Summary
Peroxisomes are involved in various metabolic processes and are important for virulence in different pathogenic fungi. How peroxisomes rapidly emerge in the appressorium during fungal infection is poorly understood. Here, we describe a gene, PEF1, which can regulate peroxisome formation in the appressorium by controlling peroxisomal fission, and is required for plant infection in the rice blast fungus Magnaporthe oryzae. Targeted deletion of PEF1 resulted in a reduction in virulence and a delay in penetration and invasive growth in host cells. PEF1 was particularly expressed during appressorial development, and its encoding protein was co‐localized with peroxisomes during appressorial development. Compared with the massive vesicle‐shaped peroxisomes formed in the wild‐type appressorium, the Δpef1 mutant could only form stringy linked immature peroxisomes, suggesting that PEF1 was involved in peroxisomal fission during appressorium formation. We also found that the Δpef1 mutant could not utilize fatty acids efficiently, which can improve significantly the expression level of PEF1 and induce peroxisomal fission. As expected, the Δpef1 mutant showed reduced intracellular production of reactive oxygen species (ROS) during appressorium formation and induced ROS accumulation in host cells during infection. Taken together, PEF1‐mediated peroxisomal fission is important for fungal infection by controlling the number of peroxisomes in the appressorium.
Keywords: appressorium, fatty acid, Magnaporthe oryzae, β‐oxidation, peroxisomal proliferation
Introduction
Peroxisomes are single‐layer, membrane‐bound organelles ubiquitously distributed in eukaryotic cells (Smith and Aitchison, 2013). They are highly dynamic structures and move along cytoskeletal tracks (Titorenko and Rachubinski, 2001). Peroxisomes contain at least 50 different enzymes, which are involved in a variety of metabolic reactions, including several aspects of energy metabolism. Peroxisomes produce a large amount of hydrogen peroxide (H2O2). Because H2O2 is harmful, peroxisomes also contain the enzyme catalase to decompose H2O2. A variety of substrates are degraded by oxidative reactions in peroxisomes, including uric acid, amino acids and fatty acids. Peroxisomes exert many activities related to lipid metabolism, including functions involving fatty acid β‐oxidation, decomposition of H2O2 and the glyoxylate cycle (Kunze et al., 2006; Poirier et al., 2006; Schrader and Fahimi, 2006; Titorenko and Rachubinski, 2001).
Eukaryotic cells may contain different numbers of peroxisomes in response to various environmental stimuli or at different developmental stages. The number of peroxisomes in a cell can be determined by several processes, including de novo formation, fission, fusion and pexophagy (Fagarasanu et al., 2007). The fusion and fission processes of peroxisomes are well balanced in order to ensure the requisite number, basic function and structures of peroxisomes for developmental needs or environmental responses. De novo formation and fission processes contribute to increase the number of peroxisomes (Fagarasanu et al., 2007). Pexophagy is a type of autophagic function in peroxisome degradation and occurs when the cells suddenly move away from peroxisome fission‐inducing conditions (Sakai et al., 2006). In Colletotrichum orbiculare, the autophagy‐related protein Atg26 is involved in pexophagy inside appressoria, and functions in the recycling of cellular components, such as amino acids, that are required for host invasion (Asakura et al., 2009; Takano et al., 2009).
In Saccharomyces cerevisiae, the peroxisomal fission process involves three key steps: peroxisomal elongation, peroxisomal membrane constriction and the fission of peroxisomes (Kaur and Hu, 2009). The peroxisomal membrane protein peroxin 11 (Pex11) may function in peroxisome elongation. Under fatty acid conditions, Pex11p‐deficient cells accumulate a few giant peroxisomes (Tam et al., 2003; Thoms and Erdmann, 2005). Dnm1p, a dynamin‐like protein, is concentrated at constrictions of the elongated peroxisomes, leading to final membrane fission through guanosine triphosphate (GTP) hydrolysis (Koch et al., 2003). Fis1p is an outer membrane protein, which recruits Dnm1p to peroxisomes with the assistance of adaptors Mdv1p or Caf4p (Koch et al., 2005). The final fission process requires the interaction among Dnm1p, Fis1p and Mdv1p or Caf4p (Saraya et al., 2010). Although extensive studies have been performed in yeasts and human cells, functions of peroxisomal fission have been addressed less well in filamentous fungi.
Magnaporthe oryzae is a fungal pathogen that causes rice blast disease, which is one of the most destructive rice diseases worldwide. During infection, the fungal pathogen produces a dome‐shaped infection structure, called an appressorium (Wilson and Talbot, 2009). In the incipient appressorium, lipid droplets are brought from conidial cells in a short time by bulk cytoplasmic streaming, and are utilized in the peroxisomes during appressorium maturation (Thines et al., 2000). Triglycerides are an abundant form of lipids, which can be decomposed by lipases to release fatty acids and glycerol. Then, the fatty acids can be utilized through peroxisomal β‐oxidation pathways to produce acetyl‐coenzyme A (acetyl‐CoA) and energy (Wang et al., 2007). A high concentration of glycerol contributes to turgor generation for appressorial penetration (Howard et al., 1991; de Jong et al., 1997). Host cell penetration is usually associated with the activation of a strong defence response [including the reactive oxygen species (ROS) burst] (Jones and Dangl, 2006). To colonize host cells, the fungus must overcome this defence response by various mechanisms (Huang et al., 2011).
It is generally accepted that β‐oxidation of fatty acids in fungi is exclusive to the peroxisomes (Hiltunen et al., 2003). Some studies have addressed the role of peroxisomal β‐oxidation, peroxisome biogenesis and peroxisomal matrix protein import in fungal pathogenesis (Bhambra et al., 2006; Goh et al., 2011; Ramos‐Pamplona and Naqvi, 2006; Wang et al., 2013, 2007; Yang et al., 2012). However, few studies have focused on peroxisome number control during infection. Recently, Wang et al. (2015) have demonstrated that M. oryzae Pex11A is required for peroxisome proliferation and virulence. Here, we show that Pef1, a WD‐repeat protein homologous to Mdv1p and Caf4p in S. cerevisiae, is also important for peroxisomal fission during appressorium formation, and prove that peroxisomal fission can be induced by fatty acids and is important for appressorium‐mediated infection of the rice blast fungus.
Results
Identification of a T‐DNA mutant with loss of pathogenicity
A T‐DNA insertional mutant library for M. oryzae was generated by the Agrobacterium tumefaciens‐mediated transformation (ATMT) of a wild‐type strain P131 with the hygromycin phosphotransferase (hph) gene as a selection marker (Chen et al., 2011). After screening 11 420 ATMT transformants, one mutant, SX4690, defective in pathogenicity was isolated. No disease lesions were generated on susceptible rice leaves when inoculated with SX4690, whereas the wild‐type strain developed numerous typical lesions (Fig. S1A, see Supporting Information). SX4690 also grew more slowly than the wild‐type P131.
To verify whether the defects in SX4690 were caused by gene disruption by the exogenous DNA insertion, a genetic cross was made between SX4690 and S1528, which is a wild‐type strain with opposite mating type. The results revealed that the hygromycin resistance was co‐segregated with the defects in pathogenicity and growth features in the mutant SX4690 (Table S1, see Supporting Information). To identify the gene disrupted in SX4690, the genomic DNA sequences flanking the T‐DNA insertion site were isolated with the thermal asymmetric interlaced‐polymerase chain reaction (TAIL‐PCR) technique (Liu and Whittier, 1995). Analysis of the flanking sequences revealed that T‐DNA was inserted in the intergenic region between two predicted genes, MGG_01711 and MGG_01710, and adjacent to their start codon sites at 130 bp and 550 bp distant, respectively (Fig. S1B). Thus, potentially the functions of both of these genes could be affected by T‐DNA insertion. Deletion of each of the genes resulted in reduced virulence, and thus both the genes are important for pathogenicity (data not shown). In this study, we focused on the gene MGG_01711.
PEF1 is important for virulence in rice and barley
A full‐length cDNA of MGG_01711 was obtained by reverse transcription‐polymerase chain reaction (RT‐PCR) method and had an open reading frame of 1986 bp for a protein with 661 amino acids. The protein showed approximately 32% identity with Caf4 and Mdv1 in S. cerevisiae (Fig. S2, see Supporting Information), which were reported to be involved in the fission of peroxisomes (Motley et al., 2008). Therefore, we named MGG_01711 PEF1 (for PEroxisomal Fission gene 1). Pef1 is well conserved among filamentous fungi (Fig. S2). To confirm the role of PEF1 in virulence, two independent PEF1 deletion mutants, PEF1KO1 and PEF1KO2, were generated by homologous recombination (Fig. S3A, see Supporting Information). These two deletion mutants were confirmed by PCR verification (Fig. S3B) and DNA gel blot analysis (Fig. S3C). As the two PEF1 deletion mutants were very similar in phenotype, PEF1KO1 was chosen for further analyses.
Similar to the mutant SX4690, colonies formed by PEF1KO1 were significantly smaller than those formed by the wild‐type on oatmeal tomato agar (OTA) medium, minimal medium (MM) and V8 medium (Fig. 1A; Table S2, see Supporting Information). When Δpef1 mycelia were stained with calcofluor white (CFW), the average lengths of apical and subapical hyphal cells were ∼75% of those of the wild‐type (Fig. 1B,C). For infection assays, only a few disease lesions were developed on leaves of rice or barley sprayed with the Δpef1 mutant. In contrast, numerous lesions were formed by the wild‐type strain. Moreover, lesions formed by the Δpef1 mutant were smaller than those of the wild‐type (Fig. 2A,B). A complementation experiment was also performed by introducing the full‐length PEF1 with a 2.0‐kb native promoter into the Δpef1 mutant. Twenty‐five transformants were obtained, and all of these transformants displayed similar phenotypes to the wild‐type strain in virulence and colony growth (Fig. 2A,B). Taken together, these results demonstrate that PEF1 plays an important role in mycelial growth and is required for full virulence of the rice blast fungus.
