Summary
The soil‐borne, plant‐pathogenic Ralstonia solanacearum strain OE1‐1 produces and secretes methyl 3‐hydroxymyristate (3‐OH MAME) as a quorum sensing (QS) signal, which contributes to its virulence. A global virulence regulator, PhcA, functioning through the QS system, positively regulates the expression of ralA, which encodes furanone synthase, to produce aryl‐furanone secondary metabolites, ralfuranones. A ralfuranone‐deficient mutant (ΔralA) is weakly virulent when directly inoculated into tomato xylem vessels. To investigate the functions of ralfuranones, we analysed R. solanacearum transcriptome data generated by RNA sequencing technology. ΔralA expressed phcB, which is associated with 3‐OH MAME production, and phcA at levels similar to those in strain OE1‐1. In addition, ΔralA exhibited down‐regulated expression of more than 90% of the QS positively regulated genes, and up‐regulated expression of more than 75% of the QS negatively regulated genes. These results suggest that ralfuranones affect the QS feedback loop. Ralfuranone supplementation restored the ability of ΔralA cells to aggregate. In addition, ralfuranones A and B restored the swimming motility of ΔralA to wild‐type levels. However, the application of exogenous ralfuranones did not affect the production of the major exopolysaccharide, EPS I, in ΔralA. Quantitative real‐time polymerase chain reaction assays revealed that the deletion of ralA results in the down‐regulated expression of vsrAD and vsrBC, which encode a sensor kinase and a response regulator, respectively, in the two‐component regulatory systems that influence EPS I production. The application of ralfuranone B restored the expression of these two genes. Overall, our findings indicate that integrated signalling via ralfuranones influences the QS and virulence of R. solanacearum.
Keywords: quorum sensing, ralfuranones, Ralstonia solanacearum, soil‐borne, plant‐pathogenic bacterium, virulence
Introduction
Cells of many bacteria communicate with each other by releasing, sensing and responding to small diffusible signalling molecules, allowing them to regulate their cooperative activities and physiological processes through quorum sensing (QS) (Ham, 2013). These QS‐controlled activities influence the virulence and pathogenic potential of bacteria. Many pathogenic bacteria use cell–cell signalling to regulate the expression of virulence factors. The signalling molecule known as diffusible signalling factor (DSF) is a cis‐unsaturated fatty acid and belongs to a novel class of QS signals in Xanthomonas campestris pv. campestris (Ham, 2013; Mills et al., 2011; Ryan and Dow, 2011; Tang et al., 1991). DSF appears to be widely conserved in diverse bacterial species. The DSF‐type QS signalling system includes a novel secondary messenger, cyclic‐di‐guanosine monophosphate (cyclic‐di‐GMP), which facilitates the coupling of QS to the bacterial intracellular regulatory networks. This system is implicated in the regulation of a wide range of bacterial functions. However, intercellular signalling between bacterial cells that are associated with QS remains unclear.
The soil‐borne, plant‐pathogenic Ralstonia solanacearum bacterial strains AW1 and K60 produce methyl 3‐hydroxypalmitate (3‐OH PAME) as a QS signal that mediates the phc QS system (Flavier et al., 1997; Kai et al., 2015). 3‐OH PAME is synthesized by PhcB, which is a methyltransferase. When the abundance of 3‐OH PAME reaches a threshold level, it decreases the ability of the histidine kinase PhcS to phosphorylate the response regulator PhcR. This results in elevated levels of functional PhcA, which is a LysR‐type transcriptional regulator (Clough et al., 1997; Flavier et al., 1997; Genin and Denny, 2012). In addition, strains OE1‐1 and GMI1000 produce methyl 3‐hydroxymyristate (3‐OH MAME) as a QS signal (Kai et al., 2015). The deduced PhcB and PhcS amino acid sequences among R. solanacearum strains are related to the productivity of QS signals. The global virulence regulator PhcA plays a central role in the phc QS system (Fig. S1, see Supporting Information) (Brumbley and Denny, 1990; Clough et al., 1994; Genin and Denny, 2012).
Ralstonia solanacearum synthesizes aryl‐furanone secondary metabolites, known as ralfuranones A, B, I, J, K and L, which are extracellularly secreted (Kai et al., 2014; Pauly et al., 2013). Ralfuranone I is a precursor of the other ralfuranones (Fig. S1). The production of transaminase and furanone synthase, which are encoded by ralD and ralA, respectively, depends on PhcA functions via the phc QS system, and they are involved in the biosynthesis of ralfuranones (Kai et al., 2014; Schneider et al., 2009; Wackler et al., 2011). Thus, ralfuranone production is dependent on the phc QS system. Kai et al. (2014) revealed that a ralfuranone‐deficient mutant (i.e. ΔralA) produces considerably less exopolysaccharide (EPS) and is less virulent than OE1‐1 on tomato plants following direct inoculations of xylem vessels. These results suggest that ralfuranones are required for the full virulence of strain OE1‐1. Furthermore, non‐phytotoxic ralfuranones were detected in the xylem fluids of tomato plants inoculated with strain OE1‐1. Thus, ralfuranones may be linked to the intercellular signalling between OE1‐1 cells required for virulence. However, the exact effects of ralfuranones on OE1‐1 virulence remain unclear.
To elucidate the exact role of ralfuranones in OE1‐1 virulence, we first examined the transcriptome profiles of the ΔralA and phc QS‐deficient mutants, as well as the wild‐type (WT) OE1‐1 strain, using RNA sequencing (RNA‐seq) technology. We also analysed the involvement of ralfuranones in phc QS‐dependent, virulence‐related phenotypes.
Results
ralA deletion affects the expression of a large set of genes also regulated by the QS system components phcA and phcB
For transcriptome analyses using RNA‐seq, total RNA was isolated from OE1‐1, phcB‐deleted mutant (ΔphcB), phcA‐deleted mutant (ΔphcA) and ΔralA mutant R. solanacearum cells cultured in one‐quarter‐strength M63 medium [to an optical density at 600 nm (OD600) = 0.3]. Cytoplasmic ribosomal RNA was removed from the total RNA, for a final RNA yield of 400 ng for each sample. The isolated RNA was subjected to Illumina RNA sequencing. The RNA samples were fragmented and ligated with adaptors prior to cDNA synthesis and polymerase chain reaction (PCR) amplifications. We obtained 41.0, 46.5, 45.3 and 44.8 million 100‐bp paired‐end reads from OE1‐1, ΔphcB, ΔphcA and ΔralA, respectively. By iterative alignment, 41.8, 42.5, 42.1 and 41.4 million 100‐bp paired‐end reads were successfully mapped to the R. solanacearum strain GMI1000 reference genome (Salanoubat et al., 2002). The mapping of the OE1‐1 RNA‐seq reads to the GMI1000 genome resulted in the identification of 4493 protein‐coding transcripts.
