Summary
Phytohormones, such as salicylic acid (SA), ethylene (ET) and jasmonic acid (JA), play key roles in plant defence following pathogen attack. The involvement of these hormones in susceptibility following Fusarium oxysporum (Fo) infection has mostly been studied in Arabidopsis thaliana. However, Fo causes vascular wilt disease in a broad range of crops, including tomato (Solanum lycopersicum). Surprisingly little is known about the involvement of these phytohormones in the susceptibility of tomato towards Fo f. sp. lycopersici (Fol). Here, we investigate their involvement by the analysis of the expression of ET, JA and SA marker genes following Fol infection, and by bioassays of tomato mutants affected in either hormone production or perception. Fol inoculation triggered the expression of SA and ET marker genes, showing the activation of these pathways. NahG tomato, in which SA is degraded, became hypersusceptible to Fol infection and showed stronger disease symptoms than wild‐type. In contrast, ACD and Never ripe (Nr) mutants, in which ET biosynthesis and perception, respectively, are impaired, showed decreased disease symptoms and reduced fungal colonization on infection. The susceptibility of the def1 tomato mutant, and a prosystemin over‐expressing line, in which JA signalling is compromised or constitutively activated, respectively, was unaltered. Our results show that SA is a negative and ET a positive regulator of Fol susceptibility. The SA and ET signalling pathways appear to act synergistically, as an intact ET pathway is required for the induction of an SA marker gene, and vice versa.
Keywords: ET, Fusarium oxysporum, JA, SA, susceptibility, tomato
Introduction
The root‐infecting fungal pathogen Fusarium oxysporum (Fo) causes vascular wilt disease in over 100 different plant species, including banana, cotton, palm, Arabidopsis and tomato (Michielse and Rep, 2009). Fo represents a species complex comprising many individual pathogenic strains, each capable of infecting one or a few host species only. Based on host specificity, strains have been grouped into formae speciales. Infection by Fo starts on attachment of fungal hyphae to the plant root surface. Subsequently, fungal hyphae enter the roots through wounds or cracks at the root tip, or at sites of lateral root formation. Ultimately, the fungus reaches the xylem vessels and proliferates, causing disease to ensue (Berrocal‐Lobo and Molina, 2008; di Pietro et al., 2003; Rep et al., 2002). In attempting to arrest pathogen spread through the vasculature, the plant blocks its infected vessels and compromises their ability to transport water and nutrients. Vascular browning, stunting, progressive wilting and, eventually, plant death are typical disease symptoms of infected plants (Agrios, 2005; di Pietro et al., 2003).
In general, plant defence responses against pathogens are controlled by complex signalling routes that often involve the classical defence phytohormones salicylic acid (SA), ethylene (ET) and jasmonic acid (JA) (Robert‐Seilaniantz et al., 2011). Usually, SA signalling triggers resistance against biotrophic and hemibiotrophic pathogens, whereas a combination of JA and ET signalling activates resistance against necrotrophs (Glazebrook, 2005).
As a result of the extensive availability of genetic and genomic resources, most studies on phytohormone involvement in defence against Fo have been performed in Arabidopsis (Edgar et al., 2006). Arabidopsis is susceptible to Fo forma specialis (f. sp.) conglutinans (Focn). Arabidopsis lines that express the salicylate hydroxylase transgene (NahG), or that carry the SA induction‐deficient 2 (sid2) mutant, are impaired in SA accumulation. Both lines show increased susceptibility to Fo, indicating the involvement of SA in reducing disease susceptibility (Berrocal‐Lobo and Molina, 2004; Diener and Ausubel, 2005).
Pretreatment of Arabidopsis seedlings with either methyl jasmonate (MeJA) or the ET precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) leads to enhanced disease symptom development on Fo inoculation, indicating that both ET and JA are involved in disease susceptibility (Trusov et al., 2009). The ET‐insensitive Arabidopsis mutants ethylene insensitive2‐1 (ein2‐1) and ethylene receptor 1 (etr1‐1) show a reduction in disease symptoms compared with Col‐0 plants when inoculated with Focn (Pantelides et al., 2013; Trusov et al., 2009). In contrast, various JA biosynthesis mutants, such as jasmonate resistant 1 (jar1‐1) and allene oxide synthase (aos), do not exhibit increased susceptibility to Fo (Thatcher et al., 2009; Trusov et al., 2009). Surprisingly, a point mutation in CORONATINE INSENSITIVE1 (COI1), an essential component of JA perception, strongly reduces disease symptom development following Fo infection (Thatcher et al., 2009; Trusov et al., 2009). In addition, disruption of MYC2, PFT1 and LBD20, transcriptional regulators of JA signalling, also results in an increased resistance to Fo (Anderson et al., 2004; Kidd et al., 2009; Thatcher et al., 2012). Taken together, ET and JA are positive regulators of susceptibility in Arabidopsis.
The role of phytohormones in determining host colonization and disease symptom development is known to vary for different formae speciales of Fo and their respective hosts (Di et al., 2016). To obtain a better insight into these processes, it is therefore crucial to investigate the role of phytohormones in defence responses to Fo in plant species other than Arabidopsis. Tomato (Solanum lycopersicum), a major and important vegetable crop (Panthee and Chen, 2010), is susceptible to Fo f. sp. lycopersici (Fol), resulting in significant yield losses each year (McGovern, 2015). The interaction between tomato and Fol has been well studied (Takken and Rep, 2010). Like other formae speciales of Fo, Fol colonizes the vasculature, and infected plants exhibit vascular browning, leaf epinasty, stunting, progressive wilting and, eventually, death (di Pietro et al., 2003). During colonization, the fungus secretes virulence factors, called effector proteins (Houterman et al., 2007). The deletion of specific effectors, such as Avr2, typically compromises fungal virulence, resulting in strains that are reduced in pathogenicity (Houterman et al., 2007, 2009).
For tomato, a large collection of lines is available which are compromised in hormone perception, metabolism or signalling. In our study, mutants affected in the biosynthesis and signalling pathway of specific defence‐related hormones were analysed for their susceptibility to Fol. The lines used in this study include the following: (i) transgenic NahG plants that express the bacterial enzyme salicylate hydroxylase, which converts SA into biologically inactive catechol, resulting in plants deficient in SA accumulation (Brading et al., 2000); (ii) Never ripe (Nr), a dominant ET‐insensitive mutant, carrying a single base substitution in the region encoding the N‐terminus of ETR3, a homologue of the Arabidopsis ETR1 receptor (Wilkinson et al., 1995); (iii) a transgenic line that expresses ACCD (1‐amino‐cyclopropane‐1‐carboxylic acid deaminase), which encodes the ACCd enzyme that catalyses the degradation of ACC; (iv) the JA‐deficient mutant defenseless‐1 (def1), which has a defect in the jasmonate pathway between 13‐hydroperoxy‐octadecatrienoic acid (13‐HPOT) and 12‐oxo‐phytodienoic acid; this mutant fails to produce JA and does not systemically accumulate proteinase inhibitors (PIs) in response to treatment with systemin or oligosaccharide elicitors (chitosan and polygalacturonide) (Li et al., 2002); and (v) a 35S::prosystemin transgenic line overexpressing prosystemin; prosystemin is a positive regulator of JA signalling, and hence these plants constitutively accumulate high levels of PI proteins (Howe and Ryan, 1999).
