on the putative Zn‐binding domain HEXXH. More precisely, the glutamic acid at position 149 was critical for the recognition of RipAX2 by the S. torvum immune system. We observed that this HEXXH domain was conserved within all the likely ‘functional’ alleles (i.e. predicted to code for a 219‐amino‐acid protein) identified in the RSSC, whereas the corresponding strains differed in virulence on AG91‐25 eggplant. In addition, we experimentally demonstrated that the mutation E149A did not change its ability to trigger resistance on AG91‐25 eggplant, indicating that this residue was not critical for this phenotype. Collectively, these results suggest that the RipAX2 elicitation of defences may involve different mechanisms in S. torvum leaves and S. melongena root infection. It will be interesting to study RipAX2 residues under selective pressure in order to test which protein features are critical for S. melongena AG91‐25 resistance.
RipAX2 is the second known RSSC effector triggering eggplant resistance, the first being RipP2 (formerly named PopP2) recognized by RE‐bw (Xi’ou et al., 2014).
It remains to be determined which plant proteins specifically recognize RipAX2. It is tempting to speculate that the direct or indirect recognition of RipAX2 involves one of the genes located within the major resistance locus EBWR9 (Salgon et al., 2017). Indeed, strains controlled by the EBWR9 locus tested so far (GMI1000, CMR134) carry a functional allele of ripAX2, whereas all EBWR9‐uncontrolled strains carry no ripAX2 (CFBP2957, CFBP3059, PSS4), or a non‐functional copy of ripAX2 (To10). EBWR9 is located in a 3‐cM zone, corresponding to an approximately 3‐Mb (Lebeau et al., 2013) interval, which has been reduced to 1.77 Mb and located on the long arm of chromosome 9 (Salgon et al., 2017). Five candidate R genes have been identified in this region. The fine genetic mapping of this quantitative trait locus (QTL) is currently in progress (Salgon, 2017) to pave the way to gene cloning. Another way to determine the candidate genes involved in the recognition of RipAX2 would be to use reverse genetics: virus‐induced gene silencing (VIGS) on eggplant (Liu et al., 2012) and RNA interference (RNAi) have been used successfully to find the R genes involved in effector recognition in Nicotiana benthamiana (Brendolise et al., 2017).
ripAX2 is a rather highly prevalent effector in the RSSC, as reflected by the observation that 68.1% of our strain collection carried an allele of this gene, but seems to be preferentially associated with phylotype I, absent in phylotype IV and rare in phylotype II. It is important to stress that future screenings of much larger collections, balanced across the different phylotypes infecting the Solanaceae, will allow a much more precise assessment of the distribution of ripAX2 in natural diversity. Moreover, RipAX2GMI1000 is the dominant allele in every region probed, suggesting that a large proportion of the pathogen population is controlled by AG91‐25 eggplant. One may thus speculate that the deployment of the AG91‐25 line or derived eggplant cultivars would provide efficient control of the prevalent populations. However, seven other potentially functional alleles have been found that may modulate the plant defence response. From our data, one strategy built by RSSC strains to bypass RipAX2‐triggered plant resistance includes the loss of ripAX2 (the PSS4 case) and the inactivation of the gene through frameshifts (the 11 probably non‐functional alleles identified), but it is plausible that point mutations within the gene coding sequence or the alteration of gene expression through mutations in the promoter region could also lead to a loss of RipAX2 recognition. A deeper understanding of the RipAX2–eggplant interaction is needed: (i) to identify the critical residues for AG91‐25 recognition and resistance, taking into account the natural variability detected; and (ii) to assess the possible variations in effector expression. Moreover, a global surveillance of RSSC populations will help to assess the respective frequencies of the different functional alleles. As this global screening considered RSSC populations that were not challenged by AG91‐25 resistance, future research should address the evolution of RipAX2‐containing populations submitted to AG91‐25 resistance selection pressure.
To conclude, we have shown that RipAX2 triggers resistance on AG91‐25 resistant eggplant, whose recognition occurs in the early infection stages and may be mediated by an R gene present in the ERs1/EBWR9 locus. This latter statement needs to be investigated further thanks to the generation of near‐isogenic lines (NILs) carrying (or not) EBWR9, or by reverse genetic methods. Both approaches could allow the cloning of this gene and a better characterization of AG91‐25 resistance to the RSSC.
