Summary
Sclerotinia sclerotiorum is a devastating necrotrophic plant pathogen with a worldwide distribution. Cell wall‐degrading enzymes and oxalic acid are important to the virulence of this pathogen. Here, we report a novel secretory protein, Ss‐Rhs1, which is essential for the virulence of S. sclerotiorum. Ss‐Rhs1 is believed to contain a typical signal peptide at the N‐terminal and eight rearrangement hotspot (Rhs) repeats. Ss‐Rhs1 exhibited a high level of expression at the initial stage of sclerotial development, as well as during the hyphal infection process. Targeted silencing of Ss‐Rhs1 resulted in abnormal colony morphology and reduced virulence on host plants. Microscopic observations indicated that Ss‐Rhs1‐silenced strains exhibited reduced efficiency in compound appressoria formation.
Keywords: appressorium, Rhs repeat, sclerotia, Sclerotinia sclerotiorum, secretory protein, virulence
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary is a devastating fungal plant pathogen with a worldwide distribution. This fungus threatens more than 400 plants, including many important crops, such as oilseed rape, sunflower, soybean, lettuce, celery and onion (Boland and Hall, 1994). Sclerotinia diseases caused by S. sclerotiorum have always posed a significant threat to crops because of the lack of effective host resistance cultivars, as well as safe and economical control measures.
Sclerotinia sclerotiorum produces sclerotia—multihyphal structures important for the long‐term survival of this pathogen (Chet and Henis, 1975; Willetts and Wong, 1980). Sclerotia may germinate myceliogenically to produce infection hyphae or carpogenically to produce millions of airborne ascospores that are critical for the maintenance and spread of the disease in the field (Steadman, 1979). Sclerotial development is a complex and multistep process that can be divided into three distinct stages: (1) initiation; (2) development; and (3) maturation (Willetts and Bullock, 1992). Many molecular components related to the cyclic adenosine monophosphate‐protein kinase A (cAMP‐PKA) and mitogen‐activated protein kinase (MAPK) cellular signal transduction pathways have been identified and are involved in the sclerotial development of S. sclerotiorum (Chen and Dickman, 2005; Chen et al., 2004; Erental et al., 2007; Harel et al., 2005). There are also several cell wall proteins that may contribute to cell–cell adhesive processes during sclerotial development (Yu et al., 2012; Zhu et al., 2013).
Sclerotinia sclerotiorum is a typical aggressive necrotrophic fungal pathogen. The fungus secretes cell wall‐degrading enzymes (CWDEs) (Martel et al., 1996; Poussereau et al., 2001; Riou et al., 1991; Yajima et al., 2009; Zuppini et al., 2005) and oxalic acid (OA) (Cessna et al., 2000; Favaron et al., 2004; Kim et al., 2008; Williams et al., 2011) to kill its hosts, and the pathogen then feeds on dead or dying cells. However, a series of recent reports have suggested that S. sclerotiorum may have a very short biotrophic phase during the early stages of infection (Kabbage et al., 2013, 2015; Williams et al., 2011). The fungus may suppress plant defence responses via different strategies during the infection phase. One strategy is the secretion of OA, which has been shown to create reducing conditions in plant cells ahead of advancing hyphae to suppress the host oxygen burst (Williams et al., 2011). Furthermore, S. sclerotiorum may also secrete small proteins that function as effectors to suppress host defence (Guyon et al., 2014; Kabbage et al., 2013; Zhu et al., 2013). This evidence shows that the interaction between S. sclerotiorum and its hosts is much more complex and unresolved than previously thought.
The Rhs (rearrangement hotspot) repeat (Pfam PF05593) was first identified in rearrangement hotspot elements that promote genomic recombination in Escherichia coli (Hill et al., 1994; Lin et al., 1984). Although the overall amino acid sequences across the family exhibit a low degree of conservation, a consensus sequence for the Rhs repeat itself has been defined: GxxxRYxYDxxGRL(I/T) (Wang et al., 1998). Rhs and related tyrosine–aspartate (YD) repeats have been found in many bacterial and vertebrate proteins. The C‐terminal part of teneurins, a type II integral membrane protein in vertebrates, harbours 26 YD repeats (Minet and Chiquet‐Ehrismann, 2000). The cell wall‐associated protein A (WapA) of Bacillus subtilis contains 31 Rhs repeats at the C‐terminal domain (Foster, 1993). The BC component of ABC toxins of Yersinia entomophaga is an Rhs repeat containing a protein encapsulation device (Busby et al., 2013). The broad distribution demonstrates that Rhs/YD repeat proteins play an important role in biology.
Although Rhs/YD repeats have been known for 30 years, the function of the Rhs repeat gene family is just now starting to be understood in bacteria and vertebrates. Rhs proteins from Gram‐negative bacteria and the related WapA from Gram‐positive bacteria mediate intercellular competition (Koskiniemi et al., 2013). The Rhs gene in Myxococcus xanthus is required for social motility (Youderian and Hartzell, 2007). In high metazoans, teneurins may help to establish neuronal cell connections during development (Hong et al., 2012; Mosca et al., 2012). These data indicate that Rhs/YD repeat proteins share a fundamental function in cell–cell contact and communication (Koskiniemi et al., 2013). However, the biological function of the Rhs repeat‐containing proteins in fungal plant pathogen still remains unknown.
A gene in S. sclerotiorum (GenBank accession No. EDO04920.1, SS1G_07404) is predicted to encode an eight Rhs repeat‐containing protein called Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1). The primary focus of this study involved the investigation of the biological role of Ss‐Rhs1 in S. sclerotiorum using genetic approaches. Our findings have the potential to advance our understanding of the role of Rhs repeats in fungal plant pathogens.
