Summary
Pseudomonas corrugata CFBP 5454 produces two kinds of cyclic lipopeptides (CLPs), cormycin A and corpeptins, both of which possess surfactant, antimicrobial and phytotoxic activities. In this study, we identified genes coding for a putative non‐ribosomal peptide synthetase and an ABC‐type transport system involved in corpeptin production. These genes belong to the same transcriptional unit, designated crpCDE. The genetic organization of this locus is highly similar to other Pseudomonas CLP biosynthetic clusters. Matrix‐assisted laser desorption ionization‐time of flight‐mass spectrometry (MALDI‐TOF‐MS) analysis revealed that transporter and synthetase genomic knock‐out mutants were unable to produce corpeptins, but continued to produce cormycin A. This suggests that CrpCDE is the only system involved in corpeptin production in P. corrugata CFBP 5454. In addition, phylogenetic analysis revealed that the CrpE ABC transporter clustered with the transporters of CLPs with a long peptide chain. Strains depleted in corpeptin production were significantly less virulent than the wild‐type strain when inoculated in tomato plants and induced only chlorosis when infiltrated into Nicotiana benthamiana leaves. Thus, corpeptins are important effectors of P. corrugata interaction with plants. Expression analysis revealed that crpC transcription occurs at high cell density. Two LuxR transcriptional regulators, PcoR and RfiA, have a pivotal role in crpC expression and thus in corpeptin production.
Keywords: ABC transporters, corpeptins, Lux R transcriptional regulators, non‐ribosomal peptide synthetase, Pseudomonas
Introduction
Pseudomonas corrugata Roberts and Scarlett 1981 emend. Sutra et al., 1997 is a ubiquitous bacterium isolated from a wide variety of sources. It was first described in the UK in the late 1970s (Scarlett et al., 1978) as the causal agent of tomato pith necrosis (TPN), and later associated worldwide with TPN in tomato (Catara, 2007). The most characteristic symptom of the disease is the discoloration and/or necrosis of the parenchymatous tissue of the stem. The injection of a high‐density inoculum within the stem of a wide range of plant species leads to pith necrosis (Catara et al., 1997, 2002; Siverio et al., 1993; Sutra et al., 1997). However, the disease is only widespread in tomato, with just a few cases of infection reported in pepper and chrysanthemum (Catara, 2007).
The hypersensitive response (HR) in tobacco has been reported as a variable test in P. corrugata identification/characterization (Catara et al., 1997; Siverio et al., 1993; Sutra et al., 1997). A number of studies by Gustine et al. (1990, 1994, 1995) have shown that P. corrugata strain 388 causes HR in tobacco leaves and elicits phytoalexin (medicarpin) biosynthesis in white clover callus, K+/H+ exchange in tobacco leaf discs and an active oxygen burst in white clover suspension cultures. However, surprisingly, in the genome of the P. corrugata strain CFBP 5454, no Hrp1 type III secretion system (T3SS) has been found (Licciardello et al., 2014).
Pseudomonas corrugata produces cyclic lipopeptide (CLP) corpeptins; two isoforms consisting of 22‐amino‐acid residues, corpeptin A and corpeptin B, have been described. Corpeptins induce chlorosis when infiltrated into tobacco leaves and show antimicrobial activity against the Gram‐positive bacterium Bacillus megaterium (Emanuele et al., 1998). Some strains also produce cormycin, a lipodepsinonapeptide, which has antimicrobial activity against B. megaterium and also against the yeast Rhodotorula pilimanae. Cormycin also exhibits phytotoxic activity, inducing chlorosis followed by necrosis in tobacco (Scaloni et al., 2004). CLPs produced by Pseudomonas spp. are composed of a fatty acid tail linked to a short oligopeptide, which is cyclized to form a lactone ring between two amino acids in the peptide chain (Raaijmakers et al., 2006). The corpeptin structure, as determined by Emanuele et al. (1998), is strictly related to peptin‐like CLPs, which are characterized by long peptide chains ranging from 18 to 25 amino acids. These CLPs include fuscopeptins, produced by the phytopathogen P. fuscovaginae (Ballio et al., 1996), syringopeptins, synthesized by phytopathogenic strains of P. syringae pv. syringae (Ballio et al., 1991), and tolaasin, produced by the mushroom‐infecting bacterium P. tolaasii (Coraiola et al., 2006). Cormycin belongs to the group of smaller nonapeptides (Scaloni et al., 2004), with syringomycin, produced by strains of P. syringae pv. syringae, having been the most extensively studied (Bender et al., 1999; Raaijmakers et al., 2006).
