Summary
Although the lifestyles and infection strategies of plant pathogens are diverse, a prevailing feature is the use of an arsenal of secreted proteins, known as effectors, which aid in microbial infection. In the case of eukaryotic filamentous pathogens, such as fungi and oomycetes, effector proteins are typically dissimilar, at the protein sequence level, to known protein families and functional domains. Consequently, we currently have a limited understanding of how fungal and oomycete effectors promote disease. Protein biochemistry and structural biology are two methods that can contribute greatly to the understanding of protein function. Both techniques are dependent on obtaining proteins that are pure and functional, and generally require the use of heterologous recombinant protein expression systems. Here, we present a general scheme and methodology for the production and characterization of small cysteine‐rich (SCR) effectors utilizing Escherichia coli expression systems. Using this approach, we successfully produced cysteine‐rich effectors derived from the biotrophic fungal pathogen Melampsora lini and the necrotrophic fungal pathogen Parastagonospora nodorum. Access to functional recombinant proteins facilitated crystallization and functional experiments. These results are discussed in the context of a general workflow that may serve as a template for others interested in understanding the function of SCR effector(s) from their plant pathogen(s) of interest.
Keywords: effector proteins, plant disease, protein biochemistry, protein expression, small cysteine‐rich proteins, structural biology
Introduction
During infection, filamentous plant pathogens, such as fungi and oomycetes, secrete proteinaceous molecules into the extracellular interface between the plant and pathogen (Oliveira‐Garcia and Valent, 2015; Tan et al., 2015). Known as effectors, these proteins either reside and function in the apoplast or are translocated into the plant cell. Collectively, effectors facilitate the infection and colonization of the host; however, they can also be recognized by plant disease resistance receptors, resulting in plant immunity (Dodds and Rathjen, 2010). The basis by which effectors facilitate disease is, in general, poorly understood.
Advances in sequencing technologies and annotation software have precipitated an explosion in the availability of sequenced genomes from plant pathogens, and candidate effector identification techniques have enabled the classification of large numbers of candidate secreted effector proteins (CSEPs) (Cantu et al., 2011, 2013; Duplessis et al., 2011a, 2011b; Fernández et al., 1997; Fernandez et al., 2012; Garnica et al., 2013, 2014; Hacquard et al., 2012; Hane et al., 2007; Nemri et al., 2014; Saunders et al., 2012). Features used to identify CSEPs include the presence of a recognizable signal peptide to facilitate their secretion from the pathogen and infection‐specific expression. Certain conserved motifs, such as RxLR and F/YxC in oomycetes and powdery mildew fungi, respectively, are also useful for particular pathogen classes (Bozkurt et al., 2012; Saunders et al., 2012; Sperschneider et al., 2015). In general, CSEPs have low sequence similarity to known proteins, domains and motifs, preventing functional inferences based on sequence alone. In addition, there often exists considerable diversity of candidate effector repertoires at the genus and species level (Nemri et al., 2014; Zheng et al., 2013). Taken together, although genetic classification and studies of plant pathogens have enhanced our global understanding of their infection strategies, it is often difficult to draw significant conclusions with regard to individual effector function from sequence alone.
Despite the diverse nature of CSEPs, small cysteine‐rich (SCR) proteins are typically highly represented in the complement of predicted effectors from filamentous pathogens (Saunders et al., 2012). For example, genomic analysis of Melampsora larici‐populina, the causative agent of poplar rust, revealed that 63% of the CSEPs contained more than four cysteine residues (Duplessis et al., 2011a). The cysteine residues of SCR effectors are often implicated in the formation of intramolecular disulfide bridges, and are thought to assist protein stability in effectors that function outside the plant cell (Saunders et al., 2012). For example, the Cladosporium fulvum SCR effector Avr2 contains four disulfide bonds, which are required for its inhibitory function against the tomato apoplast cysteine protease Rcr3 (Rooney et al., 2005). However, SCR effectors are also known to enter and function within the plant cell. For example, the Pyrenophora tritici‐repentis effector ToxA contains a disulfide‐bonded cysteine pair (Sarma et al., 2005) and is internalized into mesophyll cells in sensitive wheat lines (Manning and Ciuffetti, 2005). Similarly, the avirulence gene AvrLm4‐7 from Leptosphaeria maculans encodes a protein with four disulfide‐bonded cysteine pairs that can translocate into plant cells, where it is believed to interact with an unidentified cognate resistance (R) protein (Blondeau et al., 2015).
Structural biology has and will continue to play an important role in understanding the activity, localization and host interactions associated with plant pathogen effector proteins (reviewed by Wirthmueller et al., 2013). We are utilizing protein biochemistry, biophysics and structural biology approaches in order to understand the function of individual effectors from fungal pathogens. Here, we report a generalized workflow to facilitate SCR effector studies using recombinant protein production in Escherichia coli expression systems. Using this strategy, four of the five effector candidates studied were successfully expressed and purified. The proteins were folded and the oxidation state of the encoded cysteines was determined. Collectively, this work enabled functional and structural investigations. With the rapid increases in SCR effector identification and interest in their function in pathogenicity, we believe our approach may serve as a template for others interested in understanding the function of SCR effector(s) from their plant pathogen(s) of interest.
Results
The role of cysteines in SCR effectors
The SCR effectors selected for this study are summarized in Table 1. The AvrP and AvrP123 genes from the biotrophic fungal pathogen Melampsora lini (flax rust) are allelic variants of the AvrP123 locus of M. lini and encode proteins with 70% sequence identity (Barrett et al., 2009). The AvrP and AvrP123 proteins contain 10 conserved cysteine residues and have been reported previously to contain a cysteine signature that resembles the Kazal family of serine protease inhibitors (Catanzariti et al., 2006). The proteins are recognized by the flax intracellular resistance proteins P (AvrP) or P1, P2 and P3 (AvrP123), which leads to a localized cell death response in a plant defence process known as effector‐triggered immunity (Barrett et al., 2009; Catanzariti et al., 2006). Similarly, the unrelated M. lini protein AvrP4 is recognized by P4 (Catanzariti et al., 2006). AvrP4 encodes a predicted 67‐amino‐acid mature protein with six cysteine residues, and has been predicted previously to comply with the spacing consensus of inhibitor cysteine knot proteins (Catanzariti et al., 2006). AvrP, AvrP123 and AvrP4 induce cell death when expressed inside the plant cell, suggesting that they are translocated across the plant cell membrane (Barrett et al., 2009; Catanzariti et al., 2006).
Table 1.
Small cysteine‐rich (SCR) effectors studied.
| Protein | Organism | Molecular weight (kDa) | Residues | Cysteines | Predicted localization | ScanProsite* | Phyre2† |
|---|---|---|---|---|---|---|---|
| AvrP | Melampsora lini | 9.5 | 88 | 10 | Intracellular | Kazal serine protease inhibitor family signature | PHD domain (zinc binding) |
| AvrP123 | Melampsora lini | 10.3 | 94 | 10 | Intracellular | Kazal serine protease inhibitor family signature | 100.0 |
| AvrP4 | Melampsora lini | 7.3 | 67 | 6 | Intracellular | No hit | – |
| SnTox1 | Parastagonospora nodorum | 10.3 | 100 | 16 | Unknown | No hit | 100.0 |
| SnTox3 | Parastagonospora nodorum | 23.6 | 209 | 6 | Unknown | No hit | 100.0 |
| avrM‡ | Melampsora lini | 27.1 | 235 | 0 | Intracellular | Not applicable | Not applicable |
*ScanProsite (http://prosite.expasy.org/scanprosite/) hits matched by miniprofile (Hulo et al., 2008).
†Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id = index) hits displayed with confidence levels >80%.
‡Positive control for protein expression. The sequences of the constructs used are detailed in Table S2 (see Supporting Information).
SnTox1 and SnTox3 are two unrelated SCR effectors from the necrotrophic pathogen Parastagonospora nodorum that induce necrosis and promote disease on wheat lines harbouring the susceptibility genes Snn1 and Snn3, respectively (Liu et al., 2009, 2012). To date, the Snn1 and Snn3 susceptibility genes have not been identified and the localization of SnTox1 and SnTox3 during infection is unknown. SnTox1 is predicted to encode a mature protein of 100 residues containing 16 cysteines, and SnTox3 contains six cysteine residues in the mature protein (Liu et al., 2009, 2012). The necrotic activity of both proteins is sensitive to dithiothreitol (DTT), a disulfide bond reducing agent, suggesting that they require disulfide bonds for activity (Liu et al., 2009, 2012). Collectively, these data suggest that these SCR effectors may require disulfide bonds.
To investigate these SCR effectors further, we used the motif prediction program PrositeScan (Hulo et al., 2008) and the structural modelling program Phyre2 (Kelley et al., 2015). No significant hits were observed for AvrP4, SnTox1 and SnTox3. PrositeScan analysis of AvrP and AvrP123 identified a stretch of sequence (residues 36–59) analogous to that found in Kazal‐type serine protease inhibitor domain proteins (Fig. S1, see Supporting Information), consistent with previous reports (Catanzariti et al., 2006). Interestingly, however, AvrP structure prediction using Phyre2 generated homology models from the plant homeodomain (PHD) (Fig. S1). The top template (confidence score of 90%) was the PHD domain of the PHD Finger Protein 8 [Protein Data Bank (PDB) code 3KV4] (Horton et al., 2010). Sequence alignment revealed that seven cysteine residues are conserved among AvrP, AvrP123 and PHD domains (Fig. S1). Interestingly, PHD domains have a characteristic arrangement of cysteine and histidine residues that co‐ordinate binding to zinc ions (Aasland et al., 1995) and do not contain cysteine pairs forming disulfide bonds, and so it is possible that the cysteine residues in AvrP and AvrP123 have a metal‐binding role rather than forming disulfides.
Soluble expression of SCR effectors in E. coli
Recombinant expression in prokaryotes, particularly E. coli, remains the most attractive protein production system because of its relatively low cost, rapid application, high yield and well‐characterized genomics (Terpe, 2006). In addition, the experimental equipment and techniques required to produce proteins in E. coli are present in most molecular biology laboratories. However, large‐scale production of disulfide bond‐containing proteins in E. coli is challenging. The cytoplasmic redox potential of E. coli does not favour intracellular disulfide bond formation and disulfide bond‐containing proteins are often incorrectly folded when expressed in the cytosol of E. coli (Lobstein et al., 2012). To combat this, there are a number of strategies that researchers can employ, including the secretion of proteins into the periplasm and the use of commercially available E. coli strains that have been engineered to promote intracellular disulfide bond formation (Berkmen, 2012; Ke and Berkmen, 2014; Terpe, 2006). The strain SHuffle (New England Biolabs, Ipswich, Massachusetts, United States) is a recently developed example of an E. coli strain developed to favour cytoplasmic disulfide bond formation. SHuffle is based on the trxB gor suppressor strain SMG96, which has diminished reductive activity in the cytoplasm as a result of the disruption of the glutaredoxin and thioredoxin pathways. SHuffle is also engineered to express a cytoplasmic version of the disulfide bond isomerase protein, DsbC, collectively improving the strain's capability to produce high yields of active disulfide‐bonded protein within the cytoplasm of E. coli (Lobstein et al., 2012).
As a result of the clear advantages associated with recombinant protein expression in E. coli, we aimed to produce the selected SCR effectors in this system. At the outset of these experiments, we were still unclear whether the cysteine residues in all SCR effectors of interest were disulfide bonded. Therefore, we compared two E. coli expression strains for protein production. Expression in the BL21 (DE3) strain drives protein production in a reducing environment unconducive to disulfide bond formation, whereas the SHuffle strain presents an optimized oxidizing environment designed to facilitate the formation and maintenance of disulfide bonds (Lobstein et al., 2012). The SCR effectors were cloned without their respective signal peptides (Table S1, see Supporting Information) into the vector pMCSG7, which incorporates an N‐terminal 6 × histidine tag to aid in protein purification by metal affinity chromatography, and a Tobacco etch virus (TEV) protease cleavage site to facilitate the subsequent removal of the histidine tag in the later stages of purification (Stols et al., 2002). Small‐scale expression tests were carried out to compare the total protein expression in the BL21 and SHuffle expression strains (Fig. 1a). High expression of AvrP and AvrP123 was detected in both E. coli strains; however, stronger bands were observed from expression in SHuffle. No significant expression was identified for AvrP4, SnTox1 was only observed in SHuffle and SnTox3 showed similar expression levels in both strains. The M. lini effector avrM, which expresses well in E. coli, but does not contain any cysteines (Ve et al., 2011, 2013), was strongly expressed in both strains. To determine whether the SCR effectors were expressed in a soluble form and whether they can be purified by affinity purification using the histidine tag, we utilized the BLItz biosensor system (FortéBio, Menlo Park, California, United States) to detect histidine‐tagged proteins directly from clarified E. coli cell lysates. This system enables rapid protein detection and can be semi‐quantitative because of the directly proportional relationship between the protein concentration and protein–sensor binding rate. Strikingly, in the clarified cell lysates of AvrP and AvrP123, we observed higher levels of soluble histidine‐tagged protein in BL21 relative to SHuffle (Fig. 1b). To further validate the results from BLItz detection, the soluble fractions of the total protein were purified in a batch‐binding approach using nickel affinity beads and analysed using sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) (Fig. 1c). Both AvrP and AvrP123 showed higher soluble protein expression in BL21. In the case of SnTox3, the yields of soluble protein were almost identical between the two strains, whereas the SnTox1 protein was essentially only detected using the SHuffle strain. Histidine‐tagged protein was detected in both samples of AvrP4 using BLItz (Fig. 1b); however, protein migration on the SDS‐PAGE gel did not appear to be consistent with the expected molecular mass of AvrP4 of ∼10 kDa (including the histidine tag).
Figure 1.
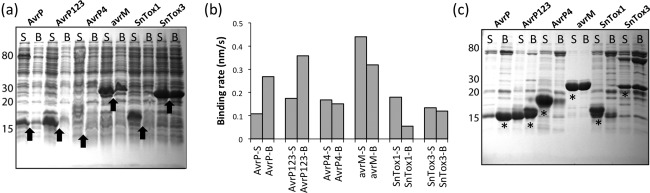
Cysteine‐rich effector protein production in Escherichia coli. (a) Coomassie brilliant blue (CBB)‐stained sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of total protein from E. coli expression experiments. S and B denote SHuffle and BL21 strains, respectively. Arrows indicate the expected migration of the overexpressed proteins. (b) Analysis of histidine‐tagged protein derived from crude cell lysates using BLItz (FortéBio). The binding rate of the histidine‐tagged protein to the sensor, which is directly proportional to the protein concentration in the sample, was measured. (c) CBB‐stained SDS‐PAGE analysis of soluble protein purified in small‐scale nickel affinity purification experiments, labelled as in (b). *Samples analysed by tryptic digest mass spectrometry.
