Summary
Autophagy, a ubiquitous intracellular degradation process, is conserved from yeasts to humans. It serves as a major survival function during nutrient depletion stress and is crucial for correct growth and differentiation. In this study, we characterized an atg1 orthologue Bcatg1 in the necrotrophic plant pathogen Botrytis cinerea. Quantitative real‐time polymerase chain reaction (qRT‐PCR) assays showed that the expression of BcATG1 was up‐regulated under carbon or nitrogen starvation conditions. BcATG1 could functionally restore the survival defects of the yeast ATG1 mutant during nitrogen starvation. Deletion of BcATG1 (ΔBcatg1) inhibited autophagosome accumulation in the vacuoles of nitrogen‐starved cells. ΔBcatg1 was dramatically impaired in vegetative growth, conidiation and sclerotial formation. In addition, most conidia of ΔBcatg1 lost the capacity to form the appressorium infection structure and failed to penetrate onion epidermis. Pathogenicity assays showed that the virulence of ΔBcatg1 on different host plant tissues was drastically impaired, which was consistent with its inability to form an appressorium. Moreover, lipid droplet accumulation was significantly reduced in the conidia of ΔBcatg1, but the glycerol content was increased. All of the defects of ΔBcatg1 were complemented by re‐introduction of an intact copy of the wild‐type BcATG1 into the mutant. These results indicate that BcATG1 plays a critical role in numerous developmental processes and is essential to the pathogenesis of B. cinerea.
Keywords: autophagy, BcATG1, Botrytis cinerea, development, pathogenesis
Introduction
Autophagy is a cellular mechanism for bulk degradation of long‐lived cytosolic or short‐lived damaged proteins and organelles within vacuoles/lysosomes to maintain cellular homeostasis (Yorimitsu and Klionsky, 2005). For many years, autophagy has been presumed to be involved in the cellular architectural changes that occur during differentiation and development, probably by the recycling of intracellular macromolecular substances (Klionsky, 2005; Reggiori and Klionsky, 2002). The analysis of the molecular mechanism of autophagy over the last two decades has mainly used the unicellular ascomycetes Saccharomyces cerevisiae and Pichia pastoris. Genetic analysis in these yeasts has identified 36 autophagy‐related (atg) genes, and most are conserved in all eukaryotes, including filamentous ascomycetes (Duan et al., 2013; Voigt and Pöggeler, 2013b). However, the autophagic mechanism has also evolved significant differences in fungi, as a consequence of adaptation to diverse fungal lifestyles (Voigt and Pöggeler, 2013b). Recent studies have shown that autophagy in filamentous fungi is involved not only in nutrient homeostasis, but also in other cellular processes, such as cell differentiation, secondary metabolism and pathogenicity (Duan et al., 2013; Kikuma et al., 2006; Liu et al., 2007; Nitsche et al., 2013; Voigt and Pöggeler, 2013a). Among the several core elements of the autophagy pathway, the serine/threonine kinase atg1 is a key member that is involved in both non‐selective and selective autophagy (Yanagisawa et al., 2013). Previous studies have shown that atg1 is involved in multiple cellular processes, including autophagy, cell growth, differentiation, lipid metabolism and virulence (Chan and Tooze, 2009). Deletion of atg1 resulted in no autophagy, mitophagy or pexophagy, decreased chronological and replicative lifespan, increased sensitivity to nitrogen starvation and failure to sporulate in S. cerevisiae (Kamada et al., 2000; Pinan‐Lucarré et al., 2005). Homologues of atg1 have also been characterized in a number of other fungi. For example, deletion of MoATG1 in Magnaporthe oryzae resulted in non‐pathogenicity, the accumulation of fewer lipid droplets, lower turgor pressure of the appressorium, slower germination of conidia and impairment of nuclei macroautophagy (He et al., 2012; Kershaw and Talbot, 2009; Liu et al., 2007). Deletion of MrATG1 in Metarhizium robertsii resulted in the impairment of aerial hyphal growth, conidiation and virulence (Yanagisawa et al., 2013). Deletion of atg1 also resulted in a reduction in conidiospores, increase in peroxisomes and over‐production of penicillin in Penicillium chrysogenum (Bartoszewska et al., 2011). Molecular interaction studies have revealed that atg1 interacts with several proteins in vitro which are specifically required for either the cytoplasm to vacuole targeting (Cvt) pathway or autophagy via Atg13 or Atg17, and the interactions can be mediated by the atg1 phosphorylation status (Cebollero and Reggiori, 2009; Kamada et al., 2000). However, the composition of the atg1 complex in vivo is elusive, which makes it difficult to uncover the molecular role of atg1.
The gray mould fungus Botrytis cinerea is a necrotrophic filamentous phytopathogen that is capable of causing disease in more than 200 crop species worldwide. As the incubation period of B. cinerea in infected tissues is long, serious damage is caused to both fresh and post‐harvest crops (Rosslenbroich and Stuebler, 2000; Williamson et al., 2007). In recent years, considerable progress has been made in understanding fungal development and pathogenesis in B. cinerea (Amselem et al., 2011). Conserved signal transduction pathways, such as the cyclic adenosine monophosphate (cAMP)‐dependent and several mitogen‐activated protein kinase (MAPK) pathways, have been shown to be important for cell function during morphogenesis, differentiation and pathogenic interactions in B. cinerea (Mehrabi et al., 2009; Schumacher et al., 2008; Tudzynski and Gronover, 2007; Zhao et al., 2007). However, none has been reported to be associated with autophagy. Here, we investigated the function of an atg1 orthologue gene from B. cinerea by the construction of BcATG1 deletion and complemented mutants, and examined its role in fungal development and pathogenesis. These results extend our knowledge of fungal autophagy in necrotrophic fungi, especially in B. cinerea.
Results
Sequence analysis of BcATG1 in B. cinerea
The serine/threonine kinase gene BcATG1 was retrieved by blastp search of the B. cinerea genome database with the S. cerevisiae atg1 protein as a query. The coding region of BcATG1 is 3061 bp in length, is predicted to have two introns and encodes a 952‐amino‐acid protein. The predicted locations and sizes of the introns in the BcATG1 gene were verified by sequencing a 2859‐bp fragment obtained from cDNA.
Phylogenetic analysis showed that the BcATG1 encoded protein contains a conserved serine/threonine kinase domain at its N‐terminal region, where the catalytic domain and ATP‐binding sites are located, and is highly homologous to ATG1 from other filamentous ascomycetes. It shows 52% identity with M. oryzae MoATG1, 51% identity with Fusarium graminearum FgATG1, 51% identity with Aspergillus oryzae AoATG1 and 34% identity with S. cerevisiae ScATG1 (Fig. S1, see Supporting Information).