Like many other biologically active secondary metabolites, CLPs are synthesized by multifunctional non‐ribosomal peptide synthetases (NRPSs) (Finking and Marahiel, 2004); Raaijmakers et al., 2006). NRPSs have a modular structure, and each module is a building block resulting in the stepwise incorporation of one amino acid in the peptide chain (Finking and Marahiel, 2004). Each amino acid activation module has a minimal set of three domains: an aminoacyl‐adenylation (A) domain, a thiolation (T) domain and a condensation (C) domain (Finking and Marahiel, 2004). The genetic organization of CLP biosynthetic loci reveals that several genes flanking the biosynthetic genes are conserved among Pseudomonas spp. Genes coding for putative CLP transporters, located downstream of the last synthetase gene, and regulatory genes positioned up‐ and downstream of the CLP biosynthesis genes, have been identified and described in a number of clusters (de Bruijn and Raaijmakers, 2009; Gross and Loper, 2009).
Our previous studies have demonstrated that P. corrugata CFBP 5454 possesses an N‐acyl‐homoserine lactone quorum sensing (AHL‐QS) system, PcoI/PcoR, consisting of an AHL synthase, PcoI, and a transcriptional sensor/regulator belonging to the LuxR family protein, PcoR (Licciardello et al., 2007). Downstream of pcoI is rfiA, which is co‐transcribed with pcoI (Licciardello et al., 2009). RfiA belongs to LuxR regulators whose particular characteristic is that they contain the typical helix–turn–helix (HTH) motif at the C‐terminus, but do not harbour the autoinducer‐binding terminus typical of QS LuxR regulators (de Bruijn and Raaijmakers, 2009). PcoR activates pcoI expression in the presence of AHL via a typical positive‐feedback regulatory loop. As pcoI and rfiA constitute an operon, the expression of rfiA is directly regulated by the PcoR–AHL complex (Licciardello et al., 2009). Pseudomonas corrugata pcoR and rfiA mutants show significantly reduced virulence when inoculated in tomatoes. CLPs were also absent in their culture filtrates (Licciardello et al., 2009, 2012). Genetically linked to rfiA is the operon pcoABC, which is under positive regulation by RfiA, and thus indirectly by the PcoI/R system (Licciardello et al., 2009). A null mutant in pcoABC retains the ability to inhibit the growth of CLP indicator microorganisms R. pilimanae and B. megaterium, although at a reduced level compared with the wild‐type (WT), and is as virulent as the WT strain. These findings indicate that other CLP secretion mechanisms must exist in P. corrugata. The aforementioned genes (pcoI/pcoR, rfiA and pcoABC) are located in a cosmid insert of approximately 20 000 bp (Licciardello et al., 2009). The insert sequence also supports the presence of genes putatively coding for an additional ABC transporter and part of an NRPS highly homologous to genes of other Pseudomonas spp. CLP biosynthetic clusters.
In this article, we demonstrate that these ABC transporter genes (crpDE) form an operon with the last gene of a putative NRPS (crpC), and that both are part of the biosynthesis cluster of P. corrugata corpeptins, which are demonstrated to have an important role in plant interaction and in virulence. A role for the transcriptional regulators PcoR and RfiA in corpeptin production and crpC gene expression is also demonstrated.
