Summary
The targeting of bacterial type III secretion systems (T3SSs), which are critical virulence factors in most Gram‐negative pathogens, is regarded as an alternative strategy for the development of novel anti‐microbial drugs. Xanthomonas oryzae pv. oryzae (Xoo) and X. oryzae pv. oryzicola (Xoc) are two of the most important bacterial pathogens on rice, which cause leaf blight and leaf streak diseases, respectively. To identify potential anti‐virulence drugs against these two pathogens, we screened a library of plant phenolic compounds and derivatives for their effects on the Xoo T3SS. Ten of 56 compounds significantly inhibited the promoter activity of a harpin gene, hpa1. These inhibitors were further tested for their impact on the hypersensitive response (HR) caused by Xoo on non‐host tobacco plants. The results showed that pretreatment of Xoo with TS006 (o‐coumaric acid, OCA), TS010, TS015 and TS018 resulted in significantly attenuated HR without affecting bacterial growth or survival. In addition, Cya translocation assays demonstrated that the translocation of two T3 effectors was suppressed by the four inhibitors. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis showed that mRNA levels of representative genes in the hrp (hypersensitive response and pathogenicity) cluster, as well as the regulatory genes hrpG and hrpX, were reduced by treatment with the four inhibitors, suggesting that expression of the Xoo T3SS was suppressed. The expression of other virulence factors was not suppressed, which indicated possible T3SS‐specific inhibition. Finally, we demonstrated that these inhibitors reduced the disease symptoms of Xoo and Xoc on the rice cultivar (Oryza sativa) IR24 to varying extents.
Keywords: bacterial leaf blight, bacterial leaf streak, harpin, HrpG, HrpX, hypersensitive response, o‐coumaric acid
Introduction
Traditional antibiotic therapy, which aims to affect the processes essential for bacterial growth and survival, has led to strong selective pressure to develop antibiotic resistance in pathogenic bacteria (Rasko and Sperandio, 2010). An alternative approach is to identify new agents that target bacterial virulence factors, rather than their growth (Barczak and Hung, 2009; Rasko and Sperandio, 2010). The type III secretion system (T3SS) is a highly conserved virulence factor in many Gram‐negative pathogenic bacteria, and is not necessary for bacterial survival in vitro (Büttner, 2012; Cornelis, 2006). Thus, it is regarded as an ideal target for the development of novel antimicrobial drugs (Charro and Mota, 2015; Marshall and Finlay, 2014). So far, several different classes of small‐molecule compounds have been identified as active T3SS inhibitors in a wide range of pathogenic bacteria, including Escherichia, Yersinia, Salmonella and Erwinia species (Felise et al., 2008; Jessen et al., 2014; Wang et al., 2011; Yang et al., 2014). These inhibitors function by directly targeting the components of the T3SS apparatus (Bowlin et al., 2014; Jessen et al., 2014), or regulating T3SS gene expression (Garrity‐Ryan et al., 2010; Yang et al., 2014), or through more indirect interactions (Bowlin et al., 2014; Wang et al., 2011).
Phenolic compounds are ubiquitous secondary metabolites in plants, which are crucial in many aspects of plant life, especially during their interactions with the environment (Lattanzio, 2013). Some phenolics play key roles in plant defence responses to pathogen or insect attacks. For example, salicylic acid (SA), a simple phenolic compound, is the signalling molecule required for the induction of systemic acquired resistance in plants (Durrant and Dong, 2004). Recently, it has been discovered that the novel phenolic compounds o‐coumaric acid (OCA), trans‐cinnamic acid (TCA) and p‐coumaric acid (PCA) modulate the expression of T3SS genes in Dickeya dadantii, a broad‐host‐range phytopathogen (Li et al., 2009; Yang et al., 2008). Some of their derivatives have been found to suppress T3SS gene expression in Pseudomonas aeruginosa and Erwinia amylovora (Khokhani et al., 2013; Yamazaki et al., 2012).
Xanthomonas oryzae pv. oryzae (Xoo) is the causal agent of bacterial blight (BB), one of the most serious diseases of rice worldwide (Nino‐Liu et al., 2006). Xoo typically invades rice vascular tissue through wounds or natural openings, such as the hydathode (Nino‐Liu et al., 2006). Xanthomonas oryzae pv. oryzicola (Xoc) is another bacterial pathogen of rice, which colonizes the mesophyll parenchyma tissue to cause bacterial leaf streak disease. Xoc enters the leaf primarily through the stomata (Nino‐Liu et al., 2006). To date, multiple virulence factors have been identified that contribute to bacterial invasion at different stages, such as adhesion‐like proteins, exopolysaccharide (EPS) and lipopolysaccharide (LPS), biofilm formation and the type II secretion system (T2SS), which is used for the secretion of extracellular enzymes, including xylanase and cellulase (Büttner and Bonas, 2010; Das et al., 2009; Kim et al., 2009; Rai et al., 2012; Rajeshwari et al., 2005). Like many other Gram‐negative plant‐pathogenic bacteria, Xoo and Xoc also possess a T3SS to inject and deliver effector proteins into host cells. It is encoded by the hypersensitive response and pathogenicity (hrp) gene locus (Cho et al., 2008; Zou et al., 2006). The T3SS and its secreted components play a critical role in conferring pathogenicity on the host and triggering the hypersensitive response (HR) on resistant or non‐host plants (Alfano and Collmer, 1997; Büttner and Bonas, 2003). The core operon consists of more than 20 genes on several transcriptional units, which contain hrp, hrc (hrp‐conserved) and hpa (hrp‐associated) genes (Cho et al., 2008; Zou et al., 2006). Two types of effector have been found in X. oryzae: TAL (transcription activator‐like) and non‐TAL effectors (Salzberg et al., 2008; White and Yang, 2009). These effectors often determine the consequence of interactions between the bacterium and different hosts.
hrp gene expression is tightly regulated, and is induced in planta or in specially prepared minimal medium, which is designed to mimic in planta conditions. Expression is suppressed in nutrient‐rich medium (Tang et al., 2006; Tsuge et al., 2002). The hrp genes in Xanthomonas spp. and Ralstonia solanacearum have been classified as hrp group II, which differ from group I of E. amylovora and Pseudomonas syringae (Alfano and Collmer, 1997; Tang et al., 2006). The expression of hrp genes in group I is activated by alternative sigma factor HrpL (Chatterjee et al., 2002; Xiao et al., 1994). In group II, hrpG and hrpX, which are spatially located away from the hrp gene cluster, are two key known regulatory genes of hrp gene expression (Wengelnik and Bonas, 1996; Wengelnik et al., 1996). HrpG, a response regulator belonging to the OmpR family of two‐component signal transduction systems (TCS), positively regulates the expression of hrpX (Wengelnik et al., 1996). HrpX is an AraC family regulator which activates the transcription of other hrp genes (hrpB to hrpF), together with the genes encoding T3 effectors (Wengelnik and Bonas, 1996). Most of the genes in the HrpG regulon are regulated by HrpX. HrpX interacts with a cis‐element within the promoter region of hrp genes, known as the plant‐inducible promoter (PIP)‐box, which is also present in the promoter of many T3 effectors (Noel et al., 2001; Wengelnik and Bonas, 1996). In addition to HrpG and HrpX, HrpD6 has been reported recently as a hrp regulator in Xoc, responsible for the control of a subgroup of hrp genes (Li et al., 2011).
In this article, a small library of phenolic compounds was screened for their effectiveness on T3SS expression of Xoo. Four inhibitors, which can suppress the HR in tobacco without killing bacterial cells, were identified and selected for further analysis. Their effects on effector translocation and expression of representative hrp genes were also examined. Virulence assays indicated that the symptoms in rice caused by both Xoo and Xoc were weakened by these four inhibitors to varying degrees.
Results
Screening of inhibitors which affect the T3SS of Xoo
In order to identify potential inhibitors of the Xoo T3SS, a library of 56 plant phenolic compounds and their derivatives (Tables 1 and S1, see Supporting Information) was screened for their effects on the promoter activity of the hpa1 gene, which encodes a harpin protein in Xoo (Furutanin et al., 2003; Wang et al., 2008). Expression of hpa1 is induced in the hrp‐inducing medium XOM2, and is controlled by the regulatory protein HrpX (Furutanin et al., 2003). The promoter region of hpa1 (Fig. S1, see Supporting Information) was inserted into the promoter‐probe vector pPROBE‐AT (Li et al., 2009), which contains a promoterless green fluorescence protein (GFP) reporter gene, resulting in pPhpa1. Xoo PXO99A strain carrying pPhpa1 was grown in XOM2 supplemented with each of the compounds at a final concentration of 200 μm for 15 h before the promoter activity of hpa1 was measured. Eleven of the 56 compounds were found to be insoluble in XOM2 medium (Table S1). Thus, their effects could not be determined. The other 45 compounds were screened for alterations in hpa1 promoter activity through the highly efficient fluorescence‐activated cell sorting (FACS) system. The mean fluorescence intensity (MFI), representing the promoter activity of hpa1 in each treatment, was recorded (Table 1). We also calculated the ratio of MFI after treatment by each compound to that of the solvent control, resulting in a number indicated by %DMSO (DMSO, dimethylsulfoxide). The results showed that 25 compounds altered hpa1 promoter activity significantly in comparison with the DMSO control and, of these, 10 compounds inhibited the hpa1 promoter activity by at least 60% (Table 1).