Complementation of yeast ATG1 deletion mutant with BcATG1
In order to determine the functional conservation of BcATG1, a full‐length BcATG1 cDNA fragment was introduced into yeast ATG1 deletion strain (YGL180W) using the pYES2 expression vector (Invitrogen Co., CA, USA). The expression of BcATG1 in the yeast ATG1 mutant was confirmed by reverse transcription‐polymerase chain reaction (RT‐PCR) (data not shown). When cultured on nitrogen starvation medium (SD–N) for 18 days, the ATG1 mutant carrying an empty pYES2 vector died, but the wild‐type strain (BY4741) and that complemented with BcATG1 survived, suggesting that BcATG1 restores the corresponding defects in these mutants, thereby ensuring their survival (Fig. 1). These results indicate that BcATG1 is presumably homologous to yeast ATG1 in structure and function.
Figure 1.
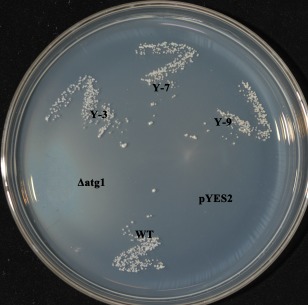
Complementation of the yeast ATG1 mutant with BcATG1. After incubation for 18 days at 30°C on nitrogen starvation medium (SD–N; 2% glucose and 0.17% yeast nitrogen base with neither amino acids nor ammonium sulfate), the yeast cells were grown on YPD (1% yeast extract, 2% peptone and 2% glucose) plates for 2 days at 30°C. In contrast with wild‐type (WT) yeast, Δatg1 and the mutant transformed with the empty pYES2 vector did not survive after nitrogen starvation; however, the mutant transformed with BcATG1 (Y‐3, Y‐7, Y‐9) survived like the WT.
The expression of BcATG1 is induced under carbon or nitrogen starvation conditions
The most typical trigger of autophagy is nutrient starvation, and several ATG genes have been shown to be induced during nitrogen or carbon starvation in yeast and plant cells (Kirisako et al., 1999; Rose et al., 2006). In order to investigate whether the same phenomenon occurs in B. cinerea, we tested BcATG1 expression of nitrogen‐ or carbon‐starved strains by qRT‐PCR. As shown in Fig. 2, after 4 and 8 h of nitrogen starvation, the expression levels of BcATG1 were up‐regulated by more than seven and ten times the initial value, respectively; however, the strains cultured in potato dextrose broth (PDB) showed no obvious change. A similar, but lesser, pattern was observed for carbon starvation. These results indicate that, as in other organisms, nutrient stress conditions are sufficient to induce the expression of BcATG1 in B. cinerea.
Figure 2.
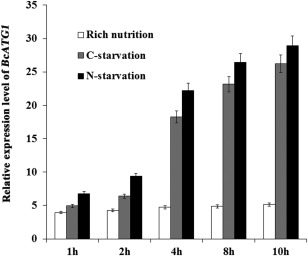
Expression levels of BcATG1 during carbon or nitrogen starvation. Relative expression levels of BcATG1 were calculated by quantitative real‐time polymerase chain reaction (qRT‐PCR). The total RNAs of the wild‐type samples were collected at the time points indicated in the graphs. Expression levels were normalized to the control gene Actin. The indicated values correspond to the means of three biological replicates; the bars represent the standard error of biological variation.
Deletion and complementation of BcATG1
To investigate the biological function of BcATG1 in B. cinerea, we generated the gene deletion mutant of BcATG1 using a homologous recombination strategy (Fig. S2A, see Supporting Information). Three deletion mutants were identified from 73 hygromycin‐resistant transformants by PCR analysis with the primer pair Bcatg1‐in‐F and Bcatg1‐in‐R (Fig. S2B and Table S1, see Supporting Information). Southern blotting patterns confirmed that ΔBcatg1 resulted from the anticipated homologous recombination events at the BcATG1 locus, and the wild‐type BcATG1 was ectopically integrated into the genome of the complemented strain with a single copy (ΔBcatg1‐C) (Fig. S2C).
BcATG1 is required for autophagy
After the mycelia of B. cinerea had been cultured in nitrogen‐limiting medium (minimal medium without NaNO3, MM–N) with 2 mm phenylmethylsulfonylfluoride (PMSF) for 6 h, they were observed by transmission electron microscopy (TEM). As shown in Fig. 3A, only the wild‐type and complemented strains accumulated autophagosomes in the vacuoles. In addition, autophagy was examined by staining the nutrient‐starved mycelia with monodansylcadaverine (MDC), which is an indicator of autophagic activity that accumulates in vacuoles (Klionsky et al., 2012). The wild‐type and complemented strains showed induction of autophagic compartments under nitrogen starvation conditions, but there were no autophagy signs of MDC staining in ΔBcatg1 (Fig. 3B). These results indicate that autophagy is blocked in ΔBcatg1.
Figure 3.
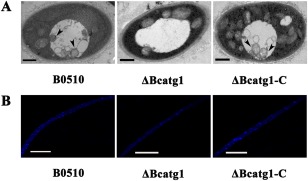
Analysis of autophagy process by transmission electron microscopy (TEM) observation and monodansylcadaverine (MDC) staining. (A) After each strain had been incubated in nitrogen‐limiting medium [minimal medium without NaNO3 (MM–N)] with 2 mm phenylmethylsulfonylfluoride (PMSF) for 6 h, autophagosomes were evident in the vacuoles of the wild‐type strain B05.10 and △Bcatg1‐C, whereas no autophagosomes were observed in the vacuoles of △Bcatg1. Arrow, autophagosome. Scale bar, 0.5 μm. (B) Mycelia were stained with the fluorescent dye MDC and observed under epifluorescence microscopy; autophagy compartments were observed in B0510 and △Bcatg1‐C, but not in △Bcatg1. Scale bar, 10 μm.
Involvement of BcATG1 in vegetative growth, conidiation and sclerotial formation
Although the mycelial radial growth rate of ΔBcatg1 was broadly similar to that of the wild‐type progenitor B05.10 on either potato dextrose agar (PDA) or MM (data not shown), ΔBcatg1 produced much fewer aerial hyphae (Fig. 4A) and included more fused hyphae (Fig. 4B). After 7 days, the hyphae of ΔBcatg1 lost their ability to extend and produced more pigments (Fig. 4A). In addition, ΔBcatg1 produced significantly fewer conidia than the wild‐type or the complemented strain after culture on PDA for 8 days (Fig. 5A,B), and most conidia showed an aberrant shape with many vacuoles (Fig. 5C). These results indicate that BcATG1 is involved in vegetative growth and conidial development.
Figure 4.
