Summary
In order to facilitate infection, the rice blast pathogen Magnaporthe oryzae secretes an abundance of proteins, including avirulence effectors, to diminish its host's defences. Avirulence effectors are recognized by host resistance proteins and trigger the host's hypersensitive response, which is a rapid and effective form of innate plant immunity. An understanding of the underlying molecular mechanisms of such interactions is crucial for the development of strategies to control disease. However, the expression and secretion of certain effector proteins, such as AVR‐Pia, have yet to be reported. Reverse transcription‐polymerase chain reaction (RT‐PCR) revealed that AVR‐Pia was only expressed during infection. Fluorescently labelled AVR‐Pia indicated that AVR‐Pia expression was induced during appressorial differentiation in the cells of both rice and onion, as well as in a penetration‐deficient (Δpls1) mutant capable of developing melanized appressoria, but unable to penetrate host cells, suggesting that AVR‐Pia expression is independent of fungal penetration. Using live‐cell imaging, we also documented the co‐localization of green fluorescent protein (GFP)‐labelled AVR‐Pia and monomeric red fluorescent protein (mRFP)‐labelled PWL2, which indicates that AVR‐Pia accumulates in biotrophic interfacial complexes before being delivered to the plant cytosol. Together, these results suggest that AVR‐Pia is a cytoplasmic effector that is expressed at the onset of appressorial differentiation and is translocated to the biotrophic interfacial complex, and then into the host's cytoplasm.
Keywords: appressorium; avirulence effector; AVR‐Pia; BIC, Pyricularia oryzae; rice
Introduction
Among the cereals, rice (Oryza sativa L.) is one of the leading food sources for humankind and, in many cultures, rice has served as a staple food for at least 10 000 years (Wani et al., 2012). However, the expanding human population is resulting in an imbalance between the supply and demand of this important crop (Jeon et al., 2011). Therefore, increasing rice production is very important, and one way to enhance net yield is to reduce the factors that cause damage to the crop, including diseases.
Rice blast disease (RBD) is one of the most serious and devastating diseases affecting rice and has been reported in 85 countries (Maciel, 2011). The disease is highly adaptable to a wide range of environmental conditions and is able to infect crops of rain‐fed upland, irrigated lowland and deep‐water rice fields. Accordingly, RBD continuously threatens global food security and presents a significant challenge to rice farmers worldwide.
The pathogen responsible for RBD is the hemibiotrophic ascomycete fungus Magnaporthe oryzae (synonym of Pyricularia oryzae; Zhang et al., 2016). During infection, M. oryzae conidia attach to the leaf surfaces of rice, develop into melanized appressoria and accumulate glycerol until they have achieved a sufficient turgor pressure to penetrate host cells. Next, the pathogen develops biotrophic invasive hyphae (IHs) to colonize host cells (Talbot, 1995) and secretes effector proteins at the host–pathogen interface to facilitate invasion. However, some of the effectors function as avirulence (AVR) effectors, which are recognized by the host's corresponding resistance (R) proteins and allow host plants to recognize the pathogen, in a classic gene‐for‐gene relationship (Flor, 1971); host recognition of AVR effectors and specific R proteins induces defence responses, such as the hypersensitive response (HR), to limit the extent of damage.
In most plants, innate immunity against microbial pathogens comprises two layers. The first layer is known as pathogen‐associated molecular pattern (PAMP)‐triggered immunity and is triggered by the recognition of PAMPs by plant pattern recognition receptors located at the host–pathogen interface. In most cases, PAMPs include essential pathogen components, such as chitin, glucan, flagellin and ergosterol (Dodds and Rathjen, 2010; Oliveira‐Garcia and Valent, 2015). Meanwhile, the second layer of defence is known as effector‐triggered immunity and involves the recognition of AVR effectors by R proteins, as mentioned previously, which belong to the nucleotide‐binding/leucine‐rich repeat family of receptors, which are known to intercept AVR effectors and induce robust resistance to pathogen attacks (Cui et al., 2015).
In order to develop effective means of preventing RBD, a thorough understanding of the plant–pathogen interaction at the molecular level is required. To date, approximately 85 RBD R genes have been identified (Ballini et al., 2008); however, only approximately 25 M. oryzae AVR genes have been genetically mapped (Dioh et al., 2000), which suggests that many more AVR genes remain to be identified. Of the known AVR genes, PWL1, PWL2, AVR‐Pita and AvrPiz‐t were isolated using a map‐based cloning strategy, and AVR‐Pia was isolated by comparing the Japanese field isolate Ina168 and its spontaneous mutant Ina168m95‐1 (Sweigard et al., 1995, Kang et al., 1995, Orbach et al., 2000, Li et al., 2009, and Miki et al., 2009). In addition, AVR‐Pia, together with two other AVR genes, AVR‐Pii and AVR‐Pik/km/kp, were identified using a polymerase chain reaction (PCR)‐based association genetics approach (Yoshida et al., 2009). Among the previously identified AVR effectors, all encode small proteins that are secreted during infection, but ACE1 encodes a large cytoplasmic non‐ribosomal peptide synthetase, which is involved in the production of secondary metabolites (Fudal et al., 2007).
The two main secretory pathways of effector proteins are the biotrophic interfacial complex (BIC)‐associated secretory pathway and the IH‐associated secretory pathway. Previous studies have demonstrated that cytoplasmic effectors, such as AvrPiz‐t, AVR‐Pita and PWL2, are delivered to the host cytoplasm via BICs and are associated with a secretory system that involves the exocyst complex (SEC5, Exo70) and t‐SNAREs, such as Sso1 (Giraldo et al., 2013). The BIC‐associated secretory pathway is thought to be the main secretion system for cytoplasmic effectors (Giraldo et al., 2013), whereas apoplastic effectors, such as Slp1 and BAS4, are secreted from IHs via the conserved endoplasmic reticulum (ER)–Golgi secretory pathway and accumulate in the extra‐invasive hyphal membrane compartment that surrounds the IH cell wall, without being secreted into the plant cytoplasm (Zhang and Xu, 2014). However, although AVR‐Pia is known to encode a small secreted protein (85 amino acids) without any known protein domains (Yoshida et al., 2009), the secretion of AVR‐Pia remains to be studied.