Results
Analysis of a putative ABC efflux system and a putative NRPS in P . corrugata
Recently, we have demonstrated that the AHL‐QS system of P. corrugate, PcoI/PcoR, and the transcriptional regulator RfiA play an important role in the regulation of virulence via CLP production (Licciardello et al., 2012). The absence of further genetic and molecular information prompted us to sequence the complete cosmid insert from which the AHL‐QS genes were identified (Fig. 1). Three complete and one partial new open reading frames (ORFs) were identified. ORF8 spans 1152 bases and codes for a putative 383‐amino‐acid protein which displays a high level of amino acid sequence similarity (81%–91%) to periplasmic membrane fusion proteins of other Pseudomonas species. Downstream of this ORF, separated by only 2 bp, and transcribed in the same direction, ORF9 spans 1962 bases and encodes a predicted 653‐amino‐acid protein. blastx analysis of P. corrugata ORF9 revealed homologies with a number of cytoplasmic ABC‐type transporters of Pseudomonas species, with a similarity ranging from 90% to 85%. Downstream of ORF9 was ORF10, which is separated by 299 bp and is transcribed in the opposite direction. ORF10 codes for a putative diaminobutyrate‐2‐oxoglutarate transaminase protein (ORF10), which displays 80% identity to P. syringae B728a (YP_235696.1). Located 78 bp upstream of ORF8 and translated in the same direction, we identified a truncated ORF (ORF7). The 6471 bases encode 2156 amino acids which show high homology (66%–50%) to members of NRPS genes involved in the CLP biosynthesis of other Pseudomonas spp., e.g. arthrofactin synthetase C (BAC67536.1), massetolide MassC (ABH06369.2) and orfamide NRPS OfaC (YP_259254.1). Analysis of the deduced amino acid sequence for the C‐terminal portion of the ORF7 protein revealed the presence of an entire amino acid activation module, containing conserved core sequences for the C, A and T domains, and a truncated amino acid activation module lacking the C domain. Two thioesterase TE domains, each containing the conserved GxSxG sequence motif involved in the linear or cyclic peptide release, were also identified (Gross and Loper, 2009). Tandem TE domains have been described in the termination modules of biosynthetic clusters of syringopeptin, arthrofactin, viscosin, massetolide, orfamide, putisolvin and entolysin (de Bruijn et al., 2008; Dubern et al., 2008; Gross et al., 2007; Roongsawang et al., 2003; Scholz‐Schroeder et al., 2003; Vallet‐Gely et al., 2010).
Figure 1.
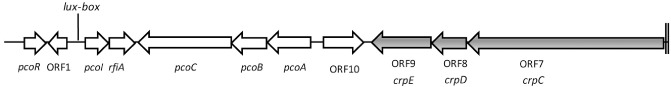
Representation of an approximately 21‐kb DNA region of Pseudomonas corrugata CFBP 5454 harbouring the N‐acyl‐homoserine lactone quorum sensing (AHL‐QS) system genes (pcoI / pcoR), the transcriptional regulator rfiA belonging to the LuxR‐type transcriptional regulators, the pcoABC operon, coding for a resistance nodulation–cell division transporter system, the crpDE operon (i.e. ORF8, 9) coding for an ATP‐binding transporter system and crpC (ORF7) which is part of a non‐ribosomal peptide synthetase.
The presence of genes flanking the terminating NRPS genes, encoding components of exporter systems, is a recurrent theme for several Pseudomonas CLP clusters. We therefore analysed the genomic context of each ORF9 homologue (ABC transporter‐encoding) within plant‐associated Pseudomonas spp. Genes coding for all of these ABC transporters were localized downstream of the last putative NRPS genes as deduced by bioinformatics (Table S1, see Supporting Information). The neighbour‐joining dendrogram of these ABC transporters aligned by ClustalW in mega 5.01 software resulted in distinct clusters (Fig. 2). Pseudomonas corrugata ORF9 formed a cluster together with ABC transporters of P. syringae pv. syringae strains 642 (ZP_07265130.1) and B728a (YP_235695.1), located downstream of genes coding for syringopeptins (Scholz‐Schroeder et al., 2003), and those of P. fuscovaginae UPB 0736 (ZP_10991311.1), P. syringae pv. avellanae ISPaVe013 (ZP_17805054) and Pseudomonas sp. CMR12a (AFH75324.1). Another well‐delineated cluster group of transporters located downstream of NRPSs includes SyfD, involved in the export of the syringafactin linear lipopeptide of P. syringae pv. tomato DC3000 (NP_792636.1) (Berti et al., 2007), and other ABC transporters in P. syringae pathovars (Baltrus et al., 2011), annotated as syringolide efflux proteins (EGH63888.1; EGH94284.1; EGH09011.1), but which, in our analysis, were located downstream of NRPS genes highly homologous to syringafactins and annotated as sifB (Table S1). The remaining ABC transporters belong to the biosynthesis gene clusters of the medium‐chain‐length peptide CLPs arthrofactin (BAF40423.1), massetolide (ABW87988.1), motilin (AFH75332.1), viscosin (YP_002872144.1) and orfamide (YP_259256.1).
Figure 2.
