Summary
During virus infection, specific viral component–host factor interaction elicits the transcriptional reprogramming of diverse cellular pathways. Alfalfa mosaic virus (AMV) can establish a compatible interaction in tobacco and Arabidopsis hosts. We show that the coat protein (CP) of AMV interacts directly with transcription factor (TF) ILR3 of both species. ILR3 is a basic helix–loop–helix (bHLH) family member of TFs, previously proposed to participate in diverse metabolic pathways. ILR3 has been shown to regulate NEET in Arabidopsis, a critical protein in plant development, senescence, iron metabolism and reactive oxygen species (ROS) homeostasis. We show that the AMV CP–ILR3 interaction causes a fraction of this TF to relocate from the nucleus to the nucleolus. ROS, pathogenesis‐related protein 1 (PR1) mRNAs, salicylic acid (SA) and jasmonic acid (JA) contents are increased in healthy Arabidopsis loss‐of‐function ILR3 mutant (ilr3.2) plants, which implicates ILR3 in the regulation of plant defence responses. In AMV‐infected wild‐type (wt) plants, NEET expression is reduced slightly, but is induced significantly in ilr3.2 mutant plants. Furthermore, the accumulation of SA and JA is induced in Arabidopsis wt‐infected plants. AMV infection in ilr3.2 plants increases JA by over 10‐fold, and SA is reduced significantly, indicating an antagonist crosstalk effect. The accumulation levels of viral RNAs are decreased significantly in ilr3.2 mutants, but the virus can still systemically invade the plant. The AMV CP–ILR3 interaction may down‐regulate a host factor, NEET, leading to the activation of plant hormone responses to obtain a hormonal equilibrium state, where infection remains at a level that does not affect plant viability
Keywords: AMV, coat protein, nucleolus, plant defense, ROS, transcription factors, virus‐interactions
Introduction
Compatible plant–virus interactions result in systemic infections that trigger many changes in host gene expression and metabolism, which may have a negative impact on normal plant development (Maule et al., 2000; Pallas and Garcia, 2011; Palukaitis et al., 2013; Rodrigo et al., 2012; Whitham et al., 2003, 2006). Gene expression changes, which occur during virus infection, can be elicited by the general accumulation of viral factors (Aparicio et al., 2005), but also by the interaction and/or interference of specific viral components with host factors (Culver and Padmanabhan, 2007; García and Pallás, 2015; Mandadi and Scholthof, 2013). Interactions between viral proteins and transcription factors (TFs) can result in the transcriptional reprogramming of different cellular pathways, making their identification interesting. For example, interactions between diverse TFs of the NAC domain family and viral proteins have been described which, depending on the virus, can either enhance or inhibit virus accumulation (Donze et al., 2014; Olsen et al., 2005; Puranik et al., 2012; Selth et al., 2005). It has been demonstrated recently that a viral protein can act as a plant TF by up‐regulating the regulator of cell proliferation upp‐L which, in turn, is a TF of the basic helix–loop–helix (bHLH) family, to cause severe leaf malformation (Lukhovitskaya et al., 2013). Diverse studies have indicated the implication of WRKY family TFs in the regulation of defence response signalling (Kim et al., 2008; Peng et al., 2012).
bHLH TFs comprise a family of transcriptional regulators that bind as homo‐ and heterodimers to specific DNA target sites, and are implicated in diverse plant metabolism and development pathways (Heim et al., 2003; Toledo‐Ortiz et al., 2003). In Arabidopsis, 147 bHLH genes have been identified and grouped into 21 families (Toledo‐Ortiz et al., 2003). Arabidopsis bHLH105/ILR3 (referred to hereafter as AtILR3) belongs to subgroup IVc (Heim et al., 2003; Toledo‐Ortiz et al., 2003), whose members are characterized by a leucine zipper domain following the bHLH domain. AtILR3 is expressed in all tissues in each plant developmental stage. Several findings have suggested that AtILR3, in combination with other regulatory proteins, may participate directly or indirectly in diverse metabolic pathways, such as iron and ROS homeostasis, auxin responsiveness and stress responses (Long et al., 2010; Nechushtai et al., 2012; Rampey et al., 2006). Thus, AtILR3 has been reported to interact directly with Arabidopsis PYE (another bHLH TF) and BRUTUS (a putative E3 ligase protein). These two proteins are implicated in iron metabolism by regulating the expression of genes involved in iron homeostasis (Long et al., 2010). Interestingly, another study, which used gain‐ and loss‐of‐function Arabidopsis ilr3 mutants (Rampey et al., 2006), has indicated that this TF may regulate the expression of a gene coding for a protein that contains an iron‐binding zinc finger CDGSH‐type domain, recently identified as the plant version of NEET proteins (AtNEET) (Nechushtai et al., 2012), and three significantly homologous genes to Arabidopsis vacuolar iron transporter 1 (Kim et al., 2006), called vacuolar iron transporter homologues (VITh) (Rampey et al., 2006). NEET proteins are involved in assisting Fe–S cluster transfer between proteins (Paddock et al., 2007). AtNEET plays a critical role in plant development, senescence, iron metabolism and reactive oxygen species (ROS) homeostasis (Nechushtai et al., 2012).
Among plant viral factors, coat proteins (CPs) are multifunctional proteins that play major roles in most virus infection steps, including the establishment of interactions with host factors (Callaway et al., 2001; Ni and Cheng‐Kao, 2013; Weber and Bujarski, 2015). Indeed, the CP of Alfalfa mosaic virus (AMV) is involved in the regulation of the replication and translation of viral RNAs, the cell‐to‐cell and systemic movement of the virus and virion formation (Sanchez‐Navarro et al., 2006; reviewed in Bol, 2005). AMV is the only member of the genus Alfamovirus in the family Bromoviridae which, with the Ilarvirus genus, requires the presence of the CP in inoculum to be infectious (for a recent review, see Pallas et al., 2013). Its genome consists of three single‐stranded RNAs of plus sense polarity. RNA1 and RNA2 encode the replicase subunits P1 and P2, respectively, whereas RNA3 encodes the movement protein (MP) and serves as a template for the synthesis of non‐replicating subgenomic RNA4 (sgRNA4), which encodes the CP (Bol, 2005). In a previous study, we identified a nucleolar localization signal (NLoS) in the AMV CP, and found that the cytoplasmic/nuclear–nucleolar balance of CP accumulation modulates viral expression (Herranz et al., 2012). It is still unknown whether CP accumulation in the nucleus/nucleolus affects general cell gene expression. A recent study has identified several Arabidopsis proteins that interact with the AMV CP. However, the effect on infection was analysed only for a component of the chloroplast oxygen‐evolving complex of Photosystem II (PsbP), whose over‐expression negatively affected virus accumulation (Balasubramaniam et al., 2014).
We report herein the interaction between the AMV CP and TF ILR3 from both Arabidopsis and Nicotiana tabacum. By comparison of AMV infection in an Arabidopsis loss‐of‐function ILR3 mutant (ilr3.2 plants) with that in wild‐type (wt) plants, we were able to link the activity of this TF with the hormone‐based plant defence system. A model is proposed in which, on infection, AMV CP–ILR3 interaction down‐regulates a host factor, NEET. This, in turn, activates ROS and salicylic acid (SA)‐ and jasmonic acid (JA)‐dependent signalling defence.
