Summary
The leucine‐rich repeat (LRR) proteins play important roles in the recognition of corresponding ligands and signal transduction networks in plant defence responses. Herein, a novel LRR protein from Capsicum annuum, CaLRR51, was identified and characterized. It was localized to the plasma membrane and transcriptionally up‐regulated by Ralstonia solanacearum infection (RSI), as well as the exogenous application of salicylic acid (SA), jasmonic acid (JA) and ethephon (ETH). Virus‐induced gene silencing of CaLRR51 significantly increased the susceptibility of pepper to RSI. By contrast, transient overexpression of CaLRR51 in pepper plants activated hypersensitive response (HR)‐like cell death, and up‐regulated the defence‐related marker genes, including PO2, HIR1, PR1, DEF1 and ACO1. Moreover, ectopic overexpression of CaLRR51 in transgenic tobacco plants significantly enhanced the resistance to RSI. Transcriptional expression of the corresponding defence‐related marker genes in transgenic tobacco plants was also found to be enhanced by the overexpression of CaLRR51, which was potentiated by RSI. These loss‐ and gain‐of‐function assays suggest that CaLRR51 acts as a positive regulator in the response of pepper to RSI. In addition, the putative signal peptide and transmembrane region were found to be required for plasma membrane targeting of CaLRR51, which is indispensable for the role of CaLRR51 in plant immunity.
Keywords: Capsicum annuum, leucine‐rich repeat (LRR) protein, plant defence response, Ralstonia solanacearum
Introduction
In their natural habitats, plants inevitably encounter a broad range of microbial pathogens, and have evolved sophisticated defence mechanisms to defend themselves against these potential threats. In addition to physical or chemical barriers, such as a waxy cuticle and preformed antimicrobial compounds, plants also employ innate immunity to actively respond to pathogen attack (Schneider, 2002). This active form of defence consists of two interconnected branches, known as pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) and effector‐triggered immunity (ETI), on perception of PAMPs and effectors by cell surface‐localized pattern recognition receptors (PRRs) and disease resistance (R) proteins in host cells, respectively (Jones and Dangl, 2006). The never‐ending arms race between plants and pathogens may drive the continuous emergence of interconnected PRRs or R proteins in plants and PAMPs or effectors in pathogens, resulting in a continuum between typical PTI and ETI (Block et al., 2008; Boller and Felix, 2009; Boller and He, 2009; Jones and Dangl, 2006; Lee et al., 2015). However, knowledge of this continuum is still very limited.
Leucine‐rich repeats (LRRs) exist widely in plant proteins and are involved in protein–protein interactions and ligand binding, and therefore in pathogen perception and signal transduction (McHale et al., 2006). Based on their subcellular localization, LRRs can be divided into extracellular (eLRR) and intracellular (iLRR) LRRs (Zhou et al., 2009). iLRRs are mainly found in NB‐LRR R proteins, named after their central nucleotide‐binding (NB) domain, with an LRR domain in the C‐terminal region and a Toll/Interleukin‐1 Receptor (TIR) homology or predicted coiled‐coil (CC) domain in the N‐terminal region (Caplan et al., 2008), in which LRRs play important roles in the determination of the recognition specificity of R proteins to pathogen effectors (Dodds et al., 2001, 2006; Hwang and Williamson, 2003; Rairdan and Moffett, 2006), or in the auto‐inhibition of the NB‐LRR R protein through intramolecular interactions with the region carrying the NB‐ARC (nucleotide‐binding adaptor shared by APAF‐1, R proteins, and CED‐4) and CC domains (Ade et al., 2007; Moffett et al., 2002; Rairdan and Moffett, 2006; Qi et al., 2012). Interactions between R proteins and other signalling components, for example SGT1, can be mediated by the LRR domain in R proteins (Bieri et al., 2004). eLRRs have been found frequently in receptor like kinases (RLKs), which contain an eLRR domain, a single membrane‐spanning helix and a cytosolic kinase domain for signal transduction (van der Hoorn and Jones, 2004), and in receptor‐like proteins (RLPs), which contain an eLRR domain, a single transmembrane helix and a small cytosolic tail (Zhou et al., 2009). Emerging data have shown that LRR proteins, such as SLRR in sorghum (Hipskind et al., 1996), LeLRP in tomato (Tornero et al., 1996), NtLRP1 in tobacco (Jacques et al., 2006), CaLRR1 in pepper (Jung et al., 2004; Jung and Hwang, 2007) and OsLRR1 in rice (Zhou et al., 2009), exhibit structural similarities to RLKs with different subcellular localizations, and may play a role in plant immunity (Zhou et al., 2009). In rice, the OsLRR1 protein can enter the endosomal pathway and act as a positive regulator in the defence response by interacting with OsHIR1 in the plasma membrane (Zhou et al., 2009, 2010). In pepper, the CaLRR1 protein has been found to act as a positive regulator in plant immunity by interacting with pathogenesis‐related protein 10 (PR10) in the cytoplasm and the apoplastic space (Choi et al., 2012); however, it suppresses hypersensitive response (HR)‐like cell death and the defence response by interacting with hypersensitive‐induced reaction 1 (HIR1) in the plasma membrane (Choi et al., 2011; Jung and Hwang, 2007), or PR4b in the plasma membrane and the apoplast (Hwang et al., 2014). Despite the difference between the sequences of CaLRR1 and OsLRR1, and their modes of action in plant immunity, both CaLRR1 and OsLRR1 can interact with HIR1, and this interaction has been found to be highly conserved among different plant species (Zhou et al., 2009). All these results indicate that eLRR‐containing proteins play important roles in plant immunity. However, the data available on the roles of eLRR‐containing proteins in plants is very limited, even though a large number of genes encoding eLRR proteins can be found in the genomes of different plant species.
In the present study, a novel LRR protein from pepper, CaLRR51, structurally distinct from the previously characterized CaLRR1, was identified and characterized. It was localized to the plasma membrane and transcriptionally up‐regulated by Ralstonia solanacearum infection (RSI), as well as by exogenously applied salicylic acid (SA), jasmonic acid (JA) and ethephon (ETH). The data of loss‐ and gain‐of‐function assays indicated that CaLRR51 acts as a positive regulator in the response of pepper to RSI. In addition, the signal peptide (SP) and transmembrane region (TR) were found to be required for plasma membrane targeting of CaLRR51, which is required for the role of CaLRR51 in pepper immunity.
Results
Cloning and sequence analysis of a novel LRR protein CaLRR51
A normalized cDNA library of pepper inbred line GZ03 was constructed previously to isolate full‐length Capsicum annuum cDNAs (our group's unpublished data). One of the positive cDNA clones was isolated by random sequencing and identified as a member of the LRR domain proteins, which is identical to the sequence of C. annuum cultivar CM334 PGAv.1.5.contig190568 (accession no: AYRZ01190568.1). The length of this cDNA was 1586 bp, including 80 bp of 5′‐untranslated region, 1287 bp of open reading frame (ORF) and 219 bp of 3′‐untranslated region. The ORF of this gene was predicted to encode a protein of 428 amino acids with three successive domains from the N‐terminus to the C‐terminus: a putative SP, an LRR domain and a TR (Fig. 1A). The putative SP was predicted with a cleavage site after the 28th amino acid. There were six tandem repeats of the LRR motif in the central region of the deduced amino acid sequence, containing a domain exhibiting similarity to LRR RLP kinases. The TR was predicted at the C‐terminus, starting at residue 402 and ending at residue 424.
