Summary
Barley net form net blotch (NFNB), caused by the necrotrophic fungus Pyrenophora teres f. teres, is a destructive foliar disease in barley‐growing regions worldwide. Little is known about the genetic and molecular basis of this pathosystem. Here, we identified a small secreted proteinaceous necrotrophic effector (NE), designated PttNE1, from intercellular wash fluids of the susceptible barley line Hector after inoculation with P. teres f. teres isolate 0–1. Using a barley recombinant inbred line (RIL) population developed from a cross between the sensitive/susceptible line Hector and the insensitive/resistant line NDB 112 (HN population), sensitivity to PttNE1, which we have named SPN1, mapped to a common resistance/susceptibility region on barley chromosome 6H. PttNE1–SPN1 interaction accounted for 31% of the disease variation when the HN population was inoculated with the 0–1 isolate. Strong accumulation of hydrogen peroxide and increased levels of electrolyte leakage were associated with the susceptible reaction, but not the resistant reaction. In addition, the HN RIL population was evaluated for its reactions to 10 geographically diverse P. teres f. teres isolates. Quantitative trait locus (QTL) mapping led to the identification of at least 10 genomic regions associated with disease, with chromosomes 3H and 6H harbouring major QTLs for resistance/susceptibility. SPN1 was associated with all the 6H QTLs, except one. Collectively, this information indicates that the barley–P. teres f. teres pathosystem follows, at least partially, an NE‐triggered susceptibility (NETS) model that has been described in other necrotrophic fungal disease systems, especially in the Dothideomycete class of fungi.
Keywords: barley, Dothidiomycete, electrolyte leakage, necrotrophic effector‐triggered susceptibility, net form net blotch, reactive oxygen species (ROS)
Introduction
Net blotch is a destructive disease of barley caused by the necrotrophic fungal pathogen Pyrenophora teres. Net blotch is divided into two forms, net form net blotch (NFNB) and spot form net blotch (SFNB), based on the formation of distinctive net‐like and spot‐like lesions, respectively, on the leaves of susceptible barley lines (Campbell et al., 2002; Rau et al., 2007; Smedegård‐Petersen, 1971). NFNB is economically important in many barley‐growing regions (Grewal et al., 2012; Murray and Brennan, 2010; Steffenson, 1997), producing yield losses of up to 40% with a moderate NFNB epidemic (Murray and Brennan, 2010; Steffenson, 1997). Cultural practices and fungicide application can be used to manage NFNB (Hysing and Wiik, 2013; Toubia‐Rahme et al., 1995), but the most desirable method of controlling NFNB is the use of resistant cultivars. Resistance to NFNB has been reported in various barley germplasm lines and the genetics and chromosomal locations of resistance genes have been investigated (reviewed by Liu et al., 2011). Early work using classical genetic analysis showed that resistance was controlled by major genes, which could be dominant, incompletely dominant, recessive, duplicated or complementary (Bockelman et al., 1977; Ho et al., 1996; Khan & Boyd 1969; Schaller, 1955). Since the 1990s, molecular genetics has been used to locate and estimate the effects of resistance genes and quantitative trait loci (QTLs). QTLs and genes for NFNB resistance have been identified on all barley chromosomes, but QTLs with major effects have been primarily located to genomic regions on chromosomes 2H, 3H, 4H and 6H (for a review, see Liu et al., 2011; also Grewal et al., 2012; König et al., 2013; O'Boyle et al., 2011). In particular, chromosome 6H has often been reported to carry major genes and QTL conditioning resistance/susceptibility to NFNB in many genetic backgrounds against a diverse group of NFNB isolates (Abu Qamar et al., 2008; Cakir et al., 2003; Emebiri et al., 2005; Friesen et al., 2006; Grewal et al., 2012; König et al., 2013; Liu et al., 2010; Ma et al., 2004; Manninen et al., 2000, 2006; Richter et al., 1998; Steffenson et al., 1996; St. Pierre et al., 2010). The exact relationship among these QTL/genes is not clear because different populations, isolates and DNA marker types were used. However, dominant (Friesen et al., 2006; O'Boyle et al., 2011) and recessive (Abu Qamar et al., 2008; Ho et al., 1996) resistances have been reported for the 6H region. The identification of dominant and recessive forms of resistance not only suggests the presence of multiple genes in the 6H region, but also indicates that some genes may in fact confer susceptibility to NFNB (Abu Qamar et al., 2008).
Necrotrophic specialist pathogens that can live on dying tissue are known to produce necrotrophic effectors (NEs) that trigger susceptibility. Sensitivity to each NE is often conferred by a single dominant gene in the plant, and the interaction of the two induces uncontrolled programmed cell death (PCD) leading to susceptibility; thus, there is no active resistance response and susceptibility is dominant (Friesen et al., 2008; Wolpert et al., 2002). Three genes conferring sensitivity to NEs have been cloned and each harbours resistance protein‐like domains, including both nucleotide‐binding (NB) and leucine‐rich repeat (LRR) domains (Faris et al., 2010; Lorang et al., 2007; Nagy and Bennetzen, 2008). In addition, it has been shown that NEs induce physiological, biochemical and transcriptional changes in the host that resemble a resistance response (Adhikari et al., 2009; Liu et al., 2012b; Lorang et al., 2012; Pandelova et al., 2009). These lines of evidence indicate that necrotrophic specialists hijack plant resistance pathways to induce cell death. The recognition of the pathogen‐produced effector leads to susceptibility as opposed to resistance, which is observed in the classical gene‐for‐gene interaction; therefore, these host–necrotroph interactions follow an inverse gene‐for‐gene or an NE‐triggered susceptibility (NETS) model (Faris et al., 2010; Friesen et al., 2007; Liu et al., 2012b). Because there are usually multiple NE–host gene pairs contributing simultaneously to the host–pathogen interaction, the reaction to necrotrophic fungal pathogens appears to be quantitatively controlled (e.g. Parastagonospora nodorum [synonym Stagonospora nodorum], causal agent of Stagonospora nodorum blotch; Friesen and Faris 2010).
Toxin production has long been associated with infection by P. teres f. teres (Keon and Hargreaves, 1983; Smedegård‐Petersen, 1977), with at least three toxins (A, B and C) having been identified (Bach et al., 1979; Smedegård‐Petersen, 1977; Weiergang et al., 2002). However, the three P. teres toxins were all characterized as non‐protein and low‐molecular‐weight metabolites (Friis et al., 1991). More recently, Sarpeleh et al. (2007, 2008) have reported the identification and initial characterization of proteinaceous toxins that were able to induce NFNB‐like brown necrotic spots or lesions on some susceptible cultivars. Although a strong correlation between toxin reaction and disease susceptibility has not been observed in these studies, they suggest that NETS probably plays a major role in the barley–P. teres f. teres interaction.
In the current work, we used a recombinant inbred population to identify and map an NE sensitivity that corresponded to a newly identified NE purified from intercellular wash fluids (IWFs) of diseased barley leaves. Inoculation data showed that the NE sensitivity contributed significantly to NFNB disease.
Results
Development of a genetic linkage map in a barley recombinant inbred population
A barley population consisting of 118 recombinant inbred lines (RILs) was developed from a cross between the Canadian barley variety Hector (highly susceptible) and the North Dakota breeding line NDB 112 (highly resistant). This population is hereafter referred to as the HN population.
A total of 701 single nucleotide polymorphism (SNP) and 77 simple sequence repeat (SSR) markers showed segregation in the HN population and were used to assemble linkage maps. The final map contained 409 markers [368 SNP + 41 SSR, with logarithm of odds (LOD) > 3.0] covering all seven chromosomes with a total map distance of 1101.6 cM (Fig. S1 and Table S1, see Supporting Information). All seven chromosomes were well represented with molecular markers; however, 3H and 6H contained gaps of 23.9 and 25.3 cM, respectively. The marker density for each chromosome was similar, ranging from an average of 0.33 (3H) to 0.46 (2H) markers per cM (Table S1). The HN maps were compared with the SNP map of Close et al. (2009) and the SSR map of Varshney et al. (2007), and were found to agree closely in marker order, position and total genetic distance.
