Summary
Rsv1, a single dominant resistance locus in soybean, confers extreme resistance to the majority of Soybean mosaic virus (SMV) strains, but is susceptible to the G7 strain. In Rsv1‐genotype soybean, G7 infection provokes a lethal systemic hypersensitive response (LSHR), a delayed host defence response. The Rsv1‐mediated LSHR signalling pathway remains largely unknown. In this study, we employed a genome‐wide investigation to gain an insight into the molecular interplay between SMV G7 and Rsv1‐genotype soybean. Small RNA (sRNA), degradome and transcriptome sequencing analyses were used to identify differentially expressed genes (DEGs) and microRNAs (DEMs) in response to G7 infection. A number of DEGs, DEMs and microRNA targets, and the interaction network of DEMs and their target mRNAs responsive to G7 infection, were identified. Knock‐down of one of the identified DEGs, the eukaryotic translation initiation factor 5A (eIF5A), diminished the LSHR and enhanced viral accumulation, suggesting the essential role of eIF5A in the G7‐induced, Rsv1‐mediated LSHR signalling pathway. This work provides an in‐depth genome‐wide analysis of high‐throughput sequencing data, and identifies multiple genes and microRNA signatures that are associated with the Rsv1‐mediated LSHR.
Keywords: degradome‐seq, eukaryotic translation initiation factor, lethal systemic hypersensitive response, Soybean mosaic virus, sRNA‐seq, transcriptome‐seq, virus–host interaction
Introduction
To protect themselves against pathogen attack, plants have evolved a complex innate immune system that consists of two interconnected tiers of receptors, i.e. pattern recognition receptors (PRRs) and nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) receptors, to modulate plant response to microbial infections (Dangl et al., 2013; Mandadi and Scholthof, 2013; Mukhtar et al., 2011; Nicaise, 2014). The extracellular PRRs recognize and respond to evolutionarily conserved pathogen‐ or microbial‐associated molecular patterns (PAMPs or MAMPs) to trigger the first layer of resistance, termed PAMP‐triggered immunity (PTI) (Boller and He, 2009). PTI is able to halt the further colonization of the majority of potential pathogens. Successful pathogens deploy variable pathogen molecules that can interfere with PTI. This process allows the pathogen to establish a successful infection, and is referred to as effector‐triggered susceptibility (ETS). In response, plants have developed a counter defence layer that is activated by single dominant resistance (R) gene products which directly or indirectly sense the presence of a specific pathogen by their effector, termed an avirulence factor (Avr). This leads to a stage called effector‐triggered immunity (ETI). Most R gene‐encoded proteins belong to the extremely polymorphic superfamily of NBS‐LRRs. ETI activates much more rapidly and strongly than PTI, and triggers a hypersensitive response (HR), which inhibits the spread of pathogen at the infection site, resulting in plant disease resistance (Boller and He, 2009; Jones and Dangl, 2006). Although it mainly derives from studies of non‐viral infections, the two‐tier immunity concept, particularly ETI, is considered to be an essential antiviral mechanism in plants, in addition to RNA silencing. It is well established that plants combat viral pathogens through their NBS‐LRR domain‐containing resistance proteins, which interact with viral effectors to activate the ETI responses (Jones and Dangl, 2006; Mandadi and Scholthof, 2013; Nicaise, 2014). Recently, PTI has also been proposed to be part of antiviral immunity in plants (Kørner et al., 2013). Regardless of the type of pathogen, the ETI responses and, to a lesser extent, the PTI responses are often associated with HR (Jones and Dangl, 2006). Perturbation of Avr or R genes, a change in the genetic background of the R gene‐carrying plant, the prevention of salicylic acid accumulation in the R‐genotype plant or a temperature shift can cause a virus to escape to distant tissues, inducing systemic HR (SHR) in resistant tissues (Chen et al., 1994; Culver et al., 1991; Delaney et al., 1994; Dinesh‐Kumar et al., 2000; Hall, 1980; Lanfermeijer et al., 2003; Moury et al., 1998). SHR may result in the death of the plant, which is termed lethal SHR (LSHR) (Hajimorad and Hill, 2001; Hajimorad et al., 2003; Seo et al., 2009). Therefore, LSHR has been suggested to be a delayed HR.
Soybean mosaic virus (SMV), belonging to the potyvirus group in the Potyviridae family, is the most prevalent viral pathogen in soybean [Glycine max (L.) Merr.]. Based on their differential responses to a set of susceptible and resistant soybean cultivars, SMV isolates in North America may be classified into seven distinct strains (G1–G7) (Cho and Goodman, 1979). Of the three independent dominant resistance loci (Rsv1, Rsv3 and Rsv4) identified from soybean thus far, Rsv1 probably comprises one or more members of the NBS‐LRR gene family (Zhang et al., 2012). Rsv1‐genotype soybean confers extreme resistance to the majority of SMV strains, except G7. Infection by G7 provokes an Rsv1‐mediated LSHR, and the symptoms usually appear 14 days after inoculation. LSHR is associated with the up‐regulation of the pathogenesis‐related 1 (PR1) gene transcript and breakdown of ARGONAUTE1 (AGO1) homeostasis (Chen et al., 2015; Chowda‐Reddy et al., 2011; Hajimorad and Hill, 2001; Hajimorad et al., 2005; Yu et al., 1994). Although the strain‐specific P3 protein of SMV has been identified as an elicitor of Rsv1‐mediated LSHR, the host components of the Rsv1‐mediated LSHR signalling pathway still largely remain unknown.
Over the past decade, genomics approaches, including the recently developed next‐generation sequencing (NGS) technologies, have been extensively employed to evaluate global gene expression changes in plants in response to viral infections (Babu et al., 2008; Bilgin et al., 2008; Du et al., 2011; Garcia‐Marcos et al., 2009; Mochizuki et al., 2014; Yang et al., 2007). In addition to the transcriptome, microRNAs (miRNAs) have also been profiled in virus‐infected plants. miRNAs are single‐stranded, non‐coding RNAs with critical functions across a wide variety of biological processes, such as the maintenance of genome integrity, development and feedback mechanisms, as well as various biotic and abiotic stress responses (Baulcombe, 2004; Ding and Voinnet, 2007; Meister, 2013; Sunkar et al., 2012). miRNA not only regulates the expression of endogenous genes, but is also indispensable for innate immunity in plants. Accumulated evidence proves that viral infections are often associated with alterations in endogenous miRNA levels and their target mRNA accumulation (Bazzini et al., 2009; Du et al., 2011; Pradhan et al., 2015; Romanel et al., 2012). For example, AGO1 mRNA and miR168 are up‐regulated by infections with several RNA viruses. MiR168 is also involved in the regulation of the virally inducible AGO18 in response to diverse viruses in rice (Chen et al., 2015; Havelda et al., 2008; Várallyay et al., 2010; Wu et al., 2015; Zhang et al., 2006). In soybean, 573 miRNA precursors and 639 mature miRNAs have been identified from different developmental stages and tissues, as well as various biotic and abiotic stress‐treated tissues (Shamimuzzaman and Vodkin, 2012; Xu et al., 2013; Yin et al., 2013; Zhai et al., 2011). Some of these miRNAs have been implicated in pathogen infections and nodule organogenesis in soybean (Wang et al., 2014; Wong et al., 2014; Yin et al., 2013). In soybean‐related Medicago, miRNA has been suggested to be a master regulator of the plant NBS‐LRR defence gene family via the production of phased, trans‐acting small interfering RNAs (siRNAs) (Zhai et al., 2011). These data suggest potential roles of miRNA in SMV G7‐provoked Rsv1‐mediated LSHR.
As LSHR is a type of delayed HR, a better understanding of LSHR would provide new insights into the HR signalling pathway that is mediated by R genes, ETI and PTI. In this study, small RNA sequencing (sRNA‐seq), degradome sequencing (degradome‐seq) and transcriptome sequencing (transcriptome‐seq) analyses were employed to identify differentially expressed genes (DEGs) and miRNAs (DEMs) that are specifically up‐regulated or down‐regulated by SMV G7 infection.
Results
An overview of deep sequencing datasets
To gain an insight into the HR signalling network, we conducted genome‐wide analyses of sRNA‐seq, degradome‐seq and transcriptome‐seq to identify genes and miRNAs that may regulate the Rsv1‐mediated LSHR provoked by G7 infection. RNA‐seq yielded a total of 5 542 824 and 6 406 830 reads from mock‐ and G7‐inoculated plants, respectively. For sRNA‐seq, the mock sample produced 5 778 554 reads and the G7 sample generated 5 937 675 reads. As shown in Table S1A,B (see Supporting Information), approximately 80% (RNA‐seq) and over 42% (sRNA‐seq) reads matched perfectly to the soybean genome. This resulted in more than 3.7 million and 1.2 million distinct genome‐matched RNA sequences and sRNA sequences per library. The RNA and sRNA sequences from each library were also mapped to the soybean chromosomes (Table S1A,B).