Results
Ss‐Rhs1 encodes a secretory Rhs repeat‐containing protein
The S. sclerotiorum Ss‐Rhs1 gene consists of eight exons and seven introns, and encodes a protein with 282 amino acids. The 20 initial N‐terminal amino acids have been predicted to encode a signal peptide with the SignalP 4.1 Server (Petersen et al., 2011). No transmembrane helices were predicted with TMHMM 2.0 (Krogh et al., 2001) or TMpred (Hofmann and Stoffel, 1993). The function of the predicted signal peptide of Ss‐Rhs1 was validated using an assay based on the requirement of yeast cells for invertase secretion to grow on raffinose medium (Gu et al., 2011; Jacobs et al., 1997). The 20 initial amino acids of Ss‐Rhs1 were fused into the invertase sequence of pSUC2 to generate pSURHS1. As shown in Fig. 1, the yeast strains transformed with pSUC2 and pSURHS1, respectively, grew on CMD‐W medium; only the strains with pSURHS1 grew on YPRAA medium (See ‘Experimental procedures’ for media constituents). These findings suggest that the signal peptide of Ss‐Rhs1 is functional and that Ss‐Rhs1 is probably a secretory protein. To test our hypothesis, a Flag‐tagged Ss‐Rhs1‐engineered S. sclerotiorum strain was obtained and cultured in liquid medium with shaking. The results of Western blot revealed that Ss‐Rhs1‐Flag could be detected in the culture filtrate (Fig. 2), which suggests that Ss‐Rhs1 is a secretory protein.
Figure 1.
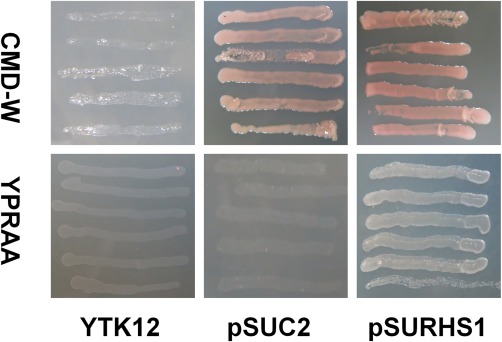
Functional validation of the predicted signal peptide of the Sclerotinia sclerotiorum Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1) protein. The 20 initial amino acids of Ss‐Rhs1 were fused into the invertase sequence of pSUC2 and the resulting vector pSURHS1 was transformed into the yeast strain YTK12. The transformed YTK12 cells were grown on CMD‐W and YPRAA media. The untransformed YTK12 strain and the YTK12 strain that transformed with pSUC2 were used as controls.
Figure 2.
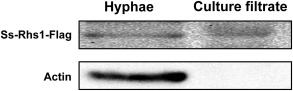
Secretion of the Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1) protein. The Ss‐Rhs1‐Flag‐engineered strain was cultured with shaking for 4 days and the proteins in the hyphae and culture filtrate were extracted. The proteins were then subjected to Western analysis using an anti‐Flag (top) or anti‐Actin (bottom) antibody.
Homologues of Ss‐Rhs1 are absent in most species of fungi and can only be found in Botryotinia fuckeliana (XP_001554189.1, E‐value: 3e‐88) and Sclerotinia borealis (ESZ96939.1, E‐value: 2e‐83); both homologues have an unknown function. A closer inspection of the sequences of Ss‐Rhs1 reveals the existence of a peptide motif that is repeated eight times (Fig. 3A). The motif is very similar with Rhs repeats, the consensus sequence of which has been defined: GXXXRYXYDXXGRL(I/T) (Wang et al., 1998) (Fig. 3B). As a result, Ss‐Rhs1 is predicted to encode a secretory Rhs repeat‐containing protein with the structure shown in Fig. 3C.
Figure 3.
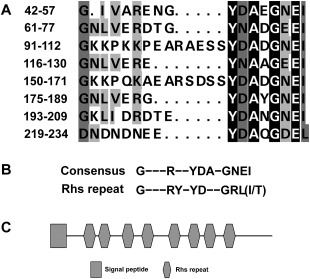
Characterization of the Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1) protein. (A) Alignment of the repeat peptide sequences of Ss‐Rhs1. (B) Comparison of the peptide repeat consensus of Ss‐Rhs1 with the Rhs repeat consensus (Wang et al., 1998). (C) Domain organization of the Ss‐Rhs1 protein.
High expression of Ss‐Rhs1 during sclerotial development and the hyphal infection process
To determine the expression levels of Ss‐Rhs1 during different stages of sclerotial development of S. sclerotiorum, a real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) approach was used. As shown in Fig. 4A, the level of Ss‐Rhs1 expression exhibited a dramatic increase at the initial stage of sclerotial development. This expression was approximately 90‐fold greater than that during hyphal growth. The expression level of Ss‐Rhs1 gradually declined as sclerotia developed and matured. These findings suggest that Ss‐Rhs1 is involved in sclerotial development. The mycelial fragments of the wild‐type strain were inoculated on the leaves of Arabidopsis thaliana or on cellophane placed on potato dextrose agar (PDA), and the expression levels of Ss‐Rhs1 were evaluated. As shown in Fig. 4B, the expression level of Ss‐Rhs1 in hyphae inoculated on plants rapidly increased (13‐fold) at 3 h post‐inoculation (hpi); the expression level was significantly higher than that in hyphae inoculated on PDA from 3 to 12 hpi. This result indicates that the expression of Ss‐Rhs1 is induced by the host plant.
Figure 4.
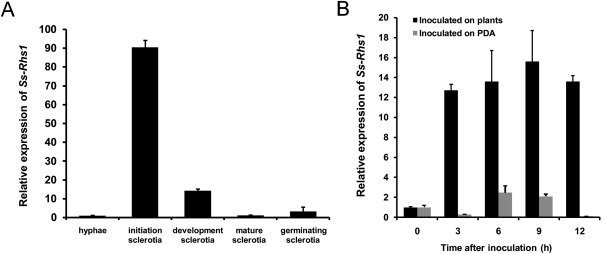
Real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of the Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1) gene transcript during different sclerotial development stages (A) and after contact with Arabidopsis thaliana (dark columns) and growing on potato dextrose agar (PDA) plates (grey columns) (B). The quantity of Ss‐Rhs1 cDNA in each sample was normalized to that of tub1 cDNA. The relative abundance of Ss‐Rhs1 cDNA in the stage of hyphal growth or in mycelium inoculated on PDA or plants at 0 h was set as unity. Bars indicate the standard error. The analyses were repeated three times. Gene expression levels in different replicates showed similar trends. One replicate is shown.
Ss‐Rhs1 gene‐silenced strains have an abnormal morphology
To study the possible roles played by Ss‐Rhs1 in S. sclerotiorum, strains with low expression of Ss‐Rhs1 were produced via RNA interference (RNAi). Two Ss‐Rhs1 coding fragments, approximately 400 bp in size, were ligated into the pCIT vector in opposite orientations between PtrpC and TtrpC to generate an Ss‐Rhs1 RNAi vector called pSIRH1 (Fig. 5A). The vector was used to transform the S. sclerotiorum wild‐type strain 1980. Real‐time RT‐PCR was then applied to examine the transcript accumulation of Ss‐Rhs1 in each transformant containing pSIRHS1. Ss‐Rhs1 expression in Sirhs‐66 and Sirhs‐93 was dramatically lower, and these strains were selected for additional analyses (Fig. 5B).