Sensitivity to the P. teres f. teres NE PttNE1 maps to barley chromosome 6H and is strongly associated with disease
To identify the NEs underlying fungal virulence, we produced fungal culture filtrates as described in Friesen et al. (2012) and infiltrated them into leaves of Hector and NDB 112; however, no reaction was induced on either parental line (data not shown). Therefore, we examined IWFs from infected leaf tissue. The extracted IWF was infiltrated into leaves of the HN RILs and parental lines. Three days after infiltration, distinct necrosis developed on Hector, and there was no visible reaction on NDB 112 (Fig. 1A). This indicated the presence of an NE in the IWF that had selective activity between Hector and NDB 112, which was designated PttNE1 (Pyrenophora teres f. teres NE 1). The HN population segregated for sensitivity to PttNE1 in a ratio of 54 sensitive : 66 insensitive, which was not significantly different from a 1:1 ratio (χ 2 = 1.22, P = 0.26), the expected ratio for a single gene controlling sensitivity. The gene, designated here as SPN1 (Sensitivity to PttNE1), mapped to chromosome 6H in the HN population, flanked by SNP markers 4191‐268 and ABC08769‐1‐1‐205 at distances of 1.8 and 1.8 cM, respectively (Fig. 1B). Analysis of sensitivity in F2 individuals of a Hector × NDB 112 cross using purified PttNE1 showed a sensitive : insensitive ratio of 91:22, which was not significantly different from a 3:1 ratio (χ 2 = 1.844, P = 0.175), indicating a single dominant gene controlling sensitivity.
Figure 1.
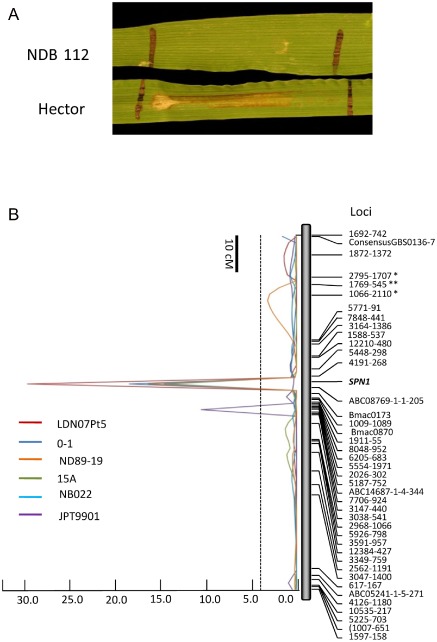
Association of sensitivity to PttNE1 (SPN1) with disease induced by multiple Pyrenophora teres f. teres isolates. (A) In planta sensitivity of Hector barley to intercellular wash fluids containing PttNE1 produced by P. teres f. teres isolate 0–1. NDB 112 is insensitive and Hector is sensitive. (B) Composite interval mapping of barley chromosome 6H which includes the phenotypic marker for PttNE1 sensitivity (SPN1). Disease quantitative trait locus (QTL) analysis for isolates 0–1, 15A, LDN07Pt5, ND89‐19 and NB022 peaked directly over SPN1. However, the disease QTL for isolate JPT9901 peaks approximately 8 cM from SPN1. A logarithm of odds (LOD) cutoff line of 4.5 is indicated by the broken line. A 10‐cM scale is shown in the top left.
Isolate 0–1 was inoculated across the HN RIL population to identify the genomic regions associated with resistance/susceptibility. A major QTL on chromosome 6H accounted for 31% of the disease variation and peaked directly over the sensitivity locus SPN1, suggesting that the SPN1–PttNE1 interaction was responsible for the effects of the QTL observed on 6H (Fig. 1B). In other words, PttNE1 serves as an important virulence factor for the pathogen. The sensitivity to PttNE1 and the susceptibility QTL at the same 6H locus were contributed by the susceptible parent Hector.
The fungal effector present in the IWF is a protein in the size range 6.5–12.5 kDa
To determine whether PttNE1 was a protein, we infiltrated 0–1 IWFs treated with Pronase into Hector barley leaves. The untreated 0–1 IWF induced necrosis on Hector, whereas 0–1 IWF treated with Pronase did not cause any obvious reaction (Fig. 2A), strongly indicating the proteinaceous nature of PttNE1 present in the IWF. We further used size‐based high‐pressure liquid chromatography (HPLC) to estimate the size of PttNE1. The infiltration bioassay on Hector indicated that a single peak was eluted in fractions 19 and 20, a region that eluted between the size standards of cytochrome C (12.3 kDa) and aprotinin (6.5 kDa), indicating that the molecular weight of the fungal effector was probably in the range 6.5–12.3 kDa (Fig. 2B).
Figure 2.
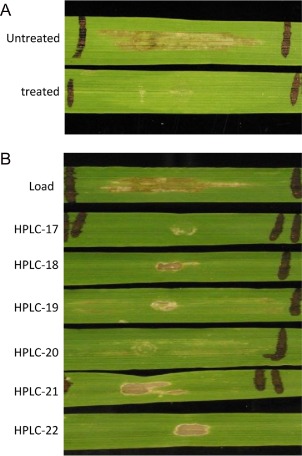
Pronase treatment and size determination of the necrotrophic effector PttNE1. (A) The reaction of Hector to infiltration with intercellular wash fluid (IWF) and the same IWF treated with Pronase for 2 h. (B) The reaction of Hector to infiltration with the IWF and fractions 17–22 from size‐exclusion high‐pressure liquid chromatography (HPLC). Necrotic centres on leaves are tissue damage from the infiltration. Sensitivity to PttNE1 is identified on the basis of tan necrosis between the infiltration point and the black marked end of the infiltrated area (i.e. fractions 19 and 20).
The compatible interaction involving isolate 0–1 was characterized by the accumulation of hydrogen peroxide (H2O2) and increased electrolyte leakage
It is well known that the plant hypersensitive response (incompatible reaction) involves an oxidative burst, a biochemical response resulting in reactive oxygen species (ROS). H2O2 is an important ROS and has been implicated in limiting or stopping the growth of a pathogen and signalling plant cell death (Tenhaken et al., 1995). Recently, it has been shown that some NEs induce ROS production during a susceptible interaction (Liu et al., 2012b; Manning et al., 2009). To investigate whether ROS accumulates during the barley–P. teres f. teres interaction, we monitored the disease progress and H2O2 production in both compatible (susceptible) and incompatible (resistant) interactions. Hector and NDB 112 were inoculated with isolate 0–1 and the infected leaves from both genotypes were photographed and stained with 3,3′‐diaminobenzidine (DAB) daily until 7 days after inoculation (DAI). In the first 2 days, reactions on Hector and NDB 112 were similar with both showing only a few pinpoint brown spots (Fig. 3A). However, we observed a weak DAB staining on Hector at 2 DAI, but not on NDB 112 (Fig. 3A). The net‐like lesions started to form on the leaves of Hector at 3 DAI, whereas only a few pinpoint spots were visible on the leaves of NDB 112 (Fig. 3A). The accumulation of DAB staining was clearly observed on the leaves of Hector surrounding the lesions, but only weak DAB staining was observed on NDB 112 (Fig. 3B). Disease rapidly progressed on Hector at 4, 5 and 6 DAI with the formation of complete net‐like lesions with surrounding chlorosis. DAB staining revealed a strong accumulation of H2O2, mostly occurring around the lesions (Fig. 3A,B). In contrast, on NDB 112, there were still only pinpoint lesions and weak accumulation of H2O2 around them. At 7 DAI, the whole leaf of Hector was dead and appeared to be bleached and, when stained with DAB, more than 90% of the leaf area was dark brown, indicating strong H2O2 production. However, the lesion size and H2O2 production on NDB 112 were not very different from the observations on the earlier samples (Fig. 3A,B).
Figure 3.
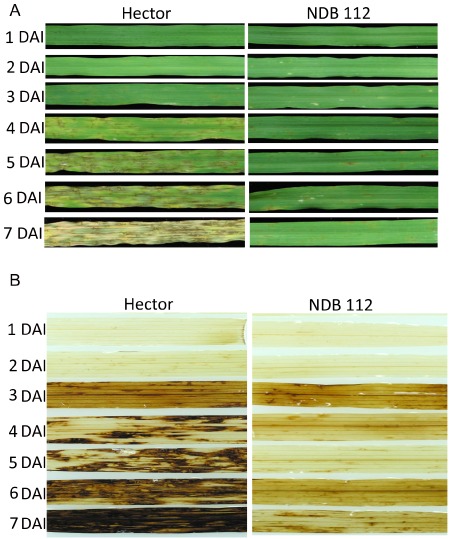
Disease progress and the accumulation of hydrogen peroxide (H2O2). (A) Hector and NDB 112 were inoculated with isolate 0–1 and the leaves of both lines were photographed daily until 7 days post‐inoculation. (B) Leaves of Hector barley and NDB 112 were stained with 3,3′‐diaminobenzidine for the detection of H2O2.