To check the identity of the protein bands, we performed tryptic digestion of the isolated gel bands (Fig. 1c) and mass spectrometry (MS) analysis. These data confirmed the soluble expression of AvrP, AvrP123, SnTox1 and SnTox3; however, the prominent band in the AvrP4 sample was shown to be a contaminating protein from E. coli (Table S3 and Fig. S2, see Supporting Information). Collectively, these data demonstrate that, in terms of soluble protein production, AvrP and AvrP123 perform better in the reducing environment of BL21, whereas SnTox1 requires the oxidizing environment provided by the expression in the SHuffle strain. SnTox3 was produced in similar quantities in both strains.
Purification and characterization of SCR effectors expressed in E. coli
Following soluble protein expression studies, AvrP and AvrP123 were expressed and purified on a large scale with BL21 cells. The proteins could be purified from the cell lysate using nickel affinity chromatography, and the histidine tag was removed before further purification using size exclusion chromatography (SEC). Yields of 1.5 and 1 mg/L (cell culture) were obtained for AvrP and AvrP123, respectively. As a result of the high sequence identity between AvrP and AvrP123 and the slightly higher yields and observed stability of AvrP, we present here further characterization of AvrP only (Fig. 2). We analysed the purified AvrP protein using circular dichroism (CD) spectropolarimetry, which revealed that AvrP did not maintain any dominant helical or sheet secondary structure elements and appeared to be generally unstructured (Fig. 2a). Despite this, AvrP eluted from SEC in a monodisperse peak, demonstrating that the protein did not form soluble protein aggregates (Fig. 2b). Secondary structure prediction of the protein was consistent with the CD data, suggesting that AvrP is dominated by a random coil structure (Fig. 2c).
Figure 2.
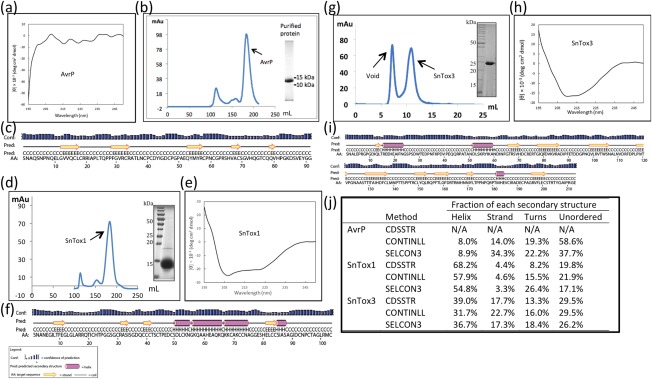
Small cysteine‐rich (SCR) effector purification and secondary structure analysis. (a) Circular dichroism (CD) analysis of the purified AvrP. (b) Size exclusion chromatography (SEC) profile of AvrP on Hiload 26/600 Superdex 75 PG. Coomassie brilliant blue (CBB)‐stained sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of protein from the identified peak (inset). (c) Predicted secondary structure analysis output from PSPIRED (http://bioinf.cs.ucl.ac.uk/psipred/) (Buchan et al., 2013; Jones, 1999). (d–f) As for (b, a, c), but performed with SnTox1. (g) SEC profile of SnTox3 on 10/300 Superdex 75HR. CBB‐stained SDS‐PAGE of protein from the identified peak (inset). (h, i) As for (a, c), but performed with SnTox3. (j) The percentage of secondary structure elements estimated using CD analysis programs CDSSTR, CONTINLL and SELCON3. CDSSTR failed to estimate the secondary structure elements of AvrP (N/A).
SnTox1 clearly favoured the disulfide bond‐supportive environment provided by E. coli SHuffle cells and was expressed and purified on a large scale using this strain. After nickel affinity chromatography, histidine tag removal and SEC, SnTox1 was purified at high purity with yields of 5 mg/L (Fig. 2d). The CD analysis of SnTox1 showed that the secondary structure was dominated by a helix, consistent across all analysis programs (Fig. 2e,j), and this was in general agreement with the secondary structure predictions (Fig. 2f). The results from the small‐scale experiments of SnTox3 were unclear, and it was subsequently expressed on a large scale using both BL21 and SHuffle cells; however, in these experiments, negligible levels of soluble protein were obtained from BL21 expression (data not shown) and low yields of ∼0.1 mg/L were obtained using SHuffle cells. SnTox3 protein produced in SHuffle cells migrated as a single species in SEC (Fig. 2g) and showed secondary structure features of both α‐helices and β‐strands when analysed by CD (Fig. 2h,j), which was again consistent with secondary structure predictions (Fig. 2i).
Oxidation state of the cysteines in the recombinant SCR effector proteins
To determine the oxidation state of the cysteine residues in the SCR effectors, we used MS to measure the intact mass of the protein (Table S4 and Figs S3–S5, see Supporting Information). MS analysis of AvrP returned a measured molecular mass of 9791.7 Da, similar to the expected molecular mass (9794 Da) associated with the reduced form of the AvrP protein. To further clarify the oxidation state, AvrP was treated with iodoacetamide (IAA). IAA alkylates free thiol (–SH) or thiolate (–S–) groups associated with reduced cysteines, adding a molecular mass of 57 Da per thiol group. IAA treatment of AvrP increased the molecular mass of AvrP by 572.2 Da, consistent with the alkylation of 10 cysteine residues, supporting our initial observation that the cysteines in AvrP are reduced. By contrast, MS analysis of SnTox1 and SnTox3 identified molecular masses of 10 574.6 Da and 24 008.9 Da, respectively, which were consistent with proteins containing disulfide‐bonded cysteine pairs (Table S4). This was further supported by IAA treatment, which did not affect the molecular masses of SnTox1 and SnTox3. The proteins were further incubated in reducing conditions (5 mm DTT at 65°C) prior to alkylation with IAA. The reduction and alkylation of SnTox3 resulted in a 348‐Da molecular mass increase in the major SnTox3 peak, indicating that the six cysteines were reduced and alkylated. A mixed molecular mass species of SnTox1 was identified in the reduced and alkylated sample, suggesting that SnTox1 was not fully reduced using this treatment. These data are consistent with the observation that the necrosis‐inducing activity of both these proteins is abolished by treatment with DTT (Liu et al., 2009, 2012). Our MS analysis also suggested that we had no additional contaminants or significant breakdown protein species associated with AvrP and SnTox3 purified proteins. However, in the case of SnTox1, MS analysis demonstrated that there was a truncated SnTox1 product in our purified sample (Fig. S4).
Zinc is required for the production and crystallization of AvrP
As the expression, purification and MS data suggest that AvrP does not contain disulfide bonds, we were interested in understanding the role of the conserved cysteine residues in this protein. The homology modelling and sequence alignment indicated that AvrP has an arrangement of cysteine residues similar to those of PHD proteins, which are involved in the binding of zinc ions (Fig. S1). Therefore, we tested whether AvrP purified from E. coli BL21 strain was bound to metals using 4‐(2‐pyridylazo)resorcinol (PAR) reagent (Fig. 3a). The uncomplexed PAR showed a maximum absorption wavelength of 416 nm, but, when incubated with purified AvrP, an additional absorption peak at a wavelength of 497 nm was observed (Fig. 3a), consistent with the formation of the PAR–metal complex and revealing that the AvrP sample contained metals.
Figure 3.
