Summary
Arabidopsis contains two proline dehydrogenase (ProDH) genes, ProDH1 and ProDH2, encoding for homologous and functional isoenzymes. Although ProDH1 has been studied extensively, especially under abiotic stress, ProDH2 has only started to be analysed in recent years. These genes display distinctive expression patterns and show weak transcriptional co‐regulation, but are both activated in pathogen‐infected tissues. We have demonstrated previously that Arabidopsis plants with silenced ProDH1/2 expression fail to trigger defences against the hemibiotrophic bacterial pathogen Pseudomonas syringae pv. tomato AvrRpm1 (Pst‐AvrRpm1), and that ProDH1 and ProDH2 are differentially regulated by salicylic acid (SA). In the current work, we used prodh1 and prodh2 single‐mutant plants to assess the particular contribution of each gene to resistance against Pst‐AvrRpm1 and the necrotrophic fungal pathogen Botrytis cinerea. In addition, we studied the sensitivity of ProDH1 and ProDH2 to the jasmonic acid (JA) defence pathway. We found that ProDH1 and ProDH2 are both necessary to achieve maximum resistance against Pst‐AvrRpm1 and B. cinerea. However, ProDH2 has a major effect on early restriction of B. cinerea growth. Interestingly, ProDH1 is up‐regulated by SA and JA, whereas ProDH2 is only activated by JA, and both genes display transcriptional inter‐regulation at basal and infection conditions. These studies provide the first evidence of the contribution of ProDH2 to disease resistance, and describe the differential regulation and non‐redundant but complementary function of both enzyme isoforms in infected tissues, providing support for a fundamental role of ProDH in the control of biotrophic and necrotrophic pathogens.
Keywords: HopX1, jasmonate, prodh1‐3 and prodh2‐2, proline dehydrogenase, Pst‐AvrRpm1, B. cinerea, salicylic acid
Introduction
Proline dehydrogenase (ProDH) catalyses the first and rate‐limiting step in the transformation of proline (Pro) into glutamic acid (Glu) that takes place at mitochondria. This enzyme oxidizes Pro into delta‐1‐pyrroline‐5‐carboxylate (P5C) using FAD as cofactor. Next, P5C is non‐enzymatically transformed into glutamate semialdehyde (GSA), which is used to generate Glu and NADH by P5C dehydrogenase (P5CDH). In higher plants, ProDH is bound to the inner mitochondrial membrane and is suspected to transfer electrons to ubiquinone in the mitochondrial electron transport chain (mETC) (Elthon and Stewart, 1981, Kiyosue et al., 1996; Liang et al., 2013).
Arabidopsis contains two genes encoding for ProDH: ProDH1 (At3g30775) and ProDH2 (At5g38710) (Funck et al., 2010; Kiyosue et al., 1996). Their products are highly homologous (75% identical), but differ at their N‐terminus and may have different subcellular localizations. Although ProDH1 and ProDH1‐GFP fusion proteins are exclusively detected at mitochondria, ProDH2‐GFP is found at mitochondria and plastids (Funck et al., 2010; Kiyosue et al., 1996; Van Aken et al., 2009). ProDH1, also called ERD5 (Early Responsive to Dehydration 5), was the first to be described and is considered to be the predominant functional isoform under most studied conditions (Kiyosue et al., 1996). ProDH2 has been recognized recently as an active enzyme, based on its ability to complement the Pro utilization defect of the Δput1 yeast strain, and the Pro hypersensitive phenotype of the prodh1‐1 Arabidopsis mutant (Funck et al., 2010). Attempts to generate transgenic plants fully silenced in ProDH1 and ProDH2 were unsuccessful (Cecchini et al., 2011a), but the first double‐mutant plant (prodh1‐4/prodh2‐2) has been described recently (Cabassa‐Hourton et al., 2016).
ProDH1 and ProDH2 genes have distinctive expression patterns and are controlled by different mechanisms. Although ProDH1 is expressed in most tissues and developmental stages with greater intensity in stigma and pollen, ProDH2 shows low expression, mostly at vascular tissues and senescent leaves (Funck et al., 2010; GENEVESTIGATOR, https://genevestigator.com). In addition, transcripts of both genes are detected in root tissues and abscission zones (Funck et al., 2010; GENEVESTIGATOR, https://genevestigator.com; Nakashima et al., 1998). The expression of ProDH1 is modulated by S1 bZIP transcription factors (bZIP1 and bZIP53; Dietrich et al., 2011) that heterodimerize with members of the C bZIP group (bZIP53/bZIP10; Weltmeier et al., 2006), whereas ProDH2 is a direct target of bZIP11 (Funck et al., 2010; Hanson et al., 2008). Both genes are repressed by sucrose and induced by exogenous Pro treatment, whereas ProDH1 is repressed and ProDH2 is induced by NaCl (Funck et al., 2010; Hanson et al., 2008; Satoh et al., 2004). Microarray analysis (GENEVESTIGATOR, https://genevestigator.com) shows poor co‐regulation of these genes, and recent studies have suggested that they play distinctive roles in plant development and stress adaptation (Funck et al., 2010). Apparently, this may also occur with the ProDH1 and ProDH2 orthologues in Nicotiana tabacum and Brassica napus (Bna). Recent studies have reported the expression of six BnaProDH1 and two BnaProDH2 genes, showing that ProDH1 is prevalent in pollen, ProDH2 in senescent leaves and both are active in roots, as in Arabidopsis (Faës et al., 2015; Ribarits et al., 2007).
In recent years, ProDH has been implicated in defences against pathogens. Arabidopsis plants show enhanced gene expression and enzyme activity on elicitation of the hypersensitive response (HR) by recognition of Pseudomonas syringae pv. tomato (Pst) AvrRpm1 (Pst‐AvrRpm1) (Cecchini et al., 2011a). ProDH1/2 genes are also induced in Nicotiana benthamiana plants establishing a non‐host interaction with Pst T1 (Senthil‐Kumar and Mysore, 2012). In both cases, silencing of ProDH genes compromises the accumulation of reactive oxygen species (ROS), the generation of cell death and the establishment of disease resistance (Cecchini et al., 2011a; Senthil‐Kumar and Mysore, 2012). In addition, ProDH activation is believed to sustain the oxidative burst and to reduce cell viability in other kingdoms (Cecchini et al., 2011b). However, the mechanism by which ProDH contributes to these responses is not understood. The consequences of enzyme activation have been analysed at the biochemical level for the Arabidopsis–Pst‐AvrRpm1 pathosystem. In this system, ProDH loses its coordination with P5CDH at the time of ROS increase, but this increases the level of the toxic metabolite P5C (Monteoliva et al., 2014). Subsequently, Pro synthesis is induced in these tissues, suggesting that ProDH triggers long‐term metabolic changes in HR (Rizzi et al., 2015).
