Summary
The plant pathogen Candidatus Phytoplasma mali (P. mali) is the causative agent of apple proliferation, a disease of increasing importance in apple‐growing areas within Europe. Despite its economic importance, little is known about the molecular mechanisms of disease manifestation within apple trees. In this study, we identified two TCP (TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR) transcription factors of Malus x domestica as binding partners of the P. mali SAP11‐like effector ATP_00189. Phytohormone analyses revealed an effect of P. mali infection on jasmonates, salicylic acid and abscisic acid levels, showing that P. mali affects phytohormonal levels in apple trees, which is in line with the functions of the effector assumed from its binding to TCP transcription factors. To our knowledge, this is the first characterization of the molecular targets of a P. mali effector and thus provides the basis to better understand symptom development and disease progress during apple proliferation. As SAP11 homologues are found in several Phytoplasma species infecting a broad range of different plants, SAP11‐like proteins seem to be key players in phytoplasmal infection.
Keywords: apple proliferation, ATP_00189, effector protein, phytohormones, TCP, transcription factor, yeast two hybrid
Infection with the biotrophic bacterial pathogen Candidatus Phytoplasma mali (P. mali), the causative agent of apple proliferation (AP), can lead to massive yield losses and economic damage in apple production regions (Strauss, 2009). The province of South Tyrol/Alto Adige in northern Italy is the largest interconnected apple‐growing region in Europe and has suffered dramatically from apple proliferation outbreaks during the last decade (Berger, 2007; Mattedi et al., 2007). Infected apple trees (Malus x domestica) develop symptoms comprising witches’ brooms, stunting, foliar reddening and undersized, colourless and tasteless fruits (Kartte and Seemüller, 1988; Seemüller and Schneider, 2007). Phytoplasma exhibit a unique life cycle that involves a reproductive phase in a phloem‐feeding insect and subsequent transmission into the plant (Christensen et al., 2005). Inside the plant phloem, the bacteria replicate and can be re‐transmitted into the phloem‐sucking insect to complete their infectious life cycle and enable their dissemination to other host plants (Sugio et al., 2011b). Much progress has been made in unravelling the molecular basis of phytoplasma infection using the Ca. P. asteris strain aster yellow‐witches’ broom (AY‐WB), mainly by the identification and characterization of the bacterial effectors that play a role in disease manifestation and symptom development in Arabidopsis thaliana (Bai et al., 2009; Kartte and Seemüller, 1988; Lu et al., 2014a, 2014b; MacLean et al., 2011, 2014; Sugawara et al., 2013; Sugio et al., 2011a, b). Although the genome has been fully sequenced (Kube et al., 2008), no functional effector protein of P. mali has been described to date. Therefore, the molecular mechanisms underlying disease manifestation and symptom development in the natural host Malus x domestica remain elusive. A study performed with P. mali identified several genes expressed in Malus x domestica during infection, amongst others a gene that encodes the protein ATP_00189 (GenBank: CAP18376.1), a protein which shares homology to the AY‐WB effector SAP11 (Siewert et al., 2014; Sugio et al., 2011a). ATP_00189 contains an N‐terminal sequence‐variable mosaic (SVM) protein signal sequence (Pfam entry: PF12113) and shares 41% identity to SAP11.
In the present study, we addressed the following questions: (i) whether the SAP11 homologue ATP_00189 of P. mali is differentially expressed in spring and autumn in naturally infected apple trees; (ii) whether sequence variants of this potential effector protein occur within South Tyrol/Alto Adige; (iii) which proteins are targeted by ATP_00189; and (iv) whether infected apple trees show differential hormonal regulation in spring and autumn.
The function of SAP11 is mainly based on three domains: the signal peptide that mediates extra‐bacterial translocation of the protein into the surrounding environment; the nuclear localization sequence that targets the protein to the plant nucleus; and the TCP (TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR) binding domain that mediates binding to TCP transcription factors (TFs) (Sugio et al., 2014). Although these domains are important for the effector function of this protein, the amino acid sequences of these domains have been described to be poorly conserved between SAP11‐like proteins of different phytoplasma species (Sugio et al., 2014). A comparison of SAP11 from AY‐WB with ATP_00189 shows that these two proteins share 40% sequence identity. At amino acid positions within the SVM signal peptide and the TCP‐binding region that are not identical in both proteins, similar hydrophobic amino acid patterns are evident in ATP_00189 and SAP11 (Fig. S1, see Supporting Information). This might indicate a functional selection based on hydrophobic parts within stretches of the proteins, rather than a selection for exact amino acid motifs. Hydrophobicity‐mediated protein functions could, for example, involve membrane binding, protein folding, and polymerization and interaction with host targets. Rümpler et al. (2015) hypothesized that the binding function of MADS‐box TFs is determined by a characteristic hydrophobicity pattern, rather than a defined amino acid sequence in the keratin‐like domain (K‐domain) of these factors. Interestingly, this K‐domain is targeted by the phytoplasmal effector SAP54/PHYL1 which, itself, mimics and binds the K‐domain of the TF, and thus primes it for ubiquitin‐mediated proteosomal degradation (MacLean et al., 2011, 2014; Maejima et al., 2014; Rümpler et al., 2015). Phytoplasmas are genetically highly dynamic bacteria (Bai et al., 2006; Jarausch et al., 2000; Sugio and Hogenhout, 2012). Sequence analysis of different loci has revealed that P. mali genotypes from different sampling sites within South Tyrol/Alto Adige and within individual trees can be highly variable (Janik