Table 1.
Screening for inhibitors of the Xanthomonas oryzae pv. oryzae (Xoo) type III secretion system (T3SS) by fluorescence‐activated cell sorting assays.
| Phenolic compound | Average MFI ± SD a | %DMSO b |
|---|---|---|
| XOM2 | 15.63 ± 1.96 | |
| Dimethylsulfoxide (DMSO) | 14.97 ± 2.33 | |
| TS001, trans‐cinnamic acid | 9.41 ± 0.10* | 62.86 |
| TS002, 2,4‐dihydroxycinnamic acid | 12.69 ± 0.10 | 84.77 |
| TS006, o‐coumaric acid | 3.58 ± 0.08** | 23.91 |
| TS007, 3‐(4‐hydroxyphenyl)propionic acid | 19.85 ± 0.74* | 132.60 |
| TS013, trans−3‐(3‐pyridyl)acrylic acid | 7.66 ± 1.06* | 51.17 |
| TS015, trans−2‐methoxycinnamic acid | 4.43 ± 0.01* | 29.59 |
| TS027, trans−4‐aminocinnamic acid | 13.59 ± 1.28 | 90.78 |
| TS028, trans‐4‐nitrocinnamic acid | 10.26 ± 1.17 | 68.54 |
| TS103, trans−4‐hydroxycinnamohydroxamic acid | 7.92 ± 0.26* | 52.91 |
| TS114, diethyl trans−2‐(4‐hydroxyphenyl)‐vinylphosphonate | 22.2 ± 3.11 | 148.30 |
| TS115, trans−2‐(4‐hydroxyphenyl)vinylphosphonic acid | 17.74 ± 1.31 | 118.50 |
| TS117, p‐coumarylamine | 14.45 ± 0.23 | 96.53 |
| TS118, N‐(4‐methoxycinnamyl)phthalimide | 16.89 ± 0.34 | 112.83 |
| TS130, trans−3‐hydroxycinnamohydroxamic acid | 5.37 ± 0.53* | 35.87 |
| TS132, trans‐cinnamohydroxamic acid | 9.10 ± 0.52* | 60.79 |
| TS139, 3‐phenylpropionohydroxamic acid | 4.85 ± 0.03* | 32.40 |
| TS141, trans−4‐fluorocinnamohydroxamic acid | 5.34 ± 0.44* | 35.67 |
| TS160, trans−2‐methylcinnamohydroxamic acid | 3.97 ± 0.04** | 26.52 |
| XOM2 | 20.26 ± 0.50 | |
| DMSO | 19.67 ± 1.45 | |
| TS003, 3,4‐dihydroxycinnamic acid | 12.54 ± 1.09* | 63.75 |
| TS004, p‐coumaric acid | 22.08 ± 2.78 | 112.25 |
| TS008, hydrocinnamic acid | 18.98 ± 0.90 | 96.49 |
| TS010, trans−2‐phenylcyclopropane‐1‐carboxylic acid | 6.68 ± 1.69** | 33.96 |
| TS011, trans−3‐(2‐thienyl)acrylic acid | 18.46 ± 1.66 | 93.85 |
| TS012, trans−3‐indoleacrylic acid | 4.41 ± 0.06** | 22.42 |
| TS014, trans−3‐(4‐imidazolyl)acrylic acid | 18.81 ± 1.28 | 95.63 |
| TS016, trans−3‐methoxycinnamic acid | 8.72 ± 0.29** | 44.33 |
| TS018, trans−2‐methylcinnamic acid | 7.26 ± 0.16** | 36.91 |
| TS019, trans−3‐methylcinnamic acid | 10.79 ± 0.46** | 54.86 |
| TS022, trans−3‐chlorocinnamic acid | 9.23 ± 0.18** | 46.92 |
| TS024, trans−2‐carboxycinnamic acid | 21.07 ± 4.16 | 107.12 |
| TS025, trans‐cinnamamide | 17.96 ± 1.87 | 91.31 |
| TS026, trans−4‐mercaptocinnamic acid | 4.48 ± 0.06** | 22.78 |
| TS029, trans−4‐formylcinnamic acid | 16.72 ± 0.72** | 85.00 |
| TS030, methyl trans‐cinnamate | 28.43 ± 5.36 | 144.53 |
| TS031, trans−4‐carboxycinnamic acid | 19.61 ± 1.09 | 99.69 |
| TS032, cinnamyl alcohol | 29.88 ± 1.49 | 151.91 |
| TS033, salicylic acid | 12.51 ± 1.36* | 63.60 |
| TS034, benzoic acid | 21.65 ± 2.306 | 110.07 |
| TS124, trans−4‐hydroxymethylcinnamic acid | 20.67 ± 0.06 | 105.08 |
| TS125, trans−4‐methoxycinnamohydroxamic acid | 20.71 ± 0.97 | 105.29 |
| TS131, trans−3,4‐dihydroxycinnamohydroxamic acid | 9.06 ± 0.61** | 46.06 |
| TS133, trans−3‐(4‐hydroxyphenyl)acrylohydrazide | 11.75 ± 0.42** | 59.74 |
| TS134, benzhydroxamic acid | 12.01 ± 1.88* | 61.06 |
| TS135, salicylhydroxamic acid | 10.21 ± 0.34** | 51.91 |
| TS136, phenylpropiolic acid | 14.80 ± 2.473 | 75.24 |
aGreen fluorescent protein (GFP) mean fluorescence intensity (MFI) was determined for gated populations of bacterial cells by flow cytometry. Values are representative of at least two independent experiments, and three replicates were used for each experiment. Asterisks indicate statistically significant differences in MFI between bacterial cells grown in XOM2 with DMSO and XOM2 supplemented with 200 μm of each compound (Student's t‐test). *P < 0.05; **P < 0.01.
b%DMSO was used to represent the relative promoter activity of hpa1 in Xoo cells grown in XOM2 supplemented with 200 μm of each compound in comparison with that in XOM2 with DMSO only, which was calculated by the formula: %DMSO = 100 × MFI(XOM2 with phenolic compounds)/MFI(XOM2 with DMSO).
Five compounds suppress the HR caused by Xoo in tobacco
To determine whether the 10 inhibitors interfered with the function of T3SS in Xoo, we performed a secondary screening by examining their effects on the HR‐inducing ability of Xoo on tobacco. Infiltration of Xoo cells into non‐host tobacco leaves can induce HR, whereas deletion of the major T3SS regulatory gene hrpX completely abolishes the HR, suggesting that a functional T3SS is required for this phenotype (Fig. 1). Therefore, the HR‐inducing ability can indicate whether the T3SS is active in the bacteria. To determine the appropriate concentration of compounds for HR assays, we first examined the dose‐dependent effects of TS006, one of the strongest inhibitors of hpa1 promoter activity. TS006 began to show an inhibitory effect at a concentration of 50 µm. The impact was significantly enhanced at 100 μm and reached a plateau at 200 μm (Fig. S2, see Supporting Information). Therefore, we chose to test the effects at concentrations of 100 and 200 μm. Bacterial cell suspensions were incubated with DMSO or each compound for 2 h before they were infiltrated into tobacco leaves. Meanwhile, the cell suspensions were plated onto peptone sucrose agar (PSA) to check whether bacterial survival was affected by these compounds. At a concentration of 100 μm, none of them showed inhibition of HR (data not shown). When the concentration was increased to 200 μm, TS006, TS010, TS012, TS015 and TS018 caused a complete loss of HR (Fig. 1), suggesting that these compounds might effectively suppress the function of the Xoo T3SS. However, the numbers of bacterial cells were significantly decreased after treatment with TS012 (Fig. S3A, see Supporting Information). Cell counting revealed that bacterial populations were reduced by at least 50% (Fig. S3B), suggesting that TS012 has strong bactericidal activity. Comparatively, the other five compounds, TS026, TS130, TS139, TS141 and TS160, did not show inhibition of the HR phenotype at 200 μm (Fig. 1). Even when the concentration was increased to 400 μm, they did not demonstrate any inhibitory effect at all (data not shown), suggesting that they might not be functional in planta.
Figure 1.

Effects of various compounds on the hypersensitive response (HR) induced by Xanthomonas oryzae pv. oryzae (Xoo) on Nicotiana benthamiana. Cell suspensions of wild‐type Xoo PXO99A (wild‐type, WT) at an optical density at 600 nm (OD600) of 0.3 were incubated with dimethylsulfoxide (DMSO) or each compound at a concentration of 200 μm for 2 h before infiltration into tobacco leaves. ΔhrpX was used as a type III secretion system (T3SS) deficiency control. Photographs were taken 24 h after infiltration. At least four independent experiments were performed with similar results.
