Summary
Xanthomonas campestris pv. campestris causes black rot, a serious disease of crucifers. Xanthomonads encode a siderophore biosynthesis and uptake gene cluster xss (Xanthomonas siderophore synthesis) involved in the production of a vibrioferrin‐type siderophore. However, little is known about the role of the siderophore in the iron uptake and virulence of X. campestris pv. campestris. In this study, we show that X. campestris pv. campestris produces an α‐hydroxycarboxylate‐type siderophore (named xanthoferrin), which is required for growth under low‐iron conditions and for optimum virulence. A mutation in the siderophore synthesis xssA gene causes deficiency in siderophore production and growth under low‐iron conditions. In contrast, the siderophore utilization ΔxsuA mutant is able to produce siderophore, but exhibits a defect in the utilization of the siderophore–iron complex. Our radiolabelled iron uptake studies confirm that the ΔxssA and ΔxsuA mutants exhibit defects in ferric iron (Fe3+) uptake. The ΔxssA mutant is able to utilize and transport the exogenous xanthoferrin–Fe3+ complex; in contrast, the siderophore utilization or uptake mutant ΔxsuA exhibits defects in siderophore uptake. Expression analysis of the xss operon using a chromosomal gusA fusion indicates that the xss operon is expressed during in planta growth and under low‐iron conditions. Furthermore, exogenous iron supplementation in cabbage leaves rescues the in planta growth deficiency of ΔxssA and ΔxsuA mutants. Our study reveals that the siderophore xanthoferrin is an important virulence factor of X. campestris pv. campestris which promotes in planta growth by the sequestration of Fe3+.
Keywords: cabbage, ferric iron uptake, siderophore, vibrioferrin, virulence, xanthoferrin
Introduction
Iron uptake and metabolism are crucial for the growth and survival of bacterial pathogens inside their hosts (Cassat and Skaar, 2013; Expert et al., 2012; Expert et al., 1996; Guerinot, 1994). Iron acquisition by both animal‐ and plant‐pathogenic bacteria inside their hosts acts as a crucial virulence‐determining factor in many infection processes in which iron is sequestered by the host proteins (Expert et al., 1996; Schaible and Kaufmann, 2004; Weinberg, 2009). Under low‐iron conditions, bacteria produce and secrete siderophores (high‐affinity, iron‐binding compounds), which chelate ferric iron (Fe3+) from the environment and facilitate its uptake (Andrews et al., 2003; Neilands, 1981). The transport of the Fe3+–siderophore complex occurs across the outer membrane through TonB‐dependent transporters (TBDTs), which are bacterial outer membrane β‐barrel proteins with 22 antiparallel β‐strands (Noinaj et al., 2010). Fe3+–siderophore‐bound outer membrane TonB‐dependent transporters couple with the cytoplasmic membrane through the protein complex TonB–ExbB–ExbD, which provides the proton motive force‐derived energy required for the conformational change of TBDTs to release the Fe3+–siderophore complex into the periplasm (Ferguson and Deisenhofer, 2004; Larsen et al., 1999; Wiener, 2005; Wiggerich et al., 1997). An ABC transporter, which is a cytoplasmic‐ATPase‐attached transmembrane permease, delivers periplasmic Fe3+–siderophores to the cytoplasm and eventually Fe3+ reduction occurs to release ferrous iron (Fe2+) (Braun and Hantke, 2013; Faraldo‐Gómez and Sansom, 2003; Krewulak and Vogel, 2008; Noinaj et al., 2010).
Siderophores exhibit substantial structural diversity and can be classified into five major groups based on the moiety involved in iron coordination: catecholates (e.g. enterobactin from enteric bacteria, Streptomyces spp.), hydroxamates (e.g. alcaligin from Alcaligenes denitrificans), phenolates (e.g. yersiniabactin from Yersinia pestis), carboxylates (e.g. staphyloferrin from Staphylococcus spp.) and mixed type (e.g. aerobactin from Enterobacter spp.) (Miethke and Marahiel, 2007).
Earlier studies have demonstrated the requirement of siderophores in the virulence of several animal‐pathogenic bacteria, such as staphyloferrin B in Staphylococcus aureus (Dale et al., 2004), yersiniabactin in Yersinia pestis and Klebsiella pneumoniae (Bearden et al., 1997; Lawlor et al., 2007), pyoverdine in Pseudomonas aeruginosa (Meyer et al., 1996) and vibriobactin in Vibrio cholerae (Henderson and Payne, 1994).
However, there are considerable differences in the contributions of siderophores to the virulence of plant‐pathogenic bacteria. The plant‐pathogenic bacterium Erwinia chrysanthemi 3937 synthesizes two types of siderophore, chrysobactin and achromobactin, which are required for the development of soft rot disease in African violets (Saintpaulia ionantha) (Enard et al., 1988; Franza et al., 2005). The fire blight‐causing bacterium Erwinia amylovora produces the siderophore desferrioxamine, which is crucial for its effective infection on apple (Malus domestica) (Dellagi et al., 1998). In contrast, siderophore‐deficient mutants of Agrobacterium tumefaciens (Rondon et al., 2004), Ralstonia solanacearum (Bhatt and Denny, 2004), Erwinia carotovora ssp. carotovora (Bull et al., 1996) and Pseudomonas syringae pv. tomato DC3000 (Jones and Wildermuth, 2011) are virulence proficient.
Xanthomonads encode an xss (Xanthomonas siderophore synthesis) operon which is homologous to the Vibrio parahaemolyticus siderophore (pvs) locus and produces a vibrioferrin‐type siderophore under iron‐restricted conditions (Pandey and Sonti, 2010; Rai et al., 2015). In Xanthomonas oryzae pv. oryzae (Xoo; a vascular pathogen of rice), it has been shown that siderophore production is not required for virulence in rice (Pandey and Sonti, 2010). Interestingly, the Xoo xss cluster is induced under low‐iron conditions; however, it is not expressed in planta (Pandey and Sonti, 2010).
In Xanthomonas campestris pv. campestris (Xcc), the TonB‐dependent Fe3+ acquisition system (exbB, exbD1 and exbD2) has been proposed to play a role in virulence. However, the role of the siderophore in iron uptake and virulence has not been demonstrated clearly in this bacterium (Wiggerich and Pühler, 2000). In order to understand the role of the siderophore in Xcc virulence and iron uptake, we characterized the siderophore biosynthesis (xssA) and utilization (xsuA) mutants of Xcc 8004. In this study, we show that Xcc produces xanthoferrin, which is similar to the α‐hydroxycarboxylate‐type siderophore vibrioferrin. We demonstrate that the production and utilization of xanthoferrin are required for Xcc growth under low‐iron conditions and contribute to its virulence and growth inside its cabbage host.
Results
Δ xssA mutant of Xcc is deficient in xanthoferrin production
The genome of strain Xcc 8004 (Qian et al., 2005) possesses a homologue of the Xanthomonas siderophore synthesis and utilization locus similar to the Xoo xss and V. parahaemolyticus pvs locus (Fig. S1; Table S1, see Supporting Information; Pandey and Sonti, 2010; Tanabe et al., 1982). The xssA (XC_1107) gene product from the Xcc 8004 strain exhibits high homology to XssA from Xoo (identity 86%, similarity 90%) and PvsA from V. parahaemolyticus (identity 46%, similarity 64%). The siderophore utilization gene product xsuA exhibits high homology to XsuA from Xoo (identity 87%, similarity 92%) and PvuA from V. parahaemolyticus (identity 45%, similarity 61%) (Table S1).
In order to understand the role of the siderophore in Xcc virulence, we constructed in‐frame marker‐free deletions of xssA (XC_1107) and xsuA (XC_1108) genes in the wild‐type Xcc 8004 strain (see Experimental Procedures; Fig. S2A; Table S2, see Supporting Information). Expression analysis by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) indicated that the expression of downstream genes is not altered in the deletion mutants relative to the wild‐type strain (Fig. S3, see Supporting Information). This result suggests that the in‐frame deletion of xssA and xsuA genes does not have a possible polar effect on the expression of genes located downstream. The ability of these mutants to produce siderophores was assessed on peptone–sucrose agar and chromeazurol S (PSA–CAS) medium containing 75 μm 2,2′‐dipyridyl (DP; specific Fe2+ chelator). In contrast with the wild‐type Xcc 8004 strain, the ΔxssA mutant failed to produce a halo on PSA–CAS–DP medium (Fig. 1A). Complementation of the ΔxssA mutant with a plasmid harbouring the entire xss cluster from Xoo (pAP15; Pandey and Sonti, 2010; Table S2) rescued the siderophore production defect.
Figure 1.