Figure 5.
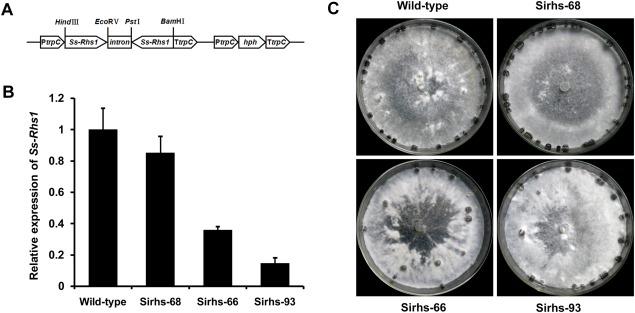
Construction of the Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1) gene RNA interference (RNAi) vector and phenotype of Ss‐Rhs1‐silenced strains. (A) Construction of the Ss‐Rhs1 RNAi vector pSIRHS1. (B) Relative expression level of Ss‐Rhs1 in different isolates containing pSIRHS1, as well as in the wild‐type strain, as determined by real‐time reverse transcription‐polymerase chain reaction (RT‐PCR). The quantity of S. sclerotiorum tub1 cDNA normalized different samples. The relative expression level of Ss‐Rhs1 in the wild‐type strain was set to unity. Bars indicate the standard error. (C) Phenotypes of the wild‐type strain, Sirhs‐66, Sirhs‐68 and Sirhs‐93. The strains were grown on PDA medium for 15 days.
Figure 5C shows the colony morphologies of the Ss‐Rhs1 gene‐silenced strains on PDA plates. The Ss‐Rhs1‐silenced strains produced fewer, but larger, sclerotia than the wild‐type strain at the late growth stage. The number and fresh weight of sclerotia produced by Sirhs‐93 were approximately 28 ± 1 and 0.24 ± 0.02 g per 9‐cm‐diameter plate; the wild‐type strains exhibited corresponding values of 20 ± 1 and 0.31 ± 0.03 g per plate. Ss‐Rhs1 did not influence the carpogenic germination of S. sclerotiorum, because the sclerotia of the Ss‐Rhs1 gene‐silenced strains produced apothecia when incubated for carpogenic germination under standard conditions. In addition to the aberrant morphology of sclerotial development, Sirhs‐93 also exhibited a slightly reduced mycelial growth rate on PDA plates (Fig. 6). The growth rate for the wild‐type strain was 2.1 cm/day, whereas, for Sirhs‐93, it was 1.8 cm/day.
Figure 6.
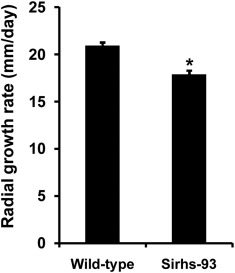
Radial growth rates of wild‐type and Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1)‐silenced strains. The experiment was repeated three times; one replicate is shown here. Bars indicate the standard error. *Significantly different from the wild‐type strain on potato dextrose agar (PDA) plates.
Ss‐Rhs1 gene‐silenced strains exhibit impaired virulence
Detached Brassica napus leaves were inoculated with agar plugs colonized with Ss‐Rhs1 gene‐silenced strains or the wild‐type strain. As shown in Fig. 7A, the lesions induced by Sirhs‐66 and Sirhs‐93 were smaller than those induced by the wild‐type strain. This test was also performed on A. thaliana, and smaller lesions were observed when Ss‐Rhs1 gene‐silenced strains were inoculated. Sirhs‐93 induced larger lesions on wounded leaves of rapeseed than on intact leaves. However, the lesions were much smaller than those caused by the wild‐type strain (Fig. 7B). These results indicate that Ss‐Rhs1 is involved in the virulence of S. sclerotiorum.
Figure 7.
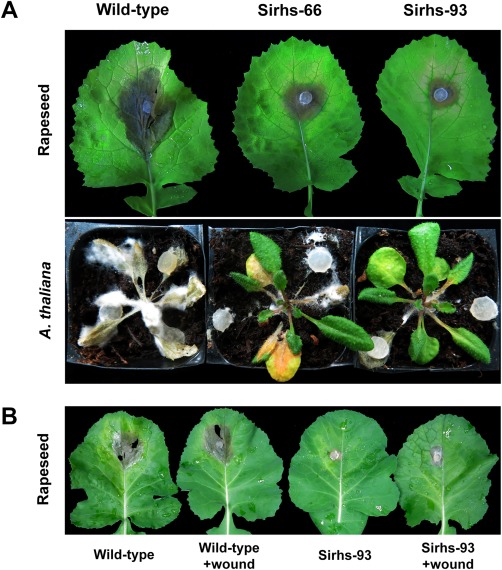
Virulence assays of Ss‐Rhs1 (Sclerotinia sclerotiorum Rearrangement hotspot repeat 1)‐silenced strains. (A) Detached leaves of rapeseed (Brassica napus) and Arabidopsis thaliana were inoculated with potato dextrose agar (PDA) plugs colonized with the wild‐type strain, Sirhs‐66 and Sirhs‐93. (B) Detached leaves of rapeseed were wounded with a dissecting needle and the wild‐type strain and Sirhs‐93 were placed over the wound. The experiment was repeated three times, and each strain was investigated with five rapeseed leaves or A. thaliana plants each time.
Ss‐Rhs1 gene‐silenced strains produce OA and CWDEs
As Ss‐Rhs1 gene‐silenced strains exhibited poor virulence on hosts, OA production and CWDE secretion by the gene‐silenced strains were evaluated. A high‐performance liquid chromatography (HPLC) assay of 3‐day‐old cultures in potato dextrose broth showed that the secreted level of OA in Sirhs‐93 (281.37 ± 11.57 mg/g of dry mycelia) was slightly lower than that secreted by the wild‐type strain (328.24 ± 13.23 mg/g). The strains were then cultured on media with various substrates to evaluate the production of CWDEs. As shown in Fig. S1 (see Supporting Information), Sirhs‐93 could secret amylases, cellulases, proteases and pectinases.
