Summary
Pectobacterium carotovorum ssp. brasiliense 1692 (Pcb1692) is an important emerging pathogen of potatoes causing blackleg in the field and soft rot during post‐harvest storage. Blackleg diseases involve the bacterial colonization of vascular tissue and the formation of aggregates, also known as biofilms. To understand the role of quorum sensing in vascular colonization by Pcb1692, we generated a Pcb1692ΔexpI mutant strain. Inactivation of expI led to the reduced production of plant cell wall‐degrading enzymes (PCWDEs), the inability to produce acyl homoserine lactone (AHL) and reduced virulence in potato tubers and stems. Complementation of the mutant strain with the wild‐type expI gene in trans successfully restored AHL and PCWDE production as well as virulence. Transmission electron microscopy and in vitro motility assays demonstrated hyperpiliation and loss of flagella and swimming motility in the mutant strain compared with the wild‐type Pcb1692. Furthermore, we noted that, in the early stages of infection, Pcb1692 wild‐type cells had intact flagella which were shed at the later stages of infection. Confocal laser microscopy of PcbΔexpI‐inoculated plants showed that the mutant strain tended to aggregate in intercellular spaces, but was unable to transit to xylem tissue. On the contrary, the wild‐type strain was often observed forming aggregates within xylem tissue of potato stems. Gene expression analyses confirmed that flagella are part of the quorum sensing regulon, whereas fimbriae and pili appear to be negatively regulated by quorum sensing. The relative expression levels of other important putative virulence genes, such as those encoding different groups of PCWDEs, were down‐regulated in the mutant compared with the wild‐type strain.
Keywords: biofilm, expI, Pectobacterium brasiliense, soft rot Enterobacteriaceae
Introduction
Blackleg and tuber soft rot are important diseases of potato plants in the field and during post‐harvest storage, respectively (Charkowski et al., 2012; Perombelon, 2002). The major causal agents of both diseases are Pectobacterium and Dickeya spp., both belonging to the soft rot Enterobacteriaceae group (Charkowski et al., 2012). Within the Pectobacterium genus, P. atrosepticum, P. carotovorum and P. wasabiae are most often isolated from infected potato plants and tubers. Although it is clear that all three Pectobacterium spp. are capable of inducing soft rot disease, not all can cause blackleg. This particular disease has often been associated with P. atrosepticum, especially in the Northern Hemisphere, including Canada, USA and Europe (De Boer et al., 2012; de Haan et al., 2008). Although generally not associated with blackleg, there have been some reports suggesting that a small subgroup of P. carotovorum is capable of causing blackleg (de Haan et al., 2008). Incidentally, P. carotovorum has now been divided into two subspecies, P. carotovorum ssp. carotovorum and P. carotovorum ssp. brasiliensis, following reports of an atypical Pectobacterium species causing blackleg in Brazil (Duarte et al., 2004; Nabhan et al., 2012).
Since its first report in Brazil, P. carotovorum ssp. brasiliense (Pcb) has emerged as a pathogen of major economic importance in countries such as Canada, USA, South Africa and New Zealand (De Boer et al., 2012; van der Merwe et al., 2010; Nunes‐Leite et al., 2014; Panda et al., 2012). In all of the countries in which it has been isolated, Pcb has been shown to be more aggressive than other Pectobacterium spp. Notable exceptions in this respect are three Canadian isolates that appear to show weak virulence on potato plants (Li et al., 2015). Reports in these countries suggest that it is a major causal agent of blackleg and, as such, a significant threat to potato production. Blackleg originates from infected seed tubers and ultimately manifests itself in the stems of infected plants. This involves the movement of bacteria from seed tubers into stems via the vascular system. Given that this is a newly identified pathogen, the mechanisms that allow colonization in the vascular system and the observed aggressiveness of Pcb in potatoes are still poorly understood. Nonetheless, we have reported recently that, in a susceptible potato cultivar, Pcb1692 colonizes host vascular systems by forming biofilm‐like aggregates that lead to the occlusion of the xylem (Kubheka et al., 2013). These biofilm‐like aggregates were not observed in the interaction of Pcb1692 with a tolerant cultivar (no disease development). This demonstrated the importance of biofilm formation in Pcb1692 infection and subsequent disease development in potato plant stems.
Quorum sensing is a density‐dependent form of communication in bacteria that plays a crucial role in the regulation of many virulence genes. In Pectobacterium spp., the LuxI homologue (ExpI) synthesizes acyl homoserine lactones (AHLs), which are small signalling molecules responsible for cell‐to‐cell communication in bacteria. In the absence of AHLs, the LuxR homologue ExpR represses the transcription of virulence genes, including those that encode plant cell wall‐degrading enzymes (PCWDEs, a major virulence factor for the soft rot Enterobacteriaceae), motility, adhesion, type III secretion system (T3SS) and others (Barnard and Salmond, 2007; Charkowski, 2009; Cui et al., 2005; Põllumaa et al., 2012). When AHLs reach threshold levels, they bind to ExpR, and the resulting ExpR–AHL complex is unable to repress the expression of these virulence factors. Biofilm formation is an important virulence factor for xylem‐dwelling phytopathogens, such as Xanthomonas, Xylella and Pantoea spp. In these pathogens, the role of cell‐to‐cell signalling in biofilm formation is well established (Barel et al., 2015; Dow et al., 2003; Koutsoudis et al., 2006). However, there are as yet no studies that have attempted to understand the role of quorum sensing in biofilm formation during the colonization of potato stem vascular tissue by the soft rot Enterobacteriaceae pathogens.
In this study, we generated a quorum sensing mutant strain and evaluated the role of quorum sensing in the virulence of Pcb on potato stems and tubers. To understand the role of quorum sensing in Pcb biofilm formation in planta, colonization and migration patterns of the quorum sensing‐defective mutant were compared with those of the wild‐type strain on potato stems. Finally, the relative expression of a selected number of virulence‐associated genes in the wild‐type strain was compared with that of the mutant strain using quantitative polymerase chain reaction (qPCR).
Results
Construction and characterization of an expI mutant strain of Pcb1692
As a shotgun genome sequence of Pcb1692 is available, we first identified an expI homologue in the genome of Pcb1692. In this respect, a P. carotovorum ssp. carotovorum expI sequence was blasted against the Pcb1692 shotgun genome sequence and a homologous gene was identified. The expI coding region is 651 bp and forms part of an operon comprising the two genes expI/R. Next, we sought to functionally characterize expI in Pcb1692. Thus, the Pcb1692ΔexpI mutant strain was generated using a combination of the lambda recombination system and triparental mating, as outlined in Experimental Procedures (Datsenko and Wanner, 2000). This resulted in a Pcb1692ΔexpI mutant strain in which the wild‐type expI gene was disrupted with a kanamycin resistance marker gene. The location of the kanamycin cassette was confirmed by PCR using primers that flank expI (InsT_up and InsT_down), and the product was sequenced. We specifically selected primers that flanked the inserted expIupstream‐kan‐expIdownstream cassette to enable us to confirm not only successful integration of the kanamycin gene, but also the exact genomic location in which integration occurred (Fig. S1, step 3, see Supporting Information). Furthermore, Southern blot analysis confirmed that a single insertion of the kanamycin cassette was present in the genome of Pcb1692 (Fig. S1B). Qualitative and semi‐quantitative assays for pectinase (pectate lyase), protease and cellulase showed that the mutant produced relatively low levels of these three enzymes in comparison with the wild‐type strain. Complementation of the mutant strain restored enzyme levels to wild‐type levels (Fig. S2, see Supporting Information). The ability of the mutant to produce AHLs was also tested using the Chromobacterium violaceum indicator strain. As expected, the mutant strain was unable to produce any AHLs (Fig. S3, see Supporting Information). However, complementation of the mutant with Pcb1692 expI in trans was able to restore AHL production to near wild‐type levels (Fig. S3). In vitro growth of Pcb1692ΔexpI was compared with that of the wild‐type over 16 h. After 16 h, there was no significant (P > 0.05) difference in the growth of the mutant and the wild‐type strains, suggesting that loss of AHL production in the mutant strain was not a result of impaired growth (Fig. S4, see Supporting Information). Finally, in planta growth assays over 72 h post‐inoculation (hpi) indicated no difference in the growth rate of the Pcb1692ΔexpI mutant compared with the wild‐type strain, suggesting that mutation of expI has no detrimental effect on the growth and multiplication of the bacteria in potato tubers and stems (results not shown).
