Summary
The lateral organ boundary domain (LBD) genes encode a group of plant‐specific proteins that function as transcription factors in the regulation of plant growth and development. Citrus sinensis lateral organ boundary 1 (CsLOB1) is a member of the LBD family and functions as a disease susceptibility gene in citrus bacterial canker (CBC). Thirty‐four LBD members have been identified from the Citrus sinensis genome. We assessed the potential for additional members of LBD genes in citrus to function as surrogates for CsLOB1 in CBC, and compared host gene expression on induction of different LBD genes. Using custom‐designed transcription activator‐like (TAL) effectors, two members of the same clade as CsLOB1, named CsLOB2 and CsLOB3, were found to be capable of functioning similarly to CsLOB1 in CBC. RNA sequencing and quantitative reverse transcription‐polymerase chain reaction analyses revealed a set of cell wall metabolic genes that are associated with CsLOB1, CsLOB2 and CsLOB3 expression and may represent downstream genes involved in CBC.
Keywords: citrus bacterial canker, lateral organ boundary domain (LBD) genes, susceptibility (S) genes
Introduction
The lateral organ boundary domain (LBD) genes encode a group of plant‐specific proteins that function as transcription factors in the regulation of plant growth and development (Husbands et al., 2007; Majer and Hochholdinger, 2011). All members contain a domain referred to as the lateral organ boundary (LOB) domain, which includes a cysteine‐rich motif (CX2CX6CX3C) that has been shown, in some cases, to mediate DNA binding (Husbands et al., 2007; Majer and Hochholdinger, 2011). The proteins also contain a conserved glycine residue and a coiled‐coil leucine zipper‐like motif (LX6LX3LX6L). Citrus sinensis lateral organ boundary 1 (CsLOB1) is a member of the LBD gene family and functions as a disease susceptibility (S) gene in citrus bacterial canker (CBC), a disease that afflicts most citrus varieties worldwide (Hu et al., 2014). The typical symptoms are erumpent water‐soaked lesions with yellow haloes on leaves, stems and fruits. Defoliation and premature fruit drop may occur if disease is severe (Gottwald et al., 2002). The causal pathogens are various species of the genus Xanthomonas. Five bacterial strain groups (group A, group A*, group Aw, group B and group C) have been identified. Group A, A* and Aw are strains of the species of Xanthomonas citri ssp. citri (Xcc), which originated from Asia, whereas groups B and C are strains of the species X. fuscans ssp. aurantifolii (Xau) (Al‐Saadi et al., 2007; Sun et al., 2004; Verniere et al., 1998). Pustule formation, regardless of the inciting strain, requires CsLOB1 expression, which is also associated with increases in bacterial populations at the site of infection (Hu et al., 2014; Pereira et al., 2014).
How CsLOB1 expression facilitates pustule formation and whether CsLOB1‐related genes can function in place of CsLOB1 are unknown. Forty‐three LBD genes have been identified in Arabidopsis (Shuai et al., 2002). The founding member of the LBD family in Arabidopsis, AtLOB, is expressed at the base of lateral organs and is involved in leaf development. AtLOB functions as a transcription activator and interacts with members of the basic helix–loop–helix (bHLH) family of transcription factors (Husbands et al., 2007; Shuai et al., 2002). Other LBD proteins have been demonstrated to regulate anthocyanin and nitrogen metabolism and to respond to phytohormones and environmental stimuli during the process of organ boundary formation (Gendron et al., 2012; Majer and Hochholdinger, 2011). An LBD gene from apple (Malus domestica B.), MdLBD11, has a similar function to the Arabidopsis gene AtAS2/AtLBD12 in terms of its role in leaf development and flowering (Wang et al., 2013). The expression of the Arabidopsis gene LBD20 is induced on infection by the pathogen Fusarium oxysporum, and silencing of the LBD20 results in increased resistance to the pathogen (Thatcher et al., 2012).
CsLOB1 induction early in the disease process is mediated by one of the six major transcription activator‐like (TAL) effectors from strains of Xcc or Xau (Hu et al., 2014; Pereira et al., 2014). PthA was the first member of the TAL effector family demonstrated to function in pathogenicity and shown to be required for pustule formation (Swarup et al., 1992). Additional pthA homologous genes have been identified from other strains of Xanthomonas and include pthA4, pthAw, pthA*, pthB and pthC (Al‐Saadi et al., 2007; Jalan et al., 2013; Sun et al., 2004; Verniere et al., 1998). All of the major TAL effectors target CsLOB1 (Hu et al., 2014; Pereira et al., 2014). TAL effectors are members of the type III secretion (T3S) effectors primarily identified in Xanthomonas species and Ralstonia solanacearum (De Feyter et al., 1993; Heuer et al., 2007; Hopkins et al., 1992; Mak et al., 2013). TAL effectors, on entry into host cells, move to the cell nuclei and bind to the specific promoter regions, designated here as effector‐binding elements (EBEs), of host genes. The expression of a variety of genes that are TAL effector‐dependent leads to either enhanced susceptibility or host resistance, or, in some cases, no clear phenotypic effect on the plant (Boch et al., 2014; Zhang et al., 2015). The cognate EBE is determined by the TAL effector central repetitive region. The amino acid sequence of the repeats is largely conserved with the exception of residues 12 and 13, which are polymorphic and referred to as repeat‐variable diresidues (RVDs) (Boch et al., 2009, 2014; Moscou and Bogdanove, 2009).
Artificially designed TAL effector (dTALe) genes can be produced by replacing the repetitive region with coding segments that target new binding sites of interest (Li et al., 2013; Morbitzer et al., 2010; Streubel et al., 2013). The targeting of novel promoter elements by dTALes has been proven to be useful for the discrimination between S genes and phenotypically silent or off‐target genes that also contain native EBEs (Cernadas et al., 2014; Cohn et al., 2014; Hu et al., 2014). For example, PthA4 and PthAw both induce CsLOB1 and a second gene CsSWEET1 (Hu et al., 2014). The construction of dTALes that targeted either gene revealed that only the induction of CsLOB1 contributed to pustule formation and enhanced population levels of the pathogen (Hu et al., 2014; Pereira et al., 2014). Furthermore, dTALes facilitate the identification of S gene surrogates, especially for the paralogues of the identified S genes. TAL effectors of field strains of X. oryzae pv. oryzae are known to target any one of three SWEET genes (OsSWEET11, OsSWEET13, OsSWEET14) encoding closely related members of clade III of sucrose transporters in rice (Antony et al., 2010; Yang et al., 2006; Zhou et al., 2015). The use of dTALes allowed the targeting of an additional 19 OsSWEET genes. However, only two additional members (OsSWEET12 and OsSWEET15) from clade III could induce the typical disease symptom of water‐soaked lesions, revealing that only members of clade III can function as S genes (Streubel et al., 2013). Here, we assessed the potential for additional members of the LBD genes in citrus to function as surrogates for CsLOB1 in CBC, and compared host gene expression on induction of different LBD genes.
Results
Phylogeny of citrus LBD gene family
Thirty‐four members in the C. sinensis LBD family were identified in the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/family.php?sp=Csi&fam=LBD) (File S1, see Supporting Information). The three citrus genes most closely related to CsLOB1 based on the C. sinensis proteome are orange1.1g047491m, orange1.1g045080m and orange1.1g036782m according to their blast scores. They share 71.0%, 67.9% and 71.9% identities to CsLOB1, respectively. None has been characterized functionally, with the exception of the function of CsLOB1 (orange1.1g026556m) in CBC (Hu et al., 2014). The constructed phylogenetic tree was divided into nine clades. The clade with CsLOB1, designated as clade I, harbours four additional members—orange1.1g047491m, orange1.1g045080m, orange1.1g047530m and orange1.1g047804m (Fig. 1). Two of the four clade I LBD proteins (orange1.1g045080m and orange1.1g047491m) and one (orange1.1g036782m) from clade III were chosen for dTALe targeting (Fig. 1, red arrows). Hereafter, the three LBD genes (orange1.1g045080m, orange1.1g047491m and orange1.1g036782m) are designated as CsLOB2, CsLOB3 and CsLOB4, respectively.
Figure 1.
