Summary
Grapevine leafroll‐associated virus 2 (GLRaV‐2) p24 has been reported to be an RNA silencing suppressor (RSS). However, the mechanisms underlying p24's suppression of RNA silencing are unknown. Using Agrobacterium infiltration‐mediated RNA silencing assays, we showed that GLRaV‐2 p24 is a strong RSS triggered by positive‐sense green fluorescent protein (GFP) RNA, and that silencing suppression by p24 effectively blocks the accumulation of small interfering RNAs. Deletion analyses showed that the region of amino acids 1–188, which contains all predicted α‐helices and β‐strands, is required for the RSS activity of p24. Hydrophobic residues I35/F38/V85/V89/W149 and V162/L169/L170, previously shown to be critical for p24 self‐interaction, are also crucial for silencing suppression, and western blotting results suggested that a lack of self‐interaction ability results in decreased p24 accumulation in plants. The mutants showed greatly weakened or a lack of RSS activity. Substitution with two basic residues at positions 2 or 86, putatively involved in RNA binding, totally abolished the RSS activity of p24, suggesting that p24 uses an RNA‐binding strategy to suppress RNA silencing. Our results also showed that W54 in the WG/GW‐like motif (W54/G55) is crucial for the RSS activity of p24, whereas p24 does not physically interact with AGO1 of Nicotiana benthamiana. Furthermore, p24 did not promote AGO1 degradation, but significantly up‐regulated AGO1 mRNA expression, and this effect was correlated with the RSS activity of p24, indicating that p24 may interfere with microRNA‐directed processes. The presented results contribute to our understanding of viral suppression of RNA silencing and the molecular mechanisms underlying GLRaV‐2 infection.
Keywords: argonaute protein, Grapevine leafroll‐associated virus 2, p24, RNA silencing suppressor, self‐interaction
Introduction
RNA silencing plays an important antiviral role in plants (Ding, 2010; Ding and Voinnet, 2007). In antiviral silencing pathways, highly structured or double‐stranded (ds) viral RNAs are processed by RNaseIII enzymes, called Dicers, into small interfering RNA (siRNA), and one strand of the siRNA duplex is then incorporated into the RNA‐induced silencing complex (RISC) which contains the argonaute (AGO) protein as a core component that initiates the sequence‐specific degradation of target RNAs (Ding and Voinnet, 2007; Malone and Hannon, 2009; Sontheimer, 2005). Several members of the AGO family are known to contribute to the biogenesis of viral siRNAs (Baulcombe, 2004; Zhang et al., 2015). In addition to Dicer and AGO proteins, RNA‐dependent RNA polymerases are also involved in reinforcing the host silencing responses through the amplification of secondary siRNAs (Díaz‐Pendón and Ding, 2008). To counter antiviral defences, most plant viruses encode an RNA silencing suppressor (RSS) to suppress RNA silencing by targeting different steps of the silencing pathway (Ding and Voinnet, 2007). For instance, the 2b protein encoded by Cucumber mosaic virus (CMV) inhibits RISC‐mediated mRNA cleavage through interaction with and inhibition of AGO1 and AGO4 (Duan et al., 2012; González et al., 2010; Hamera et al., 2012; Zhang et al., 2006), and can also suppress RNA silencing by binding directly to siRNA (Goto et al., 2007). The RSS activity of the 19‐kDa protein of tombusviruses (p19) depends on the binding and inactivation of silencing‐generated double‐stranded siRNAs to prevent their subsequent incorporation into RISC (Chapman et al., 2004; Lakatos et al., 2006; Silhavy et al., 2002), and inhibition of the translational capacity of AGO1 mRNA to alleviate the antiviral function of AGO1 protein (Várallyay et al., 2010). The helper component proteinase (HC‐Pro) of potyviruses interferes with RNA silencing through binding to siRNAs (Lakatos et al., 2006), down‐regulation of the accumulation of primary and secondary siRNAs (Zhang et al., 2008) and modulation of the accumulation of miR168, consequently interfering with AGO1‐related silencing (Várallyay et al., 2010).
In vitro structural analyses have shown that several RSSs, such as 2b of Tomato aspermy virus (Chen et al., 2008), p19 of tombusviruses (Vargason et al., 2003; Ye et al., 2003), HC‐Pro of Tobacco etch virus (Ruiz‐Ferrer et al., 2005) and p21 of Beet yellows virus (BYV) (Ye and Patel, 2005), form homodimers or oligomers. Our previous results have also shown that Grapevine leafroll‐associated virus 2 (GLRaV‐2) p24 can self‐interact and aggregate in the cytoplasm of plant cells (Liu et al., 2016). Furthermore, some viral RSSs need to be homodimers or higher order oligomers to become functional (Bragg and Jackson, 2004; Chen et al., 2008; Vargason et al., 2003; Xu et al., 2013). However, dimerization of Flock house virus B2 is not essential for interaction with the RNA silencing machinery (Seo et al., 2012).
GLRaV‐2 is a member of the genus Closterovirus in the family Closteroviridae. GLRaV‐2 virions, like those of other closteroviruses, are presumed to be limited to the phloem (Dolja, 2003), and some isolates of GLRaV‐2 may also cause systemic infection and symptoms, albeit inefficiently, in herbaceous hosts, such as Nicotiana benthamiana, via mechanical inoculation (Ghanem‐Sabanadzovic et al., 2000; Goszczynski et al., 1996). The flexuous, filamentous particles of the virus are 1400–1800 nm in length and its single‐stranded, positive‐sense RNA genome is approximately 16.5 kb in size, encoding nine open reading frames (at least 10 proteins) (Liu et al., 2009; Meng et al., 2005; Zhu et al., 1998). The genome organization is similar to that of BYV, the type member of the genus Closterovirus (Karasev, 2000; Zhu et al., 1998). In this genus, BYV and GLRaV‐2 encode RSSs p21 and p24, respectively (Chiba et al., 2006; Reed et al., 2003), and Citrus tristeza virus (CTV) encodes three distinct RSSs—p20, p23 and coat protein (Lu et al., 2004). BYV p21, the homologue of p24, has been reported to suppress dsRNA‐induced silencing of green fluorescent protein (GFP) mRNA (Reed et al., 2003), and to bind siRNAs in vitro and in vivo (Chapman et al., 2004). However, the amino acid sequence identity between BYV p21 and GLRaV‐2 p24 is only about 16%, and the mechanism underlying p24 suppression of silencing remains unknown.
In this study, we showed that GLRaV‐2 p24 is a strong RSS and that silencing suppression by p24 prevents siRNA processing. The region of amino acids 1–188, which contains predicted α‐helices and β‐strands, is required for p24's RSS activity, and self‐interaction is also important for p24 functionality. Site‐directed mutagenesis showed that R2 and R86, which may be involved in RNA binding, and W54 in the WG/GW‐like motif (W54/G55), are crucial for p24 suppression of RNA silencing. We also showed that p24 does not physically interact with N. benthamiana AGO1 (NbAGO1) or promote the degradation of Arabidopsis thaliana AGO1 (AtAGO1), but induces the expression of NbAGO1 mRNA.
Results
GLRaV‐2 p24 is a strong RSS triggered by positive‐sense GFP RNA
The gene encoding GLRaV‐2 p24 was cloned into the expression cassette of the binary vector pGD (Goodin et al., 2002) to test its suppression activity using Agrobacterium co‐infiltration assay, as described previously (Johansen and Carrington, 2001). Nicotiana benthamiana leaves infiltrated with cultures harbouring pGD–GFP and cultures harbouring either an empty vector (pGD) or a construct expressing p19 of Tomato bushy stunt virus (TBSV), a well‐characterized suppressor, were used as negative and positive controls, respectively. Leaf patches agroinfiltrated with p24 showed strong GFP fluorescence at 3 days post‐infiltration (dpi), comparable with the green fluorescence emanating from the leaf patches infiltrated with p19 (Fig. 1A). In contrast, GFP fluorescence was negligible or undetectable in tissues co‐infiltrated with pGD–GFP and the empty vector pGD (Fig. 1A) because of activation of RNA silencing. Western blot was performed for quantitative assessment of GFP accumulation, and the results showed significantly higher GFP accumulation in the sectors co‐infiltrated with the GFP construct and p24 or p19 construct than in leaf patches co‐infiltrated with the GFP construct and empty vector (Fig. 1B). This was in agreement with the fluorescence levels observed in the infiltrated tissues.
