Abstract
Summary
Pantoea ananatis, a bacterium that is well known for its phytopathogenic characteristics, has been isolated from a myriad of ecological niches and hosts. Infection of agronomic crops, such as maize and rice, can result in substantial economic losses. In the last few years, much of the research performed on P. ananatis has been based on the sequencing and analysis of the genomes of strains isolated from different environments and with different lifestyles. In this review, we summarize the advances made in terms of pathogenicity determinants of phytopathogenic strains of P. ananatis and how this bacterium is able to adapt and survive in such a wide variety of habitats. The diversity and adaptability of P. ananatis can largely be attributed to the plasticity of its genome and the integration of mobile genetic elements on both the chromosome and plasmid. Furthermore, we discuss the recent interest in this species in various biotechnological applications.
Taxonomy
Domain Bacteria; Class Gammaproteobacteria; Family Enterobacteriaceae; genus Pantoea; species ananatis.
Disease symptoms
Pantoea ananatis causes disease on a wide range of plants, and symptoms can range from dieback and stunted growth in Eucalyptus seedlings to chlorosis and bulb rotting in onions.
Disease control
Currently, the only methods of control of P. ananatis on most plant hosts are the use of resistant clones and cultivars or the eradication of infected plant material. The use of lytic bacteriophages on certain host plants, such as rice, has also achieved a measure of success.
Keywords: biotechnology, genomics, pan‐genome, pathogenicity, quorum sensing, type VI secretion system
Introduction
Pantoea ananatis is a species of Gram‐negative, rod‐shaped, aerobic or facultatively anaerobic, yellow‐pigmented bacteria belonging to the class Gammaproteobacteria and the family Enterobacteriaceae (Coutinho and Venter, 2009). Initially, it formed part of the Erwinia herbicola–Enterobacter agglomerans complex and was assigned to the genus Pantoea when it was established in 1989 (Gavini et al., 1989). Pantoea uredovora, a pathogen of Puccinia graminis, was shown to have a high level of genomic relatedness to P. ananatis, and the two species were synonymized (Mergaert et al., 1993).
Pantoea ananatis has a cosmopolitan distribution and has been found right across the globe, from South Africa in the south (Coutinho et al., 2002; Goszczynska et al., 2007; Weller‐Stuart et al., 2014) to Russia in the north (Egorova et al., 2015), Mexico in the west (Perez‐y‐Terron et al., 2009) and Australia in the east (Murrell et al., 2003). In these areas, this bacterium has an exceptional lifestyle, as it is not only associated with plants, but is also frequently isolated from a wide range of environmental sources. As a plant pathogen, it causes severe losses of many agronomic crop and tree species, such as maize (Goszczynska et al., 2007), rice (Watanabe et al., 1996), onion (Gitaitis et al., 2002) and Eucalyptus (Coutinho et al., 2002). Pantoea ananatis is also frequently isolated from a wide range of plant hosts as an epiphyte or endophyte, where it exists as a commensal without causing any disease symptoms (Coutinho and Venter, 2009). In addition to its association with plants, P. ananatis has been found to be associated with numerous insects (Dutta et al., 2016; Gitaitis et al., 2003; Murrell et al., 2003; Watanabe et al., 1996; Wells et al., 2002), as well as humans (De Baere et al., 2004), where it is capable of causing bacteraemia. As a saprophyte, it has been isolated from a plethora of different environments, ranging from soil and freshwater to aviation fuel tanks (Gasser et al., 2012; Pileggi et al., 2012; Rauch et al., 2006).
Given the ubiquitous nature of P. ananatis and its significance as an agronomic phytopathogen with potential clinical relevance, it has been well represented in the genome sequencing era, with a number of plant‐pathogenic, plant growth‐promoting and environmental isolates being sequenced in the last decade. These genome sequences and comparative genomic analyses of the sequenced genomes with one another and with those of other well‐characterized species provide a primer to unravel some of the enigmas of this species, including how it causes disease on such a wide range of plant hosts, how it can spread between hosts and what enables this bacterium to occupy such a diverse range of ecological niches.
Previously, we have reviewed some of the key concepts regarding the taxonomic background, microbiological properties, disease symptoms, control and epidemiology of P. ananatis (Coutinho and Venter, 2009). Furthermore, we have highlighted some of the gaps in our knowledge on this emerging phytopathogen, including the pathogenicity and host range determinants, as well as the ability to adapt and survive in a wide range of habitats (Coutinho and Venter, 2009). In this review, we examine the information that has been revealed by the sequencing of the genomes of P. ananatis strains isolated from numerous habitats. Key areas of the lifestyle of this important phytopathogen, including ecological diversification, pathogenicity and biotechnological applications, are discussed. We found that the plasticity of the P. ananatis genome allows it to be an adaptable, multi‐faceted bacterium that not only survives, but thrives, in numerous ecological niches and lifestyles.
Pantoea Ananatis in The Genome Sequencing era
To date, the genomes of 22 P. ananatis strains have been sequenced (Table 1). The main focus of these genome sequencing projects has been on phytopathogenic isolates, including strains from pineapple, rice, Eucalyptus, cotton, maize and onion (Adam et al., 2014; Choi et al., 2012; De Maayer et al., 2010; Medrano and Bell, 2015; Weller‐Stuart et al., 2014). However, a variety of endophytic isolates (Midha et al., 2016; Sheibani‐Tezerji et al., 2015), plant growth‐promoting strains, isolates with potential biotechnological and biological control applications (Gasser et al., 2012; Gkorezis et al., 2016; Hara et al., 2012; Kim et al., 2012; Megías et al., 2016; Shi et al., 2015; Smith et al., 2013; Wu et al., 2016) and a clinical isolate (De Maayer et al., 2012b) have also been sequenced.
Table 1.