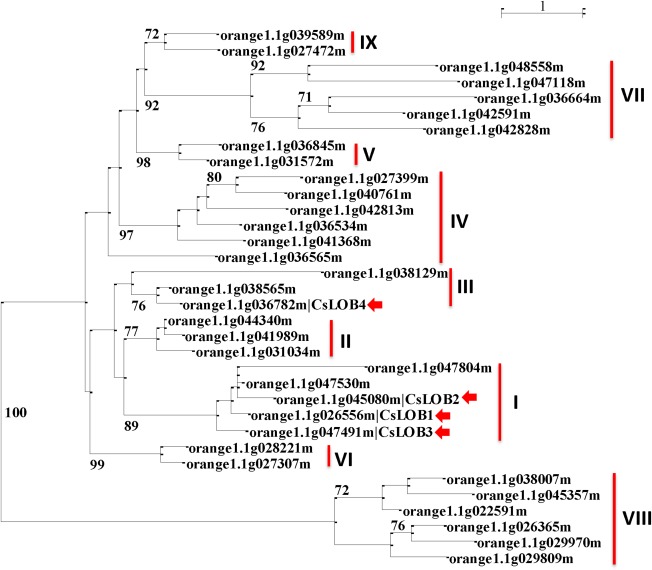
Phylogenetic tree of Citrus sinensis lateral organ boundary domain (LBD) genes. Values with a cutoff of 70 for the bootstrap support (500 replicates) are shown on the shoulders of the branches. The scale bar indicates substitutions/site, and vertical lines are used to represent different clades of C. sinensis LBD proteins.
Pustule symptoms are restored by dTALes targeting CsLOB2 and CsLOB3, but not CsLOB4
A set of dTALes was designed to target the promoters of CsLOB2, CsLOB3 and CsLOB4, which were designated as dCsLOB2, dCsLOB2L, dCsLOB3 and dCsLOB4, respectively (Fig. S1, see Supporting Information; Table 1). The functionality of each dTALe was tested by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of mRNA from grapefruit leaves after inoculation with a pthA4 mutant strain of Xcc‐306 (Xcc306ΔpthA4) and the same strain complemented by introduction of the individual dTALe genes (dCsLOB2, dCsLOB2L, dCsLOB3 and dCsLOB4). Compared with Xcc306ΔpthA4, the complemented strains induced higher fold changes for the respective gene targets (Figs 2a and S6b, see Supporting Information).
Figure 2.
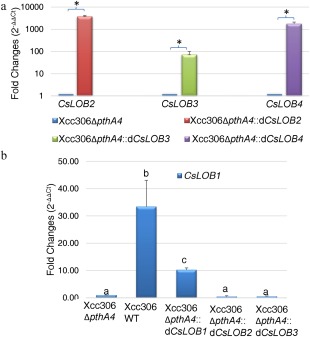
The targeted lateral organ boundary domain (LBD) genes are induced by the respective designed transcription activator‐like effectors (dTALes). (a) Validation of targeted gene induction by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). Asterisks denote significant differences in the fold changes between the treatments relative to the negative control Xcc306ΔpthA4 at P < 0.05 using a t‐test. (b) CsLOB1 is not induced by the dTALes targeting CsLOB2 or CsLOB3. Different letters indicate significant differences in fold changes among the treatments at P < 0.05 using analysis of variance (ANOVA). qRT‐PCR was conducted on mRNA extracted from grapefruit leaves inoculated with the individual Xanthomonas citri ssp. citri (Xcc) strains as indicated using LBD gene‐specific primers at 5 days post‐inoculation (dpi). Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard deviations for three independent experimental replicates. Samples were taken at 5 dpi.
The contribution of the respective strains harbouring dTALes for the respective targeted LBD genes to disease symptom development was tested on grapefruit leaves. At 15 days post‐inoculation (dpi), the inoculation sites treated with bacterial suspensions of Xcc306WT, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB2L or Xcc306ΔpthA4::dCsLOB3 showed pustule formation (Fig. 3a,b, panels 1–3, and Fig. S6a, panel 1, respectively), whereas the sites treated with bacterial suspensions of either Xcc306ΔpthA4 or Xcc306ΔpthA4::dCsLOB4 only showed water‐soaked lesions (Fig. 3b, panels 4 and 5). To determine whether induction of an alternative LBD member is also associated with an increase in the bacterial population on citrus leaves, bacterial population assays were conducted with strains of Xcc306WT (with pthA4), 306ΔpthA4 and Xcc306ΔpthA4::dCsLOB2. Indeed, Xcc306ΔpthA4::dCsLOB2 achieved higher populations than 306ΔpthA4 and was comparable with strain Xcc306WT after 3 days on host plants (Fig. 4).
Figure 3.
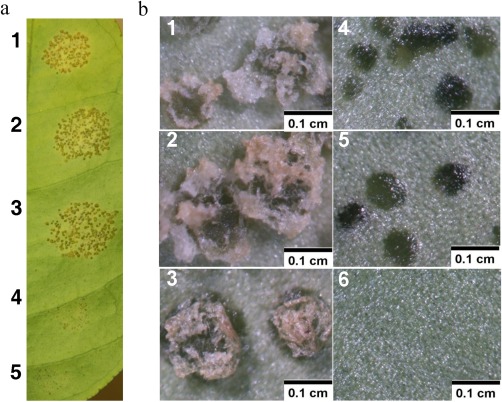
Disease symptoms induced by designed transcription activator‐like effectors (dTALes) targeting the three CsLOB1‐related genes. (a) Light photography at 1×; (b) 6.4× magnification. Panels and inoculation sites: 1, Xcc306WT; 2, Xcc306ΔpthA4::dCsLOB2; 3, Xcc306ΔpthA4::dCsLOB3; 4, Xcc306ΔpthA4::dCsLOB4; 5, Xcc306ΔpthA4; 6, healthy leaf. Photographs were taken at 15 days post‐inoculation (dpi) of various strains as shown in the figures. Inocula were 103 times dilutions of bacterial suspensions with an optical density at 600 nm (OD600) of 0.5.
Figure 4.
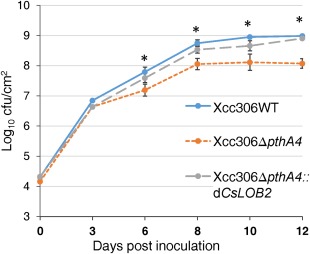
dCsLOB2 promoted an increase in the bacterial population in host plants. Bacterial populations were measured for three strains (Xcc306ΔpthA4, Xcc306WT and Xcc306ΔpthA4::dCsLOB2) on grapefruit. The x‐axis shows the days post‐inoculation. Asterisks indicate significant difference (P < 0.01) of two comparisons: Xcc306WT and Xcc306ΔpthA4::dCsLOB2 vs. Xcc306ΔpthA4. Statistical significance of the analysis was tested using t‐test. cfu, colony‐forming unit.
To exclude the possibility that pustule formation or pathogen growth conferred by CsLOB2 or CsLOB3 was caused by the coincidental expression of CsLOB1, the induction of CsLOB1 in the presence of dCsLOB2 or dCsLOB3 was also assayed. No significant induction of CsLOB1 by Xcc306ΔpthA4::dCsLOB2 or Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4 was observed (Fig. 2b).
S gene expression does not contribute to enhanced virulence of a non‐host pathogen
Xanthomonas campestris pv. vesicatoria (Xcv) Xcv85‐10, a pathogen of pepper, was used as a non‐host strain to determine whether a TAL effector gene could alter the virulence of Xcv85‐10 on a non‐host. Xcv85‐10 normally induces light water soaking on citrus (Fig. 5a). CsLOB2 was highly induced by Xcv85‐10::dCsLOB2 when inoculated on grapefruit leaves compared with the control Xcv85‐10::pHM1 (empty vector) as determined by qRT‐PCR (Fig. S2, see Supporting Information). Pustule‐like symptoms were observed at the inoculation sites with Xcv85‐10::dCsLOB2 (Fig. 5a, panels 4 and 6). However, Xcv85‐10, which grows to relatively low population levels compared with Xcc306WT, did not grow to higher populations in the presence vs. absence of dCsLOB2 (Fig. 5b).
Figure 5.
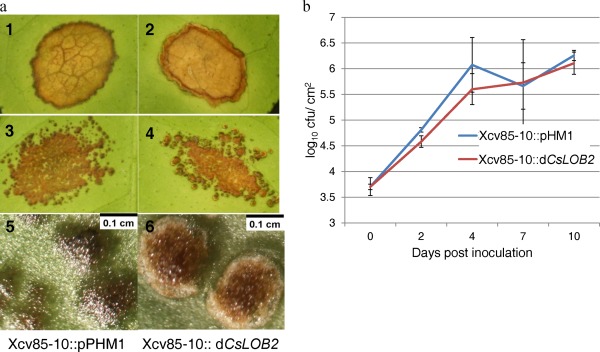
S gene expression does not contribute to enhanced virulence of a non‐host pathogen. (a) Pustule‐like symptoms were observed when DCsLOB2 was transformed into Xanthomonas euvesicatoria Xcv85‐10. Photographs were taken at 30 days post‐inoculation (dpi) of grapefruit leaves with Xcv85‐10::pHM1 or Xcv85‐10::dCsLOB2 at 1× (1–4) or 6.4× (5, 6) magnification. Inoculum with an optical density (OD600) of 0.5 (1,2) or the same inoculum diluted 103 times (3–6). (b) Bacterial population assay of the two strains Xcv85‐10::pHM1 and Xcv85‐10::dCSLOB2 on grapefruit. t‐test (P < 0.05) showed no significant difference between the two treatments. cfu, colony‐forming unit.