Figure 1.

Suppression of local silencing by Grapevine leafroll‐associated virus 2 (GLRaV‐2) p24. (A) Assessment of suppression of gene silencing in Nicotiana benthamiana leaves by co‐infiltration of an Agrobacterium strain containing pGD–GFP for the expression of green fluorescent protein (GFP), a strain containing the control pGD plasmid or strains containing pGD plasmids for the expression of p24 or p19. Photographs were taken of leaves at 3 days post‐infiltration (dpi) under long‐wavelength UV light. (B) Quantitative assessment of GFP accumulation by western blot analysis using GFP antibody. Total proteins were extracted from the agroinfiltrated regions for sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). The loading control was Rubisco stained with Coomassie blue. (C) Northern blot analysis of GFP‐specific siRNA extracted from the patches infiltrated with Agrobacterium suspensions carrying pGD–GFP/pGD or pGD–GFP/pGD–p24 at 3 dpi. U6 RNA was used as a loading control.
The presence of siRNAs is indicative of RNA silencing (Hamilton and Baulcombe, 1999). To further determine whether p24 affects GFP expression through the suppression of RNA silencing, we analysed the level of GFP‐specific siRNA. Silencing suppression by p24 resulted in a low level of GFP‐specific siRNA compared with that in the negative control (Fig. 1C).
Collectively, our data demonstrate that GLRaV‐2 p24 is a strong suppressor of RNA silencing triggered by positive‐sense GFP RNA, and that silencing suppression by p24 blocks the accumulation of silencing‐specific small RNAs.
Residues 1–188 are necessary for RSS activity of p24
Protein sequence analysis shows that p24 (205 amino acids) contains a putative conserved domain located between amino acids 31 and 150, and secondary structure analysis (http://bioinf.cs.ucl.ac.uk/psipred/) predicts that p24 possesses seven α‐helices located in the region of amino acids 10–180, and three short β‐strands (β1, amino acids 2–4; β2, amino acids 43–44; β3, amino acids 184–188). Thus, p24 deletion mutants with increasing C‐ and N‐terminal deletions, p24 with amino acids 1–188 [p24 (1–188)], p24 (1–180), p24 (1–150), p24 (10–188), p24 (10–180), p24 (31–205) and p24 (151–205), were produced to determine the functional regions necessary for suppressor activity in agroinfiltrated patch silencing suppression assays. At 3 dpi, leaves co‐infiltrated with pGD–GFP and the pGD plasmids for the expression of p24 (1–180), p24 (1–150), p24 (10–188), p24 (10–180), p24 (31–205) or p24 (151–205) failed to suppress the silencing of a GFP reporter gene. However, intense fluorescence was observed in tissues co‐infiltrated with pGD–GFP and the pGD plasmid for the expression of p24 (1–188) or wild‐type (wt) p24 (Fig. 2A), suggesting that amino acid region 1–188 retains local silencing suppression activity comparable with that of wt p24. High levels of GFP protein detected in leaves co‐infiltrated with the GFP construct and p24 (1–188) or the wt p24 construct (Fig. 2B) were in agreement with the fluorescence observations. These results demonstrate that the region of amino acids 1–188, which includes all putative α‐helices and β‐strands, is required for the RSS activity of p24.
Figure 2.
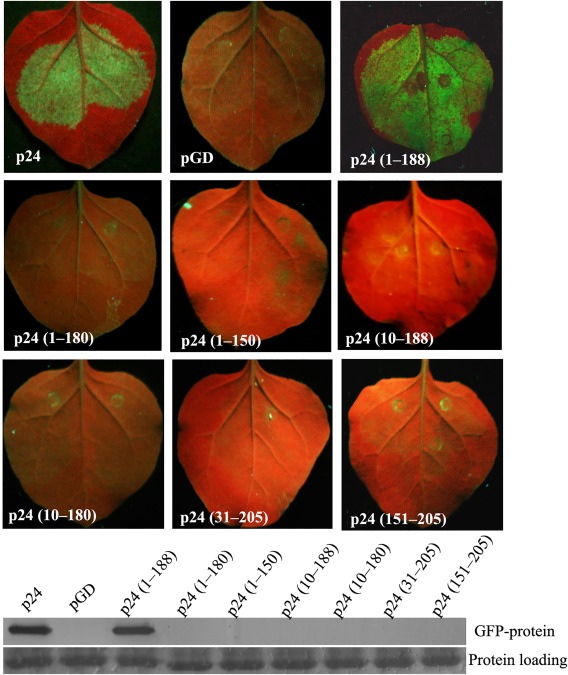
Functional region required for p24 suppression of RNA silencing. (A) Assessment of suppression of gene silencing in Nicotiana benthamiana leaves co‐infiltrated with an Agrobacterium strain containing pGD–GFP and a strain containing the control pGD plasmid, or strains containing pGD plasmids for the expression of p24 and its different deletion mutants. Photographs were taken as indicated in Fig. 1A. (B) Quantitative assessment of green fluorescent protein (GFP) accumulation by western blot analysis using GFP antibody as described in Fig. 1B.
Specific amino acids that are crucial for p24 self‐interaction are also critical for RNA silencing suppression
Hydrophobic residues I35/F38/V85/V89/W149 and V162/L169/L170 may mediate the inter‐domain interaction of the same p24 monomer and the tail‐to‐tail association, respectively (Ye and Patel, 2005). Our previous results have shown that substitution of H into each of I35, F38, V85 and V89 does not affect the self‐interaction of p24, whereas quadruple H substitutions in I35/F38/V85/V89, a single A substitution for W149 or triple H substitutions in V162/L169/L170 totally destroy the homologous interaction of p24 (Liu et al., 2016). To determine whether these hydrophobic residues are involved in the RSS activity of p24, seven p24 mutants (35H, 38H, 85H, 89H, 162/169/170H, 35/38/85/89H and 149A) were used in silencing suppression analyses. Our results showed that 35H, 38H, 85H and 89H all retain local silencing suppression activity comparable with that of wt p24 (Fig. 3A). However, mutants 162/169/170H and 35/38/85/89H showed greatly reduced GFP fluorescence and hence weakened suppressor activity, and 149A totally failed to suppress silencing of the GFP reporter gene (Fig. 3A). Western blot analyses confirmed the visual observations (Fig. 3B). These results suggest that hydrophobic residues I35/F38/V85/V89/W149 and V162/L169/L170, which are crucial for the self‐interaction of p24, are also important for RNA silencing suppression.
Figure 3.