Currently, there are many basic questions about the regulation and function of ProDH1 and ProDH2 during the activation of plant defence. In Arabidopsis and N. benthamiana, ProDH1 apparently displays a predominant role in cell death‐associated defences (Cecchini et al., 2011a; Senthil‐Kumar and Mysore, 2012). However, the role of ProDH2 in disease resistance has not been evaluated for any pathosystem. Moreover, the contribution of each isoform to resistance against pathogens with different lifestyles remains to be investigated. Defences against biotrophic and necrotrophic pathogens are mainly signalled by the salicylic acid (SA) and jasmonic acid (JA) pathways, respectively. These routes are inter‐regulated by several negative crosstalks, but may have synergistic effects on particular genes at low concentrations (Caarls et al., 2015; Mur et al., 2006). The canonical SA pathway includes the action of the isochorismate synthase SID2 enzyme (SA induction‐deficient 2), which is responsible for SA generation, as well as the SA receptor NPR1 (non‐expressor of Pathogenesis‐Related 1) (Wu et al., 2012). Two major components of the JA pathway are JAR1 (JA responsive 1), which generates the active hormone form JA‐isoleucine (JA‐Ile), and COI1 (coronatine insensitive 1), sensing JA‐Ile as part of the JA receptor (Katsir et al., 2008; Staswick and Tiryaki, 2004). Some pathogen effectors alter the JA pathway to benefit the invader. For instance, Pst DC3000 secretes the JA‐Ile analogue coronatine (COR) which down‐regulates SA‐dependent defences (Katsir et al., 2008). In the same way, Pseudomonas syringae pv. tabaci generates HopX1, which binds to and degrades jasmonate ZIM‐domain (JAZ) repressor proteins, therefore stimulating the JA route and, consequently, suppressing SA signalling (Gimenez‐Ibanez et al., 2014).
Previous observations have suggested that ProDH1 and ProDH2 genes could be differentially regulated in infected tissues (Cecchini et al., 2011a). Here, we evaluated the response of both genes to biotrophic and necrotrophic pathogens, and their contribution to disease resistance in the interaction of Arabidopsis with Pst‐AvrRpm1 and Botrytis cinerea B05.10. Our studies include the use of wild‐type, prodh1 and prodh2 single‐mutant plants to analyse pathogen growth, cytological defence markers and the regulation of ProDH genes at different infection conditions. We report a distinctive sensitivity of ProDH1 and ProDH2 to SA and JA, the requirement of both genes for resistance against both pathogens, and the predominant contribution of ProDH2 in the early control of B. cinerea infection. In addition, we provide evidence for the inter‐regulation between these genes at basal and infection conditions.
Results
Responses of prodh1 and prodh2 mutants to Pst‐AvrRpm1
Arabidopsis plants with silenced ProDH1/2 expression show enhanced sensitivity to Pst‐AvrRpm1 (Cecchini et al., 2011a). To assess how each gene individually contributes to this phenotype, we analysed pathogen growth in prodh1 and prodh2 single mutants. We used the previously characterized prodh1‐2 (SALK_081276; Col‐0), prodh1‐4 (SALK_119334; Col‐0) and prodh2‐1 (GT1788; Ler) alleles. In addition, we included the Col‐0 mutant plants prodh1‐3 (GABI_308F08) and prodh2‐2 (GABI_328G05). As the last two mutants have not been characterized at the phenotypic level, we first evaluated two traits of ProDH deficiency: Pro accumulation and hypersensitivity to exogenous Pro (Funck et al., 2010; Sharma and Verslues, 2010). Mutant and control plants were germinated on 0, 5 and 20 mm Pro and analysed at the age of 2 weeks. Both prodh mutants contained less healthy seedlings than control plants, and this was more pronounced for prodh1‐3 (Fig. 1a,b). In addition, prodh1‐3, but not prodh2‐2, increased its Pro content (Fig. 1c). This indicates that the new mutants display similar phenotypes to other prodh1 and prodh2 alleles, confirming that ProDH1 is primarily responsible for the protection of plants against exogenous Pro (Funck et al., 2010; Nanjo et al., 2003).
Figure 1.
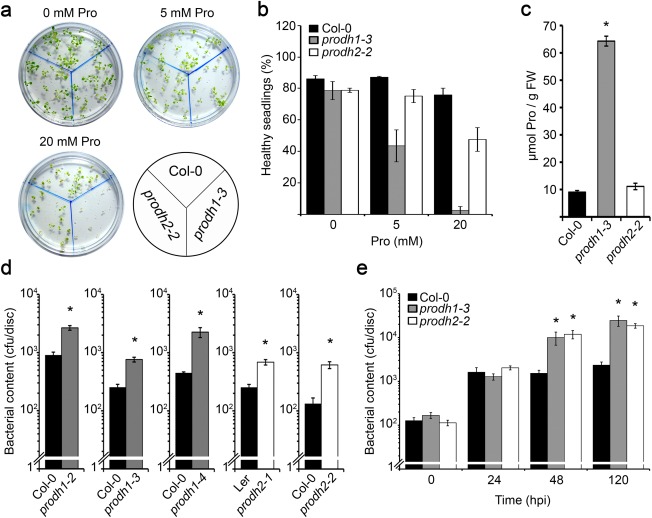
Responses of prodh1 and prodh2 mutants to exogenous proline (Pro) and Pseudomonas syringae pv. tomato AvrRpm1 (Pst‐AvrRpm1). (a) Col‐0, prodh1‐3 and prodh2‐2 seedlings were grown for 2 weeks in Gamborg's medium (GM) (1% sucrose) with 0, 5 or 20 mm Pro. (b) Percentage of healthy seedlings under the conditions described in (a). (c) Pro content in seedlings germinated in 5 mm Pro. FW, fresh weight. Values in (b) and (c) indicate mean ± standard error (SE) of two experiments. *Significant differences between mutant and wild‐type plants (P < 0.001; t‐test). (d) Pst‐AvrRpm1 content at 72 h post‐inoculation (hpi) in Col‐0, prodh1‐2, prodh1‐3, prodh1‐4, Ler, prodh2‐1 and prodh2‐2 plants. (e) Pst‐AvrRpm1 content at 24, 48 and 120 hpi in Col‐0, prodh1‐3 and prodh2‐2 plants. cfu, colony‐forming unit. Six leaf discs from at least three plants were used to determine bacterial content in each sample; one representative of three independent assays is shown (d, e). *Significant differences between mutant and wild‐type plants (P < 0.05; t‐test).
To test the involvement of ProDH1 and ProDH2 in resistance against Pst‐AvrRpm1, we quantified the bacterial content in the five prodh mutants. At late infection stage [72 h post‐inoculation (hpi)], all mutants contained higher pathogen titres than did wild‐type plants (three to eight times higher) (Fig. 1d). However, no significant differences were observed between prodh1 and prodh2 alleles. Next, we assessed Pst‐AvrRpm1 infection progression on one mutant of each type. prodh1‐3 and prodh2‐2 were selected for this purpose as they lack native transcripts (see below) and share the Col‐0 background, well characterized in the interaction with Pst. The Pst‐AvrRpm1 content was quantified at 24, 48 and 120 hpi (Fig. 1e). prodh1‐3 and prodh2‐2 contained similar pathogen concentrations at all the analysed stages and manifested their susceptibility by 48 hpi. Therefore, loss of either ProDH1 or ProDH2 reduces resistance to Pst‐AvrRpm1 in a similar manner, indicating that both isoforms help to generate defences against this hemibiotrophic pathogen.
