Summary
Bacillus amyloliquefaciens FZB42 is a type of plant growth‐promoting rhizobacterium (PGPR) which activates induced systemic resistance (ISR) in Arabidopsis. Blocking of the synthesis of cyclic lipopeptides and 2,3‐butanediol by FZB42, which have been demonstrated to be involved in the priming of ISR, results in the abolishment of the plant defence responses. To further clarify the ISR activated by PGPRs at the microRNA (miRNA) level, small RNA (sRNA) libraries from Arabidopsis leaves after root irrigation with FZB42, FZB42ΔsfpΔalsS and control were constructed and sequenced. After fold change selection, promoter analysis and target prediction, miR846‐5p and miR846‐3p from the same precursor were selected as candidate ISR‐associated miRNAs. miR846 belongs to the non‐conserved miRNAs, specifically exists in Arabidopsis and its function in the plant defence response remains unclear. The disease severity of transgenic Arabidopsis overexpressing miR846 (OEmiR846) or knockdown miR846 (STTM846) against Pseudomonas syringae DC3000 suggests that the miR846 expression level in Arabidopsis is negatively correlated with disease resistance. Moreover, miR846 in Arabidopsis Col‐0 is repressed after methyl jasmonate treatment. In addition, jasmonic acid (JA) signalling‐related genes are up‐regulated in STTM846, and the stomatal apertures of STTM846 are also less than those in Arabidopsis Col‐0 after methyl jasmonate treatment. Furthermore, the disease resistance of STTM846 transgenic Arabidopsis against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) is blocked by the addition of the JA biosynthetic inhibitor diethyldiethiocarbamic acid (DIECA). Taken together, our results suggest that B. amyloliquefaciens FZB42 inoculation suppresses miR846 expression to induce Arabidopsis systemic resistance via a JA‐dependent signalling pathway.
Keywords: Bacillus amyloliquefaciens FZB42, induced systemic resistance, jasmonic acid, miRNA, sRNA sequence
Introduction
Plant growth‐promoting rhizobacteria (PGPRs) are naturally occurring soil microorganisms that benefit plants by increasing plant growth or reducing disease. The PGPR Bacillus amyloliquefaciens FZB42 is an environmental strain that can successfully colonize the roots of plants, such as Arabidopsis, stimulate plant growth and enhance the defence response (Chowdhury et al., 2015a; Fan et al., 2012). Analysis of the whole FZB42 genome revealed a potential capability to produce several secondary metabolites, such as lipopeptides (surfactin, iturins, fengycins) and polyketides (macrolactin, bacillaene and difficidin) (Chen et al., 2007). Many secondary metabolites, such as surfactin and 2,3‐butanediol, have been confirmed to suppress pathogen growth and to activate the induced systemic resistance process (Cawoy et al., 2014; Ryu et al., 2004).
Plants have developed various mechanisms to protect themselves against bacterial, fungal and viral infection. These defence systems include pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Chisholm et al., 2006; Jones and Dangl, 2006). The outcome of PTI and ETI can initiate the hypersensitive response (HR) in local tissues and systemic acquired resistance (SAR) in distal tissues (Thomma et al., 2011). SAR is often associated with increased levels of salicylic acid (SA) which coordinate the activation of pathogenesis‐related (PR) genes (Fu and Dong, 2013). This systemic resistance can also be activated by beneficial microbes, such as Pseudomonas spp. and Bacillus spp., which is called induced systemic resistance (ISR) (Lugtenberg and Kamilova, 2009). ISR was discovered by the finding that resistance could be induced by the rhizobacterium Pseudomonas sp. strain WCS417r against Fusarium wilt of carnation (Van Peer et al., 1991). In many cases, ISR is often dependent on jasmonic acid (JA) and ethylene (ETH) signalling rather than SA signalling in plants. Several studies have shown that volatile organic compounds and cyclic lipopeptides, such as surfactin and fengycin, produced by PGPRs are capable of eliciting ISR in plants (Chowdhury et al., 2015b; Ryu et al., 2004). For example, massetolide A produced by Pseudomonas fluorescens SS101 is involved in ISR‐eliciting activity in tomato against Phytophthora infestans (Tran et al., 2007). Purified fengycins and surfactins, but not iturins, induce significant protection in bean and tomato leaves against the fungal pathogen Botrytis cinerea (Ongena et al., 2007). Treatment of tobacco cell suspensions with low concentrations of surfactin induces several early plant defence‐related events, such as Ca2+‐dependent extracellular alkalinization and oxidative burst, without causing any significant cell death or any marked toxicity (Jourdan et al., 2009). 2,3‐Butanediol released from Bacillus subtilis GB03 and B. amyloliquefaciens IN937a significantly increases plant resistance against Pectobacterium carotovorum ssp. carotovorum (Ryu et al., 2004).
MicroRNAs (miRNAs) are short, non‐coding, 20–24‐nt small RNAs (sRNAs) and have been demonstrated to play an important role in plant immunity. For example, flg22 induces the accumulation of miR393, which contributes to plant resistance against bacteria by negatively regulating the mRNA level of F‐box auxin receptors TIR1, AFB2 and AFB3 (Navarro et al., 2006). In addition to miR393, miR167 and miR160, which target auxin‐response factors (ARFs), are also induced by Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) hrcC–, a strain with a mutation in the type III secretion system (TTSS) that still triggers robust PTI in Arabidopsis (Fahlgren et al., 2007). The Arabidopsis miRNA‐deficient mutants dcl1 and hen1 display enhanced growth of the bacterium Pst DC3000 hrcG, indicating that the proteins required for sRNA biogenesis and function are also required for disease resistance to bacterial pathogens (Navarro et al., 2008). In addition, sRNAs can also trigger plant ETI by regulating resistance (R) genes, such as miR482, miR2109 and miR6019/miR6020 (Baldrich and Segundo, 2016; Li et al., 2012). Previous studies have largely focused on miRNAs involved in plant systemic resistance against pathogens. Relatively fewer studies have focused on the role of miRNAs during plant–rhizobacteria interactions. De Luis et al. (2012) reported that the legume Lotus japonicas miR171c and miR397 are specifically linked to bacterial infection and nodule function rather than organogenesis. In another study, Bacillus cereus AR156 pretreatment has been shown to prime ISR to Pst DC3000 infection by suppression of miR825 and miR825* and activation of their target toll‐interleukin‐like receptor‐nucleotide‐binding site (TIR‐NBS) and leucine‐rich repeat (LRR)‐type R genes (Niu et al., 2016). Bacillus, Arabidopsis and Pst DC3000 were introduced into the same research system; however, studies on the interaction between Bacillus and plants are needed to explore the miRNAs involved in the ISR process.
In this study, we constructed the Bacillus amyloliquefaciens FZB42ΔsfpΔalsS mutant, which is deficient in the production of cyclic lipopeptides and 2,3‐butanediol compounds. Blocking of the synthesis of these compounds resulted in the abolishment of ISR activation. To identify plant sRNAs involved in the ISR process, we analysed Arabidopsis sRNAs by Illumina Hiseq deep sequencing. The results indicated that miR846‐5p and miR846‐3p were differentially repressed by wild‐type FZB42, whereas their expression showed no significant difference between FZB42ΔsfpΔalsS treatment and control. Transgenic Arabidopsis overexpressing miR846 was more susceptible to Pst DC3000 infection, whereas miR846 knockdown in transgenic Arabidopsis showed resistance to Pst DC3000. Furthermore, miR846 was confirmed to modulate ISR through the JA‐dependent signalling pathway.