Currently sequenced Pantoea ananatis genomes.
| Strain | Assembly | Genome size (Mb) | G + C (%) | Status | No. CDSs | Interest | Source | Reference |
|---|---|---|---|---|---|---|---|---|
| P. ananatis AJ13355 | GCA_000270125.2 | 4.88 | 53.7 | Complete | 4223 | Biotechnological | Soil | Hara et al. ( 2012) |
| P. ananatis AMG521 | GCA_001465955.1 | 4.88 | 53.1 | Draft | 4315 | Plant growth promoter | Rice paddy | Megías et al. (2016) |
| P. ananatis B1‐9 | GCA_000285975.1 | 5.12 | 53.5 | Draft | 4566 | Plant growth promoter | Rhizosphere of onion root | Kim et al. ( 2012) |
| P. ananatis BD442 | GCA_000709995.1 | 4.8 | 53.6 | Draft | 4330 | Plant pathogen | Onion seed | Weller‐Stuart et al. ( 2014) |
| P. ananatis BRT175 | GCA_000475035.1 | 4.85 | 53.7 | Draft | 4336 | Antibiosis | Strawberry | Smith et al. ( 2013) |
| P. ananatis CFH 7‐1 | GCA_001187705.1 | 4.6 | 53.1 | Draft | 4094 | Plant pathogen | Cotton boll | Medrano and Bell ( 2015) |
| P. ananatis DAR 76143 | GCA_000467085.1 | 5.25 | 53.4 | Draft | * | Plant pathogen | Rice | * |
| P. ananatis LMG 20103 | GCA_000025405.2 | 4.7 | 53.7 | Complete | 4076 | Plant pathogen | Eucalyptus | De Maayer et al. ( 2010) |
| P. ananatis LMG 2665T | GCA_000661975.1 | 4.94 | 53.4 | Draft | 4403 | Plant pathogen | Pineapple fruitlet | Adam et al. ( 2014) |
| P. ananatis LMG 5342 | GCA_000283875.1 | 4.91 | 53.3 | Complete | 4345 | Clinical isolate | Human wound | De Maayer et al. ( 2012a) |
| P. ananatis NS296 | GCA_001475715.1 | 4.73 | 53.5 | Draft | 4272 | Endophyte | Rice seed | Midha et al. ( 2016) |
| P. ananatis NS303 | GCA_001476115.1 | 4.73 | 53.5 | Draft | 4266 | Endophyte | Rice seed | Midha et al. ( 2016) |
| P. ananatis NS311 | GCA_001475725.1 | 4.72 | 53.5 | Draft | 4227 | Endophyte | Rice seed | Midha et al. ( 2016) |
| P. ananatis PA13 | GCA_000233595.1 | 4.87 | 53.6 | Complete | 4403 | Plant pathogen | Rice grain | Choi et al. ( 2012) |
| P. ananatis PA4 | GCA_000710015.1 | 5.16 | 53.6 | Draft | 4698 | Plant pathogen | Maize | Weller‐Stuart et al. ( 2014) |
| P. ananatis PaMB1 | GCA_000766045.1 | 4.76 | 53.8 | Draft | 4223 | Plant pathogen | * | * |
| P. ananatis R100 | GCA_001543055.1 | 4.86 | 53.6 | Complete | 4289 | Antibiosis | Rice seed | Wu et al. ( 2016) |
| P. ananatis RSA47 | GCA_001475885.1 | 4.74 | 53.5 | Draft | 4268 | Endophyte | Rice seed | Midha et al. ( 2016) |
| P. ananatis S6 | GCA_001369355.1 | 4.34 | 54.1 | Draft | 4375 | Endophyte | Maize seed | Sheibani‐Tezerji et al. ( 2015) |
| P. ananatis S7 | GCA_001369375.1 | 4.49 | 54.0 | Draft | 4516 | Endophyte | Maize seed | Sheibani‐Tezerji et al. ( 2015) |
| P. ananatis S8 | GCA_001369395.1 | 4.48 | 54.1 | Draft | 4528 | Endophyte | Maize seed | Sheibani‐Tezerji et al. ( 2015) |
| P. ananatis Sd‐1 | GCA_000582575.1 | 4.93 | 53.3 | Draft | 4332 | Biotechnological | Rice seed | Ma et al. ( 2016) |
The strain, isolation source and reason for sequencing are indicated. The sizes of the genomes, average G + C content (%), number of proteins (CDSs) encoded on the genome and sequencing status are shown, as per the National Center for Biotechnology Information Genome Assembly and Annotation report (www.ncbi.nlm.nih.gov/genome/genomes/2606).
*Data unavailable.
The genomes of P. ananatis strains are, on average, 4.81 megabases (Mb) in size and range between 4.34 Mb (P. ananatis S6) and 5.25 Mb (P. ananatis DAR 76143). Between 4026 (P. ananatis LMG 20103) and 4698 (P. ananatis PA4) proteins are encoded on the genomes. The genome of all strains incorporates a single large chromosome and a universal large plasmid, LPP‐1, which ranges in size between 280.8 and 352.8 kilobases (kb) and codes for between 238 and 320 proteins (Choi et al., 2012; De Maayer et al., 2012a; Weller‐Stuart et al., 2014). The presence of additional plasmids cannot be excluded (Ismail et al., 2014), particularly in the strains with larger genomes, as the majority of the genomes are draft sequences. The G + C contents of the genomes are relatively similar, with an average G + C content of 53.6%.
Ecological Diversification Can Be Linked to The Extensive Genome Plasticity of P. Ananatis
The global distribution of P. ananatis strains, the wide range of sources from whence they have been isolated, the different associations with their diverse hosts as pathogens and saprophytes, and their variable metabolic and biological capacities are testament of the extreme versatility of this species. This phenotypic diversification can be linked to the extensive genomic plasticity observed within the species. A pan‐genome analysis of eight P. ananatis strains has shown that, although there is a large core genome (3876 protein coding sequences) conserved among the eight strains, there is also a sizeable accessory genome (1690 protein coding sequences). The sequencing of an additional strain would add 106 unique protein coding sequences to the open pan‐genome of the species (De Maayer et al., 2014). Although the majority of the accessory genome is derived from integrated prophages, a number of the accessory proteins are specific to bacteria that colonize animal, plant or insect hosts and include proteins involved in carbohydrate and amino acid metabolism, adherence to host tissues, protection against plant and animal defence systems and putative pathogenicity determinants (De Maayer et al., 2014). The presence of these accessory proteins hints towards the specialization of particular strains to their different ecological niches, hosts and, potentially, lifestyles.