et al., 2015). To analyse whether ATP_00189 variants found in South Tyrol/Alto Adige resemble the protein sequence published in 2008 (Kube et al., 2008), DNA from symptomatic apple trees from 20 different orchards was purified using a method described elsewhere (Schlink and Reski, 2002). In all samples, infection with P. mali was confirmed by polymerase chain reaction (PCR) using fO1/rO1 primers (Lee et al., 2000) and by real‐time PCR using the probe AP (Mehle et al., 2013). Infection with the other 16SrX group phytoplasmas P. pyri and P. prunorum was ruled out by performing the same real‐time PCR with the respective probes (Mehle et al., 2013). The atp_00189 gene was amplified, subcloned and sequenced (Methods S1, see Supporting Information). The prevalent genetic sequence of atp_00189 in South Tyrol/Alto Adige (atp_00189_STAA; Accession: KM501063), which occurred in all trees and in about 91% of the tested clones, contained three single nucleotide polymorphisms (SNPs) compared with the sequence described by Kube et al. (2008). The atp_00189 sequence published by Kube et al. (2008) was only detected in one tree, in which the prevalent sequence type atp_00189_staa was also present. Two SNPs present at the 5′‐end of atp_00189_staa lead to amino acid exchanges in the signal peptide (Fig. S1), whereas the third SNP at the 3′‐end of the gene does not lead to translational differences, leaving the amino acid sequence of the mature, i.e. signal peptide lacking ATP_00189, protein unaffected. Taking the general genetic dynamics of phytoplasma into consideration (Bai et al., 2006; Jarausch et al., 2000; Sugio and Hogenhout, 2012), the conserved nature of ATP_00189 indicates an importance of this protein for the pathogen.
Figure 1.
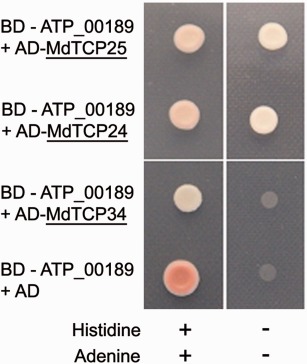
ATP_00189 binds class II TCP (TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTOR) transcription factors (TFs) MdTCP24 and MdTCP25. A yeast two‐hybrid (Y2H) screen was performed using the binding domain (BD)‐coupled ATP00189 expression plasmid (pLexA‐N‐ATP_00189) and an expression plasmid carrying the activation domain (AD) coupled to the full‐length MdTCP of Malus x domestica (pGAD‐HA‐ccdB constructs), identified in the Y2H screen. MdTCP34 was not identified as an interaction partner in the Y2H screen and serves as a negative control. Interaction between ATP_00189 and the respective TCP TF complements the auxotrophy for histidine and adenine. In the absence of interaction, co‐transformed yeast does not grow on adenine‐ and histidine‐depleted selection plates.
To determine which proteins are targeted by the SAP11 homologue of P. mali in apple trees under natural conditions, a cDNA library of the leaf transcriptome of Malus x domestica cv. ‘Golden Delicious’ was generated (see Methods S1) and a yeast two‐hybrid (Y2H) screen was performed with ATP_00189 as the bait. Sequence analyses of the positive clones in the Y2H screen revealed four different Malus x domestica binding partners of ATP_00189. Three of the four interactors found in this study are homologues of class II TCP TFs from A. thaliana: TCP4‐like (GI:657979223), which corresponds to Malus x domestica (Md) MdTCP25 (Xu et al., 2014); TCP13‐like (GI:658044279), which is a homologue of MdTCP24 (Xu et al., 2014); and an isoform of TCP18‐like (GI:657966084), similar to MdTCP16 (Xu et al., 2014). In addition to the interactions with Malus proteins containing TCP domains, interactions were found between ATP_00189 and a Malus x domestica library clone that shares partial identity with 60 amino acids of the C‐terminal part of a putative chlorophyll(ide) b reductase NYC1, chloroplastic‐like isoform X2 from Glycine max (GI:571465492). A list of all interacting cDNA fragments identified in the Y2H screen can be found below under ‘Accession numbers’.
The full‐length genes of MdTCP24 and MdTCP25 (plant transcription factor database http://planttfdb.cbi.pku.edu.cn/ id MDP0000692406 and id MDP0000442611) were de novo amplified from Malus x domestica DNA. The NYC1 and MdTCP16 genes contain introns and might have different splice variants. Thus, the identified fragments of these genes were amplified from cDNA. Using the de novo subcloned prey vectors, interaction could be shown for ATP_00189 and the full‐length MdTCP24 and MdTCP25 in co‐transformed yeast (Fig. 1). The expression of ATP_00189 was confirmed by immunoblot analysis using an antibody against the LexA‐tag, which was coupled to ATP_00189 (Fig. S2, see Supporting Information). In planta interactions of ATP_00189 with MdTCP24 and MdTCP25 were verified by bimolecular fluorescence complementation (BiFC) in Nicotiana benthamiana mesophyll protoplasts (Fig. 2). Mesophyll protoplasts were prepared as described by Sheen (2002). ATP_00189 and the MdTCP encoding cDNAs were subcloned into the BiFC vectors pE‐SPYNE and pE‐SPYCE, respectively (Walter et al., 2004). The BiFC vectors contain the information for the N‐terminal (pE‐SPYNE) or C‐terminal (pE‐SPYCE) halves of yellow fluorescent protein (YFP). An interaction of the proteins leads to the reconstitution of YFP from both halves and results in YFP fluorescence that can be visualized via fluorescence microscopy. In the analyses performed by confocal laser scanning microscopy, 19%–24% of the randomly counted protoplasts showed a strong YFP signal predominantly in the nucleus (Fig. 2 and Table S1, see Supporting Information). The putative interaction of NYC1 and the MdTCP16 isoform fragments, indicated by the initial Y2H library screen, could not be confirmed, in either ATP_00189 co‐transformed yeast carrying the cDNA of these genes or BiFC analyses. As a negative control, a member of the Malus x domestica class I TCP TF family (MdTCP34) was used as a proxy for this TF subclass (see below), for which no interactor was found in the Y2H library screen. Accordingly, no interaction between MdTCP34 and ATP_00189 was observed in the co‐transformed yeast (Fig. 1). In BiFC experiments, only 2% of the counted protoplasts showed very weak signals of ATP_00189 interaction with MdTCP34 (Table S1), whereas the majority of protoplasts did not exhibit any YFP fluorescence (Fig. 2).