Mutation of expI reduces soft rot and blackleg disease development in potato tubers and stems, respectively
To evaluate the effect of quorum sensing on the ability of Pcb1692 to macerate potato tubers, tubers were inoculated with the Pcb1692ΔexpI mutant strain, the complemented Pcb1692ΔexpI_expI mutant strain and the wild‐type strain. At 72 hpi, macerated tissue per strain was weighed and an average weight of three independent experiments was compared (Fig. 1). The mutant strain was significantly attenuated in its ability to macerate potato tubers in comparison with the wild‐type strain (Fig. 1). This phenotype, however, could be restored by introducing the wild‐type expI into the mutant strain. To evaluate blackleg caused by the wild‐type vs. mutant strain on stems of a susceptible potato cultivar, we stab inoculated the mutant, wild‐type and complemented mutant strains into potato stems and monitored disease development over 72 hpi (black arrows in Fig. 2A show stab wounds caused by the inoculation process). The experiment consisted of five stems per treatment and was repeated three times. No symptoms were observed in the first 12 h. The first signs of stem maceration by the wild‐type strain were observed at 24 hpi. At 72 hpi, a significant lesion (the white line in Fig. 2A indicates the length of the lesion) had developed in stems inoculated with either the wild‐type or the complemented mutant strain (Fig. 2A,B). On the contrary, the mutant strain was severely impaired in its ability to cause disease (Fig. 2A,B). Mock‐inoculated (control plants inoculated with MgSO4) potato stems or tubers did not show any disease symptoms.
Figure 1.
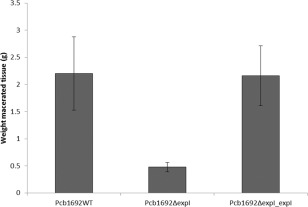
The Pectobacterium carotovorum ssp. brasiliense (Pcb1692) expI gene is required for virulence in potato tubers. Potato tubers (Solanum tuberosum cv. Mondial) were inoculated with 1 × 108 colony‐forming units (cfu)/mL of Pcb1692WT (wild‐type), Pcb1692ΔexpI and Pcb1692ΔexpI_expI complemented mutant strain. The average weight of macerated potato tissue was calculated at 72 h post‐inoculation (hpi). Error bars denote significant differences in virulence between the wild‐type and complemented strain compared with the mutant strain according to Student's t‐test (P > 0.05).
Figure 2.
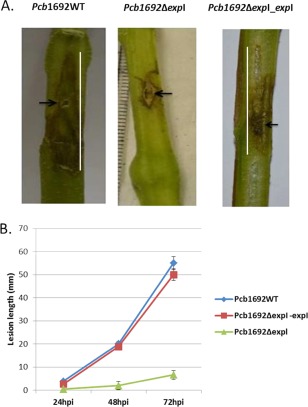
ExpIPcb1692 is important for virulence in potato stems. Stems of a susceptible potato cultivar (Solanum tuberosum cv. Valor) were inoculated with 1 × 108 colony‐forming units (cfu)/mL of Pcb1692 wild‐type, Pcb1692ΔexpI and Pcb1692ΔexpI_expI. (A) Stem lesions were captured at 72 h post‐inoculation (hpi). (B) Lesion development was monitored by measurement of the lesions (mm) over 72 hpi. The mutant strain was significantly attenuated in virulence compared with the wild‐type strain and complemented strain (Student's t‐test, P > 0.05).
Migration of the expI mutant strain up and down potato plant stems is comparable with that of the wild‐type strain
Given the importance of quorum sensing as a regulator of many virulence‐associated genes in pectobacteria, we hypothesized that a quorum sensing‐defective mutant would not be able to migrate up and down the stem from the point of inoculation (POI). To test this hypothesis, we generated wild‐type and mutant Pcb1692 strains tagged with egfp, resulting in strains Pcb1692_pMP4657 and Pcb1692ΔexpI_pMP4657, respectively. Both strains were evaluated for in vitro and in planta growth in comparison with the wild‐type strain containing an empty plasmid. There was no difference in the growth rate of these two strains, both in vitro (16 h) and in planta in potato tubers (72 h) (results not shown). To determine the rate at which each of the strains migrated up and down the stems, 6‐week‐old potato plant stems were inoculated at 6 cm above soil level. Stem sections were harvested 2 cm above and 2 cm below the POI, and bacteria per gram tissue were enumerated (Fig. 3). The results indicated that, for both Pcb1692 wild‐type and mutant strains, there were higher numbers of cells below the POI compared with those above the POI. This is interesting, given that downward‐migrating cells are migrating against the natural upward movement of water in the xylem. Furthermore, the number of bacterial cells above and below the POI was higher in the wild‐type strain compared with the mutant strain. This indicates that both the wild‐type and mutant cells are motile, although it would appear that the wild‐type moves faster than the mutant strain. Hence, these results suggest that at least some of the structures associated with motility might not be under the control of quorum sensing.
Figure 3.
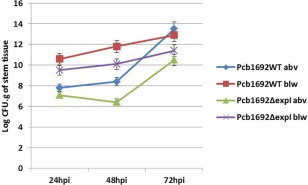
Wild‐type and mutant Pcb1692 strains migrate rapidly below (blw) the point of inoculation compared with above (abv) the point of inoculation. Stems of Solanum tuberosum cv. Valor were inoculated with 1 × 10−8 colony‐forming units (cfu)/mL of Pcb1692 wild‐type and Pcb1692ΔexpI at 6 cm above soil level. Stem sections (2 cm thick) were harvested 2 cm above and below the point of inoculation at 24, 48 and 72 h post‐inoculation (hpi), and log cfu/g stem section was recorded.
The Pcb1692 expI mutant strain is hyperpiliated but lacks flagella
In bacteria, pili and flagella are important virulence factors involved in twitching and swimming motility, respectively. Thus, we sought to determine the presence or absence of these two motility structures before and during the colonization of potato stems and tubers by the Pcb1692 wild‐type and mutant strains. Hence, a combination of swimming and twitching motility assays, together with transmission electron microscopy (TEM), was used. We started by culturing Pcb1692 wild‐type and mutant strains in Luria–Bertani (LB) broth overnight, followed by washing of the bacteria four times and viewing the cells under TEM. Both the mutant and wild‐type strains possessed pili and fimbriae. Indeed, it appeared that relatively more pili and fimbriae were present in the mutant strain compared with the wild‐type (Fig. 4), suggesting a negative regulation of these two structures by quorum sensing. In vitro twitching assays confirmed that both the wild‐type and mutant strains were positive for twitching motility (results not shown).
Figure 4.
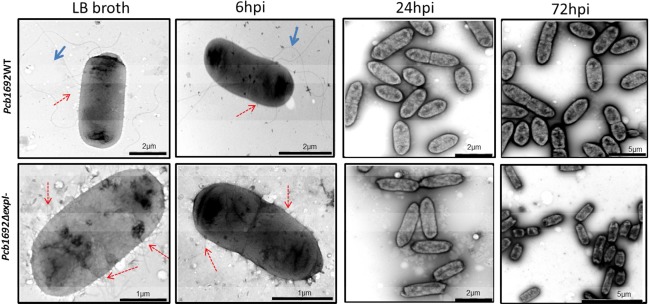
Pcb1692 wild‐type cells are flagellated, whereas Pcb1692ΔexpI cells lack flagella and are hyperpiliated. Transmission electron microscopy was used to view cells of the wild‐type and mutant strains after overnight culture in Luria–Bertani (LB) broth and stem inoculation at 6, 24 and 72 h post‐inoculation (hpi). Flagella are indicated with full blue arrows and pili/fimbriae by broken red arrows.
In contrast with the above, TEM indicated that only the wild‐type cells were flagellated, but no flagella were observed in the mutant strain (Fig. 4). This indicated that, prior to inoculation into potato stems or tubers, the mutant strain already lacked flagella and was thus unable to swim. Swimming ability and the lack thereof in the wild‐type and mutant strains, respectively, was confirmed by in vitro swimming motility assays (Fig. 5). The loss of flagella in the mutant strain could be restored by complementation of the strain with wild‐type expI in trans (Fig. S5, see Supporting Information). We subsequently inoculated 108 colony‐forming units (cfu)/mL of each bacterial strain on potato plant stems at 6 cm above soil level. Thereafter, stem sections (2 cm thick) were obtained at three time points (6, 24 and 72 hpi) and inoculated bacteria were re‐isolated and again analysed by TEM and swimming motility assay. We observed that Pcb1692 wild‐type cells harvested from stem sections that had been inoculated for 6 h possessed intact flagella, fimbriae and pili. Interestingly, we noted that, at 24 hpi onwards, Pcb1692 wild‐type cells lost their flagella (Fig. 4) and subsequently their ability to swim (Fig. 5). In the mutant strain, in keeping with our earlier observations, no flagella could be observed at these three time points post‐inoculation (Fig. 4). Consequently, in vitro swimming motility for the mutant strains was negative at all time points (Fig. 5). This was expected, given that there were no flagella in overnight cultures that were used to inoculate the plants.
Figure 5.