Figure 1.

Structural domains and comparison of amino acid sequences of CaLRR51 with its homologues. (A) The amino acid sequence of CaLRR51 was predicted with three successive domains from N‐terminus to C‐terminus: a putative signal peptide (SP), a leucine‐rich repeat (LRR) domain with six tandem repeats of the LRR motif and a transmembrane region (TR). Domains were detected by Blastp (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and smart (http://smart.embl-heidelberg.de/). (B) Sequence alignment of CaLRR51 with its homologues in some other plant species, including Arabidopsis thaliana (AtLRR51, accession no: NP_193611.1), Gossypium arboreum (GaLRR, accession no: ACD56661.1), Glycine max (GmLRR, accession no: XP_003541695.1), Nicotiana sylvestris (NsLRR, accession no: XP_009770243.1), Nicotiana tomentosiformis (NTLRR, accession no: XP_009594315.1) and Solanum lycopersicum (SlLRR, accession no: XP_004234080.1).
The deduced amino acid sequence of this isolated cDNA exhibits high similarity (56%–81% identity) with its orthologues in other plant species, including Arabidopsis thaliana (AtLRR51, accession no: NP_193611.1), Gossypium arboreum (accession no: ACD56661.1), Glycine max (accession no: XP_003541695.1), Nicotiana sylvestris (accession no: XP_009770243.1), Nicotiana tomentosiformis (accession no: XP_009594315.1) and Solanum lycopersicum (accession no: XP_004234080.1). As a result of its high sequence similarity to AtLRR51 from A. thaliana, we designated this cDNA clone as CaLRR51. Notably, CaLRR51 in pepper and its orthologues in other plant species do not seem to be functionally characterized in the existing literature.
We also compared CaLRR51 with other previously characterized eLRR proteins, such as CaLRR1 (accession number: AY237117.1), OsLRR1 (accession number: AAO85403.1) (Fig. S1, see Supporting Information), AtSERK1 (A. thaliana somatic embryogenesis receptor‐like kinase; accession number: NP_177328), AtSERK3 (accession number: NP_567920), OsSERK1 (accession number: BAD86793), rice LRR protein (Oryza sativa LRR protein; accession no: AP000815), SLRR (Sorghum bicolor LRR protein; accession no: U62279) and RLK (A. thaliana receptor‐like kinase protein; accession no: NM119198) (data not shown). This revealed that these LRR proteins share no appreciable homology with CaLRR51, except for the LRR motifs.
Up‐regulation of CaLRR51 transcript levels by RSI and exogenous phytohormone application
As LRR proteins have been implicated in plant immunity, to test whether this holds for CaLRR51, its transcriptional expression was measured by quantitative real‐time reverse transcription‐polymerase chain reaction (qRT‐PCR) on inoculation of R. solanacearum, the causal agent of pepper bacterial wilt (Fig. 2). On leaf inoculation with the highly virulent R. solanacearum strain FJC100301, the CaLRR51 transcripts were up‐regulated nearly three‐fold at 24 and 48 h post‐inoculation (hpi) in pepper plants, suggesting that CaLRR51 participates in the defence response of pepper to R. solanacearum.
Figure 2.
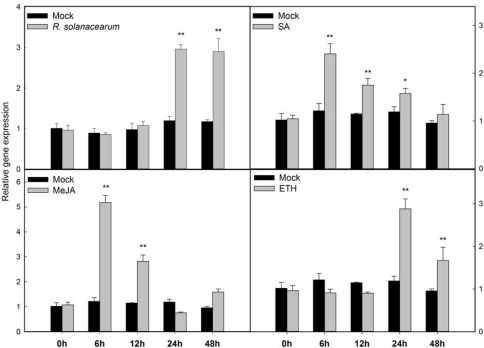
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of CaLRR51 in pepper plants during Ralstonia solanacearum infection and exogenous phytohormone application. For R. solanacearum infection assay, the third leaves from the top of pepper plants at the eight‐leaf stage were infiltrated with 10 μL of the highly virulent R. solanacearum strain FJC100301 suspension [optical density at 600 nm (OD600) = 0.8] using a syringe without a needle, and the mock was inoculated with 10 mm MgCl2. For exogenous phytohormone application, pepper plants at the four‐leaf stage were sprayed with 1 mm salicylic acid (SA), 100 µm methyl jasmonate (MeJA) or 100 µm ethephon (ETH), and the mock treatment was sprayed with a corresponding solvent or sterile double‐distilled H2O. The leaves were collected at the indicated time points for RNA extraction and qRT‐PCR analysis. The relative transcript levels of CaLRR51 were compared with those in mock‐treated control plants. Data are the means ± standard deviation from three independent experiments. Asterisks indicate statistically significant differences compared with mock treatment by the least‐significant difference (LSD) test (*P < 0.05; **P < 0.01).
Phytohormones, such as SA, JA and ethylene (ET), serve as important signalling molecules and are involved ubiquitously in the responses of plants to pathogen attack. To assess the possible association of CaLRR51 with signalling mediated by these hormones, the transcript abundance of CaLRR51 was determined by qRT‐PCR in leaves of pepper plants (at the four‐leaf stage) exogenously treated with SA, methyl jasmonate (MeJA) and ETH (Fig. 2). The results showed that the transcript levels of CaLRR51 were rapidly induced by 2.5‐fold at 6 h post‐treatment (hpt) of SA, and gradually decreased to the basal level at 48 hpt. On MeJA treatment, the transcript levels of CaLRR51 were significantly increased up to 5.3‐fold at 6 hpt and 2.8‐fold at 12 hpt. In response to ETH treatment, the transcript levels of CaLRR51 were elevated nearly three‐fold at 24 hpt and 1.6‐fold at 48 hpt. These results suggest that CaLRR51 plays a role in the response of pepper to RSI and is involved in the signalling pathways mediated by these hormones.
Silencing of CaLRR51 enhances the susceptibility of pepper to R. solanacearum
To further study the role of CaLRR51 in the response of pepper to RSI, we performed loss‐of‐function experiments in pepper seedlings by virus‐induced gene silencing (VIGS) of CaLRR51. qRT‐PCR analysis showed that CaLRR51 was effectively silenced in Tobacco rattle virus (TRV):CaLRR51 pepper plants; the transcript levels in TRV:CaLRR51 plants were reduced to approximately 28% of those in TRV:00 plants (Fig. 3A). On leaf inoculation with R. solanacearum strain FJC100301, the transcript levels in TRV:CaLRR51 plants were only 3%–18% of those in TRV:00 plants at 24 and 48 hpi, and the growth of R. solanacearum was significantly enhanced in CaLRR51‐silenced pepper plants compared with that in TRV:00 plants (Fig. 3B). Consistently, the CaLRR51‐silenced pepper plants exhibited more severe disease symptoms compared with control plants after inoculation with R. solanacearum in pepper roots. These results suggest that CaLRR51 plays a role in host defence against R. solanacearum.
Figure 3.
