Summary
Ustilago maydis is an obligate biotrophic fungal pathogen which causes common smut disease of corn. To proliferate in host tissue, U. maydis must gain access to nutrients and overcome plant defence responses, such as the production of reactive oxygen species. The elucidation of the mechanisms by which U. maydis meets these challenges is critical for the development of strategies to combat smut disease. In this study, we focused on the contributions of phospholipases (PLs) to the pathogenesis of corn smut disease. We identified 11 genes encoding putative PLs and characterized the transcript levels for these genes in the fungus grown in culture and during infection of corn tissue. To assess the contributions of specific PLs, we focused on two genes, lip1 and lip2, which encode putative phospholipase A2 (PLA2) enzymes with similarity to platelet‐activating factor acetylhydrolases. PLA2 enzymes are known to counteract oxidative damage to lipids in other organisms. Consistent with a role in the mitigation of oxidative damage, lip2 mutants were sensitive to oxidative stress provoked by hydrogen peroxide and by increased production of reactive oxygen species caused by inhibitors of mitochondrial functions. Importantly, mutants defective in lip2, but not lip1, were attenuated for virulence in corn seedlings. Finally, a comparative analysis of fatty acid and cardiolipin profiles in the wild‐type strain and a lip2 mutant revealed differences consistent with a protective role for Lip2 in maintaining lipid homeostasis and mitochondrial health during proliferation in the hostile host environment.
Keywords: corn, fungal pathogenesis, maize, mitochondria, smut disease
Introduction
An understanding of the mechanisms involved in host recognition, colonization, nutrient acquisition and survival is critical for the development of targeted strategies to combat crop losses caused by fungal phytopathogens. Phospholipases (PLs), which catalyse the hydrolysis of ester or phosphodiester bonds of glycerophospholipids, are known to contribute to the virulence of some fungal pathogens. For example, fungal pathogens of humans, such as Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus, use PLs to acquire nutrients, facilitate invasion of host cells or modulate lipid‐mediated immune responses in the host (Ghannoum, 2000; Kohler et al., 2006). PLs in fungal pathogens of plants function in morphological transitions, such as appressorium formation in Magnaporthe oryzae or conidia formation and germination in Alternaria alternata (Choi et al., 2011; Kohler et al., 2006; Rho et al., 2009; Tsai and Chung 2014).
In addition to roles in virulence and morphogenesis, fungal PLs have been shown to counteract damage caused by oxidative stress by removing oxidized fatty acids from phospholipids in membranes. For example, overexpression of the Schizosaccharomyces pombe PL plg7 in Saccharomyces cerevisiae resulted in increased resistance to oxidative stress caused by CuSO4 (Foulks et al., 2008). This PL was also identified as a suppressor of cell death in Sc. pombe strains engineered to produce high levels of ricinoleic acid (RA), a hydroxylated fatty acid capable of disrupting membrane function (Yazawa et al., 2013a,b, 2014). The PL encoded by a homologue of plg7 in Trichoderma harzianum is also a stress‐responsive protein. Deletion mutants for the gene in T. harzianum exhibit slow growth, sensitivity to oxidative stress caused by hydrogen peroxide (H2O2) and decreased levels of eicosanoids and ergosterol (Yu et al., 2014). The ability to deal with oxidative stress caused by reactive oxygen species (ROS) is an important attribute of a successful plant pathogen because the oxidative burst is an early plant defence response. This process involves the production of ROS at the site of infection via host enzymes, such as NADPH‐oxidase, superoxide dismutase, oxalate oxidase, peroxidase, lipoxygenase or amine oxidase (Jones and Dangl, 2006; Shetty et al., 2008). ROS act to strengthen the host cell wall and function as second messengers to induce programmed cell death and the synthesis of pathogen‐related proteins and phytoalexins (Bradley et al., 1992; Zurbriggen et al., 2009). ROS are also potent antimicrobial agents capable of inducing lipid peroxidation in membrane lipids, leading to cell damage (Farmer and Mueller, 2013).
The ability to suppress or overcome the host defence response, including the presence of ROS, is important for the virulence of Ustilago maydis, the causative agent of corn smut and an important model organism for the study of obligate biotrophism (Banuett and Herskowitz, 1996; Bolker et al., 1995; Dean et al., 2012; Ruiz‐Herrera et al., 1995). For example, the transcription factor Yap1 is a major regulator of the oxidative stress response in U. maydis. This protein controls the expression of a number of ROS‐detoxifying enzymes and low‐molecular‐weight antioxidants, and mutants lacking this gene exhibit both increased sensitivity to oxidative stress and reduced virulence (Molina and Kahmann, 2007). Ustilago maydis also employs the secreted effector Pep1 to suppress the host defence response. Pep1 localizes in the host apoplast and to sites of cell‐to‐cell passage, and inhibits the host peroxidase POX12 (Doehlemann et al., 2009; Hemetsberger et al., 2012).
A morphological transition from budding to filamentous growth is also a key aspect of virulence for U. maydis, and this switch occurs as a result of mating of haploid yeast cells on the plant surface to yield an infectious dikaryon capable of penetrating the host surface (Banuett and Herskowitz, 1996; Lanver et al., 2014). Other factors are also known to trigger filamentous growth, including low pH, low nitrogen, phosphate limitation and exposure to fatty acids (Boyce et al., 2006; Klose and Kronstad, 2006; Klose et al., 2004; Kretschmer et al., 2012; Lanver et al., 2014; Lovely and Perlin, 2011; Ruiz‐Herrera et al., 1995). During infection, filamentous growth occurs in and between host cells and fungal mass accumulates in parallel with the formation of large tumours on aerial parts of the plant. The tumours contain diploid teliospores which, after dissemination into the environment, germinate to yield haploid progeny (Banuett and Herskowitz, 1996).
Given the role of PLs in counteracting oxidative stress, and the importance of lipids in the morphogenesis of U. maydis, we initiated an investigation of PLs by the identification of genes for 11 candidate enzymes in the U. maydis genome. We characterized the expression of these genes and focused a detailed study on two PLs, Lip1 and Lip2, with sequence similarity to the platelet‐activating factor acetylhydrolase (PAFAH) subclass of phospholipase A2s (PLA2s). Strains containing a deletion for lip2, but not lip1, were less virulent on corn seedlings, and more sensitive to oxidative stress and ROS generated by inhibitors of mitochondrial function. These findings and a comparative analysis of fatty acid and cardiolipin profiles support the conclusion that Lip2 functions in the maintenance of lipid homeostasis and mitochondrial function during proliferation in the hostile host environment.