Responses of prodh1‐3 and prodh2‐2 mutants to B. cinerea
Next, we analysed the susceptibility of prodh1‐3 and prodh2‐2 plants to B. cinerea. Conidial suspensions (103 or 104 conidia/mL) were deposited on the adaxial surface of the leaves (three spots per side) and samples were taken before (24 and 30 hpi) and after (48 hpi) the observation of necrotic lesions. Fungal hyphae were detected by trypan blue staining of infected leaves (Fabro et al., 2008) (Fig. 2a), and mycelial development was determined by the quantification of stained areas (ImageJ program) in these samples (Fig. 2b). At 24 hpi, prodh2‐2 showed greater mycelium expansion than prodh1‐3 or control plants (Fig. 2a,b). Later (30 hpi), hyphal growth was higher in both mutants than in control plants, with a tendency of prodh2‐2 to be more susceptible than prodh1‐3 (Fig. 2b). At 48 hpi, necrotic lesions were manifested in all plants without significant differences among genotypes (Fig. 2a, bottom).
Figure 2.
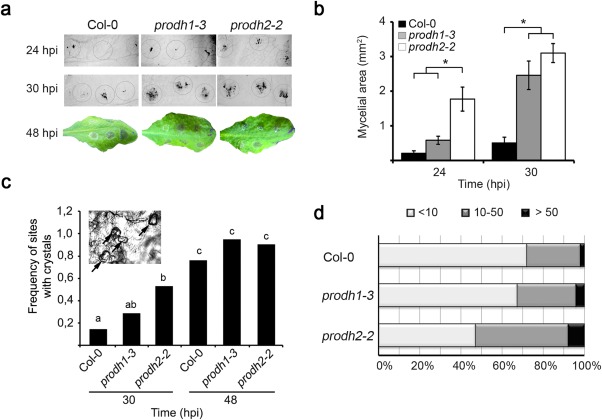
Growth of Botrytis cinerea in prodh1‐3 and prodh2‐2 mutants. (a) Leaves inoculated at different points (circles) with conidial suspension (5 μL, 104/mL) were stained with trypan blue to detect fungal mycelium at 24 or 30 h post‐inoculation (hpi). Macroscopic lesions developed at 48 hpi with 103 (top half) or 104 (botton half) conidia/mL. (b) Leaf area occupied by mycelia at 24 and 30 hpi with 104 conidia/mL (ImageJ program) based on images obtained as in (a). In (a) and (b), one representative of four independent experiments is shown (nine spots from three leaves from three plants were analysed in each case). Values indicate mean ± standard error. *Significant differences between plants (P < 0.05; t‐test). (c) Frequency of infection sites containing at least one crystal at 30 and 48 hpi in leaves treated with 103 conidia/mL. Different letters indicate significant differences among samples (P < 0.05; chi‐squared test). One representative of four independent assays is shown. (d) Number of crystals per site at 48 hpi with 104 conidia/mL. Intervals with less than 10, 10–50 or 50–100 crystals are shown, based on the analysis of four independent experiments (18–21 spots from seven leaves from at least three plants per sample). prodh2‐2 values are significantly different from those of the other two plants (P < 0.05; chi‐squared test).
Several necrotrophic fungal pathogens secret oxalic acid (OA) during plant invasion, and this appears to be critical for B. cinerea infection, as strains unable to produce OA cannot colonize Arabidopsis (Kunz et al., 2006; Prins et al., 2000). The amount of OA crystals increases during infection and is indicative of disease progression (Prins et al., 2000; Uloth et al., 2015). Thus, we used this marker to further evaluate the response of ProDH‐deficient plants to this fungus. Crystals (Fig. 2c, inset) were quantified at 30 and 48 hpi in wild‐type and mutant plants, and differences were detected among genotypes. At 30 hpi, the frequency of sites containing at least one crystal was higher for prodh2‐2 (9/17, 5/17 and 2/17 sites for prodh2‐2, prodh1‐3 and Col‐0, respectively) (Fig. 2c). At 48 hpi, most sites contained at least one crystal in all plants (19/21, 20/21 and 16/21 sites for prodh2‐2, prodh1‐3 and Col‐0, respectively) (Fig. 2c), but the number of crystals per site was also higher for prodh2‐2. Under the latter condition, sites with more than 10 crystals represented 53% of the total in prodh2‐2, and 28% or 32% of the total in wild‐type and prodh1‐3, respectively (Fig. 2d). Thus, although ProDH1 and ProDH2 are both required for full resistance against B. cinerea, ProDH2 shows a predominant role in the early control of fungal infection.
Regulation of ProDH1 and ProDH2 genes by SA and JA
We have shown that ProDH1 and ProDH2 are induced by Pst‐AvrRpm1 infection, but only the former is activated by exogenous SA (Cecchini et al., 2011a). To learn more about the response of these genes to biotic stresses, we analysed their sensitivity to other pathogens and hormones. To set up these experiments, we took into account that ProDH1 and ProDH2 have different expression levels (Fig. S1a, see Supporting Information) and regulation by light (Fig. S1b). Therefore, treatments were started 2 h after light phase onset and samples were taken 6, 24 or 48 h later in order to avoid the major effects of light (Fig. S1b).
Initially, we analysed the behaviour of ProDH2. The finding of SA‐sensitive elements in its promoter region (Table S1, see Supporting Information) prompted us to test whether the SA pathway mediates its induction on interaction with Pst‐AvrRpm1 (Cecchini et al., 2011a). With this purpose, the pathogen was inoculated in plants lacking SID2 or NPR1. Neither sid2‐2 nor npr1‐1 plants reduced ProDH2 activation by Pst‐AvrRpm1 (Fig. 3a), indicating that the canonical SA route is dispensable for this response. Arabidopsis also accumulates JA in tissues that are infected with avirulent Pst (De Vos et al., 2005). In addition, the ProDH2 promoter also contained JA‐responsive elements (Table S1). Therefore, we assessed whether ProDH2 may respond to the JA route. First, we analysed the effect of exogenous methyl jasmonate (meJA) (0.84 µm). Wild‐type plants (Col‐0, Col‐5) induced ProDH2 expression in response to this treatment (Fig. 3b). In contrast, jar1‐1 (Col‐0) mutants strongly reduced such a response (Fig. 3b, top). In turn, coi1‐1 plants induced the gene at lower level than control (Col‐5) plants (Fig. 3b). Therefore, in wild‐type plants, ProDH2 seems to be up‐regulated by JA‐Ile partially via COI1, but also through another pathway.
Figure 3.