Figure 2.
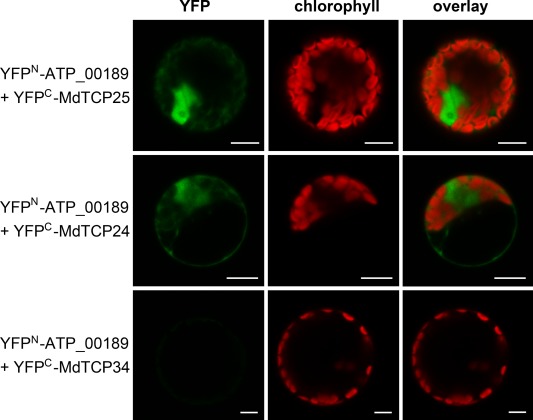
In planta interaction of ATP_00189 with MdTCP24 and MdTCP25. Mesophyll protoplasts of Nicotiana benthamiana were co‐transformed with pE‐SPYNE‐ATP_00189 and pE‐SPYCE‐MdTCP24, ‐MdTCP25 or ‐MdTCP34. Interaction of ATP_00189 with MdTCP24 and MdTCP25 leads to reconstitution of yellow fluorescent protein (YFP) in the protoplast, indicated by green fluorescence (top and middle panel), whereas an interaction with MdTCP34 was not detected (bottom panel). Bars represent 10 µm.
In a genome‐wide screen of Malus x domestica, 52 TCP genes were identified (Xu et al., 2014). These TFs share the so‐called TCP domain which mediates nuclear localization, DNA binding and protein–protein interaction (Cubas et al., 1999; Kosugi and Ohashi, 2002). TCP proteins can be divided into two classes based on sequence similarities (Cubas et al., 1999; Kosugi and Ohashi, 2002), and are involved in the regulation of diverse plant cellular processes, such as branching or floral and leaf development (Martin‐Trillo and Cubas, 2010). In analogy with the identified A. thaliana interaction partners of SAP11 (Sugio et al., 2011a), ATP_00189 binds Malus x domestica homologues of two TCP protein members of the CINCINNATA (CIN)‐related TCP group II, namely MdTCP25 (a homologue of A. thaliana TCP4) and MdTCP24 (a homologue of A. thaliana TCP13/PTF1). A TCP4 homologue of Malus x domestica was shown to bind the FLOWERING LOCUS T (MdFT1) involved in apple flowering (Kotoda et al., 2010; Mimida et al., 2011), indicating a function of TCP4 apple homologues in developmental processes, such as fruit ripening, where the expression of MdTCP25 has been shown (Xu et al., 2014). TCP4 of A. thaliana has been shown to be involved in leaf development, thereby negatively regulating cellular mitotic processes, leaf growth and abscisic acid (ABA) responses in A. thaliana (Danisman et al., 2012; Sarvepalli and Nath, 2011a, 2011b). Furthermore, it was shown that TCP4 of Arabidopsis regulates jasmonic acid (JA) biosynthesis by activating the expression of LOX2 (lipoxygenase 2) (Danisman et al., 2012; Schommer et al., 2008), an enzyme that catalyses one of the first steps of JA biosynthesis in A. thaliana (Vick and Zimmerman, 1983). SAP11 binds and destabilizes TCP4 and other CIN‐TCP proteins, and thus interferes with LOX2‐mediated JA accumulation in A. thaliana. This reduction in JA levels, in turn, enhances oviposition of the leafhopper Macrosteles quadrilineatus on the infected plant, and thus supports bacterial dissemination to other host plants (Sugio et al., 2011a). In our study, we showed that ATP_00189 binds MdTCP25 of Malus x domestica and might thus be involved in the down‐regulation of JA biosynthesis, leading to diminished JA responses (Sugio et al., 2011a, 2014) and increased ABA levels and responses (Sarvepalli and Nath, 2011b). Our results further demonstrated the binding of ATP_00189 to MdTCP24, a Malus x domestica homologue of TCP13 from A. thaliana, another CIN‐TCP group II protein, also known as PTF1. This TF is nuclear‐encoded, but located in plastids (Baba et al., 2001), thus most likely playing a role in plastid gene expression and regulation. PTF1 is involved in phosphate tolerance, carbon metabolism and root growth in maize (Li et al., 2011) and in ABA‐regulated transcriptional responses in the chloroplast in Arabidopsis (Yamburenko et al., 2015). SAP11 has been shown to degrade TCP13/PTF1 (Sugio et al., 2011a) and affects phosphate metabolism, anthocyanin accumulation and root architecture in A. thaliana (Lu et al., 2014b). Commonly observed symptoms of apple proliferation, such as reddening and altered root growth (Kunze, 1979), might thus be induced by SAP11‐mediated TCP13/PTF1 degradation in P. mali‐infected plants.
