Summary
The transcription factor MoAP1 has been shown previously to be required for pathogenicity in Magnaporthe oryzae via mediation of the oxidative stress response. In the serial analysis gene expression database, it was found that expression of MoYcp4, a homologue of the Saccharomyces cerevisiae flavodoxin‐like protein ScYcp4, was affected by MoAP1. Transcriptional analysis demonstrated that MoYCP4 was significantly up‐regulated during conidiation, appressorium formation and infection. The growth rate of a ΔMoycp4 mutant was reduced slightly, but conidial production was increased significantly (more than 10‐fold), compared with the wild‐type strain. Although the rate of appressorium formation was unaffected, the appressorial turgor was abnormal and the ability to infect rice and barley was reduced, resulting in decreased pathogenicity. In summary, MoYcp4, a target of MoAP1, is involved in the growth, conidiogenesis and pathogenicity of M. oryzae. Our studies provide a comprehensive analysis of flavodoxin‐like proteins and will aid in the study of pathogen‐related molecular mechanisms.
Keywords: conidiogenesis, growth, MoYcp4, pathogenicity
Introduction
Magnaporthe oryzae is an important pathogenic fungus which causes great damage on rice and a variety of other cereal crops. The annual grain losses caused by rice blast would be sufficient to feed 60 million people; therefore, pathogenic morphogenesis has important economic and social significance (Ou, 1985). Considering the important economic impact and genetic tractability of the pathogen and host, rice blast is considered to be an important model for the study of plant–pathogen interactions at the molecular level (Caracuel‐Rios and Talbot, 2007).
Like most other fungal pathogens, the conidia of M. oryzae play a central role in the disease cycle. When attached to the plant surface, conidia begin to germinate and form a specialized infection structure, known as an appressorium, at the end of the germ tube (Bourett and Howard, 1991). Once mature, the appressorium accumulates enormous turgor pressure (as high as 8 MPa) to help penetrate the strong leaf cuticle and to enter plant cells (Howard et al., 1991). After penetration, infectious hyphae spread and cause necrotic lesions on the plants. Once conditions are suitable, thousands of conidia are produced in the lesions, which are then released to initiate a new infection cycle on plant tissues within 3–5 days. Therefore, the characterization of the molecular mechanisms involved in conidiation and appressorium penetration‐mediated plant infection is pivotal to the production of new strategies for disease management.
We have shown previously that the transcription factor MoAP1 is required for the pathogenicity of M. oryzae via mediation of the oxidative stress response. In the serial analysis gene expression (SAGE) database, many of the genes are affected by MoAP1, including those required for pathogenicity (Guo et al., 2011). MoYcp4, a homologue of the Saccharomyces cerevisiae flavodoxin‐like protein (FLP) ScYcp4, has also been shown to be affected by MoAP1. In S. cerevisiae, ScYCP4 and its homologue ScRFS1 regulate the expression of a series of genes during late growth, and the deletion of ScYCP4 leads to the expression of genes associated with hexose transport and glycolysis, as well as altered expression of genes involved in mating and pheromone responses. In addition, the response to oxidative and osmotic stress is affected in the ΔScycp4ΔScrfs1 double mutant (Cardona et al., 2011), indicating that ScYCP4 is involved in a series of biological processes and plays an important role in the growth of S. cerevisiae. Moreover, Paracoccidioides brasiliensis PBY20, the homologue of YCP4, has been shown to play an important role in cell differentiation or even in the maintenance of the yeast form (Daher et al., 2005). Deletion of four FLPs brought about an increased sensitivity to benzoquinone and oxidants, reduced ubiquinone synthesis and loss of virulence to the host in Candida albicans (Morschhäuser et al., 2015).
Here, we define a pivotal FLP, MoYcp4, and, for the first time, elucidate its function in phytopathogenic fungi by the analysis of its role in the growth, conidiogenesis and pathogenicity of M. oryzae.
Results
Identification of MoYCP4
MoAP1 is a positive transcription factor which regulates the transcription of a series of genes important for the growth, development and pathogenicity of M. oryzae (Guo et al., 2011). To identify target genes affected by MoAP1, we screened the ΔMoap1 mutant SAGE database and identified a gene (MGG_01569) with an encoded protein that showed 55% identity and 65% similarity to the S. cerevisiae FLP ScYcp4; therefore, we named the protein MoYcp4. Considering that ScYCP4 and its homologue ScRFS1 are involved in the osmotic stress response in S. cerevisiae (Cardona et al., 2011), we expressed MoYCP4 in a ΔScycp4 mutant through the yeast expression vector pYES2 to determine whether MoYcp4 can complement ScYcp4 function. Transformants carrying the MoYCP4 gene exhibited similar growth to the wild‐type on medium containing 0.7 m NaCl and 0.6 m KCl, Growth of the transformants is much better when compared with the ΔScycp4 mutant (Fig. S1, see Supporting Information), suggesting that MoYcp4 is a functional paralogue of ScYcp4.
We have shown previously that the transcription of MoYCP4 is reduced in the ΔMoap1 mutant (Guo et al., 2011). We confirmed the expression levels of MoYCP4 using quantitative real‐time polymerase chain reaction (qRT‐PCR) and found that our result was consistent with that in the Solexa SAGE database. The expression of MoYCP4 was reduced significantly in the ΔMoap1 mutant (Fig. 1A). To further validate that MoYCP4 is regulated directly by the transcription factor MoAP1, we performed an agarose gel electrophoretic mobility shift assay (EMSA). We found that the digoxigenin (DIG)‐labelled promoter of MoYCP4 was retarded by the addition of MoAP1 protein in EMSA, but not by the addition of glutathione S‐transferase (GST) (Fig. 1B). The addition of proteinase K caused the DIG‐labelled DNA to shift back to the fast‐mobility band (Fig. 1B). Further, specificity and competition assays were performed; we found that the DIG‐labelled promoter of other gene MoLYS2 (Chen et al., 2014b) was not retarded by the addition of MoAP1 protein (Fig. S3A, see Supporting Information), and the concentrations of the binding complexes of MoAP1 and the DIG‐labelled promoter were progressively reduced with the addition of increasing amounts of unlabelled promoter of MoYCP4 (Fig. S3B), These results suggest that MoAP1 is able to bind to the promoter of MoYCP4.
Figure 1.