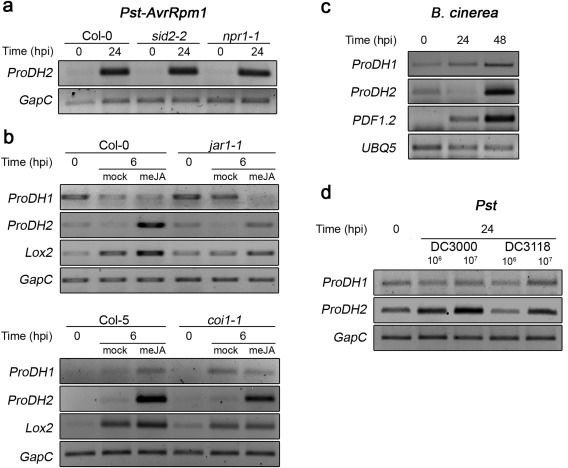
Sensitivity of ProDH1 and ProDH2 to the salicylic acid (SA) and jasmonic acid (JA) pathways. (a) ProDH2 expression in Col‐0, sid2‐2 and npr1‐1 plants at 0 and 24 h post‐inoculation (hpi) with Pseudomonas syringae pv. tomato (Pst)‐AvrRpm1. (b) ProDH1 and ProDH2 transcripts in Col‐0, jar1‐1, Col‐5 and coi1‐1 plants exposed to methyl jasmonate (meJA) (0.84 µm) or mock solution at 0 and 6 h post‐treatment (hpt). Lox2 was used as a JA‐responsive gene. (c) ProDH1 and ProDH2 expression in Col‐0 plants infected with Botrytis cinerea (six spots of 5 µL, 105 conidia/mL) at 0, 24 and 48 hpi. PDF1.2 was used as a marker of the JA pathway. (d) ProDH1 and ProDH2 transcripts in Col‐0 plants infected with Pst DC3000 or coronatine (COR)‐deficient Pst DC3118 [106 or 107 colony‐forming units (cfu)/mL] at 24 hpi. Gene expression was determined by semi‐quantitative polymerase chain reaction using GapC or UBQ5 as endogenous controls, under the conditions described in Table S2 (see Supporting Information). One representative of three biological replicates is shown.
To gain an insight into the regulation of ProDH2 by endogenous activation of the JA pathway, we evaluated its expression under infection conditions that increased the hormone levels. Plants challenged with B. cinerea accumulate high levels of JA (Liu et al., 2015). Interestingly, this treatment induced ProDH2, but triggered the maximum expression of the JA‐responsive gene marker PDF1.2 (Fig. 3c). In addition, we took advantage of the fact that Pst DC3000 synthesizes the JA‐Ile analogue COR and that the Pst DC3118 strain is deficient in this capacity (Katsir et al., 2008). Thus, we compared the responses of both strains to test the effect of this JA‐Ile analogue on ProDH2 expression. Pst DC3118 produced lower gene induction than Pst DC3000, and this was reproduced at different pathogen concentrations (Fig. 3d), indicating that COR may activate ProDH2 on infection with Pst DC3000.
Next, we analysed the sensitivity of ProDH1 to SA and JA. Responsive elements for both hormones were detected in the promoter of this gene (Table S1). As described, the SA pathway mediates early (6 hpi), but not late (24 hpi), ProDH1 activation on interaction with Pst‐AvrRpm1 (Cecchini et al., 2011a). Thus, we tested whether the latter response involved the JA pathway. ProDH1 expression was evaluated at 1 day post‐infection in control and jar1‐1 mutant tissues. jar1‐1 plants did not activate the gene under this condition (Fig. 4a), implicating JA signalling in ProDH1 regulation. JA‐Ile triggers ProDH1 induction in wild‐type tissues infected with Pst‐AvrRpm1. This was certainly surprising, as ProDH1 did not respond to exogenous meJA (Fig. 3b).
Figure 4.
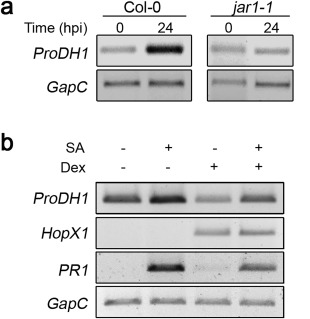
Regulation of ProDH1 expression by JAR1 and HopX1. (a) ProDH1 transcripts in Col‐0 and jar1‐1 plants infected with Pseudomonas syringae pv. tomato (Pst)‐AvrRpm1 at 24 h post‐inoculation (hpi). (b) Effect of HopX1 on ProDH1 induction by salicylic acid (SA). Transgenic plants Dex:HopX1 were initially exposed to dimethylsulfoxide (DMSO) (–) or dexamethasone (Dex) (5 µm) for 24 h, and then treated with H2O (–) or SA (1 mm) for 24 h. PR1 was used as SA‐sensitive gene and GapC as housekeeping gene. One representative from three independent assays is shown. Semi‐quantitative polymerase chain reaction conditions are described in Table S2 (see Supporting Information).
ProDH1 activation can be triggered by either SA (Cecchini et al., 2011b) or JA (Fig. 4a) pathways, suggesting that the balance between the two routes may affect the gene expression levels. To investigate this, we used transgenic plants expressing the bacterial effector HopX1 under the control of dexamethasone (Dex). As expected (Gimenez‐Ibanez et al., 2014), HopX1 suppresses SA signalling as it reduces the capacity of SA to induce the PR1 gene marker (Fig. 4b). Interestingly, HopX1 also reduces the induction of ProDH1 by SA. Therefore, non‐infected tissues expressing HopX1 override the ProDH1 activation by SA (Fig. 4b). Taken together, these results suggest that JA displays different effects on ProDH1 expression depending on whether it is exogenously applied as meJA or is generated by infected tissues. This suggests that JA does not act alone in the induction of ProDH1 by Pst‐AvrRpm1.
Inter‐regulation between ProDH1 and ProDH2
Finally, we evaluated whether ProDH1 and ProDH2 display some type of inter‐regulation. For this, we quantified gene expression in single‐mutant plants. As shown in Fig. 5, we confirmed that prodh1‐3 and prodh2‐2 are null mutants, as no transcripts were detected at basal or infection conditions for the mutant genes. In uninfected tissues, ProDH1 showed higher activation in prodh2‐2 than in control plants (4.5‐fold difference), whereas ProDH2 showed similar expression in prodh1‐3 and wild‐type plants (Fig. 5a). Thus, ProDH1 expression seems to be sensitive to ProDH2 deficiency, but not vice versa. Then, we analysed Pst‐AvrRpm1‐ and B. cinerea‐infected tissues using untreated samples as control. After bacterial infection, ProDH1 induction was marginally affected by ProDH2 deficiency, as its transcripts were increased 960 and 1200 times in prodh2‐2 and Col‐0 plants, respectively (Fig. 5b). In contrast, bacterial‐mediated ProDH2 activation was strongly reduced in prodh1‐3 plants (800‐ and 1800‐fold increase in prodh1‐3 and control plants, respectively) (Fig. 5b). Botrytis cinerea triggered lower ProDH1 and ProDH2 induction than did Pst‐AvrRpm1 treatment. However, ProDH genes displayed similar responses under both infection conditions as fungal infection generated similar ProDH1 activation in wild‐type and prodh2‐2 plants (7.9‐ and 8.6‐fold increase, respectively), and lower ProDH2 activation in prodh1‐3 plants (16.8‐ and 5.4‐fold increase in control and mutant, respectively) (Fig. 5c). Therefore, regardless of the gene induction level achieved under these infection conditions, in both cases, the absence of ProDH1 reduces ProDH2 activation, but not vice versa. This set of results reveals the inter‐regulation of the two genes, indicating that, although hyper‐activation of ProDH1 partially compensates for the absence of ProDH2 at basal conditions, such compensation is not observed in infected tissues, where ProDH1 deficiency limits ProDH2 activation.