As AY‐WB in A. thaliana affects JA biosynthesis via SAP11, we were interested in whether hormonal changes occur in Malus x domestica during apple proliferation infection. To reveal which hormonal pathways are affected by P. mali, we analysed the phytohormonal levels at two different time points. Pools of leaves from symptomatic and healthy apple trees of the variety Malus x domestica cv. ‘Golden Delicious’ were harvested in May, when phytoplasma could not be detected in the canopy, and in October, when the bacteria have colonized the leaves and express atp_00189 (Fig. 3, top panel I). Phytohormone quantification was performed using liquid chromatography‐mass spectrometry analysis (Vadassery et al., 2012). In infected and control trees, levels of cis−12‐oxo‐phytodienoic acid (OPDA), (+)−7‐iso‐jasmonoyl‐l‐isoleucine (JA‐Ile), salicylic acid (SA) and ABA were determined (Fig. 3, bottom panel II). In May, OPDA, Ja‐Ile and SA were significantly higher in AP‐infected trees than in controls (Fig. 3, bottom panel II, a–c). JA and SA levels in infected trees were not increased any further in October; indeed, they tended to be below the levels of the non‐infected control (Fig. 3, bottom panel II, b, c). From May to October, levels of SA, OPDA and JA‐Ile in the leaves of healthy controls increased, whereas the levels in P. mali‐infected apple trees did not change significantly (Fig. 3, bottom panel II, a–c). Similarly, Musetti et al. (2013) detected a reduced expression of JA marker genes in infected trees in the autumn. From May until October, ABA levels decreased in P. mali‐infected and control trees, but the amount of ABA in infected trees remained significantly higher in October compared with the control group (Fig. 3, bottom panel II, d). These results support several findings of Zimmermann et al. (2015), who showed an increased ABA accumulation for one time point after infection, which we also observed in samples harvested in October, and increased SA levels that we detected in material collected in May. In the study of Zimmermann et al., however, no effect on JA‐Ile and OPDA was observed. This could be because the authors chose another sampling time point. Our results suggest that the regular, seasonal increase of JA‐Ile, OPDA and SA is impeded by phytoplasmal infection, either by direct interference with their synthesis and/or indirectly through phytohormonal crosstalk, as antagonistic actions between ABA, SA and JA have been described (Anderson et al., 2004; Mohr and Cahill, 2007; Yasuda et al., 2008). Phytoplasma mali disappears from the apple tree crown during the winter and recolonizes the aerial parts of the tree beginning in spring (Baric et al., 2011; Loi et al., 2002; Pedrazzoli et al., 2008; Seemüller et al., 1984). Hence, the bacterial titre in the crown is low until June and increases throughout the growing season. On the one hand, this might explain the induction of SA and JA in spring when the first phytoplasmas recolonize the aerial parts and are recognized by the plant. On the other, the increasing bacterial concentration may explain their growing impact on leaf phytohormone levels in autumn. The observed hormonal changes in infected apple trees indicate a phytoplasma‐mediated effect on JA, ABA and SA signalling. It remains to be clarified whether altered JA levels and action have a similar effect on the P. mali leafhopper vectors Cacopsylla picta or C. melanoneura, as has been shown for oviposition or feeding of the leafhoppers Macrosteles quadrilineatus and Empoasca in the Arabidopsis and tobacco systems, respectively (Kallenbach et al., 2012; Sugio et al., 2011a).
Figure 3.
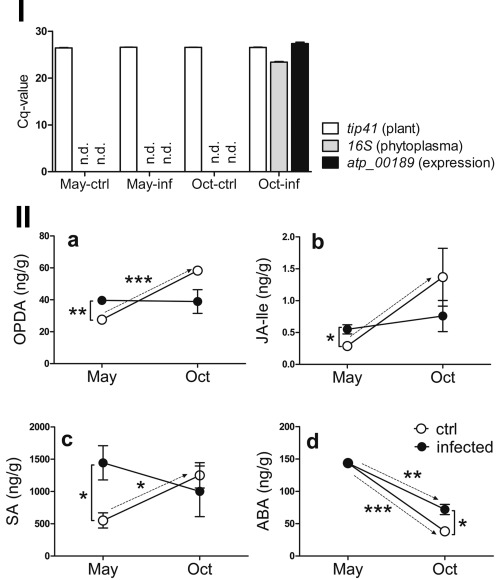
Levels of (+)−7‐iso‐jasmonoyl‐l‐isoleucine (JA‐Ile), cis‐12‐oxo‐phytodienoic acid (OPDA), salicylic acid (SA) and abscisic acid (ABA) in leaves of apple trees naturally infected with Phytoplasma mali. In infected apple trees, the presence of P. mali varies from May to October: In the top panel I, P. mali‐specific 16S and expression of atp_00189 were determined using quantitative polymerase chain reaction (PCR) with cDNA from leaves harvested in May and October (same pools as described below). As a control for equal cDNA amounts, plant‐specific tip41 was determined in parallel. Only in October could P. mali be detected and atp_00189 expression was confirmed when bacteria were present. Each pool was tested in technical triplicates. The mean Cq value of three independent pools for each condition is depicted + standard error of the mean (SEM). Results under the detection limit are designated as "not detected" (n.d.). OPDA, JA‐Ile and SA are elevated in leaves of P. mali‐infected trees in May when phytoplasma are not present in the canopy (top panel I and bottom panel II, a–c). The physiological increase in these hormones from May to October is inhibited in infected trees. Levels of ABA decrease from May to October in both control and infected trees (bottom panel II, d). In October, ABA is elevated in infected trees in comparison with the controls (bottom panel II, d). In panels I and II, for each time point, three pools of leaves from control or P. mali‐infected trees (6–11 trees/pool) were tested. The mean of three pools (n = 3) is shown ± SEM. Broken arrows in panel II indicate the trend of regulation from May to October for a simplified comparison between hormonal regulation in non‐infected vs. infected trees during these months. Differences between pools were determined with Student's t‐test. Significant differences are indicated by: *P < 0.05; **P < 0.01; ***P < 0.001.
