Summary
Sclerotinia sclerotiorum (Lib.) de Bary is a necrotrophic plant pathogen with a worldwide distribution. The sclerotia of S. sclerotiorum are pigmented multicellular structures formed from the aggregation of vegetative hyphae. These survival structures play a central role in the life and infection cycles of this pathogen. Here, we characterized an atypical forkhead (FKH)‐box‐containing protein, SsFKH1, involved in sclerotial development and virulence. To investigate the role of SsFkh1 in S. sclerotiorum, the partial sequence of SsFkh1 was cloned and RNA interference (RNAi)‐based gene silencing was employed to alter the expression of SsFkh1. RNA‐silenced mutants with significantly reduced SsFkh1 RNA levels exhibited slow hyphal growth and sclerotial developmental defects. In addition, the expression levels of a set of putative melanin biosynthesis‐related laccase genes and a polyketide synthase‐encoding gene were significantly down‐regulated in silenced strains. Disease assays demonstrated that pathogenicity in RNAi‐silenced strains was significantly compromised with the development of a smaller infection lesion on tomato leaves. Collectively, the results suggest that SsFkh1 is involved in hyphal growth, virulence and sclerotial formation in S. sclerotiorum.
Keywords: pathogenicity, RNA interference, sclerotial formation, Sclerotinia sclerotiorum, SsFkh1
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary is a notorious and ubiquitous necrotrophic fungal pathogen with a broad range of plant hosts. This pathogen attacks more than 400 plant species, including numerous economically important crops, such as sunflower, soybean, lettuce, oilseed rape, celery, onion and most species of dicots (Boland and Hall, 1994). This necrotrophic pathogen synthesizes de novo the non‐specific phytotoxin oxalic acid (OA), which is considered to be a key virulence factor which dampens host immune signalling pathways and triggers cell death in host plant tissue (Kabbage et al., 2013). Currently, genetic resistance to this fungus in host plants remains inadequate (Williams et al., 2011). Furthermore, S. sclerotiorum produces hard and long‐lived multicellular structures, sclerotia, which make it difficult to eliminate this notorious pathogen using fungicides in the field.
Sclerotia are asexual resting structures composed of condensed vegetative hyphal cells and produced by an array of phytopathogenic fungi. These structures help the fungi to survive under harsh environments, such as low temperature, desiccation, microbial infection or the absence of a host for nutrients (Smith et al., 2015). As a typical sclerotia‐producing fungus, S. sclerotiorum takes advantage of this dense aggregation of tissue to accomplish the transition from asexual to sexual development and to survive for long periods under unfavourable environmental conditions (Bolton et al., 2006). Sclerotia may germinate to either produce infectious hyphae, attacking the roots or stems near the ground directly, or to generate apothecia, which produce numerous airborne ascospores as the primary infection source in most Sclerotinia‐causing diseases. Therefore, successful sclerotial development and germination are vital biological processes in the life cycle of S. sclerotiorum (Roper et al., 2010). Sclerotial development is a complicated biological process, and many environmental changes, primary metabolism and secondary messengers affecting sclerotial development have been well characterized (Erental et al., 2008). At the molecular level, sclerotial development may be regulated by the cyclic adenosine monophosphate (cAMP)‐dependent protein kinase (Jurick and Rollins, 2007), ERK (extracellular signal‐regulated kinase)‐like mitogen‐activated protein kinase and serine/threonine (Ser/Thr) phosphatases type 2A and 2B (Kim et al., 2005), histidine kinase Shk1 (Duan et al., 2013) and secreted protein Sscaf1 (Xiao et al., 2013). However, the molecular regulatory mechanisms regulating the sclerotial development in S. sclerotiorum are still largely unknown.
Based on its macroscopic features, sclerotial formation is divided into three distinct stages: initiation, development and maturation (Erental et al., 2008). Melanin plays an important role in the formation of sclerotia because S. sclerotiorum cannot form fully formed sclerotia without the accumulation of melanin. Furthermore, the accumulation of melanin in sclerotia is of importance for S. sclerotiorum to combat the unfavourable conditions in soil, whereas the melanized outer layer of sclerotia provides protection against both biotic and abiotic stresses. The full genome of S. sclerotiorum has now been sequenced and genetic transformation systems have been established, which could facilitate the molecular manipulation and investigation of this phytopathogenic fungus.
Forkhead (FKH) box (FOX) family members encode transcription factors (TFs) containing an FKH helix‐turn‐helix DNA‐binding domain of approximately 110 amino acids, and regulate various aspects of cellular processes, such as primary metabolism, the cell cycle and morphogenesis, in animals and fungi (Lam et al., 2013; Wang et al., 2015), whereas FOX TFs are not found in plant genomes (Shimeld et al., 2010). Two TFs (FKH1 and FKH2) in Saccharomyces cerevisiae have been found to play roles in the G2 and M phases of the cell cycle, and fkh1fkh2 double mutants display abnormal morphology, such as defects in the separation of mother and daughter cells following M phase, resulting in apparent chain‐like projections and budding defects, leading to the formation of extended buds (Koranda et al., 2000; Kumar et al., 2000; Zhu et al., 2000). Three FOX TFs (fkhF, fkhE and fhpA), encoded in the genome of the filamentous fungus Aspergillus nidulans, have been reported to play critical roles during asexual development (Lee et al., 2005; Park et al., 2010), and the deletion of fkhE and fkhF resulted in impaired conidiophore formation (Park et al., 2010). In addition to their functions in development, proteins containing FOX domain have been documented to play roles in fungal pathogenicity. Magnaporthe oryzae, the causal agent of rice blast, harbours four FOX TFs encoded in its genome (MoFOX1, MoFOX2, MoFKH1 and MoHCM1) (Park et al., 2014). The knockout of MoFKH1 resulted in reduced mycelial growth and conidial germination, reduced virulence and increased sensitivity to environmental stress, whereas deletion of MoHCM1 led to reduced mycelial growth and conidial germination.
Four FOX‐like members are predicted in the S. sclerotiorum genome (Wang et al., 2016). We have recently characterized a FOX family member, SsFOXE2, which is required for the regulation of apothecial formation, but not essential for the pathogenicity and vegetative development of S. sclerotiorum (Wang et al., 2016). We have now characterized an atypical FOX‐containing protein and investigated its biological function using an RNA interference (RNAi) strategy.
Results
SsFkh1 encodes a non‐canonical FOX‐containing protein
In the search for FOX‐containing proteins in the S. sclerotiorum genome, four were found to be encoded in SsFoxE1, SsFoxE2, SsFoxE3 (Wang et al., 2016) and another non‐canonical FOX‐containing protein. A detailed multiple sequence alignment was performed based on the representative FOX domain‐containing proteins from human, fruit fly and filamentous fungi. In our alignment and sequence analysis, the non‐canonical FOX‐containing protein (SS1G_07360) encodes a 172‐amino‐acid protein containing an FKH domain (amino acids 122–170) lacking the C‐terminus conserved motif (Fig. 1A), corresponding to the third helix (H3) present in most FOX proteins (Zahiri et al., 2010). The FKH motif is conserved across animal and fungal lineages. Based on the phylogenetic analysis with other fungal FKH proteins, the fourth member of FOX proteins was named SsFKH1 (Fig. 1B).
