Summary
Rhizomania of sugar beet, caused by Beet necrotic yellow vein virus (BNYVV), is characterized by excessive lateral root (LR) formation leading to dramatic reduction of taproot weight and massive yield losses. LR formation represents a developmental process tightly controlled by auxin signaling through AUX/IAA‐ARF responsive module and LATERAL ORGAN BOUNDARIES DOMAIN (LBD) transcriptional network. Several LBD transcription factors play central roles in auxin‐regulated LR development and act upstream of EXPANSINS (EXPs), cell wall (CW)‐loosening proteins involved in plant development via disruption of the extracellular matrix for CW relaxation and expansion. Here, we present evidence that BNYVV hijacks these auxin‐regulated pathways resulting in formation LR and root hairs (RH). We identified an AUX/IAA protein (BvAUX28) as interacting with P25, a viral virulence factor. Mutational analysis indicated that P25 interacts with domains I and II of BvAUX28. Subcellular localization of co‐expressed P25 and BvAUX28 showed that P25 inhibits BvAUX28 nuclear localization. Moreover, root‐specific LBDs and EXPs were greatly upregulated during rhizomania development. Based on these data, we present a model in which BNYVV P25 protein mimics action of auxin by removing BvAUX28 transcriptional repressor, leading to activation of LBDs and EXPs. Thus, the evidence highlights two pathways operating in parallel and leading to uncontrolled formation of LRs and RHs, the main manifestation of the rhizomania syndrome.
Keywords: beet necrotic yellow vein virus, rhizomania, expansin, P25 virulence factor
Introduction
Plant viruses continue to pose a significant and constant threat to crop production affecting the yield and quality of harvested tissues. Crop losses to viruses compromise global food security and, in many cases, are attributed to virus‐reprogrammed plant morphogenesis and abnormal growth, manifested as viral disease symptoms. However, with a few exceptions, the molecular basis of symptom development in plants is still poorly understood (Jin et al., 2016; Lukhovitskaya et al., 2013; Shimura et al., 2011). Rhizomania of sugar beet, caused by Beet necrotic yellow vein virus (BNYVV), is characterized by the abnormal proliferation of lateral roots (LRs) leading to a significant decrease in taproot weight, sugar content and massive yield losses. Hence, BNYVV is able to alter the morphogenesis of the sugar beet crop to facilitate severe and dramatic changes in plant development manifested as excessive formation of LRs and root hairs (RHs).
LR emergence and formation represent a tightly controlled developmental process (Péret et al., 2009). The phytohormone auxin and several of its transport and signalling components play a crucial role during LR development (reviewed in Lavenus et al., 2013). When pumped into the cell by specialized transport machinery, the auxin signal triggers the degradation of AUXIN/INDOLE ACETIC ACID (AUX/IAA) proteins through binding to the SCFTIR1 multiprotein complex (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007). AUX/IAA proteins function as transcriptional repressors and their degradation allows interacting transcriptional activators, termed AUXIN RESPONSE FACTORs (ARFs), to alter auxin‐responsive gene expression (Dharmasiri and Estelle, 2004). In Arabidopsis thaliana, two transcriptional repressors, IAA14 and IAA28, have been demonstrated to play an essential role in LR formation (reviewed in Lavenus et al., 2013). Both IAA14 and IAA28 are capable of interacting with ARF7 and ARF19 transcription factors (TFs) (De Rybel et al., 2010; Fukaki et al., 2005). Moreover, the degradation of IAA14 allows ARF7 and ARF19 to activate the expression of auxin‐responsive genes, including LATERAL ORGAN BOUNDARIES DOMAIN (LBD) TFs. Three LBDs, LBD16, LBD18 and LBD29, play crucial and distinct roles in auxin‐regulated LR development (reviewed in Lavenus et al., 2013). LBD16 is involved in instigating the migration of nuclei and in the asymmetric division of LR founder cells to promote LR initiation (Goh et al., 2012). LBD18 plays a role in LR initiation and emergence (reviewed in Lavenus et al., 2013). LBD18 facilitates LR initiation through the transcriptional activation of a cell cycle regulator, E2Fa TF (Berckmans et al., 2011). LBD18 induces the expression of other cell cycle regulators, such as CYCLINB1;1 and CYCLIN‐DEPENDENT KINASE A1;1 (Lee et al., 2015). At later stages of LR emergence, LBD18 activates EXPANSIN A14 (EXPA14) expression by directly binding to the EXPA14 promoter, and indirectly up‐regulates EXPA17 and other EXPANSINs (EXPs) to mediate cell wall (CW) loosening and relaxation for cell growth and extension (Lee et al., 2013; Lee et al., 2015).
EXPs are non‐hydrolytic CW‐loosening proteins involved in the control of cell extension and are engaged in a variety of plant developmental processes, including leaf emergence, LR formation and RH elongation (reviewed in Cosgrove, 2015). In flowering plants, EXPs are encoded by a large multigene family comprising 29–88 members, and are phylogenetically classified into four groups, designated as EXPANSIN A (EXPA), EXPANSIN B (EXPB), EXPANSIN‐LIKE A (EXLA) and EXPANSIN‐LIKE B (EXLB) (Kende et al., 2004). The expression of most root‐specific EXPs is regulated by auxin and promoters of these genes often contain multiple auxin‐responsive factor binding elements (ARFEs) and/or conserved RH‐specific cis‐elements (RHEs) (Kim et al., 2006). Two RH‐specific A. thaliana genes, AtEXPA7 and AtEXPA18, and their functional equivalents in other species, contain RHE sequences in their promoters and are expressed specifically in RH cells immediately prior to RH initiation and elongation (Kim et al., 2006).
BNYVV is vectored in soil by zoospores of the obligate biotrophic parasite Polymyxa betae, a ubiquitous plasmodiophorid. Because BNYVV has a world‐wide geographical distribution, in sugar beet‐growing areas, the growth of BNYVV‐resistant sugar beet varieties is essential to maintain high yields. Modern varieties contain the Rz1 resistance gene (Biancardi et al., 2002), which does not provide complete resistance to infection. However, it delays the viral spread from infected LRs to the main taproot. Two different monogenic resistance genes, Rz2 and Rz3, have been identified in a sea beet: Beta vulgaris ssp. maritima (Gidner et al., 2005; Lewellen et al., 1987). Recently, the candidate gene for Rz2 has been identified by mapping‐by‐sequencing and appears to represent a typical plant R gene encoding a coiled‐coil nucleotide‐binding leucine‐rich repeat (CC‐NB‐LRR) protein (Capistrano‐Gossmann et al., 2017).
BNYVV has a multipartite genome consisting of four or five (depending on the isolate) positive‐sense, single‐stranded RNA segments (Bouzoubaa et al., 1986; Tamada et al., 1999). RNA1 and RNA2 encode genes of the ‘housekeeping module’ involved in virus replication, cell‐to‐cell movement, transmission and encapsidation, whereas the P14 gene encoded by RNA2, as well as genes encoded by RNA3, RNA4 and RNA5, represent the ‘interactive module’ comprising genes involved in virus–host interactions (Bouzoubaa et al., 1986; Tamada et al., 1999). The BNYVV RNA3‐encoded P25 protein is responsible for rhizomania symptom development in sugar beet (reviewed in Peltier et al., 2011). A short sequence of the P25 gene coding for four consecutive amino acid residues (amino acids 67–70)—the so‐called ‘tetrad’—shows great variability (Bornemann et al., 2015; Chiba et al., 2011; Schirmer et al., 2005). BNYVV strains with certain ‘tetrad’ variants harbouring specific mutations are able to overcome Rz1 and induce typical rhizomania symptoms (Acosta‐Leal et al., 1999; Koenig et al., 2009).
BNYVV RNA3 must be present in the inoculum to facilitate virus long‐distance movement in Beta species (Lauber et al., 1998). The RNA3‐encoded P25 protein is a virulence factor of the virus responsible for the ‘hairy root’ phenotype, when expressed on its own in transgenic A. thaliana (Peltier et al., 2011). At the subcellular level, the P25 protein localizes to the nucleus and to the cytoplasm (Vetter et al., 2004). The nuclear localization signal (NLS), zinc‐finger domain and nuclear export signal (NES) have been identified in P25 and characterized by mutagenesis (Vetter et al., 2004).
Previous studies in B. vulgaris and Arabidopsis on the identification of differentially expressed transcripts in response to infection or ectopic BNYVV P25 expression have suggested the importance of EXPs in rhizomania symptom development (Peltier et al., 2011; Schmidlin et al., 2008). Here, we show that, among 32 EXP genes of sugar beet, 13 were activated during rhizomania development. Moreover, three LBD TFs that act upstream of the root‐specific EXPs in model plant species, but downstream of auxin signalling and the AUX‐ARF7/19‐responsive module, were also found to be up‐regulated. Furthermore, we identified BvAUX28 as physically interacting with P25. Our results indicate that the activation of LBD TFs, and subsequently root‐specific EXPs, plays an integral role in shaping the development of the sugar beet root and suggests a link between the dysregulation of BvAUX28 and uncontrolled LR emergence. Taken together, these results support the notion that BNYVV P25 mimics the action of auxin in the induction of LRs and RHs, the phenotypic manifestations of rhizomania disease.
Results
Bioinformatics analysis identifies 32 EXP genes in the genome of B. vulgaris
Previous studies on the identification of differentially expressed transcripts in response to infection or expression of BNYVV P25 have suggested the importance of EXPs in rhizomania symptom development (Peltier et al., 2011; Schmidlin et al., 2011). As a result of unknown orthology relations, it was unclear which EXPs might be up‐regulated in B. vulgaris. Moreover, different EXPs may have a similar function (functional equivalents) in various plant species. The sugar beet EXP gene family is represented by many members. Despite the fact that the B. vulgaris genome was sequenced over 4 years ago (Dohm et al., 2014), to the best of our knowledge, there have been no systematic studies on the EXP gene family. Because of this gap, a genome‐wide analysis of B. vulgaris EXPs is required. To this end, using the Arabidopsis EXPs as queries, we identified a total of 32 putative B. vulgaris EXP gene sequences (Table S1, see Supporting Information).