Results
The U. maydis genome encodes 11 putative PLs
In this study, we focused on PLs because our previous work revealed that lipids influence the filamentous growth of U. maydis, and both mitochondrial and peroxisomal β‐oxidation contribute to the ability of the fungus to cause disease in corn seedlings (Durrenberger et al., 2001; Klose and Kronstad, 2006; Klose et al., 2004; Kretschmer et al., 2012). We initially searched the U. maydis genome to identify genes encoding potential PLs. Specifically, PLs previously identified in S. cerevisiae were used as queries for blastp searches against the predicted U. maydis proteome (Table S1, see Supporting Information). These searches identified candidate genes for the PLs of the A2, B, C and D families of enzymes, and the corresponding sequences were then used as input queries for domain‐enhanced lookup time accelerated‐blast (delta‐blast) searches of the U. maydis genome. These searches identified 11 genes encoding putative PLs, including more than one member of each main PL family. The features of these putative PLs are summarized in Table S2 (see Supporting Information), and include an analysis of the predicted localization of the enzymes. Two genes were predicted to encode PLBs (um01035/um11266 and um05871); both genes encode polypeptides with a predicted lysophospholipase catalytic domain (IPR002642), and um05871 also contains a 4′‐phosphopantetheinyl transferase domain (IPR008278). Three genes were identified which encode candidate PLCs (um00004, um02982 and um01895/um10263). A PLC‐like phosphodiesterase, TIM beta/alpha‐barrel domain (IPR017946), was identified in um00004, whereas both um02982 and um01865/um11266 contain a PLC, phosphatidylinositol‐specific X and Y domain (IPR000909 and IPR001711). The um02982 protein also contained a pleckstrin homology‐type, EF‐hand‐like domain (IPR015359) and a C2 calcium‐dependent membrane‐targeting domain (IPR000008). Three genes were identified that encode potential PLDs (um00370, um01120 and um06066). All three genes contain a PLD/transphosphatidylase domain (IPR001736); um00370 also contains a phox‐like domain (IPR001683) and pleckstrin‐like domain (IPR001849).
The three genes predicted to encode putative PLAs (um00133, um01927 and um01017) were of particular interest, given the role of PLA2 enzymes in resistance to oxidative stress in other organisms (Foulks et al., 2008; Yu et al., 2014). Domain analysis for the three candidate PLA2 enzymes identified a patatin/PLA2‐related domain (IPR002641) in the gene um01017. The other two enzymes, encoded by um00133 (designated lip1) and um01927 (designated lip2), both contain a conserved PAFAH domain (IPR005056). Lip2 also has a predicted haloacid‐dehydrogenase (HAD)‐like domain (IPR023214). Further examination revealed a conserved GXSXG lipase sequence in all three PLA2 proteins, as well as conservation of the amino acids Ser343/289 and Asp366/313 for Lip1 and Lip2, respectively; these residues are critical for enzyme activity in esterases and lipases (Arai et al., 2002; Wei et al., 1998). Overall, the domain conservation and sequence features suggest that Lip1 and Lip2 are PLA2 orthologues.
We next performed quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) measurements of transcript levels for the candidate PLs using RNA isolated from budding cells, from a mixture of mating cells in culture to represent the filamentous dikaryon, from a mixture of the cells used to inoculate plants (day 0), and from infected corn tissue at 1 and 12 days post‐inoculation (dpi) (Fig. 1). This analysis revealed low transcript levels for the PLA2 encoded by the lip1 (um00133) and lip2 (um01927) genes in the mating mixture relative to both budding cells and the day 0 time point. In contrast, the transcript levels of lip1 were high at day 1 of infection, whereas the transcript levels of lip2 were low at this time and at day 12. The transcript levels of other PLs shared similar patterns with each other, such that higher expression was observed in both cells from the mating mixture and from infected tissue, compared with budding cells; the one exception was the gene um05871 which showed low transcript levels in the mating mixture. There were no obvious trends in the transcript levels for each distinct family of PLs, but some specific candidate PL genes were more highly expressed at day 1 than at other times [um00133 and um01017 (PLAs), um01035 (PLB), um00004 and um10263 (PLCs) and um00370 and um01170 (PLDs)]. The um05871 (PLB), um02982 (PLC) and um06066 (PLD) genes showed increasing transcript levels as infection progressed. Overall, the distinct pattern of low expression in the mating mixture and increased transcript levels at the initial stage of interaction, together with the known role of PLA2 enzymes in response to oxidative stress, prompted us to begin our genetic characterization of PL genes with a focus on lip1 and lip2.
Figure 1.
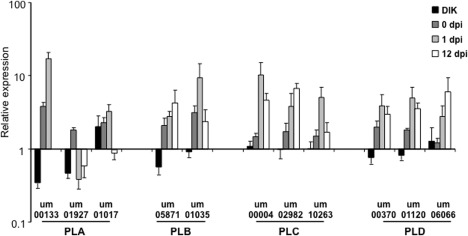
Transcript levels of candidate phospholipases (PLs) change during growth in culture and in planta for Ustilago maydis. Real‐time polymerase chain reaction analysis of putative PL genes using RNA isolated from the filamentous dikaryon (DIK), the mating culture used for infection [0 days post‐inoculation (dpi)] and infected maize tissue at early (1 dpi) and late (12 dpi) stages of infection relative to expression values in budding haploids. The control genes were actin and glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH). Three biological replicates were performed. Standard errors are shown. The 12‐dpi sample for gene um00133 did not show expression in any of the three replicates, and thus the fold change could not be determined.
Deletion of lip2, but not lip1, attenuates virulence
Single and double gene deletion mutants were constructed to assess the roles of Lip1 and Lip2. Two independent mutants were obtained for both lip1 and lip2 in each mating‐type background (001 a2 b2 and 002 a1 b1) (Fig. S1 and Table S3, see Supporting Information). In addition, a lip1 lip2 double mutant was constructed in each strain of opposite mating type. Alleles of the deletions are designated as Δlip1, Δlip2 and Δlip1 Δlip2. Furthermore, lip2 was deleted in the solopathogenic haploid strain SG200, which is capable of bypassing the requirement for mating to cause disease (Bolker et al., 1995). Correct integration of the gene deletion constructs was confirmed by PCR for all of the strains and, given that only lip2 mutants showed phenotypes, the deletion of lip2 was also confirmed via Southern hybridization to ensure integration at a single locus; the resulting strains were designated Δlip2 A3‐3, C3‐3, A2‐1 and B3‐3, and Δlip1 Δlip2 A4‐2, C5‐2, A3‐2 and C2‐1 (Fig. S1 and Table S3). We noted that strain B3‐3 had an additional ectopic band, and this mutant was therefore not used in further analyses. Deletion of lip1 or lip2 did not influence the ability of the strains to participate in mating with compatible wild‐type or mutant strains (Fig. S2, see Supporting Information). The mutants also did not show growth or morphological defects on solid or liquid media compared with the wild‐type strain (Fig. S3, see Supporting Information).
To examine the contributions of Lip1 and Lip2 to virulence, 7‐day‐old corn seedlings were infected with a mating culture containing sexually compatible crosses of wild‐type and mutant strains, and disease symptoms were assessed at 14 dpi. Seedlings infected with the mating‐compatible Δlip2 × Δlip2 mutants exhibited a reduction in the severity of disease symptoms, including a higher proportion of healthy plants and a lower proportion of severe symptoms, such as tumour formation (Fig. 2). Deletion of lip1 had no observable effect on the virulence of U. maydis because seedlings infected with mating‐compatible Δlip1 × Δlip1 strains exhibited a similar severity of disease symptoms to those infected with the wild‐type 001 × 002 mixture. Furthermore, strains containing deletions in both lip1 and lip2 exhibited a reduction in disease symptoms similar to the Δlip2 deletion mutant, indicating that lip1 does not make a contribution to virulence, even in the context of a lip2 deletion (Fig. 2). We did notice that the combination of the Δlip2 deletion mutant with the wild‐type strains (Fig. 2B) reduced virulence to a greater extent than the combination of the double mutant with the wild‐type strains. The reason for this is not clear, but may suggest a compensatory influence of the loss of lip1 in the background of a Δlip2 deletion mutant. We also found that the solopathogenic SG200 strain containing a deletion of lip2 exhibited a similar reduction in disease severity, indicating that this phenotype is independent of mating (Fig. S4, see Supporting Information). Together, these data indicate a role for Lip2 in the virulence of U. maydis, although we note that the Δlip2 mutants still cause substantial disease and this may be a result of redundant contributions of the other PLs.