An understanding of when and where AVR gene expression is induced will provide an insight into the AVR–R interactions of RBD infection and will facilitate the development of strategies for the protection of crops. Previously, Fudal et al. (2007) studied mutants related to appressorial formation and maturation, and reported that ACE1 expression was associated with the onset of appressorial‐mediated penetration. However, the gene expression of AVR‐Pia and its AVR effectors has not been well clarified. Therefore, this study investigates the timing of AVR‐Pia expression, as well as the localization of AVR‐Pia prior to its delivery to host cells. To examine AVR‐Pia expression, we used transcriptional fusion of the AVR‐Pia native promoter (PPR) and the enhanced green fluorescence protein (eGFP), and observed the fluorescence of appressoria and IHs on host surfaces.
Results
AVR‐Pia expression during differentiation of M. oryzae appressoria
Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis revealed that AVR‐Pia was not transcribed in liquid‐cultured Ina168 mycelium, but was clearly expressed in Ina168‐inoculated rice leaves at 24–60 h post‐inoculation (hpi; Fig. 1). In addition, qRT‐PCR demonstrated that AVR‐Pia expression was initiated at ∼18 hpi in compatible cultivars (Shin‐2, Nipponbare; pia) and continued until 60 hpi, whereas AVR‐Pia expression declined after 30 hpi in incompatible cultivars (Aichiasahi, Nipponbare; Pia), after which almost no expression was detected (Fig. 2). Thus, AVR‐Pia expression is likely to be induced at 18–24 hpi, which coincides with appressorial differentiation, independent of host compatibility.
Figure 1.
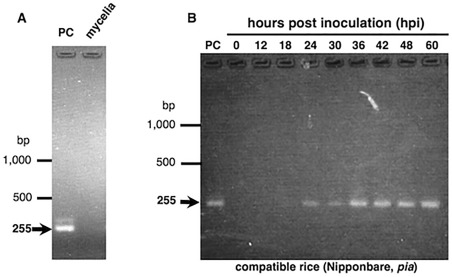
Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of AVR‐Pia expression in in vitro and in planta Magnaporthe oryzae Ina168. (A) AVR‐Pia expression in liquid‐cultured Ina168 mycelia at 22 h post‐inoculation (hpi). (B) AVR‐Pia expression in Ina168‐infected rice leaf sheaths at various time points after inoculation. PC, positive control (AVR‐Pia DNA).
Figure 2.
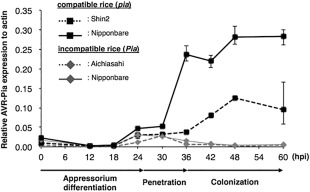
Quantification of Magnaporthe oryzae AVR‐Pia expression in compatible and incompatible rice plants. AVR‐Pia expression was quantified using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) of RNA extracted from Ina168‐infected rice leaf sheaths at various time points after inoculation. AVR‐Pia levels are shown relative to actin expression, and the data points (n = 3) and error bars indicate means ± standard deviation.
In order to monitor AVR‐Pia expression during pathogenesis‐related differentiation, the expression vector pCSN43‐DEST‐PPR::eGFP, which included the native AVR‐Pia promoter (PPR) and eGFP, was expressed in the M. oryzae strain Ina168m95‐1 (Δavr‐pia), and the resulting transformant (Ina168m95‐1‐PPR::eGFP) developed well‐melanized appressoria on the leaf sheaths of the susceptible rice cultivar Shin‐2 at 24 hpi and successfully colonized rice cells via IHs at 48 hpi. Fluorescence visualization was used to detect the eGFP signal in the transformant's appressoria (Fig. 3A), as well as in IHs that had penetrated rice cells (Fig. 3B). The eGFP signal was also detected in the appressoria and IHs of the transformant on onion epidermis at 24 hpi (Fig. 3C,D). Therefore, the results confirmed that AVR‐Pia transcription occurs during the early stage of infection that involves appressorial differentiation. In addition, fluorescence visualization of the transformant cultured in liquid medium was performed at 6 and 24 hpi (Fig. 3E,F), and no eGFP signal was detected in the hyphae of the transformant, confirming that AVR‐Pia expression is related to fungal infection.
Figure 3.
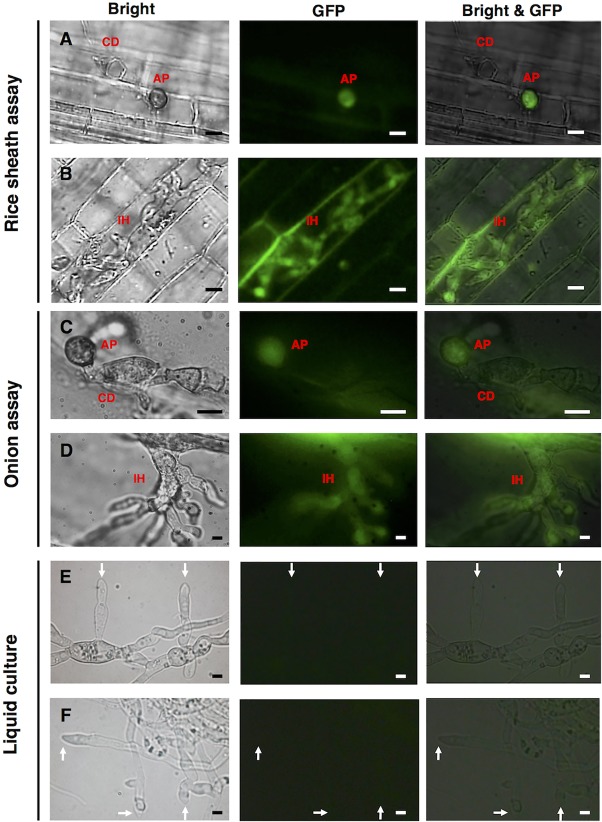
AVR‐Pia promoter activity visualized using enhanced green fluorescence protein (eGFP) fluorescence in rice leaf sheath, onion epidermis and liquid culture. PPR::eGFP‐transformed Magnaporthe oryzae Ina168m95‐1 was used to monitor AVR‐Pia expression in rice leaf sheaths at 24 h post‐inoculation (hpi) (A) and 48 hpi (B), in onion epidermis at 24 hpi (C, D) and in liquid culture at 6 hpi (E) and 24 hpi (F). AP, appressorium; CD, conidium; IH, invasive hypha; white arrow, tip of hypha. Scale bars, 10 μm.