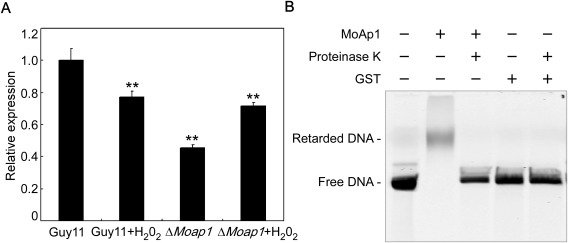
Expression analysis of MoYCP4 in the ΔMoap1 mutant and the DNA‐binding activity of MoAP1. (A) Transcriptional levels of MoYCP4 in the wild‐type and ΔMoap1 mutant before and after exposure to H2O2 measured by quantitative real‐time polymerase chain reaction (qRT‐PCR). (B) The digoxigenin (DIG)‐labelled promoter DNA of MoYCP4 was incubated in the absence or presence of purified MoAp1 protein. Proteinase K was added after the incubation of MoAp1 with the probe, and the sample was incubated for 10 min at room temperature. GST, glutathione S‐transferase. Double asterisks represent significant differences (P < 0.01).
MoYCP4 is highly expressed during various stages of infection
Before exploring the functions of MoYCP4, we evaluated the transcriptional profile in various growth and infection stages. The expression of MoYCP4 was higher in the conidial, appressorial and infection stages than in the mycelium stage, with the highest level occurring during the conidial stage (>20‐fold increase; Fig. 2). These observations suggest that MoYCP4 plays an important role in infection‐related morphogenesis.
Figure 2.
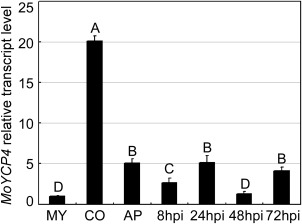
Transcription profiles of MoYCP4 at different stages of fungal development. The phase‐specific expression of MoYCP4 was quantified by quantitative real‐time polymerase chain reaction (qRT‐PCR), with the synthesis of cDNA from each sample including infectious growth, vegetative growth and conidia. MY, mycelia; CO, conidia; AP, appressoria; hpi, hour post‐inoculation. Capital letters represent significant differences (P < 0.01).
MoYCP4 deletion affects hyphal growth
A MoYCP4 deletion mutant was generated by replacing the MoYCP4 coding region with the hygromycin resistance cassette (HPH) (Fig. S2A, see Supporting Information). The putative mutant (ΔMoycp4) was screened and confirmed by Southern blot analysis (Fig. S2B). Furthermore, a complementation strain (ΔMoycp4/MoYCP4) containing the open reading frame (ORF) encoded by MoYCP4 and green fluorescent protein (GFP) under the control of the MoYCP4 native promoter was also generated (Fig. S2C). The resulting transformant showed normal growth, conidiation and infection (Figs 3, 4, 5, 6, 7, 8, 9, 10) and was considered as the complemented strain. Under epifluorescence microscopy, the GFP signal of MoYcp4‐GFP was visible in the cytoplasm of the conidia, appressoria and infectious hyphae (Fig. S4, see Supporting Information).
Figure 3.
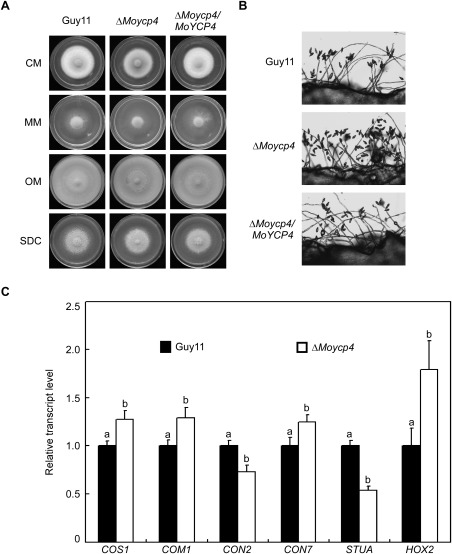
Comparison of mutants and wild‐type strains with regard to growth and conidia formation. (A) Wild‐type, complementation and mutant strains were inoculated onto complete (CM), minimal (MM), oatmeal (OM) and straw decoction and corn (SDC) media and cultured at 28 °C in the dark for 7 days. (B) Conidia formation was observed under a light microscope at 24 h at room temperature after induction of conidiation under coverslips. (C) Expression analysis of conidiation‐related genes by quantitative real‐time polymerase chain reaction (qRT‐PCR) in the ΔMoycp4 mutant. Lowercase letters represent significant differences (P < 0.05).
Figure 4.
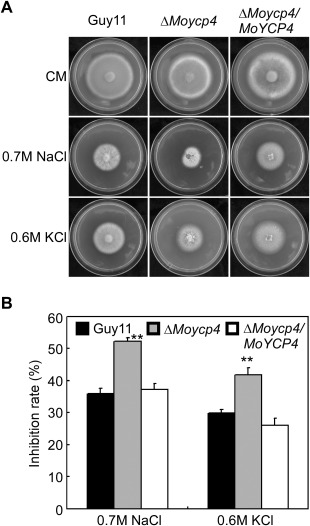
ΔMoycp4 mutant ion stress assessment. (A) The ΔMoycp4 mutant is more sensitive than Guy11 to ion stress. Colonies of the wild‐type Guy11, the ΔMoycp4 mutant and the complemented strains were grown on complete medium (CM) plates with 0.7 m NaCl or 0.6 m KCl, and cultured at 28 °C for 7 days. (B) The growth inhibition rate is estimated relative to the growth rate of each untreated control [inhibition rate = (diameter of untreated strain – diameter of treated strain)/(diameter of untreated strain × 100%)]. Three repeats were performed and similar results were obtained. Error bars represent the standard deviations and double asterisks represents significant differences (P < 0.01).
Figure 5.
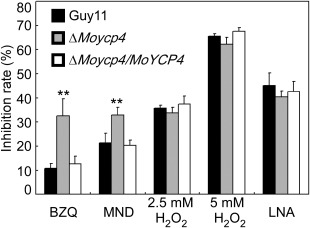
ΔMoycp4 mutant oxidation stress assessment. Equal plugs of mycelia from 5‐day‐old complete medium (CM) plates were transferred into liquid CM containing p‐benzoquinone (BZQ), menadione (MND), 2.5 mm H2O2, 5 mm H2O2 and linolenic acid (LNA); the dry weight of mycelia was determined by culture with shaking (150 rpm) at 28 °C for 2 days. The inhibition rate is estimated relative to the growth rate of each untreated control [inhibition rate = (dry weight of untreated strain – dry weight of treated strain)/(dry weight of untreated strain × 100%)]. Three repeats were performed and similar results were obtained. Error bars represent the standard deviations and double asterisks represents significant differences (P < 0.01).
Figure 6.
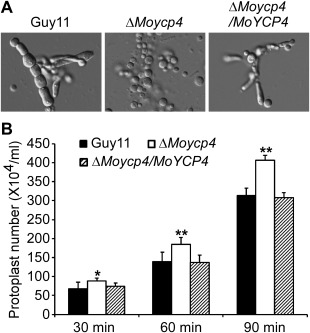
Protoplast release assay. (A) Protoplasts released on treatment with cell wall‐degrading enzymes. Light microscopic examination after 40 min. (B) The protoplasts released were quantified at 30‐min intervals. Asterisks represent significant differences (P < 0.05)
Figure 7.