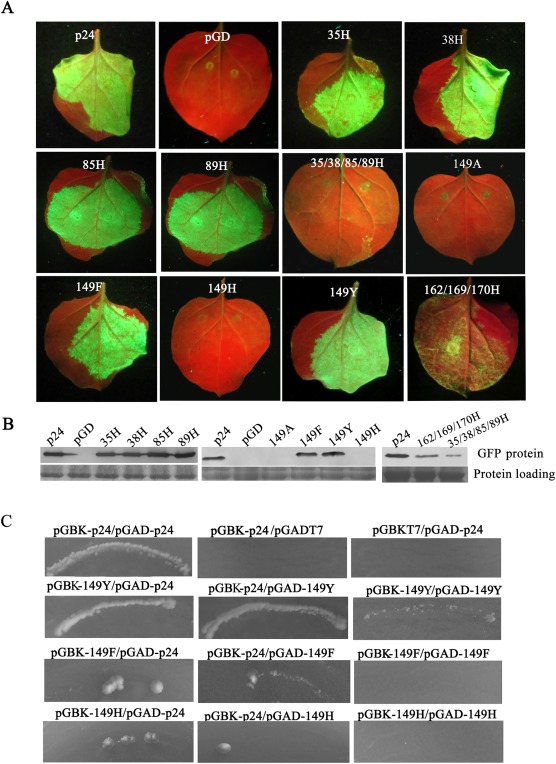
Ability of p24 mutants that can or cannot self‐interact to suppress gene silencing in Nicotiana benthamiana leaves. (A) Co‐infiltration of N. benthamiana leaves with Agrobacterium carrying pGD–GFP and the empty vector pGD, or pGD–p24, pGD–35H, pGD–38H, pGD–85H, pGD–89H, pGD–149A, pGD–149F, pGD–149H, pGD–149Y, pGD–162/169/170H or pGD–35/38/85/89H. Photographs were taken as indicated in Fig. 1A. (B) Quantitative assessment of green fluorescent protein (GFP) accumulation by western blot analysis as described in Fig. 1B. (C) Yeast two‐hybrid system (YTHS) analysis of self‐interaction of mutants as well as interactions between mutants and wild‐type (wt) p24. Yeast AH109 cells co‐transformed with the pair pGBK–p24/pGAD–p24 were used as a positive control, and cells co‐transformed with pairs pGBK–p24/pGADT7 and pGBKT7/pGAD–p24 were used as negative controls.
Ring‐structured amino acids affect the suppressor activity of Melon aphid‐borne yellows virus P0 protein (Han et al., 2010). We therefore generated three substitutions targeting W149 with F (with a benzene ring), Y (phenol ring) and H (imidazole ring), respectively, to investigate whether the ring structures are involved in the RSS activity of p24. Leaves co‐infiltrated with pGD–GFP and pGD–149F or pGD–149Y displayed strong GFP fluorescence, comparable with that of leaves co‐infiltrated with pGD–GFP and pGD–p24 at 3 dpi, whereas fluorescence in leaf patches co‐infiltrated with pGD–GFP and pGD–149H was not detectable (Fig. 3A). Western blot analyses confirmed the visual observations (Fig. 3B). In addition, we observed that mutants 149F and 149H both have weak interactions with wt p24, and that 149Y can self‐interact and interact with wt p24 (Fig. 3C). These results suggest that an aromatic amino acid at position 149 is necessary for p24 to self‐interact, as well as to suppress RNA silencing.
Two basic and polar residues at positions 2 and 86 are required for p24 suppressor activity
The putative RNA‐binding inner face of BYV p21 is composed of K2, R90, R130 and T149, and these four well‐conserved basic and polar residues in GLRaV‐2 p24 are R2, R86, R126 and K145 (Ye and Patel, 2005). We substituted each of the four residues in p24 with A to verify whether these conserved residues are related to suppression activity. The four p24 mutants, 2A, 86A, 126A and 145A, were used in agroinfiltrated patch silencing suppression assays. Mutants 126A and 145A displayed suppressive activities similar to that of wt p24 (Fig. 4A), indicating that these two residues are not necessary for the RSS activity of p24. In contrast, GFP fluorescence in leaves co‐infiltrated with pGD–GFP and pGD–2A, pGD–86A or pGD was not easily detected (Fig. 4A), suggesting that mutants 2A and 86A failed to suppress silencing of the GFP reporter gene in the patch assay. Consistent with this, western blotting showed undetectable levels of GFP protein in leaf patches co‐infiltrated with pGD–GFP and pGD–2A, pGD–86A or pGD empty vector (Fig. 4B). These results suggest that the two basic residues at positions 2 and 86 are crucial for the RSS activity of p24.
Figure 4.
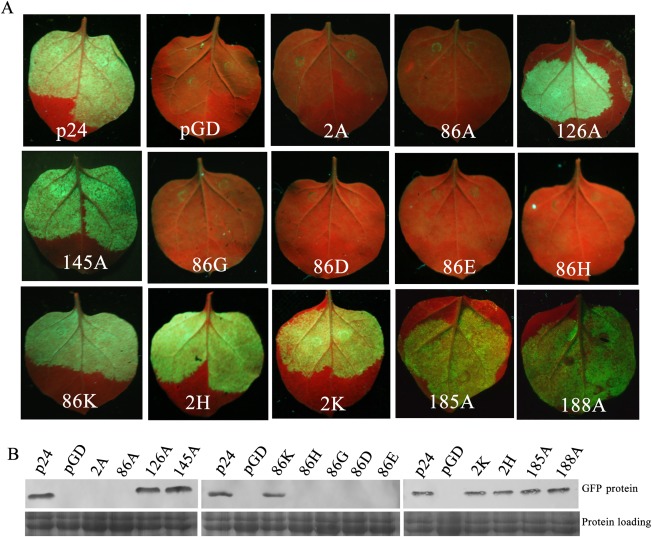
Local silencing suppression activity of substitution mutants of p24 in infiltrated patch assays. (A) Co‐infiltration of Nicotiana benthamiana leaves with Agrobacterium carrying pGD–GFP and the empty vector pGD, or pGD–p24, pGD–2A, pGD–2H, pGD–2K, pGD–86A, pGD–86D, pGD–86E, pGD–86G, pGD–86H, pGD–86K, pGD–126A, pGD–145A, pGD–185A or pGD–188A. Photographs were taken as indicated in Fig. 1A. (B) Quantitative assessment of green fluorescent protein (GFP) accumulation by western blot analysis as described in Fig. 1B.
We further engineered five amino acid substitutions targeting the positively charged residue R86 with negatively charged residues D and E, non‐polar residue G and two other positively charged residues, K and H, respectively. Mutants 86D, 86E, 86G and 86H all failed to suppress silencing, whereas mutant 86K retained suppressor activity (Fig. 4A). Then, H and K were substituted into R2, respectively, and the results showed that mutants 2H and 2K both retained RSS activity (Fig. 4A). Western blot analyses confirmed the visual observations (Fig. 4B). These results suggest that R2 can be replaced by H or K, and R86 can be replaced by K, without affecting the suppression of silencing.
As deletion mutant p24 (1–188), but not p24 (1–180), retained RSS activity (Fig. 2), and the amino acid region 181–188 contains two basic residues, H185 and R188, we tested whether these two basic residues contribute to p24 silencing suppression. We generated an A substitution for each of the two residues. Mutants 185A and 188A both displayed suppressor activity (Fig. 4), suggesting that the two basic residues are not required for the RSS activity of p24.
W in the GW/WG‐like motif (W54/G55) is crucial for p24 suppression of RNA silencing
GW/WG‐like motifs are crucial for the suppressor activity of some viral RSSs (Azevedo et al., 2010; Giner et al., 2010; Mingot et al., 2016; Untiveros et al., 2016; Zhuo et al., 2014). GLRaV‐2 p24 contains one WG/GW‐like motif (W54/G55) at its N‐terminus. To investigate the potential role of W54/G55 residues in the RSS activity of p24, we generated single mutants by substitution with A (54A and 55A), and a double mutant in which W and G were exchanged (54G/55W). At 3 dpi, sectors infiltrated with the mutants 54A and 54G/55W and with the pGD vector failed to exhibit fluorescence, but sectors infiltrated with the mutant 55A exhibited fluorescence comparable with that of wt p24 infiltration (Fig. 5A). Western blot analyses confirmed the visual observations (Fig. 5B). Hence, W54 is crucial for p24 silencing suppression.
Figure 5.