The plant defence response involves the disruption of the plant cell membrane, leading to rapid ion effluxes from plant cells into the apoplastic space (Felix et al., 1993). Furthermore, ToxA, an NE produced by Pyrenophora tritici‐repentis and P. nodorum, has been shown to induce electrolyte leakage in sensitive/susceptible genotypes (Kwon et al., 1998). Therefore, we monitored the electrolyte leakage in the compatible and incompatible barley–P. teres f. teres interactions. A continuous increase in electrolyte leakage was observed in the compatible interaction (Hector inoculated with 0–1), starting from 51.3 μS at 3 DAI to 260.0 μS at 7 DAI. In contrast, the electrolyte leakage in the incompatible interaction (NDB 112 inoculated with 0–1), as well as two controls (Hector and NDB 112 sprayed with water), remained very low. NDB 112 + 0–1 had conductivity readings of 28.2 and 34.1 μS at 6 and 7 DAI, respectively. However, these readings were significantly higher than those of the two water controls at the corresponding time points of measurement (Fig. 4), indicating that the resistance response may involve limited or controlled PCD, whereas the susceptible response results from extensive and uncontrolled PCD typical of NETS.
Figure 4.
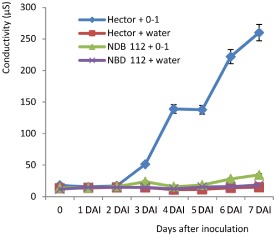
Electrolyte leakage assay in susceptible and resistant reactions. Hector and NDB 112 were inoculated with isolate 0–1 or a water control. The leaves were collected for both lines daily until 7 days post‐inoculation and were used to assay for electrolyte leakage.
QTL analysis of the HN population using a global collection of P. teres f. teres isolates
To investigate the genetic basis of host resistance/susceptibility, the HN population was evaluated for reaction to different P. teres f. teres isolates. Our hypothesis was that fungal isolates from diverse geographical regions would carry different suites of virulence/avirulence genes that would interact with the corresponding host genes to induce resistance/susceptibility. Therefore, including 0–1, we used a set of 10 P. teres f. teres isolates originating from six countries (Australia, Brazil, Canada, Denmark, Japan and the USA) representing five continents (Table 1). NDB 112 had an average reaction type ranging from 1.0 to 3.7 and was resistant to all 10 isolates tested, whereas Hector had an average disease reaction type ranging from 6.8 to 8.7 and was susceptible to all 10 isolates (Table 1). In all cases, the leaves of Hector developed extensive and large necrotic/chlorotic blotches with distinct net‐like streaks within them, whereas the leaves of NDB 112 had small dark spots or short streaks with minimal chlorosis (Fig. 5).
Table 1.
Mean disease reaction of Hector and NDB 112 to the net form net blotch isolates used in this study
| Isolate | Origin | Reference | Disease reaction | |
|---|---|---|---|---|
| Hector | NDB 112 | |||
| 0–1 | Ontario, Canada | Weiland et al. (1999) | 8.0 | 3.7 |
| 15A | CA, USA | Weiland et al. (1999) | 7.3 | 2.0 |
| LDN07Pt5 | ND, USA | Liu et al. (2012a) | 7.2 | 2.7 |
| ND89‐19 | ND, USA | Provided by B. Steffenson | 7.7 | 2 |
| NB022 | Australia | Provided by G. Platz | 6.8 | 1.7 |
| JPT9901 | Japan | Provided by J. Rasmussen | 7.7 | 3.2 |
| BB06 | Denmark | Provided by L. Jorgensen | 8.7 | 2.2 |
| NB50 | Australia | Provided by G. Platz | 8.3 | 1.8 |
| BrPteres | Brazil | Provided by F. Santana | 7.7 | 1.0 |
| 6A | CA, USA | Abu Qamar et al. (2008) | 7.7 | 3.2 |
Figure 5.

The disease reaction of barley lines Hector (susceptible) and NDB 112 (resistant) to the 10 Pyrenophora teres f. teres isolates used in this study.
QTL analysis revealed a number of genomic regions significantly (LOD ≥ 4.3) associated with the reaction to P. teres f. teres, with each isolate having two to three QTLs significantly associated with disease. The information on all the significant QTLs is summarized in Table 2 and shown graphically in Fig. 6.
Table 2.
Summary of quantitative trait loci (QTLs) in the Hector × NDB 112 population for each isolate identified by composite interval mapping
| Isolate | Chromosome location | Chromosome position (cM) | Closest marker | Peak LOD | R 2 | Source |
|---|---|---|---|---|---|---|
| 0–1 | 3HL | 164 | 2335‐1614 | 6.3 | 0.11 | NDB 112 |
| 6H | 46 | SPN1 | 19.5 | 0.31 | NDB 112 | |
| 15A | 1HL | 96–100 | 3087‐1763 | 5.1 | 0.09 | Hector |
| 2HS | 14–24 | 791–1113 | 9.4 | 0.10 | NDB 112 | |
| 6H | 46 | SPN1 | 12.1 | 0.21 | NDB 112 | |
| LDN07Pt5 | 3HS | 0–2 | ConsensusGBS0194‐1 | 19.3 | 0.20 | NDB 112 |
| 3HL | 162 | 6716‐823 | 9.7 | 0.08 | NDB 112 | |
| 6H | 46 | SPN1 | 31.6 | 0.34 | NDB 112 | |
| ND89‐19 | 3HL | 162 | 6716‐823 | 14.5 | 0.17 | NDB 112 |
| 5HS | 38 | 4570‐591 | 6.2 | 0.16 | NDB 112 | |
| 6H | 46 | SPN1 | 17.3 | 0.34 | NDB 112 | |
| NB022 | 2HS | 0–18 | 9490‐843 | 8.2 | 0.13 | NDB 112 |
| 6H | 46 | SPN1 | 15.4 | 0.28 | NDB 112 | |
| JPT9901 | 3HL | 138–144 | 1898‐580 | 7.5 | 0.07 | NDB 112 |
| 5HS | 30–36 | 4334‐482 | 9.9 | 0.18 | NDB 112 | |
| 6H | 54 | 5187‐752 | 12.4 | 0.21 | NDB 112 | |
| BB06 | 3H | 68–70 | 2804‐1832 | 36.4 | 0.64 | NDB 112 |
| 3HL | 164–166 | 2335‐1614 | 6.7 | 0.01 | NDB 112 | |
| NB50 | 3H | 68–70 | 2804‐1832 | 38.8 | 0.66 | NDB 112 |
| 3HL | 166–170 | 3718‐1026 | 9.4 | 0.04 | NDB 112 | |
| 5HS | 40–46 | 1861–2382 | 5.9 | 0.07 | NDB 112 | |
| BrPteres | 3H | 68–70 | 2804‐1832 | 38.5 | 0.73 | NDB 112 |
| 3HL | 162 | 6716‐823 | 4.5 | 0.01 | NDB 112 | |
| 5HS | 28–30 | 4977‐567 | 4.7 | 0.07 | NDB 112 | |
| 6A | 3HL | 164 | 2335‐1614 | 21.0 | 0.31 | NDB 112 |
| 5HS | 38–40 | 2664‐314 | 14.6 | 0.22 | NDB 112 |
LOD, logarithm of odds.
Figure 6.
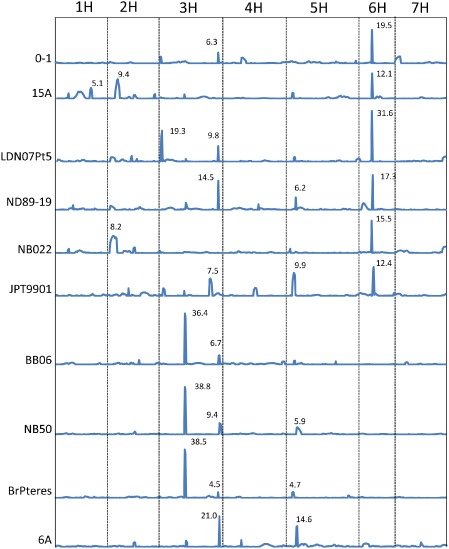
The location of quantitative trait loci (QTLs) associated with disease reaction to each isolate. Composite interval mapping of seven chromosomes was provided to show the location of all the significant QTLs with logarithm of the odds (LOD) > 4.5 (α = 0.05). The LOD value for each QTL is indicated by the number adjacent to the QTL peak.
All barley chromosomes, except 4H and 7H, harboured QTLs significantly associated with NFNB (Fig. 6). Among them, chromosome 6H carried a major QTL associated with disease caused by six of the 10 isolates, including 15A, 0–1, LDN07Pt5, ND89‐19, JPT9901 and NB022, which explained 21%–33% of the phenotypic variation. Five of the six isolates showing a 6H QTL, including 15A, 0–1, LDN07Pt5, ND89‐19 and NB022, peaked directly over the SPN1 phenotypic marker, indicating that they all probably produce PttNE1 (Fig. 1). However, the 6H QTL associated with JPT9901 peaked 8 cM from the SPN1 locus (Fig. 1), indicating the potential for a second 6H locus being involved in susceptibility/resistance.