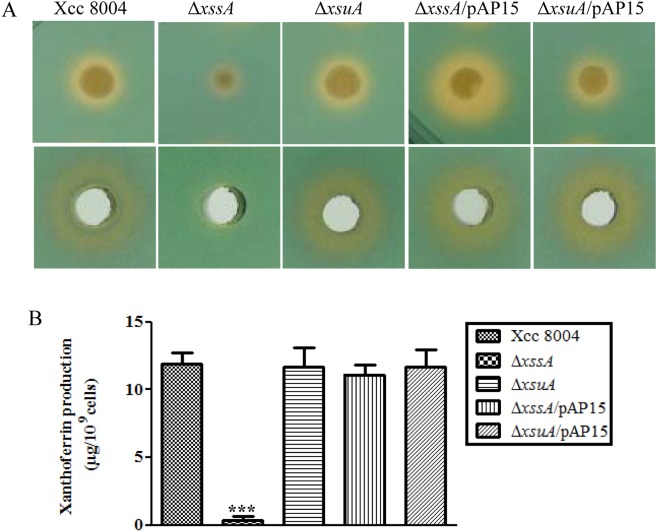
xssA gene is required for siderophore xanthoferrin biosynthesis in Xanthomonas campestris pv. campestris (Xcc). (A) Top: siderophore production by different Xcc strains on peptone–sucrose agar and chromeazurol S (PSA–CAS) plates containing 75 µm of the ferrous iron chelator 2,2′‐dipyridyl. Strains: Xcc 8004 (wild‐type strain); ΔxssA [deletion of xssA (Xanthomonas siderophore synthesis A)]; ΔxsuA [deletion of xsuA (Xanthomonas siderophore uptake A)]; ΔxssA/pAP15; and ΔxsuA/pAP15 (pAP15; wild‐type xssA and xsuA alleles). Bottom: wells on PSA–CAS plate containing siderophore isolated from the cell‐free culture supernatant of different strains of Xcc grown under low‐iron conditions [peptone–sucrose (PS) + 100 µm 2,2′‐dipyridyl] using Amberlite XAD‐16 resin column chromatography. Cell‐normalized siderophore fractions were loaded into wells prepared on a PSA–CAS indicator plate (with 75 µm 2,2′‐dipyridyl). (B) Quantification of purified xanthoferrin of different Xcc strains. Strains were grown to late exponential phase in low‐iron medium (PS + 100 μm 2,2′‐dipyridyl). Siderophores were initially purified from cell‐free culture supernatants using Amberlite XAD‐16 resin columns. The cell‐normalized active fraction of siderophore was analysed by high‐performance liquid chromatography (HPLC). Xanthoferrin was detected at 300 nm. The amount of xanthoferrin was determined by comparing the peak area (mAU × min) with the standard curve which was generated from a known concentration of pure standard vibrioferrin.
We isolated siderophores from the cell‐free culture supernatant of wild‐type Xcc 8004, ΔxssA, ΔxsuA, and mutants harbouring the complementing plasmid pAP15 (ΔxssA/pAP15 and ΔxsuA/pAP15) with Amberlite XAD‐16 resin columns (see Experimental Procedures) and checked for activity on PSA–CAS–DP plates (Fig. 1A). The siderophore was further purified from active fractions by high‐performance liquid chromatography (HPLC) and compared with the elution profile of the purified α‐hydroxycarboxylate‐type siderophore vibrioferrin (Fujita et al., 2011). The HPLC analysis indicated that the fractions corresponding to the purified active Xcc siderophore and standard synthetic vibrioferrin exhibited similar retention times (Fig. S4, see Supporting Information). We named the purified active siderophore from the Xcc 8004 strain as ‘xanthoferrin’, which is similar to vibrioferrin produced by V. parahaemolyticus and V. alginolyticus (Fujita et al., 2011; Pandey and Sonti, 2010; Rai et al., 2015; Tanabe et al., 1982). The concentration of xanthoferrin produced by different strains of Xcc was determined on the basis of the peak area and calculated from standard curves generated from known concentrations of standard vibrioferrin by HPLC analysis (Fujita et al., 2011). The wild‐type Xcc 8004 strain produced approximately 12 µg of xanthoferrin per 109 cells (Fig. 1B). In contrast, the ΔxssA mutant produced a negligible amount of xanthoferrin siderophore, which could be complemented by the plasmid harbouring the xss cluster from Xoo (Figs 1B and S4). However, the siderophore utilization ΔxsuA mutant did not exhibit any defect in xanthoferrin production (Figs 1B and S4).
Xanthoferrin‐mediated iron uptake is required for growth under low‐iron conditions
To understand the role of xanthoferrin during growth under low‐iron conditions, we compared the growth profile of wild‐type Xcc 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA/pAP15 strains in peptone–sucrose (PS) (rich medium), PS–DP (low‐iron conditions; PS medium containing 150 µm DP) and PS–DP medium supplemented with either purified xanthoferrin siderophore or Fe2+ sulfate (Fig. 2). The growth patterns of wild‐type and mutants were indistinguishable in rich medium (doubling time of approximately 2.8 h; Fig. 2A; Table 1). Under low‐iron conditions (PS–DP), the doubling time of the wild‐type Xcc 8004 strain was 3.9 h. However, the doubling times of ΔxssA and ΔxsuA mutants were significantly less (doubling time of 5.5 h) than that of the wild‐type strain (Fig. 2B; Table 1) grown under low‐iron conditions. The growth defect exhibited by the ΔxssA and ΔxsuA mutants under low‐iron conditions could be rescued by complementation with the plasmid harbouring the Xoo xss gene cluster (Fig. 2A,B; Table 1). Interestingly, supplementation of purified xanthoferrin siderophore could rescue the growth defect of ΔxssA (doubling time of approximately 4 h), but failed to restore the growth defect exhibited by ΔxsuA in low‐iron medium (doubling time 5.9 h) (Fig. 2C; Table 1). Furthermore, supplementation of purified vibrioferrin from V. parahaemolyticus also rescued the growth defect of the ΔxssA mutant, similar to xanthoferrin supplementation (Fig. S5; Table S3, see Supporting Information).
Figure 2.
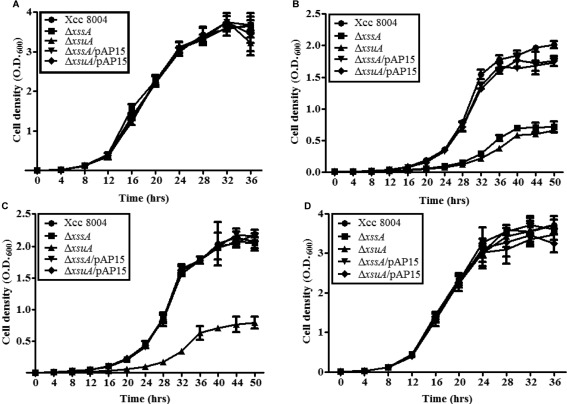
The ΔxssA and ΔxsuA mutants are growth deficient under low‐iron conditions. Growth of Xanthomonas campestris pv. campestris (Xcc) 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA/pAP15 strains under the following conditions: (A) rich peptone–sucrose (PS) medium; (B) low‐iron medium (PS + 150 µm intracellular ferrous iron chelator 2,2′‐bipyridyl); (C) xanthoferrin‐supplemented low‐iron medium (PS + 150 µm 2,2′‐dipyridyl + 20 µm xanthoferrin); (D) ferrous iron‐supplemented low‐iron medium (PS + 150 µm 2,2′‐dipyridyl + 100 µm FeSO4). Growth was monitored by determination of the optical density at 600 nm (OD600). Data are shown as mean ± standard deviation (SD) (n = 3).
Table 1.
Generation times of Xanthomonas campestris pv. campestris (Xcc) strains.
| Generation time (h)* | ||||
|---|---|---|---|---|
| PS | PS + DP† | PS + DP + FeSO4 ‡ | PS + DP + xanthoferrin§ | |
| Xcc 8004 | 2.86 ± 0.06 | 3.98 ± 0.03 | 2.84 ± 0.13 | 4.19 ± 0.14 |
| ΔxssA | 2.85 ± 0.04 | 5.56 ± 0.38¶ | 2.82 ± 0.1 | 4.06 ± 0.13 |
| ΔxsuA | 2.85 ± 0.13 | 5.51 ± 0.37¶ | 2.85 ± 0.1 | 5.91 ± 0.52¶ |
| ΔxssA/pAP15 | 2.83 ± 0.11 | 3.92 ± 0.19 | 2.8 ± 0.04 | 4.15 ± 0.11 |
| ΔxsuA/pAP15 | 2.9 ± 0.04 | 3.89 ± 0.07 | 2.88 ± 0.07 | 4.31 ± 0.19 |
*Generation times are the means of three biological replicates ± standard deviation (SD).
†150 µm 2,2′‐dipyridyl (DP) was added to peptone–sucrose (PS) medium to yield low‐iron conditions.
‡100 µm of FeSO4 was added to supplement the low‐iron medium.
§20 µm of xanthoferrin was added to supplement the low‐iron medium.
¶P < 0.01 vs. wild‐type in the same medium by two‐tailed paired Student's t‐test.
Δ xssA and Δ xsuA mutants exhibit less intracellular iron content under low‐iron conditions
The bactericidal activity of the quinone antibiotic streptonigrin is strongly dependent on the formation of oxygen radicals, which has a direct correlation with the availability of intracellular iron in bacteria. The hypersensitivity of bacterial cells towards streptonigrin indicates a higher intracellular level of iron (Cohen et al., 1987; Schmitt, 1997; Yeowell and White, 1982). Therefore, to assess the intracellular iron content of different Xcc strains, we performed streptonigrin sensitivity assay in rich PS, low‐iron and iron‐replete media (Figs 3A and S6, see Supporting Information). We observed that ΔxssA and ΔxsuA mutants exhibited a 35%–40% increase in survival rate compared with the wild‐type strain after 16 and 42 h of growth under low‐iron conditions in the presence of streptonigrin. This result suggests that the ΔxssA and ΔxsuA mutants hold less intracellular iron in the low‐iron medium relative to the wild‐type Xcc 8004, a phenotype which can be restored by complementation in trans (Fig. 3A). In rich (PS) and iron‐replete (PS + 100 µm FeSO4) media, there was no significant difference observed in the survival rate of these mutants relative to the wild‐type Xcc 8004 (Fig. S6A–D). Further, we performed atomic absorption spectroscopy to measure directly intracellular elemental iron in different Xcc strains. Iron estimation by inductively coupled plasma‐optical emission spectrometry (ICP‐OES) further indicated approximately two‐fold less intracellular iron content in the ΔxssA and ΔxsuA mutants under low‐iron conditions, which was restored by in trans expression of the wild‐type Xoo xss gene cluster in the mutant background (Fig. 3B). However, we did not observe any significant difference in the intracellular iron content of ΔxssA and ΔxsuA mutants relative to the wild‐type Xcc 8004 strain in either rich PS medium or iron‐replete conditions (PS + 100 µm FeSO4) (Fig. S6E,F).