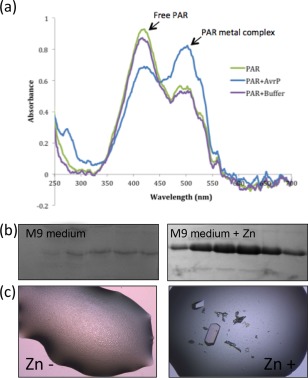
Zinc is required for the production and crystallization of AvrP. (a) Detection of metals in AvrP, using 4‐(2‐pyridylazo)resorcinol (PAR) as the reagent, demonstrated the presence of metals (Hunt et al., 1985; Sabel et al., 2009). The spectrum of PAR and AvrP mix (blue) showed an absorption peak of the PAR–metal complex at 497 nm in addition to the peak of the excess free PAR, whereas PAR and the size exclusion chromatography (SEC) buffer mix (purple) showed a similar absorption at 497 nm to the PAR‐only solution (green). (b) The histidine‐tagged AvrP fusion protein was produced in M9 minimal medium ± 50 μm ZnCl2 and purified using nickel affinity chromatography. The Coomassie brilliant blue (CBB)‐stained sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel showed the peak fractions of the nickel affinity elution without (left) and with (right) zinc supplement, demonstrating the requirement of zinc for AvrP production in M9 minimal medium. (c) Crystallization of AvrP was dependent on the addition of 50 μm ZnCl2 to the crystallization solution.
In the light of the similarities to zinc‐binding proteins and the observed metal‐binding activity, an AvrP protein expression test was performed in minimal medium (M9) with or without zinc salt as a supplement. The relative amount of AvrP was analysed after first‐step nickel affinity chromatography (Fig. 3b), which demonstrated that the introduction of zinc greatly increased the production of AvrP when expressed as a cytosolic protein in E. coli cells. Collectively, these observations suggest that the cysteine residues in AvrP bind to metals. We therefore performed crystallization experiments in the presence or absence of 50 μm zinc chloride. The AvrP protein sample with zinc removed failed to crystallize, whereas good crystals were obtained in the presence of zinc (Fig. 3c), indicating that zinc also has a role in the promotion of crystal formation.
Recombinant SnTox1 and SnTox3 proteins cause necrosis in susceptible wheat lines
Our analysis of the E. coli‐produced SnTox1 and SnTox3 proteins demonstrated that the proteins contained secondary structure and were disulphide bonded. We were interested in determining whether the E. coli‐produced proteins showed biological activity. Infiltration of purified SnTox1 and SnTox3 proteins into wheat lines harbouring the susceptibility genes Snn1 and Snn3 (Calingari and BG220), respectively, demonstrated that the proteins alone were able to induce necrosis (Fig. 4). Importantly, the necrosis was genotype specific, although we did observe mild chlorosis in the SnTox1–BG220 infiltrations. This effect appeared (by visual inspection) to be influenced by the amount of SnTox1 that was infiltrated into wheat leaves (Fig. S6, see Supporting Information), and may indicate an additional function of the protein in the absence of Snn1.
Figure 4.
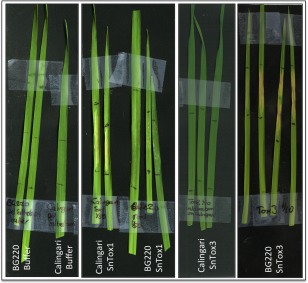
Escherichia coli‐produced SnTox1 and SnTox3 cause necrosis when infiltrated into wheat lines harbouring the susceptibility genes Snn1 and Snn3, Calingari and BG220, respectively. From left to right: buffer infiltrated into BG220 and Calingari; SnTox1 (15 μg) infiltrated into BG220 and Calingari; SnTox3 (37.5 μg) infiltrated into Calingari; SnTox3 infiltrated into BG220.
Discussion
Here, we have presented a strategy that enables the production of soluble and functional fungal SCR effectors using intracellular expression in E. coli. Of the five selected SCR effectors, four could be expressed and purified from E. coli to near homogeneity using a relatively simple purification strategy, and, of these, three were utilized in further functional and structural studies. The relatively inexpensive nature of prokaryotic expression systems and their compatibility with the equipment and expertise in most molecular biology laboratories often make them the systems of choice for initial recombinant protein expression and production trials. We believe our approach could serve as a starting point for researchers with an interest in understanding SCR effector function using recombinant protein approaches (Fig. 5). We highlight important considerations and quality checkpoints that should be considered when tailoring protein‐specific expression and purification strategies (Fig. 5).
Figure 5.
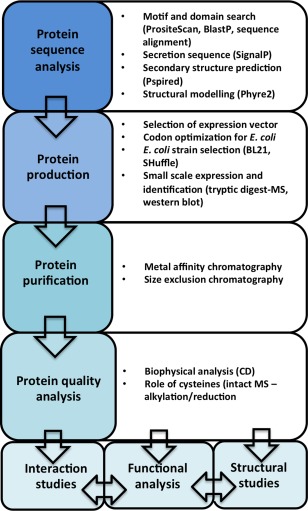
A proposed workflow for the production of small cysteine‐rich (SCR) effectors. CD, circular dichroism; MS, mass spectrometry.
Although cysteines are known to play crucial roles in the active sites of enzymes, such as proteases, and the coordination of metal ions (for example, in proteins involved in nucleic acid binding), in most cases, it is thought that the cysteines in SCR effectors are involved in the formation of disulfide bonds that function to stabilize the protein structures (Bozkurt et al., 2012; Stergiopoulos and de Wit, 2009). Although this function is likely to hold true for a large number of SCR effectors (perhaps the majority), we present data here which demonstrate that this should not be assumed to be generally true for all SCR effector proteins. Bioinformatics analysis of the M. lini SCR effector AvrP revealed two opposing hypotheses: (i) the arrangement of cysteines has features that are commonly found in the Kazal family of serine protease inhibitors; and (ii) the arrangement of cysteines has features that are found in metal‐binding PHD domains. Consequently, we used an unbiased approach in our E. coli expression studies, performing initial expression and solubility screens in E. coli strains that promote either reducing or oxidizing intracellular environments. Higher quantities of soluble AvrP and AvrP123 were produced in E. coli BL21 cells, suggesting their preference for a reducing environment. These data were consistent with the MS data for the intact AvrP and the role of metal binding in protein crystallization. The AvrP crystals diffracted X‐rays to 2.5 Å resolution at the MX1 beamline at the Australian Synchrotron and the presence of zinc was detected in the crystal by a fluorescence excitation scan (Fig. S7, see Supporting Information). We subsequently solved the structure and found that AvrP coordinates three zinc ions per protein molecule; the details of this structure will be presented elsewhere.
Despite the significant advantages of heterologous expression in E. coli, proteins that require disulfide‐bonded cysteines are challenging to produce in a soluble form. Consequently, these proteins are often produced in secreted form in eukaryotic cells, such as yeast, where trafficking through the endoplasmic reticulum and the secretion system facilitates disulfide bond formation. This method has been utilized for SCR effectors including AvrLm4‐7 from Leptosphaeria maculans and the effectors Avr2 and Avr4 from Cladosporium fulvum (Blondeau et al., 2015; Rooney et al., 2005). In the studies that identified SnTox1 and SnTox3, Pichia pastoris was used to manufacture protein that was capable of inducing necrosis in wheat leaves containing compatible susceptibility genes (Liu et al., 2009, 2012). However, the production levels reported for these proteins would not be sufficient for structural analysis. For example, SnTox1 produced in P. pastoris was observed by western blot, but could not be detected on a Coomassie‐stained gel (Liu et al., 2009). In recent years, advances in expression strains and techniques have yielded significant improvements in the utility of E. coli as a production system for disulfide‐bonded proteins (reviewed by Berkmen, 2012; Ke and Berkmen, 2014; Terpe, 2006). For example, secretion to the oxidation‐favouring environment of the periplasmic space has proved to be a successful approach for the manufacture of disulfide bond‐containing proteins (reviewed by Choi and Lee, 2004). In our study, we used the cytoplasmic disulfide bond‐supporting strain SHuffle, based on the study by Lobstein et al. (2012). In our hands, SHuffle has become a very effective strain to produce disulfide‐bonded proteins in a soluble form in E. coli. The effectiveness of the strain in protein production and structural biology is also highlighted in PDB. According to a PDB search (September 2015), SHuffle has been used to produce protein that has resulted in 45 structure entries since 2012, 43 of which were solved by X‐ray crystallography, with 19 released in 2015. Whilst our work was in progress, Maqbool et al. (2015) employed SHuffle to produce the proteins used in solving the crystal structure of the complex between the rice blast fungal SCR effector AVR‐PikD and the heavy metal‐associated (HMA) domain from the rice resistance protein Pik, providing further proof of the usefulness of this system in the study of SCR effectors.