In order to enable the large‐scale examination of miRNA‐guided cleavage products and to validate the predicted mRNA targets by miRNAs, we constructed and sequenced degradome libraries for the G7 and mock‐inoculated control samples. A total of 70 626 197 degradome sequences were obtained, and approximately 40% (over 14 million per sample) of the sequences matched perfectly to the soybean genome, resulting in more than 4.4 million genome‐matched degradome‐seq reads per library. The degradome‐seq reads were also mapped to each soybean chromosome (Table S1C).
Transcriptional response to G7 infection
To identify host components of the Rsv1‐mediated LSHR signalling pathway induced by SMV G7 infection, DEGs in response to SMV inoculation with the G7 strain were identified and compared with those of the mock control. A total of 906 genes, including two new genes, showed at least a two‐fold expression change in the G7‐infected leaf sample compared with the mock‐inoculated control [up‐regulated, log2(fold change, FC) ≥ +1.0; down‐regulated, log2(FC) ≤ −1.0; Table S2 (see Supporting Information) and Fig. 1A]. Of these DEGs, 318 were up‐regulated and 588 were down‐regulated (Fig. 1B).
Figure 1.

Global transcriptional response to G7 infection in Rsv1‐genotype soybean (PI96983). (A) At 14 days after inoculation, Soybean mosaic virus (SMV) G7‐infected plants had 906 differentially expressed genes (DEGs) compared with mock‐inoculated plants. The heat map depicts these 906 genes and presents a detailed view of the genes within the two branches. Colours indicate the log2 fold changes (FCs) in G7‐infected plants relative to mock‐inoculated plants according to the average normalized signal values. Red denotes up‐regulation and blue indicates down‐regulation. Significance was determined using a fold change threshold of at least two. (B) Venn diagram of significant up‐regulated and down‐regulated DEGs by G7 infection relative to the mock‐inoculated control. Of the 906 DEGs, 318 were up‐regulated and 588 were down‐regulated.
Gene ontology (GO) and enrichment analysis were performed using the GO database (www.pantherdb.org and www.soybase.org) to gain further insights into the biological relevance of these DEGs in response to SMV inoculation. Amongst the up‐regulated DEGs were those involved in ‘DNA replication initiation’ (GO:0006270), ‘Protein complex assembly’ (GO:0006461) and ‘Oxylipin biosynthesis/metabolism’ (GO:0031408, GO:0031408), which were all enriched by more than five‐fold (P < 0.005, Table S2). The down‐regulated genes included those involved in ‘Regulation of photosynthesis’ (GO:0010109), ‘Chloroplast localization’ (GO:0019750) and ‘Photosystem II stabilization’ (GO:0042549), which were likewise enriched by more than five‐fold (P < 0.005, Table S2). G7 infection differentially regulated 119 genes that were predicted to play a role in ‘Defence response’ (GO:0006952) (Table S3, see Supporting Information), suggesting possible roles of defence‐related genes in G7 pathogenesis in Rsv1‐genotype soybean and Rsv1‐mediated LSHR. Further breakdown of these defence genes based on their predicted molecular function revealed eight encoding proteins with transcription factor (TF) activity, 14 with ATP/ADP activity and 14 with nucleotide binding activity. Genes with protein binding and ion binding activity were the largest categories with 35 genes (Fig. S1, see Supporting Information).
miRNA regulation in response to SMV G7 infection
Genome‐wide profiling of miRNAs revealed that 84 miRNAs, including two originating from the same predicted precursor (−5p and −3p), representing 41 families, were differentially regulated by at least a two‐fold change in G7‐infected soybean compared with the mock‐inoculated control [up‐regulated, log2(FC) ≥ +1.0; down‐regulated, log2(FC) ≤ −1.0; Table S4 (see Supporting Information) and Fig. 2A]. Amongst these DEMs, 37 miRNAs were significantly up‐regulated and 47 were down‐regulated compared with the mock‐inoculated control (Fig. 2B). Numerous miRNAs are known to play a role in the plant's defence response to viral infection. miR160 and miR393 have been suggested to be involved in the soybean defence response to SMV infection by the SC7 isolate from China (Yin et al., 2013). Here, miR160a‐3p was up‐regulated, but miR393 (h/i/j/k) was significantly down‐regulated, in response to G7 infection (Table S4). Many members of miR156 (p/q/r/s/t) and miR171 (c/k/l/m/n/p/t) families were also highly responsive to SMV G7 infection (Table S4). In the miR156 family, miR156r was up‐regulated in response to infection, whereas the four other members (p/q/s/t) were remarkably down‐regulated. miR2118a/b‐5p, known to target NBS‐LRRs, was also highly up‐regulated after G7 infection (Table S4). Together, these results demonstrate that the analysis is capable of identifying known, as well as novel, miRNAs that may play a role in LSHR brought on by G7 infection in Rsv1‐genotype soybean.
Figure 2.
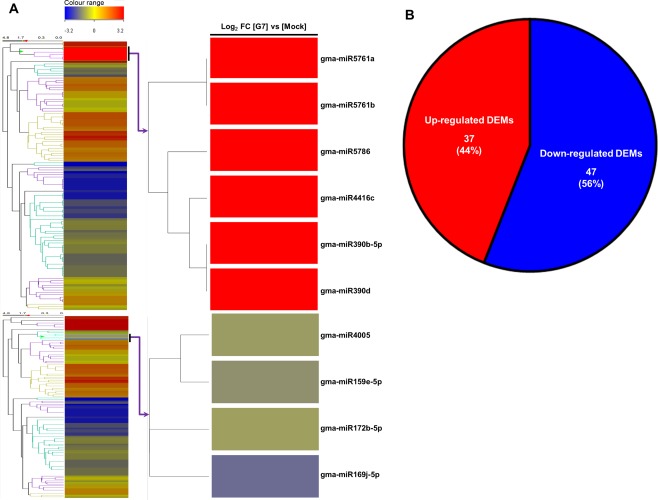
Differentially expressed microRNAs (DEMs) in response to G7 infection in Rsv1‐genotype soybean (PI96983). (A) At 14 days after inoculation, Soybean mosaic virus (SMV) G7‐infected plants had 84 DEMs compared with mock‐inoculated plants. The heat map represents these miRNAs as well as a detailed view of the DEMs from the two branches. Colours indicate the log2 fold changes (FCs) in G7‐infected plants relative to mock‐inoculated controls according to the average normalized signal values. All down‐regulations are indicated with a negative sign and are shown in blue. All up‐regulations are shown in red. Significance was determined using a fold change threshold of at least two. (B) Venn diagram of significant up‐regulated and down‐regulated DEMs by G7 infection relative to mock‐inoculated control. Of the 84 DEMs, 37 were up‐regulated and 47 were down‐regulated.
Validation of SMV‐responsive genes and miRNAs
In order to validate NGS data, 12 DEGs (nine up‐regulated and three down‐regulated), which may play a role in plant defence (Table S2), were selected for validation by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis. The nine up‐regulated DEGs included GLYMA07G01730 (stem 28‐kDa glycoprotein), GLYMA10G35871 (auxin down‐regulated protein), GLYMA11G05190 (calcium ATPase 2), GLYMA09G28310 (kunitz family trypsin and protease inhibitor protein), GLYMA08G05810 (calcium‐binding EF‐hand family protein), GLYMA03G32830 (glycine‐rich protein), GLYMA10G40990 (cyclin D3), GLYMA02G12520 (eukaryotic translation initiation factor 5A, eIF5A) and GLYMA07G01740 (HAD superfamily, subfamily IIIB acid phosphatase). The three down‐regulated DEGs included GLYMA05G08590 (carbonic anhydrase 1), GLYMA03G42240 (thylakoid membrane phosphoprotein) and GLYMA04G06970 (14‐kDa proline‐rich protein) (Fig. 3). The RT‐qPCR data for the exemplar genes largely matched the RNA‐seq data (Fig. 3 and Table S2).
Figure 3.
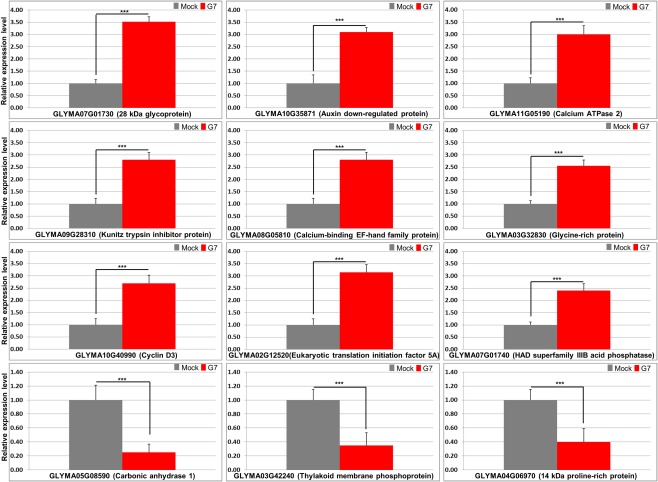
Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) validation of differentially expressed genes (DEGs) in response to G7 infection in Rsv1‐genotype soybean (PI96983). From the RNA sequencing analysis, 12 G7‐responsive DEGs (nine up‐regulated and three down‐regulated) were selected for validation by RT‐qPCR analysis. The soybean Actin (GmACT11) gene was used as an internal control. Error bars represent mean ± standard deviation (SD) and the data are averages from three biological replicates. Asterisks indicate statistically significant differences compared with the mock control (Student's t‐test): *** P < 0.001.