Ss‐Rhs1 gene‐silenced strains exhibit a reduced efficiency of compound appressoria differentiation
The production of compound appressoria was investigated in the wild‐type strain and Ss‐Rhs1 gene‐silenced strains. On parafilm‐overlaid growth media, Sirhs‐93 formed fewer cushion‐shaped appressoria than the wild‐type strain (Fig. 8A). On leaves of A. thaliana, the wild‐type strain showed complex and frequent appressoria at 6 hpi, whereas Sirhs‐93 rarely formed appressoria (Fig. 8B). These results suggest that Ss‐Rhs1 gene‐silenced strains are less efficient at compound appressoria differentiation.
Figure 8.
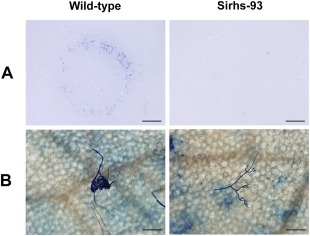
Compound appressoria formation phenotypes of wild‐type and Sirhs‐93 strain. (A) Compound appressoria formation on parafilm surrounding mycelia‐colonized agar plugs (8 h post‐inoculation, hpi). Scale bars correspond to 2 mm. (B) Compound appressoria formation on Arabidopsis thaliana leaves inoculated with mycelial plugs (6 hpi). Scale bars correspond to 100 μm.
Discussion
In this research, we characterized an Rhs repeat‐containing protein called Ss‐Rhs1 in S. sclerotiorum. Ss‐Rhs1 is a secretory protein related to sclerotial development, compound appressoria differentiation and the virulence of S. sclerotiorum. The down‐regulation of Ss‐Rhs1 leads to abnormal sclerotia and appressoria formation with poor strain virulence.
Our bioinformatics analyses showed that Ss‐Rhs1 contains eight Rhs repeats. Until recently, the function of Rhs repeat proteins in fungal pathogens has been poorly understood. Many studies have reported that Rhs repeat proteins in bacteria facilitate interactions with eukaryotic host cells. The E. coli rhsA gene is required for calf intestine colonization (van Diemen et al., 2005). The Rhs gene rhsT in Pseudomonas aeruginosa encodes a virulence determinant against mammals, and an Rhs gene in Xenorhabdus bovienii encodes a protein that is toxic to nematodes (Kung et al., 2012). In Yersinia entomophaga, the C protein of the ABC toxin contains Rhs repeats (Busby et al., 2013). Here, the expression of Ss‐Rhs1 is strongly induced via interactions with the host, and Ss‐Rhs1 gene‐silenced strains exhibit poor virulence on many hosts. To the best of our knowledge, this is the first report to show that an Rhs repeat‐containing protein is involved in fungal pathogen–host interactions.
The first 20 amino acids of Ss‐Rhs1 were predicted and functionally validated to encode a typical signal peptide, suggesting that Ss‐Rhs1 works as a secretory protein. Several secretory proteins have been confirmed to be associated with the pathogenicity of S. sclerotiorum. Zhu et al. (2013) identified an effector‐like secretory protein in S. sclerotiorum, called SSITL, which suppresses the jasmonic acid/ethylene (JA/ET) signal pathway‐mediated resistance. Another secretory protein, Ss‐Caf1, has the ability to enter host cells and trigger host cell death (Xiao et al., 2014). Interestingly, Ss‐Rhs1 is an effector candidate of S. sclerotiorum identified via bioinformatics approaches (Guyon et al., 2014). Ss‐Rhs1 contains eight Rhs repeats which are involved in the binding of carbohydrates (Krivan et al., 1986). Many effectors in fungal pathogens have been shown to contain carbohydrate‐binding domains. Cladosporium fulvum effector Ecp6 contains LysM motifs that can bind chitin oligosaccharides (Bolton et al., 2008). Ecp6 sequesters chitin oligosaccharides that are released from fungal cell walls to prevent the elicitation of the host defence response (de Jonge et al., 2010). However, the determination of the carbohydrate‐binding activity of Ss‐Rhs1 and the connection between Ss‐Rhs1 and the host defence response requires additional studies.
Many plant‐pathogenic fungi can form compound appressoria (multicellular infection structures) on the host surface (Boenisch and Schäfer, 2011; Hofman and Jongebloed, 1988; Purdy, 1958; Sharman and Heale, 1977). In S. sclerotiorum, the compound appressoria may rapidly accumulate toxins, CWDEs and defence‐suppressive factors to help penetrate into the plant tissues (Huang et al., 2008; Jamaux et al., 1995; Liang et al., 2015; Tariq and Jeffries, 1984). Until now, clues into the mechanism of compound appressoria formation in S. sclerotiorum have been rare. In this study, the efficiency of compound appressorium formation in Ss‐Rhs1 gene‐silenced strains was reduced on parafilm and leaves of A. thaliana. We suggest that Ss‐Rhs1 is associated with compound appressorium formation in S. sclerotiorum. In compound appressorium development, hyphae self‐adhere to form multicellular forms (Li et al., 2012). A very recent study has reported that a novel adhesin XadM of Xanthomonas oryzae pv. oryzae contains Rhs repeats which are essential to optimum attachment and biofilm formation (Pradhan et al., 2012). This indicates that Ss‐Rhs1 also contributes to compound appressorium formation, probably by mediating cell–cell adhesion.
Ss‐Rhs1 is related to sclerotial development in S. sclerotiorum because Ss‐Rhs1 exhibits strong expression during the sclerotial development stage; the down‐expression of Ss‐Rhs1 leads to abnormal sclerotial formation. During sclerotial development in S. sclerotiorum, hyphae adhere to form a condensed sclerotial initial body; a mucilage‐like substance is needed in this process (Erental et al., 2008). Ss‐Sl2 is an adhesive during sclerotial development and gene‐silenced strains form interwoven hyphal masses instead of mature sclerotia (Yu et al., 2012). The function of Ss‐Sl2 as an adhesive may be mediated by PAN modules that possess carbohydrate‐binding activities (Zhou et al., 1999). We hypothesized that Ss‐Rhs1 controls sclerotial development in S. sclerotiorum through the carbohydrate‐binding activity of Rhs repeats, although more evidence is needed.
In summary, Ss‐Rhs1 encodes a secretory protein that contains Rhs repeat elements and plays an important role in sclerotial development, appressorium formation and the virulence of S. sclerotiorum. Our findings suggest a conserved role of the Rhs repeat family protein in pathogenicity.