Results
AMV CP interacts with ILR3 from Arabidopsis and N. tabacum
To identify the AMV CP host interacting proteins, we performed yeast two‐hybrid (Y2H) screening using CP as bait and an Arabidopsis leaf‐specific cDNA library as prey (Németh et al., 1998). Several clones containing almost the full‐length sequence of the TF ilr3 (at3g23210) were found to grow on minimal synthetic selective interaction medium (data not shown). As we were interested in using N. tabacum as plant host, we searched the National Center for Biotechnology Information (NCBI) database for Nicotiana ILR3 homologues. We found two putative ilr3‐like genes in N. tabacum that we called NtILR3‐like1 and NtILR3‐like2, respectively. NtILR3‐like2 had a single‐nucleotide deletion at its C‐terminus, which caused a premature stop codon after amino acid 205. However, both the bHLH as well as the adjacent leucine zipper domains, characteristic of subgroup IVc, are conserved (Fig. S1, see Supporting Information). The AtILR3 protein showed 70% identity with NtILR3‐like1 and 57% identity with NtILR3‐like2 (Fig. S1). To validate the original Y2H screening of full‐length open reading frames (ORFs) of AtILR3, a homologue from subgroup IVc (AtbHLH115, at1g51070) and NtILR3‐like1 were fused to the activation domain (pAD plasmid) and transformed into yeast cells that expressed the AMV CP fused to the binding domain (pBD plasmid). After growth at 28 ºC for 5 days on interaction selective medium, we found that the CP specifically interacted with AtILR3 and NtILR3‐like1, but not with AtbHLH115 (Fig. 1A). Empty pBD vector and vector expressing the tumor protein p53 (pBD:p53) were used as negative interaction controls (Fig. 1A). The pBD:CP–pAD interaction was performed to rule out CP self‐activation (Fig. S2, see Supporting Information). Next, we determined the subcellular localization of TFs by transiently agro‐expressing the proteins fused in frame at the C‐terminus of the red fluorescent protein (dsRed). Confocal laser‐scanning microscopy (CLSM) demonstrated that all three TFs were exclusively localized throughout the nucleoplasm, except the nucleolus (Fig. 1B, only dsRed:NtILR3‐like1 and dsRed:AtbHLH115 are shown). Finally, bimolecular fluorescence complementation (BiFC) analysis was used to corroborate the in planta CP–ILR3 interactions (Aparicio et al., 2006). Reconstituted yellow fluorescent protein (YFP) fluorescence was found to form discrete granules exclusively in the nuclei of the cells infiltrated with the AMV CP and both ILR3 proteins (Fig. 2, CYFP:CP plus NYFP‐AtILR3 or NYFP‐NtILR3‐like1), whereas no interaction was detected between AtbHLH115 and the CP (Fig. 2, CYFP:AtbHLH115 plus NYFP:CP).
Figure 1.
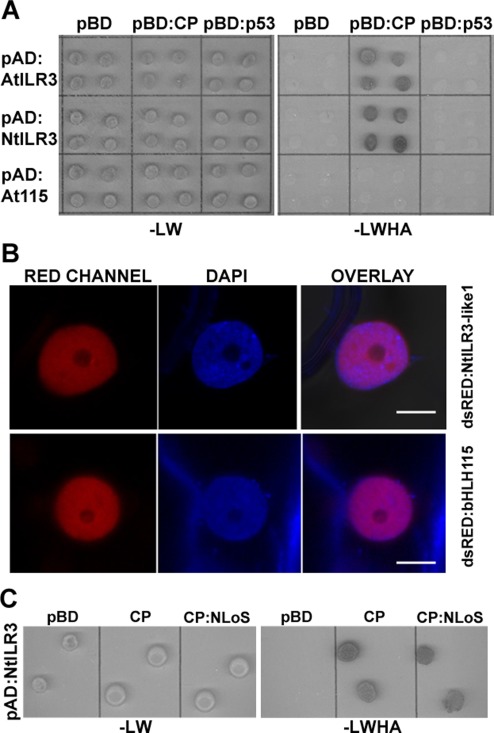
Arabidopsis and tobacco ILR3 interacts with Alfalfa mosaic virus (AMV) coat protein (CP). (A) AMV CP interaction with AtILR3, AtbHLH115 and NtILR3‐like1 transcription factors (TFs) by a yeast two‐hybrid (Y2H) system. AH109 yeast cells co‐transformed with the indicated plasmids were spotted onto minimal medium lacking leucine and tryptophan (–LW) or leucine, tryptophan, histidine and adenine (–LWHA) to confirm correct co‐transformation and positive interactions, respectively. (B) Nuclei from cells expressing NtILR3‐like1 and AtbHLH115 fused to dsRed and stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) are shown by confocal laser‐scanning microscopy (CLSM) in the red (RED) and blue (DAPI) channels. Overlay images confirmed that TFs localized exclusively in the nucleoplasm. (C) Interaction of AMV CP lacking the nucleolar localization signal (NLoS) with NtILR3‐like1 by the Y2H system. Co‐transformed AH109 yeast cells were spotted onto minimal medium lacking the corresponding amino acids (–LW or –LWHA) to confirm correct co‐transformation and positive interactions, respectively.
Figure 2.
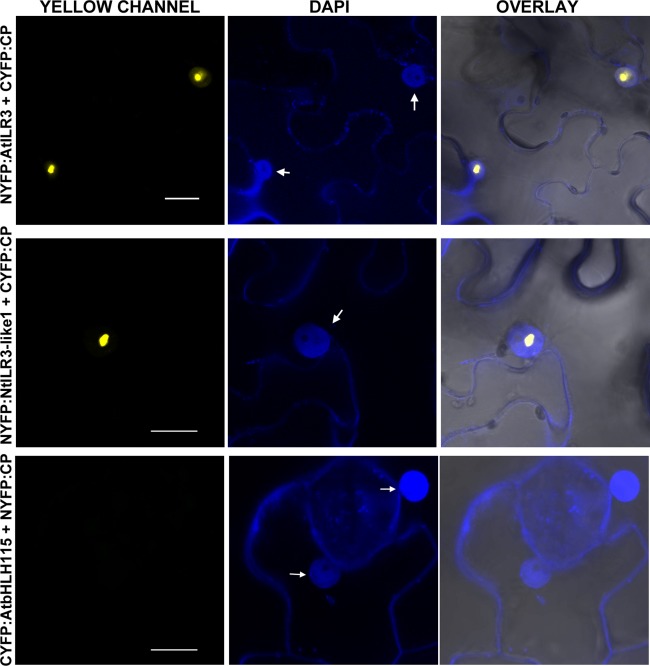
Bimolecular fluorescence complementation (BiFC) analysis of coat protein (CP)–ILR3 interaction. Confocal laser‐scanning microscopy (CLSM) images of nuclei from epidermal cells co‐infiltrated with the constructs indicated on the left and stained with 4′,6‐diamidino‐2‐phenylindole (DAPI) are shown in the yellow (YELLOW, YFP) and blue (DAPI) channels. Overlay panels are the superposition of yellow fluorescent protein (YFP) and DAPI over the bright field images. YFP reconstitution was exclusively found in the nucleus (arrows). The BiFC nomenclature of the plasmids is as follows: NYFP and CYFP refer to the N‐terminal and C‐terminal fragments of YFP and, in all constructs, the YFP tag is attached to the N‐terminus of the protein. Bars, 10 µm.
It has been shown previously that the interaction between viral and host proteins can affect their subcellular localization (e.g. Inaba et al., 2011; Ren et al., 2005; Uhrig et al., 2004). To analyse whether the ILR3–CP interaction during infection could have an effect on the subcellular location of ILR3, transgenic N. tabacum plants expressing the proteins of AMV P1 and P2 (P12 plants, Taschner et al., 1991) were inoculated with RNA transcripts from a modified AMV RNA3 clone, which expressed the green fluorescent protein (GFP) and permitted infected cells to be visualized (Fig. 3A, R3GFP‐CPwt construct) (Sanchez‐Navarro et al., 2001). At 48 h post‐inoculation (hpi), these leaves were infiltrated with Agrobacterium expressing AtILR3, NtILR3‐like1 or AtbHLH115 fused to dsRed. At 96 hpi, leaves were examined by fluorescence microscopy to localize the infection foci identified by the expression of GFP (Fig. 3B), and CLSM images were taken of the nuclei of the non‐infected and infected cells (Fig. 3B, top arrow and two bottom arrows, respectively, and Fig. 3C). In infected cells, GFP accumulated in the nucleus, except the nucleolus, whereas a fraction of dsRed:AtILR3 and dsRed:NtILR3‐like1 accumulated in the nucleolus. Their localization differed in non‐infected cells, where all three fusion proteins remained in the nucleoplasm and did not enter the nucleolus (Fig. 3C, only dsRed:AtILR3 is presented as being representative of the three TFs). In contrast, dsRed:AtbHLH115 was located outside of the nucleolus of the infected cells (Fig. 3C, bottom row). As the AMV CP accumulates in the nucleolus of the infected cells (Herranz et al., 2012), we wondered whether the ILR3 fraction found in the nucleolus was transported to this structure as a result of its interaction with the nucleolar traffic CP. To corroborate this hypothesis, we inoculated P12 leaves with an RNA3 mutant, which failed to accumulate the CP in the nucleolus as the CP lacked the NLoS (Fig. 3D, R3GFP‐CPNLoS) (Herranz et al., 2012). As before, the inoculated leaves were infiltrated with Agrobacterium expressing the TFs. Again, fluorescence microscopy was used to identify the infection foci (Fig. 3E), and CLSM images were taken of the nuclei from the infected leaves (Fig. 3F). In this case, neither of the three TFs accumulated in the nucleolus of infected cells (Fig. 3F, only dsRed:AtILR3 is shown as being a representative image of all TFs). Finally, by Y2H, we established that the CPNLoS mutant was still able to interact with NtILR3‐like1 (Fig. 1C). In summary, our results indicate that the TFs which interact with the CP by Y2H, e.g. AtILR3 and NtILR3, but not the non‐interacting AtbHLH115, are relocated towards the nucleolus of the infected cells. However, this targeting failed when a mutated CP that lacked NLoS was used.