Figure 5.
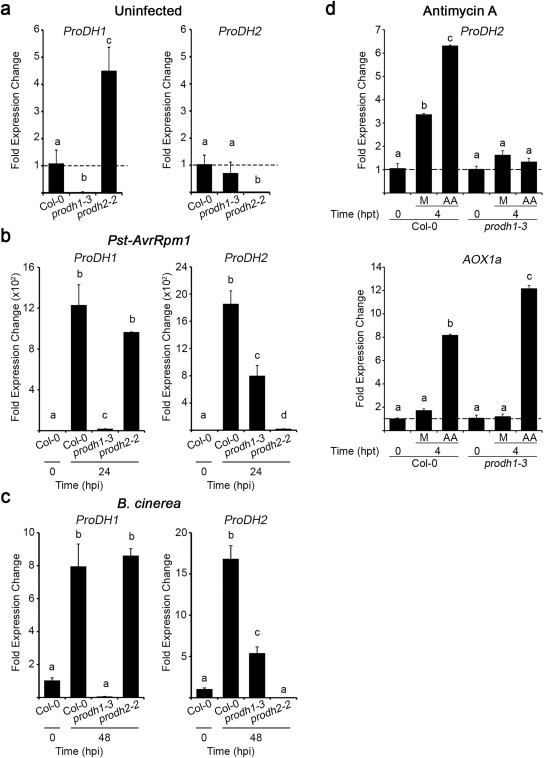
Inter‐regulation of ProDH1 and ProDH2 expression. Transcripts were quantified in Col‐0, prodh1‐3 and prodh2‐2 leaves at basal (uninfected) conditions (a), at 24 h post‐inoculation (hpi) with Pseudomonas syringae pv. tomato (Pst)‐AvrRpm1 [107 colony‐forming units (cfu)/mL] (b) or 48 hpi with Botrytis cinerea (six spots of 5 µL with 105 conidia/mL per leaf) (c). ProDH2 and AOX1a (alternative oxidase) expression in Col‐0 and prodh1‐3 tissues treated with antimycin A (AA, 10 µm) for 4 h (hpt, h post‐treatment) (d). Gene expression levels were determined by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR), applying the ΔΔCt method relative to each gene transcript level in Col‐0 at 0 hpi (a–c) or Col‐0 or prodh1‐3 at 0 hpi (d). Bars represent average ± standard deviation of three replicates. UBQ5 was used as a housekeeping gene. In each case, one representative of three independent experiments is shown. Different letters indicate significant differences among samples (P < 0.05 in a–c and P < 0.01 in d; t‐test).
Finally, we tested whether ProDH1 affects ProDH2 expression after the mitochondrial ROS burst. For this purpose, we treated wild‐type and prodh1‐3 leaf tissues with antimycin A (AA), an inhibitor of mETC that increases mitochondrial ROS levels (Fabro et al., 2016). ProDH2 was induced by AA in wild‐type, but not mutant, plants, revealing an inter‐regulation of these genes that is sensitive to ROS signalling (Fig. 5d). Mock treatments produced a lower ProDH2 activation, which was also dependent on ProDH1, suggesting that mechanical stress and/or light phase conditions affect this response.
Discussion
This work evaluates the contribution of ProDH1 and ProDH2 to defences against adapted biotrophic and necrotrophic pathogens in Arabidopsis. Both enzyme isoforms were found to be necessary to establish full resistance against Pst‐AvrRpm1 (Fig. 1d,e). This was expected for ProDH1, whose transcriptional activation requires SID2 and NPR1, which signal resistance against this biotrophic pathogen (Cecchini et al., 2011a). In contrast, ProDH2 induction does not involve this pathway (Fig. 3a), but still supports plant immunity, as prodh2 plants show enhanced susceptibility to the bacteria (Fig. 1d,e). As discussed below, ProDH2 is up‐regulated by JA. Similarly, other genes associated with defences against biotrophic pathogens are induced by COR. These include anthocyanin, phenylpropanoid, terpenoid and shikimate synthesis genes (CSH, PAL1, DHS1, etc.), as well as ELI3 (At4g37990), considered to be a marker of RPM1‐dependent resistance (Thilmony et al., 2006). Another example is the activation of JA gene markers occurring in the cpr22 (Yoshioka et al., 2001) and hrl1 (Devadas et al., 2002) mutants, which show enhanced resistance to biotrophic agents. Furthermore, in the interaction with avirulent Pst, SA and JA increase (De Vos et al., 2005), but SA signalling cannot completely suppress the JA pathway (Spoel et al., 2007). Thus, activation of ProDH2 by the JA route may persist under this condition.
The ProDH1 and ProDH2 genes show different expression levels and sensitivity to light, although their differences are small in adult leaves at the light phase stages analysed here (Fig. S1). ProDH2 maintains lower expression than ProDH1 in most conditions, but reaches maximal induction in infected tissues (GENEVESTIGATOR). These genes display distinctive responses under biotic stress. ProDH2 is up‐regulated by exogenous meJA (Fig. 3b) and COR derived from Pst DC3000 (Fig. 3d), consistent with transcriptomic analysis data of ProDH and other genes associated with amino acid metabolism (Thilmony et al., 2006). Curiously, coi1‐1 plants did not lose the ability to induce ProDH2 by meJA treatment (Fig. 3b), indicating that a COI1‐independent pathway signals gene activation. Indeed, nearly 26% of meJA‐responsive genes maintain their regulation in the absence of COI1 (Devoto et al., 2005), and components of COI1 alternative pathways have begun to be studied (Geng et al., 2014), and so the routes leading to ProDH2 induction in infected tissues could be identified soon.
ProDH1 was up‐regulated by exogenous SA, requiring SID2 and NPR1 for early stimulation by Pst‐AvrRpm1 (Cecchini et al., 2011a). Curiously, this gene is not induced by meJA (Fig. 3b), but depends on JAR1 for late activation by Pst‐AvrRpm1 (Fig. 4a). Thus, unlike ProDH2, ProDH1 is sensitive to SA and JA, and is apparently affected by the balance between these hormones, as its activation by SA is reduced by HopX1 (Fig. 4b). JA induces ProDH1 in Pst‐AvrRpm1 tissues that accumulate SA, but suppresses its up‐regulation by SA in uninfected tissues expressing HopX1. In addition, ProDH1 is induced by B. cinerea infection (Fig. 5c), where the contents of SA and JA are increased (Liu et al., 2015). Therefore, variations in SA/JA levels or hormone combinations might differently affect ProDH1 expression, as suggested for other genes sensitive to SA and JA (Mur et al., 2006).