Experimental Procedures
Fungal strains and growth conditions
The wild‐type strain of S. sclerotiorum ‘1980’ was used in this study (Godoy et al., 1990). The strain was routinely maintained on PDA (Difco Laboratories, Detroit, MI, USA). The Ss‐Rhs1 gene‐silenced strains were maintained on PDA supplemented with hygromcyin B (Calbiochem, San Diego, CA, USA) at 50 μg/mL.
Sequence analysis and alignment
The Ss‐Rhs1 gene was characterized using the publicly available genomic sequence database of S. sclerotiorum (http://www.ncbi.nlm.nih.gov/bioproject/15530). The signal peptide sequence and transmembrane domain were predicted using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/), TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). The homology analysis was based on blastp searches at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence alignments were carried out using DNAMAN software (Lynnon BioSoft, Vaudreuil, QC, Canada). Conserved amino acids are shown with a shaded background.
Functional validation of the predicted signal peptide of Ss‐Rhs1
The function of the predicted signal peptide of Ss‐Rhs1 was validated as described by Gu et al. (2011). The primer pair Rhs1spfp (5′‐GCGGAATTCATGCGTTCTTCAACAGCAAGT‐3′) and Rhs1sprp (5′‐CCGCTCGAGTGCCTGAACTGCCAACATG‐3′) was designed to amplify the initial 60‐bp fragments of Ss‐Rhs1 cDNA. The fragments were digested with EcoRI and XhoI and then ligated into yeast signal trap vector pSUC2T7M13ORI (pSUC2) (Jacobs et al., 1997). The resulting vector, pSURHS1, was transformed into yeast strain YTK12 using the lithium acetate method. Transformants were grown on CMD‐W medium (0.67% yeast N base without amino acids, 0.075% tryptophan dropout supplement, 0.1% glucose, 2% sucrose and 2% agar) and then replica plated onto YPRAA medium (2% raffinose, 2% peptone, 1% yeast extract and 2 μg/mL antimycin A) to conduct an assay for invertase secretion. The YTK12 strain transformed with pSUC2 and untransformed YTK12 strain were used as negative controls.
Western blot analysis
To generate the Ss‐Rhs1‐Flag fusion construct, the primer pairs Ss‐Rhs1‐Flagfp (CCGCTCGAGATGCGTTCTTCAACAGCAAGTTTG) and Ss‐Rhs1‐Flagrp (CGGGGTACCTTATTTGTCGTCGTCGTCTTTGTAGTCGATGTTAACCAAGTTTCCATCTTTGTCG) were designed to amplify the Ss‐Rhs1 coding sequences. The fragment was digested with XhoI and KpnI and cloned into pSilent‐1 (Nakayashiki et al., 2005). The resulting construct was transformed into the wild‐type strain of S. sclerotiorum using the polyethylene glycol (PEG) method (Rollins, 2003). The positive Ss‐Rhs1‐Flag strain was cultured in potato dextrose broth (PDB) medium for 4 days. The culture broth was filtered with four layers of Calbiochem Micracloth and concentrated by ultrafiltration with Amicon Ultra‐15 (3K) (Millipore, Bedford, MA, USA). The hyphal tissues were also harvested and the proteins were extracted as described by Jurick et al. (2004). The proteins (50 μg) were separated by 12% sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to nitrocellulose membranes. The membranes were treated with 5% nonfat dry milk in TTBS (0.1% Tween‐20, 20 mM Tris, 150 mM NaCl, pH 7.5) for 2 h at room temperature, and then incubated for 2 h with DYKDDDDK Epitope Tag Monoclonal antibody (Thermo Scientific, Waltham, MA, USA) or Monoclonal Anti‐Actin antibody produced in mouse (Sigma‐Aldrich, St Louis, MO, USA). Rabbit anti‐mouse immunoglobulin G‐horseradish peroxidase (IgG‐HRP) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used as secondary antibody. The signals were detected using a SuperSignal West Femto Trial Kit (Thermo Scientific, Rockford, IL, USA) and a Molecular Image ChemiDoc XBS system (Bio‐Rad, Hercules, CA, USA).
Construction of the Ss‐Rhs1 RNAi vector and transformation of S. sclerotiorum
Plasmid vector pCIT (Yu et al., 2012) was used to construct an RNAi gene‐silencing vector. A 400‐bp Ss‐Rhs1 coding sequence was isolated via RT‐PCR using the primer pair Sirhsfp (5′‐CGCGGATCCATCGATAACTCAAGCCGCATTTATC‐3′) and Sirhsrp (5′‐CGTCTGCAGGATATCCAGGAAATCCAATTCCAAG‐3′). The amplicons were digested with BamHI and PstI, and the excised fragment was cloned into pCIT to produce pCIT‐Rhs1. The amplicons were then digested with ClaI and EcoRV, and the excised fragment was cloned into pCIT‐Rhs1 to produce pCIT‐Rhs2. The hygromycin resistance gene cassette from pSKH (Hamid et al., 2013) was isolated as an XbaI fragment and ligated into the pCIT‐Rhs2 XbaI site. The resulting RNAi construct, pSIRHS1, was transformed into the wild‐type strain of S. sclerotiorum using the PEG method.
Nucleic acid extraction and real‐time RT‐PCR analysis
The relative quantification of Ss‐Rhs1 during the different sclerotial developmental stages and infection process was performed according to the method of Yu et al. (2015). The wild‐type strain was cultured on cellophane over PDA at 20 ºC and the mycelia were harvested at the hyphal stage (2 days post‐inoculation, dpi), the initial sclerotial stage (3 dpi), the developed sclerotial stage (5 dpi) and the mature sclerotial stage (8 dpi). The mature sclerotia from the wild‐type strain were placed on the surface of moist sand at 16 ºC. The sclerotia were collected once the stipe initials appeared.
To analyse the transcript expression levels of Ss‐Rhs1 during the infection process, the wild‐type strain was cultured in PDB for 2 days. Two grams of mycelia were collected and washed in double‐distilled H2O (ddH2O) three times. The mycelia were then ground into fragments using a mortar and pestle. The hyphal fragments were transferred to minimal medium broth and grown for 8 h at 150 rpm. The fragments were then washed with ddH2O twice and resuspended in 6 mL of ddH2O. The hyphal fragments were next sprayed onto the leaves of 10 A. thaliana plants (5‐week‐old) evenly with a small spray bottle. The plants were then placed in 90% relative humidity at 20 ºC. The leaves were collected at 0, 3, 6, 9 and 12 hpi. The hyphal fragments cultured on cellophane were placed on PDA plates and used as a control.