The normalized gene expression levels of OE1‐1, ΔphcB, ΔphcA and ΔralA were compared to detect differentially expressed transcripts. Read counts obtained for each sample were FPKM (fragments per kilobase of exon per million fragments mapped) normalized prior to being analysed for differentially expressed genes. Genes were considered to be differentially expressed if they exhibited fold changes of ≥2 or ≤–2. We detected 688, 983 and 772 genes that were expressed at significantly lower levels in ΔphcB, ΔphcA and ΔralA than in OE1‐1, respectively (Table S1, see Supporting Information, Fig. 1a). In addition, 606 genes were expressed at lower levels in both ΔphcB and ΔphcA, suggesting that the expression of these genes is positively regulated by PhcA functioning through the phc QS system (Fig. S2, see Supporting Information). Of these, 556 genes, including lecM, the major EPS (i.e. EPS I) production‐related genes, such as those in the eps operon (i.e. epsR and xpsR), the type VI secretion system‐related genes, plant cell wall degradation enzyme genes (i.e. pme, egl and pehC), two‐component system‐related genes (i.e. solI and solR) and some effector genes secreted through the type III secretion systems (i.e. RSc1800, ripG4; RSc1801, ripG5; Rsp0323, ripO1; RSp0731, ripTP5; Rsp1281, ripS; and RSp1460, ripAU) were expressed at lower levels in ΔralA than in OE1‐1 (Table S2, see Supporting Information).
Figure 1.
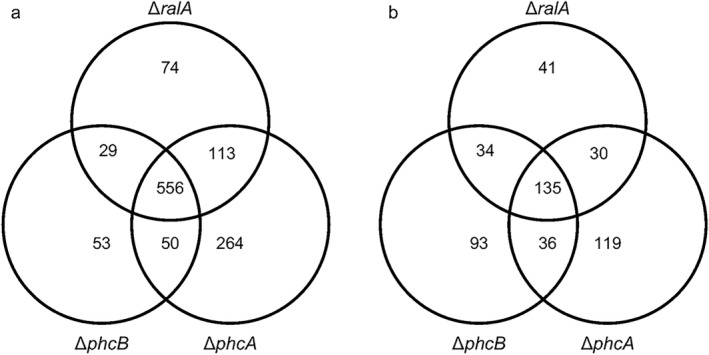
Number of genes exhibiting expression level fold changes of ≤–2 (a) or ≥2 (b) in the Ralstonia solanacearum phcB‐deleted mutant (ΔphcB), phcA‐deleted mutant (ΔphcA) and ralA‐deleted mutant (ΔralA), relative to the expression levels of strain OE1‐1. The FPKM (fragments per kilobase of exon per million fragments mapped) values of strains OE1‐1, ΔphcB, ΔphcA and ΔralA were normalized prior to the analyses of differentially expressed genes.
We also detected 298, 320 and 240 genes that were expressed at higher levels in ΔphcB, ΔphcA and ΔralA than in OE1‐1, respectively (Table S2, Fig. 1b). There were 171 genes that were expressed at higher levels in both ΔphcB and ΔphcA, suggesting that the expression of these genes is negatively regulated by PhcA functioning through the phc QS system (Fig. S2). Included among these genes were 135 genes that were more highly expressed in ΔralA than in OE1‐1, including flagellar motility‐related genes, such as fliC, type III secretion‐related genes and some type III effector genes (i.e. RSc3290, ripAX1; RSp0099, ripA2; RSp0822, ripAF1; RSp0877, ripX; RSp0876, ripAB; RSp0875, ripAC; RSp1374, ripS2; RSp0930, ripS3; RSp1277, ripQ; RSp1582, ripAZ1; and RSp1601, ripAD), and chemotaxis‐related genes (Table S3, see Supporting Information).
Expression analysis of the phc QS‐related genes and 3‐OH MAME production
The expression of ralA in strain OE1‐1 depends on the activation of PhcA in the phc QS system, with 3‐OH MAME acting as a QS signal (Kai et al., 2015). If ralfuranones control 3‐OH MAME production by feedback regulation, deletion of ralA might result in systemically decreased QS‐dependent gene expression levels. We first analysed the expression of the phc QS‐related genes, phcB and phcA, in R. solanacearum strains grown in one‐quarter‐strength M63 medium (to OD600 = 0.3) using quantitative real‐time PCR (qRT‐PCR) assays. There were no significant differences between ΔralA and OE1‐1 with regard to phcB and phcA expression levels (P > 0.05) (Fig. 2a). However, ΔralA produced slightly less 3‐OH MAME than OE1‐1 (Fig. 2b).
Figure 2.

Influence of ralA deletion on the quorum sensing of Ralstonia solanacearum. Expression of phcB and phcA in R. solanacearum OE1‐1 and ralA‐deleted mutant (ΔralA) strains (a) and methyl 3‐hydroxymyristate (3‐OH MAME) purified from the R. solanacearum OE1‐1 and ΔralA strains (b). The R. solanacearum strains were grown in one‐quarter‐strength M63 medium (to OD600 = 0.3). Total RNA was then extracted from the bacterial cells. The rpoD gene was used as an internal control for quantitative real‐time polymerase chain reaction. The gene expression levels are presented relative to the rpoD expression level. The experiment was conducted at least twice using independent samples, with similar results. Results for a single representative sample are provided. Values are presented as the mean ± standard deviation of three replicates. Asterisks indicate values that are significantly different from those of OE1‐1 cells (P < 0.05, t‐test). Synthetic 3‐OH MAME was used as a positive control. The arrows indicate the peaks corresponding to 3‐OH MAME.
Ralfuranones affect the aggregation of R. solanacearum OE1‐1 cells
The aggregation of R. solanacearum cells depends on lecM, which encodes the lectin, RS‐IIL. The expression of lecM is positively regulated by the phc QS system (Meng et al., 2015; Mori et al., 2016). Ralstonia solanacearum strains incubated in one‐quarter‐strength M63 medium for 24 h were analysed using crystal violet staining. There were significantly fewer cell aggregates in the ΔralA mutant than in the OE1‐1 or complemented ΔralA mutant strain (ralA‐comp) (P < 0.05, Fig. 3).
Figure 3.
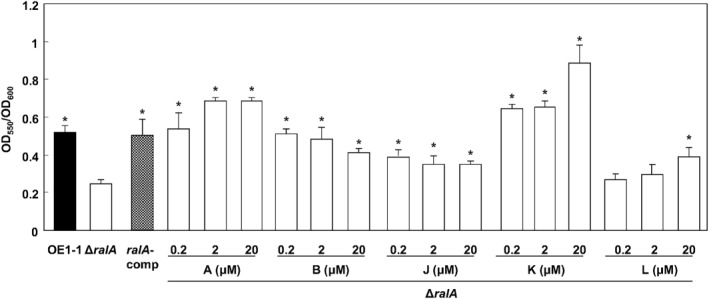
Cell aggregation results for Ralstonia solanacearum OE1‐1, the ralfuranone‐deficient mutant (ΔralA) and the complemented ΔralA mutant (ralA‐comp) strains. OE1‐1, ΔralA and ralA‐comp cells were incubated in one‐quarter‐strength M63 medium in wells of polyvinylchloride microtitre plates. ΔralA cells were also incubated in one‐quarter‐strength M63 medium supplemented with ralfuranones A, B, J, K or L at concentrations of 0.2–20 μm. The wells were stained with crystal violet. Asterisks indicate values that are significantly different from those of the ΔralA strain in apoplast fluid (P < 0.05, t‐test). OD, optical density.