Figure 1.
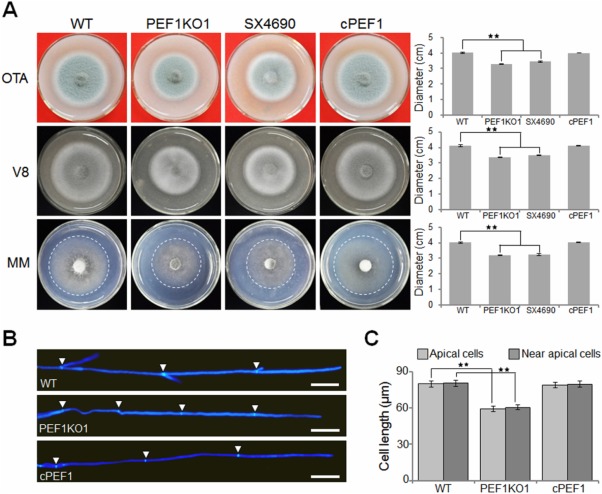
Deletion of PEF1 affects vegetative growth. (A) The Δpef1 mutants displayed reduced colony growth. The wild‐type strain P131 (WT), PEF1 deletion mutant (PEF1KO1), T‐DNA mutant (SX4690) and complemented strain (cPEF1) were cultured on oatmeal tomato agar (OTA) medium, minimal medium (MM) and V8 medium at 28°C for 5 days. (B) Hyphal tips of P131, PEF1KO1 and cPEF1 were stained with calcofluor white. The cell septa are indicated by white arrows. Bar, 20 μm. (C) Cell length of the apical and subapical cells in the hyphal tips of P131, PEF1KO1 and cPEF1. The data represent the mean ± standard error of the mean (SEM) of two independent experiments conducted in triplicate. Significant differences compared with WT are indicated by asterisks (P = 0.01).
Figure 2.
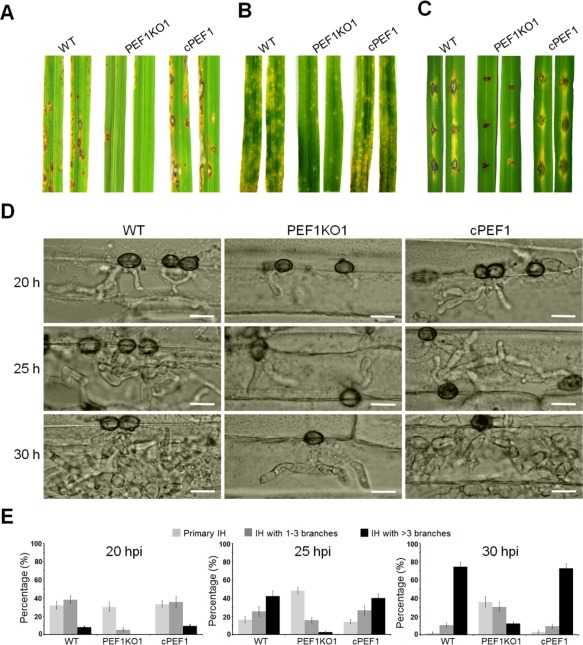
Loss of PEF1 leads to a reduction in virulence and growth of infection hyphae in host tissues. Rice leaves (A) and barley leaves (B) were inoculated with conidial suspensions of the wild‐type P131 (WT), the PEF1 deletion mutant (PEF1KO1) and one complemented transformant (cPEF1). Typical leaves were photographed at 5 days post‐inoculation (dpi). (C) Virulence test on wounded rice leaves. Rice leaves were slightly wounded by a needle before inoculation with a mycelial block of P131, PEF1KO1 or cPEF1. Typical leaves were photographed at 5 dpi. (D) Penetration and infection hyphae of P131, PEF1KO1 and cPEF1 were examined at 20, 25 and 30 h post‐inoculation (hpi). Bar, 10 μm. (E) Percentages of appressoria with distinct types of infection hyphae (IH) (primary IH, IH with one to three branches and IH with more than three branches) at 20, 25 and 30 hpi.
The Δpef1 mutant is blocked in penetration and invasive growth in planta
We also assayed the virulence of the Δpef1 mutant by inoculation of fungal blocks onto abraded rice leaves. Compared with the wild‐type, the Δpef1 mutants formed very limited lesions on wounded rice leaves (Fig. 2C), suggesting that the Δpef1 mutant was defective in invasive growth in host tissues. To confirm the role of PEF1 during the infection process, we compared major infection steps of the Δpef1 mutant with those of the wild‐type strain on barley epidermis. The Δpef1 conidia were normal in germination and appressorial formation. However, infection penetration and invasive growth of the Δpef1 mutant were blocked significantly (Table S3, see Supporting Information). At 20 h post‐inoculation (hpi), 78.1% of the wild‐type appressoria had penetrated into plant cells and 46.3% of them had developed branched infection hyphae (IH). In contrast, only 35.8% of the Δpef1 appressoria had formed primary IH, and 5.3% of them had formed branched IH (Fig. 2D,E; Table S3). At 25 hpi, 67.8% of the wild‐type primary IH had branched, and 42.3% of them had formed more than three branches, whereas only 18.6% of the Δpef1 mutant primary IH had branched, and just 2.8% of them had formed more than three branches (Fig. 2D,E; Table S3). At 30 dpi, around 74.4% of the wild‐type appressoria had formed IH with at least three branches, but only 12.5% of the Δpef1 appressoria had formed multiple branched IH (Fig. 2D,E). The Δpef1‐rescued strains exhibited similar penetration and invasive growth to the wild‐type strain (Fig. 2D,E; Table S3). Taken together, these results indicate that PEF1KO1 is able to penetrate and grow in the host cells, but its extension is delayed.
Pef1 recovers defects of the yeast Δmdv1, Δcaf4 and Δmdv1Δcaf4 mutants
In S. cerevisiae, both Mdv1 and Caf4 are involved in peroxisomal fission and mitochondrial fission processes, and the Δmdv1 and Δmdv1Δcaf4 mutants exhibit increased sensitivity to rapamycin (Guo et al., 2012). As the unique homologue, we speculated that Pef1 should facilitate the functions of both Mdv1 and Caf4. To test this possibility, we cloned PEF1 into the expression vector pYES2, and the resulting construct was transformed into the yeast Δmdv1, Δcaf4 and Δmdv1Δcaf4 mutants. All of the transformants restored the sensitivities of the Δmdv1, Δcaf4 and Δmdv1Δcaf4 mutants to 20 ng/mL rapamycin under induction conditions (Fig. 3A), indicating that M. oryzae Pef1 exerts functions of both Mdv1 and Caf4 in S. cerevisiae.
Figure 3.
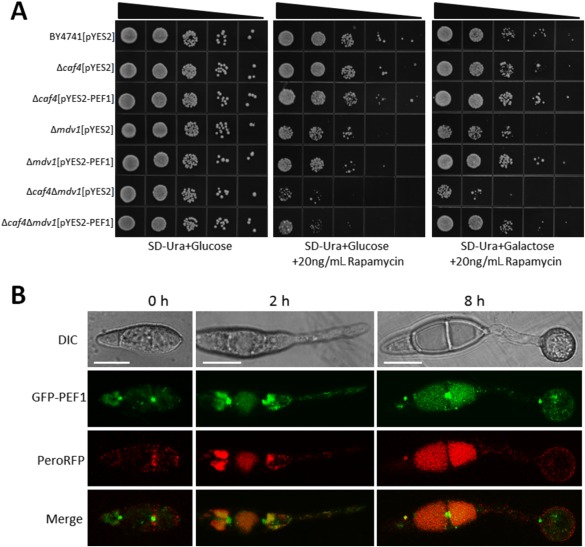
Co‐localization of Pef1‐GFP with PeroRFP during appressorial development. (A) Yeast complementation assay. Serial 10‐fold dilutions of cells from the wild‐type strain BY4741 and the null mutants Δcaf4, Δmdv1 and Δcaf4Δmdv1 transformed with pYES2 or pYES2‐PEF1 were dotted onto synthetic complete medium without uracil (SC–Ura) plates with glucose (Glu) or galactose (Gal) supplemented with 20 ng/mL rapamycin. The SC–Ura plate with Glu, but without rapamycin, was used as a control. All of the plates were incubated at 30°C for 72 h. (B) Dual colour imaging by confocal laser scanning microscopy (CLSM) of transformant PEF1DW1 expressing both PEF1‐GFP and PeroRFP. Left: conidia (0 h). Middle, germ tube (2 h). Right, appressoria (12 h). Bars, 10 μm. DIC, differential interference contrast; GFP, green fluorescent protein; RFP, red fluorescent protein.
