Summary
The perception of pathogen‐associated molecular patterns (PAMPs) by immune receptors launches defence mechanisms referred to as PAMP‐triggered immunity (PTI). Successful pathogens must suppress PTI pathways via the action of effectors to efficiently colonize their hosts. So far, plant PTI has been reported to be active against most classes of pathogens, except viruses, although this defence layer has been hypothesized recently as an active part of antiviral immunity which needs to be suppressed by viruses for infection success. Here, we report that Arabidopsis PTI genes are regulated upon infection by viruses and contribute to plant resistance to Plum pox virus (PPV). Our experiments further show that PPV suppresses two early PTI responses, the oxidative burst and marker gene expression, during Arabidopsis infection. In planta expression of PPV capsid protein (CP) was found to strongly impair these responses in Nicotiana benthamiana and Arabidopsis, revealing its PTI suppressor activity. In summary, we provide the first clear evidence that plant viruses acquired the ability to suppress PTI mechanisms via the action of effectors, highlighting a novel strategy employed by viruses to escape plant defences.
Keywords: Arabidopsis thaliana, capsid protein, effector, flg22 signalling, PAMP‐triggered immunity, plant antiviral defences, Plum pox virus
Introduction
Animal and plants possess an elaborate immune system, whose first layer enables the identification of pathogens by pattern recognition receptors (PRRs) that perceive pathogen‐associated molecular patterns (PAMPs) (Kumar et al., 2011; Zipfel, 2014). Once activated, PRRs trigger signalling cascades that launch transcriptional and physiological changes within host cells, ultimately hampering pathogen growth and establishing PAMP‐triggered immunity (PTI). To counteract this defence strategy, successful pathogens deploy a range of effectors, the primary function of which is to evade/interfere with PTI (Jones and Dangl, 2006).
The best‐studied PTI pathway in plants relies on the Arabidopsis receptor kinase FLAGELLIN‐SENSING 2 (FLS2), which perceives bacterial flagellin (or its active epitope flg22) (Gómez‐Gómez et al., 2000; Zipfel et al., 2004). FLS2 activation requires the association with the co‐receptor BRI1‐ASSOCIATED KINASE 1 (BAK1), or its closest paralog BAK1‐LIKE 1 (BKK1), within plasma membrane (PM)‐localized PRR complexes (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011; Sun et al., 2013). Activated FLS2 dissociates with the receptor‐like cytoplasmic kinases (RLCKs) BOTRYTIS‐INDUCED KINASE 1 (BIK1) and AVRPPHB SUSCEPTIBLE 1‐LIKE 1 (PBL1) (Kadota et al., 2014; Li et al., 2014; Lu et al., 2010; Zhang et al., 2010). The following downstream cascade comprises a reactive oxygen species (ROS) burst, activation of mitogen‐activated protein kinases (MAPKs) and transcriptional reprogramming (Bigeard et al., 2015).
Over the last 20 years, it has become clear that PTI mechanisms allowing both plants and animals to resist pathogen attacks follow conserved signalling strategies (Arpaia and Barton, 2011; Lester and Li, 2014; Schwessinger and Ronald, 2012; Thompson et al., 2011; Zipfel and Felix, 2005). Antiviral PRR pathways have been studied extensively in mammals, and the mechanisms whereby viral effectors manipulate PTI defences have been well characterized (Harris and Coyne, 2013; Hiscott et al., 2006; Kumar et al., 2011; Schröder and Bowie, 2007; Yokota et al., 2010). In contrast, hardly anything is known in plants, although indications concerning the existence of PTI mechanisms targeting plant viruses have emerged recently (Kørner et al., 2013; Nicaise, 2014; Zvereva and Pooggin, 2012).
Plant antiviral defences rely mainly on RNA interference, in which the cellular machinery targets virus‐derived nucleic acids, and resistance (R) proteins which recognize virus avirulence factors and trigger an array of physiological and biochemical defence processes broadly targeting pathogens (Nicaise, 2014). Interestingly, a recent model hypothesizes: (i) the action of PTI mechanisms within plant immunity against viruses in parallel with RNA interference and R proteins; and (ii) the existence of specialized effectors encoded by successful plant viruses to bypass PTI, in parallel with the well‐characterized viral silencing suppressors (Nicaise, 2014; Zvereva and Pooggin, 2012).
Potyviruses constitute one of the largest and most successful genera of plant viruses (Revers and García, 2015). Their single‐stranded RNA genome is packed into filamentous particles and encodes 11 highly multifunctional proteins (Charon et al., 2016), including the capsid protein (CP), which is primarily characterized by its structural role in forming the protective shell around the viral genome. In addition to being the causal agent of Sharka, the most damaging viral disease affecting stone fruit trees, Plum pox virus (PPV) is a representative model of RNA viruses, a dual feature that has led to its classification among the Top 10 plant viruses of scientific and economic importance (Decroocq et al., 2006; García et al., 2014; Rimbaud et al., 2015; Scholthof et al., 2011).
We address here the question of the existence of virus‐encoded effectors suppressing PTI mechanisms, using the Arabidopsis thaliana–PPV pathosystem. In this report, we show that: (i) PTI genes contribute to Arabidopsis immunity to PPV; (ii) PPV suppresses early PTI responses during plant infection; and (iii) PPV CP acts as an effector suppressing PTI mechanisms, underlining a novel strategy employed by a plant virus to counteract host defences.
Results and Discussion
Plant infection by viruses is associated with cellular perturbations, including a massive reprogramming of host gene expression (Hanley‐Bowdoin et al., 2013; Lindbo et al., 2001; Pallas and García, 2011; Whitham et al., 2006). The analysis of previously published transcriptomic data derived from Arabidopsis tissues infected with viruses (Ascencio‐Ibáñez et al., 2008; Babu et al., 2008; Espinoza et al., 2007; Fernandez‐Calvino et al., 2014; Ishihara et al., 2004; Marathe et al., 2002; Pierce and Rey, 2013; Rodrigo et al., 2012; Yang et al., 2007) has indicated that plant colonization is associated with the transcriptional regulation of genes encoding key factors from PTI pathways, such as PRRs themselves, co‐receptors, regulators, MAPKs and transcription factors (Table S1, see Supporting Information). This suggests that cellular components belonging to the PTI machinery may play a role in antiviral defences in plants.