HN progeny lines harbouring PttNE1 sensitivity (SPN1) were compared with lines insensitive to PttNE1 (spn1). Significant differences (P < 0.01) in the disease reaction were identified between SPN1 and spn1 lines only for the six isolates with a 6H QTL, with differences ranging from 0.99 to 1.87 (Table 3). Average disease reactions for PttNE1‐insensitive lines ranged from 2.97 to 5.91, whereas sensitive lines ranged from 4.32 to 6.92 (Table 3).
Table 3.
Average disease reactions of Hector × NDB 112 recombinant inbred lines sensitive to PttNE1 (SPN1) or insensitive to PttNE1 (spn1) when inoculated with 10 different P yrenophora teres f. teres isolates
| SPN1 | spn1 | Average difference | |
|---|---|---|---|
| 0–1 | 6.92 | 5.76 | 1.16* |
| 15A | 5.80 | 4.39 | 1.41* |
| LDN07Pt5 | 6.68 | 5.03 | 1.65* |
| ND89‐19 | 6.42 | 4.55 | 1.87* |
| NB022 | 4.32 | 2.97 | 1.35* |
| JPT9901 | 6.90 | 5.91 | 0.99* |
| BB06 | 5.66 | 5.57 | 0.09NS |
| NB50 | 4.46 | 4.54 | −0.08NS |
| BrPteres | 4.31 | 4.59 | −0.28NS |
| 6A | 5.84 | 5.37 | 0.47NS |
NS, not significant.
*Significantly different at the 0.01 level of probability.
Disease caused by the isolates NB50, BrPteres and BB06 was associated with a major QTL near the middle of chromosome 3H that explained as much as 73% of the disease variation, indicating that this QTL had major effects on the disease reaction. For isolate 6A, neither the 3H nor 6H QTL was associated with the disease, but two significant QTLs, one on chromosome arm 3HL and the other on 5HS, were detected, explaining 31% and 22% of disease variation, respectively.
In addition to the major QTLs on 3H and 6H, relatively minor QTLs were found to be associated with each isolate (Table 2, Fig. 6). Some of the QTLs were located on the same or similar genomic regions, indicating the involvement of the same host and pathogen genes. For example, ND89‐19, 0–1, LDN07Pt5, BB06 and BrPteres all had a relatively minor QTL located in the genetic region of 162–166 cM on chromosome 3HL, which was the same region as for the major QTL identified in the 6A inoculation. However, other QTLs were only detected for one or two isolates, which suggests unique host–pathogen gene interactions (Table 2, Fig. 6). The susceptible parent Hector conferred all susceptibility except for the 1HL QTL identified for 15A (Table 2).
Discussion
NETS in the barley–P. teres f. teres pathosystem
Previous studies have improved our understanding of the genetic basis of NFNB disease interaction in which the major gene‐for‐gene interactions between host and pathogen are involved. The findings of previous studies in which both dominant and recessive resistance genes have been identified have suggested the potential for both effector/pathogen‐associated molecular pattern‐triggered immunity (ETI/PTI) and NETS in this pathosystem. However, until now, no direct evidence at the molecular level has existed to support either model.
Necrotrophic fungal pathogens are known to produce NEs to induce host cell death, providing nutrients that benefit the growth of the pathogen. NEs have been identified from numerous necrotrophic specialists found in the order Pleosporales within the class Dothideomycetes (reviewed by Friesen et al., 2008; Stergiopoulos et al., 2012). Pyrenophora tritici‐repentis and P. nodorum, two fungal pathogens of wheat, are known to produce multiple proteinaceous NEs (reviewed by Ciuffetti et al., 2010; Oliver et al., 2012). Toxins and other metabolic compounds have previously been identified from P. teres f. teres, but these compounds have not been shown to have a strong association with disease (reviewed by Liu et al., 2011). To reveal the molecular basis of the barley–P. teres f. teres interaction and to identify NEs produced by P. teres f. teres, we followed the procedure used for the model tomato leaf mould pathogen Passalora fulva (formerly Cladosporium fulvum), where effectors were purified from apoplastic fluid (de Wit and Spikman, 1982). Using IWF from infected plant tissue, for the first time, we identified and mapped sensitivity to a proteinaceous NE (PttNE1) that shows a direct association with disease. The co‐localization of the NE sensitivity locus, designated SPN1, and the disease QTL strongly suggests that the compatible sensitivity gene–NE interaction is responsible for the effects of the QTL. Dominant sensitivity in the sensitive/susceptible parent Hector was validated by a 3:1 sensitive : insensitive ratio when F2 individuals of a Hector × NDB 112 cross were infiltrated with PttNE1 purified from IWF, indicating that the IWF contains an NE involved in the induction of necrosis and disease. This is similar to the situations for NEs that have been identified in other systems (Ciuffetti et al., 2010; Oliver et al., 2012). We further determined that the effector PttNE1 was a small protein with a molecular mass in the range 6.5–13.5 kDa, similar to the small secreted NE proteins reported in P. tritici‐repentis and P. nodorum (Liu et al., 2012b). Here, we provide the first molecular evidence that the barley NFNB disease system is, at least partially, governed by NETS. However, we cannot rule out the possibility that the ETI model might explain other aspects of the barley–P. teres f. teres interaction because several dominant resistance genes have been reported.
The accumulation of H2O2 and increased level of electrolyte leakage shed light on the necrotrophic lifestyle of P. teres f. teres
The oxidative burst characterized by the increase in ROS and PCD characterized by cell lysis are both defence responses known to inhibit biotrophic and hemibiotrophic pathogens (Tenhaken et al., 1995). H2O2 is an important ROS and has been implicated in limiting or stopping the growth of a pathogen and signalling plant cell death. However, recently, it has been shown that some NEs induce ROS production during a susceptible interaction (Liu et al., 2012b; Manning et al., 2009). The NFNB susceptible interaction was characterized by higher levels of H2O2 production and electrolyte leakage compared with the resistant reaction, as demonstrated by the dark DAB staining on leaves of the susceptible line Hector (Fig. 3) and the increased conductivity of the leakage from Hector diseased leaves (Fig. 4), respectively. These hallmarks of a resistance response are usually thought to be part of the defence response resulting from effector recognition and the resulting downstream events. However, several studies have suggested that, in addition to having a role in plant resistance, an increase in ROS concentration in planta during the early stages of infection and proliferation can be used to the advantage of necrotrophic pathogens, if the pathogen is able to survive and grow under these conditions (Bao et al., 2014; Govrin and Levine, 2000; Tiedemann, 1997). This increase and maintenance of ROS levels associated with cell lysis and nutrient leakage can be provided by pathogen‐produced ROS, plant‐produced ROS or by the pathogen's suppression of plant peroxidases (Tiedemann, 1997). It is obvious that P. teres f. teres has little difficulty surviving and proliferating under high levels of ROS, and it is likely that many necrotrophic pathogens, including P. teres f. teres, have evolved mechanisms necessary to modulate the defence‐associated oxidative burst and therefore are able to benefit from the nutrients released during the resulting cell death. The ability to induce PCD associated with the classical resistance response and not only survive, but thrive and sporulate, under these harsh conditions, has been shown to be a signature for several necrotrophic pathogens, including the closely related wheat pathogens P. tritici‐repentis and P. nodorum (Faris et al., 2010), and other NE‐producing pathogens (Lorang et al., 2007; Nagy and Bennetzen, 2008), as well as necrotrophic generalist pathogens, including some Sclerotinia, Botrytis and Fusarium species (Bao et al., 2014; Bolton et al., 2006; Govrin and Levine, 2000; Tiedemann, 1997). This ability to induce, maintain and survive the oxidative burst appears to be a major factor in the evolution of both the necrotrophic generalist and necrotrophic specialist lifestyles.
QTL detection and comparison with previous studies
Numerous studies have been conducted to map NFNB resistance genes/QTLs on barley chromosomes (see review by Liu et al., 2011). In most cases, however, these studies used a single isolate or only a few isolates from a single geographical region for disease evaluation. Unfortunately, this does not provide an indication of the scope of the pathogen virulence and host resistance. By using a geographically and pathogenically diverse set of isolates, we were able to capture more genetic loci that govern host resistance/susceptibility and to obtain a better understanding of the genetic relationship of the host and pathogen.