The RNA products were extracted using Trizol reagent (TianGen, Dalian, China). DNase treatment and first‐strand cDNA synthesis were conducted using a PrimeScript™ RT reagent kit with gDNA Eraser (Takara, Tokyo, Japan). To estimate the gene expression levels in S. sclerotiorum, real‐time RT‐PCR using SYBR Green I technology on a CFX96™ Realtime System (Bio‐Rad, Hercules, CA, USA) was performed. The primer pair Rt‐Rhs1fp (5′‐CTCGCTCGGTTTCTGGTATTG‐3′) and Rt‐Rhs1rp (5′‐TGCTTCAGGTTTCTTGGGCT‐3′) was used to evaluate the expression level of the Ss‐Rhs1 gene. Ss‐Rhs1 cDNA abundance was normalized using the β‐tubulin gene tub1 (SS1G_04652) as an internal control, which was amplified with Rt‐tubfp (5′‐GTGAGGCTGAGGGCTGTGA‐3′) and Rt‐tubrp (5′‐CCTTTGGCGATGGGACG‐3′). The amplification mixtures were composed of a 4 pmol concentration of each primer, 10 μL of SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan), 1 μL of cDNA and nuclease‐free water to a final volume of 20 μL. The amplification program was set as: 95 ºC for 2 min (one cycle), followed by 95 ºC for 20 s, 57 ºC for 15 s and 72 ºC for 20 s (40 cycles). Each sample was analysed in three biological replications, and the average cycle threshold was calculated to evaluate the relative expression. Each experiment was repeated three times.
Pathogenicity assays
A. thaliana Columbia‐0 and B. napus Zhongshuang 9 were used to assay the pathogenicity of S. sclerotiorum strains. A. thaliana was grown in a growth chamber at 25 ± 1 ºC in a 12‐h light/12‐h dark cycle for 5 weeks. B. napus was grown in a glasshouse at 20–30 ºC for approximately 10 weeks. The leaves of A. thaliana or B. napus were inoculated with 0.6‐cm mycelium‐colonized agar plugs obtained from actively growing colony edges. The inoculated plants and leaves were grown in 90% relative humidity at 20 ºC. Photographs were taken at 72 hpi for rapeseed leaves and at 96 hpi for A. thaliana. To assay the pathogenicity of strains on wounded leaves, detached leaves of rapeseed were wounded with a dissecting needle and the wild‐type strain and Sirhs‐93 were placed directly over the wound. Intact leaves inoculated with the strains were used as controls. Photographs were taken at 60 hpi. Each strain was evaluated with five leaves or plants three times.
OA assays
To evaluate the secreted level of OA in Sirhs‐93 and the wild‐type strain, three mycelium plugs (9 mm in diameter) of each strain were cultured in 50 mL of PDB for 3 days with shaking at 150 rpm. The concentration of OA was determined using HPLC (Zhang et al., 2010). The yield was expressed as milligrams of OA per gram of dry mycelia. The experiment was repeated three times.
Assays of amylases, cellulases, proteases and pectinases
The production of amylases, cellulases, proteases and pectinases for the wild‐type strain and Sirhs‐93 was analysed as described by Zhang et al. (2010) and Xiao et al. (2014). To detect the production of amylases, the strains were cultured on medium containing 0.15% amylum for 2 days. The medium was stained with Gram's iodine solution (2% iodine and 3% potassium iodide in 70% ethanol). A clear zone of the colony on a blue background indicated the amylase activity of each strain. To determine the production of cellulases, the strains were cultured on medium with 0.1% carboxymethyl cellulose for 2 days. The plates were stained with 5% Congo red and then washed with 1 m NaCl. An orange zone of the colony indicated cellulase activity. To assay the production of proteases, strains were cultured on medium with 1% gelatin for 4 days, and the plates were then stained with 15% HgCl2. Protease activity was indicated by a clear zone of the colony. To evaluate the production of pectinases, mycelial plugs were removed from the colony margin of a 7‐day‐old PDA culture of strains and placed on medium containing 0.1% polygalacturonic acid. After incubation at 45 ºC for 24 h, the mycelial plugs were removed from the medium. The medium was then stained with 0.03% ruthenium red at 4 ºC for 2 h. Pectinase activity was determined by the presence of clear zones under the area occupied by the mycelial plugs. Each assay was repeated three times.
Appressoria formation assays
To assay the appressoria formation of the wild‐type strain and Ss‐Rhs1 gene‐silenced strains, mycelial plugs were inoculated on parafilm‐overlaid PDA plates and leaves of A. thaliana Columbia‐0. For the inoculation parafilm, the plugs were removed at 8 hpi, and 5% trypan blue was added to the parafilm surface to stain the appressoria. For the inoculation leaves, the plugs were removed at 6 hpi. The leaves were cleared with 3 : 1 ethanol–acetic acid solution for 12 h and then stained with 5% trypan blue for 12 h.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Evaluation of cellulases, amylases, proteases and pectinases produced by the wild‐type strain and Sirhs‐93. Enzyme activity was visualized as a clear zone on the medium.
Acknowledgements
This research was supported by National Natural Science Foundation of China (31301612, 31671973), Fundamental Research Funds for the Central Universities, China (XDJK2013C150, XDJK2013A013), Science & Technology Innovation Funds of Shizhu Base of Southwest University (Sz201314). We thank anonymous reviewers for their kind suggestions.