In order to clarify the contribution of the PTI machinery in plant–virus interactions, we investigated Arabidopsis susceptibility to PPV in different genotypes altered in PTI signalling. For the sake of inoculation efficiency/reproducibility, whilst causing minimal injury to leaf tissues, Arabidopsis leaves were inoculated by agroinfiltration with a PPV infectious construct on a small area at the tip of each leaf, and the virus loads were quantified in the rest of the leaf by semi‐quantitative double antibody sandwich‐enzyme‐linked immunosorbent assay (DAS‐ELISA) at 11 days post‐inoculation (dpi). Wild‐type (WT) ecotypes (Columbia‐0 and Landsberg erecta) were used as susceptible controls (Fig. 1). The role of the plant PRRs FLS2, EF‐Tu receptor (EFR) and CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) is well characterized in antibacterial and antifungal immunity (Zipfel, 2014). Arabidopsis null mutants for the corresponding genes were as susceptible to PPV as WT plants (Fig. 1A), suggesting that these PRRs do not participate in anti‐PPV immunity. This is in accordance with the observation that plant PRRs are specialized in the recognition of specific classes of pathogen (Zipfel, 2014). BAK1 and BKK1 are involved in a wide array of pathways related to development and defence (Schwessinger and Rathjen, 2015). Notably, they are both key PTI activators against various non‐viral pathogens (Chaparro‐Garcia et al., 2011; Chinchilla et al., 2007; Heese et al., 2007; Kim et al., 2013; Peng and Kaloshian, 2014; Prince et al., 2014; Roux et al., 2011) and their contribution to plant resistance against viruses has been reported (Kørner et al., 2013; Yang et al., 2010). In our experiments, the single mutants bak1‐4, bak1‐5 and bkk1 were not significantly affected in PPV susceptibility (Fig. 1B), unlike the results observed previously with tobamoviruses and carmoviruses (Kørner et al., 2013; Yang et al., 2010). However, the double mutant bak1‐5 bkk1 displayed a strong increase in viral accumulation (Fig. 1B), indicating that both BAK1 and BKK1 contribute to immunity against PPV, probably in a redundant manner. In various PTI pathways, PRR downstream signalling is positively regulated by BIK1 and PBL1 (Kadota et al., 2014; Li et al., 2014; Lu et al., 2010; Zhang et al., 2010). The mutant bik1 was more susceptible to PPV (Fig. 1C), indicating that this kinase positively contributes to Arabidopsis basal resistance against PPV, whereas the loss of PBL1 failed to increase significantly the bik1 phenotype. PTI signalling is mediated by MAPK cascades comprising MPK3 and MPK6, which activate PTI responses, whereas MPK4 acts as a negative regulator of immune pathways (Rasmussen et al., 2012). Here, we observed that, although statistical analyses do not validate the role of MPK3 in our conditions, the mutants mpk6 and mpk4 are more susceptible and more resistant to PPV respectively (Fig. 1D), respectively, indicating that these two MAPKs seem to be actively involved in plant–virus interactions. Taken together, these results show that a range of host proteins previously described as key PTI factors contribute to Arabidopsis immunity to PPV.
Figure 1.
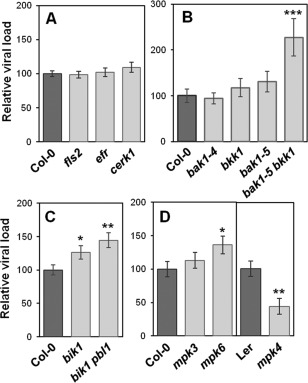
Pathogen‐associated molecular pattern‐triggered immunity (PTI) machinery contributes to Arabidopsis resistance to Plum pox virus (PPV). Arabidopsis susceptibility to PPV was evaluated at 11 days post‐inoculation (dpi) by measuring the viral loads by double antibody sandwich‐enzyme‐linked immunosorbent assay (DAS‐ELISA) in inoculated leaves from mutants affected in the expression of PTI components, such as pattern recognition receptors (PRRs) (A), co‐receptors (B), positive regulators (C) and mitogen‐activated protein kinases (MAPKs) (D). In each panel, values were normalized relative to wild‐type (WT) samples (Col‐0 for all lines, except for mpk4, where the WT ecotype used is Ler). Values presented are the average of 18–24 samples from at least three experiments ± standard error. Values labelled with asterisks are statistically significantly different from WT samples: *P < 0.05, **P < 0.01 and ***P < 0.001. Col‐0, Columbia‐0; Ler, Landsberg erecta.
Successful cellular pathogens from both animals and plants must suppress PTI pathways to efficiently colonize their hosts. In the case of acellular microorganisms, only animal viruses have so far been described as interfering with PTI pathways (Yokota et al., 2010). To determine whether plant viruses employ such a strategy during host infection, we evaluated the impact of PPV infection on Arabidopsis early PTI responses. For this purpose, Arabidopsis plants were first agroinoculated with the PPV infectious construct and systemic tissues were sampled at 4 and 11 dpi. The ability to activate PTI was then evaluated by measuring the responsiveness of infected tissues to the heterologous bacterial PAMP flg22, an efficient elicitor of PTI responses in Arabidopsis (Zipfel et al., 2004). Mock controls consisting of leaves infiltrated with WT agrobacteria were always analysed in comparison. Care was taken to collect leaf discs outside the agroinfiltrated area to avoid any impact of the presence of agrobacteria and/or agroinfiltration‐associated wound lesions on PTI assays (Fig. S1A, see Supporting Information). At an early infection stage (4 dpi), virus accumulation outside the inoculated area was highly limited and only detectable by reverse transcription‐polymerase chain reaction (RT‐PCR), whereas the 11‐dpi stage displayed full‐blown PPV infection (Fig. S1B, C). At 4 dpi, infected tissues displayed a PPV‐specific oxidative burst, whereas ROS production returned to the basal level at 11 dpi (Fig. 2A). Interestingly, PPV infection had a strong impact on the PTI‐related oxidative burst: at 4 dpi, infected tissues treated with flg22 produced a greater amount of ROS than flg22‐treated mock samples (Fig. 2A), suggesting that there is an additive effect between the PPV‐ and flg22‐induced oxidative burst. In contrast, the flg22 responsiveness of PPV‐inoculated tissues was reduced at 11 dpi compared with flg22‐treated mock samples (Fig. 2A). In consequence, our results suggest that, at a very early PPV infection stage (when only a few viral particles are present), infection triggers ROS production, confirming the previous reports on ROS release during viral infections in plants (Allan et al., 2001; Díaz‐Vivancos et al., 2008; Love et al., 2005; Manacorda et al., 2013; Nováková et al., 2015). However, at a late PPV infection stage (with full‐blown virus accumulation), PPV impairs both the PPV‐ and PTI‐related oxidative burst. With the intention of confirming a possible negative effect of PPV infection on the early PTI response, the expression of PTI‐related genes was evaluated during Arabidopsis infection by PPV at 11 dpi. Classically used as flg22‐induced marker genes (Boudsocq et al., 2010), AtFRK1 and AtNHL10 were observed to be induced upon PAMP treatment in mock samples, as expected (Fig. 2B). Interestingly, the infected tissues displayed a decrease in transcript accumulation without PAMP treatment, compared with the basal levels measured in mock samples (Fig. 2B). Moreover, PPV accumulation strongly impaired gene induction triggered by flg22 treatment (Fig. 2B), revealing that plant infection by PPV suppresses the expression of PTI‐related marker genes. These results indicate that PPV negatively regulates early PTI responses during plant infection.