TS012 severely affects the growth rate of Xoo
As shown in the previous section, except for TS012, the other four compounds did not demonstrate any apparent bactericidal effects on Xoo. In line with this, we investigated their effect on bacterial growth at different stages. The growth rate of Xoo was measured in both rich medium (M210) and hrp‐inducing medium (XOM2) for a period of 72 h. The nutrient‐scarce medium XOM2 was supplemented with 0.5% sucrose to support bacterial growth (Tian et al., 2015). TS006, TS010, TS012, TS015 and TS018 were added to the media at a concentration of 200 μm. In comparison with the non‐treatment control, bacterial growth rates with the addition of either DMSO or compounds TS006, TS010, TS015 or TS018 did not show statistically significant changes at different time points, indicating that these compounds did not affect bacterial growth (Fig. 2). In contrast, treatment with TS012 significantly reduced the growth rate of Xoo. As we were looking for compounds that suppressed bacterial virulence, but did not affect bacterial growth or survival, we focused on TS006, TS010, TS015 and TS018 for the remainder of the study. To be consistent, we used a final concentration of 200 μm of each compound in all other assays.
Figure 2.
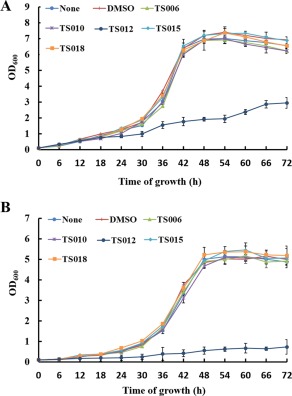
Effects of various compounds on bacterial growth rates. (A) The growth rate of Xanthomonas oryzae pv. oryzae (Xoo) PXO99A in rich medium (M210) supplemented with dimethylsulfoxide (DMSO) or 200 μm of TS006, TS010, TS012, TS015 or TS018. (B) The growth rate of Xoo PXO99A in hrp‐inducing medium (XOM2) (plus 0.5% sucrose) supplemented with DMSO or 200 μm of TS006, TS010, TS012, TS015 or TS018. The optical density at 600 nm (OD600) of the culture suspensions was measured every 6 h during the 72‐h period. Two independent tests were performed with similar results.
TS006, TS010, TS015 and TS018 suppress the translocation of PXO_04172 and PXO_03702 in rice
Translocation of T3 effectors into the host cell is the primary function of the T3SS. To evaluate directly whether this process was affected by the four inhibitors, we examined their effect on the translocation of two non‐TAL effectors, which have been confirmed previously to be translocated using a Cya‐based reporter system (Furutani et al., 2009). Two plasmids, pHM04172 and pHM03702, carrying T3 effectors XOO3803 and XOO4042 of Xoo strain MAFF31101 (Furutani et al., 2009), corresponding to effectors PXO_04172 and PXO_03702 in PXO99A, were transformed into PXO99A wild‐type and hrpX mutant strains for translocation assays. With only DMSO treatment, a considerable amount of cyclic adenosine monophosphate (cAMP) was detected in rice leaves infected with wild‐type stains carrying PXO_04172 or PXO_03702, whereas it was barely detectable after infection with the hrpX mutant strains, suggesting that these two effectors were properly translocated by the T3SS of PXO99A (Fig. 3). Compared with DMSO treatment, there was a significant reduction in cAMP accumulation in the presence of TS006, TS010, TS015 and TS018 (Fig. 3). These results indicate that translocation of PXO_04172 and PXO_03702 was suppressed by these four inhibitors, suggesting that the translocation process through the T3SS in Xoo might be repressed under these conditions.
Figure 3.
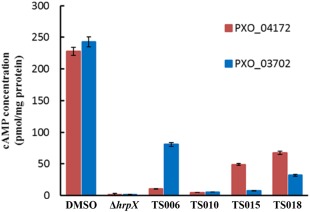
The effect of TS006, TS010, TS015 and TS018 on the translocation of the type III effectors PXO_04172 and PXO_03702 from Xanthomonas oryzae pv. oryzae (Xoo) PXO99A into rice cells, examined by Cya translocation assays. The concentration of each compound was 200 μm. An equal volume of dimethylsulfoxide (DMSO) was used as a control. The level of cyclic adenosine monophosphate (cAMP) indicates the level of the translocated T3 effectors. Three replicates were used in each experiment. At least three independent tests were performed with similar results.
Expression of representative hrp/hrc genes is suppressed by inhibitors TS006, TS010, TS015 and TS018
The data outlined above demonstrate that TS006, TS010, TS015 and TS018 suppress the Xoo T3SS without affecting bacterial growth, implying that they might specifically interfere with the regulatory pathway for T3SS gene expression. Therefore, we carried out quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) experiments to examine the effect of these inhibitors on the expression levels of T3SS genes. The first screening showed that TS006, TS010, TS015 and TS018 reduced the promoter activity of hpa1 by at least 60% (Table 1). Here, the qRT‐PCR assays demonstrated that the mRNA level of hpa1 was reduced by over 80% in Xoo cells treated with these inhibitors, which is consistent with the results obtained from the promoter assay (Fig. 4A). Next, we examined the expression levels of other hrp/hrc genes, including hrpE (encoding the hrp pilus protein), hrpF (encoding a putative translocon protein), hrcC (encoding the outer‐membrane secretin) and export apparatus genes hrcT and hrcU (Gürlebeck et al., 2006). As expected, the mRNA levels of the tested hrp/hrc genes were reduced significantly when the inhibitors were present, in comparison with the DMSO control (Fig. 4A,B). It is worth noting that TS006, among the four inhibitors, consistently showed the strongest inhibitory effect on all genes tested.
Figure 4.
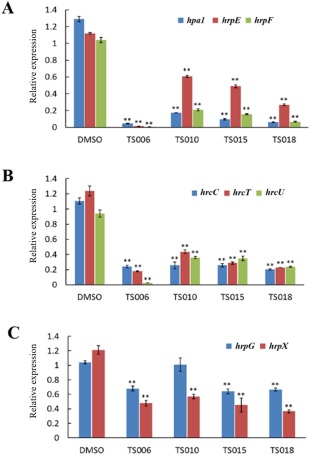
Relative mRNA levels of hrp genes in Xanthomonas oryzae pv. oryzae (Xoo) PXO99A incubated with compounds TS006, TS010, TS015 and TS018, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). (A) The mRNA levels of hpa1, hrpE and hrpF were reduced significantly after treatment with these inhibitors in comparison with the dimethylsulfoxide (DMSO) control. (B) The mRNA levels of three hrc genes, i.e. hrcC, hrcT and hrcU, were also reduced significantly after treatment with these inhibitors in comparison with the DMSO control. (C) The mRNA levels of hrpG and hrpX genes were reduced to varying extents after treatment with different inhibitors in comparison with the DMSO control. The concentration of each compound used was 200 μm. An equal volume of DMSO was used as a control. gyrB was used as an endogenous control for data analysis. Three replicates were used in each experiment. Three independent tests were performed with similar results. Asterisks indicate statistically significant differences (Student's t‐test). **P < 0.01.
The four inhibitors affect the expression of regulatory genes hrpG and hrpX
It is known that the expression of the Xanthomonas T3SS is controlled by the regulatory cascade consisting of HrpG and HrpX (Büttner and Bonas, 2010). As we observed a reduction in mRNA levels in representative hpa/hrp/hrc genes in Xoo after treatment with the four inhibitors, it was imperative to investigate whether the expression of hrpG and hrpX was affected. The mRNA levels of hrpG and hrpX were examined in Xoo cells grown in the presence of each inhibitor. In comparison with the DMSO control, the mRNA level of hrpX was reduced by approximately 50% in the presence of each inhibitor (Fig. 4C). In contrast, the mRNA level of hrpG was reduced by about 30% by TS006, TS015 and TS018, and even less by TS010 (Fig. 4C). These results suggest that the inhibitory effect of TS006, TS010, TS015 and TS018 on T3SS gene expression of Xoo exists through the HrpG–HrpX regulatory cascade, but with varying degrees of intensity.
The promoters of hrcC and hrpB respond differentially to inhibitors TS006, TS010, TS015 and TS018
To further investigate whether HrpG or HrpX played a role in relaying the impact of the inhibitors on the transcription of T3SS genes in Xoo, we examined their effect on two different promoters within their regulon. As mentioned above, most of the genes in the hrp cluster were activated by the transcriptional regulator HrpX, which recognizes the PIP‐box (TTCGC‐N15‐TTCGC) in the promoter region, such as that of hpa1 (Fig. S4A, see Supporting Information). The hrpB operon, spanning from hrpB1 to hrcT, is located within the core hrp gene cluster of Xoo (Fig. 5A) (Cho et al., 2008). hrcC, which is next to hrcT has been confirmed to be transcribed together with the hrpB operon (Cho et al., 2008). Interestingly, it has been shown recently that hrcC in Xoc also uses an internal promoter for transcription (Li et al., 2011). The internal promoter in front of hrcC, designated as PhrcC, does not contain a PIP‐box, and was regulated by HrpG, but not HrpX (Li et al., 2011). Sequence alignment of the promoter regions of hrcC in Xoo PXO99A and Xoc RS105 showed that they share over 90% identity (Fig. S4B). Therefore, we amplified this region, named PhrcC in Xoo (Fig. 5A), to test whether its activity was affected by the four inhibitors. In addition, we amplified the promoter for the hrpB operon, designated as PhrpB, which contains a perfect PIP‐box for comparison (Fig. S4C). FACS assays were performed to examine the impact of TS006, TS010, TS015 and TS018 on the activity of PhrcC and PhrpB. After calculating the ratio of compound treatment to the DMSO control (DMSO%), we found that the activity of PhrcC was not affected by these four compounds at all (Fig. 5B). In contrast, the activity of PhrpB decreased by at least 50% after treatment with these inhibitors in comparison with the DMSO control. This effect was similar to their effect on the hpa1 promoter (Fig. 5B). These results suggest that promoters containing a PIP‐box might be the main targets of these inhibitors, which also supports the statement that HrpX plays an important role in the mediation of the inhibitory effect. Although the activity of PhrcC was not affected by these inhibitors, its mRNA level was reduced significantly, as shown above (Fig. 4B). A plausible hypothesis might be that part of the mRNA transcribed from PhrpB is reduced, leading to a decrease in the total mRNA level of hrcC.