Figure 2.
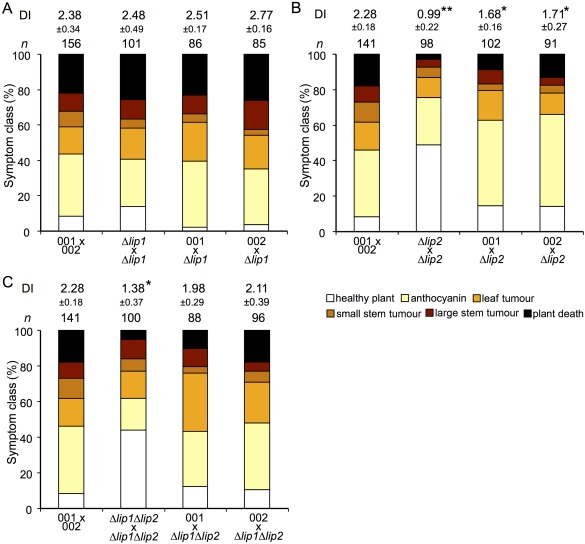
Loss of Lip2 attenuates the virulence of compatible haploid strains. Virulence assays were performed with 7‐day‐old maize seedlings (Golden Bantam) infected with sexually compatible mating cultures of the wild‐type strain or mutants for Δlip1 (A), Δlip2 (B) or the Δlip1 Δlip2 double mutant (C). Disease symptoms were scored at 14 days post‐inoculation. The disease index (DI) was calculated based on the frequency of symptoms, including plant death (score 5), large stem tumours (score 4), small stem tumours (score 3), leaf tumours (score 2), anthocyanin (score 1) and healthy plants (score 0). Biological replicates were performed for each experiment and the standard deviations for the DIs are shown. t‐tests were performed to identify significantly different DIs compared with the wild‐type strain (*P < 0.05; **P < 0.01).
Deletion of lip2 increases susceptibility to oxidative stress
We hypothesized that the virulence defect in Δlip2 mutants was caused by their increased susceptibility to oxidative stress, perhaps generated as part of the early plant defence response to infection. This would be consistent with the role of PAFAHs in the response to oxidative stress in fungi and other organisms (Foulks et al., 2008; Rosenson and Stafforini, 2012; Yazawa et al., 2013a, 2013b; Yu et al., 2014). We therefore assessed the susceptibilities of the Δlip1 and Δlip2 mutants to oxidative stress caused by H2O2. Strains containing a deletion in lip2, but not lip1, exhibited greater susceptibility to H2O2, as indicated by the increase in the size of the zone of clearing (Fig. 3). The greater susceptibility of the mutants carrying the Δlip2 deletion was also observed with two and five times lower levels of H2O2. Overall, these results indicate that Lip2 is important for the tolerance of the fungus to oxidative stress as provoked by H2O2.
Figure 3.
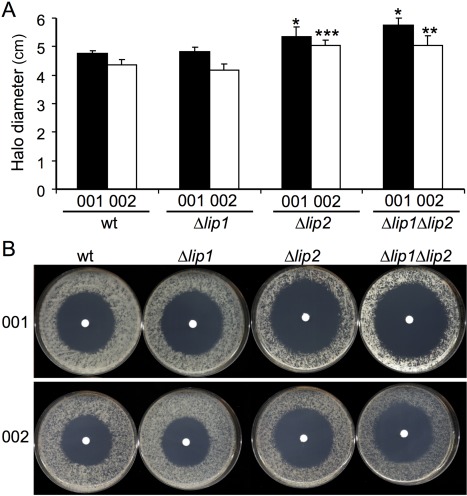
Lip2 mutants are susceptible to oxidative stress. The wild‐type (wt) strain and the indicated mutants were plated onto potato dextrose agar (PDA) and incubated with 5 µL H2O2 for 48 h. The size of the zone of clearance from the plates (B) was measured and averaged (A). Four biological replicates were performed and the standard deviations are shown. Significant differences from the respective wt strains are indicated with an asterisk (*P < 0.05, **P < 0.01, ***P < 0.001).
The increased susceptibility of the lip2 deletion mutant to H2O2 is consistent with known roles for PLA2 enzymes in other organisms (Foulks et al., 2008; Yu et al., 2014), and this observation prompted a more detailed examination of the source of the oxidative stress. However, the mutants did not show altered susceptibility to other sources of oxidative stress, including CuSO4 and tert‐butyl hydroperoxide (data not shown). The relatively minor phenotype on H2O2, compared with the much larger influence of H2O2 on other mutants of U. maydis (e.g. yap1; Molina and Kahmann, 2007), could be a result of redundancy with other enzymes, and may also suggest that the source of oxidative stress might be endogenous. In this context, the electron transport chain is known to generate ROS. We therefore tested inhibitors of mitochondrial function, including carbonyl cyanide m‐chlorophenyl hydrazone (CCCP) (an uncoupling agent that abolishes the link between respiration and phosphorylation) and rotenone (an inhibitor of the respiratory chain). With this approach, we observed increased susceptibility of the lip2 mutant to rotenone and CCCP, suggesting that the source of ROS may be endogenous and related to mitochondrial function (Fig. 4). Although we did not examine mitochondrial function directly in the lip2 mutant, our observations are consistent with the known roles of PLA2 enzymes in the maintenance of mitochondrial integrity (Ramanadham et al., 2015).
Figure 4.
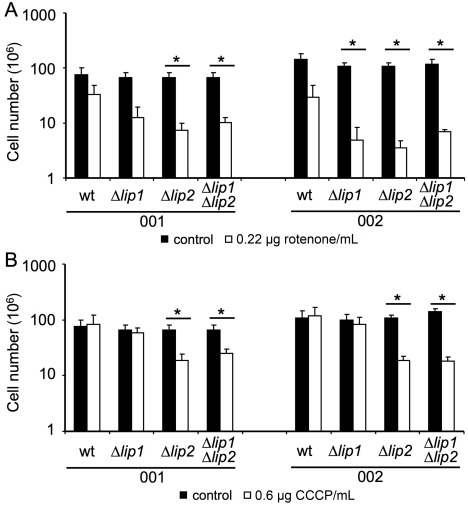
Lip2 mutants are susceptible to inhibitors of respiration. The wild‐type (wt) strains and the Δlip1, Δlip2 and Δlip1 Δlip2 mutants in both haploid backgrounds were grown in minimal medium (MM) with or without rotenone (A) or carbonyl cyanide m‐chlorophenyl hydrazone (CCCP) (B). Strains were pre‐grown in potato dextrose broth (PDB), washed, counted and 106 cells were inoculated into 5 mL of MM (pH 7) with 1% glucose as the carbon source. Rotenone (A) and CCCP (B) were added as dimethylsulfoxide (DMSO) stocks to reach final concentrations of 0.22 µg/mL and 0.6 µg/mL, respectively. Cells were incubated at 30°C for 24 h at 220 rpm, and the cell number was determined. The test was repeated three times and the standard deviations are shown. A t‐test was used to identify significantly different values between mutants and wt (*P < 0.05). The filled bars indicate cultures without drug and the open bars indicate cultures with drug.