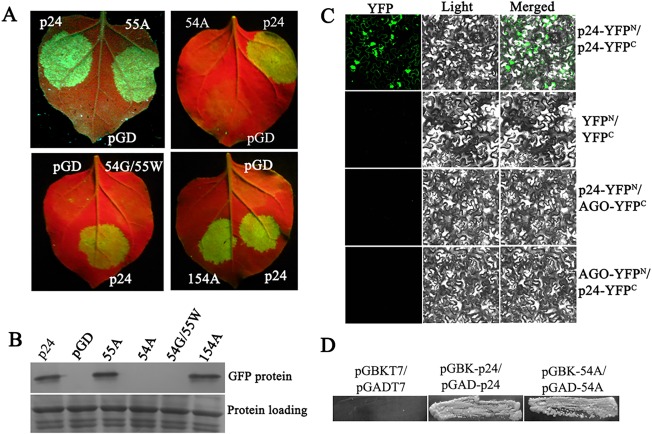
Effects of mutations in the WG/GW‐like motif of p24 on its RNA silencing suppression. (A) Co‐infiltration of Nicotiana benthamiana leaves with Agrobacterium carrying pGD–GFP and the empty vector pGD, or pGD–p24, pGD–54A, pGD–54G/55W, pGD–55A or pGD–154A. Photographs were taken as indicated in Fig. 1A. (B) Quantitative assessment of green fluorescent protein (GFP) accumulation by western blot analysis as described in Fig. 1B. (C) Bimolecular fluorescence complementation (BiFC) analysis of the interaction between p24 and AGO1 of N. benthamiana. Nicotiana benthamiana leaves were co‐infiltrated with the combination p24–YFPC/AGO–YFPN or p24–YFPN/AGO–YFPC. Pairs p24–YFPN/p24–YFPC and YFPN/YFPC were used as positive and negative controls, respectively. Yellow fluorescent protein (YFP) fluorescence images were recorded at 3 days post‐infiltration (dpi) by visualization at 488 nm. YFP, YFP fluorescence image (green); Light, bright‐field image; Merged, YFP and bright‐field overlay. (D) Yeast two‐hybrid system (YTHS) analysis of self‐interaction of the mutant 54A. Yeast AH109 cells co‐transformed with the pairs pGBK–p24/pGAD–p24 and pGBKT7/pGADT7 were used as positive and negative controls, respectively.
GW/WG repeats of some viral RSSs have been proposed to dictate their suppressor function by mediating interactions with AGO(s) (Jin and Zhu, 2010). The above results prompted us to investigate the potential interaction between p24 and AGO proteins. Plasmids AGO–YFPN and AGO–YFPC expressing AGO1 of N. benthamiana (kind gifts from Professor Chengui Han) were used in bimolecular fluorescence complementation (BiFC) analyses. Nicotiana benthamiana leaves were co‐infiltrated with Agrobacterium tumefaciens GV3101 cells harbouring the combination p24–YFPN/AGO–YFPC or p24–YFPC/AGO–YFPN. The pairs p24–YFPN/p24–YFPC (Liu et al., 2016) and YFPN/YFPC were used as positive and negative controls, respectively. Samples were examined for yellow fluorescent protein (YFP) fluorescence using spectral confocal laser scanning microscopy at 3 dpi. Strong reconstitution of YFP fluorescence was observed in N. benthamiana leaf epidermis co‐infiltrated with the positive control p24–YFPN/p24–YFPC, whereas no or only negligible fluorescence was observed in tissues infiltrated with p24–YFPN/AGO–YFPC, p24–YFPC/AGO–YFPN or the negative control (Fig. 5C). Our results show that p24 does not interact with NbAGO1 in plant cells.
As the substitution of A in W149 led to a mutant with no ability to self‐interact (Liu et al., 2016) or to suppress RNA silencing (Fig. 3), we tested whether substitution of A in W54 would also result in a loss of self‐interaction. In a yeast two‐hybrid system (YTHS), it was concluded that the mutant 54A can self‐interact (Fig. 5D).
The sequence of p24 contains three W residues, i.e. W54, W149 and W154. We substituted A into W154 to investigate its potential role in suppression activity. Mutant 154A retained the same RSS activity as the wt p24 (Fig. 5A,B), suggesting that W154 is not critical for the RSS activity of p24.
Silencing suppression‐defective mutants fail to block siRNA accumulation
To further evaluate these results, we selected representative mutants to analyse mRNA levels of GFP and GFP‐specific siRNAs. Nicotiana benthamiana leaves were co‐infiltrated with pGD–GFP and pGD–p24, pGD–2A, pGD–2K, pGD–86A, pGD–86K, pGD–149A, pGD–149Y or pGD–54A. Co‐infiltration of pGD–GFP with pGD served as the negative control. Total mRNA was then extracted from leaves co‐infiltrated with the above combinations of constructs. Northern blot analyses revealed much higher levels of GFP mRNA in leaves that expressed GFP plus wt p24, 2K, 86K or 149Y than in leaves agroinfiltrated with pGD–GFP and pGD, pGD–2A, pGD–86A, pGD–149A or pGD–54A (Fig. 6A). These results confirm those of the GFP fluorescence images and western blot results shown in Figs 3, 4, 5.
Figure 6.
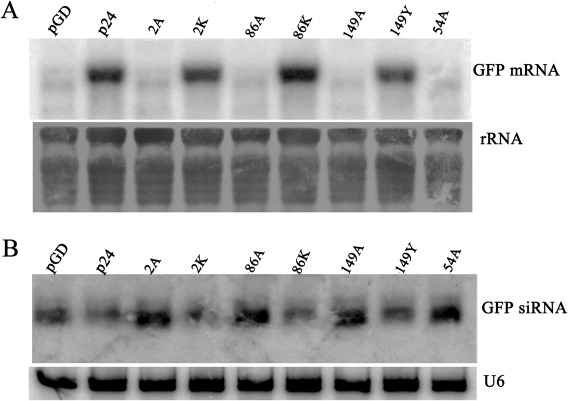
Assessment of the ability of representative p24 mutants to suppress gene silencing in Nicotiana benthamiana leaves. (A) Northern blot analysis of green fluorescent protein (GFP) mRNA extracted from the agroinfiltrated regions harvested at 3 days post‐infiltration (dpi). Blots were hybridized with probes specific for GFP mRNA. Ethidium bromide‐stained rRNA was used as an RNA loading control. (B) Northern blot analysis of siRNAs hybridized with [γ‐32P]ATP‐labelled GFP probes. U6 RNA was used as a loading control.
The levels of GFP‐specific siRNAs were also evaluated. Leaves infiltrated with pGD–GFP and pGD, pGD–2A, pGD–86A, pGD–149A or pGD–54A showed high accumulation of 21–24‐nucleotide GFP siRNA (Fig. 6B), suggesting that these silencing suppression‐defective mutants failed to affect the levels of GFP siRNA accumulation. However, the levels of the GFP siRNAs were obviously lower in leaves infiltrated with pGD–GFP plus pGD–p24, pGD–2K, pGD–86K or pGD–149Y, suggesting that wt p24 and its functional mutants 2K, 86K and 149Y can effectively block the accumulation of GFP siRNAs.
Expression of silencing suppression‐defective mutants in N. benthamiana leaves
The above silencing suppression‐defective mutants were tagged with a 3 × flag epitope at their C‐termini to investigate their expression in infiltrated N. benthamiana leaves. First, the effects of the flag tag on RSS activity were analysed. Nicotiana benthamiana leaves were co‐infiltrated with pGD–GFP and flag‐tagged wt p24 or its dysfunctional mutants 2A, 54A, 86A, 149A, 162/169/170H or 35/38/85/89H. Co‐infiltration of pGD–GFP with pGD served as the negative control. At 3 dpi, GFP fluorescence had disappeared or declined to very low levels when pGD–GFP was co‐infiltrated with flag‐tagged mutants 2A, 86A, 149A, 54A, 162/169/170H, 35/38/85/89H and the negative pGD control. In contrast, strong GFP fluorescence was observed in leaves infiltrated with the flag‐tagged p24 (Fig. 7A). Hence, the results with the flag‐tagged p24 and its mutants were entirely consistent with those of the untagged p24 and its derivatives, indicating that the flag tag does not compromise silencing suppression activity.