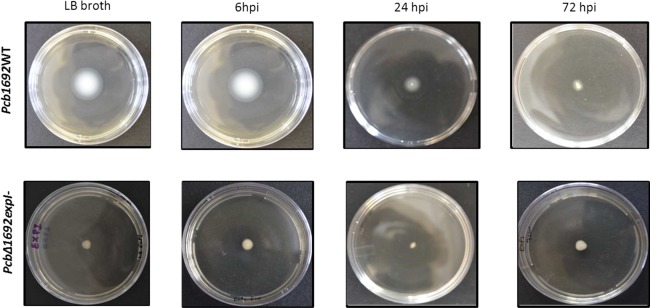
In vitro assays reveal a loss of flagella‐mediated swimming motility in the mutant compared with the wild‐type strain. Pcb1692 cells cultured overnight in Luria–Bertani (LB) broth and cells harvested from Pcb1692‐inoculated stems at 6, 24 and 72 h post‐inoculation (hpi) were obtained using a sterile pipette and stabbed in the centre of 0.4% polygalacturonic acid (PGA) plates. Photographs were taken after 48 h.
The expI mutant aggregates in intercellular spaces, but, unlike the wild‐type strain, is unable to invade xylem tissue
The colonization patterns of the Pcb1692_pMP4657 wild‐type strain and Pcb1692ΔexpI_pMP4657 mutant strain were compared in 6‐week‐old potato plant stems of a susceptible cultivar. Stems were inoculated with each of the strains 6 cm above soil level and 2‐cm‐thick stem sections were sampled for confocal imaging at different time points (Fig. 6). For both the mutant and wild‐type strains, we were able to visualize clumps of bacterial cells (green) in the intercellular spaces as early as 24 hpi. Within 48 hpi, we could see indications of bacterial aggregation for both strains. There was no clear difference in the ability of the respective strains to form aggregates in potato plant stems, albeit that we were able to visualize the wild‐type strain earlier and, consequently, observed slightly more aggregation by the wild‐type strain than the mutant strain. What was particularly remarkable was the fact that we consistently observed that, in the case of the wild‐type strain, the bacteria were able to transit from the intercellular spaces into xylem vessels, leading to the occlusion of the vascular tissue (indicated by a white arrow), whereas this was not the case for the mutant strain (Fig. 6). Thus, it appears that, although the mutant strain is able to form aggregates, these are limited to the intercellular spaces, and hence the mutant is unable to invade and occlude the xylem.
Figure 6.
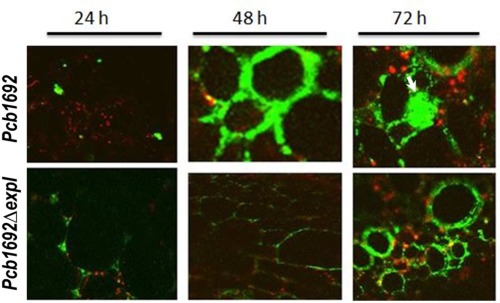
The Pcb1692ΔexpI mutant strain forms aggregates limited to intercellular spaces but, unlike the wild‐type strain, is unable to occlude xylem tissue. Potato stems were inoculated with Pcb1692_ pMP4657 and Pcb1692ΔexpI‐pMP4657 strains as outlined previously. Transverse stem sections were obtained at 2 cm above and below the point of inoculation and confocal microscopy was used to evaluate the colonization patterns of the two strains.
Differential gene expression analyses of selected genes in the expI mutant relative to the wild‐type strain
Three housekeeping genes, previously evaluated as endogenous controls for P. atrosepticum, were evaluated for their suitability as controls for Pcb1692 (Takle et al., 2007). The ffh gene, encoding a signal recognition particle subunit, was found to be most stably expressed compared with the other two housekeeping genes, and was thus used to normalize the expression values (results not shown). The expression of a few selected Pcb1692 virulence‐associated genes and transcription factors was compared in potato tuber‐inoculated mutant strain vs. the wild‐type strain. The genes selected were those encoding different PCWDEs (pelD encoding a pectate lyase, celV encoding a cellulase and prtA encoding a protease), and pilA and fimA encoding pilus and fimbriae structural proteins, respectively. Also included were rpoS and flhD, encoding a stationary phase‐inducible sigma factor and a flagellum‐specific sigma factor, respectively. At 6 hpi, there was no difference in the relative expression of pelD between wild‐type and expI mutant tuber‐inoculated bacteria. However, at 24 and 72 hpi, there was a substantial increase in the relative levels of pelD in the wild‐type strain relative to the mutant strain. A significant up‐regulation of celV and prtA (72‐ and 10‐fold, respectively) was noted at 72 hpi in the wild‐type relative to the mutant strain (three‐ and zero‐fold, respectively), indicating that the expression of these two enzymes occurs later than pectate lyase (Fig. 7). pilA was relatively higher in the mutant strain (30‐ and eight‐fold) compared with the wild‐type (15‐ and four‐fold) during the earlier time points of inoculation (at 6 and 24 hpi, respectively) (Fig. 7). Similarly, fimA was also highly induced in the mutant strain compared with the wild‐type at these two time points (Fig. 7). This was congruent with our TEM results demonstrating hyperpiliation in the mutant strain compared with the wild‐type strain. At the later stages of colonization (72 hpi), there was relatively more fimA and pilA transcripts in the wild‐type than in the mutant strain, indicating that the expression of these two genes in the wild‐type increases with the accumulation of AHLs. The relative expression of fliA (the flagella sigma factor) was more abundant in the wild‐type (23‐, 82‐ and 80‐fold) compared with the mutant strain (zero‐, two‐ and zero‐fold) at all three sampled time points. Finally, the relative expression levels of rpoS were almost double in the mutant strain at 6 hpi compared with the wild‐type strain; however, the inverse appeared to be the case at 72 hpi.
Figure 7.
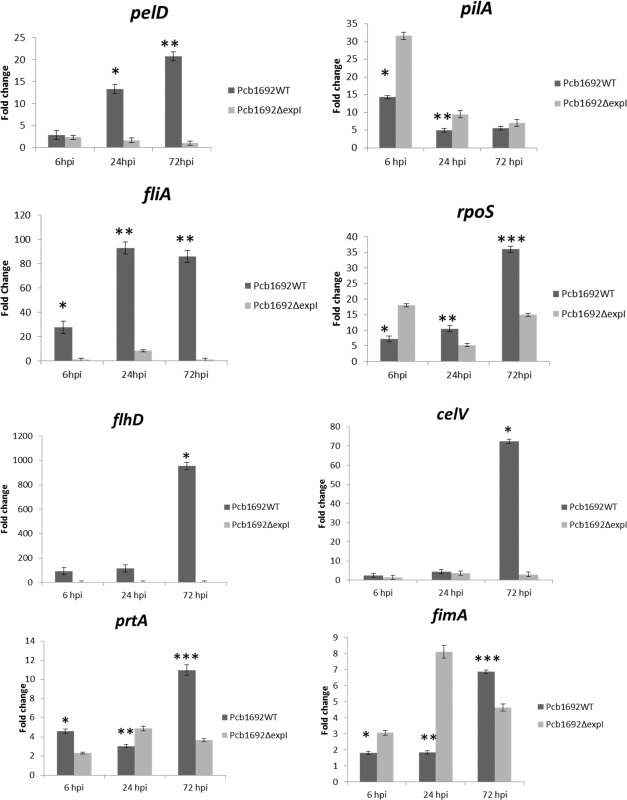
Differential gene expression analyses of candidate pathogenicity genes in the mutant compared with the wild‐type strain over 72 h post‐inoculation (hpi). Pcb1692 wild‐type and Pcb1692ΔexpI strains were harvested from inoculated potato tubers at 6, 24 and 72 hpi. The relative expression levels of putative pathogenicity genes and two transcriptional regulators were monitored. Gene expression was normalized using the ffh housekeeping gene as an endogenous control. Values of inoculated potato tubers (time 0) were used as controls for relative gene expression. Asterisks denote significant differences (P < 0.05) according to Student's t‐test.
Discussion
Quorum sensing is an important regulatory mechanism in many plant‐pathogenic bacteria (von Bodman et al., 2003). In the soft rot enterobacteria, AHL‐mediated quorum sensing has been shown to play an important role in the virulence of some of the well‐studied Pectobacterium spp., such as P. atrosepticum and P. carotovorum ssp. carotovorum (Chatterjee et al., 2005; Crépin et al., 2012a; Lui et al., 2008; Põllumaa et al., 2012; Smadja et al., 2004). On the contrary, AHLs play a minor role in the virulence of the closely related soft rot‐causing Dickeya spp. (Charkowski, 2009; Charkowski et al., 2012). In recent times, there has been a global emergence of new soft rot‐ and blackleg‐causing bacteria, posing a significant threat to the production of potato and other crops (Ma et al., 2007; Pitman et al., 2008; Toth et al., 2011). Amongst these new soft rot bacteria are P. wasabiae and Pcb, both recently identified in South Africa, and D. solani causing problems in Europe (van der Merwe et al., 2010; Moleleki et al., 2012; Toth, 2011). Recently, Crépin et al. (2012b) characterized the production and diversity of AHLs in some of these emerging bacteria. However, the role played by AHLs in potato stem vascular colonization of these emerging strains remains largely uncharacterized. Moreover, studies on the role of AHLs in virulence, even in the well‐studied P. atrosepticum and P. carotovorum ssp. carotovorum, have mainly been either in vitro or on potato tubers. Hence, there still remains a gap in our understanding of the underlying mechanisms that allow blackleg‐causing soft rot Enterobacteriaceae to colonize stem vasculature and form biofilms.