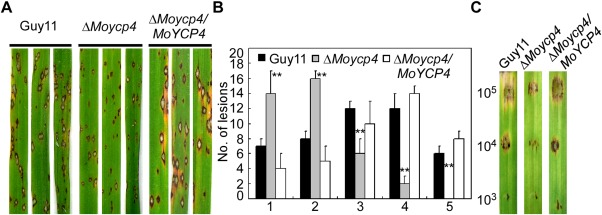
Pathogenicity assay of the mutant. (A) Leaf spraying assay. Four millilitres of conidial suspension (5 × 104 spores/mL) of each strain were sprayed onto 2‐week‐old rice seedlings. Diseased leaves were photographed at 7 days after inoculation. (B) Quantification of lesion type (0, no symptom; 1, pinhead‐sized brown specks; 2, 1.5‐mm brown spots; 3, 2–3‐mm grey spots with brown margins; 4, many elliptical grey spots longer than 3 mm; 5, coalesced lesions infecting 50% or more of the leaf area). Lesions were photographed and measured or scored at 7 days post‐inoculation (dpi) and experiments were repeated twice with similar results. (C) Conidia of different concentrations were drop inoculated onto barley; the ΔMoycp4 mutant showed a virulence defect compared with the wild‐type and complemented strains, as manifested by smaller lesions at 5 dpi. Double asterisks represent significant differences (P < 0.01).
Figure 8.
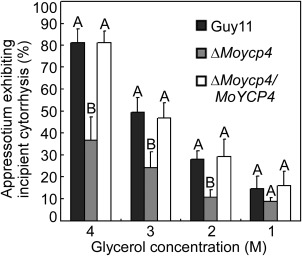
Collapsed appressoria were observed in the mutant strain. For each glycerol concentration, at least 100 appressoria were observed and the numbers of collapsed appressoria were counted. Capital letters represent significant differences (P < 0.01).
Figure 9.
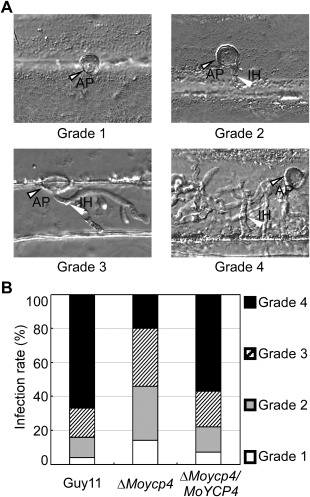
Infectious hyphal growth on barley leaves. (A) Excised barley leaves from 7‐day‐old barley seedlings were inoculated with conidial suspension (5 × 104 spores/mL). Infectious growth was observed at 24 h post‐inoculation (hpi). (B) Statistical analysis of each type of infectious hyphal shape for each tested strain; 100 infecting hyphae were counted per replicate and the experiment was repeated three times. AP, appressorium; IH, infected hypha. Grade 1, no penetration; grade 2, with a penetration peg; grade 3, with a single invasive hypha; grade 4, with extensive hyphal growth.
Figure 10.
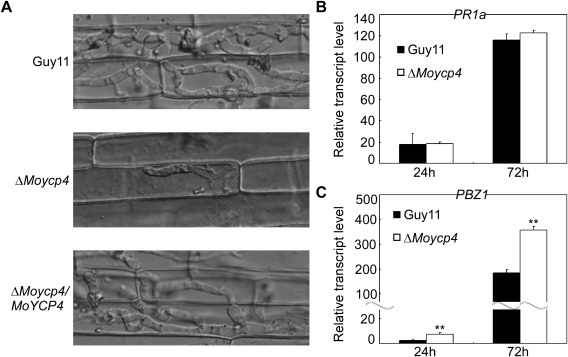
Close observation of infectious growth. (A) Excised rice sheath from 4‐week‐old rice seedling was inoculated with conidial suspension (5 × 104 spores/mL). Infectious growth was observed 48 h after inoculation. (B, C) The transcription of PR1a and PBZ1 in the infected host was assayed using quantitative real‐time polymerase chain reaction (qRT‐PCR). Three independent biological experiments with three replicates each time were performed, and similar results were obtained each time. Double asterisks represent significant differences (P < 0.01).
We evaluated the growth of the ΔMoycp4 mutant on complete (CM), minimal (MM), oatmeal (OM), and straw decoction and corn (SDC) media. The ΔMoycp4 mutants showed slightly smaller colony diameter than the wild‐type strain (Guy11) and the complemented strain ΔMoycp4/MoYCP4 on all media types, except for CM (Fig. 3A). These results were further supported by statistical analysis (Table 1), indicating that MoYcp4 plays a role in hyphal growth.
Table 1.
Comparison of mycological characteristics among strains.
| Strain | Mycelial growth* (cm) | Conidiation † (×104/cm2) | Appressorial formation ‡ (%) |
|---|---|---|---|
| CM MM OM SDC | |||
| Guy11 | 3.90 ± 0.10A 3.53 ± 0.05A 3.87 ± 0.15A 4.03 ± 0.15A | 3.72 ± 0.39A | 96.00 ± 2.79A |
| ΔMoycp4 | 3.60 ± 0.10A 3.1 ± 0.10B 3.03 ± 0.15B 3.70 ± 0.10B | 50.22 ± 2.79B | 92.67 ± 2.79A |
| ΔMoycp4/MoYCP4 | 4.00 ± 0.20A 3.63 ± 0.12A 3.63 ± 0.12A 4.10 ± 0.10A | 4.34 ± 0.80A | 93.33 ± 2.36A |
*Diameter of hyphal radii at day 7 after incubation on complete (CM), minimal (MM), oatmeal (OM) and straw decoction and corn (SDC) media plates at room temperature.
†Number of conidia harvested from a 9‐cm SDC plate at day 10 after incubation at room temperature.
‡Percentage of appressorium formation on an artificial surface at 24 h post‐incubation at room temperature.
Capital letters represent significant differences (P < 0.01).
MoYcp4 is important in stress responses
To investigate whether ΔMoycp4 exhibited any defects under different conditions of stress, the ΔMoycp4 mutant was exposed to 0.7 m NaCl and 0.6 m KCl. The ΔMoycp4 mutant showed stronger growth inhibition than Guy11 and the complemented strain (Fig. 4A). Inhibition of the ΔMoycp4 mutant was 17% and 12% higher than Guy11 in NaCl‐ and KCl‐containing CM, respectively (Fig. 4B). These findings suggest that MoYcp4 contributes to the osmotic stress response of the fungus.