Figure 5.
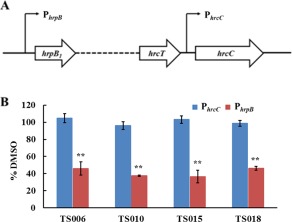
Impact of TS006, TS010, TS015 and TS018 on the promoter activity of hrpB and hrcC. (A) Schematic diagram of PhrpB and PhrcC. (B) Promoter activities of hrpB and hrcC in Xanthomonas oryzae pv. oryzae (Xoo) PXO99A grown in XOM2 supplemented with 200 μm of TS006, TS010, TS015 and TS018 for 15 h, measured by fluorescence‐activated cell sorting (FACS). Dimethylsulfoxide (DMSO) was used as a negative control. Green fluorescent protein (GFP) mean fluorescence intensity (MFI) was determined by flow cytometry. %DMSO = 100 × MFI(XOM2 with each compound)/MFI(XOM2 with equal volume of DMSO). Three independent tests were performed with similar results.
The expression of other virulence factors of Xoo is not suppressed by T3SS inhibitors
Previously, it has been shown that several compounds related to salicylidene acylhydrazides suppress both T3SS and the EPS amylovoran in E. amylovora (Yang et al., 2014). Therefore, we wanted to investigate whether the effects of the Xoo T3SS inhibitors identified above were specific or effective on multiple virulence factors. The disruption of the T2SS structural genes xpsE and xpsF led to reduced secretion of extracellular enzymes and impaired virulence of Xoo (Ray et al., 2000; Sun et al., 2005). The mRNA levels of xpsE and xpsF were measured by qRT‐PCR in Xoo cells treated with the T3SS inhibitors TS006, TS010, TS015 and TS018. In comparison with the DMSO control, the four compounds did not reduce the mRNA levels of xpsE and xpsF (Fig. 6A). The expression of representative gum genes in Xoo cells treated with the four inhibitors was also examined. The results showed that the mRNA levels of gumB, gumD, gumK and gumM were not reduced by these T3SS inhibitors (Fig. 6B).
Figure 6.
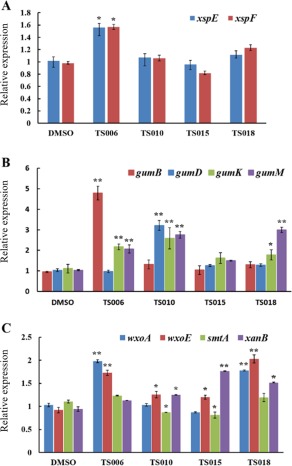
Impact of 200 μm of TS006, TS010, TS015 and TS018 on the mRNA levels of genes encoding the type II secretion system (T2SS) (A), exopolysaccharide (EPS) (B) and lipopolysaccharide (LPS) (C) in Xanthomonas oryzae pv. oryzae (Xoo) PXO99A, measured by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The gyrB gene was used as an endogenous control for data analysis. Three replicates were used in each experiment. Three independent tests were performed with similar results. Asterisks indicate statistically significant differences (Student's t‐test). *P < 0.05; **P < 0.01. DMSO, dimethylsulfoxide.
The impact of the four inhibitors on the mRNA levels of wxoE, smtA and wxoA from cluster I, and xanB from cluster II, was similarly examined. Except for smtA, which was slightly down‐regulated by TS010 and TS015, the other three genes did not show reduced expression after treatment by the four inhibitors. Overall, these results clearly demonstrate that the four T3SS inhibitors do not suppress major gene expression of T2SS, EPS and LPS. Instead, their expression is enhanced in some cases (Fig. 6A–C), which is probably a result of negative cross‐talk between different virulence factors. Therefore, we conclude that the inhibitory effect of these inhibitors is specific to T3SS, and not expanded to other virulence factors.
TS006, TS010, TS015 and TS018 suppress the water‐soaking and disease symptoms of Xoo on rice
Our focus in this work was to demonstrate that these inhibitors can suppress the virulence phenotypes of Xoo on rice. On the seedlings of susceptible rice cultivar IR24, Xoo PXO99A induced water‐soaked lesions after infiltration of bacterial cells into the leaves (Fig. 7A). The water‐soaking symptoms were reduced to various levels by treatment of the bacterial cells with TS006, TS010, TS015 and TS018 (Fig. 7A). Moreover, the yellowish disease symptoms on adult IR24 plants were also weakened by these inhibitors (Fig. 7B), and the lesion lengths were significantly reduced in comparison with DMSO treatment (Fig. 7C). In addition, we also measured the bacterial population in the diseased leaf tissue. After treatment with each inhibitor, the bacterial numbers isolated from the leaves were significantly reduced (Fig. S5, see Supporting Information). These results indicate that TS006, TS010, TS015 and TS018 do indeed suppress the virulence of Xoo cells, and reflect their propagation in planta.
Figure 7.
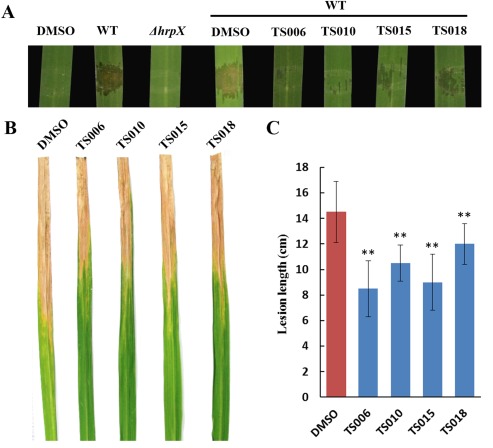
Compounds TS006, TS010, TS015 and TS018 suppress the virulence of Xanthomonas oryzae pv. oryzae (Xoo) PXO99A on rice cultivar IR24. (A) The effect of TS006, TS010, TS015 and TS018 on the water‐soaking symptoms caused by Xoo PXO99A (wild‐type, WT) on IR24 seedling. ΔhrpX was used as a type III secretion system (T3SS) deficiency control. Photographs were taken 3 days after infiltration. The disease symptoms (B) and lesion lengths (C) of Xoo PXO99A on adult plants of rice cultivar IR24 were reduced after pretreatment with TS006, TS010, TS015 and TS018. Photographs were taken 14 days after infiltration. At least three independent tests were performed with similar results. Asterisks indicate statistically significant differences (Student's t‐test). **P < 0.01. DMSO, dimethylsulfoxide.
TS006, TS010, TS015 and TS018 suppress the virulence of Xoc on rice
As the hrp clusters among Xanthomonas spp. are highly conserved (Büttner and Bonas, 2010; Nino‐Liu et al., 2006), and the major regulatory proteins HrpG and HrpX are also functional in Xoc (Zou et al., 2006), we speculated that it might be possible for these inhibitors to also suppress the T3SS of Xoc. Therefore, it was reasonable to test whether they suppressed the virulence of Xoc on rice. As shown in Fig. 8A, the disease symptoms caused by Xoc RS105 on rice seedlings were significantly reduced by TS006 and TS010 (Fig. 8A). On adult plants with only DMSO treatment, the lesion length can reach 4–5 cm, whereas, with the addition of the four compounds, lesion lengths were below 1 cm (Fig. 8B,C), suggesting that the virulence of Xoc was significantly suppressed by these inhibitors. As the four compounds exhibited stronger inhibitory effects on the virulence of Xoc than Xoo at the same concentration (200 µm), it is necessary to include the possibility that they might affect the growth or survival of Xoc. Therefore, we measured the growth rate of Xoc in both rich medium (nutrient broth, NB) and hrp‐inducing medium (XOM2) (plus 0.5% sucrose) during a period of 72 h. TS006, TS010, TS015 and TS018 were added to the media at a final concentration of 200 μm. In comparison with the non‐treatment control, bacterial growth rates with the addition of either DMSO or each compound did not show significant alterations at different time points, indicating that these compounds did not affect the bacterial growth of Xoc (Fig. S6, see Supporting Information). Thus, we conclude that TS006, TS010, TS015 and TS018 suppress the virulence of Xoc, but do not affect its growth.
Figure 8.