Interestingly, increased LOX activity in P. mali‐infected apple tree leaves correlates with the recovery phenomenon, characterized by a type of resilience against P. mali in the tree canopy (Carraro et al., 2004; Musetti et al., 2013; Patui et al., 2013; Seemüller et al., 1984; Schmid, 1975); this emphasizes the importance of decreased JA levels for the success of phytoplasma infection. However, biotrophic pathogens are generally sensitive to SA‐mediated responses (Glazebrook, 2005; Thomma et al., 2001). SA induction can lead to systemic acquired resistance, which results in a broad‐spectrum resistance against pathogens and thus plays an important role in plant immunity (Ryals et al., 1996). The latest results of a study with SAP11‐transgenic A. thaliana plants indicate a role of SAP11 in the down‐regulation of SA responses (Lu et al., 2014b). As SA accumulation in P. mali‐infected apple trees is reduced, as shown in this study, it is plausible to consider a similar function for ATP_00189 as well.
In our study, we focused on the characterization of the Malus x domestica binding partners of P. mali ATP_00189, a homologue of the well‐characterized AY‐WB effector SAP11, to elucidate the molecular processes underlying the disease progress of AP. Interestingly, we found that ATP_00189 shares targets with SAP11, supporting the idea that the principles of SAP11‐mediated TCP factor degradation found in A. thaliana might also be valid during P. mali infection in apple trees. Infected apple trees show altered hormonal levels compared with controls, indicating an effect of P. mali on the hormone system of Malus x domestica. Integrating our data with results achieved in AY‐WB SAP11 studies in A. thaliana, we hypothesize that, during infection of its natural host, the apple tree, ATP_00189 of P. mali has similar functions to SAP11 revealed in its host, the model plant A. thaliana. Given the fact that SAP11 homologues are found in many different phytoplasma species and that the motifs of this protein seem to be conserved throughout different phytoplasma species, it is likely that SAP11 and its relatives play a pivotal role in phytoplasmal infection. Targeting this protein and its derivatives might be a suitable approach for future infection prevention or therapy of economically important phytoplasmal diseases.
Accession Numbers
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Comparison of amino acid sequences of SAP11 from aster yellow‐witches’ broom (AY‐WB) phytoplasma (WP_011412651.1) and ATP_00189 (CAP18376.1). Sequences of the SAP11 protein from AY‐WB, the SAP11‐like protein (AP) from apple proliferation phytoplasma as described by Kube et al. (2008) and the SAP11‐like protein AP_STAA (the main variant found in northern Italy, South Tyrol/Alto Adige) share stretches of identical amino acid motifs or amino acids of similar hydrophobicity. This indicates similar functions of the proteins. The colour and height of the bars in the respective panels indicate the relative levels of hydrophobicity of adjacent amino acids within the sequence (Hydrophobicity). Sequence identities of AP and AP_STAA with the AY‐WB reference protein are highlighted by green boxes above the sequences. Differences in the signal peptide between AP and AP_STAA are indicated by red letters in a box. Analysis was performed using Geneious software (http://www.geneious.com; Kearse et al., 2012).
Fig. S2 Expression of the ATP_00189 protein in co‐transformed Saccharomyces cerevisiae NMY51. The expression of ATP_00189 was analysed with an antibody against the LexA‐tag [i.e. the yeast two‐hybrid (Y2H) transcriptional activator binding domain] fused to the ATP_00189 protein. Cell lysates from: non‐transformed NMY51 (1), NMY51 co‐transformed with the empty bait (only expressing the LexA‐tag) vector pLexA and the empty prey vector pGAD‐HA‐ccdB (2), co‐transformed with pLexA‐ATP_00189 + pGAD‐HA‐ccdB‐MdTCP25 (3) or co‐transformed with pLexA‐ATP_00189 + pGAD‐HA‐ccdB‐MdTCP24 (4). The left lane contains the protein marker (M) with the corresponding fragment sizes. LexA has a size of ∼24 kDa and ATP_00189 + LexA has a size of ∼35 kDa.
Table S1 In planta interaction of ATP_00189 with MdTCP24, MdTCP25 or MdTCP34. Mesophyll protoplasts of Nicotiana benthamiana were co‐transfected with the binary bimolecular fluorescence complementation (BiFC) vectors pE‐SPYNE and pE‐SPYCE harbouring ATP_00189 [fused to the N‐terminal subunit of yellow fluorescent protein (YFP)] and MdTCP24, MdTCP25 or MdTCP34 (each fused to the C‐terminal subunit of YFP), respectively. Interaction leads to the generation of YFP fluorescence. Transformation with pE‐SPYNE‐ATP_00189 alone served as a negative control. Protoplasts were randomly counted and the number and percentage of YFP‐positive protoplasts for each co‐transfection were determined.