AVR‐Pia expression is independent of penetration
Using the penetration‐deficient (Δpls1) mutant Ina86‐137Δlig4, in which the gene (i.e. Pls1) that encodes the tetraspanin‐like protein required for the formation of penetration pegs (Clergeot et al., 2001) was disrupted, we found that the mutant was unable to develop penetration pegs on either rice leaves or onion epidermis (data not shown). However, introduction of the pBlastr‐PPR::eGFP vector (Δpls1‐PPR::eGFP) resulted in the detection of eGFP in Δpls1‐PPR::eGFP appressoria on rice leaf sheaths at 24 hpi (Fig. 4A), clearly indicating that AVR‐Pia expression is induced before fungal penetration. Infection‐induced AVR‐Pia expression was checked by fluorescence observation of Δpls1‐PPR::eGFP in liquid culture at 24 hpi (Fig. 4B), and showed that there was no detected eGFP in Δpls1‐PPR::eGFP, confirming that AVR‐Pia expression is induced by infection.
Figure 4.
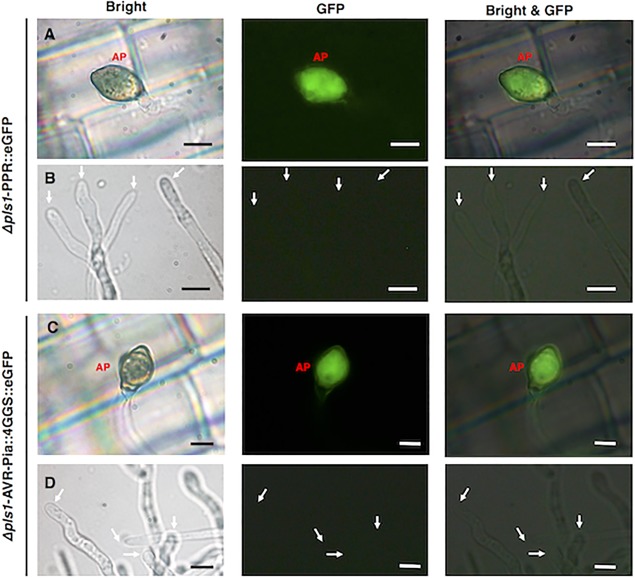
AVR‐Pia expression in a penetration‐deficient Magnaporthe oryzae mutant. Rice sheath assays of Δpls1‐PPR::eGFP (A) and Δpls1‐PPR::AVR‐Pia::4GGS::eGFP (C) were conducted at 24 h post‐inoculation (hpi), and fluorescence observation of liquid culture of Δpls1‐PPR::eGFP (B) and Δpls1‐PPR::AVR‐Pia::4GGS::eGFP (D) was performed at 24 hpi. AP, appressorium; GFP, green fluorescence protein; white arrow, tip of hypha. Scale bars, 5 μm.
Construction of GFP‐fused AVR‐Pia expression vector
To check the function of AVR‐Pia::eGFP, we produced PPR::AVR‐Pia::eGFP by fusing eGFP to the C‐terminus of AVR‐Pia, and conducted a virulence assay using Ina168m95‐1‐PPR::AVR‐Pia::eGFP. We found that the introduction of AVR‐Pia::eGFP could not complement the avirulence of Ina168m95‐1 in inoculated Pia+ rice leaf sheaths (Aichiasahi), suggesting that AVR‐Pia::eGFP cannot function as an AVR‐Pia effector.
We hypothesized that the fusion protein failed to function, as eGFP is a relatively bulky protein (239 amino acids) compared with AVR‐Pia (85 amino acids), and therefore inserted a spacer linker between AVR‐Pia and eGFP. Once the glycine–glycine–serine (GGS) spacer was inserted, virulence assays of Ina168m95‐1 transformed with vectors containing two, four or six GGS repeats demonstrated that 2GGS and 4GGS, but not 6GGS, could complement the function of AVR‐Pia as an AVR effector (Fig. 5).
Figure 5.
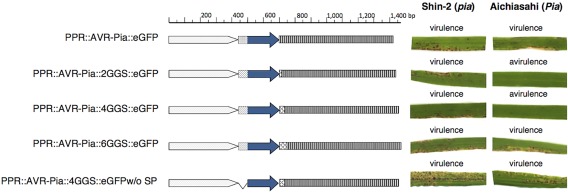
Illustration of five enhanced green fluorescence protein (eGFP)‐fused AVR‐Pia constructs and the virulence of Magnaporthe oryzae transformed with each. The spacer sequence consisted of two, four or six glycine–glycine–serine repeats (2GGS, 4GGS and 6GGS, respectively) and was inserted between AVR‐Pia and eGFP. White arrows, native promoter region; spotted boxes, secreted signal; blue arrows, AVR‐Pia; latticed boxes, GGS spacer sequence; striped boxes, eGFP.
AVR‐Pia translation in appressoria
We introduced pBlastr‐PPR::AVR‐Pia::4GGS::eGFP into the Δpls1 mutant and observed that the fusion protein was localized in the transformant's appressoria (Fig. 4C), indicating that the AVR‐Pia protein is produced independent of fungal penetration. Figure 4D showed that AVR‐Pia translation was not induced in liquid culture of Δpls1‐PPR::AVR‐Pia::4GGS::eGFP.
Localization of AVR‐Pia after fungal penetration
Examination of Ina168m95‐1‐PPR::AVR‐Pia::4GGS::eGFP‐inoculated leaf sheaths revealed that eGFP‐labelled AVR‐Pia was localized in the BIC‐like structures of the first IHs at 27 and 30 hpi (Fig. 6A). Furthermore, the fluorescence signal was detected in the tip of the third IH in the third invaded rice cells at 60 and 61 hpi and in the BIC‐like structures at 63 and 64 hpi (Fig. 6B), indicating that AVR‐Pia was localized in the BIC‐like structures of IHs in both the first invaded rice cells and afterwards. Moreover, our examination of fluorescence in rice leaf sheaths inoculated with Ina168m95‐1‐PPR::AVR‐Pia::4GGS::eGFP‐w/oSP, which did not contain the signal peptide sequence, revealed that the protein was localized in IHs (Fig. S1, see Supporting Information), indicating that the AVR‐Pia signal peptide sequence (57 bp) was necessary for AVR‐Pia secretion.
Figure 6.

Preferential localization of enhanced green fluorescence protein (eGFP)‐labelled AVR‐Pia in Magnaporthe oryzae biotrophic interfacial complex (BIC) structures. White arrows indicate eGFP fluorescence. Scale bars, 10 μm. (A) The first invasive hyphae in the first invaded sheath epidermal cells at 27 and 30 h post‐inoculation (hpi) are indicated by eGFP‐labelled AVR‐Pia in the BIC‐like structures. (B) The third invasive hyphae in the third invaded sheath epidermal cells at 60 and 63 hpi are indicated by eGFP‐labelled AVR‐Pia in the BIC‐like structures. The boxes in the top right‐hand corners of the images are magnifications of the smaller corresponding boxes.
