Summary
Hibiscus green spot virus (HGSV) is a recently discovered and so far poorly characterized bacilliform plant virus with a positive‐stranded RNA genome consisting of three RNA species. Here, we demonstrate that the proteins encoded by the ORF2 and ORF3 in HGSV RNA2 are necessary and sufficient to mediate cell‐to‐cell movement of transport‐deficient Potato virus X in Nicotiana benthamiana. These two genes represent a specialized transport module called a ‘binary movement block’ (BMB), and ORF2 and ORF3 are termed BMB1 and BMB2 genes. In agroinfiltrated epidermal cells of N. benthamiana, green fluorescent protein (GFP)‐BMB1 fusion protein was distributed diffusely in the cytoplasm and the nucleus. However, in the presence of BMB2, GFP‐BMB1 was directed to cell wall‐adjacent elongated bodies at the cell periphery, to cell wall‐embedded punctate structures co‐localizing with callose deposits at plasmodesmata, and to cells adjacent to the initially transformed cell. Thus, BMB2 can mediate the transport of BMB1 to and through plasmodesmata. In general, our observations support the idea that cell‐to‐cell trafficking of movement proteins involves an initial delivery to membrane compartments adjacent to plasmodesmata, subsequent entry of the plasmodesmata cavity and, finally, transport to adjacent cells. This process, as an alternative to tubule‐based transport, has most likely evolved independently in triple gene block (TGB), double gene block (DGB), BMB and the single gene‐coded transport system.
Keywords: cell‐to‐cell movement, Hibiscus green spot virus, movement protein, plant virus, plasmodesmata
Introduction
Studies of plant virus cell‐to‐cell movement carried out in the last 35 years have revealed several types of viral transport system exhibiting remarkable differences in their gene organization, as well as in the structure and biochemical properties of their encoded movement proteins (MPs), with the movement functions being performed by a single protein or distributed among several MPs (Heinlein, 2015; Lucas, 2006; Tilsner et al., 2014).
The most‐studied model MP, which represents a single gene‐coded cell‐to‐cell transport system, is the 30‐kDa protein (30K) of Tobacco mosaic virus (TMV) and other viruses of the genus Tobamovirus (Heinlein, 2015; Liu and Nelson, 2013; Niehl and Heinlein, 2011; Niehl et al., 2013, 2014). The TMV 30K MP is capable of intercellular movement and modification of plasmodesmata (PD) independently of other virus proteins and virus genomic RNA (Oparka et al., 1995; Tomenius et al., 1987; Waigmann et al., 1994). However, in the context of TMV infection, cell‐to‐cell movement is tightly linked to virus replication (Guenoune‐Gelbart et al., 2008; Hirashima and Watanabe, 2001; Peña et al., 2014). The 30K MP has several activities, such as viral RNA‐binding, interaction with endomembranes and associated cytoskeleton, and PD gating (Heinlein, 2015, 2016; Liu and Nelson, 2013). The TMV MP belongs to the so‐called ‘30K superfamily’ of structure‐related MPs with no enzymatic functions (Melcher, 2000; Mushegian and Elena, 2015), which includes, in addition to the MPs of tobamoviruses, the MPs encoded by diverse viruses with positive‐ and negative‐stranded RNA genomes, including viruses of the family Bromoviridae, genera Furovirus, Tobravirus, Umbravirus, Ourmiavirus, Cytorhabdovirus and Tospovirus, and many representatives of the order Tymovirales (Martelli et al., 2007; Mushegian and Elena, 2015).
Among the transport systems of plant RNA viruses encoding more than one MP, the double gene block (DGB) is the shortest gene module, which encodes two small non‐enzymatic MPs (Hull, 2002). The DGB is characteristic of the many Tombusviridae genera, including Carmovirus, Machlomovirus, Alphanecrovirus, Betanecrovirus, Panicovirus, Avenavirus, Macanavirus, Galantivirus and Pelarspovirus (Scheets et al., 2015). The first DGB gene encodes the RNA‐binding protein believed to form transport ribonucleoproteins (RNPs), whereas the second DGB gene codes for a transmembrane protein, which is targeted to PD in a Golgi‐ and actin‐dependent manner (Genovés et al., 2010; Navarro et al., 2006). Apparently, the most complex transport system is found in closteroviruses (viruses of the family Closteroviridae). It consists of five proteins encoded by a quintuple gene block (QGB), namely a small transmembrane protein, a heat shock protein 70 (Hsp70) chaperone‐like protein (Hsp70h) with enzymatic ATPase activity, coat protein (CP) and two distant coat protein homologues, p64 and CPm (Dolja et al., 2006). Coordinated incorporation of the last four proteins (CP, CPm, Hsp70h and p64) into filamentous virions, which are targeted to PD with the aid of the small transmembrane protein, is required for virus cell‐to‐cell movement (Dolja et al., 2006; Kiss et al., 2013). The Hsp70h ATPase activity is essential for Beet yellows virus (BYV) transport through PD. Moreover, when expressed alone, this protein is incorporated into granular structures capable of actin‐dependent transport to PD (Alzhanova et al., 2001; Prokhnevsky et al., 2005), suggesting that Hsp70h can serve as a ‘driving force’ for virion intracellular trafficking. Yet another multicomponent movement system including an enzymatic protein is found in potyviruses (the family Potyviridae). The enzymatic protein, designated as a cylindrical inclusion (CI) protein, is a replicative superfamily II (SFII) RNA helicase capable of targeting to PD, forming PD‐associated cylindrical structures, and guiding potyviral filamentous virions to and through PD (Revers and García, 2015; Sorel et al., 2014). In addition, four more potyviral proteins, namely the CP, the helper component proteinase (HC‐Pro), the viral genome‐linked protein (VPg) and P3N‐PIPO are involved in viral intercellular movement (Revers and García, 2015). The VPg protein of encapsidated genomic RNA binds to HC‐Pro (Torrance et al., 2006), which, in turn, can interact with the CI protein (Gabrenaite‐Verkhovskaya et al., 2008) targeted to PD by P3N‐PIPO, the protein capable of directed trafficking to PD and translocation through PD to adjacent cells (Vijayapalani et al., 2012). In addition, the transmembrane 6K2 protein induces the formation of replication vesicles, which can move from cell to cell during Turnip mosaic potyvirus infection (Grangeon et al., 2013; Jiang et al., 2015). Thus, the 6K2 membrane protein involved in directed transport to PD makes the potyvirus transport system reminiscent of the closterovirus QGB and the DGB of Tombusviridae.
A distinct type of plant virus transport system is represented by the triple gene block (TGB) found in many virus genera of the Alphaflexiviridae, Betaflexiviridae and Virgaviridae families and the genus Benyvirus. Three partially overlapping TGB genes encode proteins essential for virus cell‐to‐cell movement. In addition to the TGB proteins, some of these viruses (possessing ‘potex‐like’ TGB in Alphaflexiviridae and Betaflexiviridae) depend on the CP in both cell‐to‐cell and long‐distance transport (Morozov and Solovyev, 2003; Park et al., 2014; Solovyev et al., 2012; Verchot‐Lubicz et al., 2010). TGB1 belongs to a divergent lineage of ‘accessory’ viral SFI helicases that evolved after a duplication of the replicative RNA helicase domain (Koonin and Dolja, 1993; Morozov and Solovyev, 2012). It is a multifunctional protein, which, in addition to its RNA helicase activity, is able to bind single‐stranded RNA. This protein is essential for the formation of the transport form of the viral genome, either RNPs formed by TGB1 and viral genomic RNA in viruses with a ‘hordei‐like’ TGB, or filamentous virions modified by binding of TGB1 molecules to the virion end containing the 5′‐terminus of viral genomic RNA, as found for viruses with a ‘potex‐like’ TGB (Morozov and Solovyev, 2003; Obraztsova et al., 2015; Verchot‐Lubicz et al., 2010). TGB2 and TGB3 are small proteins integrated into cell membranes by hydrophobic sequence segments and are required for the delivery of TGB1 to PD. Several studies have suggested that TGB3 contains specific signals for PD targeting, and that therefore this protein can serve as a ‘driving force’ for the targeting of TGB2‐ and TGB1‐containing movement‐competent complexes (RNPs or virions) to PD‐associated membrane domains (Lim et al., 2009; Park et al., 2014; Schepetilnikov et al., 2005; Shemyakina et al., 2011; Zamyatnin et al., 2004).
Some recently discovered plant viruses have been found to encode putative transport modules including ‘accessory’ helicase genes and small hydrophobic protein genes (Morozov and Solovyev, 2012). Among these viruses is Hibiscus green spot virus (HGSV, proposed genus Higrevirus). This virus forms bacilliform particles and carries a TGB‐like gene module (Melzer et al., 2012). The HGSV genome consists of three polyadenylated RNAs, designated as RNA1 (8.35 kb), RNA2 (3.17 kb) and RNA3 (3.11 kb). HGSV RNA1 has a single open reading frame (ORF) for viral replicase protein, RNA2 possesses four ORFs and encodes a protein of unknown function (ORF1) and three TGB‐like proteins (ORF2, ORF3 and ORF4), and RNA3 codes for the viral CP and two other proteins of unknown function (Melzer et al., 2012). Similarly to TGB1 proteins, the HGSV RNA2 ORF2‐encoded protein comprises a helicase sequence domain, which, however, shows stronger similarity to SFI replicative helicases of the genus Benyvirus than to those of TGB1 proteins. Both HGSV ORF3‐ and ORF4‐encoded proteins, like TGB2 and TGB3 proteins, contain long hydrophobic segments, but show only marginal similarity to other TGB proteins (Morozov and Solovyev, 2012, 2015).