To investigate the evolutionary relationships of B. vulgaris EXPs, we performed multiple sequence alignment and phylogenetic analysis. The results showed that the B. vulgaris genes were clearly divided into two large subfamilies, EXPA and EXPB, and two smaller subfamilies, EXLA and EXLB (Fig. 1a). This clustering confirmed previous classification of EXPs in Arabidopsis, rice and other plant species (Cosgrove, 2015). Notably, compared with other plant species, the EXLA subfamily is expanded in B. vulgaris to seven members (Fig. 1a,b). A previous analysis of the 26 EXPA gene sequences in Arabidopsis allowed their classification into a total of 12 clades (Sampedro and Cosgrove, 2005). To define orthologous groups, we performed multiple sequence alignment and phylogenetic analysis by comparing the 32 EXP gene sequences of sugar beet and the 36 EXP gene sequences of Arabidopsis. Most BvEXPA genes clustered together with the corresponding Arabidopsis genes into ten defined clades (Fig. 1d). Notably, two small clades—clades II and VIII—as well as clade VI were exclusively represented by Arabidopsis genes, of which AtEXPA14 and AtEXPA17 are involved in LR development (Lee and Kim, 2013). The lack of AtEXPA14 and AtEXPA17 orthologues in the sugar beet genome suggests that their function in LR development is taken over by other EXPs.
Figure 1.
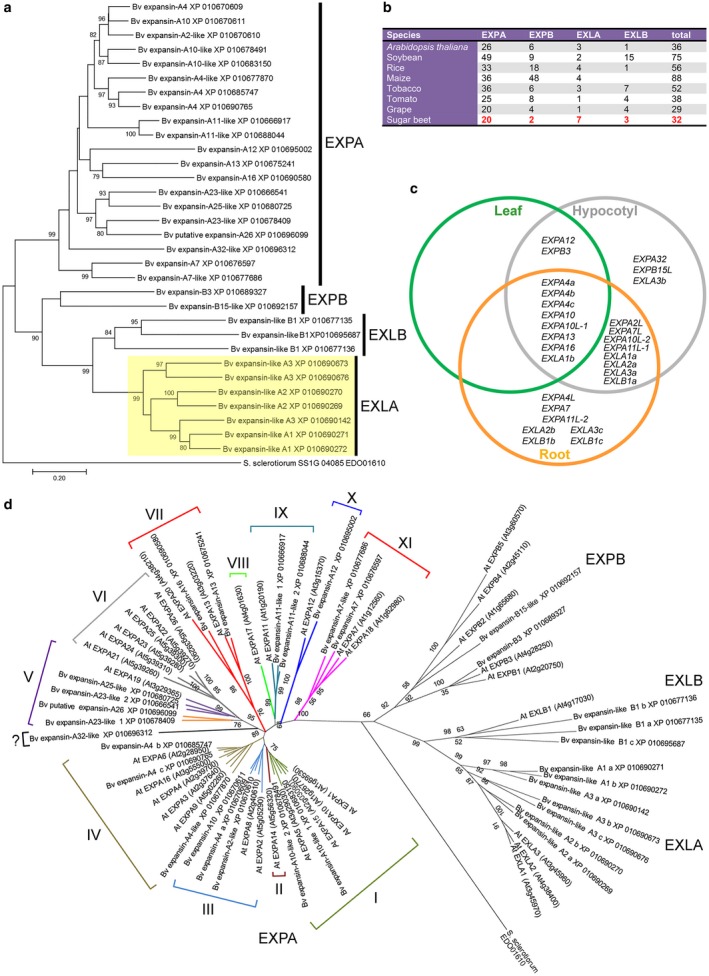
Expansin gene family in Beta vulgaris. (a) Phylogenetic analysis of the EXPANSIN (EXP) family in B. vulgaris. EXP of Sclerotinia sclerotiorum was used as outgroup. (b) Minimum gene numbers reported for the four EXP subfamilies of representative plant genomes compared with those in B. vulgaris. (c) Expression of EXP genes in root, hypocotyl and leaf of B. vulgaris. Data in the Venn diagram are compiled from our reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) results shown in their entirety in Table S2 (see Supporting Information). (d) A phylogenetic tree of the EXP family, including protein sequences from Arabidopsis thaliana and B. vulgaris. This tree does not correctly resolve clade EXPA‐V, possibly because of changes in amino acid usage between Arabidopsis and sugar beet EXPs. The clades of the EXPA subfamily are numbered according to a previous classification in Sampedro and Cosgrove (2005).
To characterize the pattern of EXP gene expression, roots and hypocotyl samples were collected from 3‐week‐old healthy seedlings and from young fully expanded leaves of 4‐week‐old healthy B. vulgaris plants. RNA was extracted and the abundance of individual EXP transcripts was assayed by reverse transcription‐quantitative polymerase chain reactions (RT‐qPCRs) (Table S2, see Supporting Information). The RT‐qPCR data indicated that a total of 23 EXP genes are expressed to some extent in B. vulgaris root tissues (Fig. 1c).
Identification of EXP genes induced in response to rhizomania development
To identify EXP genes and investigate the abundance change of their transcript levels in response to rhizomania development, seedlings of a BNYVV‐susceptible sugar beet variety were grown in different soils infested with BNYVV and root samples were collected for RNA extraction from the infected and mock‐inoculated plants at two time points: 4 and 6 weeks post‐sowing (wps). The soils used in these experiments were collected from three locations in Europe [Germany, Sweden and the Netherlands (Holland)], and were characterized previously as containing the B‐strain of BNYVV (Germany and Sweden) and the A‐strain of BNYVV (Holland) (Koenig and Lennefors, 2000; Lennefors et al., 2008). The presence of BNYVV and P. betae in the sampled roots of the plants grown in infested soils was confirmed by RT‐PCR and sequencing (Fig. S1a,b,d, see Supporting Information).
To determine which EXPs showed altered expression in response to rhizomania development, RT‐qPCRs were conducted for 26 EXP genes using total RNAs of mock‐inoculated and virus‐infected plants. The mRNA levels of 13 EXPs were significantly higher (Student’s two‐tailed t‐test, P < 0.05) in virus‐infected relative to mock‐inoculated plants at 4 wps (Fig. S2a, Table S3, see Supporting Information). A similar set of EXPs was up‐regulated at 6 wps, but the levels of induction were, depending on the gene, either higher or lower compared with the 4‐wps time point, and mRNA levels for three EXPs (BvEXPA7L, BvEXPA13 and BvEXLA2a) were lower than those in mock‐inoculated plants. Collectively, these data suggest highly dynamic changes in EXP expression during rhizomania development.
Three LBD TFs are up‐regulated by natural soil‐mediated infection with BNYVV
The expression of at least three expansins (AtEXPA4, AtEXPA14 and AtEXPA17), which mediate the loosening and remodelling of CW during LR development in Arabidopsis, is induced directly or indirectly by AtLBD16, AtLBD18 and AtLBD29. To determine whether natural soil‐mediated infection with BNYVV also affects BvLBD16, BvLBD18 and BvLBD29 expression, RT‐qPCRs were conducted. In infected plants displaying typical rhizomania symptoms, the mRNA levels of BvLBD16, BvLBD18 and BvLBD29 were significantly higher than those in the roots of healthy plants, as measured at two time points (Fig. S2b). Similar to previous observations with EXPs, the induction of the LBD TFs during rhizomania disease development was highly dynamic with mRNA expression levels noticeably higher at the early stage of virus infection (4 wps) relative to a later sampling date (6 wps) (Fig. S2b). These results indicate that the expression of LBD TFs, presumably involved in LR formation in B. vulgaris (Table S4, see Supporting Information), is indeed up‐regulated by soil‐mediated infection.
Identification of EXPs and LBD TFs specifically involved in rhizomania development through the use of Rz1 and Rz2 resistant varieties, benyvirus infectious clones and virus reassortants
We identified 13 EXPs and three LBDs as being activated in response to rhizomania development by growing plants in BNYVV‐infested soils. In principle, some of these EXPs/LBDs may be induced by P. betae and/or by other unidentified pathogens present in the soil, and thus may not directly contribute to rhizomania development. To resolve this issue, we took advantage of the availability of two sugar beet varieties carrying the resistance genes Rz1 or Rz2. Although resistant varieties are infected with BNYVV, they do not develop the disease (Fig. 2a). Therefore, we reasoned that the genes which are induced in susceptible, but not resistant, varieties could be directly linked to rhizomania development.
Figure 2.