To validate the unique DEMs in response to G7 infection, the accumulation of 12 top DEMs (six up‐regulated and six down‐regulated) was validated by stem‐loop RT‐qPCR (Table S4 and Fig. 4). G7 infection enhanced the accumulation of miR5761a, miR4416c, miR390b‐5p, miR5786, miR397b‐5p and miR1535b, whereas miR5778, miR319i, miR169j‐5p, miR159e‐5p, miR4364a and miR4405 were specifically down‐regulated during G7 infection (Fig. 4). As with the RT‐qPCR results and RNA‐seq, these results matched those obtained from sRNA‐seq and thus lent validity to the data obtained from the pooled sequencing. Together, these data demonstrate the involvement of a group of genes and miRNAs in G7 infection and the Rsv1‐mediated LSHR.
Figure 4.
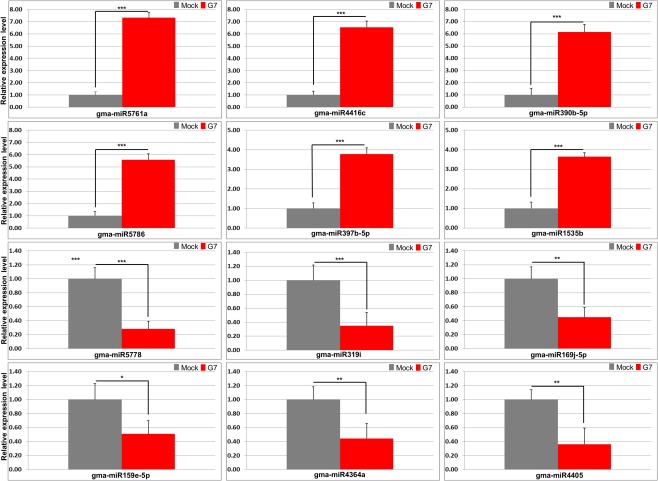
Stem‐loop reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis of differentially expressed microRNAs (DEMs) in response to G7 infection in Rsv1‐genotype soybean (PI96983). The accumulation of 12 top DEMs (six up‐regulated and six down‐regulated) was validated by stem‐loop RT‐qPCR. Soybean 18S rRNA was used as the internal control. Error bars represent mean ± standard deviation (SD) and the data are averages from three biological replicates. Asterisks indicate statistically significant differences compared with the mock control (Student's t‐test): * P < 0.05; ** P < 0.01; *** P < 0.001; ns, not significant.
Validation of the predicted target genes of miRNAs by degradome‐seq
To understand the potential regulatory roles of the detected SMV‐responsive miRNAs, G7 and mock‐treated control degradome libraries were constructed to identify the target genes of all SMV‐responsive miRNAs. The identified targets were classified into five categories (0–4) based on the strength of the degradome signals at the miRNA target sites, as described previously (Folkes et al., 2012). A total of 169 transcripts were identified as potential cleavage targets by 27 miRNA families (131 members) in which two, 12, 83, 35 and 146 transcripts (including multiple targets) fell into categories 0–4, respectively (Table S5, see Supporting Information).
Among these potential targets, many were associated with various signalling pathways in the plant response to biotic and abiotic stress. Glyma13g29320 (auxin response factor 8), Glyma08g04770 (jasmonate‐zim‐domain protein 3), Glyma09g24900 (HXXXD‐type acyl‐transferase family protein), Glyma12g07780 (cytosolic ascorbate peroxidase 2), Glyma05g09440 {disease resistance protein [coiled coil (CC)‐NBS‐LRR class] family} and Glyma18g14410 (photosystem II subunit P‐1) transcripts were targeted by miR167a, miR169a, miR171o, miR396b‐3p, miR5374 and miR5668, respectively. These were also experimentally validated by RNA ligase‐mediated rapid amplification of 5′·cDNA ends (RLM‐5′ RACE) assay to confirm the precise cleavage sites (Fig. S2, see Supporting Information). These data support the predicted cleavage sites from the degradome‐seq results.
Genome‐wide data analysis reveals differentially regulated genes that are possibly involved in the Rsv1‐mediated LSHR
RNA‐seq analysis and RT‐qPCR experimental results revealed the association of a group of DEGs with G7 infection and the Rsv1‐mediated LSHR. Many of these DEGs may have roles as pivotal regulators of the plant HR to pathogen infection (Table S2 and Fig. 3). Based on their potential biological functions, some may participate in the LSHR signalling pathway. The soybean eIF5A, referred to as GmeIF5A in this study, showed strong up‐regulation after G7 infection at 14 days post‐inoculation (dpi) in Rsv1‐genotype soybean. In Arabidopsis, three isoforms of elF5A (AtelF5A‐1, AtelF5A‐2 and AtelF5A‐3) have been identified (Hopkins et al., 2008; Wang et al., 2001), but very limited information about eIF5A is available in soybean. We therefore examined the possible involvement of eIF5A in G7 infection.
There are seven potential GmeIF5A genes in the soybean genome (Fig. 5A). To determine whether the expression of other GmeIF5A family members is also affected in response to G7 infection, a heat map was obtained using a log scale of the values of read density (normalized read count per kilobase of exon model per million reads) from the RNA‐seq data (Fig. 5B). The expression levels of these seven GmeIF5A genes were quantified by the read density value (computed as the normalized read count per million reads) from RNA‐seq (Fig. 5C). All seven genes were up‐regulated, with GmeIF5A (GLYMA02G12520) showing the highest expression in response to G7 infection (Fig. 5C). RT‐PCR was performed to monitor the expression levels of GmeIF5A in systemic leaves at 0, 7, 14 and 21 dpi after SMV inoculation. The results showed that GmeIF5A (GLYMA02G12520) was significantly induced at 14 and 21 dpi (Fig. 5D).
Figure 5.
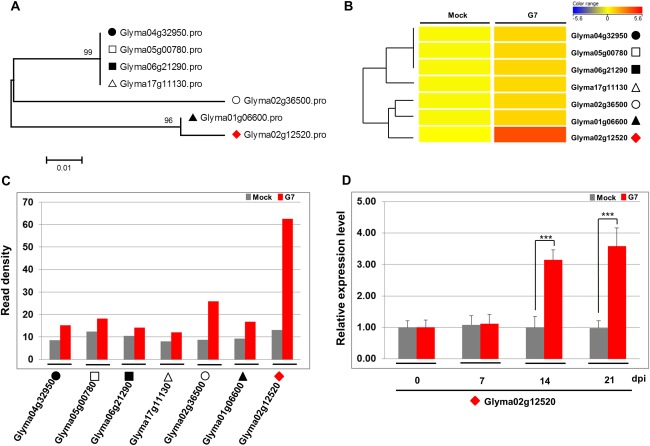
Expression levels of GmelF5A induced by G7 infection. (A) Phylogenetic analysis of seven homologous soybean GmeIF5A genes. The numbers at the nodes are percentages of bootstrap values (1000 replicates). The scale bar indicates 0.01 substitutions per nucleotide position. (B) Heat map displaying the hierarchical clustering of the expression patterns of seven soybean eIF5A genes induced by Soybean mosaic virus (SMV) inoculation at 14 days post‐inoculation (dpi). Colours indicate the log‐scaled value of the normalized read count per kilobase of exon model per million reads. (C) The RNA‐seq read density of seven soybean GmeIF5A genes induced by SMV inoculation compared with mock‐inoculated plants. The read density value is the normalized read count per million reads. (D) The related expression level of Glyma02g12520 in systemic leaves was validated by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis at 0, 7, 14 and 21 dpi after SMV inoculation. The soybean Actin (GmACT11) gene was used as an internal control. Error bars represent mean ± standard deviation (SD) and the data are averages from three biological replicates. Asterisks indicate statistically significant differences compared with the mock control (Student's t‐test): *** P < 0.001.