Figure 3.
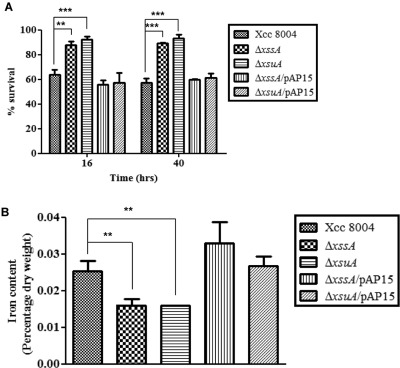
The ΔxssA and ΔxsuA mutants exhibit less intracellular iron content under low‐iron conditions. (A) Streptonigrin (SNG) sensitivity assay in broth. Percentage survival of different Xanthomonas campestris pv. campestris (Xcc) strains in peptone–sucrose (PS) + 150 µm 2,2′‐dipyridyl + 0.5 μg/mL SNG + 0.01 m sodium citrate after 16 and 42 h of growth. Percentage survival in SNG was calculated by comparison with growth in PS + 150 µm 2,2′‐dipyridyl as described in Experimental Procedures. (B) Intracellular iron content quantification determined by inductively coupled plasma‐optical emission spectrometry (ICP‐OES). Different Xcc strains were grown to late exponential phase in low‐iron medium (PS + 150 µm intracellular ferrous iron chelator 2,2′‐dipyridyl). Cells were harvested by centrifugation, freeze dried and solubilized in 30% HNO3 as described in Experimental Procedures. The data shown in the graphs are the mean ± standard error (SE) (n = 3). **P < 0.01; ***P < 0.001; statistical significance by paired Student's t‐test.
Expression of the xss operon is induced under low‐iron conditions and during in planta growth
To investigate the role of xanthoferrin during the in planta growth of Xcc, we performed expression analysis inside the plant using a chromosomal fusion of the upstream putative promoter of the xss cluster with the β‐glucuronidase (gusA) reporter gene in the wild‐type Xcc 8004 background (Fig. S2; Table S2). The Xcc 8004 Pxss::gusA reporter strain exhibited phenotypes similar to the wild‐type Xcc strain when grown under low‐iron conditions (Fig. S7, see Supporting Information). Cabbage leaves were infiltrated with the Xcc 8004 Pxss::gusA strain with and without FeSO4 or DP. GUS activity was monitored by staining the leaves with the chromogenic substrate X‐Gluc (5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronide; see Experimental Procedures). The histochemical blue–green GUS staining was detected in leaves inoculated with the Xcc 8004 Pxss::gusA reporter strain at 5 days post‐inoculation (dpi). Leaves co‐infiltrated with DP and Xcc 8004 Pxss::gusA reporter strain exhibited slightly more intense GUS staining relative to leaves infiltrated with the Xcc 8004 Pxss::gusA strain alone (Fig. 4A). The intensity of GUS staining decreased strongly in leaves co‐infiltrated with Xcc 8004 Pxss::gusA strain with Fe2+ (Fig. 4A). GUS activity was measured from the extracts of cabbage leaves inoculated with the Xcc 8004 Pxss::gusA reporter strain from 0 to 8 dpi. GUS assay indicated significant in planta expression of the xss operon (Fig. 4B). There was approximately 100‐fold decrease in GUS activity in leaves co‐infiltrated with Xcc 8004 Pxss::gusA and iron compared with those infiltrated with Xcc 8004 Pxss::gusA strain alone. In addition, we performed in vitro expression analysis with the chromosomal fusion of Pxss::gusA grown in rich (PS), low‐iron (PS + DP) and low‐iron medium supplemented with Fe2+. There was approximately three‐ to four‐fold induction in the expression of the xss gene cluster under low‐iron conditions relative to rich PS medium. The addition of exogenous iron to the low‐iron medium suppressed the expression of the xss operon (Fig. 4C).
Figure 4.
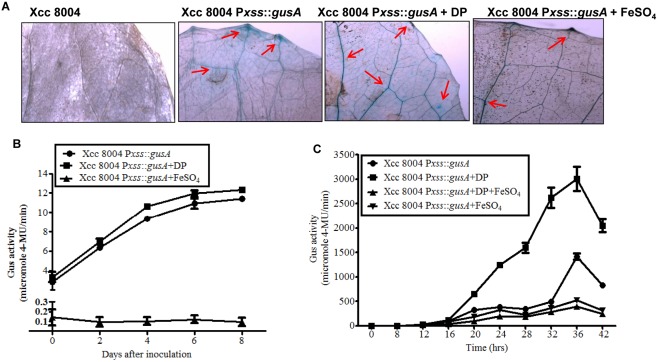
The xss (Xanthomonas siderophore synthesis) gene cluster is expressed during the in planta growth of Xanthomonas campestris pv. campestris (Xcc). (A) Histochemical β‐glucuronidase (GUS) staining with the chromogenic substrate X‐Gluc (5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐glucuronide) of cabbage leaves infiltrated with reporter strain Xcc 8004 Pxss::gusA with water, 100 µm 2,2′‐dipyridyl or 200 µm FeSO4. Control is a GUS‐negative Xcc 8004 strain. Arrows indicate GUS staining in veins. (B) In planta expression analysis of xss gene cluster in wild‐type (Xcc 8004 Pxss::gusA). GUS activity was monitored from 1 cm2 of cabbage leaves inoculated with reporter strain Xcc 8004 Pxss::gusA with water, 100 µm 2,2′‐dipyridyl or 200 µm FeSO4. (C) Cell‐normalized in vitro GUS assay of reporter strain Xcc 8004 Pxss::gusA in rich peptone–sucrose (PS), low‐iron (PS + 150 µm 2,2′‐dipyridyl), iron‐supplemented low‐iron (PS + 150 µm 2,2′‐dipyridyl + 100 µm FeSO4) and excess iron (PS + 100 µm FeSO4) media. 4‐MU, 4‐methylumbelliferone.
Δ xssA and Δ xsuA mutants are defective in Fe3+ uptake and exogenous xanthoferrin restores Fe3+ uptake in Δ xssA
To understand the contribution of xanthoferrin in iron uptake, we performed in vitro iron uptake assay to measure the iron transport ability of wild‐type Xcc 8004, ΔxssA, ΔxsuA and the mutants harbouring the wild‐type Xoo xss cluster (pAP15) using radiolabelled iron, as described previously (Ardon et al., 1997; Velayudhan et al., 2000) with a few modifications (see Experimental Procedures). The total amounts of radiolabelled Fe3+ incorporated into the ΔxssA and ΔxsuA mutants were significantly less (approximately two‐fold lower) than in the wild‐type Xcc 8004, ΔxssA/pAP15 and ΔxsuA/pAP15 over the 10‐min time course of the uptake assay experiment (Fig. 5A). In addition, we performed the Fe3+ uptake assay with the Fe3+–xanthoferrin complex. The xanthoferrin‐mediated incorporation of radiolabelled Fe3+ was of a similar level in ΔxssA and wild‐type Xcc 8004, but the ΔxsuA mutant was defective in xanthoferrin‐mediated Fe3+ uptake (Fig. 5B). However, we did not observe any significant difference in radiolabelled Fe2+ uptake in the ΔxssA and ΔxsuA mutants compared with the wild‐type Xcc 8004 (data not shown).
Figure 5.
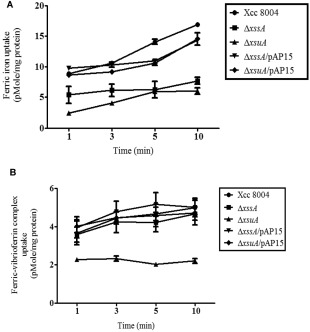
The ΔxssA and ΔxsuA mutants are defective in ferric iron uptake and xanthoferrin supplementation restores ferric iron uptake in the ΔxssA mutant. (A) The ΔxssA and ΔxsuA mutants show deficiency in ferric iron uptake under low‐iron conditions. The ferric iron transport was initiated by the addition of 0.4 μm 55FeCl3 to cell suspensions of wild‐type Xcc 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA//pAP15 strains. (B) Xanthoferrin supplementation restores the ferric iron uptake deficiency of ΔxssA. The ferric‐xanthoferrin complex transport was initiated by the addition of 0.4 μm 1 : 1 ratios of 55FeCl3 and xanthoferrin to cell suspensions of various Xanthomonas campestris pv. campestris (Xcc) strains grown under low‐iron conditions. Data are shown as mean ± standard error (SE) (n = 3).
Xanthoferrin synthesis and uptake are required for the optimum virulence of Xcc 8004 on cabbage
To understand the role of xanthoferrin in the virulence of Xcc, we performed infection studies with wild‐type Xcc 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA/pAP15 strains on cabbage plants. We inoculated cabbage leaves with bacterial cell suspensions by the leaf clip method and monitored lesion development, bacterial migration inside the leaves and in planta bacterial growth (see Experimental Procedures; Fig. S8, see Supporting Information). The ΔxssA and ΔxsuA mutants were significantly compromised in lesion development, which was rescued by plasmid harbouring the Xoo xss cluster (pAP15) (Fig. 6A,B). The lesions caused by the wild‐type Xcc 8004 strain were approximately 3 cm in length at 21 dpi. In contrast, the lesions caused by ΔxssA and ΔxsuA mutants were approximately 1 cm in length at 21 dpi (Fig. 6B). To monitor bacterial migration inside cabbage leaves, surface‐sterilized infected leaves (5 dpi) were cut into 1‐cm pieces from the bottom of the leaf to the top (site of inoculation) with sterile scissors, and were incubated on PSA medium containing appropriate antibiotics (Fig. S8A). Migration was estimated by observing the colonies formed after 1–3 days by bacterial ooze from the cut ends of the cabbage leaf pieces. Migration assay indicated that the ΔxssA and ΔxsuA mutants exhibited reduced in planta migration relative to the wild‐type Xcc 8004 strain (Fig. 6C). In planta growth assay (0–14 dpi) indicated that the ΔxssA and ΔxsuA mutants exhibited significantly reduced growth (approximately four‐fold less) than that exhibited by the wild‐type Xcc 8004 strain or the mutants harbouring the complementing plasmid (Fig. 6D).