Figure 3.
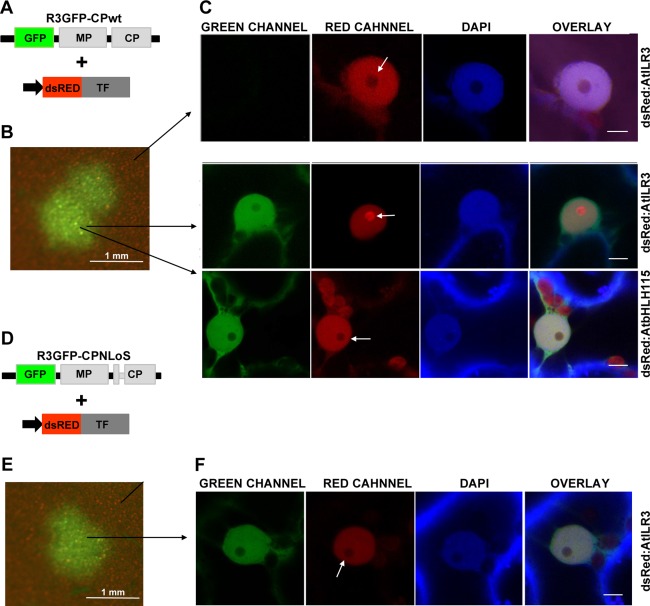
Alfalfa mosaic virus (AMV) infection promotes nucleolar relocalization of AtILR3. (A, D) Schematic representation showing the modified AMV RNA3 with the wild‐type (wt) coat protein (CP) (R3GFP‐CPwt) or the mutated CP lacking the nucleolar localization signal (NLoS) (R3GFP‐CPNLoS) and expressing green fluorescent protein (GFP). (B, E) Images of one infection focus denoted by the accumulation of GFP. (C) Confocal laser‐scanning microscopy (CLSM) images in the green (GREEN), red (RED) and blue (4′,6‐diamidino‐2‐phenylindole, DAPI) channels of nuclei from non‐infected and infected cells with R3GFP‐CPwt and transiently expressing the transcription factors (TFs) indicated on the right of the panels. White arrows indicate the nucleolus. (F) CLSM images of a nucleus of a cell infected with R3GFP‐CPNLoS and transiently expressing dsRed:AtILR3. In this case, the TF does not accumulate in the nucleolus (white arrow). In all images, bar = 5 µm.
Loss of ILR3 activity activates plant defence responses
A previous transcriptomic analysis of the Arabidopsis ILR3 loss‐of‐function mutant ilr3.2, identified by Rampey et al. (2006), has shown altered mRNA levels of AtNEET. This suggests that ILR3 may be implicated directly or indirectly in the transcriptional regulation of this gene. Another study has reported that Arabidopsis RNA interference (RNAi) lines with low AtNEET mRNA present enhanced ROS accumulation, indicating that this protein is involved in ROS homeostasis (Nechushtai et al., 2012).
As reported previously, reverse transcription‐polymerase chain reaction (RT‐PCR) analysis did not detect intact ILR3 mRNA in homozygous ilr3.2 (Fig. 4A) (Rampey et al., 2006). Under our plant growth conditions, northern blot analysis showed that, in AtNEET, mRNA was reduced slightly in the ilr3.2 mutant (Fig. 4A, the NEET panel). Next, we wondered whether reduced NEET mRNA accumulation would be accompanied by a deregulation of ROS levels. We analysed ROS by visualization of H2O2 accumulation using 3,3′‐diaminobenzidine (DAB) staining. DAB polymerizes in contact with H2O2, which produces a visible reddish‐brown precipitate (Fryer et al., 2003). We found a brown precipitate in ilr3.2 leaves, which correlated with the relative decrease in AtNEET mRNA (Fig. 4B). As ROS has been reported to be implicated in the plant response against pathogen attack, we examined whether other defence pathways could also be challenged in ilr3.2 plants. We found that pathogenesis‐related protein 1 (PR1) mRNA, a hallmark of SA signalling activation, was also clearly induced (Fig. 4A, the PR1 panel). Finally, quantification of fresh weight demonstrated that ilr3.2 presented an approximately 30% reduction in weight compared with wt plants under our growth conditions (Fig. 4C). Altogether, these data show that interference with ILR3 function activates defence responses.
Figure 4.
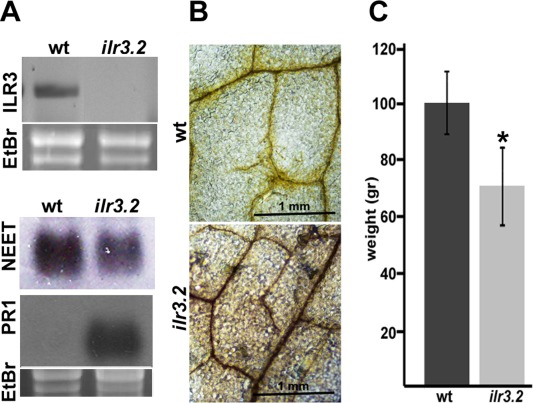
Characterization of Arabidopsis ilr3.2 mutant plants. (A) Northern blots to detect mRNA accumulation of several genes (indicated on the left) in wild‐type (wt) and ilr3.2 plants. Ethidium bromide (EtBr) staining of ribosomal RNAs was used as RNA loading control. PR1, pathogenesis‐related protein 1. (B) 3,3′‐Diaminobenzidine (DAB) staining of leaves from wt and ilr3.2 plants to reveal H2O2 accumulation (dark brown precipitate). (C) Plant weight of wt and ilr3.2 plants. Rosettes from 21‐day‐old plants were cut from the roots and weighed. Asterisk indicates significant difference from wt (*P < 0.05) using the paired t‐test (n = 25). Error bars represent the standard error of the mean.
AMV infection interferes with the putative activity of ILR3 and increases SA and JA biosynthesis
An open question was whether AMV infection would induce, in wt plants, the same metabolic effects as found in the ilr3.2 mutant. The fact that NEET mRNA expression could be regulated by ILR3 led us to identify putative NEET proteins in tobacco. The NCBI database search identified a putative protein sequence in tobacco (Accession number EB680812), which showed an identity of 70% with AtNEET (at5g51720). The N‐terminal part was the most dissimilar domain (Fig. S1). We examined its subcellular localization by fusing the protein to the N‐terminus of GFP (NtNEET:GFP). As AtNEET has been described previously to accumulate in both chloroplast and mitochondria (Nechushtai et al., 2012), we co‐infiltrated NbNEET:GFP and a mitochondrial marker (mt‐rk CD3‐991) in N. benthamiana leaves (Nelson et al., 2007). Unlike AtNEET, CLSM showed that the tobacco protein exclusively accumulated in chloroplasts (Fig. 5, arrowheads and arrows indicate chloroplasts and mitochondria, respectively).