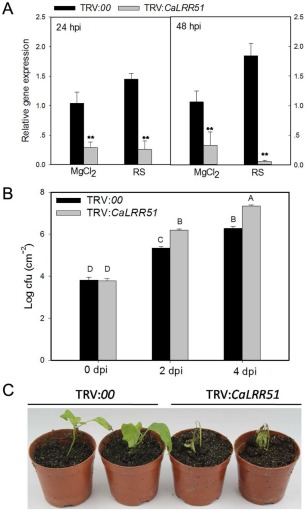
Virus‐induced gene silencing (VIGS) of CaLRR51 enhances the susceptibility of pepper to Ralstonia solanacearum. (A) Relative transcriptional expression of CaLRR51 in leaves of CaLRR51‐silenced pepper plants (TRV:CaLRR51) versus empty vector control plants (TRV:00) by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) at 24 and 48 h post‐inoculation (hpi) with R. solanacearum or MgCl2 (mock). (B) Bacterial growth (colony‐forming units, cfu) in CaLRR51‐silenced or empty vector control pepper leaves inoculated with R. solanacearum at 2 and 4 days post‐inoculation (dpi). Data are the means ± standard deviation of three independent experiments. Different letters indicate statistically significant differences compared with the empty vector control plants by the least‐significant difference (LSD) test (P < 0.01). (C) Disease symptoms of CaLRR51‐silenced and empty vector control plants at 7 dpi after root inoculation with R. solanacearum.
Transient overexpression of CaLRR51 induces cell death and defence‐related marker gene expression in pepper plants
To further confirm that CaLRR51 acts as a positive regulator in the defence response of pepper to R. solanacearum, CaLRR51 was transiently expressed in pepper leaves by infiltration with Agrobacterium tumefaciens strain GV3101 carrying 35S:CaLRR51 or 35S:00 (empty vector). As shown in Fig. 4A, transient overexpression of CaLRR51 triggered an obvious HR‐like cell death response at 4 days post‐inoculation (dpi), but not in empty vector control plants. The HR‐like cell death response was then assessed by trypan blue staining to identify necrotic cells. There was no or a very weak HR‐mediated necrotic response in leaves infiltrated with Ag. tumefaciens carrying the empty vector, whereas transient overexpression of 35S:CaLRR51 strongly induced the necrotic response in pepper leaves. Hydrogen peroxide production in pepper leaves was also detected by diaminobenzidine (DAB) staining. There was visible DAB staining in leaves with transient expression of 35S:CaLRR51, but not in leaves infiltrated with GV3101 strain containing the empty vector. We also performed an ion leakage test to analyse the severity of cell necrosis caused by plasma membrane damage in leaves expressing 35S:CaLRR51 (Fig. 4B). Greater ion leakage at 48 and 72 h after agroinfiltration was observed in leaves expressing 35S:CaLRR51 than in the empty vector control.
Figure 4.
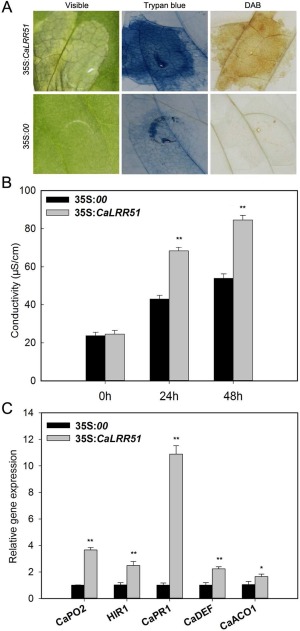
Transient overexpression of CaLRR51 induces hypersensitive response (HR)‐like cell death and defence‐related gene expression in pepper plants. (A) Pepper leaves were infiltrated with Agrobacterium GV3101 carrying the 35S:00 (empty vector) or 35S:CaLRR51 construct. Phenotypes of infiltrated pepper leaves at 4 days after agroinfiltration (left), by trypan blue staining (middle) and by diaminobenzidine (DAB) staining (right). (B) Electrolyte leakage assay of pepper leaves after agroinfiltration with the 35S:00 or 35S:CaLRR51 construct at 24 and 48 h post‐inoculation (hpi). (C) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of defence‐related gene expression in pepper leaves transiently overexpressing 35S:CaLRR51 or 35S:00 at 48 hpi. The relative transcript levels were normalized to the pepper CaActin gene. Data are the means ± standard deviation of three independent experiments. Asterisks indicate statistically significant differences compared with the empty vector controls by the least‐significant difference (LSD) test (*P < 0.05; **P < 0.01).
To test whether transient overexpression of CaLRR51 could alter defence‐related gene expression in pepper plants, we also examined the transcript abundances of defence‐related marker genes, including the HR‐associated gene CaHIR1, reactive oxygen species (ROS) detoxification‐associated gene CaPO2, SA‐responsive gene CaPR1, JA‐responsive gene CaDEF1 and ET‐associated gene CaACO1 (Fig. 4C). The results showed that the relative transcription levels of these defence‐related genes were increased in plants by transient overexpression of CaLRR51. In particular, the transcript level of CaPR1 was increased over 10‐fold compared with that in control plants.
Stable overexpression of CaLRR51 enhances resistance of tobacco to R. solanacearum inoculation
As pepper is very recalcitrant to genetic transformation, we generated stable transgenic tobacco plants to determine the in vivo function of CaLRR51. We obtained 15 independent T0 transgenic tobacco lines that constitutively expressed CaLRR51 driven by the Cauliflower mosaic virus (CaMV) 35S promoter. Two T2 transgenic lines, CaLRR51‐OX‐5 and CaLRR51‐OX‐6, which exhibited the highest levels of CaLRR51 transcripts, displayed visible normal morphology and set viable seeds, were selected for further analyses. Eight‐week‐old plants of the two transgenic lines and wild‐type (WT) line were inoculated in roots with the highly virulent R. solanacearum strain FJC100301. Seven days after inoculation, severe wilting symptoms had developed on wild‐type plants; however, the transgenic lines displayed less severe disease symptoms (Fig. 5A). To quantify the extent of disease in R. solanacearum‐infected plants, bacterial growth in the third leaves of WT and transgenic tobacco plants was assessed at 36 hpi. Significantly decreased bacterial growth was observed in transgenic tobacco plants compared with wild‐type plants (Fig. 5B). We also examined local defence responses by DAB (as an indicator of H2O2 accumulation) and trypan blue (as an indicator of cell death or necrosis) staining of R. solanacearum‐inoculated leaves. The infected leaves of CaLRR51‐OX transgenic lines exhibited clearly increased intensities of H2O2 accumulation (dark brown) and HR‐like cell death (dark blue) compared with wild‐type plants (Fig. 5C).
Figure 5.
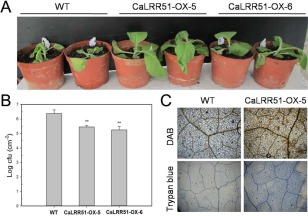
Transgenic tobacco plants overexpressing CaLRR51 show increased resistance to Ralstonia solanacearum infection. (A) Disease symptoms of 8‐week‐old transgenic tobacco lines (CaLRR51‐OX‐5 and CaLRR51‐OX‐6) and the wild‐type (WT) line at 7 days after root inoculation with the highly virulent R. solanacearum strain FJC100301. (B) Bacterial growth (colony‐forming units, cfu) in the third leaves of WT and transgenic tobacco plants at 36 h after leaf inoculation with R. solanacearum. Data are the means ± standard deviation of three independent experiments. Asterisks indicate statistically significant differences compared with the WT controls by the least‐significant difference (LSD) test (**P < 0.01). (C) Increased H2O2 accumulation and hypersensitive response (HR)‐like cell death in CaLRR51‐OX‐5 transgenic tobacco leaves compared with WT leaves inoculated with R. solanacearum at 24 h post‐inoculation. DAB, diaminobenzidine.