Figure 1.

SsFkh1 encodes a non‐canonical forkhead box‐containing protein in Sclerotinia sclerotiorum. (A) Multiple alignment of the forkhead box domain from different species. The selected forkhead box domains are from: S. sclerotiorum, SsFoxE2 (XP_001592912.1) and SsFKH1 (XP_001591914.1); Homo sapiens, HsFOXK1 (NP_001032242.1); Danio rerio, DrFOXI1 (NP_944600.1); Saccharomyces cerevisiae, ScFKH2 (NP_014331.3) and ScFKH1 (NP_012135.1); Magnaporthe oryzae, MoFKH1 (XP_003717334.1); Acremonium chrysogenum, AcFKH1 (AAP35674.1); Ustilago maydis, UmFox1 (XP_011387558.1). The conserved motif is marked with a red box. (B) Phylogenetic analysis of putative forkhead box‐containing transcription factors based on the conserved forkhead box domain. The phylogenetic tree was constructed using the neighbour‐joining method and forkhead box‐containing proteins in S. sclerotiorum are indicated with black triangles. The selected forkhead box domains are from: Botrytis cinerea, BcFox (XP_001555067.1); Aspergillus flavus, AfFox (XP_002374794.1); Aspergillus terreus, AtFoxC2 (EAU35186.1) and AtFoxL2 (EAU30606.1); Madurella mycetomatis, MmFoxO (KOP42143.1); Neurospora crassa, NcFKH1 (EAA27770.1); Fusarium oxysporum, FoFoxO4 (ENH61621.1); S. sclerotiorum, SsFoxE1 (XP_001598491.1), SsFoxE2 (XP_001592912.1), SsFoxE3 (XP_001588166.1) and SsFKH1 (XP_001591914.1); Saccharomyces cerevisiae, ScFKH2 (NP_014331.1) and ScFKH1 (NP_012135.1); Acremonium chrysogenum, AcFKH1 (AAP35674.1); Ustilago maydis, UmFox1 (XP_011387558.1); Aspergillus nidulans, AnFkhF (XP_682218.1), AnFkhE (XP_659629.1) and AnFhpA (AKN52397.1); Magnaporthe oryzae, MoFKH1 (XP_003717334.1), MoHCM1 (XP_003717145.1), MoFOX2 (XP_003712110.1) and MoFOX1 (ELQ36733.1).
To examine the expression profiles of SsFkh1 in S. sclerotiorum at various developmental stages, we examined the SsFkh1 transcriptional changes using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). The results showed that, in sclerotia (stages SI, SII and SIII) and most apothecium stages (AI–AII), SsFkh1 showed a lower abundance than in the hyphal stage. However, the expression of SsFkh1 was maintained at a relatively stable level in S. sclerotiorum at various developmental stages (Fig. S1, see Supporting Information).
Expression of SsFkh1 is decreased by RNAi
SsFKH1 is phylogenetically related to well‐characterized FKH1 proteins from Acremonium chrysogenum (Schmitt et al., 2004), Sa. cerevisiae (Ostrow et al., 2014) and M. oryzae (Park et al., 2014), which have been demonstrated previously to be required for development. We hypothesized that SsFkh1 was also associated with development and possibly virulence in S. sclerotiorum. SsFkh1‐silenced strains were therefore generated using RNAi to assess the role of SsFkh1 in development and virulence in S. sclerotiorum. Two SsFkh1 RNA‐silenced constructs, pS1‐SsFkh1 and pSD‐SsFkh1, were introduced into wild‐type S. sclerotiorum via polyethylene glycol (PEG)‐mediated protoplast transformation. The transformation efficiency of S. sclerotiorum protoplasts was approximately 1 in every 6.0 × 104 protoplasts. A total of 120 transformants was obtained using potato dextrose agar (PDA) medium amended with 100 μg/mL hygromycin B or geneticin. Amongst all of the silenced transformants, 10 transformants with common phenotypic changes and PCR‐positive results (Fig. 2) were designated as pSl‐1, pSl‐2, pSl‐3, pSl‐4 and pSl‐5 (originating from pS1‐SsFkh1 transformation) and pSD‐1, pSD‐2, pSD‐3, pSD‐4 and pSD‐5 (originating from pSD‐SsFkh1 transformation). These 10 SsFkh1‐silenced strains were employed for further experiments. The altered phenotypes included a reduced hyphal growth rate and condensed colony development (Fig. 2). Consistent with the phenotypic alterations, the expression level of SsFkh1 was reduced significantly in RNAi mutants (Fig. 2C), but the reduced levels varied among different RNA‐silenced strains. For example, strains pS1‐3 and pSD‐3 exhibited less SsFkh1 transcription reduction and fewer defects in hyphal development.
Figure 2.
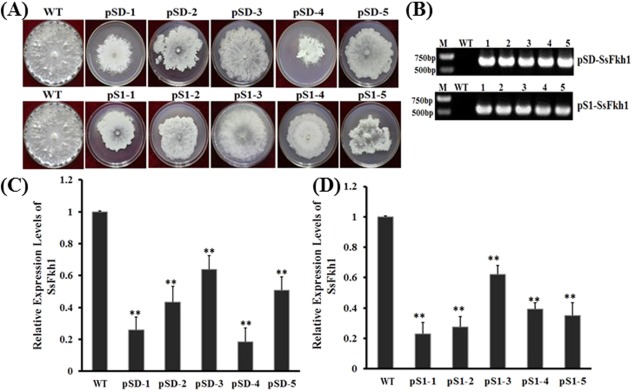
Generation and functional analysis of RNA interference (RNAi)‐mediated SsFkh1‐silenced strains. (A) Morphological observation of wild‐type (WT) and RNAi mutant strains. Selected transformants were cultured on potato dextrose agar (PDA) medium and grown at 25°C. Photographs were taken 7 days after inoculation. (B) Identification of selective marker insertions by polymerase chain reaction (PCR). RNAi mutants were inoculated on PDA medium for 2 days and hyphae were collected for DNA preparation. Conventional PCR was performed using primers specific for hygromycin and geneticin resistance genes, and produced 646‐ and 650‐bp PCR products from RNAi mutants, respectively. (C, D) Expression levels of SsFkh1 in RNAi mutants. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed using cDNA template generated from each RNAi mutant and WT. Each data point represents the mean ± standard deviation (SD) from three biological repeats (n = 3). Asterisks indicate significant difference compared with the WT strain [**P < 0.01, one‐way analysis of variance (ANOVA) in SPSS].