Results
Construction of ISR‐deficient FZB42ΔsfpΔalsS mutant
Cyclic lipopeptides, such as surfactin, and volatile compounds, such as 2,3‐butanediol, produced by Bacillus have been confirmed to be elicitors of ISR in plants (Chowdhury et al., 2015b; Ryu et al., 2004). The genes sfp (encoding phosphopantetheinyl transferase) and alsS (encoding acetolactate dehydrogenase) are involved in the synthesis of cyclic lipopeptides and 2,3‐butanediol, respectively. In order to prepare an ISR‐deficient mutant, we constructed a site‐directed mutant of sfp and alsS genes in FZB42 by double‐crossover homologous recombination. The disruption of the sfp gene abolishes the production of cyclic lipopeptides, such as surfactin, which has haemolytic activity. The haemolytic activity of the FZB42ΔsfpΔalsS mutant disappeared as we expected, and polymerase chain reaction (PCR) amplification also validated that the double mutant constructed was successful (Fig. S1, see Supporting Information).
We then tested the ISR‐eliciting activity of the FZB42ΔsfpΔalsS mutant through the oxidative burst, stomatal closure, disease severity and disease resistance‐related gene expression. FZB42 root inoculation caused Arabidopsis stomatal closure and H2O2 accumulation, whereas FZB42ΔsfpΔalsS root inoculation showed no difference compared with the control (Fig. 1A,B). Moreover, FZB42ΔsfpΔalsS‐treated Arabidopsis showed the same disease severity as the control, and FZB42‐treated Arabidopsis exhibited an increased disease resistance against Pst DC3000 (Fig. 1C). The expression of pathogenesis‐related protein (PR1), plant defence factor 1.2 (pdf1.2) and ETH response factor 1 (ERF1) genes, involved in the SA‐, JA‐ and ETH‐dependent defence signalling pathways in Arabidopsis, were also analysed. The expression of PR1, pdf1.2 and ERF1 in Arabidopsis was enhanced after root inoculation with FZB42 for 12 and 24 h; in contrast their expression levels showed no significant changes in response to FZB42ΔsfpΔalsS compared with the control (Fig. 1D). These results indicate that the FZB42ΔsfpΔalsS mutant is deficient in ISR activation.
Figure 1.
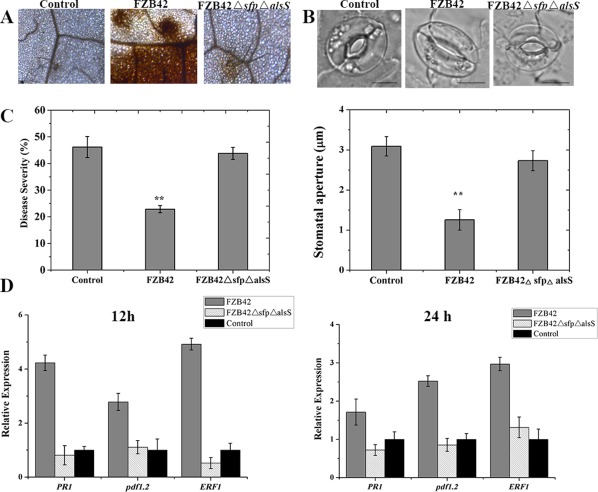
Hydrogen peroxide (A), stomatal aperture (B), disease severity (C) and expression of disease resistance‐related genes (D) in Arabidopsis Col‐0 leaves after inoculation with Bacillus amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. (A) Detection of hydrogen peroxide in Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control by 3,3′‐diaminobenzidine (DAB) staining. Brown precipitates represent hydrogen peroxide generation. (B) Stomatal apertures of Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. Scale bars indicate 10 μm. Values represent the means of 50 random selected stomata. All experiments were repeated three times. **Highly significant difference (P < 0.01). (C) Disease severity of Arabidopsis against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control. Four‐week‐old Arabidopsis Col‐0 plants were inoculated with cell suspensions of FZB42, FZB42ΔsfpΔalsS and water control. Five days later, leaves were sprayed with a cell suspension of Pst DC3000 at 108 colony‐forming units (CFU)/mL. The disease severity was determined according to the disease index measured at 7 days post‐inoculation (dpi). Each treatment had 12 plants and the experiment was repeated three times. (D) Expression levels of disease resistance‐related genes in Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, the FZB42ΔsfpΔalsS mutant and control for 12 and 24 h. Pathogenesis‐related protein (PR1), plant defence factor 1.2 (PDF 1.2) and ethylene response factor 1 (ERF1) genes, involved in the salicylic acid‐, jasmonic acid‐ and ethylene‐dependent defence signalling pathways in Arabidopsis. Data are expressed as the relative expression of the respective mRNAs normalized to the endogenous actin 2. Error bars represent significant differences according to Fisher's least‐significant difference test (P = 0.05) using SPSS software. The experiment was repeated three times.
Identification of candidate ISR‐associated miRNAs
To identify ISR‐associated miRNAs, three sRNA libraries were constructed: Arabidopsis leaves after root irrigation with FZB42, FZB42ΔsfpΔalsS (mutant deficient in ISR activation) or control. sRNAs ranging from 18 to 30 nucleotides (nt) were isolated from these tissues and sequenced using Illumina. Deep sequencing data revealed 1158k, 1117k and 1141k total reads from the FZB42‐, FZB42ΔsfpΔalsS‐ and control‐treated Arabidopsis leaves, respectively (Table S1, see Supporting Information). In addition, the class of sRNA with a length of 21 nt was the most abundant amongst the three sRNA libraries (Fig. S2, see Supporting Information). Most sRNAs ranging from 18 to 30 nt predominantly favoured a U (uridine) at the 5′ end (Fig. S3, see Supporting Information). Moreover, 252 known miRNAs and 19 novel miRNAs were identified in Arabidopsis.
We first screened the differentially expressed miRNAs regarding ISR following the selection criteria Q value < 0.01 and |log2(fold change)| > 1. Using a two‐fold change cut‐off, we found that 29 miRNAs were differentially expressed (Table 1). Amongst these differentially expressed miRNAs, three miRNAs were selected for expression analysis using stem‐loop quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) (Chen et al., 2005). The expression of miR173‐3p, miR846‐5p and miR846‐3p was repressed, consistent with the sRNA sequencing data, which confirmed the reliability of the sRNA sequence (Fig. S4, see Supporting Information). The cross‐sections of differentially expressed miRNAs between FZB42 versus FZB42ΔsfpΔalsS and FZB42 versus control were analysed and profiled, because we speculated that ISR‐associated miRNAs would be differentially expressed in FZB42‐treated Arabidopsis versus FZB42ΔsfpΔalsS‐ and control‐treated Arabidopsis. According to the criteria, six suppressed miRNAs (miR173‐3p, miR1888a, miR5651, miR781a, miR846‐5p and miR846‐3p) and one induced miRNA (miR850) were identified (Fig. 2A).
Table 1.