The role of accessory proteins in determining the lifestyle of a bacterium was demonstrated in a study comparing the genomes of three P. ananatis strains isolated from maize seeds. These strains showed 85%–87% similarity in their core genomes. The strains selected were pathogenic, commensal and beneficial to the host, respectively (Sheibani‐Tezerji et al., 2015). These phenotypic differences were attributed to the integration of mobile genetic elements, such as phages and transposases, as well as differences in the type VI secretion systems (T6SSs) and type IV pili (Sheibani‐Tezerji et al., 2015). Eukaryotic‐like protein domains were identified in these strains and included enzymes that increased mutation frequency, allowing bacteria to adapt to changing environmental conditions. Other eukaryotic‐like protein domains included proteins that play a role in adhesion and virulence (Sheibani‐Tezerji et al., 2015).
A major contributing factor in the adaptability of P. ananatis strains may be the presence of the universal plasmid (LPP‐1) which is derived from a plasmid that is ancestral to the genus Pantoea (De Maayer et al., 2012a). Within both the genus and species, LPP‐1 has undergone extensive genetic diversification with the insertion of several phage and mobile genetic elements (De Maayer et al., 2012a). Many of the genes encoded on LPP‐1 play a role in determining the phenotype of this bacterium. These various phenotypes include metabolic diversity, iron and nitrogen assimilation, resistance to selected antibiotics and heavy metals, and host–microbe interactions (De Maayer et al., 2012a).
Another key factor in the diversification of P. ananatis is the presence of an Integrative and Conjugative Element (ICEPan), an integrating and excising mobile genetic element, which was observed in a subset of strains [five of eight genome sequenced strains and 24 of 46 strains as determined by polymerase chain reaction (PCR) amplification of conserved ICEPan genes] isolated from a wide variety of hosts and geographical origins (De Maayer et al., 2015). In silico analyses showed that ICEPan encodes various proteins that may play a role in antibiosis and the stress response, which probably enables P. ananatis to occupy diverse environmental niches (De Maayer et al., 2015). For example, orthologues of alternative RNA polymerase σ factors that play a role in overcoming stress factors, such as starvation, heat and cold shock, pH and oxidative stress and DNA damage, are present on ICEPan (De Maayer et al., 2015). A putative novel antibiotic and bacteriocins encoded on ICEPan may also confer a competitive advantage over other organisms occupying the same environmental niche (De Maayer et al., 2015).
New Insights Into The Pathogenicity of P. Ananatis
Early genomic evidence revealed that many of the typical pathogenicity determinants found in well‐known plant pathogens, such as Pseudomonas, Xanthomonas, Ralstonia and the more closely related enterobacterial phytopathogens Erwinia amylovora and Pectobacterium spp., such as the type II secretion system, Hrp type III secretion system and phytotoxins, such as coronatine, are absent from the genomes of P. ananatis strains (Coutinho and Venter, 2009; De Maayer et al., 2014). However, orthologues of other predicted pathogenicity determinants (Fig. 1), such as flagella and fimbriae biosynthetic proteins, non‐fimbrial adhesins, amylovoran/stewartan‐like exopolysaccharide and potential cell wall‐degrading enzymes (CWDEs), are encoded on the genomes of many P. ananatis strains (De Maayer et al., 2014; Ma et al., 2016; Miller et al., 2016). Recent genetic and mutagenic analyses have also contributed to our understanding of the mechanisms underlying P. ananatis phytopathogenesis. For example, P. ananatis strains encode up to three T6SSs, which may play a role in pathogenesis in both plant and animal hosts (De Maayer et al., 2011; Shyntum et al., 2014). The first and second T6SSs appear to be universal among P. ananatis strains and are hypothesized to play a role in antibiosis, fitness and niche adaptation (Shyntum et al., 2014). It is likely that the second T6SS locus, which is incomplete, arose through the duplication of the first locus, whereas the third locus, which is plasmid borne, was acquired through horizontal gene transfer (De Maayer et al., 2011). In a study conducted by Shyntum et al. (2015), it was shown that the first locus is fully functional and plays a role in pathogenicity in plant hosts, specifically onion seedlings, and is also an important factor in intra‐ and interspecies bacterial competition.
Figure 1.
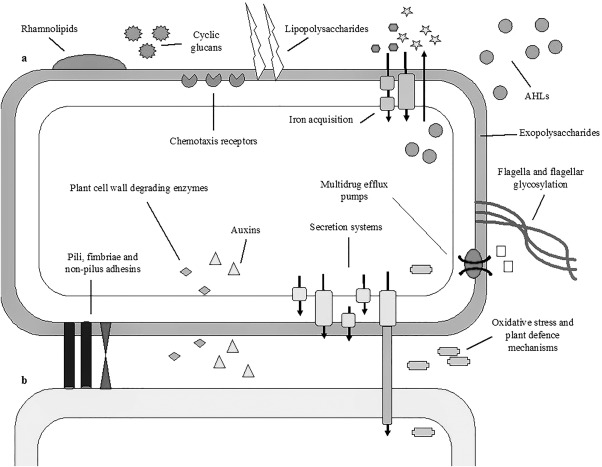
Putative plant‐pathogenicity factors of Pantoea ananatis. The pathogenicity determinants of P. ananatis (a) when infecting a host plant cell (b). The secretion systems include Tat and Sec secretion systems, as well as the type I secretion system (T1SS), T5SS and T6SS. AHLs, acyl‐homoserine lactones.