References
- Boenisch, M.J. and Schäfer, W. (2011) Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 11, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Can. J. Plant Pathol. 16, 93–108. [Google Scholar]
- Bolton, M.D. , van Esse, H.P. , Vossen, J.H. , de Jonge, R. , Stergiopoulos, I. , Stulemeijer, I.J. , van den Berg, G.C. , Borrás‐Hidalgo, O. , Dekker, H.L. , de Koster, C.G. , de Wit, P.J. , Joosten, M.H. and Thomma, B.P. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69, 119–136. [DOI] [PubMed] [Google Scholar]
- Busby, J.N. , Panjikar, S. , Landsberg, M.J. , Hurst, M.R. and Lott, J.S. (2013) The BC component of ABC toxins is an RHS‐repeat‐containing protein encapsulation device. Nature, 501, 547–550. [DOI] [PubMed] [Google Scholar]
- Cessna, S.G. , Sears, V.E. , Dickman, M.B. and Low, P.S. (2000) Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell, 12, 2192–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. and Dickman, M.B. (2005) cAMP blocks MAPK activation and sclerotial development via Rap‐1 in a PKA‐independent manner in Sclerotinia sclerotiorum . Mol. Microbiol. 55, 299–311. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Harel, A. , Gorovoits, R. , Yarden, O. and Dickman, M.B. (2004) MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and cAMP sensing. Mol. Plant–Microbe. Interact. 17, 404–413. [DOI] [PubMed] [Google Scholar]
- Chet, I. and Henis, Y. (1975) Sclerotial morphogenesis in fungi. Annu. Rev. Phytopathol. 13, 169–192. [Google Scholar]
- van Diemen, P.M. , Dziva, F. , Stevens, M.P. and Wallis, T.S. (2005) Identification of enterohemorrhagic Escherichia coli O26: H‐genes required for intestinal colonization in calves. Infect. Immun. 73, 1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erental, A. , Harel, A. and Yarden, O. (2007) Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum . Mol. Plant–Microbe. Interact. 20, 944–954. [DOI] [PubMed] [Google Scholar]
- Erental, A. , Dickman, M.B. and Yarden, O. (2008) Sclerotial development in Sclerotinia sclerotiorum: awakening molecular analysis of a “Dormant” structure. Fungal Biol. Rev. 22, 6–16. [Google Scholar]
- Favaron, F. , Sella, L. and D'Ovidio, R. (2004) Relationships among endo‐polygalacturonase, oxalate, pH, and plant polygalacturonase‐inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant–Microbe. Interact. 17, 1402–1409. [DOI] [PubMed] [Google Scholar]
- Foster, S.J. (1993) Molecular analysis of three major wall‐associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two‐domain ligand‐binding protein. Mol . Microbiol. 8, 299–310. [DOI] [PubMed] [Google Scholar]
- Godoy, G. , Steadman, J.R. , Dickman, M.B. and Dam, R. (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris . Physiol. Mol. Plant Pathol. 37, 179–191. [Google Scholar]
- Gu, B. , Kale, S.D. , Wang, Q. , Wang, D. , Pan, Q. , Cao, H. , Meng, Y. , Kang, Z. , Tyler, B.M. and Shan, W. (2011) Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR‐like motif sufficient for translocation into plant cells. PLoS One, 6, e27217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon, K. , Balagué, C. , Roby, D. and Raffaole, S. (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum . BMC Genomics, 15, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid, M.I. , Zeng, F. , Cheng, J. , Jiang, D. and Fu, Y. (2013) Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans . Fungal Genet. Biol. 53, 42–49. [DOI] [PubMed] [Google Scholar]
- Harel, A. , Gorovits, R. and Yarden, O. (2005) Changes in protein kinase A activity accompany sclerotial development in Sclerotinia sclerotiorum . Phytopathology, 95, 397–404. [DOI] [PubMed] [Google Scholar]
- Hill, C.W. , Sandt, C.H. and Vlazny, D.A. (1994) Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12, 865–871. [DOI] [PubMed] [Google Scholar]
- Hofman, T.W. and Jongebloed, P.H. (1988) Infection process of Rhizoctonia solani on Solanum tuberosum and effects of granular nematicides. Neth. J. Plant Pathol. 94, 243–252. [Google Scholar]
- Hofmann, K. and Stoffel, W. (1993) TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe‐Seyler, 374, 166. [Google Scholar]
- Hong, W. , Mosca, T.J. and Luo, L. (2012) Teneurins instruct synaptic partner matching in an olfactory map. Nature, 484, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Buchenauer, H. , Han, Q. , Zhang, X. and Kang, Z. (2008) Ultrastructural and cytochemical studies on the infection process of Sclerotinia sclerotiorum in oilseed rape. J. Plant Dis. Protect. 115, 9. [Google Scholar]
- Jacobs, K.A. , Collins‐Racie, L.A. , Colbert, M. , Duckett, M. , Golden‐Fleet, M. , Kelleher, K. , Kriz, R. , LaVallie, E.R. , Merberg, D. , Spaulding, V. , Stover, J. , Williamson, M.J. and McCoy, J.M. (1997) A genetic selection for isolating cDNAs encoding secreted proteins. Gene, 198, 289–296. [DOI] [PubMed] [Google Scholar]
- Jamaux, I. , Gelie, B. and Lamarque, C. (1995) Early stages of infection of rapeseed petals and leaves by Sclerotinia sclerotiorum revealed by scanning electron microscopy. Plant Pathol. 44, 22–30. [Google Scholar]
- de Jonge, R. , van Esse, H.P. , Kombrink, A. , Shinya, T. , Desaki, Y. , Bours, R. , van der Krol, S. , Shibuya, N. , Joosten, M.H. and Thomma, B.P. (2010) Conserved fungal LysM effector Ecp6 prevents chitin‐triggered immunity in plants. Science, 329, 953–955. [DOI] [PubMed] [Google Scholar]
- Jurick, W.M. , Dickman, M.B. and Rollins, J.A. (2004) Characterization and functional analysis of a cAMP‐dependent protein kinase. A catalytic subunit gene (pka1) in Sclerotinia sclerotiorum . Physiol. Mol. Plant Pathol. 64, 155–163. [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbage, M. , Yarden, O. and Dickman, M.B. (2015) Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. [DOI] [PubMed] [Google Scholar]
- Kim, K.S. , Min, J.Y. and Dickman, M.B. (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant–Microbe. Interact. 21, 605–612. [DOI] [PubMed] [Google Scholar]