We also tested the mutants for a wide range of additional phenotypes, including the effects of the vacuole‐accumulating drugs chloroquine and quinacrine, because of speculation that the mode of action of chloroquine against Plasmodium falciparum involves ROS (although this idea has been controversial) (Lehane et al., 2012; Monti et al., 2002). These drugs have been reported to have broad‐spectrum toxicity against viruses, bacteria and fungi, and are reported to be inhibitors of PLA2s (Farooqui et al., 2006; Rolain et al., 2007). We noted that strains containing deletions in lip2 were more susceptible to both chloroquine and quinacrine, as indicated by a reduction in growth. However, this phenotype was most obvious in the 001 (a2 b2) background, but not for mutants in the 002 (a1 b1) background (data not shown). We also examined the susceptibility of Δlip2 mutants to stress caused by the presence of 1 m NaCl or 10 mm LiCl, and found that strains lacking Lip2 showed increased resistance to these stressors (data not shown). Taken together, these results suggest that Lip2 may function more broadly in counteracting ionic stress.
Analysis of the influence of Lip2 on lipid composition
The lip2 mutant phenotypes associated with oxidative stress and mitochondrial functions led us to speculate that Lip2 may be involved, either indirectly or directly, in the maintenance of lipid homeostasis and membrane integrity in response to endogenous ROS. In addition, defects in PAFAHs in other fungi cause alterations to lipid profiles, such as a reduction in eicosanoic acid and ergosterol in T. harzianum, as well as a reduction in membrane phospholipid‐associated RA moieties in Sc. pombe strains overexpressing plg7 (Yazawa et al., 2014; Yu et al., 2014). We therefore hypothesized that the deletion of lip2 would affect the lipid composition of the cell. As shown in Fig. 5, analysis of fatty acid methyl esters (FAMEs) in the wild‐type strain and the lip2 mutant revealed differences in lipid composition, including a notable increase in the proportion of 18:2 (linoleic acid) as a percentage of total fatty acids in the mutant. Furthermore, changes in the proportion of additional FAMEs were detected in the Δlip1 Δlip2 mutant relative to the wild‐type strain, and these included a decrease in 18:0, 18:1 and 20:0 fatty acids, and an increased proportion of 18:2. These results support the conclusion that Lip2 influences fatty acid composition and suggest that Lip1 may also have a minor influence on lipid homeostasis in U. maydis.
Figure 5.
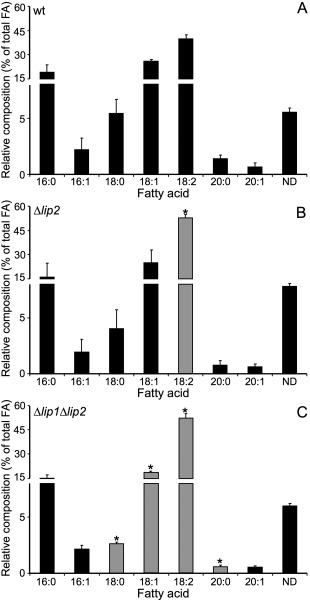
Loss of Lip2 alters the fatty acid composition of Ustilago maydis cells. Lipid profiles for cells of the wild‐type (wt) strain (A), the Δlip2 mutant (B) and the Δlip1 Δlip2 mutant (C) were measured by gas chromatography with flame ionization detection of the fatty acid methyl ester derivatives. The analysis was repeated three times with multiple injections. The standard deviations are shown and statistically significant differences compared with the wt strain are indicated with an asterisk (* P < 0.05) and highlighted with grey bars. A t‐test with Holm–Sidak correction was used for statistical analysis. FA, fatty acid; ND, not identified.
Given the influence of CCCP and rotenone on the growth of the lip2 mutant, we hypothesized that Lip2 may also participate in the maintenance of fatty acyl chain homeostasis and remodelling of cardiolipins in mitochondria. We therefore performed a targeted analysis of cardiolipin composition in the wild‐type strain and the lip2 deletion mutant, and observed an increase in the proportion of the 72:8 cardiolipin molecular species and a decrease in the proportion of the 72:7 cardiolipin species in the mutant (Fig. 6). Interestingly, this result is consistent with the increase in the 18:2 fatty acyl group detected in the FAME analysis of total lipids, given that the 72:8 cardiolipin would be expected to contain four 18:2 fatty acyl groups (compared with three 18:2 and one 18:1 for the 72:7 species). Taken together, these analyses reveal an impact of lip2 deletion on fatty acid and cardiolipin composition consistent with a role in supporting mitochondrial function.
Figure 6.
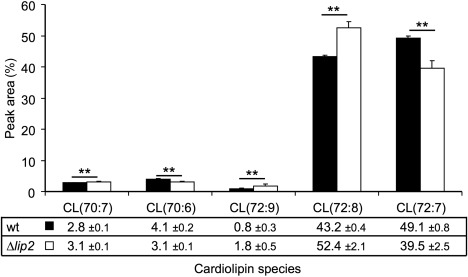
Loss of Lip2 alters the cardiolipin (CL) composition of Ustilago maydis cells. CL profiles were obtained for the wild‐type (wt) strain and the Δlip2 mutant by liquid chromatography‐mass spectrometry (LC‐MS) analysis. The predicted compositions of the CLs are labelled below the graph based on the predicted acyl chains and double bonds (e.g. 72:7). For the two most abundant species, 72:7 is predicted to have three linoleic acid chains (18:2) and one oleic acid chain (18:1), whereas the 72:8 species is predicted to have four linoleic acid chains. The percentage of each of the indicated CL species is also indicated below the graph. The analysis was repeated with multiple injections. Injections were analysed in the (M + H)+ and (M + NH4)+ modes, and the average of the technical replicates was calculated for each mode and each biological replicate. The overall averages for the wt strain and the mutant and the standard deviations are shown. A t‐test was used to identify significant differences between wt (filled bars) and mutant (open bars) (**P < 0.01).
Discussion
Biotrophic fungal pathogens, such as smut, bunt and rust fungi, are economic threats to cereal crop production (Fisher et al., 2012). An understanding of the pathogen requirements for proliferation in host tissue and subsequent disease is critical for the design of strategies to protect crops. In this study, we predicted that PLs play important roles in biotrophic disease based on previous studies on lipid signalling and metabolism in U. maydis and studies in other fungal pathogens (Choi et al., 2011; Klose and Kronstad, 2006; Klose et al., 2004; Kohler et al., 2006; Kretschmer et al., 2012; Rho et al., 2009; Tsai and Chung, 2014). We identified candidate PLs and focused specifically on two PLA2 enzymes, Lip1 and Lip2, to establish their roles in virulence. Our main findings were that Lip1 had little influence, but that Lip2 played a role in virulence, resistance to ROS, lipid homeostasis and the maintenance or support of mitochondrial function. These roles for Lip2 are supported by the following phenotypes of the lip2 mutant: (i) increased susceptibility to H2O2; (ii) increased susceptibility to inhibitors of mitochondrial respiration and oxidative phosphorylation; and (iii) altered ratio of 18:1 versus 18:2 fatty acids, including changes in cardiolipins (presumably associated with mitochondria and possibly influencing respiration).