SsFkh1 is essential for S. sclerotiorum hyphal growth
Because RNAi‐mediated SsFkh1‐silenced mutants exhibited impaired growth phenotypes, we further investigated the significance of SsFkh1 in S. sclerotiorum hyphal growth and development. Wild‐type strains and SsFkh1 RNAi mutants were inoculated on PDA and incubated at 25°C. The colony diameters of these strains were measured every 12 h. The RNAi mutant strains exhibited slower hyphal growth rates than the wild‐type strain (Fig. 3A,B), with both pS1‐1 and pSD‐4 mutants showing the slowest hyphal growth rates. The hyphal tip morphology observed under light microscopy demonstrated that both the RNAi mutants and wild‐type strains had similar hyphal branching patterns and hyphal tip projection (Fig. 3C). These data reveal that SsFkh1 is essential for hyphal growth and development, but not for correct hyphal branching.
Figure 3.
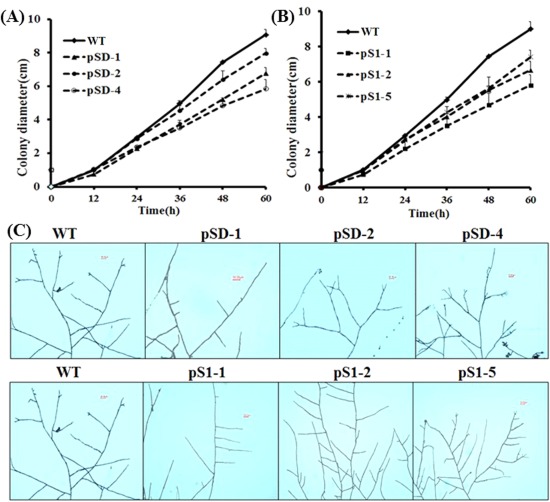
SsFkh1 is essential for hyphal growth and development. (A, B) Knockdown of the expression of SsFkh1 impaired hyphal growth and development. Both RNA interference (RNAi) mutants and wild‐type (WT) strains were inoculated on potato dextrose agar (PDA) and placed in an incubator at 25 ºC. The colony diameters were measured every 12 h after inoculation. (C) Hyphal branching patterns of WT and RNAi mutants.
SsFkh1 RNAi mutants exhibit impaired pathogenicity
To determine whether a transcriptional reduction of SsFkh1 influences the pathogenicity of S. sclerotiorum, we inoculated healthy detached tomato leaves with agar plugs with SsFkh1 RNAi mutants or the wild‐type strain. The pathogenicity of SsFkh1 mutants was strongly impaired. Consistent with the RNA silencing efficiency, six RNAi strains (pSl‐1, pSl‐2, pS1‐4, pSD‐1, pSD‐2 and pSD‐4) showed reduced ability to infect tomato leaves and caused smaller necrotic lesions than those resulting from the wild‐type strain. The other four strains (pSl‐3, pS1‐5, pSD‐3 and pSD‐5) also produced significant disease symptoms, but manifested reduced symptoms compared with the fully pathogenic wild‐type strain (Fig. 4). The necrotic lesion diameters of pSl‐3 and pSD‐3 mutants were larger than those of the other mutants, but were significantly smaller than those of the wild‐type. These data demonstrate that SsFkh1 is involved in the pathogenicity of S. sclerotiorum, and that infection capacity and necrotic lesion development are largely impaired when the expression level of SsFkh1 is reduced by RNA silencing.
Figure 4.
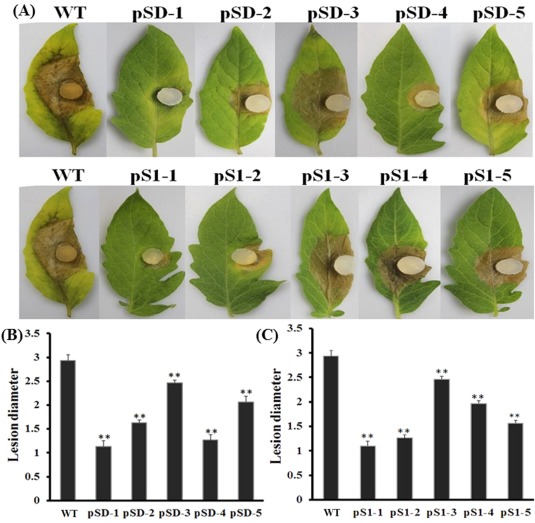
RNA interference (RNAi)‐mediated SsFkh1 silencing compromised the pathogenicity of Sclerotinia sclerotiorum. (A) RNAi mutants and wild‐type (WT) strains were inoculated on detached 45‐day‐old tomato leaves and placed in a high‐humidity growth chamber. Photographs were taken 6 days after inoculation. (B, C) Diameters of infectious necrotic lesions were measured from (A). Each data point represents the mean ± standard deviation (SD) from three biological repeats (n = 3). Asterisks indicate significant difference compared with the WT strain [**P < 0.01, one‐way analysis of variance (ANOVA) in SPSS].
SsFkh1 is necessary for sclerotial formation and is involved in melanin biosynthesis
As FOX family members have been documented to be critical factors for the cell cycle and development, we performed sclerotial formation assay to examine whether SsFkh1 functions in this vital biological process. Both wild‐type and mutants were placed in the same autoclaved plates (containing smashed potato with 1.5% agar) for 7 days to induce sclerotial formation. Numerous normal sclerotial formations were observed in the wild‐type strain, whereas no sclerotia were produced by the RNAi strains (Fig. 5). Melanin accumulation is closely associated with the production of sclerotia. Therefore, we profiled the gene expression patterns of the polyketide synthase‐encoding gene (SsPks13) and three laccase genes (lac1, lac2 and lac3), which are probably involved in melanin biosynthesis. Consistent with the deficient phenotype in sclerotial formation, the expression levels of these genes were down‐regulated significantly in the RNAi strains (Fig. 6). These data demonstrate that the lack of sclerotial development also blocks putative biosynthesis genes.
Figure 5.
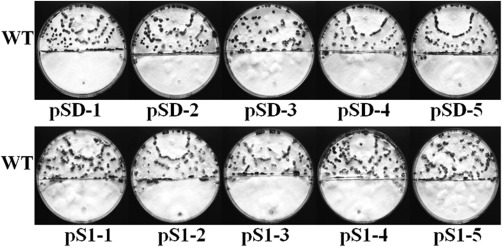
Reduced expression of SsFkh1 abolished sclerotial formation of Sclerotinia sclerotiorum. Wild‐type (WT) and RNA interference (RNAi) mutant strains were inoculated on the same autoclaved smashed potato medium plates, and the inoculated plates were incubated at 25 ºC for 7 days; sclerotial formation was observed and photographs were taken.
Figure 6.