Silencing of GmeIF5A in Rsv1‐genotype soybean inhibits LSHR induced by G7 infection
To investigate whether GmeIF5A is a key element involved in the signal transduction pathway of Rsv1‐mediated LSHR, we modified a Bean pod mottle virus (BPMV)‐based virus‐induced gene silencing (VIGS) vector to silence the GmeIF5A gene in Rsv1‐genotype soybean (Kachroo and Ghabrial, 2012). The modified vector, introduced into Rsv1‐genotype soybean, contained a 320‐bp specific fragment of the open reading frame of GmeIF5A. At 21 dpi (Fig. 6A), infection by the recombinant BPMV was verified by RT‐PCR (Fig. 6B). RT‐qPCR results suggested that GmeIF5A mRNA was significantly reduced in the soybean plants infected with the BPMV vector containing a GmeIF5A VIGS construct, in comparison with the plants bombarded with the empty vector control (Fig. 6C). Both GmeIF5A‐silenced and vector control plants were then challenged with G7. Unlike the vector control plants, necrosis symptoms and LSHR were diminished in GmeIF5A‐silenced soybean (Fig. 7A). In order to gauge viral accumulation and HR, the expression levels of the SMV coat protein (SMV‐CP) and pathogen‐responsive HR marker gene PR1, respectively, were measured with RT‐qPCR. In contrast with the vector control plants, the accumulation of SMV G7 RNA largely increased in the GmeIF5A‐silenced plants (Fig. 7B), indicating that silencing of GmeIF5A enhanced susceptibility to infection by G7. Conversely, the expression of PR1, a pathogen‐responsive marker for HR, was significantly reduced in the GmeIF5A‐silenced soybean at 14 and 21 dpi (Fig. 7B).
Figure 6.
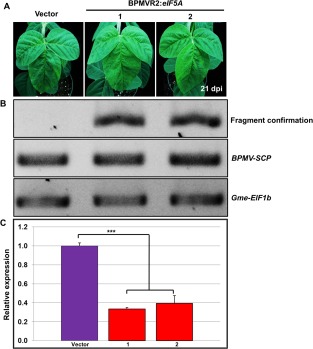
Efficient silencing of GmeIF5A in Rsv1‐genotype soybean (PI96983) by a Bean pod mottle virus (BPMV)‐based virus‐induced gene silencing (VIGS) vector. (A) Modified BPMV‐based VIGS vector, containing a 320‐bp fragment specific to GmeIF5A, was used to silence the GmeIF5A gene in Rsv1 soybean. At 21 days post‐inoculation (dpi), mild BPMV viral symptoms developed on the systemic leaves of plants inoculated with the vector containing the GmeIF5A fragment. Vector, inoculated with the empty vector; BPMVR2:eIF5A, inoculated with the BPMV‐based VIGS vector targeting the GmeIF5A gene. (B) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis confirms vector infection in all samples. The eIF5a fragment is absent from the vector‐only control. The soybean Gme‐EIF1b served as an internal control for fragment specificity. (C) Following VIGS infection, the level of GmeIF5A mRNA was detected by RT‐qPCR in both the vector control and GmeIF5A‐silenced plant (1, 2). The soybean Actin (GmACT11) gene was used as an internal control. Error bars represent mean ± standard deviation (SD) and the data are averages from three biological replicates. Asterisks indicate statistically significant differences compared with the vector control (Student's t‐test): *** P < 0.001.
Figure 7.
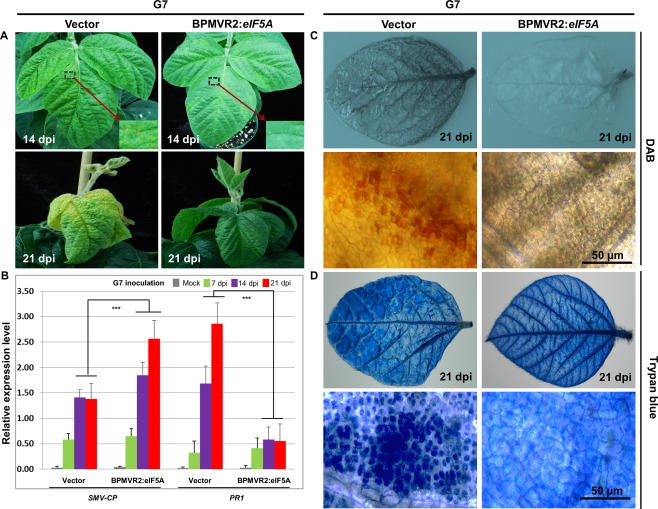
Effects of silencing of GmeIF5A on lethal systemic hypersensitive response (LSHR) severity and Soybean mosaic virus (SMV) G7 viral RNA accumulation in Rsv1‐genotype soybean (PI96983). (A) Vector‐only control plants and GmeIF5A virus‐induced gene‐silenced plants were inoculated with SMV G7. LSHR symptoms developed on leaves of both control and GmeIF5A‐silenced plants at 14 and 21 days post‐inoculation of G7. Vector, inoculated with the empty vector; BPMVR2:eIF5A, inoculated with the eIF5A‐BPMV vector. (B) Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analyses of the accumulated viral RNA (SMV coat protein, SMV‐CP) and the expression of pathogenesis‐related 1 (PR1) transcript in the control and GmeIF5A‐silenced plants at 7, 14 and 21 dpi of G7. The soybean Actin (GmACT11) gene was used as an internal control. Error bars represent mean ± standard deviation (SD) and the data are averages from three independent experiments. Asterisks indicate statistically significant differences between the vector control plant and the GmeIF5A‐silenced plant at 14 and 21 dpi after G7 inoculation (Student's t‐test): *** P < 0.001. The oxidative burst and LSHR formation in response to G7 infection were observed by staining with diaminobenzidine (DAB) (C) and trypan blue (D) in control and GmeIF5A‐silenced leaves at 21 dpi after G7 inoculation. Bars, 50 µm.
To further examine the contribution of GmeIF5A to hydrogen peroxide (H2O2) production during LSHR following G7 infection, the infected systemic leaves on both GmeIF5A‐silenced plants and vector control plants were stained with diaminobenzidine (DAB) and trypan blue at 21 dpi with G7. In contrast with the strong, brown precipitate in the control plants, GmeIF5A‐silenced leaves were DAB negative (Fig. 7C). Trypan blue staining performed on additional leaves from the plants showed a similar result (Fig. 7D). These data suggest that GmeIF5A is an essential component of the G7‐induced, Rsv1‐mediated LSHR signalling pathway in soybean.
Discussion
Viral infection induces vast transcriptional alterations in plants, leading to diverse morphological changes and symptoms. In general, defence‐related genes, including the NBS‐LRR gene family, are up‐regulated in infected plants, whereas photosynthesis‐related genes are often suppressed (Li et al., 2016). To gain an insight into the Rsv1‐mediated LSHR response initiated by G7 infection and to mine potential candidate targets for future study, we conducted a genome‐wide study of mRNA and miRNA regulatory changes following viral infection. In soybean carrying the Rsv1 locus (where one or more NBS‐LRR genes are located), infection with the G7 isolate of SMV induced the down‐regulation of 588 genes and the up‐regulation of 318 genes (Table S2). As expected, genes down‐regulated in response to G7 infection were strongly and significantly enriched in biological processes associated with photosynthesis (GO:0042549, 0042548, 0043467, 0010109, 0019750, etc.; Table S2). In contrast, up‐regulated DEGs were similarly strongly enriched for DNA replication (GO:0006270, 0006261, 0071103) as well as oxylipin biosynthesis (GO:0031408) and metabolism (GO:0031407). Oxylipins are acyclic or cyclic oxidation products, derived from the catabolism of fatty acids, which regulate many defence and developmental pathways in plants (Creelman and Mulpuri, 2002). These data support the previously described general transcriptional responses to viral infection and lend credibility to the data obtained by our approach (Li et al., 2016). More specifically, 119 genes with potential roles in plant defence (GO:006952) were identified. In view of the essential role of this gene group in viral pathogenesis, these DEGs responsive to G7 infection are probably involved in Rsv1‐mediated LSHR, and some key DEGs warrant more detailed studies. For instance, a kunitz family trypsin and protease inhibitor protein (GLYMA09G28310) and a chloroplast carbonic anhydrase (GLYMA05G08590) were specifically up‐regulated and down‐regulated by G7 infection, respectively, and both genes play a key role in the signalling pathway involved in the HR during host–pathogen interactions (Li et al., 2008; Slaymaker et al., 2002).
Thirty‐seven miRNAs were up‐regulated and 47 were down‐regulated (Fig. 2B,C) in response to G7 infection. Many miRNAs identified from this study have been implicated in plant–microbe interactions in previous studies. For instance, miR160 (up‐regulated by G7 infection, Table S4) and miR393 (down‐regulated, Table S4) have also been shown to be involved in the soybean defence response to SMV infection (SC7 isolate) (Yin et al., 2013). In Arabidopsis, miR156 is induced by expression of a viral silencing suppressor, such as P1/HC‐Pro of Turnip mosaic virus (TuMV) (Kasschau et al., 2003) and p69 of Turnip yellow mosaic virus (TYMV) (Chen et al., 2004). TuMV infection also up‐regulates miR171, which directs the cleavage of several mRNAs coding for Scarecrow‐like TFs (Kasschau et al., 2003). In the current study, many members of the miR156 (p/q/r/s/t) and miR171 (c/i/k/l/m/n/o/p/q/t) families were highly responsive to SMV inoculation (Table S4). The miR482/miR2118 superfamily in tomato, soybean and Medicago truncatula typically triggers phased siRNA generation and targets numerous NB‐LRR defence genes at the conserved P‐loop‐encoding motif (Shivaprasad et al., 2012; Zhai et al., 2011; Zhao et al., 2015). In chickpea (Cicer arietinum), miR2118 is up‐regulated in response to wilt infection with the fungus Fusarium oxysporum, but is down‐regulated after infection with the fungus Verticillium dahliae in cotton (Kohli et al., 2014; Yin et al., 2012). miR482 is down‐regulated in the inoculated leaves of tomato plants infected with Cucumber mosaic virus (CMV), Turnip crinkle virus (TCV) and Tobacco rattle virus (TRV) (Shivaprasad et al., 2012). In this study, miR2118a/b‐5p was found to be highly up‐regulated by G7 infection (Tables S4). It would be interesting to determine how these miRNAs are involved in G7 pathogenesis and the Rsv1‐mediated LSHR.