Based on our findings, we speculate that Lip2 normally functions, at least in part, to remove polyunsaturated acyl chains from phospholipids and cardiolipins to maintain the appropriate stoichiometry of monounsaturated versus polyunsaturated fatty acyl moieties in these compounds. It is known that phospholipids with sn‐2 polyunsaturated fatty acyl residues are targets of oxidizing radicals that can be pro‐apoptotic or perturb membranes (Foulks et al., 2008). In the absence of Lip2, polyunsaturated fatty acids may accumulate in membrane lipids and serve as targets of oxidative radicals, leading to membrane damage. This would account for the increased susceptibility of the lip2 deletion mutant to H2O2, as well as the potential challenges to mitochondrial function by the inhibitors that may provoke ROS accumulation to damage polyunsaturated acyl chains in cardiolipins. In particular, rotenone promotes ROS accumulation through the inhibition of complex I of the mitochondrial respiratory chain, and the oxidative phosphorylation uncoupler CCCP is also known to generate ROS (Izeradjene et al., 2005; Li et al., 2003).
In addition to internally generated ROS, it is also possible that Lip2 plays a protective role against the oxidative stress encountered on host colonization by removing detrimental oxidized polyunsaturated fatty acids from the cell membrane, mitigating the damage caused by plant ROS‐triggered lipid peroxidation. Repair functions of PLA2s have been reported and lipid peroxidation is known to stimulate PLA2 activity (McLean et al., 1993; Rosenson and Stafforini, 2012; Sevanian et al., 1981; Tjoelker et al., 1995). For example, plg7 in Sc. pombe has been shown to protect the cell by cleaving hydroxylated fatty acid moieties, such as RA, that are known to damage cell membranes (Yazawa et al., 2013a,b, 2014). We noted that the lip2 mutant, however, does not exhibit as severe a phenotype as previously identified factors that appear to play protective roles in U. maydis to withstand the host oxidative burst. These factors include the transcription factor Yap1, which is a major regulator of the oxidative stress response in U. maydis, and the secreted effector Pep1, which inhibits the host peroxidase POX12 (Doehlemann et al., 2009; Hemetsberger et al., 2012; Molina and Kahmann, 2007). We also found that the virulence defect of the lip2 mutant was not restored by the addition of ascorbate or diphenyleneiodonium (DPI), and pretreatment of cells with H2O2 prior to inoculation did not exacerbate the virulence defect (data not shown). It appears to be more likely that Lip2 is important for supporting lipid homeostasis during the metabolic requirements needed by U. maydis to proliferate in host tissue. Of course, it is possible that Lip2 makes a contribution at both the initial stages of infection and during later proliferation.
Given the increased susceptibility of the lip2 mutant to inhibitors of respiration, Lip2 may indirectly support a role for mitochondrial function by influencing the integrity of cardiolipins in the inner membrane. In turn, part or all of the attenuated virulence of the lip2 mutant may result from mitochondrial dysfunction that impairs pathogen growth in host tissue. It is known that mitochondria contribute to the virulence of fungal pathogens of animals, and mitochondrial defects also influence the ability of phytopathogenic fungi to cause disease (An et al., 2015; Chatre and Ricchetti, 2014). A mitochondrial location is not predicted for the Lip2 protein, but the enzyme may act by influencing the fatty acid composition in the cytoplasm or in the remodelling of cardiolipins (Baile et al., 2013).
In conclusion, our results suggest that the initial adaptation of U. maydis during the early stages of proliferation in host tissues involves a requirement to maintain lipid homeostasis. The growth of the fungus in the host may pose a specific challenge to lipid integrity related to the generation of endogenous ROS via mitochondrial activity. The influence of Lip2 on lipid homeostasis provides a novel insight into the metabolic functions required for the virulence of U. maydis that may reflect a conserved feature of fungal biotrophy.
Experimental Procedures
Strains and growth conditions
Mutants were derived from the U. maydis strains 001 (also designated 518, mating type a2 b2) and 002 (also designated 521, mating type a1 b1) (Table S3). Strains were grown at 30°C in potato dextrose broth (PDB), minimal medium (MM) or double complete medium (DCM) with 2% agar in solid plates when applicable.
Bioinformatic and statistical analyses
Putative PLs in the U. maydis genome were identified using a blastx search employing sequences from S. cerevisiae, and the results were then used as query in a reciprocal delta‐blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Table S1).
All analyses were carried out in biological triplicates unless stated otherwise and standard deviations are shown unless stated otherwise. The data were analysed for statistical differences with t‐tests. Significance levels are *P < 0.05, **P < 0.01 and ***P < 0.001.
Construction of deletion mutants
An overlap PCR strategy was used to delete the genomic sequence coding for the genes lip1 (um00133) and lip2 (um01927) employing the primers listed in Table S4 (see Supporting Information) to replace the wild‐type open reading frame with a resistance marker. Briefly, ∼1‐kb sequences of upstream and downstream sequence, together with a selectable marker, were amplified and fused using overlap PCR. The deletion construct was PCR amplified using nested primers and used to transform U. maydis cells via biolistic transformation, as described in Toffaletti et al. (1993). Transformants were grown on DCM supplemented with 0.75 m sorbitol, followed by multiple rounds of selection on DCM containing 250 µg/mL hygromycin B or 100 µg/mL nourseothricin. Transformants were confirmed by PCR and Southern hybridization. Two independent transformants in each mating type were used for subsequent experiments. Gel electrophoresis, restriction enzyme digestion and Southern hybridization were performed using standard procedures (Sambrook et al., 1989).
Virulence assays
The ability of U. maydis strains to cause disease was assessed by inoculation of 7‐day‐old maize seedlings (Golden Bantam) with a mixture of sexually compatible haploid pairs of wild‐type and mutant strains at a cell concentration of 1 × 107 cells/mL (5 × 106 cells/mL of each haploid strain). Disease symptoms were evaluated at 14 dpi as described previously (Klose and Kronstad, 2006).
RNA extraction and quantitative real‐time PCR
To isolate RNA from budding yeast cells, cells grown overnight in PDB were centrifuged, washed in sterile water and approximately 108 cells were homogenized using glass beads in a bead mill for 3 min in 60‐s intervals, followed by cooling on ice. RNA from dikaryotic cells was obtained by co‐spotting equal numbers of 001 and 002 cells onto DCM agar plates containing 1% activated charcoal and scraping filamentous colonies after 48 h of growth at room temperature. Dikaryotic cells were homogenized by grinding in liquid nitrogen with a mortar and pestle. For the infection time course, cells of the 001 and 002 strains were grown overnight in PDB, washed twice in water and diluted in equal numbers to 108 cells/mL. Cells for the 0‐dpi time point were treated with the same methods as used for RNA isolation for the budding yeast cells described above. The 1‐dpi time point was obtained after spotting 100 µL of mating culture on 22‐day‐old leaves, and 7‐day‐old seedlings were inoculated with the mating culture as described above, and subsequently harvested at the 12‐dpi time point. Approximately 100 mg of infected tissue (fresh weight) were collected, immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation.
RNA for RT‐PCR was isolated using a low‐pH RNA extraction method (Kretschmer et al., 2014). Briefly, for 50 mg of tissue or 108 cells, samples were homogenized in 1 mL of cell lysis reagent [2% sodium dodecylsulfate (SDS), 68 mm tri‐sodium citrate dehydrate, 132 mm citric acid, 10 mm ethylenediaminetetraacetic acid (EDTA)], followed by the addition of 340 µL of protein‐DNA precipitation reagent (4 m NaCl, 17 mm tri‐sodium citrate dehydrate, 33 mm citric acid). Samples were incubated on ice for 5 min and centrifuged for 10 min at maximum speed (Gentra Systems, Inc., Qiagen, Toronto, Ontario, Canada). The supernatant was transferred to a clean tube and RNA was extracted using phenol–chloroform, as described previously (Sambrook et al., 1989). RNA was DNase treated using TURBO DNase following the manufacturer's instructions (Life Technologies, Burlington, Ontario, Canada).