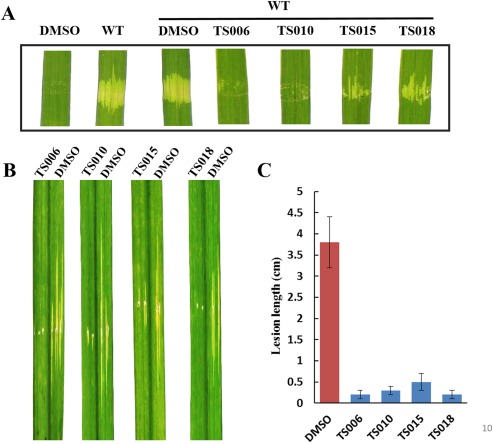
Compounds TS006, TS010, TS015 and TS018 suppress the virulence of Xanthomonas oryzae pv. oryzicola (Xoc) RS105 on rice cultivar IR24. (A) The impact of TS006, TS010, TS015 and TS018 on disease symptoms caused by infiltration of Xoc RS105 (wild‐type, WT) in IR24 seedlings. Photographs were taken 3 days after infiltration. (B) The impact of TS006, TS010, TS015 and TS018 on disease symptoms cause by needling infiltration of Xoc RS105 in IR24 adult plants. Photographs were taken 14 days after infiltration. (C) Lesion lengths on adult plants. At least three independent tests were performed with similar results. DMSO, dimethylsulfoxide.
Discussion
Rice is the staple food of the world, feeding more than one‐half of its population. However, rice is vulnerable to pathogen infection, which often leads to devastating diseases, resulting in severe yield losses. Bacterial leaf blight and leaf streak diseases caused by the two pathovars of X. oryzae are the most important bacterial diseases around the world, especially in Asia and Africa (Sundaram et al., 2014). Here, we report, for the first time, that phenolic compounds TS006, TS010, TS015 and TS018 are able to suppress the disease symptoms of Xoo and Xoc on rice by specifically inhibiting the function of the T3SS. The compounds were identified by screening a small library of natural phenolics and their derivatives, some of which have been shown previously to affect the T3SS gene expression of several bacterial pathogens, including D. dadantii (Li et al., 2009; Yang et al., 2008), P. aeruginosa (Yamazaki et al., 2012) and E. amylovora (Khokhani et al., 2013; Li et al., 2015).
A whole‐cell‐based high‐throughput screening (HTS) approach has often been used to identify T3SS inhibitors from compound libraries (Marshall and Finlay, 2014). Here, the Xoo strain PXO99A, harbouring a reporter plasmid with a gfp gene transcriptionally fused to the hpa1 promoter, was constructed for screening. Purposely, the hpa1 promoter was used because Hpa1 has been confirmed to be a T3SS‐secreted harpin protein in both Xoo and Xoc (Furutanin et al., 2003; Zhu et al., 2000; Zou et al., 2006). Twenty‐five of 45 compounds showed significant inhibitory effects on hap1 promoter activity (Table 1). This efficiency is much higher than that of HTS using large libraries containing thousands of natural or synthetic small molecules (Charro and Mota, 2015; Felise et al., 2008). A secondary screening was performed to identify HR inhibitors from the 10 strongest compounds showing inhibition of promoter activity. At a concentration of 200 μm, TS006, TS010, TS012, TS015 and TS018 showed suppression of HR of Xoo in tobacco (Fig. 1). Interestingly, the other five compounds (TS026, TS130, TS139, TS141 and TS160) did not demonstrate any inhibitory effects at all, even at higher concentrations (Fig. 1). There could be many reasons for their inability to suppress HR in planta. One possible explanation is that they might not be stable in planta to perform their functions. Of course, more work needs to be performed in the future to fully elucidate the exact cause.
As a result of their possible positive impact on agriculture, it is important to understand the functional mechanism of these T3SS inhibitors. Our results indicated that TS006, TS010, TS015 and TS018 suppressed hrp/hrc gene expression, probably through the major regulatory proteins HrpG and HrpX (Fig. 4). In addition, we examined the activity of PhrcC, which does not contain a PIP‐box like Phpa1 (Fig. 5). In Xoc, PhrcCP was positively regulated by HrpG, but not HrpX (Li et al., 2011). The activity of PhrcC was not suppressed by the four T3SS inhibitors, suggesting that the regulatory role of HrpX might be indispensable for the function of these inhibitors. We speculated that these inhibitors may exert their effects upstream of HrpG, as the expression of hrpG and hrpX was affected (Fig. 4). Presumably, phosphorylated HrpG activates the expression and production of HrpX, and HrpX regulates the downstream genes (Büttner and Bonas, 2010). However, the signalling transduction upstream of HrpG remains elusive. Recently, a putative histidine kinase of HrpG has been reported in X. campestris pv. campestris (Li et al., 2014). In addition, the RNA‐binding protein RsmA has been shown to positively regulate the T3SS by stabilizing HrpG mRNA in X. citri ssp. citri (Andrade et al., 2014). It will be of great interest to study whether these components play similar roles in regulating the T3SS in X. oryzae, and whether they mediate the inhibitory effects of these compounds on the T3SS.
By comparison with previous screening results of these phenolic compounds in other bacteria, we observed some differences in their activities. For example, TS006 (OCA), the inhibitor of the Xoo T3SS, which has also been shown to inhibit the T3SS of E. amylovora (Khokhani et al., 2013), was initially identified to induce T3SS gene expression in D. dadantii (Li et al., 2009; Yang et al., 2008). In contrast, TS004 (PCA), a strong inhibitor of the T3SS in D. dadantii and E. amylovora, did not show significant alteration of hpa1 promoter activity in Xoo (Table 1). In D. dadantii, PCA inhibited the T3SS gene expression through the HrpX/HrpY TCS (Li et al., 2009), which is a pathway that does not exist in Xoo. Therefore, different T3SS regulatory pathways might be a major reason for the distinctive activities detected for these compounds.
Given the possibility that the T3SS inhibitors might also affect the expression of other virulence factors, it was important to observe the behaviour of these types of T3SS inhibitor. Here, we analyzed the effects of TS006, TS010, TS015 and TS018 on the expression of other important virulence factors in Xoo, including the T2SS, EPS and LPS (Büttner and Bonas, 2010; Das et al., 2009; Kim et al., 2009; Rai et al., 2012; Rajeshwari et al., 2005). We found that most genes encoding their products were not affected by the four T3SS inhibitors (Fig. 6). These results are consistent with the proposed mechanism that the T3SS inhibitors act through the HrpG–HrpX cascade, as the transcription of these genes is most probably not controlled by HrpX. Meanwhile, it was intriguing that some genes were up‐regulated by the addition of the inhibitors. A similar phenomenon has also been reported previously. For example, in E. amylovora, transcriptome analysis demonstrated that a large number of genes were up‐regulated when treated with T3SS inhibitors (Yang et al., 2014). In Salmonella, when a compound was used to inhibit the secretion of the T3 substrate SipA, a significantly increased amount of the flagellin protein FliC was detected (Felise et al., 2008). This might be a result of cross‐talk between virulence factors in bacteria, which would warrant further investigation.
Water soaking is a symptom specifically related to the function of the AvrBs3/PthA family of effectors in Xanthomonads, which were more recently renamed as ‘TAL (transcription activator‐like)’ effectors (Verdier et al., 2012; White and Yang, 2009). In Xoo strain PXO99A, there are multiple TAL effectors, which confer gene‐specific resistance in host plants (White and Yang, 2009). After syringe infiltration in the susceptible rice cultivar IR24, where no corresponding R genes exist, the PXO99A strain caused strong water‐soaked lesions (Verdier et al., 2012; Fig. 7A). We found that, after performing a treatment protocol with TS006 and TS010, the water‐soaking phenotypes on rice were almost completely abolished (Fig. 7A). TS015 and TS018 also suppressed the water‐soaking phenotype to varying levels (Fig. 7A). Using the translocation assays including two non‐TAL effectors as representatives (Fig. 3), these results demonstrated that the primary function of the T3SS, which is the delivery of effectors into plant cells, was suppressed by TS006, TS010, TS015 and TS018 with different intensities.
Finally, virulence assays were performed to evaluate the effect of the four inhibitors in preventing the disease symptoms of Xoo and Xoc on rice. Unexpectedly, TS006 and TS010, which almost completely abolished the water‐soaking symptoms, only reduced the disease lesion lengths of Xoo by 20%–30% (Fig. 7C). This might be because other virulence factors were still functional or even enhanced by these inhibitors (Fig. 6). In comparison, the reduction in disease lesion lengths of Xoc on rice was more evident (Fig. 8B,C). Although these T3SS inhibitors were screened using Xoo as the reporter strain, it was not surprising to observe their functions in Xoc, as the major T3SS regulatory pathways are very similar between Xoo and Xoc (Cho et al., 2008; Zou et al., 2006). With regard to why the inhibitors were more efficient in suppressing the virulence of Xoc, different infection methods between the two pathovars might be a possible reason.