Methods S1 Supplementary Material and Methods Section. Several methods mentioned a legend is thus not intended.
Acknowledgements
We thank the South Tyrolean Extension Service for Fruit and Wine Growing for samples from South Tyrol, Sabine Oettl and Andrea Lehr for sample collection and preparation, Silvia Schmidt and Thomas Letschka for participation in the establishment and selection of trees for phytohormone analyses, and Christine Kerschbamer, Michael Reichelt and Mirelle Borges Dias Schnetzer for technical support. This work was performed as part of the APPL2.0 project and was partially funded by the Autonomous Province of Bozen/Bolzano, Italy and the South Tyrolean Apple Consortium.
[The copyright line for this article was changed on October 13, 2017 after original online publication]
References
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , et al (2004) Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, K. , Nakano, T. , Yamagishi, K. and Yoshida, S. (2001) Involvement of a nuclear‐encoded basic helix‐loop‐helix protein in transcription of the light‐responsive promoter of psbD1. Plant Physiol. 125, 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Zhang, J. , Ewing, A. , Miller, S.A. , Jancso Radek, A. , Shevchenko, D.‐V. , Tsukerman, K., Walunas, T., Lapidus, A., Campbell, J.‐W., Hogenhout, S.‐A. (2006) Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. In: J. Bacteriol. 188, 3682–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, X. , Correa, V.R. , Toruño, T.Y. , Ammar, E.‐D. , Kamoun, S. and Hogenhout, S.A. (2009) AY‐WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant–Microbe Interact. 22, 18–30. [DOI] [PubMed] [Google Scholar]
- Baric, S. , Berger, J. , Cainelli, C. , Kerschbamer, C. , Letschka, T. and Dalla Via, J. (2011) Seasonal colonisation of apple trees by ‘Candidatus Phytoplasma mali’ revealed by a new quantitative TaqMan real‐time PCR approach. Eur. J. Plant Pathol. 129, 455–467. [Google Scholar]
- Berger, H. (2007): Apple proliferation in South Tyrol. In: Apfel aktuell 21 (2), p. 2 Online available at http://www.vog.it/en/news/archive/apple-proliferation-in-south-tyrol.html. [Google Scholar]
- Carraro, L. , Ermacora, P. , Loi, N. and Osler, R. (2004) The recovery phenomenon in apple proliferation‐infected apple trees. J. Plant Pathol. 86, 141–146. [Google Scholar]
- Christensen, N.M. , Axelsen, K.B. , Nicolaisen, M. and Schulz, A. (2005) Phytoplasmas and their interactions with hosts. Trends Plant Sci. 10, 526–535. [DOI] [PubMed] [Google Scholar]
- Cubas, P. , Lauter, N. , Doebley, J. and Coen, E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- Danisman, S. , van der Wal, F. , Dhondt, S. , Waites, R. , de Folter, S. , Bimbo, A. , et al (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Janik, K. , Oettl, S. and Schlink, K. (2015) Local distribution of ‘Candidatus Phytoplasma mali’ genetic variants in South Tyrol (Italy) based on a MLST study. Phytopathogenic Mollicutes, 5, S29. [Google Scholar]
- Jarausch, W. , Saillard, C. , Helliot, B. , Garnier, M. and Dosba, F. (2000) Genetic variability of apple proliferation phytoplasmas as determined by PCR‐RFLP and sequencing of a non‐ribosomal fragment. Mol. Cell Probes, 14, 17–24. [DOI] [PubMed] [Google Scholar]
- Kallenbach, M. , Bonaventure, G. , Gilardoni, P.A. , Wissgott, A. and Baldwin, I.T. (2012) Empoasca leafhoppers attack wild tobacco plants in a jasmonate‐dependent manner and identify jasmonate mutants in natural populations. Proc . Natl. Acad. Sci. USA, 109, 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartte, S. and Seemüller, E. (1988) Variable response within the genus Malus to the apple proliferation disease. J. Plant Dis. Protect. 95, 25–34. [Google Scholar]
- Kosugi, S. and Ohashi, Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. [DOI] [PubMed] [Google Scholar]
- Kotoda, N. , Hayashi, H. , Suzuki, M. , Igarashi, M. , Hatsuyama, Y. , Kidou, S. , et al (2010) Molecular characterization of FLOWERING LOCUS T‐like genes of apple (Malus x domestica Borkh.). Plant Cell Physiol. 51, 561–575. [DOI] [PubMed] [Google Scholar]
- Kube, M. , Schneider, B. , Kuhl, H. , Dandekar, T. , Heitmann, K. , Migdoll, A.M. , et al (2008) The linear chromosome of the plant‐pathogenic mycoplasma 'Candidatus Phytoplasma Mali'. BMC Genomics, 9, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, L. (1979) Wurzelschäden durch Triebsuchtbefall an Apfelbäumen. Mitt. Biol. Bundesanstalt, 191, p. 204–205. [Google Scholar]
- Lee, I.M. , Davis, R.E. and Gundersen‐Rindal, D.E. (2000) Phytoplasma: phytopathogenic mollicutes. Annu. Rev. Microbiol. 54, 221–255. [DOI] [PubMed] [Google Scholar]
- Li, Z. , Gao, Q. , Liu, Y. , He, C. , Zhang, X. and Zhang, J. (2011) Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta, 233, 1129–1143. [DOI] [PubMed] [Google Scholar]