In total, we identified more than 10 genomic regions significantly (P < 0.05) associated with the reaction to the 10 P. teres f. teres isolates, with each isolate having two to three QTLs associated with the disease (Fig. 6, Table 2). Two genomic regions, one on chromosome 6H and the other on 3H, had the largest effects and were associated with disease induced by multiple isolates. This suggests that isolates with different geographical origins may carry similar NE genes that interact with the gene products of these barley loci. In addition to these two major QTLs, there were other genomic regions associated with disease induced by only one or two isolates, indicating that these isolates may harbour virulence/avirulence factors that are less common in the global population. This result agrees well with previous reports on NFNB where one to three major genes, in addition to minor genes, have been shown to control NFNB resistance/susceptibility in barley in any given biparental population (reviewed in Liu et al., 2011). Current and previous genetic studies using fungal mapping populations have also shown that a few major genes in each P. teres f. teres isolate condition virulence/avirulence towards a particular host genotype (Afanasenko et al., 2007; Beattie et al., 2007; Lai et al., 2007; Shjerve et al., 2014).
The 6H QTL region has been reported in several previous studies using a variety of host populations and pathogen isolates (see review by Liu et al., 2011). It is likely that multiple resistance/susceptibility genes are present at the 6H locus, not only because of the number of reports of the 6H region, but also because of the report of this region harbouring both dominant and recessive forms of resistance (Abu Qamar et al., 2008; Friesen et al., 2006; Gupta et al., 2010; St. Pierre et al., 2010). In the current study, we found that the 6H association with susceptibility was identified for six of the 10 isolates tested, with these six being geographically diverse. Five of the six 6H QTLs peaked directly above SPN1, highlighting the importance of PttNE1 in virulence. The data presented in Table 3 show that, when comparing PttNE1‐sensitive and PttNE1‐insensitive lines, there is a significant difference between the two groups, as expected on the basis of QTL analysis. However, it is interesting to note that the average disease reactions of the insensitive lines for three of the five isolates that have a QTL peak directly over SPN1 is still above 5.0, indicating that the other relatively minor QTLs are probably a result of undetected NE–susceptibility gene interactions. The peak for the 6H QTL of JPT9901 did not align with SPN1, but instead appeared to be a distinct locus. This also suggests that, even within the HN population, different genes may exist in this region, conferring resistance/susceptibility depending on the isolate used.
Several studies using various RIL and doubled haploid (DH) mapping populations have identified a region on chromosome 3H which, on the basis of common markers, appears to be in the same region as the major 3H QTLs reported here (Graner et al., 1996; Gupta et al., 2004, 2010; Raman et al., 2003; Yun et al., 2005). At least three of these studies also used the Australian isolate NB50 (Gupta et al., 2004, 2010; Raman et al., 2003). Using this same NB50 isolate on the HN population, we also detected a major QTL on 3H (R 2 = 0.66) in addition to two minor effect QTLs on 3HL and 5HS. Gupta et al. (2004) used NB50 to evaluate four DH populations, all using the same susceptible parent, and a 3H QTL alone was identified in two of the populations, but the major QTL mapped to either chromosome 6H or 4H in the other two populations. In the DH population of Sloop/Halcyon, seedling resistance to NB50 mapped to chromosomes 4H and 3H with R 2 = 0.64 and 0.09, respectively (Raman et al., 2003). Similarly, the Californian isolate 6A has been shown previously to have virulence towards a dominant susceptibility gene on chromosome 6H in the DH population of Rika × Kombar (Abu Qamar et al., 2008; Liu et al., 2010). However, in the current study, there was no obvious 6H QTL detected for 6A. Instead, two new QTLs were identified, one on the distal end of the long arm of 3H and the other on the short arm of 5H, each having a similar size of effect (Table 2). Collectively, these studies indicate that the interaction of barley–P. teres f. teres is determined by both resistance and susceptibility genes from the host and avirulence and virulence genes in the pathogen, which act in a gene‐for‐gene (ETI/PTI) or inverse gene‐for‐gene (NETS) manner, respectively. The cloning and functional characterization of the host and pathogen genes will help to expose the mechanism or mechanisms of pathogen virulence and host resistance.
Gene action of NFNB resistance
NFNB resistance has been described as dominant, recessive and incompletely dominant (Liu et al., 2011), but many studies in which genes or QTLs have been localized to genomic regions have been performed using RIL or DH populations, neither of which allows dominance to be measured. In two previous studies, we evaluated F2 individuals of crosses in which a single major gene was mapped on chromosome 6H using DH populations. In one case, the ratios of the F2 individuals indicated dominant resistance (Friesen et al., 2006) and in the other indicated dominant susceptibility (Abu Qamar et al., 2008). Ho et al. (1996) analyzed the reactions of F1 and F2 lines to NFNB and found resistance to be recessive (dominant susceptibility).
O'Boyle et al. (2011) reported a single dominant resistance gene in NDB 112 based on the analysis of F2 individuals of a Hector × NDB 112 cross, the same cross as used in this article. Using the same ND89‐19 isolate, our analysis of the HN RIL population did not indicate a single resistance gene, but rather multiple genes. Several reasons for this discrepancy are possible; however, based on QTL analysis of the HN RIL population, it is clear that multiple loci are involved in conferring resistance/susceptibility to isolate ND89‐19. It is also probable that ND89‐19 produces PttNE1, because the SPN1 locus defined the peak of the 6H QTL associated with disease caused by ND89‐19. In addition, inoculation of Hector × NDB 112 F2 individuals showed a continuum of reaction types which made it impossible for us to confidently call a cutoff between resistance and susceptibility, which also indicated that genetic control of resistance was more complex than a single dominant gene (data not shown). Extensive effort has been made to transfer the source of resistance in NDB 112 for SFNB and NFNB. This has resulted in many resistant cultivars with SFNB resistance, but little success has been met in transferring NFNB resistance into released varieties (O'Boyle et al., 2011; Steffenson et al., 1996). This is probably a result of the fact that breeders are often looking to incorporate dominant resistance genes. Our study suggests that, when dealing with necrotrophic specialist pathogens, breeders may need to focus on the removal of the dominant susceptibility gene or genes inducing NETS.
The current study has identified the NE sensitivity locus SPN1, which corresponds to sensitivity to the NE PttNE1. SPN1 was also shown to be highly associated with disease when the HN population was inoculated with 0–1, the isolate from which PttNE1 was isolated. In addition, four other isolates had major 6H QTLs that peaked directly over SPN1, indicating that PttNE1 is also produced in planta by these isolates. Inoculation of the PttNE1‐sensitive and highly susceptible barley line Hector with the PttNE1‐producing P. teres f. teres isolate 0–1 induced cellular disruption (PCD), as indicated by electrolyte leakage and a significant increase in ROS, both hallmarks of resistance in a biotrophic or hemibiotrophic interaction.
Experimental Procedures
Plant materials
A barley population of 118 RILs was developed from a cross between the barley variety Hector (CIho 15514), developed in Alberta, Canada (Wells, 1973), and the North Dakota breeding line NDB 112. Hector is highly susceptible to all NFNB isolates tested by us and NDB 112 has been used as a source of resistance to both NFNB and SFNB (caused by Cochliobolus sativus) (Rasmusson et al., 1999; Steffenson et al., 1996; Wilcoxson et al., 1990). The Hector × NDB 112 (HN) RIL population was advanced to the F7 generation from F2 individuals by single seed descent.
Marker development
SSR and SNP markers were used to genotype the 118 lines of the HN population. The sequences for barley SSR primers were obtained from Varshney et al. (2007) and SSR genotyping was performed in the US Department of Agriculture‐Agricultural Research Service (USDA‐ARS) genotyping center at Fargo, ND, USA using a high‐throughput procedure described by Tsilo et al. (2009). Eighty‐seven SSR primer pairs were tested on the parental lines and 77 pairs were selected to run on the whole population. For SNP genotyping, we used the barley OPA1 (BOPA1) platform containing 1536 SNPs (Close et al., 2009). SNP genotype calling was performed using Illumina's GenomeStudio software (Illumina, Inc., San Diego, CA, USA). The genotype calls were then manually inspected to ensure call accuracy. Of the 1536 SNPs genotyped, 701 SNPs that were found to be polymorphic and segregating in the HN population were used in the mapping analysis.
Map construction
A total of 778 markers (701 SNPs and 77 SSRs) was used to assemble the map in the HN population. The Microsoft Excel‐based computer software MapDisto (Lorieux, 2007) was utilized to build the genetic map with reference to the previously published barley maps (Close et al., 2009; Varshney et al., 2007). Linkage groups were determined using the ‘Find groups’ command with LODmin = 3.0 and r max = 0.3. Linkage groups were then broken down into smaller groups by increasing LODmin to 5.0 and r max = 0.3 for individual chromosome mapping. Markers in each linkage group were ordered using the ‘order sequence’ command that implements a seriation algorithm with the Sum of Adjacent Recombination Frequencies as criteria. ‘Ripple order’ and ‘Check inversions’ commands were repeatedly executed to refine the marker order and to eliminate any co‐segregating or low‐quality markers. After the map was determined, map data were saved in a QGene file format for QTL analysis.