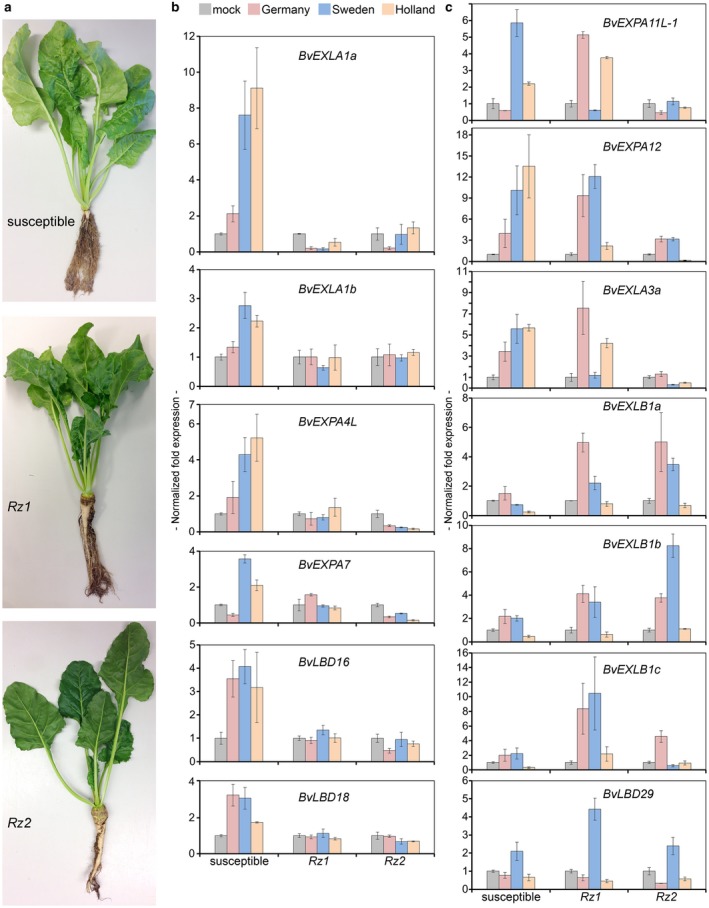
Four EXPANSIN (EXPs) and two LATERAL ORGAN BOUNDARIES DOMAIN (LBDs) genes are activated in Beet necrotic yellow vein virus (BNYVV)‐susceptible, but not BNYVV‐resistant, sugar beet varieties, whereas BvLBD29 and six EXPs are up‐regulated in both susceptible and resistant varieties, by natural soil‐mediated infection with BNYVV. (a) Phenotypes of three different varieties (BNYVV‐susceptible, as well as varieties harbouring Rz1 or Rz2 resistance genes) grown in soil infested with BNYVV for 6 weeks. (b) Expression of BvEXLA1a, BvEXLA1b, BvEXPA4L, BvEXPA7, BvLBD16 and BvLBD18 in sugar beet BNYVV‐susceptible vs. BNYVV‐resistant varieties harbouring Rz1 or Rz2 resistance genes at 6 weeks post sowing (wps). (c) Expression of six EXPs and BvLBD29 induced by BNYVV in both susceptible and resistant varieties at 6 wps. Bars represent means ± standard deviation (n ≥ 4).
To compare the induction of LBDs and EXPs in susceptible vs. resistant varieties, RT‐qPCRs were conducted using total RNAs from roots sampled at 6 wps. BvLBD16, BvLBD18, BvEXP4L, BvEXPA7, BvEXLA1a and BvEXLA1b were significantly (Student’s two‐tailed t‐test, P < 0.05) up‐regulated in susceptible (Fig. 2b), but not in resistant, varieties (Student’s two‐tailed t‐test, P > 0.05; Fig. 2b), suggesting that the activation of these genes contributes to rhizomania development. There was not much change detected in the expression of BvEXPA7L, BvEXPA13 and BvEXLA2a (Fig. S3, see Supporting Information). On the other hand, six EXPs and BvLBD29 were activated in both susceptible and resistant varieties, although to a different extent depending on the type of infested soil (Fig. 2c), suggesting that the expression of these genes does not directly contribute to rhizomania symptoms, but might synergistically enhance the severity of the disease.
To determine whether the presence of BNYVV RNA3, which encodes the P25 virulence factor, is required for the activation of EXP and LBD genes (identified through the use of Rz1 and Rz2 resistant varieties as being important for rhizomania development), we performed infection experiments with recombinant viruses reassorted for RNA3 genomic segments. To this end, we took advantage of the availability of the infectious cDNA clones for BNYVV and Beet soil‐borne mosaic virus (BSBMV) (Laufer et al., 2018). As for BNYVV, BSBMV belongs to the genus Benyvirus, has a genome organization similar to BNYVV and infects sugar beet, but does not cause the ‘hairy root’ phenotype typical of rhizomania (Workneh et al., 2003). Notably, the BSBMV RNA3‐encoded orthologue of P25, the P29 protein, has a very low similarity to P25, namely 23% identity (Lee et al., 2001). Moreover, genomic components can be exchanged between the viruses, as they can be trans‐replicated (Ratti et al., 2009). To inoculate the roots of sugar beet seedlings, four different inocula were assembled, each comprising RNA1 + RNA2 of either BNYVV or BSBMV, and RNA3 genomic components reassorted between the two viruses (Fig. 3a). The RT‐qPCR analysis performed on total RNA isolated at 32 days post‐inoculation (dpi) clearly showed that BvLBD16, BvEXPA4L and BvEXPA7 were up‐regulated by the presence of BNYVV RNA3, but not by the presence of BSBMV RNA3 (Fig. 3b). Interestingly, we did not observe changes in the level of BvLBD18, BvEXLA1a and BvEXLA1b transcript accumulation, suggesting that, compared with the experiments involving Rz1 and Rz2 resistant varieties, the expression of these genes was not induced at this time point.
Figure 3.
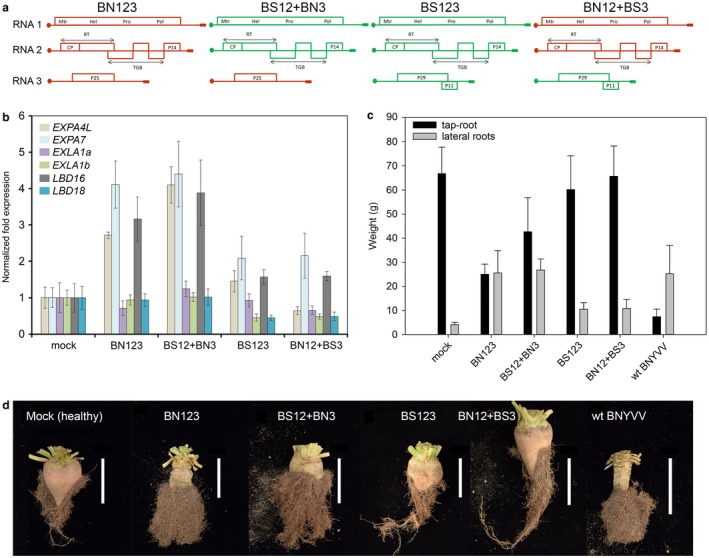
Differences in the abundance of EXPANSIN (EXPs) and LATERAL ORGAN BOUNDARIES DOMAIN (LBDs) gene transcripts, and rhizomania development, in Beta vulgaris roots on infection with Beet necrotic yellow vein virus (BNYVV), Beet soil‐borne mosaic virus (BSBMV) and interspecies RNA3 reassortants. (a) Schematic representation of the inocula, consisting of BNYVV and BSBMV RNA1, RNA2 and RNA3 genomic segments (BN123 and BS123, respectively) and reassortants for the RNA3 segment (BS12+BN3 and BN12+BS3, respectively), used in the virus infection experiments. (b) Expression of EXPs and LBDs in roots of B. vulgaris infected with BNYVV (BN123), BSBMV (BS123) and reassortants for the RNA3 segment (BS12+BN3 and BN12+BS3, respectively) at 32 days post‐inoculation (dpi). Bars represent means ± standard deviation (n ≥ 6). (c) Fresh weight of B. vulgaris lateral roots (LRs) infected with BNYVV (BN123), BSBMV (BS123) and reassortants for the RNA3 segment at 111 dpi. (d) Symptom appearance in roots of B. vulgaris infected with BNYVV (BN123), BSBMV (BS123) and reassortants for the RNA3 segment. Images were taken at 111 dpi. Bars, 5 cm.
In another experiment, we evaluated the symptom appearance and fresh weight of LRs infected with reassortant viruses following a growth period that allows the formation of taproot and rhizomania symptoms (111 dpi). Consistent with the RT‐qPCR data, the occurrence of symptoms and the increase in fresh weight of LRs correlated with the presence of BNYVV RNA3 in the inoculum (Figs 3c,d and S1c).
LBDs and EXPs can be induced by auxin treatment
As the expression of LBD16/18/29 TFs and some EXPs in Arabidopsis is controlled through auxin signalling, we asked whether their orthologues in sugar beet could be induced by auxin. To address this question, roots of 3‐week‐old B. vulgaris seedlings were treated with auxin (indole‐3‐acetic acid, 3‐IAA) at low non‐toxic concentrations of 10 µm for 2 h. The RT‐qPCRs of RNA samples isolated from the roots challenged with 3‐IAA revealed significant up‐regulation of BvLBD16, BvLBD18 and BvLBD29, with BvLBD29 being up‐regulated up to 16‐fold relative to mock‐treated samples (Fig. 4). Moreover, two of ten EXPs tested in these experiments—BvEXLA1a and BvEXPA4L—were also slightly up‐regulated—up to 2.5‐fold (Student’s one‐tailed t‐test, P < 0.05). Thus, these experiments linked auxin signalling to the induction of LBDs and EXPs in sugar beet and prompted us to analyse auxin signalling components acting upstream of LBD16/18/29.
Figure 4.
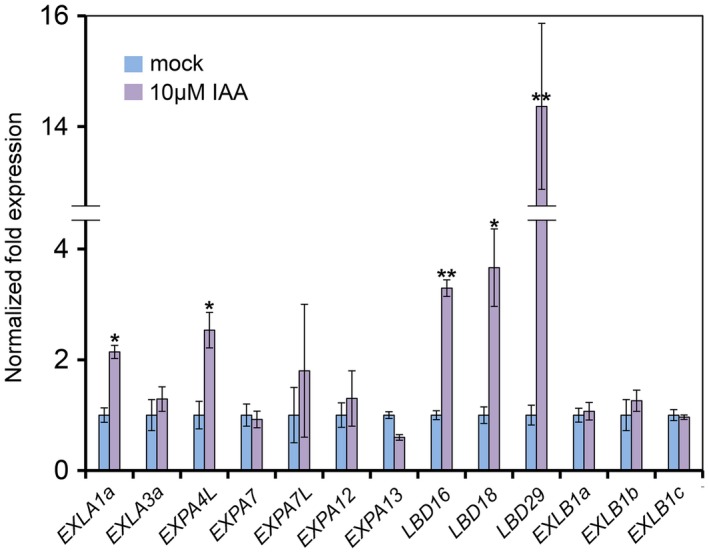
Auxin modulates the expression of EXPANSIN (EXPs) and LATERAL ORGAN BOUNDARIES DOMAIN (LBDs) genes in the roots of Beta vulgaris. Roots of B. vulgaris seedlings were treated with auxin and samples were collected 2 h after treatment with indole‐3‐acetic acid (3‐IAA) or 10% ethanol (mock). Bars represent means ± standard deviation (n = 3). *P < 0.05, Student’s one‐tailed t‐test; **P < 0.05, Student’s two‐tailed t‐test.