EXPERIMENTAL PROCEDURES
Plant material
Two eggplant accessions (S. melongena) were used: the resistant eggplant AG91‐25, also called MM960, and the susceptible eggplant MM738 (Ano et al., 1991, 2002; Doganlar et al., 2002; Wu et al., 2009). Eggplant seeds were sown 4 weeks before inoculation and were then transplanted into 7 × 7 × 6‐cm3 black plastic pots at 1 week after sowing. Four‐week‐old plants were transferred to a growth chamber at 90% relative humidity, with a day/night thermoperiod of 28 °C/24 °C and a 12‐h light/12‐h dark photoperiod.
Bacterial strains
The distribution of the ripAX2 coding sequence within the RSSC was assessed on a world collection of 91 bacterial strains, gathering representatives of the species R. pseudosolanacearum (phylotypes I and III) and R. solanacearum (phylotypes IIA and IIB) (see Table S1 in Supporting Information). Because of its limited epidemiological importance in Solanaceae, R. syzygii ssp. indonesiensis (phylotype IV) was not considered in this screening. Strains were chosen following two criteria: (i) they were sampled from 13 geographical areas of agronomic importance, and/or containing international breeding stations, in Africa (Burkina Faso, Cameroon, Ivory Coast, South Africa), Asia and Indian Ocean (Reunion Island, India, Indonesia, Thailand, Philippines, Taiwan, Australia), and Caribbean and South America (French Guiana, Martinique); and (ii) they were mostly isolated from the Solanaceae. Some of these strains were screened for pathogenicity on MM738 and AG91‐25, following the methodology described in N’Guessan et al (2012). All were able to trigger disease on MM738, but displayed different interaction profiles on AG91‐25 resistant eggplant, ranging from being unable to colonize and wilt AG91‐25 to highly pathogenic strains (Table S1).
Bacterial cultures and DNA extraction
From stock tubes stored at –80 °C, strains were revived in nutrient broth (one to two beads per 3 mL of broth) for 48 h at 28 °C, and then quadrant streaked (50 µL of broth) on Kelman’s medium. Cultures were then multiplied from a single colony and suspended (one 10‐µL loopful) in 1 mL of NaCl (0.5 m) for rinsing. Suspensions were then centrifuged (6000 g for 3 min) and pellets were stored at –20 °C until DNA extraction. Genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Charbonnières, France) following the manufacturer’s recommendations. DNA concentration and quality were assessed using a NanoDrop® 8000 device. Bacterial DNAs were then diluted to 2 ng/μL and stored at –20 °C until use.
ripAX2 cloning and complementation constructs
The strain GMI1000 ripAX2::pCZ367 has been obtained previously (Cunnac et al., 2004). The complementation was generated by a double crossing‐over insertion using a plasmid derived from the pRC toolkit (Monteiro et al., 2012). The pNP329 plasmid (Wang et al., 2016) was modified (Acc65i‐BsrGI fragment was removed) to generate pNP380. The ripAX2 gene [coding sequence (CDS) flanked in 5′ by 300 bp and in 3′ by 949 bp] was subcloned from GMI1000 genomic DNA into pNP380 to generate the pNP384 plasmid. Starting from pNP384, the ripAX2 CDS was point mutated to generate the equivalent of Rip36E149A (Nahar et al., 2014) by site‐directed mutagenesis using the following oligonucleotides: oAM277 (GGGCATGGATCAGTGCATGGGCGAGGACA) and oAM278 (TGTCCTCGCCCATGCACTGATCCATGCCC). This generated the plasmid pAM123. These plasmids were used to transform both GMI1000 ripAX2::pCZ367 and the PSS4 strain.
Plant infection
Bacterial strains were grown overnight in BG medium and then diluted to an optical density at 600 nm (OD600) of 0.1 (~108 CFU/mL) with water to obtain the inocula. For each plant, a deep ‘L’‐shaped cut was made in the soil with a sharp scalpel in order to cut the roots. Ten millilitres of inoculum were poured into the ‘L’‐shaped open soil surface. The symptoms were scored daily on a 0–4 scale (no wilting to complete wilting). For stem bacterial load quantification, 1 cm of stem was sampled above the cotyledons, weighed and the surface was disinfected in a 70% alcohol solution, followed by rinsing in water. It was then cut into six parts and placed into 1 mL of water for a minimum of 30 min to allow the bacteria to diffuse. Appropriate dilutions were then plated in order to evaluate the bacterial load.