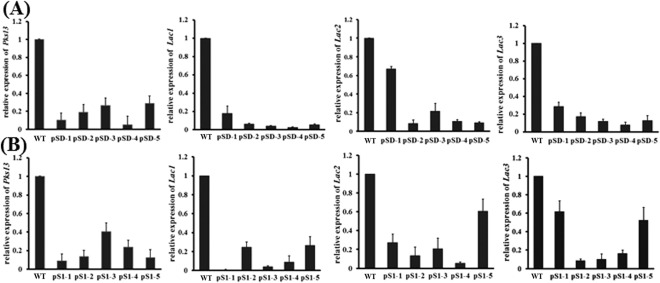
Expression profiles of melanin biosynthesis‐related genes. (A, B) Expression levels of the melanin biosynthesis‐associated genes, polyketide synthase‐encoding gene (SsPks13) and laccase genes (SsLac1, SsLac2 and SsLac3), were investigated in hyphal tissue in RNA interference (RNAi) and wild‐type (WT) strains. cDNA was normalized based on actin cDNA in extracts from each strain. Each data point represents the mean ± standard deviation (SD) from three biological repeats (n = 3).
SsFkh1 is a key factor in the maintenance of hyphal cell integrity
When cultured on PDA supplemented with hyperosmotic and oxidative factors, such as NaCl, KCl or H2O2, SsFkh1 RNAi strains exhibited significant hyphal growth inhibition compared with the wild‐type strain (Fig. 7). Under the same concentration of osmotic stress, both wild‐type and RNAi mutant strains were more sensitive to KCl than NaCl.
Figure 7.
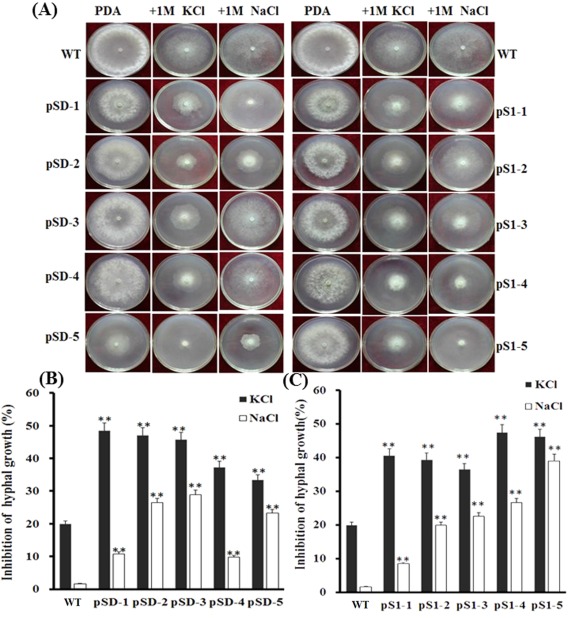
Decreased expression of SsFkh1 increased the sensitivity to osmotic stress of Sclerotinia sclerotiorum. (A) Hyphae from either wild‐type (WT) or RNA interference (RNAi) mutant strains were inoculated on potato dextrose agar (PDA) containing osmotic factors (1 m NaCl or 1 m KCl) and incubated at 25 ºC for growth. Photographs were taken and colony diameters were measured 3 days after inoculation. (B, C) Inhibition rates were calculated based on (A). Values are means ± standard deviation (SD) from three biological repeats (n = 3). Asterisks indicate significant difference compared with the WT strain [**P < 0.01, one‐way analysis of variance (ANOVA) in SPSS].
Similar phenotypes were also observed when the PDA medium was amended with varying concentrations of H2O2 from 5 to 25 mm, indicating that fungal hyphal growth was greatly inhibited by oxidative stress (Fig. 8). Moreover, no hyphal growth was observed in most RNAi strains with 25 mm H2O2, whereas neither wild‐type nor RNAi mutants survived the oxidative stress treatment when the concentration of H2O2 was increased to 30 mm on PDA. These data indicate that silencing of SsFkh1 causes impaired cellular integrity and increases sensitivity to both osmotic and oxidative stresses.
Figure 8.
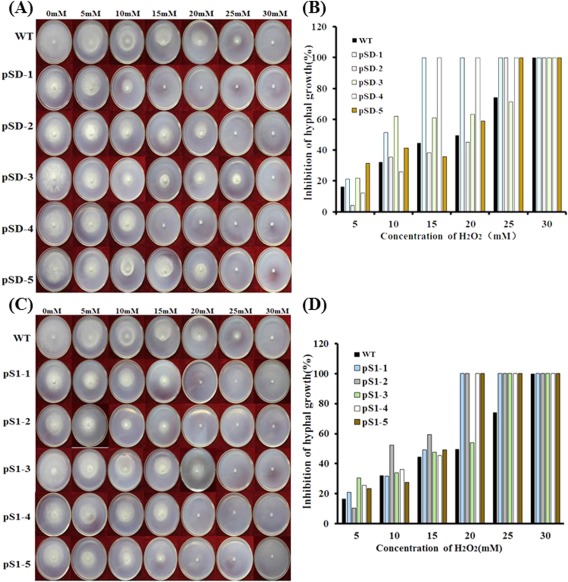
Knockdown of the expression of SsFkh1 enhanced the sensitivity to oxidative stress of Sclerotinia sclerotiorum. (A, C) Hyphae from either wild‐type (WT) or RNA interference (RNAi) mutant strains were inoculated on potato dextrose agar (PDA) supplemented with sequential concentrations of H2O2 mimicking oxidative stress. Photographs were taken and colony diameters were measured after incubation at 25 ºC for 3 days. (B, D) Inhibition rates were calculated based on (A) and (C). Values are means ± standard deviation (SD) from three biological repeats (n = 3).
Discussion
TFs control and regulate multiple biological processes by the manipulation of gene expression networks. FKH proteins constitute a small proportion of TF families, but display a remarkable functional diversity involved in a wide variety of biological processes (Carlsson and Mahlapuu, 2002). In fungi, there are several cases reporting the roles of FOX proteins, and most are FKH proteins. FKH proteins play roles in the regulation of development, growth rate, pathogenicity and the stress response in fungi (Lee et al., 2005; Park et al., 2014, 2010), which are important for fungal adaptation to the environment (Wang et al., 2015). In this study, we characterized an atypical FOX‐containing TF, designated as SsFKH1, which lacks the conserved ‘RHNLS’ motif (Zahiri et al., 2010). Phylogenetically, this protein is closely related to a group of FKH proteins. Four FKH‐containing proteins (SsFoxE1, SsFoxE2, SsFoxE3 and SsFKH1) are encoded in the S. sclerotiorum genome, and SsFoxE2 has been demonstrated previously to be essential for sexual reproduction through apothecial development (Wang et al., 2016). At the transcriptional level, transcripts of SsFkh1 fluctuate in different developmental stages, whereas SsFoxE2 is expressed exclusively in apothecial developmental stages. Both phylogenetic analysis and transcriptional profiling showed that SsFoxE2 exhibited distinct expression patterns from SsFkh1, indicating divergent roles played by these FKH‐containing proteins in S. sclerotiorum.