To further investigate the possible mode of action of CaLRR51, the expression of defence‐related marker genes was analysed in CaLRR51‐OX transgenic tobacco plants and wild‐type plants by qRT‐PCR. We examined the transcript levels of the HR‐associated gene NtHSR201, ROS detoxification‐associated gene NtCAT1, SA‐responsive genes NtPR1a and NtCHN50, JA‐responsive gene NtLOX1 and ET‐associated gene NtACO1. The results showed that the relative transcription levels of these defence‐related genes were constitutively increased in CaLRR51‐OX transgenic plants, except for NtPR1a and NtACO1 (Fig. 6A). We also detected the transcript levels of these defence‐related genes in tobacco plants after leaf inoculation with R. solanacearum at 24 and 48 hpi. All the defence marker genes were strongly induced in the transgenic lines in at least one of the two tested time points during R. solanacearum infection (Fig. 6B). Taken together, the overexpression of CaLRR51 in tobacco plants appears to enhance disease resistance against R. solanacearum by elevated HR‐like cell death and H2O2 levels, accompanied by increased transcript levels of HR‐associated genes and other defence genes, which is consistent with the positive role of CaLRR51 observed in pepper plants.
Figure 6.
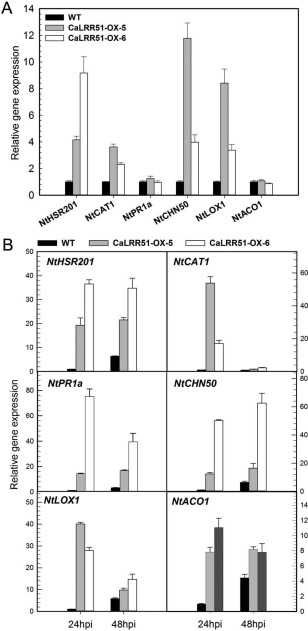
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of defence‐related gene expression in transgenic tobacco plants overexpressing CaLRR51, with or without Ralstonia solanacearum inoculation. (A) The relative transcript levels of defence‐related genes in the third leaves of CaLRR51‐OX transgenic tobacco plants and wild‐type plants without R. solanacearum inoculation. (B) The relative transcript levels of defence‐related genes in the third leaves of CaLRR51‐OX transgenic tobacco plants and wild‐type plants at 24 and 48 h after leaf inoculation with R. solanacearum. NtHSR201, hypersensitive response (HR)‐associated gene; NtCAT1, reactive oxygen species (ROS) detoxification‐associated gene; NtPR1a and NtCHN50, salicylic acid (SA)‐responsive genes; NtLOX1, jasmonic acid (JA)‐responsive gene; NtACO1, ethylene (ET)‐associated gene. The relative transcript levels were normalized to the pepper CaActin gene. Data were the means ± standard deviation of three independent experiments.
The SP/TR deletion mutants of CaLRR51 attenuate HR‐like cell death in pepper plants
To determine the subcellular localization of CaLRR51, we generated a CaLRR51‐GFP fusion construct (35S:CaLRR51‐GFP) driven by the constitutive CaMV 35S promoter. As computational analysis of the CaLRR51 protein highlighted putative SP and TR at the N‐terminus and C‐terminus, respectively (Fig. 1), we also constructed vectors of SP and/or TR deletion mutants of CaLRR51 fused with the green fluorescent protein (GFP). Nicotiana benthamiana leaves were used to express these constructs by co‐agroinfiltration with a 35S:CBL1n‐RFP fusion construct (as an additional control targeting to the plasma membrane), and were visualized at 48 hpi using a confocal microscope (Batistic et al., 2008) (Fig. 7A). The green fluorescent signals of 35S:GFP control construct in N. benthamiana leaves were observed in the cytoplasm and nuclei. However, CaLRR51‐GFP was localized primarily at the plasma membrane, similar to our observation with CBL1n‐RFP. The subcellular localization of ΔSP‐CaLRR51‐GFP was shown to be similar to that of the 35S:GFP control construct, except that the intensity of green fluorescence was discontinuous in the plasma membrane. In N. benthamiana leaves expressing the CaLRR51‐ΔTR‐GFP fusion protein, the GFP signals were observed in multiple subcellular compartments, including the cytoplasm and nuclei. Significantly, the fluorescent signals of CaLRR51‐ΔTR‐GFP exhibited a punctate pattern in the cytoplasm. For the ΔSP‐CaLRR51‐ΔTR‐GFP deletion mutant construct, the subcellular localization was similar to that of ΔSP‐CaLRR51‐GFP and also exhibited some punctate signals in the cytoplasm.
Figure 7.
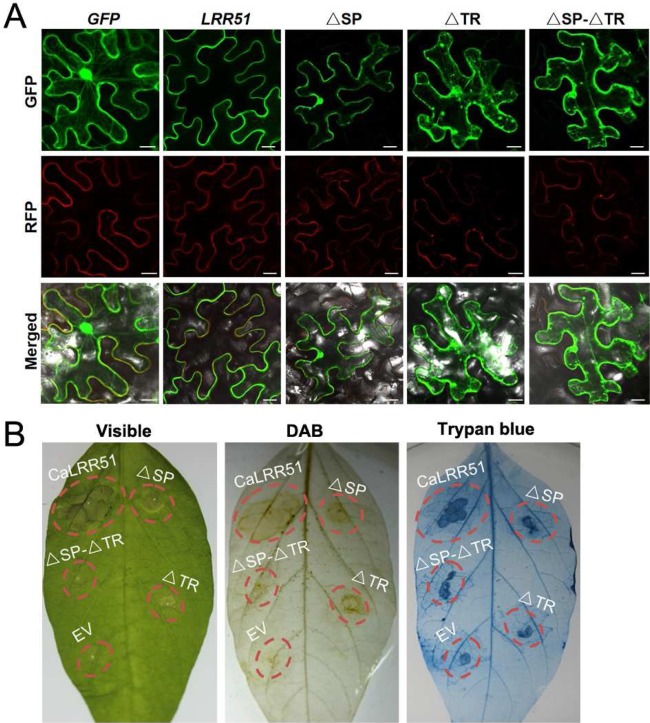
Subcellular localization and hypersensitive response (HR)‐like cell death responses of CaLRR51 and its signal peptide/transmembrane region (SP/TR) deletion mutants. (A) Nicotiana benthamiana leaves were co‐infiltrated with Agrobacterium tumefaciens strains containing 35S:CBL1n‐RFP and 35S:CaLRR51‐GFP or their ΔSP/ΔTR‐GFP fusion constructs (35S:ΔSP‐CaLRR51‐GFP, 35S:CaLRR51‐ΔTR‐GFP or 35S:ΔSP‐CaLRR51‐ΔTR‐GFP). Fluorescence was imaged using a confocal microscope at 48 h post‐inoculation (hpi). GFP, green fluorescent protein; RFP, red fluorescent protein. Bars, 30 μm. (B) Pepper leaves were infiltrated with Agrobacterium GV3101 carrying the 35S:CaLRR51, 35S:ΔSP‐CaLRR51, 35S:CaLRR51‐ΔTR, 35S:ΔSP‐CaLRR51‐ΔTR or 35S:00 construct. Phenotypes of infiltrated pepper leaves 4 days after agroinfiltration (left), by diaminobenzidine (DAB) staining (middle) and by trypan blue staining (right).