For stem injection, 10 µL of a bacterial suspension at 106 CFU/mL were injected above the cotyledons with a microsyringe. The microsyringe was sterilized between each plant by pumping up and down with ethanol and then four times with water. For sampling, 1 cm of stem surrounding the injection point was taken, weighed, surface sterilized, cut into pieces and placed into 1 mL of water to diffuse. Appropriate dilutions were plated.
ripAX2 prevalence and allele diversity in RSSC
The prevalence and diversity of the ripAX2 gene were addressed on a 91‐strain RSSC collection described above, taking advantage of the available genomic sequences (Ailloud et al., 2015; Guinard et al., 2016; Remenant et al., 2010; Salanoubat et al., 2002), as well as using PCR screening.
The distribution of the ripAX2 coding sequence within the RSSC was also assessed on a world collection of 74 whole‐genome sequences (Table S2, see Supporting Information), publicly available and harboured in the RalstoT3E database (https://iant.toulouse.inra.fr/bacteria/annotation/site/prj/T3Ev3/). ripAX2 was assessed as ‘present in the strain as one copy’, ‘present but frameshifted in the strain’, ‘missing in the strain’ or ‘present as a pseudogene copy in the strain’.
Design of ripAX2 PCR primers
The ripAX2 GMI1000 coding sequence was downloaded from the website ‘RalstoT3E’ (https://iant.toulouse.inra.fr/bacteria/annotation/site/prj/T3Ev3/) and BLAST‐queried on the MaGe interface (Magnifying Genomes, https://genoscope.cns.fr/microscope/mage) to search for gene orthologues and to extract the 400‐bp upstream and downstream regions. Orthology search was performed using the following parameters: (i) sequence identity above 80%; and (ii) minimal (MinLrap) and maximal (MaxLrap) ratios of alignment lengths above 90%.
Gene and flanking regions of ripAX2 GMI1000, ripAX2 Y45 and ripAX2 MOLK2 were concatenated using Geneious v5.5 (https://www.geneious.com, Kearse et al., 2012) and then aligned using ClustalW (Thompson et al., 1994). External primers were designed on the consensus sequences of the flanking regions of ripAX2 GMI100 and ripAX2 Y45 only (phylotype I) using the Primer3 algorithm under Geneious, with the following parameters: annealing temperature around 60 °C and primer sizes ranging from 18 to 25 nucleotides. Details of the primer sequences are given in Table S4 (see Supporting Information).
Gene amplification and sequencing
For each strain, putative effectors were PCR amplified on 20 ng of sample DNA template. PCRs (total volume of 25 µL) consisted of 1 U of Red Goldstar Taq DNA polymerase, 25 pmol of each primer, 1 × PCR buffer, 1.5 mm MgCl2, 0.2 mm of each deoxynucleotide (dNTP) and 1 × Q‐solution. The reaction was cycled in an Eppendorf (Montesson, France) Mastercycler Gradient thermocycler or an Applied Biosystems (Villebon‐sur‐Yvette, France) GenAmp PCR System 9700 thermocycler with a first denaturation step at 96 °C for 5 min, followed by 30 cycles of 30 s at 95 °C, 60 s at 55–57 °C (see Table S2) and 60 s at 72 °C, and a final elongation step of 10 min at 72 °C. All PCR products were resolved on a 2% agarose gel and visualized with UV light after ethidium bromide staining (5 µg/mL); fragment sizes were estimated in comparison with a 100‐bp DNA ladder (New England BioLabs , Evry, France).
PCR products were vacuum dehydrated and sent to Beckman Coulter Genomics (Takeley, Essex, UK) for further purification and double‐strand sequencing using PCR primers as sequencing primers. Raw sequences from both strands were assembled using Geneious v5.5 to generate consensus sequences; all ambiguous sequences were reamplified and resequenced. Alignments were carried out using the ClustalW alignment tool under Geneious, together with the reference sequences (GMI1000, RS1000, MOLK2).
Sequence extremities were trimmed on the basis of the sequence quality and reading frame, and to match the gene entire length. In some cases, the entire gene sequence was obtained by assembling sequences obtained from both external and internal primers. Each strain was amplified at least twice.
Accession numbers
The DNA sequences described in this article were deposited in GenBank under the numbers MF055715–MF055746. Details are given in Table S3.
Conflict of interest
The authors declare no conflicts of interest.
Supporting information
Fig. S1 Kaplan–Meier survival curves representing the wilting of AG91‐25 plants inoculated with the strains GMI1000, GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, PSS4 and PSS4::ripAX2 in two supplemental replicates (12 and 22 plants per strain, respectively). Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S2 Kaplan–Meier survival curves observed on MM738 plants (susceptible control) inoculated with strains GMI1000, GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, PSS4 and PSS4::ripAX2 in one supplemental replicate. Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S3 Alignment of the RipAX2 protein alleles from GMI1000, RS1000 and MOLK2, and their percentages of identity.