Localization of AVR‐Pia in BIC before being delivered into plant cells
Examination of leaf sheaths inoculated with pCSN‐DEST‐PPR::AVR‐Pia::4GGS::eGFP‐ and pPWL2::PWL2::mRFP‐transformed Ina168m95‐1 revealed that the fluorescently labelled AVR‐Pia and PWL2, which were under the control of their respective native promoters, were co‐localized; the mRFP‐labelled PWL2 clearly accumulated in the BICs of IHs, as did the eGFP‐labelled AVR‐Pia (Fig. 7). Therefore, AVR‐Pia appears to accumulate in the BIC structures prior to its translocation into rice host cells. A clear fluorescence signal was detected in rice cell walls and was confirmed not to be caused by rice autofluorescence, but by AVR‐Pia translocated to cell walls, from the result of inoculation assay with mCherry‐labelled AVR‐Pia (Fig. S2, see Supporting Information).
Figure 7.
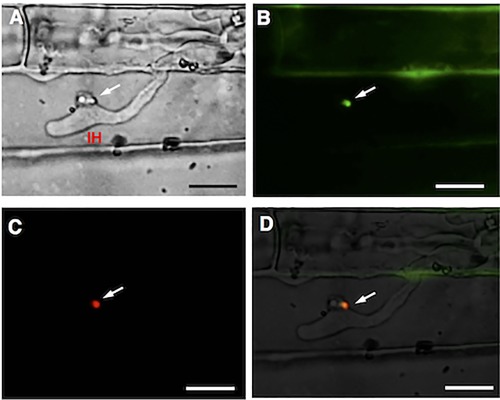
Co‐localization of AVR‐Pia and PWL2 in the biotrophic interfacial complex (BIC) structures of Magnaporthe oryzae. Microscopic observation of rice sheath cells infected with Ina168m95‐1‐pCSN43‐DEST‐PPR::AVR‐Pia::4GGS::eGFP‐pPWL2::PWL2::mRFP‐transformed M. oryzae was performed at 57 h post‐inoculation (hpi). IH, invasive hypha; white arrows, BICs. Scale bars, 10 μm. (A) Bright‐field image. (B) Green enhanced green fluorescence protein (eGFP)‐labelled AVR‐Pia fluorescence in BICs. (C) Red monomeric red fluorescent protein (mRFP)‐labelled PWL2 fluorescence in BIC. (D) Merged bright‐field, eGFP::AVR‐Pia (green) and mRFP::PWL2 (red) images.
Translocation of AVR‐Pia protein into rice cytoplasm
In order to verify that AVR‐Pia was translocated into rice cytoplasm, we examined the cytoplasmic fluorescence of sucrose‐plasmolysed rice cells that had been inoculated with a transformant expressing eGFP‐fused AVR‐Pia and mRFP‐fused PWL2. Both the red‐fluorescent PWL2 and green‐fluorescent AVR‐Pia were detected in the plasmolysed cytoplasm of infected cells (Fig. 8), indicating that AVR‐Pia was secreted into rice cytoplasm after its accumulation in the BIC structures.
Figure 8.
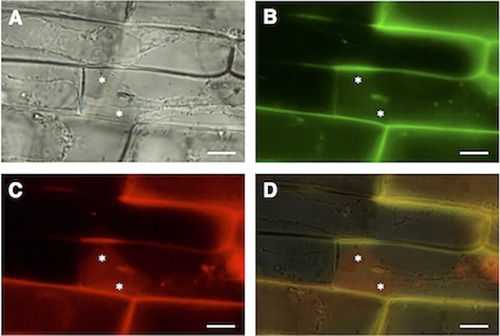
Translocation of AVR‐Pia and PWL2 in the plasmolysed cytoplasm of Magnaporthe oryzae‐infected rice sheath cells at 61 h post‐inoculation (hpi). Asterisks, plasmolysed and concentrated rice cytoplasm. Scale bars, 10 μm. (A) Bright‐field image. (B) Green enhanced green fluorescence protein (eGFP)‐labelled AVR‐Pia fluorescence. (C) Red monomeric red fluorescent protein (mRFP)‐labelled PWL2 fluorescence. (D) Merged bright‐field, eGFP::AVR‐Pia (green) and mRFP::PWL2 image (red) images.
Discussion
AVR‐Pia expression is independent of penetration
One of the objectives of this study was to determine the timing of AVR‐Pia expression. RT‐PCR analysis indicated that AVR‐Pia was only expressed during fungal infection, and qRT‐PCR indicated that AVR‐Pia expression decreased at 30 hpi in incompatible rice cultivars. This suggests that the AVR‐Pia effector is recognized by the Pia resistance protein, leading to the induction of a host defence response. However, it remains unclear whether the decreased AVR‐Pia expression is the result of fungal cell death or whether the recognition of AVR‐Pia and Pia induces the suppression of AVR‐Pia expression. Furthermore, our visualization of Ina168m95‐1‐PPR::eGFP‐inoculated leaf sheaths and onion epidermis suggested that AVR‐Pia transcription is induced during appressorial differentiation.
Previously, Clergeot et al. (2001) have reported that the turgor, melanization and glycogen accumulation of appressoria in Δpls1 mutants are similar to those of mature wild‐type appressoria before penetration, but the mutant appressoria fail to develop penetration pegs. We clearly demonstrated that fungal penetration is not required for AVR‐Pia expression, as it is induced before penetration and also occurs in the appressoria of Δpls1. Previously, AVR‐Pia expression has been reported to occur in well‐melanized appressoria of M. oryzae inoculated onto glass slides (Sornkom et al., 2015), when the ability of fungal penetration was blocked by impenetrable artificial membranes. Therefore, AVR‐Pia expression is clearly independent of fungal penetration.
Previously, the induction of ACE1 expression has been reported to occur during appressorial‐mediated penetration, independent of fungal penetration (Fudal et al., 2007). Although ACE1 differs from AVR genes, in that it encodes a cytoplasmic enzyme for secondary metabolite biosynthesis, the expression pattern of ACE1 is similar to that of AVR‐Pia. Therefore, it is possible that ACE1 and AVR‐Pia share the same regulatory expression system. In addition, Bok and Keller (2004) reported that LaeA is a global regulator of secondary metabolite production in fungi. Therefore, we speculate that LaeA may also regulate the expression of AVR‐Pia. However, the study of the expression regulatory system remains to be clarified to understand the factors involved in AVR‐Pia expression.