The use of effectors has become a very important tool in plant breeding in recent years, in order to speed up the process of R gene or susceptibility gene identification (Vleeshouwers and Oliver, 2014). For example, effectors from the hemibiotrophic oomycete Phytophthora infestans have been used to identify R genes in wild Solanum plants for introduction into commercial potato species (Vleeshouwers and Oliver, 2014). Allele mining using AVRblb1 identified the R gene Rpi‐sto1 from Solanum stoloniferum, which can now be integrated into cultivated potato by direct crossing (Vleeshouwers et al., 2008). The potato system has the advantage that it can utilize Agrobacterium‐based methods to deliver effectors; however, other plant species, such as wheat, are not compatible with this approach. Nevertheless, the injection of purified protein has been proven to be very useful for the ToxA effector, which occurs in two necrotrophic fungi that infect wheat: Pyrenophora tritici‐repentis and Parastagonospora nodorum (Ballance et al., 1996; Friesen et al., 2006). In 2009, semi‐pure ToxA was first given to plant breeders to test wheat progeny for the presence of the corresponding susceptibility gene Tsn1 and, by 2012, sufficient ToxA was produced to test 30 000 cultivars (Vleeshouwers and Oliver, 2014). Likewise, SnTox1 and SnTox3 proteins sufficient for 6000 doses have been produced using a P. pastoris system to screen for the presence of the susceptibility genes Snn1 and Snn3. In the current study, we used a dose of approximately 0.015 mg of SnTox1 for each infiltration and achieved yields of 5 mg/L of E. coli culture at an estimated culture cost of ∼20$/L, corresponding to a cost per infiltration of a very modest $0.06. This dose rate could potentially be reduced, as the visible signs of necrosis were still very strong at this protein level (Fig. 4). For the SnTox3 infiltrations (Fig. 4), we required ∼0.030 mg to induce visible necrosis. Our yields were much lower for SnTox3 and, at this stage, we can only perform approximately three infiltrations with the protein obtained per litre of E. coli cells grown, with an estimated cost of ∼$7 per infiltration. At these current levels, E. coli‐produced SnTox1 may provide a viable alternative for plant breeders testing susceptibility in wheat, broadening the utility of our E. coli‐produced proteins beyond structural and biochemical studies.
Based on our experience in heterologous protein production, we recommend the workflow depicted in Fig. 5 as a reference point for the investigation of SCR effectors. We stop short of calling this a pipeline, as the individual properties of the proteins of interest will almost certainly require the optimization of protein‐specific protocols at the expression and purification levels. We find structural modelling (in our case, we used Phyre2; Kelley et al., 2015) to be a very useful first step when investigating a new SCR effector and, in our hands, we benefited from an unbiased approach in our choice of E. coli expression systems to help guide further strategies for large‐scale protein production. We and others have found SHuffle to be an effective strain for the production of disulfide‐bonded SCR effectors (Maqbool et al., 2015), and this is our method of choice before embarking on more expensive and time‐consuming expression hosts. The inclusion of SEC, MS and CD is critical to assess protein quality, purity and folding, respectively, and we recommend the inclusion of similar characterization techniques whenever possible. Collectively, this study provided the protein material for subsequent structural and functional experiments that will assist our understanding of their function.
In conclusion, we believe that this work will provide a guide to others interested in the production of SCR effectors to complement recent technical reports which, collectively, will assist a multi‐disciplined approach towards the understanding of the role of effectors from filamentous plant pathogens (Gong et al., 2015; Hughes and Banfield, 2014; Petre et al., 2015).
Experimental Procedures
Vectors and gene constructs
The DNA sequence that encodes SnTox3 was generated by polymerase chain reaction (PCR)‐based gene synthesis. Codon optimization for expression in E. coli and automatic oligonucleotide design were carried out using the webserver DNAWorks (http://helixweb.nih.gov/dnaworks/) (Hoover and Lubkowski, 2002), and synthesized by overlapping PCR. The DNA sequence that encodes SnTox1 was codon optimized for expression in E. coli and synthesized by GeneArt (Thermofisher Scientific, Waltham, Massachusetts, United States). Both genes were subcloned into pMCSG7 (Stols et al., 2002) for expression studies in E. coli. The cDNAs for AvrP, AvrP123 and AvrP4 (Catanzariti et al., 2006) were cloned into the pMCSG7 vector using ligation‐independent cloning (LIC) (Stols et al., 2002). The resulting constructs contained an N‐terminal 6 × histidine tag followed by a TEV protease cleavage site. The avrM expression construct was that used previously (Ve et al., 2011, 2013). The oligonucleotides used in this study are summarized in Table S1.
Small‐scale protein expression and analysis
For small‐scale expression experiments, E. coli strains BL21(DE3) and SHuffle were transformed with relevant expression vectors and grown in 500 mL of Luria–Bertani (LB) medium at 37°C, supplemented with 100 μg/mL ampicillin, with shaking until the cultures reached an optical density at 600 nm (OD600) of 0.6–0.8. The temperature was then reduced to 20°C and isopropyl β‐d−1‐thiogalactopyranoside (IPTG) was added to a final concentration of 1 mm. The cultures were grown for a further 16 h before harvesting by centrifugation. The cell pellets were resuspended in 4 mL of lysis buffer [50 mm N−2‐hydroxyethylpiperazine‐N'‐2‐ethanesulfonic acid (Hepes), pH 8, 300 mm NaCl] with 1 mm phenylmethylsulfonyl fluoride (PMSF) added. The cells were lysed by sonication and the lysate was centrifuged at 25 000 g for 10 min to collect the supernatant. The histidine‐tagged proteins in the supernatant were detected and quantified using a BLItz system (FortéBio) employing Ni2+‐nitrilotriacetate (Ni‐NTA) as biosensor. The sensor was immersed in the clarified crude lysate for 120 s and the rate of histidine‐tagged protein binding to the sensor was measured. The concentration of the histidine‐tagged protein in the sample is directly proportional to the binding rate. To purify the desired protein, the lysate was incubated with 500 μL of Ni Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) for 1 h. The beads were transferred to a gravity‐flow column and washed with 20 mL of wash buffer (50 mm Hepes, pH 8, 300 mm NaCl, 20 mm imidazole) to remove the unbound proteins, and the proteins of interest were eluted in 5 mL of elution buffer (50 mm Hepes, pH 8, 300 mm NaCl, 250 mm imidazole). The eluted proteins were further concentrated using Amicon Ultra centrifugal filter devices (Millipore, Billerica, Massachusetts, United States).