- Koskiniemi, S. , Lamoureux, J.G. , Nikolakakis, K.C. , de Roodenbeke, C. , Kaplan, M.D. , Low, D.A. and Hayes, C.S. (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. USA, 110, 7032–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan, H.C. , Clark, G.F. , Smith, D.F. and Wilkins, T.D. (1986) Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Galα1‐3Galβ1‐4GlcNAc. Infect. Immun. 53, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A. , Larsson, B. , Von Heijne, G. and Sonnhammer, E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kung, V.L. , Khare, S. , Stehlik, C. , Bacon, E.M. , Hughes, A.J. and Hauser, A.R. (2012) An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc. Natl. Acad. Sci. USA, 109, 1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. , Liang, X. and Rollins, J.A. (2012) Sclerotinia sclerotiorum γ‐glutamyl transpeptidase (Ss‐Ggt1) is required for regulating glutathione accumulation and development of sclerotia and compound appressoria. Mol. Plant–Microbe. Interact. 25, 412–420. [DOI] [PubMed] [Google Scholar]
- Liang, X. , Moomaw, E.W. and Rollins, J.A. (2015) Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16, 825–836. doi: 10.1111/mpp.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R.J. , Capage, M. and Hill, C.W. (1984) A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K‐12 chromosome. J. Mol. Biol. 177, 1–18. [DOI] [PubMed] [Google Scholar]
- Martel, M.B. , Létoublon, R. and Fěvre, M. (1996) Purification of endo polygalacturonases from Sclerotinia sclerotiorum: multiplicity of the complex enzyme system. Curr. Microbiol. 33, 243–248. [DOI] [PubMed] [Google Scholar]
- Minet, A.D. and Chiquet‐Ehrismann, R. (2000) Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli . Gene, 257, 87–97. [DOI] [PubMed] [Google Scholar]
- Mosca, T.J. , Hong, W. , Dani, V.S. , Favaloro, V. and Luo, L. (2012) Trans‐synaptic teneurin signalling in neuromuscular synapse organization and target choice. Nature, 484, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Quoc, N.B. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42, 275–283. [DOI] [PubMed] [Google Scholar]
- Petersen, T.N. , Brunak, S. , von Heijne, G. and Nielsen, H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods, 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Poussereau, N. , Gente, S. , Rascle, C. , Billon‐Grand, G. and Fèvre, M. (2001) aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol. Lett. 194, 27–32. [DOI] [PubMed] [Google Scholar]
- Pradhan, B.B. , Ranjan, M. and Chatterjee, S. (2012) XadM, a novel adhesin of Xanthomonas oryzae pv. oryzae, exhibits similarity to Rhs family proteins and is required for optimum attachment, biofilm formation, and virulence. Mol. Plant–Microbe. Interact. 25, 1157–1170. [DOI] [PubMed] [Google Scholar]
- Purdy, L.H. (1958) Some factors affecting penetration and infection by Sclerotinia sclerotiorum . Phytopathology, 48, 605–609. [Google Scholar]
- Riou, C. , Freyssinet, G. and Fevre, M. (1991) Production of cell wall‐degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum . Appl. Environ. Microb. 57, 1478–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins, J.A. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant–Microbe. Interact. 16, 785–795. [DOI] [PubMed] [Google Scholar]
- Sharman, S. and Heale, J.B. (1977) Penetration of carrot roots by the grey mould fungus Botrytis cinerea Pers. ex Pers. Physiol. Plant Pathol. 10, 63–71. [Google Scholar]
- Steadman, J.R. (1979) Control of plant diseases caused by Sclerotinia species. Phytopathology, 69, 904–907. [Google Scholar]
- Tariq, V.N. and Jeffries, P. (1984) Appressorium formation by Sclerotinia sclerotiorum: scanning electron microscopy. Trans. Br. Mycol. Soc. 82, 645–651. [Google Scholar]
- Wang, Y.D. , Zhao, S. and Hill, C.W. (1998) Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli . J. Bacteriol. 180, 4102–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts, H.J. and Bullock, S. (1992) Developmental biology of sclerotia. Mycol. Res. 96, 801–816. [Google Scholar]
- Willetts, H.J. and Wong, J.A. (1980) The biology of Sclerotinia sclerotiorum, Sclerotinia trifoliorum, and Sclerotinia minor with emphasis on specific nomenclature. Bot. Rev. 46, 101–165. [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X. , Xie, J. , Cheng, J. , Li, G. , Yi, X. , Jiang, D. and Fu, Y. (2014) Novel secretory protein Ss‐Caf1 of the plant‐pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant–Microbe. Interact. 27, 40–55. [DOI] [PubMed] [Google Scholar]
- Yajima, W. , Liang, Y. and Kav, N.N. (2009) Gene disruption of an arabinofuranosidase/β‐xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Mol. Plant–Microbe. Interact. 22, 783–789. [DOI] [PubMed] [Google Scholar]
- Youderian, P. and Hartzell, P.L. (2007) Triple mutants uncover three new genes required for social motility in Myxococcus xanthus . Genetics, 177, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Jiang, D. , Xie, J. , Cheng, J. , Li, G. , Yi, X. and Fu, Y. (2012) Ss‐Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum . PLoS One, 7, e34962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Xiao, J. , Yang, Y. , Bi, C. , Qing, L. and Tan, W. (2015) Ss‐Bi1 encodes a putative BAX inhibitor‐1 protein that is required for full virulence of Sclerotinia sclerotiorum . Physiol. Mol. Plant Pathol. 90, 115–122. [Google Scholar]
- Zhang, L. , Wu, M. , Li, G. , Jiang, D. and Huang, H. (2010) Effect of mitovirus infection on formation of infection cushions and virulence of Botrytis cinerea . Physiol. Mol. Plant Pathol. 75, 71–80. [Google Scholar]
- Zhou, H. , Casas‐Finet, J.R. , Heath, Coats, R. , Kaufman, J.D. , Stahl, S.J. , Wingfield, P.T. , Rubin, J.S. , Bottaro, D.P. and Byrd, R.A. (1999) Identification and dynamics of a heparin‐binding site in hepatocyte growth factor. Biochemistry, 38, 14 793–14 802. [DOI] [PubMed] [Google Scholar]
- Zhu, W. , Wei, W. , Fu, Y. , Chen, J. , Xie, J. , Li, G. , Yi, X. , Kang, Z. , Dickman, M.B. and Jiang, D. (2013) A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS One, 8, e53901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuppini, A. , Navazio, L. , Sella, L. , Castiglioni, C. , Favaron, F. and Mariani, P. (2005) An endopolygalacturonase from Sclerotinia sclerotiorum induces calcium‐mediated signaling and programmed cell death in soybean cells. Mol. Plant–Microbe. Interact. 18, 849–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Evaluation of cellulases, amylases, proteases and pectinases produced by the wild‐type strain and Sirhs‐93. Enzyme activity was visualized as a clear zone on the medium.