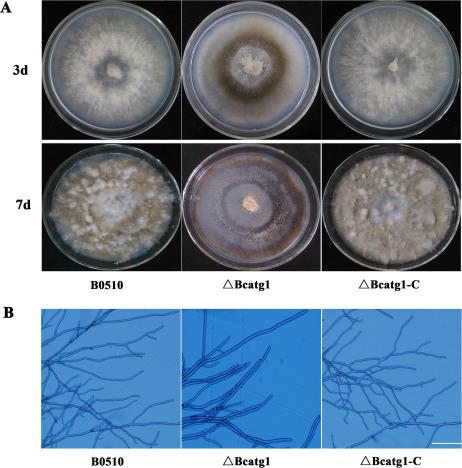
Morphological comparisons of the wild‐type B0510, △Bcatg1 and △Bcatg1‐C. (A) Colony morphology of B0510, △Bcatg1 and △Bcatg1‐C on potato dextrose agar (PDA). The photographs of the plates were taken after 3 and 7 days of incubation on PDA. (B) Hyphal tip growth and branching patterns of B0510, △Bcatg1 and △Bcatg1‐C growing on PDA plates. Hyphal branching was decreased and fused hyphae were increased in the BcATG1 deletion mutant △Bcatg1. Scale bar, 100 μm.
Figure 5.
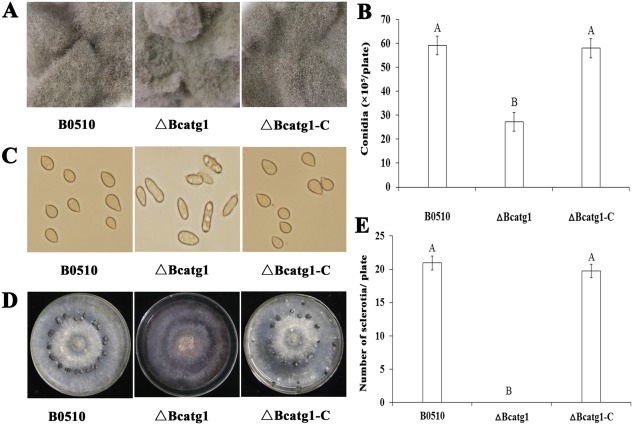
Impact of BcATG1 deletion on conidiation and sclerotial formation. (A) Comparison of conidiation among the wild‐type B0510, ΔBcatg1 and ΔBcatg1‐C after 7 days of incubation on sterilized potato fragments. (B) The number of conidia produced by the wild‐type B0510, ΔBcatg1 and ΔBcatg1‐C on potato dextrose agar (PDA) plates (diameter, 9 cm). (C) Conidial morphology of the wild‐type B0510, ΔBcatg1 and ΔBcatg1‐C. (D) Comparison of sclerotial formation among the wild‐type B05.10, ΔBcatg1 and ΔBcatg1‐C after 4 weeks of incubation on PDA at 25°C in the dark. (E) The numbers of sclerotia produced by the wild‐type B0510, ΔBcatg1 and ΔBcatg1‐C were quantified. The bars in each column denote the standard errors of three experiments. Values on the bars followed by the same letter are not significantly different at P = 0.05.
Sclerotial formation is an important survival mechanism for B. cinerea to overwinter (Williamson et al., 2007). We therefore investigated the effect of BcATG1 on sclerotial formation. After 4 weeks of incubation on PDA in the dark, sclerotia were observed only on the wild‐type and the complemented strains (Fig. 5D,E), indicating that BcATG1 is essential for sclerotial development in B. cinerea.
BcATG1 is required for pathogenesis and appressorium formation
To analyse the role of BcATG1 in virulence, infection tests on different host plant tissues were performed. ΔBcatg1 failed to infect wounded leaves of cucumber after inoculation for 60 h, whereas the wild‐type progenitor B05.10 and the complemented strain ΔBcatg1‐C caused serious disease lesions (Fig. 6). Interestingly, ΔBcatg1 showed slight virulence on wounded apple, grape, tomato and onion. After incubation for 72 h, ΔBcatg1 caused very small disease lesions (<0.5 cm in diameter), whereas the disease lesions caused by the wild‐type and complemented strains were very obvious (>1.5 cm) (Fig. 7A–E).
Figure 6.
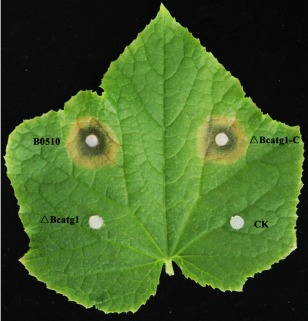
Pathogenicity assays on wounded cucumber leaf following inoculation with the wild‐type B0510, ΔBcatg1 and ΔBcatg1‐C. Agar plugs without fungal mycelia were used as negative controls (CK). Disease symptoms were analysed at 60 h post‐inoculation (hpi).
Figure 7.
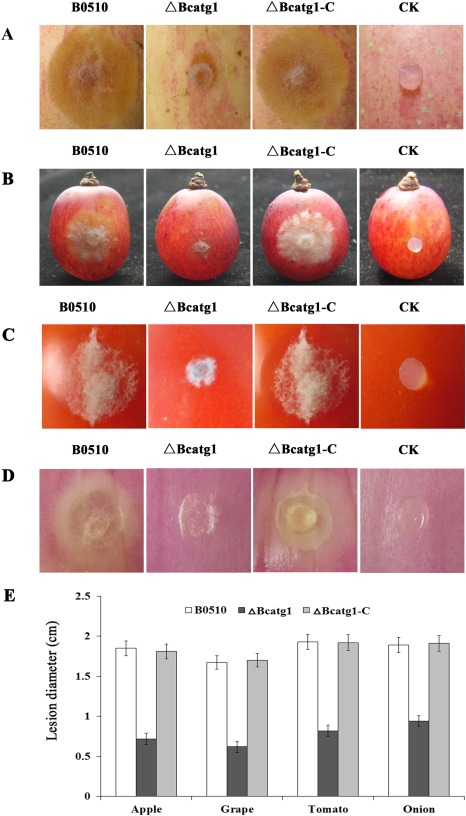
Pathogenicity assays on different plant tissues following inoculation with the wild‐type B0510, △Bcatg1 and △Bcatg1‐C. Agar plugs without fungal mycelia were used as negative controls (CK). Disease symptoms on wounded apple fruits (A), wounded grape fruits (B), wounded tomato fruits (C) and wounded onion (D) were phtographed at 72 h post‐inoculation (hpi). (E). Diameter of disease lesions on various host tissues caused by each strain at 72 hpi. Bars denote standard errors of three experiments.
To further analyse the virulence defects of ΔBcatg1, the formation of the appressorium infection structure was investigated. There was no significant difference in conidial germination between ΔBcatg1 and the wild‐type strain (data not shown). After inoculation for 8 h, 63.2% of germinated conidia in the wild‐type formed terminal thickenings representing appressoria‐like structures, whereas only 9.7% of germinated conidia in the ΔBcatg1 mutant produced appressoria‐like structures (Fig. 8A,B). In addition, onion penetration assay was performed. As shown in Fig. 8C, after 12 h of incubation, most wild‐type germlings, but only a few germlings of the mutants, successfully penetrated onion epidermal cells. These results indicate that BcATG1 is involved in the virulence of B. cinerea.
Figure 8.