To determine whether the location sites of CaLRR51 and its SP/TR deletion mutants are important for function, we investigated HR‐like cell death responses by transient overexpression of a set of vectors, including 35S:CaLRR51, 35S:ΔSP‐CaLRR51, 35S:CaLRR51‐ΔTR and 35S:ΔSP‐CaLRR51‐ΔTR, in pepper leaves (Fig. 7B). At 4 days after agroinfiltration, the transient overexpression of CaLRR51 triggered a severe visible cell death response. However, all three SP/TR deletion mutants of CaLRR51 attenuated HR‐like cell death, in accordance with the results by trypan blue and DAB staining.
Discussion
Although conserved LRRs have been found to exist widely in plant proteins and have been implicated in pathogen perception and signal transduction, these studies have focused on iLRRs (Zhou et al., 2009), and eLRRs remain largely uncharacterized. The data in this study provide evidence that CaLRR51, a novel eLRR protein, acts as a positive regulator in the response of pepper to RSI.
The evidence that CaLRR51 acts as a positive regulator in the response of pepper to RSI comes from the following aspects. First, the transcriptional expression of CaLRR51 was found to be significantly enhanced by inoculation of R. solanacearum into pepper leaves compared with that in control plant leaves, implying that CaLRR51 may play a role in the response of pepper to RSI. As shown by previous studies, the responses of plants to pathogens are largely regulated at the transcriptional level (Bartsch et al., 2006; Buscaill and Rivas, 2014; Eulgem, 2005; Katagiri, 2004; Lewis et al., 2015; Tsuda and Somssich, 2015), and genes up‐regulated by pathogen challenge are frequently found to play important roles in plant immunity (Bartsch et al., 2006; Cai et al., 2015; Dang et al., 2014, 2013; Ramonell et al., 2005). As CaLRR1, which has been found previously to be involved in pepper immunity (Choi et al., 2012, 2011; Hwang et al., 2014; Jung and Hwang, 2007), was also found to be up‐regulated by RSI in the present study (Fig. S2, see Supporting Information), this suggests that the response of CaLRR51 to RSI is not specific. Second, the silencing of CaLRR51 by VIGS significantly enhanced the susceptibility of pepper plants to RSI relative to the observation in control plants. By contrast, HR‐like cell death was found to be significantly triggered by transient overexpression of CaLRR51 in pepper plants, manifested by the coupled darker trypan blue staining and DAB staining, as HR‐like cell death has been found previously to be coupled with the accumulation of H2O2, a signalling molecule leading to HR (Torres et al., 2006). We speculate that the HR‐like cell death and H2O2 accumulation in pepper plants by local transient assays may be closely related because of the high expression levels of CaLRR51. Moreover, transient overexpression of CaLRR51 in pepper plants triggered the expression of defence‐related marker genes, including CaHIR1 (Jung and Hwang, 2007), CaPO2 (Choi et al., 2007), CaPR1 (Kim and Hwang, 2014), CaDEF1 (Hwang and Hwang, 2010) and CaACO1 (Aizat et al., 2013; Lee et al., 2014), which have been found previously to be transcriptionally up‐regulated by pathogens and to play important roles in plant immunity. Third, ectopic overexpression of CaLRR51 significantly enhanced the resistance of transgenic tobacco plants to RSI, with more intensive trypan blue and DAB staining compared with that in control plants. Transcriptional expression of defence‐related genes, including NtHSR201 (Takahashi et al., 2004), NtCAT1 (Takahashi et al., 1997), NtPR1a (Sohn et al., 2007), NtCHN50 (Menke et al., 2005), NtLOX1 (Fammartino et al., 2007, 2010) and NtACO1 (Kim et al., 2003), was also found to be enhanced by the overexpression of CaLRR51, which was potentiated by RSI. All these data strongly suggest that CaLRR51 acts as a positive component in the response of pepper to RSI.
Although CaLRR51 acts as a positive regulator in the HR‐like response by local transient assays, the CaLRR51‐OX transgenic tobacco lines described here did not display visible abnormal morphology. This may be because the expression levels of CaLRR51 in transgenic tobacco plants were too low to trigger visible HR‐like responses. One explanation may be that, despite the random nature of insertion of the 35S:CaLRR51 construct into the tobacco genome, high‐expression‐level CaLRR51 transgenic plants failed to grow because of severe growth retardation or lethal effects. Thus, only those transgenic plants with lower CaLRR51 expression levels would be recoverable. Of note, the transformation efficiency of CaLRR51‐OX tobacco lines was very low in this study.
It was predicted that CaLRR51 contains an LRR domain with six tandem repeats of LRR motifs and a TR domain, which are distinct from that in previously characterized eLRR proteins, such as OsLRR1 (Zhou et al., 2009), CaLRR1 (Jung et al., 2004), NtLRP1 (Jacques et al., 2006), LeLRP (Tornero et al., 1996) and SLRR (Hipskind et al., 1996). These eLRR proteins contain LRR domains with only four to five LRR motifs and no TR domain. The function of a given protein is generally related closely to its structurally dependent subcellular localization, and data from previous studies have shown diverse subcellular localizations of eLRR proteins. For example, CaLRR1 localizes to the extracellular matrix (Choi et al., 2011), NtLRP1 localizes to the endoplasmic reticulum (Jacques et al., 2006) and OsLRR1 targets the plasma membrane, clathrin‐coated vesicles, early endosome and late endosome (Zhou et al., 2009). Unlike these LRR proteins, our data showed that CaLRR51 was exclusively localized to the plasma membrane. Both TM and SP in CaLRR51 are required for plasma membrane targeting. The SP deletion mutant of CaLRR51 was localized to whole cells including nuclei; however, the intensity of green fluorescent signals was discontinuous in the plasma membrane. The TR deletion mutant of CaLRR51 was localized to the cytoplasm with punctate GFP signals, similar to that of OsLRR1, which lacks the TR domain (Zhou et al., 2009). The membrane targeting of CaLRR51 was found to be crucial for its function in immunity, as the blocking of its membrane targeting by either SP or TR deletion triggered lower levels of HR‐like cell death, manifested by lower levels of DAB and trypan blue staining, compared with that of the wild‐type CaLRR51. As it has been generally established that PTI is triggered by the perception of PAMPs by cell surface‐localized PRRs (Jones and Dangl, 2006), we speculate that CaLRR51 might act as a cell surface‐localized PRR. As CaLRR51 is predicted to be an LRR RLP kinase, it might fulfil its function in plant immunity via interaction with certain unidentified ligands, as LRR proteins generally serve as protein interaction platforms; for example, CaLRR1 has been implicated via interaction with HIR1 (Choi et al., 2011; Jung and Hwang, 2007), PR10 (Choi et al., 2012) and PR4b (Hwang et al., 2014).To elucidate the underlying mechanism of immunity mediated by CaLRR51, further studies to identify and characterize its possible interacting proteins from either pathogens or pepper are required.