Figure 7.
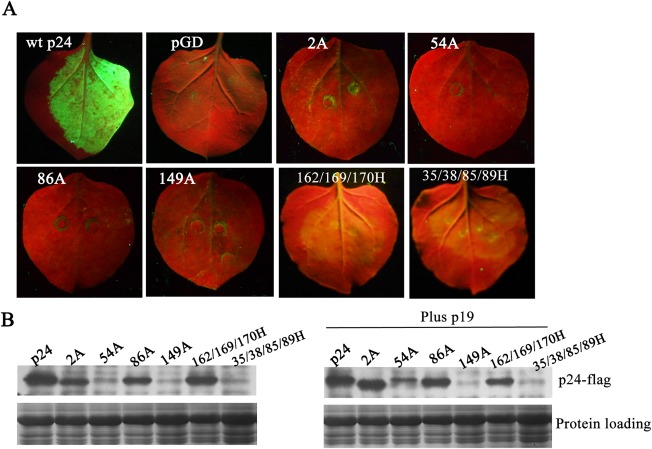
Accumulation of 3 × flag‐tagged p24 and p24 mutant derivatives in infiltrated tissues at 3 days post‐infiltration (dpi) in the presence or absence of p19 protein. (A) Tests of RNA silencing suppression for flag‐tagged p24 and p24 mutant derivatives. Nicotiana benthamiana leaves were co‐infiltrated with pGD–GFP and flag‐tagged wild‐type (wt) p24 or its dysfunctional mutants 2A, 54A, 86A, 149A, 162/169/170H and 35/38/85/89H. Photographs were taken as indicated in Fig. 1A. (B) Assessment of the accumulation of flag‐tagged p24 and p24 mutant derivatives by western blot analysis using a flag antibody. Plus p19, in the presence of Tomato bushy stunt virus (TBSV) p19.
The accumulation of the above mutants was then examined at 3 dpi in the presence or absence of TBSV p19. The accumulated protein levels of 2A and 86A were lower than those of wt p24 in the absence of p19, but reached comparable levels with those of wt p24 in the presence of p19 (Fig. 7B). The accumulation of 54A increased markedly in the presence of p19, but was still lower than that of wt p24. However, p19 did not have any beneficial effect on the accumulation of the other dysfunctional mutants—149A, 162/169/170H and 35/38/85/89H—without self‐interaction ability; among them, the accumulation of 162/169/170H was lower than that of wt p24, whereas 149A and 35/38/85/89H were only present in trace amounts (Fig. 7B).
GLRaV‐2 p24 up‐regulates the AGO1 mRNA level
To test the effects of p24 on AGO1 accumulation, a 6 × myc‐tagged version of AtAGO1 (6myc–AtAGO1, a gift from Huishan Guo) was introduced into N. benthamiana leaves by infiltration with the pGD–p24 or pGD control vector. At 2, 4, 5 and 6 dpi, extracts from co‐infiltrated leaves were analysed by western blotting. In time‐course analyses, the accumulation of 6myc–AtAGO1 in pGD–p24‐infiltrated tissues did not decrease with time (Fig. 8A), suggesting that p24 does not promote AtAGO1 degradation.
Figure 8.

Effects of p24 on protein and transcript levels of AGO1. (A) p24 does not promote argonaute 1 (AGO1) of Arabidopsis thaliana (AtAGO1). Nicotiana benthamiana leaves were co‐infiltrated with 6 × myc‐tagged AtAGO1 and pGD or pGD–p24. The accumulation of AtAGO1 was evaluated by western blotting in agroinfiltrated regions harvested at 2, 4, 5 and 6 days post‐infiltration (dpi) using anti‐myc antibody. (B) Effects of p24 and its dysfunctional mutant on the transcript level of N. benthamiana AGO1 (NbAGO1). Nicotiana benthamiana leaves were agroinfiltrated with pGD–p24–flag, pGD–2A–flag or pGD–GFP. Total RNA and protein were extracted from agroinfiltrated regions harvested at 3 and 5 dpi for the detection of the mRNA expression level of NbAGO1 by quantitative real‐time reverse transcription‐polymerase chain reaction, testing the accumulation of p24 and 2A by western blotting. Bars represent grand means ± standard deviation (SD).
We then determined the expression level of NbAGO1 mRNA in N. benthamiana leaves infiltrated with 3 × flag‐tagged p24 (pGD–p24–flag) or pGD–2A–flag by quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR), and compared it with the level in pGD–GFP‐infiltrated leaves. In parallel with the increase in p24 accumulation, NbAGO1 mRNA expression levels in p24‐expressing leaves were about 450% and 1160% of those in GFP‐expressing leaves at 3 and 5 dpi (Fig. 8B), respectively. This indicates that p24 induces the expression of NbAGO1 mRNA. The expression level of NbAGO1 mRNA in leaves expressing the dysfunctional mutant 2A showed only a moderate increase (Fig. 8B). Our results indicate that p24 induces the expression of AGO1 mRNA and that this effect is correlated with its RSS activity.
Discussion
In this study, using Agrobacterium infiltration‐mediated RNA silencing assays, we showed that p24 of GLRaV‐2 can suppress positive‐sense GFP RNA‐induced silencing, with activity comparable with the strong p19 suppressor protein of TBSV (Fig. 1). Silencing suppression by p24 effectively blocks siRNA accumulation (Fig. 1C), and the effect is correlated with its RSS activity (Fig. 6).
Binding of siRNA is a general strategy for viral suppression of RNA silencing (Lakatos et al., 2006; Mérai et al., 2006; Silhavy et al., 2002). Several unrelated viral RSSs, such as Peanut clump virus p15, Barley stripe mosaic virus γB, Carnation Italian ringspot virus p19, Tobacco etch virus HC‐Pro, CMV 2b, Influenza A virus NS1, Flock house virus B2 (Bucher et al., 2004; Goto et al., 2007; Lakatos et al., 2006; Lu et al., 2005; Mérai et al., 2006) and BYV p21 (Chapman et al., 2004), the homologue of p24, have been reported to show siRNA‐ and/or microRNA (miRNA)‐binding properties. Using substitution analyses, we found that p24 may share this strategy, because substitution of A in R2 or R86, both of which may be involved in RNA binding (Ye et al., 2003), abolished the suppressor function (Fig. 4), and additional western blotting results showed that the accumulation of the two mutants was relatively lower than that of wt p24 in the absence of p19, but reached comparable levels in the presence of p19 (Fig. 7). This suggests that the suppression inability of 2A and 86A is not a result of degradation. In addition, we observed that R2 can be replaced by basic residues H and K, and R86 can be replaced by K, suggesting that p24–siRNA interactions may be primarily electrostatic.
Deletion mutant p24 (1–188), but not p24 (1–180), showed suppressor activity (Fig. 2), and the two basic residues H and R, at positions 185 and 188, respectively, were not required for p24 suppression of RNA silencing (Fig. 4), suggesting that all predicted α‐helices and β‐strands are required for RSS activity. Our results agree with crystal structure predictions that α‐helices and β‐strands are important for the suppressive activity of some viral RSSs (Chen et al., 2008; Vargason et al., 2003; Ye et al., 2003).