Identified microRNAs (miRNAs) with significant differential expression.
| miR name | miR sequence |
|
|
|
|||
|---|---|---|---|---|---|---|---|
| miR158a‐5p | CUUUGUCUACAAUUUUGGAAA | −1.6922 | 1.1791 | ||||
| miR159c | UUUGGAUUGAAGGGAGCUCCU | 1.2171 | 2.2209 | ||||
| miR166e‐5p | GGAAUGUUGUCUGGCACGAGG | 1.0465 | |||||
| miR167d | UGAAGCUGCCAGCAUGAUCUGG | 1.0657 | |||||
| miR172a | AGAAUCUUGAUGAUGCUGCAU | −1.3662 | |||||
| miR172b‐5p | GCAGCACCAUUAAGAUUCAC | 1.106 | |||||
| miR172c | AGAAUCUUGAUGAUGCUGCAG | −1.1988 | |||||
| miR173‐3p | UGAUUCUCUGUGUAAGCGAAA | −2.0867 | −1.4266 | ||||
| miR1888a | UAAGUUAAGAUUUGUGAAGAA | −1.7632 | −2.1491 | ||||
| miR393a‐5p | UCCAAAGGGAUCGCAUUGAUCC | −1.6796 | |||||
| miR3932b‐5p | UUUGACGUGCUCGAUCUGCUC | 1.5155 | 1.4638 | ||||
| miR396a‐5p | UUCCACAGCUUUCUUGAACUG | 1.0794 | |||||
| miR398b‐3p | UGUGUUCUCAGGUCACCCCUG | 1.4839 | |||||
| miR399d | UGCCAAAGGAGAUUUGCCCCG | −1.0753 | |||||
| miR447a.2–3p | UAUGGAAGAAAUUGUAGUAUU | −1.2288 | |||||
| miR5651 | UUGUGCGGUUCAAAUAGUAAC | −1.2507 | −1.5276 | ||||
| miR5996 | UGACAUCCAGAUAGAAGCUUUG | −2.0714 | |||||
| miR781a | UUAGAGUUUUCUGGAUACUUA | −1.5449 | −1.7001 | ||||
| miR8183 | UUUAGUUGACGGAAUUGUGGC | 2.3745 | |||||
| miR824‐3p | CCUUCUCAUCGAUGGUCUAGA | 1.0624 | |||||
| miR824‐5p | UAGACCAUUUGUGAGAAGGGA | −1.1461 | |||||
| miR828 | UCUUGCUUAAAUGAGUAUUCCA | −1.3346 | |||||
| miR840‐5p | ACACUGAAGGACCUAAACUAAC | −1.4847 | |||||
| miR843 | UUUAGGUCGAGCUUCAUUGGA | 1.2199 | |||||
| miR844‐3p | UUAUAAGCCAUCUUACUAGUU | −1.4455 | |||||
| miR846‐5p | CAUUCAAGGACUUCUAUUCAG | −1.1414 | −1.4858 | ||||
| miR846‐3p | UUGAAUUGAAGUGCUUGAAUU | −1.1029 | −1.0363 | ||||
| miR850 | UAAGAUCCGGACUACAACAAAG | 1.2239 | 1.0209 | ||||
| novel_36 | AUGAAGUGAUGAUUGAACU | −1.4007 |
Figure 2.
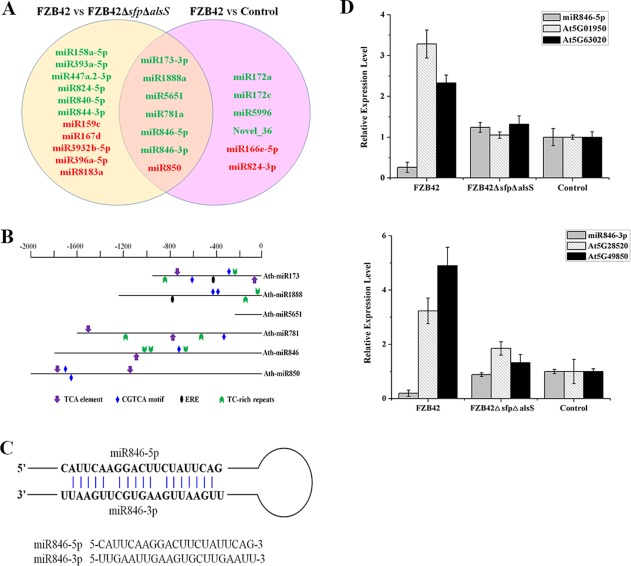
miR846‐5p and miR846‐3p are induced systemic resistance (ISR)‐associated microRNAs (miRNAs) after inoculation with Bacillus amyloliquefaciens FZB42. (A) Venn diagram of differentially expressed miRNAs between FZB42 and FZB42ΔsfpΔalsS and FZB42 and control. Red represents up‐regulated miRNAs and green represents down‐regulated miRNAs. (B) Cis‐elements in the promoter regions of differentially expressed miRNAs. The defence‐responsive cis‐elements distributed on the sense strand and reverse strand are shown above and below the black lines, respectively. TCA, CGTCA and ERE elements represent cis‐acting elements involved in salicylic acid, jasmonic acid and ethylene responsiveness, respectively. TC‐rich repeat represents cis‐acting element involved in defence responsiveness. (C) Secondary stem‐loop structure for miR846‐5p and miR846‐3p with mature miRNA sequences indicated. (D) Expression levels of miR846‐5p, miR846‐3p and their target genes (At5G01950 and At5G63020 for miR846‐5p; At5G49850 and At5G28520 for miR846‐3p) from Arabidopsis leaves after inoculation with B. amyloliquefaciens FZB42, FZB42ΔsfpΔalsS and control. Data are expressed as the relative expression of the respective mRNAs normalized to the endogenous actin 2. The experiment was repeated three times. Error bars represent significant differences according to Fisher's least‐significant difference test (P = 0.05) using SPSS software.
Next, the potential defence‐responsive elements in the promoters of ISR‐associated miRNAs were also investigated. First, the promoters of pri‐miRNA were obtained according to Ding et al. (2011). Cis‐acting elements in the miRNA promoters were then searched for by PlantCARE. The TCA element, CGTCA motif and ERE element involved in SA, JA and ETH responsiveness, as well as TC‐rich repeats involved in defence responsiveness. were recruited, and their distribution patterns were characterized (Fig. 2B). These defence‐related cis‐acting elements were present in the promoters of miR173, miR1888, miR781, miR846 and miR850.
We further predicted the putative targets of the above miRNAs by psRobot (Table 2). LRR genes (AT5G01950 and AT5G63020) and jacalin lectin genes (At5G49850 and At5G28520) were predicted as the targets of miR846‐5p and miR846‐3p, respectively. Interestingly, miR846‐5p and miR846‐3p originated from the same precursor (Fig. 2C). Thus, miR846‐5p and miR846‐3p were the most outstanding miRNAs that might play a role in ISR activation. The expression of miR846‐5p and miR846‐3p was suppressed in FZB42‐treated Arabidopsis, whereas their expression levels were not altered in FZB42ΔsfpΔalsS‐ and control‐treated Arabidopsis (Fig. S4). Jacalin lectin family genes (At5G49850 and At5G28520) were confirmed as the target genes of miR846‐3p (Fahlgren et al., 2007; Jia and Rock, 2013a). The expression levels of jacalin‐related lectin (JRL) genes (At5G49850 and At5G28520) and LRR genes (At5G01950 and At5G63020) were accordingly induced in FZB42‐treated Arabidopsis (Fig. 2D). Many JRL genes have been shown to be associated with disease resistance, abiotic stress signalling, wounding insect damage or multiple stresses (Song et al., 2014). Hence, our functional study of ISR‐associated miRNAs focused mostly on miR846.