Here, we report our inoculation assays, using both wild‐type Fol and a FolΔAvr2 mutant, of the various tomato lines affected in SA, ET or JA signalling. In contrast with JA signalling, both SA and ET play major and opposing roles in disease susceptibility and development. The SA and ET signalling pathways appear to act synergistically, as an intact ET pathway is required for the induction of an SA reporter gene, and vice versa. A model for the role of SA, ET and JA signalling in tomato in the susceptibility to Fol is proposed and compared with that in Arabidopsis.
Results
NahG tomato plants show enhanced disease symptom development on Fol infection
To assess the potential role of SA in the modulation of susceptibility to Fol, 3‐week‐old wild‐type tomato plants (cultivar Moneymaker) and transgenic NahG plants impaired in SA accumulation were inoculated with either water (mock) or wild‐type Fol, notably a race 2 isolate called Fol007. In addition, to allow the assessment of hypersusceptibility, a Fol007 Avr2 knockout strain (FolΔAvr2) was included. This mutant is compromised in virulence and causes fewer disease symptoms than wild‐type Fol on susceptible plants (Houterman et al., 2009). As shown in Fig. 1a, NahG plants inoculated with Fol007 exhibited stronger disease symptoms than did wild‐type plants. These symptoms included extensive wilting and a greater stunting 3 weeks after inoculation. Consistent with this, the fresh weight of Fol007‐infected NahG tomato plants was significantly lower than that of corresponding wild‐type plants (Fig. 1b). Moreover, all vascular bundles of infected NahG plants had turned brown, and plants were either dead or very small and wilted. On a scale from 0–4 (Rep et al., 2005), infected NahG plants scored the maximal disease index (Fig. 1c). As expected, FolΔAvr2‐inoculated plants developed weaker disease symptoms (Fig. 1a). Similar to Fol007, FolΔAvr2‐infected NahG plants showed a significant reduction in fresh weight and a higher disease index relative to infected wild‐type plants (Fig. 1b,c).
Figure 1.
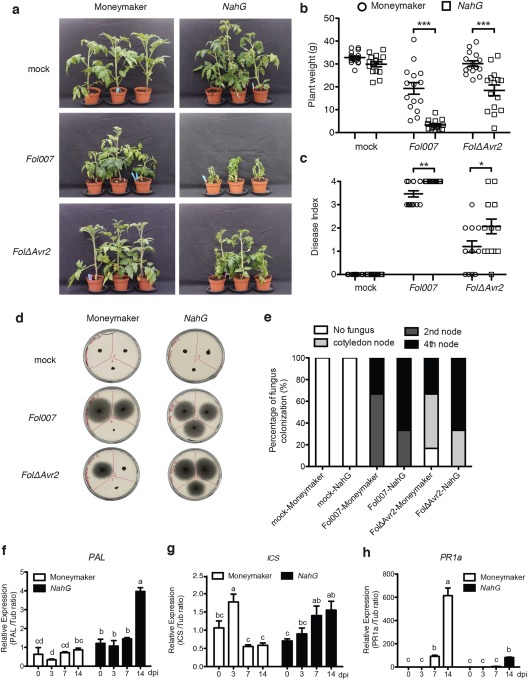
Impaired salicylic acid (SA) signalling enhances Fusarium oxysporum f. sp. lycopersici (Fol) disease symptom development in tomato. (a) Three‐week‐old wild‐type Moneymaker and NahG tomato plants inoculated with water (mock), Fol007 or FolΔAvr2 at 21 days post‐infection (dpi). Disease development was scored by measuring the fresh plant weight (b) and determining the disease index (range, 0–4) (c) of 20 plants per treatment/genotype combination. Circles and squares indicate Moneymaker and NahG plants, respectively. Plant weight was subjected to a pairwise comparison with a Student's t‐test, whereas the disease index was analysed by a non‐parametric Mann–Whitney U‐test (*P < 0.05; **P < 0.01; ***P < 0.001). The bioassay was repeated three times with similar results. (d) Representative stem sections taken from the cotyledon node (top left), second node (top right) and fourth node (bottom) of individual treated plants (n = 6) after incubation for 5 days on potato dextrose agar (PDA) plates. (e) Percentage of infected slices showing fungal outgrowth. Fungal progression in the stem was expressed as the infected percentage of all stem pieces. The experiment was performed twice with similar results. Transcription patterns of phenylalanine ammonia‐lyase (PAL) (f), isochorismate (ICS) (g) and pathogenesis‐related 1a (PR1a) (h) in Fol007‐inoculated wild‐type Moneymaker and NahG plants at 0, 3, 7 and 14 dpi. Gene expression levels relative to the internal control tubulin genes were quantified by quantitative polymerase chain reaction (qPCR). The data are expressed as the mean ± standard deviation (SD). Three biological replicates for each line per time point were analysed. The different letters show significant difference at P < 0.05 as determined by Duncan's multiple‐range test. The experiment was performed twice with similar results.
To investigate whether the augmented disease symptom development in NahG plants correlated with increased host colonization, a fungal recovery assay was performed. Sections were taken from Fol‐inoculated wild‐type and NahG plants at different heights of the stem, notably at the position of the cotyledon node, second node and fourth node. Following sterilization, the sections were placed on potato dextrose agar (PDA) plates and incubated for 5 days at 25 °C. As shown in Fig. 1d,e, wild‐type Fol colonized the stems more extensively than did the FolΔAvr2 strain. In all cases, the wild‐type fungus was able to reach the second or even fourth node, whereas, in the Avr2 knockout, only 30% reached the second node and the majority of the fungus was contained at the cotyledon node or below. Notably, the FolΔAvr2 strain was able to more effectively colonize NahG plants, and fungal outgrowth was observed in the fourth node in 70% of the inoculations (Fig. 1e). The NahG plants were also hypersusceptible to the wild‐type fungus, as depicted by the higher percentage of plants in which fungal outgrowth was seen from the fourth node. Together, these data suggest that NahG plants are hypersusceptible to Fusarium infection, and that the increased disease symptoms correlate with increased fungal colonization of the transgenic plants.