Figure 2.
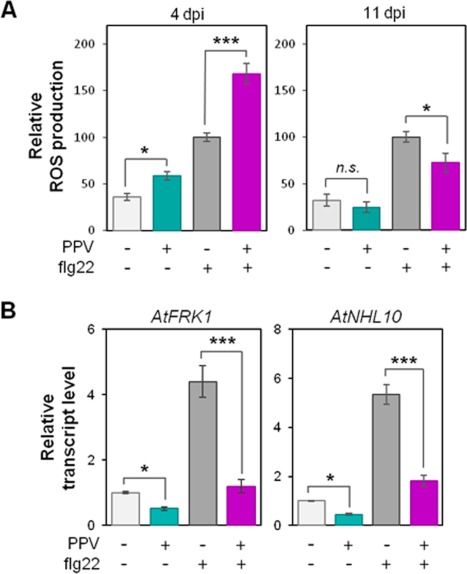
Plum pox virus (PPV) suppresses early pathogen‐associated molecular pattern‐triggered immunity (PTI) responses during Arabidopsis infection. (A) The PTI‐related oxidative burst is affected upon PPV infection. Reactive oxygen species (ROS) production was measured in PPV‐inoculated (+) or mock‐inoculated (–) leaves at 4 days post‐inoculation (dpi)/11 dpi in response to treatment with 200 nm flg22. The results are presented as the total photon count during 40 min of treatment, normalized in comparison with mock‐inoculated leaves treated with flg22. The values presented are the average of 24–30 samples from at least three experiments ± standard error. Connecting lines with asterisks indicate two statistically significantly different values: *P < 0.05 and ***P < 0.001; n.s., not significant. (B) PTI marker gene expression is suppressed upon PPV infection. The transcript accumulation of Arabidopsis PTI marker genes AtFRK1 and AtNHL10 was assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) in PPV‐inoculated (+) or mock‐inoculated (–) leaves at 11 dpi, 30 min after treatment with 1 µm flg22. Values are the average of 12 samples from three experiments ± standard error presented as fold induction compared with untreated mock‐inoculated samples. Connecting lines with asterisks indicate two statistically significantly different values: *P < 0.05 and ***P < 0.001.
We hypothesized that one (or several) virus genome‐encoded protein(s) may act as PTI‐suppressing effectors. Overlapping immune signalling induced by different classes of pathogen enables the successful identification of effectors suppressing PTI responses triggered by heterologous PAMPs (Bos et al., 2010; Chen et al., 2013; Jaouannet et al., 2013; Park et al., 2012; Pel et al., 2014; Zheng et al., 2014). Thus, we sought to determine the effects of the expression of PPV proteins on early PTI responses triggered by flg22. For this purpose, agrobacteria expressing green fluorescent protein (GFP)‐tagged versions of PPV‐encoded proteins were infiltrated into Nicotiana benthamiana leaves. At 2 days post‐agroinfiltration (dpa), leaf discs were collected inside the agroinfiltrated area (Fig. S2A, see Supporting Information) and flg22 responsiveness was evaluated. We found that in planta‐expressed CP‐GFP (detected in Fig. S2C, E) strongly reduced the flg22‐triggered oxidative burst relative to GUS‐GFP (Fig. 3A). The genes NbACRE31 and NbACRE132 are rapidly up‐regulated upon flg22 treatment in N. benthamiana (Heese et al., 2007; Segonzac et al., 2011). Although flg22 responsiveness was not affected in leaves overexpressing GUS‐GFP and those infiltrated with WT agrobacteria, the induction of the flg22 marker genes was suppressed in CP‐expressing samples (Fig. 3B). Transient expression experiments performed in Arabidopsis seedlings (Fig. S2B, D) confirmed these results, as the induction of flg22 marker genes, AtFRK1 and AtNHL10, was also inhibited upon PPV CP expression, compared with the negative controls (Fig. 3C). Therefore, our results show that PPV CP suppresses early PTI responses, revealing, for the first time, the existence of a plant virus PTI‐suppressing effector.
Figure 3.
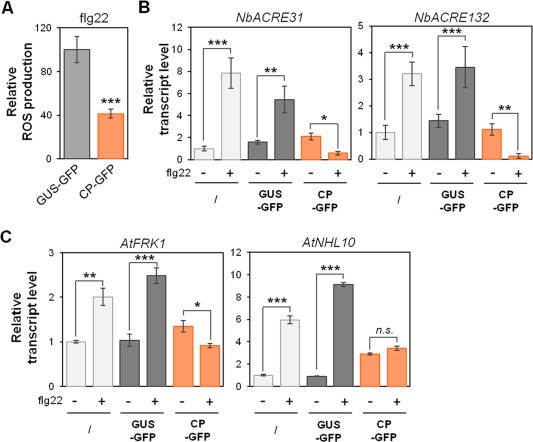
Plum pox virus (PPV) capsid protein (CP) suppresses early pathogen‐associated molecular pattern‐triggered immunity (PTI) responses in Nicotiana benthamiana and Arabidopsis. (A) In planta‐expressed PPV CP impairs the PTI‐associated oxidative burst in Nicotiana benthamiana. Reactive oxygen species (ROS) production in response to treatment with 200 nm flg22 was measured on leaves transiently overexpressing CP‐green fluorescent protein (CP‐GFP) or β‐glucuronidase‐GFP (GUS‐GFP) at 2 days post‐agroinfiltration (dpa). ROS production is presented as the total photon count during 40 min of treatment, normalized relative to GUS‐GFP‐expressing leaves. Values presented are the average of 24 samples from three biological experiments ± standard error. Values labelled with asterisks are statistically significantly different: ***P < 0.001. (B) In planta‐expressed PPV CP suppresses PTI‐associated gene expression in Nicotiana benthamiana. Transcript accumulation of PTI marker genes NbACRE31 and NbACRE132 was assessed by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) on leaf tissues overexpressing CP‐GFP and GUS‐GFP or infiltrated with wild‐type agrobacteria (control samples represented by ‘/’) at 2 dpa, 1 h after treatment with 1 µm flg22. Values are the average of 12 samples from three biological experiments ± standard error presented as fold induction compared with mock‐treated control samples. Connecting lines with asterisks indicate two statistically significantly different values: *P < 0.05, **P < 0.01 and ***P < 0.001. (C) In planta‐expressed PPV CP suppresses flg22‐induced gene expression in Arabidopsis seedlings. Transcript accumulation of PTI marker genes AtFRK1 and AtNHL10 was assessed at 2 dpa by quantitative RT‐PCR on seedlings overexpressing CP‐GFP or GUS‐GFP or on control seedlings treated with wild‐type agrobacteria (represented by ‘/’), 30 min after treatment with 1 µm flg22. Values are the average of eight samples from two biological experiments ± standard error presented as fold induction compared with mock‐treated control samples. Connecting lines with asterisks indicate two statistically significantly different values: *P < 0.05, **P < 0.01 and ***P < 0.001; n.s., not significant.