Motility plays a crucial role in the infection process of many well‐characterized plant pathogens, such as Ralstonia solanacearum, Pseudomonas syringae and Xanthomonas oryzae pv. oryzae (Kang et al., 2002; Meng et al., 2011; Shen and Ronald, 2002), and it has been shown to play a similar role in P. ananatis (Weller‐Stuart et al., 2016). Research on P. ananatis–onion seedling interactions demonstrated the need for functional flagella in initial attachment and pathogenicity. Both the structure and function of the flagella were disrupted in separate, site‐specific mutations, as the actual flagellar filament may play a role in adhesion, whereas swimming motility allows the bacterial cell to overcome the natural repulsive forces that exist between itself and a surface, and also allows the cell to respond to chemotactic signals in the environment (O'Toole and Kolter, 1998; Ramos et al., 2004). The flagellar filament has also been shown to be subject to flagellar glycosylation, which may play a role in the avoidance of host recognition during infection, host specificity, attachment and virulence (De Maayer and Cowan, 2016; Logan, 2006). Flagellar glycosylation occurs when N‐ and O‐linked carbohydrates are added to flagellin, the repeating major subunit of the filament, to change both its antigenic properties and function (Takeuchi et al., 2003). Twitching motility also plays a pivotal role in allowing P. ananatis to spread and colonize the surface of the leaf once attachment has occurred (Weller‐Stuart et al., 2016). Deletion mutations in the type IV pilin major subunit, as well as the ATPase responsible for retracting type IV pili during twitching motility, demonstrated that, although the role of type IV pili in attachment may be complemented by other pili, such as type I and type III pili, twitching motility is instrumental in the spread of P. ananatis across the surface of a leaf (Weller‐Stuart et al., 2016). Motility is thus crucial to the infection process in P. ananatis.
Plant CWDEs break down one of the primary defences of the plant cell, namely the cell wall. In order to cause disease, bacteria commonly need to breach this barrier and gain entry into the plant cell (Esquerre‐Tugaye et al., 2000). All analysed P. ananatis strains have several CWDEs, including a putative xylanase, pectin acetylesterase, cellulase and two polygalacturonases (De Maayer et al., 2014). The lignocellulose‐degrading strain P. ananatis Sd‐1 has an unusually high number of carbohydrate‐active enzymes among members of the species, including enzymes required for the degradation of cellulose and hemicellulose (Ma et al., 2016). Included in its arsenal of CWDEs are 59 glycoside hydrolases, 25 carbohydrate esterases, 10 cellulases and two polysaccharide lyases (Ma et al., 2016). Pantoea ananatis is thus well equipped to degrade the integrity of the plant cell wall and cause disease in its host.
Little is known about how P. ananatis causes disease in animal hosts. In humans, infections by P. ananatis usually occur in immunocompromised individuals or neonates, where infections can result in bacteraemia (De Baere et al., 2004; Van Rostenberghe et al., 2006). Genomic factors which have been suggested to play a role in animal colonization and infection include adhesins, the T6SS and type I fimbriae (De Maayer et al., 2014; Shyntum et al., 2014). All analysed P. ananatis strains also have rhlA and rhlB genes which are required for the biosynthesis of a rhamnolipid (Smith et al., 2016). This glycolipid is involved in the production of a biosurfactant that not only enables swarming motility, but is also cytotoxic to the bacteriovorous grazing amoeba Dictyostelium discoideum, and may thus enable P. ananatis to establish infections in humans (Smith et al., 2016). Dictyostelium discoideum is often employed in mammalian pathogenicity trials as many of the D. discoideum genes encoded on its genome are homologous to human genes and, as a result, its cellular response to pathogenicity factors is similar to that of mammalian cells (Pan et al., 2011).
Quorum sensing is a highly regulated population density‐dependent method of regulation of gene expression within a bacterial cell (Morohoshi et al., 2011). Generally speaking, quorum sensing comprises a LuxI homologue, which is an autoinducer synthase and is responsible for the synthesis of the signalling molecules, namely acyl‐homoserine lactones (AHLs), and a LuxR homologue, which is the AHL receptor (von Bodman et al., 2003). Pantoea ananatis has two LuxI/R homologues, namely EanI/R and RhlI/R (Sibanda et al., 2016). The specific AHLs that are synthesized by P. ananatis are N‐hexanoyl‐HL, N‐heptanoyl‐HL and N‐octanoyl‐HL, with the most abundant of the three being N‐hexanoyl‐HL (Pomini et al., 2006). N‐Heptanoyl‐HL and N‐octanoyl‐HL are uncommon in that they are used both as signalling substances and as antimicrobials against Gram‐positive bacteria. This may give P. ananatis a competitive advantage in its natural environment (Pomini and Marsaioli, 2008). It has been shown that quorum sensing in P. ananatis also plays a role in exopolysaccharide biosynthesis, biofilm formation, pathogenicity, cell aggregation and the biosynthesis of hydrolytic enzymes, such as extracellular alkaline phosphatase (Jatt et al., 2015; Morohoshi et al., 2007, 2011; Sibanda et al., 2016).
Pantoea Ananatis in The Age of Biotechnology
Although much P. ananatis research has focused on pathogenicity on various plant hosts, this bacterium also provides us with an array of beneficial characteristics that we have only just begun to investigate. For example, several strains are known to aid the growth of their host plants, such as poplar (Gkorezis et al., 2016), onions (Kim et al., 2012), papaya (Thomas et al., 2007) rice (Megías et al., 2016; Sanchez‐Matamoros et al., 2013) and red pepper, and can increase crop yield up to three times (Kim et al., 2012). Several P. ananatis strains are capable of phosphate solubilization (Kim et al., 2012; Megías et al., 2016), whereby organic and inorganic forms of phosphorus are solubilized and converted into a bioavailable form for plant root growth, seed formation and major metabolic processes, such as photosynthesis and respiration (Sharma et al., 2013). Pantoea ananatis can also promote plant growth through the production of cellulose (Megías et al., 2016) and indole acetic acid (IAA) (Kim et al., 2012; Megías et al., 2016). The production of bacterial cellulose aids in inter‐domain attachments and biofilm formation, allowing growth‐promoting bacteria to effectively deliver growth‐promoting agents to their host plant (Augimeri et al., 2015). IAA is a phytohormone that can have both beneficial and deleterious effects on the host plant (Spaepen et al., 2007). When produced in the correct concentrations, IAA aids the plant in cell division and enlargement, tissue differentiation, root proliferation and responses to light and gravity (Spaepen et al., 2007).