Considering the involvement of FLPs in the response to quinone and oxidants in C. albicans, and the ubiquinone reduction of the FLP deletion mutant (Morschhäuser et al., 2015), we examined the sensitivity of the ΔMoycp4 mutant to these agents. Consistent with the previous study, the ΔMoycp4 mutant showed a higher inhibition rate on exposure to p‐benzoquinone (BZQ) and menadione (MND) than did Guy11 and the complemented strain (Fig. 5), indicating that the ΔMoycp4 mutant is more sensitive to quinone. However, when treated with 2.5 or 5 mm H2O2, the growth inhibition rates were not significantly different between the ΔMoycp4 mutant and Guy11 (Fig. 5). Consistently, the transcription level of MoYCP4 was also down‐regulated in Guy11 in the presence vs. absence of H2O2 treatment (Fig. 1A). We also examined the sensitivity of the ΔMoycp4 mutant to linolenic acid (LNA), a polyunsaturated fatty acid which induces lipid peroxidation (Do et al., 1996); no significant difference was observed between the ΔMoycp4 mutant and Guy11 (Fig. 5). These results indicate that the deletion of MoYCP4 does not affect the oxidation stress response in M. oryzae.
MoYcp4 is involved in cell wall integrity
To examine the role of MoYcp4 in cell wall integrity, mycelial growth was measured on CM containing sodium dodecylsulfate (SDS), calcofluor white (CFW) and Congo red (CR), all of which are cell wall‐perturbing agents. The sensitivity of the ΔMoycp4 mutant to SDS, CFW and CR at low concentrations was not significantly different from that of the wild‐type strain Guy11 (Table 2); however, the mutant was more sensitive to these agents at high concentrations (Table 2). We further examined the effects of lytic enzymes (10 mg/mL) on the ΔMoycp4 mutant. More protoplasts were found in the ΔMoycp4 mutant than in the controls after incubation for 30, 60 and 90 min (Fig. 6A,B). Mycelial fragments were still found after 60 min of incubation (Fig. 6A). In contrast, no mycelial fragments were found in the ΔMoycp4 mutant (Fig. 6A). These results indicate that MoYcp4 plays a role in cell wall integrity.
Table 2.
ΔMoycp4 mutant cell wall stress response (inhibition rate).
| Guy11 | ΔMoycp4 | ΔMoycp4/MoYCP4 | |
|---|---|---|---|
| 0.05% SDS | 34.69 ± 0.59A | 35.04 ± 3.37A | 35.01 ± 1.71A |
| 0.08% SDS | 45.54 ± 1.46A | 54.99 ± 1.67B | 47.55 ± 2.17A |
| 0.10% SDS | 54.33 ± 1.77A | 69.48 ± 1.31B | 55.35 ± 2.10A |
| 200 µg/mL CFW | 5.70 ± 0.51A | 7.94 ± 1.48A | 5.39 ± 1.37A |
| 400 µg/mL CFW | 9.04 ± 0.42A | 10.69 ± 1.31A | 9.20 ± 0.48A |
| 600 µg/mL CFW | 15.05 ± 0.85A | 23.59 ± 2.04B | 17.54 ± 1.06A |
| 200 µg/mL CR | 10.78 ± 0.61A | 11.11 ± 1.42A | 10.12 ± 2.25A |
| 400 µg/mL CR | 15.80 ± 0.25A | 23.94 ± 1.66B | 15.87 ± 1.22A |
| 600 µg/mL CR | 21.00 ± 1.04A | 27.24 ± 1.90B | 21.27 ± 1.58A |
CFW, calcofluor white; CR, Congo red; SDS, sodium dodecylsulfate.
Capital letters represent significant differences (P < 0.01)
MoYcp4 is involved in conidiogenesis, but not in appressorium formation
As conidia play an important role during M. oryzae infection, we measured the conidial production of the ΔMoycp4 mutant. We found that conidia production was significantly increased on SDC medium (Fig. 3B). The ΔMoycp4 mutant produced normal‐shaped conidia; the production was increased by over 10‐fold compared with the wild‐type Guy11 (Table 1). We further examined the expression of six conidiation‐related genes. The expression levels of MoCON2 and MoSTUA (Nishimura et al., 2009; Shi et al., 1998) were significantly lower in the ΔMoycp4 mutant than in the Guy11 strain (Fig. 3C). In contrast, the transcript levels of MoCOS1, MoCOM1, MoCON7 and MoHOX2 (Kim et al., 2009; Liu et al., 2010; Shi et al., 1998; Yang et al., 2010; Zhou et al., 2009) were significantly increased in the ΔMoycp4 mutant (Fig. 3C), indicating that MoYcp4 is involved in the regulation of the expression of conidiation‐related genes. Next, we examined appressorium formation in the ΔMoycp4 mutant. The ΔMoycp4 mutant formed normal germ tubes. Microscopic examination revealed that the rate of appressorium formation in ΔMoycp4 was not significantly different from that in Guy11, as well as in the complemented strain ΔMoycp4/MoYCP4 (Table 1). These results suggest that MoYcp4 negatively regulates the conidiogenesis of M. oryzae.
MoYcp4 is required for full virulence
To determine whether MoYcp4 is involved in pathogenicity, conidial suspensions of the ΔMoycp4 mutant, wild‐type and complemented strain were sprayed onto 2‐week‐old rice seedlings (cv. CO‐39) and detached barley leaves. When observed at 7 days post‐infection in rice, the ΔMoycp4 mutant produced disease lesions, but a remarkable reduction was observed when compared with the lesions caused by the control strain in both size and number (Fig. 7A). Signs of disease on rice plants were also quantified using a ‘lesion‐type’ scoring assay (Valent et al., 1991). The ΔMoycp4 mutant mainly produced type 1–3 lesions with fewer lesions of types 4 and 5 (severe, coalescing) (Fig. 7B). On detached barley leaves, which were inoculated with three different concentrations of a conidial suspension (105, 104 and 103 spores/mL), the lesions caused by the ΔMoycp4 mutant were obviously less severe (Fig. 7C). Taken together, these results indicate that MoYcp4 is involved in pathogenicity.
MoYcp4 is required for normal appressorium turgor pressure
To penetrate the rice leaf cuticle and cause infection, a high appressorium internal turgor pressure is required (Talbot and Foster, 2001). Appressorium turgor can be measured using an incipient cytorrhysis assay, which applies hyperosmotic concentrations of a solute to collapse the appressoria (Howard et al., 1991; de Jong et al., 1997). To further elucidate the mechanism underlying virulence in the ΔMoycp4 mutant, we examined the appressorium turgor in the ΔMoycp4 mutant and wild‐type strain. Surprisingly, the appressoria of the ΔMoycp4 mutant showed a reduced collapse rate in 2, 3 and 4 m glycerol compared with the appressoria of the wild‐type (Fig. 8), indicating that the appressoria of the ΔMoycp4 mutant produced an abnormal turgor pressure.