RNA‐seq also revealed a number of genes with potential roles in the LSHR signalling pathway. As mentioned above, GLYMA09G28310 codes for a kunitz family trypsin and protease inhibitor protein (KTI) which is known to be involved in the modulation of programmed cell death (PCD) or apoptosis in the HR of the plant response to pathogens (Li et al., 2008). KTI was up‐regulated by G7 infection. GLYMA05G08590, encoding a chloroplast carbonic anhydrase that may play a key role in the hypersensitive defence response (Slaymaker et al., 2002), was down‐regulated by G7 infection. GLYMA02G12520 encodes eIF5A, which is critical for the signalling transduction pathway of cell death (Hopkins et al., 2008). A soybean elF5A gene (GLYMA02G12520) was specifically up‐regulated by G7 infection (Fig. 5A). eIF5A is a highly conserved protein found in different kingdoms of eukaryotic organisms (Jenkins et al., 2001). Of the three eIF5A isoforms in Arabidopsis, AteIF5A‐2 is a key element of the signal transduction pathway of cell death, shown to be involved in the development of disease symptoms induced by infection with virulent Pseudomonas syringae pv. tomato DC3000 (Hopkins et al., 2008). eIF5A has been suggested to play a crucial role in plant growth and development by the regulation of cell division, cell growth and cell death (Feng et al., 2007; Hopkins et al., 2008; Thompson et al., 2004; Wang et al., 2001). Despite this growing evidence, the precise way in which this protein functions in the cell is not yet known, and no study has been performed on elF5A in soybean.
There are seven loci which may encode elF5A homologues in soybean (Fig. 5A). The soybean genome has undergone two whole‐genome duplication events, occurring approximately 59 and 13 million years ago, followed by gene diversification and loss, and numerous chromosome rearrangements, leading to the relatively large number of eIF5A homologues compared with Arabidopsis (Schmutz et al., 2010). Among these seven eIF5A homologues, GmeIF5A (GLYMA02G12520) was most highly induced by G7 infection (Fig. 5C,D). Silencing of GmelF5A (GLYMA02G12520) diminished G7‐induced necrosis symptoms and the LSHR (Fig. 7A), and was accompanied by the accumulation of a higher level of G7 viral RNA (Fig. 7B), suggesting that silencing of GmeIF5A enhanced the susceptibility to infection by G7. Conversely, the expression levels of the PR1 gene were significantly reduced and oxidative bursts were hardly detectable in the GmeIF5A‐silenced, G7‐infected soybean compared with the G7‐infected control plants (Fig. 7B–D). LSHR, a type of necrosis, is elicited by reactive oxygen species (ROS) (Hernández et al., 2015). Our data are consistent with dual roles of ROS (including H2O2) during viral infection: the elicitation of localized cell death, which may lead to the containment of the invading viruses in local cells, and a function as a diffusible signal to induce antioxidant and pathogenesis‐related defence responses in adjacent plant tissues. As GmeIF5A shares a high nucleotide sequence identity (97%) with Glyma01g06600 and, to a lesser extent (∼80%), with five other eIF5A homologues, we could not exclude the possibility that other eIF5A homologous genes (particularly Glyma01g06600) were knocked down and thus also contributed to the Rsv1‐mediated LSHR. Overall, these results suggest that the GmeIF5A gene family plays a crucial and complex role, upstream of ROS and PR1 in the LSHR signalling pathway. Future studies examining the impact of increased GmeIF5A levels during G7 infection in Rsv1 plants could provide further insight into the precise role of eIF5A within the cell.
In this study, sRNA‐seq, degradome‐seq and transcriptome‐seq analyses were employed to identify DEGs and DEMs that are specifically up‐regulated or down‐regulated by SMV G7 infection. Our results identified multiple defence genes and miRNA candidates that may be involved in SMV G7‐induced Rsv1‐mediated LSHR. Further investigation of one of these candidates, eIF5A, suggests a central role of this gene family in the Rsv1‐mediated LSHR signalling pathway, and future studies will be directed towards the elucidation of the exact mechanism of GmeIF5A in the LSHR pathway. This finding strengthens our belief that the bulk data gathered in this study represent a fertile resource for future studies on the molecular mechanism of Rsv1‐mediated LSHR as well as a means to prevent losses in crop species.
Experimental Procedures
Soybean cultivar, virus strains, inoculation and detection
Rsv1‐genotype soybean (PI96983) was used in this study. Plants were grown in a growth chamber under a 16‐h light at 22°C/8‐h dark at 18°C cycle. Plants were inoculated by biolistic bombardment with SMV infectious clones (G7), and mechanical inoculation with infected tissues as an inoculum was carried out as described previously (Gagarinova et al., 2008a, 2008b; Hajimorad and Hill, 2001; Hajimorad et al., 2003). The nearly fully expanded unifoliate leaves of Rsv1‐genotype soybean (PI96983) were mechanically inoculated with the infectious sap of SMV G7 (a virulent strain). As a control, soybean leaves were inoculated with a mock solution (buffer only). The systemic leaves of G7‐infected plants were sampled at 14 dpi, showing symptoms of LSHR.
At 14 dpi, symptomatic (in the case of virus‐infected plants) leaf tissue was collected from mock and SMV G7‐inoculated plants. Systemic viral infection was subsequently determined by RT‐PCR analysis as described previously (Chen et al., 2015). Three biological replicates (five plants in each replicate) for each treatment (G7‐infected and mock) were performed; 15 plants from each treatment in three replicates were pooled for RNA extraction and all library construction.
Transcriptome, sRNA and degradome library construction, sequencing and data analysis
Transcriptome libraries were constructed as described previously (Jones and Vodkin, 2013; Mortazavi et al., 2008; Severin et al., 2010). sRNA libraries were constructed as described previously with minor modifications (Chen et al., 2015; Lu et al., 2007). Degradome libraries were constructed as described previously with some modified procedures (German et al., 2008, 2009; Zhai et al., 2014). RNA was extracted using a mirVana miRNA isolation kit (Ambion, Austin, TX, USA). The quantity, size and integrity of the RNAs used in these analyses were assessed using an RNA 6000 Nano kit on a Bioanalyzer 2100 system (Agilent Technologies, Palo Alto, CA, USA). For transcriptome analysis, an input of 1 µg of total RNA was employed to construct cDNA libraries using a TruSeq Stranded Total RNA Sample Prep Kit (Illumina, San Diego, CA, USA) with a Ribo‐Zero rRNA depletion kit, following the manufacturer's instructions. For sRNA analysis, an input of 50 ng of sRNA fractions was employed to construct cDNA libraries using a TruSeq sRNA Sample Prep Kit (Illumina), following the manufacturer's instructions. For degradome sequencing, poly(A)+ RNA was isolated using a NucleoTrap mRNA purification kit (Machery‐Nagel, St. Neumann Neander, Düren, Germany) according to the instructions of the supplier. A 5′ RNA adapter was ligated to the cleavage product, which contains a 5′ monophosphate, and the ligated product was reverse transcribed into cDNA using an oligo(dT) primer with a 3′ adapter sequence via SuperScript III RTase (Invitrogen, Carlsbad, CA, USA), and amplified by PCR [(98°C for 30 s, 58°C for 30 s and 72°C for 5 min, eight cycles), 72°C for 7 min] with a pair of cDNA primers using Phusion polymerase (NEB, Woburn, MA, USA). The resulting product was digested with restriction enzyme MmeI (NEB, Woburn, MA, USA) to capture ∼20‐bp fragments from the 5´ end of double‐stranded cDNA. The digested products were ligated with an annealed duplex DNA adapter using T4 DNA ligase (NEB, Woburn, MA, USA). Then, the ligated double‐stranded DNA products were isolated by running a 12% polyacrylamide (PAGE) gel based on size (∼62 bp), and the purified products were amplified by PCR [98°C for 30 s, (98°C for 10 s, 58°C for 30 s and 72°C for 20 s, 20 cycles), 72°C for 10 min] with a set of indexed TruSeq primers (forward, RP1 and reverse, RPI 1–4 for each library, Illumina). The final PCR products were purified by running a 6% PAGE gel based on size (∼128 bp).