Comparison of genes induced in association with CsLOB1, CsLOB2, CsLOB3 or CsLOB4 expression
CsLOB1, CsLOB2 and CsLOB3 might induce CBC development by the regulation of common downstream genes and pathways. RNA sequencing (RNA‐Seq) was performed to investigate the transcriptional profile of infected tissues with strains of the pathogen that induce any one of the genes CsLOB1, CsLOB2, CsLOB3 or CsLOB4. Grapefruit leaves were separately inoculated with Xcc306ΔpthA4 (negative control), Xcc306WT, which harbours pthA4 and induces CsLOB1, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB3 and Xcc306ΔpthA4::dCsLOB4 (data uploaded at Sequence Read Archive with accession number SRP066665). Comparisons of gene expression were conducted between Xcc306ΔpthA4 and each of Xcc306WT, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB3 and Xcc306ΔpthA4::dCsLOB4, resulting in 3748, 3892, 4749 and 466 significantly differentially expressed genes (DEGs), respectively, with a control of 5% false discovery rate (FDR) for each comparison. Furthermore, 1797 common DEGs were identified, with 737 up‐regulated and 977 down‐regulated, in all three comparisons: Xcc306WT vs. Xcc306ΔpthA4, Xcc306ΔpthA4::dCsLOB2 vs. Xcc306ΔpthA4 and Xcc306ΔpthA4::dCsLOB3 vs. Xcc306ΔpthA4. Among the 1797 genes, 163 genes were significantly up‐ or down‐regulated in all four comparisons. Thus, 1634 genes were associated with the expression of CsLOB1, CsLOB2 and CsLOB3, but not with the expression of CsLOB4 (Fig. 6).
Figure 6.
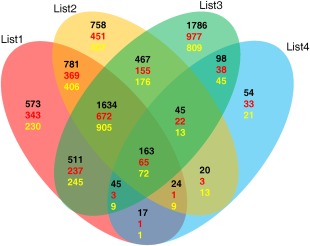
RNA‐sequencing (RNA‐Seq) results of the gene expression profile induced by CsLOB1, CsLOB2, CsLOB3 or CsLOB4. Venn diagrams show the number of genes that were significantly differentially expressed in each comparison (List1, Xcc306WT vs. 306ΔpthA4; List2, 306ΔpthA4::dCsLOB2 vs. 306ΔpthA4; List3, 306ΔpthA4::dCsLOB3 vs. 306ΔpthA4; List4, 306ΔpthA4::dCsLOB4 vs. 306ΔpthA4). The comparisons are represented by blue, yellow, green or red ovals. Genes that show significant differences in more than one comparison are plotted in the overlapping areas. The black numbers denote the total significantly regulated genes at a false discovery rate (FDR) < 0.05, the red numbers denote the up‐regulated genes and the yellow numbers denote the down‐regulated genes.
Mercator data analysis was used to predict the pathways associated with the expression of each one of the four LBD genes. For the up‐regulated genes, a threshold of fold change greater than two was applied as a filter to the 737 genes associated with the expression of CsLOB1, CsLOB2 and CsLOB3, which yielded 466 up‐regulated common genes. The gene annotations were clustered into categories of cell, cell wall, protein, RNA and signalling (Fig. S3a, File S2, see Supporting Information). Cell and cell wall were the two categories apportioned to most of the genes. Genes in the cell category were mostly classified into cell organization, cell division and cell cycle. The cell wall genes were mostly apportioned into cell wall degradation and cell wall modification (File S2).
A cutoff of fold change greater than two was also utilized to filter the 977 significantly down‐regulated genes associated with the expression of CsLOB1, CsLOB2 and CsLOB3, and 564 genes were identified. Mercator analysis of these 564 transcripts showed that most of the transcripts were classified into categories of signalling, stress, protein, hormone metabolism and secondary metabolism (Fig. S3b, File S3, see Supporting Information). Signalling and stress were the groups with the largest number of genes. Most of the signalling category genes were receptor kinase signalling and calcium signalling genes, and genes in the stress category were mostly grouped into biotic stress (File S3).
Expression assays were conducted using qRT‐PCR to validate the RNA‐Seq data for a selected set of genes. Most of the induced cell wall‐related genes were categorized into cell wall degradation and cell wall modification according to the Mercator analysis results (Table S1, see Supporting Information). Eight representative genes for cell wall degradation and modification with high fold changes were selected for validation by qRT‐PCR analysis. Five were cell wall degradation genes, including an endo‐β‐1,4‐glucanase gene (orange1.1g009690m), a glycosyl hydrolase family 5 gene (orange1.1g014426m), one polygalacturonase gene (orange1.1g043061m) and two pectate lyase genes (orange1.1g015623m and orange1.1g011814m). Three were cell wall modification genes, including two β‐expansin 3 genes (orange1.1g023962m and orange1.1g024303m) and an α‐expansin 9 gene (orange1.1g024916m). RNA samples were prepared from independent inoculations of grapefruit leaves treated with Xcc306ΔpthA4, Xcc306WT, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB3 or Xcc306ΔpthA4::dCsLOB4. The selected cell wall‐related genes were up‐regulated in response to inoculations by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 or Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4; five were also up‐regulated by Xcc306ΔpthA4::dCsLOB4 relative to Xcc306ΔpthA4, but the remaining three were not (Fig. 7). The results indicated that the expression of CsLOB1, CsLOB2 and CsLOB3 was associated with cell wall‐related gene expression. The eight selected cell wall‐related genes were all induced by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 or Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4 at significance levels of P < 0.05. Although the expression levels of the eight genes increased in association with CsLOB4 expression, the levels were, at a maximum, elevated to one‐fifth of the level associated with CsLOB3 expression (Fig. 7). Three of the selected cell wall‐related genes, which were exclusively up‐regulated by expression of CsLOB1, CsLOB2 and CsLOB3, included one polygalacturonase gene (orange1.1g043061m), one pectate lyase gene (orange1.1g011814m) and one β‐expansin gene (orange1.1g024303m) (Fig. 7).
Figure 7.
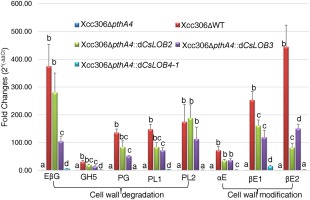
Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) validation of cell wall‐related genes induced by the expression of CsLOB1, CsLOB2, CsLOB3 and CsLOB4. RNA samples were grapefruit leaves treated with various Xanthomonas citri ssp. citri (Xcc) strains as indicated in the figure, and were taken at 5 days post‐inoculation (dpi). Inoculum had an optical density at 600 nm (OD600) of 0.5. Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard error for three independent experimental replicates. Different letters indicate significant differences among the various treatments for each test gene at the significance level of P < 0.05 according to analysis of variance (ANOVA). Different genes are represented by abbreviations of the annotations of various transcripts. Gene IDs are from Phytozome. EβG, endo β‐1,4‐glucanase (orange1.1g009690m); GH5, glycosyl hydrolase family 5 (orange1.1g014426m); PG, polygalacturonase (orange1.1g043061m); PL1, pectate lyase1 (orange1.1g015623m); PL2, pectate lyase2 (orange1.1g011814m); αE, α‐expansin (orange1.1g024916m); βE1, β‐expansin1 (orange1.1g023962m); βE2, β‐expansin2 (orange1.1g024303m).
To exclude the possibility that the induction of genes by Xcc306ΔpthA4::dCsLOB4 relative to Xcc306ΔpthA4 might be a result of direct induction by dCsLOB4, a second dTALe, dCsLOB4‐2, was constructed, targeting a different EBE in the promoter region of CsLOB4 (Fig. S1). After introduction of dCsLOB4‐2 into Xcc306ΔpthA4, CsLOB4 was efficiently induced, and no pustule formation was observed (Fig. S5a,b). Nine genes whose log2 fold changes (Xcc306ΔpthA4::dCsLOB4 vs. Xcc306ΔpthA4) were greater than two were selected to perform the qRT‐PCR validation using both Xcc306ΔpthA4::dCsLOB4 and Xcc306ΔpthA4::dCsLOB4‐2. qRT‐PCR results revealed that four of the nine genes were significantly induced by both Xcc306ΔpthA4::dCsLOB4 and Xcc306ΔpthA4::dCsLOB4‐2 relative to Xcc306ΔpthA4 (Fig. 8).
Figure 8.
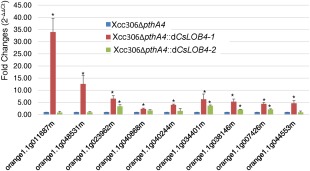
Quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) validation of genes up‐regulated by the expression of CsLOB4, which was induced by either dCsLOB4 or dCsLOB4‐2. RNA samples were grapefruit leaves treated with various Xanthomonas citri ssp. citri (Xcc) strains as indicated in the figure, and were taken at 5 days post‐inoculation (dpi). Inoculum had an optical density at 600 nm (OD600) of 0.5. Expression values were normalized to the housekeeping gene orange1.1g001725m. Different genes are represented by the transcript ID from Phytozome. Error bars represent the standard error for three independent experimental replicates. Single asterisks indicate a significant difference relative to 306ΔpthA4 at the significance level of P < 0.05 according to t‐test.