Plant hormones, such as SA, JA and ET, have been implicated ubiquitously in defence signalling networks that are recruited on perception of an invader (Clarke et al., 2000; Klessig et al., 2000; Pieterse et al., 2012). Although signalling pathways mediated by these hormones act as common signalling cascades in both PTI and ETI (Tsuda and Katagiri, 2010), the relationships among the signalling sectors in PTI and ETI are different. Synergistic relationships are evident in PTI, which may amplify the signal; compensatory relationships among the sectors dominate in ETI (Tsuda et al., 2009). The aforementioned membrane targeting of CaLRR51 and its high sequence similarity to LRR RLP kinases strongly suggest that CaLRR51 acts as a probable receptor of unidentified PAMPs to trigger PTI. Consistent with this, our data showed that exogenously applied SA, MeJA and ETH significantly induced the transcriptional expression of CaLRR51. Moreover, the ectopic overexpression of CaLRR51 in tobacco plants and transient overexpression in pepper plants synergistically activated the transcriptional expression of SA‐, JA‐ and ET‐dependent immunity‐associated marker genes, suggesting that CaLRR51 might be involved in PTI. Notably, in addition to the above‐mentioned structural differences between CaLRR1 and CaLRR51, there are significant differences in the responses of CaLRR1 and CaLRR51 to the exogenous application of SA, MeJA and ETH (Jung et al., 2004). It can be speculated that CaLRR1 and CaLRR51 have evolved in pepper's co‐evolution with pathogens with different lifestyles or invasion strategies. Further investigations on the evolution and roles of different LRR proteins as well as their relationships would provide new insights into plant immunity.
Experimental procedures
Plant materials and growth conditions
Seeds of pepper (C. annuum) cultivar GZ03, tobacco (Nicotiana tabacum) cultivar Honghuadajinyuan and N. benthamiana were sown in a soil mix [peat moss : perlite, 2 : 1 (v/v)] in plastic pots and placed in a growth room at 25°C, 60–70 mmol photons/m2/s, relative humidity of 70% and a 16‐h light/8‐h dark photoperiod.
Pathogens and R. solanacearum inoculation
A highly virulent R. solanacearum strain FJC100301 was isolated from wilted samples of pepper in Fujian province (China) by our laboratory and amplified according to the method described previously (Dang et al., 2013). The R. solanacearum strain was cultured at 28°C, 200 rpm in PSA medium (200 g/L potato, 20 g/L sucrose, 3 g/L beef extract, 5 g/L tryptone) and resuspended in 10 mm MgCl2 solution. The bacterial cell solution used for inoculation was diluted to 108 colony‐forming units (cfu)/mL [optical density at 600 nm (OD600) = 0.8]. For root inoculation, pepper or tobacco plants at the eight‐leaf stage were irrigated with 1 mL of the resulting R. solanacearum suspension, and the disease symptoms were scored at 7 dpi. For leaf inoculation, the third leaves from the top of pepper or tobacco plants at the eight‐leaf stage were infiltrated with 10 μL of the R. solanacearum suspension described above using a syringe without a needle, and the mock was inoculated with 10 mm MgCl2. The leaves were collected at the indicated time points for further analysis.
Treatment of plants with exogenous hormones
Pepper plants at the four‐leaf stage were sprayed with 1 mm SA (in 10% distilled ethanol), 100 µm MeJA (in 10% distilled ethanol) or 100 µm ETH (in sterile double‐distilled H2O). Mock treatment was performed by spraying with the corresponding solvent or sterile double‐distilled H2O.
Vector construction
To construct vectors for overexpression, the full‐length ORF of CaLRR51, or SP and/or TR deletion mutants of CaLRR51 (ΔSP‐CaLRR51, CaLRR51‐ΔTR and ΔSP‐CaLRR51‐ΔTR), were cloned into the entry vector pDONR207 by BP reaction, and then cloned into destination vectors pEarleyGate201 by LR reaction using a Gateway cloning technique (Invitrogen, Carlsbad, CA, USA). To construct the vector for VIGS, a specific 351‐bp fragment in the ORF of CaLRR51 was searched for by blast against the genome sequence in the databases of CM334 (http://peppergenome.snu.ac.kr/) and Zunla‐1 (http://peppersequence.genomics.cn/page/species/blast.jsp). The specific fragment was cloned into the entry vector pDONR207, and then cloned into the PYL279 vector. For subcellular localization, CaLRR51 and its SP/TR deletion mutants (ΔSP‐CaLRR51, CaLRR51‐ΔTR and ΔSP‐CaLRR51‐ΔTR) were fused with GFP. These GFP fusion constructs were driven by the constitutive CaMV 35S promoter. As the N‐terminal 12‐amino‐acid peptide of CBL1 (CBL1n) had been demonstrated to confer plasma membrane targeting (Batistic et al., 2008), a CBL1n‐RFP fusion construct (35S:CBL1n‐RFP) was also generated for use as an additional control.
VIGS of CaLRR51 in pepper plants
For VIGS of CaLRR51 in pepper plants, the TRV‐based VIGS system was employed according to previous studies (Dang et al., 2013). Agrobacterium tumefaciens strains GV3101 harbouring PYL192 and PYL279‐CaLRR51, or PYL279 as a negative control, were resuspended in induction medium [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), 10 mm MgCl2, 200 µm acetosyringone, pH 5.6; OD600 = 0.8] at a 1 : 1 ratio, and then co‐infiltrated into cotyledons of 2‐week‐old pepper plants. The agroinfiltrated pepper plants were grown in the dark at 16°C for 56 h, and then transferred into a growth room at 25°C, 60–70 mmol photons/m2/s, relative humidity of 70% and a 16‐h light/8‐h dark photoperiod for 3–4 weeks for further analysis.
Transient expression of CaLRR51 in pepper leaves
For transient expression analysis, Ag. tumefaciens strains GV3101 harbouring the 35S:CaLRR51, 35S:ΔSP‐CaLRR51, 35S:CaLRR51‐ΔTR, 35S:ΔSP‐CaLRR51‐ΔTR or 35S:00 vector (empty vector used as a control) were grown overnight, and then resuspended in induction medium. A bacterial suspension (OD600 = 0.8) was infiltrated into leaves of pepper plants at the eight‐leaf stage using a syringe without a needle, and the injected leaves were collected at the indicated time points for further use.
Generation of transgenic CaLRR51‐overexpressing tobacco plants
Tobacco cultivar Honghuadajinyuan was used to generate CaLRR51‐overexpressing tobacco plants by the transformation of leaf discs with Ag. tumefaciens strain GV3101 harbouring the 35S:CaLRR51 vector. Fifteen independent T0 transgenic tobacco lines were selected by hygromycin, and further confirmed by PCR and qRT‐PCR. Two T2 transgenic lines exhibiting the highest levels of CaLRR51 transcripts were selected for analyses in this study.
Subcellular localization
Agrobacterium tumefaciens strains GV3101 containing 35S:CaLRR51‐GFP, 35S:ΔSP‐CaLRR51‐GFP, 35S:CaLRR51‐ΔTR‐GFP, 35S:ΔSP‐CaLRR51‐ΔTR‐GFP, 35S:GFP (used as a control) or 35S:CBL1n‐RFP (used as an additional control targeting to the plasma membrane) were grown overnight, and then resuspended in induction medium (OD600 = 0.8). Agrobacterium tumefaciens strains harbouring each of the GFP fusion constructs and the 35S:CBL1n‐RFP construct were mixed in a 1 : 1 ratio and co‐infiltrated into N. benthamiana leaves using a syringe without a needle. At 48 hpi, fluorescence was imaged using a laser scanning confocal microscope (TCS SP8, Leica, Solms, Germany).