The constructed libraries were analysed for size distribution and quality assessment on an Agilent 2100 Bioanalyzer and were quantified by running a library quantification qPCR kit (KAPA Biosystems, Woburn, MA, USA) on a CFX96 real‐time PCR detection system (Bio‐Rad, Hercules, CA, USA) in order to determine the concentration of libraries and the loading volume for deep sequencing on an Illumina Miseq platform.
For RNA‐seq and sRNA‐seq analysis, after the low‐quality reads and the adapter sequences had been trimmed, the sequences were mapped to the Glycine max reference genome (Glyma1, Ensembl) and aligned to the known miRNA gene of Glycine max (miRBase 21) using Strand NGS software (Strand Life Sciences, Strand genomics, San Francisco, CA, USA, version 2.1) following RNA/sRNA alignment and the RNA/sRNA analysis pipeline with standard parameters. Only reads with 100% match to the genome were used for further analysis. For degradome‐seq analysis, the adapter sequences, tRNA/rRNA sequences and low‐complexity sequences were removed from the raw reads, and the length of the degradome sequences and sRNA sequences were trimmed to 20–21 and 19–24 nucleotides, respectively. Any sequences without a match to the soybean genome (Glyma1, Ensembl) were also removed from further analysis. The potential targets of sRNAs were identified and validated using the UEA sRNA workbench following the PAREsnip pipeline under a high‐stringency setting with the Glycine max reference genome (Glyma1, Ensembl) and the known miRNAs of Glycine max (miRBase 21).
Transcriptome‐seq, sRNA‐seq and degradome‐seq data used for this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) under accession number GSE77796.
qRT‐PCR
Total RNA collected from mock‐inoculated and SMV‐infected plants was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed with a Superscript III Reverse Transcriptase kit (Life Technologies, Rockville, MD, USA) using gene‐specific reverse primer according to the manufacturer's instructions. qPCR was performed using the respective forward and reverse primer pairs, as shown in Table S6 (see Supporting Information). The soybean Actin (GmACT11) gene was used as an internal control. Three independent experiments were performed with five soybean plants.
Stem‐loop RT‐qPCR
The stem‐loop RT‐qPCR assay was carried out as described previously (Mestdagh et al., 2008, 2009). Briefly, RT was performed using a TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer's protocol with a stem‐loop RT primer bound to the 3′ portion of the miRNAs. The RT product was amplified using TaqMan® Universal PCR Master Mix (Applied Biosystems) with an miRNA‐specific forward primer and a universal reverse primer. Soybean 18S rRNA was used as the internal control. Primer sequences are shown in Table S6. The experiments were repeated at least three times.
RLM‐5′ RACE
RLM‐5′ RACE was performed using a FirstChoice RLM‐RACE Kit (Ambion) essentially as described previously (German et al., 2008; Llave et al., 2011). Poly(A)+ mRNAs were purified from total RNA using a NucleoTrap mRNA purification kit (Machery‐Nagel) according to the manufacturer's protocol. Approximately 150 ng of mRNAs were ligated to the 5′ RACE adapter. The ligated products were purified using spin‐column chromatography (Roche, Indianapolis, IN, USA) and reverse transcribed into first‐strand cDNA using an antisense gene‐specific (GSP) outer primer. PCR amplification was carried out using the 5′ RACE outer primer and GSP outer primer. The initial PCR was diluted (1 : 50) and used for nested PCR with the 5′ RACE inner primer and GSP inner primer. The primers are shown in Table S6. RACE fragments were cloned into pCR‐BluntII‐TOPO vector (Invitrogen) and 10 positive clones were sequenced according to standard methods.
Phylogenetic analysis of GmeIF5a homologues
A phylogenetic tree was produced using the maximum likelihood method with the deduced amino acid sequences with mega5.1 software (Tamura et al., 2011) with 1000 replications.
BPMV VIGS construction and challenging inoculation by G7 on GmeIF5A‐silenced Rsv1‐genotype soybean
To increase silencing efficiency, a DNA‐based BPMV vector was developed based on a previous version of the RNA‐based BPMV vector (Kachroo and Ghabrial, 2012). In brief, a 307‐bp fragment of the NOS terminator was amplified by PCR with pCAMBIA3301 as a template. PCR products were double digested with BamHI and PmeI, and ligated into pCass4‐Rz previously digested with the same enzymes (Annamalai and Rao, 2005), resulting in pCass4‐NOS. The cDNAs of BPMV genomic RNA1 and RNA2 were PCR amplified with RNA‐based BPMV vector pGHoR1 and pGG7R2‐M, respectively, as a template (Diaz‐Camino et al., 2011). PCR products were digested with BamHI and XbaI, respectively, and placed in between the 35S promoter and NOS terminator of pCass4‐NOS, which was previously digested with StuI/BamHI and StuI/XbaI, respectively, resulting in pCass4R1‐NOS and pCass4R2‐NOS. An overlapping PCR was used to introduce StuI and AvrII restriction sites after the stop codon of the BPMV RNA2 polyprotein coding sequence to generate the VIGS vector pCass4R2OL‐NOS. For the generation of an eIF5A VIGS vector, a 320‐bp GmeIF5A fragment was amplified with cDNA as a template, which transcribed with total RNA extracted from leaves of Rsv1‐genotype soybean (PI96983). The PCR product was cloned into StuI‐digested pCass4R2OL‐NOS to generate eIF5A VIGS vector pCass4R2OL‐eIF5A‐NOS. The insert in the construct was verified by sequencing. For inoculation, biolistic bombardment with the constructed BPMV VIGS vectors was carried out as described previously (Chen et al., 2015). The inoculated plants were grown in a growth chamber at 20°C. At 3 weeks post‐inoculation with BPMV, the systemic leaves on BPMV‐infected Rsv1‐genotype soybean plants were rub inoculated with SMV G7‐infected tissues as an inoculum. At 14 dpi with SMV G7, the SMV‐infected leaves were detached for qRT‐PCR analysis and histochemical assay. Three independent tests for each construct were performed. In total, 10 eIF5A‐silenced plants from each test were used to evaluate G7‐provoking Rsv1‐mediated LSHR. Primer sequences are included in Table S6.
Histochemical assays
DAB staining was performed as described previously (Daudi et al., 2012; Thordal‐Christensen et al., 1997). Trypan blue staining was carried out as described previously (Koch and Slusarenko, 1990). The representative phenotypes were photographed with a microscope EVOS (Life Technologies, Grand Island, NY, USA).
Author Contributions
H.C. and A.W. conceived the project and designed the work. H.C. performed the experiments. H.C. and A.A.A. conducted bioinformatics analyses. All authors analysed and reviewed the experimental data. K.Y. provided soybean materials. H.C., A.A. and A.W. wrote the paper.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Gene ontology (GO) and enrichment analysis of the differentially expressed genes (DEGs) in response to G7 infection using the GO Database (Soybase) in Rsv1‐genotype soybean (PI96983). G7 infection resulted in a significant change in expression of eight genes with transcription factor (TF) activity, 14 genes with ATP/ADP activity and 14 genes with nucleotide binding activity. Genes with protein and ion binding activity were the largest categories with 35 combined genes.
Fig. S2 Target plots (t‐plots) of identified microRNA (miRNA) targets by degradome sequencing (degradome‐seq). t‐plots are shown at the top and sequence alignments of miRNA and their targets are shown at the bottom for miR167a, miR169a, miR171o, miR396b‐3p, miR5374 and miR5668. In the t‐plots, the degradome sequence corresponding to the miRNA‐directed cleaved transcript is represented by a red diamond and black arrow. The x axis indicates the nucleotide position on the targeted transcript (nt, nucleotide). The y axis indicates the normalized read abundance (TPM, transcripts per million) of cleaved transcript detected in degradome‐seq. In the alignments, the vertical lines, missing lines and circles indicate matches, mismatches and G:U wobble pairs, respectively. The black arrows (red coloured nucleotide) above the target transcript indicate the cleavage site detected in degradome‐seq. The numbers of clones sequenced show the cleavage frequencies detected by RNA ligase‐mediated rapid amplification of 5′·cDNA ends (RLM‐5′ RACE) assay.
Table S1 Summary of deep sequencing library datasets from mock‐ and virus‐infected Rsv1‐genotype soybean (PI96983).
Table S2 List of differentially expressed genes (DEGs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S3 List of defence‐related differentially expressed genes (DEGs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S4 List of differentially expressed microRNAs (DEMs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S5 List of identified microRNA targets by degradome sequencing in Rsv1‐genotype soybean (PI96983).
Table S6 Primer sequences used in this study.
Acknowledgements
The authors are very grateful to Jamie McNeil [Agriculture and Agri‐Food Canada (AAFC), Ottawa, ON, Canada] for expert technical assistance and Alex Molnar (AAFC) for artwork. The SMV G7 infectious clone was kindly provided by Professor John Hill (Iowa State University, Ames, IA, USA). The BPMV vector was a kind gift from Professor Said Ghabrial (University of Kentucky, Lexington, KY, USA).
This work was supported in part by AAFC Genomics R and D Initiative (GRDI), the Natural Sciences and Engineering Research Council of Canada (NSERC) and Grain Farmers Ontario (GFO).