Some viral RSSs have been shown to require self‐interaction to be functional (Chapman et al., 2004; Chen et al., 2008; Vargason et al., 2003; Xu et al., 2013; Ye et al., 2003). We observed that the RSS activity of p24 mutants without self‐interaction ability (Liu et al., 2016) was abolished (149A) or substantially weakened (162/169/170H and 35/38/85/89H), whereas mutants 35H, 38H, 85H and 89H, which can self‐interact, all retained RSS activity comparable with that of wt p24 (Fig. 3). Western blotting results showed that the accumulation of these three dysfunctional mutants (149A, 162/169/170H and 35/38/85/89H) does not increase when co‐expressed with p19, in contrast with the beneficial effects of p19 on the accumulation of dysfunctional mutants 2A, 86A and 54A, which can self‐interact (Fig. 7). GLRaV‐2 p24 aggregates in the cytoplasm of plant cells (Liu et al., 2016). Therefore, our results suggest that the aggregation of p24 is important for its stability and RSS function. Moreover, p24 maintained its RSS activity when W149 was substituted by two other aromatic amino acids, F or Y, but not by H (with an imidazole ring), although the three mutants (149F, 149Y and 149H) could all interact with wt p24 (Fig. 3C). These results indicate that aromatic amino acids at position 149 are essential for p24 to self‐interact, as well as to suppress RNA silencing.
Repeated GW/WG motifs are well‐known AGO‐binding platforms used by some cellular proteins, and viral RSSs p38 of Turnip crinkle virus and P1 of Sweet potato mid mottle potyvirus also use GW motifs as an AGO hook (Azevedo et al., 2010; Giner et al., 2010). However, Zhuo et al. (2014) reported that mutations in the GW/WG‐like motifs substantially weaken or abolish RSS activity of PLRV P0, although P0 does not interact with AGO1. We also found that W54 in the GW/WG‐like motif (W54/G55) is critical for p24 suppressor function, whereas p24 does not physically interact with NbAGO1 (Fig. 5C). This suggests that the important role of W54 in the silencing suppression of p24 may not be caused by its binding to AGO1. However, we cannot eliminate the possibility that W54 participates in interactions with other AGO proteins. In addition, we observed that the substitution of A into W54 does not lead to the loss of self‐interacting ability (Fig. 5D), and western blot analysis showed the beneficial effects of p19 on the accumulation of this mutant (Fig. 7B), suggesting that the involvement of W54 in the RSS activity of p24 is different from that of W149. Szabó et al. (2012) reported that the only WG/GW‐like motif in the N‐terminal domain of Sweet potato feathery mottle virus P1 does not confer RSS activity. In addition, by sequence analysis, we also noted that GW residues are not present in p24 homologues from members of the Closterovirus genus, such as BYV p21 and CTV p20. Therefore, the specific mechanism of W54/55G involvement in silencing suppression by p24 warrants further investigation.
Polerovirus silencing suppressor P0 has been reported to target AGO1 for degradation (Baumberger et al., 2007; Csorba et al., 2010; Zhuo et al., 2014). In the time‐course analysis, p24 did not promote AtAGO1 degradation (Fig. 8A). However, ectopic expression of p24 significantly up‐regulated the expression of NbAGO1 mRNA, and the effect was correlated with its RSS activity (Fig. 8B). Other viral RSSs, such as P0 of Beet western yellows virus (Bortolamiol et al., 2007) and p19 of Cymbidium ringspot virus (Várallyay et al., 2010), also have similar effects on the expression levels of miRNA‐targeted mRNAs, including AGO1 mRNA. Chapman et al. (2004) have suggested that miRNA interference may be a general property of viral RSSs, such as BYV p21, TBSV p19 and P1/HC‐Pro of Turnip mosaic virus. The increased accumulation of AGO1 mRNA in the presence of p24 suggests that p24 may also interfere with miRNA‐directed processes.
Collectively, to the best of our knowledge, this is the first report characterizing the function of GLRaV‐2 p24 protein as an RSS. However, the mechanisms whereby substitutions affect suppressor function need to be further investigated. Additional experiments should address whether p24 interacts directly with other AGOs. Moreover, transgenic plants expressing GLRaV‐2 p24 need to be produced to further investigate the effect of p24 on miRNA‐directed processes.
Experimental Procedures
Preparation of plasmids
To generate plasmids for agroinfiltrated patch silencing suppression assays, pGBK–p24 (Liu et al., 2016) was used to amplify the coding sequences of p24 and its derivatives [p24 (1–188) (contains amino acids 1–188), p24 (1–180), p24 (1–150), p24 (10–188), p24 (10–180), p24 (31–205) and p24 (151–205)] with primer pairs F1/R1, F1/R2, F1/R3, F1/R4, F2/R2, F2/R3, F3/R5 and F4/R5 (Table S1, see Supporting Information), respectively. The sequences of mutants 86A, 149A, 35H, 38H, 85H, 89H, 162/169/170H and 35/38/85/89H were PCR amplified using pGBK–86A, pGBK–35H, pGBK–38H, pGBK–85H, pGBK–89H, pGBK–149A, pGBK–162/169/170H and pGBK–35/38/85/89H (Liu et al., 2016), respectively, as templates and the primer pair F1/R1 (Table S1). To amplify the sequence of 2A, template pGBK–2A was used with primer pair F5/R1 (Table S1). The PCR products were digested with HindIII/BamHI and cloned into the binary vector pGD (Zhuo et al., 2014).
The sequences of the other p24 substitution mutants 2H, 2K, 86G, 86D, 86H, 126A, 145A, 185A, 188A, 149F, 149Y, 149H, 54A, 55A and 54G/55W were produced by reverse PCR using pGBK–p24 as the template with primer pairs F6/R6, F6/R7, F7/R8, F8/R8, F9/R8, F10/R9, F11/R10, F12/R11, F13/R12, F14/R13, F15/R13, F16/R13, F17/R14, F18/R15 and F18/R16 (Table S1), respectively. The PCR products were cloned into PMD18‐T (simple) to produce pMD–2H, pMD–2K, pMD–86G, pMD–86D, pMD–86H, pMD–126A, pMD–145A, pMD–185A, pMD–188A, pMD–149F, pMD–149H, pMD–54A, pMD–55A, pMD–54G/55W and pMD–154A, respectively. The PCR products amplified from the above recombinant plasmids with primer pair F1/R1 (except for sequences amplified from pMD–2H with F19/R1 and pMD–2K using F20/R1) were cloned into pGD.
To generate plasmids for YTHS, sequences of 54A, 149H, 149F and 149Y were PCR amplified with primer pair F21/R17 (Table S1). Products were digested with EcoRI/BamHI and cloned into pGBKT7 and pGADT7, respectively.
For western blotting analyses, sequences of p24 and its substitution mutants 149A, 86A, 54A, 162/169/170H and 35/38/85/89H were PCR amplified with primer pair F22/R18 (except 2A which was amplified using primer pair F23/R18). Products were digested with XhoI/ApaI and cloned into 3 × flag–pGAD (Zhuo et al., 2014).
Agroinfiltration and GFP imaging
Agrobacterium tumefaciens GV3101 was transformed with each plasmid. Each A. tumefaciens culture was grown to an optical density at 600 nm (OD600) of unity and mixed in equal volumes prior to infiltration. Plants were illuminated with a 100‐W hand‐held long‐wave ultraviolet lamp (UV Products, Upland, CA, USA; Black Ray model B 100AP/R) for photography, and images were taken with a Nikon (Tokyo, Japan) 4500 digital camera. All reported experiments were repeated at least three times.
Protein extraction and western analysis
Infiltrated N. benthamiana leaves were ground in liquid nitrogen and mixed with 2 × SDS sample buffer containing 10% β‐mercaptoethanol. The samples were then boiled at 100 ºC for 5 min, and centrifuged for 5 min at 13 000 g before loading on a gel. To detect GFP expression, proteins were separated by 12% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and western blot analysis was performed by probing first with a mouse anti‐GFP antibody (diluted 1 : 1000), and then incubating with anti‐mouse alkaline phosphatase (Sigma, St. Louis, USA). Finally, GFP was detected with the substrates nitroblue tetrazolium (NBT) and 5‐bromo‐4‐chloro‐3‐indolyl phosphate (BCIP) (Sigma).