Expression of AvrP in M9 minimal medium
The method for AvrP protein expression using M9 minimal medium was adapted from Klint et al. (2013). The cells were grown at 37°C in LB medium with shaking until the cultures reached an OD600 of 0.7–0.8. The cells were then harvested by centrifugation for 15 min at 5000 g and washed gently in 1 L M9 salts solution (22 mm KH2PO4, 90 mm Na2HPO4, 17 mm NaCl). The cells were pelleted again by centrifugation for 15 min at 5000 g and resuspended in M9 minimal medium (M9 salts supplied with 1.6 mm MgSO4, 80 nm CaCl2, 18 nm NH4Cl, 22 mm d‐glucose, 2 µg/mL thiamine, 0.2% (v/v) vitamin solution). The volume of M9 medium equalled one‐quarter of the volume of LB medium used to grow the cells. The cultures were grown at 37°C for 1–1.5 h with shaking before cooling to 20°C. To induce protein expression, IPTG was added to the cultures to a final concentration of 1 mm. The cultures were then grown for a further 16 h before harvesting by centrifugation.
Protein expression and purification
A number of optimization experiments were performed to arrive at the conditions used for large‐scale protein production. Details of the final expression and purification conditions are provided in Methods S1 (see Supporting Information).
Tryptic digest and intact MS
To determine whether the partially purified proteins from small‐scale expression/purification experiments represented the SCR effectors, tryptic digest MS was performed on the predominant protein bands (Fig. 1c). Details of the tryptic digest, peptide extraction and MS analysis are described in Methods S2 (see Supporting Information). MS was also performed on the purified AvrP, SnTox1 and SnTox3 proteins to understand the oxidation state of the cysteines. Methods for the production of native, alkylated and reduced/alkylated samples and MS analysis are described in Methods S3 (see Supporting Information).
CD spectropolarimetry
The CD spectra were recorded on a Jasco, Easton, Maryland, United States J710 spectropolarimeter at 25°C. Purified protein samples of SnTox1, SnTox3 and AvrP were analysed at protein concentrations of 0.25, 0.2 and 0.1 mg/mL, respectively, in phosphate‐buffered saline. Measurements were taken at 0.2‐nm wavelength increments from 195 to 250 nm at 100 nm/min using a cell with a path length of 1 mm, bandwidth of 2 nm, response time of 1 s and five accumulations, and corrected for buffer baseline contribution. The secondary structure was estimated by the programs SELCON3 (Sreerama and Woody, 1993), CDSSTR (Johnson, 1999) and CONTIN (Provencher and Glockner, 1981) in the CDPro software package (Sreerama and Woody, 2000).
Metal‐binding assay
The PAR metal‐binding assay was used to detect the presence of metals in the AvrP protein samples (Hunt et al., 1985; Sabel et al., 2009). The PAR solution was prepared in reaction buffer (50 mm Hepes, pH 7.4, 4 m guanidine‐HCl) to a PAR concentration of 1 mm. The AvrP protein was denatured in the reaction buffer by boiling. The denatured AvrP protein was mixed with the PAR solution to a final concentration of 50 μm PAR and 14 μm AvrP. The absorbance of the samples at wavelengths between 250 and 700 nm was measured using a NanoDrop (Thermo Scientific, Waltham, Massachusetts, United States). The uncomplexed PAR displays an absorption peak at 416 nm and the PAR–metal complex has an absorption peak at 497 nm.
AvrP crystallization
To prepare homogeneous AvrP protein for crystallization, the purified proteins were treated with ethylenediaminetetraacetic acid (EDTA) to remove metals captured during expression and purification, and buffer exchanged into SEC buffer containing 50 μm ZnCl2. Eight commercial screens were utilized: Index, PEG/Ion and PEGRx (Hampton Research, Aliso Viejo, California, United States), Morpheus, ProPlex, JCSG+ and Pact Premier (Molecular Dimensions, Newmarket, Suffolk, United Kingdom), and Precipitant Synergy (Emerald Biosystems, Bainbridge Island, Washington, United States). Hanging drops consisting of 100 nL of protein solution and 100 nL of reservoir solution were prepared using a Mosquito robot (TTP LabTech, Royston, Cambridgeshire, United Kingdom) and equilibrated against 100 μL of reservoir solution. Crystals of AvrP (5 mg/mL) appeared after 1 day under several different screening conditions, including Index condition A9 consisting of 0.1 m Bis‐Tris, pH 5.5, and 3 m NaCl, and Index condition F12 consisting of 0.1 m Hepes, pH 7.5, 25% (w/v) PEG 3350 and 0.2 m NaCl. Optimization, including pH and precipitant changes, was carried out and the best quality crystals were obtained in 0.1 m Bis‐Tris, pH 7.0, 22% (w/v) PEG 3350 and 0.2 m NaCl. Crystals were back soaked in well solution that included 25% (v/v) ethylene glycol as a cryoprotectant and flash cooled in liquid nitrogen for data collection on the MX1 beamline at the Australian Synchrotron.
Wheat infiltration assays
Wheat plants were grown in a controlled environment chamber with a 16‐h day/8‐h night cycle, with 20°C day temperature and 12°C night temperature. The light intensity was 250 µE/m2/s with 85% relative humidity. Purified SnTox1 and SnTox3 were diluted in 10 mm Hepes, pH 7.5, and 150 mm NaCl to 0.1 and 0.25 mg/mL, respectively. Syringe infiltrations of the second leaf of 2‐week‐old wheat seedlings were conducted with 150 μL of protein (or buffer) solution and the plants were monitored from 2 days post‐infiltration (dpi). Leaves were harvested and images were taken at 4 dpi.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Protein expression and purification of AvrP, SnTox1 and SnTox3.
Methods S2 Tryptic digest mass spectrometry.
Methods S3 Intact mass spectrometry.
Fig. S1 In silico analysis of AvrP.
Fig. S2 Tryptic digest mass spectrometry MASCOT search results.
Fig. S3 Intact AvrP mass spectrometry analysis.
Fig. S4 Intact SnTox1 mass spectrometry analysis.
Fig. S5 Intact SnTox3 mass spectrometry analysis.
Fig. S6 Concentration‐dependent necrosis activity of SnTox1.
Fig. S7 Diffraction image and excitation scan of an AvrP crystal.
Table S1 List of oligonucleotides used in this work.
Table S2 Amino acid sequences of small cysteine‐rich (SCR) effectors.
Table S3 MASCOT results from tryptic digest mass spectrometry.
Table S4 Intact mass analysis of untreated, iodoacetamide (IAA)‐treated and dithiothreitol (DTT)/IAA‐treated small cysteine‐rich (SCR) effectors.
Acknowledgements
This research was supported by the Australian Research Council (ARC) Discovery Projects DP120100685 and DP120103558. X.Z. was a recipient of an ANZ Trustee's PhD Scholarship for Medical Research in Queensland. B.K. is a National Health and Medical Research Council (NHMRC) Research Fellow (1003325 and 1110971). The mass spectrometry analysis was carried out at the Mass Spectrometry Facility in the School of Chemistry and Molecular Biosciences, University of Queensland. We thank Peter Josh and Amanda Nouwens for their technical assistance with the mass spectrometry experiments, and Mehdi Mobli for suggesting the use of SHuffle. We acknowledge the use of the University of Queensland Remote Operation Crystallization and X‐ray Diffraction Facility (UQ ROCX). The X‐ray diffraction data collection was undertaken on the MX beamlines at the Australian Synchrotron. We declare that we have no conflicts of interest.
Contributor Information
Bostjan Kobe, Email: b.kobe@uq.edu.au.
Simon J. Williams, Email: s.williams8@uq.edu.au.