Figure 6.
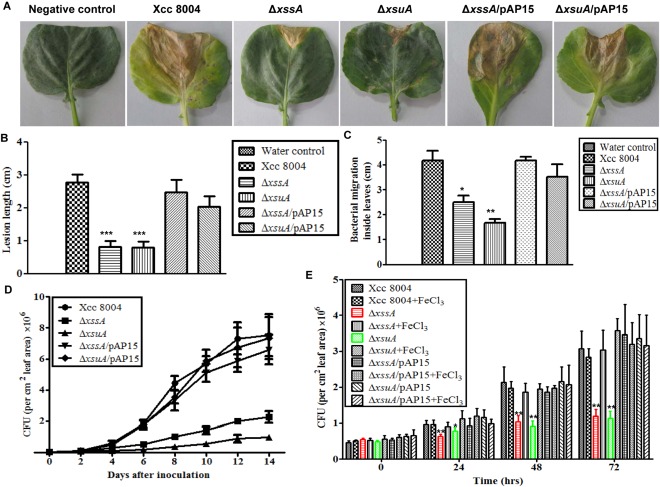
The ΔxssA and ΔxsuA mutants are deficient in virulence and growth inside cabbage. (A) Cabbage leaves (Indian Super Hybrid variety) infected with wild‐type Xcc 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA//pAP15 strains showing lesion symptoms at 21 days post‐inoculation. Bacterial cultures (1 × 109 cells/mL suspension) were inoculated into 30‐day‐old plants by the clip method. (B) Quantification of lesion length at 21 days post‐inoculation. Twenty‐five leaves were inoculated per strain. (C) Bacterial migration at 5 days post‐inoculation in host leaves was assayed by inoculating 1‐cm pieces of infected leaf, cut from the base to the tip, on a peptone–sucrose agar (PSA) plate with the respective antibiotics (see Table S2). Migration was estimated by observing colonies formed after 1–3 days by bacterial ooze from the cut ends of the cabbage leaf pieces. For each experiment, six leaves were used (three independent experiments). (D) In planta growth assays of wild‐type Xcc 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA//pAP15 strains. Bacterial populations were measured by crushing leaves (1 cm2) followed by serial dilution plating at the indicated post‐inoculation days. For each experiment, six leaves were used (three independent experiments). (E) Detached leaf assay with exogenous iron supplementation. Different Xcc strains were inoculated on detached cabbage leaves by the clip method. The leaves were maintained in 1 μg/mL of benzyl amino purine (BAP; first‐generation synthetic cytokinin) with or without 50 µm FeCl3 supplementation. Bacterial populations were determined from a leaf area of 1 cm2 at the indicated post‐inoculation days. The data shown in the graphs are the mean ± standard error (SE) (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; significant difference between the data obtained from mutants and the data obtained from the wild‐type and complemented strains by paired Student's t‐test. CFU, colony‐forming unit.
Exogenous iron supplementation promotes the growth of Δ xssA and Δ xsuA mutants in cabbage leaves
To investigate whether iron supplementation can rescue the growth defect of ΔxssA and ΔxsuA mutants inside host leaves, detached leaf assays were performed as described previously (Chatterjee and Sonti, 2002) with modifications (see Experimental Procedures; Fig. S8B). The detached cabbage leaves were maintained in beakers with or without iron (50 µm FeCl3) supplementation. Benzyl amino purine (BAP), a first‐generation synthetic cytokinin, was added to each beaker to maintain the freshness of the leaves. The detached cabbage leaves were inoculated with different Xcc strains and bacterial growth within the leaves was estimated at 0, 24, 48 and 72 h post‐inoculation. In planta growth assay indicated that exogenous iron supplementation rescued the growth defect exhibited by the ΔxssA and ΔxsuA mutants (Fig. 6E). In contrast, iron supplementation in cabbage leaves did not significantly affect the growth of wild‐type Xcc 8004, ΔxssA/pAP15 and ΔxsuA/pAP15 strains (Fig. 6E).
Discussion
In this study, we have characterized the role of the siderophore in the virulence of Xcc, a pathogen of crucifers. The results of this study have established the following: Xcc produces xanthoferrin, an α‐hydroxycarboxylate‐type siderophore similar to vibrioferrin, under low‐iron conditions; xanthoferrin production is required for growth under low‐iron conditions and for virulence; the xanthoferrin production and uptake mutants ΔxssA and ΔxsuA exhibit defects in Fe3+ uptake; and exogenous supplementation of iron promotes the in planta growth of ΔxssA and ΔxsuA mutants in cabbage.
Xanthomonas axonopodis pv. citri (Xac) and Xcc possess non‐ribosomal peptide synthetase (NRPS) enzymes encoding the genes Xac3922 and Xcc3867, respectively, which show similarity to enterobactin, a catecholate‐type siderophore, synthase component F (EntF) in Escherichia coli (Etchegaray et al., 2004; Ryan et al., 2011; da Silva et al., 2002). It has been proposed that the siderophores produced by Xac and Xcc do not belong to the catechol or hydroxamate family, but might be assigned to an unknown novel category (Etchegaray et al., 2004). The genome sequence of Xcc 8004 (Qian et al., 2005) indicated that it possesses an xss gene cluster, homologous to the Xoo xss locus (Lee et al., 2005; Pandey and Sonti, 2010) and V. parahaemolyticus pvs locus (Tanabe et al., 1982), which is involved in siderophore synthesis and uptake (Fig. S1). Siderophore purified from the cell‐free culture supernatant by Amberlite XAD‐16 resin columns and HPLC analysis suggests that Xcc 8004 synthesizes a xanthoferrin siderophore (similar to the vibrioferrin of V. parahaemolyticus) under iron‐limited conditions, which is an α‐hydroxycarboxylate‐type siderophore (Figs 1A and S4). The homology of the xssA gene to pvsA, the vibrioferrin synthesis gene of V. parahaemolyticus, and the drastic reduction in xanthoferrin production in the ΔxssA mutant suggest that xssA encodes a protein which is involved in xanthoferrin synthesis (Fig. 1B; Table S1). Furthermore, rescue of the growth defect exhibited by the ΔxssA mutant of Xcc by the exogenous supplementation of purified vibrioferrin from V. parahaemolyticus strongly suggests that the xanthoferrin form Xcc and vibrioferrin are structurally related (Fig. S5).
In Xoo, a xylem‐specific pathogen of rice, it has been demonstrated that siderophore production is not required for virulence and the xss cluster is not expressed in planta (Pandey and Sonti, 2010). In contrast, it has been shown that, in Xanthomonas oryzae pv. oryzicola (Xoc; a pathogen of rice parenchyma), siderophore production is required for virulence and is induced in planta (Rai et al., 2015). These studies suggest that the contribution of the xss locus in pathogenicity seems to be variable among closely related xanthomonads. Our results indicate that siderophore‐mediated iron uptake is required for the virulence and colonization of Xcc. We speculate that differences in host iron storage and forms (Fe2+ vs. Fe3+) may contribute to differences in iron utilization strategies and need for closely related pathogens.
Many bacteria acquire iron through energy‐independent, low‐affinity iron uptake systems in iron‐rich environments; however, in iron‐limited environments, siderophore‐mediated iron uptake occurs, which is a highly energy‐dependent process (Jones and Niederweis, 2010; Noinaj et al., 2010). We observed that Xcc 8004 produces siderophores under low‐iron conditions, which ceases on iron supplementation (data not shown). Although, iron or xanthoferrin supplementation restores the impaired growth of the ΔxssA mutant under iron‐limited conditions, the addition of xanthoferrin fails to restore the compromised growth of the ΔxsuA mutant in low‐iron medium (Fig. 2C). Similarly, xanthoferrin supplementation restores the impaired Fe3+ uptake of the ΔxssA mutant, but fails to restore the Fe3+ uptake deficiency of the ΔxsuA mutant (Fig. 5B). These results suggest that xsuA, a homologue of V. parahaemolyticus pvuA (a receptor for the Fe3+–siderophore complex), encodes an outer membrane receptor protein required for xanthoferrin‐mediated Fe3+ uptake. Quantitative GUS reporter assay suggests that the expression of the xss operon is induced under low‐iron conditions and is suppressed to the basal level after iron supplementation (Fig. 4C). The ΔxssA and ΔxsuA mutants possess less intracellular iron than the wild‐type Xcc 8004 grown under iron‐limited conditions (Fig. 3). These results indicate that Xcc 8004 requires xanthoferrin‐mediated iron uptake, particularly in the iron‐limited environment of the plant. Siderophore‐independent, low‐affinity iron uptake systems maintain iron homeostasis under iron‐replete conditions in many microorganisms (Andrews et al., 2003). The porins, Msp in Mycobacterium smegmatis (Jones and Niederweis, 2010) and the SFU system in Serratia marcescens (Angerer et al., 1990; Zimmermann et al., 1989), have been reported to be involved in siderophore‐independent Fe3+ uptake. The FeoB system is involved in the uptake of the less commonly available, but soluble, Fe2+ form of iron (Cartron et al., 2006; Kammler et al., 1993). Extracellular or cell‐associated Fe3+ reductases have been reported in many bacteria, which assist in iron uptake by increasing the availability of the soluble form of iron by reducing Fe3+ to Fe2+ (Schröder et al., 2003). In our study, we detected Fe3+ reductase activity in Xcc strains, but no significant difference was observed among wild‐type Xcc 8004, ΔxssA and ΔxsuA mutants (Fig. S9, see Supporting Information). It is pertinent to note that the iron uptake facilitated by Fe3+ reductases and low‐affinity iron uptake systems might be sufficient to fulfil the iron requirement under rich or iron‐replete conditions. However, under iron‐depleted conditions, Xcc 8004 may depend particularly on xanthoferrin‐mediated iron uptake. This might be the reason why the xss cluster is induced under iron‐limiting conditions.