Table 2.
Predicted targets of microRNAs (miRNAs) in Arabidopsis.
| miR name | Accession no. of miRNA targets | Protein annotation |
|---|---|---|
| miR173‐3p | AT1G53600 | Pentatricopeptide repeat‐containing protein |
| AT4G38960 | B‐box type zinc finger family protein | |
| miR1888a | AT1G18090 | 5′–3′ exonuclease family protein |
| AT1G28620 | Pseudo | |
| AT1G35740 | Pseudo | |
| AT1G53630 | Pseudo | |
| miR781a | AT1G44900 | Minichromosome maintenance protein 2 |
| AT1G52820 | 2‐Oxoglutarate‐dependent dioxygenase | |
| AT1G69490 | NAC transcription factor protein family | |
| AT2G37760 | Aldo–keto reductase family 4 member C8 | |
| AT5G23480 | SWIB/MDM2, Plus‐3 and GYF domain‐containing protein | |
| miR846‐5p | AT5G01950 | Leucine‐rich repeat protein kinase‐like protein |
| AT5G63020 | CC‐NBS‐LRR class disease resistance protein | |
| miR846‐3p | AT1G33790 | Jacalin lectin family protein |
| AT1G52050 | Jacalin‐like lectin domain‐containing protein | |
| AT1G52060 | Jacalin‐like lectin domain‐containing protein | |
| AT1G52070 | Jacalin‐like lectin domain‐containing protein | |
| AT1G52110 | Jacalin‐like lectin domain‐containing protein | |
| AT1G52130 | Mannose‐binding lectin‐like protein | |
| AT1G57570 | Jacalin‐like lectin domain‐containing protein | |
| AT1G60095 | Jacalin‐like lectin domain‐containing protein | |
| AT1G60110 | Jacalin‐like lectin domain‐containing protein | |
| AT1G61230 | Jacalin‐like lectin domain‐containing protein | |
| AT2G25980 | Myrosinase‐binding protein‐like protein | |
| AT2G42270 | U5 small nuclear ribonucleoprotein helicase | |
| AT3G60730 | Putative pectinesterase/pectinesterase inhibitor 36 | |
| AT5G28520 | Mannose‐binding lectin‐like protein | |
| AT5G38540 | Mannose‐binding lectin superfamily protein | |
| AT5G38550 | Jacalin lectin family protein | |
| AT5G49850 | Jacalin lectin family protein | |
| AT5G49870 | Mannose‐binding lectin superfamily protein | |
| miR850 | AT3G28007 | Nodulin MtN3 family protein |
| AT2G43240 | Nucleotide‐sugar transporter family protein |
CC‐NBS‐LRR, coiled coil‐nucleotide‐binding site‐leucine‐rich repeat.
Expression level of miR846 is negatively correlated with plant resistance ability
To investigate the function of miR846, we generated transgenic Arabidopsis overexpressing miR846 (OEmiR846) or with knockdown of miR846 (STTM846). We first investigated the miR846 expression level in OEmiR846 and STTM846 to validate transgenic Arabidopsis. The miR846 expression levels in OEmiR846 and STTM846 were higher and lower, respectively, than in wild‐type Col‐0, which confirmed the correctness of the constructed OEmiR846 and STTM846 transgenic Arabidopsis. In addition, the expression of target genes (At5G49850 and At5G28520 for miR846‐3p; At5G01950 and At5G63020 for miR846‐5p) was repressed in OEmiR846 transgenic Arabidopsis, and the target genes were up‐regulated in STTM846 transgenic Arabidopsis (Fig. 3A). STTM846 transgenic Arabidopsis had a smaller stature than the wild‐type, whereas OEmiR846 transgenic Arabidopsis had a larger stature (Fig. 3B). We further detected the disease resistance of transgenic Arabidopsis against Pst DC3000. The disease severity and Pst DC3000 density in OEmiR846 were significantly higher than those in the wild‐type. In contrast, the disease severity and Pst DC3000 density in STTM846 were less than those in the wild‐type (Fig. 3C,D). Evans blue staining showed similar results (Fig. 3E). We also detected the expression levels of disease resistance‐related genes in OEmiR846 and STTM846. The disease resistance‐related gene pdf1.2 was significantly induced in STTM846 (Fig. 3F). Taken together, these results indicate that the expression level of miR846 in Arabidopsis is negatively correlated with plant disease resistance.
Figure 3.
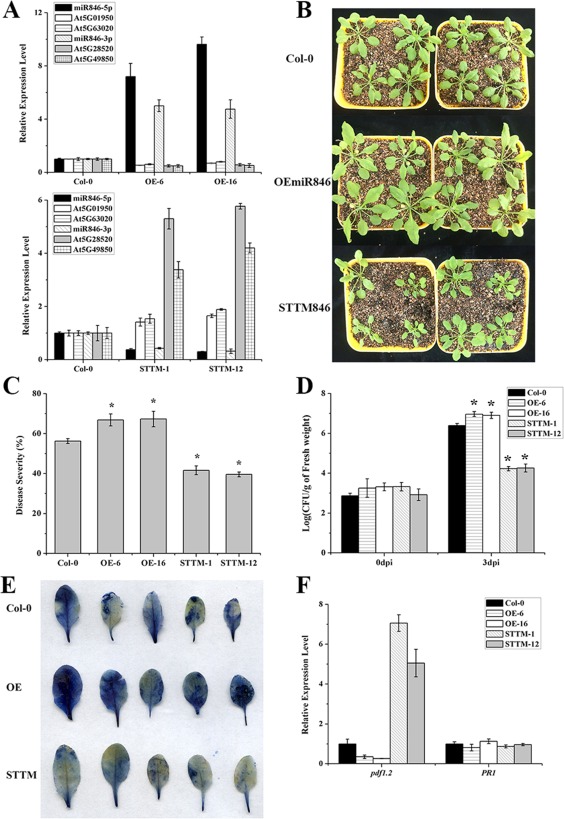
The miR846 expression level in Arabidopsis is negatively correlated with disease resistance. (A) Expression levels of miR846‐5p/miR846‐3p and their target genes were detected in transgenic Arabidopsis overexpressing miR846‐5p/miR846‐3p (OEmiR846) or with knockdown of miR846‐5p/miR846‐3p (STTM846) by stem‐loop quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The leucine‐rich repeat genes (AT5G01950 and AT5G63020) were predicted as the targets of miR846‐5p. The jacalin lectin genes (At5G49850 and At5G28520) were predicted as the targets for miR846‐3p. (B) Phenotypes of transgenic Arabidopsis OEmiR846 and STTM846. Photographs were taken 4 weeks after germination. (C) Disease severity of transgenic Arabidopsis OEmiR846 and STTM846 after Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) infection for 7 days. *Highly significant difference (P < 0.05). (D) Quantification of Pst DC3000 growth in transgenic Arabidopsis OEmiR846 and STTM846. *Highly significant difference (P < 0.05). CFU, colony‐forming unit; dpi, days post‐inoculation. (E) Cell death in transgenic Arabidopsis OEmiR846 and STTM846 leaves after Pst DC3000 infection for 7 days. Blue areas represent cell death. (F) Expression levels of disease resistance‐associated genes in transgenic Arabidopsis OEmiR846 and STTM846 were detected by qRT‐PCR. Pathogenesis‐related protein (PR1) and plant defence factor 1.2 (PDF 1.2) represent marker genes involved in the salicylic acid‐ and jasmonic acid‐dependent defence signalling pathways in Arabidopsis. Error bars represent significant differences according to Fisher's least‐significant difference test (P = 0.05) using SPSS software. Each experiment was repeated three times.