Pef1 can be localized to peroxisomes and mitochondria
In S. cerevisiae, Mdv1 and Caf4 were found to be involved in the peroxisomal fission process and co‐localized to peroxisomes (Motley et al., 2008); we wondered whether Pef1 played a similar role in M. oryzae. We first generated a construct that contained the enhanced green fluorescent protein‐encoding gene (eGFP) fused with PEF1 under a 1.5‐kb PEF1 native promoter. The eGFP‐PEF1 fusion construct was then co‐transferred with the widely used peroxisome marker PeroRFP into the Δpef1 mutant. All the resulting transformants were recovered to the wild‐type levels in mycelial growth and virulence. One, PEF1DW1, was randomly selected for further confocal laser scanning microscopy (CLSM) observation. In conidia, the GFP‐PEF1 protein was not well co‐localized with the peroxisome marker PeroRFP protein. However, together with the development of germ tubes and appressoria, punctuated structures of the GFP‐PEF1 protein were observed, which were partially co‐localized with the PeroRFP protein (Fig. 3B). As the yeast proteins Mdv1 and Caf4, orthologues of Pef1, are also involved in the fission process of the mitochondria (Griffin et al., 2005; Tieu and Nunnari, 2000), we performed co‐localization observation of Pef1 and mitochondria. CLSM observation demonstrated that the GFP‐PEF1 protein was also partially co‐localized with mitochondria in the strain PEF1DW1 stained with Mitotracker (Fig. S4, see Supporting Information). Importantly, the GFP‐PEF1 protein was strongly co‐localized with M. oryzae MoFis1 (Khan et al., 2015), the orthologue of S. cerevisiae Fis1 (Fig. S5, see Supporting Information). In S. cerevisiae, the Fis1 protein was also found to be involved in both peroxisomal fission and mitochondrial fission by combination with Mdv1 or Caf4 (Motley et al., 2008). These data indicate that Pef1 could be functional in the fission processes of both peroxisomes and mitochondria.
Deletion of PEF1 results in fewer numbers of peroxisomes in appressoria
Gene expression profiling showed that the expression levels of PEF1 in conidia and mycelia were similar. However, when conidia were germinated on a hydrophobic surface, the expression level of PEF1 was greatly elevated in germ tubes, reached a maximum in unmelanized appressoria (3.5‐fold that in mycelia) and then decreased during the maturation of appressoria. Interestingly, the expression level of PEF1 was much lower during invasive growth (Fig. 4A). These results suggest that PEF1 may play an important role during appressorial formation and maturation.
Figure 4.
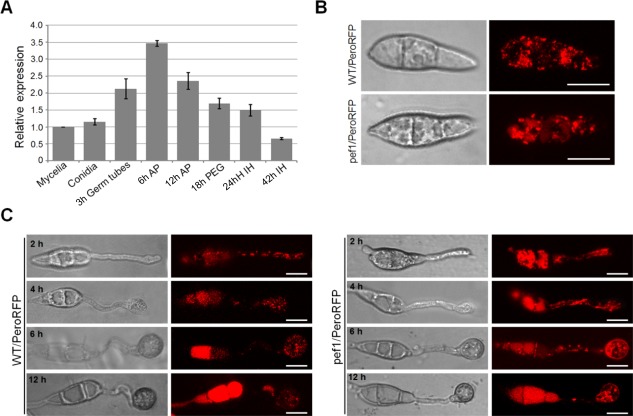
Peroxisome fission defects of the Δpef1 mutant during appressorium formation. (A) Expression levels of PEF1 in mycelia, conidia, germ tubes, 6‐h appressoria and 12‐h appressoria. Mean and standard errors were calculated from three independent replicates. AP, appressoria; IH, infection hyphae; PEG, penetration pegs. (B) Peroxisomal morphologies in conidia of WT/PeroRFP and pef1/PeroRFP. WT/PeroRFP and pef1/PeroRFP are transformants of the wild‐type (WT) and Δpef1 strains expressing pPeroRFP, respectively. Bars, 10 μm. (C) Peroxisomal morphology during development. WT/PeroRFP and pef1/PeroRFP were inoculated onto an inductive surface and assessed for peroxisomal morphology by confocal laser scanning microscopy (CLSM) at 2, 4, 6 and 12 h post‐inoculation (hpi). Bars, 10 µm.
In S. cerevisiae, Mdv1 and Caf4 are two fission adaptors that can help Fis1 to recruit Dnm1 to the peroxisome for fission (Motley et al., 2008). To clarify whether peroxisomal fission was also regulated by PEF1 in M. oryzae, we first compared peroxisome production in conidia and during appressorial formation of the Δpef1 mutant and the wild‐type strain. The vector expressing PeroRFP was transformed into the wild‐type strain and the Δpef1 mutant, and the resulting transformants were named WT/PeroRFP and pef1/PeroRFP, respectively. CLSM observation showed that many independent punctuated structures of PeroRFP were formed in conidia, germ tubes and appressoria of the WT/PeroRFP strain (Fig. 4B,C). In contrast, in the Δpef1 mutant, string‐shaped structures of PeroRFP were more commonly observed; punctuated structures were also found together with the development of appressoria, and fluorescence was much brighter than that in the wild‐type (Fig. 4C). These results indicate that PEF1 is an important factor controlling the number of peroxisomes in conidia and appressoria in M. oryzae.
PEF1 affects fatty acid‐induced formation of peroxisomes
The number of cellular peroxisomes is known to be variable in environments with different carbon sources. We therefore compared the number and morphology of peroxisomes in mycelia of the wild‐type strain and Δpef1 mutant cultured on MM with different carbon sources. When glucose was used as the sole carbon source (normal conditions), both the wild‐type strain and Δpef1 mutant showed a number of peroxisomes that appeared as punctuated structures and were evenly distributed in mycelia (Fig. 5A). When ethanol or acetate was added as the sole carbon source, greater numbers of punctuated peroxisomes were formed, and the Δpef1 mutant was not significantly different from the wild‐type strain with regard to the number and morphology of peroxisomes (Fig. 5A,B). However, when oleic acid (C18), myristic acid (C14) or lauric acid (C12) was added as the sole carbon source, the Δpef1 mutant seldom formed punctuated peroxisomes. In contrast, the Δpef1 mutant generated sheet or string‐like structures, whereas the wild‐type strain formed many more punctuated peroxisomes than formed under ethanol or acetate (Fig. 5A). These results show that fatty acids can induce the formation of punctuated peroxisomes in mycelia of M. oryzae, and that the PEF1 deletion mutant is deficient in the induction of peroxisome formation by fatty acids.
Figure 5.
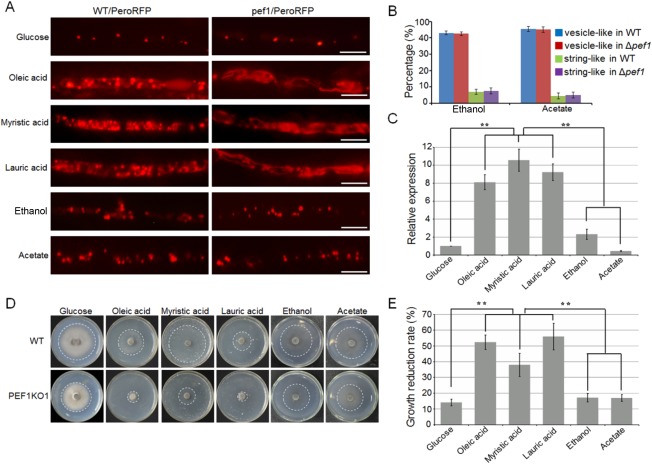
Peroxisomal fission is induced by fatty acids. (A) The wild‐type (WT) and Δpef1 strains expressing PeroRFP were incubated on plates with different fatty acids or carbon sources, and assessed for peroxisome morphology by epifluorescence microscopy with red fluorescent protein (RFP) optics. Bars, 10 μm. (B) Statistical analysis of vesicle‐like or string‐like peroxisomes formed in the WT and Δpef1 strains expressing PeroRFP under ethanol or acetate conditions. Means and standard errors were calculated from three independent replicates. (C) Expression profiling of PEF1 in minimal medium with different fatty acids or carbon sources as sole carbon source. Means and standard errors were calculated from three independent replicates. Significant differences are indicated by asterisks (P = 0.01). (D) Utilization of different fatty acids and carbon sources by the WT and Δpef1 strains. The strains were cultured on minimal medium with glucose, fatty acids (C18, oleic acid; C14, myristic acid; C12, lauric acid) or alternative carbon sources (ethanol and acetate) as sole carbon source. The colonies were photographed at 5 days post‐inoculation (dpi). (E) Statistical analysis of growth reduction rates of colonies on different media. Means and standard errors were calculated from three independent replicates. Significant differences are indicated by asterisks (P = 0.01).
We further determined the relative expression levels of PEF1 in mycelia of the wild‐type strain cultured in MM amended with different carbon sources, and found that the expression levels of PEF1 were significantly elevated when fatty acids were added as the sole carbon source relative to glucose, ethanol or acetate as the sole carbon source (Fig. 5C). This result suggests that the fatty acid‐induced expression of PEF1 is involved in the induced formation of punctuated peroxisomes in mycelia of M. oryzae.
Δpef1 is deficient in fatty acid utilization
The above data clearly showed that PEF1 could be significantly induced by fatty acids and that the induced expression of PEF1 was important for peroxisomal fission. We thus speculated that fatty acid utilization would be defective in the PEF1 deletion mutant as a result of a lesser formation of peroxisomes. To test the hypothesis, we cultured the wild‐type and Δpef1 mutant strains and compared their growth on MM supplemented with glucose and different fatty acids, including oleic acid, myristic acid or lauric acid, as the sole carbon source. After 5 days of culture at 28 ºC, colony diameters of the Δpef1 mutant were slightly reduced when grown on glucose (14.1% reduction) compared with those of the wild‐type strain. In contrast, the reduction rates were significantly higher under the oleic acid, myristic acid or lauric acid condition, which reached 52.3%, 37.9% and 56.0%, respectively (Fig. 5D,E). This suggests that the utilization of fatty acids is affected, and the defects in peroxisomal fission may lead to a β‐oxidation block in the peroxisomes. We also tested the utilization of non‐fermentable carbon sources (ethanol and acetate), but no significant differences were detected between the wild‐type strain and the Δpef1 mutant when grown under these conditions (Fig. 5D,E), suggesting that peroxisomal fission does not respond to the non‐fermentable carbon sources, consistent with a previous report in S. cerevisiae (Kuravi et al., 2006).