Fig. S4 Kaplan–Meier survival curves observed on AG91‐25 plants inoculated with strains GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, GMI1000 ripAX2::pCZ367::ripAX2‐E149A, PSS4, PSS4::ripAX2 and PSS4::ripAX2‐E149A in two supplemental replicates (22 plants and 18 plants per strain for each respective replicate). Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S5 Kaplan–Meier survival curves observed on MM738 plants inoculated with strains GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, GMI1000 ripAX2::pCZ367::ripAX2‐E149A, PSS4, PSS4::ripAX2 and PSS4::ripAX2‐E149A in three replicates. Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S6 DNA alignment of the reference ripAX2 alleles (GMI1000 and RS1000) with the long ripAX2 alleles amplified within the phylotype I Ivorian strains RUN1546 (sequevar 13), RUN1740 and RUN1743 (sequevar 31), and in the Guianese phylotype IIA RUN1994 (sequevar 41). Alignments were performed using MUSCLE under GENEIOUS v5.5. Each strain is represented by its nucleotide sequence (upper line) and proteic sequence (lower line). Black squares indicate stop codons.
Fig. S7 Geographical distribution of the 19 RipAX2 alleles, either functional (PROT) or non‐functional (NF), across regions within the phylotype I strains. The heatmap was built with the R package pheatmap, based on Euclidean distances between observations; the clusters were calculated using the Ward method. For each region, the number of RipAX2 alleles (G) and phylotype I strains (N) are presented. ‘Africa’ gathers Burkina‐Faso, Cameroon, Ivory Coast, Reunion Island and South Africa; ‘Asia’ gathers Australia, India, Indonesia, Philippines, Taiwan and Thailand; ‘South America’ gathers French Guiana and Martinique. The heatmap values correspond to the numbers of strains.
Table S1 Bacterial strains of the Ralstonia solanacearum species complex (RSSC) tested for the presence of the ripAX2 coding sequence. Phylotype I and III strains belong to the species R. pseudosolanacearum, and phylotype IIA and IIB strains belong to the species R. solanacearum.
Table S2 Distribution of ripAX2 within the 74 Ralstonia solanacearum species complex (RSSC) genomes harboured in the RalstoT3E database. Strains are ranked by phylotype and code. Phylotypes I and III are grouped within R. pseudosolanacearum, phylotypes IIA and IIB within R. solanacearum and phylotype IV within R. syzygii. The ripAX2 type III effector is present as a complete sequence (PROT), frameshifted (FS), pseudogenized (PS) or absent (NO).
Table S3 Comparison of ripAX2 distribution, assessed by polymerase chain reaction (PCR) amplification or genome sequencing, in the 15 strains common to our collection and the RalstoT3E database. The gene was present (1), frameshifted (FS) or absent (0).
Table S4 External and internal primers designed to amplify the ripAX2 coding sequence. Both external and internal primers were designed on the alignment of GM1000 and RS1000 ripAX2 gene and neighbouring regions.
Table S5 Bacterial strains of the Ralstonia solanacearum species complex (RSSC) with their host of isolation, nucleotidic and proteic RipAX2 alleles, and GenBank accession numbers.
Table S6 Specificities in distributions of the RipAX2 protein alleles (Hap_AA) across regions, as assessed by χ 2 tests. (A) Distribution of each allele across regions. Status is ‘absence’ (ABS), ‘functional’ (PROT) or ‘non‐functional’ (NF). (B) Distribution of each region across alleles.
ACKNOWLEDGEMENTS
Edith Lallemand‐Mamosa, Sylvain Lebon and Jean‐Michel Baptiste are greatly acknowledged for their excellent technical support. We wish to thank Professors Yuki Ichinose (Okayama University, Okayama, Japan) and Takafumi Mukaihara [Research Institute for Biological Sciences (RIBS), Okayama, Japan] for sharing their anti‐RipAX2 antibody. We also thank Sebastien Cunnac for critical reading of the first version of this article.