Discussion
The results indicate that more than one member of the LBD genes could potentially serve as an S gene in CBC. Two LBD family members, CsLOB2 and CsLOB3, were capable of directing pustule formation when strains of Xcc containing LBD‐specific dTALe genes were inoculated on citrus leaves. TAL effector‐mediated induction of CsLOB4 failed to support pustule formation, indicating that at least one member was incapable of supporting pustule formation. CsLOB2 and CsLOB3 both encode members of the same clade as CsLOB1, whereas CsLOB4 lies in a close, but distinct, clade of LBD family members. At the same time, it is premature to conclude that pustule formation is limited to clade I members, as only one member outside of clade I was tested. The induction of CsLOB1, CsLOB2 and CsLOB3 by the respective TAL effectors facilitated symptom development and, at least in the case of CsLOB1 and CsLOB2, enhanced pathogen population levels in leaves. The results may reflect a common biochemical function of the genes within a specific clade, as observed for the SWEET genes in bacterial blight of rice (Streubel et al., 2013). In contrast with bacterial blight of rice, where three SWEET genes are targeted by field strains, only one member of LBD clade I has been shown to be targeted for CBC in the field (Antony et al., 2010; Hu et al., 2014; Pereira et al., 2014; Yang et al., 2006; Zhou et al., 2015).
The lack of alternative S gene targeting in the field might be attributed to the breeding histories of cultivated rice and citrus varieties. Genetic breeding for rice can be traced back 10 000 years (Asano et al., 2011). The repeated deployment of genes for resistance in rice to bacterial blight for specific TAL effector genes in X. oryzae pv. oryzae, and the long breeding history, undoubtedly produced allelic S gene targets and selective pressure for new TAL effector specificities (Garris et al., 2005; Hutin et al., 2015; Shinada et al., 2014). The documented breeding history of citrus is for a much shorter time, perhaps 200 years, and, as a tree species, breeding cycles are considerably longer (Khan, 2007). Therefore, the genetic complexity of the bacterial–plant interactions may be much lower in citrus than in rice, putting less selective pressure on the pathogen population and TAL effector evolution. An alternative model is that the disease itself is rather young, resulting from the recent horizontal transfer of a TAL effector gene. A variety of Xanthomonas strains do not induce a strong non‐host hypersensitive reaction in citrus and cause bacterial spot‐like symptoms (Gent et al., 2005). CBC strains, in addition to a single S gene target, have relatively fewer copies of TAL effector genes within a single strain in comparison with X. oryzae pv. oryzae, which can have as many as 19 genes in a single strain, and the genes are often plasmid‐borne, providing further evidence of recent acquisition (Al‐Saadi et al., 2007; Salzberg et al., 2008; da Silva et al., 2002).
CBC pustule formation has been observed previously on transfer of pthA from X. citri pv. citri to Xanthomonas pathovars, although the growth within a new host was not assessed (Swarup et al., 1991). Here, the dCsLOB2 gene was transferred into X. euvesicatoria Xcv85‐10, which is a pathogen of pepper and causes slight water‐soaking symptoms in Duncan grapefruit. Xcv85‐10 with dCsLOB2 also induced pustules in grapefruit leaves. However, no differences were observed in pathogen leaf populations compared with populations of Xcv85‐10 without dCsLOB2, indicating that the presence of pustule‐associated TAL effector genes does not, in itself, override other requirements for host adaptation. From a practical standpoint, the results also indicate that the CBC causal strains are broadly at risk, at least in the short term, of recessive resistance strategies involving engineering polymorphisms in the CsLOB1 promoter, similar to changes in the EBEs of rice S gene promoters (Antony et al., 2010; Li et al., 2012; Yang et al., 2006; Zhou et al., 2015)
Little is known about the biological functions of LBD members in citrus. The constructed phylogenetic tree (Fig. S4a, see Supporting Information) shows that CsLOB1, CsLOB2 and CsLOB3 are most closely related to AtLBD1 and AtLBD11, whereas CsLOB4 is more closely related to AtLBD12. The protein sequence alignment reveals the consensus LOB domain that contains the three conserved motifs: a cysteine‐rich domain CX2CX6CX3C, a conserved glycine residue and a coiled‐coil leucine zipper‐like motif LX6LX3LX6L (Fig. S4b). AtLBD12 and CsLOB4 have much shorter N‐terminals, which might account for the dysfunction of CsLOB4 as an S gene in CBC. However, reports on the function of AtLBD1 or AtLBD11 are few (Matsui et al., 2008). AtLBD12 mutants have been reported to show reduced leaf size, apical dominance and epinastic leaves, and the mutant plants show increased sterility (Nakazawa et al., 2003). Therefore, the prediction of the biological functions of CsLOB1, CsLOB2 and CsLOB3 is premature based on homology.
Citrus LBD genes potentially function in the regulation of downstream pathways for normal plant development. RNA‐Seq analysis of the expression profiles associated with CsLOB1, CsLOB2 and CsLOB3 exposed a common set of downstream genes; genes in categories of cell organization, cell division, cell cycle, cell wall degradation and cell wall modification were significantly up‐regulated. However, genes in classes of receptor kinase signalling, calcium signalling and biotic stress were mainly down‐regulated, which indicates that CsLOB1, CsLOB2 and CsLOB3 might regulate this set of downstream genes which ultimately trigger pustule development.
The reason why the induction of CsLOB4 could not restore the pustule formation ability of the pthA4 mutant strain is not clear. First, the possibility that CsLOB4 protein is dysfunctional cannot be excluded. Four hundred and sixty‐six genes were significantly induced by dCsLOB4. Nine genes showed a log2 fold change (Xcc306ΔpthA4::dCsLOB4 vs. Xcc306ΔpthA4) greater than two, some of which were also associated with the expression of the other three LBD genes (Table 2). Furthermore, four genes were induced by both Xcc306ΔpthA4::dCsLOB4 and Xcc306ΔpthA4::dCsLOB4‐2 relative to Xcc306ΔpthA4, as revealed by qRT‐PCR analysis, which reduces the likelihood that the induced genes might be off‐targets of dTALes and indicates that CsLOB4 is not dysfunctional. However, the low efficiency of CsLOB4 as a transcription factor might be the reason why the expression of CsLOB4 could not induce pustule formation. Expression of CsLOB4 was shown to be associated with a much smaller set of downstream genes compared with those related to the expression of the other three pustule‐inducible LBD members. Although there are 163 genes that are commonly associated with the expression of all four LBD genes, for most of the genes, the absolute log2 fold changes in the comparison of Xcc306ΔpthA4::dCsLOB4 vs. Xcc306ΔpthA4 are lower than those in the other three comparisons (File S4, see Supporting Information).
Table 2.
Genes whose log2 fold changes are greater than two in a comparison between Xcc306ΔpthA4::dCsLOB4 and Xcc306ΔpthA4, and the differential expression levels in three other comparisons: Xcc306WT vs. Xcc306ΔpthA4, Xcc306ΔpthA4::dCsLOB2 vs. Xcc306ΔpthA4, and Xcc306ΔpthA4::dCsLOB3 vs. Xcc306ΔpthA4.
| Gene ID (Phytozome) | log2 fold change (Xcc306ΔpthA4::dCsLOB4 vs. Xcc306ΔpthA4) | log2 fold change (Xcc306WT vs. Xcc306ΔpthA4) | log2 fold change (Xcc306ΔpthA4::dCsLOB2 vs. Xcc306ΔpthA4) | log2 fold change (Xcc306ΔpthA4::dCsLOB3 vs. Xcc306ΔpthA4) |
|---|---|---|---|---|
| orange1.1g036782m | Infinite | |||
| orange1.1g011687m | 5.7325 | |||
| orange1.1g048531m | 3.8539 | |||
| orange1.1g023962m | 3.6642 | 2.8367 | 2.9486 | 6.5571 |
| orange1.1g040868m | 2.7413 | 2.7184 | 3.4253 | 6.2149 |
| orange1.1g040244m | 2.4699 | 3.9254 | ||
| orange1.1g034401m | 2.2665 | 1.8605 | 1.9224 | 4.3398 |
| orange1.1g038146m | 2.2480 | 2.6675 | 2.5362 | 4.7505 |
| orange1.1g007426m | 2.1797 | 0.7902 | 4.7797 | |
| orange1.1g044553m | 2.0393 | 1.3147 | 2.2238 |
Blank cells indicate that the differential expression levels are not significant at a false discovery rate (FDR) < 0.05.