We used an RNAi‐mediated gene silencing technique to elucidate the roles of SsFkh1 in development, pathogenicity and response to stress in S. sclerotiorum. qRT‐PCR data revealed that SsFkh1 was silenced efficiently, and this was also confirmed by observations of the hyphal development phenotypes. RNA‐silenced mutants exhibited impaired developmental phenotypes, such as reduced hyphal growth rates and abolished sclerotial formation, but hyphal branching patterns did not differ appreciably from those of the wild‐type. These results also agree with the observations in other fungi. When MoFKH1 was depleted from the M. oryzae genome, the mutants showed reduced mycelial growth and conidial germination, abnormal septation and stress response, as well as reduced infection capacity (Park et al., 2014). When ScFkh1 and ScFkh2 were double deleted from Sa. cerevisiae, the resulting yeast mutants exhibited a series of abnormal phenotypes, such as a defect in the separation of mother and daughter cells following M phase, resulting in obvious chain‐like projections and budding defects, leading to the formation of extended buds (Koranda et al., 2000; Kumar et al., 2000; Zhu et al., 2000). Sep1+ is the homologue of the FKH TFs in Schizosaccharomyces pombe and has been shown to prevent growth at cell tips (Ribár et al., 1999). FKH TFs have also been shown to influence morphogenesis in both Sa. cerevisiae and Sz. pombe (Bensen et al., 2002). In Ustilago maydis, the causal agent of corn smut disease, the deletion of a FOX protein resulted in tumour development and reduced virulence on corn leaves (Zahiri et al., 2010). Similar to the phenotypes recorded in U. maydis and M. oryzae, silencing of SsFkh1 also resulted in reduced pathogenicity when the RNAi mutants were inoculated on tomato leaves (Fig. 4). These results indicate that some members of FKH proteins not only play roles in development but may also participate in pathogenicity.
Sclerotia are considered to be important asexual resting structures for both the fungal life cycle and pathogenicity (Smith et al., 2015). Melanin is known to be involved in the process of sclerotial formation. SsFkh1 RNAi strains cannot produce sclerotia and therefore fail to accomplish the transition from asexual to sexual reproduction, and subsequently generate no ascospores as infecting agents. Sclerotial formation is a complicated process controlled by several determining factors, such as ERK‐like mitogen‐activated protein kinase and Ser/Thr phosphatases type 2A and 2B (Kim et al., 2005), histidine kinase Shk1 (Duan et al., 2013) and secreted protein Sscaf1 (Xiao et al., 2013). These proteins may function in signalling transduction or the regulation of gene expression. As a TF, SsFkh1 may act as a regulator orchestrating the gene expression network, controlling the biosynthesis of sclerotial components, such as melanin (Liu and Free, 2016). Melanin accumulation is related to sclerotial biosynthesis (Yu et al., 2012) and is affected by polyketide synthase (SsPks13) and laccases (SsLac) (Liu and Free, 2016). The expression patterns of these genes were largely affected by reducing the expression of SsFkh1, which is probably the cause of the deficient phenotypes of sclerotia.
Hyphal cell integrity is important for fungi to respond to abiotic stress (Yu et al., 2012), When S. sclerotiorum was challenged with osmotic or oxidative stress, the RNAi mutants demonstrated increased sensitivity to both types of stress, indicating that SsFkh1 plays a role in the maintenance of cell integrity. As a TF, SsFKH1 might function as a regulator in this process. Moreover, in the initial interface established between the pathogen and a host plant, reactive oxygen species (ROS) are generated to resist the establishment of infection (Apel and Hirt, 2004). Therefore, reduced oxidative and osmotic stress resistance and impaired hyphal development could contribute to the reduced infection capacity and ROS endurance in RNAi mutant strains, which, in turn, impairs the infection capacity.
In conclusion, we have characterized SsFKH1, an atypical FOX TF in S. sclerotiorum. Phylogenetically, SsFKH1 is closely related to a group of FKH proteins and is structurally distinct from the previously characterized SsFoxE2. FKH proteins play divergent roles in S. sclerotiorum. Based on the RNA‐mediated SsFkh1 silencing results, SsFkh1 is involved in hyphal development and cell integrity, sclerotial development and pathogenicity in S. sclerotiorum. However, as a TF, the regulating signalling components upstream of SsFKH1 are still unknown.
Experimental Procedures
Fungal strains and media
Wild‐type S. sclerotiorum isolate 1980 (Godoy et al., 1990) and RNA‐silenced mutants were cultured, unless otherwise stated, on PDA medium at 25°C. PDA medium supplemented with 100 µg/mL hygromycin B (Roche, Indianapolis, IN, USA) or 100 µg/mL geneticin (Roche) was utilized to culture the transformation mutants. To test the osmotic and oxidative tolerance of RNAi mutants, PDA medium was supplemented with various concentrations of salts (NaCl and KCl) and H2O2.
Bioinformatics analysis of SsFKH1
In a previous study, based on PFAM analysis, we found that there were four FOX proteins encoded in the S. sclerotiorum genome, namely SsFoxE1, SsFoxE2, SsFoxE3 and another non‐canonical FOX‐containing protein (Wang et al., 2016). To analyse the relationships among the documented FOX proteins from human, fruit fly and filamentous fungi, multiple sequence alignments and phylogenetic analyses were performed. Amino acid sequences of FOX proteins were recovered by blast from GenBank and aligned by ClustalX software (version 1.83). A phylogenetic tree was constructed using mega software (version 6.06) with the maximum likelihood method using 1000 bootstrap replicates to ascertain the reliability of a given branch pattern in the neighbor‐joining tree. Meanwhile, detailed protein sequence alignment using S. sclerotiorum FoxE2 (XP_001592912.1) and SsFKH1 (XP_001591914.1), Homo sapiens HsFOXK1 (NP_001032242.1), Danio rerio FOXI1 (NP_944600.1), Sa. cerevisiae ScFKH2 (NP_014331.3) and ScFKH1 (NP_012135.1), M. oryzae MoFKH1 (XP_003717334.1), Acremonium chrysogenum AcFKH1 (AAP35674.1) and U. maydis UmFox1 (XP_011387558.1) was performed using ClustalX software.
Expression levels of SsFkh1 in the life cycle of S. sclerotiorum
The expression levels of SsFkh1 at various developmental stages were investigated to capture the expression profile of this gene. Fungal tissues from major developmental stages [i.e. hyphae, sclerotial stages I–VI (Li and Rollins, 2009) and apothecial stages I–III] were collected, and RNA was isolated using Trizol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). One microgram of total RNA was subjected to cDNA synthesis using PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). qRT‐PCR was performed with the primer pair qSsFkh1‐F and qSsFkh1‐R (Table 1). Total cDNA abundance in samples was normalized using the actin gene (XP_001589969.1; SS1G_08733) as an internal reference. Amplification conditions were set up as follows: 30 s at 95 ºC, then 40 cycles consisting of 5 s at 95 ºC, 40 s at 60 ºC and 15 s at 95 ºC, followed by 1 min at 60 ºC and 15 s at 95 ºC. The 2–ΔΔCT method was used to analyse the relative quantification of gene expression (Schmittgen et al., 2000). The actin gene was used as internal control. Each RT‐PCR experiment was performed at least three times.