miR846 exerts its regulatory function in ISR through JA‐dependent signalling in Arabidopsis
SAR primed by pathogens is often dependent on the SA signalling pathway, and many miRNAs have been confirmed to play a role in SAR, such as miR393 and miR482 (González et al., 2015; Navarro et al., 2006). ISR elicited by beneficial bacteria is often dependent on the JA signalling pathway; however, the function of miRNA in the ISR‐activating process remains unclear. The up‐regulated expression of pdf1.2 in STTM846 suggests that miR846 may be involved in the JA signalling pathway. To further confirm this, we first detected the miR846 expression level in wild‐type Arabidopsis Col‐0 after spraying with SA and methyl jasmonate (MeJA). miR846‐5p and miR846‐3p were both repressed by spraying with MeJA, as expected (Fig. 4A). In addition, the expression levels of AOS, LOX2 and LOX3, associated with JA biosynthesis, in OEmiR846 and STTM846 were also observed. The three JA biosynthesis‐related genes were all up‐regulated in STTM846 (Fig. 4B). The stomatal apertures of OEmiR846 and STTM846 transgenic Arabidopsis showed no difference under light conditions. However, the addition of MeJA led to stomatal closure in STTM846, and the stomatal apertures of OEmiR846 were larger than those of wild‐type Col‐0 (Fig. 4C). Furthermore, the disease resistance of STTM846 transgenic Arabidopsis against Pst DC3000 was blocked by spraying with the JA biosynthetic inhibitor diethyldiethiocarbamic acid (DIECA) (Fig. 4D). All of these results suggest that miR846 modulates ISR through the JA‐dependent signalling pathway.
Figure 4.
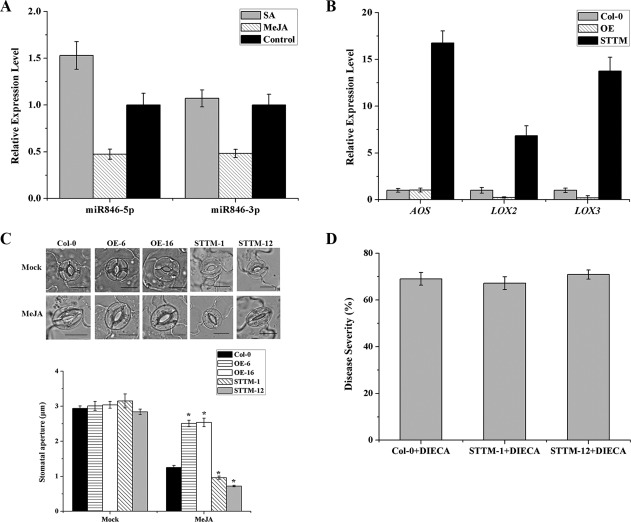
miR846 modulates induced systemic resistance in Arabidopsis through the jasmonic acid (JA)‐dependent signalling pathway. (A) Expression levels of miR846‐5p/miR846‐3p were detected in wild Arabidopsis Col‐0 after spraying with 1 mm salicylic acid (SA) or 0.1 mm methyl jasmonate (MeJA). (B) Expression levels of JA biosynthesis genes (AOS, LOX2 and LOX3) in transgenic Arabidopsis OEmiR846 and STTM846. (C) Stomatal apertures in transgenic Arabidopsis OEmiR846 and STTM846 after the addition of 0.1 mm MeJA. Scale bars indicate 10 μm. Values represent the means of 50 random selected stomata. (D) Disease resistance of STTM846 against Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) after spraying with the JA biosynthetic inhibitor diethyldiethiocarbamic acid (DIECA). Four‐week‐old STTM846 transgenic Arabidopsis and wild‐type Col‐0 were previously sprayed with 200 μm DIECA containing 0.02% (v/v) Tween‐20. Twenty‐four hours later, DIECA‐treated plants were inoculated with 108 colony‐forming units (CFU)/mL Pst DC3000. Disease severity was determined according to the disease index measured at 7 days post‐inoculation (dpi). Each treatment had 12 plants and the experiment was repeated three times. Error bars represent significant differences according to Fisher's least‐significant difference test (P = 0.05) using SPSS software. *Highly significant difference (P < 0.05). All experiments were repeated three times.
Discussion
Numerous reports have demonstrated that plant sRNAs are involved in the evolution of plant immune systems in response to pathogens (Padmanabhan et al., 2009). However, whether miRNAs contribute to the ISR‐eliciting process in response to biocontrol bacteria still remains unclear. In the present study, we successfully constructed an ISR‐deficient mutant of B. amyloliquefaciens (FZB42ΔsfpΔalsS) by blocking cyclic lipopeptide and 2,3‐butanediol synthesis. This provides the foundation for the analysis of the Arabidopsis sRNA profile in response to FZB42 (triggers ISR), FZB42ΔsfpΔalsS (deficient in ISR) and control to identify miRNAs involved in the ISR process. After fold change selection, promoter analysis and target prediction, miR846‐5p and miR846‐3p from the same precursor were selected as candidate ISR‐associated miRNAs. miR846 is only present in Arabidopsis, and belongs to the non‐conserved miRNAs. Non‐conserved miRNAs can be distinguished from conserved miRNAs by the feature of existing only in limited plant species (Qin et al., 2014). Several non‐conserved miRNAs have been confirmed to be involved in plant defence responses. For instance, tomato miR482 and tobacco miR6019 are typical species‐specific miRNAs that modulate plant immunity by cleaving LRR disease resistance (Li et al., 2012). Another non‐conserved miR472 also influences PTI via the mediation of secondary siRNAs (Boccara et al., 2014). These observations show the significant importance of non‐conserved miRNAs in plant defence, and that different types of plant may adopt different strategies at the sRNA level to elicit resistance against various microbes, including pathogens and PGPRs, during their interaction process.miR846 was first reported in the miRNA diversity analysis of Arabidopsis thaliana (Rajagopalan et al., 2006). In addition, miR846 has also been mentioned in several reports. For example, the expression of miR846 was repressed by the application of exogenous abscisic acid (ABA) to Arabidopsis seedlings, whereas miR846 was dramatically up‐regulated under nitrogen starvation conditions (Jia and Rock, 2013b; Liang et al., 2012). In addition, miR846 was also induced in transgenic Arabidopsis overexpressing rGRF3 or purple acid phosphatase 2 (Liang et al., 2014; Liu et al., 2014). However, these studies only reported the expression level of miR846 under different conditions; the function of miR846, especially in plant defence responses, remains unknown.miR846 was predicted to target several jacalin lectin genes; two jacalin lectin genes (At5G49850 and At5G28520) have been confirmed as the targets of miR846 through 5′ RNA ligase‐mediated rapid amplification of cDNA ends (5′RLM‐RACE) (Fahlgren et al., 2007; Jia and Rock, 2013a). The family of JRL genes is named after jacalin, an 18‐kDa T‐antigen disaccharide‐binding lectin domain first isolated from the seeds of jackfruit (Artocarpus integrifolia). Many JRL genes have been shown to be associated with disease resistance, abiotic stress signalling, wounding insect damage or multiple stresses (Song et al., 2014). Transgenic tobacco plants overexpressing the JRL gene Ta‐JA1 show increased resistance to bacterial, fungal and viral pathogens (Ma et al., 2010). Arabidopsis jacalin‐related JAX1 confers broad but specific resistance to potex viruses by the inhibition of viral RNA accumulation (Yamaji et al., 2012). Hence, we predicted that B. amyloliquefaciens FZB42 activated Arabidopsis ISR by the suppression of miR846.