In this work, we used the HGSV ORF2, ORF3 and ORF4 coding sequences for transient expression assays to analyse the subcellular localization of the HGSV TGB‐like proteins and their possible involvement in virus cell‐to‐cell movement.
Results
HGSV proteins mediate cell‐to‐cell transport of Potato virus X (PVX)
To experimentally verify that the proteins encoded by ORF2, ORF3 and ORF4 in the HGSV genomic RNA2 (Fig. 1A) represent viral MPs corresponding to the TGB proteins in other plant viruses (Melzer et al., 2012), a virus movement complementation assay was used. For these experiments, the HGSV ORF2, ORF3 and ORF4 coding sequences were cloned into a binary vector under the control of the Cauliflower mosaic virus 35S promoter (Fig. 1B). Agrobacterial cultures carrying the constructed vectors for the expression of ORF2, ORF3 and ORF4 were used, individually or in combination, for the infiltration of Nicotiana benthamiana leaves, together with an agrobacterial culture carrying a binary vector with the 35S promoter‐driven construct PVX‐POL‐GFP (Komarova et al., 2006). This clone encodes a PVX genome derivative containing only the viral replicase gene and an inserted green fluorescent protein (GFP) gene (Fig. 1C). The transcribed RNA of this virus is able to replicate and express GFP in initially infected cells, but is deficient in cell‐to‐cell movement because of the absence of TGB and CP genes. In co‐infiltration experiments, the HGSV constructs were agroinfiltrated at a high density of bacterial culture, resulting in the expression of proteins in most cells of the infiltrated leaf areas, whereas the PVX‐POL‐GFP culture was 2000‐fold diluted prior to infiltration (see Experimental Procedures) to ensure the initiation of PVX‐POL‐GFP infection in individual cells located rather far from each other.
Figure 1.
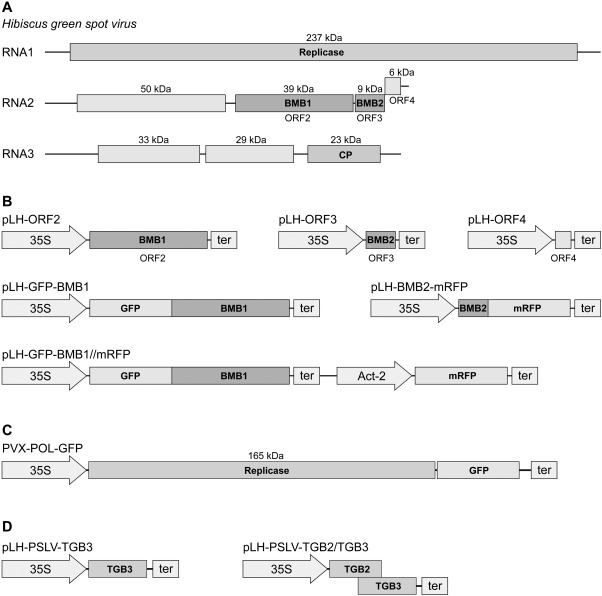
The genome of Hibiscus green spot virus (HGSV) and constructs with HGSV genes used in this study. (A) Schematic representation of the HGSV genome. Boxes indicate genes; the molecular weights of encoded HGSV proteins are indicated. CP, coat protein. (B) Expression cassettes in the binary vectors used. Arrows indicate promoters. 35S, Cauliflower mosaic virus 35S promoter; Act‐2, Arabidopsis thaliana Actin‐2 promoter; ter, transcriptional terminator. (C) Schematic representation of the transport‐deficient Potato virus X (PVX) derivative PVX‐POL‐GFP cloned in a binary vector under the control of the 35S promoter. (D) Expression cassettes in binary vectors for the expression of Poa semilatent virus (PSLV) TGB2 and TGB2/TGB3 proteins. BMB, binary movement block; GFP, green fluorescent protein; ORF, open reading frame; TGB, triple gene block.
Infiltrated N. benthamiana leaves were examined under UV light at 4 days post‐infiltration (dpi). Leaf areas infected with PVX‐POL‐GFP produced low overall fluorescence signals visible to the unaided eye as tiny bright fluorescent spots (Fig. 2A). Similar fluorescence patterns were observed in areas in which PVX‐POL‐GFP was co‐expressed with individual HGSV proteins (ORF2, ORF3 or ORF4 products), the combination of ORF2‐ and ORF4‐encoded proteins, and the combination of ORF3‐ and ORF4‐encoded proteins (Fig. 2A and data not shown). As revealed by microscopy, fluorescent spots in these infiltration areas in most cases corresponded to single cells or, rarely, pairs of neighbouring cells (Fig. 2B,C). By contrast, infection with PVX‐POL‐GFP in the presence of co‐expressed ORF2 and ORF3 proteins resulted in substantially larger fluorescent foci visible in the infiltrated area, and similar larger foci were also observed for infection with PVX‐POL‐GFP in the presence of co‐expressed ORF2, ORF3 and ORF4 constructs (Fig. 2A). Fluorescence microscopy revealed that, in both cases, these foci represented groups of fluorescent cells (Fig. 2D,E). The larger foci sizes indicate the ability of movement‐deficient PVX‐POL‐GFP to move from initially infected cells and to form multicellular infection sites. Therefore, these observations revealed that the expression of two combinations of HGSV proteins, namely ORF2 + ORF3 and ORF2 + ORF3 + ORF4, complemented the cell‐to‐cell transport of PVX‐POL‐GFP. Therefore, the ORF2 + ORF3 combination was functional in movement complementation in both the presence and absence of the ORF4 protein. In leaves co‐agroinfiltrated with ORF2 + ORF3 + ORF4 constructs, the expression of the ORF4 gene was confirmed by reverse transcription‐polymerase chain reaction (RT‐PCR) with ORF4‐specific primers (data not shown). To analyse quantitatively whether the ORF4 product had any influence on the efficiency of PVX‐POL‐GFP movement complementation by the ORF2 + ORF3 construct combination, the sizes of the fluorescent foci were measured in leaf areas infected with PVX‐POL‐GFP co‐expressed with either ORF2 + ORF3 or ORF2 + ORF3 + ORF4 proteins. The mean foci sizes calculated for 541 and 488 measured loci, respectively, demonstrated the absence of a statistically significant difference between these two complementation experiments (Fig. 2F).
Figure 2.
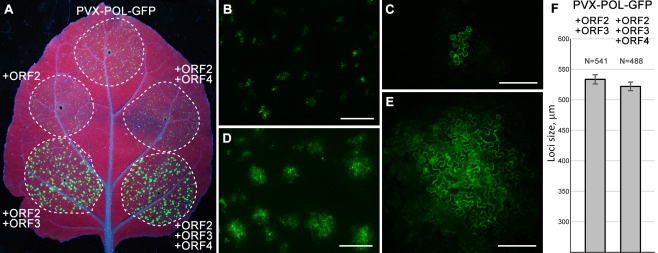
Complementation of cell‐to‐cell movement of transport‐deficient Potato virus X (PVX) derivative PVX‐POL‐GFP by Hibiscus green spot virus (HGSV) proteins in Nicotiana benthamiana leaves. (A) Green fluorescent protein (GFP) fluorescence in the leaf infiltrated with PVX‐POL‐GFP or PVX‐POL‐GFP in combination with the indicated HGSV protein‐encoding constructs. The leaf was imaged under UV light at 4 days post‐infiltration (dpi). Broken lines encircle infiltrated areas. (B–E) Fluorescence microscopy of infiltrated areas at low magnification (B, D) and higher magnification (C, E). (B, C) Images taken for PVX‐POL‐GFP. (D, E) Images taken for PVX‐POL‐GFP + ORF2 + ORF3. Scale bars: (B, D) 500 μm; (C, E) 200 μm. (F) The mean sizes of fluorescent foci observed in leaf areas in which PVX‐POL‐GFP was co‐agroinfiltrated with either ORF2 + ORF3 or ORF2 + ORF3 + ORF4 construct. ORF, open reading frame.
These data demonstrate that the HGSV proteins encoded by ORF2 and ORF3 are necessary and sufficient to mediate cell‐to‐cell movement of PVX‐POL‐GFP in N. benthamiama. Thus, we conclude that ORF2 and ORF3 of HGSV genomic RNA2 represent a specialized transport module, which is called hereafter the ‘binary movement block’ (BMB) to distinguish it from DGB found in Tombusviridae (Hull, 2002), which does not encode a helicase and consists of two small non‐enzymatic MPs. Accordingly, the HGSV RNA2 ORF2 and ORF3 are termed BMB1 and BMB2 genes, respectively.