The ΔralA mutant was then grown in one‐quarter‐strength M63 medium containing 0.2–20 μm of each ralfuranone. The ΔralA cell aggregation level increased with increasing concentrations of ralfuranones A, K and L. When the growth medium was supplemented with 20 μm ralfuranones A or K, more ΔralA cells than OE1‐1 cells aggregated (Fig. 3). In addition, ralfuranone L restored the aggregation of ΔralA cells to approximately 75.5% of that of WT cells. Supplementation with ralfuranone B restored the aggregation of ΔralA cells to >80% of the OE1‐1 levels, regardless of concentration. In contrast, supplementation with 20 μm ralfuranone J resulted in only a slight increase in the aggregation of ΔralA cells. These results suggest that ralfuranones, especially ralfuranones A and K, influence the aggregation of OE1‐1 cells. Based on these assay results, we used 20 μm ralfuranones in subsequent experiments.
ΔralA produces significantly less EPS I than does OE1‐1
We quantified the EPS I produced by R. solanacearum strains growing on one‐quarter‐strength M63 solid medium using an enzyme‐linked immunosorbent assay. The ΔralA mutant produced significantly less EPS I than did the OE1‐1 and ralA‐comp strains (P < 0.05, Fig. 4a). Supplementation with individual ralfuranones did not affect EPS I production by ΔralA.
Figure 4.
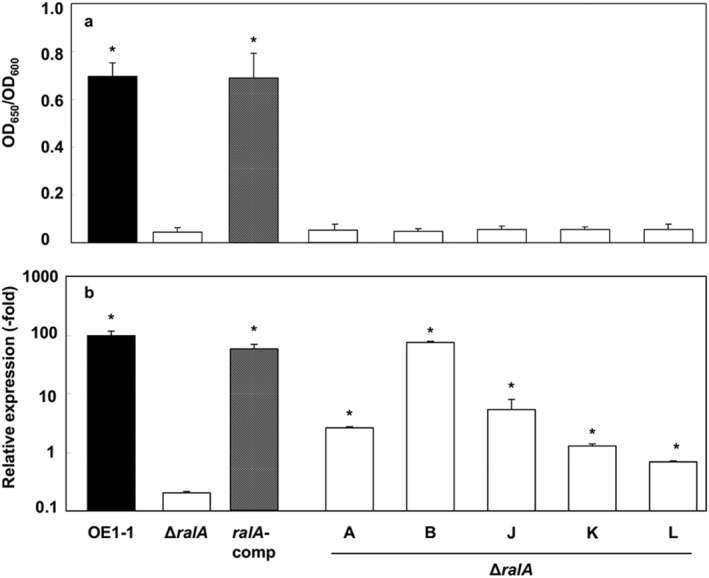
Influence of ralA deletion on exopolysaccharide I (EPS I) production by Ralstonia solanacearum. (a) Quantification of EPS I in supernatants using an enzyme‐linked immunosorbent assay with anti‐R. solanacearum EPS I antibodies. The R. solanacearum OE1‐1, ralfuranone‐deficient mutant (ΔralA) and ΔralA complemented mutant (ralA‐comp) strains were incubated on one‐quarter‐strength M63 medium in plates. The ΔralA mutant was also incubated on one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L. (b) epsB expression levels in three R. solanacearum strains. Strains OE1‐1, ΔralA and ralA‐comp were cultured in one‐quarter‐strength M63 medium. The ΔralA mutant was also cultured in one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L. Total RNA was extracted from a bacterial culture [optical density at 600 nm (OD600) = 0.3]. The rpoD gene was used as an internal control for quantitative real‐time polymerase chain reaction. The gene expression levels are presented relative to the rpoD expression level. The experiment was conducted at least twice using independent samples, with similar results. Results for a single representative sample are provided. Values are presented as the mean ± standard deviation of three replicates. Asterisks indicate values that are significantly different from those of the ΔralA cells (P < 0.05, t‐test).
We then analysed the expression of epsB in R. solanacearum strains grown in one‐quarter‐strength M63 medium using qRT‐PCR assays. This gene is included in the eps operon and is thought to be important for EPS I biosynthesis (Huang and Schell, 1995). The epsB expression level was significantly lower in ΔralA than in OE1‐1 or ralA‐comp (P < 0.05, Fig. 4b). Supplementation with ralfuranones A, J, K or L resulted in slight increases in epsB expression in the ΔralA mutant, whereas ralfuranone B supplementation increased epsB expression in the mutant to 76.0% of the level in OE1‐1.
ΔralA cells exhibit greater swimming motility than OE1‐1 cells
Flagella biogenesis is negatively regulated by the phc QS system, and swimming motility is essential for biofilm formation by R. solanacearum (Tans‐Kersten et al., 2001). Thus, we analysed the swimming motility of the R. solanacearum strains. The ΔralA mutant exhibited greater swimming motility than the WT strain OE1‐1 on one‐quarter‐strength M63 medium solidified with 0.25% agar, similar to the phcA‐deleted mutant. The swimming motility of the ralA‐comp mutant strain was similar to that of the WT strain (Fig. 5a).
Figure 5.
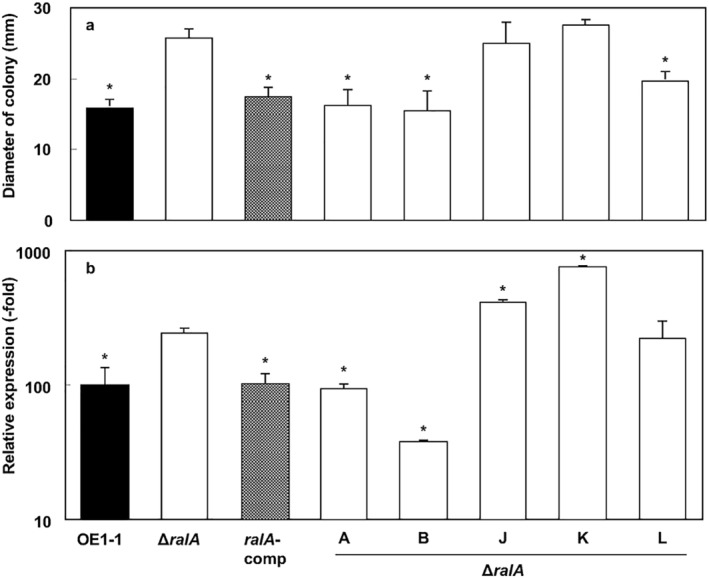
Swimming motility and fliC expression in Ralstonia solanacearum strains. (a) To analyse swimming motility, R. solanacearum OE1‐1, ralfuranone‐deficient mutant (ΔralA) and complemented ΔralA mutant (ralA‐comp) strains were grown on one‐quarter‐strength M63 medium solidified with 0.25% agar. The ΔralA mutant was also incubated on agar‐solidified, one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L. (b) To examine fliC expression, R. solanacearum strains OE1‐1, ΔralA and ralA‐comp were cultured in one‐quarter‐strength M63 medium. The ΔralA mutant was also cultured in one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L. Total RNA was extracted from a bacterial culture [optical density at 600 nm (OD600) = 0.3]. The rpoD gene was used as an internal control for quantitative real‐time polymerase chain reaction. The gene expression levels are presented relative to the rpoD expression level. The experiment was conducted at least twice using independent samples, with similar results. Results for a single representative sample are provided. Values are presented as the mean ± standard deviation of three replicates. Asterisks indicate values that are significantly different from those of the ΔralA strain (P < 0.05, t‐test).