For the detection of 6myc–AtAGO1 or 3 × flag–p24, proteins were separated by10% SDS‐PAGE and probed with an anti‐c‐myc antibody (1 : 1000; Sigma) or an anti‐flag antibody (1 : 1000; Sigma) followed by an alkaline phosphatase‐goat anti‐rabbit immunoglobulin G (IgG) (1 : 3000, Bio‐Rad, Hercules, CA, USA). Antibody–protein interactions were visualized using an enhanced chemiluminescence detection kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) according to the manufacturer's instructions. All experiments were repeated at least three times.
YTHS assay
YTHS tests were performed using the BD Matchmaker Library Construction and Screening kits (Clontech, Mountain View, CA, USA). The small‐scale lithium acetate transformation method was used to transform pairs of constructs simultaneously into yeast AH109 cells according to the manufacturer's protocol. Transformed yeast cells were plated on a high‐stringency synthetic dropout (SD) selection medium lacking leucine, tryptophan, adenine and histidine. Protein interaction was determined by colony growth on this medium. The plasmid combination pGBK–p24/pGAD–p24 (Liu et al., 2016) served as a positive control. All experiments were repeated at least three times, and identical results were obtained.
BiFC assay
Leaves of 4‐week‐old N. benthamiana plants were cultured in growth chambers (16 h light/8 h dark at 25–26°C) and used for agroinfiltration. Agrobacterium tumefaciens strain GV3101 carrying p24–YFPC, p24–YFPN, NbAGO1–YFPC or NbAGO1–YFPN was cultured separately and the cells were resuspended to an OD600 of 0.4–0.5 in MMA buffer (10 mm MES/NaOH, pH 5.6, 10 mm MgCl2, 150 μm acetosyringone). For co‐infiltration, equal volumes of the combinations were mixed. At 3 dpi, epidermal cells of agroinfiltrated leaves were observed for fluorescence emission under a confocal laser scanning microscope (Olympus FLUOVIEW FV1000, Tokyo, Japan) at a wavelength of 488 nm. Experiments were repeated three times.
RNA extraction and RNA gel blot analysis
Total RNA was extracted from agroinfiltrated leaves of N. benthamiana using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. For the detection of high‐molecular‐weight RNA, 15 µg of total RNA were separated on a 1% agarose–formaldehyde gel, transferred to Hybond‐N+ membranes (GE Healthcare) and hybridized with GFP probes randomly labelled with [α‐32P]dCTP using the Random Primer DNA Labelling Kit Ver. 2 (TaKaRa Bio Inc., Dalian, China). Blots were hybridized using hybridization buffer (Sigma) and washed as instructed by the manufacturer. For the detection of small RNAs, 30 µg of total RNA were separated on 15% polyacrylamide–8 m urea gels and transferred onto Hybond‐NX membrane (GE Healthcare). GFP‐specific probes were radiolabelled with [γ‐32P]ATP, and hybridizations were performed as described previously (Han et al., 2010). Experiments were repeated three times.
qRT‐PCR analysis
SYBR Green qRT‐PCR was performed with a SYBR® PrimeScript™ RT‐PCR Kit (TaKaRa Bio Inc.) according to the manufacturer's instructions. Synthesis of cDNA was primed with a mix of random primers and oligo dT provided by the kit using 500 ng of total RNA. The primer pair F25/R20 (Table S1) was used for the analysis of NbAGO1 mRNA. Nicotiana benthamiana actin mRNA served as an internal control with the primer pair F24/R19 (Table S1). qPCR analysis was conducted in an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, USA). The qPCR consisted of 10 μL of 2 × SYBR Premix Ex Taq DNA polymerase, 0.2 μL (200 nm) of each specific primer pair, 2 μL of diluted reverse‐transcribed cDNA and 0.4 μL of ROX Dye II, in a total reaction volume of 20 μL, as instructed by the manufacturer (TaKaRa Bio Inc.). Quantification was conducted according to the method described previously (Pfaffl, 2001). The experiments were conducted independently at least three times.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Table S1 Oligonucleotide primers used for polymerase chain reaction (PCR).
Acknowledgements
We thank Professor Huishan Guo (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China) for providing plasmid pCAMBIA–35S–6myc–AGO1, Professor Chengui Han (Department of Plant Pathology, China Agricultural University, Beijing, China) for providing plasmids AGO–YFPC and AGO–YFPN, and Professor Jörg Kudla (Institut für Biologie und Biotechnologie der Pflanzen, University of Münster, Münster, Germany) for providing the BiFC vectors pSPYNE–35S and pSPYCE–35S. This work was supported by the earmarked fund for Modern Agro‐Industry Technology Research System (CARS‐30‐bc‐1).
References
- Azevedo, J. , Garcia, D. , Pontier, D. , Ohnesorge, S. , Yu, A. , Garcia, S. , Braun, L. , Bergdoll, M. , Hakimi, M.A. , Lagrange, T. and Voinnet, O. (2010) Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen‐encoded GW repeat protein. Genes Dev. 24, 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baumberger, N. , Tsai, C.H. , Lie, M. , Havecker, E. and Baulcombe, D.C. (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614. [DOI] [PubMed] [Google Scholar]
- Bortolamiol, D. , Pazhouhandeh, M. , Marrocco, K. , Genschik, P. and Ziegler‐Graff, V. (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621. [DOI] [PubMed] [Google Scholar]
- Bragg, J.N. and Jackson, A.O. (2004) The C‐terminal region of the Barley stripe mosaic virus gamma protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5, 465–481. [DOI] [PubMed] [Google Scholar]
- Bucher, E. , Hemmes, H. , De Haan, P. , Goldbach, R. and Prins, M. (2004) The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85, 983–991. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J. , Prokhnevsky, A.I. , Gopinath, K. , Dolja, V.V. and Carrington, J.C. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.Y. , Yang, J. , Lin, C. and Yuan, Y.A. (2008) Structural basis for RNA‐silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, M. , Reed, J.C. , Prokhnevsky, A.I. , Chapman, E.J. , Mawassi, M. , Koonin, E.V. , Carrington, J.C. and Dolja, V.V. (2006) Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology, 346, 7–14. [DOI] [PubMed] [Google Scholar]
- Csorba, T. , Lózsa, R. , Hutvágner, G. and Burgyán, J. (2010) Polerovirus protein P0 prevents the assembly of small RNA‐containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472. [DOI] [PubMed] [Google Scholar]
- Díaz‐Pendón, J.A. and Ding, S.W. (2008) Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. (2010) RNA‐based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja, V.V. (2003) Beet yellows virus: the importance of being different. Mol. Plant Pathol. 4, 91–98. [DOI] [PubMed] [Google Scholar]
- Duan, C.G. , Fang, Y.Y. , Zhou, B.J. , Zhao, J.H. , Hou, W.N. , Zhu, H. , Ding, S.W. and Guo, H.S. (2012) Suppression of Arabidopsis ARGONAUTE1‐mediated slicing, transgene‐induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell, 24, 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem‐Sabanadzovic, N.A. , Sabanadzovic, S. , Castellano, M.A. , Boscia, D. and Martelli, G.P. (2000) Properties of a new isolate of grapevine leafroll‐associated virus 2. Vitis, 39, 119–121. [Google Scholar]
- Giner, A. , Lakatos, L. , García‐Chapa, M. , López‐Moya, J.J. and Burgyán, J. (2010) Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6, e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, I. , Martínez, L. , Rakitina, D.V. , Lewsey, M.G. , Atencio, F.A. , Llave, C. , Kalinina, N.O. , Carr, J.P. , Palukaitis, P. and Canto, T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant–Microbe Interact. 23, 294–303. [DOI] [PubMed] [Google Scholar]