AVR‐Pia is secreted into rice cytoplasm via BIC structures
In this study, we have demonstrated that AVR‐Pia is co‐localized with PWL2, suggesting that AVR‐Pia preferentially localizes to BIC structures before secretion. In addition, we have also confirmed that the AVR‐Pia signal peptide is important for BIC localization, and have demonstrated that AVR‐Pia is translocated into rice cytoplasm, which is similar to most other BIC‐localized effectors.
In addition, it has been reported that AVR‐Pia binds directly and physically to RGA5, which is one of the two nucleotide‐binding/leucine‐rich repeat coding genes (RGA4 and RGA5) in Pia (Okuyama et al., 2011). The recognition of AVR‐Pia and RGA5 results in the induction of cell death and HR, activated by RGA4. It has also been revealed that RGA4 and RGA5 are localized in the cytoplasm (Césari et al., 2014). Consequently, it is suggested that the recognition of Pia and AVR‐Pia occurs in the plant cytoplasm.
AvrPiz‐t has been identified as a cytoplasmic effector that functions as a suppressor of PAMP‐triggered immunity and thus facilitates pathogenesis (Park et al., 2012). Moreover, it has been reported that the protein structure of AvrPiz‐t is similar to that of AVR‐Pia (Ose et al., 2015). Nevertheless, the effector activity of AVR‐Pia has not yet been clarified, but there is a strong possibility that AVR‐Pia is a cytoplasmic effector, similar to AvrPiz‐t.
A detectable amount of AVR‐Pia was found in rice cell walls. The mechanism for this has yet to be elucidated. Recently, Satoh et al. (2014) have reported that recombinant AVR‐Pia protein (rAVR‐Pia) infiltration can induce an HR‐like reaction in Pia rice leaf, without the inoculation of AVR‐Pia blast fungus. They indicated the possibility of rAVR‐Pia protein movement through the apoplast for HR induction in a site distal to infiltration. The cell wall localization of AVR‐Pia observed in this study might represent evidence for AVR‐Pia movement in the apoplast, but additional intensive study is required to confirm this possibility.
Conclusions
This study is the first to report that AVR‐Pia expression is induced during early fungal infection (before penetration) and confirms that AVR‐Pia is delivered to rice cytoplasm via BIC, where it is recognized by Pia and induces the host's defence response. However, the factors that control AVR‐Pia expression (transcription factors, cis‐elements, etc.) and AVR‐Pia activity (effector targets) remain to be identified. The AVR‐Pia expression reported in this study suggests plant–pathogen communication during early infection. The study of the mechanism of AVR‐Pia expression in future work will provide clues to an understanding of the nature of AVR gene expression.
Experimental Procedures
Fungal and bacterial strains
Japanese field isolate Ina168 (AVR‐Pia+) and its spontaneous mutant Ina168m95‐1 (avr‐pia−; Miki et al., 2009), and the mutant strain Ina86‐137Δlig4, which lacked non‐homologous recombination, were used as recipient strains for transformation. Fungal isolates were stored at −20 °C as dried mycelium on pieces of filter paper, and were cultured on oatmeal agar before use. Escherichia coli TOP10 (Invitrogen, Carlsbad, CA, USA) was used for recombinant DNA experiments and plasmid construction.
DNA extraction and manipulation
Plasmid DNA was extracted using NucleoSpin Plasmid EasyPure or NucleoBond Xtra Midi Kits (MACHEREY‐NAGEL GmbH & Co. KG, Düren, Germany), and fungal DNA was isolated from M. oryzae using the protocol described previously (Sone et al., 1997). DNA digestion was performed according to the manufacturer's instructions for restriction enzymes, and PCR amplifications were performed using GoTaq Green Master Mix (Promega, Madison, WI, USA) or KOD FX Neo (TOYOBO, Osaka, Japan), according to the manufacturers’ instructions, and gene‐specific primers (Table 1). Capillary blotting for Southern hybridization was performed using a Hybond‐N+ nylon transfer membrane (GE Healthcare, Amersham, Buckinghamshire, UK) following the manufacturer's instructions, and the labelling and detection of DNA probes were performed using an AlkPhos Direct Labelling system and CDP‐Star detection reagent (GE Healthcare).
Table 1.
Synthetic oligonucleotides used in this study.
| Primer name | Sequence (5′–3′) | Target gene | Application |
|---|---|---|---|
| ORF‐F (21509)2 | TAAACCTAGTAAGGCTCGGCAGC | AVR‐Pia ORF | RT‐PCR for AVR‐Pia transcript |
| ORF‐R (21771 + 1)5 | ATGCATTTTTCGACAATTTTCATCCC | AVR‐Pia ORF | RT‐PCR for AVR‐Pia transcript |
| mgactin‐F | GACGACATGGAGAAGATCTGGC | Magnaporthe oryzae actin ORF | RT‐PCR for actin transcript |
| mgactin‐R | CGTCGTACTCCTGCTTCGAGAT | M. oryzae actin ORF | RT‐PCR for actin transcript |
| AVR‐Pia‐ANYM1 | ACGGCCACCTTCCCGC | AVR‐Pia ORF | Taqman probe for AVR‐Pia qRT‐PCR |
| AVR‐Pia‐ANY F | CTGCGCCAGCTAGATTTTGC | AVR‐Pia ORF | qRT‐PCR for AVR‐Pia transcript |
| AVR‐Pia‐ANY R | GCTGTAGTGCCGATTCTAACGTA | AVR‐Pia ORF | qRT‐PCR for AVR‐Pia transcript |
| Actin probe | CTACGAGCTTCCCGACGG | M. oryzae actin ORF | Taqman MGB probe for actin qRT‐PCR |
| actqF | CTGCCCAGAGCTCCAGCTT | M. oryzae actin ORF | qRT‐PCR for actin transcript |