Δ Moycp4 mutant is defective in infectious hyphal growth on plants
To further explore why the ΔMoycp4 mutant showed reduced virulence on host plants, we performed an infection assay to examine infectious hyphal growth on barley leaves and rice leaf sheaths. Infectious hyphal growth on barley was also evaluated using an ‘invasive hypha type’ assay (Wang et al., 2013) at 48 h post‐inoculation (hpi) employing spore suspensions; four grades (grade 1, no penetration; grade 2, with a penetration peg; grade 3, with a single invasive hypha; grade 4, with extensive hyphal growth) of invasive hyphae were observed in barley tissues (Fig. 9A). In the wild‐type and complemented strains, about 60% of the cells showed grade 4 growth; few strains showed grades 1 and 2 invasive hyphal growth. In contrast, 20% of the cells showed grade 4 and more than 40% of the cells showed grades 1 and 2 invasive hyphal growth in the ΔMoycp4 mutant (Fig. 9B). A similar result was observed in rice leaf sheaths. In the ΔMoycp4 mutant, invasive hyphae were mostly restricted to the primary infected leaf sheath cells, in contrast with the free spread of invasive hyphae in the wild‐type Guy11 (Fig. 10A). We also examined the expression of the plant defence genes PR1a and PBZ1, and observed higher transcription levels of PBZ1 in rice infected with the ΔMoycp4 mutant when compared with that infected with wild‐type Guy11 (Fig. 10B,C). Finally, 3,3′‐diaminobenzidine (DAB) staining was performed to detect the reactive oxygen species (ROS) burst at the infection site of Guy11 and the ΔMoycp4 mutant; however, no distinguishable difference between Guy11 and the ΔMoycp4 mutant was observed (data not shown).
Discussion
Our previous studies have shown that the transcription factor MoAP1 mediates the oxidative stress response and is required for M. oryzae pathogenicity (Guo et al., 2011). In the ΔMoap1 mutant SAGE database, many MoAP1‐regulated genes, including MoYCP4 encoding an FLP, play important roles in infection‐related morphogenesis. This study increases our understanding of the mechanism involved in the pleiotropic functions of MoAP1 and the molecular mechanisms involved in rice blast.
MoYcp4 is involved in vegetative growth and conidiation in M. oryzae. In S. cerevisiae, the expression of metabolism genes related to hexose transport and glycolysis is affected, suggesting that the ΔScycp4 mutant possesses defects in nutrient absorption and glycometabolism (Cardona et al., 2011). In this study, we found that the growth rate of the ΔMoycp4 mutant was decreased in CM, MM, OM and SDC media; in particular, the colony diameter of the ΔMoycp4 mutant was significantly lower than that of wild‐type Guy11, suggesting that the disruption of MoYCP4 reduces the ability of the fungus to absorb and utilize nutrients. Further, the ΔMoycp4 mutant showed defects in cell wall integrity, as it exhibited increased sensitivity to cell wall stress agents and lysis. It is well known that the fungal cell wall plays an important role in the maintenance of cell morphogenesis and adaptation to the environment (Levin, 2005). Moreover, previous studies have shown that cell wall integrity‐defective mutants exhibit multiple defects in growth and stress responses (Li et al., 2006; Qi et al., 2012). Therefore, it is reasonable to speculate that the defects of growth and osmotic response of the ΔMoycp4 mutant result from a breach in cell wall integrity.
Previous studies have indicated that FLPs are involved in the oxidative stress response, and YCP4, a member of the FLP family, is up‐regulated under oxidative stress (Daher et al., 2005; Gasch et al., 2001; Kudo et al., 1999). However, our data showed that the expression level of MoYCP4 was decreased and the inhibition rate was no different between the ΔMoycp4 mutant and wild‐type when treated with H2O2. In addition, MoYcp4 was dispensable for the elimination of ROS of host cells, indicating that MoYcp4 was not sensitive to oxidation stress and such a function might be differentiated in M. oryzae. Considering that MoAP1 functioned as a redox sensor and was involved in the regulation of the oxidative stress response (Guo et al., 2011), our further study will focus on the elucidation of why MoYcp4 does not participate in the oxidation response.
The ΔMoycp4 mutant showed markedly increased conidial production. Combined with our transcript level analysis, MoYCP4 was obviously up‐regulated during the conidial stage, indicating that MoYcp4 is a negative regulator of conidiation. These results are similar to those obtained for MoPac2, which is also regulated by MoAP1. The ΔMopac2 mutant produced normal conidia, but the number was increased by about two‐fold when compared with wild‐type Guy11 (Chen et al., 2014a). Although ΔMoap1 showed reduced production of conidia and affected conidial morphology, MoYCP4 and MoPAC2, both of which function downstream of MoAP1, showed a different regulatory mechanism to MoAP1. These results indicate that other genes positively control conidiation downstream of MoAP1; a number of genes are known to be involved in this process, including MoSSADH, MoACT1 and MoGTI1 (Chen et al., 2014a; Guo et al., 2011).
Disruption of MoYCP4 leads to a defect in infection‐related morphogenesis in M. oryzae. The ΔMoycp4 mutant showed reduced pathogenicity in rice and barley. To elucidate the mechanism of reduced pathogenicity, we examined appressorium turgor pressure. We found that the collapse rate of appressoria of the ΔMoycp4 mutant was less than that of Guy11, suggesting that appressorium turgor in the ΔMoycp4 mutant was increased. Previous studies have shown that the broken balance of appressorium turgor decreases the infectivity of a fungus (Zhang et al., 2009). As expected, invasive growth of the ΔMoycp4 mutant was inhibited in rice sheaths, and the expansion ratio was markedly lower than that for wild‐type Guy11 in barley epidermis. Moreover, PBZ1, a key component in the jasmonic acid (JA) pathway and involved in JA‐induced plant defence (Mei et al., 2006), showed a stronger activation in mutant‐inoculated plants compared with controls. We conclude that these factors contribute to the decreased pathogenicity of the ΔMoycp4 mutant.
Collectively, we have identified an important pathogenic factor, MoYcp4, which functions downstream of MoAP1. Our results indicate that MoYcp4 plays an important role in growth, conidiation, infection and pathogenicity in M. oryzae.
Experimental Procedures
Strains and culture conditions
Magnaporthe oryzae Guy11 strain was used as the wild‐type in this study. All strains were cultured on CM agar plates for 3–15 days at 28 °C (Talbot et al., 1993). Fungal mycelia were harvested from liquid CM and used for genomic DNA and RNA extractions. Protoplasts were prepared and transformed as described by Sweigard et al. (1992). Transformants were selected on TB3 medium (3 g of yeast extract, 3 g of casamino acids, 200 g of sucrose, 7.5 g of agar in 1 L of distilled water) with 300 µg/mL hygromycin B (Roche, San Francisco, California, USA) or 200 µg/mL zeocin (Invitrogen, Shanghai, China). For conidiation, strain blocks were maintained on RDC medium at 28 °C for 7 days in the dark, followed by 3 days of continuous illumination under fluorescent light (Zhang et al., 2009).