Ultrastructural defects in the Δpef1 mutant
Transmission electron microscopy (TEM) observation demonstrated that, in appressoria at 12 hpi on plant cells, many lipid bodies were found in the wild‐type, but only a few in the Δpef1 mutant (Fig. 6A). Interestingly, in lipid bodies of the wild‐type strains, no detectable content was observed, whereas massive content could be found in the lipid bodies of the Δpef1 mutant (Fig. 6A,B). This observation further confirms that the fatty acids may not be efficiently oxidized because of defects in peroxisome fission in the Δpef1 mutant.
Figure 6.
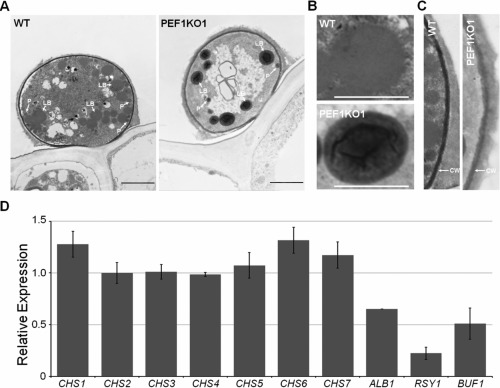
Transmission electron microscopy (TEM) observation of the appressorium. (A) Ultrastructural analysis of the appressorium; 12‐h appressoria of the wild‐type (WT) and Δpef1 mutant strains were processed for TEM. LB, lipid body; P, peroxisome. Bars, 5 μm. (B) Lipid bodies in appressoria of the WT and Δpef1 mutant strains. Bars, 2.5 μm. (C) Cell wall structure in appressoria of the WT and Δpef1 mutant strains. CW, cell wall. (D) Expression level changes of chitin synthase genes and melanin biosynthesis‐related genes. Means and standard errors were calculated from three independent replicates.
From the TEM images, we also found that the cell walls of wild‐type appressoria were darker and more condensed than those of the Δpef1 mutant (Fig. 6C), suggesting that PEF1 might be associated with melanin biosynthesis and cell wall integrity. Subsequent real‐time quantitative PCR analyses revealed that the expression levels of the key genes involved in melanin biosynthesis, ALB1, RSY1 and BUF1 (Chumley and Valent, 1990), were significantly reduced in the Δpef1 mutant (Fig. 6D). However, the expression levels of the seven chitin synthase genes (Kong et al., 2012) were not evidently changed (Fig. 6D).
In order to test whether the mycelial cell wall integrity of the Δpef1 mutant was also affected, we assayed the sensitivity of the Δpef1 mutant to cell wall‐perturbing agents, including CFW, Congo red (CR) and sodium dodecylsulfate (SDS). As expected, the Δpef1 mutant exhibited increased sensitivity to all of the cell wall‐perturbing reagents (Fig. S6, see Supporting Information) compared with the wild‐type strain. These results show that peroxisomal fission can affect the cell wall integrity of both mycelia and appressoria.
The Δpef1 mutant is reduced in intracellular ROS production during appressorium maturation
In M. oryzae, intracellular ROS are crucial for virulence, and the deletion mutants of NADPH oxidases lose virulence on the seedlings of susceptible rice cultivars because of obstructed intracellular ROS generation (Egan et al., 2007). As peroxisomes play important roles in ROS detoxification, we investigated whether defects in peroxisomal fission resulted in the disruption of intracellular ROS homeostasis. The intracellular ROS levels in the wild‐type and Δpef1 mutant strains were detected by the nitroblue tetrazolium (NBT) staining assay. As expected, significant reduction in intracellular ROS was detected in the Δpef1 mutant from conidia to mature appressoria (Fig. 7A). These findings confirm that defects in peroxisomal fission may lead to a reduction in intracellular ROS generation.
Figure 7.
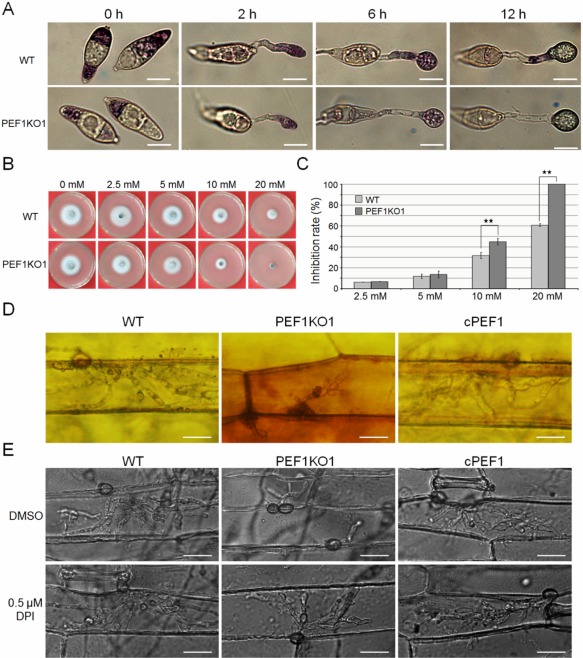
Detection of superoxide and sensitivity to hydrogen peroxide (H2O2) pressure. (A) Conidia of the wild‐type (WT) and Δpef1 mutant strains were inoculated on plastic cover slips and incubated in a moist chamber at 28°C for 0, 2, 6 and 12 h before being stained with a 0.3 mm nitroblue tetrazolium (NBT) aqueous solution for 20 min and viewed by bright‐field microscopy. Representative bright‐field images at each time point are shown. Bar, 10 μm. (B) Sensitivity of P131, the Δpef1 mutant and the complemented strain cPEF1 to different concentrations of H2O2. Four‐day‐old complete medium (CM) cultures were photographed. (C) Statistical analysis of growth reduction rates of mycelia on CM with different concentrations of H2O2. Mean and standard errors were calculated from triplicates. Significant differences are indicated by asterisks (P = 0.01). (D) 3,3′‐Diaminobenzidine (DAB) staining assays of the penetrated plant cells. Barley epidermis was stained with DAB at 30 h post‐inoculation (hpi) and observed under a Nikon microscope. Bar, 20 μm. (E) Invasive growth of infection hyphae restored by diphenyleneiodonium (DPI) treatment. Barley leaves were treated with or without 0.5 μm DPI dissolved in dimethylsulfoxide (DMSO) before inoculation. Penetration and invasive growth were observed at 30 hpi. Bar, 20 μm.
The Δpef1 mutant induces host cellular ROS accumulation during infection
Reduced fungal intracellular ROS levels could lead to attenuated suppression of the host defence system, especially ROS accumulation (Jones and Dangl, 2006). To investigate whether deletion of PEF1 resulted in sensitivity to oxidative oxygen, the wild‐type strain, Δpef1 mutant and complemented strain cPEF1 were exposed to H2O2 stress. There were no differences in colony growth when these strains were exposed to 2.5 and 5 mm H2O2. However, compared with the wild‐type strain, growth inhibition for the Δpef1 mutant was significantly increased when exposed to 10 mm H2O2. Moreover, the Δpef1 mutant, but not the wild‐type strain, failed to grow on 20 mm H2O2 (Fig. 7B,C). These results indicate that deletion of PEF1 indeed leads to increased sensitivity to oxidative pressure.
We subsequently detected ROS accumulation in the host cells by staining with 3,3′‐diaminobenzidine (DAB) at 30 hpi. Barley cells infected by the wild‐type strain were not well stained by DAB, and infrequent reddish brown precipitates were detected. In contrast, the primary barley cells infected by Δpef1 were intensely stained by DAB, and abundant reddish brown precipitates around the infected cells were observed (Fig. 7D). The accumulation of ROS functions as a second signal mediating plant defence (Gechev and Hille, 2005; Nurnberger et al., 2004). The activation of ROS scavenging by fungal pathogens is a mechanism to cope with plant defences and to allow infection (Molina and Kahmann, 2007). We hypothesized that the blocked hyphal growth in the infected host cell and attenuated virulence of the Δpef1 mutant were caused by defects in the scavenging of ROS generated by the host plant. To test this hypothesis, we inhibited the plant cell NADPH oxidase activity using diphenyleneiodonium (DPI), one of the most important generators of ROS (Torres and Dangl, 2005). Without DPI treatment, the Δpef1 infectious hyphae were restricted and only one to two branches formed at 30 hpi. Under the same conditions, the wild‐type strain developed many more branches of infectious hyphae (Fig. 7E). Interestingly, when treated with 0.5 μm DPI to inhibit plant‐generated ROS, the previously inhibited infectious hyphae of the Δpef1 mutant were almost rescued and developed comparable numbers of branches to the wild‐type strain (Fig. 7E). Moreover, the expression levels of the rice pathogenesis‐related (PR) genes, including OsPR1a, OsPR3 and OsPBZ1, were significantly elevated when rice was infected by the Δpef1 mutant (Fig. S7, see Supporting Information). Taken together, scavenging of host‐derived ROS at the infection sites is essential for virulence of the Δpef1 mutant, and deletion of PEF1 is a vital reason for the delay in invasive growth as a result of a failure to produce sufficient fungal intracellular ROS.