To estimate the efficiency of the two BMB proteins in mediating viral movement in the experimental system used, we compared the transport of PVX‐POL‐GFP in the presence of HGSV BMB1 + BMB2 and in the presence of the well‐studied TMV MP known to complement in trans the cell‐to‐cell movement of PVX (Fedorkin et al., 2001; Morozov et al., 1997). Examination of infiltrated leaves under UV light carried out at 4 dpi revealed that the infection foci formed in the presence of the TMV MP were slightly brighter and larger, but comparable in size, with those formed in the same leaves in the presence of BMB1/BMB2 (Fig. 3). This observation indicates that the effect of BMB1/BMB2 on PVX‐POL‐GFP is, as in the case of the TMV MP, a result of movement complementation rather than an influence of BMB proteins on the physiological state of the leaves.
Figure 3.
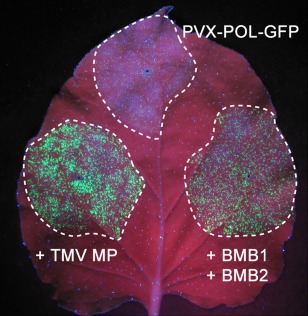
Comparison of PVX‐POL‐GFP movement complementation by Hibiscus green spot virus (HGSV) BMB1 + BMB2 and Tobacco mosaic virus (TMV) 30K movement protein (MP). The leaf was agroinfiltrated for infection with PVX‐POL‐GFP or PVX‐POL‐GFP in the presence of the indicated constructs. Green fluorescent protein (GFP) fluorescence was detected under UV light at 4 days post‐infiltration (dpi). Broken lines encircle infiltrated areas. BMB, binary movement block; PVX, Potato virus X.
Subcellular localization of individual BMB proteins
To analyse the subcellular localization of BMB proteins, the BMB1 and BMB2 genes were translationally fused to the genes of GFP and monomeric red fluorescent protein (mRFP) (Fig. 1B). In the two BMB1 constructs GFP‐BMB1 and BMB1‐GFP, GFP was fused to the N‐ and C‐termini of the encoded BMB1 protein, respectively. Similarly, mRFP‐BMB2 and BMB2‐mRFP constructs were obtained. Agrobacterial cultures carrying binary vectors with the fusion genes cloned in a 35S promoter‐based expression cassette were used for the infiltration of N. benthamiana leaves. The subcellular localization of the fusion proteins in leaf epidermal cells was analysed by confocal laser scanning microscopy at 2 or 3 dpi.
GFP‐BMB1 and BMB1‐GFP localized similarly; therefore, only the data for GFP‐BMB1 are presented below. This fusion protein exhibited diffuse localization in the cytoplasm and the nucleus (Fig. 4A), and was therefore similar to the localization of non‐fused fluorescent proteins in plant cells. However, unlike non‐fused mRFP, which is excluded from the nucleolus (Fig. 4B), GFP‐BMB1 was uniformly distributed in the nucleoplasm (Fig. 4A). In addition, GFP‐BMB1 formed a ring around the nucleus (Fig. 4A).
Figure 4.
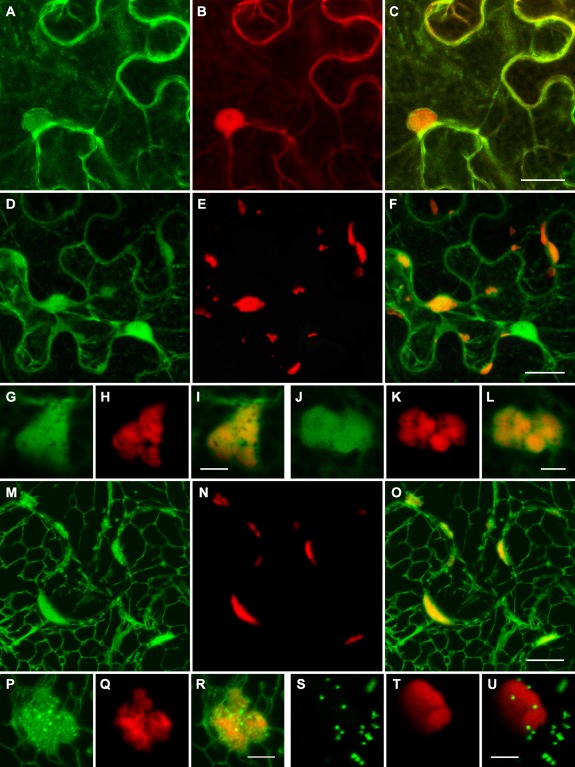
Subcellular localization of GFP‐BMB1 and BMB2‐mRFP. (A–C) Co‐expression of GFP‐BMB1 and mRFP. (D–L) Co‐expression of BMB2‐mRFP and GFP. (M–R) Co‐expression of BMB2‐mRFP and ER‐GFP. (S–U) Co‐expression of BMB2‐mRFP and the Golgi marker ST‐GFP. Higher magnification images in (G–L) show the localization of the BMB2‐mRFP bodies in GFP‐containing cytoplasmic areas, and in (P–R) show fine granular ER‐GFP‐containing structures associated with the BMB2‐mRFP bodies. GFP and mRFP fluorescence signals were imaged independently. Merged images are shown in (C), (F), (I), (L), (O), (R) and (U). All images are reconstructed from Z‐series of optical sections. Scale bars: (C, F) 20 μm; (O) 10 μm; (I, L, R, U) 5 μm. BMB, binary movement block; ER, endoplasmic reticulum; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
BMB2‐mRFP was found in bodies of different shapes and sizes, located mostly close to the cell wall of epidermal cells and, in addition, in the cytoplasm (Fig. 4E). A similar localization was observed for mRFP‐BMB2 (data not shown). Higher magnification imaging revealed that the BMB2‐containing bodies had a complex structure, having many holes and cavities (Fig. 4H,K). In cells co‐expressing BMB2‐mRFP and GFP, the BMB2 bodies were visible within cytoplasmic sack regions containing diffusely localized GFP (Fig. 4D,G,J). Co‐expression of BMB2‐mRFP with the endoplasmic reticulum (ER) marker m‐GFP5‐ER (Zamyatnin et al., 2002) demonstrated that the BMB2 bodies co‐localized with m‐GFP5‐ER‐containing ER subdomains connected to the cortical ER network (Fig. 4M–O). Interestingly, a part of the ER marker co‐localized with BMB2‐mRFP was visible in fine granular substructures and seemed to surround the BMB2‐containing bodies (Fig. 4P–R). In addition, BMB2‐mRFP did not co‐localize with the Golgi marker (Fig. 4S–U).
To verify the integrity of GFP‐BMB1 and BMB2‐mRFP fusion proteins expressed in plants, Western blotting was carried out with GFP‐ and mRFP‐specific antibodies, respectively. The GFP‐BMB1 fusion protein migrated in gel as a single band of the expected mobility (Fig. S1A, see Supporting Information), whereas BMB2‐mRFP was detected as a band of molecular mass of more than 100 kDa (Fig. S1B). Similarly, a hordeivirus TGB3 protein has been found previously in high‐molecular‐mass protein complexes (Shemyakina et al., 2011). In an additional experiment, we found that the HGSV ORF4‐GFP fusion protein had the expected mobility in gel and was localized to the cortical ER (Fig. S2, see Supporting Information), which was in agreement with the presence of hydrophobic sequence segments in this protein (Morozov and Solovyev, 2012).
BMB2 directs trafficking of BMB1 to subdomains of cortical ER, PD and neighbouring cells
To analyse whether the HGSV BMB1 and BMB2 can influence the subcellular localization of each other, these proteins were co‐expressed in N. benthamiana leaves by agroinfiltration.
Co‐expression of GFP‐BMB1 with BMB2 dramatically changed the localization of GFP‐BMB1. Instead of the diffuse distribution throughout the cytoplasm and the nucleus observed for GFP‐BMB1 expressed alone, GFP‐BMB1 co‐expressed with the BMB2 protein was found in heterogeneous bodies, often being elongated structures appressed against the cell wall, similar to the BMB2‐mRFP‐containing bodies (Fig. 5A). To test directly whether BMB1 can be targeted to BMB2‐containing structures, GFP‐BMB1 was co‐expressed with BMB2‐mRFP. Indeed, both fusion proteins co‐localized (Fig. 5D–F). Higher magnification imaging demonstrated perfect co‐localization of GFP‐BMB1 and BMB2‐mRFP in the observed bodies (Fig. 5G–L). Thus, the HGSV BMB2 protein is able to direct the intracellular targeting of BMB1.
Figure 5.