SA is synthesized through both the isochorismate (ICS) and phenylalanine ammonia‐lyase (PAL) pathways (Lee et al., 1995; Wildermuth et al., 2001). To assess the expression of the PAL and ICS genes during Fol infection, a reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) analysis was carried out on hypocotyls. Samples were taken at 0, 3, 7 and 14 days post‐infection (dpi) of wild‐type and NahG plants. PAL and ICS expression levels were measured and normalized to tubulin. We found that, in NahG plants, only at 14 dpi transcript levels of PAL were significantly up‐regulated relative to that at 0 dpi (Fig. 1f). In the wild‐type Moneymaker plants, no significant induction of PAL was observed at any time point. ICS expression in NahG plants was significantly induced at 7 and 14 dpi relative to that at 0 dpi (Fig. 1g). In the wild‐type plants, a significant, but only transient, induction of ICS was observed at 3 dpi. Together, these data indicate the involvement of SA biosynthesis genes in the Fol–tomato interaction. Possibly, the high SA turnover in NahG plants elicits a feedback mechanism activating the PAL and ICS SA biosynthesis pathways following Fol infection.
Pathogenesis‐related 1a (PR1a) expression is often used as a reporter for SA‐dependent defence signalling (Kunkel and Brooks, 2002). To assess whether PR1a expression is altered during Fol infection, its expression at 0, 3, 7 and 14 dpi was measured. Transcript levels of PR1a were significantly induced in wild‐type plants at 7 and 14 dpi, suggesting that SA signalling is activated late during Fol infection (Fig. 1h). Compared with wild‐type plants, the expression of PR1a in infected NahG plants was less strongly induced. This weaker induction correlates with the loss of SA accumulation in NahG plants, confirming the proposed of the transgenic line. Overall, these data show that impaired SA signalling enhances susceptibility to Fol and disease symptom development in tomato.
ET enhances susceptibility to Fol in tomato plants
Pretreatment of Arabidopsis seedlings with the ET precursor ACC leads to enhanced disease symptom development on Focn inoculation, indicating that ET is involved in disease susceptibility (Trusov et al., 2009).
To investigate the role of ET in disease symptom development in tomato, transgenic plants impaired in ET biosynthesis were analysed for their susceptibility to Fol. In the transgenic ACD line, constitutively expressing a bacterial ACC deaminase gene, ET production is reduced by 90% relative to the wild‐type (Klee et al., 1991). As the transgene is present in cultivar UC82B, this cultivar was used as wild‐type control. Although wild‐type UC82B showed severe wilting and stunting following Fol infection, most ACD plants showed only mild disease symptoms (Fig. 2a). The weight of infected ACD plants was also significantly higher than that of infected wild‐type plants (Fig. 2b). In addition, the disease index in ACD plants was significantly attenuated relative to that in wild‐type plants (Fig. 2c). A similar reduction in symptom development was also observed in ACD lines inoculated with FolΔAvr2. To monitor fungal colonization, stem sections were taken and incubated on PDA plates. The fungal recovery assay showed that Fol007 grew‐out from most stem sections of both wild‐type and ACD plants, whereas much less fungal growth was observed in FolΔAvr2‐inoculated ACD plants. A typical example of a plate assay is shown in Fig. 2d and the data from the fungal recovery assay are summarized in Fig. 2e. FolΔAvr2 was found to efficiently colonize wild‐type UC82B plants, as fungal outgrowth was often observed up to the fourth node. In contrast, colonization of ACD plants was much reduced: in 80% of cases, the fungus was only observed in stem sections collected at the cotyledon node. These data indicate that the ACD line exerts a reduced susceptibility towards Fol infection concomitant with a reduction in symptom development.
Figure 2.
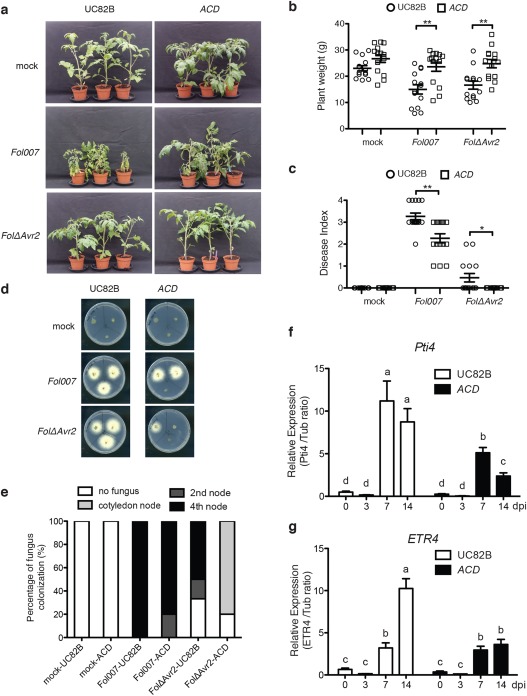
Impaired ethylene (ET) biosynthesis and production in tomato reduces disease susceptibility to Fusarium oxysporum f. sp. lycopersici (Fol). (a) Three‐week‐old wild‐type UC82B and ACD tomato plants inoculated with water (mock), Fol007 or FolΔAvr2 at 21 days post‐infection (dpi). Disease development was scored by measuring the fresh weight (b) and disease index (range, 0–4) (c) of 20 plants per treatment/genotype combination. Circles and squares indicate UC82B and ACD plants, respectively. Plant weight was subjected to a pairwise comparison using a Student's t‐test, whereas the disease index was analysed by a non‐parametric Mann–Whitney U‐test (*P < 0.05; **P < 0.01; ***P < 0.001). The bioassay was repeated three times with similar results. (d) Representative stem sections taken from the cotyledon node (top left), second node (top right) and fourth node (bottom) of individual treated plants (n = 6) after incubation for 5 days on potato dextrose agar (PDA) plates. (e) Colonization is expressed as the percentage of infected slices of all stem pieces (n = 6). The experiment was repeated twice with similar results. (f, g) Transcription patterns of ET‐regulated marker genes Pti4 and ETR4 in Fol007‐inoculated UC82B and ACD at 0, 3, 7 and 14 dpi. Gene expression levels relative to the internal control tubulin genes were quantified by quantitative polymerase chain reaction (qPCR). The data are expressed as the mean ± standard deviation (SD). Three biological replicates for each line per time point were analysed. The different letters show the significant difference at P < 0.05 as determined by Duncan's multiple‐range test. The experiment was performed twice with similar results.
It has been reported that the ET receptor gene ETR and ET‐responsive factors ERFs are induced following Fo infection (Berrocal‐Lobo and Molina, 2008; Pantelides et al., 2013). A well‐characterized ERF family member in tomato is Pti4, which has been shown to specifically bind to the GCC‐box cis‐element present in the promoter of PR genes (Wu et al., 2002). GCC‐box binding by ERFs induces PR gene expression and Pti4 could be a functional tomato homologue of ERF1 (Wu et al., 2002). In Arabidopsis, ERF1 overexpression enhances resistance to Fo (Berrocal‐Lobo and Molina, 2004). As shown in Fig. 2f,g, the transcription of Pti4 and ETR4 was strongly induced in wild‐type plants at 7 and 14 days, respectively, following Fol inoculation. Compared with wild‐type plants, Fol‐infected ACD plants showed a weaker increase in expression of the two ET marker genes, which confirms the proposed plant genotypes. In addition, the weaker induction of ETR4 and Pti4 expression in the ACD line is a further indication that ET‐mediated signalling is activated during Fol infection.