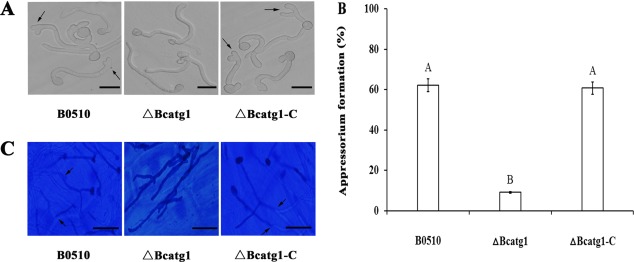
Formation assays of appressoria‐like structures on cellophane. (A) Detection of appressoria‐like structures of the conidia in the wild‐type B0510, △Bcatg1 and △Bcatg1‐C after inoculation on cellophane paper for 8 h. Arrows, appressoria‐like structures. Scale bars, 20 μm. (B) Bar chart showing the percentage of appressoria‐like structures of conidia in the wild‐type B0510, △Bcatg1 and △Bcatg1‐C. Error bars represent standard deviations. Values on the bars followed by the same letter are not significantly different at P = 0.05. (C) Onion penetration assays with the wild‐type B0510, △Bcatg1 and △Bcatg1‐C. Infection structures (stained) indicated by arrows and hyphae growing in planta (not stained) were observed clearly. Scale bars, 20 μm.
Involvement of BcATG1 in lipid metabolism
It has been reported that autophagy regulates lipid metabolism (Singh et al., 2009).
To assess the effect of BcATG1 on lipid droplet synthesis in B. cinerea, conidia stained with Nile red were observed by light microscopy. As shown in Fig. 9A, the conidia of ΔBcatg1 contained less abundant lipid bodies than those of the wild‐type or of the complemented strain.
Figure 9.
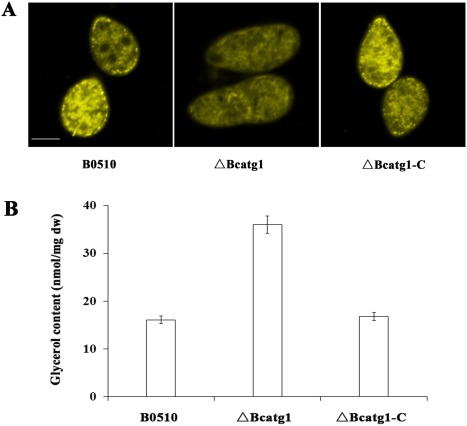
Effects of BcATG1 on lipid metabolism. (A) Histochemical analyses of lipid metabolism within conidia. Lipid droplets within conidia of the wild‐type B0510, △Bcatg1 and △Bcatg1‐C were stained with Nile red and examined under an episcopic fluorescence microscope. Scale bar, 10 μm. (B) Effects of BcATG1 on glycerol biosynthesis. The intracellular glycerol concentration (nmol/mg dried mycelia) in mycelia of the wild‐type B0510, △Bcatg1 and △Bcatg1‐C was analysed after incubation in potato dextrose broth (PDB) for 3 days. Bars denote the standard errors from three repeated experiments. dw, dry weight.
It is well known that glycerol is a product of lipolysis (Chopra, 1984). We therefore analysed the glycerol content in mycelia of the mutant ΔBcatg1. As shown in Fig. 9B, the glycerol content in ΔBcatg1 was significantly higher than that in the wild‐type B0510 and the complemented strain ΔBcatg1‐C. These results suggest that lipid metabolism is influenced by the BcATG1 gene.
Discussion
Although autophagy has been well documented in both biotrophic and hemibiotrophic fungi in recent decades, few have reported this mechanism in necrotrophic fungi. The mechanism of autophagy has evolved significant differences in fungi, as a consequence of adaptation to diverse fungal lifestyles (Voigt and Pöggeler, 2013b), which led us to perform this work. In this study, we reveal a tight relationship between autophagy and several biological aspects of B. cinerea through a reverse genetic approach. To our knowledge, this is the first report on autophagy in the necrotrophic plant‐pathogenic fungus B. cinerea.
BcATG1 is involved in the vegetative growth process of B. cinerea
A distinguishing feature of filamentous ascomycetes is growth by tip extension, and the extended hyphal tips are filled with cytoplasm from basal hyphal compartments. Thus, a mycelium has young, actively growing hyphae at the colony margin, surrounding an older, inner hyphal network that recycles nutrients to fuel the actively growing tips (Glass et al., 2004). In this study, deletion of BcATG1 in B. cinerea led to reduced vegetative growth: the colony margin was fresh and active during the first 3 days, whereas it became older and pigmented, but still alive, and failed to extend on the seventh day. In addition, the BcATG1 deletion mutant displayed more fused hyphae. In a previous study, autophagy has been shown to regulate hyphal fusion and post‐fusion nuclear degradation in Fusarium oxysporum (Corral‐Ramos et al., 2015), and autophagy is essential for filamentous fusion in Podospora anserina (Pinan‐Lucarré et al., 2003). These results indicate that autophagy is required for B. cinerea to recycle material and for morphogenesis during the vegetative growth process.
BcATG1 is essential for fungal reproductive development in the necrotrophic pathogen B. cinerea
Defective sporulation is a known effect of blocked autophagy in yeast (Matsuura et al., 1997; Straub et al., 1997; Tsukada and Ohsumi, 1993). As in yeast, deletion of MgATG1 in M. oryzae significantly reduced the production of conidia (Liu et al., 2007). Our results showed that the deletion of BcATG1 also had a negative effect on sporulation, and, moreover, most conidia of the mutant showed an abnormal morphology and many vacuoles, which is consistent with the phenomenon in M. oryzae (Liu et al., 2007). Sclerotium development within dying host tissues under a freezing environment is necessary for B. cinerea to complete the whole life cycle (Nitsche et al., 2013; Yang et al., 2013). In this study, the BcATG1 mutant completely lost the ability to form sclerotia. In previous studies, the PaATG1 mutant of P. anserina (Pinan‐Lucarré et al., 2005) and the MgATG1 mutant of M. oryzae (Liu et al., 2007) produced no perithecia, suggesting that autophagy is required for the differentiation of the fruiting body. Thus, autophagy is essential to fungal reproductive development in necrotrophic pathogens, although its exact role varies with the fungal species.
BcATG1 is required for the full virulence of B. cinerea on host plants
Genes affecting the virulence of B. cinerea have been functionally identified from those mediating secretion, penetration and host defence (Nakajima and Akutsu, 2014). Similar to other plant‐pathogenic fungi (Pollack et al., 2009; Soanes et al., 2012), which require autophagy for infection, blocking of autophagy in B. cinerea also led to a significantly reduced virulence on different host plants. Moreover, most conidia of the BcATG1 mutant lost the ability to form infection structures, thus almost losing the capacity to penetrate the host cuticle. These results further confirm that autophagy is involved in the pathogenicity of B. cinerea.