Discussion
The utilization of lipid stores in peroxisomes during appressorial maturation is vital for infection by the rice blast fungus, through the provision of energy and biosynthetic requirements. Recent studies have shown that peroxisomes play important roles in lipid degradation in plant‐pathogenic fungi (Kimura et al., 2001; Wang et al., 2007). Several genes have been found to maintain normal functions of peroxisomes in fungal pathogens, such as M. oryzae, Colletotrichum lagenarium and C. orbiculare. Their functions include the β‐oxidation of fatty acids (MFP1/FOX2, PTH2, CRC1) (Wang et al., 2007; Yang et al., 2012), the assembly of peroxisomes (PEX6 and PEX13) (Fujihara et al., 2010; Kimura et al., 2001; Ramos‐Pamplona and Naqvi, 2006; Wang et al., 2007), peroxisomal protein import (PEX5 and PEX7) (Goh et al., 2011; Wang et al., 2013), glyoxylate cycle processes (ICL1 and MLS1) (Solomon et al., 2004; Wang et al., 2003) and pexophagy (ATG26) (Asakura et al., 2009). However, studies on the control of peroxisome numbers are relatively scarce. Recently, M. oryzae MoPEX11A was found to be a main factor controlling peroxisomal division and is required for full virulence (Wang et al., 2015). MoFis1, orthologue of S. cerevisiae Fis1, was found to be important for the mitochondrial fission process in M. oryzae (Khan et al., 2015), but whether MoFis1 is important for peroxisomal fission remains unclear. Here, we showed that PEF1 is another gene involved in peroxisomal fission in M. oryzae, which is important for the rapid increase in the number of appressorial peroxisomes and is required for appressorium‐mediated infection.
Pef1 is involved in the peroxisomal fission process
The mechanism of peroxisomal fission is best understood in S. cerevisiae, involving the genes Dnm1, Fis1 and Mdv1 or Caf4. During peroxisomal fission, Fis1 recruits the dynamin‐related GTPase Dnm1, and this recruitment requires Mdv1 or Caf4, either of which functions as a molecular adaptor to link Dnm1 and Fis1 (Kaur and Hu, 2009). Here, we found that Pef1 was the sole counterpart of Mdv1/Caf4 in M. oryzae. Similar to Mdv1 or Caf4 (Zhang et al., 2012), Pef1 has a modular structure consisting of an N‐terminal extension (NTE) that binds to Fis1, a central coiled‐coil region for dimerization and a C‐terminal WD40 repeat region binding to Dnm1 (Fig. S8, see Supporting Information). In S. cerevisiae, the functions of Mdv1 and Caf4 were not equivalent, because the mdv1 mutant exhibited a clear fission defect, but the caf4 mutant did not (Griffin et al., 2005). As no genes with high identity to PEF1 could be found in the M. oryzae genome, we inferred that Pef1 can play the roles of both Mdv1 and Caf4. Deletion of PEF1 resulted in an obvious defect in peroxisomal fission under inducing conditions with fatty acids, which caused the formation of string‐linked peroxisomes, but not massive small particles in the mycelia, as found with the mdv1caf4 double mutant or the fis1 and dnm1 mutants in yeast (Motley et al., 2008). This function is also similar to that of a mammal protein Mff1, which shows no evident similarity to Pef1 at the amino acid level (Gandre‐Babbe and van der Bliek, 2008).
As the yeast peroxisomal fission machinery is also functional in mitochondrial fission (Hoppins et al., 2007; Schrader, 2006), further research is needed on whether Pef1 controls or influences mitochondrial fission. Moreover, whether the other components, such as the counterpart of yeast Dnm1, are also involved in peroxisomal fission must be clarified in M. oryzae.
The role of PEF1 in appressorium‐mediated host infection
Deletion of PEF1 significantly reduced the virulence towards both rice and barley. The Δpef1 mutant showed normal conidial germination and appressorium formation, but delayed penetration and invasive growth. Why is PEF1 important for penetration and invasive growth? First, deletion of PEF1 led to a reduction in the β‐oxidation of fatty acids. As the acetyl‐CoA generated from peroxisomal lipid oxidation is an important resource for glycerol accumulation in appressoria, its deficiency will lead to a reduction in appressorial turgor in the Δpef1 mutant. Acetyl‐CoA can also be used in the biosynthesis of glucan and chitin, which are mediated by the glyoxylate cycle and required for cell wall generation (Howard and Valent, 1996). Moreover, acetyl‐CoA can also be used in melanin biosynthesis (Fujii et al., 2000). Thus, the defects of turgor generation, cell wall integrity and melanin accumulation in the appressoria of the Δpef1 mutant may lead to a block in penetration. In addition, a reduction in peroxisomal lipid oxidation may lead to a lack of nutrients before biotrophic growth. Second, deletion of PEF1 results in a lack of intracellular ROS in appressoria, which is important for counteracting host defence systems. There may also be other possible causes of virulence reduction.
Peroxisomal fission is induced by appressorial fatty acids
In this study, the peroxisomal fission process was found to be activated in the presence of fatty acids. This phenomenon coincides with the process of appressorium formation, during which fatty acids are produced. The expression level of PEF1 was significantly increased during appressorial maturation, which could be induced by appressorial fatty acids. Together with conidial germination, abundant fatty acids formed in a short time, and peroxisomal fission could also be activated under these conditions (Wilson and Talbot, 2009). However, how the peroxisomal fission process can be activated by PEF1 is still unclear. Future studies should focus on this intriguing process.
Fission, de novo formation and autophagy of the peroxisomes
The number of peroxisomes in a cell is regulated by several processes, including de novo formation, fission, fusion and pexophagy (Fagarasanu et al., 2007). De novo formation and fission processes contribute to increase the number of peroxisomes. However, the role of peroxisomal fission and endoplasmic reticulum (ER)‐dependent peroxisome formation (de novo formation) to the total number of peroxisomes remains unclear. It is possible that the peroxisome formation process from both the ER and fission of the pre‐existing organelles might occur in cells of most organisms. However, when abundant fatty acids must be utilized in a short time, the de novo formation process may not be sufficient of efficient enough, whereas the fission process can produce large numbers of organelles in a limited time. Therefore, the fission process may be more significant during fungal infection when lipid stores need to be utilized in a short time (less than 12 h) for functional appressorium formation. Pexophagy occurs when cells are released from fission‐inducing conditions, such as the presence of fatty acids. This autophagic process is also found to be important for fungal infection (Takano et al., 2009). In C. orbiculare, CoAtg26‐dependent pexophagy functions in the recycling of cellular components, including amino acids, inside the appressoria, which might promote protein synthesis required for host invasion (Asakura et al., 2009). Thus, peroxisomal fission and pexophagy comprise an in‐and‐out of peroxisome burst stage during the appressoria development.
In summary, we provide a hypothesis of the life cycle of peroxisomes during appressorium‐mediated infections (Fig. 8). In this model, lipid bodies are mobilized by the Pmk1‐mitogen‐activated protein kinase (Pmk1‐MAPK) and cyclic adenosine monophosphate/protein kinase A (cAMP/PKA) signalling pathways, and are decomposed by lipolysis. The resulting products include fatty acids and glycerol. The emergence of fatty acids can induce the Pef1‐mediated peroxisomal fission process, which leads to a rapid increase in peroxisome number. After peroxisome biogenesis and the import of peroxisomal matrix proteins, the mature peroxisomes can digest the fatty acids by β‐oxidation. When the fatty acids have been totally utilized, the peroxisomes are degraded by ATG26‐mediated pexophagy.
Figure 8.
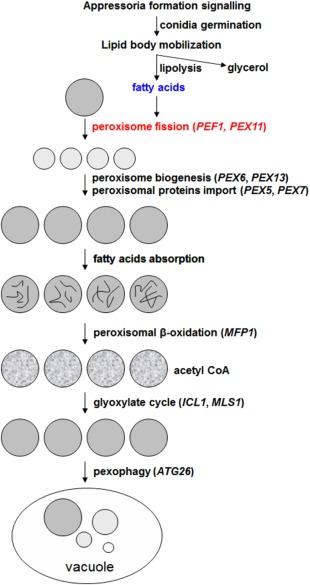
Working model for the role of peroxisomes in fungal infection. During infection, lipid bodies in fungal conidia are mobilized by the Pmk1‐mitogen‐activated protein kinase (Pmk1‐MAPK) signalling pathway and cyclic adenosine monophosphate/protein kinase A (cAMP/PKA) signalling pathway. After lipolysis, lipid bodies are degraded into fatty acids and glycerol. Fatty acids can induce the peroxisomal fission process, through which the peroxisomal number is rapidly increased. After peroxisome biogenesis and the peroxisomal protein import process, the newly formed peroxisomes become mature. Then, fatty acids can be degraded by peroxisomal β‐oxidation. The main product of fatty acid oxidation is acetyl‐coenzyme A (acetyl‐CoA), which can be used by the glyoxylate cycle for structural and nutrient material during host infection. When fatty acids are consumed, the peroxisomes are degraded by the pexophagy process.
Experimental Procedures
Strains and culture conditions
Two M. oryzae wild‐type strains were used. One was the field isolate P131 (Yang et al., 2010), which was used for diverse transformations and assays. The other was S1528 (Yang et al., 2010), which was used for crossing with mutants of P131. The wild‐type strains and transformants generated in this study (Table S4, see Supporting Information) were maintained on OTA plates at 28°C. For the extraction of genomic DNA, RNA, protein and isolation of protoplasts, mycelia were collected from liquid CM cultures (180 rpm) incubated at 28°C for 36 h. For the measurement of colony sizes, hyphal blocks (5 mm in diameter) were inoculated onto the centre of OTA plates and cultured for 120 h. Conidiation was assayed as described previously (Yang et al., 2010). Conidia harvested from 7‐day‐old OTA cultures were used for the assay of virulence, penetration and invasive growth, as described previously (Chen et al., 2014).