To further validate our hypothesis, we constructed transgenic Arabidopsis overexpressing OEmiR846 and the knockdown mutant STTM846. The expression level of miR846 in Arabidopsis was negatively correlated with plant disease resistance, indicating that miR846 negatively modulates ISR. On the other hand, jacalin lectins, the target genes of miR846, can be subdivided into two subgroups (galactose‐specific JRLs and mannose‐specific JRLs) based on differences in molecular structure, subcellular localization and carbohydrate‐binding properties. A mannose‐specific JRL TaJRLL1, consisting of two jacalin‐like domains, is considered to be a component of SA‐ and JA‐dependent plant defence signalling, A. thaliana transformed with TaJRLL1 displays increased resistance to Fusarium graminearum and Botrytis cinerea (Xiang et al., 2011). The phytohormones SA, JA and ETH play important roles in plant defence responses. ISR is often regulated by JA/ETH‐dependent signalling pathways. To understand the signalling pathway in which miR846 is involved, the miR846 expression level in wild‐type Arabidopsis Col‐0 in response to SA and MeJA, as well as the expression levels of JA biosynthesis‐related genes in transgenic OEmiR846 and STTM846, were detected. Our results suggest that miR846 may negatively regulate ISR via the JA‐dependent signalling pathway.
Previous studies investigating the functions of miRNAs in plant defence have focused on SAR elicited by pathogens. The involvement of miRNAs in beneficial bacteria‐primed ISR processes has been ignored. Our study has revealed that lipopeptides and volatile compounds produced by B. amyloliquefaciens FZB42 repress Arabidopsis miR846 to induce defence‐related gene expression and stomatal closure, thus leading to ISR. Moreover, miR846 modulates ISR via the JA‐dependent signalling pathway.
Experimental Procedures
Bacterial strains, plant and growth conditions
Bacillus amyloliquefaciens FZB42 wild‐type and FZB42Δsfp mutant were provided by Professor Gao from Nanjing Agricultural University, Nanjing, China. Bacillus amyloliquefaciens FZB42 wild‐type and FZB42ΔsfpΔalsS were grown in Luria–Bertani (LB) medium at 37 °C for 12 h. The antibiotics 5 μg/ml chloramphenicol and 10 μg/ml erythromycin were added to culture FZB42ΔsfpΔalsS. Subsequently, bacterial cells were harvested by centrifugation, resuspended in distilled water and adjusted to a final concentration of 108 colony‐forming units (CFU)/mL.
Pseudomonas syringae pv. tomato DC3000 was grown in King's B (KB) medium at 28 °C for 24 h. Cells were then collected by centrifugation and resuspended in 10 mm MgSO4 containing 0.01% (v/v) surfactant Silwet L‐77 with a final concentration of 108 CFU/mL.
Arabidopsis thaliana Col‐0 seeds were surface sterilized with 0.12% NaClO for 15 min, washed three times with sterile distilled water, spread on solid Murashige and Skoog (MS) medium and vernalized for 3 days at 4 °C in the absence of light. Seedlings were then placed in growth cabinets under a 16‐h/8‐h light/dark condition at 22 ºC.
Construction of mutants
To generate the site‐directed mutant FZB42ΔsfpΔalsS, two partial sequence fragments of alsS genes were amplified from FZB42 chromosomal DNA; the sequences of chloramphenicol were obtained from plasmid pAD43‐25. The recombinant fragments were fused by overlap extension PCR and transformed into the FZB42Δsfp strain to generate the site‐directed mutant FZB42ΔsfpΔalsS.
To generate the overexpression construct OEmiR846, the miR846 precursor sequence was cloned from the A. thaliana genome, and then inserted into the plant overexpression vector pCAMBIA1301. Short tandem target mimic (STTM) was used to inactivate miR846 (Yan et al., 2012). The STTM846 construct carries two tandem miR846 binding sites linked by a 48‐nt spacer. The miR846 binding site contains perfect complementary sequences of miR846, with the exception of the ‘CTA’ bulges, to prevent miR846‐mediated cleavage. The construct was then inserted into the vector pCAMBIA1301. The constructs OEmiR846 and STTM846 were electroporated into Agrobacterium tumefaciens GV3101, and used to transform Arabidopsis by the floral dipping method.
Reactive oxygen species (ROS) assay
Arabidopsis leaves collected 6 h after treatment were incubated in phosphate‐buffered saline (PBS) containing 1 mg/mL 3,3′‐diaminobenzidine (DAB) for 8 h. The leaves were then boiled in 95% ethanol for 15 min, and brown precipitates were observed (Samuel et al., 2005).
Stomatal aperture measurement
Arabidopsis Col‐0 leaf epidermis was excised from Bacillus‐inoculated plants after 5 h of exposure to light within the growth chamber and floated in MES buffer (10 mM MES‐Tris, 5 mM KCl, and 50 mM CaCl2, pH 6.15). The leaf epidermis from transgenic Arabidopsis leaves was first soaked in MES buffer under light for 6 h to open the stomata, followed by the addition of 0.1 mm MeJA for 4 h. The stomatal aperture was detected using a Leica microscope (wetzlar, Germany), and the diameters were measured from 50 randomly selected stomata using ImageJ software. Each assay was repeated three times.
Cell death measurement
Cell death was measured by Evans blue staining. Arabidopsis leaves which had been inoculated with Pst DC3000 for 7 days were soaked in 0.5% Evans blue for 24 h, and then soaked in 1 g/mL chloral hydrate for 24 h. The blue areas representing cell death were observed.