PTI governs a fast and powerful defence line that has been reported to be active in many eukaryotic organisms. In plants, pathogens from various lifestyle classes have been shown to be controlled by PTI, most models excluding viruses (Boller and Felix, 2009; Dangl et al., 2013; Schwessinger and Ronald, 2012). The contribution of key PTI components in Arabidopsis resistance against PPV suggests that plants, similar to animals, defend themselves against viruses using PTI machinery. Since the submission of this work, viral double‐stranded RNAs (dsRNAs) have been reported to act as PAMPs that trigger PTI responses and protection against viral infection in Arabidopsis (Niehl et al., 2016), confirming the existence of antiviral PTI in plants. In this context, the discovery of the first PTI‐targeting effector encoded by a plant virus illustrates further the biological significance of PTI for antiviral defences in plants.
Most animal PRRs involved in virus perception are intracellular (Kumar et al., 2011). Whether plant PRRs specialized in virus perception fulfil similar canonical structures and subcellular localizations has yet to be determined. Notwithstanding, the role of PM‐localized PTI actors (such as BAK1 and BKK1) in plant–virus interactions raises the question of the way in which intracellular pathogens could be perceived extracellularly. Although this point remains unknown, a similar situation occurring in animal cells indicates that extracellular treatment with viral PAMPs could activate immune pathways via a clathrin‐dependent endocytic pathway and/or the activation of PRRs located at the cell surface, in addition to their classical inner membrane‐associated localization (Itoh et al., 2008; Pohar et al., 2013).
It is reasonable to consider that antiviral PTI pathways may display specificity relative to the mechanisms identified in other plant pathosystems. However, the fact that key PTI components involved in defence against non‐viral pathogens contribute to defence against viruses confirms that immune pathways against various classes of pathogen share certain signalling components. Nonetheless, it is worth mentioning that BAK1, BKK1 and BIK1, in addition to their role in PTI, are involved in signalling pathways associated with BRI1 (Brassinosteroid‐insensitive 1), a steroid phytohormone receptor involved in plant development (He et al., 2007; Lin et al., 2013), which could suggest that brassinosteroid (BR) signalling has an impact on plant defence against PPV, as reported for other plant viruses (Ali et al., 2014; Baebler et al., 2009; Deng et al., 2016; Nakashita et al., 2003; Zhang et al., 2015). Although the use of the mutant bak1‐5 [carrying a point mutation impairing PTI responses, but not BR signalling (Schwessinger et al., 2011)] favours the role of PTI on PPV infection, the possible cross‐talk between PTI and BR pathways in plant–virus interactions has yet to be clarified. In addition, future investigations will need to focus on possible interconnections between immune pathways triggered by viral PAMPs and host DAMPs (danger‐associated molecular patterns) released by cells during the infection process.
In this work, we hypothesize that PPV infection triggers PTI mechanisms, which are afterwards suppressed by the viral CP to enable plant colonization (hypothesis developed in Fig. S3, see Supporting Information). PPV is a representative model of potyviruses and more broadly of RNA viruses. Hence, it is likely that other plant viruses have evolved strategies to suppress PTI mechanisms. Remarkably, the fact that the expression of PTI genes seems to be mostly down‐regulated during plant infection (Table S1) could suggest that DNA and RNA viruses from different families share a common feature that involves the targeting of plant PTI pathways, notably at the gene expression level.
In accordance with the current concept of plant innate immunity, effector proteins can act as both a virulence factor (suppressing PTI) and avirulence factor [triggering effector‐triggered immunity (ETI)]. Although many proteins from all cellular pathogens meet this definition (Dangl et al., 2013), only the avirulence factor side has so far been reported for viruses, including many CPs recognized by plant R genes (De Ronde et al., 2014). The fact that PPV CP displays PTI suppressor activity emphasizes, for the first time, that plant viruses integrate the host–pathogen conceptual arms race illustrated by the zig–zag model (Jones and Dangl, 2006).
Although CPs from animal and plant viruses were initially characterized for their role as structural proteins in forming protective shells around viral genomes, they possess numerous non‐encapsidation activities, including the regulation of host immune defences (Ni and Cheng Kao, 2013; Weber and Bujarski, 2015). Here, our findings provide evidence that PPV CP possesses PTI suppressor activity. Remarkably, potyviral CPs are intrinsically disordered (Baratova et al., 2001; Charon et al., 2016; Ksenofontov et al., 2013; Rantalainen et al., 2008), a structural feature shared by many pathogen effectors to efficiently bypass the immune system (Marín et al., 2013).
Plant viruses have been known for some time to successfully evade/manipulate host defences via specific proteins, the most widely known being the virus‐encoded silencing suppressors (Csorba et al., 2015; Nakahara and Masuta, 2014; Pumplin and Voinnet, 2013). The present work reports the existence of a plant virus‐encoded PTI suppressor, and therefore a novel strategy employed by a plant virus to escape host defences. By providing evidence that viruses belong to the list of plant pathogens that suppress PTI, our findings raise considerable questions about the tight molecular dialogue underlying plant–virus interactions.
Experimental Procedures
All primers used in this work are described in Methods S1 (see Supporting Information).
Plant material
All plants were grown in a glasshouse at 20–22 ºC with a 16‐h light/8‐h dark photoperiod. Arabidopsis genotypes were in the Columbia‐0 background, except for the mpk4‐1 mutant, which was in the Landsberg erecta background. Arabidopsis mutants have been published previously: fls2 (=fls2c; Zipfel et al., 2004); efr (=efr‐1; Zipfel et al., 2006); cerk1 (=cerk1‐2; Gimenez‐Ibanez et al., 2009); bak1‐4 (Chinchilla et al., 2007); bkk1 (He et al., 2007); bak1‐5 and bak1‐5 bkk1 (Schwessinger et al., 2011); bik1 and bik1 pbl1 (Zhang et al., 2010); mpk3 (=mpk3‐1; Bartels et al., 2009); mpk4 (=mpk4‐1; Petersen et al., 2000); mpk6 (mpk6‐2; Bartels et al., 2009).