| actqR | CGTTGCCGATGGTGATAACC | M. oryzae actin ORF | qRT‐PCR for actin transcript |
| cacc22212R | CACCCCAGTCGCTTGAGATTCTTTG | PPR |
PPR::AVR‐Pia fragment amplification PPR::eGFP fragment amplification |
| 21517F | GTAAGGCTCGGCAGCAAGCC | ORF end of AVR‐Pia |
PPR::AVR‐Pia fragment amplification Inverse PCR for GGS spacer insertion |
| eGFP‐F | ATGGTGAGCAAGGGCGAGGA | ORF start of eGFP |
eGFP amplification Inverse PCR for GGS spacer insertion |
| eGFP‐R | TTACTTGTACAGCTCGTCCATGCCG | ORF end of eGFP |
eGFP amplification PPR::eGFP PPR::AVR‐Pia::4GGS::eGFP |
| 2GGS‐F | GGCGGCAGCGGCGGCAGC | GGS spacer | 2GGS spacer construction (sense strand) |
| 2GGS‐R | GCTGCCGCCGCTGCCGCC | GGS spacer | 2GGS spacer construction (antisense strand) |
| 4GGS‐F | GGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGC | GGS spacer | 4GGS spacer construction (sense strand) |
| 4GGS‐R | GCTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCC | GGS spacer | 4GGS spacer construction (antisense strand) |
| 6GGS‐F | GGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGC | GGS spacer | 6GGS spacer construction (sense strand) |
| 6GGS‐R | GCTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCC | GGS spacer | 6GGS spacer construction (antisense strand) |
| 21714R w/o SP +ATG | atgCCGCCAGCTAGATTTTGCGTC | AVR‐Pia | Inverse PCR for signal peptide deletion |
| pAVRR | TTTGAGAATTTTCGTGTATGGGGC | AVR‐Pia promoter | Inverse PCR for signal peptide deletion |
| pls1homo‐F | CATGACTGAAACCCGCGTGC | 5′ flanking region of Pls1 | Pls1 fragment amplification |
| pls1homo‐R | CATGTTGGATGTTGACCAAGGGG | 3′ flanking region of Pls1 | Pls1 fragment amplification |
| pls1inv‐F | CACCGTGCCGCTTGTATTAGGCAG | Internal region of Pls1 | Pls1 disruption vector construction |
| pls1inv‐R | TGAAGGAGAGGATTCCATGGCTC | Internal region of Pls1 | Pls1 disruption vector construction |
| PWL2P‐F | GGGTACCGCGTCAGTGAACAAACC | PWL2 promoter | PWL2::mRFP fragment amplification |
| Term‐R | CTCTAGATCATCATCATGCAACATGCA | Terminator of PWL2 | PWL2::mRFP fragment amplification |
| 591‐mCherry‐F | ATGGTGAGCAAGGGCGAGG | ORF start of mCherry | mCherry::NLS‐pBAS4::BAS4 amplification |
| BAS4‐R | AGCAGGGGGGATAGACGAGC | ORF end of BAS4 | mCherry::NLS‐pBAS4::BAS4 amplification |
| mCherry‐F‐69.4 | ATGGTGAGCAAGGGCGAGGAGGATAA | ORF start of mCherry | Inverse PCR for AVR‐Pia::mCherry::NLS |
| AVR‐Pia‐R‐68 | GTAAGGCTCGGCAGCAAGCCAATC | ORF end of AVR‐Pia | Inverse PCR for AVR‐Pia::mCherry::NLS |
eGFP, enhanced green fluorescent protein; GGS, glycine–glycine–serine; mRFP, monomeric red fluorescent protein; ORF, open reading frame; qRT‐PCR, quantitative reverse transcription‐polymerase chain reaction.
Plasmid construction
To construct the pCSN43‐DEST‐PPR::AVR‐Pia::eGFP plasmid, AVR‐Pia, with its putative promoter, and eGFP were amplified from pCSN43‐DEST‐Vm (Miki et al., 2009) and pPCG664 (provided by Dr T. Kamakura, Tokyo University of Science, Tokyo, Japan), respectively, and the two fragments were ligated into the pENTR/D‐TOPO vector to produce pENTR‐AVR‐Pia::eGFP, which was then transferred to pCSN43‐DEST (S. Miki et al., unpublished data) via an LR reaction using Gateway LR Clonase II Enzyme mix (Invitrogen, Life Technologies Japan Ltd., Tokyo, Japan). This vector was subsequently used as a template for inverse PCR, to construct pCSN43‐DEST‐PPR::eGFP by self‐ligation and for the insertion of GGS spacers.
To insert the GGS spacers, the plasmid was opened at the end of the AVR‐Pia sequence using inverse PCR, phosphorylated and ligated using synthesized GGS spacers, thus producing pCSN43‐DEST‐PPR::AVR‐Pia::eGFP with two, four or six repeats of the GGS spacer. In addition, pCSN43‐DEST‐PPR::AVR‐Pia::4GGS::eGFP‐w/oSP was constructed using inverse PCR and self‐ligation, and both PPR::eGFP and PPR::AVR‐Pia::4GGS::eGFP were introduced into the blasticidin S‐resistant pBLASTR vector to produce the pBLASTR‐PPR::eGFP and pBLASTR‐PPR::AVR‐Pia::4GGS::eGFP plasmids, respectively.
To construct Δpls1 mutants, homologous recombination was used to disrupt the Pls1 gene, the sequence of which was obtained from the GenBank and Broad Institute databases (accession nos. AX058235 and MGG_12594). In order to facilitate homologous recombination, Ina86‐137Δlig4 was used to knock out Pls1, and a Pls1 disruption vector was constructed using pBARST, as reported previously (Abe et al., 2006). Briefly, genomic DNA fragments containing Pls1 were amplified and digested with EcoRV (TAKARA Bio Inc., Shiga, Japan) and, following self‐ligation, inverse PCR was performed using Pls1‐inverse primers (Table 1). The resulting fragment was then inserted into the pENTR‐D‐TOPO vector (Life Technologies Japan Ltd.), and the resulting pBARST‐pls1homo::bia construct was used to disrupt Pls1.
In order to investigate AVR‐Pia localization, a plasmid was constructed that contained fused eGFP and AVR‐Pia and fused mRFP and PWL2. A linear pCSN43‐DEST‐PPR::AVR‐Pia::4GGS::eGFP fragment was prepared using DNA digestion with SmaI, and the pPWL2::PWL2::mRFP fragment was PCR‐amplified using specific primers and the pBV377 plasmid (provided by Dr Barbara Valent, Kansas State University, Manhattan, KS, USA). Then, pCSN43‐DEST‐PPR::AVR‐Pia::eGFP and pPWL2::PWL2::mRFP were ligated.