B. vulgaris AUX‐ARF7/19‐responsive module genes are not transcriptionally activated by rhizomania development
To determine whether the genes acting upstream of LBDs and EXPs, but downstream of auxin signalling, are activated by natural soil‐mediated infection with BNYVV, total RNA isolated at 6 wps was subjected to RT‐qPCR analysis. RT‐qPCR results clearly demonstrated that the mRNA levels of BvIAA14, BvARF7 and BvARF19 either did not show a significant alteration in expression or were slightly down‐regulated in comparison with control mock‐inoculated plants (Fig. S4, see Supporting Information). These findings are consistent with the notion that the mode of regulation of the AUX‐ARF7/19 module occurs through protein–protein interactions and protein stability/turnover.
P25 interacts with BvAUX28 and the interaction involves BvAUX28 domains I and II
The finding that several EXPs and LBDs belonging to the LR and RH developmental pathways (in Arabidopsis) are up‐regulated by rhizomania, and, moreover, that a similar set of genes is induced by auxin treatment of sugar beet roots, raises the possibility that LBDs and EXPs may be induced downstream of auxin signalling via the AUX‐ARF7/19‐responsive module. Interestingly, a previous study, employing yeast two‐hybrid (Y2H) screens, identified a root‐specific transcriptional repressor BvAUX28 as an interacting partner of P25 (Thiel and Varrelmann, 2009), although the biological role of this interaction has not been addressed. To confirm the interactions between BvAUX28 and P25, we carried out three independent assays. We cloned BvAUX28 full‐length cDNA into a prey plasmid and validated its interactions with P25 in yeast (Fig. 5a,b).
Figure 5.
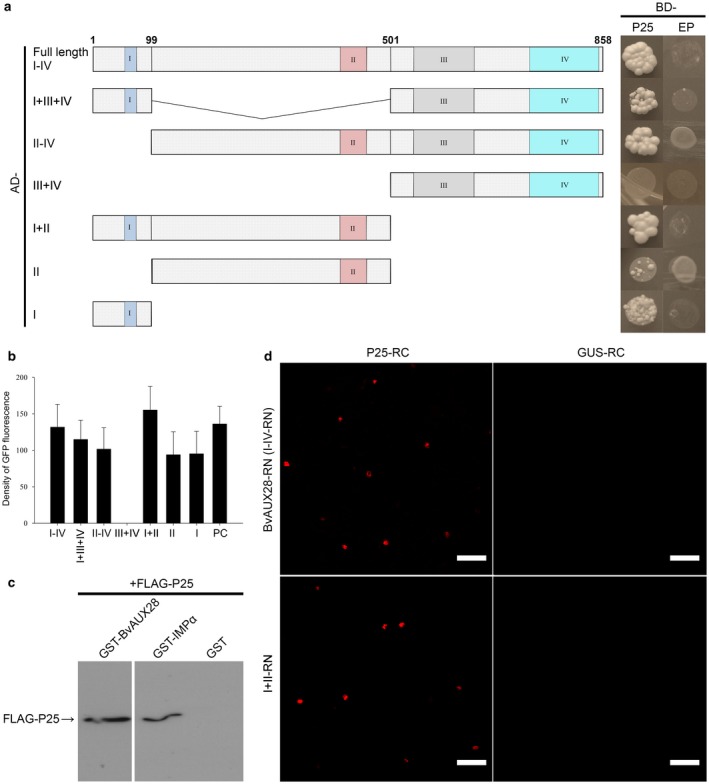
The Beet necrotic yellow vein virus (BNYVV) P25 protein interacts with BvAUX28. (a) Evaluation of P25 interaction with BvAUX28 in yeast and identification of the interacting domain of BvAUX28. Left: schematic diagram showing the BvAUX28 domains; conserved domains are represented by coloured rectangles. Right: yeast transformants were spotted onto a high‐stringency synthetic dropout (SD/‐Trp/‐Leu/‐His/‐Ade) medium. Empty BD vector/plasmid (EP) co‐transformed with corresponding AD constructs tested for transcriptional autoactivation (negative control). AD, activation domain. BD, binding domain. (b) Relative quantification of leucine‐driven green fluorescent protein (GFP) (mean total area density) in diploid yeast expressing the combination of P25 and BvAUX28 or its truncated derivatives as shown in (a). Bars represent means ± standard error of the mean. (c) In vitro pull‐down assay to confirm P25 interaction with BvAUX28. FLAG epitope‐tagged P25 was incubated with glutathione‐S‐transferase (GST)‐tagged BvAUX28, GST‐tagged OsIMPα (importin α from rice) or with GST tag. GST pull‐down products were visualized by immunoblotting with anti‐P25 antiserum. (d) Bimolecular fluorescence complementation (BiFC) conformation of the BNYVV P25 and BvAUX28 interaction. P25‐RC and BvAUX28‐RN (or I+II‐RN) were co‐expressed in Nicotiana benthamiana leaf epidermal cells via agroinfiltration. Co‐expression of GUS‐RC and BvAUX28‐RN or I+II‐RN was used as negative control. Images were taken at 4 days post‐inoculation (dpi). Scale bars, 50 μm.
To identify which domain of BvAUX28 is required for the interaction with P25, six constructs expressing various sets of the domains were tested by Y2H assays. The interactions were detected with constructs expressing domains I, II, I+II, II–IV, I+III+IV, as well as full‐length BvAUX28 (domains I–IV), whereas a derivative expressing domains III+IV did not show interaction (Fig. 5a). The strength of the P25–BvAUX28 interaction was evaluated by an indirect method involving the quantification of the leucine‐driven expression of plasmid‐encoded green fluorescent protein (GFP) in yeast (Fig. 5b). When expressed on their own, domains I and II, as well as the II–IV construct, showed weak interaction activity relative to the construct containing both domains I and II (Fig. 5b). Together, these results indicate that the interaction between BvAUX28 and P25 is contributed by domains I and II.
To verify the BvAUX28–P25 interaction, we expressed a recombinant FLAG‐tagged P25 protein from a Baculovirus vector in Sf9 insect cells and glutathione‐S‐transferase (GST)‐tagged BvAUX28 or GST (as a negative control) proteins in Escherichia coli, and performed in vitro pull‐down assays. As shown in Fig. 5c, FLAG‐P25 was detected in GST‐BvAUX28, but not in GST pull‐down complexes.
To further verify the BvAUX28–P25 interaction in vivo, we performed bimolecular fluorescence complementation (BiFC) assay. First, we examined the subcellular localization of BvAUX28 by transiently co‐expressing it as a GFP‐tagged fusion together with the DsRed‐tagged NLS of Simian virus 40 (SV40) T‐antigen in Nicotiana benthamiana using Agrobacterium tumefaciens. As expected, this experiment showed that GFP‐BvAUX28 localized exclusively to the nucleus (Fig. S5, see Supporting Information). Next, we verified the interaction between BvAUX28 and P25 in planta using BiFC. P25‐RC and BvAUX28‐RN (or I+II‐RN) were transiently co‐expressed in N. benthamiana leaf epidermal cells and the positive signals of red fluorescent protein (RFP) were detected in the nucleus (Fig. 5d). We did not find any RFP signal in the nucleus when we co‐expressed the BvAUX28‐RN or I+II‐RN fusions together with GUS‐RC (Fig. 5d). Taken together, these experiments demonstrate that P25 physically interacts with BvAUX28 in vitro and in vivo.
P25 inhibits BvAUX28 nuclear localization
We then investigated whether P25 affects the nuclear accumulation of BvAUX28. As expected, the GFP‐fused P25 localized to the nucleus and cytoplasm (Fig. 6a). Consistent with its role in the regulation of transcription, the RFP‐fused BvAUX28 was exclusively localized to the nucleus in 100% of examined cells (Fig. 6a). To examine the effect of the P25–BvAUX28 interaction on the subcellular localization of BvAUX28, GFP‐P25 and RFP‐BvAUX28 were transiently co‐expressed in N. benthamiana. Surprisingly, RFP‐BvAUX28 fluorescence, besides localizing in the nucleus, accumulated in the cytoplasm (Fig. 6b). In general, RFP‐BvAUX28 fluorescence co‐localized with the green fluorescence of GFP‐P25 by being detected in the nucleus and cytoplasm (Fig. 6b). Together, these results demonstrate that P25 disrupts the function of BvAUX28 in nuclear accumulation.
Figure 6.
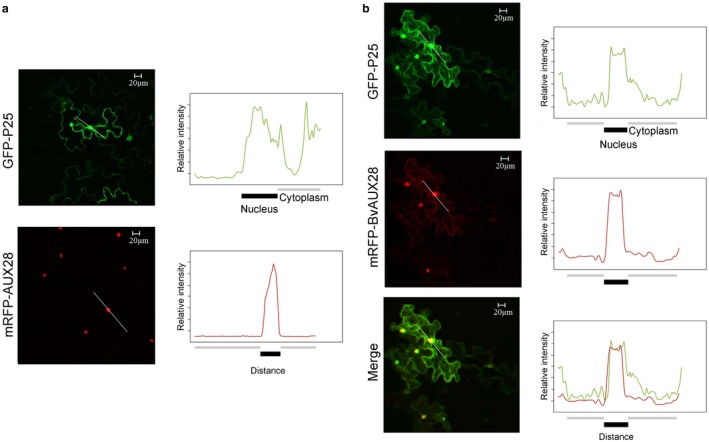
Beet necrotic yellow vein virus (BNYVV) P25 inhibits BvAUX28 nuclear localization. (a) Intracellular localization of the BNYVV P25 protein and BvAUX28. BNYVV P25‐GFP and mRFP‐BvAUX28 were transiently expressed in Nicotiana benthamiana. (b) Co‐expression of BNYVV P25 and BvAUX28. (a, b) Densitometry scans were performed along the lines indicated in white. Different colours indicate P25 (green) and BvAUX28 (red) intensities. Bars below the densitometry scans indicate nucleus (black) and cytoplasm including plasma membrane (grey). GFP, green fluorescent protein; RFP, red fluorescent protein. Scale bars, 20 μm.