- Loi, N. , Ermacora, P. , Carraro, L. , Osler, R. and Chen, T. A. (2002) Production of monoclonal antibodies against apple proliferation phytoplasma and their use in serological detection. Eur. J. Plant Pathol. 108, 81–86. [Google Scholar]
- Lu, Y.‐T. , Cheng, K.‐T. , Jiang, S.‐Y. and Yang, J.‐Y. (2014a) Post‐translational cleavage and self‐interaction of the phytoplasma effector SAP11. Plant Signal. Behav. 9, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y.‐T. , Li, M.‐Y. , Cheng, K.‐T. , Tan, C.M. , Su, L.‐W. , Lin, W.‐Y. , et al (2014b) Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 164, 1456–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, A.M. , Sugio, A. , Makarova, O.V. , Findlay, K.C. , Grieve, V.M. , Tóth, R. , et al (2011) Phytoplasma effector SAP54 induces indeterminate leaf‐like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean, A.M. , Orlovskis, Z. , Kowitwanich, K. , Zdziarska, A.M. , Angenent, G.C. , Immink, R.G.H. and Hogenhout, S.A. (2014) Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS‐box proteins and promotes insect colonization in a RAD23‐dependent manner. PLoS Biol. 12, e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima, K. , Iwai, R. , Himeno, M. , Komatsu, K. , Kitazawa, Y. , Fujita, N. , et al (2014) Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J. 78, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Trillo, M. and Cubas, P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Mattedi L., Forno F. and Varner M. (eds) (2007) Scopazzi del Melo: Conoscenze ed Osservazioni in Campo. San Michele All'Adige: Fondazione Edmund Mach. [Google Scholar]
- Mehle, N. , Nikolić, P. , Gruden, K. , Ravnikar, M. and Dermastia, M. (2013) Real‐time PCR for specific detection of three phytoplasmas from the apple proliferation group. Methods Mol. Biol. 938, 269–281. [DOI] [PubMed] [Google Scholar]
- Mimida, N. , Kidou, S.‐I. , Iwanami, H. , Moriya, S. , Abe, K. , Voogd, C. , et al (2011) Apple FLOWERING LOCUS T proteins interact with transcription factors implicated in cell growth and organ development. Tree Physiol. 31, 555–566. [DOI] [PubMed] [Google Scholar]
- Mohr, P.G. and Cahill, D.M. (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes in Arabidopsis infected with Pseudomonas syringae pv. tomato . Funct. Integr. Genomics, 7, 181–191. [DOI] [PubMed] [Google Scholar]
- Musetti, R. , Farhan, K. , de Marco, F. , Polizzotto, R. , Paolacci, A. , Ciaffi, M. , et al (2013) Differentially‐regulated defence genes in Malus domestica during phytoplasma infection and recovery. Eur. J. Plant Pathol. 136, 13–19. [Google Scholar]
- Patui, S. , Bertolini, A. , Clincon, L. , Ermacora, P. , Braidot, E. , Vianello, A. and Zancani, M. (2013) Involvement of plasma membrane peroxidases and oxylipin pathway in the recovery from phytoplasma disease in apple (Malus domestica). Physiol Plant. 148, 200–213. [DOI] [PubMed] [Google Scholar]
- Pedrazzoli, F. , Cicotti, A.M. , Bianchedi, P.L. , Salvadori, A. and Zorer, R. (2008) Seasonal colonisation behaviour of Candidatus Phytoplasma mali in apple trees in Trentino. Acta Hortic. 781, 483–489. [Google Scholar]
- Rümpler, F. , Gramzow, L. , Theißen, G. and Melzer, R. (2015) Did convergent protein evolution enable phytoplasmas to generate ‘zombie plants’? Trends Plant Sci. 20, 1–9. [DOI] [PubMed] [Google Scholar]
- Ryals, J.A. , Neuenschwander, U.H. , Willits, M.G. , Molina, A. , Steiner, H.Y. and Hunt, M.D. (1996) Systemic acquired resistance. Plant Cell, 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli, K. and Nath, U. (2011a) Interaction of TCP4‐mediated growth module with phytohormones. Plant Signal. Behav. 6, 1440–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli, K. and Nath, U. (2011b) Hyper‐activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67, 595–607. [DOI] [PubMed] [Google Scholar]
- Schlink, K. and Reski, R. (2002) Preparing high‐quality DNA from moss (Physcomitrella patens). Plant Mol. Biol. Rep. 20, 423a–423f. [Google Scholar]
- Schmid, G. (1975) Prolonged observations on spread and behaviour of proliferation disease in apple orchards. Acta Hortic. 44, 183–192. [Google Scholar]
- Schommer, C. , Palatnik, J.F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E.E. , et al (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemüller, E. and Schneider, B. (2007) Differences in virulence and genomic features of strains of ‘Candidatus Phytoplasma mali’, the apple proliferation agent. Phytopathology, 97, 964–970. [DOI] [PubMed] [Google Scholar]
- Seemüller, E. , Kunze, L. and Schaper, U. (1984) Colonization behaviour of MLO, and symptom expression of proliferation‐diseased apple trees and decline‐diseased pear trees over a period of several years In: Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz, 91, 371–382. [Google Scholar]
- Sheen, J. (2002). A transient expression assay using Arabidopsis mesophyll protoplasts. Harvard Medical School, Department of Molecular Biology. Boston, MA (USA). Available at: http://genetics.mgh.harvard.edu/sheenweb/. last update 22nd March 2004.