Table 1.
Primers used in this study.
| Primer | Primer sequence | Comments |
|---|---|---|
| SsFkh1‐F | CCATCTTCCGGTAAGAGGGCCC | SsFkh1 (Forward) |
| SsFkh1‐R | ATAACTTACTTGCCAACCGTGGG | SsFkh1 (Reverse) |
| pSD‐1 | CTCTAGAGACGTCGAGAACCAATGATGAA(XbaI) | SsFkh1 (Forward) |
| pSD‐2 | CTCTAGAGCGAACATCATCTCAACTCCA(XbaI) | SsFkh1 (Reverse) |
| pSl‐L‐1 | CCTCGAGGACGTCGAGAACCAATGATGAA(XhoI) | SsFkh1 (Forward) |
| pSl‐L‐2 | CAAGCTTGCGAACATCATCTCAACTCCA(HindIII) | SsFkh1 (Reverse) |
| pSl‐R‐1 | GGCATGCCACGTCGAGAACCAATGATGAA(SphI) | SsFkh1 (Forward) |
| pSl‐R‐2 | CAGATCTGCGAACATCATCTCAACTCCA(BglII) | SsFkh1 (Reverse) |
| gene‐F | TGTCCGGTGCCCTGAATGAACT | Geneticin (Forward) |
| gene‐R | GCCGCCAAGCTCTTCAGCAATAT | Geneticin (Reverse) |
| hyg‐F | CGTCTGCTGCTCCATACAAG | Hygromycin (Forward) |
| hyg‐R | GCGAAGAATCTCGTGCTTTC | Hygromycin (Reverse) |
| qSsFkh1‐F | CCTCAAATCAACTGCCACGC | RT‐PCR for SsFkh1 (Forward) |
| qSsFkh1‐R | AGAGTGACGCCCAACATTGA | RT‐PCR for SsFkh1 (Reverse) |
| Actin‐F | GAATGTGTAAGGCCGGTTTCGC | Actin (Forward) |
| Actin‐R | CATCCCAGTTGGTGACGACACC | Actin (Reverse) |
| qSsPks1‐F | ACTGCTACGCCGAAACCATC | RT‐PCR for SsPks1 (Forward) |
| qSsPks1‐R | CGCATAGGACCTGCCAACTC | RT‐PCR for SsPks1 (Reverse) |
| qSsLac1‐F | ACCGCAGTCGCTTCTCAATA | RT‐PCR for SsLac1 (Forward) |
| qSsLac1‐R | TGCATGATCGGAGGGAACAC | RT‐PCR for SsLac1 (Reverse) |
| qSsLac2‐F | ACACTGTGAGAGCGGAATGG | RT‐PCR for SsLac2 (Forward) |
| qSsLac2‐R | CGACGTAGTGCTAGATCCCG | RT‐PCR for SsLac2 (Reverse) |
| qSsLac3‐F | ACCACACCGTTACCCAATCC | RT‐PCR for SsLac3 (Forward) |
| qSsLac3‐R | ACCACATTGGTGTAGCGACC | RT‐PCR for SsLac3 (Reverse) |
Italic indicates the restriction enzyme digestion sites.
Generation of SsFkh1 RNAi constructs
To collect hyphae from cultures grown on solid medium, a sterilized cellophane membrane was placed on the medium before inoculation. Hyphae were collected and frozen in liquid nitrogen and stored at −80 ºC until needed. Fungal genomic DNA was extracted with a modified cetyltrimethylammonium bromide (CTAB) protocol (Proctor et al., 1997). Total RNA was prepared with Trizol reagent according to the manufacturer's instructions (Invitrogen). cDNA synthesis has been described above. To amplify the full‐length cDNA of SsFkh1 (XP_001591914.1; SS1G_07360), specific primers SsFkh1‐F and SsFkh1‐R (Table 1) were designed, and the synthesized cDNA was used as the PCR template. The purified PCR product was ligated into pMD18‐T cloning vector (TaKaRa) and verified by DNA sequencing.
To silence the SsFkh1 gene, a 246‐bp cDNA fragment was amplified with two sets of primers (pSl‐L‐1/pSl‐L‐2 and pSl‐R‐1/pSl‐R‐2) using pMD18T‐SsFkh1 as template. The amplified DNA was inserted into two widely used fungal RNA silencing vectors, pSilent1 and pSilent‐Dual1 (Nakayashiki et al., 2005; Nguyen et al., 2008), to generate pS1‐SsFkh1 and pSD‐SsFkh1 constructs, respectively. The resulting RNAi constructs were verified by DNA sequencing. The resulting constructs were used for the transformation of S. sclerotiorum.
Transformation and evaluation of RNAi mutants
Protoplasts of S. sclerotiorum were prepared and the quality of protoplasts was examined by microscopy, as described previously (Qu et al., 2014). The RNAi constructs were introduced into the prepared protoplasts by a PEG‐mediated transformation method (Rollins, 2003). Colonies regenerated through the selective medium were transferred to PDA with either 100 mg/mL hygromycin B (pSilent1) or 100 mg/mL geneticin (pSilent‐Dual1). Transformants were cultured and purified at least three times on PDA containing the corresponding antibiotics using hyphal tips. The transformants containing the transformation vectors were verified by PCR with specific primers (hyg‐F/hyg‐R or gene‐F/gene‐R) using genomic DNA as template. Five transformants from each vector were used for phenotypic assay.
Evaluation of the expression levels of SsFkh1 in RNA‐silenced mutants
To evaluate the accumulation level of SsFkh1 transcripts in transformants containing either pS1‐SsFkh1 or pSD‐SsFkh1, the transformants and wild‐type strain were cultured on PDA for 2 days. Total RNA of these strains was isolated and cDNA was synthesized as described above. qRT‐PCR was performed as described above with the primer pair qSsFkh1‐F/qSsFkh1‐R. The PCR was conducted as follows: 30 s at 95 ºC, then 40 cycles each consisting of 5 s at 95 ºC, 40 s at 60 ºC and 15 s at 95 ºC, followed by 1 min at 60 ºC and 15 s at 95 ºC. The 2–ΔΔCT method was used to analyse the relative quantification of gene expression as described above.
Hyphal morphology and growth rate observation
To investigate the influence of silencing SsFkh1 at the RNA level on hyphal growth rates and morphology, wild‐type and selected RNAi mutant strains with the lowest levels of SsFkh1 expression were cultured on PDA medium at 25°C for 2 days. Colony diameters of each strain, inoculated in the centre of a Petri dish with an agar mycelium plug derived from the growing margin of the PDA culture, were measured over time to determine the radial mycelial growth on PDA plates. Hyphal morphology was observed by light microscopy (Nikon Eclipse 80i digital microscopy, Melville, NY, USA).