References
- Annamalai, P. and Rao, A.L. (2005) Replication‐independent expression of genome components and capsid protein of Brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338, 96–111. [DOI] [PubMed] [Google Scholar]
- Babu, M. , Gagarinova, A.G. , Brandle, J.E. and Wang, A. (2008) Association of the transcriptional response of soybean plants with Soybean mosaic virus systemic infection. J. Gen. Virol. 89, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bazzini, A.A. , Almasia, N.I. , Manacorda, C.A. , Mongelli, V.C. , Conti, G. , Maroniche, G.A. , Rodriguez, M.C. , Distéfano, A.J. , Hopp, H.E. and del Vas, M. (2009) Virus infection elevates transcriptional activity of miR164a promoter in plants. BMC Plant Biol. 9, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin, D.D. , Aldea, M. , O'Neill, B.F. , Benitez, M. , Li, M. , Clough, S.J. and DeLucia, E.H. (2008) Elevated ozone alters soybean–virus interaction. Mol. Plant–Microbe Interact. 21, 1297–1308. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He, S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Zhang, L. , Yu, K. and Wang, A. (2015) Pathogenesis of Soybean mosaic virus in soybean carrying Rsv1 gene is associated with miRNA and siRNA pathways, and breakdown of AGO1 homeostasis. Virology, 476, 395–404. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Li, W.X. , Xie, D. , Peng, J.R. and Ding, S.W. (2004) Viral virulence protein suppresses RNA silencing‐mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell, 16, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. , Buss, G.R. , Roane, C.W. and Tolin, S.A. (1994) Inheritance in soybean of resistant and necrotic reactions to Soybean mosaic virus strains. Crop Sci. 34, 414–422. [Google Scholar]
- Cho, E.K. and Goodman, R.M. (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology, 69, 467–470. [Google Scholar]
- Chowda‐Reddy, R.V. , Sun, H. , Hill, J.H. , Poysa, V. and Wang, A. (2011) Simultaneous mutations in multi‐viral proteins are required for Soybean mosaic virus to gain virulence on soybean genotypes carrying different R genes. PLoS One, 6, e28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A. and Mulpuri, R. (2002) The oxylipin pathway in Arabidopsis. Arabidopsis Book, 1, e0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver, J.N. , Lindbeck, A.G.C. and Dawson, W.O. (1991) Virus–host interactions: induction of chlorotic and necrotic responses in plants by tobamoviruses. Annu. Rev. Phytopathol. 29, 193–217. [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi, A. , Cheng, Z. , O'brien, J.A. , Mammarella, N. , Khan, S. , Ausubel, F.M. and Bolwell, G.P. (2012) The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern‐triggered immunity. Plant Cell, 24, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Diaz‐Camino, C. , Annamalai, P. , Sanchez, F. , Kachroo, A. and Ghabrial, S.A. (2011) An effective virus‐based gene silencing method for functional genomics studies in common bean. Plant Methods, 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh‐Kumar, S.P. , Tham, W.H. and Baker, B.J. (2000) Structure–function analysis of the Tobacco mosaic virus resistance gene N . Proc. Natl. Acad. Sci. USA, 97, 14 789–14 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, P. , Wu, J. , Zhang, J. , Zhao, S. , Zheng, H. , Gao, G. , Wei, L. and Li, Y. (2011) Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 7, e1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H. , Chen, Q. , Feng, J. , Zhang, J. , Yang, X. and Zuo, J. (2007) Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A‐2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant . Physiol. 144, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkes, L. , Moxon, S. , Woolfenden, H.C. , Stocks, M.B. , Szittya, G. , Dalmay, T. and Moulton, V. (2012) PAREsnip: a tool for rapid genome‐wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res. 40, e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagarinova, A.G. , Babu, M. , Poysa, V. , Hill, J.H. and Wang, A. (2008a) Identification and molecular characterization of two naturally occurring Soybean mosaic virus isolates that are closely related but differ in their ability to overcome Rsv4 resistance. Virus Res. 138, 50–56. [DOI] [PubMed] [Google Scholar]
- Gagarinova, A.G. , Babu, M. , Strömvik, M.V. and Wang, A. (2008b) Recombination analysis of Soybean mosaic virus sequences reveals evidence of RNA recombination between distinct pathotypes. Virol. J. 5, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Marcos, A. , Pacheco, R. , Martianez, J. , Gonzalez‐Jara, P. , Diaz‐Ruiz, J.R. and Tenllado, F. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant–Microbe Interact. 22, 1431–1444. [DOI] [PubMed] [Google Scholar]
- German, M.A. , Pillay, M. , Jeong, D.H. , Hetawal, A. , Luo, S. , Janardhanan, P. , Kannan, V. , Rymarquis, L.A. , Nobuta, K. and German, R. (2008) Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. [DOI] [PubMed] [Google Scholar]
- German, M.A. , Luo, S. , Schroth, G. , Meyers, B.C. and Green, P.J. (2009) Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4, 356–362. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. and Hill, J.H. (2001) Rsv1‐mediated resistance against Soybean mosaic virus‐N is hypersensitive response‐independent at inoculation site, but has the potential to initiate a hypersensitive response‐like mechanism. Mol. Plant–Microbe Interact. 14, 587–598. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2003) Evolution of Soybean mosaic virus‐G7 molecularly cloned genome in Rsv1‐genotype soybean results in emergence of a mutant capable of evading Rsv1‐mediated recognition. Virology, 314, 497–509. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2005) Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1‐mediated lethal systemic hypersensitive response maps to P3. J. Virol. 79, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T.J. (1980) Resistance at the Tm‐2 locus in the tomato to Tomato mosaic virus . Euphytica, 29, 189–197. [Google Scholar]
- Havelda, Z. , Várallyay, É. , Valoczi, A. and Burgyán, J. (2008) Plant virus infection‐induced persistent host gene downregulation in systemically infected leaves. Plant J. 55, 278–288. [DOI] [PubMed] [Google Scholar]
- Hernández, J.A. , Gullner, G. , Clemente‐Moreno, M.J. , Künstler, A. , Juhász, C. , Díaz‐Vivancos, P. and Király, L. (2015) Oxidative stress and antioxidative responses in plant–virus interactions. Physiol. Mol. Plant Pathol. doi: 10.1016/j.pmpp.2015.09.001. [DOI] [Google Scholar]
- Hopkins, M.T. , Lampi, Y. , Wang, T.W. , Liu, Z. and Thompson, J.E. (2008) Eukaryotic translation initiation factor 5A is involved in pathogen‐induced cell death and development of disease symptoms in Arabidopsis . Plant Physiol. 148, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, Z.A. , Haag, P.G. and Johansson, H.E. (2001) Human eIF5A2 on chromosome 3q25‐q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue‐specific expression. Genomics, 71, 101–109. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jones, S.I. and Vodkin, L.O. (2013) Using RNA‐Seq to profile soybean seed development from fertilization to maturity. PLoS One, 8, e59270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A. and Ghabrial, S. (2012) Virus‐induced gene silencing in soybean. Methods Mol. Biol. 894, 287–297. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D. , Xie, Z. , Allen, E. , Llave, C. , Chapman, E.J. , Krizan, K.A. and Carrington, J.C. (2003) P1/HC‐Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell, 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, D. , Joshi, G. , Deokar, A.A. , Bhardwaj, A.R. , Agarwal, M. , Katiyar‐Agarwal, S. , Srinivasan, R. and Jain, P.K. (2014) Identification and characterization of wilt and salt stress‐responsive microRNAs in chickpea through high‐throughput sequencing. PLoS One, 9, e108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C.J. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. , Heinlein, M. and Hann, D.R. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant–Microbe Interact. 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer, F.C. , Dijkhuis, J. , Sturre, M.J. , de Haan, P. and Hille, J. (2003) Cloning and characterization of the durable Tomato mosaic virus resistance gene Tm‐2(2) from Lycopersicon esculentum . Plant Mol. Biol. 52, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2008) Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis . Mol. Plant. 1, 482–495. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Cui, H. , Cui, X. and Wang, A. (2016) The altered photosynthetic machinery during compatible virus infection. Curr. Opin. Virol. 17, 19–24. [DOI] [PubMed] [Google Scholar]
- Llave, C. , Franco‐Zorrilla, J.M. , Solano, R. and Barajas, D. (2011) Target validation of plant microRNAs. Methods Mol. Biol. 732, 187–208. [DOI] [PubMed] [Google Scholar]
- Lu, C. , Meyers, B.C. and Green, P.J. (2007) Construction of small RNA cDNA libraries for deep sequencing. Methods, 43, 110–117. [DOI] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.B. (2013) Plant immune responses against viruses: how does a virus cause disease? Plant Cell, 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G. (2013) Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genetics, 14, 447–459. [DOI] [PubMed] [Google Scholar]