Fungal transformation
All fungal transformations were performed using the protoplast‐polyethylene glycol (PEG) method, as described previously (Abe et al., 2006; Miki et al., 2009). For hygromycin selection, 500 μg/mL of hygromycin B were used to select transformants and, for bialaphos or blasticidin selection, 5 μg/mL of bialaphos or blasticidin were employed. Single conidia were isolated from each transformant, as described previously (Sone et al., 1997), and both PCR and Southern hybridization were performed to check and confirm the transformants.
RNA extraction, RT‐PCR and qRT‐PCR
A piece of M. oryzae Ina168 mycelium was inoculated into 2YEG (yeast extract, 2 g/L; glucose, 10 g/L) and cultured with shaking for 72 h, and the same strain was also spray inoculated onto the rice cultivars Shin‐2 (pia), Nipponbare‐pia (pia, JP222429), Aichiasahi (Pia) and Nipponbare‐Pia (Pia, JP222431), according to Miki et al. (2009). Inoculated leaves were sampled at various time points (i.e. 0, 12, 18, 24, 30, 36, 42, 48 and 60 hpi) and stored at −80 °C, and RNA was extracted from the samples using RNAiso Plus reagent (TAKARA Bio Inc.), following the manufacturer's instructions, and kept at −80 °C. The RT‐PCR experiments were performed using the Superscript III One‐step RT‐PCR System with Platinum Taq DNA Polymerase (Invitrogen) and gene‐specific primers (Table 1). For qRT‐PCR of AVR‐Pia and actin (internal control), primers and probes (Table 1) were designed using Primer Express software (Applied Biosystems, Foster City, CA, USA). The expression of AVR‐Pia was quantified using the TaqMan expression detection kit (Applied Biosystems) and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems), with plasmid DNA harbouring AVR‐Pia or actin as standard DNA, following the manufacturer's instructions. Expression of the AVR‐Pia gene was calculated using the following formula: eAVR‐Pia = cAVR‐Pia/cactin, where cAVR‐Pia and cactin are the copy numbers of AVR‐Pia cDNA and actin cDNA, respectively.
In planta leaf sheath assay and fluorescence microscopy
Seeds of Shin‐2 rice, which lacks the Pia resistance gene, were sown in plastic seedling pots containing soil with fertilizer (Honens seedling soil; Honen Agri, Co., Niigata, Japan) and grown in a glasshouse for 60 days, or until the seventh leaf stage, when intact leaf sheaths were inoculated by injecting a conidial suspension, as described previously (Sornkom et al., 2015). The inoculated leaf sheaths were prepared for microscopy by excising thin layers of sheath epidermal cells that had been in contact with the conidial suspension, and the excised leaf sheath pieces were placed on slides and visualized using a BX‐50 fluorescence microscope (Olympus, Tokyo, Japan) equipped with a U‐MNIBA3 filter set.
Onion epidermis assay
Onion epidermis was prepared as described previously (Sornkom et al., 2015), inoculated with diluted conidial suspensions from each transformant (104−105 cells/mL) by dropping about 10 drops (1 μL/drop) of conidial suspension over the membranes and incubated at 27 °C for 24 h. Fluorescence visualization was then performed as in the rice sheath assay.
Spray inoculation
Virulence assays were conducted using the rice cultivars Shin‐2 (pia –) and Aichiasahi (Pia+) with spray inoculation, as described previously (Miki et al., 2009).
Plasmolysis assay for fluorescence observation in rice cytoplasm
Plasmolysis assay was performed as described previously (Khang et al., 2010) with some modifications to concentrate the cytoplasm and separate it from the cell wall. Plasmolysis was performed slowly by sequential incubation of inoculated rice tissue in 0.25, 0.5, 0.75 and 1.0 m sucrose to avoid causing damage to the host cells. The incubation time of each concentration of sucrose was 2 min. Microscopic observation was performed immediately after plasmolysis.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Localization of enhanced green fluorescence protein (eGFP)‐labelled AVR‐Pia fluorescence in the invasive hyphae of Ina168m95‐1‐PPR::AVR‐Pia::4GGS::eGFP‐w/oSP at 52 h post‐inoculation (hpi). Scale bars, 10 μm.
Fig. S2 Autofluorescence control of green fluorescence protein (GFP)‐labelled AVR‐Pia. Transformant expressing mCherry‐NLS‐labelled AVR‐Pia and GFP‐labelled BAS4 was inoculated on rice sheath at 33 h post‐inoculation (hpi). This control strain does not express GFP‐labelled AVR‐Pia, which was used for study in our research. No green fluorescence was found in the cell wall of rice cells. White arrows indicate the biotrophic interfacial complex (BIC). Bars, 10 μm.
Acknowledgements
This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio‐oriented Industry (BRAIN) and Grants‐in‐Aid for Scientific Research (C‐20580043 and B‐23380024) by the Japan Society for the Promotion of Science (JSPS).
The authors have no conflicts of interest to declare.