- Goodin, M.M. , Dietzgen, R.G. , Schichnes, D. , Ruzin, S. and Jackson, A.O. (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383. [DOI] [PubMed] [Google Scholar]
- Goszczynski, D.E. , Kasdorf, G.G.F. , Pietersen, G. and van Tonder, H. (1996) Detection of two strains of Grapevine leafroll‐associated virus 2 . Vitis, 35, 133–135. [Google Scholar]
- Goto, K. , Kobori, T. , Kosaka, Y. , Natsuaki, T. and Masuta, C. (2007) Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA‐binding abilities. Plant Cell Physiol. 48, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Hamera, S. , Song, X. , Su, L. , Chen, X. and Fang, R. (2012) Cucumber mosaic virus suppressor 2b binds to AGO4‐related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J. and Baulcombe, D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Han, Y.H. , Xiang, H.Y. , Wang, Q. , Li, Y.Y. , Wu, W.Q. , Han, C.G. , Li, D.W. and Yu, J.L. (2010) Ring structure amino acids affect the suppressor activity of melon aphid‐borne yellows virus P0 protein. Virology, 406, 21–27. [DOI] [PubMed] [Google Scholar]
- Jin, H. and Zhu, J.K. (2010) A viral suppressor protein inhibits host RNA silencing by hooking up with Argonautes. Genes Dev. 24, 853–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, L.K. and Carrington, J.C. (2001) Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium‐mediated transient expression system. Plant Physiol. 126, 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasev, A.V. (2000) Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38, 293–324. [DOI] [PubMed] [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y.P. , Dolja, V.V. , Calvino, L.F. , López‐Moya, J.J. and Burgyán, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Guo, R. , Li, M. , Feng, M. , Wang, X. , Wang, Q. and Cheng, Y. (2016) Critical regions and residues for self‐interaction of grapevine leafroll‐associated virus 2 protein p24. Virus Res. 15, 57–63. [DOI] [PubMed] [Google Scholar]
- Liu, Y.P. , Peremyslov, V.V. , Medina, V. and Dolja, V.V. (2009) Tandem leader proteases of Grapevine leafroll‐associated virus 2: host‐specific functions in the infection cycle. Virology, 383, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Folimonov, A. , Shintaku, M. , Li, W.X. , Falk, B.W. , Dawson, W.O. and Ding, S.W. (2004) Three distinct suppressors of RNA silencing encoded by a 20‐kb viral RNA genome. Proc. Natl. Acad. Sci. USA, 101, 15 742–15 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Maduro, M. , Li, F. , Li, H.W. , Broitman‐Maduro, G. , Li, W.X. and Ding, S.W. (2005) Animal virus replication and RNAi‐mediated antiviral silencing in Caenorhabditis elegans . Nature, 436, 1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C.D. and Hannon, G.J. (2009) Small RNAs as guardians of the genome. Cell, 136, 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, B. , Li, C. , Goszczynski, D.E. and Gonsalves, D. (2005) Genome sequences and structures of two biologically distinct strains of Grapevine leafroll‐associated virus 2 and sequence analysis. Virus Genes, 31, 31–41. [DOI] [PubMed] [Google Scholar]
- Mérai, Z. , Kerényi, Z. , Kertész, S. , Magna, M. , Lakatos, L. and Silhavy, D. (2006) Double‐stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 80, 5747–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot, A. , Valli A., Rodamilans, B. , San León, D. , Baulcombe, D.C. , García, J.A. and López‐Moya, J.J. (2016) The P1N‐PISPO trans‐Frame Gene of Sweet Potato Feathery Mottle Potyvirus Is Produced during Virus Infection and Functions as an RNA Silencing Suppressor. J. Virol. 90, 3543–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.C. , Kasschau, K.D. , Prokhnevsky, A.I. , Gopinath, K. , Pogue, G.P. , Carrington, J.C. and Dolja, V.V. (2003) Suppressor of RNA silencing encoded by Beet yellows virus. Virology, 306, 203–209. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ferrer, V. , Boskovic, J. , Alfonso, C. , Rivas, G. , Llorca, O. , López‐Abella, D. and López‐Moya, J.J. (2005) Structural analysis of tobacco etch potyvirus HC‐pro oligomers involved in aphid transmission. J. Virol. 79, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Kwon, S.J. and Rao, A.L. (2012) Molecular dissection of Flock house virus protein B2 reveals that electrostatic interactions between N‐terminal domains of B2 monomers are critical for dimerization. Virology, 25, 296–305. [DOI] [PubMed] [Google Scholar]
- Silhavy, D. , Molnár, A. , Lucioli, A. , Szittya, G. , Hornyik, C. , Tavazza, M. and Burgyán, J. (2002) A viral protein suppresses RNA silencing and binds silencing‐generated, 21‐ to 25‐nucleotide double‐stranded RNAs. EMBO J. 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E.J. (2005) Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell. Biol. 6, 127–138. [DOI] [PubMed] [Google Scholar]
- Szabó, E.Z. , Manczinger, M. , Göblös, A. , Kemény, L. and Lakatos, L. (2012) Switching on RNA silencing suppressor activity by restoring argonaute binding to a viral protein. J. Virol. 86, 8324–8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untiveros, M. , Olspert, A. , Artola, K. , Firth, A.E. , Kreuze, J.F. and Valkonen, J.P. (2016) A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol. Plant Pathol. 17, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várallyay, E. , Válóczi, A. , Agyi, A. , Burgyán, J. and Havelda, Z. (2010) Plant virus‐mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M. , Szittya, G. , Burgyán, J. and Hall, T.M. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 26, 799–811. [DOI] [PubMed] [Google Scholar]
- Xu, A. , Zhao, Z. , Chen, W. , Zhang, H. , Liao, Q. , Chen, J.P. and Du, Z. (2013) Self‐interaction of the cucumber mosaic virus 2b protein plays a vital role in the suppression of RNA silencing and the induction of viral symptoms. Mol. Plant Pathol. 14, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K. and Patel, D.J. (2005) RNA silencing suppressor p21 of Beet yellows virus forms an RNA binding octameric ring structure. Structure, 13, 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K. , Malinina, L. and Patel, D.J. (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature, 18, 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Wu, Z. , Li, Y. and Wu, J. (2015) Biogenesis, function, and applications of virus‐derived small RNAs in plants. Front Microbiol. 6, 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Du, P. , Lu, L. , Xiao, Q. , Wang, W. , Cao, X. , Ren, B. , Wei, C. and Li, Y. (2008) Contrasting effects of HC‐Pro and 2b viral suppressors from Sugarcane mosaic virus and Tomato aspermy cucumovirus on the accumulation of siRNAs. Virology, 374, 351–360. [DOI] [PubMed] [Google Scholar]
- Zhu, H.Y. , Ling, K.S. , Goszczynski, D.E. , McFerson, J.R. and Gonsalves, D. (1998) Nucleotide sequence and genome organization of grapevine leafroll‐associated virus‐2 are similar to beet yellows virus, the closterovirus type member. J. Gen. Virol. 79, 1289–1298. [DOI] [PubMed] [Google Scholar]
- Zhuo, T. , Li, Y.Y. , Xiang, H.Y. , Wu, Z.Y. , Wang, X.B. , Wang, Y. , Zhang, Y.L. , Li, D.W. , Yu, J.L. and Han, C.G. (2014) Amino acid sequence motifs essential for P0‐mediated suppression of RNA silencing in an isolate of Potato leafroll virus from Inner Mongolia. Mol. Plant–Microbe Interact. 27, 515–527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Table S1 Oligonucleotide primers used for polymerase chain reaction (PCR).