Discussion
BNYVV continues to pose a significant threat to sugar beet production, as exemplified by the emergence of Rz1 resistance‐breaking isolates in different areas of the world (Bornemann et al., 2015; Koenig et al., 2008; Liu and Lewellen, 2007; Liu et al., 2005; Pferdmenges et al., 2009). Therefore, there is a growing need to understand the molecular basis of rhizomania disease and to identify the pathways of the host involved in order to develop sustainable solutions for disease management. Although progress is being made in understanding the molecular details of plant–virus interactions in different pathosystems, our knowledge about how viruses interfere with plant development and growth, culminating in diseases, is still insufficient to provide genetic resources for resistance‐based breeding in various crops and to develop sustainable methods to control the viruses. Recent advances in the genome sequencing of sugar beet, combined with progress made in understanding the mechanisms and genetic basis of LR emergence and RH induction in the model plant Arabidopsis, prompted us to analyse similar pathways operating in the sugar beet crop and their potential contribution to the development of rhizomania disease.
Previous studies examining the transcriptome changes in response to BNYVV in sugar beet (Schmidlin et al., 2008) or transgenic expression of P25 in Arabidopsis (Peltier et al., 2011) have identified several differentially expressed EXP genes, including AtEXPA14 and AtEXPA17, as well as AtEXPA7 and AtEXPA18, which were later shown to be involved in LR development and RH elongation in Arabidopsis, respectively (Lee et al., 2015). However, it remained unclear which of the functional equivalents of Arabidopsis EXPs are up‐regulated in sugar beet plants, because different EXPs may play similar roles and converge on similar pathways in various plant species. Therefore, we sought to characterize the EXP gene family in B. vulgaris, as well as changes in EXP expression during rhizomania development. These analyses highlighted at least two virus‐reprogrammed pathways, operating in parallel and leading to the development of typical symptoms of rhizomania, namely excessive formation of LRs and RHs.
Among the factors studied during rhizomania development, EXPs are primarily known for their role in plant CW remodelling by breaking the hydrogen bonds between cellohexaose and pectin; they thus participate in various aspects of plant development involving cell division and expansion (Cosgrove, 2015), and as potential pro‐viral factors (Chen et al., 2018; Park et al., 2017). The data accumulated from studies in various plant pathosystems, including nematodes, fungi and viruses, suggest that the down‐regulation of EXPs may represent a defence response (Marowa et al., 2016). In Arabidopsis, the down‐regulation or knockdown of AtEXLA2 enhances resistance to the necrotrophic fungus Alternaria brassicicola (Abuqamar et al., 2013). The knockdown of NbEXPA1 in N. benthamiana induces resistance to Turnip mosaic virus (TuMV) (Park et al., 2017). Furthermore, it has been shown that NbEXPA1 is a plasmodesmata‐specific protein recruited by TuMV to the virus replication complex through its interaction with the viral RNA‐dependent RNA polymerase, the NIb protein, and transient expression of NbEXPA1 promotes TuMV replication and cell‐to‐cell movement (Park et al., 2017). Similarly, the knockdown of NbEXPA4 expression reduces Tobacco mosaic virus (TMV) accumulation in systemically infected plants, whereas the overexpression of NbEXPA4 accelerates virus replication (Chen et al., 2018). The pro‐viral role of EXPs is further supported (this study) by the up‐regulation of 13 EXP genes by natural soil‐mediated infection with BNYVV, among which four root‐specific EXPs are up‐regulated in susceptible, but not resistant, varieties, linking the activation of these EXPs to rhizomania development.
By integrating RT‐qPCR data with the results of Y2H screening into a broader picture, we were able to obtain a model for rhizomania development in sugar beet, which revealed a prominent role of LBD TFs and EXPs in shaping the abnormal growth of the sugar beet taproot. By comparing the expression signatures of LBDs and EXPs in susceptible and resistant varieties of sugar beet, as well as in B. vulgaris roots infected with virus reassortants, for a genomic component encoding the virulence factor of the virus, we identified two LBD TFs and four EXPs as putative downstream targets, the ectopic expression of which contributes to rhizomania development. Indeed, there was a strong correlation between BvLBD16, BvLBD18, BvEXPA7, BvEXPA4L, BvEXLA1a and BvEXLA1b expression in susceptible varieties and the development of typical rhizomania symptoms. Furthermore, the pro‐rhizomania LBDs and EXPs were either unchanged or down‐regulated in Rz1 and Rz2 resistant varieties. These findings provide new insights into the pathways that lead to rhizomania. For example, the activation of BvEXPA7 and BvEXPA4L, both of which have RHE signature motives in their promoters (data not shown) and seem to be involved in RH initiation and elongation, on the one hand, and the up‐regulation of BvLBD16, BvLBD18, BvEXLA1a and BvEXLA1b, presumably involved in LR development, on the other, suggests that two pathways operate in parallel: one leading to LR emergence and the other leading to RH induction and elongation. Although the question of the increase in root hair number on rhizomania development has not yet been addressed properly, our data support this hypothesis. It also remains to be determined whether there is a cross‐talk between these pathways. Intriguingly, some of these genes are induced by the auxin treatment of sugar beet roots, indicating that both pathways are regulated by auxin signalling. Indeed, it has been shown that LR emergence and RH development are auxin‐dependent developmental processes (Lee et al., 2015).
Previous studies have reported that plant viruses can hijack the auxin signalling pathway to promote virus infection and virus movement in the host plants (Collum et al., 2016; Jin et al., 2016). The capsid protein of Rice dwarf virus (RDV) interferes with auxin signalling by binding to the rice AUX/IAA protein OsIAA10, thereby protecting it from degradation by the 26S proteasome and resulting in typical RDV symptoms in infected plants (Jin et al., 2016). The symptoms include dwarfism, excessive tillering and stunted crown roots, and are phenocopied in transgenic rice overexpressing OsIAA10 or its degradation‐resistant derivative (Jin et al., 2016). The TMV replicase physically interacts with three vascular‐expressed AUX/IAA proteins, including IAA26 (Padmanabhan et al., 2006). These transcriptional repressors have been suggested to regulate genes involved in phloem loading. TMV infection inhibits the nuclear localization of AUX/IAA proteins, resulting in the transcriptional reprogramming of mature vascular tissue, which is correlated with enhanced TMV spread in the phloem of older leaves (Collum et al., 2016). Taken together, these data are consistent with our findings showing that BNYVV P25 interacts with BvAUX28, presumably inactivating its function as a transcriptional repressor through the disruption of its nuclear localization. Moreover, hormonal content assays measuring levels of auxin in BNYVV‐infected sugar beet plants, as well as in Arabidopsis plants transgenic for P25, have demonstrated significantly elevated levels of auxin relative to those in healthy plants, suggesting a positive feedback loop, presumably leading to further acceleration of LR/RH formation (Peltier et al., 2011; Pollini et al., 1990).
Therefore, here, we propose a regulatory model for rhizomania development integrating our present data in sugar beet and previous reports on LR and RH development in Arabidopsis (Fig. 7). In healthy plants, LR and RH formation is tightly regulated. Auxin activates the signalling pathways, inducing LR founder cell priming, through the degradation of root‐specific AUX/IAA transcriptional repressors (Fig. 7; left panel). Another auxin‐dependent pathway also controls RH formation. In BNYVV‐infected plants, the P25 virulence factor physically interacts with BvAUX28 and disrupts its function in nuclear accumulation, perhaps as a result of nuclear to cytoplasmic shuttling of P25, thus preventing BvAUX28 interaction with ARFs. Hence, ARFs are derepressed and activate the expression of LBDs, promoting further LR development. In turn, LBDs activate the expression of EXPs for CW loosening and remodelling, facilitating cell separation for LR formation. Another pathway must be operating in parallel and leads to the up‐regulation of BvEXPA4L and BvEXPA7, presumably needed for RH formation and elongation.
Figure 7.
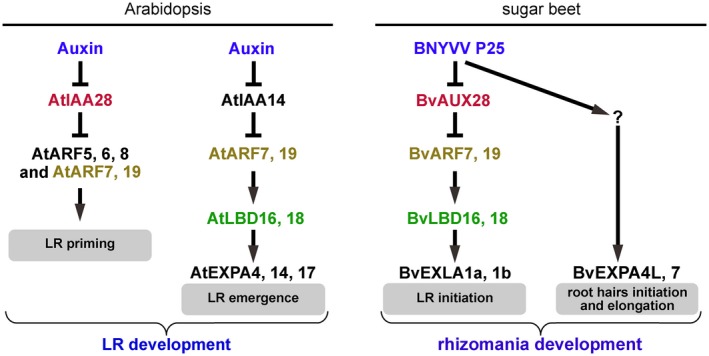
Proposed regulatory model for rhizomania development. In healthy Arabidopsis plants, auxin controls lateral root (LR) development through multiple auxin signalling modules. Two modules involved in LR development are shown in the left panel (modified from Lavenus et al., 2013 and Lee et al., 2015). LR founder cell priming involves the AtIAA28‐AtARF5, 6, 7, 8, 19 module. LR emergence is controlled by the AtIAA14‐AtARF7, 19 module. AtLBD16, 18 regulate LR emergence downstream of AtARF7, 19. In sugar beet, following Beet necrotic yellow vein virus (BNYVV) infection, the P25 virulence factor is produced. P25 inactivates BvAUX28 transcriptional repressor leading to the derepression of BvARF7, 19 transcriptional activators. Subsequently, the expression of a number of their target genes is turned on, including BvLBD16 and BvLBD18 transcription factors (TFs) involved in LR formation. BvLBD16, 18 activate the expression of two EXLA1 genes to facilitate cell wall (CW) relaxation and remodelling during LR initiation and emergence. The up‐regulation of the positive regulators of LR formation, BvLBD16 and BvLBD18, as well as EXPANSIN genes (EXPs), leads to the uncontrolled formation of LR, giving a ‘hairy root’ appearance of rhizomania‐afflicted sugar beet taproot. Meanwhile, another pathway is turned on leading to the activation of root hair (RH)‐specific EXPs, BvEXPA4L and BvEXPA7, to promote RH initiation and elongation. The formation of LRs and RHs is beneficial for the propagation of Polymyxa betae, the formation of zoospores and, ultimately, the efficient transmission of BNYVV. Auxin and P25 are shown in blue. The proteins encoded by homologous genes in Arabidopsis and sugar beet are shown by similar colours.