To further test how ET signalling contributes to increased susceptibility, the involvement of ET perception by the host was investigated. Bioassays were performed with wild‐type tomato cultivar Pearson and the ET‐insensitive Pearson mutant Nr. Following the inoculation of wild‐type Pearson with Fol007, the older leaves of infected plants became chlorotic and the plants showed mild wilting symptoms (Fig. 3a). On inoculation with FolΔAvr2, wild‐type Pearson plants showed hardly any symptoms (Fig. 3a). Notably, no obvious disease symptoms were observed in Nr plants inoculated with either Fol007 or FolΔAvr2. Although the fresh weight of Nr plants was identical to that of Pearson plants after Fol007 infection (Fig. 3b), the Nr plants exhibited a significantly lower disease index than Pearson plants; fewer brown vessels were observed in the stems (Fig. 3c). Fungal recovery assays revealed that FolΔAvr2 either completely failed to colonize Nr plants or, in the rare cases it did, it only reached the basal part of the stem that forms the hypocotyl (Fig. 3d,e). Taken together, the data suggest that both the ability to synthesize ET and the ability to perceive the hormone are essential for disease development and the ability of the fungus to colonize the plant.
Figure 3.
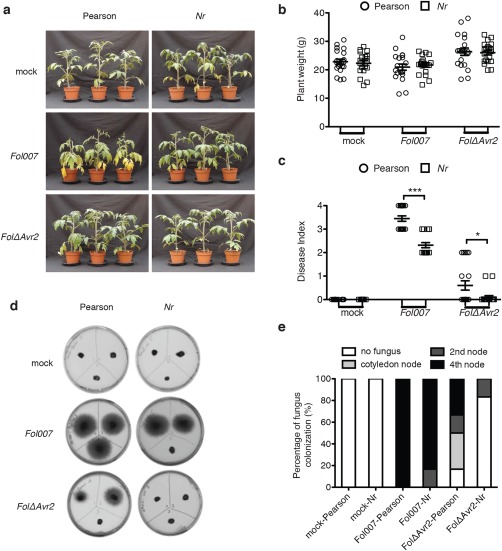
Ethylene (ET) perception is required for Fusarium oxysporum f. sp. lycopersici (Fol) disease symptom development in tomato. (a) Three‐week‐old wild‐type and Never ripe (Nr) Pearson tomato plants inoculated with water (mock), Fol007 or FolΔAvr2 at 21 days post‐infection (dpi). Disease symptoms were scored by measuring the fresh weight (b) and disease index (range, 0–4) (c) of 20 plants per treatment/genotype combination. Circle and square indicate Pearson plant and Nr plant, respectively. Plant weight was subjected to a pairwise comparison using a Student's t‐test, whereas the disease index was analysed by a non‐parametric Mann–Whitney U‐test (*P < 0.05; **P < 0.01; ***P < 0.001). The bioassay was repeated three times with similar results. (d) Representative stem sections taken from the cotyledon node (top left), second node (top right) and fourth node (bottom) of individual treated plants (n = 6) after incubation for 5 days on potato dextrose agar (PDA) plates. (e) Colonization is expressed as the percentage of infected slices of all stem pieces (n = 6). The experiment was repeated twice with similar results.
Perturbation of JA signalling has no detectable effect on plant susceptibility and disease symptom development
In Arabidopsis, various JA biosynthesis mutants, such as jasmonate resistant 1 (jar1‐1) and allene oxide synthase (aos), do not exhibit increased susceptibility to Focn (Thatcher et al., 2009; Trusov et al., 2009). To investigate the role of JA biosynthesis in the susceptibility of tomato to Fol, we used the def1 mutant. This line has a defect in its octadecanoid biosynthesis pathway, which provides precursors for JA synthesis, making the plant hypersusceptible to herbivores because of its impaired accumulation of PIs I and II in response to wounding (Lightner et al., 1993). In addition to def1, 35S::prosystemin plants were included in our assays. In these plants, the prosystemin gene is constitutively overexpressed by the 35S cauliflower mosaic virus promoter. Prosystemin is the precursor of systemin, which initiates a signalling pathway that leads to the synthesis of JA from linolenic acid (Ryan, 2000). The constitutive induction of the JA pathway in 35S::prosystemin plants results in the systemic accumulation of high levels of PIs in these plants (McGurl et al., 1994).
To confirm the genotype of def1 and 35S::prosystemin plants, the transcript levels of PI‐I were examined in leaves of wild‐type Castlemart, def1 and 35S::prosystemin plants in response to wounding. As shown in Fig. S1 (see Supporting Information), wounding of wild‐type plants triggered the induction of PI‐I expression. In contrast, no PI‐I accumulation was observed on wounding of def1 plants, whereas a constitutive expression of PI‐I was observed in 35S::prosystemin plants irrespective of treatment. These results confirm the plant genotypes and the involvement of def1 and 35S::prosystemin in JA signalling.
To test whether JA signalling affects the susceptibility to Fol, the JA mutant lines were inoculated with the fungus. As shown in Fig. 4a, Fol007‐infected def1 and 35S::prosystemin lines became equally chlorotic as the wild‐type Castlemart. Inoculation with the less pathogenic FolΔAvr2 strain did not result in obvious disease symptoms in any of the lines. No significant differences in fresh weight or disease index were observed between Castlemart and its derivatives following either Fol007 or FolΔAvr2 inoculation (Fig. 4b,c).
Figure 4.
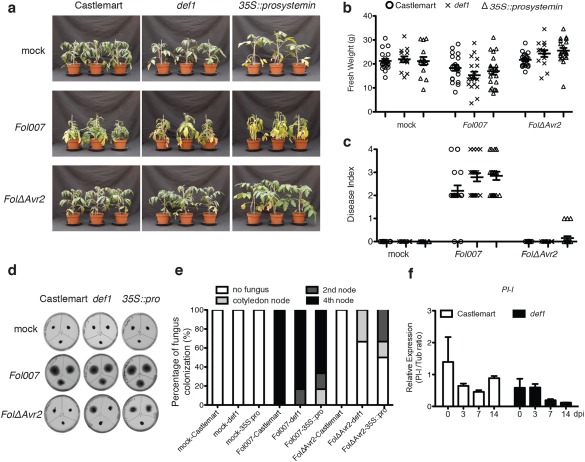
Perturbation of jasmonic acid (JA) signalling has no detectable effect on plant susceptibility. (a) Three‐week‐old wild‐type Castlemart, def1 and 35S::prosystemin tomato plants inoculated with water (mock), Fol007 or FolΔAvr2 at 21 days post‐infection (dpi). Disease symptoms were scored by measuring the fresh weight (b) and disease index (range, 0–4) (c) of 20 plants per treatment/genotype combination. Circle, × and Δ indicate Castlemart, def1 and 35S::prosystemin, respectively. The bioassay was repeated three times with similar results. (d) Representative stem sections taken from the cotyledon node (top left), second node (top right) and fourth node (bottom) of individual treated plants (n = 6) after incubation for 5 days on potato dextrose agar (PDA) plates. (e) Colonization is expressed as the percentage of infected slices of all stem pieces (n = 6). The experiment was repeated twice with similar results. (f) Transcription patterns of proteinase inhibitor I (PI‐I) in Fol007‐inoculated wild‐type Castlemart and def1 at 0, 3, 7 and 14 dpi. Three biological replicates for each line were analysed. The data are expressed as the mean ± standard deviation (SD). The experiment was performed twice with similar results.