Virus inoculation and detection
The PPV isolate used in this work was PPV‐R3‐GFP (D strain). Rosette leaves from 6–7‐week‐old plants were inoculated on a small area at the tip of each leaf (Fig. S1) by infiltration with Agrobacterium tumefaciens C58C1 cells [optical density at 600 nm (OD600) = 0.15; ∼80 µL per leaf tip] carrying the infectious clone pBIN‐PPV‐NK‐GFP construct, as described previously (Jiménez et al., 2006; Nicaise et al., 2007). DAS‐ELISA experiments were performed using anti‐PPV polyclonal antibodies on an EPOCH microplate spectrophotometer (Biotek, Colmar, France). RT‐PCRs were performed on cDNAs synthesized using Superscript® II Reverse Transcriptase (Invitrogen, Paisley, UK) from total RNAs isolated using the TRI‐Reagent® method (Sigma‐Aldrich, Saint Louis, USA).
ROS measurement and PTI marker gene expression
ROS production was measured using an Infinite® 200 PRO photon counting reader (TECAN, Mannedorf, Switzerland), as described previously (Zipfel et al., 2004), in the presence of 17 mm L‐012 (Wako Chemical Inc., Richmond, USA), 1 µm horseradish peroxidase (Sigma‐Aldrich) and 200 nm flg22 (Peptron Inc., Deajeon, South Korea).
Total RNAs were isolated using the TRI‐Reagent® method (Sigma‐Aldrich) and treated with Turbo DNA‐free DNase (Ambion, Austin, USA). First‐strand cDNA was synthesized using Superscript® II Reverse Transcriptase (Invitrogen) and an oligo(dT) primer, according to the manufacturer's instructions. cDNAs were amplified in triplicate or quadruplicate by quantitative PCR using SYBR Green JumpStart Taq ReadyMix (Sigma‐Aldrich) and the LightCycler® 480 System (Roche, Meylan, France). AtUbox and NbEF1α genes were used as internal controls (in Arabidopsis and N. benthamiana experiments, respectively). The expression in control samples was used to normalize with the expression level set to unity. Relative expression was determined using the comparative Ct method (2–ΔΔ Ct).
DNA constructs
The PPV capsid coding sequence was cloned into the pENTR/D‐TOPO vector (Invitrogen). A control pENTR/D‐TOPO‐GUS was also obtained in parallel. Entry constructs were recombined by LR reaction into the Gateway‐compatible pK7FWG2.0 (Ghent University, Belgium).
In planta transient expression
Transient expression in N. benthamiana was performed as described previously (Bos et al., 2010) on leaves from 5‐week‐old plants. Transient expression in Arabidopsis efr seedlings was performed as described previously (Wu et al., 2014).
Protein extraction, purification and western blotting
In planta‐expressed tagged proteins were extracted and immunoprecipitated with GFP‐Trap® agarose beads (Chromotek, Planegg‐Martinsried, Germany), as described previously (Nicaise et al., 2013). Proteins were fractionated by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), transferred onto poly(vinylidene difluoride) (PVDF) membrane and detected using anti‐GFP antibodies (Ambion, Abingdon, UK) and peroxidase‐conjugated anti‐rabbit antibodies (Sigma‐Aldrich). Immunodetection was performed using the reagent SuperSignal™ West‐Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, USA).
Statistical analyses
Statistical significance based on one‐way analysis of variance (ANOVA) was determined with InStat 3.10 software (GraphPad, La Jolla, CA, USA).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Plum pox virus (PPV) accumulation in inoculated leaves during Arabidopsis infection.
Fig. S2 In planta transient over‐expression of capsid protein‐green fluorescent protein (CP‐GFP) and β‐glucuronidase‐GFP (GUS‐GFP) in Nicotiana benthamiana leaves and Arabidopsis seedlings.
Fig. S3 Schematic model depicting capsid protein (CP)‐mediated pathogen‐associated molecular pattern‐triggered immunity (PTI) suppression during Plum pox virus (PPV) infection. ETS, effector‐triggered susceptibility.
Table S1 Pathogen‐associated molecular pattern‐triggered immunity (PTI) machinery‐related genes are regulated upon virus infection in Arabidopsis. The table represents the PTI‐related genes up‐regulated (in red) or down‐regulated (in green) by RNA and DNA viruses during Arabidopsis infection. The absence of colour indicates that the corresponding genes are not up‐ or down‐regulated. Data were extracted from transcriptomic analyses published previously (Ascencio‐Ibáñez et al., 2008; Babu et al., 2008; Espinoza et al., 2007; Fernandez‐Calvino et al., 2014; Ishihara et al., 2004; Marathe et al., 2002; Pierce and Rey, 2013; Rodrigo et al., 2012; Yang et al., 2007). ER‐QC, endoplasmic reticulum‐quality control; CDPK, calcium‐dependent protein kinase; MAPK, mitogen‐activated protein kinase; PRR, pattern recognition receptor; RBP, RNA‐binding protein; SERK, somatic embryogenesis receptor kinase (protein family containing BAK1/SERK3 and BKK1/SERK4). DNA viruses: CaLCuV, Cabbage leaf curl begomovirus; SACMV, South African cassava mosaic begomovirus. RNA viruses: CMV, Cucumber mosaic bromovirus; PPV, Plum pox potyvirus; TCV, Turnip crinkle carmovirus; TEV, Tobacco etch potyvirus; TMV, Tobacco mosaic tobamovirus; TRV, Tobacco rattle tobravirus; TuMV, Turnip mosaic potyvirus.
Methods S1 Primers used in this study.
Acknowledgements
We thank Drs Scott Peck, Morten Petersen, Jian‐Min Zhou and Cyril Zipfel for sharing biological material. This research was funded by an FP7 Marie‐Curie Intra‐European Fellowship allocated to V.N. (Grant number ⋕327341).