Infinite denotes that the fold change is positive infinite. The table is a derivative from the RNA‐Seq analysis.
The results revealed that a subset of cell wall‐related genes, especially those involving cell wall degradation and cell wall modification, were induced when the pustule‐associated LBD genes were present. Induction of CsLOB4, one member incapable of forming pustules, was associated with much lower levels of expression or no induction of the cell wall‐related genes, which indicates that these cell wall‐related genes may be functionally important for pustule formation and symptom development. The plant cell wall acts as a physical barrier in interactions with plant pathogens, and pathogens employ various strategies to manipulate their host plant cell wall metabolism to favour infection (Bellincampi et al., 2014). In tomato, several cell wall‐related genes, including Cel1 and Cel2 (endo‐β‐1,4‐glucanase), LePG (polygalacturonase) and LeExp1 (expansin), were up‐regulated during infection by Botrytis cinerea. Silencing of the genes attenuated the susceptibility of tomato fruits to B. cinerea (Cantu et al., 2009; Flors et al., 2007). The expression of CsLOB1, CsLOB2 and CsLOB3 may reflect the role played by a set of cell wall metabolic genes as susceptibility factors. Targeting of individual downstream citrus genes may allow the identification of specific critical genes using the dTALe strategy.
The clade I LBD genes of citrus join a limited club of TAL effector‐dependent S genes that have been shown to contribute major phenotypic features to the respective disease processes. Three other examples have been characterized. The only one for bacterial spot of pepper is upa20, encoding a bHLH family transcription factor, the S gene targeted by TAL effector AvrBs3 from X. campestris pv. vesicatoria. UPA20 has been revealed to play possible roles in cell enlargement in pepper bacterial leaf spot disease (Kay et al., 2007). OsSULTR3;6, a putative sulfate transporter gene, is targeted by Tal2g of X. oryzae pv. oryzicola in rice bacterial leaf streak disease, where the gene facilitates lesion expansion and bacterial exudation (Cernadas et al., 2014). The largest group is the SWEET genes of rice and cassava (Antony et al., 2010; Cohn et al., 2014; Streubel et al., 2013; Yang et al., 2006; Zhou et al., 2015). These SWEET proteins have been demonstrated to function in the transport of sucrose or glucose during pathogenesis (Chen et al., 2010, 2012; Zhou et al., 2015). The range of genes that have been identified to date indicates that a variety of host genes can serve as S genes. At the same time, little is known about the function of TAL effector‐dependent S genes from a physiological perspective. The characterization of additional pathosystems that involve TAL effector‐mediated virulence may ultimately reveal common pathways for the enhancement of host susceptibility.
Experimental Procedures
Plant material, bacterial strains and plasmids
Grapefruit trees were grown at 28 ºC, 80% humidity, at a setting of 16 h of daylight and 8 h of darkness. All Xanthomonas strains were incubated at 28 ºC, and Escherichia coli strains were cultured at 37 ºC. All the bacterial strains and plasmids are shown in Table 1. The E. coli competent cells used were Stellar™ Competent Cells from Clontech Mountain View, CA , USA or NEB 5‐alpha Competent E. coli (High Efficiency) from New England Biology (Ipswich, MA, USA). The Xanthomonas competent cells were prepared by washing the cultured bacteria with 10% glycerol three times, and transformed using the electroporation method (Alexandre et al., 2005).
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Feature | Source |
|---|---|---|
| Xanthonomans citri ssp. citri | ||
| Xcc306WT | Xcc306 Group A, wild‐type, Rif | DPI* |
| Xcc306ΔpthA4 | pthA4 deletion mutant | Jeffery Johne's laboratory (Gainesville, FL, USA) |
| Xcc306ΔpthA4::dCsLOB1 |
Artificial TALE targeting CsLOB1 complement Xcc306ΔpthA4, Gm |
This study |
| Xcc306ΔpthA4::dCsLOB2 |
Artificial TALE targeting CsLOB2 complement Xcc306ΔpthA4, Gm |
This study |
| Xcc306ΔpthA4::dCsLOB2L |
Artificial TALE targeting CsLOB2 complement Xcc306ΔpthA4, Gm |
This study |
| Xcc306ΔpthA4::dCsLOB3 |
Artificial TALE targeting CsLOB3 complement Xcc306ΔpthA4, Gm |
This study |
| Xcc306ΔpthA4::dCsLOB4/Xcc306ΔpthA4::dCsLOB4‐2 |
Artificial TALE targeting CsLOB4 complement Xcc306ΔpthA4, Gm |
This study |
| Xanthomonas campestris pv. vesicatoria | ||
| Xcv8510 | Wild‐type | Jeffery Johne's laboratory (Gainesville, FL, USA) |
| Xcv8510::pHMI | Empty vector pHMI complement Xcv8510 | This study |
| Xcv8510:: dCsLOB2 |
Artificial TALE targeting CsLOB2 complement Xcv8510, Gm |
This study |
| Escherichia coli | ||
| DH5α | F−recAϕ80dlacZΔM15 | New England Biology (NEB) |
| Plasmid | ||
| pBluescript KS(+) | Phagemid, pUC derivative, Ampr | Stratagene |
| pHMI | Broad‐host‐range resistance to Sp | Hopkins et al. (1992) |
| pUFR053 | repW, Mob+, LacZα+, Par+, Gmr | Clontech |
Ampr, ampicillin‐resistant; Gm, gentamicin; Sp, spectinomycin; Rif, rifamycin.
*DPI, Division of Plant Industry of the Florida Department of Agriculture and Consumer Services (Gainesville, FL, USA).
†BRL, Bethesda Research Laboratories (Gaithersburg, MD, USA).
Bacterium inoculation
Xanthomonas strains were streaked on tryptone sucrose agar (TSA) medium incubated at 28 ºC for 48 h. Single colonies were picked, spread on new TSA plates and incubated at 28 ºC for 2 days. The bacteria were suspended in sterilized double‐distilled water to an optical density at 600 nm (OD600) of 0.5 and, where required, the suspensions were diluted 103 times in double‐distilled water. The bacterial suspensions were delivered to the underside (abaxial) of young grapefruit leaves with a needleless 1‐mL syringe.
Imaging of grapefruit leaves inoculated with various bacterial strains
Grapefruit leaves were removed from the trees when the phenotypes were ready for imaging; 1× photographs were taken using a digital SLR camera; 6.4× photographs were imaged using a Nikon SMZ800 dissecting microscope, which was equipped with a Nikon DS‐Vi1 camera.
Phylogenetic analysis of C. sinensis LBD proteins
LBD protein sequences were downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/family.php?sp = Csi&fam = LBD). Amino acid sequences were aligned using SeaView version 4 with the default parameters. The unrooted phylogenetic tree was computed using the setting of PhyML‐3.1 with the LG amino acid substitution model and the non‐parametric bootstrap support test. SeaView 4.1 was used to display and edit the phylogenetic trees (Gouy et al., 2009).
Phylogenetic analysis of LBD proteins of CsLOB1‐4 and the closely related LBD members in A. thaliana
Sequences were aligned with the online ClustalW server (http://www.ch.embnet.org/software/ClustalW.html) using the default values. mega6.0 was used to generate a tree on the basis of ClustalW output. Phylogenetic calculations were based on the maximum likelihood method, and bootstrap analysis was used to evaluate the reliability of the nodes of the phylogenetic trees.
dTALe design and construction
Promoter sequences were the 1000‐bp upstream DNA sequences from the initiation codon ATG of each CsLOB gene retrieved from the Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Csinensis) according to the respective ID. The promoter regions of CsLOB2, CsLOB3 and CsLOB4 were scanned for potential EBEs using TAL Effector Nucleotide Targeter 2.0 (https://tale-nt.cac.cornell.edu/node/add/talef-off). Sites closest to the TATA box were selected as the targeted EBEs (Fig. S1A).
Four types of repeat (NI, NN, NG and HD), which correspond to the respective nucleotides A, G, T and C, were used to assemble the middle repeat domains of the artificial dTALes according to the TAL effector assembly method of Li et al. (2011). The restriction enzymes BstAPI and AatII were used to replace the repeat region of the TAL effector gene pthAw. Each gene was sequenced to verify each construct.
RNA extraction, reverse transcription and qRT‐PCR analysis
Grapefruit leaves were syringe infiltrated with bacterial suspensions at OD600 = 0.5, and the leaf tissues were harvested at 5 dpi. Total RNA was extracted using TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer's instructions. RNA was subjected to LiCl re‐precipitation and DNase treatment to remove the remaining DNA. First‐strand cDNA synthesis was achieved using a Verso cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Two‐step real‐time PCR was performed using iQ™ SYBR® Green Supermix (Bio‐Rad, Hercules, CA, USA). The gene‐specific primer sequences are listed in Table S2 (see Supporting Information). The gene with transcript ID number orange1.1g001725m (Phytozome) was used as an endogenous control. The 2–ΔΔCt method was used for relative quantification (Livak and Schmittgen, 2001).