Figure 5.
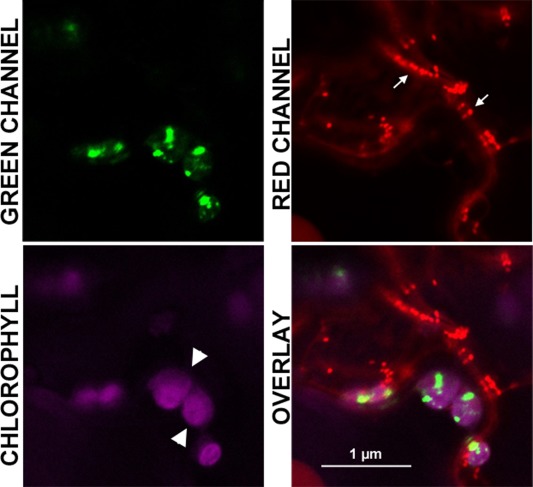
Subcellular localization of NtNEET protein. Magnified image of a cell co‐expressing NtNEET:GFP (GREEN CHANNEL) and the mitochondrial marker mt‐rk CD3‐991 (RED CHANNEL). Autofluorescence of chlorophyll in chloroplasts is shown in magenta (CHLOROPHYLL). Overlay image reveals that NtNEET accumulates in the chloroplast (white arrowhead), but not in mitochondria (arrows). GFP, green fluorescent protein.
To analyse the effect of AMV infection, N. tabacum cv. Xanthi and Arabidopsis wt and ilr3.2 plants were inoculated with the AMV PV0196 isolate (DSMZ GmbH, Plant Virus Collection, Braunschweig, Germany). In tobacco, this isolate induces reduced plant size, accompanied by a chlorotic pattern in the inoculated and upper leaves, whereas it is asymptomatic in Arabidopsis. Total RNA was extracted from inoculated and upper systemic tobacco leaves [at 2 and 4 days post‐inoculation (dpi), respectively] and from inoculated Arabidopsis leaves (at 4 dpi). In tobacco, northern blot analyses showed that NtILR3 mRNA accumulation was similar in mock and infected plants, whereas NtNEET and NtPR1 mRNAs were clearly down‐ and up‐regulated, respectively, in both inoculated and systemically infected leaves (Fig. 6A, compare lanes M and A). The same effect was found in infected Arabidopsis wt plants (Fig. 6B, compare lanes M and A in wt plants). In ilr3.2 plants, AtNEET was induced slightly, whereas AtPR1 mRNA was reduced in infected vs. mock‐inoculated plants (Fig. 6B, compare lanes M and A in ilr3.2 plants). In contrast, northern blot analysis using a digoxigenin‐labelled probe to detect AMV RNA3 corroborated that the virus accumulated in inoculated plants (Fig. 6A,B, bottom panels). We wondered whether the observed reduction in NEET in infected plants would also be accompanied by the deregulation of ROS. DAB staining revealed a strong brown precipitate, indicative of increased H2O2 (Fryer et al., 2003), in both tobacco and Arabidopsis wt infected leaves, which was absent in non‐infected material (Fig. 7, compare the AMV and mock panels, respectively).
Figure 6.
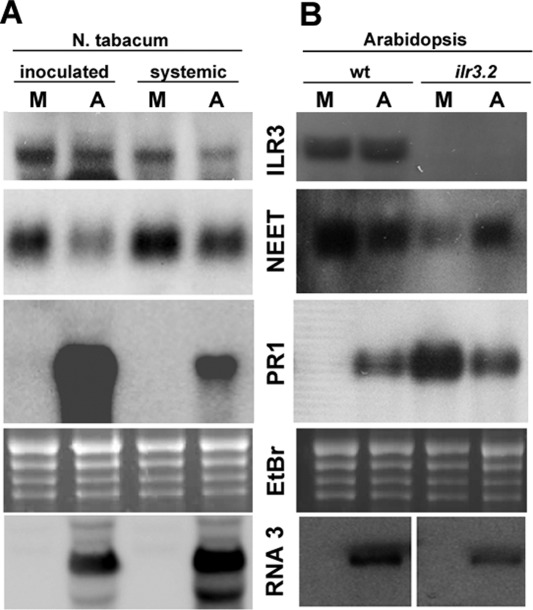
Alfalfa mosaic virus (AMV) infection activates plant defence responses. Northern blots to detect mRNA accumulation of several genes (indicated in the middle of the panels) from mock‐ and AMV‐inoculated plants (M and A, respectively) in Nicotiana tabacum (A) and Arabidopsis (B). Inoculated and upper systemic leaves in tobacco and inoculated leaves in Arabidopsis wt and ilr3.2 plants were analysed (indicated at the top). Ethidium bromide (EtBr) staining of ribosomal RNAs was used as RNA loading control. Northern blot to detect the AMV RNA3 (bottom panel) was used to corroborate viral accumulation.
Figure 7.
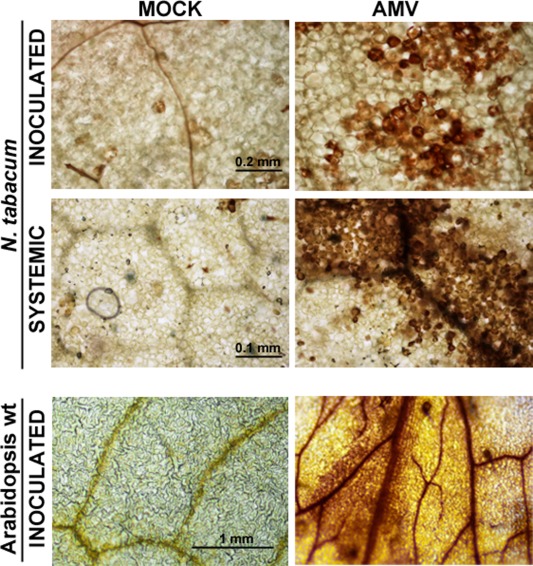
3,3′‐Diaminobenzidine (DAB) staining of Nicotiana tabacum and Arabidopsis wild‐type (wt) plants. Inoculated or upper systemic leaves (indicated on the left) from healthy (mock) and infected (Alfalfa mosaic virus, AMV) plants were infiltrated with DAB solution to visualize H2O2 accumulation.
As AMV infection led to the activation of defence responses, we measured the content of two hormones that modulate plant immunity, i.e. SA and JA. Phytohormone content was quantified in mock and inoculated leaves at 4 dpi from both Arabidopsis wt and ilr3.2 plants. In Arabidopsis wt, SA and JA contents increased in infected vs. healthy plants (Fig. 8, wt plants, AMV and mock, respectively). The same response was observed in mock ilr3.2 plants compared with wt plants (Fig. 8, compare mock bars of wt and ilr3.2 plants), which indicates that a loss of ILR3 activity correlates with the induction of SA and JA biosynthesis. The SA content decreased, whereas the JA content increased more than 10‐fold in infected vs. mock‐inoculated ilr3.2 plants (Fig. 8, ilr3.2 plants). This indicates that AMV infection can also induce JA biosynthesis, irrespective of ILR3 activity. As found in other virus–host interactions, the considerable JA accumulation in ilr3.2‐infected plants was able to antagonize the SA pathway, which explains the reduction in SA and AtPR1 mRNA found in infected vs. mock ilr3.2 plants (reviewed in Alazem and Lin, 2015; Collum and Culver, 2016).
Figure 8.