References
- Ali, S.S. , Gunupuru, L.R. , Kumar, G.B. , Khan, M. , Scofield, S. , Nicholson, P. and Doohan, F. (2014) Plant disease resistance is augmented in uzu barley lines modified in the brassinosteroid receptor BRI1. BMC Plant Biol. 14, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan, A.C. , Lapidot, M. , Culver, J.N. and Fluhr, R. (2001) An early tobacco mosaic virus‐induced oxidative burst in tobacco indicates extracellular perception of the virus coat protein. Plant Physiol. 126, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia, N. and Barton, G.M. (2011) Toll‐like receptors: key players in antiviral immunity. Curr. Opin. Virol. 1, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio‐Ibáñez, J.T. , Sozzani, R. , Lee, T.J. , Chu, T.M. , Wolfinger, R.D. , Cella, R. and Hanley‐Bowdoin, L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, M. , Griffiths, J.S. , Huang, T.S. and Wang, A. (2008) Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to plum pox virus infection. BMC Genomics, 9, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler, Š. , Krečič‐Stres, H. , Rotter, A. , Kogovsek, P. , Cankar, K. , Kok, E.J. , Gruden, K. , Kovac, M. , Zel, J. , Pompe‐Novak, M. and Ravnikar, M. (2009) PVYNTN elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol. Plant Pathol. 10, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratova, L.A. , Efimov, A.V. , Dobrov, E.N. , Fedorova, N.V. , Hunt, R. , Badun, G.A. , Ksenofontov, A.L. , Torrance, L. and Järvekülg, L. (2001) In situ spatial organization of Potato virus A coat protein subunits as assessed by tritium bombardment. J. Virol. 75, 9696–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, S. , Anderson, J.C. , González Besteiro, M.A. , Carreri, A. , Hirt, H. , Buchala, A. , Métraux, J.P. , Peck, S.C. and Ulm, R. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1‐mediated responses in Arabidopsis. Plant Cell, 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard, J. , Colcombet, J. and Hirt, H. (2015) Signaling mechanisms in pattern‐triggered immunity (PTI). Mol. Plant, 8, 521–539. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bos, J.I.B. , Prince, D. , Pitino, M. , Maffei, M.E. , Win, J. and Hogenhout, S.A. (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6, e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. , Willmann, M.R. , McCormack, M. , Lee, H. , Shan, L. , He, P. , Bush, J. , Cheng, S.H. and Sheen, J. (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature, 464, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro‐Garcia, A. , Wilkinson, R. , Gimenez‐Ibanez, S. , Findlay, K. , Coffey, M. , Zipfel, C. , Rathjen, J. , Kamoun, S. and Schornack, S. (2011) The receptor‐like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana . PLoS One, 6, e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon, J. , Theil, S. , Nicaise, V. and Michon, T. (2016) Protein intrinsic disorder within the Potyvirus genus: from proteome‐wide analysis to functional annotation. Mol. BioSyst. 12, 634–652. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Chronis, D. and Wang, X. (2013) The novel GrCEP12 peptide from the plant‐parasitic nematode Globodera rostochiensis suppresses flg22‐mediated PTI. Plant Signal. Behav. 8, 22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nürnberger, T. , Jones, J.D.G. , Felix, G. and Boller, T. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Csorba, T. , Kontra, L. and Burgyán, J. (2015) Viral silencing suppressors: tools forged to fine‐tune host–pathogen coexistence. Virology, 480, 85–103. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L. , Horvath, D.M. and Staskawicz, B.J. (2013) Pivoting the plant immune system from dissection to deployment. Science, 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq, V. , Sicard, O. , Alamillo, J.M. , Lansac, M. , Eyquard, J.P. , García, J.A. , Candresse, T. , Gall, O. and , Le Revers, F. (2006) Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana . Mol. Plant–Microbe. Interact. 19, 541–549. [DOI] [PubMed] [Google Scholar]
- Deng, X.G. , Zhu, T. , Peng, X.J. , Xi, D.H. , Guo, H. , Yin, Y. , Zhang, D.W. and Lin, H.H. (2016) Role of brassinosteroid signaling in modulating Tobacco mosaic virus resistance in Nicotiana benthamiana . Sci. Rep. 6, 20 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ronde, D. , Butterbach, P. and Kormelink, R. (2014) Dominant resistance against plant viruses. Front. Plant Sci. 5, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Vivancos, P. , Clemente‐Moreno, M.J. , Rubio, M. , Olmos, E. , García, J.A. , Martínez‐Gómez, P. and Hernández, J.A. (2008) Alteration in the chloroplastic metabolism leads to ROS accumulation in pea plants in response to plum pox virus. J. Exp. Bot. 59, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza, C. , Medina, C. , Somerville, S. and Arce‐Johnson, P. (2007) Senescence‐associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J. Exp. Bot. 58, 3197–3212. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Calvino, L. , Osorio, S. , Hernandez, M.L. , Hamada, I.B. , del Toro, F.J. , Donaire, L. , Yu, A. , Bustos, R. , Fernie, A.R. , Martínez‐Rivas, J.M. and Llave, C. (2014) Virus‐induced alterations in primary metabolism modulate susceptibility to tobacco rattle virus in Arabidopsis. Plant Physiol. 166, 1821–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García, J.A. , Glasa, M. , Cambra, M. and Candresse, T. (2014) Plum pox virus and sharka: a model potyvirus and a major disease. Mol. Plant Pathol. 15, 226–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Ntoukakis, V. and Rathjen, J.P. (2009) The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 4, 539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Hanley‐Bowdoin, L. , Bejarano, E.R. , Robertson, D. and Mansoor, S. (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. [DOI] [PubMed] [Google Scholar]
- Harris, K.G. and Coyne, C.B. (2013) Enter at your own risk: how enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine, 63, 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K. , Gou, X. , Yuan, T. , Lin, H. , Asami, T. , Yoshida, S. , Russell, S.D. and Li, J. (2007) BAK1 and BKK1 regulate brassinosteroid‐dependent growth and brassinosteroid‐independent cell‐death pathways. Curr. Biol. 17, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Heese, A. , Hann, D.R. , Gimenez‐Ibanez, S. , Jones, A.M.E. , He, K. , Li, J. , Schroeder, J.I. , Peck, S.C. and Rathjen, J.P. (2007) The receptor‐like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA, 104, 12 217–12 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott, J. , Nguyen, T.L.A. , Arguello, M. , Nakhaei, P. and Paz, S. (2006) Manipulation of the nuclear factor‐kappaB pathway and the innate immune response by viruses. Oncogene, 25, 6844–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, T. , Sakurai, N. , Sekine, K.T. , Hase, S. , Ikegami, M. , Shibata, D. and Takahashi, H. (2004) Comparative analysis of expressed sequence tags in resistant and susceptible ecotypes of Arabidopsis thaliana infected with cucumber mosaic virus. Plant Cell Physiol. 45, 470–480. [DOI] [PubMed] [Google Scholar]