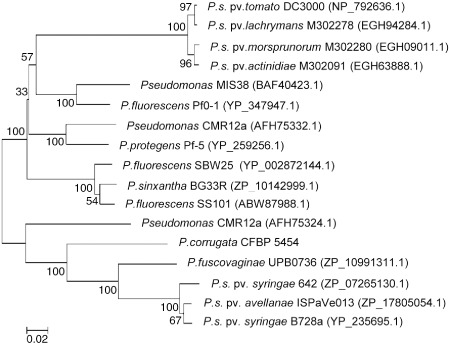
Phylogenetic analysis of amino acid sequences of 17 ABC‐type transporters identified in cyclic lipopeptide (CLP) biosynthetic clusters of plant‐associated Pseudomonas spp. The numbers at the nodes indicate the level of bootstrap support, based on neighbour‐joining analysis of 500 re‐sampled datasets. The bar indicates the relative number of substitutions per site. The protein Accession numbers are given in parentheses.
crpCDE is part of the corpeptin biosynthetic cluster
In order to examine the role of these newly identified ORFs in CLP biosynthesis, P. corrugata ORF7 and ORF8 were insertionally inactivated. The two genomic mutant strains were designated as PCONRPS and PCOMFP, respectively (Table 1). Matrix‐assisted laser desorption ionization‐time of flight‐mass spectrometry (MALDI‐TOF‐MS) analysis of CFBP 5454 cell‐free culture filtrates used as controls highlighted the presence of cormycin and corpeptins, corresponding to specific peaks at m/z 1274, 2095.3 and 2121.2, respectively (Licciardello et al., 2012). Analysis of the cell‐free culture filtrates of the aforementioned knock‐out mutants grown in CLP‐inducing conditions revealed that the two mutants no longer produced corpeptins, whereas they continued to produce cormycin, suggesting that the knocked‐out genes are involved in corpeptin production (Fig. 3A). This also suggested that CrpDE was probably involved in the secretion of corpeptins and that CrpC was part of the NRPS complex for the biosynthesis of this CLP. The bioactivity of the culture filtrates of the two mutants was tested using an assay based on the inhibition of the in vitro growth of the Gram‐positive bacterium B. megaterium and the yeast R. pilimanae. The inhibitory activity of PCOMFP and PCONRPS strain cell‐free culture filtrates was significantly reduced against both microorganisms compared with that of the parent strain (Fig. 3B).
Table 1.
Bacterial strains, plasmids and oligonucleotides used in this study
| Strain | Genotype/relevant characteristics | Reference or source |
|---|---|---|
| Pseudomonas corrugata | ||
| CFBP 5454 | Wild‐type, source of crpC and crpD | CFBP |
| PCOMFP | crpD:: pKnock, Kmr | This study |
| PCONRPS | crpC:: pknock, Kmr | This study |
| PCOMFPC | PCOMFP complemented with pBBR‐CrpDE | This study |
| GL2 | pcoR76::Tn5, Kmr | Licciardello et al. (2007) |
| GLRFIA | rfiA:: pKnock, Kmr | Licciardello et al. (2009) |
| Escherichia coli | ||
| pLC3.34 | pLAFR3 containing P. corrugata CFBP 5454 DNA, Tcr | DISPA |
| DH5α | F2 f80dlaczZDM15 D(lacZYA‐argF)U169 endA1 recA1 hsdR17 deoR gyrA96 thi‐1 relA1 supE44 | Sambrook et al. (1989) |