We argue that the formation of LRs and RHs is beneficial for P. betae, the vector of BNYVV. The life cycle of P. betae and other plasmodiophorids starts with a primary zoospore that attaches to the wall of an RH often in the zone of elongation (Ahm and Buchenauer, 1999; Barr and Asher, 1999). Moreover, during the vegetation period, sporosori of plasmodiophorids could be observed in RHs of the infected plants, suggesting that RHs are the main sites of plasmodiophorid propagation. Hence, P. betae propagation and the formation of sporosori are beneficial for BNYVV transmission to increase the number of zoospores and the probability of infecting further neighbouring plants.
To conclude, our study further extends the range of plant developmental processes affected by viruses through interference with phytohormone signalling. Our work identifies at least two pathways for the development of rhizomania and paves the way for follow‐up studies, e.g. on the detailed characterization of the molecular mechanism of AUX28 inactivation by P25 and its contribution to rhizomania development. It remains to be determined whether nuclear export or import of AUX28 is affected by P25, and whether interaction of AUX28 with P25 leads to AUX28 degradation via the 26S proteasome. Although these aspects of P25 functioning are fascinating, they are beyond the scope of this article and will be addressed in future studies.
Experimental procedures
Virus isolates and plant inoculation
The sources of BNYVV isolates used in this study were soil samples from Thurnhof, Germany (B‐type BNYVV), Landskrona, Sweden (B‐type) and Holland (A‐type). The B‐type isolates have been partially sequence characterized (Koenig and Lennefors, 2000; Lennefors et al., 2006). Sugar beet inbred pollinator lines from the MariboHilleshog breeding pool (susceptible, line 13012502; Rz1, line 15014536; Rz2, line 14109005) were germinated in sterile sand and 1‐week‐old seedlings were transplanted into 0.25‐L plastic tubes, which contained BNYVV‐infested soil mixed with sterile sand (1 : 10 ratio). Each treatment included at least ten plants. The experiment was repeated twice.
Plant material and growth conditions
Tubes with sugar beet plants were cultivated in a quarantine glasshouse under controlled conditions (day, 22 °C/16 h; night, 20 °C/8 h). Beta macrocarpa, B. vulgaris and N. benthamiana plants were cultivated under glasshouse conditions (day, 24 °C/14 h; night, 18 °C/10 h).
Sequence alignments and phylogenetic analysis
Full‐length protein sequences of B. vulgaris and A. thaliana EXPs were retrieved from GenBank (Benson et al., 2013). Using MEGA version 7 (Kumar et al., 2016), multiple alignments were made with the algorithm MUSCLE with default parameters (Edgar, 2004), followed by phylogenetic reconstruction by neighbour joining. The topology of the tree was tested using bootstrap (1000 replications) and a Poisson model was used as substitution model.
RT‐qPCR and detection of BNYVV and P. betae by RT‐PCR
Total RNA from leaves was extracted with a MagJET RNA Purification Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Total RNA (1 µg) was transcribed into cDNA using an iScript cDNA Synthesis Kit (Bio‐Rad, Hercules, California, USA). Four microlitres of one‐tenth‐diluted cDNA were used for qPCR with the DyNAmo Flash SYBR Green qPCR Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). This reaction was supplemented with 0.125 µm of each primer. The BvICDH (isocitrate dehydrogenase) gene was used as reference. The resulting qPCR data were analysed using the 2‐ΔΔCt method. The sequences of the primers used for qRT‐PCR are provided in Table S5 (see Supporting Information).
For pathogen detection, BNYVV‐ and P. betae‐specific primers were used (Table S5). The reaction was set with initial denaturation of 95 °C for 1 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s, and a final extension of 72 °C for 5 min.
Plant inoculation with recombinant viruses
Reassortment experiments in B. vulgaris were conducted using infectious full‐length cDNA clones of BNYVV and BSBMV (Laufer et al., 2018). Different combinations of viral RNAs were first inoculated into B. macrocarpa leaves using agroinfiltration with A. tumefaciens. Then, leaves of B. macrocarpa displaying symptoms of systemic infection were used for vortex inoculation of 7‐day‐old B. vulgaris seedlings, as reported previously (Bornemann and Varrelmann, 2011). BNYVV‐ and BSBMV‐specific enzyme‐linked immunosorbent assay (ELISA) of infected LRs was performed to determine the virus content, as described previously (Pferdmenges et al., 2009).
Auxin treatment of sugar beet seedlings
Auxin treatment was carried out as follows: 21‐day‐old sugar beet seedlings (KWS03) were treated with 10 µm 3‐IAA (Sigma‐Aldrich, St. Louis, Missouri, USA) applying vacuum infiltration for 2 min (González‐Lamothe et al., 2012); seedlings were kept in infiltration solution for a further 2 h in the dark at room temperature; subsequently, RNA was extracted and subjected to RT‐qPCR analysis.
Y2H assays
The Grow’n’Glow Y2H system (MoBiTec, Göttingen, Germany) used is a modified version of the LexA (pEG202 as bait) and B42 (as prey) yeast two‐hybrid system (Gyuris et al., 1993). In addition, it contains a LexA operator‐controlled GFP reporter plasmid pGNG1 (Cormack, 1998). The analysis of the optical density of the total area of GFP fluorescence was carried out by epifluorescence microscopy of yeast cells using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The yeast vector pJG4‐5 (MoBiTec) carrying a partial cDNA clone of BvAUX28 with domains I–II was named pJG4‐5‐I‐II, and was obtained from a P25 Y2H screen of a sugar beet cDNA library (Thiel and Varrelmann, 2009). The complete BvAUX28 open reading frame (ORF) was cloned from sugar beet breeding line MS150 (Syngenta, Landskrona, Sweden). To obtain constructs carrying full‐length as well as truncated derivatives of the BvAUX28 ORF, PCR products were cloned into EcoRI‐XhoI sites giving rise to plasmids named pJG4‐5‐I, pJG4‐5‐II, pJG4‐5‐II‐IV, pJG4‐5‐III‐IV and pJG4‐5‐I+III‐IV. The truncated derivative constructs contain BvAUX28 sequences encoding the conserved domains I (amino acids 1–99), II (amino acids 100–501), II–IV (amino acids 100–858), III–IV (amino acids 502–858) and I + III–IV (amino acids 1–99 + amino acids 502–858).
GST pull‐down assay
FLAG epitope‐tagged P25 was produced in Sf9 insect cells infected with recombinant baculovirus. The E. coli BL21‐codon Plus (DE3) RIL strain (Stratagene, La Jolla, California, USA) was used to express the GST fusion proteins. The in vitro GST pull‐down experiments were performed as described previously (Vetter et al., 2004). The expression of the protein from pGEX‐BvAUX28 was performed at 28 °C. Aliquots of the supernatants were analysed using 12% sodium dodecylsulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE), and the proteins were immunodetected using anti‐P25 immunoglobulin G (IgG).
Transient expression and live cell imaging of fluorescent proteins
For subcellular localization and BiFC studies, all constructs were delivered by agroinfiltration to N. benthamiana leaves. Agrobacterium tumefaciens (strain C58C1/pGV2260) cells were electro‐transformed with the plasmid constructs. Agrobacterium tumefaciens cultures carrying the plasmids were grown overnight (16 h) at 28 °C. Bacterial cells were pelleted by centrifugation at 1200 × g for 10 min, and then resuspended in infiltration buffer (10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulfonic acid, 150 μm acetosyringone) prior to infiltration. Confocal imaging of GFP‐ and mRFP‐expressing leaf tissues was performed with a Leica, Wetzlar, Germany TCS SP5 confocal imaging system applying excitation and emission wavelengths of 488 and 510–515 nm for GFP, and 514–561 and 600–630 nm for mRFP.
BiFC assay
A BiFC system with an optimized mRFP was chosen for protein–protein analysis in N. benthamiana epidermal leaf cells (Zilian and Maiss, 2011). Therefore, P25, BvAUX28 and BvAUX28‐I+II were amplified with specific oligonucleotides (Table S5) and cloned into the vectors pES264 and pES265 using BamHI or BglII and SalI. The plasmids obtained were named pES264‐P25, pES265‐I‐IV and pES265‐I‐II. The coat protein (CP) of Plum pox virus (PPV) displaying self‐interaction served as a positive control, whereas β‐glucuronidase (GUS) was used as a negative control.
Conflicts of interest
The authors have no conflicts of interest to declare.
Author contributions
EIS and MV conceived the research plans; JFG, SL, B‐LL, TK, DG, EM, MV and EIS designed the research; EIS, JFG, SL and HT performed the experiments; JFG, SL, B‐LL, TK, DG, MV and EIS analysed the data; EIS wrote the paper with contributions from all authors.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Detection of Beet necrotic yellow vein virus (BNYVV) and Polymyxa betae, and comparison of P25 sequences from BNYVV strains used in our experiments.
Fig. S2 Thirteen EXPANSIN (EXP) and three LATERAL ORGAN BOUNDARIES DOMAIN (LBD) genes are up‐regulated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV).