Pathogenicity assay
To evaluate the impact of RNA silencing on pathogenicity, 7‐week‐old tomato leaves were detached and placed in Petri dishes with moist filter paper. They were inoculated with a PDA‐colonized agar plug of either wild‐type or SsFkh1 RNA‐silenced mutants, and placed in a high‐humidity chamber at 20°C for 6 days. The diameters of the necrotic lesions were measured and the pathogenicity assays were performed three times. The photographs were taken with a Canon EOS 550D (Canon, Tokyo, Japan) camera in this study.
Sclerotial production assay
Wild‐type and RNA‐silenced mutants were cultured on PDA medium at 25°C for 2 days. Agar plugs taken from the colony from either the wild‐type or RNAi mutant strains using a cork borer (5 mm in diameter) were inoculated at opposite sides of the same autoclaved medium (smashed potato with 1.5% agar). The inoculated plates were incubated at 25 ºC for 7 days or longer for sclerotial production.
Expression profiling of melanin biosynthesis‐associated genes
Melanin is required for sclerotial biosynthesis. To investigate whether SsFkh1 is involved in the biosynthesis of sclerotia, the expression profiles of a set of melanin biosynthesis‐associated genes (Liu and Free, 2016) (polyketide synthase‐encoding gene SsPks13, XP_001585805.1, SS1G_13322) (Amselem et al., 2011) and three laccases (lac1, NXP_001595941, SS1G_02155; lac2, XP_001591159, SS1G_07784; and lac3, XP_001598720, SS1G_00809) in S. sclerotiorum were analysed by qRT‐PCR. The transformants and wild‐type strains were cultured on PDA overlaid with cellophane for 2 days. Total RNA isolation and cDNA synthesis were conducted as described above. qRT‐PCR was performed using the primer pairs listed in Table 1. The expression level of actin in S. sclerotiorum was used as an internal reference. PCRs were conducted as follows: 30 s at 95 ºC, then 40 cycles each consisting of 5 s at 95 ºC, 40 s at 60 ºC and 15 s at 95 ºC, followed by 1 min at 60 ºC and 15 s at 95 ºC. The relative expression levels were calculated as described above.
Osmotic and oxidative stress assays
The wild‐type strain and RNA‐silenced mutants were inoculated on medium containing H2O2, or grown under hyperosmotic stress, for the examination of cell integrity maintenance in hyphae. The hyphal growth rates of each strain on PDA supplemented with H2O2 (0–30 mm), NaCl (1 m) or KCl (1 m) were measured 48 h after inoculation. Each experiment was repeated at least three times.
Statistical analysis
All statistical analyses and plots were performed using SigmaPlot 12.0, Microsoft Excel 2010 or SPSS 19.0 (SPSS Science, Chicago, IL, USA). The data are expressed as the mean ± standard deviation (SD).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 SsFkh1 is expressed differentially at different developmental stages. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed to profile the SsFkh1 expression of Sclerotinia sclerotiorum at different developmental stages: H, hyphae; SI–SIII, sclerotial stages I – stage III; AI – AIII, sclerotial stages I–III. Gene expression values are normalized relative to the constitutively expressed actin gene. Values are the means ± standard deviation (SD) (n = 3). Asterisks indicate significant differences compared with the wild‐type strain [*P < 0.05, **P < 0.01, one‐way analysis of variance (ANOVA) in SPSS].
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31271991, 31471730), the Ministry of Education Fund for the Doctoral of China (No.20120061110082) and the Special Program of the Ministry of Agriculture for Public Profession (201103016). All authors declare that there are no conflicts of interest.
References
- Amselem, J. , Cuomo, C.A. , van Kan, J.A.L. , Viaud, M. , Benito, E.P. , Couloux, A. , Coutinho, P.M. , de Vries, R.P. , Dyer, P.S. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K.M. , Pradier, J.M. , Quévillon, E. , Sharon, A. , Simon, A. , ten Have, A. , Tudzynski, B. , Tudzynski, P. , Wincker, P. , Andrew, M. , Anthouard, V. , Beever, R.E. , Beffa, R. , Benoit, I. , Bouzid, O. , Brault, B. , Chen, Z. , Choquer, M. , Collémare, J. , Cotton, P. , Danchin, E.G. , Da Silva, C. , Gautier, A. , Giraud, C. , Giraud, T. , Gonzalez, C. , Grossetete, S. , Güldener, U. , Henrissat, B. , Howlett, B.J. , Kodira, C. , Kretschmer, M. , Lappartient, A. , Leroch, M. , Levis, C. , Mauceli, E. , Neuvéglise, C. , Oeser, B. , Pearson, M. , Poulain, J. , Poussereau, N. , Quesneville, H. , Rascle, C. , Schumacher, J. , Ségurens, B. , Sexton, A. , Silva, E. , Sirven, C. , Soanes, D.M. , Talbot, N.J. , Templeton, M. , Yandava, C. , Yarden, O. , Zeng, Q. , Rollins, J.A. , Lebrun, M.H. and Dickman M. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel, K. and Hirt, H. (2004) REACTIVE OXYGEN SPECIES: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. [DOI] [PubMed] [Google Scholar]
- Bensen, E.S. , Filler, S.G. and Berman, J. (2002) A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans . Eukaryot. Cell, 1, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland, G.J. and Hall, R. (1994) Index of plant hosts of Sclerotinia sclerotiorum . Can. J. Plant Pathol. 16, 93–108. [Google Scholar]
- Bolton, M.D. , Thomma, B.P.H.J. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Carlsson, P. and Mahlapuu, M. (2002) Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250, 1–23. [DOI] [PubMed] [Google Scholar]
- Duan, Y. , Ge, C. , Liu, S. , Wang, J. and Zhou, M. (2013) A two‐component histidine kinase Shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum . Mol. Plant Pathol. 14, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erental, A. , Dickman, M.B. and Yarden, O. (2008) Sclerotial development in Sclerotinia sclerotiorum: awakening molecular analysis of a “Dormant” structure. Fungal Biol. Rev. 22, 6–16. [Google Scholar]
- Godoy, G. , Steadman, J.R. , Dickman, M.B. and Dam, R. (1990) Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris . Physiol. Mol. Plant Pathol. 37, 179–191. [Google Scholar]
- Jurick II, W.M. and Rollins, J.A. (2007) Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum . Fungal Genet. Biol. 44, 521–530. [DOI] [PubMed] [Google Scholar]
- Kabbage, M. , Williams, B. and Dickman, M.B. (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum . PLoS Pathog. 9, e1003287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.S. , Park, S.H. , Kwon, S.B. , Youn, S.W. , Park, E.S. and Park, K.C. (2005) Heat treatment decreases melanin synthesis via protein phosphatase 2A inactivation. Cell. Signal. 17, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Koranda, M. , Schleiffer, A. , Endler, L. and Ammerer, G. (2000) Forkhead‐like transcription factors recruit Ndd1 to the chromatin of G2/M‐specific promoters. Nature, 406, 94–98. [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Reynolds, D.M. , Shevchenko, A. , Shevchenko, A. , Goldstone, S.D. and Dalton, S. (2000) Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M‐phase. Curr. Biol. 10, 896–906. [DOI] [PubMed] [Google Scholar]