Yeast ΔScycp4 mutant complementation
The full lengths of MoYCP4 and ScYCP4 cDNA were amplified using primers FL13475/FL13476 and FL13477/FL13478 (Table S1, see Supporting Information), respectively. The PCR products were digested with EcoRI/SphI and XhoI/SphI, respectively, cloned into the pYES2 vector (Invitrogen) and transformed into the ΔScycp4 mutant. Colonies were selected on synthetic drop‐out (SD) medium without uracil. As a control, the wild‐type strain BY4742 and the ΔScycp4 mutant were transformed with the empty pYES2 vector. Yeast cells were incubated on liquid YPD medium (2% glucose, 2% peptone and 1% yeast extract) and aliquots (5 µL) of a 10‐fold serial dilution were grown on SD‐NaCl (galactose + 0.7 m NaCl) and SD‐KCl (galactose + 0.6 m KCl) plates at 30 °C for 4 days and photographed.
MoYCP4 gene disruption and ΔMoycp4 mutant complementation
The ligation PCR approach (Zhao et al., 2004) was used to generate the MoYCP4 gene replacement constructs. Approximately 1‐kb upstream and downstream flanking sequences of the MoYCP4 gene were amplified by PCR with the primer pairs FL12705/FL12706 and FL12707/FL12708 (Table S1), respectively. The resulting PCR products of primer pairs FL12705/FL12706 and primer pairs FL12707/FL12708 were digested with XhoI/HindIII and SpeI/SacI, respectively, and then purified and orderly ligated to vector pCX62. The MoYCP4 gene replacement constructs were transformed into protoplasts of Guy11. Putative ΔMoycp4 mutants were identified by PCR and further confirmed by Southern blot analyses. For complementation assays, the full length (except stop codon) of the MoYCP4 gene, including the native promoter, was amplified and cloned into the bleomycin‐resistant vector pYF11 by the yeast in vivo recombination approach (Bruno et al., 2004; Zhou et al., 2011) and transformed into the ΔMoycp4 mutant.
Vegetative growth, stress response and protoplast release assay
Vegetative growth of ΔMoycp4 and Guy11 was measured on CM (50 mL 20 × nitrate salts, 1 mL trace elements, 10 g glucose, 2 g peptone, 1 g yeast extract, 1 g casamino acids, 1 mL vitamin solution and 15 g agar in 1 L distilled water), MM (6 g NaNO3, 0.52 g KCl, 0.52 g MgSO4, 1.52 g KH2PO4, 10 g glucose, 0.5% biotin and 15 g agar in 1 L distilled water), OM (30 g oatmeal and 10 g agar in 1 L distilled water) and SDC medium (100 g straw, 40 g corn powder and 15 g agar in 1 L distilled water) for 7 days. Mycelial plugs of equal size, from 5‐day‐old CM plates, were transferred into liquid CM. The mycelia were cultured with shaking (150 rpm) at 28 °C for 2 days. All growth assays were repeated three times, with three replicates each time.
Mycelial plugs (5 mm × 5 mm) were placed onto freshly prepared CM agar plates with NaCl (0.7 m), KCl (0.6 m), SDS (0.05%, 0.08% and 0.1%), CFW (200, 400 and 600 µg/mL) and CR (200, 400 and 600 µg/mL) and cultured in the dark at 28 °C to determine their effects on fungal growth. The size of the colonies were measured and photographed after 7 days of incubation. The inhibition rate was determined by the percentage decrease in the colony diameter (Zhang et al., 2014). The experiment was repeated three times with three replicates each time.
For protoplast release assays, mycelia were cultured in liquid CM for 48 h and harvested by filtration; they were then resuspended in 0.7 m NaCl with lysing enzyme (7.5 mg/mL; Sigma‐Aldrich, St. Louis, MO, USA) and placed in a shaker (70 rpm) at 28 °C. Lysis activity was stopped after 30, 60 and 90 min of incubation, and protoplasts were counted under a light microscope using a haemocytometer.
Nucleic acid manipulation, Southern blotting and EMSA
The standard Southern blot protocol was utilized (Sambrook and Russell, 2001). The target gene probe and HPH probe were amplified with primer pairs FL6084/FL6085 (for MoYCP4) (Table S1) and FL1111/FL1112 (for HPH), respectively. Probe labelling, hybridization and detection were performed with the DIG High Prime DNA Labelling and Detection Starter Kit (Roche Applied Science, Penzberg, Germany). Total RNA was isolated from frozen fungal mycelia using the RNA extraction kit (Invitrogen, Carlsbad, California, USA). To measure the relative abundance of gene transcripts, RNAs were extracted from mycelia grown in CM liquid medium for 2 days at 28 °C in a 150 rpm orbital shaker.
The MoAP1 protein was expressed and purified from Escherichia coli strain Rosetta using the pGEX4T‐2 construct. The DNA fragment from the MoYCP4 promoter was end labelled with Alex660 by PCR amplification using the 5′ Alex660‐labelled primer. The purified protein was mixed with Alex660‐labelled DNA and incubated for 20 min at 25 °C in binding buffer, and then 10× loading buffer was added. The reaction mixtures were separated by electrophoresis in 1% agarose with 1 × TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.0). Gels were visualized using a LI‐COR Odyssey scanner (Pleasanton, California, USA) with excitation at 700 nm.
Conidiation, appressorium formation, turgor analysis and examination of the pathogenesis‐related (PR) gene transcription level
For conidiation, 10‐day‐old conidia were collected with 5 mL of distilled water, filtered through three layers of lens paper and counted with a haemocytometer under a microscope. Conidial germination and appressorium formation were measured on a hydrophobic surface. Conidial suspensions of 30 µL (5 × 104 spores/mL) were dropped onto a hydrophobic surface and placed in a moistened box at 28 °C (Zhang et al., 2011). The appressorium formation rate was counted at 24 hpi under the microscope; more than 200 appressoria were counted for each strain. Photographs were taken at 24 hpi. The appressorium turgor was measured using an incipient cytorrhysis (cell collapse) assay and a 1–4 m glycerol solution (Howard et al., 1991). Droplets (20 µL) of the conidial suspensions (5 × 104 spores/mL) were placed on plastic coverslips and incubated in a humid chamber for 24 h at room temperature. The water surrounding the conidia was removed carefully and then replaced with an equal volume (20 µL) of glycerol in the concentration range 1–4 m. The number of appressoria that had collapsed after 10 min was recorded. The experiment was repeated three times, and >100 appressoria were observed for each replicate. To measure the relative abundance of rice PR gene transcripts, conidial suspensions of the wild‐type and ΔMoycp4 mutant were inoculated onto rice leaves for 24 and 72 h, respectively, and plant total RNA samples were extracted according to a previous method (Chen et al., 2014b).