When ΔralA was supplemented with ralfuranones A or B, the observed swimming motility was similar to that of the OE1‐1 and ralA‐comp strains (Fig. 5a). Supplementation with ralfuranone L resulted in slightly decreased ΔralA swimming motility, whereas ralfuranones J and K had no effect.
We analysed the fliC expression level in R. solanacearum strains grown on one‐quarter‐strength M63 medium using qRT‐PCR assays. We observed higher fliC expression levels in the ΔralA mutant than in the WT and ralA‐comp strains (Fig. 5b). When grown on one‐quarter‐strength M63 medium containing ralfuranones A or B, the fliC expression level in strain ΔralA decreased to a level similar to that of the WT OE1‐1 strain. Supplementation with ralfuranone L resulted in a slightly decreased fliC expression level in ΔralA, whereas ralfuranones J and K had the opposite effect.
Ralfuranones affect the expression of two‐component, system‐encoding vsrA/vsrD and vsrB/vsrC
Although supplementation with ralfuranone B restored epsB expression in the ΔralA mutant strain, EPS I production was unaffected. The VsrAD and VsrBC two‐component sensor kinase/response regulatory systems are reportedly involved in the regulation of EPS I production dependent on the phc QS (Garg et al., 2000; Huang et al., 1998). To elucidate the role of ralfuranones in these two‐component systems, we analysed the expression of vsrA/vsrD and vsrB/vsrC in the R. solanacearum strains using qRT‐PCR assays. The expression levels of all four genes were significantly lower in the ΔralA mutant than in the WT and ralA‐comp strains (P < 0.05, Fig. 6). When grown in one‐quarter‐strength M63 medium containing ralfuranone B, the expression of vsrA/vsrD and vsrB/vsrC was restored to WT levels in the ΔralA mutant. Supplementation with ralfuranone J resulted in partially recovered gene expression levels (Fig. 6), whereas ralfuranones K and L did not influence gene expression in ΔralA. Supplementation with ralfuranone A resulted in slightly decreased vsrA, vsrD and vsrB expression levels in the ΔralA mutant.
Figure 6.
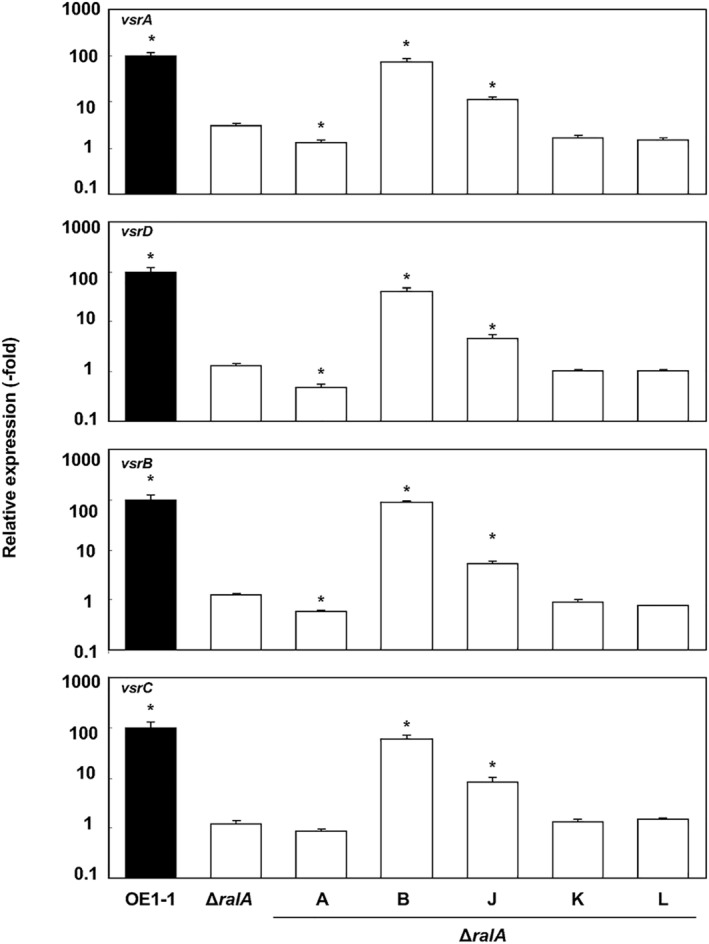
Expression of vsrA/vsrD and vsrB/vsrC two‐component, system‐encoding genes in Ralstonia solanacearum. Strain OE1‐1 and the ralfuranone‐deficient mutant ΔralA were cultured in one‐quarter‐strength M63 medium. The ΔralA mutant was also cultured in one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L. Total RNA was extracted from a bacterial culture [optical density at 600 nm (OD600) = 0.3]. The rpoD gene was used as an internal control for quantitative real‐time polymerase chain reaction. The gene expression levels are presented relative to the rpoD expression level. The experiment was conducted at least twice using independent samples, with similar results. Results for a single representative sample are provided. Values are presented as the mean ± standard deviation of three replicates. Asterisks indicate values that are significantly different from those of the ΔralA mutant (P < 0.05, t‐test).
Ralfuranones affect the expression of genes encoding sigma factors
RpoN (i.e. sigma factor σ54) regulates flagellar motility, EPS biosynthesis and biofilm formation in plant‐pathogenic bacteria (Dong and Mekalanos, 2012; Hao et al., 2013; Kazmierczak et al., 2005; O'Toole et al., 1997; Reitzer and Schneider, 2001; da Silva Neto et al., 2008). Ralstonia solanacearum has two genes encoding σ54 (i.e. rpoN1 and rpoN2). rpoN1 also influences the virulence of strain GMI1000 (Ray et al., 2015). Furthermore, the expression of solI and solR, which are associated with the SolI/R acyl‐homoserine lactone QS system, depends on PhcA and RpoS (Flavier et al., 1997). To clarify the involvement of ralfuranones in the regulation of sigma factors, we compared the expression of rpoN1, rpoN2 and rpoS in strains ΔralA and OE1‐1 incubated in one‐quarter‐strength M63 medium. We observed significantly decreased rpoN2 expression levels in the ΔralA mutant, whereas the expression of rpoN1 was not significantly affected (P < 0.05, Fig. 7a). The rpoS expression level was 2.5 times higher in ΔralA than in OE1‐1. We subsequently analysed the effects of ralfuranones on the expression of rpoN2. Although supplementation with ralfuranones A, K and L did not influence the expression of rpoN2 in ΔralA, ralfuranones B and J partially restored rpoN2 expression (Fig. 7b).