In planta GUS reporter assay indicated that the xss operon is expressed in the iron‐limited host environment, but is suppressed on co‐infiltration of the reporter strain and iron in cabbage leaves (Fig. 4). These results suggest that Xcc 8004 requires xanthoferrin‐mediated iron uptake in the iron‐limiting environment of the host. The disruption of either xanthoferrin synthesis or the xanthoferrin‐mediated iron uptake system diminishes the ability of Xcc to grow, colonize and migrate inside cabbage leaves (Fig. 6). The results of the detached leaf assay suggested that the in planta growth deficiency of ΔxssA and ΔxsuA mutants can be rescued by the exogenous supplementation of a moderate amount of iron (Fig. 6E). Thus, there is a correlation between xanthoferrin‐mediated iron uptake and the ability of Xcc to cause disease. These data suggest that the ability to acquire iron by the energy‐dependent xanthoferrin synthesis and uptake system may have evolved in Xcc as a mechanism to take up iron under iron‐depleted conditions, such as in the host environment.
This study is also important from the disease perspective, as targeting of the iron uptake system in these bacteria could help to prevent disease development in the near future. Further, a thorough understanding of iron uptake systems in different bacteria may aid in the development of novel strategies to control various animal and plant diseases, which are mainly dependent on the iron uptake ability of pathogens.
Experimental Procedures
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S2. Xcc 8004 strains were grown at 28 ºC in PS medium (Tsuchia et al., 1982) at 200 rpm (New Brunswick Scientific, Innova 43, Edison, NJ, USA). The concentrations of antibiotics used were as follows: rifampicin (Rif), 50 µg/mL; kanamycin (Kan), 50 µg/mL; tetracycline (Tet), 5 µg/mL; gentamycin (Gent), 5 µg/mL; ampicillin (Amp), 50 µg/mL. We used an iron chelator, DP (Fluka Analytical, Steinheim, Westphalia, Germany), to create the low‐iron conditions.
Molecular biology and microbiology techniques
All standard molecular biology techniques, including plasmid isolations, genomic DNA isolations, gel extractions and PCR purifications, were performed as described by Sambrook et al. (1989) or using kits provided by Qiagen Inc., (Valencia, CA, USA). PCR amplifications were carried out with high‐fidelity accu taq polymerase (Sigma‐Aldrich, St. Louis, MO, USA), KAPA HiFi HotStart DNA Polymerase (Kappa Biosystems Inc., Wilmington, MA, USA) and Taq polymerase (Thermo Fisher Scientific, Waltham, MA, USA) according to the user manuals provided by the manufacturers. Restriction digestions and ligations were performed with enzymes provided by New England Biolabs (Ipswich, MA, USA) according to the user manual provided by the manufacturer. Transformations were performed by electroporation, the heat shock method or conjugation. The primers used in this study are listed in Table S4 (see Supporting Information).
Generation of Δ xssA and Δ xsuA deletion mutants in the wild‐type Xcc 8004 background
In‐frame marker‐free deletion strains were prepared as described previously by Schäfer et al. (1994) with a few modifications. The chromosomal deletion of the xssA and xsuA genes in the Xcc 8004 background was obtained by allelic exchange and homologous recombination using the suicide vector pK18mobsacB harbouring 5′ and 3′ flanking regions of the gene to be deleted. The 5′ flanking regions of ΔxssA and ΔxsuA were amplified with the primers (listed in Table S4) SCPsidelxc1F & SCPsidelxc2R and SCPxsuAdel 1F & SCPxsuA1R, respectively, whereas the 3′ flanking regions were amplified with the primers SCPsidelxc2F & SCPsidelxc2R and SCPxsuAdel2F & SCPxsuAdel2R, respectively. The 5′ and 3′ flanking PCR products were digested with a common restriction enzyme (XbaI) and ligated. The ligation product was PCR amplified with end primers to select the deletion construct. The PCR products and pK18mobsacB were then digested with appropriate restriction enzymes (BamH1 & HindIII for the deletion of xssA and EcoRI & HindIII for the deletion of xsuA), ligated and transformed in E. coli DH5α cells. Appropriate clones were selected on nalidixic acid and kanamycin‐containing Luria–Bertani (LB) agar plates and confirmed by nucleotide sequencing. pK18mobsacB carrying deletion constructs (Table S2) was transformed into the wild‐type Xcc 8004 strain by electroporation, and transformants with single crossover were selected on nutrient agar (NA) plates containing rifampicin and kanamycin. Colonies were passaged in antibiotic‐free nutrient broth medium for the second recombination to occur for the removal of the plasmid background, and eventually selected on PSA containing rifampicin and 5% sucrose. Colonies growing on PSA, but not on NA with kanamycin, were confirmed by the sequencing of PCR products amplified using the outward primers (Table S4): SCP54F Sid del out 2 & SCP54R Sid del out 2 and SCPxsuA del out F and SCPxsuAdel2R for deletion in the xssA and xsuA genes, respectively. For complementation, the cosmid clone with a 38.2‐kb genomic insert of Xoo containing the entire xss cluster (pAP15) (Table S2; Pandey and Sonti, 2010) was mobilized in the ΔxssA and ΔxsuA background by biparental mating with E. coli strain S17′‐1 harbouring pAP15.
Generation of chromosomal promoter fusions to GUS
Chromosomal fusions of the promoter of the xss gene cluster with GUS were created using the suicidal plasmid pVO155 containing the promoterless gusA gene, as described previously (Oke and Long, 1999; Pandey and Sonti, 2010) with a few modifications. Briefly, a 971‐bp sequence (upstream of the xsuA gene) containing the putative promoter of the xss gene cluster (Fig. S2; Table S2) was amplified with SCP_sid_prom F & SCP_sid_prom R primers (listed in Table S4) and cloned at the HindIII‐XbaI site of the promoterless suicidal plasmid pVO155. The GUS reporter construct was mobilized from E. coli DH5α to E. coli S17‐1 by triparental mating, and subsequently introduced into wild‐type Xcc 8004 by biparental mating with the S17′‐1 construct. Insertion of the gus reporter cassette was confirmed by PCR amplification and sequencing with primers specific for gusA and the neighbouring flanking sequence (data not shown).
Growth under low‐iron conditions
We used different concentrations of DP (75, 100 and 150 µm) to create low‐iron conditions in PS medium in order to identify the conditions inducing siderophore production without affecting the growth of different Xcc strains, and also to identify the low‐iron conditions in which the siderophore‐deficient mutants exhibit growth deficiency (Figs 2 and S10, see Supporting Information). We found that 75 μm DP induced siderophore production in the wild‐type Xcc 8004 strain, but did not cause significant growth defects in the wild‐type or the ΔxsuA and ΔxssA mutants (Fig. S10B). To induce siderophore production and purification from liquid PS medium, we used 100 μm DP. We found that 100 μm DP did not cause severe growth deficiency in the ΔxsuA and ΔxssA mutants compared with the wild‐type Xcc 8004 strain (Fig. S10C). At 150 μm DP, we observed that the ΔxsuA and ΔxssA mutants exhibited growth deficiency which could be rescued by supplementation of either iron or purified siderophores. Therefore, for growth assay under low‐iron conditions, we used 150 µm DP in this study (Fig. 2).
CAS plate assays for the assessment of siderophore production
CAS agar plate assays for the assessment of siderophore production were performed as described previously (Schwyn and Neilands, 1987) with a few modifications (Chatterjee and Sonti, 2002). Xanthomonas grew quite slowly on minimal medium MM9; therefore, we used rich PSA medium supplemented with 75 μm DP to create low‐iron conditions. The different Xcc strains were spotted onto the CAS agar plate and incubated at 28 °C.
Purification and quantification of xanthoferrin
Primary cultures of different Xcc strains were grown up to 1.0 × 109 cells/mL in PS broth with the respective antibiotics (see Table S2); 0.3% of inoculum was transferred to 500 mL of PS medium supplemented with 100 μm DP and grown to an optical density at 600 nm (OD600) of unity at 28 °C in a shaking incubator at 200 rpm (New Brunswick Scientific). The cultures were centrifuged at 13201 g for 1 h. The supernatants from different cultures were collected and further filtered through a 0.22‐µm membrane to remove the remaining cells. Initially, the siderophore was isolated from cell‐free supernatants using Amberlite XAD‐16 resin columns, as described by Wright (2010) with a few modifications. Briefly, the different cell‐free supernatants were acidified up to pH 2 with concentrated HCl and allowed to pass through the prepared XAD‐16 resin column (2.4 × 30 cm2). Elution was performed with methanol and the flow‐through was collected into 100 fractions. Each fraction was tested for siderophores on CAS agar plates supplemented with 75 µm DP. As a negative control, empty buffer eluate from the column run was spotted onto a PSA–CAS + 75 µm DP plate to detect any cross‐contaminating CAS‐reactive agent during the separation of xanthoferrin (Fig. S11, see Supporting Information). Further, we processed the samples with an Agilent 1100series HPLC system (Agilent, Santa Clara, CA, USA). The data were recorded and analysed using chemstation software (Agilent 1100). We compared the chromatogram of xanthoferrin with the chromatogram of standard purified vibrioferrin (a kind gift from M. J. Fujita). The estimation of siderophore production was performed as described previously (Amin et al., 2009) with a few modifications. The xanthoferrin production by different Xcc strains was estimated by comparing the peak area at a particular retention time with standard curves generated from a known concentration of pure standard vibrioferrin.