Fungal recovery assay revealed that, in wild‐type Castlemart, Fol007 was present in all probed stem sections, showing that the fungus colonized the stem until the fourth node (Fig. 4d,e). In most of the inoculated def1 and 35S::prosystemin plants, Fol007 had also colonized the entire stem until the fourth node. In contrast with Fol007, FolΔAvr2 completely failed to colonize wild‐type Castlemart plants, which is consistent with the lack of disease symptoms observed in these plants. However, in the 35S::prosystemin plants and def1 mutants, the FolΔAvr2 strain was detected at a limited number of stem sections collected at either the position of the cotyledon node or the second node (Fig. 4e). The latter data suggest that disturbed JA signalling might facilitate fungal colonization.
To assess whether JA signalling is induced following Fol007 inoculation, the expression of PI‐I was monitored at 0, 3, 7 and 14 dpi. As shown in Fig. 4f, no significant difference in PI‐I expression was observed in Fol‐inoculated wild‐type plants at any time point. The same low levels of PI‐I expression were detected in the def1 mutant, and these levels did not change during infection. Taken together, these data imply that perturbation of JA signalling has no detectable effect on the susceptibility to Fol.
SA and ET signalling pathways act synergistically in tomato susceptibility to Fol infection
The degradation of SA was found to enhance the susceptibility to Fol, whereas impaired ET signalling reduced the susceptibility. We therefore wanted to test whether these pathways interact following Fol infection. The expression of PR1a was monitored over time (0–14 dpi) in both the ACD ET synthesis mutant and in wild‐type UC82B plants on Fol007 infection. As shown in Fig. 5a, expression of the SA marker gene PR1a was strongly induced in wild‐type plants, but much less so in ACD lines. This finding indicates that the induction of PR1a following Fol infection requires an intact ET pathway. Subsequently, the expression of Pti4 and ETR4 was assessed in the NahG line and its parental Moneymaker background following Fol007 inoculation. As shown in Fig. 5b,c, expression of both ET marker genes was strongly induced in wild‐type plants, but not in NahG plants, following Fol infection. This finding suggests that an intact SA signalling pathway is required for Pti4 and ETR4 induction.
Figure 5.
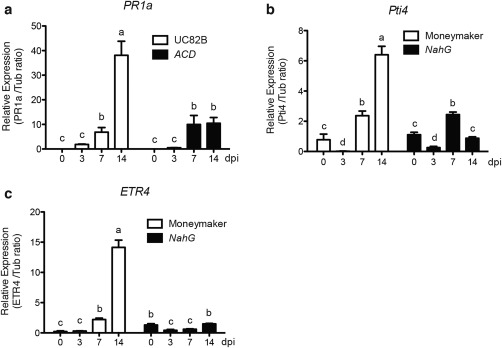
Time course of transcription patterns of salicylic acid (SA) and ethylene (ET) marker genes on Fusarium oxysporum f. sp. lycopersici (Fol) inoculation. (a) Expression of the SA marker gene pathogenesis‐related 1a (PR1a) in wild‐type UC82B and the transgenic ACD line at 0, 3, 7 and 14 days post‐infection (dpi). (b, c) Transcription patterns of the ET marker genes Pti4 and ETR4 in NahG and the Moneymaker progenitor at 0, 3, 7 and 14 dpi. Gene expression levels relative to the internal control tubulin genes were quantified by quantitative polymerase chain reaction (qPCR). The data are expressed as the mean ± standard deviation (SD). Three biological replicates for each line per time point were analysed. The different letters show the significant difference at P < 0.05 as determined by Duncan's multiple‐range test. The experiment was performed twice with similar results.
Discussion
Here, the role of SA, ET and JA in the modulation of the susceptibility of tomato plants to Fol was investigated. NahG plants that failed to accumulate SA were hypersusceptible to Fol infection and showed severe disease symptoms and extensive fungal colonization of their xylem vessels. Together with the strong induction of the SA marker gene PR1a in wild‐type plants, which was not strongly induced in NahG plants, these data show that SA plays a positive role in reducing disease susceptibility. This conclusion is in agreement with chemical studies, in which exogenous application of SA to tomato through root feeding or foliar sprays reduced vascular browning, leaf yellowing and wilting following Fol inoculation (Mandal et al., 2009). The positive role of SA in Fol resistance in tomato is consistent with studies with NahG Arabidopsis, showing an increased susceptibility to Fo (Berrocal‐Lobo and Molina, 2004; Diener and Ausubel, 2005; Thatcher et al., 2009; Trusov et al., 2009). Therefore, the role of SA in Fo susceptibility seems to be conserved in both plant species. Interestingly, in several studies, elevated SA has been reported to enhance the susceptibility to necrotrophic pathogens, but to promote resistance to hemibiotrophs (Bari and Jones, 2009; El Oirdi et al., 2011). Our findings, in which SA reduces the susceptibility of tomato plants to Fol, are in line with the hemibiotrophic lifestyle of the latter (Di et al., 2016).
The ACD tomato line, in which ET biosynthesis is compromised, showed a reduced susceptibility to Fol infection. On Fol infection, the transgenic line showed fewer disease symptoms and a reduced fungal colonization relative to the wild‐type UC82B cultivar (Fig. 2). The ET marker genes ETR4 and Pti4 were highly induced in wild‐type plants, but not in the ACD line, indicating that ET signalling is induced in response to Fol infection and is important for disease development. The ET‐insensitive Nr mutant was also found to be less susceptible than wild‐type Pearson plants to infection with Fol007 (Fig. 3). These data are consistent with studies by Lund et al. (1998) and Francia et al. (2007), which also reported a reduction in Fol disease symptoms in the Nr mutant, The fact that both ET synthesis and perception were found to be important for disease development and fungal colonization suggests that ET signalling is important for full susceptibility to Fol.