DNase‐treated total RNA was used as template for cDNA synthesis employing a Verso cDNA first strand synthesis kit (Thermo Fisher Scientific, Burlington, Ontario, Canada) and oligo(dT) primers. SYBR green PCR master mix (Applied Biosystems, Burlington, Ontario, Canada) was used for real‐time PCRs in 20‐µL volumes employing the primers listed in Table S4. Gene expression was calculated relative to the constitutive expression of U. maydis glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) and actin genes, as described previously (Kretschmer et al., 2014).
Susceptibility of PLA2 mutants to inhibitors and agents that provoke stress
To assess susceptibility to oxidative stress, 1 × 104 cells were plated onto potato dextrose agar (PDA) together with a 5‐mm piece of Whatmann paper (Fisher Scientific, Burlington, Ontario, Canada) soaked with 5 µL of H2O2. Plates were incubated at 30°C for 48 h and the zone of clearance was measured. Susceptibility to rotenone and CCCP was tested by inoculating 1 × 106 cells into 5 mL of MM (pH 7.0) containing either 0.22 μg/mL of rotenone or 0.60 μg/mL of CCCP, and monitoring growth after 24 h at 30°C with shaking. Rotenone and CCCP were dissolved in dimethylsulfoxide (DMSO). DMSO was added to the control samples at the same concentration.
Fatty acid analysis
For fatty acid analysis, cell pellets were resuspended in 2 mL of 1 M methanolic hydrochloric acid and heated at 80°C for 2 h to transmethylate fatty acids. FAMEs were then extracted in 2 mL of hexane, using 1 mL of 0.9% sodium chloride to improve phase separation, and the hexane phase was dried under a nitrogen stream. To facilitate the separation of hydroxylated FAMEs by gas chromatography, hydroxyl groups were silylated by heating FAMEs in 10 μL pyridine and 10 μL N,O‐bis(trimethylsilyl)trifluoroacetamide with 1% trimethylchlorosilane at 80°C for 1 h. The derivatized FAMEs were again dried under a nitrogen stream, and then resuspended in 50 μL of hexane. FAMEs were separated and relative quantities were determined using gas chromatography coupled with flame ionization detection (FID). For this analysis, an Agilent 6890 Plus System (Mississauga, Ontario, Canada) with a DB‐23 capillary column was employed, based on the protocol described by Kunst et al. (1992). The injection was made with a 10 : 1 split. Separation was carried out at an initial oven temperature of 180°C for 5 min, followed by an increase at 0.5°C/min to 190°C, then an increase at 10°C/min to 240°C, and maintained for a further 5 min. The injector was set to 280°C, and the FID was set to 300°C. H2 was used as a carrier gas with a flow rate of 40 mL/min, air flow was 450 mL/min and He was supplied as a make‐up gas for FID at a rate of 20 mL/min. Peaks were identified based on the comparison of retention times with standards and seed oil of Arabidopsis thaliana. The analysis was repeated three times with independent samples.
Cardiolipin analysis
Cells were resuspended in 200 µL of cold 50% methanol (containing 0.5 mg/mL butylated hydroxytoluene) and lysed by shaking on a MM400 mill mixer with 5‐mm metal balls (2 × 1 min). Then, 600 µL of methanol and 200 µL of chloroform were added, followed by sonication for 5 min on ice. The mixture was centrifuged, the supernatant was collected and the pellet was resuspended in 400 µL of chloroform for further extraction. The supernatants were pooled and partitioned into aqueous and organic phases with a water–methanol–chloroform mixture (1 : 2 : 0.75, v/v/v). The organic phase was dried and subsequently dissolved in 200 µL of methanol for liquid chromatography‐mass spectrometry (LC‐MS) analysis on a Dionex Ultimate 3400 RSLC coupled to a Thermo Linear Trap Quadrupole (LTQ) Orbitrap Velos mass spectrometer operated with electrospray ionization (ESI) and in the positive‐ion detection mode (Thermo Fisher, Burlington, Ontario, Canada). The LC‐MS analysis was performed over m/z 200–1800 with detection in positive‐ion Fourier‐transformed mass spectrometry (FTMS) mode at 60 000 full width at half‐maximum (FWHM) (at m/z 400). Lock‐mass internal calibration was employed during LC‐MS to ensure mass accuracy. Separation of cardiolipin from other lipids was achieved with a hydrophilic interaction chromatography‐ultraperformance liquid chromatography (HILIC‐UPLC) column using ammonium acetate (solvent A) and methanol (solvent B) as the mobile phase for binary solvent gradient elution.
The identification of cardiolipins on the LC‐MS chromatograms was achieved by: (i) comparison of retention times with that of cardiolipin (18:1); (ii) searching the LIPID MAPS database with the accurately measured m/z values (mass errors < 3 ppm) of the detected lipids; and (iii) confirmation of the assignments with the aid of MS/MS acquired with on‐line collision‐induced dissociation. No oxidized cardiolipins were detected in any of the samples. The linear range of MS detection was over a concentration range of >1000‐fold concentrations. A standard curve for quantification was prepared as the total peak areas of the (M + H)+ and (M + NH4)+ ions of cardiolipin (18:1) versus molar concentrations (nmol/mL). The samples from each of two independent cultures per strain were processed in duplicate and the cardiolipin concentrations were calculated as the total amount of all cardiolipins in the (M + H)+ and (M + NH4)+ ion forms over the mass range of m/z 1424 to m/z 1512.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1. Saccharomyces cerevisiae phospholipase genes used to identify putative Ustilago maydis phospholipases.
Table S2. Summary of putative phospholipases in Ustilago maydis.
Table S3. Strains used in this study.
Table S4. Primers used in this study.
Fig. S1. Southern blot hybridization confirms the genotypes of the lip2 deletion mutants. (A) HindIII‐digested DNA from the wild‐type (wt) strain and the mutants carrying the lip2 deletion was hybridized with a radiolabelled probe to test for the correct integration of the lip2 deletion construct. (B) A schematic diagram indicating HindIII cut sites and expected fragment sizes. Note that the lip2 mutant B3‐3 in strain 002 had a second ectopic integration of the construct, and was therefore not used for further analysis.
Fig. S2. Deletion of lip1 and lip2 does not affect mating in Ustilago maydis. Cultures of haploids or sexually compatible haploid pairs of the wild‐type strains (001 or 002) and the lip1 (A), lip2 (B) or lip1 lip2 (C) deletion mutants in the 001 or 002 backgrounds (M001 or M002) were spotted onto charcoal‐containing double complete medium (DCM) and incubated at room temperature for 48 h. Successful mating is indicated by filamentous growth seen as fuzzy white colonies.
Fig. S3. The lip1 and lip2 mutants are similar to the wild‐type (wt) strains in growth and morphology. (A) The growth rates of wt and lip1 or lip2 deletion strains were measured in both the 001 (bottom panel) and 002 (top panel) strain backgrounds. The strains were grown in 50 mL of minimal medium (MM) + 1% glucose and readings of the optical density at 600 nm (OD600) were taken at 12‐h intervals for 144 h. (B) Spot assays of the mutants and wt strains on MM with 1% glucose for 48 h at 30°C. Two independent mutants were tested for each strain. Note that one lip2 mutant in the 002 background (*) had an additional ectopic integration of the deletion construct and this strain was not used for further experiments. (C) Examples of the cell morphologies of the wt strain and a lip2 mutant.