Iron uptake assay
We performed the iron uptake assays using radiolabelled iron as described previously (Ardon et al., 1997; Velayudhan et al., 2000) with a few modifications. Primary cultures of different Xcc strains were grown to 1.0 × 109 cells/mL in rich PS medium with the respective antibiotics (Table S2); 0.3% of inoculum was transferred to fresh PS medium containing 150 µm DP and the antibiotic required to maintain cosmids, and grown to the late stationary phase. Further bacterial cells were harvested and washed twice in 50 mm sodium phosphate buffer (pH 7.4). Bacterial pellets were resuspended in 50 mm sodium phosphate buffer and subsequently diluted with chelex‐100 (Sigma, St. Louis, MO, USA)‐treated PS to an OD600 of 1.0 and incubated at 28 °C for 5 min. The Fe3+ uptake was initiated by the addition of 0.4 µm of 55FeCl3 (specific activity of 10.18 mCi/mg; American Radiolabeled Chemicals, Inc., St. Louis, MO, USA). For the uptake assay of the Fe3+–xanthoferrin complex, the uptake was initiated after combining vibrioferrin (7.6 mm stock) and 55FeCl3 in a 1 : 1 ratio. Uptake was stopped at different time points by layering onto di‐butyl phthalate and di‐octyl phthalate (1 : 1) solution. Immediately, the cultures were centrifuged at 17968 g for 2 min; eventually, the pellets were resuspended in 100 µL of 1% (v/v) Triton X‐100. The suspensions were transferred to 5 mL of scintillation cocktail and counted in the 3H channel of a scintillation counter (Perkin‐Elmer, Waltham, MA, USA, Liquid Scintillation Analyzer, Tri‐Carb 2910 TR, USA). As a control, iron uptake was performed in the presence of carbonyl cyanide p‐trifluoromethoxy phenylhydrazone (FCCP; 50 µm), a proton motive force uncoupler. There was no significant increase in the incorporation of radiolabelled iron in the presence of FCCP, indicating that iron uptake is an energy‐dependent process (data not shown).
Streptonigrin sensitivity assay
To assess intracellular iron content, we performed streptonigrin sensitivity assay as described previously (Wilson et al., 1998) with a few modifications. Briefly, overnight‐grown cultures of different Xcc strains were harvested by centrifugation at 3300 g for 5 min. Pellets were resuspended in fresh PS medium at OD600 = 0.6; 2% of inoculum was transferred to 4 mL of fresh PS medium with and without streptonigrin (Sigma‐Aldrich) and sodium citrate in six‐well plates. The plates were incubated in a static incubator at 28 ºC. Absorbance at 600 nm was measured after 16 and 40 h of incubation. The percentage survival was calculated by comparing the bacterial growth in streptonigrin relative to that in rich PS medium. This assay was also performed on a PSA plate containing streptonigrin and sodium citrate by spotting serial dilutions of cell‐normalized bacterial cultures.
Estimation of intracellular iron level
The intracellular iron content was estimated using atomic absorption spectrophotometry as described previously (Velayudhan et al., 2000) with a few modifications. Different Xcc strains were grown to stationary phase culture in rich PS, PS with 150 µm FeSO4 (Fe‐replete) and PS with 100 µm DP (Fe‐restricted) media. Cells were harvested by centrifugation, followed by washing twice with phosphate‐buffered saline (50 mm PBS, pH 7.4). Pellets were lyophilized and their dry weights were determined. A further equal amount of lyophilized cells was solubilized in 30% HNO3 at 80 ºC overnight and subsequently diluted 10‐fold with sterile milliQ water. The iron content was determined using ICP‐OES (JY 2000 sequential ICP‐OES spectrometer, Jobin Yvon, Horiba, France).
GUS reporter assays
The GUS reporter strain was grown in rich PS medium with the required antibiotics at 28 ºC and 200 rpm overnight; 0.2% of primary inoculum was transferred to rich PS, PS with 100 µm FeSO4 (Fe‐replete), PS with 100 µm DP (Fe‐restricted) and PS with 100 µm DP + 100 µm FeSO4 (FeSO4 supplementation to Fe‐restricted) media. The absorbance at 600 nm and GUS expression were measured at regular time intervals. GUS expression assays were performed as described previously (Jefferson et al., 1987) with a few modifications. Briefly, cells were harvested from 1 mL of culture and washed with sterile milliQ water. Pellets were resuspended in 250 μL extraction buffer [50 mm sodium dihydrogen phosphate (pH 7.0), 10 mm ethylenediaminetetraacetic acid (EDTA), 10 mm β‐mercaptoethanol, 0.1% Triton X‐100 and 0.1% sodium lauryl sarcosine] with added 1 mm MUG (4‐methylumbelliferyl β‐d‐glucuronide) and incubated at 37 °C. After a definite time interval, reactions were terminated by the addition of 675 μL of 0.2 m Na2CO3 into 75 μL of reaction mixture. Fluorescence was measured with 4‐methylumbelliferone (4‐MU; Sigma) as standard at an excitation wavelength of 365 nm and emission wavelength of 455 nm. GUS activity was presented as nanomoles of 4‐MU produced per minute.
In planta GUS expression assay
The GUS reporter strain was grown to 1.0 × 109 cells/mL in the presence or absence of 100 µm DP or 100 µm FeSO4. Cells were harvested, washed with sterile milliQ water and infiltrated into cabbage leaves (Indian Super Hybrid variety) with and without 100 µm DP or 100 µm FeSO4. The leaves were harvested at 0, 2, 4, 6 and 8 dpi and crushed in 1 mL of extraction buffer [50 mm sodium dihydrogen phosphate (pH 7.0), 0.1% Triton X‐100, 10 mm EDTA, 0.1% sodium lauryl sarcosine and 10 mm β‐mercaptoethanol] without substrate. Subsequently, 250 μL of extraction buffer with 1 mm MUG substrate was added to the plant extract and incubated at 37 °C. Reactions were stopped by the addition of 675 μL of 0.2 m Na2CO3 into 75 μL of reaction mixture. Fluorescence was measured with 4‐MU (Sigma) as standard at an excitation wavelength of 365 nm and emission wavelength of 455 nm. GUS activity was expressed as nanomoles of MU produced per minute per square centimetre of leaf area.
Histochemical staining
Thirty‐day‐old cabbage leaves (Indian Super Hybrid variety) were infiltrated with GUS reporter strains in the presence or absence of 100 µm DP or 100 µm FeSO4. At 5 dpi, the leaves were stained with 1 mm X‐Gluc in GUS assay buffer [50 mm sodium dihydrogen phosphate (pH 7.0), 10 mm EDTA, 0.1% sodium lauryl sarcosine, 0.1% Triton X‐100 and 10 mm β‐mercaptoethanol] to determine β‐d‐glucuronidase activity. A vacuum was applied for 1 h to facilitate X‐Gluc penetration into the leaves and then incubated at 37 °C for 2 h. Subsequently, chlorophyll was removed from the leaves by incubation in absolute alcohol for 72 h at 37 °C, and then observed under a compound microscope. The experiment was performed with a minimum of five infiltrated leaves for each strain and repeated three times.
Assay for Fe3+ reductase activity
The Fe3+ reductase activity of Xcc strains was measured using ferrozine (Sigma‐Aldrich), a chromogenic Fe2+ chelator, as described previously (Deneer et al., 1995; Velayudhan et al., 2000; Worst et al., 1998) with slight modifications. Briefly, 1 mm ferrozine and 100 µm FeCl3 were added to different Xcc strains, grown to a density of 1.0 × 106 cells/mL and incubated at 28 °C. Cell‐free PS medium was incubated under the same conditions as a blank; 1‐mL aliquots were taken at each time interval and centrifuged at 17968 g for 2 min. The absorbance of the magenta‐coloured Fe2+–ferrozine complex in the supernatant was measured at 562 nm.
Virulence assays on cabbage plants
Virulence assay was conducted by inoculation of Xcc strains onto 30‐day‐old Indian cabbage (Indian Super Hybrid variety) using the scissor‐clipping method. In brief, different Xcc strains were grown to a density of 1.0 × 109 cells/mL in PS medium with the required antibiotics. Cells were pelleted down at 3300 g for 5 min and resuspended in sterile milliQ water. Sterile scissors were dipped in bacterial cultures and cabbage leaves were gently incised at the apex. The lesion length, number of colony‐forming units (CFU) and migration of bacteria inside host leaves were recorded. A plant inoculated with sterile milliQ water was used as a control (Robinson and Callow, 1986).