| CC118λpir | Δ(ara, leu)7697 araD139 ΔlacX74 galE galK phoA20 thi‐1 rpsE rpoB (Rfr) argE(Am) recA1 λpir | Herrero et al. (1990) |
| Chromobacterium violaceum | ||
| CV026 | ATCC 31532 derivative, cviI::Tn5xylE Kmr Smr | McClean et al. (1997) |
| Plasmids | ||
| pCR2.1 | Cloning vector TA, Ampr | Invitrogen |
| pGEM‐T | Cloning vector TA, Ampr | Promega |
| pKNOCK‐Kmr | Mobilizable suicide vector, Kmr | Alexeyev (1999) |
| pRK2013 | Kmr100, Tra+ Mob+ ColE1 replicon | Figurski and Helinski (1979) |
| pKMMfp | pKNOCK containing an internal fragment of P. corrugata CFBP 5454 corpD gene | This study |
| pKMNrps | pKNOCK containing an internal fragment of P. corrugata CFBP 5454 corpC gene | This study |
| pBBR‐CrpDE | pBBR1MCS‐5 containing the full‐length P. corrugata CFBP 5454 crpDE genes | This study |
| Oligonucleotides | Sequence | |
| MFPkn‐fw | 5′‐AAGGATCCAGTGGCTGGCGGAAATC‐3′ | This study |
| MFPkn‐rev | 5′‐GGTCTAGAGGATGGTGAAATACACT‐3′ | This study |
| NRPSkn‐fw | 5′‐CAGGATCCGGATCTATCTGCTCGAC‐3′ | This study |
| NRPSkn‐rev | 5′‐AATCTAGAGCCGATAGTGCCGAGGG‐3′ | This study |
| PCR1‐fw | 5′‐ACCGCAACATCAATACAGCG‐3′ | This study |
| PCR1‐rev | 5′‐ACCGACATCAACCTGCTTGAC‐3′ | This study |
| PCR2‐fw | 5′‐CATCGCCTGCGTATCTCGAT‐3′ | This study |
| PCR2‐rev | 5′‐CAACTCATGGTCGTCCATCG‐3′ | This study |
| Pco16S fw | 5′‐TGTAGCGGTGAAATGCGTAGAT‐3′ | Conte et al. (2006) |
| Pco16S rev | 5′‐CCTCAGTGTCAGTATCAGTCCAG‐3′ | Conte et al. (2006) |
| Abc1‐fw | 5′‐CAAAATCGCTATCGTGCTTGTC‐3′ | This study |
| Abc1‐rev | 5′‐CGACCGTAGCGGTCAGGTA‐3′ | This study |
| Nrps‐fw | 5′‐ACGGGCCACCCGAAAG‐3′ | This study |
| Nrps rev | 5′‐GAGGCGAAAGCCACGTGAT‐3′ | This study |
| ABC‐fw | 5′‐AAGCTTTGGGCACACCCATG‐3′ | This study |
| ABC‐rev | 5′‐GGATCCCACAGAGGACAGTG‐3′ | This study |
| Primerseq‐fw | 5′‐GATCCATCGACGGACTGTC‐3′ | This study |
| Primerseq‐rev | 5′‐CATCGTGTTCGGTTTCGTAC‐3′ | This study |
CFBP, Collection Francaise de Bacteries Phytopathogenes, Angers, France; DISPA, Dipartimento di Scienze delle Produzioni Agrarie ed Alimentari, University of Catania, Catania, Italy.
Figure 3.
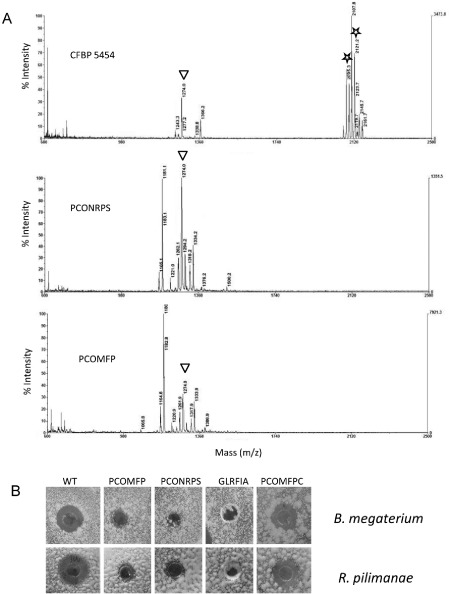
Analysis of cell‐free extracts of Pseudomonas corrugata strains after 96 h of incubation at 28 °C: CFBP 5454 (wild‐type, WT); PCONRPS (crpC::pKnock); PCOMFP (crpD::pKnock). (A) Representative matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectra. The triangles indicate the peaks corresponding to cormycin and the stars indicate those corresponding to corpeptins. (B) Antimicrobial assay to evaluate the bioactivity of cell‐free culture filtrates; the GLRFIA mutant was included as a negative control; PCOMFPC, PCOMFP strain complemented in trans with intact crpDE.