References
- Aasland, R. , Gibson, T.J. and Stewart, A.F. (1995) The PHD finger: implications for chromatin‐mediated transcriptional regulation. Trends Biochem. Sci. 20, 56–59. [DOI] [PubMed] [Google Scholar]
- Ballance, G.M. , Lamari, L. , Kowatsch, R. and Bernier, C.C. (1996) Cloning, expression and occurrence of the gene encoding the Ptr necrosis toxin from Pyrenophora tritici‐repentis . Mol. Plant Pathol. (online first: http://www.bspp.org.uk/mppol/1996/1209ballance). [Google Scholar]
- Barrett, L.G. , Thrall, P.H. , Dodds, P.N. , van der Merwe, M. , Linde, C.C. , Lawrence, G.J. and Burdon, J.J. (2009) Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini . Mol. Biol. Evol. 26, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkmen, M. (2012) Production of disulfide‐bonded proteins in Escherichia coli . Protein Expr. Purif. 82, 240–251. [DOI] [PubMed] [Google Scholar]
- Blondeau, K. , Blaise, F. , Graille, M. , Kale, S.D. , Linglin, J. , Ollivier, B. , Labarde, A. , Lazar, N. , Daverdin, G. , Balesdent, M.‐H. , Choi, D.H. , Tyler, B.M. , Rouxel, T. , van Tilbeurgh, H. and Fudal, I. (2015) Crystal structure of the effector AvrLm4‐7 of Leptosphaeria maculans reveals insights into its translocation into plant cell and recognition by resistance proteins. Plant J. 83, 610–624. [DOI] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Banfield, M.J. and Kamoun, S. (2012) Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. [DOI] [PubMed] [Google Scholar]
- Buchan, D.W.A. , Minneci, F. , Nugent, T.C.O. , Bryson, K. and Jones, D.T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, D. , Govindarajulu, M. , Kozik, A. , Wang, M. , Chen, X. , Kojima, K.K. , Jurka, J. , Michelmore, R.W. and Dubcovsky, J. (2011) Next generation sequencing provides rapid access to the genome of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS One, 6, e24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, D. , Segovia, V. , Maclean, D. , Bayles, R. , Chen, X. , Kamoun, S. , Dubcovsky, J. , Saunders, D.G.O. and Uauy, C. (2013) Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics, 14, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A.‐M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J.H. and Lee, S.Y. (2004) Secretory and extracellular production of recombinant proteins using Escherichia coli . Appl. Microbiol. Biot. 64, 625–635. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Duplessis, S. , Cuomo, C.A. , Lin, Y.C. , Aerts, A. , Tisserant, E. , Veneault‐Fourrey, C. , Joly, D.L. , Hacquard, S. , Amselem, J. , Cantarel, B.L. , Chiu, R. , Coutinho, P.M. , Feau, N. , Field, M. , Frey, P. , Gelhaye, E. , Goldberg, J. , Grabherr, M.G. , Kodira, C.D. , Kohler, A. , Kues, U. , Lindquist, E.A. , Lucas, S.M. , Mago, R. , Mauceli, E. , Morin, E. , Murat, C. , Pangilinan, J.L. , Park, R. , Pearson, M. , Quesneville, H. , Rouhier, N. , Sakthikumar, S. , Salamov, A.A. , Schmutz, J. , Selles, B. , Shapiro, H. , Tanguay, P. , Tuskan, G.A. , Henrissat, B. , Van de Peer, Y. , Rouze, P. , Ellis, J.G. , Dodds, P.N. , Schein, J.E. , Zhong, S. , Hamelin, R.C. , Grigoriev, I.V. , Szabo, L.J. , Martin, F. (2011a) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA, 108, 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Hacquard, S. , Delaruelle, C. , Tisserant, E. , Frey, P. , Martin, F. and Kohler, A. (2011b) Melampsora larici‐populina transcript profiling during germination and timecourse infection of poplar leaves reveals dynamic expression patterns associated with virulence and biotrophy. Mol. Plant–Microbe Interact. 24, 808–818. [DOI] [PubMed] [Google Scholar]
- Fernández, C. , Szyperski, T. , Bruyère, T. , Ramage, P. , Mösinger, E. and Wüthrich, K. (1997) NMR solution structure of the pathogenesis‐related protein P14a. J. Mol. Biol. 266, 576–593. [DOI] [PubMed] [Google Scholar]
- Fernandez, D. , Tisserant, E. , Talhinhas, P. , Azinheira, H. , Vieira, A. , Petitot, A.S. , Loureiro, A. , Poulain, J. , Da Silva, C. , Silva Mdo, C. , Duplessis, S. , (2012) 454‐pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta‐expressed pathogen‐secreted proteins and plant functions in a late compatible plant–rust interaction. Mol. Plant Pathol. 13, 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Garnica, D.P. , Upadhyaya, N.M. , Dodds, P.N. and Rathjen, J.P. (2013) Strategies for wheat stripe rust pathogenicity identified by transcriptome sequencing. PLoS One, 8, e67150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnica, D.P. , Nemri, A. , Upadhyaya, N.M. , Rathjen, J.P. and Dodds, P.N. (2014) The ins and outs of rust haustoria. PLoS Pathog. 10, e1004329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, X. , Hurtado, O. , Wang, B. , Wu, C. , Yi, M. , Giraldo, M. , Valent, B. , Goodin, M. and Farman, M. (2015) pFPL vectors for high‐throughput protein localization in fungi: detecting cytoplasmic accumulation of putative effector proteins. Mol. Plant–Microbe Interact. 28, 107–121. [DOI] [PubMed] [Google Scholar]
- Hacquard, S. , Joly, D.L. , Lin, Y.‐C. , Tisserant, E. , Feau, N. , Delaruelle, C. , Legué, V. , Kohler, A. , Tanguay, P. , Petre, B. , Frey, P. , Van de Peer, Y. , Rouzé, P. , Martin, F. , Hamelin, R.C. , Duplessis, S. (2012) A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici‐populina (poplar leaf rust). Mol. Plant–Microbe Interact. 25, 279–293. [DOI] [PubMed] [Google Scholar]
- Hane, J.K. , Lowe, R.G.T. , Solomon, P.S. , Tan, K.‐C. , Schoch, C.L. , Spatafora, J.W. , Crous, P.W. , Kodira, C. , Birren, B.W. , Galagan, J.E. , Torriani, S.F.F. , McDonald, B.A. , Oliver, R.P. (2007) Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum . Plant Cell, 19, 3347–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, D.M. and Lubkowski, J. (2002) DNAWorks: an automated method for designing oligonucleotides for PCR‐based gene synthesis. Nucleic Acids Res. 30, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, J.R. , Upadhyay, A.K. , Qi, H.H. , Zhang, X. , Shi, Y. and Cheng, X. (2010) Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17, 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, R.K. and Banfield, M.J. (2014) Production of RXLR effector proteins for structural analysis by X‐ray crystallography. Methods Mol. Biol. (Clifton, NJ) 1127, 231–253. [DOI] [PubMed] [Google Scholar]
- Hulo, N. , Bairoch, A. , Bulliard, V. , Cerutti, L. , Cuche, B.A. , de Castro, E. , Lachaize, C. , Langendijk‐Genevaux, P.S. and Sigrist, C.J.A. (2008) The 20 years of PROSITE. Nucleic Acids Res. 36, 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, J.B. , Neece, S.H. and Ginsburg, A. (1985) The use of 4‐(2‐pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 146, 150–157. [DOI] [PubMed] [Google Scholar]
- Johnson, W.C. (1999) Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins‐Struct. Funct. Genet. 35, 307–312. [PubMed] [Google Scholar]
- Jones, D.T. (1999) Protein secondary structure prediction based on position‐specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Ke, N. and Berkmen, M. (2014) Production of Disulfide‐Bonded Proteins in Escherichia coli. Current Protocols in Molecular Biology. 108:I:16.1B:16.1B.1–16.1B.21. [DOI] [PubMed]