Histochemical staining
Leaves were stained with trypan blue and DAB according to the method described by Choi et al. (2012), following the detailed process reported previously (Cai et al., 2015; Dang et al., 2013; Liu et al., 2015).
qRT‐PCR
To determine the relative transcription levels of selected genes, real‐time PCR was performed with specific primers (Table S1, see Supporting Information) according to the manufacturer's instructions for the BIO‐RAD Real‐time PCR system (Foster City, CA, USA) and the SYBR Premix Ex Taq II system (TaKaRa, Dalian, China). Total RNA preparation and real‐time RT‐PCR were carried out following the procedures described in our previous studies (Cai et al., 2015; Dang et al., 2013). Four independent biological replicates of each treatment were performed. Data were analysed by the Livak method (Livak and Schmittgen, 2001) and expressed as a normalized relative expression level (2–ΔΔCT) of the respective genes. The relative transcript level of each sample was normalized to CaActin.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Comparison of the amino acid sequence of CaLRR51 with that of other representative related leucine‐rich repeat (LRR) proteins. Sequence alignment of CaLRR51 with Arabidopsis thaliana AtLRR51 (accession no: NP_193611.1), Capsicum annuum CaLRR1 (accession number: AY237117.1) and Oryza sativa OsLRR1 (accession number: AAO85403.1).
Fig. S2 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of CaLRR1 in pepper plants during Ralstonia solanacearum infection. The third leaves from the top of pepper plants at the eight‐leaf stage were infiltrated with 10 μL of the highly virulent R. solanacearum strain FJC100301 suspension [optical density at 600 nm (OD600) = 0.8] using a syringe without a needle, and the mock was inoculated with 10 mm MgCl2. The leaves were collected at 24 and 48 h after leaf inoculation with R. solanacearum or MgCl2 for RNA extraction and qRT‐PCR analysis.
Table S1 Primers for polymerase chain reaction (PCR) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) used in this study.
Acknowledgements
We thank Mark D. Curtis for kindly providing the Gateway destination vectors. This work was supported by grants from the National Natural Science Foundation of China (31372061, 31401890, 31401312, 31501767 and 31572136).
References
- Ade, J. , DeYoung, B.J. , Golstein, C. and Innes,R.W. (2007) Indirect activation of a plant nucleotide binding site‐leucine‐rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA, 104, 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizat, W.M. , Able, J.A. , Stangoulis, J.C.R. and Able,A.J. (2013) Characterisation of ethylene pathway components in non‐climacteric capsicum. BMC Plant Biol. 13, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker,J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic, O. , Sorek, N. , Schultke, S. , Yalovsky, S. and Kudla,J. (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell, 20, 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri, S. , Mauch, S. , Shen, Q.H. , Peart, J. , Devoto, A. , Casais, C. , Ceron, F. , Schulze, S. , Steinbiss, H.H. , Shirasu, K. and Schulze‐Lefert,P. (2004) RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell, 16, 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. , Li, G. , Fu, Z.Q. and Alfano,J.R. (2008) Phytopathogen type III effector weaponry and their plant targets. Curr. Opin. Plant Biol. 11, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix,G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Boller, T. and He,S.Y. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill, P. and Rivas,S. (2014) Transcriptional control of plant defence responses. Curr. Opin. Plant. Biol. 20, 35–46. [DOI] [PubMed] [Google Scholar]
- Cai, H. , Yang, S. , Yan, Y. , Xiao, Z. , Cheng, J. , Wu, J. , Qiu, A. , Lai, Y. , Mou, S. , Guan, D. , Huang, R. and He,S. (2015) CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high‐temperature and high‐humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. [DOI] [PubMed] [Google Scholar]
- Caplan, J. , Padmanabhan, M. and Dinesh‐Kumar,S.P. (2008) Plant NB‐LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe, 3, 126–135. [DOI] [PubMed] [Google Scholar]
- Choi, D.S. , Hwang, I.S. and Hwang,B.K. (2012) Requirement of the cytosolic interaction between PATHOGENESIS‐RELATED PROTEIN10 and LEUCINE‐RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell, 24, 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.W. , Kim, Y.J. , Lee, S.C. , Hong, J.K. and Hwang,B.K. (2007) Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 145, 890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H.W. , Kim, Y.J. and Hwang,B.K. (2011) The hypersensitive induced reaction and leucine‐rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant–Microbe Interact. 24, 68–78. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D. , Volko, S.M. , Ledford, H. , Ausubel, F.M. and Dong,X. (2000) Roles of salicylic acid, jasmonic acid, and ethylene in cpr‐induced resistance in Arabidopsis. Plant Cell, 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, F. , Wang, Y. , She, J. , Lei, Y. , Liu, Z. , Eulgem, T. , Lai, Y. , Lin, J. , Yu, L. , Lei, D. , Guan, D. , Li, X. , Yuan, Q. and He,S. (2014) Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant, 150, 397–411. [DOI] [PubMed] [Google Scholar]
- Dang, F.F. , Wang, Y.N. , Yu, L. , Eulgem, T. , Lai, Y. , Liu, Z.Q. , Wang, X. , Qiu, A.L. , Zhang, T.X. , Lin, J. , Chen, Y.S. , Guan, D.Y. , Cai, H.Y. , Mou, S.L. and He,S.L. (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. and Ellis,J.G. (2001) Six amino acid changes confined to the leucine‐rich repeat beta‐strand/beta‐turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell, 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. , Lawrence, G.J. , Catanzariti, A.M. , Teh, T. , Wang, C.I. , Ayliffe, M.A. , Kobe, B. and Ellis,J.G. (2006) Direct protein interaction underlies gene‐for‐gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA, 103, 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem,T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. [DOI] [PubMed] [Google Scholar]
- Fammartino, A. , Cardinale, F. , Gobel, C. , Mene‐Saffrane, L. , Fournier, J. , Feussner, I. and Esquerre‐Tugaye,M.T. (2007) Characterization of a divinyl ether biosynthetic pathway specifically associated with pathogenesis in tobacco. Plant Physiol. 143, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fammartino, A. , Verdaguer, B. , Fournier, J. , Tamietti, G. , Carbonne, F. , Esquerre‐Tugaye, M.T. and Cardinale,F. (2010) Coordinated transcriptional regulation of the divinyl ether biosynthetic genes in tobacco by signal molecules related to defense. Plant Physiol. Biochem. 48, 225–231. [DOI] [PubMed] [Google Scholar]
- Hipskind, J.D. , Nicholson, R.L. and Goldsbrough,P.B. (1996) Isolation of a cDNA encoding a novel leucine‐rich repeat motif from Sorghum bicolor inoculated with fungi. Mol. Plant–Microbe Interact. 9, 819–825. [DOI] [PubMed] [Google Scholar]
- van der Hoorn, R.A. and Jones,J.D. (2004) The plant proteolytic machinery and its role in defence. Curr. Opin Plant Biol. 7, 400–407. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F. and Williamson,V.M. (2003) Leucine‐rich repeat‐mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J. 34, 585–593. [DOI] [PubMed] [Google Scholar]
- Hwang, I.S. and Hwang,B.K. (2010) The pepper 9‐lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 152, 948–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I.S. , Choi Du, S. , Kim, N.H. , Kim, D.S. and Hwang,B.K. (2014) Pathogenesis‐related protein 4b interacts with leucine‐rich repeat protein 1 to suppress PR4b‐triggered cell death and defense response in pepper. Plant J. 77, 521–533. [DOI] [PubMed] [Google Scholar]