The role of ET is multifaceted in Arabidopsis (Di et al., 2016). Similar to Nr in tomato, soil‐grown ein2‐1 and etr1‐1 mutants of Arabidopsis showed a reduction in disease symptoms relative to wild‐type Col‐0 plants when inoculated with Focn (Pantelides et al., 2013; Trusov et al., 2009). Fo inoculation of Arabidopsis plants carrying the ein2‐5 allele revealed a markedly enhanced susceptibility to Fo in plate assays (Berrocal‐Lobo and Molina, 2004). These differences might be explained by the different mutations, or by the different inoculation methods. As inoculation of soil‐grown plants best mimics the natural infection process, which resembles that of our tomato assays, it seems that, in both plant species, the role of ET is conserved, in that its absence reduces disease symptom development (Pantelides et al., 2013; Trusov et al., 2009).
No significant difference in disease index and fresh weight between the wild‐type and JA‐deficient def1 line was found after Fol007 or FolΔAvr2 inoculation (Fig. 4). These findings contrast those of Thaler et al. (2004), who showed that the weight of def1, but not wild‐type tomato plants, was reduced on inoculation with a race 1 Fol isolate. The reason for the discrepancy might be that a different Fusarium race or different assay conditions were used. On Fol007 infection, 35S::prosystemin plants also did not show a significant difference in disease index or fresh weight relative to wild‐type tomato. Thus, under our assay conditions, JA does not appear to play a major role in the development of disease symptoms or disease susceptibility.
In different host–Fo pathosystems, JA can promote either resistance or susceptibility (Di et al., 2016). A point mutation in COI1 in Arabidopsis, an essential component for JA perception, strongly reduces the susceptibility to Fo (Thatcher et al., 2009; Trusov et al., 2009). In contrast, jar1 mutants, which are defective in the synthesis of the bioactive JA–isoleucine (JA‐Ile) conjugate, show wild‐type‐like symptoms or only a slight increase in susceptibility (Thatcher et al., 2009; Trusov et al., 2009). In addition, Cole et al. (2014) have reported that infection by Focn and Fo f. sp. matthioli, which produce JA‐Ile and leucine‐conjugated JA (JA‐Leu), respectively, is suppressed in coi1. In contrast, the coi1 mutation has no effect on infection by Fo f. sp. raphani, which produces no detectable JA‐Ile/JA‐Leu. Furthermore, the JA‐insensitive mutation (jai1) has no effect on the infection of tomato plants by Fol, which produces no detectable jasmonates. Therefore, different formae speciales may adopt different strategies to infect their host and to cause disease symptoms.
The SA, ET and JA signalling pathways are entangled in a complex network in which the different pathways influence each other through positive and negative regulatory interactions (Grant and Jones, 2009). We observed that, relative to wild‐type plants, the expression of PR1a was less strongly induced in the ET biosynthesis mutant ACD (Fig. 5). Similarly, following Fol inoculation, ET signalling, as monitored by ETR4 and Pti4 expression, was strongly induced in wild‐type plants, but less so in NahG plants. Collectively, these results indicate that, in tomato, SA and ET signalling might act synergistically during Fol infection, as an intact ET pathway is required for the induction of the SA reporter gene, and vice versa. In addition, for Xanthomonas campestris pv. vesicatoria infection of tomato, accumulation of SA was found to require ET synthesis, suggesting that ET positively regulates SA‐induced defences (O'Donnell et al., 2003).
Altogether, these data allow us to propose a model for the involvement of SA, ET and JA signalling in tomato in susceptibility to Fol (Fig. 6a). SA and ET signalling interact and have opposite roles in disease susceptibility. The infection of tomato plants by Fol activates both the ET and SA pathways. The ET response enhances the susceptibility to Fol infection and disease development, whereas SA responses restrict colonization. No apparent role for JA was shown in the interaction, as disease symptoms in JA mutants were indistinguishable from those of the wild‐type plants. A comparison with the reported roles of these phytohormones in Arabidopsis in Fo infection reveals shared and unique effects between Arabidopsis and tomato. As shown in Fig. 6b, SA signalling also negatively regulates susceptibility to Fo, whereas ET signalling likewise positively enhances susceptibility. Notably, JA can be hijacked by the fungus to enhance pathogenicity in Arabidopsis, but apparently does not play an important role in tomato.
Figure 6.
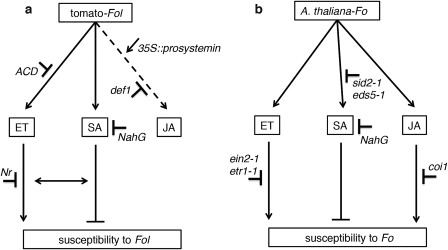
Proposed models for the involvement of jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) signalling in tomato (a) and Arabidopsis (b) on Fusarium oxysporum (Fo) infection. Compromised ET biosynthesis and perception reduce disease susceptibility, whereas compromised SA signalling promotes hypersusceptibility to Fo f. sp. lycopersici (Fol) infection in tomato. The SA and ET pathways act synergistically, in that induction of one pathway requires the intactness of the other. Although, in Arabidopsis, the JA pathway is required for disease susceptibility, it is not in tomato. By convention, the arrowhead implies positive regulation (stimulation) and the T‐bar implies negative regulation.
Although the Arabidopsis–Fo system serves as a useful model for some plant–Fo interactions, the tomato system can be used to study Fo–plant interactions in which JA does not play a major role. It will be interesting to also investigate other plant–Fo systems to identify whether there may be more species‐specific aspects that differ from either model. These insights are relevant to allow the translation of molecular mechanisms obtained in these models into various crops to aim for a reduced susceptibility to wilt disease. The molecular mechanisms underlying susceptibility, however, are currently unknown and their elucidation is a challenge for future studies.
Experimental Procedures
Ten different tomato (Solanum lycopersicum) genotypes were used in these studies, including the four wild‐type cultivars from which these mutants were derived: Moneymaker, UC82B, Pearson and Castlemart. The transgenic NahG line, which is compromised in SA accumulation, is in a Moneymaker background (Brading et al., 2000). The ET‐impaired mutant ACD (Klee et al., 1991) and the Nr mutation (Lanahan et al., 1994) are in a UC82B and Pearson background, respectively. The JA‐impaired mutant def1 (Howe et al., 1996) and the prosystemin‐overexpressing line (35S::prosystemin) are in the Castlemart background (McGurl et al., 1994). Tomato seedlings were grown in a conditioned glasshouse with day–night temperatures of 23 °C–18 °C and a 16‐h light/8‐h dark regime.
Fusarium inoculation assay
Wild‐type Fusarium strain Fol007 (race 2) and the derived FolΔAvr2 mutant have been described previously (Houterman et al., 2009). Fol strains were grown on PDA (Oxoid Ltd., Basingstoke, Hampshire, UK) at 25 °C for 7–10 days. Subsequently, a piece of agar carrying the fungus was transferred to 100 mL minimal medium (100 mm KNO3, 3% sucrose and 0.17% yeast nitrogen base without amino acids or ammonia). Conidial spores were harvested after 3–5 days of cultivation at 25 °C with shaking. After washing with sterilized water, the spores were diluted to 106 spores/mL. For bioassay, 3‐week‐old tomato seedlings were uprooted from the soil. The seedlings were placed for 5 min in the Fol spore suspension (106 spores/mL) and subsequently potted. Disease progression was evaluated after 3 weeks. Plant weight and disease index (Rep et al., 2005) were scored for 20 plants/treatment.