Based on genotypic and phenotypic evidence, we designated ORF7, ORF8 and ORF9 as crpC, crpD and crpE, respectively, and PCONRPS and PCOMFP as crpC::pKnock and crpD::pKnock, respectively. In addition, the three genes constitute an operon, designated crpCDE, as determined by a reverse transcription approach with two sets of primers: PCR1 to capture the intervening region between crpC and crpD, upstream of the knock‐out inactivation site of PCOMFP, and PCR2 which amplifies the intervening region between crpD and crpE (Fig. 4). This result was also confirmed by quantitative real‐time polymerase chain reaction (Q‐PCR) assays, which showed that crpC transcription occurred in the PCOMFP strain, whereas crpD expression was undetectable in the PCONRPS strain (data not shown).
Figure 4.
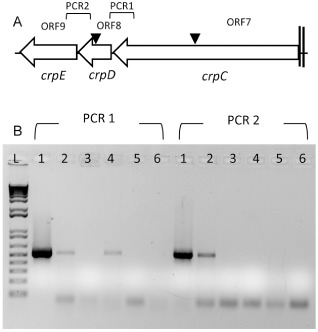
Polycistronic organization of crpCDE cluster identified in Pseudomonas corrugata CFBP 5454. (A) Triangles indicate the positions of insertional mutagenesis. The brackets indicate the positions of primer sets overlapping crpC and crpD (PCR1) and crpD and crpE (PCR2). (B) Agarose gel electrophoresis of reverse transcription‐polymerase chain reaction (RT‐PCR) analysis performed with total RNA isolated from P. corrugata CFBP 5454 and mutant strains incubated on Improved Minimal Medium (IMM) for 4 days at 28 °C. Two sets of primers (PCR1 and PCR2) were designed for the amplification of approximately 500‐bp DNA fragments at each inter‐gene locus gap. Lanes: L, 1‐kb plus ladder (Life Technologies, Carlsbad, CA, USA); 1, wild‐type (WT) genomic DNA; 2, WT cDNA; 3, crpC::pKnock; 4, crpD::pKnock; 5, negative control of the reverse transcription step; 6, PCR negative control, water.
Corpeptins contribute to P . corrugata–plant interaction
To investigate the role of crpCDE in P. corrugata–plant interaction, we set up in planta assays. The tests were performed with the PCOMFP and PCONRPS mutant strains (producing only cormycin) and, as controls, the parent strain CFBP 5454 (which produces both cormycin and corpeptins) and the GLRFIA mutant strain (rfiA –) (which is no longer able to produce both cormycin and corpeptins) (Licciardello et al., 2012).
Fifteen days after inoculation, stem pith‐infected tissues of tomato plants inoculated with the parent strain showed dark brown lesions ranging in length from 3 to 6 cm, necrotic in the inner part and at times hollow (Figs 5 and 6A). Again, the GLRFIA mutant strain did not cause any lesions on tomato stem pith other than a pale discoloration at the inoculation site. The plants inoculated with PCOMFP and PCONRPS mutant strains showed a reduced brown discoloration of the stem pith (approximately 1–2 cm) (Figs 5 and 6A). At 3 and 7 days post‐inoculation (dpi), CFBP 5454 showed a bacterial titre of approximately 1012 colony‐forming units (cfu)/cm of stem at the inoculation site, whereas the two derivative strains showed a titre of approximately 109 cfu/cm of stem (Table 2). Monitoring at different distances above and below the inoculation site always revealed the presence of all three strains. Three‐way analysis of variance (ANOVA) showed significant effects of strains (P < 0.001) and distances (P < 0.001), but not of positions or interaction effects. The population concentration of the three strains varied significantly at each sampling time and with the following order CFBP 5454 > PCONRPS > GLRFIA (Table 2). The bacterial titre declined significantly with increasing distance from the inoculation site both below and above the inoculation site ( Table 2).
Figure 5.
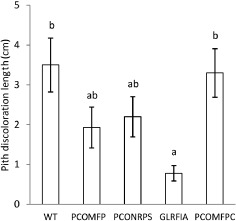
Length of pith discoloration in tomato plants inoculated with Pseudomonas corrugata CFBP 5454 (wild‐type, WT), PCOMFP (crpD::pKnock), PCONRPS (crpC::pKnock), GLRFIA (rfiA mutant) and PCOMFPC (PCOMFP strain complemented in trans with intact crpDE). Values are means ± standard error (SE) of 20 replicates. Values followed by the same letters do not differ significantly at P ≤ 0.05 according to Student–Newman–Keuls test. The results presented are representative of two independent experiments.
Figure 6.