In summary, we have demonstrated the inhibitory effect of phenolic compounds TS006, TS010, TS015 and TS018 on the T3SS of Xoo both in vitro and in planta. Furthermore, these compounds have been proven to be effective in suppressing the disease symptoms caused by Xoo and Xoc on rice leaves. These findings are important because they may provide potential anti‐virulence drugs, which might be used to prevent infection in agricultural production in the future.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. Escherichia coli was grown in Luria–Bertani (LB) medium at 37°C. Xoo wild‐type strain PXO99A and the derived strains were grown in M210 medium (0.8% casein enzymatic hydrolysate, 0.5% sucrose, 0.4% yeast extract, 17.2 mm K2HPO4, 1.2 mm MgSO4·7H2O) or on PSA plates. XOM2 medium (0.18% d‐(+) xylose, 670 µm l‐methionine, 10 mm sodium l‐(+) glutamate, 14.7 mm KH2PO4, 40 µm MnSO4, 240 µm Fe(III)‐EDTA and 5 mm MgCl2, the pH was adjusted to 6.5 with KOH) was used for hrp‐inducing conditions (Tsuge et al., 2002). Xoc wild‐type strain RS105 was grown on NA (0.5% peptone, 0.1% yeast, 1% sucrose, 0.3% beef extract and 1.5% agar) or NB (NA without agar) medium at 28°C. Antibiotics were used at the following final concentrations (μg/mL) when required: ampicillin (Ap), 100; spectinomycin (Sp), 50; kanamycin (Kan), 50; cephalexin (Cp), 25.
Table 2.
Strains and plasmids used in this study.
| Strains and plasmids | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| Xanthomonas oryzae pv. oryzae | ||
| PXO99A | Wild‐type strain, Philippine race 6, Cpr | Laboratory collection |
| ΔhrpX | hrpX gene deletion mutant of PXO99A, Cpr | This study |
| Xanthomonas oryzae pv. oryzicola | ||
| RS105 | Wild‐type, Chinese race 2 | Dr Fengquan Liu |
| Plasmids | ||
| pPROBE‐AT | Promoter‐probe vector, Apr | |
| pKMS1 | Suicidal vector carrying sacB gene for non‐marker mutagenesis, Kmr | Dr Gong‐you Chen (Li et al., 2011) |
| pPhpa1 | pProbe‐AT derivative with PCR fragment containing hpa1 promoter region, Apr | This study |
| pPhrcC | pProbe‐AT derivative with PCR fragment containing hrcC promoter region, Apr | This study |
| pPhrpB | pProbe‐AT derivative with PCR fragment containing hrpB promoter region, Apr | This study |
| pKHrpX | pKMS1 derivative carrying an in‐frame hrpX mutation, Kmr | This study |
| pHM04172 | The putative promoter region and the 5'‐coding region of PXO_04172 fused with a cya‐tag, Spr | Dr Seiji Tsuge (Furutani et al., 2009) |
| pHM03702 | The putative promoter region and the 5'‐coding region of PXO_03702 fused with a cya‐tag, Spr | Dr Seiji Tsuge (Furutani et al., 2009) |
Apr, ampicillin resistance; Cpr, cephalexin resistance; Kmr, kanamycin resistance; Spr, spectinomycin resistance.
Sources of the screened compounds
Compounds TS001 to TS035, TS108 to TS113, and TS134 to TS136 were purchased from commercial sources Aldrich (St. Louis, MO, USA), Alfa Aesar (Ward Hill, MA, USA), and TCI (Tokyo, Japan). The remaining compounds were synthesized via the routes described in our previously published papers (Khokhani et al., 2013; Yamazaki et al., 2012). All compounds were dissolved in DMSO.
Construction of the hrpX gene deletion mutant
An in‐frame deletion mutation of the hrpX gene was constructed in PXO99A through homologous recombination using the suicide vector pKMS1, as described previously (Li et al., 2011). The sacB gene (sucrose sensitivity counter‐selectable marker) on pKMS1 confers suicide ability to the host bacterium during growth on high‐concentration sucrose‐containing medium. Briefly, approximately 600 bp of the upstream and 900 bp of the downstream region of the hrpX gene were amplified from PXO99A genomic DNA using the primer pairs hrpXUSF/R and hrpXDSF/R, respectively. The primers used in this study are listed in Table S2 (see Supporting Information). The two fragments were ligated into the suicide vector pKMS1, and introduced into PXO99A by electroporation. The transformants were first selected on NAN medium (consisting of kanamycin, 1% tryptone, 0.1% yeast extract, 0.3% peptone, 1.5% agar) and after continuous transfer culture in NBN broth (1% tryptone, 0.1% yeast extract, 1% sucrose, 0.3% peptone) four times. The potential mutants were selected on NAS medium (1% tryptone, 0.1% yeast extract, 10% sucrose, 0.3% peptone and 1.5% agar). The mutant candidates that grew on NAS, but were sensitive to kanamycin, were further confirmed by PCR.
Construction of reporter strains and flow cytometry analysis
To screen compounds that induce or inhibit the expression of the PXO99A T3SS, a 246‐bp fragment containing the promoter region of hpa1 was PCR amplified using the primers Phpa1‐F and Phpa1‐R. The amplified fragment was digested with BamHI and EcoRI, and ligated to pPROBE‐AT, a broad‐host‐range vector carrying a promoter‐less gfp gene (Miller et al., 2000), resulting in pPhpa1. This plasmid was then transferred to PXO99A by electroporation. PXO99A carrying the pPhpa1 or promoterless pPROBE‐AT was grown in M210 overnight and transferred to XOM2 or XOM2 supplemented with 200 μm of each compound. The promoter activity of hpa1 was analysed using a FACS‐Caliber flow cytometer (BD Bioscience, San Jose, CA, USA) as described previously (Yamazaki et al., 2012). An equivalent volume of DMSO was added as a negative control. Three independent experiments were performed, and three replicates were used in each experiment. The promoter activities of hrcC and hrpB were analysed by a similar method. PhrcC‐F/R and PhrcT‐F/R were used to amplify the promoter regions of hrcC and hrpB, respectively (Li et al., 2011).
RNA extraction and qPCR analysis
Xoo cells were cultured in M210 medium overnight at 28°C and subcultured to XOM2 at an optical density at 600 nm (OD600) of 0.3, supplemented with DMSO or 200 μm of each compound for 15 h. Total RNA was isolated using an RNAprep Pure Bacteria Kit (Tiangen, Beijing, China). cDNA was synthesized using an HiScriptII Q RT SuperMix Kit (Vazyme, Nanjing, China). The cDNA levels of different samples were quantified by real‐time PCR using a SYBR Green Master Mix (Vazyme). The relative levels of gene expression were determined using the 2–ΔΔCT method (Livak and Schmittgen, 2001), with the DNA gyrase subunit B (gyrB) gene as the internal control (Tsuge et al., 2002). Three technical replicates were used each time.
HR assay
Xoo cells were cultured in M210 medium overnight at 28°C and resuspended in sterile distilled water. The bacterial suspensions were adjusted to OD600 = 0.3. Nicotiana benthamiana plants were used for HR assays. Cell suspensions were mixed with 200 μm of each compound or DMSO, and incubated at 28°C for 2 h, before infiltration into tobacco using a needleless syringe. The HR symptoms were observed and photographed at 24 h after inoculation. Bacterial cell numbers were counted after serial dilutions and plating on PSA plates.
Measurement of the growth rate
Xoo or Xoc cells were grown overnight in M210 or NB at 28°C. The cells were then resuspended in M210, NB or XOM2 (plus 0.5% sucrose) medium supplemented with DMSO or 200 μm of each compound, starting at an OD600 of 0.05. The growth rates were monitored every 6 h during the 72‐h period using a Synergy 4 multimode microplate reader (BioTek, Winooski, VT, USA). Two independent experiments were performed, and three replicates were used in each experiment.
Adenylate cyclase translocation assays
Plasmids expressing C‐terminal Cya fusions of type III effectors PXO_04172 and PXO_03702, which were named as pHM04172 and pHM03702, were kindly provided by Dr Seiji Tsuge (Furutani et al., 2009). PXO99A strain carrying pHM04172 or pHM03702 was grown in M210 medium overnight at 28°C, and resuspended in sterile distilled water to an OD600 of 0.6. The bacterial cell suspensions were incubated with DMSO or 200 μm of each compound for 2 h before infiltration into the leaves of rice (Oryza sativa ssp. indica cultivar IR24) for translocation assays. Rice leaf sections (length, 1 cm) that included the inoculation sites were collected 3 days after inoculation, and homogenized in 300 μL of 0.1 m HCl with stainless beads using a vibration‐ball mill (GRINOER, Beijing, China) (1000 rpm for 1 min). Then, cell debris was removed by centrifugation (13 400 g) for 2 min. The supernatant was collected in new tubes and stored at −80°C until use. The level of cAMP was quantified using the cAMP ELISA kit (Enzo Life Sciences, Lausen, Switzerland) system according to the manufacturer's instructions. The protein concentration of each sample was determined with a BCA Protein Assay kit (CWBIO, Beijing, China).
Pathogenicity assays
Xoo and Xoc cells were cultured in M210 or NB medium overnight at 28°C and resuspended in sterile distilled water. Cell suspensions at an OD600 of 0.8 were mixed with 200 μm of the compounds or DMSO and incubated at 28°C for 2 h. Oryza sativa ssp. indica rice cultivar IR24 was used for pathogenicity assays. On 2‐week‐old seedlings, bacterial cells of Xoo or Xoc were inoculated using a needleless syringe. On 2‐month‐old adult plants, bacterial cells of Xoo or Xoc were inoculated by the leaf clipping or leaf needling method, respectively. Plants were scored at 3 days post‐inoculation (dpi) for symptoms in seedlings, and at 14 dpi for lesion lengths in adult rice plants. Plants were maintained in a glasshouse at 25°C (16 h of light and 8 h of darkness) during the experiments.