Figure 7.
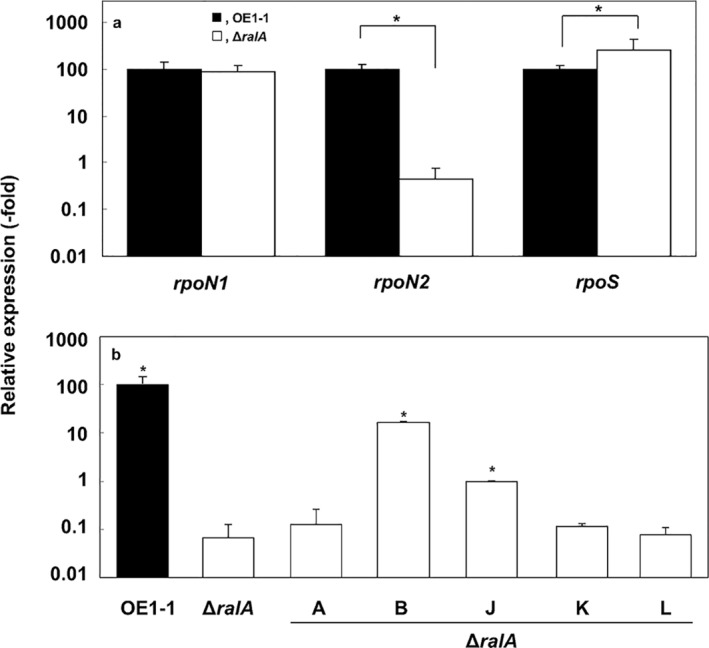
Expression of rpoN1, rpoN2 and rpoS in Ralstonia solanacearum strain OE1‐1 and the ralfuranone‐deficient mutant (ΔralA). Total RNA was extracted from cells grown in one‐quarter‐strength M63 medium [optical density at 600 nm (OD600) = 0.3] (a) or in one‐quarter‐strength M63 medium containing 20 μm ralfuranones A, B, J, K or L (b). The rpoD gene was used as an internal control for quantitative real‐time polymerase chain reaction. The gene expression levels are presented relative to the rpoD expression level. The experiment was conducted at least twice using independent samples, with similar results. Results for a single representative sample are provided. Values are presented as the mean ± standard deviation of three replicates. Asterisks indicate values that are significantly different from those of OE1‐1 (a) and ΔralA (b) (P < 0.05, t‐test).
Discussion
The QS system in R. solanacearum consists of phc regulatory elements, and PhcA functioning via the phc QS system plays a central role, leading to the virulence of this bacterial species (Clough et al., 1997; Flavier et al., 1997; Genin and Denny, 2012). The results of our transcriptome analysis using RNA‐seq (Fig. 1, Tables S1–S3) and qRT‐PCR assays revealed that the deletion of ralA leads to significant changes in the expression levels of the majority of the phc QS‐dependent genes, but not phcB and phcA (Fig. 2a). These results suggest that a lack of ralA may have implications for transcriptional regulation by the phc QS system. It is likely that the ralfuranones influence the phc QS‐mediated functionalization of PhcA. It is tempting to speculate that ralfuranones affect the regulation of 3‐OH MAME production at the post‐transcriptional level based on the decreased production of 3‐OH MAME in the ΔralA mutant. The expression of ralfuranone production‐related genes (i.e. ralA and ralD) is positively regulated by PhcA functions through the phc QS system (Kai et al., 2014; Schneider et al., 2009; Wackler et al., 2011). These results suggest that ralfuranones may be associated with the feedback loop of the phc QS system (Fig. S3, see Supporting Information).
Kai et al. (2014) reported that the ΔralA mutant exhibits significantly decreased virulence following direct inoculations of tomato xylem vessels. This implies that ralfuranones contribute to the full virulence of strain OE1‐1. Furthermore, Kai et al. (2014) also detected ralfuranones in the xylem fluids of tomato plants following the direct inoculation of xylem vessels with strain OE1‐1 using the wounded petiole inoculation method. These observations indicate that ralfuranones, produced by strain OE1‐1 infecting xylem vessels, affect the expression of phc QS‐regulated genes, leading to the full virulence of strain OE1‐1.
The production of EPS I by OE1‐1 is positively regulated by the phc QS system (Brumbley and Denny, 1990; Clough et al., 1994; Genin and Denny, 2012). Although all of the ralfuranones used in this study positively regulated epsB expression in the ΔralA mutant (Fig. 4b), EPS I production was not affected by any of the ralfuranones (Fig. 4a). Ralfuranones may thus affect the expression of unknown additional crucial EPS I biosynthesis‐related genes. Furthermore, ralfuranones B and J were involved in the positive regulation of vsrA, vsrD, vsrB and vsrC expression (Fig. 6). In contrast, ralfuranone A may help to negatively regulate the expression of these genes. The PhcA and VsrAD two‐component sensor/response regulatory systems are necessary for the full activation of the transcription of xpsR. In addition, both a transcriptional regulator XpsR and a response regulator VsrC up‐regulate the expression of the eps operon (Garg et al., 2000; Huang et al., 1998). EPS I production is thus influenced by the VsrAD and VsrBC two‐component systems. Therefore, the integrated regulation of vsrAD and vsrBC expression by ralfuranones A, B and J may also contribute to EPS I production (Fig. S3).
Flagella biogenesis in R. solanacearum is negatively regulated by the phc QS system (Tans‐Kersten et al., 2001). Interestingly, ralfuranones A and B negatively regulated fliC expression (Fig. 5b) and swimming motility (Fig. 5a) in the current study. Furthermore, the VsrAD and VsrBC two‐component systems affect the regulation of flagella biogenesis (Genin and Denny, 2012). Therefore, ralfuranones A, B and J may also influence flagellar motility by regulating the expression of vsrAD and vsrBC. Furthermore, Schneider et al. (2009) reported that VsrAD is upstream of PhcA and is involved in the biosynthesis of ralfuranones. These observations suggest that the expression of vsrAD may be feedback regulated through ralfuranones A, B and J, leading to the regulation of PhcA function (Fig. S3).