- Siewert, C. , Luge, T. , Duduk, B. , Seemüller, E. , Büttner, C. , Sauer, S. and Kube, M. (2014) Analysis of expressed genes of the bacterium 'Candidatus phytoplasma Mali' highlights key features of virulence and metabolism. PLoS One, 9, e94391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, E. (2009) Microbiology. Phytoplasma research begins to bloom. Science, 325, 388–390. [DOI] [PubMed] [Google Scholar]
- Sugawara, K. , Honma, Y. , Komatsu, K. , Himeno, M. , Oshima, K. and Namba, S. (2013) The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant. Physiol. 162, 2005–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. and Hogenhout, S.A. (2012) The genome biology of phytoplasma: modulators of plants and insects. Curr. Opin. Microbiol. 15, 247–254. [DOI] [PubMed] [Google Scholar]
- Sugio, A. , Kingdom, H.N. , MacLean, A.M. , Grieve, V.M. and Hogenhout, S.A. (2011a) Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA, 108, E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio, A. , MacLean, A.M. , Kingdom, H.N. , Grieve, V.M. , Manimekalai, R. and Hogenhout, S.A. (2011b) Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu. Rev. Phytopathol. 49, 175–195. [DOI] [PubMed] [Google Scholar]
- Sugio, A. , MacLean, A.M. and Hogenhout, S.A. (2014) The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN‐TCP binding and destabilization. New Phytol. 202, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P. , Penninckx, I.A. , Broekaert, W.F. and Cammue, B.P. (2001) The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Vadassery, J. , Reichelt, M. , Hause, B. , Gershenzon, J. , Boland, W. and Mithofer, A. (2012) CML42‐mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick, B.A. and Zimmerman, D.C. (1983) The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem. Biophys. Res. Commun. 111, 470–477. [DOI] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. , et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Sun, P. , Jia, F. , Lu, L. , Li, Y. , Zhang, S. and Huang, J. (2014) Genomewide analysis of TCP transcription factor gene family in Malus domestica . J. Genet. 93, 733–746. [DOI] [PubMed] [Google Scholar]
- Yamburenko, M.V. , Zubo, Y.O. and Börner, T. (2015) Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C‐dependent activation of nuclear genes: repression by guanosine‐30‐50‐bisdiphosphate and activation by sigma factor 5. Plant J. 82, 1030–1041. [DOI] [PubMed] [Google Scholar]
- Yasuda, M. , Ishikawa, A. , Jikumaru, Y. , Seki, M. , Umezawa, T. , Asami, T. , et al (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid‐mediated abiotic stress response in Arabidopsis. Plant Cell, 20, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, M.R. , Schneider, B. , Mithöfer, A. , Reichelt, M. , Seemüller, E. and Furch, C.U. (2015) Implications of Candidatus Phytoplasma mali infection on phloem function of apple trees. J. Endocytobiosis Cell Res. 26, 67–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Comparison of amino acid sequences of SAP11 from aster yellow‐witches’ broom (AY‐WB) phytoplasma (WP_011412651.1) and ATP_00189 (CAP18376.1). Sequences of the SAP11 protein from AY‐WB, the SAP11‐like protein (AP) from apple proliferation phytoplasma as described by Kube et al. (2008) and the SAP11‐like protein AP_STAA (the main variant found in northern Italy, South Tyrol/Alto Adige) share stretches of identical amino acid motifs or amino acids of similar hydrophobicity. This indicates similar functions of the proteins. The colour and height of the bars in the respective panels indicate the relative levels of hydrophobicity of adjacent amino acids within the sequence (Hydrophobicity). Sequence identities of AP and AP_STAA with the AY‐WB reference protein are highlighted by green boxes above the sequences. Differences in the signal peptide between AP and AP_STAA are indicated by red letters in a box. Analysis was performed using Geneious software (http://www.geneious.com; Kearse et al., 2012).
Fig. S2 Expression of the ATP_00189 protein in co‐transformed Saccharomyces cerevisiae NMY51. The expression of ATP_00189 was analysed with an antibody against the LexA‐tag [i.e. the yeast two‐hybrid (Y2H) transcriptional activator binding domain] fused to the ATP_00189 protein. Cell lysates from: non‐transformed NMY51 (1), NMY51 co‐transformed with the empty bait (only expressing the LexA‐tag) vector pLexA and the empty prey vector pGAD‐HA‐ccdB (2), co‐transformed with pLexA‐ATP_00189 + pGAD‐HA‐ccdB‐MdTCP25 (3) or co‐transformed with pLexA‐ATP_00189 + pGAD‐HA‐ccdB‐MdTCP24 (4). The left lane contains the protein marker (M) with the corresponding fragment sizes. LexA has a size of ∼24 kDa and ATP_00189 + LexA has a size of ∼35 kDa.
Table S1 In planta interaction of ATP_00189 with MdTCP24, MdTCP25 or MdTCP34. Mesophyll protoplasts of Nicotiana benthamiana were co‐transfected with the binary bimolecular fluorescence complementation (BiFC) vectors pE‐SPYNE and pE‐SPYCE harbouring ATP_00189 [fused to the N‐terminal subunit of yellow fluorescent protein (YFP)] and MdTCP24, MdTCP25 or MdTCP34 (each fused to the C‐terminal subunit of YFP), respectively. Interaction leads to the generation of YFP fluorescence. Transformation with pE‐SPYNE‐ATP_00189 alone served as a negative control. Protoplasts were randomly counted and the number and percentage of YFP‐positive protoplasts for each co‐transfection were determined.
Methods S1 Supplementary Material and Methods Section. Several methods mentioned a legend is thus not intended.