BcATG1 participates in lipid metabolism
Autophagy is believed to be associated with changes in cellular architecture during differentiation and development, presumably via its role in organic material recycling (Levine and Klionsky, 2004). Recent research has shown that lipid droplets serve as a substrate for macroautophagy, and the ability to undergo autophagy in response to changes in nutrient supply allows the cell to alter lipid droplet metabolism to meet cellular energy demands (Dong and Czaja, 2011). In M. oryzae, the accumulated content of lipid droplets in conidia is affected by autophagy, and lipid droplets in conidia are much fewer in the MoATG1 mutant (Liu et al., 2007). In this study, deletion of BcATG1 dramatically reduced the lipid droplets in conidia, which was also consistent with the results in the MrATG8 mutant of Metarhizum robertsii (Duan et al., 2013) and the MoATG1 mutant (Liu et al., 2007).
Deletion of BcATG1 leads to dramatically increased glycerol accumulation in the mycelia of B. cinerea
In M. oryzae, blocking of autophagy resulted in a reduction in glycerol accumulation in the appressorium (Liu et al., 2012). A previous study has shown that an autophagic inhibitor potently stimulates lipolysis in adipocytes (Heckmann et al., 2013). In addition, glycerol prevents the triggering of autophagy induced by sucrose starvation in plant cells (Aubert et al., 1994). In the current study, however, deletion of BcATG1 led to the inhibition of autophagy and the augmentation of glycerol accumulation, which was totally different from that in appressoria when autophagy was blocked in M. oryzae. These results indicate that the effects of autophagy on glycerol accumulation may be different in necrotrophic fungi. We suspect that lipid metabolism is regulated by autophagy and thus influences the accumulation of glycerol. Further study is necessary to elucidate the relationship between autophagy, lipid metabolism and glycerol biosynthesis.
Experimental Procedures
Fungal strains, media and culture conditions
Botrytis cinerea strain B05.10, isolated from grape in California, USA, was used as a recipient strain for the transformation experiments. The wild‐type progenitor and its derived mutants were grown on PDA (200 g potato, 20 g glucose, 20 g agar and 1 L water) and MM (0.5 g KCl, 2 g NaNO3, 1 g K2HPO4, 0.5 g MgSO4.7H2O, 0.01 g FeSO4.7H2O, trace elements, 30 g glucose and 1 L water, pH 6.9) for the examination of colony morphology, conidiation and sclerotial formation. MM–C (MM without glucose) and MM–N were employed for autophagy induction. MM–N, employed for the evaluation of autophagosome accumulation, was amended with 2 mm PMSF (Sigma, St. Louis, MO, USA) to inhibit autophagosome degradation by hydrolysis. Yeast wild‐type strain BY4741 and ATG1 deletion mutant YJL180W were cultured in YPD (1% yeast extract, 2% peptone and 2% glucose) and SD–N (2% glucose and 0.17% yeast nitrogen base with neither amino acids nor ammonium sulfate).
Sequence analysis of BcATG1
BcATG1 (BC1G_05432) was originally identified by homology search of the B. cinerea genome database (http://www.broadinstitute.org/annotation/genome/botrytis_cinerea/) using the blastp algorithm with ATG1 from S. cerevisiae (Klionsky et al., 2003) as a query. To verify the existence and sizes of the introns, RNA was extracted from mycelia of the wild‐type strain B05.10 with the RNAsimple Total RNA Kit (Tiangen Biotech. Co., Beijing, China) and used for reverse transcription (RT) with the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China). RT‐PCR amplification of the cDNA was conducted with the YES2‐Bcatg1‐F and YES2‐Bcatg1‐R primer pair (Table S1). The resulting PCR product was purified, cloned and sequenced.
Complementation of the yeast ATG1 mutant
For functional complementation of the yeast ATG1‐defective mutant, the full‐length cDNA of BcATG1 was amplified from the total RNA of wild‐type strain B0510 with primers YES2‐Bcatg1‐F and YES2‐Bcatg1‐R (Table S1), and cloned into the KpnI and EcoRI sites of the yeast expression vector pYES2 (Invitrogen, V825‐20). The resulting vector, namely pYES2‐BcATG1, was introduced into the yeast ATG1 mutant using a small‐scale yeast transformation protocol (Invitrogen, V825‐20). The uracil prototrophic transformants were selected and verified by PCR. The complementation effect of the BcATG1 gene was tested in a starvation challenge, as described previously. (Liu et al., 2007). Briefly, the yeast cells were streaked on YPD plates and cultured for 2 days at 30 °C, and then moved to SD–N medium for 18 days at 30 °C.
Construction of BcATG1 deletion and complemented mutants
The BcATG1 deletion and complemented mutants were generated according to the previously described method (Duan et al., 2013) with some modifications. Briefly, 1.4‐kb upstream and 0.8‐kb downstream sequences of BcATG1 were amplified from the genome of the wild‐type strain B05.10 with the Bcatg1‐up‐F/Bcatg1‐up‐R and Bcatg1‐down‐F/Bcatg1‐down‐R primer pairs (Table S1, Fig. S2), respectively. The HPH cassette containing the Aspergillus nidulans trpC promoter (Mullins et al., 2001) was amplified using the HPH‐F/HPH‐R primer pair. Then, the upstream, downstream and HPH cassette sequences were fused using the Bcatg1‐N‐F/Bctg1‐N‐R primer pair (Yu et al., 2004) (Table S1, Fig. S2). The resulting PCR product was purified and used to transform protoplasts of the B05.10 strain. Protoplasts were prepared and transformed using the previously described method (Gronover et al., 2001).
The BcATG1 deletion mutant was complemented with the full‐length BcATG1 gene. The full‐length Bcatg1 gene containing the 630‐bp upstream and 372‐bp terminator regions was amplified from the genome of the wild‐type strain B05.10 with the Bcatg1‐com‐F/Bcatg1‐com‐R primer pair (Table S1, Fig. S2). The PCR product was cloned into the SmaI and PstI sites of pNEO and used for protoplast transformation (Duan et al., 2013).
Nucleic acid manipulations
Fungal genomic DNA was isolated as described previously (McDonald and Martinez, 1990). Plasmid DNA was isolated using a Plasmid Mini Kit I (OMEGA bio‐tek Co., Shanghai, China). Southern blot hybridization analysis of the BcATG1 transformants was performed using a 1098‐bp downstream fragment of BcATG1 as a probe. The probe was labelled with digoxigenin (DIG) using the High Prime DNA Labelling and Detection Starter Kit II, according to the protocol of the manufacturer (Roche Diagnostics, Mannheim, Germany). DNA extracted from B. cinerea was digested with NcoI and used for Southern hybridization analysis.