Fig. S3 BvEXPA7L, BvEXPA13 and BvEXLA2a are not transcriptionally activated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV) in both susceptible and resistant varieties.
Fig. S4 BvIAA14, BvARF7 and BvARF19 are not transcriptionally activated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV).
Fig. S5 BvAUX28 localizes to the nucleus.
Table S1 The EXPANSIN genes in sugar beet (Beta vulgaris).
Table S2 Relative expression of Beta vulgaris EXPANSINs in different organs measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
Table S3 Beet necrotic yellow vein virus (BNYVV)‐dependent changes (by natural soil‐mediated infection with BNYVV) in EXPANSIN gene expression at 4 and 6 weeks post‐sowing (wps) assayed by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
Table S4 Beta vulgaris orthologues of Arabidopsis genes involved in lateral root development used in our analysis.
Table S5 Sequences of all oligonucleotides used in this study.
Acknowledgements
This work was supported by the Swedish Foundation for Strategic Research (SSF), Carl Tryggers Foundation and German Research Foundation (project numbers 165716485 and 278522005).
These authors contributed equally to this work.
References
- Abuqamar, S. , Ajeb, S. , Sham, A. , Enan, M.R. and Iratni, R. (2013) A mutation in the expansin‐like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana . Mol. Plant Pathol. 14, 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta‐Leal, R. , Bryan, B.K. and Rush, C.M. (2010) Host effect on the genetic diversification of beet necrotic yellow vein virus single‐plant populations. Phytopathology 100, 1204–1212. [DOI] [PubMed] [Google Scholar]
- Ahm, H.D. and Buchenauer, H. (1999) Studies on the biology of Polymyxa betae, the vector of beet necrotic yellow vein virus. J. Phytopathology. 139, 329–338. [Google Scholar]
- Barr, K.J. and Asher, M.J.C. (1996) Studies on the life‐cycle of Polymyxa betae in sugar beet roots. Mycological Research 100, 203–208. [Google Scholar]
- Benson, D.A. , Cavanaugh, M. , Clark, K. , Karsch‐Mizrachi, I. , Lipman, D.J. , Ostell, J. and Sayers, E.W. (2013) GenBank. Nucleic Acids Res. 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans, B. , Vassileva, V. , Schmid, S.P.C. , Maes, S. , Parizot, B. , Naramoto, S. , Magyar, Z. , Kamei, C.L.A. , Koncz, C. , Bögre, L. , Persiau, G. , De Jaeger, G. , Friml, J. , Simon, R. , Beeckman, T. and De Veylder, L. (2011) Auxin‐dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell, 23, 3671–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancardi, E. , Lewellen, R.T. , De Biaggi, M. , Erichsen, A.W. and Stevanato, P. (2002) The origin of rhizomania resistance in sugar beet. Euphytica 127, 383–397. [Google Scholar]
- Bornemann, K. , Hanse, B. , Varrelmann, M. and Stevens, M. (2015) Occurrence of resistance‐breaking strains of Beet necrotic yellow vein virus in sugar beet in northwestern Europe and identification of a new variant of the viral pathogenicity factor P25. Plant Pathol. 64, 25–34. [Google Scholar]
- Bornemann, K. and Varrelmann, M. (2011) Analysis of the resistance‐breaking ability of different Beet necrotic yellow vein virus isolates loaded into a single Polymyxa betae population in soil. Phytopathology 101, 718–724. [DOI] [PubMed] [Google Scholar]
- Bouzoubaa, S. , Ziegler, V. , Beck, D. , Guilley, H. , Richards, K. and Jonard, G. (1986) Nucleotide sequence of Beet Necrotic Yellow Vein Virus RNA‐2. J. Gen. Virol 67, 1689–1700. [Google Scholar]
- Capistrano‐Gossmann, G.G. , Ries, D. , Holtgräwe, D. , et al (2017) Crop wild relative populations of Beta vulgaris allow direct mapping of agronomically important genes. Nat. Commun. 8, 15 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐J. , Zou, W.‐S. , Wu, G. , Lin, H.‐H. and Xi, D.‐H. (2018) Tobacco alpha‐expansin EXPA4 plays a role in Nicotiana benthamiana defence against Tobacco mosaic virus. Planta 247, 355–368. [DOI] [PubMed] [Google Scholar]
- Chiba, S. , Kondo, H. , Miyanishi, M. , Andika, I.B. , Han, C. and Tamada, T. (2011) The evolutionary history of Beet necrotic yellow vein virus deduced from genetic variation, geographical origin and spread, and the breaking of host resistance. Mol. Plant–Microbe Interact. 24, 207–218. [DOI] [PubMed] [Google Scholar]
- Collum, T.D. , Padmanabhan, M.S. , Hsieh, Y.‐C. and Culver, J.N. (2016) Tobacco mosaic virus‐directed reprogramming of auxin/indole acetic acid protein transcriptional responses enhances virus phloem loading. Proc. Natl. Acad. Sci. USA, 113, E2740–E2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack, B. (1998) Green fluorescent protein as a reporter of transcription and protein localization in fungi. Curr. Opin. Microbiol. 1, 406–410. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2015) Plant expansins: diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 25, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel, B. , Vassileva, V. , Parizot, B. , Demeulenaere, M. , Grunewald, W. , Audenaert, D. , Van Campenhout, J. , Overvoorde, P. , Jansen, L. , Vanneste, S. , Möller, B. , Wilson, M. , Holman, T. , Van Isterdael, G. , Brunoud, G. , Vuylsteke, M. , Vernoux, T. , DeVeylder, L. , Inzé, D. , Weijers, D. , Bennett, M.J. and Beeckman, T. (2010) A novel Aux/IAA28 signaling cascade activates GATA23‐dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N. , Dharmasiri, S. and Estelle, M. (2005) The F‐box protein TIR1 is an auxin receptor. Nature 435, 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N. and Estelle, M. (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci. 9, 302–308. [DOI] [PubMed] [Google Scholar]
- Dohm, J.C. , Minoche, A.E. , Holtgräwe, D. , Capella‐Gutiérrez, S. , Zakrzewski, F. , Tafer, H. , Rupp, O. , Sörensen, T.R. , Stracke, R. , Reinhardt, R. , Goesmann, A. , Kraft, T. , Schulz, B. , Stadler, P.F. , Schmidt, T. , Gabaldón, T. , Lehrach, H. , Weisshaar, B. and Himmelbauer, H. (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Sun, X. , Wang, G. , Liu, H. and Zhu, J. (2012) LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana . Ann. Bot. 110, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki, H. , Nakao, Y. , Okushima, Y. , Theologis, A. and Tasaka, M. (2005) Tissue‐specific expression of stabilized SOLITARY‐ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395. [DOI] [PubMed] [Google Scholar]
- Gidner, S. , Lennefors, B.‐L. , Nilsson, N.‐O. , Bensefelt, J. , Johansson, E. , Gyllenspetz, U. and Kraft, T. (2005) QTL mapping of BNYVV resistance from the WB41 source in sugar beet. Genome 48, 279–285. [DOI] [PubMed] [Google Scholar]
- Goh, T. , Kasahara, H. , Mimura, T. , Kamiya, Y. and Fukaki, H. (2012) Multiple AUX/IAA‐ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3‐mediated auxin signalling. Philos. Trans. R. Soc. London B: Biol. Sci. 367, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Lamothe, R. , Oirdi, M.El , Brisson, N. and Bouarab, K. (2012) The conjugated auxin indole‐3‐acetic acid‐aspartic acid promotes plant disease development. Plant Cell, 24, 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris, J. , Golemis, E. , Chertkov, H. and Brent, R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Jin, L. , Qin, Q. , Wang, Y. , Pu, Y. , Liu, L. , Wen, X. , Ji, S. , Wu, J. , Wei, C. , Ding, B. and Li, Y. (2016) Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 12, e1005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende, H. , Bradford, K. , Brummell, D. , Cho, H. , Cosgrove, D. , Fleming, A. , Gehring, C. , Lee, Y. , Mcqueen‐mason, S. , Rose, J. and Voesenek, L. (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 55, 311–314. [DOI] [PubMed] [Google Scholar]
- Kepinski, S. and Leyser, O. (2005) The Arabidopsis F‐box protein TIR1 is an auxin receptor. Nature 435, 446–451. [DOI] [PubMed] [Google Scholar]
- Kim, D.W. , Lee, S.H. , Choi, S.B. , Won, S.K. , Heo, Y.K. , Cho, M. , Park, Y.I. and Cho, H.T. (2006) Functional conservation of a root hair cell‐specific cis‐element in angiosperms with different root hair distribution patterns. Plant Cell, 18, 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig, R. , Kastirr, U. , Holtschulte, B. , Deml, G. and Varrelmann, M. (2008) Distribution of various types and P25 subtypes of Beet necrotic yellow vein virus in Germany and other European countries. Arch. Virol. 153, 2139–2144. [DOI] [PubMed] [Google Scholar]
- Koenig, R. , Holtschulte, B. , Deml, G. , Lüddecke, P. , Schuhmann, S. , Maaß, C. and Richert‐Pöggeler, K. (2009) Beet necrotic yellow vein virus genome reassortments in a resistant sugar beet variety showing–in a small area in France–strong rhizomania symptoms. J. Plant Dis. Prot. 116, 7–9. [Google Scholar]
- Koenig, R. and Lennefors, B.‐L. (2000) Molecular analyses of European A, B and P type sources of Beet necrotic yellow vein virus and detection of the rare P type in Kazakhstan. Arch. Virol. 145, 1561–1570. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, E. , Guilley, H. , Tamada, T. , Richards, K.E. and Jonard, G. (1998) Vascular movement of beet necrotic yellow vein virus in Beta macrocarpa is probably dependent on an RNA 3 sequence domain rather than a gene product. J. Gen. Virol. 79, 385–393. [DOI] [PubMed] [Google Scholar]