Potential applications of P. ananatis in bioremediation and biofuel production have also been suggested. Strains of P. ananatis have been isolated from aquatic environments near maize fields that were treated with mesotrione, a recalcitrant herbicide commonly used in Europe, Brazil and the USA (Alferness and Wiebe, 2002; Pileggi et al., 2012). Pantoea ananatis strains proved effective in the complete degradation of mesotrione in a relatively short period of time (Pileggi et al., 2012). The secretome of P. ananatis Sd‐1 was analysed and found to encode 154 putative carbohydrate‐active enzymes. These enzymes are predominantly responsible for the degradation of cellulose and hemicellulose in plant cell wall polymers (Ma et al., 2016). The secretome also revealed the presence of ligninolytic and lignocellulolytic enzymes and laccases, which have great potential in the lignocellulosic bioenergy industry for the production of biofuels, as well as the decolorization of synthetic dyes (Ma et al., 2016; Shi et al., 2015).
There is increasing evidence that P. ananatis can be applied as an effective biological control agent against a range of bacterial and fungal plant pathogens. The sequenced genomes of P. ananatis BRT175 and R100 have shed light on the mechanisms of antagonism (Smith et al., 2013; Wu et al., 2016). Pantoea ananatis BRT175 produces PNP‐1, which is a compound inhibitory against E. amylovora (Walterson et al., 2014) and provides protection against Xanthomonas axonopodis pv. vesicatoria in pepper (Kang et al., 2007). Both E. amylovora and X. axonopodis pv. vesicatoria are causal agents of destructive diseases that have severe economic implications (Johnson, 2000; Ritchie, 2000). The P. ananatis R100 genome encodes oxazolomycin and chalcomycin biosynthetic genes, which are antibiotics capable of inhibiting bacterial growth, such as Agrobacterium tumefaciens, Staphylococcus aureus and Streptococcus pyogenes (Kanzaki et al., 1998; Ward et al., 2004; Wu et al., 2016).
Conclusions
Pantoea ananatis has been well represented in the genome sequencing era with the genomes of 22 strains sequenced to date. The data that we now have available to us have delineated some of the complexities of this species. The plasticity of its genome enables P. ananatis to proliferate in numerous environmental niches and to cause disease in a wide variety of hosts. It can be envisaged that, with the availability of the complete gene sets of many different P. ananatis strains isolated from different hosts and with distinct lifestyles, and the application of cutting edge molecular tools, we will gain deeper insights into the pathogenicity determinants employed, host range determinants and biology of this important enterobacterium. Future investigations may be able to shed light on how the host genotype may affect the pathogenicity of P. ananatis, and what mechanisms this pathogen uses to avoid recognition and the defence response by the broad range of plant hosts it infects. Questions such as these may be answered by combining the wealth of genomic data available with other state‐of‐the‐art molecular tools, such as transcriptomic and proteomic approaches, which could identify differences in the gene expression and protein complement of both the bacterium and host under various circumstances, or with population genetic studies, where the genetic variation within strains is highlighted. We have only just begun to investigate the biotechnological applications of P. ananatis, and this is undoubtedly a field of research that merits further investigation.
Acknowledgements
The authors would like to thank the National Research Foundation (NRF) of South Africa, the University of Pretoria, the Forestry and Agricultural Biotechnology Institute (FABI), the Tree Protection Cooperative Program (TPCP) and the Centre of Excellence in Tree Health Biotechnology (CTHB) for supporting this research.
References
- Adam, Z. , Tambong, J.T. , Lewis, C.T. , Levesque, C.A. , Chen, W. , Bromfield, E.S.P. , Khan, I.U.H. and Xu, R. (2014) Draft genome sequence of Pantoea ananatis strain LMG 2665T, a bacterial pathogen of pineapple fruitlets. Genome Announc. doi: 10.1128/genomeA.00489-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alferness, P. and Wiebe, L. (2002) Determination of mesotrione residues and metabolites in crops, soil, and water by liquid chromatography with fluorescence detection. J. Agric. Food Chem. 50, 3926–3934. [DOI] [PubMed] [Google Scholar]
- Augimeri, R.V. , Varley, A.J. and Strap, J.L. (2015) Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm‐producing Proteobacteria . Front. Microbiol. doi: 10.3389/fmicb.2015.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman, S.B. , Bauer, W.D. and Coplin, D.L. (2003) Quorum sensing in plant‐pathogenic bacteria. Annu. Rev. Phytopathol. 41, 455–482. [DOI] [PubMed] [Google Scholar]
- Choi, O. , Lim, J.Y. , Seo, Y.‐S. , Hwang, I. and Kim, J. (2012) Complete genome sequence of the rice pathogen Pantoea ananatis strain PA13. J. Bacteriol. 194, 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho, T.A. , Preisig, O. , Mergaert, J. , Cnockaert, M.C. , Riedel, K.H. , Swings, J. and Wingfield, M.J. (2002) Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis. 86, 20–25. [DOI] [PubMed] [Google Scholar]