Fungal recovery assay
Fungal colonization in tomato plants was assessed at 21 dpi. Representative stem sections taken from the cotyledon node, second node and fourth node were collected separately. The stem pieces were surface sterilized in 70% ethanol and rinsed in sterile distilled water, after which the ends of the stems were removed with a sterile scalpel. Stem sections about 5 mm thick were cut and placed on PDA supplemented with 200 mg/L streptomycin and 100 mg/L penicillin at 25 °C, allowing the fungus to grow out of the stem sections. Photographs were taken after 5 days of incubation at 25 °C. Data were expressed as a percentage of slices showing fungal outgrowth.
Wounding assay
Wounding experiments have been descripted previously (Howe et al., 1996). Briefly, a haemostat was used to inflict damage to the primary leaf of 15‐day‐old tomato plants. Twenty‐four hours later, the wounded leaf and an upper unwounded leaf were dissected and snap frozen in liquid N2 to use for expression analysis.
Analysis of gene expression by RT‐qPCR
RNA isolation and cDNA synthesis were performed as described previously (Gawehns et al., 2014). Briefly, total RNA from tomato stem beneath the cotyledon was extracted using Trizol‐Reagent (Invitrogen, Life Technologies, Grand Island, NY, USA) according to the manufacturer's instructions. The RNA was subsequently purified with an RNeasy Mini kit (Qiagen, Düsseldorf, Germany) and DNA was removed by on‐column treatment with RNase‐free DNase (Qiagen). cDNA was synthesized using the M‐MulV reverse‐transcriptase RNase H minus kit (Fermentas, Thermo Scientific, Pittsburgh, PA, USA). Stem tissue was collected from tomato plants at 0, 3, 7 and 14 dpi. The conditions of the RT‐qPCR experiments and the relative quantification of specific mRNA levels were performed according to Lopez‐Raez et al. (2010) and using the gene‐specific primers described in Table S1 (see Supporting Information). PCRs were performed in an ABI 7500 Real‐Time PCR system (Applied Biosystems, http://www.appliedbiosystems.com Foster City CA, USA) using the Platinum SYBR Green qPCR SuperMix‐UDG kit (Invitrogen). The 20‐μL PCRs contained 0.25 μm of each primer, 0.1 μL ROX reference dye and 1 μL of cDNA. The cycling programme was set to 5 min at 50 °C, 5 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C, followed by a melting curve analysis. The expression levels of selected genes were normalized to tomato α‐tubulin (Solyc04g077020.2) expression. Relative gene expression was calculated using the 2–Δ CT method. Three biological replicates for each of the selected genes were performed.
Statistical analyses
The statistical significance of the results was determined by performing PRISM 5.0 (GraphPad, http://www.graphpad.com). The data on plant weight were subjected to a pairwise comparison with Student's t‐test. For data on the disease index in tomato plants, a Mann–Whitney test was performed for each genotype. Gene expression data were statistically evaluated, and bars annotated with different letters were significantly different according to Duncan's multiple‐range test.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 The expression of proteinase inhibitor I (PI‐I) was examined by reverse transcription‐polymerase chain reaction (RT‐PCR) in unwounded (u) and wounded (w) leaves of Castlemart, def1 and 35S::prosystemin. Two biological replicates for each line were analysed.
Table S1 Primer sequences used in the gene expression analysis.
Acknowledgements
We thank Jonathan D. G. Jones (Sainsbury Laboratory, Norwich, Norfolk, UK) for providing the transgenic NahG line. ACD and Nr mutants were obtained from M. Mudgett, Stanford University, Stanford, CA, USA (provided by H. Klee, University of Florida, Gainesville, FL, USA). def1 was obtained from M. Mudgett (provided by G. Howe, Michigan State University, East Lansing, MI, USA). We also thank M. Kant (University of Amsterdam, Amsterdam, the Netherlands) for providing 35S::prosystemin. The authors wish to thank Ben Cornelissen for critical reading and comments on the manuscript. X.D. was financially supported by the China Scholarship Council program grant number 201206180034 and by the ZonMw Program ‘Enabling Technologies Hotels’ file number 435000001. F.L.W.T. received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skodowska‐Curie grant agreement No. 676480 (Bestpass), and from the NWO‐Earth and Life Sciences funded VICI project No. 865.14.003.
References
- Agrios, G.N. (2005) Plant Pathology. St. Louis, MO: Academic Press. [Google Scholar]
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari, R. and Jones, J.D.G. (2009) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum . Mol. Plant–Microbe Interact. 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal‐Lobo, M. and Molina, A. (2008) Arabidopsis defense response against Fusarium oxysporum . Trends Plant Sci. 13, 145–150. [DOI] [PubMed] [Google Scholar]
- Brading, P.A. , Hammond‐Kosack, K.E. , Parr, A. and Jones, J.D.G. (2000) Salicylic acid is not required for Cf‐2‐ and Cf‐9‐dependent resistance of tomato to Cladosporium fulvum . Plant J. 23, 305–318. [DOI] [PubMed] [Google Scholar]
- Cole, S.J. , Yoon, A.J. , Faull, K.F. and Diener, A.C. (2014) Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine‐ and leucine‐conjugated jasmonates. Mol. Plant Pathol. 15, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, X. , Takken, F.L.W. and Tintor, N. (2016) How phytohormones shape interactions between plants and the soil‐borne fungus Fusarium oxysporum . Front. Plant Sci. 7, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, A.C. and Ausubel, F.M. (2005) Resistance to Fusarium oxysporum 1, a dominant Arabidopsis disease‐resistance gene, is not race specific. Genetics, 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, C.I. , McGrath, K.C. , Dombrecht, B. , Manners, J.M. , Maclean, D.C. , Schenk, P.M. and Kazan, K. (2006) Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana . Australas. Plant Pathol. 35, 581–591. [Google Scholar]
- El Oirdi, M. , Abd El Rahman, T. , Rigano, L. , El Hadrami, A. , Rodriguez, M.C. , Daayf, F. , Vojnov, A. and Bouarab, K. (2011) Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell, 23, 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia, D. , Demaria, D. , Calderini, O. , Ferraris, L. , Valentino, D. , Arcioni, S. , Tamietti, G. and Cardinale, F. (2007) Wounding induces resistance to pathogens with different lifestyles in tomato: role of ethylene in cross‐protection. Plant Cell Environ. 30, 1357–1365. [DOI] [PubMed] [Google Scholar]