- Laufer, M. , Mohammad, H. , Maiss, E. , Richert‐Pöggeler, K. , Dall’Ara,M. , Ratti,C. , Gilmer,D. , Liebe,S. and Varrelmann,M. (2018) Biological properties of Beet soil‐borne mosaic virus and Beet necrotic yellow vein virus cDNA clones produced by isothermal in vitro recombination: insights for reassortant appearance. Virology 518, 25–33. [DOI] [PubMed] [Google Scholar]
- Lavenus, J. , Goh, T. , Roberts, I. , Guyomarc’h, S. , Lucas, M. , De Smet, I. , Fukaki, H. , Beeckman, T. , Bennett, M. and Laplaze, L. (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. [DOI] [PubMed] [Google Scholar]
- Lee, H.W. , Cho, C. and Kim, J. (2015) Lateral Organ Boundaries Domain16 and 18 act downstream of the AUXIN1 and LIKE‐AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 168, 1792–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W. , Cho, C. and Kim, J. (2015) LBD16 and LBD18 act downstream of the AUX1 and LAX3 auxin influx carriers to control lateral root development in Arabidopsis thaliana . Plant Physiol. 168, 1792–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W. and Kim, J. (2013) EXPANSINA17 up‐regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 54, 1600–1611. [DOI] [PubMed] [Google Scholar]
- Lee, H.W. , Kim, N.Y. , Lee, D.J. and Kim, J. (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.W. , Kim, M.J. , Kim, N.Y. , Lee, S.H. and Kim, J. (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 73, 212–224. [DOI] [PubMed] [Google Scholar]
- Lee, L. , Telford, E.B. , Batten, J.S. , Scholthof, K.B.G. and Rush, C.M. (2001) Complete nucleotide sequence and genome organization of Beet soilborne mosaic virus, a proposed member of the genus Benyvirus. Arch. Virol. 146, 2443–2453. [DOI] [PubMed] [Google Scholar]
- Lennefors, B.‐L. , Savenkov, E.I. , Bensefelt, J. , Wremerth‐Weich, E. , van Roggen, P. , Tuvesson, S. , Valkonen, J.P.T. and Gielen, J. (2006) dsRNA‐mediated resistance to Beet Necrotic Yellow Vein Virus infections in sugar beet (Beta vulgaris L. ssp. vulgaris). Mol. Breed. 18, 313–325. [Google Scholar]
- Lennefors, B.‐L. , van Roggen, P.M. , Yndgaard, F. , Savenkov, E.I. and Valkonen, J.P.T. (2008) Efficient dsRNA‐mediated transgenic resistance to Beet necrotic yellow vein virus in sugar beets is not affected by other soilborne and aphid‐transmitted viruses. Transgenic Res. 17, 219–228. [DOI] [PubMed] [Google Scholar]
- Lewellen, R. , Skoyen, I. and Erichsen, A. (1987) Breeding sugarbeet for resistance to rhizomania: evaluation of host‐plant reactions and selection for and inheritance of resistance. In: 50th Winter Congress of the IIRB. International Institute of Sugar Beet Research (IIRB), Brussels, Belgium: pp. 139–156. [Google Scholar]
- Liu, H.‐Y. and Lewellen, R.T. (2007) Distribution and molecular characterization of resistance‐breaking isolates of Beet necrotic yellow vein virus in the United States. Plant Dis. 91, 847–851. [DOI] [PubMed] [Google Scholar]
- Liu, H.‐Y. , Sears, J.L. and Lewellen, R.T. (2005) Occurrence of resistance‐breaking Beet necrotic yellow vein virus of sugar beet. Plant Dis. 89, 464–468. [DOI] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Solovieva, A.D. , Boddeti, S.K. , Thaduri, S. , Solovyev, A.G. and Savenkov, E.I. (2013) An RNA virus‐encoded zinc‐finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell, 25, 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marowa, P. , Ding, A. and Kong, Y. (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 35, 949–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y. , Fukaki, H. , Onoda, M. , Theologis, A. and Tasaka, M. (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell, 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan, M.S. , Shiferaw, H. and Culver, J.N. (2006) The Tobacco mosaic virus replicase protein disrupts the localization and function of interacting Aux/IAA proteins. Mol. Plant–Microbe Interact. 19, 864–873. [DOI] [PubMed] [Google Scholar]
- Park, S.‐H. , Li, F. , Renaud, J. , Shen, W. , Li, Y. , Guo, L. , Cui, H. , Sumarah, M. and Wang, A. (2017) NbEXPA1, an α‐expansin, is plasmodesmata‐specific and a novel host factor for potyviral infection. Plant J. 92, 846–861. [DOI] [PubMed] [Google Scholar]
- Peltier, C. , Schmidlin, L. , Klein, E. , Taconnat, L. , Prinsen, E. , Erhardt, M. , Heintz, D. , Weyens, G. , Lefebvre, M. , Renou, J.P. and Gilmer, D. (2011) Expression of the Beet necrotic yellow vein virus p25 protein induces hormonal changes and a root branching phenotype in Arabidopsis thaliana . Transgenic Res. 20, 443–466. [DOI] [PubMed] [Google Scholar]
- Péret, B. , De Rybel, B. , Casimiro, I. , Benková, E. , Swarup, R. , Laplaze, L. , Beeckman, T. and Bennett, M.J. (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408. [DOI] [PubMed] [Google Scholar]
- Pferdmenges, F. , Korf, H. and Varrelmann, M. (2009) Identification of rhizomania‐infected soil in Europe able to overcome Rz1 resistance in sugar beet and comparison with other resistance‐breaking soils from different geographic origins. Eur. J. Plant Pathol. 124, 31–43. [Google Scholar]
- Pollini, C.P. , Masia, A. and Giunchedi, L. (1990) Free indole‐3‐acetic acid in sugar‐beet root of rhizomania‐susceptible and moderately resistant cultivars. Phytopathol. Mediterr. 29, 191–195. [Google Scholar]
- Ratti, C. , Hleibieh, K. , Bianchi, L. , Schirmer, A. , Autonell, C.R. and Gilmer, D. (2009) Beet soil‐borne mosaic virus RNA‐3 is replicated and encapsidated in the presence of BNYVV RNA‐1 and ‐2 and allows long distance movement in Beta macrocarpa . Virology 385, 392–399. [DOI] [PubMed] [Google Scholar]
- Sampedro, J. and Cosgrove, D.J. (2005) The expansin superfamily. Genome Biol. 6, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin, L. , De Bruyne, E. , Weyens, G. , Lefebvre, M. and Gilmer, D. (2008) Identification of differentially expressed root genes upon rhizomania disease. Mol. Plant Pathol. 9, 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura, H. , Pantaleo, V. , Ishihara, T. , Myojo, N. , Inaba, J. , Sueda, K. , Burgyán, J. and Masuta, C. (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery Ding, S.‐W., ed. PLoS Pathog. 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada, T. , Uchino, H. , Kusume, T. and Saito, M. (1999) RNA 3 deletion mutants of beet necrotic yellow vein virus do not cause rhizomania disease in sugar beets. Phytopathology 89, 1000–1006. [DOI] [PubMed] [Google Scholar]
- Tan, X. , Calderon‐Villalobos, L.I.A. , Sharon, M. , Zheng, C. , Robinson, C.V. , Estelle, M. and Zheng, N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Thiel, H. and Varrelmann, M. (2009) Identification of Beet necrotic yellow vein virus P25 pathogenicity factor–interacting sugar beet proteins that represent putative virus targets or components of plant resistance. Mol. Plant–Microbe Interact. 22, 999–1010. [DOI] [PubMed] [Google Scholar]
- Vetter, G. , Hily, J.‐M. , Klein, E. , Schmidlin, L. , Haas, M. , Merkle, T. and Gilmer, D. (2004) Nucleo‐cytoplasmic shuttling of the beet necrotic yellow vein virus RNA‐3‐encoded p25 protein. J. Gen. Virol. 85, 2459–2469. [DOI] [PubMed] [Google Scholar]
- Wang, J.‐J. and Guo, H.‐S. (2015) Cleavage of INDOLE‐3‐ACETIC ACID INDUCIBLE28 mRNA by MicroRNA847 upregulates auxin signaling to modulate cell proliferation and lateral organ growth in Arabidopsis. Plant Cell, 27, 574–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workneh, F. , Villanueva, E. , Steddom, K. and Rush, C.M. (2003) Spatial association and distribution of Beet necrotic yellow vein virus and Beet soilborne mosaic virus in sugar beet fields. Plant Dis. 87, 707–711. [DOI] [PubMed] [Google Scholar]
- Zilian, E. and Maiss, E. (2011) An optimized mRFP‐based bimolecular fluorescence complementation system for the detection of protein–protein interactions in planta. J. Virol. Methods, 174, 158–165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Detection of Beet necrotic yellow vein virus (BNYVV) and Polymyxa betae, and comparison of P25 sequences from BNYVV strains used in our experiments.
Fig. S2 Thirteen EXPANSIN (EXP) and three LATERAL ORGAN BOUNDARIES DOMAIN (LBD) genes are up‐regulated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV).
Fig. S3 BvEXPA7L, BvEXPA13 and BvEXLA2a are not transcriptionally activated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV) in both susceptible and resistant varieties.
Fig. S4 BvIAA14, BvARF7 and BvARF19 are not transcriptionally activated by natural soil‐mediated infection with Beet necrotic yellow vein virus (BNYVV).
Fig. S5 BvAUX28 localizes to the nucleus.
Table S1 The EXPANSIN genes in sugar beet (Beta vulgaris).
Table S2 Relative expression of Beta vulgaris EXPANSINs in different organs measured by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
Table S3 Beet necrotic yellow vein virus (BNYVV)‐dependent changes (by natural soil‐mediated infection with BNYVV) in EXPANSIN gene expression at 4 and 6 weeks post‐sowing (wps) assayed by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
Table S4 Beta vulgaris orthologues of Arabidopsis genes involved in lateral root development used in our analysis.
Table S5 Sequences of all oligonucleotides used in this study.