J.G.’s PhD grant was financially supported by a fellowship from both CIRAD and the University of la Réunion, by the projects ‘GENETOM3’ and ‘BSV2.1’ (FEDER project financed by the European Union and Région Réunion), by the CASDAR project RESAUBER (‘Sustainable management of eggplant resistances to bacterial wilt’, French Ministry of Agriculture) and by the Agreenium Foundation through the EIR‐A program. A.M. and F.L. were funded by a grant from the French Ministry of National Education and Research. This work was also supported by the French Laboratory of Excellence project ‘TULIP’ (ANR‐10‐LABX‐41).
†Present address: IPME, Université de Montpellier, CIRAD, IRD, F‐34394, Montpellier, France.
Contributor Information
Nemo Peeters, Email: nemo.peeters@inra.fr.
Emmanuel Wicker, Email: wicker@cirad.fr.
REFERENCES
- Ailloud, F. , Lowe, T. , Cellier, G. , Roche, D. , Allen, C. and Prior, P. (2015) Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genomics, 16(1), 270 10.1186/s12864-015-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano, G. , Hebert, Y. , Prior, P. and Messiaen, C.M. (1991) A new source of resistance to bacterial wilt of eggplants obtained from a cross – Solanum‐aethiopicum L × Solanum‐melongena L. Agronomie, 11, 555–560. [Google Scholar]
- Ano, G. , Anais, G. and Chidiac, A. (2002) Creation and use of disease‐resistant varieties in Guadeloupe [France]. ‐ Creation de varietes resistantes aux maladies en Guadeloupe [France]. Elements indispensables de diversification agricole. Phytoma‐La Défense des Végétaux, 551, 36–37. [Google Scholar]
- Ben, C. , Debelle, F. , Berges, H. , Bellec, A. , Jardinaud, M. , Anson, P. , Huguet, T. , Gentzbittel, L. and Vailleau, F. (2013) MtQRRS1, an R‐locus required for Medicago truncatula quantitative resistance to Ralstonia solanacearum . New Phytol. 199, 758–772. [DOI] [PubMed] [Google Scholar]
- Bernoux, M. , Timmers, T. , Jauneau, A. , Briere, C. , de Wit, P. , Marco, Y. and Deslandes, L. (2008) RD19, an Arabidopsis cysteine protease required for RRS1‐R‐mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell, 20, 2252–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, L.A. , Ridout, C. , O'Sullivan, D.M. , Leach, J.E. and Leung, H. (2013) Plant–pathogen interactions: disease resistance in modern agriculture. Trends Genet. 29, 233–240. [DOI] [PubMed] [Google Scholar]
- Brendolise, C. , Montefiori, M. , Dinis, R. , Peeters, N. , Storey, R.D. and Rikkerink, E.H. (2017) A novel hairpin library‐based approach to identify NBS‐LRR genes required for effector‐triggered hypersensitive response in Nicotiana benthamiana. Plant Methods, 13, 32 10.1186/s13007-017-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, D. (2016) Behind the lines—actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, C.R. , Studholme, D.J. , Hayes, B. , Runde, B. , Weisberg, A. , Cai, R. , Wroblewski, T. , Daunay, M.C. , Wicker, E. , Castillo, J.A. and Vinatzer, B.A. (2015) Genome‐enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum Race 3 Biovar 2 and screening for sources of resistance against its core effectors. Phytopathology, 105, 597–607. [DOI] [PubMed] [Google Scholar]
- Cunnac, S. , Occhialini, A. , Barberis, P. , Boucher, C. and Genin, S. (2004) Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. [DOI] [PubMed] [Google Scholar]
- Daverdin, G. , Rouxel, T. , Gout, L. , Aubertot, J.N. , Fudal, I. , Meyer, M. , Parlange, F. , Carpezat, J. and Balesdent, M.H. (2012) Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathog. 8(11), e1003020 10.1371/journal.ppat.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, T.P. (2006) Plant pathogenic Ralstonia species In: Plant‐associated bacteria (Gnanamanickam, S.S. ed.), pp. 573–644. Dordrecht: Springer. [Google Scholar]
- Deslandes, L. and Genin, S. (2014) Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 20C, 110–117. [DOI] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulieres, F. , Hirsch, J. , Feng, D. , Bittner, E.P, Beynon, J. , Marco, Y. , Feng, D. . (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar, S. , Frary, A. , Ku, H.M. and Tanksley, S.D. (2002) Mapping quantitative trait loci in inbred backcross lines of Lycopersicon pimpinellifolium (LA1589). Genome, 45(6), 1189–1202. [DOI] [PubMed] [Google Scholar]