Induction of systemic resistance
The roots of 4‐week‐old Arabidopsis were inoculated with a cell suspension of FZB42, FZB42ΔsfpΔalsS or water control. Five days later, Arabidopsis was challenge inoculated by spraying the leaves with Pst DC3000 at 108 CFU/mL and then kept in a dew chamber at 100% relative humidity for 3 days. After challenge inoculation for 7 days, the disease index was assessed according to Hossain and associates (Hossain et al., 2007). Each treatment had 12 plants and the experiment was repeated three times.
sRNA library construction and deep sequencing
After 12 h of root inoculation with FZB42, FZB42ΔsfpΔalsS or control, 24 Arabidopsis leaves per sample were collected. sRNA extraction and library construction were carried out by Novogene (Beijing, China). A total amount of 3 µg of total RNA per sample was used as input material for the sRNA library. Sequencing libraries were generated using a NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, Ipswich, MA) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. sRNAs (18–30 nt) were excised and ligated to 5′‐ and 3′‐RNA adaptors using T4 RNA ligase, followed by RT‐PCR and gel purification, as instructed by Illumina. The sRNA libraries were sequenced on an Illumina Hiseq 2500/2000 platform.
Sequence data processing
Raw data in fastq format were first processed through custom perl and python scripts; clean data were then obtained by removing reads containing poly‐N, with 5′ adapter contaminants, without 3′ adapter or the insert tag, containing poly A or T or G or C, and low‐quality reads from raw data. Meanwhile, the sRNA sequences were mapped to the genome using A. thaliana without mismatch to analyse their expression and distribution on the reference. P values were adjusted using the Q value. Q value < 0.01 and |log2(fold change)| > 1 was set as the threshold for significantly differential expression by default. Prediction of the target gene of miRNA was performed by psRobot_tar in psRobot for plants (Wu et al., 2012).
miRNA promoter selection and cis‐acting element analysis
The promoter sequences of pri‐miRNAs were obtained as follows (Ding et al., 2011). In general, if pri‐miRNA and its closest upstream gene had the same transcription, and the distance between them was less than 2400 bp, the region from the site 400 bp downstream of the upstream gene to pri‐miRNA was obtained. If the pri‐miRNA and its upstream gene had the opposite transcription, and their distance was less than 4000 bp, the region between pri‐miRNA and their middle point was used. Otherwise, the 2000‐bp upstream area of pri‐miRNA was used. PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was then used to analyse these miRNA promoters. Cis‐acting elements involved in SA, MeJA, ETH and defence responsiveness were retrieved.
qRT‐PCR
Stem‐loop qRT‐PCR was used to measure the expression of miRNA. Total RNA was extracted from Arabidopsis leaves using TRizol reagent. The reverse transcription reaction was performed according to a PrimeScript™ RT kit (TaKaRa, Dalian, China) with stem‐loop reverse transcription primer. Stem‐loop qRT‐PCR was then carried out on an ABI 7300 Fast Real‐time PCR System (Applied Biosystems, Foster City, CA, USA). 18S rRNA was used as the internal control.
Statistical analysis
Three independent experiments were performed for each assay. The data were analysed by Fisher's least‐significant difference test (P < 0.05) with SPSS software (SPSS Inc. Chicago, USA).
Competing Interests
The authors declare no conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Construction of the induced systemic resistance (ISR)‐deficient mutant FZB42ΔsfpΔalsS. (A) Haemolytic activity detection of the FZB42ΔsfpΔalsS mutant: 1, FZB42; 2, FZB42ΔsfpΔalsS mutant; 3, methyl alcohol control. (B) Polymerase chain reaction (PCR) amplification verification of the FZB42ΔsfpΔalsS mutant: 1, FZB42; 2, FZB42Δsfp mutant; 3–5, FZB42ΔsfpΔalsS transformants; 6, sterilized water control.
Fig. S2 Length distribution of small RNAs from the sequencing of the three Arabidopsis libraries. (A) Arabidopsis leaves treated by wide‐type FZB42. (B) Arabidopsis leaves treated by mutant FZB42ΔsfpΔalsS. (C) Arabidopsis leaves treated by control.
Fig. S3 Relative nucleotide bias at the first position of the known Arabidopsis microRNAs (miRNAs). (A) Arabidopsis leaves treated by wide‐type FZB42. (B) Arabidopsis leaves treated by mutant FZB42ΔsfpΔalsS. (C) Arabidopsis leaves treated by control.
Fig. S4 The expression levels of three differentially expressed microRNAs (miRNAs) were examined by stem‐loop quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Table S1 Summary of small RNA (sRNA) sequencing.
Acknowledgements
We gratefully acknowledge Professor Xuewen Gao for supplying the Bacillus amyloliquefaciens FZB42 and FZB42Δsfp mutants. This study was funded by grants from China Postdoctoral Science Foundation (2016M592036) and Key Science and Technology Program of Anhui Province (15czz03119).
References
- Baldrich, P. and Segundo, B.S. (2016) MicroRNAs in rice innate immunity. Rice, 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccara, M. , Sarazin, A. , Thiébeauld, O. , Jay, F. , Voinnet, O. , Navarro, L. and Colot, V. (2014) Correction: the Arabidopsis miR472‐RDR6 silencing pathway modulates PAMP‐ and effector‐triggered immunity through the post‐transcriptional control of disease resistance genes. PLoS Pathog. 10, e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy, H. , Mariutto, M. , Henry, G. , Fisher, C. , Vasilyeva, N. , Thonart, P. , Dommes, J. and Ongena, M. (2014) Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant–Microbe Interact. 27, 87–100. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Ridzon, D.A. , Broomer, A.J. , Zhou, Z. , Lee, D.H. , Nguyen, J.T. , Barbisin, M. , Xu, N.L. , Mahuvakar, V.R. , Andersen, M.R. , Lao, K.Q. , Livak, K.J. and Guegler, K.J. (2005) Real‐time quantification of microRNAs by stem‐loop RT–PCR. Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.H. , Koumoutsi, A. , Scholz, R. , Eisenreich, A. , Schneider, K. , Heinemeyer, I. , Morgenstern, B. , Voss, B. , Hess, W.R. , Reva, O. , Junge, H. , Voigt, B. , Jungblut, P.R. , Vater, J. , Süssmuth, R. , Liesegang, H. , Strittmatter, A. , Gottschalk, G. and Borriss, R. (2007) Comparative analysis of the complete genome sequence of the plant growth‐promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 25, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S.P. , Hartmann, A. , Gao, X. and Borriss, R. (2015a) Biocontrol mechanism by root‐associated Bacillus amyloliquefaciens FZB42 – a review. Front. Microbiol. 6, 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S.P. , Uhl, J. , Grosch, R. , Alquéres, S. , Pittroff, S. , Dietel, K. , Schmitt‐Kopplin, P. , Borriss, R. and Hartmann, A. (2015b) Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani . Mol. Plant–Microbe Interact. 28, 984–995. [DOI] [PubMed] [Google Scholar]