Bacterial population assay in citrus plants
Each strain was inoculated using a bacterial suspension solution of OD600 = 0.5 diluted 103 times in double‐distilled water. Six 1‐cm2 pieces of leaf disc in the inoculation area for each treatment were taken and ground in 10 mL of sterile water; after serial dilution, 50 µL were plated on TSA medium and incubated at 28 °C for 2 days. The colony number was counted to determine the internal populations. Each treatment was repeated three times.
RNA‐Seq assay and analysis
Two RNA‐Seq assays were performed. For the first, three treatments of Xcc306WT, Xcc306ΔpthA4 and Xcc306ΔpthA4::dCsLOB2 with three replications for each strain were executed. For the second, three treatments of Xcc306ΔpthA4, Xcc306ΔpthA4::dCsLOB3 and Xcc306ΔpthA4::dCsLOB4 were performed, also with three replications for each strain. Libraries were constructed using the TruSeq™ RNA and DNA Sample Prep Kits (Illumina, San Diego, CA, USA). Nine samples were pooled into one lane for sequencing. For the first, Illumina High Output 100‐cycle single‐end (HO‐SR100) sequencing was used; for the second, an Illumina NextSeq500:MID Throughput 2 × 75 cycle run was performed.
The quality of raw reads was analysed with fastQC software (Andrews, 2010). Adapter trimming and filtering were conducted using FastX‐toolKit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Reads were mapped to the reference genome sequence of C. sinensis obtained from the hytozome database V10 (https://phytozome.jgi.doe.gov/pz/portal.html) using TopHat V2.1 (Trapnell et al., 2009). Gene expression quantification was conducted with HTSeq‐count software (Anders et al., 2015) using the gene annotation file from the C. sinensis genome project at Phytozome database v10. Differential gene expression analysis was conducted using DESeq (Anders and Huber, 2010). Genes were considered to be differentially expressed with a cutoff of the adjusted P value of 0.05. DEGs from both assays were compared to generate a Venn diagram using the downloaded software VennPlex version 1.0.0.2 (Cai et al., 2013).
Gene annotation and enrichment were analysed using the online software Mercator employing the amino acid sequences with parameters all selected, except IS_DNA, and a BLAST_CUTOFF value was set as 50 (Lohse et al., 2013).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
File S1 The accession ID (Phytozome) and protein sequences of the 34 lateral organ boundary domain (LBD) members in Citrus sinensis downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/family.php?sp=Csi&fam=LBD).
File S2 Mercator analysis of the genes that were up‐regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. A log2(fold change) larger than unity was applied as the filtering threshold.
File S3 Mercator analysis of the genes that were down‐regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. A log2(fold change) smaller than −1 was applied as the filtering threshold.
File S4 RNA‐sequencing (RNA‐Seq) analysis results of genes that are commonly regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB3 and Xcc306ΔpthA4::dCsLOB4 relative to Xcc306ΔpthA4.
Fig. S1 Schematic diagrams of designed transcription activator‐like effectors (dTALes) for CsLOB2, CsLOB3 and CsLOB4. (a) Promoter regions of three targeted lateral organ boundary domain (LBD) genes in Citrus sinensis. The yellow highlight shows the effector‐binding elements (EBEs) targeted by dTALes, and the red letters indicate the predicted TATA boxes and start codons for each gene. (b) The repeat variable diresidues (RVDs) of three dTALes and the corresponding EBEs in the promoters of CsLOB2, CsLOB3 and CsLOB4. (c) The transcription activator‐like (TAL) code based on Grau et al. (2013).
Fig. S2 dCsLOB2 induces CsLOB2 expression when transformed into Xanthomonas euvesicatoria strain Xcv85‐10. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of RNA samples of grapefruit leaves inoculated with Xcv85‐10::pHM1 and Xcv85‐10::dCsLOB2 [5 days post‐inoculation (dpi)]. Inoculum was a bacterial suspension with an optical density at 600 nm (OD600) of 0.5. Samples were taken at 5 dpi. Error bars represent the standard errors for three independent experimental replicates. *Significant difference between the two treatments (t‐test, P < 0.05).
Fig. S3 Functional category assignments of common genes that were significantly regulated in grapefruit by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. (a) Mercator analysis of the up‐regulated genes was performed using the 466 genes related to the expression of CsLOB1, CsLOB2 and CsLOB3 in grapefruit at 5 days post‐inoculation (dpi). A log2(fold change) larger than unity was applied as the filtering threshold. (b) Mercator analysis of the down‐regulated genes was performed using the 564 genes related to the expression of CsLOB1, CsLOB2 and CsLOB3 in grapefruit at 5 dpi. A log2(fold change) smaller than −1 was applied as the infiltration threshold.
Fig. S4 Phylogenetic analysis of lateral organ boundary domain (LBD) proteins of CsLOB1, CsLOB2, CsLOB3, CsLOB4 and closely related LBD members in Arabidopsis thaliana. (a) Phylogenetic tree of LBD proteins of CsLOB1, CsLOB2, CsLOB3, CsLOB4 and closely related LBD members in A. thaliana. A monophyletic group containing CsLOB1, CsLOB2 and CsLOB3 is boxed in red. The group containing CsLOB4 is boxed in blue. Bootstrap values were based on 500 replications. The branch lengths of the tree are proportional to divergence. The 0.1 scale represents 10% change. (b) Protein sequence alignment of LBD genes from Citrus sinensis (Cs) and Arabidopsis thaliana (At). Three conserved motifs (CX2CX6CX3C, glycine residue and LX6LX3LX6L) are highlighted in red, pink and blue, respectively. The alignment was conducted using ClustalW with the default settings.
Fig. S5 A second designed transcription activator‐like effector (dTALe) targeting CsLOB4 did not induce pustule formation on citrus. (a) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) validation of CsLOB4 induction by dCsLOB4‐2. Asterisk denotes a significant difference in the fold change between the treatments relative to the negative control Xcc306ΔpthA4 at P < 0.05 using t‐test. qRT‐PCR was conducted on mRNA extracted from grapefruit leaves inoculated with the individual Xanthomonas citri ssp. citri (Xcc) strains as indicated, using CsLOB4 gene‐specific primers at 5 days post‐inoculation (dpi). Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard deviations for three independent experimental replicates. Samples were taken at 5 dpi. (b) Pustule was not induced by the two dTALes targeting CsLOB4: 1, Xcc306ΔpthA4; 2, Xcc306ΔpthA4::dCsLOB4; 3, Xcc306ΔpthA4::dCsLOB4‐2. Photographs were taken at 7 dpi of various strains as shown in the figures. Inocula were bacterial suspensions with an optical density at 600 nm (OD600) of 0.5.
Fig. S6 A second longer version of designed transcription activator‐like effector (dTALe) targeting CsLOB2 induced pustule formation on citrus. (a) Pustule was induced by the two dTALes targeting CsLOB2: 1, Xcc306ΔpthA4::dCsLOB2L; 2, Xcc306ΔpthA4. Photographs were taken at 7 days post‐inoculation (dpi) of various strains as shown in the figures. Inocula were bacterial suspensions with an optical density at 600 nm (OD600) of 0.5. (b) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) validation of CsLOB2 induction by dCsLOB2L. Asterisk denotes significant difference in the fold change between the treatments relative to the negative control Xcc306ΔpthA4 at P < 0.05 using t‐test. qRT‐PCR was conducted on mRNA extracted from grapefruit leaves inoculated with the individual Xanthomonas citri ssp. citri (Xcc) strains as indicated, using CsLOB2 gene‐specific primers at 5 dpi. Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard deviations for three independent experimental replicates. Samples were taken at 5 dpi.
Table S1 Cell wall‐related genes clustered by Mercator analysis. The yellow highlighted genes are those tested by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Table S2 Primers used in this study
Acknowledgements
Thanks are due to the Agriculture and Food Research Initiative National Institute of Food and Agriculture (AFRI NIFA, grant 2011‐04153) and the Plant Genome Research Program of the National Science Foundation (grant IOS‐1238189).