- Fegan, M. and Prior, P. (2005) How complex is the "Ralstonia solanacearum species complex" In: Bacterial wilt disease and the Ralstonia solanacearum species complex ( Allen, C. , Prior, P. and Hayward, A.C. eds.), pp. 449–462. Madison, WI: APS Press. [Google Scholar]
- Guinard, J. , Vinatzer, B.A. , Poussier, S. , Lefeuvre, P. and Wicker, E. (2016) Draft genome sequences of nine strains of Ralstonia solanacearum differing in virulence to eggplant (Solanum melongena). Genome Announc. 4(1), e01415–15. 10.1128/genomeA.01415-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward, A.C. (1991) Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum . Annu. Rev. Phytopathol. 29, 65–87. [DOI] [PubMed] [Google Scholar]
- Hayward, A.C. (1994) The hosts of Pseudomonas solanacearum In: Bacterial wilt – the disease and its causative agent, Pseudomonas solanacearum (Hayward A.C. and Hartman G.L., ed.), pp. 9–24. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , Buxton, S. , Cooper, A. , Markowitz, S. , Duran, C. , Thierer, T. , Ashton, B. , Meintjes, P. and Drummond, A. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa, S. (1982) Genetics and epidemiological modeling of breakdown of plant disease resistance. Annu. Rev. Phytopathol. 20, 93–117. [Google Scholar]
- Le Roux, C. , Huet, G. , Jauneau, A. , Camborde, L. , Tremousaygue, D. , Kraut, A. , Zhou, B. , Levaillant, M. , Adachi, H. , Yoshioka, H. , Raffaele, S. , Berthome, R. , Coute, Y. , Parker, J.E. and Deslandes, L. (2015) A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell, 161, 1074–1088. [DOI] [PubMed] [Google Scholar]
- Lebeau, A. (2010) Résistance de la tomate, l’aubergine et le piment à Ralstonia solanacearum: interactions entre les géniteurs de résistance et la diversité bactérienne, caractérisation et cartographie des facteurs génétiques impliqués chez l'aubergine. PhD Thesis, Université de la Réunion. Saint Denis de la Réunion, Reunion Island, France.
- Lebeau, A. , Daunay, M.C. , Frary, A. , Palloix, A. , Wang, J.F. , Dintinger, J. , Chiroleu, F. , Wicker, E. and Prior, P. (2011) Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology, 101, 154–165. [DOI] [PubMed] [Google Scholar]
- Lebeau, A. , Gouy, M. , Daunay, M. , Wicker, E. , Chiroleu, F. , Prior, P. , Frary, A. and Dintinger, J. (2013) Genetic mapping of a major dominant gene for resistance to Ralstonia solanacearum in eggplant. Theor. Appl. Genet. 126, 143–158. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Fu, D. , Zhu, B. , Yan, H. , Shen, X. , Zuo, J. , Zhu, Y. and Luo, Y. (2012) Virus‐induced gene silencing in eggplant (Solanum melongena). J. Integr. Plant Biol. 54, 422–429. [DOI] [PubMed] [Google Scholar]
- Lohou, D. , Turner, M. , Lonjon, F. , Cazalcb, A. , Peeters, N. , Genin, S. and Vailleau, F. (2014) HpaP modulates type III effector secretion in Ralstonia solanacearum and harbours a substrate specificity switch domain essential for virulence. Mol. Plant Pathol. 15, 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B.A. and Linde, C. (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- Montarry, J. , Corbiere, R. , Lesueur, S. , Glais, I. and Andrivon, D. (2006) Does selection by resistant hosts trigger local adaptation in plant–pathogen systems? J. Evol. Biol. 19, 522–531. [DOI] [PubMed] [Google Scholar]
- Monteiro, F. , Sole, M. , van Dijk, I. and Valls, M. (2012) A chromosomal insertion toolbox for promoter probing, mutant complementation, and pathogenicity studies in Ralstonia solanacearum . Mol. Plant–Microbe Interact. 25, 557–568. [DOI] [PubMed] [Google Scholar]
- Moury, B. (2010) A new lineage sheds light on the evolutionary history of Potato virus Y . Mol. Plant Pathol. 11, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar, K. , Matsumoto, I. , Taguchi, F. , Inagaki, Y. , Yamamoto, M. , Toyoda, K. , Shiraishi, T. , Ichinose, Y. and Mukaihara, T. (2014) Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum . Mol. Plant Pathol. 15, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N'Guessan, A.C. (2013) Phylogénie, structure génétique et diversité de virulence de Ralstonia solanacearum, agent du flétrissement bactérien, en Côte d'Ivoire. PhD Thesis, Université Félix Houphouët‐Boigny (Cocody)‐Abidjan. Abidjan, Côte d'Ivoire.