This is the first report of the contribution of ProDH to resistance against necrotrophic pathogens. The B. cinerea strain used here is adapted to suppress host defences. Nevertheless, the prodh mutants displayed enhanced susceptibility to the fungus. At early infection stages, mycelial expansion was faster in prodh2‐2 than in wild‐type plants, and this was also observed, albeit less noticeable, in the prodh1‐3 mutant. Therefore, ProDH and, mostly, ProDH2 seem to act early, probably counteracting fungal germination, penetration, hyphal development or other initial infection events. We did not evaluate whether the enzyme also strengthen late defences, as this should be tested with a less virulent fungal strain. Under the analysed conditions, lack of ProDH had no obvious effects on the development of necrotic lesions.
A key finding of this study was that ProDH1 and ProDH2 provided resistance against pathogens with different lifestyles, suggesting their effect on a primary or fundamental process required to cope with infection. Consistently, both isoforms contributed to generate the oxidative burst after the perception of flagellin (Fabro et al., 2016). Similarly, in animal cells, ProDH plays protective roles in different adverse conditions, such as genotoxic processes, inflammation or metabolic stress (Phang and Liu, 2011).
ProDH is associated with the inner mitochondrial membrane and has the capacity to charge electrons into the respiratory chain at ubiquinone. It is expected that this enzyme affects respiration, energy production and the redox balance (Hancock et al., 2015; Schertl and Braun, 2014). Its coordination with P5CDH yields 30 moles of ATP per mole of Pro. Its coupling with P5CR has been suggested to activate the Pro/P5C cycle, which may either increase mitochondrial ROS or enhance reducing power at mitochondria (Ben Rejeb et al., 2014). However, there is no conclusive evidence that this cycle operates in plants at present. Currently, the exact consequences of ProDH activation in different infection conditions are unknown. The same applies for other mitochondrial enzymes controlling basic cellular functions that support defences against biotrophs and necrotrophs. For instance, mutations impairing the activity of mitochondrial succinate dehydrogenase (complex II) slow respiration and reduce ROS, weakening resistance against Pst, Rhizoctonia solani and Alternaria brassicicola (Gleason et al., 2011). Deficiency in hydroxymethyltransferase serine (SHMT1), involved in photorespiration, generates redox alterations and increases susceptibility to Pst‐AvrRpm1, Alternaria brassicicola and B. cinerea (Moreno et al., 2005).
It was interesting that in infected tissues the healthy gene present in the prodh single mutants did not compensate for the lack of the second gene. This suggests that ProDH1 and ProDH2 provide non‐redundant functions in these tissues. These studies were conducted with prodh1‐3 and prodh2‐2 plants which were found to be null mutants (Fig. 5). Both plants were hypersusceptible to exogenous Pro (Fig. 1a,b), something new for prodh2 alleles, as prodh2‐1 plants previously analysed in this sense were in the Ler background which is Pro sensitive (Funck et al., 2010). At present, we do not know how ProDH1 and ProDH2 are coordinated in infected tissues. One possibility is that both isoenzymes work in different cells or tissues, and their combination leads to full resistance. This is consistent with the induction of both genes in tissues infected with Pst‐AvrRpm1 or B. cinerea. Prior knowledge of these genes suggests a ubiquitous function for ProDH1 and a predominant role of ProDH2 in perivascular tissues (Faës et al., 2015; Funck et al., 2010; GENEVESTIGATOR; https://genevestigator.com). Alternatively, both isoforms may coexist in the same cells either in the same or different organelles (mitochondria and chloroplast; Funck et al., 2010; Van Aken et al., 2009). Adding complexity to this issue, ProDH1 and ProDH2 are inter‐regulated. Lack of ProDH2 increases ProDH1 expression in uninfected, but not infected, tissues (Fig. 5). Probably, Pro‐treated prodh2‐2 plants do not accumulate Pro as a result of ProDH1 compensation (Fig. 1c). In turn, lack of ProDH1 prevents maximum ProDH2 activation on infection, having no effect on basal gene expression. The induction of ProDH2 also requires ProDH1 in tissues treated with AA, an inducer of the mitochondrial ROS burst (Fabro et al., 2016), suggesting that ProDH1 contributes to ProDH2 transcriptional control under oxidative stress conditions.
The results described herein provide novel findings on ProDH action under biotic stress which are useful for the further evaluation of the circuits responsible for the inter‐regulation and balance of both ProDH genes, the pathways regulating their expression, and the subcellular and tissue locations of each enzyme isoform under different infection conditions.
Experimental Procedures
Plant material
The Arabidopsis (Arabidopsis thaliana) Col‐0 plants used in this study include sid2‐2, npr1‐1 and jar1‐1 (Arabidopsis Biological Resource Center, Columbus, OH, USA), as well as prodh1‐2 (SALK_081276), prodh1‐3 (GABI_308F08), prodh1‐4 (SALK_119334) and prodh2‐2 (GABI_328G05). prodh2‐1 (GT1788), coi1‐1 (Katsir et al., 2008) and the HopX1 transgenic line (Gimenez‐Ibanez et al., 2014) are Ler, Col‐5 and Aa‐0 plants, respectively. Seeds were germinated on Gamborg's medium (GM) plates for 10 days and transferred to soil to be grown under an 8‐h light/16‐h dark cycle at 22 ºC. For infection studies, plants were used at the age of 6 weeks. Sensitivity to Pro (5, 10 or 20 mm) was assayed on GM plates with 1% sucrose using 2‐week‐old plants. The Pro content was determined according to Bates et al. (1973).
Treatments with pathogens, JA and AA
Pst DC3000 (virulent), Pst‐AvrRpm1 (avirulent) and Pst DC3118 (COR‐deficient mutant; Thilmony et al., 2006) were grown on King's B medium supplemented with antibiotics. Pathogens were infiltrated into leaf tissues at 5 × 105 colony‐forming units (cfu)/mL for bacterial growth curves and 106 or 107 cfu/mL for expression analyses (Pavet et al., 2005). Botrytis cinerea B05.10 was grown in potato dextrose agar (PDA) for 10 days, exposed to black light for 2 days and maintained under normal growth conditions for 4 additional days. At this time, conidia were harvested, washed once in 0.1% Tween 20, twice in water, resuspended and maintained for 2 h in 10 mm K3PO4 with 10 mm sucrose, and deposited onto the adaxial side of the leaf using 5‐µL aliquots. Fungal hyphae were detected by trypan blue staining (Fabro et al., 2008) and infection areas were quantified with the ImageJ program. meJA (95%; Sigma Aldrich Buenos Aires, Argentina) was applied at 0.84 µm concentration as described previously (Fabro et al., 2008). Samples contained a set of three leaves collected from different plants. Treatments with AA (10 µm; Sigma Aldrich, Buenos Aires, Argentina, A8674) were performed as described previously (Fabro et al., 2016) using leaf discs incubated overnight in water and then exposed to water (mock) or AA for 4 h.