The key to effective vascular colonization by bacteria is the ability to switch from a planktonic to a sessile mode of growth (formation of biofilms). In our previous work, we have shown that the difference between planktonic and sessile behaviour in Pcb1692 determines its ability to cause disease or inability thereof in a susceptible and a tolerant cultivar, respectively (Kubheka et al., 2013). Hence, in the current study, we evaluated the role of quorum sensing in the emerging plant pathogen Pcb1692, particularly with respect to biofilm formation and colonization in xylem tissue of potato stems. To this end, we identified a homologue of the homoserine synthase (expI) in the Pcb1692 shotgun genome sequence. Thereafter, we generated a mutation of Pcb1692expI by replacing the expI gene with a kanamycin cassette. Loss of the expI gene in Pcb1692 resulted in the loss of the ability to synthesize AHLs. As AHLs are key signalling molecules that mediate cell density‐dependent communication in bacteria, loss of AHLs led to the attenuation in virulence of the mutant strain in both stems and tubers (Figs 1 and 2), as well as a reduction in the production of PCWDEs (Fig. S2). Similar to other Pectobacterium spp., these data confirm the central role of cell‐to‐cell communication in the pathogenesis of Pcb1692. Before studying the effect of quorum sensing on biofilm formation in the vascular system of susceptible potato plant stems, we first studied the wild‐type colonization patterns in potato plant stems. We hypothesized that Pcb1692 wild‐type cells would be planktonic and highly motile before inoculation and during the early stages of host colonization. In the early stages of colonization, bacterial cells are localized in nutrient‐deficient environments, such as the apoplast or xylem tissue. Indeed, our TEM assays showed that overnight cultures of Pcb1692 wild‐type are typically flagellated and appear able to swim in vitro and in planta (Figs 4 and 5). Similarly, at 6 hpi, Pcb1692 cells appear to be planktonic with the presence of flagella. Interestingly, we observed that Pcb1692 cells lose their flagella at 24 and 72 hpi and start to aggregate (Fig. 4). Incidentally, this aggregation coincides with the increased expression of the stress alarmone, rpoS, as well as genes encoding PCWDEs (Fig. 7). Furthermore, this is the time at which we observed a sizeable lesion in potato stems and rot in potato tubers (48–72 hpi) (Figs 1 and 2). Thus, it is conceivable that degradation of tissue by PCWDEs leads to the release of nutrients for the otherwise starved cells.
Contrary to our observations with respect to the wild‐type, our phenotypic assays showed that overnight cultures of the Pcb1692ΔexpI mutant did not have flagella and were impaired in their ability to swim (Fig. 4). Indeed, we observed loss of swimming motility for Pcb1692ΔexpI mutant cells before inoculation and in those cells that were isolated from potato stems at 6, 24 and 72 hpi (Fig. 5). This suggests that flagella in Pcb1692 are regulated by quorum sensing. Rather unexpectedly, using TEM, we observed that the Pcb1692ΔexpI mutant appeared to have more fimbriae and pili than the wild‐type (Fig. 4). This suggests that both pili and fimbriae are negatively regulated by quorum sensing. This could indicate that the hyperpiliated mutant probably switches from using swimming motility and relies strongly on twitching motility. In this respect, we further tested the ability of the mutant and wild‐type strains to migrate up and down stems and to colonize the vascular system of potato plant stems. As we had already observed the loss of flagella and hyperpiliation in the mutant strain, we wanted to determine whether the mutant would be able to move up and down the stems during colonization at a similar rate as the flagellated wild‐type. Our results showed that both the mutant and wild‐type strains were capable of movement up and down the stems of potato plants (Fig. 3). However, the wild‐type strain appeared to move faster than the mutant strain, possibly because of the presence of both flagella and pili.
Remarkably, we noted that both strains moved much faster in a downward rather than upward direction. All things considered, downward translocation, which occurs against the general direction of the transpiration stream, appears to be a rather energy costly process for bacteria. However, both Pcb and D. solani have been shown previously to localize in the vascular tissues of potato stems and to move downwards using the vascular system (Czajkowski et al., 2010a, 2010b2010a, b; Kubheka et al., 2013). Several studies have also shown that this downward translocation is mediated by twitching motility using the type 4 pilus (T4P) and is important for the virulence of vascular pathogens, such as Xylella fastidiosa and Ralstonia (Liu et al., 2001; Meng et al., 2005; Wairuri et al., 2012). Furthermore, knock‐out mutants of selected pil and fim genes in Xanthomonas citri ssp. citri resulted in a loss of twitching motility and adherence (Dunger et al., 2014).
We observed that both the mutant and wild‐type Pcb1692 strains were able to form aggregates in vitro as well as in intercellular spaces of plant tissue (Fig. 6). Incidentally, both fimbriae and pili have been implicated in adhesion and biofilm formation (Dunger et al., 2014). Hence, it is not surprising that both strains were able to form aggregates in the intercellular spaces. However, we noted two differences between the colonization patterns of the two strains. First, in comparison with the wild‐type, there was a delay in the visibility of the mutant strain within the plant apoplast (Fig. 6, 48 h). This delay can be attributed to the lack of flagella and thus limited swimming ability in the mutant strain. Second, the wild‐type Pcb1692 strain, because of its ability to produce PCWDEs, was able to transit to xylem and other tissues. However, unlike the wild‐type, the mutant strain tended to remain in the intercellular spaces, generally unable or inefficient in its ability to transit into the xylem tissue. This is consistent with the fact that the mutant strain, at this time point, does not produce PCWDEs which are required for maceration of the cell wall.
The expression patterns of several genes associated with virulence were determined in the mutant strain compared with the wild‐type strain. Similar to our earlier phenotypic observations, we noted a relative up‐regulation of both the pil and fim genes in the mutant strain compared with the wild‐type strain. In our study, we observed that the expression of pelD, one of the major pectinases, was low in the wild‐type strain during the early planktonic stages of colonization, but peaked at later time points of 24 and 72 hpi. This coincided with symptom appearance on stems and tubers. The two PCWDEs known to play no or, at best, a minor role in the pathogenesis of soft rot Enterobacteriaceae, cellulase and proteases, were induced at a much later time point of 72 hpi. These data confirm the proposed hierarchy in expression of PCWDEs in the pathology of soft rot Enterobacteriaceae, whereby pectinases are first and most important, followed by the less important cellulases and proteases (Perombelon, 2002; Toth et al., 2003). We noted an increase in the expression of rpoS in the wild‐type strain at the same time point at which we observed extensive lesions on stems and maceration of potato tubers (72 hpi). The stress/stationary phase transcriptional factor rpoS is responsive to the intercellular alarmone (p)ppGpp induced by nutrient limitation in the cell (Hengge, 2011; Lange and Hengge‐Aronis, 1991). Furthermore, up‐regulation of rpoS leads to the induction of biofilm‐associated phenotypes and the suppression of flagella‐mediated motility.
Monson et al. (2013) have shown previously that, in P. atrosepticum, genes such as those encoding motility and PCWDEs are direct targets of VirR. In their study, they proposed that, in the absence of AHLs, VirR binds to promoter regions of these genes and represses them. At above threshold concentrations, VirR is bound to AHLs and, subsequently, de‐repression of virulence‐associated genes, such as the pil gene cluster and those encoding PCWDEs, occurs. In our study, we observed that, in the Pcb1692 wild‐type strain, the expression of genes encoding different PCWDEs increases over time (possibly as a result of increasing AHLs). However, as no AHLs are produced in the mutant strain, these genes remain repressed. This therefore suggests that, as in P. atrosepticum, Pcb1692 genes encoding PCWDEs are probable targets of VirR. However, we observed the opposite for pil and fim genes. At an early time point of 6 hpi (presumably associated with low levels of AHLs), we observed that the expression of these genes is relatively higher in both the mutant and wild‐type strains compared with later time points (24 and 72 hpi) (Fig. 7). What is remarkable is that the expression of these genes is relatively higher in the AHL‐deficient mutant compared with the wild‐type strain. This suggests that, unlike in P. atrosepticum, these genes might not be part of the virR regulon in Pcb1692. Alternatively, they might be negatively regulated by quorum sensing or there may be alternative regulators. Incidentally, a negative relationship between the diffusible signal factor (DSF) and T4Ps has also been demonstrated for Xylella fastidiosa (Chatterjee et al., 2008). To this end, in their microarray experiments, Wang et al. (2012) observed the up‐regulation of T4Ps in a DSF‐deficient (rpf –) mutant compared with the wild‐type X. fastidiosa strain. However, Nykyri et al. (2013) have suggested previously that the tight adherence (Tad) and fimbrial low molecular (flp) gene clusters, encoding the T4Ps, are not under quorum sensing regulation, but, rather, they identified a novel two‐component system as a regulator of that system. In addition, recent studies have pointed to the second messenger, Cyclic diguanylate monophosphate (c‐di‐GMP), as an important player in the regulation of fimbriae and pili, subsequently affecting the transition from planktonic to sessile behaviour of many prokaryotes (Bordeleau et al., 2015; Romling, 2012; Skotnicka et al., 2015).