- N'Guessan, C.A. , Abo, K. , Fondio, L. , Chiroleu, F. , Lebeau, A. , Poussier, S. , Wicker, E. and Kone, D. (2012) So near and yet so far: the specific case of Ralstonia solanacearum populations from Cote d'Ivoire in Africa. Phytopathology, 102, 733–740. [DOI] [PubMed] [Google Scholar]
- Palloix, A. , Ayme, V. and Moury, B. (2009) Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 183, 190–199. [DOI] [PubMed] [Google Scholar]
- Pensec, F. , Lebeau, A. , Daunay, M. , Chiroleu, F. , Guidot, A. and Wicker, E. (2015) Towards the identification of type III effectors associated with Ralstonia solanacearum virulence on tomato and eggplant. Phytopathology, 105, 1529–1544. [DOI] [PubMed] [Google Scholar]
- Remenant, B. , Coupat‐Goutaland, B. , Guidot, A. , Cellier, G. , Wicker, E. , Allen, C. , Fegan, M. , Pruvost, O. , Elbaz, M. , Calteau, A. , Salvignol, G. , Mornico, D. , Mangenot, S. , Barbe, V. , Medigue, C. and Prior, P. (2010) Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics, 11(379), 1–16. 10.1186/1471-2164-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safni, I. , Cleenwerck, I. , De Vos, P. , Fegan, M. , Sly, L. and Kappler, U. (2014) Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp.nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov . Int. J. Syst. Evol. Microbiol. 64, 3087–3103. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , and Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Salgon, S. (2017) Déterminisme génétique de la résistance au flétrissement bactérien chez l'aubergine et application en sélection variétale [Genetic determinism of resistance to bacterial wilt in eggplant and applications in plant breeding]. PhD Thesis, Université de la Réunion. Saint Pierre, Reunion Island.
- Salgon, S. , Jourda, C. , Sauvage, C. , Daunay, M.‐C. , Reynaud, B. , Wicker, E. and Dintinger, J. (2017) Eggplant resistance to the Ralstonia solanacearum species complex involves both broad‐spectrum and strain‐specific quantitative trait loci. Front. Plant Sci. 8, 828 10.3389/fpls.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock, E.H. and McDonald, B.A. (2008) The origins of plant pathogens in agro‐ecosystems. Annu. Rev. Phytopathol. 46, 75–100. [DOI] [PubMed] [Google Scholar]
- Tasset, C. , Bernoux, M. , Jauneau, A. , Pouzet, C. , Briere, C. , Kieffer‐Jacquinod, S. , Rivas, S. , Marco, Y. and Deslandes, L. (2010) Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1‐R‐mediated immunity in Arabidopsis . Plos Pathogens, 6(11), e1001202 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, M. , Jauneau, A. , Genin, S. , Tavella, M.J. , Vailleau, F. , Gentzbittel, L. and Jardinaud, M.F. (2009) Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 150, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau, F. , Sartorel, E. , Jardinaud, M.F. , Chardon, F. , Genin, S. , Huguet, T. , Gentzbittel, L. and Petitprez, M.A. (2007) Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula . Mol. Plant–Microbe Interact. 20, 159–167. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. and Oliver, R.P. (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant–Microbe Interact. 27, 196–206. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Remigi, P. , Anisimova, M. , Lonjon, F. , Kars, I. , Kajava, A. , Li, C.H. , Cheng, C.P. , Vailleau, F. , Genin, S. and Peeters, N. (2016) Functional assignment to positively selected sites in the core type III effector RipG7 from Ralstonia solanacearum. Mol. Plant Pathol. 17, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, E. , Lefeuvre, P. , Lemaire, C. , de Cambiaire, J.C. , Poussier, S. , and Prior, P. (2012) Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J. 6, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F.N. , Eannetta, N.T. , Xu, Y.M. and Tanksley, S.D. (2009) A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor. Appl. Genet. 118, 927–935. [DOI] [PubMed] [Google Scholar]
- Xi’ou, X., Bihao, C., Guannan, L., Jianjun, L., Qinghua, C., Jin, J. and Yujing, C. (2014) Functional characterization of a putative bacterial wilt resistance gene (RE‐bw) in eggplant. Plant Mol. Biol. Rep. 33, 1058–1073. [Google Scholar]
- Zeigler, R.S., Tohme, J., Nelson, R., Levy, M. and Correavictoria, F.J. (1994) Lineage exclusion – a proposal for linking blast population analysis to resistance breeding In: (Zeigler, R.S., Leong, S.A. and Teng, P.S., eds.), pp. 267–292. Wallingford, Oxfordshire: CAB International. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Kaplan–Meier survival curves representing the wilting of AG91‐25 plants inoculated with the strains GMI1000, GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, PSS4 and PSS4::ripAX2 in two supplemental replicates (12 and 22 plants per strain, respectively). Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S2 Kaplan–Meier survival curves observed on MM738 plants (susceptible control) inoculated with strains GMI1000, GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, PSS4 and PSS4::ripAX2 in one supplemental replicate. Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S3 Alignment of the RipAX2 protein alleles from GMI1000, RS1000 and MOLK2, and their percentages of identity.