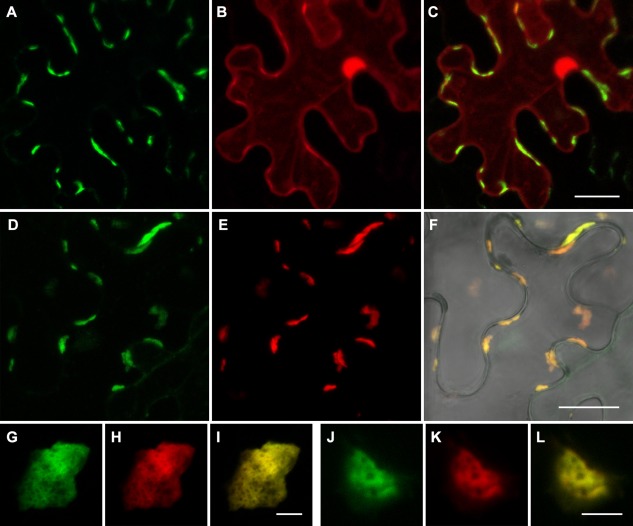
BMB2‐directed trafficking of BMB1 to cell peripheral compartments. (A–C) Co‐expression of GFP‐BMB1, BMB2 and mRFP. (D–L) Co‐expression of GFP‐BMB1 and BMB2‐mRFP. (F) Merged (D) and (E) images superimposed on a bright‐field image to show the position of the cell walls. Higher magnification images in (G–L) show perfect co‐localization of GFP‐BMB1 and BMB2‐mRFP. All images are reconstructed from Z‐series of optical sections. Scale bars: (C, F) 20 μm; (I, L) 5 μm. BMB, binary movement block; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
Next, the potential of BMB2 to mediate the cell‐to‐cell transport of BMB1 was analysed. To enable the visualization of BMB1‐expressing cells, we constructed a binary vector carrying an additional fluorescent marker gene. To this end, the binary vector carrying the GFP‐BMB1 fusion gene under the control of the 35S promoter and 35S terminator, which was used in the experiments described above, was modified by insertion of an additional expression cassette with the mRFP gene under the control of the Arabidopsis thaliana Actin‐2 promoter and the nopaline synthase transcriptional terminator (pLH‐GFP‐BMB1//mRFP; Fig. 1B). This construct was agroinfiltrated into N. benthamiana leaves at low density of bacterial culture (dilution 1 : 200) to achieve protein expression in individual cells located distantly from each other. In these conditions, when examined at 3 dpi, GFP‐BMB1 exhibited the localization typical for this protein and was confined to cells expressing mRFP (Fig. 6A–C), showing that GFP‐BMB1 itself is incapable of cell‐to‐cell movement. When the agrobacterial culture at low density was co‐infiltrated with the BMB2 culture at high density, GFP‐BMB1 was observed in the individual cells expressing mRFP, as well as in surrounding cells, being localized to discrete sites in the cell walls (Fig. 6D–F). As the BMB2 protein was expressed in most leaf cells, GFP‐BMB1 was transported not only to cells immediately adjacent to the cell in which it was expressed, but also to more distant epidermal cells. These observations demonstrate the ability of BMB2 to mediate cell‐to‐cell transport of BMB1. It should be noted that the mRFP fused to BMB2 interfered with the ability of BMB2 to mediate the cell‐to‐cell transport of GFP‐BMB1 (data not shown), although BMB2‐mRFP and mRFP‐BMB2 were competent in targeting GFP‐BMB1 to cell peripheral structures, thus indicating that these fusion proteins were only partially functional.
Figure 6.
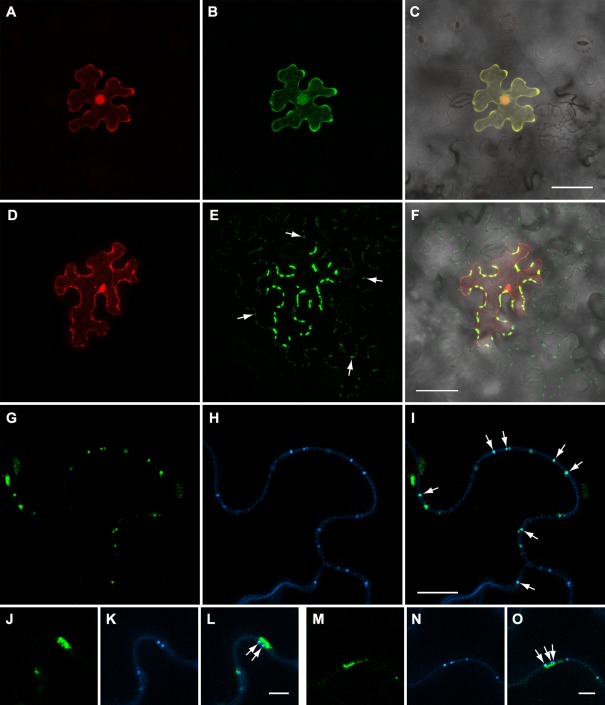
BMB2‐directed transport of BMB1 to plasmodesmata (PD) and neighbouring cells. (A–C) Epidermal cell expressing GFP‐BMB1 and mRFP in a leaf agroinfiltrated at low bacterial culture density with GFP‐BMB1//mRFP, a vector with two expression cassettes, to ensure agrotransformation of individual cells located distantly from each other. (D, E) Epidermal cell of a leaf co‐agroinfiltrated with GFP‐BMB1//mRFP at low bacterial culture density and the BMB2 construct at high culture density. (F) Merged (D) and (E) images superimposed on a bright‐field image to show the localization of GFP‐BMB1 along the cell walls of cells surrounding the primary cell. Arrows in (E) point to some of the discrete sites of GFP‐BMB1 localization in cells adjacent to the primary cell. (G–O) Callose staining with aniline blue in leaves co‐expressing GFP‐BMB1//mRFP and BMB2. (G–I) Cell walls between secondary cells are shown; arrows point to co‐localization of the GFP‐BMB1‐containing sites in cell walls with stained callose deposits. (J–O) Cell wall‐embedded BMB1‐containing punctate bodies located in close proximity to elongated membrane bodies and co‐localized with stained callose (shown with arrows). (J–L) Images taken for a primary cell. (M–O) Images taken for a secondary cell. Images in (A–F) are reconstructed from Z‐series of optical sections. Images in (G–O) represent single optical sections. Scale bars: (C, F) 50 μm; (I) 10 μm; (L, O) 5 μm. BMB, binary movement block; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
To determine whether the GFP‐BMB1‐containing foci in cell walls could represent PD‐associated sites, specimens of leaves co‐expressing GFP‐BMB1//mRFP and BMB2 were stained for PD‐associated callose with aniline blue. In cells to which GFP‐BMB1 had been transported from primary cells expressing the mRFP marker, the GFP‐BMB1‐containing sites in cell walls were found to co‐localize with stained callose deposits (Fig. 6G–I). In primary cells, where the BMB1 protein mostly accumulated in cortical ER subdomains, it was also observed in cell wall‐embedded punctate bodies co‐localized with stained callose (Fig. 6J–O). These observations show that BMB2 can direct the BMB1 protein to cortical ER subdomains in close vicinity to PD and to PD themselves.
Heterologous TGB2/TGB3 proteins cannot functionally replace BMB2
To analyse whether BMB2 could be functionally replaced with TGB2 and TGB3 proteins, we tested whether TGB2 and TGB3 would complement BMB1 functions and allow PVX‐POL‐GFP cell‐to‐cell movement. Thus, a bacterial culture with the PVX‐POL‐GFP construct was diluted into pre‐mixed high‐density cultures of BMB1, BMB1 and Poa semilatent virus (PSLV, genus Hordeivirus) TGB2, or BMB1 and PSLV TGB2/TGB3, and infiltrated into leaves. The latter construct contained the PSLV genome region, which encompassed the overlapping TGB2 and TGB3 genes, under the control of the 35S promoter (Fig. 1D). This construct could ensure the expression of TGB3 at a relatively low, compared with TGB2, level and avoid the TGB3 overexpression known to be detrimental for hordeivirus infections (Lim et al., 2009). As controls, the diluted PVX‐POL‐GFP construct was either agroinfiltrated alone, or co‐infiltrated with BMB1 and BMB2 constructs. Examination of infiltrated N. benthamiana leaves under UV light at 4 dpi revealed that the combination of HGSV BMB1 with PSLV TGB2/TGB3 was unable to complement the cell‐to‐cell transport of PVX‐POL‐GFP (Fig. 7A). In addition, the potential of PSLV TGB2 and TGB3 to influence the subcellular localization of GFP‐BMB1 was analysed in co‐agroinfiltration experiments. Confocal microscopy revealed that the localization of GFP‐BMB1 remained largely unchanged in the presence of PSLV TGB2 and TGB3 (Fig. 7B,C). These data indicate that the BMB2‐dependent targeting of BMB1 depends on a specific interaction between BMB1 and BMB2, and that BMB2 cannot be functionally replaced by TGB proteins.
Figure 7.