- Gawehns, F. , Houterman, P.M. , Ichou, F.A. , Michielse, C.B. , Hijdra, M. , Cornelissen, B.J.C. , Rep, M. and Takken, F.L.W. (2014) The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I‐2‐mediated cell death. Mol. Plant–Microbe Interact. 27, 336–348. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Grant, M.R. and Jones, J.D.G. (2009) Hormone (dis)harmony moulds plant health and disease. Science, 324, 750–752. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Speijer, D. , Dekker, H.L. , de Koster, C.G. , Cornelissen, B.J.C. and Rep, M. (2007) The mixed xylem sap proteome of Fusarium oxysporum‐infected tomato plants. Mol. Plant Pathol. 8, 215–221. [DOI] [PubMed] [Google Scholar]
- Houterman, P.M. , Ma, L. , van Ooijen, G. , de Vroomen, M.J. , Cornelissen, B.J.C. , Takken, F.L.W. and Rep, M. (2009) The effector protein Avr2 of the xylem‐colonizing fungus Fusarium oxysporum activates the tomato resistance protein I‐2 intracellularly. Plant J. 58, 970–978. [DOI] [PubMed] [Google Scholar]
- Howe, G.A. and Ryan, C.A. (1999) Suppressors of system in signaling identify genes in the tomato wound response pathway. Genetics, 153, 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A. , Lightner, J. , Browse, J. and Ryan, C.A. (1996) An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell, 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd, B.N. , Edgar, C.I. , Kumar, K.K. , Aitken, E.A. , Schenk, P.M. , Manners, J.M. and Kazan, K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate‐dependent defense in Arabidopsis. Plant Cell, 21, 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee, H.J. , Hayford, M.B. , Kretzmer, K.A. , Barry, G.F. and Kishore, G.M. (1991) Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell, 3, 1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B. , Yen, H.C. , Giovannoni, J.J. and Klee, H.J. (1994) The Never Ripe mutation blocks ethylene perception in tomato. Plant Cell, 6, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.I. , Leon, J. and Raskin, I. (1995) Biosynthesis and metabolism of salicylic‐acid. Proc. Natl. Acad. Sci. USA, 92, 4076–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.Y. , Williams, M.M. , Loh, Y.T. , Lee, G.I. and Howe, G.A. (2002) Resistance of cultivated tomato to cell content‐feeding herbivores is regulated by the octadecanoid‐signaling pathway. Plant Physiol. 130, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner, J. , Pearce, G. , Ryan, C.A. and Browse, J. (1993) Isolation of signaling mutants of tomato (Lycopersicon esculentum). Mol. Gen. Genet. 241, 595–601. [DOI] [PubMed] [Google Scholar]
- Lopez‐Raez, J.A. , Kohlen, W. , Charnikhova, T. , Mulder, P. , Undas, A.K. , Sergeant, M.J. , Verstappen, F. , Bugg, T.D. , Thompson, A.J. , Ruyter‐Spira, C. and Bouwmeester, H. (2010) Does abscisic acid affect strigolactone biosynthesis? New Phytol. 187, 343–354. [DOI] [PubMed] [Google Scholar]
- Lund, S.T. , Stall, R.E. and Klee, H.J. (1998) Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell, 10, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S. , Mallick, N. and Mitra, A. (2009) Salicylic acid‐induced resistance to Fusarium oxysporum f. sp lycopersici in tomato. Plant Physiol. Biochem. 47, 642–649. [DOI] [PubMed] [Google Scholar]
- McGovern, R.J. (2015) Management of tomato diseases caused by Fusarium oxysporum . Crop Prot. 73, 78–92. [Google Scholar]
- McGurl, B. , Orozcocardenas, M. , Pearce, G. and Ryan, C.A. (1994) Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase‐inhibitor synthesis. Proc. Natl. Acad. Sci. USA, 91, 9799–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse, C.B. and Rep, M. (2009) Pathogen profile update: Fusarium oxysporum . Mol. Plant Pathol. 10, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, P.J. , Schmelz, E. , Block, A. , Miersch, O. , Wasternack, C. , Jones, J.B. and Klee, H.J. (2003) Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 133, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides, I.S. , Tjamos, S.E. , Pappa, S. , Kargakis, M. and Paplomatas, E.J. (2013) The ethylene receptor ETR1 is required for Fusarium oxysporum pathogenicity. Plant Pathol. 62, 1302–1309. [Google Scholar]
- Panthee, D.R. and Chen, F. (2010) Genomics of fungal disease resistance in tomato. Curr. Genomics, 11, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pietro, A. , Madrid, M.P. , Caracuel, Z. , Delgado‐Jarana, J. and Roncero, M.I. (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Rep, M. , Dekker, H.L. , Vossen, J.H. , de Boer, A.D. , Houterman, P.M. , Speijer, D. , Back, J.W. , de Koster, C.G. and Cornelissen, B.J.C. (2002) Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus‐infected tomato. Plant Physiol. 130, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep, M. , Meijer, M. , Houterman, P.M. , van der Does, H.C. and Cornelissen, B.J.C. (2005) Fusarium oxysporum evades I‐3‐mediated resistance without altering the matching avirulence gene. Mol. Plant–Microbe Interact. 18, 15–23. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim. Biophys. Acta, 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Takken, F.L.W. and Rep, M. (2010) The arms race between tomato and Fusarium oxysporum . Mol. Plant Pathol. 11, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler, J.S. , Owen, B. and Higgins, V.J. (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher, L.F. , Manners, J.M. and Kazan, K. (2009) Fusarium oxysporum hijacks COI1‐mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58, 927–939. [DOI] [PubMed] [Google Scholar]
- Thatcher, L.F. , Powell, J.J. , Aitken, E.A.B. , Kazan, K. and Manners, J.M. (2012) The Lateral Organ Boundaries Domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 160, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov, Y. , Sewelam, N. , Rookes, J.E. , Kunkel, M. , Nowak, E. , Schenk, P.M. and Botella, J.R. (2009) Heterotrimeric G proteins‐mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid‐, jasmonic acid/ethylene‐ and abscisic acid‐mediated defense signaling. Plant J. 58, 69–81. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Wilkinson, J.Q. , Lanahan, M.B. , Yen, H.C. , Giovannoni, J.J. and Klee, H.J. (1995) An ethylene‐inducible component of signal‐transduction encoded by Never‐Ripe . Science, 270, 1807–1809. [DOI] [PubMed] [Google Scholar]
- Wu, K. , Tian, L. , Hollingworth, J. , Brown, D.C. and Miki, B. (2002) Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128, 30–37. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 The expression of proteinase inhibitor I (PI‐I) was examined by reverse transcription‐polymerase chain reaction (RT‐PCR) in unwounded (u) and wounded (w) leaves of Castlemart, def1 and 35S::prosystemin. Two biological replicates for each line were analysed.
Table S1 Primer sequences used in the gene expression analysis.