- Itoh, K. , Watanabe, A. , Funami, K. , Seya, T. and Matsumoto, M. (2008) The clathrin‐mediated endocytic pathway participates in dsRNA‐induced IFN‐beta production. J. Immunol. 181, 5522–5529. [DOI] [PubMed] [Google Scholar]
- Jaouannet, M. , Magliano, M. , Arguel, M.J. , Gourgues, M. , Evangelisti, E. , Abad, P. and Rosso, M.N. (2013) The root‐knot nematode calreticulin Mi‐CRT is a key effector in plant defense suppression. Mol. Plant–Microbe Interact. 26, 97–105. [DOI] [PubMed] [Google Scholar]
- Jiménez, I. , López, L. , Alamillo, J.M. , Valli, A. and García, J.A. (2006) Identification of a plum pox virus CI‐interacting protein from chloroplast that has a negative effect in virus infection. Mol. Plant–Microbe. Interact. 19, 350–358. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kadota, Y. , Sklenar, J. , Derbyshire, P. , Stransfeld, L. , Asai, S. , Ntoukakis, V. , Jones, J.D. , Shirasu, K. , Menke, F. , Jones, A. and Zipfel, C. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR‐associated kinase BIK1 during plant immunity. Mol. Cell, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Kim, B.H. , Kim, S.Y. and Nam, K.H. (2013) Assessing the diverse functions of BAK1 and its homologs in Arabidopsis, beyond BR signaling and PTI responses. Mol. Cells, 35, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kørner, C.J. , Klauser, D. , Niehl, A. , Domínguez‐Ferreras, A. , Chinchilla, D. , Boller, T. , Heinlein, M. and Hann, D.R. (2013) The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant–Microbe. Interact. 26, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Ksenofontov, A.L. , Paalme, V. , Arutyunyan, A.A.M. , Semenyuk, P.P.I. , Fedorova, N.N.V. , Rumvolt, R. , Baratova, L.L.A. , Järvekülg, L. and Dobrov, E.E.N. (2013) Partially disordered structure in intravirus coat protein of potyvirus potato virus A. PLoS One, 8, e67830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, H. , Kawai, T. and Akira, S. (2011) Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. [DOI] [PubMed] [Google Scholar]
- Lester, S.N. and Li, K. (2014) Toll‐like receptors in antiviral innate immunity. J. Mol. Biol. 426, 1246–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Li, M. , Yu, L. , Zhou, Z. , Liang, X. , Liu, Z. , Cai, G. , Gao, L. , Zhang, X. , Wang, Y. , Chen, S. and Zhou, J.M. (2014) The FLS2‐associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe, 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Lin, W. , Lu, D. , Gao, X. , Jiang, S. , Ma, X. , Wang, Z. , Mengiste, T. , He, P. and Shan, L. (2013) Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor‐like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA, 110, 12 114–12 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo, J.A. , Fitzmaurice, W.P. and Della‐Cioppa, G. (2001) Virus‐mediated reprogramming of gene expression in plants. Curr. Opin. Plant Biol. 4, 181–185. [DOI] [PubMed] [Google Scholar]
- Love, A.J. , Yun, B.W. , Laval, V. , Loake, G.J. and Milner, J.J. (2005) Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense‐signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol. 139, 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D. , Wu, S. , Gao, X. , Zhang, Y. , Shan, L. and He, P. (2010) A receptor‐like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA, 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manacorda, C.A. , Mansilla, C. , Debat, H.J. , Zavallo, D. , Sánchez, F. , Ponz, F. and Asurmendi, S. (2013) Salicylic acid determines differential senescence produced by two Turnip mosaic virus strains involving reactive oxygen species and early transcriptomic changes. Mol. Plant–Microbe. Interact. 26, 1486–1498. [DOI] [PubMed] [Google Scholar]
- Marathe, R. , Anandalakshmi, R. , Liu, Y. and Dinesh‐Kumar, S.P. (2002) The tobacco mosaic virus resistance gene, N. Mol. Plant Pathol. 3, 167–172. [DOI] [PubMed] [Google Scholar]
- Marín, M. , Uversky, V.N. and Ott, T. (2013) Intrinsic disorder in pathogen effectors: protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell, 25, 3153–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. and Masuta, C. (2014) Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr. Opin. Plant Biol. 20, 88–95. [DOI] [PubMed] [Google Scholar]
- Nakashita, H. , Yasuda, M. , Nitta, T. , Asami, T. , Fujioka, S. , Arai, Y. , Sekimata, K. , Takatsuto, S. , Yamaguchi, I. and Yoshida, S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. [DOI] [PubMed] [Google Scholar]
- Ni, P. and Cheng Kao, C. (2013) Non‐encapsidation activities of the capsid proteins of positive‐strand RNA viruses. Virology, 446, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. (2014) Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise, V. , Gallois, J.L. , Chafiai, F. , Allen, L.M. , Schurdi‐Levraud, V. , Browning, K.S. , Candresse, T. , Caranta, C. , Le Gall, O. and German‐Retana, S. (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana . FEBS Lett. 581, 1041–1046. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Joe, A. , Jeong, B. , Korneli, C. , Boutrot, F. , Westedt, I. , Staiger, D. , Alfano, J.R. and Zipfel, C. (2013) Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 32, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl, A. , Wyrsch, I. , Boller, T. and Heinlein, M. (2016) Double‐stranded RNAs induce a pattern‐triggered immune signaling pathway in plants. New Phytol. 211, 1008–1019. [DOI] [PubMed] [Google Scholar]
- Nováková, S. , Flores‐Ramírez, G. , Glasa, M. , Danchenko, M. , Fiala, R. and Skultety, L. (2015) Partially resistant Cucurbita pepo showed late onset of the Zucchini yellow mosaic virus infection due to rapid activation of defense mechanisms as compared to susceptible cultivar. Front. Plant Sci. 6, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallas, V. and García, J.A. (2011) How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92, 2691–2705. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. , Bellizzi, M. , Valent, B. and Wang, G.L. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel, M.J.C. , Wintermans, P.C.A. , Cabral, A. , Robroek, B.J.M. , Seidl, M.F. , Bautor, J. , Parker, J.E. , Ackerveken, G. and Van den Pieterse, C.M.J. (2014) Functional analysis of Hyaloperonospora arabidopsidis RXLR effectors. PLoS One, 9, e110624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, H.C.H. and Kaloshian, I. (2014) The tomato leucine‐rich repeat receptor‐like kinases SlSERK3A and SlSERK3B have overlapping functions in bacterial and nematode innate immunity. PLoS One, 9, e93302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M. , Brodersen, P. , Naested, H. , Andreasson, E. , Lindhart, U. , Johansen, B. , Nielsen, H.B. , Lacy, M. , Austin, M.J. , Parker, J.E. , Sharma, S.B. , Klessig, D.F. , Martienssen, R. , Mattsson, O. , Jensen, A.B. and Mundy, J. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell, 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pierce, E.J. and Rey, M.E.C. (2013) Assessing global transcriptome changes in response to South African cassava mosaic virus [ZA‐99] infection in susceptible Arabidopsis thaliana . PLoS One, 8, e67534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohar, J. , Pirher, N. , Benčina, M. , Manček‐Keber, M. and Jerala, R. (2013) The role of UNC93B1 protein in surface localization of TLR3 receptor and in cell priming to nucleic acid agonists. J. Biol. Chem. 288, 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, D.C. , Drurey, C. , Zipfel, C. and Hogenhout, S.A. (2014) The leucine‐rich repeat receptor‐like kinase BRASSINOSTEROID INSENSITIVE1‐ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 164, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin, N. and Voinnet, O. (2013) RNA silencing suppression by plant pathogens: defence, counter‐defence and counter‐counter‐defence. Nat. Rev. Microbiol. 11, 745–760. [DOI] [PubMed] [Google Scholar]