For bacterial population assay, the top 20 cm of each leaf was ground in sterilized distilled water and the cell suspensions were plated onto PSA after serial dilutions, and then incubated at 28°C. Bacterial colonies were counted after 72 h. At least 10 leaves were used for each compound treatment, and all experiments were repeated at least three times.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 DNA sequence of hpa1 gene and its promoter region. Sequences for a plant‐inducible promoter (PIP)‐box, start codon, stop codon and the primers used for amplification of the promoter region are indicated in bold.
Fig. S2 Effects of TS006 on promoter activity of hpa1 at different concentrations. The growth of Xanthomonas oryzae pv. oryzae (Xoo) in XOM2 supplemented with different concentrations of TS006 was recorded by measuring the absorbance of the bacterial suspension at 600 nm. DMSO, dimethylsulfoxide; OD, optical density.
Fig. S3 Effects of various compounds on bacterial survival. Bacterial cells were incubated with dimethylsulfoxide (DMSO) or various compounds at a concentration of 200 μm for 2 h at 28°C before being plated on a peptone sucrose agar (PSA) plate after serial dilutions. Three independent tests were performed with similar results. CFU, colony‐forming unit.
Fig. S4 Sequence alignment of the promoter regions of hpa1 (A), hrcC (B) and hrpB (C) between Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc).
Fig. S5 Effects of TS006, TS010, TS015 and TS018 on the bacterial population of Xanthomonas oryzae pv. oryzae (Xoo) PXO99A in leaves of IR24. Bacterial populations were measured in 20‐cm leaf segments at 14 days after inoculation. CFU, colony‐forming unit; DMSO, dimethylsulfoxide.
Fig. S6 Effects of various compounds on bacterial growth rates. (A) Growth rate of Xanthomonas oryzae pv. oryzicola (Xoc) RS105 in rich medium (NB, nutrient broth) supplemented with dimethylsulfoxide (DMSO) or 200 μm of TS006, TS010, TS015 or TS018. (B) Growth rate of Xoc RS105 in hrp‐inducing medium XOM2 (plus 0.5% sucrose) supplemented with DMSO or 200 μm of TS006, TS010, TS015 or TS018. The optical density at 600 nm (OD600) of the culture suspensions was measured every 6 h during the 72‐h period. Two independent tests were performed with similar results.
Table S1 Phenolic compounds precipitated when added to XOM2.
Table S2 Primers used in this study.
Acknowledgements
We would like to thank Dr Seiji Tsuge for providing us with plasmids pHM04172 and pHM03702. We also thank Dr Fengquan Liu for providing the Xoc RS105 strain. This work was supported by grants from the Special Fund for Agro‐Scientific Research in the Public Interest of China (201303015), the National Key Project for Basic Research (973 Project, 2015CB150600), National Science Foundation of China (31370160) for CH, Beiing Natural Science Foundation (5142017) for FT, and the Research Growth Initiative of the University of Wisconsin‐Milwaukee and the Catalyst Grant in Advanced Automation of UWM Research Foundation for C‐HY.
Contributor Information
Fang Tian, Email: ftian@ippcaas.cn.
Ching‐Hong Yang, Email: chyang@uwm.edu.
References
- Alfano, J.R. and Collmer, A. (1997) The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179, 5655–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.O. , Farah, C.S. and Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5′ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas . PLoS Pathog. 10, e1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczak, A.K. and Hung, D.T. (2009) Productive steps toward an antimicrobial targeting virulence. Curr. Opin. Microbiol. 12, 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowlin, N.O. , Williams, J.D. , Knoten, C.A. , Torhan, M.C. , Tashjian, T.F. , Li, B. , Aiello, D. , Mecsas, J. , Hauser, A.R. , Peet, N.P. , Bowlin, T.L. and Moir, D.T. (2014) Mutations in the Pseudomonas aeruginosa needle protein gene pscF confer resistance to phenoxyacetamide inhibitors of the type III secretion system. Antimicrob. Agents Chemother. 58, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol. Mol. Biol. Rep. 76, 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2003) Common infection strategies of plant and animal pathogenic bacteria. Curr. Opin. Microbiol. 6, 312–319. [DOI] [PubMed] [Google Scholar]
- Büttner, D. and Bonas, U. (2010) Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 34, 107–133. [DOI] [PubMed] [Google Scholar]
- Charro, N. and Mota, L.J. (2015) Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin. Drug. Discov. 10, 373–387. [DOI] [PubMed] [Google Scholar]
- Chatterjee, A. , Cui, Y. and Chatterjee, A.K. (2002) Regulation of Erwinia carotovora hrpL(Ecc) (sigma‐L(Ecc)), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant–Microbe Interact. 15, 971–980. [DOI] [PubMed] [Google Scholar]
- Cho, H.J. , Park, Y.J. , Noh, T.H. , Kim, Y.T. , Kim, J.G. , Song, E.S. , Lee, D.H. and Lee, B.M. (2008) Molecular analysis of the hrp gene cluster in Xanthomonas oryzae pathovar oryzae KACC10859. Microb. Pathog. 44, 473–483. [DOI] [PubMed] [Google Scholar]
- Cornelis, G.R. (2006) The type III secretion injectisome. Nat. Rev. 4, 811–825. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E. , and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Das, A. , Rangaraj, N. and Sonti, R.V. (2009) Multiple adhesin‐like functions of Xanthomonas oryzae pv. oryzae are involved in promoting leaf attachment, entry, and virulence on rice. Mol. Plant–Microbe Interact. 22, 73–85. [DOI] [PubMed] [Google Scholar]
- Felise, H.B. , Nguyen, H.V. , Pfuetzner, R.A. , Barry, K.C. , Jackson, S.R. , Blanc, M.P. , Bronstein, P.A. , Kline, T. and Miller, S.I. (2008) An inhibitor of gram‐negative bacterial virulence protein secretion. Cell Host Microbe, 4, 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutanin, A. , Tsuge, S. , Oku, T. , Tsuno, K. , Inoue, Y. , Ochiai, H. , Kaku, H. and Kubo, Y. (2003) Hpa1 secretion via type III secretion system in Xanthomonas oryzae pv. oryzae . J. Gen. Plant Pathol. 69, 271–275. [Google Scholar]
- Furutani, A. , Takaoka, M. , Sanada, H. , Noguchi, Y. , Oku, T. , Tsuno, K. , Ochiai, H. and Tsuge, S. (2009) Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 22, 96–106. [DOI] [PubMed] [Google Scholar]
- Garrity‐Ryan, L.K. , Kim, O.K. , Balada‐Llasat, J.M. , Bartlett, V.J. , Verma, A.K. , Fisher, M.L. , Castillo, C. , Songsungthong, W. , Tanaka, S.K. , Levy, S.B. , Mecsas, J. and Alekshun, M.N. (2010) Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect . Immun. 78, 4683–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürlebeck, D. , Thieme, F. and Bonas, U. (2006) Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163, 233–255. [DOI] [PubMed] [Google Scholar]
- Jessen, D.L. , Bradley, D.S. and Nilles, M.L. (2014) A type III secretion system inhibitor targets YopD while revealing differential regulation of secretion in calcium‐blind mutants of Yersinia pestis . Antimicrob. Agents Chemother. 58, 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhani, D. , Zhang, C. , Li, Y. , Wang, Q. , Zeng, Q. , Yamazaki, A. , Hutchins, W. , Zhou, S.S. , Chen, X. and Yang, C.H. (2013) Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora . Appl. Environ. Microb. 79, 5424–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , Kim, J.G. , Lee, B.M. and Cho, J.Y. (2009) Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv oryzae . Biotechnol. Lett. 31, 265–270. [DOI] [PubMed] [Google Scholar]
- Lattanzio, V. (2013) Phenolic compounds: introduction In: Handbook of Natural Products (Ramawat K.G. and Mérillon J.‐M, eds), pp. 1543–1580. Berlin, Heidelberg: Springer. [Google Scholar]
- Livak, K. J. , and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Li, R.F. , Lu, G.T. , Li, L. , Su, H.Z. , Feng, G.F. , Chen, Y. , He, Y.Q. , Jiang, B.L. , Tang, D.J. and Tang, J.L. (2014) Identification of a putative cognate sensor kinase for the two‐component response regulator HrpG, a key regulator controlling the expression of the hrp genes in Xanthomonas campestris pv. campestris . Environ. Microbiol. 16, 2053–2071. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Peng, Q. , Selimi, D. , Wang, Q. , Charkowski, A.O. , Chen, X. and Yang, C.H. (2009) The plant phenolic compound p‐coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl. Environ. Microb. 75, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Hutchins, W. , Wu, X. , Liang, C. , Zhang, C. , Yuan, X. , Khokhani, D. , Chen, X. , Che, Y. , Wang, Q. and Yang, C.H. (2015) Derivative of plant phenolic compound inhibits the type III secretion system of Dickeya dadantii via HrpX/HrpY two‐component signal transduction and Rsm systems. Mol. Plant Pathol. 16, 150–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.R. , Zou, H.S. , Che, Y.Z. , Cui, Y.P. , Guo, W. , Zou, L.F. , Chatterjee, S. , Biddle, E.M. , Yang, C.H. and Chen, G.Y. (2011) A novel regulatory role of HrpD6 in regulating hrp‐hrc‐hpa genes in Xanthomonas oryzae pv. oryzicola . Mol. Plant–Microbe Interact. 24, 1086–1101. [DOI] [PubMed] [Google Scholar]