σ54 (rpoN) is involved in the regulation of nitrogen metabolism, and subsequently affects many other biological activities in diverse Proteobacteria (Buck et al., 2000). In R. solanacearum, RpoN1, but not RpoN2, helps to regulate the transcription of twitching motility‐related genes, and affects the nitrate assimilation pathway and virulence (Ray et al., 2015). Furthermore, rpoN2 expression is dependent on rpoN1. In the current study, the inability to produce ralfuranones resulted in significantly decreased rpoN2 expression levels, whereas rpoN1 expression was relatively unaffected (Fig. 7a). Ralfuranones B and J positively regulated rpoN2 expression (Fig. 7b), suggesting that these two ralfuranones may directly influence rpoN2 expression. Ray et al. (2015) revealed the broad conservation and stability of rpoN2 in the R. solanacearum species complex. They speculated that rpoN2 might be involved in the adaptation of the bacterium to a specific niche or environmental condition during its life cycle. Based on these observations, we propose that ralfuranones also regulate the expression of virulence‐related genes via the regulation of rpoN2 expression.
A novel transcriptional regulator (i.e. EfpR) in R. solanacearum strain GMI1000 acts as a global catabolic repressor that directly or indirectly down‐regulates the expression of multiple metabolic pathway genes (Perrier et al., 2016). Furthermore, EfpR also controls virulence traits, such as EPS production and motility (i.e. swimming or twitching). However, the expression of efpR is not significantly altered in the phcA mutant. In addition, phcA does not appear to be differentially regulated in the efpR mutant background. Our transcriptome analyses revealed differences between the ΔphcA (Table S2) and ΔefpR (Perrier et al., 2016) mutants with regard to the expression profiles of genes related to EPS I production and flagella motility. Interestingly, EfpR suppressed ralD expression, which regulates ralfuranone production. Furthermore, the phcA mutant was observed to exhibit increased metabolic versatility, with the ability to metabolize a wider repertoire of metabolic substrates than the efpR mutant (Peyraud et al., 2016). Thus, there is probably some interplay between the two central regulators (i.e. PhcA and EfpR) that is mediated through ralfuranones. EPS biosynthesis represents a significant cost for R. solanacearum (Peyraud et al., 2016) and PhcA controls multiple virulence functions encoded by hundreds of genes, including the eps gene cluster (Genin and Denny, 2012). The expression of efpR in the phc QS‐deficient mutants, ΔphcB and ΔphcA, not only ΔralA, did not differ significantly from that in strain OE1‐1 (Table S1). These results may lead to the complementation of epsB expression in the ΔralA mutant, but not EPS I production by the ΔralA strain, supplemented with individual ralfuranones.
Ralstonia solanacearum produces and extracellularly secretes ralfuranones. Ralfuranone I is a precursor for the other ralfuranones (Fig. S1) (Kai et al., 2016; Pauly et al., 2013), and is non‐enzymatically converted into ralfuranone B in the supernatant (Kai et al., 2016). The non‐enzymatic elimination of benzaldehyde from ralfuranone B produces ralfuranone A, whereas ralfuranones J and K are the products of the enzymatic oxidation of ralfuranone B. Ralfuranone L is enzymatically synthesized from ralfuranone I. One obvious question is why does R. solanacearum produce so many types of ralfuranone. It is thought that the extracellular secretion of each ralfuranone by OE1‐1 changes over time (Fig. S1; Kai et al., 2016). Interestingly, the application of ralfuranone B and other ralfuranones led to recovered phenotypes of ΔralA. Therefore, during the early stages of infection, 3‐OH MAME‐mediated intercellular signalling activates phc QS, leading to the production and secretion of ralfuranones. Each ralfuranone may then mediate intercellular signalling between OE1‐1 cells in association with the feedback loop of the phc QS system. Overall, the integrated intracellular/intercellular signalling of OE1‐1 cells via each ralfuranone, coupled with phc QS, may contribute to the elaborate and tunable regulation of R. solanacearum virulence (Fig. S3). This may enable R. solanacearum to infect many plant species and remain virulent during infections.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
We used the following R. solanacearum strains: OE1‐1 (Kanda et al., 2003), ΔralA (i.e. ralfuranone‐deficient mutant) (Kai et al., 2014), ralA‐comp (i.e. native ralA‐expressing complemented ΔralA mutant) (Kai et al., 2014) and the phc QS‐deficient mutants, ΔphcB (Kai et al., 2015) and ΔphcA (Mori et al., 2016). The R. solanacearum strains were routinely grown in one‐quarter‐strength M63 medium (Cohen and Rickenberg, 1956) at 30 °C. Escherichia coli strains were grown in Luria–Bertani medium (Hanahan, 1983) at 37 °C. Gentamycin (50 μg/mL) was used in selective media.
Synthesis of ralfuranones
Ralfuranones A, B, J, K and L were synthesized and analysed as described previously (Kai et al., 2014).
Analysis of 3‐OH MAME produced by R. solanacearum strains
Ralstonia solanacearum cultures grown in B medium (Clough et al., 1994) at 30 °C for 4–6 h were diluted in fresh medium (until OD600 = 1.0). A 50‐µL aliquot of the cell suspension was transferred onto a Brilliant Green (BG) agar plate (diameter, 90 mm; capacity, 25 mL; Kai et al., 2015) and incubated for 24 h at 30 °C. The BG agar was then cut into small pieces and soaked twice in ethyl acetate (50 mL) for 2 h each. The combined extracts were dried over Na2SO4 and concentrated. The residue was dissolved in 2 mL of ethyl acetate and analysed using a gas chromatography–mass spectrometry system (Kai et al., 2015).
RNA extraction, elimination of ribosomal RNA and sequencing
Total RNA was isolated from R. solanacearum strains grown in one‐quarter‐strength M63 medium (until OD600 = 0.3) using a High Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany). Ribosomal RNA was removed from the extracted total RNA using a Ribo‐Zero rRNA Removal Kit (Gram‐negative bacteria) (Illumina, Madison, WI, USA). Oriented paired‐end RNA sequencing (2 × 100 bp) was conducted by Hokkaido System Science (Sapporo, Japan) using an Illumina Hiseq 2000 system and the procedures recommended by Illumina. The adaptors and primers were designed by Hokkaido System Science. The selected inserts were 100 bp. We conducted paired‐end sequencing of the libraries.
Mapping and analysis of RNA‐seq data
Reads were trimmed using Cutadapt (version 1.1; http://code.google.com/p/cutadapt/) and Trimmomatic (version 0.32; http://www.usadellab.org/cms/?page=trimmomatic), and then mapped with TopHat (version 2.0.10; http://tophat.cbcb.umd.edu/). Read counts obtained for each of the samples are presented as FPKM, which was calculated with Cufflinks (version 2.2.1; http://cole-trapnell-lab.github.io/cufflinks/).
qRT‐PCR assay
A 500‐ng total RNA template was reverse transcribed using a PrimeScript RT Reagent Kit (Takara, Otsu, Japan). A qRT‐PCR assay was conducted with a 20‐μL reaction mixture containing 1 μL cDNA stock and 10 pm primers (Table S4, see Supporting Information) using a SYBR GreenER qPCR Reagent System (Invitrogen, Tokyo, Japan). Reactions were completed in an Applied Biosystems 7300 Real‐time PCR system (Applied Biosystems, Foster City, CA, USA). The cycling parameters for all primers were as follows: 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 31 s. Melting curve runs were completed at the end of each reaction to verify the specificity of the primers (i.e. presence of a single product). The relative gene expression quantities were calculated using the comparative cycle threshold method. All values were normalized against the rpoD expression level (i.e. internal standard for each cDNA sample). There were no significant differences in the rpoD expression level between R. solanacearum strains.