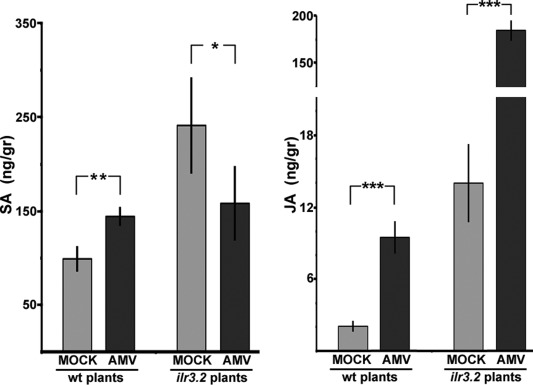
Salicylic acid (SA) and jasmonic acid (JA) contents in Arabidopsis wild‐type (wt) and ilr3.2 plants. Hormone content was measured in mock and Alfalfa mosaic virus (AMV)‐inoculated leaves at 4 days post‐inoculation (dpi). Asterisks indicate the statistical significance of AMV infection with respect to mock‐inoculated plants within each plant group: *P < 0.1; **P < 0.05; ***P < 0.01. Error bars represent the standard error of the mean obtained from three biological replicates.
ILR3 activity influences AMV accumulation
Finally, we analysed whether the absence of ILR3 had an effect on AMV infection. Arabidopsis wt and ilr3.2 plants were inoculated with AMV PV0196 isolate virion particles. A northern blot analysis using a digoxigenin‐labelled AMV CP (Fig. 9A) ORF, plus quantification of the blot signals by Image J software, was carried out to measure virus accumulation. Figure 9A shows the northern blot of the upper non‐inoculated leaves, whereas the graph in Fig. 9B illustrates how the accumulation of AMV RNAs decreased by 40% in ilr3.2 relative to wt inoculated leaves at 4 dpi, and by around 30% in upper non‐inoculated leaves at 10 dpi. This experiment was repeated three times and similar results were obtained. Our results indicate that loss of ILR3 function activates the plant defence signalling response in Arabidopsis. However, the virus escaped the defence response, still being able to move systemically, infecting the whole plant, but at a lower accumulation level.
Figure 9.
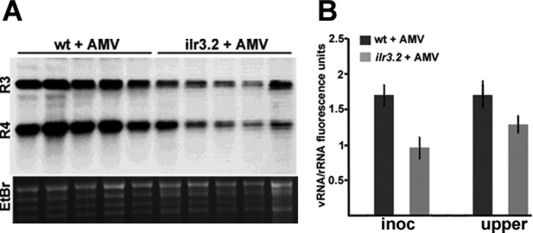
ILR3 activity affects Alfalfa mosaic virus (AMV) accumulation in Arabidopsis. (A) Northern blot corresponding to five Arabidopsis wild‐type (wt) and ilr3.2 plants to detect viral RNA3 and RNA4 (R3 and R4). Ethidium bromide (EtBr) staining of ribosomal RNAs was used as RNA loading control. (B) Graph showing the average viral RNA accumulation measured in inoculated and upper systemic leaves (inoc and upper, respectively) in wt and ilr3.2 plants (*P < 0.05 using paired t‐test, n = 5).
Discussion
Plant viruses usurp a large number of host factors/resources to their own benefit to survive, whereas the infected plant activates a whole series of responses to protect itself from the negative impact of the pathogen (Culver and Padmanabhan, 2007; García and Pallás, 2015; Mandadi and Scholthof, 2013). Herein, we have demonstrated that the interaction of a viral CP with a TF can participate in the activation of the plant defence response in infected plants, although this host response does not prevent the virus from invading the plant.
The CP of AMV is a multifunctional protein that plays essential roles in the viral cycle: from the regulation of the replication and translation of viral RNAs to cell‐to‐cell and systemic movement and encapsidation (reviewed in Bol, 2005 and Pallas et al., 2013). In the present work, we report the interaction between CP and ILR3, a TF that belongs to the bHLH family. This interaction leads to the relocation of one ILR3 protein pool to the nucleolus. This interaction was observed with ILR3 from two plant species, suggesting that this phenomenon may be a general feature of AMV infection. We have shown previously that AMV CP accumulates in the cytoplasm and nucleus/nucleolus of infected cells (Herranz et al., 2012). Here, we provide evidence that ILR3 nucleolar translocation is probably driven by viral protein transport towards the nucleolus. Subcellular localization changes in host factors, induced by interactions with viral proteins, have been reported previously. For example, in Arabidopsis infected with Cucumber mosaic virus, cytoplasmic catalase 3 is relocated in the nucleus through its interaction with the 2b protein (Inaba et al., 2011), whereas the p19 protein of Tomato bushy stunt virus interacts with three proteins of the AYL family to activate transport from the nucleus to the cytoplasm (Uhrig et al., 2004). The CP of Turnip crinkle virus interacts with TF TIP and prevents its nuclear localization (Ren et al., 2004).
One relevant question was raised: how does the CP–ILR3 interaction affect TF function. Although it has been established that bHLH TFs function as homo‐ and heterodimers (Heim et al., 2003; Toledo‐Ortiz et al., 2003), and several findings have led to the hypothesis that ILR3 might function in combination with other bHLH TFs (Long et al., 2010), no direct targets of ILR3 have yet been identified. Experiments conducted to determine AtILR3 function with gain‐ and loss‐of‐function Arabidopsis mutants have reported that the mRNA levels of AtNEET are altered in both mutant types, suggesting that this TF may directly or indirectly influence NEET expression (Rampey et al., 2006). Our northern blot analysis showed that mRNA accumulation of NEET in both tobacco and Arabidopsis was slightly down‐regulated during the course of infection, and resembled Arabidopsis loss‐of‐function ilr3‐2 (Rampey et al., 2006). However, taking into account the critical role of NEET in Arabidopsis metabolism, we cannot rule out the possibility that other TFs might function redundantly to regulate NEET mRNA transcription.
AtNEET has been localized previously in both mitochondria and chloroplasts, and has been found to be a key regulator in plant development, senescence, iron metabolism and ROS homeostasis (Nechushtai et al., 2012). Contradictory results on its subcellular localization have also been reported, as a recent study concluded that AtNEET translocates in the chloroplast, but not in mitochondria (Su et al., 2013). With NtNEET, we found that the protein was localized only in the chloroplast. The transport of proteins to mitochondria and chloroplasts can be achieved through N‐terminal transit peptides. Accordingly, AtNEET and NtNEET contain the proposed N‐terminal chloroplast transit peptide cleavage motif V‐R/K‐A‐E (Su et al., 2013). However, this N‐terminal region presents most of the differences observed in their sequences. The absence of NtNEET in mitochondria can be explained by assuming that the N‐terminal region of NtNEET lacks the mitochondrial transit peptide.
Our results link chloroplast function with viral infection. The chloroplast is a key organelle that generates plant defence signalling molecules, including ROS and SA (Spoel and Dong, 2012; Torres, 2010). Indeed, several viruses encode proteins that interact with photosynthetic machinery components, and it has been found that defence host response activation is prevented through these interactions (Abbink et al., 2002; Bhat et al., 2013; Jimenez et al., 2006). A recent report has shown that the AMV CP, through its interaction with chloroplast‐targeted PsbP, can also favour AMV replication by sequestering the host protein in the cytosol, which impedes the activation of plant defence responses (Balasubramaniam et al., 2014).
ROS, SA and JA concentrations are high in healthy ilr3.2 plants, which indicates that ILR3 activity participates in plant defence response modulation (Fig. 10). Our results suggest that, through the CP–ILR3 interaction, AMV can interfere with TF function to bring about NEET expression down‐regulation. This, in turn, leads to the activation of ROS and SA defence responses. Furthermore, JA biosynthesis activation is partially independent of ILR3 function (Fig. 10). Increased ROS and SA signalling pathway activation has been found in diverse compatible virus–host interactions (Whitham et al., 2006). In compatible host–virus interactions, the expression of the majority of defence‐related genes is induced by an SA‐dependent signalling pathway (Huang et al., 2005). The high JA concentration in ilr3.2‐infected plants might antagonize the SA pathway and lead to the strong down‐regulation of AtPR1 mRNA observed in these plants. Different studies have reported that, depending on the concentration of each hormone, the SA and JA pathways display antagonistic or synergistic effects (reviewed in Collum and Culver, 2016; Pieterse et al., 2012). In this sense, it has been found that, in some compatible virus–host interactions, early components of the SA pathway may be regulated by JA (reviewed in Alazem and Lin, 2015).
Figure 10.