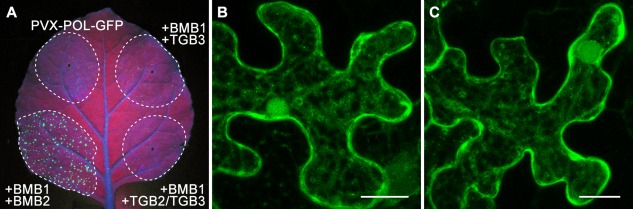
The Poa semilatent virus (PSLV) TGB2 and TGB3 proteins are unable to functionally replace the Hibiscus green spot virus (HGSV) BMB2 protein. (A) Complementation of cell‐to‐cell movement of PVX‐POL‐GFP by combinations of BMB1 with BMB2, TGB3 and TGB2 + TGB3. The leaf was imaged under UV light at 4 days post‐infiltration (dpi). Broken lines encircle infiltrated areas. (B) Co‐expression of GFP‐BMB1 with PSLV TGB3. (C) Co‐expression of GFP‐BMB1 with PSLV TGB2 and TGB3. Images in (B) are reconstructed from Z‐series of optical sections. Scale bars: (B, C) 20 μm. BMB, binary movement block; GFP, green fluorescent protein; PVX, Potato virus X; TGB, triple gene block.
Discussion
Studies of plant virus movement have revealed several types of viral transport system, which radically differ in terms of the number and properties of MPs involved. In this article, we describe yet another new transport gene module found in the HGSV genome.
In a transient complementation assay, the combination of proteins encoded by ORF2 and ORF3 in the HGSV genomic RNA2 was necessary and sufficient for cell‐to‐cell movement of transport‐deficient PVX in N. benthamiana. We refer to this transport module as the ‘binary movement block’ (BMB) composed of the BMB1 gene (ORF2) and the BMB2 gene (ORF3). Previously, the HGSV BMB1 and BMB2 genes, together with the ORF4 located just downstream of the BMB2 gene, have been suggested to correspond to the TGB transport module, consisting of genes TGB1, TGB2 and TGB3, in other plant viruses (Melzer et al., 2012; Morozov and Solovyev, 2012, 2015). However, our experiments reveal that the HGSV ORF4 product is not necessary for the complementation of PVX movement, and that this protein has no influence on the efficiency of PVX movement mediated by the BMB1 and BMB2 proteins, thus showing a clear difference between the TGB and BMB transport systems. Furthermore, as noted previously, the BMB1 and BMB2 proteins are only distantly related to the TGB1 and TGB2 proteins, respectively, as the BMB1 helicase domain is more closely related to viral replicative helicases than to TGB1 proteins, and BMB2, although having two highly hydrophobic sequence regions as all known TGB2 proteins, has only a limited sequence similarity to the most conserved central hydrophilic region of TGB2 (Morozov and Solovyev, 2012). These observations substantiate our conclusion that BMB represents a novel movement gene module different from TGB. We presume that BMB originated independently of TGB, i.e. by autonomization of a replicative helicase domain and subsequent acquisition of the BMB2 gene.
The conclusion that the HGSV BMB transport system includes only two essential proteins is consistent with our previous sequence analyses of numerous available plant transcriptomes, which sometimes contain, in addition to true plant mRNA sequences, sequences of RNA plant viruses infecting the analysed plant. We have revealed virus‐like RNAs encoding proteins closely related to viral replicases and containing ORFs for two proteins highly similar to HGSV BMB1 and BMB2. In such virus‐like RNAs found in Lathyrus sativus and Litchi chinensis, no ORF is present downstream of the BMB2‐related gene (Morozov and Solovyev, 2015). Although these identified virus‐like RNAs have not been characterized experimentally, this observation indirectly confirms that BMB1 and BMB2 proteins are sufficient for virus movement.
It should be noted that the analysis of the movement functions of HGSV proteins was carried out in N. benthamiana, whereas the known HGSV plant hosts are Citrus volkameriana and Hibiscus arnottianus. We cannot exclude the possibility that the RNA2 ORF4 protein, although not required for cell‐to‐cell movement in N. benthamiana, facilitates the transport of HGSV in its natural hosts. An auxiliary host‐specific p33 MP has been described recently for Citrus tristeza virus (CTV, genus Closterovirus) (Bak and Folimonova, 2015). To verify this prediction, we attempted to carry out PVX movement complementation tests in hibiscus plants, but failed to achieve transient protein expression in infiltrated hibiscus leaves with any Agrobacterium strain used (data not shown). Therefore, the function of the polypeptide encoded by the HGSV RNA2 ORF4 awaits further experimental investigations. Interestingly, HGSV was originally isolated from citrus plants infected also with CTV (Melzer et al., 2012), suggesting that some HGSV movement functions in this particular host may rely on CTV MPs. Nevertheless, irrespective of ORF4 function, complementation experiments demonstrated that the BMB1 and BMB2 proteins have all the necessary functional activities and are competent for functional interactions required for intracellular transport to PD and translocation through PD microchannels into neighbouring cells. This conclusion is further supported by the observed BMB1 and BMB2 subcellular localization patterns.
BMB1 was localized diffusely in the cytoplasm and the nucleus when expressed alone. In co‐expression experiments, BMB2 was able to direct BMB1 to sites of BMB2 localization in cortical ER bodies and, in addition, to PD identified by callose staining. Furthermore, BMB2 directed the transport of BMB1 to neighbouring cells, where BMB1 was found in PD. These findings are consistent with the subcellular localization and transport of Potato mop‐top virus (PMTV, genus Pomovirus), in which the TGB1 protein is directed by two smaller TGB proteins, TGB2 and TGB3, from cytoplasmic TGB1 locations to peripheral membrane bodies, PD and neighbouring cells (Zamyatnin et al., 2004). Thus, our HGSV findings support the hypothesis, which has been proposed previously on the basis of PMTV experiments, that the process of MP transport includes two stages: (i) delivery to membrane structures in close vicinity to PD; and (ii) translocation into the PD internal cavity and further to neighbouring cells (Zamyatnin et al., 2004). Recently, a similar two‐stage transport strategy has been reported for the 30K TMV MP, for which the delivery to PD, but not further ingress into PD and transport through PD, is specified by the protein N‐terminal region and can be uncoupled from other protein functions (Yuan et al., 2016). As shown for TMV movement, both stages of translocation depend on the functions of myosins (Amari et al., 2014). Importantly, as for the TMV MP, the two stages of BMB1 translocation can be uncoupled. Both BMB2‐mRFP and mRFP‐BMB2 fusion proteins are able to direct BMB1 to peripheral membrane bodies, but do not support either BMB1 translocation into PD and neighbouring cells, or the cell‐to‐cell movement of transport‐deficient PVX in combination with BMB1 (data not presented). As BMB2 is distantly sequence related to TGB2 proteins, which have, as shown for the PVX TGB2 protein, the capability for PD gating (Tamai and Meshi, 2001), we presume that a similar function can be performed by the HGSV BMB2 protein, and that this function can be blocked in BMB2 fusions with fluorescent protein markers. Whatever the reason for the partial functionality of fused BMB2, these observations confirm the idea that MP delivery to membrane compartments adjacent to PD is an independent function in at least TGB, BMB and single gene‐coded transport systems.
Based on co‐expression experiments, we conclude that the HGSV BMB2 protein appears to be functionally equivalent to two TGB proteins, possibly combining the functions of TGB2 and TGB3. However, in its subcellular localization, HGSV BMB2 is similar to the TGB3 protein of PMTV and other viruses (Ju et al., 2008; Lim et al., 2009; Schepetilnikov et al., 2005; Zamyatnin et al., 2004). According to the current TGB‐mediated transport model, the TGB3 protein contains signals of PD targeting and is required for the trafficking of two other TGB proteins and, presumably, transport‐competent RNPs containing TGB1 and viral genomic RNA to PD (Lim et al., 2009; Shemyakina et al., 2011; Zamyatnin et al., 2004). Previously, we have found that TGB2 proteins and even non‐TGB membrane‐associated proteins can be targeted by heterologous TGB3 proteins to cell wall‐associated peripheral cell compartments; moreover, heterologous combinations of the TGB3 and TGB2 proteins can direct PMTV TGB1 to the peripheral ER bodies, but not to PD and the adjacent cells (Zamyatnin et al., 2002, 2004). However, our finding that the TGB2 and TGB3 proteins of PSLV do not target GFP‐BMB1 to peripheral compartments or complement the PVX cell‐to‐cell transport in the presence of BMB1 suggest a more specific functional interaction between BMB proteins in comparison with TGB proteins.
Although the biochemical properties of BMB1 have not been studied, we hypothesize that this protein, by analogy with TGB1, might be the RNA‐binding protein forming transport RNPs that are delivered to PD by means of the BMB2 protein. Indeed, taking into account that the size and bacilliform shape of HGSV virions are not compatible with translocation through PD, the transport form of the HGSV genome is most probably an RNP rather than a virion, or a modified virion derivative. It should also be noted that our functional complementation experiments were performed in the absence of the virion‐forming CP, which is required for the movement of the PVX genome believed to be transported between cells in the form of a virion (Santa Cruz et al., 1998). Thus, given the absence of CP, BMB2 facilitated movement of the PVX genome as an RNP, probably formed by the BMB1 protein. In a similar manner, the TMV 30K MP is also sufficient to complement the cell‐to‐cell movement of transport‐deficient PVX in a non‐virion form.