Expression analysis of BcATG1 under nitrogen or carbon starvation stress
The expression levels of the BcATG1 gene in the wild‐type strain B05.10 were measured by a qRT‐PCR method. Briefly, the strain was cultured in PDB at 25 °C for 2 days in a 180‐rpm shaker. After incubation in MM–C or MM–N for 1, 2, 4, 8 or 10 h, mycelia of each treatment were harvested and ground in liquid nitrogen. RNA extraction and reverse transcription were performed using the protocol described previously (Yan et al., 2010). The real‐time PCR amplifications were performed as described in Ren et al (2015). The expression of the measured genes in each sample was normalized to actin gene expression, and relative changes in gene expression levels were analysed with ABI 7500 SDS software (Applied Biosystems, Foster City, CA, USA), which automatically set the baseline (Zheng et al., 2014). Data from three biological replicates were used to calculate the mean and standard deviation.
Analysis of autophagy by TEM observation and MDC staining
The analysis of autophagy by TEM was performed as described in Liu et al. (2007). Briefly, the conidia of each strain were cultured in PDB for 24 h at 25 °C in a 180‐rpm shaker. The young mycelia were collected, thoroughly washed in distilled water, transferred to MM–N medium with 2 mm PMSF and incubated at 25 °C for 6 h in a 180‐rpm shaker. The fungal mass was then collected and fixed overnight at 4 °C in fixative containing 2% paraformaldehyde and 2.5% (v/v) glutaraldehyde in 0.1 m phosphate buffer (pH 7.2). The fixed samples were washed three times, for 10 min each time, with 0.1 m phosphate buffer (pH 7.2). The samples were post‐fixed in 1% OsO4 for 2 h at 25 °C, washed three times with phosphate buffer as before, dehydrated in a graded ethanol series, embedded in resin and stained with 2% uranyl acetate and Reynold's lead solution before sectioning. The ultrathin sections were examined under a JEM‐1230 electron microscope (JEOL, Tokyo, Japan) operating at 70 kV.
For the analysis of autophagy by MDC staining, the conidia of each strain were added to 10 mL PDB. They were incubated at 25 °C and 180 rpm for 24 h, washed with sterile water and transferred into MM–N in the presence of 2 mm PMSF for 6 h, prior to staining with MDC at a final concentration of 50 mm for 30 min in the dark. After washing with water, samples were observed under differential interference contrast (DIC) and epifluorescence microscopy using a 4',6‐diamidino‐2‐phenylindole (DAPI) filter.
Conidiation and sclerotial formation assays
For conidiation assays, a mycelial plug (5 mm in diameter), taken from the periphery of a 3‐day‐old colony, was inoculated on a PDA plate. After the plates had been cultured at 25 °C with a 12‐h photophase for 7 days, conidia taken from three mycelial discs were suspended in sterile water and counted with a haemocytometer under a microscope.
For sclerotial formation assays, a mycelial plug (5 mm in diameter), taken from the periphery of a 3‐day‐old colony, was inoculated on a PDA plate. After the plates had been incubated at 25 °C for 4 weeks in the dark, the sclerotia formed on each plate were examined. All the experiments were repeated three times with three replicates each time, independently.
Assays for infection‐related morphogenesis, penetration and pathogenicity
For infection cushion formation assays, conidia of each strain were collected from 7‐day‐old PDA plates and resuspended with sterile water to a concentration of 1 × 105 conidia/mL. Sterile cellophane paper was placed on PDA medium, inoculated with 10 μL of conidial suspension and kept in a moist chamber at 25 °C for 8 h. The infection structures were observed at 40× magnification using an Olympus IX–71 microscope (Tokyo, Japan).
For penetration assays, onion epidermis, washed with H2O and incubated at 70 °C for 1 h in a humid chamber to kill the living cells prior to inoculation, was placed on glass slides. The onion epidermis was inoculated with 10 μL of conidial suspension (1 × 105 conidia/mL) and kept in a moist chamber at 25 °C for 10 h. The infection behaviour was observed under a microscope after staining with lactophenol blue.
For pathogenicity assays on host plant leaves, mycelial plugs taken from the periphery of a 3‐day‐old colony were used to inoculate primary leaves of host plants. Before inoculation, leaves were wounded with a sterilized needle to facilitate the penetration of the fungus into plant tissue. For the pathogenicity assays on fruits, apple, grape, tomato and onion were harvested at the commercial maturity stage, and were wounded with a sterilized needle before inoculation. Inoculated leaves and fruits were incubated in high relative humidity (about 95%) at 25 °C with 16 h of daylight. The diameters of disease lesions were recorded after the indicated times in the figures. The experiment was repeated three times.
Histochemical analysis of lipid droplets
After incubation on PDA for 7 days, conidia of each strain were harvested and stained directly with Nile red solution consisting of 20 mg/mL polyvinylpyrrolidone and 2.5 μg/mL Nile red oxazone (9‐diethylamino‐5H‐benzo[a]phenoxazine‐5‐one, Sigma, St. Louis, MO, USA) in 50 mm Tris‐maleate buffer (pH 7.5) (Weber et al., 1999). Within a few seconds in the dark, lipid droplets in conidia began to fluoresce when viewed under an episcopic fluorescence microscope. The experiment was repeated independently three times.
Determination of intracellular glycerol accumulation
Each strain was grown in PDB for 3 days at 25 °C in a 180‐rpm shaker. Mycelia of each strain were harvested and ground in liquid nitrogen. Then, mycelial powder (50 mg) was transferred to a 2‐mL microcentrifuge tube containing 0.1 mL of glycerol extraction buffer (Applygen Technologies Inc., Beijing, China). After mixing by a vortex shaker three times for 30 s each, the tubes were centrifuged at 5000 g for 10 min. The resulting supernatant was transferred to a new tube, and 10 μL of each supernatant was mixed with 190 μL detection buffer of a glycerol assay kit (Applygen Technologies Inc.). After the mixture had been incubated at 37 °C for 15 min, the glycerol concentration was determined by a SpectraMax M5 microplate reader (Molecular Devices Inc., Sunnyvale, CA, USA) at 550 nm. The experiment was repeated three times.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Oligonucleotide primers used in this study.
Fig. S1 Sequence alignment of serine‐threonine kinase domains from the predicted autophagy‐related BcATG1, MgATG1 of Magnaporthe oryzae (MGG_06393), FgATG1 of Fusarium graminearum (FGSG_05547), AoATG1 of Aspergillus oryzae (AOR_1_1116144) and ScATG1 of Saccharomyces cerevisiae (YGL180W). The ‘overline’ indicates the serine‐threonine kinase domain and asterisks indicate ATP‐binding sites.
Fig. S2 Generation and identification of the BcATG1 deletion mutant. (A) Gene deletion strategy of BcATG1. Primer (codes 3–8) binding sites are indicated by arrows (see Table S1 for the primer sequences). (B) Identification of BcATG1 deletion and complemented mutants by polymerase chain reaction (PCR) analyses with the primer pair 9 and 10. (C) Southern blot hybridization analysis for the identification of the BcATG1 deletion mutant and complemented strain using the upstream fragment of BcATG1 as a probe. Genomic DNA preparations from the wild‐type strain B0510, the mutant ΔBcatg1 and the complemented strain ΔBcatg1‐C were digested with NcoI.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31171880 and 31201543), Special Fund for Agro‐scientific Research in the Public Interest (201303023 and 201303025) and the Agricultural Science and Technology Innovation Fund Project of Jiangsu Province, China (CX (14) 2054).