Exogenous iron supplementation and bacterial growth assay in cabbage leaves
Exogenous iron supplementation and bacterial growth assays in detached leaves were performed as described previously (Chatterjee and Sonti, 2002) with a few modifications. Leaves, together with the petiole, of 30‐day‐old cabbage plants were cut with sterile scissors and dipped in 250‐mL beakers (four to six leaves per beaker) containing 100 mL of 1.0 µg/mL of BAP with or without 50 μm FeCl3 (Standard Reagents, Hyderabad, India) in sterile milliQ water. The addition of BAP, a first‐generation synthetic cytokinin, helped to maintain the detached cabbage leaves in a fresh condition. In a control experiment, we added red‐coloured safranin dye to a beaker containing BAP to determine the conductance of water in the leaves. After overnight incubation, the spread of red colour into the leaves indicated water conductance (Fig. S5B). The leaves were clip inoculated with cultures of different Xcc strains at a density of 1.0 × 109 cells/mL. Bacterial growth was determined by the number of CFU obtained after crushing the surface‐sterilized 1‐cm2 leaf area surrounding the inoculation site, followed by dilution plating.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Schematic presentation of the xss (Xanthomonas siderophore synthesis) gene cluster of Xanthomonas campestris pv. campestris (Xcc) 8004, Xanthomonas oryzae pv. oryzae and Vibrio parahaemolyticus.
Fig. S2 Genetic organization of the Xanthomonas campestris pv. campestris (Xcc) 8004 xss (Xanthomonas siderophore synthesis) cluster indicating the location of the deletions in the knockout strains ΔxssA and ΔxsuA, and pSSP70 insertion in the β‐glucuronidase (GUS) reporter strain Xcc 8004 Pxss::gusA.
Fig. S3 Relative quantification of the expression of downstream genes of the xss (Xanthomonas siderophore synthesis) operon in ΔxssA and ΔxsuA mutants compared with the wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004 strain by real‐time quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Fig. S4 High‐performance liquid chromatography (HPLC) chromatograms of purified xanthoferrin from different Xanthomonas campestris pv. campestris (Xcc) strains.
Fig. S5 Purified vibrioferrin from Vibrio parahaemolyticus rescues the growth deficiency of the ΔxssA mutant under low‐iron conditions.
Fig. S6 Streptonigrin sensitivity assay to assess the intracellular iron content among wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA//pAP15 strains grown in peptone–sucrose (PS) medium and iron‐replete conditions.
Fig. S7 The Xanthomonas campestris pv. campestris (Xcc) 8004 Pxss::gusA reporter strain exhibits wild‐type Xcc 8004‐like phenotypes under low‐iron conditions.
Fig. S8 Assay for bacterial migration inside cabbage leaves and control experiment for conductance.
Fig. S9 Ferric reductase assay of wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004, ΔxssA and ΔxsuA strains.
Fig. S10 The growth of Xanthomonas campestris pv. campestris (Xcc) 8004 (wild‐type), ΔxssA and ΔxsuA strains in different concentrations of 2,2′‐dipyridyl.
Fig. S11 Column buffer control and active fraction of wild‐type Xanthomonas campestris pv. campestris (Xcc) xanthoferrin on peptone–sucrose agar and chromeazurol S (PSA–CAS) plates containing 75 µm 2,2′‐dipyridyl.
Table S1 Homology of proteins encoded in the xss (Xanthomonas siderophore synthesis; involved in siderophore biosynthesis, putative export and uptake) gene cluster of Xanthomonas campestris pv. campestris (Xcc) with those of Vibrio parahaemolyticus, and X. oryzae pv. oryzae (Xoo) KACC103331.
Table S2 List of strains and plasmids used in this study.
Table S3 Generation times of different Xanthomonas campestris pv. campestris (Xcc) strains.
Table S4 List of the primers used in this study.
Acknowledgements
We are grateful to Dr Masaki J. Fujita (Hokkaido University, Japan) for providing the pure vibrioferrin standard. S.S.P. and R.R were recipients of Senior Research Fellowships from the Council of Scientific and Industrial Research (CSIR), India. This study was supported by funding to S.C. from the Department of Biotechnology (DBT), Council of Scientific & Industrial Research ‐ Human Resource Development Group (CSIR‐HRDG), Department of Science and Technology ‐ Science & Engineering Research Board (DST‐SERB), Government of India and core funding from Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad, India.
References
- Amin, S.A. , Green, D.H. , Küpper, F.C. and Carrano, C.J. (2009) Vibrioferrin, an unusual marine siderophore: iron binding, photochemistry, and biological implications. Inorg. Chem. 48, 11 451–11 458. [DOI] [PubMed] [Google Scholar]
- Andrews, S.C. , Robinson, A.K. and Rodríguez‐Quiñones, F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. [DOI] [PubMed] [Google Scholar]
- Angerer, A. , Gaisser, S. and Braun, V. (1990) Nucleotide sequences of the sfuA, sjiiB, and sfiC genes of Serratia marcestens suggest a periplasmic‐binding protein‐dependent iron transport mechanism. J. Bacteriol. 172, 572–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardon, O. , Weizman, H. , Libman, J. , Shanzer, A. , Chen, Y. and Hadar, Y. (1997) Iron uptake in Ustilago maydis: studies with fluorescent ferrichrome analogues. Microbiology, 143, 3625–3631. [DOI] [PubMed] [Google Scholar]
- Bearden, S.W. , Fetherston, J.D. and Perry, R.D. (1997) Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis . Infect. Immun. 65, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, G. and Denny, T.P. (2004) Ralstonia solanacearum iron scavenging by the siderophore staphyloferrin B is controlled by PhcA, the global virulence regulator. J. Bacteriol. 186, 7896–7904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, V. and Hantke, K. (2013) The tricky ways bacteria cope with iron limitation. In Iron Uptake in Bacteria with Emphasis on E. coli and Pseudomonas. (Chakraborty, R., Braun, V., Hantke, K., and Cornelis, P., eds), pp. 31–66. Netherlands: Springer.
- Bull, C.T. , Carnegie, S.R. and Loper, E.J. (1996) Pathogenicity of mutants of Erwinia carotovora subsp. carotovora deficient in aerobactin and catecholate siderophore production. Mol. Plant Pathol. 86, 260–266. [Google Scholar]
- Cartron, M.L. , Maddocks, S. , Gillingham, P. , Craven, C.J. and Andrews, S.C. (2006) Feo – transport of ferrous iron into bacteria. BioMetals, 19, 143–157. [DOI] [PubMed] [Google Scholar]
- Cassat, J.E. and Skaar, E.P. (2013) Iron in infection and immunity. Cell Host Microbe, 13, 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, S. and Sonti, R.V. (2002) rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant–Microbe. Interact. 15, 463–471. [DOI] [PubMed] [Google Scholar]
- Cohen, M.S. , Chai, Y. , Britigan, B.E. , McKenna, W. , Adams, J. , Svendsen, T. , Bean, K. , Hassett, D.J. and Sparling, P.F. (1987) Role of extracellular iron in the action of the quinone antibiotic streptonigrin: mechanisms of killing and resistance of Neisseria gonorrhoeae . Antimicrob. Agents Chemother. 31, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, S.E. , Doherty‐Kirby, A. , Lajoie, G. , Heinrichs, D.E. , Al, D.E.T. and Mmun, I.N.I. (2004) Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72, 29–37. [DOI] [PMC free article] [PubMed]
- Dellagi, A. , Brisset, M.N. , Paulin, J.P. and Expert, D. (1998) Dual role of desferrioxamine in Erwinia amylovora pathogenicity. Mol. Plant–Microbe. Interact. 11, 734–742. [DOI] [PubMed] [Google Scholar]
- Deneer, H.G. , Healey, V. and Boychuk, I. (1995) Reduction of exogenous ferric iron by a surface‐associated ferric reductase of Listeria spp. Microbiology, 141, 1985–1992. [DOI] [PubMed] [Google Scholar]
- Enard, C. , Diolez, A. and Expert, D. (1988) Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J. Bacteriol. 170, 2419–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray, A. , Silva‐Stenico, M.E. , Moon, D.H. and Tsai, S.M. (2004) In silico analysis of nonribosomal peptide synthetases of Xanthomonas axonopodis pv. citri: identification of putative siderophore and lipopeptide biosynthetic genes. Microbiol. Res. 159, 425–437. [DOI] [PubMed] [Google Scholar]
- Expert, D. , Enard, C. and Masclaux, C. (1996) The role of iron in plant host–pathogen interactions. Trends Microbiol. 4, 232–237. [DOI] [PubMed] [Google Scholar]
- Expert, D. , Franza, T. and Dellagi, A. (2012) Molecular aspects of iron metabolism in pathogenic and symbiotic plant–microbe associations. In SpringerBriefs in Biometals. (Expert, D. and O'Brian, M.R., eds), pp. 7–39. London WC1X 8HB, UK: Springer.