- Coutinho, T.A. and Venter, S.N. (2009) Pantoea ananatis: an unconventional plant pathogen. Mol. Plant. Pathol. 10, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baere, T. , Verhelst, R. , Labit, C. , Verschraegen, G. , Wauters, G. , Claeys, G. and Vaneechoutte, M. (2004) Bacteremic infection with Pantoea ananatis . J. Clin. Microbiol. 42, 4393–4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. and Cowan, D.A. (2016) Flashy flagella: flagellin modification is relatively common and highly versatile among the Enterobacteriaceae . BMC Genomics, doi: 10.1186/s12864-016-2735-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Chan, W.‐Y. , Venter, S.N. , Toth, I.K. , Birch, P.R.J. , Joubert, F. and Coutinho, T.A. (2010) Genome sequence of Pantoea ananatis LMG 5342. J. Bacteriol. 194, 1615–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Venter, S.N. , Kamber, T. , Duffy, B. , Coutinho, T.A. and Smits, T.H.M. (2011) Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics, 12, 576–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Chan, W.‐Y. , Blom, J. , Venter, S.N. , Duffy, B. , Smits, T.H. and Coutinho, T.A. (2012a) The large universal Pantoea plasmid LPP‐1 plays a major role in biological and ecological diversification. BMC Genomics, doi: 10.1186/1471-2164-13-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Chan, W.‐Y. , Rezzonica, F. , Buhlmann, A. , Venter, S.N. , Blom, J. , Goesmann, A. , Frey, J.E. , Smits, T.H.M. , Duffy, B. and Coutinho, T.A. (2012b) Complete genome sequence of clinical isolate Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J. Bacteriol. 192, 2936–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Chan, W.‐Y. , Rubagotti, E. , Venter, S.N. , Toth, I.K. , Birch, P.R.J. and Coutinho, T.A. (2014) Analysis of the Pantoea ananatis pan‐genome reveals factors underlying its ability to colonise and interact with plant, insect and vertebrate hosts. BMC Genomics, 15, 404–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maayer, P. , Chan, W.‐Y. , Martin, D.A.J. , Blom, J. , Venter, S.N. , Duffy, B. , Cowan, D.A. , Smits, T.H. and Coutinho, T.A. (2015) Integrative conjugative elements of the ICEPan family play a potential role in Pantoea ananatis ecological diversification and antibiosis. Front. Microbiol. doi: 10.3389/fmicb.2015.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, B. , Gitaitis, R. , Barman, A. , Avci, U. , Marasigan, K. and Srinivasan, R. (2016) Interactions between Frankliniella fusca and Pantoea ananatis in the center rot epidemic on onion (Allium cepa). Phytopathology, 106, 956–962. [DOI] [PubMed] [Google Scholar]
- Egorova, M. , Mazurin, E. and Ignatov, A.N. (2015) First report of Pantoea ananatis causing grain discolouration and leaf blight of rice in Russia. New Dis. Rep. 32, 21. [Google Scholar]
- Esquerre‐Tugaye, M.‐T. , Boudart, G. and Dumas, B. (2000) Cell wall degrading enzymes, inhibitory proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol. Biochem. 38, 157–163. [Google Scholar]
- Gasser, F. , Cardinale, M. , Schildberger, B. and Berg, G. (2012) Biocontrol of Botrytis cinerea by successful introduction of Pantoea ananatis in the grapevine phyllosphere. Int. J. Wine Res. 4, 53–63. [Google Scholar]
- Gavini, F. , Mergaert, J. , Beji, A. , Mielcarek, C. , Izard, D. , Kersters, K. and De Ley, J. (1989) Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 39, 337–345. [Google Scholar]
- Gitaitis, R.D. , Walcott, R.D. , Culpepper, S. , Sanders, F.H. , Zolobowska, L. and Langston, D. (2002) Recovery of Pantoea ananatis, causal agent of center rot of onion, from weeds and crops in Georgia, USA. Crop Prot. 21, 983–989. [Google Scholar]
- Gitaitis, R.D. , Walcott, R.R. , Wells, M.L. , Diaz Perez, J.C.D. and Sanders, F.H. (2003) Transmission of Pantoea ananatis, causal agent of center rot of onion, by tobacco thrips, Frankliniella fusca . Plant Dis. 87, 675–678. [DOI] [PubMed] [Google Scholar]
- Gkorezis, P. , Van Hamme, J.D. , Bottos, E.M. , Thijs, S. , Balseiro‐Romero, M. , Monterroso, C. , Kidd, P.S. , Rineau, F. , Weyens, N. and Vangronsveld, J. (2016) Draft genome sequence of Pantoea ananatis GB1, a plant‐growth‐promoting hydrocarbonoclastic root endophyte, isolated at a diesel fuel phytoremediation site planted with Populus . Genome Announc. doi: 10.1128/genomeA.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goszczynska, T. , Venter, S.N. and Coutinho, T.A. (2007) Isolation and identification of the causal agent of brown stalk rot, a new disease of corn in South Africa. Plant Dis. 91, 711–718. [DOI] [PubMed] [Google Scholar]
- Hara, Y. , Kadotani, N. , Izui, H. , Katshkina, J.I. , Kuvaeva, T.M. , Andreeva, I.G. , Golubeva, L.I. , Malko, D.B. , Makeev, V.J. , Mashko, S.V. and Kozlov, Y.I. (2012) The complete genome sequence of Pantoea ananatis AJ13355, an organism with great biotechnological potential. Appl. Microbiol. Biotechnol. 93, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, E. , Blom, J. , Bultreys, A. , Ivanović, M. , Obradović, A. , van Doorn, J. , Bergsma‐Vlami, M. , Maes, M. , Willems, A. , Duffy, B. , Stockwell, V.O. , Smits, T.H.M. and Pulawska, J. (2014) A novel plasmid pEA68 of Erwinia amylovora and the description of a new family of plasmids. Arch. Microbiol. 196, 891–899. [DOI] [PubMed] [Google Scholar]
- Jatt, A.N. , Tang, K. , Liu, J. , Zhang, J. and Zhang, X.‐H. (2015) Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol. Ecol. 91, 1–13. [DOI] [PubMed] [Google Scholar]
- Johnson, K.B. (2000) Fire blight of apple and pear. Plant Health Instruct. doi: 10.1094/PHI-I-2000-0726-01. [DOI] [Google Scholar]