References
- Amselem, J. , Cuomo, C.A. , van Kan, J.A.L. , Viaud, M. , Benito, E.P. , Couloux, A. , Coutinho, P.M. , de Vries, R.P. , Dyer, P.S. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K.M. , Pradier, J.M. , Quévillon, E. , Sharon, A. , Simon, A. , ten Have, A. , Tudzynski, B. , Tudzynski, P. , Wincker, P. , Andrew, M. , Anthouard, V. , Beever, R.E. , Beffa, R. , Benoit, I. , Bouzid, O. , Brault, B. , Chen, Z. , Choquer, M. , Collémare, J. , Cotton, P. , Danchin, E.G. , Da Silva, C. , Gautier, A. , Giraud, C. , Giraud, T. , Gonzalez, C. , Grossetete, S. , Güldener, U. , Henrissat, B. , Howlett, B.J. , Kodira, C. , Kretschmer, M. , Lappartient, A. , Leroch, M. , Levis, C. , Mauceli, E. , Neuvéglise, C. , Oeser, B. , Pearson, M. , Poulain, J. , Poussereau, N. , Quesneville, H. , Rascle, C. , Schumacher, J. , Ségurens, B. , Sexton, A. , Silva, E. , Sirven, C. , Soanes, D.M. , Talbot, N.J. , Templeton, M. , Yandava, C. , Yarden, O. , Zeng, Q. , Rollins, J.A. , Lebrun, M.H. and Dickman, M. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, S. , Gout, E. , Bligny, R. and Douce, R. (1994) Multiple effects of glycerol on plant cell metabolism. J. Biol. Chem. 269, 21 420–21 427. [PubMed] [Google Scholar]
- Bartoszewska, M. , Kiel, J.A.K.W. , Bovenberg, R.A.L. , Veenhuis, M. and van der Klei, I.J. (2011) Autophagy deficiency promotes β‐lactam production in Penicillium chrysogenum . Appl. Environ. Microb. 77, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero, E. and Reggiori, F. (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae . Biochim. Biophys. Acta, 1793, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Chan, E.Y. and Tooze, S.A. (2009) Evolution of Atg1 function and regulation. Autophagy, 5, 758–765. [DOI] [PubMed] [Google Scholar]
- Chopra, A. (1984) Lipid metabolism in Fungi. Crit. Rev. Microb. 11, 209–271. [DOI] [PubMed] [Google Scholar]
- Corral‐Ramos, C. , Roca, M.G. , Di Pietro, A. , Roncero, M.I.G. and Ruiz‐Roldán, C. (2015) Autophagy contributes to regulation of nuclear dynamics during vegetative growth and hyphal fusion in Fusarium oxysporum . Autophagy, 11, 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. and Czaja, M.J. (2011) Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. 22, 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Z. , Chen, Y. , Huang, W. , Shang, Y. , Chen, P. and Wang, C. (2013) Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii . Autophagy, 9, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, N.L. , Rasmussen, C. , Roca, M.G. and Read, N.D. (2004) Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12, 135–141. [DOI] [PubMed] [Google Scholar]
- Gronover, C.S. , Kasulke, D. , Tudzynski, P. and Tudzynski, B. (2001) The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea . Mol. Plant–Microbe Interact. 14, 1293–1302. [DOI] [PubMed] [Google Scholar]
- He, M. , Kershaw, M.J. , Soanes, D.M. , Xia, Y. and Talbot, N.J. (2012) Infection‐associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non‐selective macro‐autophagy. PLoS One, 7, e33270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann, B.L. , Yang, X. , Zhang, X. and Liu, J. (2013) The autophagic inhibitor 3‐methyladenine potently stimulates PKA‐dependent lipolysis in adipocytes. Br. J. Pharmacol. 168, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, Y. , Funakoshi, T. , Shintani, T. , Nagano, K. , Ohsumi, M. and Ohsumi, Y. (2000) Tor‐mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw, M.J. and Talbot, N.J. (2009) Genome‐wide functional analysis reveals that infection‐associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA, 106, 15 967–15 972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuma, T. , Ohneda, M. , Arioka, M. and Kitamoto, K. (2006) Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae . Eukaryot. Cell, 5, 1328–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T. , Baba, M. , Ishihara, N. , Miyazawa, K. , Ohsumi, M. , Yoshimori, T. , NodaT. and OhsumiY. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in Yeast. J. Cell Biol, 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J. (2005) The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D.J. , Cregg, J.M. , Dunn Jr, W.A. , Emr, S.D. , Sakai, Y. , Sandoval, I.V. , Sibirny, A. , Subramani, S. , Thumm, M. , Veenhuis, M. and Ohsumi, Y. (2003) A unified nomenclature for yeast autophagy‐related genes. Dev. Cell, 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J. , Abdalla, F.C. , Abeliovich, H. , Abraham, R.T. , Acevedo‐Arozena, A. , Adeli, K. , et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy, 8, 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B. and Klionsky, D.J. (2004) Development by self‐digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell, 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Liu, X.‐H. , Lu, J.‐P. , Zhang, L. , Dong, B. , Min, H. and Lin, F.‐C. (2007) Involvement of a Magnaporthe oryzae serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell, 6, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.‐H. , Gao, H.‐M. , Xu, F. , Lu, J.‐P. , Devenish, R.J. and Lin, F.‐C. (2012) Autophagy vitalizes the pathogenicity of pathogenic fungi. Autophagy, 8, 1415–1425. [DOI] [PubMed] [Google Scholar]
- Matsuura, A. , Tsukada, M. , Wada, Y. and Ohsumi, Y. (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae . Gene, 192, 245–250. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A. and Martinez, J.P. (1990) Restriction fragment length polymorphisms in Septoria tritici occur at a high frequency. Curr. Genet. 17, 133–138. [Google Scholar]
- Mehrabi, R. , Zhao, X. , Kim, Y. and Xu, J.‐R. (2009) The cAMP signaling and MAP kinase pathways in plant pathogenic fungi In: Plant Relationships (Deising H., ed.), pp. 157–172. Berlin, Heidelberg: Springer. [Google Scholar]
- Mullins, E.D. , Chen, X. , Romaine, P. , Raina, R. , Geiser, D.M. and Kang, S. (2001) Agrobacterium‐mediated transformation of Fusarium oxysporum: An efficient tool for insertional mutagenesis and gene transfer. Phytopathology, 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Nakajima, M. and Akutsu, K. (2014) Virulence factors of Botrytis cinerea . J. Gen. Plant Pathol. 80, 15–23. [Google Scholar]