To investigate the utilization of carbon sources by the Δpef1 mutant, colony diameter was measured at 5 days after growth on MM agar plates amended with 1% glucose, 5 mm sodium acetate or ethanol, or 2.5 mm oleic acid, myristic acid or lauric acid as sole carbon source. The diameters of the colonies were recorded and used for the calculation of the growth reduction rates compared with the diameters of the wild‐type strain (Kong et al., 2012).
Molecular manipulations with DNA and RNA
Genomic DNA was extracted from mycelia using the cetyltrimethylammonium bromide (CTAB) protocol, as described previously (Xu and Hamer, 1996). Total RNA was extracted with the Trizol kit (Invitrogen, Carlsbad, CA, USA). Standard molecular manipulation procedures were followed for plasmid isolation, DNA gel blot analysis and enzymatic digestion with DNA (Sambrook and Russell, 2001). Probes used for DNA gel blots were labelled with the Random Primer Labelling Kit (Takara, Dalian, China). TAIL‐PCR was performed as described previously (Liu and Whittier, 1995).
Gene disruption and complementation of PEF1
For the PEF1 gene replacement construct, 1.5‐kb upstream and downstream flanking sequence segments were amplified from the genomic DNA of P131 with the primer pairs PEF1LF/PEF1LR and PEF1RF/PEF1RR, respectively (Table S6, see Supporting Information). The two flanking sequences were cloned into pKNH (Table S5, see Supporting Information) (Chen et al., 2014) as the deletion vector pKNH‐PEF1, which was linearized by NotI and transformed into P131 protoplasts with the polyethylene glycol (PEG)/CaCl2 approach (Chen et al., 2014). For the selection of hygromycin‐ or neomycin‐resistant transformants, CM plates were supplemented with 250 μg/mL hygromycin B (Roche, Indianapolis, IN, USA) or 400 μg/mL neomycin (Ameresco, Solon, OH, USA). For complementation assays, the PEF1 gene containing 1.5‐kb native promoter and 0.5‐kb terminator regions was amplified with the primer pairs PEF1CF/PEF1CR and cloned into pKN (Table S5, see Supporting Information) (Yang et al., 2010). The resulting construct pKN‐PEF1 was digested with NotI and transformed into the Δpef1 mutant. Neomycin‐resistant transformants were isolated and verified by PCR.
Yeast complementation
Saccharomyces cerevisiae strains were derivatives of BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0). The full‐length cDNA of PEF1 from M. oryzae was amplified and cloned into plasmid pYES2 (Invitrogen). The subsequent construct was transformed into Δcaf4, Δmdv1 and Δcaf4Δmdv1 mutants. Positive transformants were selected on synthetic complete medium without uracil (SC‐Ura). For complementation assays, the cells of the tested strains were diluted by a ten‐fold series and spotted onto SC‐Ura plates containing 2% glucose or 2% galactose with and without 20 ng/mL rapamycin. The cells were grown for 3 days at 30°C. The wild‐type strain BY4741, and Δcaf4, Δmdv1 and Δcaf4Δmdv1 mutants expressing empty pYES2 vector, were used as controls.
Assay for co‐localization
For co‐localization assay, the PEF1‐eGFP and RFP‐SKL fusion constructs were generated by overlapping PCR and cloned into pKN. The resulting constructs, pKN‐PEF1GFP and pPeroRFP, were digested with NotI and co‐transformed into the Δpef1 null mutant. Neomycin‐resistant transformants were obtained, and the growth complementary strains were used to generate strains with red fluorescent protein (RFP). Co‐localization of Pef1‐GFP and PeroRFP was analysed by confocal laser scanning microscopy (CLSM) (Olympus, Shinjuku, Tokyo, Japan). A similar approach was performed to co‐localize GFP‐PEF1 with Fis1‐RFP. In order to co‐localize GFP‐PEF1 with the mitochondria, the eGFP‐PEF1 fusion construct was transformed into the Δpef1 mutant, and the resulting transformants were stained with Mitotracker (Invitrogen); co‐localization of GFP‐PEF1 and mitochondria was analysed using CLSM.
Virulence test and observation of the infection process
To investigate the infection process, a conidial suspension of 1 × 105 conidia/mL in 0.025% Tween‐20 was spotted onto the lower epidermis of barley leaves and incubated in a moist dark chamber at 28°C. After inoculation, microscopy observations were performed at 2, 12, 20, 25 and 30 hpi. Rice seedlings (Oryza sativa cv. LTH) at the fifth leaf stage and 8‐day‐old barley seedlings (Hordeum vulgare cv. E9) were sprayed with suspensions of 5 × 104 conidia/mL of M. oryzae and incubated as described previously (Yang et al., 2010). Lesion formation was examined at 5 days post‐inoculation (dpi).
For the extraction of RNAs during different developmental stages, samples of the germ tubes and appressoria were harvested on a hydrophobic plastic surface at 3, 6 and 12 hpi with suspensions of 5 × 105 conidia/mL, whereas penetration pegs and IH were harvested by collecting the barley epidermis after inoculation by spraying (with suspensions of 5 × 105 conidia/mL) at 18, 24 and 42 hpi.
For the evaluation of the growth of IH in ROS‐suppressed barley cells, a 0.5‐µL conidial suspension supplemented with 0.5 μm DPI was placed on barley leaves (Chen et al., 2014). At different time points after inoculation (20 and 30 hpi), the lower epidermis of the inoculated leaves was removed and used to observe the growth of IH by microscopy. The infection process was observed by differential interference contrast microscopy imaging using a Nikon Ni90 epifluorescence microscope (Nikon, Tokyo, Japan).
Quantitative real‐time PCR analysis
In order to test the expression levels of PEF1 at different developmental stages and for different types of carbon source, total RNA was extracted using TRIzol (Invitrogen) from conidia inoculated onto a hydrophobic glass surface and incubated for different lengths of time, or from mycelia cultured for 2 days in liquid MM supplemented with different carbon sources. Quantitative real‐time PCR was performed on an ABI 7500 real‐time PCR system (Applied Biosystems, Foster City, CA, U.S.A.) according to the manufacturer's instructions.
Staining assays
CFW staining used Fluorescent Brightener 28 (10 μg/mL, Sigma‐Aldrich) for the microscopy of mycelial cells. Both the deletion mutants and the wild‐type were inoculated onto cover slips that contained a thin layer of agar medium, and cultured for 48 h. Agar pieces with hyphal tips were removed and stained with 10 μg/mL CFW for 10 min in the dark, rinsed twice with phosphate‐buffered saline (PBS) and viewed under a fluorescence microscope (Nikon).
Host‐derived ROS was observed by staining with DAB (Sigma‐Aldrich, St. Louis, MO, USA), as described previously (Chen et al., 2014). Barley leaves inoculated with the mutant and wild‐type strains at 30 hpi were incubated in 1 mg/mL DAB solution (pH 3.8) at room temperature for 8 h, de‐stained with a clearing solution (ethanol–acetic acid; 94 : 4, v/v) for 1 h, and observed with an epifluorescence microscope.
Tem
Conidia and 12‐h‐old appressoria formed on barley leaves were processed for TEM, with processing and imaging of the TEM samples performed as described previously (Soundararajan et al., 2004).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Virulence test of Agrobacterium tumefaciens‐mediated transformation (ATMT) mutant SX4690 and T‐DNA insertion site. (A) Rice seedlings were sprayed with conidial suspensions (5 × 104 conidia/mL) of P131 (wild‐type, WT) and SX4690. Typical leaves were photographed at 5 days post‐inoculation (dpi). (B) Schematic diagram of the integration site of T‐DNA in the ATMT mutant SX4690.
Fig. S2 Alignment of the amino acid sequences of Caf4 and Mdv1 from Saccharomyces cerevisiae and Pef1 from Magnaporthe oryzae and several selected filamentous fungi. Amino acid sequences were obtained by blast and aligned with ClustalW (http://www.ch.embnet.org/software/ClustalW.html). Identical and similar residues are indicated by red regions and red characters, respectively. Sequences aligned were the predicted products of S. cerevisiae CAF4 (CAA82110), MDV1 (DAA08688), M. oryzae PEF1 (EHA54899) and Pef1 orthologues from Neurospora crassa (XP_961974), Fusarium oxysporum (EWZ42274), Botrytis cinerea (CCD52776), Sclerotinia sclerotiorum (XP_001590828) and Aspergillus nidulans (AN6867). The five putative WD repeat regions were predicted by InterProscan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) and are indicated by blue rectangles.
Fig. S3 Gene disruption strategy and verification of disruption mutants. (A) Schematic diagram of the PEF1 deletion strategy. HPH indicates the hygromycin B phosphotransferase gene cassette. ‘P’ indicates PstI, which was used for genomic DNA digestion. The theoretical calculated sizes for Southern blot analysis of the deletion mutants and the wild‐type strain are indicated. (B) Products of polymerase chain reaction (PCR) replication of PEF1 in the wild‐type and PEF1 disruption strains. An actin encoding gene was used as a control. (C) Southern blot confirmation of the PEF1 deletion mutants. PstI‐digested genomic DNAs were hybridized with a 1.5‐kb 3′‐flanking fragment of PEF1. WT, the wild‐type strain P131; PEF1KO1 and PEF1KO2, the PEF1 deletion mutants. Compared with the wild‐type, both deletion mutants lacked a 5.3‐kb band, but showed a 6.9‐kb band in its place, as demonstrated by probing with the 1.5‐kb 3′‐flanking sequence of PEF1.
Fig. S4 Co‐localization of GFP‐PEF1 with mitochondria stained by Mitotracker during appressorial development. Dual colour imaging by confocal laser scanning microscopy (CLSM) of transformant PEF1DW2 expressing GFP‐PEF1, which was stained with the mitochondrial marker Mitotracker. Left, conidia (0 h). Middle, germ tube (2 h). Right, appressoria (12 h). Bars, 10 μm. DIC, differential interference contrast; GFP, green fluorescent protein.