Plant infection and penetration assays
Plant infection assays were performed on 4‐week‐old susceptible rice seedlings (Oryza sativa) CO‐39 by spraying 4 mL of a conidial suspension (5 × 104 conidia/mL in 0.2% gelatin) with a sprayer. Inoculated plants were placed in a moist chamber at 28 °C for the first 24 h in darkness, and then transferred to another moist chamber with a photoperiod of 12 h under fluorescent light. The disease severity was assessed at 7 days after inoculation. Approximately 6‐cm‐long diseased rice blades were photographed to evaluate the virulence of the mutants (Chen et al., 2014b). For microscopic observation of penetration and infectious hyphal expansion on rice and barley tissue, rice was inoculated with 100 µL of conidial suspension (5 × 104 spores/mL) on inner leaf sheath cuticle cells and barley was inoculated with 30 µL of conidial suspension (5 × 104 spores/mL) on the underside of the leaves; after 48 h (rice) and 24 h (barley) of incubation under humid conditions at 28 °C, the leaf sheaths and barley leaves were observed under a microscope (Chen et al., 2015).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The MoYCP4 gene rescued the defect of the ΔScycp4 mutant. The ΔScycp4 mutant was transformed with the pYES2::MoYCP4 construct encoding MoYcp4 and pYES2::ScYCP4 construct encoding ScYcp4. Serial dilutions of cultures were grown overnight on SD (galactose+0.7M NaCl) and SD (galactose+0.6M KCl) plates, and grown at 30 °C for 4 days and photographed. The experiment was repeated three times and representative results were obtained.
Fig. S2 Targeted gene replacement and complementation. (A) A 955‐bp fragment of the MoYCP4 coding region were replaced by a 1.4‐kb fragment containing the hygromycin B resistance cassette to create the ΔMoycp4 a alleles, respectively. (B) Southern hybridization analysis was used to validate the deletion of MoYCP4 gene and the addition of a single copy integration of the HPH gene. (C) Semiquantitative RT‐PCR was carried out to confirm the deletion and reintroduction of MoYCP4 gene.
Fig. S3 Specificity and competition assays of MoAP1. (A) The DIG‐labeled promoter DNA of MoYCP4 and other gene MoLYS2 was incubated in the absence or presence of purified MoAp1 protein, respectively. (B) Increasing amounts of unlabeled competitor (promoter DNA of MoYCP4) were mixed with the DIG‐labeled promoter DNA of MoYCP4, then the complex were incubated in the purified MoAP1 protein.
Fig. S4 Expression and localization of MoYCP4‐GFP fusion gene. Conidium, appressoria and infectious hyphae of the ΔMoycp4/MoYCP4‐GFP transformant examined under an epifluorescence microscope.
Table S1 Primers used in this study.
Acknowledgements
This research was supported by the Natural Science Foundation of China (Grant No. 31271998 to Z.Z. and Grant No. 31501599 to Y.C.). We thank Yan Wang of Nanjing Agricultural University for critical review of the manuscript and for providing helpful suggestions.
Contributor Information
Yong Liu, Email: haoasliu@163.com.
Zhengguang Zhang, Email: zhgzhang@njau.edu.cn.
References
- Bourett, T.M. and Howard, R.J. (1991) In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea . Can. J. Bot. 68, 329–342. [Google Scholar]
- Bruno, K.S. , Tenjo, F. , Li, L. , Hamer, J.E. and Xu, J.R. (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea . Eukaryot. Cell, 3, 1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuel‐Rios, Z. and Talbot, N.J. (2007) Cellular differentiation and host invasion by the rice blast fungus Magnaporthe grisea . Curr. Opin. Microbiol. 10, 339–345. [DOI] [PubMed] [Google Scholar]
- Cardona, F. , Orozco, H. , Friant, S. , Aranda, A. and del Olmo, M. (2011) The Saccharomyces cerevisiae flavodoxin‐like proteins Ycp4 and Rfs1 play a role in stress response and in the regulation of genes related to metabolism. Arch. Microbiol. 193, 515–525. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Zhai, S. , Zhang, H. , Zuo, R. , Wang, J. , Guo, M. , Zheng, X. , Wang, P. and Zhang, Z. (2014a) Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae . Environ. Microbiol. 16, 788–801. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Zuo, R. , Zhu, Q. , Sun, Y. , Li, M. , Dong, Y. , Ru, Y. , Zhang, H. , Zheng, X. and Zhang, Z. (2014b) MoLys2 is necessary for growth, conidiogenesis, lysine biosynthesis, and pathogenicity in Magnaporthe oryzae . Fungal Genet. Biol. 67, 51–57. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Zhai, S. , Sun, Y. , Li, M. , Dong, Y. , Wang, X. , Zhang, H. , Zheng, X. , Wang, P. and Zhang, Z. (2015) MoTup1 is required for growth, conidiogenesis and pathogenicity of Magnaporthe oryzae . Mol. Plant Pathol. 16, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher, B.S. , Venancio, E.J. , de Freitas, S.M. , Bao, S.N. , Vianney, P.V. , Andrade, R.V. , Dantas, A.S., Soares, C.M., Silva‐Pereira, I. and Felipe, M.S. (2005) The highly expressed yeast gene pby20 from Paracoccidioides brasiliensis encodes a flavodoxin‐like protein. Fungal Genet. Biol. 42, 434–443. [DOI] [PubMed] [Google Scholar]
- Do, T.Q. , Schultz, J.R. and Clarke, C.F. (1996) Enhanced sensitivity of ubiquinone‐deficient mutants of Saccharomyces cerevisiae to products of autoxidized polyunsaturated fatty acids. Proc. Natl. Acad. Sci. USA, 93, 7534–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A.P. , Spellman, P.T. , Kao, C.M. , Carmelharel, O. , Eisen, M.B. , Storz, G. , Botstein, D. and Brown, P.O. (2001) Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell, 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Chen, Y. , Du, Y. , Dong, Y. , Guo, W. , Zhai, S. , Zhang, H. , Dong, S. , Zhang, Z. , Wang, Y. , Wang, P. and Zheng, X. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae . PLoS Pathog. 7, e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, R.J. , Ferrari, M.A. , Roach, D.H. and Money, N.P. (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA, 88, 11 281–11 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, J.C. , McCormack, B.J. , Smirnoff, N. and Talbot, N.J. (1997) Glycerol generates turgor in rice blast. Nature, 389, 244–245. [Google Scholar]
- Kim, S. , Park, S.Y. , Kim, K.S. , Rho, H.S. , Chi, M.H. , Choi, J. , Park, J. , Kong, S. , Park, J. , Goh, J. and Lee, Y.H. (2009) Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae . PLoS Genet. 5, e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo, N. , Taoka, H. , Toda, T. , Yoshida, M. and Horinouchi, S. (1999) A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274, 15 151–15 158. [DOI] [PubMed] [Google Scholar]