- Nitsche, B. , Burggraaf‐van Welzen, A.‐M. , Lamers, G. , Meyer, V. and Ram, A.J. (2013) Autophagy promotes survival in aging submerged cultures of the filamentous fungus Aspergillus niger . Appl. Microbiol. Biotechnol. 97, 8205–8218. [DOI] [PubMed] [Google Scholar]
- Pinan‐Lucarré, B. , Paoletti, M. , Dementhon, K. , Coulary‐Salin, B. and Clave, C. (2003) Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina . Mol. Microbiol. 47, 321–333. [DOI] [PubMed] [Google Scholar]
- Pinan‐Lucarré, B. , Balguerie, A. and Clavé, C. (2005) Accelerated cell death in Podospora autophagy mutants. Eukaryot. Cell, 4, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack, J.K. , Harris, S.D. and Marten, M.R. (2009) Autophagy in filamentous fungi. Fungal Genet. Biol, 46, 1–8. [DOI] [PubMed] [Google Scholar]
- Reggiori, F. and Klionsky, D.J. (2002) Autophagy in the eukaryotic cell. Eukaryot. Cell, 1, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, W. , Zhao, H. , Shao, W. , Ma, W. , Wang, J. , Zhou, M. and Chen, C. (2015) Identification of a novel phenamacril‐resistance‐related gene by the cDNA‐RAPD method in Fusarium asiaticum . Pest Manag. Sci. doi: 10.1002/ps.4186. [DOI] [PubMed] [Google Scholar]
- Rose, T.L. , Bonneau, L. , Der, C. , Marty‐Mazars, D. and Marty, F. (2006) Starvation‐induced expression of autophagy‐related genes in Arabidopsis. Biol. Cell, 98, 53–67. [DOI] [PubMed] [Google Scholar]
- Rosslenbroich, H.‐J. and Stuebler, D. (2000) Botrytis cinerea—history of chemical control and novel fungicides for its management. Crop Prot. 19, 557–561. [Google Scholar]
- Schumacher, J. , Kokkelink, L. , Huesmann, C. , Jimenez‐Teja, D. , Collado, I.G. , Barakat, R. , Tudzynski, P. and Tudzynski, B. (2008) The cAMP‐dependent signaling pathway and its role in conidial germination, growth, and virulence of the gray mold Botrytis cinerea . Mol. Plant–Microbe Interact. 21, 1443–1459. [DOI] [PubMed] [Google Scholar]
- Singh, R. , Kaushik, S. , Wang, Y. , Xiang, Y. , Novak, I. , Komatsu, M. , Tanaka, K. , Cuervo, A.M. and Czaja, M.J. (2009) Autophagy regulates lipid metabolism. Nature, 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes, D.M. , Chakrabarti, A. , Paszkiewicz, K.H. , Dawe, A.L. and Talbot, N.J. (2012) Genome‐wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae . PLoS Pathog, 8, e1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, M. , Bredschneider, M. and Thumm, M. (1997) AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae . J. Bacteriol. 179, 3875–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M. and Ohsumi, Y. (1993) Isolation and characterization of autophagy‐defective mutants of Saccharomyces cerevisiae . FEBS Lett. 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Tudzynski, B. and Gronover, C. (2007) Signalling in Botrytis cinerea In: Botrytis: Biology, Pathology and Control (Elad Y., Williamson B., Tudzynski P. and Delen N., eds.), pp. 85–97. Dordrecht: Springer. [Google Scholar]
- Voigt, O. and Pöggeler, S. (2013a) Autophagy genes Smatg8 and Smatg4 are required for fruiting‐body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora . Autophagy, 9, 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, O. and Pöggeler, S. (2013b) Self‐eating to grow and kill: autophagy in filamentous ascomycetes. Appl. Microbiol. Biotechnol. 97, 9277–9290. [DOI] [PubMed] [Google Scholar]
- Weber, R.W.S. , Wakley, G.E. and Pitt, D. (1999) Histochemical and ultrastructural characterization of vacuoles and spherosomes as components of the lytic system in hyphae of the fungus Botrytis cinerea . Histochem. J. 31, 293–301. [DOI] [PubMed] [Google Scholar]
- Williamson, B. , Tudzynski, B. , Tudzynski, P. and Van Kan, J.A.L. (2007) Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. [DOI] [PubMed] [Google Scholar]
- Yan, L. , Yang, Q. , Sundin, G.W. , Li, H. and Ma, Z. (2010) The mitogen‐activated protein kinase kinase BOS5 is involved in regulating vegetative differentiation and virulence in Botrytis cinerea . Fungal Genet. Biol. 47, 753–760. [DOI] [PubMed] [Google Scholar]
- Yanagisawa, S. , Kikuma, T. and Kitamoto, K. (2013) Functional analysis of Aoatg1 and detection of the Cvt pathway in Aspergillus oryzae . FEMS Microbiol. Lett. 338, 168–176. [DOI] [PubMed] [Google Scholar]
- Yang, Q. , Yu, F. , Yin, Y. and Ma, Z. (2013) Involvement of protein tyrosine phosphatases BcPtpA and BcPtpB in regulation of vegetative development, virulence and multi‐stress tolerance in Botrytis cinerea . PLoS One, 8, e61307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu, T. and Klionsky, D.J. (2005) Autophagy: molecular machinery for self‐eating. Cell Death Differ, 12, 1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.H. , Hamari, Z. , Han, K.H. , Seo, J.A. , Reyes‐Dominguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations infilamentous fungi. Fungal Genet. Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Mehrabi, R. and Xu, J.‐R. (2007) Mitogen‐activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell, 6, 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z. , Gao, T. , Zhang, Y. , Hou, Y. , Wang, J. and Zhou, M. (2014) FgFim, a key protein regulating resistance to the fungicide JS399‐19, asexual and sexual development, stress responses and virulence in Fusarium graminearum . Mol. Plant Pathol. 15, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1 Oligonucleotide primers used in this study.
Fig. S1 Sequence alignment of serine‐threonine kinase domains from the predicted autophagy‐related BcATG1, MgATG1 of Magnaporthe oryzae (MGG_06393), FgATG1 of Fusarium graminearum (FGSG_05547), AoATG1 of Aspergillus oryzae (AOR_1_1116144) and ScATG1 of Saccharomyces cerevisiae (YGL180W). The ‘overline’ indicates the serine‐threonine kinase domain and asterisks indicate ATP‐binding sites.
Fig. S2 Generation and identification of the BcATG1 deletion mutant. (A) Gene deletion strategy of BcATG1. Primer (codes 3–8) binding sites are indicated by arrows (see Table S1 for the primer sequences). (B) Identification of BcATG1 deletion and complemented mutants by polymerase chain reaction (PCR) analyses with the primer pair 9 and 10. (C) Southern blot hybridization analysis for the identification of the BcATG1 deletion mutant and complemented strain using the upstream fragment of BcATG1 as a probe. Genomic DNA preparations from the wild‐type strain B0510, the mutant ΔBcatg1 and the complemented strain ΔBcatg1‐C were digested with NcoI.