Results of intercellular movement studies employing individual MP genes transiently expressed in the absence of virus infection should be interpreted in view of the recently accepted idea that virus replication complexes (and the replicase itself) are involved in the cell‐to‐cell spread of viral genomic RNA. The data supporting this model have been reported for both single gene‐based transport systems (Amari et al., 2014; Guenoune‐Gelbart et al., 2008; Heinlein, 2015; Kaido et al., 2014; Kawakami et al., 2004) and more complex transport systems (Grangeon et al., 2013; Park et al., 2014; Tilsner et al., 2013). However, viral transport may not involve specific protein–protein binding of MPs to replication complexes (Tilsner et al., 2013). For example, viruses of the family Betaflexiviridae with closely related replicases encode dissimilar MPs, either single MPs of the 30K superfamily (genera Capillovirus, Citrivirus, Tepovirus, Trichovirus and Vitivirus) or TGB (genera Carlavirus and Foveavirus) (Martelli et al., 2007). Moreover, the PVX intercellular movement can be trans‐complemented by single gene‐coded MPs (Fedorkin et al., 2001; Morozov et al., 1997; this study). Therefore, functional interactions between MPs and the replication complexes may result from a common compartmentalization in membrane sites (Tilsner and Oparka, 2012). These specific sites are proposed to be microtubule associated and are targeted by viruses as general platforms for the formation of membrane‐based macromolecular complexes involved in signalling and trafficking (Heinlein, 2015). Thus, assuming that interaction with cognate MPs is not required for the coupling of viral replication and transport and, on the other hand, RNA binding by MPs is known to be sequence‐unspecific, heterologous transient expression of individual MPs can be used as a tool not only for the analysis of intracellular and cell‐to‐cell transport of the MPs themselves, but also for the molecular dissection of viral RNA trafficking and the role of replication in plant virus transport.
In conclusion, it is not unexpected that new types of transport systems will be present in so far unstudied plant viruses. In addition to the HGSV BMB described in this article, a new single gene‐coded transport system has been revealed recently in Rice dwarf virus (genus Phytoreovirus) encoding the single Pns6 MP with both enzymatic ATPase activity and several trans‐membrane domains (Ji et al., 2011). Therefore, ongoing studies of virus diversity can further disclose the general principles of functioning of plant virus movement systems and their evolution.
Experimental Procedures
Recombinant constructs
PVX‐POL‐GFP has been described previously under the designation PVXdt‐GFP (Komarova et al., 2006). The Golgi marker ST‐GFP was constructed by substitution of the GFP gene for the yellow fluorescent protein (YFP) gene in the previously described ST‐YFP construct (Schepetilnikov et al., 2005) The m‐GFP5‐ER construct has been described previously (Zamyatnin et al., 2002).
To obtain pLH‐PSLV‐TGB3 and pLH‐PSLV‐TGB2/TGB3, regions coding for the PSLV proteins were cut out by NcoI–XbaI digestion from the previously described constructs pRT‐18K and pRT‐15K/18K (Solovyev et al., 2000) and cloned into the similarly digested binary vector pLH* (Solovyev et al., 2013).
Coding regions of the HGSV RNA2 ORF2, ORF3 and ORF4 were chemically synthesized according to published sequences (Melzer et al., 2012) and cloned into the binary vector pLH* (Solovyev et al., 2013) to give pLH‐ORF2, pLH‐ORF3 and pLH‐ORF4, respectively. To obtain pLH‐GFP‐BMB1, the GFP gene was amplified with primers 5′‐CGCTCGAGAACATGGTGAGCAAGGGCGAGGA and 5′‐CGCCATGGACTTGTACAGCTCGTCCATGC, and the product was digested with XhoI–NcoI; the BMB1 gene was cut out from pLH‐ORF2 as an NcoI–XbaI fragment and then both fragments were ligated into pLH* digested with XhoI and XbaI. To obtain pLH‐BMB1‐GFP, the BMB1 gene was amplified with primers 5′‐GACCATGGAGAGTTTTAATTATGTGACT and 5′‐GCGGATCCAGATGAAAGTACGTAAAAATCACCTCTCA and digested with NcoI–BamHI; the GFP gene was cut out of the construct pLH‐ShVX‐TGB2/TGB3‐GFP (Lezzhov et al., 2015) as a BamHI–XbaI fragment and then both fragments were ligated into pLH* digested with NcoI and XbaI. To obtain pLH‐mRFP‐BMB2, the mRFP gene was amplified with primers 5′‐CGCTCGAGAACATGGCCTCCTCCGAGGACGTCAT and 5′‐GCGGATCCTGAAGAGGCGCCGGTGGAGTGGCGGC; the BMB2 gene was amplified with primers 5′‐GCGGATCCTCTTCAGGACCGATATTTCAAGGTCAAGCGA and 5′‐GCTCTAGATTACATATGCGTAACAACAACCAAC; the resulting products were cut with XhoI–BamHI and BamHI–XbaI, respectively, and cloned into pLH* digested with NcoI and XbaI. To obtain pLH‐BMB2‐mRFP, the BMB2 gene was amplified with primers 5′‐CGCTCGAGACCATGCCGATATTTCAA and 5′‐GCGGATCCAGATGACATATGCGTAACAACAACCAACA; the mRFP gene was amplified with primers 5′‐GCGGATCCTCAGTGGCCTCCTCCGAGGACGTCAT and 5′‐GCTCTAGATTAGGCGCCGGTGGAGTGGCGGC; the resulting products were cut with XhoI–BamHI and BamHI–XbaI, respectively, and cloned into pLH* digested with NcoI and XbaI. pLH‐GFP‐BMB1//mRFP was constructed in two steps. First, the expression cassette including the A. thaliana Actin‐2 promoter, mRFP and Nos transcriptional terminator from pBin19 was assembled in pGEM‐7Zf(+). For this purpose, the Actin‐2 promoter was amplified on the template of A. thaliana genomic DNA with specific primers 5′‐GCGAATTCACGCGTTTCGACAAAATTTAGAAC and 5′‐GCCCATGGAGCTCGGATCCTTCAAAGCGGAGAGGAAAATATATG. Second, the assembled cassette was cut out from the resulting plasmid by SalI digestion and cloned into pLH‐GFP‐BMB1 linearized with SalI.
Plant material
Nicotiana benthamiana plants were grown and maintained in growth chambers under 16‐h/8‐h light/dark cycles, 24/20°C day/night temperatures and approximately 50% humidity. Five‐ to 6‐week‐old plants were used for transient expression and movement complementation assay.
Plant agroinfiltration
Binary vectors were transformed into Agrobacterium tumefaciens (C58C1). Agrobacterial cultures were prepared for infiltration as described previously (Solovyev et al., 2013). Briefly, agrobacterial cultures were grown in Luria–Bertani (LB) medium with 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), 20 μm acetosyringone and antibiotics at 28°C, pelleted and resuspended in infiltration medium (10 mm MES, pH 5.5, 10 mm MgCl2, 150 μm acetosyringone). After 3 h of incubation, A. tumefaciens suspensions were diluted to a final optical density at 600 nm (OD600) = 0.3 prior to infiltration. For BMB1 cell‐to‐cell movement assay virus movement complementation test, GFP‐BMB1//mRFP and PVX‐POL‐GFP constructs were agroinfiltrated at OD600 = 0.0015 (1 : 200 dilution) and OD600 = 0.0001 (1 : 2500 dilution), respectively. Agrobacterial cultures were infiltrated onto the abaxial surface of Nicotiana benthamiana leaves using a needle‐less syringe.
Aniline blue staining
To visualize PD‐associated callose, leaf discs were vacuum infiltrated with aniline blue solution (0.1% aniline blue in 67 mm phosphate buffer, pH 8). Leaf discs were incubated in the dark at room temperature for 15 min before microscopy (Amari et al., 2014).
Confocal microscopy
To visualize the subcellular localization of proteins, imaging was performed 2–3 days after agroinfiltration. Leaf discs were vacuum infiltrated with water and imaged using a Zeiss (Oberkochen, Germany) LSM780 confocal laser scanning microscope under a multitrack mode. Excitation wavelengths were 488 nm for GFP, 543 nm for RFP and 405 nm for aniline blue. For dual‐colour imaging of GFP and mRFP, acquisition windows were 493–550 nm and 582–640 nm, respectively. For dual‐colour imaging of GFP and aniline blue‐stained callose, acquisition windows were 493–554 nm and 416–490 nm, respectively. Confocal images were processed using Zeiss Zen software and ImageJ (1.47s) software. In complementation assays, the cell‐to‐cell movement of PVX‐POL‐GFP was observed using a Zeiss AxioZoom V16 microscope. Foci sizes were measured using Zeiss Zen software and analysed by Student's t‐test.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot detection of the GFP‐BMB1 and BMB2‐mRFP fusion proteins in agroinfiltrated Nicotiana benthamiana leaves. (A) Detection of GFP‐BMB1 with GFP‐specific antibodies. (B) Detection of BMB2‐mRFP with mRFP‐specific antibodies. Arrow points to the position of a BMB2‐containing high‐molecular‐mass complex. Control, non‐infiltrated leaf. The positions of molecular weight markers are shown on the right; their molecular masses are indicated in kilodaltons. BMB, binary movement block; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
Fig. S2 Expression of the Hibiscus green spot virus (HGSV) ORF4‐GFP fusion protein. The ORF4‐GFP protein was transiently expressed in Nicotiana benthamiana leaves by agroinfiltration. (A) Subcellular localization of ORF4‐GFP imaged at 3 days post‐infiltration by confocal laser scanning microscopy. The image was reconstructed from a Z‐series of optical sections. Scale bar, 10 μm. (B) Detection of ORF4‐GFP expression by Western blotting with GFP‐specific antibodies. GFP, non‐fused GFP; control, non‐infiltrated leaf. The expected molecular mass of the fusion protein is 32.8 kDa. The positions of molecular weight markers are shown on the right; their molecular masses are indicated in kilodaltons. GFP, green fluorescent protein; ORF, open reading frame.