- Kang, S.H. , Cho, H.S. , Cheong, H. , Ryu, C.M. , Kim, J.F. and Park, S.H. (2007) Two bacterial endophytes eliciting both plant growth promotion and plant defense on pepper (Capsicum annuum L.). J. Microbiol. Biotechnol. 17, 96–103. [PubMed] [Google Scholar]
- Kang, Y. , Liu, H. , Genin, S. , Schell, M.A. and Denny, T.P. (2002) Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 2, 427–437. [DOI] [PubMed] [Google Scholar]
- Kanzaki, H. , Wada, K. , Nitoda, T. and Kawazu, K. (1998) Novel bioactive oxazolomycin isomers produced by Streptomyxes albus JA3453. Biosci. Biotechnol. Biochem. 62, 438–442. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Lee, J.H. , Kang, B.R. , Rong, X. , McSpadden Gardener, B.B. , Ji, H.J. , Park, C.‐S. and Kim, Y.C. (2012) Draft genome sequence of Pantoea ananatis B1‐9, a non‐pathogenic plant growth‐promoting bacterium. J. Bacteriol. 194, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan, S.M. (2006) Flagellar glycosylation – a new component of the motility repertoire? Microbiology, 152, 1249–1262. [DOI] [PubMed] [Google Scholar]
- Ma, J. , Zhang, K. , Liao, H. , Hector, S.B. , Shi, X. , Li, J. , Liu, B. , Xu, T. , Tong, C. , Liu, X. and Zhu, Y. (2016) Genomic and secretomic insight into lignocellulolytic system of an endophytic bacterium Pantoea ananatis Sd‐1. Biotechnol. Biofuels, doi: 10.1186/s13068-016-0439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano, E.G. and Bell, A.A. (2015) Genome sequence of Pantoea ananatis strain CFH 7‐1, which is associated with a vector‐borne cotton fruit disease. Genome Announc. doi: 10.1128/genomeA.01029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megías, E. , Megías, M. , Ollero, F.J. and Hungria, M. (2016) Draft genome sequence of Pantoea ananatis strain AMG521, a rice plant growth‐promoting bacterial endophyte isolated from the Guadalquivir marshes in southern Spain. Genome Announc. doi: 10.1128/genomeA.01681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, F. , Yao, J. and Allen, C. (2011) A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J. Bacteriol. 193, 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert, J. , Verdonck, L. and Kersters, K. (1993) Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int. J. Syst. Bacteriol. 43, 162–173. [Google Scholar]
- Midha, S. , Bansal, K. , Sharma, S. , Kumar, N. , Patil, P.P. , Chaudhry, V. and Patil, P.B. (2016) Genomic resource of rice seed associated bacteria. Front. Microbiol. doi: 10.3389/fmicb.2015.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A.M. , Figueiredo, J.E.F. , Linde, G.A. , Colauto, N.B. and Paccola‐Meirelles, L.D. (2016) Characterization of the inaA gene and expression of ice nucleation phenotype in Pantoea ananatis isolates from maize white spot disease. Genet. Mol. Res. doi: 10.4238/gmr.15017863 [DOI] [PubMed] [Google Scholar]
- Morohoshi, T. , Nakamura, Y. , Yamazaki, G. , Ishida, A. , Kato, N. and Ikeda, T. (2007) The plant pathogen Pantoea ananatis produces N‐acylhomoserine lactone and causes center rot disease of onion by quorum sensing. J. Bacteriol. 189, 8333–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi, T. , Ogata, Y. and Ikeda, T. (2011) Cell aggregation is negatively regulated by N‐acylhomoserine lactone‐mediated quorum sensing in Pantoea ananatis SK‐1. J. Biosci. Bioeng. 112, 566–569. [DOI] [PubMed] [Google Scholar]
- Murrell, A. , Dobson, S.J. , Yang, X. , Lacey, E. and Barker, S.C. (2003) A survey of bacterial diversity in ticks, lice and fleas from Australia. Parasitol. Res. 89, 326–334. [DOI] [PubMed] [Google Scholar]
- O'Toole, G.A. and Kolter, R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. [DOI] [PubMed] [Google Scholar]
- Pan, Y. , Lin, T. , Hsu, C. and Wang, J. (2011) Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae . Infect. Immun. 79, 997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐y‐Terron, R. , Villegas, M.C. , Cuellar, A. , Munoz‐Rojas, R. , Castaneda‐Lucio, M. , Hernandez‐Lucas, I. , Bustillos‐Cristales, R. , Bautista‐Sosa, L. , Munive, J.A. , Caicedo‐Rivas, R. and Fuentes‐Ramirez, L.E. (2009) Detection of Pantoea ananatis, causal agent of leaf spot disease of maize, in Mexico. Australas. Plant Dis. Notes, 4, 96–99. [Google Scholar]
- Pileggi, M. , Pileggi, S.A.V. , Olchanheski, L.R. , da Silva, P.A.G. , Gonzalez, A.M.M. , Koskinen, W.C. , Barber, B. and Sadowsky, M.J. (2012) Isolation of mesotrione‐degrading bacteria from aquatic environments in Brazil. Chemosphere, 86, 1127–1132. [DOI] [PubMed] [Google Scholar]
- Pomini, A.M. and Marsaioli, A.J. (2008) Absolute configuration and antimicrobial activity of acylhomoserine lactones. J. Nat. Prod. 71, 1032–1036. [DOI] [PubMed] [Google Scholar]
- Pomini, A.M. , Araujo, W.L. and Marsaioli, A.J. (2006) Structural elucidation and biological activity of acyl‐homoserine lactones from the phytopathogen Pantoea ananatis Serrano 1928. J. Chem. Ecol. 32, 1769–1778. [DOI] [PubMed] [Google Scholar]
- Ramos, H.G. , Rumbo, M. and Sirard, J.C. (2004) Bacterial flagellins: mediators of pathogenicity, and host immune responses in mucosa. Trends Microbiol. 12, 509–517. [DOI] [PubMed] [Google Scholar]
- Rauch, M.E. , Graef, H.W. , Rozenzhak, S.M. , Jones, S.E. , Bleckmann, C.A. , Kruger, R.L. , Naik, R.R. and Stone, M.O. (2006) Characterization of microbial contamination in United States Air Force aviation fuel tanks. J. Ind. Microbiol. Biotechnol. 33, 29–36. [DOI] [PubMed] [Google Scholar]
- Ritchie, D.F. (2000) Bacterial spot of pepper and tomato. Plant Health Instruct. doi: 10.1094/PHI-I-2000-1027-01. [DOI] [Google Scholar]