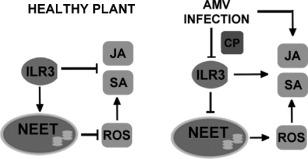
Hypothetical model to illustrate the effect of the coat protein (CP)–ILR3 interaction on the plant defence response. In healthy plants, ILR3 activity may regulate NEET levels, contributing to the repression of reactive oxygen species (ROS) and salicylic acid (SA) signalling pathways. In addition, ILR3 participates in the regulation of jasmonic acid (JA) biosynthesis. In Alfalfa mosaic virus (AMV) infection, interaction between CP and ILR3 may reduce ILR3 activity, leading to the repression of NEET mRNA accumulation and the activation of ROS and SA signalling. At the same time, AMV infection induces JA biosynthesis independent of ILR3 activity.
In summary, our results reveal that AMV infection activates plant defence pathways, which can negatively affect virus accumulation without impeding systemic host infection. A model is proposed (Fig. 10) by which, on infection, the AMV CP–ILR3 interaction modifies the subcellular location of the TF by down‐regulating a host factor, NEET; in turn, this activates ROS and SA‐ and JA‐dependent signalling defence pathways, with the latter being partially independent of ILR3.
Experimental Procedures
Y2H screening
The Arabidopsis cDNA library fused to the GAL4 AD in pGADT7 has been described previously (Németh et al., 1998). Plasmids pBD:CP and pBD:CPNLoS containing the full‐length AMV CP variants fused to the GAL4 BD in pGBKT7 plasmid have been described by Herranz et al. (2012). Y2H screening was performed with the Matchmaker Gal4 Two‐hybrid System 3 Clontech (Saint‐Germain‐en‐Laye, France) following the manufacturer's recommendations. Briefly, yeast reporter strain AH109 (Clontech) was sequentially transformed with pBD:CP and pGAD:cDNA, and co‐transformants were selected by culture on minimal synthetic medium lacking leucine and tryptophan (–LW). Positive interactions were selected by culture on medium lacking leucine, tryptophan, adenine and histidine (–LWHA). Cultures were kept at 28 ºC for 5 days. pAD:cDNA clones from positive interactions were rescued and subjected to DNA sequencing. Full‐length ORFs of AtILR3, AtbHLH115 and NtILR3‐like1 were PCR amplified with specific primers (Table S1, see Supporting Information), cloned into pGADT7 plasmid and transformed into AH109 carrying the empty pGBKT7 (pBD), pBD:CP or pBD:p53 plasmids. Reconfirmation of the interactions was carried out by growing cells in –LWHA medium for 5 days at 28 ºC.
BiFC and subcellular localization analysis
AtILR3, AtbHLH115, NtILR‐like1 and NtNEET ORFs were amplified with specific primers (Table S1) designed for cloning using the Gateway System Invitrogen (Waltham, MA, USA). Amplified products were recombined into a donor vector using the BP reaction, and then transferred into binary destination vectors expressing the full‐length dsRed or GFP for subcellular localization studies, and the N‐terminal and C‐terminal parts of YFP for BiFC analysis, following the manufacturer's recommendations. The fusion proteins generated were as follows: dsRed:AtILR3, dsRed:AtbHLH115, dsRed:NtILR3‐like1, NtNEET:GFP, NYFP:AtILR3, NYFP:NtILR3 and CYFP:AtbHLH115. Plasmids expressing the AMV CP fused to the N‐terminal and C‐terminal parts of YFP (NYFP:CP and CYFP:CP) and the mitochondrial marker mt‐rk CD3‐991 have been described previously (Aparicio et al., 2006; Nelson et al., 2007). All binary vectors were transformed into Agrobacterium tumefaciens C58 cells. Cultures were diluted at an optical density at 600 nm (OD600) of 0.2 in infiltration solution [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.5, 10 mm MgCl2] and infiltrated into 3‐week‐old N. benthamiana plants. For nuclei staining, 10 mg/mL 4′,6‐diamidino‐2‐phenylindole (DAPI) Sigma‐Aldrich (St. Louis, MO, USA) was infiltrated into N. benthamiana leaves, 1 h before observation.
Confocal images were taken at 48 h after agro‐infiltration with a Zeiss (Oberkochen, Germany) LSM 780 AxiObserver microscope. All images correspond to single slices, 1.8 μm thick, of epidermal cells. Excitation and emission wavelengths were 359 and 457 nm for DAPI, 488 and 508 nm for GFP, 514 and 527 nm for YFP, 545 and 572 nm for dsRed, and 488 and 750 nm for chloroplast visualization, respectively.
Plant growth conditions and virus inoculation
Arabidopsis ecotype Col‐0 wt and il3.2, N. tabacum cv. Xanthi, N. benthamiana and N. tabacum cv. Samsun P12 plants (Taschner et al., 1991) were grown in 12‐cm pots in a growth chamber with a photoperiod of 24 ºC, 16 h light/20 ºC, 8 h dark.
To analyse AMV infection effects on Arabidopsis or N. tabacum cv. Xanthi, two leaves of 3‐week‐old plants were mechanically inoculated with purified virions (1 mg/mL) of AMV PV0196 isolate (DSMZ GmbH, Plant Virus Collection) in 30 mm sodium phosphate buffer, pH 7, or with buffer alone (mock plants).
Plasmids px032/GFP‐MP‐CP and px032/GFP‐MP‐CP(K5‐13:A) (Herranz et al., 2012; Sanchez‐Navarro et al., 2001) (here labelled R3CPwt‐GFP and R3CPNLoS‐GFP, respectively) were used to perform ILR3 relocalization studies in P12 plants. For inoculation purposes, PstI‐linearized plasmids were transcribed with T7 RNA polymerase Takara (Saint‐Germain‐en‐Laye, France) following the manufacturer's recommendations. Leaves were mechanically inoculated with 1 µg/leaf of the corresponding transcripts and A. tumefaciens C58 cultures expressing the corresponding fusion proteins were prepared in infiltration buffer at OD600 = 0.1 and infiltrated at 48 hpi. Images of infection foci were taken at 4 dpi with a Zeiss LSM 780 AxiObserver microscope.
Cloning of host genes for northern blot analysis
NCBI and http://sydney.edu.au/science/molecular_bioscience/sites/benthamiana/database were searched for tobacco NEET and PR1 genes. Multiple sequence alignment by the clustalw program (Thompson et al., 1994) was used to analyse homologies between Arabidopsis and tobacco genes. RT‐PCRs were carried out from RNA extractions with specific primers (Table S1). Amplified products were cloned into pTZ57R/T plasmid (ThermoFisher Scientific). After digestion with XbaI restriction enzyme ThermoFisher Scientific (Waltham, MA, USA), linearized plasmids were used as templates to transcribe digoxigenin‐labelled probes. Transcriptions were carried out with T7 RNA polymerase (Takara) following the manufacturer's recommendations.
The detection of host mRNAs and viral RNAs was carried out by northern blot analysis as described (Aparicio et al., 2003). In the case of N. tabacum, inoculated leaves were harvested at 2 dpi and upper systemic leaves were collected at 4 dpi. In the case of Arabidopsis, inoculated leaves were harvested at 4 dpi and upper systemic leaves were collected at 10 dpi. Leaves were ground in liquid nitrogen with a mortar and pestle, and total RNA was extracted from 0.1 g of leaf material using Trizol Reagent (Sigma); 10 µg and 1 µg of total RNA, for mRNA and viral RNA detection, respectively, were denatured by formaldehyde treatment and analysed by northern blot hybridization, as described previously (Sambrook et al., 1989). Viral RNAs were visualized on blots using a digoxigenin‐labelled riboprobe corresponding to the AMV CP gene. Synthesis of the digoxigenin‐labelled riboprobe, hybridization and digoxigenin detection procedures were carried out as described previously (Pallas et al., 1998).
DAB staining
DAB staining was carried out as described previously (Liu et al., 2012) with some modifications. Samples were analysed at the same time as northern blot studies. To prepare DAB solution, DAB (Sigma) was diluted at 1 mg/mL in H2O at pH 3.6 by vigorous shaking at 37 ºC for at least 30 min. Leaves were infiltrated with DAB solution with a syringe and placed in a Petri dish containing paper towels saturated with a fixative solution (ethanol–acetic acid–glycerol, 3 : 1 : 1, v/v/v) for 3–4 days at room temperature until the green colour disappeared. Bright field images were recorded with a Nikon (Tokyo, Japan) Eclipse E600.