IWF extraction and plant infiltration, Pronase treatment and HPLC separation
For IWF extraction, Hector plants were inoculated at the two‐ to three‐leaf stage with isolate 0–1 using the method described in Friesen et al. (2006). After incubation in 100% humidity for 24 h, the plants were placed in a growth chamber under a 12‐h photoperiod at 21 °C. Infected leaves were collected to extract IWFs when distinctive net‐like symptoms appeared, but before chlorosis developed around the lesions. The leaves were cut into 3–4‐cm‐long fragments and placed in 50‐mL conical centrifuge tubes containing distilled water, followed by vacuum infiltration for 5 min, and incubation for another 30 min in the vacuum chamber at a pressure of 1500 Pa. The leaf sections were then air dried briefly to remove surface water. Approximately 30 leaf fragments were bundled and placed in the bottom of a 10‐mL syringe with the plunger removed and placed in an empty 50‐mL conical centrifuge tube. The tubes were then centrifuged at 3000 × g in a swinging bucket rotor for 15 min to collect the fluids from the intercellular space. The raw IWF from ∼120 leaf fragments was concentrated to ∼500 μL using a 4‐mL, 3‐kDa, Amicon Ultra (EMD/Millipore, Billerica, MA, USA), regenerated cellulose centrifugal filter. The concentrated IWF was used fresh for downstream applications, including plant infiltration, protease treatment and chromatography.
The IWF infiltration was conducted as described in Liu et al. (2004) and the reactions were evaluated on the basis of the presence or absence of necrosis (Fig. 2). The HN population was evaluated for reaction to IWF in two independent experiments and the results were used as phenotypic trait data to map the chromosomal location of the Hector sensitivity. Protease treatments were performed as described in Liu et al. (2004). The 0–1 IWF was incubated at room temperature for 4 h in the presence of Pronase (EMD Biosciences, Inc., San Diego, CA, USA) at 1 mg/mL. Samples were incubated with water alone as an untreated control. The untreated and treated samples were infiltrated into Hector at the second‐leaf stage. The activity was assayed by scoring plant leaves as sensitive or insensitive as mentioned previously. The size of the NE was determined by HPLC using a Phenomenex BioSep‐SEC‐S 2000 (600 × 7.8 mm2, 5 μm) size exclusion column (Phenomenex, Torrance, CA, USA). After chromatography, individual HPLC fractions were infiltrated on Hector and rated for biological activity as stated above.
Monitoring of disease progress and DAB staining
To monitor disease progress and H2O2 accumulation in the resistant and susceptible reactions, NDB 112 and Hector plants were inoculated with isolate 0–1 and leaves of both genotypes were collected each day until 7 days post‐inoculation for photographing and DAB staining. The preparation of the DAB staining buffer (1 mg/mL) and the process for staining followed the procedure described by Liu et al. (2012b).
Electrolyte leakage assay
To monitor the electrolyte leakage in compatible and incompatible barley–P. teres f. teres interactions, Hector and NDB 112 plants were inoculated with P. teres f. teres isolate 0–1 or a water control and the leaves were collected at the designated time points and placed in water for the measurement of electrical conductivity. For each measurement, two leaves were collected and cut into eight 3‐cm‐long fragments. Leaves were immersed in 10 mL of distilled water in a 15‐mL centrifuge tube and vacuum infiltrated for 30 min. After 1 h of incubation at room temperature, the solution was mixed thoroughly and measured for the electrical conductivity with an Orion 3 Star portable conductivity meter (Thermo Scientific, Waltham, MA, USA). The reading for distilled water was around 1.0. The mean and standard error from three biological replications were calculated using Microsoft Excel (Microsoft, Redmond, WA, USA). To detect whether there were significant differences among different treatments, a Fisher's protected least‐significant difference (LSD) at α < 0.05 was calculated using SAS 9.03 (SAS Institute, Cary, IN, USA, 2011).
Disease evaluation
Ten fungal strains (Table 1) from six different countries representing five continents were used in the evaluation. Plant materials were prepared as described in Friesen et al. (2006) and inoculated at the two‐ to three‐leaf stage. For fungal inoculum, conidia were grown and harvested as described by Weiland et al. (1999). The conidia concentration was adjusted to 3000 spores/mL and two drops of Tween 20 (polyoxyethylene sorbitan monolaurate) per 100 mL were added to the inoculum solution. Inoculation and plant incubation post‐inoculation were performed as described by Friesen et al. (2006). The 1–10 scale described by Tekauz (1985) was used to score disease reaction types. Three replications were completed for each isolate and the disease mean from three replications was used in QTL mapping.
QTL mapping
The computer program QGene 4.3 (http://www.qgene.org/qgene/index.php) (Joehanes and Nelson, 2008) was used to conduct QTL analysis on the HN population for disease response. A permutation test with 1000 permutations indicated that a LOD threshold of 4.5 in this population yields an experiment‐wise significance level of 0.05. The location and LOD value of each QTL were determined using composite interval mapping (CIM) which was implemented in the software. The R2 for each QTL was determined using simple interval mapping.
Supporting information
Fig. S1 Genetic maps of the seven chromosomes (1H–7H) in the Hector × NDB 112 recombinant inbred population.
Table S1 Summary of genetic maps in the Hector × NDB 112 population.
Acknowledgements
The authors thank Drs Melvin Bolton and Jared LeBoldus for critical review of the manuscript. This research was supported by funding from USDA‐NIFA‐AFRI grant #2011‐68002‐30029 (T‐CAP), the North Dakota Barley Council and USDA‐ARS CRIS Project no. 5442‐22000‐048‐00D.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture (USDA). USDA is an equal opportunity provider and employer.
References
- Abu Qamar, M. , Liu, Z.H. , Faris, J.D. , Chao, S. , Edwards, M.C. , Lai, Z. , Franckowiak, J.D. and Friesen, T.L. (2008) A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor. Appl. Genet. 117, 1261–1270. [DOI] [PubMed] [Google Scholar]
- Adhikari, T.B. , Bai, J. , Meinhardt, S.W. , Gurung, S. , Myfield, M. , Patel, J. , Ali, S. , Gudmestad, N.C. and Rasmussen, J.B. (2009) Tsn1‐mediated host responses to ToxA from Pyrenophora tritici‐repentis . Mol. Plant–Microbe Interact. 22, 1056–1068. [DOI] [PubMed] [Google Scholar]
- Afanasenko, O. , Mironenko, N. , Filatova, O. , Kopahnke, D. , Krämer, I. and Ordon, F. (2007) Genetics of host–pathogen interactions in the Pyrenophora teres f. teres (net form)–barley (Hordeum vulgare) pathosystem. Eur. J. Plant Pathol. 117, 267–280. [Google Scholar]
- Bach, E. , Christensen, S. , Dalgaard, L. , Larsen, P.O. and Olsen, C.E. (1979) Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres . Physiol. Plant Pathol. 14, 41–46. [Google Scholar]
- Bao, G. , Bi, Y. , Li, Y. , Kou, Z. , Hu, L. , Ge, Y. , Wang, Y. and Wang, D. (2014) Overproduction of reactive oxygen species involved in the pathogenicity of Fusarium in potato tubers. Physiol. Mol. Plant Pathol. 86, 35–42. [Google Scholar]
- Beattie, A.D. , Scoles, G.J. and Rossnagel, B.G. (2007) Identification of molecular markers linked to a Pyrenophora teres avirulence gene. Phytopathology, 97, 842–849. [DOI] [PubMed] [Google Scholar]
- Bockelman, H.E. , Sharp, E.L. and Eslick, R.F. (1977) Trisomic analysis of genes for resistance to scald and net blotch in several barley cultivars. Can. J. Bot. 55, 2142–2148. [Google Scholar]
- Bolton, M.D. , Thomma, B.P.H.J. and Nelson, B.D. (2006) Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. [DOI] [PubMed] [Google Scholar]