In silico gene promoters and gene expression analysis
Gene promoter sequences were analysed with PLACE (Higo et al., 1999), Agris (Davuluri et al., 2003) and Plant CARE (Lescot et al., 2002) programs after defining the promoter region according to Agris (2481 and 2562 bp for ProDH1 and ProDH2 promoters, respectively). Elements recognized by at least two programs were selected. Gene expression was determined by semi‐quantitative (Monteoliva et al., 2014) or quantitative (Fabro and Alvarez, 2012) reverse transcription‐polymerase chain reaction (RT‐PCR) using GapC or UBQ5 as control genes. The RT‐PCR conditions are described in Table S2 (see Supporting Information). For quantitative RT‐PCR, the gene expression values were determined by the ΔΔCt method using UQB5 as housekeeping gene, and primers at 200 nm, except for ProDH2 Rv, which was applied at 300 nm.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Expression of ProDH1 and ProDH2 at different stages of development (a) and during the light/dark cycle (b). (a) Comparison of ProDH1 (red), ProDH2 (blue) and GapC (green) expression using GENEVESTIGATOR tools. (b) Gene expression levels of ProDH1 and ProDH2 in 7‐day‐old seedlings grown under the light/dark cycle used in this study (8 h light, white bar; 16 h dark, black bar), according to data informed by the DIURNAL website (http://diurnal.mocklerlab.org/). Black arrows indicate the time points at which samples were analysed [0, 6 and 24 h post‐treatment (hpt)]. Differences in ProDH1 and ProDH2 expression are low in adult leaves (a) at the light phase stages selected for this study (b).
Table S1 cis‐regulatory elements in the ProDH1 and ProDH2 gene promoters.
Table S2 Primers and conditions used in reverse transcription‐polymerase chain reaction (RT‐PCR) assays.
Acknowledgements
This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica (PICT 2012‐2117; PICT 2014‐3255) and Secretaría de Ciencia y Tecnología, Universidad Nacional de Córdoba to M.E.A. Y.S.R. is a CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas) fellow. G.F. and M.E.A. are senior Career Investigators of CONICET. We thank Drs A. Savouré (UPMC, Université Paris 06, Jussieu, France), D. Funck (University of Konstanz, Germany) and S. Gimenez‐Ibañez (CNB‐CSIC, Madrid, Spain) for providing seeds, and Dr F. Pieckenstain (IIB‐INTECH, Argentina) for the Botrytis cinerea B05.10 strain.
References
- Bates, L.S. , Waldren, R.P. and Teare, I.D. (1973) Rapid determination of free proline for water‐stress studies. Plant Soil, 39, 205–207. [Google Scholar]
- Ben Rejeb, K. , Abdelly, C. and Savouré, A. (2014) How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 80, 278–284. [DOI] [PubMed] [Google Scholar]
- Caarls, L. , Pieterse, C.M. and Van Wees, S.C. (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabassa‐Hourton, C. , Schertl, P. , Bordenave‐Jacquemin, M. , Saadallah, K. , Guivarc'h, A. , Lebreton, S. , Planchais, S. , Klodmann, J. , Eubel, H. , Crilat, E. , Lefebvre‐De Vos, D. , Ghelis, T. , Richard, L. , Abdelly, C. , Hans‐Peter Braun, P. and Savoure, A. (2016) Proteomic and functional analysis of ProDH1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. Biochem. J. 473, 2623–2634. [DOI] [PubMed] [Google Scholar]
- Cecchini, N.M. , Monteoliva, M.I. and Alvarez, M.E. (2011a) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol. 155, 1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, N.M. , Monteoliva, M.I. and Alvarez, M.E. (2011b) Proline dehydrogenase is a positive regulator of cell death in different kingdoms. Plant Signal. Behav. 6, 1195–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri, R.V. , Sun, H. , Palaniswamy, S.K. , Matthews, N. , Molina, C. , Kurtz, M. and Grotewold, E. (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis‐regulatory elements and transcription factors. BMC Bioinformatics, 4, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas, S.K. , Enyedi, A. and Raina, R. (2002) The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. [DOI] [PubMed] [Google Scholar]
- De Vos, M. , Van Oosten, V.R. , Van Poecke, R.M. , Van Pelt, J.A. , Pozo, M.J. , Mueller, M.J. , Buchala, A.J. , Metraux, J.P. , Van Loon, L.C. , Dicke, M. and Pieterse, C.M. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant–Microbe Interact. 18, 923–937. [DOI] [PubMed] [Google Scholar]
- Devoto, A. , Ellis, C. , Magusin, A. , Chang, H.S. , Chilcott, C. , Zhu, T. and Turner, J.G. (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound‐ and methyl jasmonate‐induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 58, 497–513. [DOI] [PubMed] [Google Scholar]
- Dietrich, K. , Weltmeier, F. , Ehlert, A. , Weiste, C. , Stahl, M. , Harter, K. and Droge‐Laser, W. (2011) Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell, 23, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon, T.E. and Stewart, C.R. (1981) Submitochondrial location and electron transport characteristics of enzymes involved in proline oxidation. Plant Physiol. 67, 780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. and Alvarez, M.E. (2012) Loss of compatibility might explain resistance of the Arabidopsis thaliana accession Te‐0 to Golovinomyces cichoracearum . BMC Plant Biol. 12, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. , Di Rienzo, J.A. , Voigt, C.A. , Savchenko, T. , Dehesh, K. , Somerville, S. and Alvarez, M.E. (2008) Genome‐wide expression profiling of Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol. 146, 1421–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. , Rizzi, Y.S. and Alvarez, M.E. (2016) Arabidopsis proline dehydrogenase contributes to flagellin‐mediated PAMP‐triggered immunity by affecting RBOHD. Mol. Plant–Microbe Interact. 29, 620–628. [DOI] [PubMed] [Google Scholar]
- Faës, P. , Deleu, C. , Ainouche, A. , Le Caherec, F. , Montes, E. , Clouet, V. , Gouraud, A.M. , Albert, B. , Orsel, M. , Lassalle, G. , Leport, L. , Bouchereau, A. and Niogret, M.F. (2015) Molecular evolution and transcriptional regulation of the oilseed rape proline dehydrogenase genes suggest distinct roles of proline catabolism during development. Planta, 241, 403–419. [DOI] [PubMed] [Google Scholar]