- Mestdagh, P. , Feys, T. , Bernard, N. , Guenther, S. , Chen, C. , Speleman, F. and Vandesompele, J. (2008) High‐throughput stem‐loop RT‐qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 36, e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh, P. , Van Vlierberghe, P. , De Weer, A. , Muth, D. , Westermann, F. , Speleman, F. and Vandesompele, J. (2009) A novel and universal method for microRNA RT‐qPCR data normalization. Genome Biol. 10, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, T. , Ogata, Y. , Hirata, Y. and Ohki, S.T. (2014) Quantitative transcriptional changes associated with chlorosis severity in mosaic leaves of tobacco plants infected with Cucumber mosaic virus . Mol. Plant Pathol. 15, 242–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐Seq. Nat. Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Selassie, K.G. , Marchoux, G. , Daubeze, A.M. and Palloix, A. (1998) High temperature effects on hypersensitive resistance to Tomato spotted wilt Tospovirus (TSWV) in pepper (Capsicum chinense Jacq.). Eur. J. Plant Pathol. 104, 489–498. [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. and Nishimura, M.T. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. (2014) Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan, B. , Naqvi, A.R. , Saraf, S. , Mukherjee, S.K. and Dey, N. (2015) Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum . Virus Res. 195, 183–195. [DOI] [PubMed] [Google Scholar]
- Romanel, E. , Silva, T.F. , Corrêa, R.L. , Farinelli, L. , Hawkins, J.S. , Schrago, C.E.G. and Vaslin, M.F.S. (2012) Global alteration of microRNAs and transposon‐derived small RNAs in cotton (Gossypium hirsutum) during Cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol. Biol. 80, 443–460. [DOI] [PubMed] [Google Scholar]
- Schmutz, J. , Cannon, S.B. , Schlueter, J. , Ma, J. , Mitros, T. , Nelson, W. , Hyten, D.L. , Song, Q. , Thelen, J.J. and Cheng, J. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Severin, A.J. , Woody, J.L. , Bolon, Y.T. , Joseph, B. , Diers, B.W. , Farmer, A.D. , Muehlbauer, G.J. , Nelson, R.T. , Grant, D. , Specht, J.E. , Graham, M.A, Cannon, S.B., May, G.D., Vance, C.P. and Shoemaker, R.C. (2010) RNA‐Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Lee, S.H. and Kim, K.H. (2009) Strain‐specific cylindrical inclusion protein of soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant–Microbe Interact. 22, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Shamimuzzaman, M. and Vodkin, L. (2012) Identification of soybean seed developmental stage‐specific and tissue‐specific miRNA targets by degradome sequencing. BMC Genomics, 13, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Chen, H.M. , Patel, K. , Bond, D.M. , Santos, B.A. and Baulcombe, D.C. (2012) A microRNA superfamily regulates nucleotide binding site‐leucine‐rich repeats and other mRNAs. Plant Cell, 24, 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker, D.H. , Navarre, D.A. , Clark, D. , del Pozo, O. , Martin, G.B. and Klessig, D.F. (2002) The tobacco salicylic acid‐binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc . Natl. Acad. Sci. USA, 99, 11 640–11 645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R. , Li, Y.F. and Jagadeeswaran, G. (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol . Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.E. , Hopkins, M.T. , Taylor, C. and Wang, T.W. (2004) Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 9, 174–179. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. , Zhang, Z. and Wei, Y. , Collinge, D.B. (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. Plant J. 11, 1187–1194. [Google Scholar]
- Várallyay, É. , Válóczi, A. , Ágyi, Á. , Burgyán, J. and Havelda, Z. (2010) Plant virus‐mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 29, 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.W. , Lu, L. , Wang, D. and Thompson, J.E. (2001) Isolation and characterization of senescence‐induced cDNAs encoding deoxyhypusine synthase and eukaryotic translation initiation factor 5A from tomato. J. Biol. Chem. 276, 17 541–17 549. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, L. , Zou, Y. , Chen, L. , Cai, Z. , Zhang, S. , Zhao, F. , Tian, Y. , Jiang, Q. , Ferguson, B.J. , Gresshoff, P.M. and Li, X. (2014) Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell, 26, 4782–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J. , Gao, L. , Yang, Y. , Zhai, J. , Arikit, S. , Yu, Y. , Duan, S. , Chan, V. , Xiong, Q. , Yan, J. , Li, S., Liu, R., Wang, Y., Tang, G., Meyers, B.C., Chen, X. and Ma, W. (2014) Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, Z. , Wang, Y. , Zheng, L. , Ye, R. , Ji, Y. , Zhao, S. , Ji, S. , Liu, R. , Xu, L. , Zheng, H., Zhou, Y., Zhang, X., Cao, X., Xie, L., Wu, Z., Qi, Y. and Li, Y. (2015) Viral‐inducible Argonaute18 confers broad‐spectrum virus resistance in rice by sequestering a host microRNA. eLife, 4, e05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Liu, Q. , Chen, L. , Kuang, J. , Walk, T. , Wang, J. and Liao, H. (2013) Genome‐wide identification of soybean microRNAs and their targets reveals their organ‐specificity and responses to phosphate starvation. BMC Genomics, 14, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Guo, R. , Jie, F. , Nettleton, D. , Peng, J. , Carr, T. , Yeakley, J.M. , Fan, J.B. and Whitham, S.A. (2007) Spatial analysis of Arabidopsis thaliana gene expression in response to Turnip mosaic virus infection. Mol. Plant–Microbe Interact. 20, 358–370. [DOI] [PubMed] [Google Scholar]
- Yin, X. , Wang, J. , Cheng, H. , Wang, X. and Yu, D. (2013) Detection and evolutionary analysis of soybean miRNAs responsive to Soybean mosaic virus . Planta, 237, 1213–1225. [DOI] [PubMed] [Google Scholar]
- Yin, Z. , Li, Y. , Han, X. and Shen, F. (2012) Genome‐wide profiling of miRNAs and other small non‐coding RNAs in the Verticillium dahliae‐inoculated cotton roots. PLoS One, 7, e35765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.G. , Saghai Maroof, M.A. , Buss, G.R. , Maughan, P.J. and Tolin, S.A. (1994) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology, 84, 60–64. [Google Scholar]
- Zhai, J. , Jeong, D.H. , De Paoli, E. , Park, S. , Rosen, B.D. , Li, Y. , González, A.J. , Yan, Z. , Kitto, S.L. and Grusak, M.A. (2011) MicroRNAs as master regulators of the plant NB‐LRR defense gene family via the production of phased, trans‐acting siRNAs. Genes Dev. 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, J. , Arikit, S. , Simon, S.A. , Kingham, B.F. and Meyers, B.C. (2014) Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods, 67, 84–90. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Grosic, S. , Whitham, S.A. and Hill, J.H. (2012) The requirement of multiple defense genes in soybean Rsv1‐mediated extreme resistance to Soybean mosaic virus . Mol. Plant–Microbe Interact. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.R. , Pei, Y. , Lin, S.S. , Tuschl, T. , Patel, D.J. and Chua, N.H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. , Meyers, B.C. , Cai, C. , Xu, W. and Ma, J. (2015) Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell, 27, 546–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Gene ontology (GO) and enrichment analysis of the differentially expressed genes (DEGs) in response to G7 infection using the GO Database (Soybase) in Rsv1‐genotype soybean (PI96983). G7 infection resulted in a significant change in expression of eight genes with transcription factor (TF) activity, 14 genes with ATP/ADP activity and 14 genes with nucleotide binding activity. Genes with protein and ion binding activity were the largest categories with 35 combined genes.
Fig. S2 Target plots (t‐plots) of identified microRNA (miRNA) targets by degradome sequencing (degradome‐seq). t‐plots are shown at the top and sequence alignments of miRNA and their targets are shown at the bottom for miR167a, miR169a, miR171o, miR396b‐3p, miR5374 and miR5668. In the t‐plots, the degradome sequence corresponding to the miRNA‐directed cleaved transcript is represented by a red diamond and black arrow. The x axis indicates the nucleotide position on the targeted transcript (nt, nucleotide). The y axis indicates the normalized read abundance (TPM, transcripts per million) of cleaved transcript detected in degradome‐seq. In the alignments, the vertical lines, missing lines and circles indicate matches, mismatches and G:U wobble pairs, respectively. The black arrows (red coloured nucleotide) above the target transcript indicate the cleavage site detected in degradome‐seq. The numbers of clones sequenced show the cleavage frequencies detected by RNA ligase‐mediated rapid amplification of 5′·cDNA ends (RLM‐5′ RACE) assay.
Table S1 Summary of deep sequencing library datasets from mock‐ and virus‐infected Rsv1‐genotype soybean (PI96983).
Table S2 List of differentially expressed genes (DEGs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S3 List of defence‐related differentially expressed genes (DEGs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S4 List of differentially expressed microRNAs (DEMs) in response to Soybean mosaic virus (SMV) G7 infection in Rsv1‐genotype soybean (PI96983).
Table S5 List of identified microRNA targets by degradome sequencing in Rsv1‐genotype soybean (PI96983).
Table S6 Primer sequences used in this study.