- Sanchez‐Matamoros, R.C. , Serrano, A.M.G. , Tejero‐Mateo, P. , Ollero, J. , Saavedra, E.M. and Rodriguez‐Carvajal, M.A. (2013) Structure of the O‐antigen of the lipopolysaccharide isolated from Pantoea ananatis AEP17, a rhizobacterium associated with rice. Carbohydr. Res. 369, 25–30. [DOI] [PubMed] [Google Scholar]
- Sharma, S.B. , Sayyed, R.Z. , Trivedi, M.H. and Gobi, T.A. (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus, doi: 10.1186/2193-1801-2-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheibani‐Tezerji, R. , Naveed, M. , Jehl, M.‐A. , Sessitsch, A. , Rattei, T. and Mitter, B. (2015) The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front. Microbiol. doi: 10.3389/fmicb.2015.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. and Ronald, P. (2002) Molecular determinants of disease and resistance in interactions of Xanthomonas oryzae pv. oryzae and rice. Microb. Infect. 4, 1361–1367. [DOI] [PubMed] [Google Scholar]
- Shi, X. , Liu, Q. , Ma, J. , Liao, H. , Xiong, X. , Zhang, K. , Wang, T. , Liu, X. , Xu, T. , Yuan, S. , Zhang, X. and Zhu, Y. (2015) An acid‐stable bacterial laccase identified from the endophyte Pantoea ananatis Sd‐1 genome exhibiting lignin degradation and dye decolorization abilities. Biotechnol. Lett. 37, 2279–2288. [DOI] [PubMed] [Google Scholar]
- Shyntum, D.Y. , Venter, S.N. , Moleleki, L.N. , Toth, I. and Coutinho, T.A. (2014) Comparative genomics of type VI secretion systems in strains of Pantoea ananatis from different environments. BMC Genomics, 15, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyntum, D.Y. , Theron, J. , Venter, S.N. , Moleleki, L.N. , Toth, I. and Coutinho, T.A. (2015) Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. Plant–Microbe Interact. 28, 420–431. [DOI] [PubMed] [Google Scholar]
- Sibanda, S. , Theron, J. , Shyntum, D.Y. , Moleleki, L. and Coutinho, T.A. (2016) Characterization of two Lux I/R homologs in Pantoea ananatis LMG 2665T . Can. J. Microbiol. doi: 10.1139/cjm-2016-0143. [DOI] [PubMed] [Google Scholar]
- Smith, D.D.N. , Kirzinger, M.W.B. and Stavrinides, J. (2013) Draft genome sequence of the antibiotic‐producing epiphytic isolate Pantoea ananatis BRT175. Genome Announc. doi: 10.1128/genomeA.00902-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.D.N. , Nickzad, A. , Deziel, E. and Stavrinides, J. (2016) A novel glycolipid biosurfactant confers grazing resistance upon Pantoea ananatis BRT175 against the social amoeba Dictyostelium discoideum . mSphere, doi: 10.1128/mSphere.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. , Vanderleyden, J. and Remans, R. (2007) Indole‐3‐acetic acid in microbial and microorganism‐plant signalling. FEMS Microbiol. Rev. 4, 425–448. [DOI] [PubMed] [Google Scholar]
- Takeuchi, K. , Taguchi, F. , Inagaki, Y. , Toyoda, K. , Shiraishi, T. and Ichinose, Y. (2003) Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185, 6658–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P. , Kumari, S. , Swarna, G.K. and Gowda, T.K.S. (2007) Papaya shoot tip associated endophytic bacteria isolated from in vitro cultures and host–endophyte interaction in vitro and in vivo. Can. J. Microbiol. 53, 380–390. [DOI] [PubMed] [Google Scholar]
- Van Rostenberghe, H. , Noraida, R. , Wan Pauzi, W.I. , Habash, H. , Zeehaida, M. , Rosliza, A.R. , Fatimah, I. , Nik Sharimah, N.Y. and Maimunah, H. (2006) The clinical picture of neonatal infection with Pantoea species. Jpn. J. Infect. Dis. 59, 120–121. [PubMed] [Google Scholar]
- Walterson, A.M. , Smith, D.D.N. and Stavrinides, J. (2014) Identification of a Pantoea biosynthetic cluster that directs the synthesis of an antimicrobial natural product. PLoS One, doi: 10.1371/journal/pone.0096208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S.L. , Hu, Z. , Schirmer, A. , Reid, R. , Revill, W.P. , Reeves, C.D. , Petrakovsky, O.V. , Dong, S.D. and Katz, L. (2004) Chalcomycin biosynthesis gene cluster from Streptomyces bikinienesis: novel features of an unusual ketolide produced through expression of the chm polyketide synthase in Streptomyces fradiae . Antimicrob. Agents Chemother. 48, 4703–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K. , Kawakita, H. and Sato, M. (1996) Epiphytic bacterium, Erwinia ananas, commonly isolated from rice plants and brown plant hoppers (Nilaparvata lugens) in hopperburn patches. Appl. Entomol. Zool. 31, 459–462. [Google Scholar]
- Weller‐Stuart, T. , Chan, W.‐Y. , Coutinho, T.A. , Venter, S.N. , Smits, T.H.M. , Duffy, B. , Goszczynska, T. , Cowan, D.A. and De Maayer, P. (2014) Draft genome sequences of the onion center rot pathogen Pantoea ananatis PA4 and maize brown stalk rot pathogen P. ananatis BD442. Genome Announc. doi: 10.1128/genomeA.00750-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller‐Stuart, T. , Toth, I.D. , Maayer, P. and Coutinho, T. (2016) Swimming and twitching motility are essential for attachment and virulence of Pantoea ananatis in onion seedlings. Mol. Plant Pathol. doi: 10.1111/mpp.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, J.M. , Gitaitis, R.D. and Sanders, F.H. (2002) Association of tobacco thrips, Frankliniella fusca (Thysanoptera: Thripidae) with two species of bacteria of the genus Pantoea . Ann. Entomol. Soc. Am. 95, 719–723. [Google Scholar]
- Wu, L. , Liu, R. , Niu, Y. , Lin, H. , Ye, W. , Guo, L. and Hu, X. (2016) Whole genome sequence of Pantoea ananatis R100, an antagonistic bacterium isolated from rice seed. J. Biotechnol. doi: 10.1016/j.jbiotec.2016.03.007. [DOI] [PubMed] [Google Scholar]