In planta inoculations of Pseudomonas corrugata CFBP 5454, PCOMFP (crpD mutant), PCONRPS (crpC mutant), GLRFIA (rfiA mutant) and PCOMFPC (PCOMFP strain complemented in trans). (A) Stem pith necrosis symptoms in prick‐inoculated tomato plants. (B, C) Hypersensitivity reaction on tobacco leaves at 24 h (B) and 96 h (C) post‐inoculation with bacterial suspension. These photographs are representative of at least two experiments with replicates.
Table 2.
Pseudomonas corrugata strain CFBP 5454 and derivative mutant strain PCONRPS and GLRFIA population concentrations (log cfu/cm) in tomato stem 7 days after inoculation at different distances above and below the inoculation sites*
| Strain | Position | Strain population concentration at different distances from the inoculation site (0)† | Average position/strains† | Average strains† | |||
|---|---|---|---|---|---|---|---|
| 0 | 1 cm | 2 cm | 3 cm | ||||
| CFBP5454 | Above | 12.44 ± 0.01 | 7.73 ± 0.57 | 6.50 ± 0.40 | 5.27 ± 0.37 | 6.50 c | |
| Below | 7.29 ± 0.17 | 7.64 ± 0.38 | 6.49 ± 0.34 | 7.14 c | |||
| Mean | 7.51 c | 7.07 c | 5.88 c | 6.81 c | |||
| PCONRPS | Above | 9.86 ± 0.20 | 6.34 ± 0.36 | 5.90 ± 0.14 | 4.81 ± 0.14 | 5.68 b | |
| Below | 6.72 ± 0.13 | 5.61 ± 0.10 | 4.40 ± 0.07 | 5.58 b | |||
| Mean | 6.53 b | 5.75 b | 4.60 b | 5.63 b | |||
| GLRFIA | Above | 9.42 ± 0.19 | 5.30 ± 0.30 | 4.11 ± 0.37 | 3.20 ± 0.30 | 4.20 a | |
| Below | 5.36 ± 0.09 | 4.56 ± 0.11 | 3.39 ± 0.33 | 4.44 a | |||
| Mean | 5.33 a | 4.33 a | 3.29 a | 4.32 a | |||
| Average distances‡ | 6.46 c | 5.72 b | 4.59 a | ||||
| Average positions† | Above | 5.46 a | |||||
| Below | 5.72 a | ||||||
*A three‐way analysis of variance (ANOVA) was performed to investigate differences in bacterial population concentration of the three strains and expressed as log cfu/cm of stem sampled at different distances (1, 2 and 3 cm) from and positions (above and below) the inoculation sites (values are means of six replicates ± standard error). Three‐way ANOVA showed significant effects of strains (n = 36; P < 0.001), distances (n = 36; P < 0.001) but not positions (n = 54; P = 0.072).
†Averages followed by the same letter within columns are not significantly different according to Student–Newman–Keuls test (P = 0.01).
‡Averages of strain bacterial populations at different distances are compared within the row. This experiment was repeated on two separate dates with similar results.
We have observed previously that both GL2 (pcoR–) (Licciardello et al., 2007) and GLRFIA (V. Catara, University of Catania, Catania, unpublished data) mutant strains do not cause any necrosis symptoms when inoculated in Nicotiana tabacum and N. benthamiana leaf mesophyll, in contrast with the WT CFBP 5454 strain, which causes the collapse of the infiltrated panels within 24 h, followed by necrosis. To investigate the possible influence of corpeptin production in such leaf necrosis, we infiltrated N. benthamiana leaf panels with a bacterial suspension of the parent strain CFBP 5454 and derivative mutants. Twenty‐four hours after inoculation, the leaf panels inoculated with the parent strain showed a collapsed mesophyll, which turned necrotic within the following 24 h (Fig. 6B). At the same scoring times, leaf panels inoculated with PCOMFP and PCONRPS showed a healthy turgid appearance (Fig. 6B), comparable with the water‐inoculated control (data not shown). Four days after inoculation, chlorosis was evident in the leaf panels inoculated with PCOMFP and PCONRPS, but not with GLRFIA (Fig. 6C). In a time course experiment, we monitored the bacterial population titres of strain CFBP 5454 and the PCONRPS derivative mutant in infiltrated N. benthamiana leaf panels (Fig. 7). We included the GLRFIA mutant and a P. syringae pv. tabaci strain (DAPP‐PG632) in the trial as pathogenic controls. In tissues infiltrated with P. syringae pv. tabaci, the bacterial population increased drastically to 1010 cfu/leaf disc, whereas the P. corrugata strain CFBP 5454 population began to decay from the first dpi to 104 cfu/leaf disc. The bacterial population in leaf panels inoculated with PCONRPS and GLRFIA mutants maintained the same bacterial population titres (e.g. 106 cfu/leaf disc) from the first sampling at 0 dpi throughout the trial. At all sampling dates from 1 dpi these two strain titres were significantly different from those of strain CFBP 5454 (P < 0.01). Expression in trans of crpDE in the PCOMFP mutant by the introduction of pBBR‐CrpDE restored both virulence and antimicrobial activity similar to the WT strain level, as well as the HR‐positive phenotype (Figs 5 and 6).