References
- Alexandre, M. , Do, A. , Toledo, C.P. , Baptista, J.C. and Machado, M.A. (2005) Transformation of Xanthomonas axonopodis pv. citri by electroporation. Trop. Plant Pathol. 30, 292. doi: 10.1590/S0100-41582005000300013. [DOI] [Google Scholar]
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q. and Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats on citrus, but none determine host‐range pathogenicity variation. Mol. Plant–Microbe Interact. 20, 934–943. doi: 10.1094/MPMI-20-8-0934. [DOI] [PubMed] [Google Scholar]
- Anders, S. and Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. and Huber, W. (2015) HTSeq – a python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Antony, G. , Zhou, J. , Huang, S. , Li, T. , Liu, B. , White, F. and Yang, B. (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os11N3 . Plant Cell, 22, 3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, K. , Yamasaki, M. , Takuno, S. , Miura, K. , Katagiri, S. , Ito, T. , Doi, K. , Wu, J. , Ebana, K. , Matsumoto, T. , Innan, H. , Kitano, H. , Ashikari, M. and Matsuoka, M. (2011) Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA, 108, 11 034–11 039. doi: 10.1073/pnas.1019490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellincampi, D. , Cervone, F. and Lionetti, V. (2014) Plant cell wall dynamics and wall‐related susceptibility in plant‐pathogen interactions. Front. Plant Sci. 5, 228. doi: 10.3389/fpls.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye. T, Nickstadt , and A., Bonas, U . (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Boch, J. , Bonas, U. and Lahaye, T. (2014) TAL effectors ‐ pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015. [DOI] [PubMed] [Google Scholar]
- Cai, H. , Chen, H. , Yi, T. , Daimon, C.M. , Boyle, J.P. , Peers, C. , Maudsley, S. and Martin, B. (2013) A novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PLoS One, 8, e53388. doi: 10.1371/journal.pone.0053388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantu, D. , Blanco‐Ulate, B. , Yang, L. , Labavitch, J.M. , Bennett, A.B. and Powell, A.L.T. (2009) Ripening‐regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 150, 1434–1449. doi: 10.1104/pp.109.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas, R.A. , Doyle, E.L. , Niño‐Liu, D.O. , Wilkins, K.E. , Bancroft, T. , Wang, L. , Schmidt, C.L., Caldo, R., Yang, B., White, F.F., Nettleton, D., Wise, R,P., and Bogdanove, A.J. (2014) Code‐assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10, e1003972. doi: 10.1371/journal.ppat.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Hou, B. , Lalonde, S. , Takanaga, H. , Hartung, M.L. , Qu, X. , Guo, W.J. , Kim, J.G. , Underwood, W. , Chaudhuri, B. , Chermak, D. , Antony, G. , White, F.F. , Somerville, S.C. , Mudgett, M.B. and Frommer, W.B. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature, 468, 527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Qu, X. , Hou, B. , Sosso, D. , Osorio, S. , Fernie, A.R. and Frommer, W.B. (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science, 335, 207. [DOI] [PubMed] [Google Scholar]
- Cohn, M. , Bart, R.S. , Shybut, M. , Dahlbeck, D. , Gomez, M. , Morbitzer, R. , Hou, B.H. , Frommer, W.B. , Lahaye, T. and Staskawicz, B.J. (2014) Xanthomonas axonopodis virulence is promoted by a transcription activator‐like effector mediated induction of a SWEET sugar transporter in cassava. Mol. Plant‐Microbe Interact. 27(11), 1186–1198. doi: 10.1094/MPMI-06-14-0161-R. [DOI] [PubMed] [Google Scholar]
- De Feyter, R. , Yang, Y. and Gabriel, D.W. (1993) Gene‐for‐gene interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant‐Microbe Interact. 6, 225. [DOI] [PubMed] [Google Scholar]
- Flors, V. , Leyva, M.D.L.O. , Vicedo, B. , Finiti, I. , Real, M.D. , Garcia‐Agustin, P. , Bennett, A.B. and González‐Bosch C. (2007) Absence of the endo‐beta‐1,4‐glucanases Cel1 and Cel2 reduces susceptibility to Botrytis cinerea in tomato. Plant J. 52, 1027–1040. doi: 10.1111/j.1365-313X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Garris, A. , Tai, T. , Coburn, J. , Kresovich, S. and McCouch, S. (2005) Genetic structure and diversity in Oryza sativa L. Genetics, 169, 1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron, J.M. , Liu, J. , Fan, M. , Bai, M. , Wenkel, S. , Springer, P.S. , Barton, M.K. and Wang, Z.Y. (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis . Proc. Natl. Acad. Sci. USA, 109, 21 152–21 157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent, D.H. , Al‐Saadi, A. , Gabriel, D.W. , Louws, F.J. , Ishimaru, C.A. and Schwartz, H.F. (2005) Pathogenic and genetic relatedness among Xanthomonas axonopodis pv. allii and other pathovars of X. axonopodis . Phytopathology, 95, 918–925. doi: 10.1094/PHYTO-95-0918. [DOI] [PubMed] [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002) Citrus canker: the pathogen and its impact. Plant Health Prog. 1–32, doi: 10.1094/PHP-2002-0812-01-RV. [DOI] [Google Scholar]
- Gouy, M. , Guindon, S. and Gascuel, O. (2009) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grau, J. , Wolf, A. , Reschke, M. , Bonas, U. , Posch, S. and Boch, J. (2013) Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol. 9, e1002962. doi: 10.1371/journal.pcbi.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer, H. , Yin, Y. , Xue, Q. , Smalla, K. and Guo, J. (2007) Repeat domain diversity of avrBs3‐like genes in Ralstonia solanacearum strains and association with host preferences in the field. Appl. Environ. Microbiol. 73, 4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, C.M. , White, F.F. , Choi, S.H. , Guo, A. and Leach, J.E. (1992) Identification of a family of avirulence genes from Xanthomonas oryzae pv oryzae . Mol. Plant‐Microbe Interact. 5, 451–459. doi: 10.1094/MPMI-5-451. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. , Yang, B. , White, F.F. , Wang, N. and Jones, J.B. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111, E521–E529. doi: 10.1073/pnas.1313271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husbands, A. , Bell, E.M. , Shuai, B. , Smith, H.M.S. and Springer, P.S. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA‐binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin, M. , Pérez‐Quintero, A. , Lopez, C. and Szurek, B. (2015) MorTAL kombat: the story of defense against TAL effectors through loss‐of‐susceptibility. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan, N. , Kumar, D. Andrade, M.O. , Yu, F. , Jones, J.B. , Graham, J.H. , White, F.F. , Setubal, J.C. and Wang, N. (2013) Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics, 14, 551. doi: 10.1186/1471-2164-14-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, S. , Hahn, S. , Marois, E. , Hause, G. and Bonas, U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science, 318, 648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- Khan, I.A. (2007) In: Citrus Genetics, Breeding and Biotechnology (Khan I.A. ed). pages 1–8. Wallingford, Oxfordshire: CAB International. doi: 10.1079/9780851990194.000 ER. [DOI] [Google Scholar]
- Li, T. , Huang, S. , Zhao, X. , Wright, D.A. , Carpenter, S. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2011) Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2013) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Li, T. , Huang, S. , Zhou, J. and Yang, B. (2013) Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Mol. Plant, 6, 781–789. doi: 10.1093/mp/sst034. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2-ΔΔCT method. Methods, 25, 402–408. doi:10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lohse, M. , Nagel, A. , Herter, T. , May, P. , Schroda, M. , Zrenner, R. , Tohge, T. , Fernie, A.R. , Stitt, M. and Usadel, B. (2013) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37, 1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- Majer, C. and Hochholdinger, F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Mak, A.N. , Bradley, P. , Bogdanove, A.J. and Stoddard, B.L. (2013) TAL effectors: function, structure, engineering and applications. Curr. Opin. Struct. Biol. 23, 93–99. doi: 10.1016/j.sbi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, A. , Ishida, J. , Morosawa, T. , Mochizuki, Y. , Kaminuma, E. , Endo, T.A. , Okamoto, M. , Nambara, E. , Nakajima, M. , Kawashima, M. , Satou, M. , Kim, J.M. , Kobayashi, N. , Toyoda, T. , Shinozaki, K. and Seki, M. (2008) Arabidopsis transcriptome analysis under drought, cold, high‐salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Morbitzer, R. , Roemer, P. , Boch, J. and Lahaye, T. (2010) Regulation of selected genome loci using de novo‐engineered transcription activator‐like effector (TALE)‐type transcription factors. Proc. Natl. Acad. Sci. USA, 107, 21 617–21 622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M. , Ichikawa, T. , Ishikawa, A. , Kobayashi, H. , Tsuhara, Y. , Kawashima, M. , Suzuki, K. , Muto, S. and Matsui, M. (2003) Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 34, 741–750. doi: 10.1046/j.1365-313X.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- Pereira, A.L.A. , Carazzolle, M.F. , Abe, V.Y. , de Oliveira, M.L.P. , Domingues, M.N. , Silva, J.C. , Cernadas, R.A. and Benedetti, C.E. (2014) Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics, 15, 157. doi: 10.1186/1471-2164-15-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. and Szurek, B. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics, 9, 534. doi: 10.1186/1471-2164-9-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinada, H. , Yamamoto, T. , Yamamoto, E. , Hori, K. , Yonemaru, J. , Matsuba, S. and Fujino, K. (2014) Historical changes in population structure during rice breeding programs in the northern limits of rice cultivation. Theor. Appl. Genet. 127, 995–1004. doi: 10.1007/s00122-014-2274-2. [DOI] [PubMed] [Google Scholar]