- Rantalainen, K.I. , Uversky, V.V.N. , Permi, P. , Kalkkinen, N. , Dunker, A.K. and Mäkinen, K. (2008) Potato virus A genome‐linked protein VPg is an intrinsically disordered molten globule‐like protein with a hydrophobic core. Virology, 377, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, M.W. , Roux, M. , Petersen, M. and Mundy, J. (2012) MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Rimbaud, L. , Dallot, S. , Gottwald, T. , Jacquot, E. and Soubeyrand, S. (2015) Sharka epidemiology and worldwide management strategies: learning lessons to optimize disease control in perennial plants. Annu. Rev. Phytopathol. 53, 357–378. [DOI] [PubMed] [Google Scholar]
- Rodrigo, G. , Carrera, J. , Ruiz‐Ferrer, V. , Toro, F.J. , , del Llave, C. , Voinnet, O. and Elena, S.F. (2012) A meta‐analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PLoS One, 7, e40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, M. , Schwessinger, B. , Albrecht, C. , Chinchilla, D. , Jones, A. , Holton, N. , Malinovsky, F.G. , Tör, M. , de Vries, S. and Zipfel, C. (2011) The Arabidopsis leucine‐rich repeat receptor‐like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell, 23, 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof, K.B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G.D. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, M. and Bowie, A.G. (2007) An arms race: innate antiviral responses and counteracting viral strategies. Biochem. Soc. Trans. 35, 1512–1514. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Rathjen, J.P. (2015) Changing SERKs and priorities during plant life. Trends Plant Sci. 20, 531–533. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Ronald, P.C. (2012) Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. , Roux, M. , Kadota, Y. , Ntoukakis, V. , Sklenar, J. , Jones, A. and Zipfel, C. (2011) Phosphorylation‐dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor‐like kinase BAK1. PLoS Genet. 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.M. and Chai, J. (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Thompson, M.R. , Kaminski, J.J. , Kurt‐Jones, E.A. and Fitzgerald, K.A. (2011) Pattern recognition receptors and the innate immune response to viral infection. Viruses, 3, 920–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, P.H. and Bujarski, J.J. (2015) Multiple functions of capsid proteins in (+) stranded RNA viruses during plant–virus interactions. Virus Res. 196, 140–149. [DOI] [PubMed] [Google Scholar]
- Whitham, S.A. , Yang, C. and Goodin, M.M. (2006) Global impact: elucidating plant responses to viral infection. Mol. Plant–Microbe. Interact. 19, 1207–1215. [DOI] [PubMed] [Google Scholar]
- Wu, H.Y. , Liu, K.H. , Wang, Y.C. , Wu, J.F. , Chiu, W.L. , Chen, C.Y. , Wu, S.H. , Sheen, J. and Lai, E.M. (2014) AGROBEST: an efficient Agrobacterium‐mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods, 10, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Guo, R. , Jie, F. , Nettleton, D. , Peng, J. , Carr, T. , Yeakley, J.M. , Fan, J.B. and Whitham, S.A. (2007) Spatial analysis of Arabidopsis thaliana gene expression in response to turnip mosaic virus infection. Mol. Plant–Microbe. Interact. 20, 358–370. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Gou, X. , He, K. , Xi, D. , Du, J. , Lin, H. and Li, J. (2010) BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to turnip crinkle virus. Eur. J. Plant Pathol. 127, 149–156. [Google Scholar]
- Yokota, S.I. , Okabayashi, T. and Fujii, N. (2010) The battle between virus and host: modulation of Toll‐like receptor signaling pathways by virus infection. Mediators Inflamm. 2010, 184323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.W. , Deng, X.G. , Fu, F.Q. and Lin, H.H. (2015) Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana . Planta, 241, 875–885. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X. , Zou, Y. , Gao, M. , Zhang, X. , Chen, S. , Mengiste, T. , Zhang, Y. and Zhou, J.M. (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , McLellan, H. , Fraiture, M. , Liu, X. , Boevink, P.C. , Gilroy, E.M. , Chen, Y. , Kandel, K. , Sessa, G. , Birch, P.R. and Brunner, F. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22‐triggered immunity. PLoS Pathog. 10, e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. (2014) Plant pattern‐recognition receptors. Trends Immunol. 35, 345–351. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. and Felix, G. (2005) Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 8, 353–360. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
- Zvereva, A.S. and Pooggin, M.M. (2012) Silencing and innate immunity in plant defense against viral and non‐viral pathogens. Viruses, 4, 2578–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Plum pox virus (PPV) accumulation in inoculated leaves during Arabidopsis infection.
Fig. S2 In planta transient over‐expression of capsid protein‐green fluorescent protein (CP‐GFP) and β‐glucuronidase‐GFP (GUS‐GFP) in Nicotiana benthamiana leaves and Arabidopsis seedlings.
Fig. S3 Schematic model depicting capsid protein (CP)‐mediated pathogen‐associated molecular pattern‐triggered immunity (PTI) suppression during Plum pox virus (PPV) infection. ETS, effector‐triggered susceptibility.
Table S1 Pathogen‐associated molecular pattern‐triggered immunity (PTI) machinery‐related genes are regulated upon virus infection in Arabidopsis. The table represents the PTI‐related genes up‐regulated (in red) or down‐regulated (in green) by RNA and DNA viruses during Arabidopsis infection. The absence of colour indicates that the corresponding genes are not up‐ or down‐regulated. Data were extracted from transcriptomic analyses published previously (Ascencio‐Ibáñez et al., 2008; Babu et al., 2008; Espinoza et al., 2007; Fernandez‐Calvino et al., 2014; Ishihara et al., 2004; Marathe et al., 2002; Pierce and Rey, 2013; Rodrigo et al., 2012; Yang et al., 2007). ER‐QC, endoplasmic reticulum‐quality control; CDPK, calcium‐dependent protein kinase; MAPK, mitogen‐activated protein kinase; PRR, pattern recognition receptor; RBP, RNA‐binding protein; SERK, somatic embryogenesis receptor kinase (protein family containing BAK1/SERK3 and BKK1/SERK4). DNA viruses: CaLCuV, Cabbage leaf curl begomovirus; SACMV, South African cassava mosaic begomovirus. RNA viruses: CMV, Cucumber mosaic bromovirus; PPV, Plum pox potyvirus; TCV, Turnip crinkle carmovirus; TEV, Tobacco etch potyvirus; TMV, Tobacco mosaic tobamovirus; TRV, Tobacco rattle tobravirus; TuMV, Turnip mosaic potyvirus.
Methods S1 Primers used in this study.