Fig. S4. Deletion of lip2 reduces the virulence of a solopathogenic haploid strain. Virulence assays were performed with 7‐day‐old maize seedlings (Golden Bantam) infected with the SG200 wild‐type (wt) strain or mutants for Lip2. Disease symptoms were scored at 14 days post‐inoculation. The disease index (DI) was calculated based on the frequency of symptoms, including plant death (score 5), large stem tumours (score 4), small stem tumours (score 3), leaf tumours (score 2), anthocyanin (score 1) and healthy plants (score 0). The experiment was repeated twice with wt and two independent lip2 mutants. The overall DI for each set of infections is shown with standard deviations.
Acknowledgements
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (to J.W.K.). J.W.K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology. The authors thank Jun Han of The Metabolomics Innovation Centre and the UVic Proteomics Centre for cardiolipin analysis. The strain SG200 was kindly provided by Dr Gunther Döhlemann (University of Cologne, Cologne, Germany).
References
- An, B. , Li, B. , Qin, G. and Tian, S. (2015) Function of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea . Fungal Genet. Biol. 75, 46–55. [DOI] [PubMed] [Google Scholar]
- Arai, H. , Koizumi, H. , Aoki, J. and Inoue, K. (2002) Platelet‐activating factor acetylhydrolase (PAF‐AH). J. Biochem. 131, 635–640. [DOI] [PubMed] [Google Scholar]
- Baile, M.G. , Lu, Y.‐W. and Claypool, S.M. (2013) The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chem. Phys. Lipids, 179, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett, F. and Herskowitz, I. (1996) Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis . Development, 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Bolker, M. , Genin, S. , Lehmler, C. and Kahmann, R. (1995) Genetic regulation of mating and dimorphism in Ustilago maydis . Can. J. Bot. 73, S320–S325. [Google Scholar]
- Boyce, K.J. , Kretschmer, M. and Kronstad, J.W. (2006) The vtc4 gene influences polyphosphate storage, morphogenesis, and virulence in the maize pathogen Ustilago maydis . Eukaryot. Cell, 5, 1399–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D.J. , Kjellbom, P. and Lamb, C.J. (1992) Elicitor and wound induced oxidative cross linking of a proline rich plant cell wall protein: a novel, rapid defense response. Cell, 70, 21–30. [DOI] [PubMed] [Google Scholar]
- Chatre, L. and Ricchetti, M. (2014) Are mitochondria the Achilles' heel of the kingdom fungi? Curr. Opin. Microbiol. 20, 49–54. [DOI] [PubMed] [Google Scholar]
- Choi, J. , Kim, K.S. , Rho, H.S. and Lee, Y.H. (2011) Differential roles of the phospholipase C genes in fungal development and pathogenicity of Magnaporthe oryzae . Fungal Genet. Biol. 48, 445–455. [DOI] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann, G. , van der Linde, K. , Assmann, D. , Schwammbach, D. , Hof, A. , Mohanty, A. , Jackson, D. and Kahmann, R. (2009) Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5, e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrenberger, F. , Laidlaw, R.D. and Kronstad, J.W. (2001) The hgl1 gene is required for dimorphism and teliospore formation in the fungal pathogen Ustilago maydis . Mol. Microbiol. 41, 337–348. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. and Mueller, M.J. (2013) ROS‐mediated lipid peroxidation and RES‐activated signaling. Annu. Rev. Plant Biol. 64, 429–450. [DOI] [PubMed] [Google Scholar]
- Farooqui, A.A. , Ong, W.Y. and Horrocks, L.A. (2006) Inhibitors of brain phospholipase A2 activity: their neuropharmacological effects and therapeutic importance for the treatment of neurologic disorders. Pharmacol. Rev. 58, 591–620. [DOI] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr, S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulks, J.M. , Weyrich, A.S. , Zimmerman, G.A. and McIntyre, T.M. (2008) A yeast PAF acetylhydrolase ortholog suppresses oxidative death. Free Radic. Biol. Med. 45, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum, M.A. (2000) Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13, 122–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemetsberger, C. , Herrberger, C. , Zechmann, B. , Hillmer, M. and Doehlemann, G. (2012) The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8, e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeradjene, K. , Douglas, L. , Tillman, D.M. , Delaney, A.B. and Houghton, J.A. (2005) Reactive oxygen species regulate caspase activation in tumor necrosis factor‐related apoptosis‐inducing ligand‐resistant human colon carcinoma cell lines. Cancer Res. 65, 7436–7445. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Klose, J. and Kronstad, J.W. (2006) The multifunctional beta‐oxidation enzyme is required for full symptom development by the biotrophic maize pathogen Ustilago maydis . Eukaryot. Cell, 5, 2047–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, J. , Moniz de Sa, M. and Kronstad, J.W. (2004) Lipid‐induced filamentous growth in Ustilago maydis . Mol. Microbiol. 52, 823–835. [DOI] [PubMed] [Google Scholar]
- Kohler, G.A. , Brenot, A. , Haas‐Stapleton, E. , Agabian, N. , Deva, R. and Nigam, S. (2006) Phospholipase A2 and phospholipase B activities in fungi. Biochim. Biophys. Acta, 1761, 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer, M. , Klose, J. and Kronstad, J.W. (2012) Defects in mitochondrial and peroxisomal beta‐oxidation influence virulence in the maize pathogen Ustilago maydis . Eukaryot. Cell, 11, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer, M. , Reiner, E. , Hu, G. , Tam, N. , Oliveira, D.L. , Caza, M. , Yeon, J.H. , Kim, J. , Kastrup, C.J. , Jung, W.H. and Kronstad J.W. (2014) Defects in phosphate acquisition and storage influence the virulence of Cryptococcus neoformans . Infect. Immun. 82, 2697–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst, L. , Taylor, D.C. and Underhill, E.W. (1992) Fatty acid elongation in developing seeds of Arabidopsis thaliana . Plant Physiol. Biochem. 30, 425–434. [Google Scholar]
- Lanver, D. , Berndt, P. , Tollot, M. , Naik, V. , Vranes, M. , Warmann, T. , Munch, K. , Rossel, N. and Kahmann, R. (2014) Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 10, e1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane, A.M. , McDevitt, C.A. , Kirk, K. and Fidock, D.A. (2012) Degrees of chloroquine resistance in Plasmodium – is the redox system involved? Int. J. Parasitol. Drugs Drug Resist. 2, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Ragheb, K. , Lawler, G. , Sturgis, J. , Rajwa, B. , Melendez, J.A. and Robinson, J.P. (2003) Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 278, 8516–8525. [DOI] [PubMed] [Google Scholar]
- Lovely, C.B. and Perlin, M.H. (2011) Cla4, but not Rac1, regulates the filamentous response of Ustilago maydis to low ammonium conditions. Commun. Integr. Biol. 4, 670–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean, L.R. , Hagaman, K.A. and Davidson, W.S. (1993) Role of lipid structure in the activation of phospholipase A2 by peroxidized phospholipids. Lipids, 28, 505–509. [DOI] [PubMed] [Google Scholar]