Acknowledgements
This work was supported by the Russian Foundation for Basic Research (grant 15‐04‐03922 to A.G.S.) and the Agence Nationale de la Recherche (grant ANR‐13‐KBBE‐0005‐01 to M.H.). E.A.L. gratefully acknowledges receipt of a FEBS short‐term fellowship. We thank Mikhail Schepetilnikov, Khalid Amari and Jérôme Mutterer for helpful assistance during the visits of E.A.L. and A.G.S. to the Institut de Biologie Moléculaire des Plantes (IBMP) (Strasbourg, France) and for excellent support in confocal microscopy. The authors declare no conflicts of interest.
Contributor Information
Sergey Y. Morozov, Email: morozov@genebee.msu.su
Andrey G. Solovyev, Email: solovyev@genebee.msu.su
References
- Alzhanova, D.V. , Napuli, A.J. , Creamer, R. and Dolja, V.V. (2001) Cell‐to‐cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20, 6997–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amari, K. , Donato, M. , Di Dolja, V.V. and Heinlein, M. (2014) Myosins VIII and XI play distinct roles in reproduction and transport of tobacco mosaic virus. PLoS Pathog. 10, e1004448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak, A. and Folimonova, S.Y. (2015) The conundrum of a unique protein encoded by citrus tristeza virus that is dispensable for infection of most hosts yet shows characteristics of a viral movement protein. Virology, 485, 86–95. [DOI] [PubMed] [Google Scholar]
- Dolja, V.V. , Kreuze, J.F. and Valkonen, J.P.T. (2006) Comparative and functional genomics of closteroviruses. Virus Res. 117, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorkin, O.N. , Solovyev, A.G. , Yelina, N.E. , Zamyatnin, A.A. , Zinovkin, R.A. , Mäkinen, K. , Schiemann, J. and Morozov, S.Y. (2001) Cell‐to‐cell movement of Potato virus X involves distinct functions of the coat protein. J. Gen. Virol. 82, 449–458. [DOI] [PubMed] [Google Scholar]
- Gabrenaite‐Verkhovskaya, R. , Andreev, I.A. , Kalinina, N.O. , Torrance, L. , Taliansky, M.E. and Mäkinen, K. (2008) Cylindrical inclusion protein of potato virus A is associated with a subpopulation of particles isolated from infected plants. J. Gen. Virol. 89, 829–838. [DOI] [PubMed] [Google Scholar]
- Genovés, A. , Navarro, J.A. and Pallás, V. (2010) The intra‐ and intercellular movement of Melon necrotic spot virus (MNSV) depends on an active secretory pathway. Mol. Plant–Microbe Interact. 23, 263–272. [DOI] [PubMed] [Google Scholar]
- Grangeon, R. , Jiang, J. , Wan, J. , Agbeci, M. , Zheng, H. and Laliberté, J.F. (2013) 6K2‐induced vesicles can move cell to cell during turnip mosaic virus infection. Front. Microbiol. 4, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenoune‐Gelbart, D. , Elbaum, M. , Sagi, G. , Levy, A. and Epel, B.L. (2008) Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana . Mol. Plant–Microbe Interact. 21, 335–345. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2015) Plant virus replication and movement. Virology, 479–480, 657–671. [DOI] [PubMed] [Google Scholar]
- Heinlein, M. (2016) Viral transport and interaction with the host cytoskeleton. In: Plant–Virus Interactions Molecular Biology, Intra‐ and Intercellular Transport (Kleinow T., ed), pp. 39–66. Cham: Springer International Publishing. [Google Scholar]
- Hirashima, K. and Watanabe, Y. (2001) Tobamovirus replicase coding region is involved in cell‐to‐cell movement. J. Virol. 75, 8831–8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2002) Induction of disease 1: virus movement through the plant and effects on plant metabolism In: Matthews' Plant Virology, pp. 373–436. London: Elsevier. [Google Scholar]
- Ji, X. , Qian, D. , Wei, C. , Ye, G. , Zhang, Z. , Wu, Z. , Xie, L. and Li, Y. (2011) Movement protein Pns6 of rice dwarf phytoreovirus has both ATPase and RNA binding activities. PLoS One, 6, e24986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. , Patarroyo, C. , Garcia Cabanillas, D. , Zheng, H. and Laliberté, J.F. (2015) The vesicle‐forming 6K2 protein of turnip mosaic virus interacts with the COPII coatomer Sec24a for viral systemic infection. J. Virol. 89, 6695–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, H.J. , Ye, C.M. and Verchot‐Lubicz, J. (2008) Mutational analysis of PVX TGBp3 links subcellular accumulation and protein turnover. Virology, 375, 103–117. [DOI] [PubMed] [Google Scholar]
- Kaido, M. , Abe, K. , Mine, A. , Hyodo, K. , Taniguchi, T. , Taniguchi, H. , Mise, K. and Okuno, T. (2014) GAPDH‐a recruits a plant virus movement protein to cortical virus replication complexes to facilitate viral cell‐to‐cell movement. PLoS Pathog. 10, e1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, S. , Watanabe, Y. and Beachy, R.N. (2004) Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA, 101, 6291–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, Z.A. , Medina, V. and Falk, B.W. (2013) Crinivirus replication and host interactions. Front. Microbiol. 4, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova, T.V. , Skulachev, M.V. , Zvereva, A.S. , Schwartz, A.M. , Dorokhov, Y.L. and Atabekov, J.G. (2006) New viral vector for efficient production of target proteins in plants. Biochemistry (Moscow), 71, 846–850. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. and Dolja, V.V. (1993) Evolution and taxonomy of positive‐strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28, 375–430. [DOI] [PubMed] [Google Scholar]
- Lezzhov, A.A. , Gushchin, V.A. , Lazareva, E.A. , Vishnichenko, V.K. , Morozov, S.Y. and Solovyev, A.G. (2015) Translation of the shallot virus X TGB3 gene depends on non‐AUG initiation and leaky scanning. J. Gen. Virol. 96, 3159–3164. [DOI] [PubMed] [Google Scholar]
- Lim, H.S. , Bragg, J.N. , Ganesan, U. , Ruzin, S. , Schichnes, D. , Lee, M.Y. , Vaira, A.M. , Ryu, K.H. , Hammond, J. and Jackson, A.O. (2009) Subcellular localization of the barley stripe mosaic virus triple gene block proteins. J. Virol. 83, 9432–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. and Nelson, R.S. (2013) The cell biology of Tobacco mosaic virus replication and movement. Front. Plant Sci. 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J. (2006) Plant viral movement proteins: agents for cell‐to‐cell trafficking of viral genomes. Virology, 344, 169–184. [DOI] [PubMed] [Google Scholar]
- Martelli, G.P. , Adams, M.J. , Kreuze, J.F. and Dolja, V.V. (2007) Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45, 73–100. [DOI] [PubMed] [Google Scholar]
- Melcher, U. (2000) The “30K” superfamily of viral movement proteins. J. Gen. Virol. 81, 257–266. [DOI] [PubMed] [Google Scholar]
- Melzer, M.J. , Sether, D.M. , Borth, W.B. and Hu, J.S. (2012) Characterization of a virus infecting Citrus volkameriana with citrus leprosis‐like symptoms. Phytopathology, 102, 122–127. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2003) Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84, 1351–1366. [DOI] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2012) Did silencing suppression counter‐defensive strategy contribute to origin and evolution of the triple gene block coding for plant virus movement proteins? Front. Plant Sci. 3, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, S.Y. and Solovyev, A.G. (2015) Phylogenetic relationship of some “accessory” helicases of plant positive‐stranded RNA viruses: toward understanding the evolution of triple gene block. Front. Microbiol. 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, S.Y. , Fedorkin, O.N. , Jüttner, G. , Schiemann, J. , Baulcombe, D.C. and Atabekov, J.G. (1997) Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J. Gen. Virol. 78, 2077–2083. [DOI] [PubMed] [Google Scholar]
- Mushegian, A.R. and Elena, S.F. (2015) Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology, 476, 304–315. [DOI] [PubMed] [Google Scholar]
- Navarro, J.A. , Genovés, A. , Climent, J. , Saurí, A. , Martínez‐Gil, L. , Mingarro, I. and Pallás, V. (2006) RNA‐binding properties and membrane insertion of Melon necrotic spot virus (MNSV) double gene block movement proteins. Virology, 356, 57–67. [DOI] [PubMed] [Google Scholar]