- Funck, D. , Eckard, S. and Muller, G. (2010) Non‐redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biol. 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, X. , Jin, L. , Shimada, M. , Kim, M.G. and Mackey, D. (2014) The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae . Planta, 240, 1149–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernandez‐Barbero, G. , Chini, A. , Rathjen, J.P. and Solano, R. (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason, C. , Huang, S. , Thatcher, L.F. , Foley, R.C. , Anderson, C.R. , Carroll, A.J. , Millar, A.H. and Singh, K.B. (2011) Mitochondrial complex II has a key role in mitochondrial‐derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA, 108, 10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, C.N. , Liu, W. , Alvord, W.G. and Phang, J.M. (2015) Co‐regulation of mitochondrial respiration by proline dehydrogenase/oxidase and succinate. Amino Acids, 48, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, J. , Hanssen, M. , Wiese, A. , Hendriks, M.M. and Smeekens, S. (2008) The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53, 935–949. [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir, L. , Schilmiller, A.L. , Staswick, P.E. , He, S.Y. and Howe, G.A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA, 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T. , Yoshiba, Y. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1996) A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell, 8, 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, C. , Vandelle, E. , Rolland, S.P. , Poinssot, B. , Bruel, C. , Cimerman, A.S. , Zotti, C. , Moreau, E. , Vedel, R. , Pugin, A. and Boccara, M. (2006) Characterization of a new, nonpathogenic mutant of Botrytis cinerea with impaired plant colonization capacity. New Phytol. 170, 537–550. [DOI] [PubMed] [Google Scholar]
- Lescot, M. , Dehais, P. , Thijs, G. , Marchal, K. , Moreau, Y. , Van de Peer, Y. , Rouze, P. and Rombauts, S. (2002) PlantCARE, a database of plant cis‐acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Zhang, L. , Natarajan, S.K. and Becker, D.F. (2013) Proline mechanisms of stress survival. Antioxid. Redox Signal. 19, 998–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kracher, B. , Ziegler, J. , Birkenbihl, R.P. and Somssich, I.E. (2015) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife, 4, e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteoliva, M.I. , Rizzi, Y.S. , Cecchini, N.M. , Hajirezaei, M.R. and Alvarez, M.E. (2014) Context of action of proline dehydrogenase (ProDH) in the Hypersensitive Response of Arabidopsis. BMC Plant Biol. 14, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, J.I. , Martin, R. and Castresana, C. (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 41, 451–463. [DOI] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Atzorn, R. , Miersch, O. and Wasternack, C. (2006) The outcomes of concentration‐specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K. , Satoh, R. , Kiyosue, T. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1998) A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiol. 118, 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo, T. , Fujita, M. , Seki, M. , Kato, T. , Tabata, S. and Shinozaki, K. (2003) Toxicity of free proline revealed in an arabidopsis T‐DNA‐tagged mutant deficient in proline dehydrogenase. Plant Cell Physiol. 44, 541–548. [DOI] [PubMed] [Google Scholar]
- Pavet, V. , Olmos, E. , Kiddle, G. , Mowla, S. , Kumar, S. , Antoniw, J. , Alvarez, M.E. and Foyer, C.H. (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 139, 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang, J.M. and Liu, W. (2011) Proline metabolism and cancer. Front. Biosci. 17, 1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins, T.W. , Tudzynski, P. , von Tiedemann, A. , Tudzynski, B. , Ten Have, A. , Hansen, M.E. , Tenberge, K. and van Kan, J.A.L. (2000) Infection strategies of Botrytis cinerea and related necrotrophic pathogens In: Fungal Pathology (Krostand J. W., ed.), pp. 33–64. XXXX: Springer. [Google Scholar]
- Ribarits, A. , Abdullaev, A. , Tashpulatov, A. , Richter, A. , Heberle‐Bors, E. and Touraev, A. (2007) Two tobacco proline dehydrogenases are differentially regulated and play a role in early plant development. Planta, 225, 1313–1324. [DOI] [PubMed] [Google Scholar]
- Rizzi, Y.S. , Monteoliva, M.I. , Fabro, G. , Grosso, C.L. , Larovere, L.E. and Alvarez, M.E. (2015) P5CDH affects the pathways contributing to Pro synthesis after ProDH activation by biotic and abiotic stress conditions. Front. Plant Sci. 6, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, R. , Fujita, Y. , Nakashima, K. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2004) A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity‐responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45, 309–317. [DOI] [PubMed] [Google Scholar]
- Schertl, P. and Braun, H.P. (2014) Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil‐Kumar, M. and Mysore, K.S. (2012) Ornithine‐delta‐aminotransferase and proline dehydrogenase genes play a role in non‐host disease resistance by regulating pyrroline‐5‐carboxylate metabolism‐induced hypersensitive response. Plant Cell Environ. 35, 1329–1343. [DOI] [PubMed] [Google Scholar]
- Sharma, S. and Verslues, P.E. (2010) Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ. 33, 1838–1851. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Johnson, J.S. and Dong, X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA, 104, 18 842–18 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. and Tiryaki, I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell, 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony, R. , Underwood, W. and He, S.Y. (2006) Genome‐wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53. [DOI] [PubMed] [Google Scholar]
- Uloth, M.B. , Clode, P.L. , You, M.P. and Barbetti, M.J. (2015) Calcium oxalate crystals: an integral component of the Sclerotinia sclerotiorum/Brassica carinata pathosystem. PLoS One, 10, e0122362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken, O. , Zhang, B. , Carrie, C. , Uggalla, V. , Paynter, E. , Giraud, E. and Whelan, J. (2009) Defining the mitochondrial stress response in Arabidopsis thaliana . Mol. Plant. 2, 1310–1324. [DOI] [PubMed] [Google Scholar]
- Weltmeier, F. , Ehlert, A. , Mayer, C.S. , Dietrich, K. , Wang, X. , Schutze, K. , Alonso, R. , Harter, K. , Vicente‐Carbajosa, J. and Droge‐Laser, W. (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 25, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Zhang, D. , Chu, J.Y. , Boyle, P. , Wang, Y. , Brindle, I.D. , De Luca, V. and Despres, C. (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. [DOI] [PubMed] [Google Scholar]
- Yoshioka, K. , Kachroo, P. , Tsui, F. , Sharma, S.B. , Shah, J. and Klessig, D.F. (2001) Environmentally sensitive, SA‐dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Expression of ProDH1 and ProDH2 at different stages of development (a) and during the light/dark cycle (b). (a) Comparison of ProDH1 (red), ProDH2 (blue) and GapC (green) expression using GENEVESTIGATOR tools. (b) Gene expression levels of ProDH1 and ProDH2 in 7‐day‐old seedlings grown under the light/dark cycle used in this study (8 h light, white bar; 16 h dark, black bar), according to data informed by the DIURNAL website (http://diurnal.mocklerlab.org/). Black arrows indicate the time points at which samples were analysed [0, 6 and 24 h post‐treatment (hpt)]. Differences in ProDH1 and ProDH2 expression are low in adult leaves (a) at the light phase stages selected for this study (b).
Table S1 cis‐regulatory elements in the ProDH1 and ProDH2 gene promoters.
Table S2 Primers and conditions used in reverse transcription‐polymerase chain reaction (RT‐PCR) assays.