Bacterial cell aggregation assay
The aggregation of R. solanacearum cells was measured in vitro using a slightly modified polyvinylchloride (PVC) microtitre plate assay (O'Toole and Kolter, 1998). Briefly, 5 μL of overnight cultures of R. solanacearum adjusted to OD600 = 0.005 were used to inoculate 95 μL of one‐quarter‐strength M63 medium in the wells of a PVC microtitre plate (Nunc MicroWell plate; Thermo Fisher Scientific Inc., Waltham, MA, USA). Tomato apoplast fluid was added to the wells, and the plate was incubated at 30 °C for 24 h without shaking. To quantify the cell aggregation, 25 μL of 1.0% (w/v) crystal violet solution was added to the wells. After a 15‐min incubation, the unbound crystal violet stain was gently removed with a pipette, and the wells were washed with distilled water, 70% ethanol and then distilled water. The remaining crystal violet in each well was solubilized with 100 μL of 100% ethanol, and then quantified by measurement of the absorbance at 550 nm. The resulting value was normalized according to the number of cells. This value was considered to represent the relative cell aggregation (OD550/OD600).
Measurement of EPS I production
Overnight cultures of R. solanacearum strains were washed with distilled water and then diluted to a cell density of 1.0 × 102 colony‐forming units (CFU)/mL. A 100‐μL aliquot of these cell suspensions was spread on one‐quarter‐strength M63 agar plates and incubated for 2 days at 30 °C. Cells were then resuspended to a concentration of 1.0 × 105 CFU/mL, and the cell density was confirmed through dilution plating. EPS was quantified using anti‐R. solanacearum EPS antibodies (Agdia Inc., Elkhart, IN, USA) in an enzyme‐linked immunosorbent assay (Agdia Inc.). The assay was conducted using a 100‐μL cell suspension (1.0 × 104 CFU). Three technical replicates were assessed. The production of EPS I was quantified on the basis of the absorbance at 650 nm.
Swimming motility
Overnight cultures of R. solanacearum strains were washed with distilled water and then diluted to a cell density of 5.0 × 105 CFU/mL. For the swimming assay, 5‐μL aliquots of cell suspensions were added to the centre of one‐quarter‐strength M63 medium solidified with 0.25% agar. Motility was examined using three plates per strain. All plates were incubated at 30 °C. The diameter of the swimming areas was measured at 48 h post‐inoculation.
Nucleotide sequence accession numbers
The nucleotide sequences of the genes analysed by qRT‐PCR were deposited in the DDBJ/GenBank/EMBL databases (Table S5, see Supporting Information).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Schematic diagram of the quorum sensing and conversion of ralfuranone I to various ralfuranones in Ralstonia solanacearum strain OE1‐1.
Fig. S2 Relationships between gene expression level changes in the phcB‐deleted (ΔphcB, a), phcA‐deleted (ΔphcA, b) and ralfuranone‐deficient (ΔralA) Ralstonia solanacearum mutants, and between expression level fold changes (i.e. ≥2 or ≤–2) of genes regulated by PhcB and PhcA in the ΔphcA and ΔralA mutants (c). The FPKM (fragments per kilobase of exon per million fragments mapped) values for R. solanacearum strains OE1‐1, ΔphcB, ΔphcA and ΔralA were normalized prior to the analyses of differentially expressed genes.
Fig. S3 Model of the regulation of the phc quorum sensing (QS) system mediated through ralfuranones in Ralstonia solanacearum strain OE1‐1.
Table S1 RNA‐sequencing data for all transcripts in Ralstonia solanacearum strain OE1‐1, phcB‐deleted mutant (ΔphcB), phcA‐deleted mutant (ΔphcA), and ralfuranones‐deficient mutant (ΔralA) grown in 1/4 × M63 medium.
Table S2 Predicted function of proteins encoded by genes whose expression is positively regulated by both the phc quorum sensing systems and ralfuranones in Ralstonia solanacearum strain OE1‐1 grown in one‐quarter‐strength M63 medium.
Table S3 Predicted function of proteins encoded by genes whose expression is negatively regulated by both the phc quorum sensing systems and ralfuranones in Ralstonia solanacearum strain OE1‐1 grown in one‐quarter‐strength M63 medium.
Table S4 Primers used in this study.
Table S5 Genes analysed by quantitative real‐time polymerase chain reaction.
Acknowledgements
This work was supported by Grants‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, a research grant from Sumitomo Chemical Co. and the Sasakawa Scientific Research Grant from The Japan Science Society to Y.M.
The authors declare that they have no conflicts of interest.
References
- Brumbley, S.M. and Denny, T.P. (1990) Cloning of wild‐type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J. Bacteriol. 172, 5677–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M. , Gallegos, M.T. , Studholme, D.J. , Guo, Y. and Gralla, J.D. (2000) The bacterial enhancer‐dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 182, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. , Schell, M.A. and Denny, T.P. (1994) Evidence for involvement of a volatile extracellular factor in Pseudomonas solanacearum virulence gene expression. Mol. Plant–Microbe. Interact. 7, 621–630. [Google Scholar]
- Clough, S.J. , Lee, K.E. , Schell, M.A. and Denny, T.P. (1997) A two‐component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA‐regulated virulence factors in response to 3‐hydroxypalmitic acid methyl ester. J. Bacteriol. 179, 3639–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, G.N. and Rickenberg, H.V. (1956) La galactoside‐perméase d'Escherichia coli. Ann . Inst. Pasteur (Paris), 91, 693–720. [PubMed] [Google Scholar]
- Dong, T.G. and Mekalanos, J.J. (2012) Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40, 7766–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavier, A.B. , Clough, S.J. , Schell, M.A. and Denny, T.P. (1997) Identification of 3‐hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum . Mol. Microbiol. 26, 251–259. [DOI] [PubMed] [Google Scholar]
- Garg, R.P. , Huang, J. , Yindeeyoungyeon, W. , Denny, T.P. and Schell, M.A. (2000) Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum . J. Bacteriol. 182, 6659–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin, S. and Denny, T.P. (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. [DOI] [PubMed] [Google Scholar]
- Ham, J.H. (2013) Intercellular and intracellular signalling systems that globally control the expression of virulence genes in plant pathogenic bacteria. Mol. Plant Pathol. 14, 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hao, B. , Mo, Z.L. , Xiao, P. , Pan, H.J. , Lan, X. and Li, G.Y. (2013) Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl. Microbiol. Biotechnol. 97, 2575–2585. [DOI] [PubMed] [Google Scholar]