- Faraldo‐Gómez, J.D. and Sansom, M.S.P. (2003) Acquisition of siderophores in gram‐negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116. [DOI] [PubMed] [Google Scholar]
- Ferguson, A.D. and Deisenhofer, J. (2004) Metal import through microbial membranes. Cell, 116, 15–24. [DOI] [PubMed] [Google Scholar]
- Franza, T. , Mahé, B. and Expert, D. (2005) Erwinia chrysanthemi requires a second iron transport route dependent on the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55, 261–275. [DOI] [PubMed] [Google Scholar]
- Fujita, M.J. , Kimura, N. , Sakai, A. , Ichikawa, Y. , Hanyu, T. and Otsuka, M. (2011) Cloning and heterologous expression of the vibrioferrin biosynthetic gene cluster from a marine metagenomic library. Biosci. Biotechnol. Biochem. 75, 2283–2287. [DOI] [PubMed] [Google Scholar]
- Guerinot, M.L. (1994) Microbial iron transport. Annu. Rev. Microbiol. 48, 743–772. [DOI] [PubMed] [Google Scholar]
- Henderson, D.P. and Payne, S.M. (1994) Vibrio‐cholerae iron transport‐systems – roles of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport‐systems. Infect. Immun. 62, 5120–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.M. and Wildermuth, M.C. (2011) The phytopathogen Pseudomonas syringae pv. tomato DC3000 has three high‐affinity iron‐scavenging systems functional under iron limitation conditions but dispensable for pathogenesis. J. Bacteriol. 193, 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, C.M. and Niederweis, M. (2010) Role of porins in iron uptake by Mycobacterium smegmatis . J. Bacteriol. 192, 6411–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammler, M. , Schön, C. and Hantke, K. (1993) Characterization of the ferrous iron uptake system of Escherichia coli . J. Bacteriol. 175, 6212–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewulak, K.D. and Vogel, H.J. (2008) Structural biology of bacterial iron uptake. Biochim. Biophys. Acta, 1778, 1781–1804. [DOI] [PubMed] [Google Scholar]
- Larsen, R.A. , Thomas, M.G. and Postle, K. (1999) Proton motive force, ExbB and ligand‐bound FepA drive conformational changes in TonB. Mol. Microbiol. 31, 1809–1824. [DOI] [PubMed] [Google Scholar]
- Lawlor, M.S. , O'Connor, C. and Miller, V.L. (2007) Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75, 1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.M. , Park, Y.J. , Park, D.S , Kang, H.W. , Kim, J.G. , Song, E.S. , Park, I.C. , Yoon, U.H. , Hahn, J.H. , Koo, B.S. , Lee, G.B. , Kim, H. , Park, H.S. , Yoon, K.O. , Kim, J.H. , Jung, C.H. , Koh, N.H. , Seo, J.S. and Go, S.J. (2005) The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J.M. , Neely, A. , Stintzi, A. , Georges, C. and Holder, I.A. (1996) Pyoverdin is essential for virulence of Pseudomonas aeruginosa . Infect. Immun. 64, 518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke, M. and Marahiel, M.A. (2007) Siderophore‐based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands, J.B. (1981) Microbial iron compounds. Annu. Rev. Biochem. 50, 715–731. [DOI] [PubMed] [Google Scholar]
- Noinaj, N. , Guillier, M. , Barnard, T.J. and Buchanan, S.K. (2010) TonB‐dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke, V. and Long, S.R. (1999) Bacterial genes induced within the nodule during the Rhizobium ± legume symbiosis. Mol. Microbiol. 32, 837–849. [DOI] [PubMed] [Google Scholar]
- Pandey, A. and Sonti, R.V. (2010) Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. J. Bacteriol. 192, 3187–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W. , Jia, Y. , Ren, S.X. , He, Y.Q. , Feng, J.X. , Lu, L.F. , Sun, Q. , Ying, G. , Tang, D.J. , Tang, H. , Wu, W. , Hao, P. , Wang, L. , Jiang, B.L. , Zeng, S. , Gu, W.Y. , Lu, G. , Rong, L. , Tian, Y. , Yao, Z. , Fu, G. , Chen, B. , Fang, R. , Qiang, B. , Chen, Z. , Zhao, G.P. , Tang, J.L. and He, C. (2005) Comparative and functional genomic analyses of the pathogenicity of phytopathogen. Genome Res. 15, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, R. , Javvadi, S. and Chatterjee, S. (2015) Cell–cell signaling promotes ferric iron uptake in Xanthomonas oryzae pv. oryzicola that contributes to its virulence and growth inside rice. Mol. Microbiol. 96, 789–801. [DOI] [PubMed] [Google Scholar]
- Robinson, J.N. and Callow, J.A. (1986) Multiplication and spread of pathovars of Xanthomonas campestris in host and non‐host plants. Plant Pathol. 35, 169–177. [Google Scholar]
- Rondon, M.R. , Ballering, K.S. and Thomas, M.G. (2004) Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology, 150, 3857–3866. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.J. , Potnis, N. , Jones, J.B. , Van Sluys, M.A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T.A. (1989) Molecular Cloning: A Laboratory Manua 2nd ed., Cold Spring Harbor, NY, U.S.A:Cold Spring Harbor Laboratory Press.
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Schaible, U.E. and Kaufmann, S.H.E. (2004) Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.P. (1997) Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, I. , Johnson, E. and de Vries, S. (2003) Microbial ferric iron reductases. FEMS Microbiol. Rev. 27, 427–447. [DOI] [PubMed] [Google Scholar]
- Schwyn, B. and Neilands, J.B. (1987) Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Tanabe, T. , Funahashi, T. , Nakao, H. , Miyoshi, S. , Shinoda, S. and Yamamoto, S. (2003) Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus . J. Bacteriol., 185, 6938–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchia, K. , Mew, T.W. and Wakimoto, S. (1982) Bacteriological and pathological charecteristics of wild types and induced mutants of Xanthomonas campestris pv. oryzae. Phytopathology, 72, 43–46. [Google Scholar]
- Velayudhan, J. , Hughes, N.J. , McColm, A.A. , Bagshaw, J. , Clayton, C.L. , Andrews, S.C. and Kelly, D.J. (2000) Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high‐affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286. [DOI] [PubMed] [Google Scholar]
- Weinberg, E.D. (2009) Iron availability and infection. Biochim. Biophys. Acta, 1790, 600–605. [DOI] [PubMed] [Google Scholar]
- Wiener, M.C. (2005) TonB‐dependent outer membrane transport: going for Baroque? Curr. Opin. Struct. Biol. 15, 394–400. [DOI] [PubMed] [Google Scholar]
- Wiggerich, H.G. and Pühler, A. (2000) The exbD2 gene as well as the iron‐uptake genes tonB, exbB and exbD1 of Xanthomonas campestris pv. campestris are essential for the induction of a hypersensitive response on pepper (Capsicum annuum). Microbiology, 146, 1053–1060. [DOI] [PubMed] [Google Scholar]
- Wiggerich, H.G. , Klauke, B. , Köplin, R. , Priefer, U.B. and Pühler, A. (1997) Unusual structure of the tonB‐exb DNA region of Xanthomonas campestris pv. campestris: tonB, exbB, and exbD1 are essential for ferric iron uptake, but exbD2 is not. J. Bacteriol. 179, 7103–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.J. , Bertrand, N. , Tang, J.L. , Feng, J.X. , Pan, M.Q. , Barber, C.E. , Dow, J.M. and Daniels, M.J. (1998) The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol. Microbiol. 28, 961–970. [DOI] [PubMed] [Google Scholar]
- Worst, D.J. , Gerrits, M.M. , Vandenbroucke‐Grauls, C.M.J.E. and Kusters, J.G. (1998) Helicobacter pylori ribBA‐mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, W.H. IV (2010) “Isolation and Identification of the Siderophore “Vicibactin” Produced by Rhizobium leguminosarum ATCC 14479.” Electronic Theses and Dissertations. Paper 1690. http://dc.etsu.edu/etd/1690
- Yeowell, H.N. and White, J.R. (1982) Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob Agents Chemother. 22, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L. , Angerer, A. and Braun, V. (1989) Mechanistically novel iron(III) transport system in Serratia marcescens . J. Bacteriol. 171, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Schematic presentation of the xss (Xanthomonas siderophore synthesis) gene cluster of Xanthomonas campestris pv. campestris (Xcc) 8004, Xanthomonas oryzae pv. oryzae and Vibrio parahaemolyticus.
Fig. S2 Genetic organization of the Xanthomonas campestris pv. campestris (Xcc) 8004 xss (Xanthomonas siderophore synthesis) cluster indicating the location of the deletions in the knockout strains ΔxssA and ΔxsuA, and pSSP70 insertion in the β‐glucuronidase (GUS) reporter strain Xcc 8004 Pxss::gusA.
Fig. S3 Relative quantification of the expression of downstream genes of the xss (Xanthomonas siderophore synthesis) operon in ΔxssA and ΔxsuA mutants compared with the wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004 strain by real‐time quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR).
Fig. S4 High‐performance liquid chromatography (HPLC) chromatograms of purified xanthoferrin from different Xanthomonas campestris pv. campestris (Xcc) strains.
Fig. S5 Purified vibrioferrin from Vibrio parahaemolyticus rescues the growth deficiency of the ΔxssA mutant under low‐iron conditions.
Fig. S6 Streptonigrin sensitivity assay to assess the intracellular iron content among wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004, ΔxssA, ΔxsuA, ΔxssA/pAP15 and ΔxsuA//pAP15 strains grown in peptone–sucrose (PS) medium and iron‐replete conditions.
Fig. S7 The Xanthomonas campestris pv. campestris (Xcc) 8004 Pxss::gusA reporter strain exhibits wild‐type Xcc 8004‐like phenotypes under low‐iron conditions.
Fig. S8 Assay for bacterial migration inside cabbage leaves and control experiment for conductance.
Fig. S9 Ferric reductase assay of wild‐type Xanthomonas campestris pv. campestris (Xcc) 8004, ΔxssA and ΔxsuA strains.
Fig. S10 The growth of Xanthomonas campestris pv. campestris (Xcc) 8004 (wild‐type), ΔxssA and ΔxsuA strains in different concentrations of 2,2′‐dipyridyl.
Fig. S11 Column buffer control and active fraction of wild‐type Xanthomonas campestris pv. campestris (Xcc) xanthoferrin on peptone–sucrose agar and chromeazurol S (PSA–CAS) plates containing 75 µm 2,2′‐dipyridyl.
Table S1 Homology of proteins encoded in the xss (Xanthomonas siderophore synthesis; involved in siderophore biosynthesis, putative export and uptake) gene cluster of Xanthomonas campestris pv. campestris (Xcc) with those of Vibrio parahaemolyticus, and X. oryzae pv. oryzae (Xoo) KACC103331.
Table S2 List of strains and plasmids used in this study.
Table S3 Generation times of different Xanthomonas campestris pv. campestris (Xcc) strains.
Table S4 List of the primers used in this study.