- Kelley, L.A. , Mezulis, S. , Yates, C.M. , Wass, M.N. and Sternberg, M.J.E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klint, J.K. , Senff, S. , Saez, N.J. , Seshadri, R. , Lau, H.Y. , Bende, N.S. , Undheim, E.A.B. , Rash, L.D. , Mobli, M. and King, G.F. (2013) Production of recombinant disulfide‐rich venom peptides for structural and functional analysis via expression in the periplasm of E. coli . PLoS One, 8, e63865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Faris, J.D. , Oliver, R.P. , Tan, K.‐C. , Solomon, P.S. , McDonald, M.C. , McDonald, B.A. , Nunez, A. , Lu, S. , Rasmussen, J.B. , Friesen, T.L . (2009) SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 5, e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Zhang, Z. , Faris, J.D. , Oliver, R.P. , Syme, R. , McDonald, M.C. , McDonald, B.A. , Solomon, P.S. , Lu, S. , Shelver, W.L. , Xu, S. , Friesen, T.L. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 8, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobstein, J. , Emrich, C.A. , Jeans, C. , Faulkner, M. , Riggs, P. and Berkmen, M. (2012) SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, V.A and Ciuffetti, L.M. (2005) Localization of Ptr ToxA produced by Pyrenophora tritici‐repentis reveals protein import into wheat mesophyll cells. Plant Cell, 17, 3203–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool, A. , Saitoh, H. , Franceschetti, M. , Stevenson, C. , Uemura, A. , Kanzaki, H. , Kamoun, S. , Terauchi, R. and Banfield, M.J. (2015) Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife, 4, e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemri, A. , Saunders, D.G.O. , Anderson, C. , Upadhyaya, N.M. , Win, J. , Lawrence, G.J. , Jones, D.A. , Kamoun, S. , Ellis, J.G. and Dodds, P.N. (2014) The genome sequence and effector complement of the flax rust pathogen Melampsora lini . Front. Plant Sci. 5, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira‐Garcia, E. and Valent, B. (2015) How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 26, 92–101. [DOI] [PubMed] [Google Scholar]
- Petre, B. , Saunders, D.G.O. , Sklenar, J. , Lorrain, C. , Win, J. , Duplessis, S. and Kamoun, S. (2015) Candidate effector proteins of the rust pathogen Melampsora larici‐populina target diverse plant cell compartments. Mol. Plant–Microbe Interact. 28, 689–700. [DOI] [PubMed] [Google Scholar]
- Provencher, S.W. and Glockner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry, 20, 33–37. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C.E. , Van't Klooster, J.W. , van der Hoorn, R.A.L. , Joosten, M.H.A.J. , Jones, J.D.G. and de Wit, P.J.G.M. (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf‐2‐dependent disease resistance. Science, 308, 1783–1786. [DOI] [PubMed] [Google Scholar]
- Sabel, C.E. , Shepherd, J.L. and Siemann, S. (2009) A direct spectrophotometric method for the simultaneous determination of zinc and cobalt in metalloproteins using 4‐(2‐pyridylazo)resorcinol. Anal. Biochem. 391, 74–76. [DOI] [PubMed] [Google Scholar]
- Sarma, G.N. , Manning, V.A. , Ciuffetti, L.M. and Karplus, P.A. (2005) Structure of Ptr ToxA: an RGD‐containing host‐selective toxin from Pyrenophora tritici‐repentis . Plant Cell, 17, 3190–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, D. , Win, J. , Cano, L.M. , Szabo, L.J. and Kamoun, S. (2012) Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One, 7, e29847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider, J. , Dodds, P.N. , Gardiner, D.M. , Manners, J.M. , Singh, K.B. and Taylor, J.M. (2015) Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11, e1004806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama, N. and Woody, R.W. (1993) A self‐consistent method for the analysis of protein secondary structure from circular‐dichroism. Anal. Biochem. 209, 32–44. [DOI] [PubMed] [Google Scholar]
- Sreerama, N. and Woody, R.W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stols, L. , Gu, M. , Dieckman, L. , Raffen, R. , Collart, F.R. and Donnelly, M.I. (2002) A new vector for high‐throughput, ligation‐independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15. [DOI] [PubMed] [Google Scholar]
- Tan, K.C. , Phan, H.T. , Rybak, K. , John, E. , Chooi, Y.H. , Solomon, P.S. and Oliver, R.P. (2015) Functional redundancy of necrotrophic effectors – consequences for exploitation for breeding. Front. Plant Sci. 6, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe, K. (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biot. 72, 211–222. [DOI] [PubMed] [Google Scholar]
- Ve, T. , Williams, S.J. , Stamp, A. , Valkov, E. , Dodds, P.N. , Anderson, P.A. and Kobe, B. (2011) Crystallization and X‐ray diffraction analysis of the C‐terminal domain of the flax rust effector protein AvrM. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 67, 1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ve, T. , Williams, S.J. , Catanzariti, A.‐M. , Rafiqi, M. , Rahman, M. , Ellis, J.G. , Hardham, A.R. , Jones, D.A. , Anderson, P.A. , Dodds, P.N. , Kobe, B. (2013) Structures of the flax‐rust effector AvrM reveal insights into the molecular basis of plant‐cell entry and effector‐triggered immunity. Proc. Natl. Acad. Sci. USA, 110, 17 594–17 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.K. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G. , Jacobsen, E. , Govers, F. , Kamoun, S. , Van der Vossen, E.A. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A.A. and Oliver, R.P. (2014) Effectors as tools in disease resistance breeding against biotrophic, hemi‐biotrophic and necrotrophic plant pathogens. Mol. Plant–Microbe Interact. 27, 196–206. [DOI] [PubMed] [Google Scholar]
- Wirthmueller, L. , Maqbool, A. and Banfield, M.J. (2013) On the front line: structural insights into plant–pathogen interactions. Nat. Rev. Microbiol. 11, 761–776. [DOI] [PubMed] [Google Scholar]
- Zheng, W. , Huang, L. , Huang, J. , Wang, X. , Chen, X. , Zhao, J. , Guo, J. , Zhuang, H. , Qiu, C. , Liu, J. , Liu, H. , Huang, X. , Pei, G. , Zhan, G. , Tang, C. , Cheng, Y. , Liu, M. , Zhang, J. , Zhao, Z. , Zhang, S. , Han, Q. , Han, D. , Zhang, H. , Zhao, J. , Gao, X. , Wang, J. , Ni, P. , Dong, W. , Yang, L. , Yang, H. , Xu, J.R. , Zhang, G. , Kang, Z. (2013) High genome heterozygosity and endemic genetic recombination in the wheat stripe rust fungus. Nat. Commun. 4, 2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Methods S1 Protein expression and purification of AvrP, SnTox1 and SnTox3.
Methods S2 Tryptic digest mass spectrometry.
Methods S3 Intact mass spectrometry.
Fig. S1 In silico analysis of AvrP.
Fig. S2 Tryptic digest mass spectrometry MASCOT search results.
Fig. S3 Intact AvrP mass spectrometry analysis.
Fig. S4 Intact SnTox1 mass spectrometry analysis.
Fig. S5 Intact SnTox3 mass spectrometry analysis.
Fig. S6 Concentration‐dependent necrosis activity of SnTox1.
Fig. S7 Diffraction image and excitation scan of an AvrP crystal.
Table S1 List of oligonucleotides used in this work.
Table S2 Amino acid sequences of small cysteine‐rich (SCR) effectors.
Table S3 MASCOT results from tryptic digest mass spectrometry.
Table S4 Intact mass analysis of untreated, iodoacetamide (IAA)‐treated and dithiothreitol (DTT)/IAA‐treated small cysteine‐rich (SCR) effectors.