In conclusion, we generated an expI mutant in the emerging blackleg/soft rot pathogen Pcb. Comparison of the mutant with the wild‐type strain demonstrated attenuated virulence, reduced expression of genes encoding PCWDEs and abolition of flagella. On the contrary, hyperpiliation and an abundance of fimbriae were observed in the mutant strain compared with the wild‐type strain.
Experimental Procedures
Bacterial strains
The bacterial strains and plasmids used in this study are listed in Table 1. Pcb1692, Chromobacterium violaceum CV026 and Escherichia coli strains were cultured overnight in LB broth or on nutrient agar at 37 °C. Where required, media were supplemented with kanamycin, streptomycin, ampicillin and tetracycline to final concentrations of 100 µg/mL, 100 µg/mL, 50 µg/mL and 10 µg/mL, respectively. All antibiotics were obtained from Sigma‐Aldrich (St. Louis, Missouri, United States).
Table 1.
Bacterial strains and plasmids used in this study.
| Bacterial strain | Description | Origin |
|---|---|---|
| Pectobacterium carotovorum ssp. brasiliense 1692 (Pcb1692) | Type strain of P. carotovorum ssp. brasiliense, isolated from potato in Brazil, sequenced strain | Professor A. Charkowski, Wisconsin University (Duarte et al., 2004) |
| Pcb1692ΔexpI | P. carotovorum ssp. brasiliense 1692ΔexpI, Kanr | This study |
| Pcb1692ΔexpI_expI | P. carotovorum ssp. brasiliense 1692ΔexpI with pKNG101ΔexpI– complementation plasmid; Kanr, Ampr | This study |
| Pcb1692ΔexpI – (pempty) | P. carotovorum ssp. brasiliense 1692ΔexpI with empty pGEM‐T Easy plasmid; Ampr | This study |
| Pcb1692ΔexpI_pMP4657 | Pcb1692ΔexpI mutant strain harbouring plasmid pMP4657 | This study |
| Pcb1692_pMP4657 | Pcb1692 harbouring plasmid pMP4657 | This study |
| Escherichia coli DH5α | supE44 ΔlacU169 (Δ80lacZΔM15) hsdR17 recA1EnA1 gyrA96 thi‐1 relA1 | Invitrogen |
| E. coli CC118λpir | araD D (ara, leu) DlacZ74 phoA20 galK thi‐1 rspE rpoB argE recA1 λ pir | Herrero et al. (1990) |
| E. coli HH26 | Mobilizing strain for conjugal transfer of pKNG101‐derived marker exchange plasmids | Herrero et al. (1990) |
| Chromobacterium violaceum CV026 | Acyl homoserine lactone (AHL)‐deficient, AHL reporter strain | McClean et al. (1997) |
| Plasmids | ||
| pGEM‐T Easy | Commercial cloning vector, f1 ori, E. coli pMB1 ori, lacZ', Ampr | Promega Corp. |
| pNJ5000 | Mobilizing plasmid used in marker exchange, Tetr | Hepburn et al. (1985) |
| pKNG101 | Suicide vector for allelic marker exchange, mobRK2, oriT, oriR6 K, sacBR, Smr | Kaniga et al. (2011) |
| pKD13 | Vector containing a Kanr cassette | Datsenko and Wanner (2000) |
| pKNG101ΔexpI | Derivative of pKNG101 plasmid containing the mutant expI allele | This study |
| pMP4657 | pME6010 derivative, egfp, TetR, Ptac | Bloemberg et al. (2000) |
Kanr, Ampr, Smr, Tetr, resistant to kanamycin, ampicillin, streptomycin and tetracycline, respectively.
Sequence analysis
The expI gene in the completely sequenced genome of P. carotovorum ssp. carotovorum was employed to identify an expIPcb1692 homologue using the blastn alignment tool available at the National Center for Biotechnology Information (NCBI) blast. The Pcb1692expI sequence reference is Pcb1692 ID: gi|198439928|gb|ABVX01000038.1| and the expI location is on Contig: (gi|198439928|gb|ABVX01000038.1|0003_0002_Sequence01346) Start: 17572 End: 18222.
Manipulation of bacterial strains
Genomic DNA was extracted from Pcb1692 (Promega, (Madison, Wisconsin, United States) Wizard Genomic DNA Purification Kit). To generate a Pcb1692ΔexpI mutant (for detailed outline of the strategy and PCR products, please refer to Fig. S1), upstream and downstream expI flanking regions were PCR amplified from Pcb1692 genomic DNA using the primer combinations (expI_Fup and expI_Rup) and (expI_Fdown and expIRdown), respectively (Table 2). The kanamycin resistance cassette was amplified from plasmid DNA pKD13 using primers Kan_F and Kan_R. The expI_Rup and exp_Fdown primers were modified to contain a 50‐bp region which is homologous to the upstream (5′) and downstream (3′) regions, respectively, of the kanamycin gene found on pKD13, whereas the Kan_F and Kan_R primers each had a 50‐bp homology to the downstream and upstream expI flanking amplicons. PCRs were conducted using a High Fidelity PCR kit (Life Technologies, Carlsbad, California, United States). All primers were synthesized by Integrated DNA Technologies (IDT), (Coralville, Iowa, United States) (Table 2). The three PCR fragments were purified (Promega Wizard Genomic DNA Purification Kit) and then mixed together (15 ng of each fragment) and amplified using the expI_Fup and ExpI_Rdown primers to generate a hybrid PCR amplicon consisting of expI upstream, kanamycin and expI downstream (Fig. S1, Step 2). The pKNG101 plasmid was obtained from E. coli CC118λ, digested with SmaI and treated with alkaline phosphatase (Thermo Scientific, Waltham, Massachusetts, United States). The hybrid amplicon was then ligated to pKNG101 and transformed into E. coli CC118λ, generating strain E. coli CC118λ pKNG101_ΔexpI. Triparental mating of strains E. coli CC118λ pKNG101_ΔexpI (donor), E. coli HH26 habouring pNJ5000 (helper) and Pcb1692 (recipient) was undertaken on minimal medium (20% glucose as sole carbon source) supplemented with streptomycin. Three transformants were selected and streaked on minimal medium (50% sucrose as sole carbon source) to cure the plasmid. Colonies positive on kanamycin, but negative on streptomycin, were selected for further analysis. Disruption of the expI gene with kanamycin was validated by PCR amplication using combinations of expI gene flanking primers (InsT_up and InsT_down), followed by sequencing of the amplicon. These primers were also used in combination with kanamycin‐specific internal primers (Kan_T_F and Kan_T_R), as illustrated in Fig. S1A, step 3. In addition to PCR, Southern blot analysis was performed following standard procedures to determine the integration and copy number of the kanamycin cassette. Primers Comp F and R were used to amplify the expI gene with its native promoter and ribosomal binding site (RBS) from Pcb1692, and then purified (Qiagen, Hilden, Germany) and cloned into pGEMT‐Easy (Promega) (strain Pcb1692ΔexpI_expI) for complementation studies. Strains Pcb1692_pMP4657 and Pcb1692ΔexpI_pMP4657 were generated by electroporation of plasmid pMP4657 into the Pcb1692 and Pcb1692ΔexpI strains, respectively.
Table 2.