Hormone analysis
For SA and JA quantification, mock‐ and AMV‐inoculated leaves from Arabidopsis wt and ilr3.2 plants were collected at 4 dpi. Each sample, containing a mixture of leaves from three plants, was ground in liquid nitrogen with a mortar and pestle. Material (100 mg fresh weight) was suspended in 80% methanol–1% acetic acid containing internal standards and mixed by shaking for 1 h at 4 ºC. The extract was kept at −20 ºC overnight and then centrifuged, and the supernatant was dried in a vacuum evaporator. The dry residue was dissolved in 1% acetic acid and passed through an Oasis (Waters Cromatografia, Cerdanyola del Vallès, Barcelona, Spain) HLB (reverse‐phase) column, as described in Seo et al. (2011).
The dried eluate was dissolved in 5% acetonitrile–1% acetic acid; the hormones were separated using an autosampler and reverse‐phase ultrahigh‐performance liquid chromatography (UHPLC) (2.6‐µm Accucore RP‐MS column; 50 mm length × 2.1 mm i.d.; ThermoFisher Scientific) with an acetonitrile gradient of 5%–50%, containing 0.05% acetic acid, at 400 µL/min over 14 min. For SA and JA quantification, the dried eluate was dissolved in 5% acetonitrile–1% acetic acid, and the hormones were separated using an autosampler and reverse‐phase UHPLC (2.6‐µm Accucore RP‐MS column; 50 mm length × 2.1 mm i.d.; ThermoFisher Scientific) with an acetonitrile gradient of 5%–50%, containing 0.05% acetic acid, at 400 µL/min over 14 min.
The internal standard for SA quantification was the deuterium‐labelled hormone, whereas, for JA, the compound dhJA (dihydrojasmonic acid) was used. The hormones were analysed with a Q‐Exactive mass spectrometer (Orbitrap detector; ThermoFisher Scientific) by targeted selected ion monitoring (SIM). The concentrations of the hormones in the extracts were determined using embedded calibration curves and the Xcalibur 2.2 SP1 build 48 and TraceFinder programs. Measurements were performed in triplicate.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of primers and plasmids used in this study.
Fig. S1 Amino acid sequence alignments of ILR3 and NEET proteins of Arabidopsis and Nicotiana tabacum.
Fig. S2 Analysis of putative yeast two‐hybrid (Y2H) self‐activation by Alfalfa mosaic virus (AMV) coat protein (CP). AH109 yeast cells co‐transformed with the indicated plasmids were spotted onto minimal medium lacking leucine and tryptophan (–LW) or leucine, tryptophan, histidine and adenine (–LWHA). Colony growth was only detected with the pair pBD:CP and pAD:CP, confirming that AMV CP does not self‐activate yeast growth.
Acknowledgements
F.A. was the recipient of a contract Ramón y Cajal (RYC‐2010‐06169) program of the Ministerio de Educación, Cultura y Deporte of Spain. We thank L. Corachan for excellent technical assistance. This work was supported by Grants BIO2014‐54862‐R from the Spanish grant agency Dirección General de Investigación Científica y Técnica (DGICT) the Prometeo Program GV2015/010 from the Generalitat Valenciana and PAID‐06‐10‐1496 from the Universitat Politecnica de Valencia (Spain). Arabidopsis T‐DNA insertion mutant ilr3.2 seeds were kindly provided by Dr Bonnie Bartel. We thank Dr Isabel Lopez‐Diaz and Dr Esther Carrera for the hormone quantification carried out at the Plant Hormone Quantification Service, IBMCP, Valencia, Spain. We declare that we have no conflicts of interest.
Contributor Information
Frederic Aparicio, Email: vpallas@ibmcp.upv.es.
Vicente Pallás, Email: faparici@ibmcp.upv.es.
References
- Abbink, T.E. , Peart, J.R. , Mos, T.N. , Baulcombe, D.C. , Bol, J.F. and Linthorst, H.J. (2002) Silencing of a gene encoding a protein component of the oxygen‐evolving complex of photosystem II enhances virus replication in plants. Virology, 295, 307–319. [DOI] [PubMed] [Google Scholar]
- Alazem, M. and Lin, N.S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, F. , Vilar, M. , Pérez‐Payá, E. and Pallas, V. (2003) The coat protein of Prunus necrotic ringspot virus specifically binds and regulates the conformation of its genomic RNA. Virology, 313, 213–223. [DOI] [PubMed] [Google Scholar]
- Aparicio, F. , Thomas, C.L. , Lederer, C. , Niu, Y. , Wang, D. , and Maule, A.J. (2005) Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 138, 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, F. , Sánchez‐Navarro, J.A. and Pallás, V. (2006) In vitro and in vivo mapping of the Prunus necrotic ringspot virus coat protein C‐terminal dimerization domain by bimolecular fluorescence complementation. J. Gen. Virol. 87, 1745–1750. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam, M. , Kim, B.S. , Hutchens‐Williams, H.M. and Loesch‐Fries, L.S. (2014) The photosystem II oxygen evolving complex protein, PsbP, interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant–Microbe Interact. 27, 1107–1118. [DOI] [PubMed] [Google Scholar]
- Bhat, S. , Folimonova, S.Y. , Cole, A.B. , Ballard, K.D. , Lei, Z. , Watson, B.S. , Sumner, L.W. and Nelson, R.S. (2013) Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 161, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol, J.F. (2005) Replication of alfamo‐ and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43, 39–62. [DOI] [PubMed] [Google Scholar]
- Callaway, A. , Giesman‐Cookmeyer, D. , Gillock, E.T. , Sit, T.L. and Lommel S.A. (2001) The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39, 419–460. [DOI] [PubMed] [Google Scholar]
- Collum, T.D. and Culver, J.N. (2016). The impact of phytohormones on virus infection and disease. Curr. Opin Virol. 17, 25–31. [DOI] [PubMed] [Google Scholar]
- Culver, J.N. and Padmanabhan, M.S. (2007) Virus‐induced disease: altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 45, 221–243. [DOI] [PubMed] [Google Scholar]
- Donze, T. , Qu, F. and Morris, T.J. (2014). Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology, 449, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer, M.J. , Ball, L. , Oxborough, K. , Karpinski, S. , Mullineaux, P.M. and Baker, N.R. (2003) Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis‐leaves. Plant J. 33, 691–705. [DOI] [PubMed] [Google Scholar]
- García, J.A. and Pallás, V. (2015) Viral factors involved in plant pathogenesis. Curr. Opin. Virol. 11, 21–30. [DOI] [PubMed] [Google Scholar]
- Heim, M , A., Jakoby , M., Werber , M., Martin , C., Weisshaar , B. and Bailey, P.C. (2003) The basic helix–loop–helix transcription factor family in plants: a genome‐wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Herranz, M.C. , Pallas, V. and Aparicio, F. (2012) Multifunctional roles for the n‐terminal basic motif of Alfalfa mosaic virus coat protein: nucleolar/cytoplasmic shuttling, modulation of RNA‐binding activity, and virion formation. Mol. Plant–Microbe Interact. 25, 1093–1103. [DOI] [PubMed] [Google Scholar]
- Huang, Z. , Yeakley, J.M. , Garcia, E.W. , Holdridge, J.D. , Fan, J.‐F. and Whitham, S.A. (2005). Salicylic acid‐dependent expression of host genes in compatible Arabidopsis–virus interactions. Plant Physiol. 137, 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, J.‐I , Kim, B.M. , Shimura, H. and Masuta, C. (2011) Virus‐induced necrosis is a consequence of direct protein–protein interaction between a viral RNA‐silencing suppressor and a host catalase. Plant Physiol. 156, 2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, I. , Lopez, L. , Alamillo, J.M. , Valli, A. and Garcia, J.A. (2006) Identification of a Plum pox virus CI‐interacting protein from chloroplast that has a negative effect in virus infection. Mol. Plant–Microbe Interact. 19, 350–358. [DOI] [PubMed] [Google Scholar]