- Huang, J. and Schell, M. (1995) Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol. Microbiol. 16, 977–989. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Yindeeyoungyeon, W. , Garg, R.P. , Denny, T.P. and Schell, M.A. (1998) Joint transcriptional control of xpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR‐type transcriptional activator. J. Bacteriol. 180, 2736–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai, K. , Ohnishi, H. , Mori, Y. , Kiba, A. , Ohnishi, K. and Hikichi, Y. (2014) Involvement of ralfuranone production in the virulence of Ralstonia solanacearum OE1‐1. ChemBioChem, 15, 2590–2597. [DOI] [PubMed] [Google Scholar]
- Kai, K. , Ohnishi, H. , Shimatani, M. , Ishikawa, S. , Mori, Y. , Kiba, A. , Ohnishi, K. , Tabuchi, M. and Hikichi, Y. (2015) Methyl 3‐hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum . ChemBioChem, 16, 2309–2318. [DOI] [PubMed] [Google Scholar]
- Kai, K. , Ohnishi, H. , Kiba, A. , Ohnishi, K. and Hikichi, Y. (2016) Studies on the biosynthesis of ralfuranones in Ralstonia solanacearum . Biosci. Biotechnol. Biochem. 80, 440–444. [DOI] [PubMed] [Google Scholar]
- Kanda, A. , Yasukohchi, M. , Ohnishi, K. , Kiba, A. , Okuno, T. and Hikichi, Y. (2003) Ectopic expression of Ralstonia solanacearum effector protein PopA early in invasion results in loss of virulence. Mol. Plant–Microbe Interact. 16, 447–455. [DOI] [PubMed] [Google Scholar]
- Kazmierczak, M.J. , Wiedmann, M. and Boor, K.J. (2005) Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F. , Babujee, L. , Jacobs, J.M. and Allen, C. (2015) Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum race 3 biovar 2. PloS One, 10, e0139090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, E. , Pultz, I.S. , Kulasekara, H.D. and Miller, S.I. (2011) The bacterial second messenger c‐di‐GMP: mechanisms of signalling. Cell Microbiol. 13, 1122–1129. [DOI] [PubMed] [Google Scholar]
- Mori, Y. , Inoue, K. , Ikeda, K. , Nakayashiki, H. , Higashimoto, C. , Ohnishi, K. , Kiba, A. and Hikichi, Y. (2016) The vascular plant‐pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 17, 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G.A. and Kolter, R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. [DOI] [PubMed] [Google Scholar]
- O'Toole, R. , Milton, D.L. , Horstedt, P. and Wolf‐Watz, H. (1997) RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water‐borne but intraperitoneal route of inoculation. Microbiology, 143, 3849–3859. [DOI] [PubMed] [Google Scholar]
- Pauly, J. , Spiteller, D. , Linz, J. , Jacobs, J. , Allen, C. , Nett, M. and Hoffmeister, D. (2013) Ralfuranone thioether production by the plant pathogen Ralstonia solanacearum . ChemBioChem, 14, 2169–2178. [DOI] [PubMed] [Google Scholar]
- Perrier, A. , Peyraud, R. , Rengel, D. , Barlet, X. , Lucasson, E. , Gouzy, J. , Peeters, N. , Genin, S. and Guidot, A. (2016) Enhanced in planta fitness through adaptive mutations in EfpR, a dual regulator of virulence and metabolic functions in the plant pathogen Ralstonia solanacearum . PLOS Pathog. 12, e1006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyraud, R. , Cottret, L. , Marmiesse, L. , Gouzy, J. and Genin, S.A. (2016) A resource allocation trade‐off between virulence and proliferation drives metabolic versatility in the plant pathogen Ralstonia solanacearum . PLOS Pathog. 12, e1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S.K. , Kumar, R. , Peeters, N. , Boucher, C. and Genin, S. (2015) rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum . Front. Microbiol. 6, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer, L. and Schneider, B.L. (2001) Metabolic context and possible physiological themes of sigma(54)‐dependent genes in Escherichia coli . Microbiol. Mol. Biol. Rev. 65, 422–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. and Dow, J.M. (2011) Communication with a growing family: diffusible signal factor (DSF) signaling in bacteria. Trends. Microbiol. 19, 145–152. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebaul, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 145, 497–502. [DOI] [PubMed] [Google Scholar]
- Schneider, P. , Jacobs, J.M. , Neres, J. , Aldrich, C.C. , Allen, C. , Nett, M. and Hoffmeister, D. (2009) The global virulence regulators VsrAD and PhcA control secondary metabolism in the plant pathogen Ralstonia solanacearum . ChemBioChem, 10, 2730–2732. [DOI] [PubMed] [Google Scholar]
- da Silva Neto, J.F. , Koide, T. , Abe, C.M. , Gomes, S.L. and Marques, M.V. (2008) Role of sigma 54 in the regulation of genes involved in type I and type IV pili biogenesis in Xylella fastidiosa . Arch. Microbiol. 189, 249–261. [DOI] [PubMed] [Google Scholar]
- Tang, J.L. , Liu, Y.N. , Barber, C.E. , Dow, J.M. , Wootton, J.C. and Daniels, M.J. (1991) Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris . Mol. Gen. Genet. 226, 409–417. [DOI] [PubMed] [Google Scholar]
- Tans‐Kersten, J. , Huang, H. and Allen, C. (2001) Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183, 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackler, B. , Schneider, P. , Jacobs, J.M. , Pauly, J. , Allen, C. , Nett, M. and Hoffmeister, D. (2011) Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem. Biol. 18, 354–360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Schematic diagram of the quorum sensing and conversion of ralfuranone I to various ralfuranones in Ralstonia solanacearum strain OE1‐1.
Fig. S2 Relationships between gene expression level changes in the phcB‐deleted (ΔphcB, a), phcA‐deleted (ΔphcA, b) and ralfuranone‐deficient (ΔralA) Ralstonia solanacearum mutants, and between expression level fold changes (i.e. ≥2 or ≤–2) of genes regulated by PhcB and PhcA in the ΔphcA and ΔralA mutants (c). The FPKM (fragments per kilobase of exon per million fragments mapped) values for R. solanacearum strains OE1‐1, ΔphcB, ΔphcA and ΔralA were normalized prior to the analyses of differentially expressed genes.
Fig. S3 Model of the regulation of the phc quorum sensing (QS) system mediated through ralfuranones in Ralstonia solanacearum strain OE1‐1.
Table S1 RNA‐sequencing data for all transcripts in Ralstonia solanacearum strain OE1‐1, phcB‐deleted mutant (ΔphcB), phcA‐deleted mutant (ΔphcA), and ralfuranones‐deficient mutant (ΔralA) grown in 1/4 × M63 medium.
Table S2 Predicted function of proteins encoded by genes whose expression is positively regulated by both the phc quorum sensing systems and ralfuranones in Ralstonia solanacearum strain OE1‐1 grown in one‐quarter‐strength M63 medium.
Table S3 Predicted function of proteins encoded by genes whose expression is negatively regulated by both the phc quorum sensing systems and ralfuranones in Ralstonia solanacearum strain OE1‐1 grown in one‐quarter‐strength M63 medium.
Table S4 Primers used in this study.
Table S5 Genes analysed by quantitative real‐time polymerase chain reaction.