- Levin, D.E. (2005) Cell wall integrity signaling in Saccharomyces cerevisiae . Microbiol. Mol. Biol. Rev. 69, 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Myung, K. , Guse, D. , Donkin, B. , Proctor, R.H. , Grayburn, W.S. and Calvo, A.M. (2006) FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides . Mol. Microbiol. 62, 1418–1432. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Xie, S. , Zhao, X. , Chen, X. , Zheng, W. , Lu, G. , Xu, J.R. and Wang, Z. (2010) A homeobox gene is essential for conidiogenesis of the rice blast fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 366–375. [DOI] [PubMed] [Google Scholar]
- Mei, C. , Qi, M. , Sheng, G. and Yang, Y. (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant–Microbe Interact. 19, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Morschhäuser, J. , Li, L. , Naseem, S. , Sharma, S. and Konopka, J.B. (2015) Flavodoxin‐like proteins protect Candida albicans from oxidative stress and promote virulence. PLOS Pathog. 11, e1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M. , Fukada, J. , Moriwaki, A. , Fujikawa, T. , Ohashi, M. , Hibi, T. and Hayashi, N. (2009) Mstu1, an APSES transcription factor, is required for appressorium‐mediated infection in Magnaporthe grisea . Biosci. Biotechnol. Biochem. 73, 1779–1786. [DOI] [PubMed] [Google Scholar]
- Ou, S.H. (1985) Rice Diseases. Kew, Surrey: Commonwealth Mycological Institute. [Google Scholar]
- Qi, Z. , Wang, Q. , Dou, X. , Wang, W. , Zhao, Q. , Lv, R. , Zhang, H. , Zheng, X. , Wang, P. and Zhang, Z. (2012) MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae . Mol. Plant Pathol. 13, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D. (2001) Molecular Cloning – A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Shi, Z. , Christian, D. , Fau‐Leung, H. and Leung, H. (1998) Interactions between spore morphogenetic mutations affect cell types, sporulation, and pathogenesis in Magnaporthe grisea . Mol. Plant–Microbe Interact. 11, 199–207. [DOI] [PubMed] [Google Scholar]
- Sweigard, J.A. , Chumley, F.G. and Valent, B. (1992) Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet. 232, 183–190. [PubMed] [Google Scholar]
- Talbot, N.J. and Foster, A.J. (2001) Genetics and genomics of the rice blast fungus Magnaporthe grisea: developing an experimental model for understanding fungal diseases of cereals. In: Advances in Botanical Research Incorporating Advances in Plant Pathology (Jacqot, J.‐P., ed.) Vol.34, pp. 263–287, Elsevier. [Google Scholar]
- Talbot, N.J. , Ebbole, D.J. and Hamer, J.E. (1993) Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell, 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, B. , Farral, L. and Chumley, F.G. (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics, 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Du, Y. , Zhang, H. , Zhou, C. , Qi, Z. , Zheng, X. , Wang, P. and Zhang, Z. (2013) The actin‐regulating kinase homologue MoArk1 plays a pleiotropic function in Magnaporthe oryzae . Mol. Plant Pathol. 14, 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Zhao, X. , Sun, J. , Kang, Z. , Ding, S. , Xu, J.R. and Peng, Y.L. (2010) A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 23, 112–123. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Tang, W. , Liu, K. , Huang, Q. , Zhang, X. , Yan, X. , Chen, Y., Wang, J., Qi, Z., Wang, Z., Zheng, X., Wang, P. and Zhang, Z. (2011) Eight RGS and RGS‐like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae . PLoS Pathog. 7, e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhao, Q. , Guo, X. , Guo, M. , Qi, Z. , Tang, W. , Dong, Y. , Ye, W. , Zheng, X. , Wang, P. and Zhang, Z. (2014) Pleiotropic function of the putative zinc‐finger protein MoMsn2 in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 27, 446–460. [DOI] [PubMed] [Google Scholar]
- Zhang, H.F. , Zhao, Q. , Liu, K.Y. , Zhang, Z.G. , Wang, Y.C. and Zheng, X.B. (2009) MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol. Lett. 293, 160–169. [DOI] [PubMed] [Google Scholar]
- Zhao, X.H. , Xue, C. , Kim, Y. and Xu, J.R. (2004) A ligation‐PCR approach for generating gene replacement constructs in Magnaporthe grisea . Fungal Genet. Newsl. 51, 17–18. [Google Scholar]
- Zhou, X. , Liu, W. , Wang, C. , Xu, Q. , Wang, Y. , Ding, S. and Xu, JR. (2011) A MADS‐box transcription factor MoMcm1 is required for male fertility, microconidium production and virulence in Magnaporthe oryzae . Mol Microbiol. 80, 33–53. [DOI] [PubMed] [Google Scholar]
- Zhou, Z. , Li, G. , Lin, C. and He, C. (2009) Conidiophore stalk‐less1 encodes a putative zinc‐finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 22, 402–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 The MoYCP4 gene rescued the defect of the ΔScycp4 mutant. The ΔScycp4 mutant was transformed with the pYES2::MoYCP4 construct encoding MoYcp4 and pYES2::ScYCP4 construct encoding ScYcp4. Serial dilutions of cultures were grown overnight on SD (galactose+0.7M NaCl) and SD (galactose+0.6M KCl) plates, and grown at 30 °C for 4 days and photographed. The experiment was repeated three times and representative results were obtained.
Fig. S2 Targeted gene replacement and complementation. (A) A 955‐bp fragment of the MoYCP4 coding region were replaced by a 1.4‐kb fragment containing the hygromycin B resistance cassette to create the ΔMoycp4 a alleles, respectively. (B) Southern hybridization analysis was used to validate the deletion of MoYCP4 gene and the addition of a single copy integration of the HPH gene. (C) Semiquantitative RT‐PCR was carried out to confirm the deletion and reintroduction of MoYCP4 gene.
Fig. S3 Specificity and competition assays of MoAP1. (A) The DIG‐labeled promoter DNA of MoYCP4 and other gene MoLYS2 was incubated in the absence or presence of purified MoAp1 protein, respectively. (B) Increasing amounts of unlabeled competitor (promoter DNA of MoYCP4) were mixed with the DIG‐labeled promoter DNA of MoYCP4, then the complex were incubated in the purified MoAP1 protein.
Fig. S4 Expression and localization of MoYCP4‐GFP fusion gene. Conidium, appressoria and infectious hyphae of the ΔMoycp4/MoYCP4‐GFP transformant examined under an epifluorescence microscope.
Table S1 Primers used in this study.