References
- Abe, A. , Elegado, E. and Sone, T. (2006) Construction of pDESTR, a GATEWAY vector for gene disruption in filamentous fungi. Curr. Microbiol. 52, 210–215. [DOI] [PubMed] [Google Scholar]
- Ballini, E. , Morel, J.B. , Droc, G. , Price, A. , Courtois, B. , Notteghem, J.L. and Tharreau, D. (2008) A genome‐wide meta‐analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant–Microbe Interact. 21, 859–868. [DOI] [PubMed] [Google Scholar]
- Bok, J.W. and Keller, N.P. (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell, 3, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari, S. , Kanzaki, H. , Fujiwara, T. , Bernoux, M. , Chalvon, V. , Kawano, Y. , Shimamoto, K. , Dodds, P. , Terauchi, R. and Kroj, T. (2014) The NB‐LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot, P.H. , Gourgues, M. , Cots, J. , Laurans, F. , Latorse, M.P. , Pepin, R. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2001) PLS1, a gene encoding a tetraspanin‐like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea . Proc. Natl. Acad. Sci. USA, 98, 6963–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dioh, W. , Tharreau, D. , Notteghem, J.L. , Orbach, M. and Lebrun, M.H. (2000) Mapping of avirulence genes in the rice blast fungus, Magnaporthe grisea, with RFLP and RAPD markers. Mol. Plant–Microbe Interact. 13, 217–227. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fudal, I. , Collemare, J. , Böhnert, H.U. , Melayah, D. and Lebrun, M.H. (2007) Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium‐mediated penetration. Eukaryot. Cell, 6, 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo, M.C. , Dagdas, Y.F. , Gupta, Y.K. , Mentlak, T.A. , Yi, M. , Martinez‐Rocha, A.L. , Saitoh, H. , Terauchi, R. , Talbot, N.J. and Valent, B. (2013) Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae . Nat. Commun. 4, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, J.S. , Jung, K.H. , Kim, H.B. , Suh, J.P. and Khush, G.S. (2011) Genetic and molecular insights into the enhancement of rice yield potential. J. Plant Biol. 54, 1–9. [Google Scholar]
- Kang, S. , Sweigard, J.A. and Valent, B. (1995) The PWL host specificity gene family in the blast fungus Magnaporthe grisea . Mol. Plant–Microbe Interact. 8, 939–948. [DOI] [PubMed] [Google Scholar]
- Khang, C.H. , Berruyer, R. , Giraldo, M.C. , Kankanala, P. , Park, S.Y. , Czymmek, K. , Kang, S. and Valent, B. (2010) Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell‐to‐cell movement. Plant Cell, 22, 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Wang, B. , Wu, J. , Lu, G. , Hu, Y. , Zhang, X. , Zhang, Z. , Zhao, Q. , Feng, Q. , Zhang, H. , Wang, Z. , Wang, G. , Han, B. , Wang, Z. and Zhou, B. (2009) The Magnaporthe oryzae avirulence gene AvrPiz‐t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz‐t. Mol. Plant–Microbe Interact. 22, 411–420. [DOI] [PubMed] [Google Scholar]
- Maciel, J.L.N. (2011) Magnaporthe oryzae, the blast pathogen: current status and options for its control. Plant Sci. Rev. 6, 264. [Google Scholar]
- Miki, S. , Matsui, K. , Kito, H. , Otsuka, K. , Ashizawa, T. , Yasuda, N. , Fukiya, S. , Junko, S. , Hirayae, K. , Fujita, Y. , Nakajima, T. , Tomita, F. and Sone, T. (2009) Molecular cloning and characterization of the AVR‐Pia locus from a Japanese field isolate of Magnaporthe oryzae . Mol. Plant Pathol. 10, 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama, Y. , Kanzaki, H. , Abe, A. , Yoshida, K. , Tamiru, M. , Saitoh, H. , Fujibe, T. , Matsumura, H. , Shenton, M. , Galam, D.C. , Undan, J. , Ito, A. , Terauchi, R. and Sone, T. (2011) A multifaceted genomics approach allows the isolation of the rice Pia‐blast resistance gene consisting of two adjacent NBS‐LRR protein genes. Plant J. 66, 467–479. [DOI] [PubMed] [Google Scholar]
- Oliveira‐Garcia, E. and Valent, B. (2015) How eukaryotic filamentous pathogens evade plant recognition. Curr. Opin. Microbiol. 26, 92–101. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J. , Farrall, L. , Sweigard, J.A. , Chumley, F.G. and Valent, B. (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ose, T. , Oikawa, A. , Nakamura, Y. , Maenaka, K. , Higuchi, Y. , Satoh, Y. , Fujiwara, S. , Demura, M. , Sone, T. and Kamiya, M. (2015) Solution structure of an avirulence protein, AVR‐Pia, from Magnaporthe oryzae . J. Biomol. NMR, 63, 229–235. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. , Valent, B. and Wang, G.L. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, Y. , Miki, S. , Ose, T. , Oikawa, A. , Maenaka, K. , Terauchi, R. , Asano, K. and Sone, T. (2014) Heterologous production, purification, and immunodetection of Magnaporthe oryzae avirulence protein AVR‐Pia. Biosci. Biotechnol. Biochem. 78, 680–686. [DOI] [PubMed] [Google Scholar]
- Sone, T. , Abe, T. , Yoshida, N. , Suto, M. and Tomita, F. (1997) DNA fingerprinting and electrophoretic karyotyping of Japanese isolates of rice blast fungus. Jpn. J. Phytopathol. 63, 155–163. [Google Scholar]
- Sornkom, W. , Asano, K. and Sone, T. (2015) Use of native promoter‐eGFP as a gene reporter on onion epidermis to analyze gene expression of AVR‐Pia, an avirulence effector of rice blast pathogen. Eng. J. 19, 85–94. [Google Scholar]
- Sweigard, J.A. , Carroll, A.M. , Kang, S. , Farrall, L. , Chumley, F.G. and Valent, B. (1995) Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell, 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J. (1995) Having a blast: exploring the pathogenicity of Magnaporthe grisea . Trends Microbiol. 3, 9–16. [DOI] [PubMed] [Google Scholar]
- Wani, A.A. , Singh, P. , Shah, M.A. , Schweiggert‐Weisz, U. , Gul, K. and Wani, I.A. (2012) Rice starch diversity: effects on structural, morphological, thermal, and physicochemical properties—a review Compr. Rev. Food Sci. Food Saf. 11, 417–436. [Google Scholar]
- Yoshida, K. , Saitoh, H. , Fujisawa, S. , Kanzaki, H. , Matsumura, H. , Yoshida, K. , Tosa, Y. , Chuma, I. , Takano, Y. , Win, J. , Kamoun, S. and Terauchi, R. (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae . Plant Cell, 21, 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. , Luo, J. , Rossman, A.Y. , Aoki, T. , Chuma, I. , Crous, P.W. , Dean, R. , Vries, R.P. , Donofrio, N. , Hyde, K.D. , Lebrun, M.H. , Talbot, N.J. , Tharreau, D. , Tosa, Y. , Valent, B. , Wang, Z. and Xu, J.R. (2016) Generic names in Magnaporthales. IMA Fungus, 7, 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. and Xu, J.R. (2014) Effectors and effector delivery in Magnaporthe oryzae . PLoS Pathog. 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Localization of enhanced green fluorescence protein (eGFP)‐labelled AVR‐Pia fluorescence in the invasive hyphae of Ina168m95‐1‐PPR::AVR‐Pia::4GGS::eGFP‐w/oSP at 52 h post‐inoculation (hpi). Scale bars, 10 μm.
Fig. S2 Autofluorescence control of green fluorescence protein (GFP)‐labelled AVR‐Pia. Transformant expressing mCherry‐NLS‐labelled AVR‐Pia and GFP‐labelled BAS4 was inoculated on rice sheath at 33 h post‐inoculation (hpi). This control strain does not express GFP‐labelled AVR‐Pia, which was used for study in our research. No green fluorescence was found in the cell wall of rice cells. White arrows indicate the biotrophic interfacial complex (BIC). Bars, 10 μm.