List of primers used in this study.
| Primer name | Sequence (5′–3′) | Use | Origin |
|---|---|---|---|
| ExpI_Fup | ATAGCCCCGGGCTGTCTTTTTGTGTATGAACTCTTG | Mutant expI cassette construction | This study |
| ExpI_Rup | AAAGTATAGGAACTTCGAAGCAGCTCCAGCCTACACCGACAAAGAACCGACTAGATT | Mutant expI cassette construction | This study |
| ExpI_Fdown | TAGGAACTTCGAACTGCAGGTCGACGGATCCCCGGAATATTAGTTCGATGTTGTTTCTTTC | Mutant expI cassette construction | This study |
| ExpI_Rdown | ATAGCCCCGGGATTATTCTCGGCATCAAAACCA | Mutant expI cassette construction | This study |
| Kan_F | AAGGGAATTACCTGGAATCTAGTCGGTTCTTTGTCGGTGTAGGCTGGAGCTGCTTCG | Kanamycin resistance gene amplification | This study |
| Kan_R | GTAATTAATCATTGAAAGAAACAACATCGAACTAATATTCCGGGGATCCGTCGACCT | Kanamycin resistance gene amplification | This study |
| Ins_Tup | GGTAGGGCAACTTCAGGATAA | Verification of mutation | This study |
| Ins_Tdown | AATGAAATCCTTTACTGGGC | Verification of mutation | This study |
| Comp_F | GCATCGGGGCCCGCGCTGTCTTTTTGTGTATG | Wild‐type allele amplification | This study |
| Comp_R | GCATCGGGGCCCGAAAAAACTTGGTGTCCTG | Wild‐type allele amplification | This study |
| GyrA –F | ACGAACGTCGTACGGAAAT | DNA gyrase subunit A (housekeeping gene) | Takle et al. (2007) |
| GyrA –R | GGCTGGAGAAGCACAGAATC | ||
| Ffh –F | TGGCAAGCCAATTAAATTCC | Signal recognition particle subunit (housekeeping gene) | Takle et al. (2007) |
| Ffh –R | TCCAGGAAGTCGGTCAAATC | ||
| ProC‐F | TAAGCTGGTGCTGTCGATTG | Pyrroline‐5‐carboxylate reductase (housekeeping gene) | Takle et al. (2007) |
| ProC‐R | ACGCCGTTGATACCTTGTTC | ||
| Pect –F | GGACTCAAGCGGTAAAGCAG | Pectate lyase 1 precursor | This study |
| Pect –R | ATGGTGATCCCTTTGGTGAA | ||
| RpoS –F | AACGGACCTGAAGACACCAC | RNA polymerase sigma factor RpoS | This study |
| RpoS –R | GCCGATTTCACGACCTACAT | ||
| FlhD –F | TTCTGCGATGTTTCGTCTTG | Flagellar transcriptional activator FlhD | This study |
| FlhD –R | CGACAACAAAATCCCCGTAT | ||
| FliA –F | TATCAACAGCGCGAAAGATG | Flagellin protein | This study |
| FliA –R | ATGCGCTGAGTCATTTCCTT | ||
| Cel‐F | CGTTAAACCGGAACCAACTG | Putative cellulase | This study |
| Cel‐R | AACCACCGTACTGCCTTTTG | ||
| Pro‐F | GCCGCACCAAGTCATTCTAT | Putative protease | This study |
| Pro‐R | CTTCCAGCGTTTCCAGTAGC | ||
| Fim‐F | GCTTTCACACTCGCCATCTT | Putative fimbrial protein | This study |
| Fim‐R | GCCTTGCTCTCCGTACTCAC |
AHL detection
Chromobacterium violaceum CV026, Pcb1692 wild‐type, Pcb1692ΔexpI mutant and complemented Pcb1692ΔexpI mutant strains were cultured overnight, and 1.5 mL of the overnight cultures were centrifuged for 6 min at 11904.464g. The filter‐sterilized cultures of CV026 and Pcb1692 strains were mixed in a ratio of 3 : 1 and incubated at 25 °C for 48 h. A purple colour change was used as a positive indication of AHL presence. LB broth was included as a negative control.
Virulence assays
To assess the virulence and symptom development caused by Pcb1692 and Pcb1692ΔexpI strains, susceptible potato tubers (Solanum tuberosum cv. Mondial) were used. Potato tubers were surface sterilized in 10% (v/v) sodium hypochlorite solution containing Tween‐20 for 10 min, washed and then air dried. Tubers were inoculated with 10 μL of Pcb1692 or Pcb1692ΔexpI mutant and Pcb1692ΔexpI_expI strain [optical density at 600 nm (OD600) of 1.0, equivalent to 108 cfu/mL] and incubated at 25 °C with high humidity. Inoculated tubers were examined for visible symptom development and samples were collected at 6, 12, 24 and 72 hpi by scooping the macerated tissue from each inoculated spot. MgSO4 buffer was inoculated into tubers as a negative control. The macerated tissue was weighed to quantify the extent of tuber maceration at each time point.
To monitor lesion development in inoculated potato stems, 4‐week‐old potato stems of S. tuberosum cv. Valor were inoculated with 10 µL (OD600 of 1.0, equivalent to 108 cfu/mL) of Pcb1692, Pcb1692ΔexpI mutant and Pcb1692ΔexpI_expI strains by stabbing with a sterile pipette tip and injecting 10 µL of the bacterial suspension (1.0 × 10−8 cfu/mL). As a negative control, stems were mock‐inoculated with MgSO4 buffer. The inoculated spot was sealed with parafilm, and lesion development and length were measured. Lesion development and size were monitored for 72 hpi. The mean lesion length (mm) of three biological replicates was calculated. Each experiment consisted of three stem and three biological replicates.
In vitro and in planta growth
In vitro growth of Pcb1692, Pcb1692_plasmid, Pcb1692ΔexpI Pcb1692ΔexpI_expI, Pcb1692 _pMP4657 and Pcb1692ΔexpI‐_pMP4657 strains was evaluated by growing in LB broth with incubation at 37 °C overnight (16 h) with shaking at 200 rpm. The optical densities of the cultures were adjusted to OD600 = 0.1 and diluted 1 : 200 in fresh LB broth in a total volume of 200 mL. OD600 was measured using a spectrophotometer every hour for 16 h to determine bacterial growth. Three biological experiments were performed. For in planta growth, bacterial strains were inoculated into either potato stems or tubers, as outlined under the virulence assays. Rotting potato tuber tissue (g) was scooped out and stem segments (2 cm thick) were cut out using a sterile scalpel, weighed and macerated in 2 mL of MgSO4 buffer, followed by serial dilutions and plated onto LB agar plates supplemented with the appropriate antibiotics.
Confocal laser scanning microscopy (CLSM) and bacterial migration on potato stems
Colonization patterns of enhanced green fluorescent protein (eGFP)‐tagged Pcb1692 strains in potato stems of a susceptible potato cultivar S. tuberosum cv. Valor were investigated to study the bacterial cell–host tissue colonization pattern over a period of 72 h. Stem inoculations were performed as described in Kubheka et al. (2013). The experiment was repeated three times with each repeat having 12 plants with a minimum of three stems. Control plants were inoculated with 10 mm MgSO4. Stem fragments (2 cm thick) from inoculated stems were sampled at 24, 48 and 72 dpi at 2 cm above and 2 cm below the POI. The presence and pattern of colonization by Pcb1692_pMP4657 and Pcb1692ΔexpI_pMP4657 strains were evaluated using a Zeiss 510 META CSLM (Jena, Germany), with bacterial cells fluorescing green against the red autofluorescence of the plant material. To determine the rate of migration, 2 g of combined stem tissue was ground in 2 mL of MgSO4 with a mortar and pestle, serially diluted and the amount of cfu/g tissue was enumerated.
TEM
Potato stems or tubers of S. tuberosum cv. Valor were inoculated with 10 µL (OD600 of 1.0, equivalent to 108 cfu/mL) each of Pcb1692, Pcb1692ΔexpI or Pcb1692ΔexpI_expI, as outlined previously. Approximately 0.2 g of inoculated potato stem or tuber tissue was harvested at 6, 24 and 72 hpi. In each case, the tissue was cut into small pieces using a sterile scalpel and then gently soaked in 1 mL of double‐distilled water. After 15 min, 500 μL of the bacterial suspension were pipetted into a sterilized Eppendorf tube and centrifuged for 3 min at 6296.576g. A drop of re‐suspended strain was placed on a carbon‐coated grid and incubated for about 30 s. The carbon grid was negatively stained using 0.1% (w/v) uranyl acetate for approximately 30 s, followed by viewing under TEM.
Swimming motility assays
Overnight cultures of potato tuber‐ or stem‐inoculated Pcb1692 and Pcb1692ΔexpI mutant strains were assayed for swimming motility at zero (LB broth), 6, 24 and 72 h. In vitro motility was determined by dipping a sterile 200‐µL pipette into each of the bacterial suspensions of both strains harvested from overnight cultures at 6, 24 and 72 hpi and then stabbing the pipette tip into 0.3% (w/v) of agar plate medium containing 0.4% (w/v) polygalacturonic acid (PGA) as a carbon source.