- Niehl, A. and Heinlein, M. (2011) Cellular pathways for viral transport through plasmodesmata. Protoplasma, 248, 75–99. [DOI] [PubMed] [Google Scholar]
- Niehl, A. , Peña, E.J. , Amari, K. and Heinlein, M. (2013) Microtubules in viral replication and transport. Plant J. 75, 290–308. [DOI] [PubMed] [Google Scholar]
- Niehl, A. , Pasquier, A. , Ferriol, I. , Mély, Y. and Heinlein, M. (2014) Comparison of the oilseed rape mosaic virus and Tobacco mosaic virus movement proteins (MP) reveals common and dissimilar MP functions for tobamovirus spread. Virology, 456–457, 43–54. [DOI] [PubMed] [Google Scholar]
- Obraztsova, E.A. , Makhotenko, A.V. , Makarova, S.S. , Makarov, V.V. and Kalinina, N.O. (2015) In vitro properties of hordeivirus TGB1 protein forming ribonucleoprotein complexes. J. Gen. Virol. 96, 3422–3431. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. , Roberts, A.G. , Prior, D.A.M. , Chapman, S. , Baulcombe, D. and Santa Cruz, S. (1995) Imaging the green fluorescent protein in plants – viruses carry the torch. Protoplasma, 189, 133–141. [Google Scholar]
- Park, M.R. , Jeong, R.D. and Kim, K.H. (2014) Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 5, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña, E.J. , Ferriol, I. , Sambade, A. , Buschmann, H. , Niehl, A. , Elena, S.F. , Rubio, L. and Heinlein, M. (2014) Experimental virus evolution reveals a role of plant microtubule dynamics and TORTIFOLIA1/SPIRAL2 in RNA trafficking. PLoS One, 9, e105364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhnevsky, A.I. , Peremyslov, V.V. and Dolja, V.V. (2005) Actin cytoskeleton is involved in targeting of a viral Hsp70 homolog to the cell periphery. J. Virol. 79, 14 421–14 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Santa Cruz, S. , Roberts, A. , Prior, D. , Chapman, S. and Oparka, K. (1998) Cell‐to‐cell and phloem‐mediated transport of potato virus X. The role of virions. Plant Cell, 10, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheets, K. , Jordan, R. , White, K.A. and Hernández, C. (2015) Pelarspovirus, a proposed new genus in the family Tombusviridae. Arch. Virol. 160, 2385–2393. [DOI] [PubMed] [Google Scholar]
- Schepetilnikov, M.V. , Manske, U. , Solovyev, A.G. , Zamyatnin, A.A. , Schiemann, J. and Morozov, S.Y. (2005) The hydrophobic segment of Potato virus X TGBp3 is a major determinant of the protein intracellular trafficking. J. Gen. Virol. 86, 2379–2391. [DOI] [PubMed] [Google Scholar]
- Shemyakina, E.A. , Erokhina, T.N. , Gorshkova, E.N. , Schiemann, J. , Solovyev, A.G. and Morozov, S.Y. (2011) Formation of protein complexes containing plant virus movement protein TGBp3 is necessary for its intracellular trafficking. Biochimie, 93, 742–748. [DOI] [PubMed] [Google Scholar]
- Solovyev, A.G. , Stroganova, T.A. , Zamyatnin, A.A. , Fedorkin, O.N. , Schiemann, J. and Morozov, S.Y. (2000) Subcellular sorting of small membrane‐associated triple gene block proteins: TGBp3‐assisted targeting of TGBp2. Virology, 269, 113–127. [DOI] [PubMed] [Google Scholar]
- Solovyev, A.G. , Kalinina, N.O. and Morozov, S.Y. (2012) Recent advances in research of plant virus movement mediated by triple gene block. Front. Plant Sci. 3, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev, A.G. , Minina, E.A. , Makarova, S.S. , Erokhina, T.N. , Makarov, V.V. , Kaplan, I.B. , Kopertekh, L. , Schiemann, J. , Richert‐Pöggeler, K.R. and Morozov, S.Y. (2013) Subcellular localization and self‐interaction of plant‐specific Nt‐4/1 protein. Biochimie, 95, 1360–1370. [DOI] [PubMed] [Google Scholar]
- Sorel, M. , Garcia, J.A. and German‐Retana, S. (2014) The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Mol. Plant–Microbe Interact. 27, 215–226. [DOI] [PubMed] [Google Scholar]
- Tamai, A. and Meshi, T. (2001) Cell‐to‐cell movement of Potato virus X: the role of p12 and p8 encoded by the second and third open reading frames of the triple gene block. Mol. Plant–Microbe Interact. 14, 1158–1167. [DOI] [PubMed] [Google Scholar]
- Tilsner, J. and Oparka, K.J. (2012) Missing links? – the connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2, 705–711. [DOI] [PubMed] [Google Scholar]
- Tilsner, J. , Linnik, O. , Louveaux, M. , Roberts, I.M. , Chapman, S.N. and Oparka, K.J. (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201, 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner, J. , Taliansky, M.E. and Torrance, L. (2014) Plant virus movement. eLS, 1–12. [Google Scholar]
- Tomenius, K. , Clapham, D. and Meshi, T. (1987) Localization by immunogold cytochemistry of the virus‐coded 30K protein in plasmodesmata of leaves infected with tobacco mosaic virus. Virology, 160, 363–371. [DOI] [PubMed] [Google Scholar]
- Torrance, L. , Andreev, I.A. , Gabrenaite‐Verhovskaya, R. , Cowan, G. , Mäkinen, K. and Taliansky, M.E. (2006) An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357, 1–8. [DOI] [PubMed] [Google Scholar]
- Verchot‐Lubicz, J. , Torrance, L. , Solovyev, A.G. , Morozov, S.Y. , Jackson, A.O. and Gilmer, D. (2010) Varied movement strategies employed by triple gene block‐encoding viruses. Mol. Plant–Microbe Interact. 23, 1231–1247. [DOI] [PubMed] [Google Scholar]
- Vijayapalani, P. , Maeshima, M. , Nagasaki‐Takekuchi, N. and Miller, W.A. (2012) Interaction of the trans‐frame potyvirus protein P3N‐PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8, e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann, E. , Lucas, W.J. , Citovsky, V. and Zambryski, P. (1994) Direct functional assay for tobacco mosaic virus cell‐to‐cell movement protein and identification of a domain involved in increasing plasmodesmal permeability. Proc . Natl. Acad. Sci. USA, 91, 1433–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, C. , Lazarowitz, S.G. and Citovsky, V. (2016) Identification of a functional plasmodesmal localization signal in a plant viral cell‐to‐cell movement protein. MBio. 7, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatnin, A.A. , Solovyev, A.G. , Sablina, A.A. , Agranovsky, A.A. , Katul, L. , Vetten, H.J. , Schiemann, J. , Hinkkanen, A.E. , Lehto, K. and Morozov, S.Y. (2002) Dual‐colour imaging of membrane protein targeting directed by Poa semilatent virus movement protein TGBp3 in plant and mammalian cells. J. Gen. Virol. 83, 651–662. [DOI] [PubMed] [Google Scholar]
- Zamyatnin, A.A. , Solovyev, A.G. , Savenkov, E.I. , Germundsson, A. , Sandgren, M. , Valkonen, J.P.T. and Morozov, S.Y. (2004) Transient coexpression of individual genes encoded by the triple gene block of potato mop‐top virus reveals requirements for TGBp1 trafficking. Mol. Plant–Microbe Interact. 17, 921–930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot detection of the GFP‐BMB1 and BMB2‐mRFP fusion proteins in agroinfiltrated Nicotiana benthamiana leaves. (A) Detection of GFP‐BMB1 with GFP‐specific antibodies. (B) Detection of BMB2‐mRFP with mRFP‐specific antibodies. Arrow points to the position of a BMB2‐containing high‐molecular‐mass complex. Control, non‐infiltrated leaf. The positions of molecular weight markers are shown on the right; their molecular masses are indicated in kilodaltons. BMB, binary movement block; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein.
Fig. S2 Expression of the Hibiscus green spot virus (HGSV) ORF4‐GFP fusion protein. The ORF4‐GFP protein was transiently expressed in Nicotiana benthamiana leaves by agroinfiltration. (A) Subcellular localization of ORF4‐GFP imaged at 3 days post‐infiltration by confocal laser scanning microscopy. The image was reconstructed from a Z‐series of optical sections. Scale bar, 10 μm. (B) Detection of ORF4‐GFP expression by Western blotting with GFP‐specific antibodies. GFP, non‐fused GFP; control, non‐infiltrated leaf. The expected molecular mass of the fusion protein is 32.8 kDa. The positions of molecular weight markers are shown on the right; their molecular masses are indicated in kilodaltons. GFP, green fluorescent protein; ORF, open reading frame.