- Jacques, A. , Ghannam, A. , Erhardt, M. , de Ruffray, P. , Baillieul, F. and Kauffmann,S. (2006) NtLRP1, a tobacco leucine‐rich repeat gene with a possible role as a modulator of the hypersensitive response. Mol. Plant–Microbe Interact. 19, 747–757. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl,J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jung, E.H. , Jung, H.W. , Lee, S.C. , Han, S.W. , Heu, S. and Hwang,B.K. (2004) Identification of a novel pathogen‐induced gene encoding a leucine‐rich repeat protein expressed in phloem cells of Capsicum annuum . Biochim. Biophys. Acta, 1676, 211–222. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. and Hwang,B.K. (2007) The leucine‐rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8, 503–514. [DOI] [PubMed] [Google Scholar]
- Katagiri,F. (2004) A global view of defense gene expression regulation–a highly interconnected signaling network. Curr. Opin. Plant Biol. 7, 506–511. [DOI] [PubMed] [Google Scholar]
- Kim, C.Y. , Liu, Y.D. , Thorne, E.T. , Yang, H.P. , Fukushige, H. , Gassmann, W. , Hildebrand, D. , Sharp, R.E. and Zhang,S. (2003) Activation of a stress‐responsive mitogen‐activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell, 15, 2707–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.S. and Hwang,B.K. (2014) An important role of the pepper phenylalanine ammonia‐lyase gene (PAL1) in salicylic acid‐dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 65, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig, D.F. , Durner, J. , Noad, R. , Navarre, D.A. , Wendehenne, D. , Kumar, D. , Zhou, J.M. , Shah, J. , Zhang, S. , Kachroo, P. , Trifa, Y. , Pontier, D. , Lam, E. and Silva,H. (2000) Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA, 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.J. , Chi, Y.T. , Kim, D.M. , Choi, S.H. , Lee, J.Y. and Choi,G.W. (2014) Ectopic expression of CaRLK1 enhances hypoxia tolerance with increasing alanine production in Nicotiana spp. Plant Mol. Biol. 86, 255–270. [DOI] [PubMed] [Google Scholar]
- Lee, J. , Manning, A.J. , Wolfgeher, D. , Jelenska, J. , Cavanaugh, K.A. , Xu, H. , Fernandez, S.M. , Michelmore, R.W. , Kron, S.J. and Greenberg,J.T. (2015) Acetylation of an NB‐LRR plant immune‐effector complex suppresses immunity. Cell Rep. 13, 1670–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, L.A. , Polanski, K. , de Torres‐Zabala, M. , Jayaraman, S. , Bowden, L. , Moore, J. , Penfold, C.A. , Jenkins, D.J. , Hill, C. , Baxter, L. , Kulasekaran, S. , Truman, W. , Littlejohn, G. , Prusinska, J. , Mead, A. , Steinbrenner, J. , Hickman, R. , Rand, D. , Wild, D.L. , Ott, S. , Buchanan‐Wollaston, V. , Smirnoff, N. , Beynon, J. , Denby, K. and Grant,M. (2015) Transcriptional dynamics driving MAMP‐triggered immunity and pathogen effector‐mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell, 27, 3038–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.Q. , Qiu, A.L. , Shi, L.P. , Cai, J.S. , Huang, X.Y. , Yang, S. , Wang, B. , Shen, L. , Huang, M.K. , Mou, S.L. , Ma, X.L. , Liu, Y.Y. , Lin, L. , Wen, J.Y. , Tang, Q. , Shi, W. , Guan, D.Y. , Lai, Y. and He,S.L. (2015) SRC2‐1 is required in PcINF1‐induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698 [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen,T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- McHale, L. , Tan, X. , Koehl, P. and Michelmore,R.W. (2006) Plant NBS‐LRR proteins: adaptable guards. Genome Biol. 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F.L.H. , Kang, H.G. , Chen, Z.X. , Park, J.M. , Kumar, D. and Klessig,D.F. (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR‐like cell death in tobacco. Mol. Plant–Microbe Interact. 18, 1027–1034. [DOI] [PubMed] [Google Scholar]
- Moffett, P. , Farnham, G. , Peart, J. and Baulcombe,D.C. (2002) Interaction between domains of a plant NBS‐LRR protein in disease resistance‐related cell death. EMBO J. 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees,S.C. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Qi, D. , DeYoung, B.J. and Innes,R.W. (2012) Structure–function analysis of the coiled‐coil and leucine‐rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158, 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan, G.J. and Moffett,P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell, 18, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell, K. , Berrocal‐Lobo, M. , Koh, S. , Wan, J. , Edwards, H. , Stacey, G. and Somerville,S. (2005) Loss‐of‐function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum . Plant Physiol. 138, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider,D.S. (2002) Plant immunity and film Noir: what gumshoe detectives can teach us about plant–pathogen interactions. Cell, 109, 537–540. [DOI] [PubMed] [Google Scholar]
- Sohn, S. , Kim, Y. , Kim, B. , Lee, S. , Lim, C.K. , Hur, J.H. and Lee,J.Y. (2007) Transgenic tobacco expressing the hrpN(EP) gene from Erwinia pyrifoliae triggers defense responses against Botrytis cinerea . Mol. Cell, 24, 232–239. [PubMed] [Google Scholar]
- Takahashi, H. , Chen, Z. , Du, H. , Liu, Y. and Klessig,D.F. (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 11, 993–1005. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y. , Uehara, Y. , Berberich, T. , Ito, A. , Saitoh, H. , Miyazaki, A. , Terauchi, R. and Kusano,T. (2004) A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J. 40, 586–595. [DOI] [PubMed] [Google Scholar]
- Tornero, P. , Mayda, E. , Gomez, M.D. , Canas, L. , Conejero, V. and Vera,P. (1996) Characterization of LRP, a leucine‐rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J. 10, 315–330. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. , Jones, J.D. and Dangl,J.L. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Katagiri,F. (2010) Comparing signaling mechanisms engaged in pattern‐triggered and effector‐triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. and Somssich,I.E. (2015) Transcriptional networks in plant immunity. New Phytol. 206, 932–947. [DOI] [PubMed] [Google Scholar]
- Tsuda, K. , Sato, M. , Stoddard, T. , Glazebrook, J. and Katagiri,F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L. , Cheung, M.Y. , Zhang, Q. , Lei, C.L. , Zhang, S.H. , Sun, S.S. and Lam,H.M. (2009) A novel simple extracellular leucine‐rich repeat (eLRR) domain protein from rice (OsLRR1) enters the endosomal pathway and interacts with the hypersensitive‐induced reaction protein 1 (OsHIR1). Plant Cell Environ. 32, 1804–1820. [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Cheung, M.Y. , Li, M.W. , Fu, Y. , Sun, Z. , Sun, S.M. and Lam,H.M. (2010) Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol. 10, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Comparison of the amino acid sequence of CaLRR51 with that of other representative related leucine‐rich repeat (LRR) proteins. Sequence alignment of CaLRR51 with Arabidopsis thaliana AtLRR51 (accession no: NP_193611.1), Capsicum annuum CaLRR1 (accession number: AY237117.1) and Oryza sativa OsLRR1 (accession number: AAO85403.1).
Fig. S2 Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of CaLRR1 in pepper plants during Ralstonia solanacearum infection. The third leaves from the top of pepper plants at the eight‐leaf stage were infiltrated with 10 μL of the highly virulent R. solanacearum strain FJC100301 suspension [optical density at 600 nm (OD600) = 0.8] using a syringe without a needle, and the mock was inoculated with 10 mm MgCl2. The leaves were collected at 24 and 48 h after leaf inoculation with R. solanacearum or MgCl2 for RNA extraction and qRT‐PCR analysis.
Table S1 Primers for polymerase chain reaction (PCR) and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) used in this study.