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) assays
The Pcb1692 wild‐type and Pcb1692ΔexpI strain were harvested from inoculated potato tubers for total RNA extraction at 6, 24 and 72 hpi. To harvest inoculated bacteria, potato tissue from each inoculated site was scooped out and homogenized in double‐distilled water. Bacterial cells were separated from starch material by centrifugation at 9838.4g for 1 min. The supernatant was carefully removed and transferred into sterile 50‐mL Falcon tubes containing RNA stabilization buffer (Qiagen). Total RNA was extracted from the stabilized bacterial cells using the RNeasy mini kit (Qiagen). Extracted total RNA was treated with DNaseI (Qiagen) to remove contaminating genomic DNA. Total RNA was quantified using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). First‐strand cDNA was synthesized from 1 μg of each total RNA sample using a Superscript III First‐Strand Synthesis SuperMix cDNA synthesis kit (Invitrogen, Carlsbad, California, United States) in a final volume of 20 μL. Primers for putative virulence and internal normalization genes used for qPCR were designed using Primer3 online software and synthesized by IDT (Table 2). The cDNA was subjected to real‐time PCR using SYBR green master mix (Applied Biosystems, Foster City, California, United States) in a final volume of 25 μL. Reactions were carried out in a Quantstudio 12 flex thermo cycler (Applied Biosystems). All reactions were performed in triplicate and three biological replicates for each gene were performed. Values of inoculated potato tubers (time 0) were used as controls for relative gene expression. The data were analysed using the comparative 2ΔΔCt method.
Statistical analysis
When required, the comparison of the mean values of three biological experiments was statistically analysed using Student's t‐test. P < 0.05 was considered to be statistically significant. Analyses were performed using JMP Statistical Discovery Software version 5.0. SAS (Statistical Analysis Software) Institute ( North Carolina, USA).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Generation of Pcb1692ΔexpI mutant. (A) Schematic diagram of polymerase chain reaction (PCR) strategy. (B) PCR products obtained for the generation of the expI mutant and validation of insertion and integration of the kanamycin gene by sequencing and Southern blot analysis, respectively.
Fig. S2 The activity of different plant cell wall‐degrading enzymes was reduced in the Pcb1692ΔexpI mutant compared with the Pcb1692 wild‐type strain.
Fig. S3 Loss of acyl homoserine lactone (AHL) production in the Pcb1692ΔexpI mutant compared with the Pcb1692 wild‐type strain. Complementation of the mutant strain restored AHL production.
Fig. S4 Growth curves of the Pcb1692 wild‐type (WT), Pcb1692ΔexpI mutant, wild‐type strain harbouring an empty plasmid (Pcb1692_plasmid) and complemented Pcb1692ΔexpI_expI mutant strain. Cell density was measured at an optical density (OD) at 600 nm.
Fig. S5 Complementation of the Pcb1692ΔexpI mutant with the wild‐type expI gene restores flagella in vitro. LB, Luria–Bertani.
Acknowledgements
This study was partially supported by the following grants: NRF Thuthuka 69362; Research Development Grant for Y‐Rated Researchers 93357; Research Technology Fund 92098; Bioinformatics and Functional Genomics (BFG 93685); and The Genomics Research Institute, University of Pretoria. The NRF is acknowledged for CKT, RGP and GM student bursaries. The Potatoes Transformation Fund also funded RGP's MSc studentship. Any findings and/or recommendations expressed here are those of the author(s), and the NRF does not accept any liability in this regard.
References
- Barel, V. , Chalupowicz, L. , Barash, I. , Sharabani, G. , Reuven, M. , Dror, D. , Burdman, S. and Manulis‐Sasson, S. (2015) Virulence and in planta movement of Xanthomonas hortonum pv pelargonii are affected by the diffusible signal factor (DSF)‐dependent quorum sensing system. Mol. Plant Pathol. 16, 710–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, A.M. and Salmond, G.P. (2007) Quorum sensing in Erwinia species. Anal. Bioanal. Chem. 387, 415–423. [DOI] [PubMed] [Google Scholar]
- Bloemberg, G.V. , Wijfjies, A.H.M. , Lamers, G.E.M. , Stuurman, N. and Lugtenberg, B.J.J. (2000) Simultaneous imaging of Pseudomonas fluorescens WCS 365 populations expressing three different autofluorescence proteins in the rhizosphere: new perspectives for studying communities. Mol. Plant–Microbe Interact. 13, 1170–1176. [DOI] [PubMed] [Google Scholar]
- Bordeleau, E. , Purcell, E.B. , Lafontaine, D.A. , Fortier, L.C. , Tamayo, R. and Burrus, V. (2015) Cyclic di‐GMP riboswitch‐regulated type IV pili contribute to aggregation of Clostridium difficile . J. Bacteriol. 197: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A.O. (2009) Decaying signals: will understanding bacterial plant communications lead to control of soft rot? Curr. Opin. Biotechnol. 20, 178–184. [DOI] [PubMed] [Google Scholar]
- Charkowski, A.O. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. , Hugouvieux‐Cotte‐Pattat, N. , Solanilla, E.L. , Low, D. , Moleleki, L. , Pirhonen, M. , Palenzuela, P.R. , Francisco, M.S. , Toth, I. , Tsuyumu, S. , van der Waals, J. , van der Wolf, J. , Gijsegem, F.V. , Yang, C.‐H. and Yedidia, I. (2012) The role of secretion systems and small molecules in soft rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A. , Cui, Y. , Hasegawa, H. , Leigh, N. , Dixit, V. and Chatterjee, A.K. (2005) Comparative analysis of two classes of quorum‐sensing signaling systems that control production of extracellular proteins and secondary metabolites in Erwinia carotovora subspecies. J. Bacteriol. 187, 8026–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. , Wilstrom, C. and Lindow, S.E. (2008) A cell–cell signalling sensor is required for virulence and insect transmission of Xylella fastidiosa . Proc. Natl. Acad. Sci. USA, 105, 2670–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crépin, A. , Beury‐Cirou, A. , Barbey, C. , Farmer, C. , Hélias, V. , Burini, J.‐F. , Faure, D. and Latour, X. (2012a) N‐acyl homoserine lactones in diverse Pectobacterium and Dickeya plant pathogens: diversity, abundance and involvement in virulence. Sensors, 12, 3484–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crépin, A. , Barbey, C. , Beury‐Cirou, A. , Hélias, V. , Taupin, L. , Revershon, S. , Nasser, W. , Dufour, A. , Orange, N. , Feuilloley, M. , Heurlier, K. , Burini, J.‐F. and Latour, X. (2012b) Quorum sensing signalling molecules produced by reference and emerging soft rot bacteria (Dickeya and Pectobacterium spp.). PLoS One, 7, e35176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Chatterjee, A. , Hasegawa, H. , Dixit, V. , Leigh, N. and Chatterjee, A.R. (2005) ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA binding protein. J. Bacteriol. 184, 4792–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski, R. , de Boer, W.J. , Velvis, H. and van der Wolf, J.M. (2010a) Systemic colonization of potato plants by a soilborne green fluorescent protein‐tagged strain of Dickeya sp. biovar 3. Phytopathology, 100, 134–142. [DOI] [PubMed] [Google Scholar]
- Czajkowski, R. , de Boer, W.J. , van Veen, J.A. and van der Wolf, J.M. (2010b) Downward vascular translocation of a green fluorescent protein‐tagged strain of Dickeya sp. (biovar 3) from stem and leaf inoculation sites on potato. Phytopathology, 100, 1128–1137. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A. and Wanner, B.L. (2000). One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. Proc. Natl. Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, S.H. , Li, X. and Ward, L.J. (2012) Pectobacterium spp. associated with bacterial stem rot syndrome of potato in Canada. Phytopathology, 102, 937–947. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Crossman, L. , Findlay, K. , He, Y.Q. , Feng, J.X. and Tang, J.L. (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell to cell signalling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA, 100, 10 995–11 000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, V. , De Boer, S.H. , Ward, L.J. and de Oliviera, A.M.R. (2004) Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 96, 535–545. [DOI] [PubMed] [Google Scholar]
- Dunger, G. , Guzzo, C.R. , Andrade, M.O. , Jones, J.B. and Farah, C.S. (2014) Xanthomonas citri subsp. citri type IV pilus is required for twitching motility, biofilm development and adherence. Mol. Plant–Microbe Interact. 27, 1132–1147. [DOI] [PubMed] [Google Scholar]