Fig. S5 Co‐localization of GFP‐PEF1 with Fis1‐RFP during appressorial development. Dual colour imaging by confocal laser scanning microscopy (CLSM) of transformant PEF1DW3 expressing both GFP‐PEF1 and Fis1‐RFP. Left, conidia (0 h). Middle, germ tube (2 h). Right, appressoria (12 h). Bars, 10 μm. DIC, differential interference contrast; GFP, green fluorescent protein; RFP, red fluorescent protein.
Fig. S6 Cell wall integrity test. (A) Colony growth of the wild‐type P131, Δpef1 mutant and complementation strain cPEF1 on complete medium (CM) supplemented with the cell wall‐disturbing agents: 0.1 mg/mL calcofluor white (CFW), 0.2 mg/mL Congo red and 0.005% sodium dodecylsulfate (SDS). The cultures were incubated at 28°C for 4 days before being photographed. (B) Statistical analysis of the growth reduction rates of mycelia on CM supplemented with the three cell wall‐disturbing agents. Light grey bars, the wild type; dark grey bars, the Δpef1 mutant. Means and standard errors were calculated from three independent replicates. Significant differences are indicated by asterisks (P = 0.01).
Fig. S7 Expression pattern of the rice pathogenesis‐related (PR) genes. The expression of representative PR genes (OsPR1a, OsPR3, OsPBZ1) in rice inoculated with the wild‐type strain, the deletion mutant PEF1KO1 and the complemented strain cPEF1 was analysed using quantitative real‐time polymerase chain reaction at 48 h post‐inoculation (hpi). The gene expression levels were normalized by OsUBQ1. Asterisks indicate statistically significant differences vs. the control (Mock) (P = 0.01).
Fig. S8 Domain architecture of Pef1. Pef1 contains an NH2‐terminal domain, a central coiled‐coil domain and a COOH‐terminal WD40 repeat domain.
Table S1 Co‐segregation test of colony growth and hygromycin B sensitivity in F1 progeny.
Table S2 Phenotype characterization.
Table S3 Intracellular growth in barley epidermal cells.
Table S4 Fungal strains used in this study.
Table S5 Plasmids used in this study.
Table S6 Primers used in this study.
Acknowledgements
This work was performed in Professor You‐Liang Peng's lab, and we thank him for experiment design and critical reading of the manuscript. We thank Professor Tom Hsiang from the University of Guelph, Guelph, ON, Canada for critical reading of the manuscript. The yeast null mutant srains were kindly provided by Professor Ewald H. Hettema from the University of Sheffield, Sheffield, UK. This work was supported by the 973 Project (2012CB114000) from the Ministry of Sciences and Technology, China.
References
- Asakura, M. , Ninomiya, S. , Sugimoto, M. , Oku, M. , Yamashita, S. , Okuno, T. , Sakai, Y. and Takano, Y. (2009) Atg26‐mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell, 21, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhambra, G.K. , Wang, Z.Y. , Soanes, D.M. , Wakley, G.E. and Talbot, N.J. (2006) Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea . Mol. Microbiol. 61, 46–60. [DOI] [PubMed] [Google Scholar]
- Chen, X.L. , Yang, J. and Peng, Y.L. (2011) Large‐scale insertional mutagenesis in Magnaporthe oryzae by Agrobacterium tumefaciens‐mediated transformation. Methods Mol. Biol. 722, 213–224. [DOI] [PubMed] [Google Scholar]
- Chen, X.L. , Shi, T. , Yang, J. , Shi, W. , Gao, X. , Chen, D. , Xu, X. , Xu, J.R. , Talbot, N.J. and Peng, Y.L. (2014). N‐glycosylation of effector proteins by an alpha‐1,3‐mannosyltransferase is required for the rice blast fungus to evade host innate immunity. Plant Cell, 26, 1360–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley, F.G. and Valent, B. (1990) Genetic‐analysis of melanin‐deficient, nonpathogenic mutants of Magnaporthe grisea . Mol. Plant–Microbe Interact. 3, 135–143. [Google Scholar]
- Egan, M.J. , Wang, Z.Y. , Jones, M.A. , Smirnoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA, 104, 11 772–11 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu, A. , Fagarasanu, M. and Rachubinski, R.A. (2007) Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23, 321–344. [DOI] [PubMed] [Google Scholar]
- Fujihara, N. , Sakaguchi, A. , Tanaka, S. , Fujii, S. , Tsuji, G. , Shiraishi, T. , O'Connell, R. and Kubo, Y. (2010) Peroxisome biogenesis factor PEX13 is required for appressorium‐mediated plant infection by the anthracnose fungus Colletotrichum orbiculare . Mol. Plant–Microbe Interact. 23, 436–445. [DOI] [PubMed] [Google Scholar]
- Fujii, I. , Mori, Y. , Watanabe, A. , Kubo, Y. , Tsuji, G. and Ebizuka, Y. (2000) Enzymatic synthesis of 1,3,6,8‐tetrahydroxynaphthalene solely from malonyl coenzyme A by a fungal iterative type I polyketide synthase PKS1. Biochemistry, 39, 8853–8858. [DOI] [PubMed] [Google Scholar]
- Gandre‐Babbe, S. and van der Bliek, A.M. (2008) The novel tail‐anchored membrane protein Mff controls mitochondrial and Peroxisomal Fission in mammalian cells. Mol. Biol. Cell, 19, 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev, T.S. and Hille, J. (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J. Cell Biol. 168, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, J. , Jeon, J. , Kim, K.S. , Park, J. , Park, S.Y. and Lee, Y.H. (2011) The PEX7‐mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae . PLoS One, 6, e28220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, E.E. , Graumann, J. and Chan, D.C. (2005) The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Q. , Koirala, S. , Perkins, E.M. , McCaffery, J.M. and Shaw, J.M. (2012) The mitochondrial fission adaptors Caf4 and Mdv1 are not functionally equivalent. PLoS One, 7, e53523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltunen, J.K. , Mursula, A.M. , Rottensteiner, H. , Wierenga, R.K. , Kastaniotis, A.J. and Gurvitz, A. (2003) The biochemistry of peroxisomal beta‐oxidation in the yeast Saccharomyces cerevisiae . FEMS Microbiol. Rev. 27, 35–64. [DOI] [PubMed] [Google Scholar]
- Hoppins, S. , Lackner, L. and Nunnari, J. (2007) The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780. [DOI] [PubMed] [Google Scholar]
- Howard, R.J. and Valent, B. (1996) Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea . Annu. Rev. Microbiol. 50, 491–512. [DOI] [PubMed] [Google Scholar]
- Howard, R.J. , Ferrari, M.A. , Roach, D.H. and Money, N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA, 88, 11 281–11 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigar, J.A.D and Donofrio, N.M. (2011) HYR1‐mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7, e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006). The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- de Jong, J.C. , McCormack, B.J. , Smirnoff, N. and Talbot, N.J. (1997). Glycerol generates turgor in rice blast. Nature, 389, 244–245. [Google Scholar]
- Kaur, N. and Hu, J. (2009) Dynamics of peroxisome abundance: a tale of division and proliferation. Curr. Opin. Plant Biol. 12, 781–788. [DOI] [PubMed] [Google Scholar]
- Khan, I.A. , Ning, G. , Liu, X. , Feng, X. , Lin, F. and Lu, J. (2015) Mitochondrial fission protein MoFis1 mediates conidiation and is required for full virulence of the rice blast fungus Magnaporthe oryzae . Microbiol. Res. 178, 51–58. [DOI] [PubMed] [Google Scholar]
- Kimura, A. , Takano, Y. , Furusawa, I. and Okuno, T. (2001) Peroxisomal metabolic function is required for appressorium‐mediated plant infection by Colletotrichum lagenarium . Plant Cell, 13, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Thiemann, M. , Grabenbauer, M. , Yoon, Y. , McNiven, M.A. and Schrader, M. (2003) Dynamin‐like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. [DOI] [PubMed] [Google Scholar]
- Koch, A. , Yoon, Y. , Bonekamp, N.A. , McNiven, M.A. and Schrader, M. (2005) A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell, 16, 5077–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L.A. , Yang, J. , Li, G.T. , Qi, L.L. , Zhang, Y.J. , Wang, C.F. , Zhao, W.S. , Xu, J.R. and Peng, Y.L. (2012). Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae . PLoS Pathog. 8, e1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, M. , Pracharoenwattana, I. , Smith, S.M. and Hartig, A. (2006) A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim. Biophys. Acta, 1763, 1441–1452. [DOI] [PubMed] [Google Scholar]
- Kuravi, K. , Nagotu, S. , Krikken, A.M. , Sjollema, K. , Deckers, M. , Erdmann, R. , Veenhuis, M. and van der Klei, I.J. (2006) Dynamin‐related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae . J. Cell Sci. 119, 3994–4001. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G. and Whittier, R.F. (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Molina, L. and Kahmann, R. (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell, 19, 2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley, A.M. , Ward, G.P. and Hettema, E.H. (2008) Dnm1p‐dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J. Cell Sci. 121, 1633–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Poirier, Y. , Antonenkov, V.D. , Glumoff, T. and Hiltunen, J.K. (2006) Peroxisomal beta‐oxidation—a metabolic pathway with multiple functions. Biochim. Biophys. Acta, 1763, 1413–1426. [DOI] [PubMed] [Google Scholar]