- Marshall, N.C. and Finlay, B.B. (2014) Targeting the type III secretion system to treat bacterial infections. Expert Opin. Ther. Targets, 18, 137–152. [DOI] [PubMed] [Google Scholar]
- Miller, W.G. , Leveau, J.H. and Lindow, S.E. (2000) Improved gfp and inaZ broad‐host‐range promoter‐probe vectors. Mol. Plant–Microbe Interact. 13, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Nino‐Liu, D.O. , Ronald, P.C. and Bogdanove, A.J. (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. [DOI] [PubMed] [Google Scholar]
- Noel, L. , Thieme, F. , Nennstiel, D. and Bonas, U. (2001) cDNA‐AFLP analysis unravels a genome‐wide hrpG‐regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria . Mol. Microbiol. 41, 1271–1281. [DOI] [PubMed] [Google Scholar]
- Rai, R. , Ranjan, M. , Pradhan, B.B. and Chatterjee, S. (2012) Atypical regulation of virulence‐associated functions by a diffusible signal factor in Xanthomonas oryzae pv. oryzae . Mol. Plant–Microbe Interact. 25, 789–801. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. and Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant–Microbe Interact. 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Rasko, D.A. and Sperandio, V. (2010) Anti‐virulence strategies to combat bacteria‐mediated disease. Nat. Rev. Drug Discov. 9, 117–128. [DOI] [PubMed] [Google Scholar]
- Ray, S.K. , Rajeshwari, R. and Sonti, R.V. (2000) Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol. Plant–Microbe Interact. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L. , Sommer, D.D. , Schatz, M.C. , Phillippy, A.M. , Rabinowicz, P.D. , Tsuge, S. , Furutani, A. , Ochiai, H. , Delcher, A.L. , Kelley, D. , Madupu, R. , Puiu, D. , Radune, D. , Shumway, M. , Trapnell, C. , Aparna, G. , Jha, G. , Pandey, A. , Patil, P.B. , Ishihara, H. , Meyer, D.F. , Szurek, B. , Verdier, V. , Koebnik, R. , Dow, J.M. , Ryan, R.P. , Hirata, H. , Tsuyumu, S. , Won Lee, S. , Seo, Y.S. , Sriariyanum, M. , Ronald, P.C. , Sonti, R.V. , Van Sluys, M.A. , Leach, J.E. , White, F.F. and Bogdanove, A.J. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q.H. , Hu, J. , Huang, G.X. , Ge, C. , Fang, R.X. and He, C.Z. (2005) Type‐II secretion pathway structural gene xpsE, xylanase‐ and cellulase secretion and virulence in Xanthomonas oryzae pv. oryzae . Plant Pathol. 54, 15–21. [Google Scholar]
- Sundaram, R.M. , Chatterjee, S. , Oliva, R. , Laha, G.S. , Cruz, C.V. , Leach, J.E. and Sonti, R.V. (2014) Update on Bacterial Blight of Rice: Fourth International Conference on Bacterial Blight. Rice, 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, X. , Xiao, Y. and Zhou, J.M. (2006) Regulation of the type III secretion system in phytopathogenic bacteria. Mol. Plant–Microbe Interact. 19, 1159–1166. [DOI] [PubMed] [Google Scholar]
- Tian, F. , Yu, C. , Li, H. , Wu, X. , Li, B. , Chen, H. , Wu, M. and He, C. (2015) Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae . Microbiol. Res. 170, 177–183. [DOI] [PubMed] [Google Scholar]
- Tsuge, S. , Furutani, A. , Fukunaka, R. , Oku, T. , Tsuno, K. , Ochiai, H. , Inoue, Y. , Kaku, H. and Kubo, Y. (2002) Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen . Plant Pathol. 68, 363–371. [Google Scholar]
- Verdier, V. , Triplett, L.R. , Hummel, A.W. , Corral, R. , Cernadas, R.A. , Schmidt, C.L. , Bogdanove, A.J. and Leach, J.E. (2012) Transcription activator‐like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector‐deficient strain of Xanthomonas oryzae . New Phytol. 196, 1197–1207. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Zetterström, C.E. , Gabrielsen, M. , Beckham, K.S.H. , Tree, J.J. , Macdonald, S.E. , Byron, O. , Mitchell, T.J. , Gally, D.L. , Herzyk, P. , Mahajan, A. , Uvell, H. , Burchmore, R. , Smith, B.O. , Elofsson, M. and Roe, A.J. (2011) Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence‐blocking compounds. J. Biol. Chem. 286, 29 922–29 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Y. , Song, C.F. , Miao, W.G. , Ji, Z.L. , Wang, X. , Zhang, Y. , Zhang, J.H. , Hu, J.S. , Borth, W. and Wang, J.S. (2008) Mutations in the N‐terminal coding region of the harpin protein Hpa1 from Xanthomonas oryzae cause loss of hypersensitive reaction induction in tobacco. Appl. Microbiol. Biotechnol. 81, 359–369. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- White, F.F. and Yang, B. (2009) Host and pathogen factors controlling the rice–Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Heu, S. , Yi, J. , Lu, Y. and Hutcheson, S.W. (1994) Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, A. , Li, J. , Zeng, Q. , Khokhani, D. , Hutchins, W.C. , Yost, A.C. , Biddle, E. , Toone, E.J. , Chen, X. and Yang, C.H. (2012) Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS–GacA two‐component signal transduction system. Antimicrob. Agents Chemother. 56, 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Korban, S.S. , Pusey, P.L. , Elofsson, M. , Sundin, G.W. and Zhao, Y. (2014) Small‐molecule inhibitors suppress the expression of both type III secretion and amylovoran biosynthesis genes in Erwinia amylovora . Mol. Plant Pathol. 15, 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Peng, Q. , San Francisco, M. , Wang, Y. , Zeng, Q. and Yang, C.H. (2008) Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS One, 3, e2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, W. , MaGbanua, M.M. and White, F.F. (2000) Identification of two novel hrp‐associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae . J. Bacteriol. 182, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L.F. , Wang, X.P. , Xiang, Y. , Zhang, B. , Li, Y.R. , Xiao, Y.L. , Wang, J.S. , Walmsley, A.R. and Chen, G.Y. (2006) Elucidation of the hrp clusters of Xanthomonas oryzae pv. oryzicola that control the hypersensitive response in nonhost tobacco and pathogenicity in susceptible host rice. Appl. Environ. Microbiol. 72, 6212–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 DNA sequence of hpa1 gene and its promoter region. Sequences for a plant‐inducible promoter (PIP)‐box, start codon, stop codon and the primers used for amplification of the promoter region are indicated in bold.
Fig. S2 Effects of TS006 on promoter activity of hpa1 at different concentrations. The growth of Xanthomonas oryzae pv. oryzae (Xoo) in XOM2 supplemented with different concentrations of TS006 was recorded by measuring the absorbance of the bacterial suspension at 600 nm. DMSO, dimethylsulfoxide; OD, optical density.
Fig. S3 Effects of various compounds on bacterial survival. Bacterial cells were incubated with dimethylsulfoxide (DMSO) or various compounds at a concentration of 200 μm for 2 h at 28°C before being plated on a peptone sucrose agar (PSA) plate after serial dilutions. Three independent tests were performed with similar results. CFU, colony‐forming unit.
Fig. S4 Sequence alignment of the promoter regions of hpa1 (A), hrcC (B) and hrpB (C) between Xanthomonas oryzae pv. oryzae (Xoo) and Xanthomonas oryzae pv. oryzicola (Xoc).
Fig. S5 Effects of TS006, TS010, TS015 and TS018 on the bacterial population of Xanthomonas oryzae pv. oryzae (Xoo) PXO99A in leaves of IR24. Bacterial populations were measured in 20‐cm leaf segments at 14 days after inoculation. CFU, colony‐forming unit; DMSO, dimethylsulfoxide.
Fig. S6 Effects of various compounds on bacterial growth rates. (A) Growth rate of Xanthomonas oryzae pv. oryzicola (Xoc) RS105 in rich medium (NB, nutrient broth) supplemented with dimethylsulfoxide (DMSO) or 200 μm of TS006, TS010, TS015 or TS018. (B) Growth rate of Xoc RS105 in hrp‐inducing medium XOM2 (plus 0.5% sucrose) supplemented with DMSO or 200 μm of TS006, TS010, TS015 or TS018. The optical density at 600 nm (OD600) of the culture suspensions was measured every 6 h during the 72‐h period. Two independent tests were performed with similar results.
Table S1 Phenolic compounds precipitated when added to XOM2.
Table S2 Primers used in this study.