- de Haan, E.G. , Dekker‐Nooren, C.E.M. , van den Bovenkamp, G.W. , Speksnijder, A.G.C.L. , Van den Zouwen, P.S. and Van der Wolf, J.M. (2008) Pectobacterium carotovorum subsp. carotovorum can cause blackleg in temperate climates. Eur. J. Plant Pathol. 122, 561–569. [Google Scholar]
- Hengge, R. (2011) The general stress response in gram negative bacteria In: Bacterial stress responses (Storz G. and Hengge E., eds), pp. 251–289. Washington DC: ASM Press. [Google Scholar]
- Hepburn, A.G. , White, J. , Pearson, L. , Maunders, M.J. , Clarke, L.E. , Prescott, A.G. and Blundy, K.S. (1985) The use of pNJ5000 as an intermediate vector for the genetic manipulation of Agrobacterium Ti‐plasmids. J. Gen. Microbiol. 131, 2961–2969. [DOI] [PubMed] [Google Scholar]
- Herrero, M. , de Lorenzo, V. and Timmis, K.N. (1990). Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram‐negative bacteria. J. Bacteriol. 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga, K. , Delor, I. and Cornelis, G.R. (1991). A wide‐host‐range suicide vector for improving reverse genetics in gram‐negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica . Gene, 109, 137–141. [DOI] [PubMed] [Google Scholar]
- Koutsoudis, M.D. , Tsaltas, D. , Minogue, T.D. and von Bodman, S.B. (2006) Quorum‐sensing regulation governs bacterial adhesion, biofilm development and host colonisation in Pantoea stewartii subspecies stewartii . Proc. Natl. Acad. Sci. USA, 103, 5983–5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubheka, G.C. , Coutinho, T.A. , Moleleki, N. and Moleleki, L.N. (2013) Colonisation patterns of a mCherry‐tagged Pectobacterium carotovorum subsp. brasiliense in potato. Phytopathology, 103, 1269–1279. [DOI] [PubMed] [Google Scholar]
- Lange, R. and Hengge‐Aronis, R. (1991) Identification of a central regulator of stationary phase gene expression in Escherichia coli . Mol. Microbiol. 5, 49–59. [DOI] [PubMed] [Google Scholar]
- Li, X. , Yuan, K. , Cullis, J. , Lévesque, A. , Chen, W. , Lewis, C.T. and De Boer, S.H. (2015) Draft genome sequence for Canadian isolates of Pectobacterium carotovorum subsp. brasiliense with weak virulence on potato. Genome Announc. 3, e00240–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H.L. , Kang, Y.W. , Genin, S. , Schell, M.A. and Denny, T.P. (2001) Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology, 147, 3215–3229. [DOI] [PubMed] [Google Scholar]
- Lui, H. , Coulthurst, S.J. , Pritchard, L. , Hedley, P.E. , Ravensdale, M. , Humprhis, S. , Burr, T. , Takle, G. , Brunberg, M.‐B. , Birch, P.R.B. , Salmond, G.P. and Toth, I.K. (2008) Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum . PLoS Pathog. 4, e10000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, B. , Hibbing, M.E. , Kim, H.S. , Reedy, R.M. , Yedidia, I. , Breuer, J. , Breuer, J. , Glasner, J.D. , Perna, N.T. , Kelman, A. and Charkowski, A.O. (2007) Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya . Phytopathology, 97, 1150–1163. [DOI] [PubMed] [Google Scholar]
- McClean, K.H. , Winson, M.K. , Fish, L. , Taylor, A. , Chhabra, S.R. , Camara, M. , Daykin, M. , Lamb, J.H. , Swift, S. and Bycroft, B.W. (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N‐acylhomoserine lactones. Microbiology, 143, 3703–3711. [DOI] [PubMed] [Google Scholar]
- Meng, Y. , Li, Y. , Galvani, C.D. , Hao, G. , Turner, J.N. , Burr, T.J. and Hoch, H.C. (2005) Upstream migration of Xylella fastidiosa via pilus‐driven twitching motility. J. Bacteriol. 187, 5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe, J.J. , Coutinho, T.A. , Korsten, L. and van der Waals, J.E. (2010). Pectobacterium carotovorum subsp. brasiliensis causing blackleg on potatoes in South Africa. Eur. J. Plant Pathol. 126, 175–185. [Google Scholar]
- Moleleki, L.N. , Onkendi, E.M. , Mongae, A. and Kubheka, G.C. (2012) Characterisation of Pectobacterium wasabiae causing blackleg and soft rot diseases in South Africa. Eur. J. Plant Pathol. 135, 279–288. [Google Scholar]
- Monson, R ., Burr, T ., Carlton, T ., Liu, H ., Hedley, P ., Toth, I . and Salmond, G. P . (2013). Identification of genes in the VirR regulon of Pectobacterium atrosepticum and characterization of their roles in quorum sensing‐dependent virulence. Environmental microbiology, 15, 687–701. [DOI] [PubMed] [Google Scholar]
- Nabhan, S. , De Boer, S.H. , Maiss, E. and Wydra, K. (2012) Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J. Appl. Microbiol. 113, 904–913. [DOI] [PubMed] [Google Scholar]
- Nunes‐Leite, L. , de Haan, E.G. , Krijger, M. , Kastelein, P. , van der Zouwen, P.S. , van den Bovenkamp, G.W. , Tebaldi, N.D. and van der Wolf, J.M. (2014) First report of potato blackleg caused by Pectobacterium carotovorum subsp. brasiliensis in the Netherlands. New Dis. Rep. 29, 24. [Google Scholar]
- Nykyri, J. , Mattinen, L. , Niemi, O. , Adhikari, S. , Kõiv, V. , Somervuo, P. , Fang, X. , Auvinen, P. , Mäe, A. , Palva, T. and Pirhonen, M. (2013) Role and regulation of the Flp/Tad pilus in the virulence of Pectobacterium atrosepticum SCRI 1043 and P. wasabiae Scc193. PLoS One, 8, e73718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, P. , Fiers, M.A.W.J. , Armstrong, K. and Pitman, A.R. (2012) First report of blackleg and soft rot of potato caused by Pectobacterium carotovorum subsp. brasiliensis in New Zealand. New Dis. Rep. 29, 15. [Google Scholar]
- Perombelon, M.C.M. (2002) Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 51, 1–12. [Google Scholar]
- Pitman, A.R. , Wright, P.J. , Gailbraith, M.D. and Harrow, S.A. (2008) Biochemical and genetic diversity of pectolytic enterobacteria causing soft rot disease of potatoes in New Zealand. Australas. Plant Pathol. 37, 559–568. [Google Scholar]
- Põllumaa, L. , Alamäe, T. and Mäe, A. (2012) Quorum sensing and expression of virulence in Pectobacteria. Sensors, 12, 3327–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling, U. (2012) Cyclic di‐GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14, 1817–1829. [DOI] [PubMed] [Google Scholar]
- Skotnicka, D. , Petters, T. , Heering, J. , Hoppert, M. , Kaever, V. and Søgaard‐Andersen, L. (2015) c‐di‐GMP regulates type IV pili‐dependent‐motility in Myxococcus xanthus . J. Bacteriol. 198, 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja, B. , Latour, X. , Faure, D. , Chevalier, S. , Dessaux, Y. and Orange, N. (2004) Involvement of N‐acylhomoserine lactones throughout the plant infection by Erwinia carotovora subsp. atrosepticum (Pectobacterium atrosepticum). Mol. Plant–Microbe Interact. 17, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Takle, G.W. , Toth, I.K. and Brurberg, M.B. (2007) Evaluation of reference genes for real‐time RT‐PCR expression studies in the plant pathogen Pectobacterium atrosepticum BMC plant biology 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, I.K. , Bell, K.S. , Holeva, M.C. and Birch, P.R.J. (2003) Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4, 17–30. [DOI] [PubMed] [Google Scholar]
- Toth, I.K. , van der Wolf, J.M. , Saddler, G. , Lojkowska, E. , Helias, V. , Pirhonen, M. , Tsror (Lahkim), L. and Elphinstone, J.G. (2011) Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 60, 385–399. [Google Scholar]
- von Bodman, S.B. , Bauer, W.D. and Coplin, D.L. (2003) Quorum sensing in plant pathogenic bacteria. Annu. Rev. Phytopathol. 41, 455–482. [DOI] [PubMed] [Google Scholar]
- Wairuri, C.K. , van der Waals, J.E. , van Schalkwyk, A. and Theron, J.E. (2012) Ralstonia solanacearum needs Flp pili for virulence on potato. Mol. Plant–Microbe Interact. 25, 546–556. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Li, J.L. and Lindow, S.E. (2012) RpfF dependent regulon of Xylella fastidiosa . Phytopathology, 102, 1045–1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Generation of Pcb1692ΔexpI mutant. (A) Schematic diagram of polymerase chain reaction (PCR) strategy. (B) PCR products obtained for the generation of the expI mutant and validation of insertion and integration of the kanamycin gene by sequencing and Southern blot analysis, respectively.
Fig. S2 The activity of different plant cell wall‐degrading enzymes was reduced in the Pcb1692ΔexpI mutant compared with the Pcb1692 wild‐type strain.
Fig. S3 Loss of acyl homoserine lactone (AHL) production in the Pcb1692ΔexpI mutant compared with the Pcb1692 wild‐type strain. Complementation of the mutant strain restored AHL production.
Fig. S4 Growth curves of the Pcb1692 wild‐type (WT), Pcb1692ΔexpI mutant, wild‐type strain harbouring an empty plasmid (Pcb1692_plasmid) and complemented Pcb1692ΔexpI_expI mutant strain. Cell density was measured at an optical density (OD) at 600 nm.
Fig. S5 Complementation of the Pcb1692ΔexpI mutant with the wild‐type expI gene restores flagella in vitro. LB, Luria–Bertani.