- Cakir, M. , Gupta, S. , Platz, G.J. , Ablett, G.A. , Loughman, R. , Emebiri, L.C. , Poulsen, D. , Li, C.D. , Lance, R.C.M. , Galway, N.W. , Jones, M.G.K. and Appels, R. (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Aust. J. Agric. Res. 54, 1369–1377. [Google Scholar]
- Campbell, G.F. , Lucas, J.A. and Crous, P.W. (2002) Evidence of recombination between net‐ and spot‐type populations of Pyrenophora teres as determined by RAPD analysis. Mycol. Res. 106, 602–608. [Google Scholar]
- Ciuffetti, L.M. , Manning, V.A. , Pandelova, I. , Betts, M.F. and Martinez, J.P. (2010) Host‐selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici‐repentis–wheat interaction. New Phytol. 187, 911–919. [DOI] [PubMed] [Google Scholar]
- Close, T.J. , Bhat, P.R. , Lonardi, S. , Wu, Y. , Rostoks, N. , Ramsay, L. , Druka, A. , Stein, N. , Svensson, J.T. , Wanamaker, S. , Bozdag, S. , Roose, M.L. , Moscou, M.J. , Chao, S. , Varshney, R.K. , Szucs, P. , Sato, K. , Hays, P.M. , Matthews, D.E. , Kleinhofs, A. , Muehlbauer, G.J. , DeYoung, J. , Marshall, D.F. , Madishetty, K. , Fenton, R.D. , Condamine, P. , Graner, A. and Waugh, R. (2009) Development and implementation of high‐throughput SNP genotyping in barley. BMC Genomics, 10, 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emebiri, L.C. , Platz, G. and Moody D.B. (2005) Disease resistance genes in a doubled haploid population of two‐rowed barley segregating for malting quality attributes. Aust. J. Agric. Res, 56, 49–56. [Google Scholar]
- Faris, J.D. , Zhang, Z. , Lu, H.J. , Lu, S.W. , Reddy, L. , Cloutier, S. , Fellers, J.P. , Meinhardt, S.W. , Rasmussen, J.B. , Xu, S.S. , Oliver, R.P. , Simons, K.J. and Friesen, T.L. (2010) A unique wheat disease resistance‐like gene governs effector‐triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA, 107, 13 544–13 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, G. , Regenass, M. and Boller, T. (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein‐phosphorylation, and establishment of a refractory state. Plant J. 4, 307–316. [Google Scholar]
- Friesen, T.L. and Faris, J.D. (2010) Characterization of the wheat–Stagonospora nodorum disease system: what is the molecular basis of this quantitative necrotrophic disease interaction? Can. J. Plant Pathol. 32, 20–28. [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Lai, Z. and Steffenson, B.J. (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled‐haploid barley population. Genome, 49, 855–859. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Meinhardt, S.W. and Faris, J.D. (2007) The Stagonospora nodorum–wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51, 681–692. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Solomon, P.S. and Oliver, R.P. (2008) Host‐specific toxins, effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. [DOI] [PubMed] [Google Scholar]
- Friesen, T.L. , Faris, J.D. , Chu, C. and Xu, S.S. (2012) SnTox5‐Snn5: a novel Stagonospora nodorum effector–wheat gene interaction and its relationship with the SntoxA–Tsn1 and SnTox3–Snn3‐B1 interactions. Mol. Plant Pathol. 13, 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis, P. , Olsen, C.E. and Møller, B.L. (1991) Toxin production in Pyrenophora teres, the Ascomycete causing the net‐spot blotch disease of barley (Hordeum vulgare L.). J. Biol. Chem. 266, 13 329–13 335. [PubMed] [Google Scholar]
- Govrin, E.M. and Levine, A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea . Curr. Biol. 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Graner, A. , Foroughi‐Wehr, B. and Tekauz, A. (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica, 91, 229–234. [Google Scholar]
- Grewal, T.S. , Rossnagel, B.G. and Scoles, G.J. (2012) Mapping quantitative trait loci associated with spot blotch and net blotch resistance in a doubled haploid barley population. Mol. Breed. 30, 267–279. [Google Scholar]
- Gupta, S. , Wielinga, C. , Li, C. , Cakir, M. , Platz, G. , Loughman, R. , Lance, R. and Appels, R. (2004) Gene distribution and SSR markers linked with net type net blotch resistance in barley. In: Proceedings of the 9th International Barley Genetics Symposium (Spunar, J. and Janikova, J., eds.) Agricultural Research Institute Kromeriz, Ltd., Brno, Czech Republic, June 20–26, 2004. pp. 668–673.
- Gupta, S. , Li, C. , Loughman, R. , Cakir, M. , Platz, G. , Westcott, S. , Bradley, J. , Broughton, S. and Lance, R. (2010) Quantitative trait loci and epistatic interactions in barley conferring resistance to net type net blotch Pyrenophora teres f. teres isolates. Plant Breed. 129, 362–368. [Google Scholar]
- Ho, K.M. , Tekauz, A. , Choo, T.M. and Martin, R.A. (1996) Genetic studies on net blotch resistance in a barley cross. Can. J. Plant Sci. 76, 715–720. [Google Scholar]
- Hysing, S.‐C. and Wiik, L. (2013) The role of seed infection level and fungicide seed treatments in control of net blotch in barley. Eur. J. Plant Pathol. 137, 169–180. [Google Scholar]
- Joehanes, R. and Nelson, J.C. (2008) QGene 4.0, an extensible Java QTL analysis platform. Bioinformatics, 24, 2788–2789. doi: 10.1093/bioinformatics/btn523. [DOI] [PubMed] [Google Scholar]
- Keon, J.P.R. and Hargreaves, J.A. (1983) A cytological study of the net blotch disease of barley caused by Pyrenophora teres . Physiol. Plant Pathol. 22, 321–329. [Google Scholar]
- Khan, T.N. and Boyd, W.J.R. (1969) Inheritance of resistance to net blotch in barley. II. Genes conditioning resistance against race W.A.‐2. Can. J. Genet. Cytol. 11, 592–597. [Google Scholar]
- König, J. , Perovic, D. , Kopahnke, D. and Ordon, F. (2013) Development of an efficient method for assessing resistance to the net type of net blotch Pyrenophora teres f. teres in winter barley and mapping of quantitative trait loci for resistance. Mol. Breed. 32, 641–650. [Google Scholar]
- Kwon, C.Y. , Rasmussen, J.B. and Meinhardt, S.W. (1998) Activity of Ptr ToxA from Pyrenophora tritici‐repentis requires host metabolism. Physiol. Mol. Plant Pathol. 52, 201–212. [Google Scholar]
- Lai, Z. , Faris, J.D. , Weiland, J.J. , Steffenson, B.J. and Friesen, T.L. (2007) Genetic mapping of Pyrenophora teres f. teres genes conferring avirulence on barley. Fungal Genet. Biol. 44, 323–329. [DOI] [PubMed] [Google Scholar]
- Liu, Z.H. , Faris, J.D. , Meinhardt, S.W. , Ali, S. , Rasmussen, J.B. and Friesen, T.L. (2004) Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host selective toxin produced by Stagonospora nodorum . Phytopathology, 94, 1056–1060. [DOI] [PubMed] [Google Scholar]
- Liu, Z.H. , Faris, J.D. , Edwards, M.C. and Friesen, T.L. (2010) Development of expressed sequence tag EST‐based markers for genomic analysis of a barley 6H region harboring multiple net form net blotch resistance genes. Plant Genome, 3, 41–52. [Google Scholar]
- Liu, Z.H. , Ellwood, S.R. , Oliver, R.P. and Friesen, T.L. (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 121, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.H. , Zhong, S. , Stasko, A.K. , Edwards, M.C. and Friesen, T.L. (2012a) Virulence profile and genetic structure of a North Dakota population of Pyrenophora teres f. teres, the causal agent of net form net blotch of barley. Phytopathology, 102, 539–546. [DOI] [PubMed] [Google Scholar]
- Liu, Z.H. , Zhang, Z. , Faris, J.D. , Oliver, R.P. , Syme, R. , McDonald, M.C. , McDonald, B.M. , Solomon, P.S. , Lu, S. , Shelver, W.L. , Xu, S.S. and Friesen, T.L. (2012b) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 81, e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl. Acad. Sci. USA, 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M. , Kidarsa, T. , Bradford, C.S. , Gilbert, B. , Curtis, M. , Tzeng, S.‐C. , Maier, C.S. and Wolpert, T.J. (2012) Tricking the guard: exploiting plant defense for disease susceptibility. Science, 338, 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux, M. (2007) MapDisto, a free user‐friendly program for computing genetic maps. computer demonstration. P958 given at the Plant and Animal Genome XV Conference, 13–17 January 2007, San Diego, CA, USA. Available at http://mapdisto.free.fr/ (Accessed August 1, 2014).