- Kim, K.C. , Lai, Z. , Fan, B. and Chen, Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell, 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.A. , Punshon, T. , Lanzirotti, A. , Li, L. , Alonso, J.M. , Ecker, J.R. , Kaplan, J. and Guerinot, M.L. (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science, 314, 1295–1298. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Zhang, Z. , Faris, J.D. , Oliver, R.P. , Syme, R. , McDonald, M.C. , McDonald, B.A. , Solomon, P.S. , Lu, S. , Shelver, W.L. , Xu, S. and Friesen, T.L. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 8, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, T.A. , Tsukagoshi, H. , Busch, W. , Lahner, B. , Salt, D.E. and Benfey, P.N. (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell, 22, 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhovitskaya, N.I. , Solovieva, A.D. , Boddeti, S.K. , Thaduri, S. , Solovyev, A.G. and Savenkova, E.I. (2013) An RNA virus‐encoded zinc‐finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. Plant Cell, 25, 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi, K.K. and Scholthof, K.‐B.G. (2013) Plant immune responses against viruses: how does a virus cause disease? Plant Cell, 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule, A.J. , Escaler, M. and Aranda, M.A. (2000) Programmed responses to virus replication in plants. Mol. Plant Pathol. 1, 9–15. [DOI] [PubMed] [Google Scholar]
- Nechushtai, R. , Conlan, A.R. , Harir, Y. , Song, L. , Yogev, O. , Eisenberg‐Domovich, Y. , Livnah, O. , Michaeli, D. , Rosen, R. , Ma, V. , Luo, Y. , Zuris, J.A. , Paddock, M.L. , Cabantchik, Z.I. , Jennings, P.A. and Mittler, R. (2012) Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell, 24, 2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K. , Cai, X. and Nebenführ, A. (2007) A multi‐color set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Németh, K. , Salchert, K. , Putnoky, P. , Bhalerao, R. , Koncz‐Kálmán, Z. , Stankovic‐Stangeland, B. , Bakó, L. , Mathur, J. , Okrész, L. , Stabel, S. , Geigenberger, P. , Stitt, M. , Rédei, GP. , Schell, J. and Koncz, C. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12, 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, P. and Cheng‐Kao, C. (2013) Non‐encapsidation activities of the capsid proteins of positive‐strand RNA viruses. Virology, 446, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A.N. , Ernst, H.A. , Leggio, L.L. and Skriver K. (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. [DOI] [PubMed] [Google Scholar]
- Paddock, M.L. , Wiley, S.E. , Axelrod, H.L. , Cohen, A.E. , Roy, M. , Abresch, E.C. , Capraro, D. , Murphy, A.N. , Nechushtai, R. , Dixon, J.E. and Jennings, P.A. (2007) MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc . Natl. Acad. Sci. USA, 104, 14 342–14 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas, V. and Garcia, J.A. (2011) How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92, 2691–2705. [DOI] [PubMed] [Google Scholar]
- Pallas, V. , Mas, P. and Sanchez‐Navarro, J.A. (1998) Detection of plant RNA viruses by nonisotopic dot‐blot hybridization. Methods Mol. Biol. 81, 461–468. [DOI] [PubMed] [Google Scholar]
- Pallas, V. , Aparicio, F. , Herranz, M.C. , Sanchez‐Navarro, J.A. and Scott, S.W. (2013) The molecular biology of ilarviruses. Adv. Virus Res. 87, 139–181. [DOI] [PubMed] [Google Scholar]
- Palukaitis, P. , Groen, S.C. and Carr, J.P. (2013) The Rumsfeld paradox: some of the things we know that we don't know about plant virus infection. Curr. Opin. Plant Biol. 16, 513–519. [DOI] [PubMed] [Google Scholar]
- Peng, X. , Hu, Y. , Tang, X. , Zhou, P. , Deng, X. , Wang, H.H. and Guo, Z.J. (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236, 1485–1498. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012). Hormonal regulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Puranik, S. , Sahu, P.P. , Srivastava, P.S. and Prasad, M. (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 1–13. [DOI] [PubMed] [Google Scholar]
- Rampey, R.A , Woodward, A.W. , Hobbs, B.N. , Tierney, M.P. , Lahner, B. , Salt, D.E. and Bartel, B. (2006) An Arabidopsis basic helix‐loop‐helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics, 174, 1841–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, T. , Qu, F. and Morris. T.J. (2005) The nuclear localization of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crinkle virus. Virology, 331, 316–324. [DOI] [PubMed] [Google Scholar]
- Rodrigo, G. , Carrera, J. , Ruiz‐Ferrer, V. , del Toro, F.J. , Llave, C. , Voinnet, O. and Elena, S.F. (2012) A meta‐analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PLoS One, 7, e40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sanchez‐Navarro, J.A. , Miglino, R. , Ragozzino, A. and Bol, J.F. (2001) Engineering of Alfalfa mosaic virus RNA 3 into an expression vector. Arch. Virol. 146, 923–939. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Navarro, J.A. , Herranz, M.C. and Pallás, V. (2006) Cell‐to‐cell movement of Alfalfa mosaic virus can be mediated by the movement proteins of Ilar‐, bromo‐, cucumo‐, tobamo‐, and comoviruses and does not require virion formation. Virology, 346, 66–73. [DOI] [PubMed] [Google Scholar]
- Selth, L.A. , Dogra, S.C. , Rasheed, M.S. , Healy, H. , Randles, J.W. and Rezaian. M.A. (2005) A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell, 17, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, M. , Jikumaru, Y. and Kamiya, Y. (2011) Profiling of hormones and related metabolites in seed dormancy and germination studies. Methods Mol. Biol. 773, 99–111. [DOI] [PubMed] [Google Scholar]
- Su, L.‐W. , Chang, S.H. , Li, M.‐Y. , Huang, H.‐Y. , Jane, W.‐N. and Yang, J.‐Y. (2013) Purification and biochemical characterization of Arabidopsis At‐NEET, an ancient iron–sulfur protein, reveals a conserved cleavage motif for subcellular localization. Plant Sci. 213, 46–54. [DOI] [PubMed] [Google Scholar]
- Taschner, P.E. , Van der Kuyl, A.C. , Neeleman, L. and Bol, J.F. (1991) Replication of an incomplete alfalfa mosaic virus genome in plants transformed with viral replicase genes. Virology, 181, 445–450. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D. , Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res. 11, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo‐Ortiz, G. , Huq, E. and Quail, P.H. (2003) The Arabidopsis basic/helix‐loop‐helix transcription factor family. Plant Cell, 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. (2010) ROS in biotic interactions. Physiol. Plant. 138, 414–429. [DOI] [PubMed] [Google Scholar]
- Uhrig, J.F. , Canto, T. , Marshall, D. and MacFarlane, S.A. (2004) Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol. 135, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, P.H. and Bujarski, J.J. (2015) Multiple functions of capsid proteins in (+) stranded RNA viruses during plant–virus interactions. Virus Res. 196, 140–149. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Quan, S. , Chang, H.S. , Cooper, B. , Estes, B. , Zhu, T. , Wang, X. and Hou, Y.M. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Yang, C. and Goodin, M. (2006) Global impact: elucidating plant responses to viral infection. Mol. Plant–Microbe Interact. 19, 1207–1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 List of primers and plasmids used in this study.
Fig. S1 Amino acid sequence alignments of ILR3 and NEET proteins of Arabidopsis and Nicotiana tabacum.
Fig. S2 Analysis of putative yeast two‐hybrid (Y2H) self‐activation by Alfalfa mosaic virus (AMV) coat protein (CP). AH109 yeast cells co‐transformed with the indicated plasmids were spotted onto minimal medium lacking leucine and tryptophan (–LW) or leucine, tryptophan, histidine and adenine (–LWHA). Colony growth was only detected with the pair pBD:CP and pAD:CP, confirming that AMV CP does not self‐activate yeast growth.