- Molina, L. and Kahmann, R. (2007) An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell, 19, 2293–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti, D. , Basilico, N. , Parapini, S. , Pasini, E. , Olliaro, P. and Taramelli, D. (2002) Does chloroquine really act through oxidative stress? FEBS Lett. 522, 3–5. [DOI] [PubMed] [Google Scholar]
- Ramanadham, S. , Ali, T. , Ashley, J.W. , Bone, R.N. , Hancock, W.D. and Lei, X. (2015) Calcium‐independent phospholipases A2 and their roles in biological processes and diseases. J. Lipid Res. 56, 1643–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho, H.S. , Jeon, J. and Lee, Y.H. (2009) Phospholipase C‐mediated calcium signalling is required for fungal development and pathogenicity in Magnaporthe oryzae . Mol. Plant Pathol. 10, 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolain, J.M. , Colson, P. and Raoult, D. (2007) Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int. J. Antimicrob. Agents, 30, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenson, R.S. and Stafforini, D.M. (2012) Modulation of oxidative stress, inflammation, and atherosclerosis by lipoprotein‐associated phospholipase A2. J. Lipid Res. 53, 1767–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Herrera, J. , Leon, C. , Guevara‐Olvera, L. and Carabez‐Trejo, A. (1995) Yeast–mycelial dimorphism of haploid and diploid strains of Ustilago maydis . Microbiology, 141, 695–703. [Google Scholar]
- Sambrook, J. , Fritsch, E. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sevanian, A. , Stein, R.A. and Mead, J.F. (1981) Metabolism of epoxidized phosphatidylcholine by phospholipase A2 and epoxide hydrolase. Lipids, 16, 781–789. [DOI] [PubMed] [Google Scholar]
- Shetty, N.P. , Lyngs Jorgensen, H.L. , Due Jensen, J. , Collinge, D.B. and Shetty, H.S. (2008) Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280. [Google Scholar]
- Tjoelker, L.W. , Wilder, C. , Eberhardt, C. , Stafforini, D.M. , Dietsch, G. , Schlmpf, B. , Hooper, S. , Le Trong, H. , Cousens, L.S. , Zimmerman, G.A. , Yamada, Y. , McIntyre, T.M. , Prescott, S.M. and Gray, P.W. (1995) Anti‐inflammatory properties of a platelet activating factor acetylhydrolase. Nature, 374, 549–553. [DOI] [PubMed] [Google Scholar]
- Toffaletti, D. , Rude, T. , Johnston, S. , Durack, D. and Perfect, J. (1993) Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, H.C. and Chung, K.R. (2014) Calcineurin phosphatase and phospholipase C are required for developmental and pathological functions in the citrus fungal pathogen Alternaria alternata . Microbiology, 160, 1453–1465. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Swenson, L. , Castro, C. , Derewenda, U. , Minor, W. , Arai, H. , Aoki, J. , Inoue, K. , Servin‐Gonzalez, L. and Derewenda, Z.S. (1998) Structure of a microbial homologue of mammalian platelet‐activating factor acetylhydrolases: Streptomyces exfoliatus lipase at 1.9 Å resolution. Structure, 6, 511–519. [DOI] [PubMed] [Google Scholar]
- Yazawa, H. , Holic, R. , Kumagai, H. and Uemura, H. (2013a) Toxicity of ricinoleic acid production in fission yeast Schizosaccharomyces pombe is suppressed by the overexpression of plg7, a phospholipase A2 of a platelet‐activating factor (PAF) family homolog. Appl. Microbiol. Biotechnol. 97, 8193–8203. [DOI] [PubMed] [Google Scholar]
- Yazawa, H. , Kumagai, H. and Uemura, H. (2013b) Secretory production of ricinoleic acid in fission yeast Schizosaccharomyces pombe . Appl. Microbiol. Biotechnol. 97, 8663–8671. [DOI] [PubMed] [Google Scholar]
- Yazawa, H. , Ogiso, M. , Kumagai, H. and Uemura, H. (2014) Suppression of ricinoleic acid toxicity by ptl2 overexpression in fission yeast Schizosaccharomyces pombe . Appl. Microbiol. Biotechnol. 98, 9325–9337. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Fan, L. , Wu, Q. , Fu, K. , Gao, S. , Wang, M. , Gao, J. , Li, Y. and Chen, J. (2014) Biological role of Trichoderma harzianum‐derived platelet‐activating factor acetylhydrolase (PAF‐AH) on stress response and antagonism. PLoS One, 9, e100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurbriggen, M.D. , Carrillo, N. , Tognetti, V.B. , Melzer, M. , Peisker, M. , Hause, B. and Hajirezaei, M.R. (2009) Chloroplast‐generated reactive oxygen species play a major role in localized cell death during the non‐host interaction between tobacco and Xanthomonas campestris pv. vesicatoria . Plant J. 60, 962–973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Table S1. Saccharomyces cerevisiae phospholipase genes used to identify putative Ustilago maydis phospholipases.
Table S2. Summary of putative phospholipases in Ustilago maydis.
Table S3. Strains used in this study.
Table S4. Primers used in this study.
Fig. S1. Southern blot hybridization confirms the genotypes of the lip2 deletion mutants. (A) HindIII‐digested DNA from the wild‐type (wt) strain and the mutants carrying the lip2 deletion was hybridized with a radiolabelled probe to test for the correct integration of the lip2 deletion construct. (B) A schematic diagram indicating HindIII cut sites and expected fragment sizes. Note that the lip2 mutant B3‐3 in strain 002 had a second ectopic integration of the construct, and was therefore not used for further analysis.
Fig. S2. Deletion of lip1 and lip2 does not affect mating in Ustilago maydis. Cultures of haploids or sexually compatible haploid pairs of the wild‐type strains (001 or 002) and the lip1 (A), lip2 (B) or lip1 lip2 (C) deletion mutants in the 001 or 002 backgrounds (M001 or M002) were spotted onto charcoal‐containing double complete medium (DCM) and incubated at room temperature for 48 h. Successful mating is indicated by filamentous growth seen as fuzzy white colonies.
Fig. S3. The lip1 and lip2 mutants are similar to the wild‐type (wt) strains in growth and morphology. (A) The growth rates of wt and lip1 or lip2 deletion strains were measured in both the 001 (bottom panel) and 002 (top panel) strain backgrounds. The strains were grown in 50 mL of minimal medium (MM) + 1% glucose and readings of the optical density at 600 nm (OD600) were taken at 12‐h intervals for 144 h. (B) Spot assays of the mutants and wt strains on MM with 1% glucose for 48 h at 30°C. Two independent mutants were tested for each strain. Note that one lip2 mutant in the 002 background (*) had an additional ectopic integration of the deletion construct and this strain was not used for further experiments. (C) Examples of the cell morphologies of the wt strain and a lip2 mutant.
Fig. S4. Deletion of lip2 reduces the virulence of a solopathogenic haploid strain. Virulence assays were performed with 7‐day‐old maize seedlings (Golden Bantam) infected with the SG200 wild‐type (wt) strain or mutants for Lip2. Disease symptoms were scored at 14 days post‐inoculation. The disease index (DI) was calculated based on the frequency of symptoms, including plant death (score 5), large stem tumours (score 4), small stem tumours (score 3), leaf tumours (score 2), anthocyanin (score 1) and healthy plants (score 0). The experiment was repeated twice with wt and two independent lip2 mutants. The overall DI for each set of infections is shown with standard deviations.