- Ma, Z.Q. , Lapitan, N.L.V. and Steffenson, B. (2004) QTL mapping of net blotch resistance genes in a doubled‐haploid population of six‐rowed barley. Euphytica, 137, 291–296. [Google Scholar]
- Manninen, O. , Kalendar, R. , Robinson, J. and Schulman, A.H. (2000) Application of BARE‐1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol. Gen. Genet. 264, 325–334. [DOI] [PubMed] [Google Scholar]
- Manninen, O.M. , Jalli, M. , Kalendar, R. , Schulman, A. , Afanasenko, O. and Robinson, J. (2006) Mapping of major spot‐type and net‐type net blotch resistance genes in the Ethiopian barley (Hordeum vulgare) line CI 9819. Genome, 49, 1564–1571. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Chu, A. , Steeves, J.E. , Wolpert, T.J. and Ciuffetti, L.M. (2009) A host‐selective toxin of Pyrenophora tritici‐repentis, Ptr ToxA, induces photosystem changes and reactive oxygen species accumulation in sensitive wheat. Mol. Plant–Microbe Interact. 22, 665–676. [DOI] [PubMed] [Google Scholar]
- Murray, G.M. and Brennan, J.P. (2010) Estimating disease losses to the Australian barley industry. Australas. Plant Pathol. 39, 85–96. [Google Scholar]
- Nagy, E.D. and Bennetzen, J.L. (2008) Pathogen corruption and site directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle, P. , Brooks, W. , Steffenson, B. , Stromberg, E. and Griffey, C. (2011) Genetic characterization of barley net blotch resistance genes. Plant Dis. 95, 19–23. [DOI] [PubMed] [Google Scholar]
- Oliver, R. , Friesen, T.L. , Faris, J.D. and Solomon, P.S. (2012) Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–44. [DOI] [PubMed] [Google Scholar]
- Pandelova, I. , Betts, M.F. , Manning, V.A. , Wilhelm, L.J. , Mockler, T.C. and Ciuffetti, L.M. (2009) Analysis of transcriptome changes induced by Ptr ToxA in wheat provides insights into the mechanisms of plant susceptibility. Mol. Plant, 2, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Raman, H. , Platz, G.J. , Chalmers, K.J. , Raman, R. , Read, B.J. , Barr, A.R. and Moody, D.B. (2003) Mapping of genetic regions associated with net form of net blotch resistance in barley. Aust. J. Agric. Res. 54, 1359–1367. [Google Scholar]
- Rasmusson, D.C. , Wilcoxson, R.D. , Dill‐Macky, R. , Schiefelbein, E.L. and Wiersma, J.V. (1999) Registration of MNBrite barley. Crop Sci. 391, 290. [Google Scholar]
- Rau, D. , Attene, G. , Brown, A.H.D. , Nanni, L. , Maier, F.J. , Balmas, V. , Saba, E. , Schäfer, W. and Papa, R. (2007) Phylogeny and evolution of mating‐type genes from Pyrenophora teres, the causal agent of barley ‘net blotch’ disease. Curr. Genet. 51, 377–392. [DOI] [PubMed] [Google Scholar]
- Richter, K. , Schondelmaier, J. and Jung, C. (1998) Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor. Appl. Genet. 97, 1225–1234. [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Catcheside, D.E.A. , Tate, M.E. and Able, A.J. (2007) Evidence of involvement of proteinaceous toxins from Pyrenophora teres in symptom development of net blotch of barley. Phytopathology, 97, 907–915. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Tate, M.E. , Catcheside, D.E.A. and Able, A.J. (2008) Initial characterization of phytotoxic proteins isolated from Pyrenophora teres . Physiol. Mol. Plant Pathol. 72, 73–79. [Google Scholar]
- SAS Institute Inc. (2011) The SAS system for Windows. Release 9.3. SAS Institute Cary, NC. [Google Scholar]
- Schaller, C.W. (1955) Inheritance of resistance to net blotch of barley. Phytopathology, 45, 174–176. [Google Scholar]
- Shjerve, R.A. , Faris, J.D. , Brueggeman, R.S. , Yan, C. , Zhu, Y. , Koladia, V. and Friesen, T.L. (2014) Evaluation of a Pyrenophora teres f. teres mapping population reveals multiple independent interactions with the barley 6H chromosome region. Fungal Genet. Biol. Doi: 10.1016/j.fgb.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Smedegård‐Petersen, V. (1971) Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on barley in Denmark. K. Vet. Landbohojsk Arsskr. 1971, 124–144. [Google Scholar]
- Smedegård‐Petersen, V. (1977) Isolation of two toxins produced by Pyrenophora teres and their significance in disease development of net‐spot blotch of barley. Physiol. Plant Pathol. 10, 203–211. [Google Scholar]
- St. Pierre, S. , Gustus, C. , Steffenson, B. , Dill‐Macky, R. and Smith, K.P. (2010) Mapping net form net blotch and Septoria speckled leaf blotch resistance loci in barley. Phytopathology, 100, 80–84. [DOI] [PubMed] [Google Scholar]
- Steffenson, B.J. (1997) Net blotch In: Compendium of Barley Diseases (Mather D.E., ed.), 2nd edn, pp. 28–31. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Steffenson, B.J. , Hayes, P.M. and Kleinhofs, A. (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor. Appl. Genet. 92, 552–558. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. and de Wit, P.J.G.M. (2009) Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Collemare, J. , Mehrabi, R. and de Wit, P.J.G.M. (2012) Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 37, 67–93. [DOI] [PubMed] [Google Scholar]
- Tekauz, A. (1985) A numerical scale to classify reactions of barley to Pyrenophora teres . Can. J. Plant Pathol. 7, 181–183. [Google Scholar]
- Tenhaken, R. , Levine, A. , Brisson, L.F. , Dixon, R.A. and Lamb, C. (1995) Function of the oxidative burst in hypersensitive disease resistance. Proc. Natl. Acad. Sci. USA, 92, 4158–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedemann, A.V. (1997) Evidence for a primary role of active oxygen species in induction of host cell death during infection of bean leaves with Botrytis cinerea . Physiol. Mol. Plant Pathol. 50, 151–166. [Google Scholar]
- Toubia‐Rahme, H. , Ali‐Haimoud, D.E. , Barrault, G. and Albertini, L. (1995) Effect of four fungicides on barley net blotch caused by Drechslera teres . J. Phytopathol. 143, 335–339. [Google Scholar]
- Tsilo, T.J. , Chao, S. , Jin, Y. and Anderson, J.A. (2009) Identification and validation of SSR markers linked to the stem rust resistance gene Sr6 on the short arm of chromosome 2D in wheat. Theor. Appl. Genet. 118, 515–524. [DOI] [PubMed] [Google Scholar]
- Varshney, R.K. , Marcel, T.A. , Ramsay, L. , Russel, J. , Roder, M.S. , Stein, N. , Waugh, R. , Langridge, P. , Niks, R.E. and Graner, A. (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 114, 1091–1103. [DOI] [PubMed] [Google Scholar]
- Weiergang, I. , Jørgensen, H.J.L. , Møller, I.M. , Friis, P. and Smedegård‐Petersen, V. (2002) Correlation between sensitivity of barley to Pyrenophora teres toxins and susceptibility to the fungus. Physiol. Mol. Plant Pathol. 60, 121–129. [Google Scholar]
- Weiland, J.J. , Steffenson, B.J. , Cartwright, R.D. and Webster, R.K. (1999) Identification of molecular genetic markers in Pyrenophora teres f. teres associated with low virulence on ‘Harbin’ barley. Phytopathology, 89, 176–181. [DOI] [PubMed] [Google Scholar]
- Wells, S.A. (1973) Hector barley. Can. J. Plant Sci. 53, 497. [Google Scholar]
- Wilcoxson, R.D. , Rasmusson, D.C. and Miles, M.R. (1990) Development of barley resistant to spot blotch and genetics of resistance. Plant Dis. 74, 207–210. [Google Scholar]
- de Wit, P.J.G.M. and Spikman, G. (1982) Evidence for the occurrence of race and cultivar‐specific elicitors of necrosis in intercellular fluids of compatible interactions of Cladosporium fulvum and tomato. Physiol. Mol. Plant Pathol. 21, 1–11. [Google Scholar]
- Wolpert, T.J. , Dunkle, L.D. and Ciuffetti, L.M. (2002) Host‐selective toxins and avirulence determinants: what's in a name. Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Yun, S.J. , Gyenis, L. , Bossolini, E. , Hayes, P.M. , Matus, I. , Smith, K.P. , Steffenson, B.J. , Tuberosa, R. and Muehlbauer, G.J. (2005) Validation of quantitative trait loci for multiple disease resistance in barley using advanced backcross lines developed with a wild barley. Crop Sci. 46, 1179–1186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Genetic maps of the seven chromosomes (1H–7H) in the Hector × NDB 112 recombinant inbred population.
Table S1 Summary of genetic maps in the Hector × NDB 112 population.