Fig. S4 Kaplan–Meier survival curves observed on AG91‐25 plants inoculated with strains GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, GMI1000 ripAX2::pCZ367::ripAX2‐E149A, PSS4, PSS4::ripAX2 and PSS4::ripAX2‐E149A in two supplemental replicates (22 plants and 18 plants per strain for each respective replicate). Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S5 Kaplan–Meier survival curves observed on MM738 plants inoculated with strains GMI1000 ripAX2::pCZ367, GMI1000 ripAX2::pCZ367::ripAX2, GMI1000 ripAX2::pCZ367::ripAX2‐E149A, PSS4, PSS4::ripAX2 and PSS4::ripAX2‐E149A in three replicates. Log‐rank (Mantel–Cox) tests are given with P value.
Fig. S6 DNA alignment of the reference ripAX2 alleles (GMI1000 and RS1000) with the long ripAX2 alleles amplified within the phylotype I Ivorian strains RUN1546 (sequevar 13), RUN1740 and RUN1743 (sequevar 31), and in the Guianese phylotype IIA RUN1994 (sequevar 41). Alignments were performed using MUSCLE under GENEIOUS v5.5. Each strain is represented by its nucleotide sequence (upper line) and proteic sequence (lower line). Black squares indicate stop codons.
Fig. S7 Geographical distribution of the 19 RipAX2 alleles, either functional (PROT) or non‐functional (NF), across regions within the phylotype I strains. The heatmap was built with the R package pheatmap, based on Euclidean distances between observations; the clusters were calculated using the Ward method. For each region, the number of RipAX2 alleles (G) and phylotype I strains (N) are presented. ‘Africa’ gathers Burkina‐Faso, Cameroon, Ivory Coast, Reunion Island and South Africa; ‘Asia’ gathers Australia, India, Indonesia, Philippines, Taiwan and Thailand; ‘South America’ gathers French Guiana and Martinique. The heatmap values correspond to the numbers of strains.
Table S1 Bacterial strains of the Ralstonia solanacearum species complex (RSSC) tested for the presence of the ripAX2 coding sequence. Phylotype I and III strains belong to the species R. pseudosolanacearum, and phylotype IIA and IIB strains belong to the species R. solanacearum.
Table S2 Distribution of ripAX2 within the 74 Ralstonia solanacearum species complex (RSSC) genomes harboured in the RalstoT3E database. Strains are ranked by phylotype and code. Phylotypes I and III are grouped within R. pseudosolanacearum, phylotypes IIA and IIB within R. solanacearum and phylotype IV within R. syzygii. The ripAX2 type III effector is present as a complete sequence (PROT), frameshifted (FS), pseudogenized (PS) or absent (NO).
Table S3 Comparison of ripAX2 distribution, assessed by polymerase chain reaction (PCR) amplification or genome sequencing, in the 15 strains common to our collection and the RalstoT3E database. The gene was present (1), frameshifted (FS) or absent (0).
Table S4 External and internal primers designed to amplify the ripAX2 coding sequence. Both external and internal primers were designed on the alignment of GM1000 and RS1000 ripAX2 gene and neighbouring regions.
Table S5 Bacterial strains of the Ralstonia solanacearum species complex (RSSC) with their host of isolation, nucleotidic and proteic RipAX2 alleles, and GenBank accession numbers.
Table S6 Specificities in distributions of the RipAX2 protein alleles (Hap_AA) across regions, as assessed by χ 2 tests. (A) Distribution of each allele across regions. Status is ‘absence’ (ABS), ‘functional’ (PROT) or ‘non‐functional’ (NF). (B) Distribution of each region across alleles.