- Shuai, B. , Reynaga‐Pena, C.G. and Springer, P.S. (2002) The LATERAL ORGAN BOUNDARIES gene defines a novel, plant‐specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- Streubel, J. , Pesce, C. , Hutin, M. , Koebnik, R. , Boch, J. and Szurek, B. (2013) Five phylogenetically close rice SWEET genes confer TAL effector‐mediated susceptibility to Xanthomonas oryzae pv. oryzae . New Phytol. 200, 808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- Sun, X.A. , Stall, R.E. , Jones, J.B. , Cubero, J. , Gottwald, T.R. , Graham, J.H. , Dixon, W.N. , Schubert, T.S. , Chaloux, P.H. , Stromberg, V.K. , Lacy, G.H. and Sutton, B.D. (2004) Detection and characterization of a new strain of citrus canker bacteria from key Mexican lime and alemow in south Florida. Plant Dis. 88, 1179–1188. doi: 10.1094/PDIS.2004.88.11.1179. [DOI] [PubMed] [Google Scholar]
- Swarup, S. , De Feyter, R. , Brlansky, R.H. and Gabriel, D.W. (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit cankerlike lesions on citrus. Phytopathology, 81, 802–809. doi: 10.1094/Phyto-81-802. [DOI] [Google Scholar]
- Swarup, S. , Yang, Y.N. , Kingsley, M.T. and Gabriel, D.W. (1992) An Xanthomonas citri pathogenicity gene, ptha, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant–Microbe Interact. 5, 204–213. doi: 10.1094/MPMI-5-204. [DOI] [PubMed] [Google Scholar]
- Thatcher, L.F. , Powell, J.J. , Aitken, E.A.B. , Kazan, K. and Manners, J.M. (2012) The lateral organ boundaries domain transcription factor LBD20 functions in fusarium wilt susceptibility and jasmonate signaling in Arabidopsis . Plant Physiol. 160, 407–418. doi: 10.1104/pp.112.199067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. and Salzberg, S.L. (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics, 25, 1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verniere, C. , Hartung, J.S. , Pruvost, O.P. , Civerolo, E.L. , Alvarez, A.M. , Maestri, P. and Luisetti, J. (1998) Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from southwest Asia. Eur. J. Plant Pathol. 104, 477–487. doi: 10.1023/A:1008676508688. [DOI] [Google Scholar]
- Wang, X. , Zhang, S. , Su, L. , Liu, X. and Hao, Y. (2013) A genome‐wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS One, 8, e57044. doi: 10.1371/journal.pone.0057044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Sugio, A. and White, F.F. (2006) Os8N3 is a host disease‐susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA, 103, 10 503–10 508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Yin, Z. and White, F. (2015) TAL effectors and the executor R genes. Front. Plant Sci. 6, 641. doi: 10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Peng, Z. , Long, J. , Sosso, D. , Liu, B. , Eom, J. , Huang, S. , Liu, S. , Vera Cruz, C. , Frommer, W.B. , White, F.F. and Yang, B. (2015) Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
File S1 The accession ID (Phytozome) and protein sequences of the 34 lateral organ boundary domain (LBD) members in Citrus sinensis downloaded from the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/family.php?sp=Csi&fam=LBD).
File S2 Mercator analysis of the genes that were up‐regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. A log2(fold change) larger than unity was applied as the filtering threshold.
File S3 Mercator analysis of the genes that were down‐regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. A log2(fold change) smaller than −1 was applied as the filtering threshold.
File S4 RNA‐sequencing (RNA‐Seq) analysis results of genes that are commonly regulated by Xcc306WT, Xcc306ΔpthA4::dCsLOB2, Xcc306ΔpthA4::dCsLOB3 and Xcc306ΔpthA4::dCsLOB4 relative to Xcc306ΔpthA4.
Fig. S1 Schematic diagrams of designed transcription activator‐like effectors (dTALes) for CsLOB2, CsLOB3 and CsLOB4. (a) Promoter regions of three targeted lateral organ boundary domain (LBD) genes in Citrus sinensis. The yellow highlight shows the effector‐binding elements (EBEs) targeted by dTALes, and the red letters indicate the predicted TATA boxes and start codons for each gene. (b) The repeat variable diresidues (RVDs) of three dTALes and the corresponding EBEs in the promoters of CsLOB2, CsLOB3 and CsLOB4. (c) The transcription activator‐like (TAL) code based on Grau et al. (2013).
Fig. S2 dCsLOB2 induces CsLOB2 expression when transformed into Xanthomonas euvesicatoria strain Xcv85‐10. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of RNA samples of grapefruit leaves inoculated with Xcv85‐10::pHM1 and Xcv85‐10::dCsLOB2 [5 days post‐inoculation (dpi)]. Inoculum was a bacterial suspension with an optical density at 600 nm (OD600) of 0.5. Samples were taken at 5 dpi. Error bars represent the standard errors for three independent experimental replicates. *Significant difference between the two treatments (t‐test, P < 0.05).
Fig. S3 Functional category assignments of common genes that were significantly regulated in grapefruit by Xcc306WT, Xcc306ΔpthA4::dCsLOB2 and Xcc306ΔpthA4::dCsLOB3 relative to Xcc306ΔpthA4. (a) Mercator analysis of the up‐regulated genes was performed using the 466 genes related to the expression of CsLOB1, CsLOB2 and CsLOB3 in grapefruit at 5 days post‐inoculation (dpi). A log2(fold change) larger than unity was applied as the filtering threshold. (b) Mercator analysis of the down‐regulated genes was performed using the 564 genes related to the expression of CsLOB1, CsLOB2 and CsLOB3 in grapefruit at 5 dpi. A log2(fold change) smaller than −1 was applied as the infiltration threshold.
Fig. S4 Phylogenetic analysis of lateral organ boundary domain (LBD) proteins of CsLOB1, CsLOB2, CsLOB3, CsLOB4 and closely related LBD members in Arabidopsis thaliana. (a) Phylogenetic tree of LBD proteins of CsLOB1, CsLOB2, CsLOB3, CsLOB4 and closely related LBD members in A. thaliana. A monophyletic group containing CsLOB1, CsLOB2 and CsLOB3 is boxed in red. The group containing CsLOB4 is boxed in blue. Bootstrap values were based on 500 replications. The branch lengths of the tree are proportional to divergence. The 0.1 scale represents 10% change. (b) Protein sequence alignment of LBD genes from Citrus sinensis (Cs) and Arabidopsis thaliana (At). Three conserved motifs (CX2CX6CX3C, glycine residue and LX6LX3LX6L) are highlighted in red, pink and blue, respectively. The alignment was conducted using ClustalW with the default settings.
Fig. S5 A second designed transcription activator‐like effector (dTALe) targeting CsLOB4 did not induce pustule formation on citrus. (a) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) validation of CsLOB4 induction by dCsLOB4‐2. Asterisk denotes a significant difference in the fold change between the treatments relative to the negative control Xcc306ΔpthA4 at P < 0.05 using t‐test. qRT‐PCR was conducted on mRNA extracted from grapefruit leaves inoculated with the individual Xanthomonas citri ssp. citri (Xcc) strains as indicated, using CsLOB4 gene‐specific primers at 5 days post‐inoculation (dpi). Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard deviations for three independent experimental replicates. Samples were taken at 5 dpi. (b) Pustule was not induced by the two dTALes targeting CsLOB4: 1, Xcc306ΔpthA4; 2, Xcc306ΔpthA4::dCsLOB4; 3, Xcc306ΔpthA4::dCsLOB4‐2. Photographs were taken at 7 dpi of various strains as shown in the figures. Inocula were bacterial suspensions with an optical density at 600 nm (OD600) of 0.5.
Fig. S6 A second longer version of designed transcription activator‐like effector (dTALe) targeting CsLOB2 induced pustule formation on citrus. (a) Pustule was induced by the two dTALes targeting CsLOB2: 1, Xcc306ΔpthA4::dCsLOB2L; 2, Xcc306ΔpthA4. Photographs were taken at 7 days post‐inoculation (dpi) of various strains as shown in the figures. Inocula were bacterial suspensions with an optical density at 600 nm (OD600) of 0.5. (b) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) validation of CsLOB2 induction by dCsLOB2L. Asterisk denotes significant difference in the fold change between the treatments relative to the negative control Xcc306ΔpthA4 at P < 0.05 using t‐test. qRT‐PCR was conducted on mRNA extracted from grapefruit leaves inoculated with the individual Xanthomonas citri ssp. citri (Xcc) strains as indicated, using CsLOB2 gene‐specific primers at 5 dpi. Expression values were normalized to the housekeeping gene orange1.1g001725m. Error bars represent the standard deviations for three independent experimental replicates. Samples were taken at 5 dpi.
Table S1 Cell wall‐related genes clustered by Mercator analysis. The yellow highlighted genes are those tested by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Table S2 Primers used in this study