- Lam, E.W.F. , Brosens, J.J. , Gomes, A.R. and Koo, C.Y. (2013) Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer, 13, 482–495. [DOI] [PubMed] [Google Scholar]
- Lee, B.Y. , Han, S.Y. , Choi, H.G. , Kim, J.H. , Han, K.H. and Han, D.M. (2005) Screening of growth‐ or development‐related genes by using genomic library with inducible promoter in Aspergillus nidulans . J. Microbiol. 43, 523–528. [PubMed] [Google Scholar]
- Li, M. and Rollins, J.A. (2009) The development‐specific protein (Ssp1) from Sclerotinia sclerotiorum is encoded by a novel gene expressed exclusively in sclerotium tissues. Mycologia, 101, 34–43. [DOI] [PubMed] [Google Scholar]
- Liu, L. and Free, S.J. (2016) Characterization of the Sclerotinia sclerotiorum cell wall proteome. Mol. Plant Pathol. 17, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayashiki, H. , Hanada, S. , Quoc, N.B. , Kadotani, N. , Tosa, Y. and Mayama, S. (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 42, 275–283. [DOI] [PubMed] [Google Scholar]
- Nguyen, Q.B. , Kadotani, N. , Kasahara, S. , Tosa, Y. , Mayama, S. and Nakayashiki, H. (2008) Systematic functional analysis of calcium‐signalling proteins in the genome of the rice‐blast fungus, Magnaporthe oryzae, using a high‐throughput RNA‐silencing system. Mol. Microbiol. 68, 1348–1365. [DOI] [PubMed] [Google Scholar]
- Ostrow, A.Z. , Nellimoottil, T. , Knott, S.R.V. , Fox, C.A. , Tavaré, S. and Aparicio, O.M. (2014) Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS One, 9, e87647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kong, S. , Kim, S. , Kang, S. and Lee, Y.H. (2014) Roles of forkhead‐box transcription factors in controlling development, pathogenicity, and stress response in Magnaporthe oryzae . Plant Pathol. J. 30, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.H. , Kim, H.Y. , Kim, J.H. and Han, K.H. (2010) Gene structure and function of fkhE, a forkhead gene in a filamentous fungus Aspergillus nidulans . Korean J. Mycol. 38, 160–166. [Google Scholar]
- Proctor, R.H. , Hohn, T.M. and McCormick, S.P. (1997) Restoration of wild‐type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology, 143, 2583–2591. [DOI] [PubMed] [Google Scholar]
- Qu, X. , Yu, B. , Liu, J. , Zhang, X. , Li, G. , Zhang, D. , Li, L. , Wang, X. , Wang, L. , Chen, J. , Mu, W. , Pan, H. and Zhang, Y. (2014) MADS‐box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum . Int. J. Mol. Sci. 15, 8049–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribár, B. , Grallert, A. , Oláh, É. and Szállási, Z. (1999) Deletion of the sep1+ forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe . Biochem. Biophys. Res. Commun. 263, 465–474. [DOI] [PubMed] [Google Scholar]
- Rollins, J.A. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant–Microbe Interact. 16, 785–795. [DOI] [PubMed] [Google Scholar]
- Roper, M. , Seminara, A. , Bandi, M.M. , Cobb, A. , Dillard, H.R. and Pringle, A. (2010) Dispersal of fungal spores on a cooperatively generated wind. Proc. Natl. Acad. Sci. USA, 107, 17 474–17 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, E.K. , Hoff, B. and Kück, U. (2004) AcFKH1, a novel member of the forkhead family, associates with the RFX transcription factor CPCR1 in the cephalosporin C‐producing fungus Acremonium chrysogenum . Gene, 342, 269–281. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. , Zakrajsek, B.A. , Mills, A.G. , Gorn, V. , Singer, M.J. and Reed, M.W. (2000) Quantitative reverse transcription‐polymerase chain reaction to study mRNA decay: comparison of endpoint and real‐time methods. Anal. Biochem. 285, 194–204. [DOI] [PubMed] [Google Scholar]
- Shimeld, S.M. , Degnan, B. and Luke, G.N. (2010) Evolutionary genomics of the Fox genes: origin of gene families and the ancestry of gene clusters. Genomics, 95, 256–260. [DOI] [PubMed] [Google Scholar]
- Smith, M.E. , Henkel, T.W. and Rollins, J.A. (2015) How many fungi make sclerotia? Fungal Ecol. 13, 211–220. [Google Scholar]
- Wang, J.J. , Qiu, L. , Cai, Q. , Ying, S.H. and Feng, M.G. (2015) Transcriptional control of fungal cell cycle and cellular events by Fkh2, a forkhead transcription factor in an insect pathogen. Sci. Rep. 5, 10 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Liu, Y. , Liu, J. , Zhang, Y. , Zhang, X. and Pan, H. (2016) The Sclerotinia sclerotiorum FoxE2 gene is required for apothecial development. Phytopathology, 106, 484–490. [DOI] [PubMed] [Google Scholar]
- Williams, B. , Kabbage, M. , Kim, H.J. , Britt, R. and Dickman, M.B. (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X. , Xie, J. , Cheng, J. , Li, G. , Yi, X. , Jiang, D. and Fu, Y. (2013) Novel secretory protein Ss‐Caf1 of the plant‐pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant–Microbe Interact. 27, 40–55. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Jiang, D. , Xie, J. , Cheng, J. , Li, G. , Yi, X. and Fu, Y. (2012) Ss‐Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum . PLoS One, 7, e34962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahiri, A. , Heimel, K. , Wahl, R. , Rath, M. and Kämper, J. (2010) The Ustilago maydis forkhead transcription factor fox1 is involved in the regulation of genes required for the attenuation of plant defenses during pathogenic development. Mol. Plant–Microbe Interact. 23, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Zhu, G. , Spellman, P.T. , Volpe, T. , Brown, P.O. , Botstein, D. , Davis, T.N. and Futcher, B. (2000) Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature, 406, 90–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 SsFkh1 is expressed differentially at different developmental stages. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) was performed to profile the SsFkh1 expression of Sclerotinia sclerotiorum at different developmental stages: H, hyphae; SI–SIII, sclerotial stages I – stage III; AI – AIII, sclerotial stages I–III. Gene expression values are normalized relative to the constitutively expressed actin gene. Values are the means ± standard deviation (SD) (n = 3). Asterisks indicate significant differences compared with the wild‐type strain [*P < 0.05, **P < 0.01, one‐way analysis of variance (ANOVA) in SPSS].