- De Luis, A. , Markmann, K. , Cognat, V. , Holt, D.B. , Charpentier, M. , Parniske, M. , Stougaard, J. and Voinnet, O. (2012) Two microRNAs linked to nodule infection and nitrogen‐fixing ability in the legume Lotus japonicus . Plant Physiol. 160, 2137–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Chen, Z. and Zhu, C. (2011) Microarray‐based analysis of cadmium‐responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 62, 3563–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Howell, M.D. , Kasschau, K.D. , Chapman, E.J. , Sullivan, C.M. , Cumbie, J.S. , Givan, S.A. , Law, T.F. , Grant, S.R. , Dangl, J.L. and Carrington, J.C. (2007) High‐throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One, 2, e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, B. , Borriss, R. , Bleiss, W. and Wu, X. (2012) Gram‐positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J. Microbiol. 50, 38–44. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. and Dong, X. (2013) Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. [DOI] [PubMed] [Google Scholar]
- González, V.M. , Müller, S. , Baulcombe, D. and Puigdomènech, P. (2015) Evolution of NBS‐LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol. Plant. 8, 329–331. [DOI] [PubMed] [Google Scholar]
- Hossain, M. , Sultana, F. , Kubota, M. , Koyama, H. and Hyakumachi, M. (2007) The plant growth‐promoting fungus Penicillium simplicissimum GP17‐2 induces signals. Plant Cell Physiol. 48, 1724–1736. [DOI] [PubMed] [Google Scholar]
- Jia, F. and Rock, C.D. (2013a) Jacalin lectin At5g28520 is regulated by ABA and miR846. Plant Signal. Behav. 8, e24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, F. and Rock, C.D. (2013b) MiR846 and miR842 comprise a cistronic miRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis . Plant Mol. Biol. 81, 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jourdan, E. , Henry, G. , Duby, F. , Dommes, J. , Barthelemy, J.‐P. , Thonart, P. and Ongena, M. (2009) Insights into the defense‐related events occurring in plant cells following perception of surfactin‐type lipopeptide from Bacillus subtilis . Mol. Plant–Microbe Interact. 22, 456–468. [DOI] [PubMed] [Google Scholar]
- Li, F. , Pignatta, D. , Bendix, C. , Brunkard, J.O. , Cohn, M.M. , Tung, J. , Sun, H. , Kumar, P. and Baker, B. (2012) MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA, 109, 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C. , Liu, X. , Sun, Y. , Yiu, S.‐M. and Lim, B.L. (2014) Global small RNA analysis in fast‐growing Arabidopsis thaliana with elevated concentrations of ATP and sugars. BMC Genomics, 15, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, G. , He, H. and Yu, D. (2012) Identification of nitrogen starvation‐responsive microRNAs in Arabidopsis thaliana . PLoS One, 7, e48951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Rice, J.H. , Chen, N. , Baum, T.J. and Hewezi, T. (2014) Synchronization of developmental processes and defense signaling by growth regulating transcription factors. PLoS One, 9, e98477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg, B. and Kamilova, F. (2009) Plant‐growth‐promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. [DOI] [PubMed] [Google Scholar]
- Ma, Q.‐H. , Tian, B. and Li, Y.‐L. (2010) Overexpression of a wheat jasmonate‐regulated lectin increases pathogen resistance. Biochimie, 92, 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, L. , Dunoyer, P. , Jay, F. , Arnold, B. , Dharmasiri, N. , Estelle, M. , Voinnet, O. and Jones, J.D.G. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science, 312, 436–439. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Jay, F. , Nomura, K. , He, S.Y. and Voinnet, O. (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science, 321, 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, D. , Xia, J. , Jiang, C. , Qi, B. , Ling, X. , Lin, S. , Zhang, W. , Guo, J. , Jin, H. and Zhao, H. (2016) Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825* and activating defense related genes in Arabidopsis . J. Integr. Plant Biol. 58, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Adam, A. , Paquot, M. , Brans, A. , Joris, B. , Arpigny, J.L. and Thonart, P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Padmanabhan, C. , Zhang, X. and Jin, H. (2009) Host small RNAs are big contributors to plant innate immunity. Curr. Opin. Plant Biol. 12, 465–472. [DOI] [PubMed] [Google Scholar]
- Qin, Z. , Li, C. , Mao, L. and Wu, L. (2014) Novel insights from non‐conserved microRNAs in plants. Front. Plant Sci. 5, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, R. , Vaucheret, H. , Trejo, J. and Bartel, D.P. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes Dev. 20, 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.‐H. , Reddy, M.S. , Kloepper, J.W. and Paré, P.W. (2004) Bacterial volatiles induce systemic resistance in Arabidopsis . Plant Physiol. 134, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel, M.A. , Hall, H. , Krzymowska, M. , Drzewiecka, K. , Hennig, J. and Ellis, B.E. (2005) SIPK signaling controls multiple components of harpin‐induced cell death in tobacco. Plant J. 42, 406–416. [DOI] [PubMed] [Google Scholar]
- Song, M. , Xu, W. , Xiang, Y. , Jia, H. , Zhang, L. and Ma, Z. (2014) Association of jacalin‐related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 84, 95–110. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Nürnberger, T. and Joosten, M.H. (2011) Of PAMPs and effectors: the blurred PTI–ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H. , Ficke, A. , Asiimwe, T. , Höfte, M. and Raaijmakers, J.M. (2007) Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens . New Phytol. 175, 731–742. [DOI] [PubMed] [Google Scholar]
- Van Peer, R. , Niemann, G. and Schippers, B. (1991) Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology, 81, 728–734. [Google Scholar]
- Wu, H.‐J. , Ma, Y.‐K. , Chen, T. , Wang, M. and Wang, X.‐J. (2012) PsRobot: a web‐based plant small RNA meta‐analysis toolbox. Nucleic Acids Res. 40, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, Y. , Song, M. , Wei, Z. , Tong, J. , Zhang, L. , Xiao, L. , Ma, Z. and Wang, Y. (2011) A jacalin‐related lectin‐like gene in wheat is a component of the plant defence system. J. Exp. Bot. 62, 5471–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji, Y. , Maejima, K. , Komatsu, K. , Shiraishi, T. , Okano, Y. , Himeno, M. , Sugawara, K. , Neriya, Y. , Minato, N. , Miura, C. , Hashimoto, M. and Namba, S. (2012) Lectin‐mediated resistance impairs plant virus infection at the cellular level. Plant Cell, 24, 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J. , Gu, Y. , Jia, X. , Kang, W. , Pan, S. , Tang, X. , Chen, X. and Tang, G. (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis . Plant Cell, 24, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Construction of the induced systemic resistance (ISR)‐deficient mutant FZB42ΔsfpΔalsS. (A) Haemolytic activity detection of the FZB42ΔsfpΔalsS mutant: 1, FZB42; 2, FZB42ΔsfpΔalsS mutant; 3, methyl alcohol control. (B) Polymerase chain reaction (PCR) amplification verification of the FZB42ΔsfpΔalsS mutant: 1, FZB42; 2, FZB42Δsfp mutant; 3–5, FZB42ΔsfpΔalsS transformants; 6, sterilized water control.
Fig. S2 Length distribution of small RNAs from the sequencing of the three Arabidopsis libraries. (A) Arabidopsis leaves treated by wide‐type FZB42. (B) Arabidopsis leaves treated by mutant FZB42ΔsfpΔalsS. (C) Arabidopsis leaves treated by control.
Fig. S3 Relative nucleotide bias at the first position of the known Arabidopsis microRNAs (miRNAs). (A) Arabidopsis leaves treated by wide‐type FZB42. (B) Arabidopsis leaves treated by mutant FZB42ΔsfpΔalsS. (C) Arabidopsis leaves treated by control.
Fig. S4 The expression levels of three differentially expressed microRNAs (miRNAs) were examined by stem‐loop quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Table S1 Summary of small RNA (sRNA) sequencing.