- Ramos‐Pamplona, M. and Naqvi, N.I. (2006) Host invasion during rice‐blast disease requires carnitine‐dependent transport of peroxisomal acetyl‐CoA. Mol. Microbiol. 61, 61–75. [DOI] [PubMed] [Google Scholar]
- Sakai, Y. , Oku, M. , van der Klei, I.J. and Kiel, J.A. (2006) Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta, 1763, 1767–1775. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Saraya, R. , Veenhuis, M. and van der Klei, I.J. (2010) Peroxisomes as dynamic organelles: peroxisome abundance in yeast. FEBS J. 277, 3279–3288. [DOI] [PubMed] [Google Scholar]
- Schrader, M. (2006) Shared components of mitochondrial and peroxisomal division. Biochim. Biophys. Acta, 1763, 531–541. [DOI] [PubMed] [Google Scholar]
- Schrader, M. and Fahimi, H.D. (2006) Peroxisomes and oxidative stress. Biochim. Biophys. Acta, 1763, 1755–1766. [DOI] [PubMed] [Google Scholar]
- Smith, J.J. and Aitchison, J.D. (2013) Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, P.S. , Lee, R.C. , Wilson, T.J. and Oliver, R.P. (2004) Pathogenicity of Stagonospora nodorum requires malate synthase. Mol. Microbiol. 53, 1065–1073. [DOI] [PubMed] [Google Scholar]
- Soundararajan, S. , Jedd, G. , Li, X. , Ramos‐Pamplona, M. , Chua, N.H. and Naqvi, N.I. (2004) Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell, 16, 1564–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, Y. , Asakura, M. and Sakai, Y. (2009) Atg26‐mediated pexophagy and fungal phytopathogenicity. Autophagy, 5, 1041–1042. [DOI] [PubMed] [Google Scholar]
- Tam, Y.Y. , Torres‐Guzman, J.C. , Vizeacoumar, F.J. , Smith, J.J. , Marelli, M. , Aitchison, J.D. and Rachubinski, R.A. (2003) Pex11‐related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae . Mol. Biol. Cell, 14, 4089–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, E. , Weber, R.W. and Talbot, N.J. (2000) MAP kinase and protein kinase A‐dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea . Plant Cell, 12, 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms, S. and Erdmann, R. (2005) Dynamin‐related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272, 5169–5181. [DOI] [PubMed] [Google Scholar]
- Tieu, Q. and Nunnari, J. (2000) Mdv1p is a WD repeat protein that interacts with the dynamin‐related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I. and Rachubinski, R.A. (2001) The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2, 357–368. [DOI] [PubMed] [Google Scholar]
- Torres, M.A. and Dangl, J.L. (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhang, Z. , Wang, Y. , Li, L. , Chai, R. , Mao, X. , Jiang, H. , Qiu, H. , Du, X. , Lin, F.C. and Sun, G. (2013) PTS1 peroxisomal import pathway plays shared and distinct roles to PTS2 pathway in development and pathogenicity of Magnaporthe oryzae . PLoS One, 8, e55554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.Y. , Li, L. , Zhang, Z. , Qiu, H.P. , Li, D.M. , Fang, Y. , Jiang, H. , Chai, R.Y. , Mao, X.Q. , Wang, Y.L. and Sun, G.C. (2015) One of three Pex11 family members is required for peroxisomal proliferation and full virulence of the rice blast fungus Magnaporthe oryzae . PLoS One, 10, e0134249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y. , Thornton, C.R. , Kershaw, M.J. , Li, D.B. and Talbot, N.J. (2003) The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea . Mol. Microbiol. 47, 1601–1612. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y. , Soanes, D.M. , Kershaw, M.J. and Talbot, N.J. (2007) Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta‐oxidation during appressorium‐mediated plant infection. Mol. Plant–Microbe Interact. 20, 475–491. [DOI] [PubMed] [Google Scholar]
- Wilson, R.A. and Talbot, N.J. (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae . Nat. Rev. Microbiol. 7, 185–195. [DOI] [PubMed] [Google Scholar]
- Xu, J.R. and Hamer, J.E. (1996) MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea . Genes Dev. 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Zhao, X. , Sun, J. , Kang, Z. , Ding, S. , Xu, J.R. and Peng, Y.L. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 112–123. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Kong, L.A. , Chen, X.L. , Wang, D.W. , Qi, L.L. , Zhao, W.S. , Zhang, Y. , Liu, X. and Peng, Y.L. (2012) A carnitine‐acylcarnitine carrier protein, MoCrc1, is essential for pathogenicity in Magnaporthe oryzae . Curr. Genet. 58, 139–148. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Chan, N.C. , Ngo, H.B. , Gristick, H. and Chan, D.C. (2012) Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (Mdv1) coiled coil. J. Biol. Chem. 287, 9855–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Virulence test of Agrobacterium tumefaciens‐mediated transformation (ATMT) mutant SX4690 and T‐DNA insertion site. (A) Rice seedlings were sprayed with conidial suspensions (5 × 104 conidia/mL) of P131 (wild‐type, WT) and SX4690. Typical leaves were photographed at 5 days post‐inoculation (dpi). (B) Schematic diagram of the integration site of T‐DNA in the ATMT mutant SX4690.
Fig. S2 Alignment of the amino acid sequences of Caf4 and Mdv1 from Saccharomyces cerevisiae and Pef1 from Magnaporthe oryzae and several selected filamentous fungi. Amino acid sequences were obtained by blast and aligned with ClustalW (http://www.ch.embnet.org/software/ClustalW.html). Identical and similar residues are indicated by red regions and red characters, respectively. Sequences aligned were the predicted products of S. cerevisiae CAF4 (CAA82110), MDV1 (DAA08688), M. oryzae PEF1 (EHA54899) and Pef1 orthologues from Neurospora crassa (XP_961974), Fusarium oxysporum (EWZ42274), Botrytis cinerea (CCD52776), Sclerotinia sclerotiorum (XP_001590828) and Aspergillus nidulans (AN6867). The five putative WD repeat regions were predicted by InterProscan (http://www.ebi.ac.uk/Tools/pfa/iprscan/) and are indicated by blue rectangles.
Fig. S3 Gene disruption strategy and verification of disruption mutants. (A) Schematic diagram of the PEF1 deletion strategy. HPH indicates the hygromycin B phosphotransferase gene cassette. ‘P’ indicates PstI, which was used for genomic DNA digestion. The theoretical calculated sizes for Southern blot analysis of the deletion mutants and the wild‐type strain are indicated. (B) Products of polymerase chain reaction (PCR) replication of PEF1 in the wild‐type and PEF1 disruption strains. An actin encoding gene was used as a control. (C) Southern blot confirmation of the PEF1 deletion mutants. PstI‐digested genomic DNAs were hybridized with a 1.5‐kb 3′‐flanking fragment of PEF1. WT, the wild‐type strain P131; PEF1KO1 and PEF1KO2, the PEF1 deletion mutants. Compared with the wild‐type, both deletion mutants lacked a 5.3‐kb band, but showed a 6.9‐kb band in its place, as demonstrated by probing with the 1.5‐kb 3′‐flanking sequence of PEF1.
Fig. S4 Co‐localization of GFP‐PEF1 with mitochondria stained by Mitotracker during appressorial development. Dual colour imaging by confocal laser scanning microscopy (CLSM) of transformant PEF1DW2 expressing GFP‐PEF1, which was stained with the mitochondrial marker Mitotracker. Left, conidia (0 h). Middle, germ tube (2 h). Right, appressoria (12 h). Bars, 10 μm. DIC, differential interference contrast; GFP, green fluorescent protein.
Fig. S5 Co‐localization of GFP‐PEF1 with Fis1‐RFP during appressorial development. Dual colour imaging by confocal laser scanning microscopy (CLSM) of transformant PEF1DW3 expressing both GFP‐PEF1 and Fis1‐RFP. Left, conidia (0 h). Middle, germ tube (2 h). Right, appressoria (12 h). Bars, 10 μm. DIC, differential interference contrast; GFP, green fluorescent protein; RFP, red fluorescent protein.
Fig. S6 Cell wall integrity test. (A) Colony growth of the wild‐type P131, Δpef1 mutant and complementation strain cPEF1 on complete medium (CM) supplemented with the cell wall‐disturbing agents: 0.1 mg/mL calcofluor white (CFW), 0.2 mg/mL Congo red and 0.005% sodium dodecylsulfate (SDS). The cultures were incubated at 28°C for 4 days before being photographed. (B) Statistical analysis of the growth reduction rates of mycelia on CM supplemented with the three cell wall‐disturbing agents. Light grey bars, the wild type; dark grey bars, the Δpef1 mutant. Means and standard errors were calculated from three independent replicates. Significant differences are indicated by asterisks (P = 0.01).
Fig. S7 Expression pattern of the rice pathogenesis‐related (PR) genes. The expression of representative PR genes (OsPR1a, OsPR3, OsPBZ1) in rice inoculated with the wild‐type strain, the deletion mutant PEF1KO1 and the complemented strain cPEF1 was analysed using quantitative real‐time polymerase chain reaction at 48 h post‐inoculation (hpi). The gene expression levels were normalized by OsUBQ1. Asterisks indicate statistically significant differences vs. the control (Mock) (P = 0.01).
Fig. S8 Domain architecture of Pef1. Pef1 contains an NH2‐terminal domain, a central coiled‐coil domain and a COOH‐terminal WD40 repeat domain.
Table S1 Co‐segregation test of colony growth and hygromycin B sensitivity in F1 progeny.
Table S2 Phenotype characterization.
Table S3 Intracellular growth in barley epidermal cells.
Table S4 Fungal strains used in this study.
Table S5 Plasmids used in this study.
Table S6 Primers used in this study.