Figure 7.
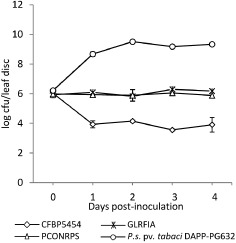
Pseudomonas corrugata strain CFBP 5454 and derivative mutant strain PCONRPS and GLRFIA population concentrations in Nicotiana benthamiana leaves Three N. benthamiana leaf discs were collected independently and processed at each time point (0, 1, 2, 3 and 4 days post‐inoculation). In this trial, Pseudomonas syringae pv. tabaci strain DAPP‐PG632 was used as the pathogenic control strain. In both trials, samples were homogenized with 1 mL of distilled water; serial dilutions were plated onto NDA medium (nutrient agar plus 1% dextrose) plus antibiotics as described in Experimental procedures. Values are means ± standard error (SE) of multiple replicates. The results presented are representative of two independent experiments.
LuxR transcriptional regulators PcoR and RfiA are involved in crpCDE expression
Pseudomonas corrugata mutants in the genes coding for the transcriptional regulators pcoR (GL2 mutant) and rfiA (GLRFIA mutant) no longer produced CLPs (Licciardello et al., 2012). By investigating the expression of crpC and crpDE in both the GL2 and GLRFIA mutant strains, we observed that transcript levels were significantly and consistently reduced in both mutant strains (data not shown). These results suggest that, under the conditions tested, both transcriptional regulators play a role in corpeptin biosynthesis/secretion in P. corrugata.
CLP production, evaluated on the basis of antimicrobial activity against the two CLP‐sensitive bioindicators R. pilimanae and B. megaterium, occurs in a cell density‐dependent manner with a trend similar to AHL production (Licciardello et al., 2009). Consequently, we speculated whether crpC transcriptional levels would follow this trend in a time‐course experiment. Bacterial cell samples were recovered from still cultures grown in CLP‐inducing medium at 17, 24, 48, 72 and 96 h post‐inoculation (hpi) (i.e. T17, T24, T48, T72 and T96). During growth of the WT strain, transcript levels of crpC were similar between 17 and 48 h, whereas they reached a maximum after 72 h of growth and decreased consistently 96 h after inoculation (Fig. 8A). In cell‐free culture filtrates sampled at the same time points, MALDI‐TOF analysis detected small amounts of corpeptins (but also cormycin) starting from T48, when the cell concentration was approximately as low as 7 × 108 cfu/mL (data not shown). At the same sampling time (T48), the presence of AHL was detected for the first time by indirect violacein production by the biosensor Chromobacterium violaceum CV026 cultures (Fig. 8B). The sampling of 72‐h‐old cultures highlighted high CLP peak signals in MALDI‐TOF mass spectra and the first evidence of in vitro antimicrobial activity (Fig. 8C).
Figure 8.
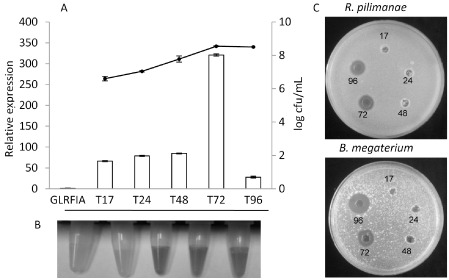
Cyclic lipopeptide (CLP) production in Pseudomonas corrugata CFBP 5454 during bacterial growth. (A) Time‐course expression (17, 24, 48, 72 and 96 h post‐inoculation) of crpC in P. corrugata CFBP 5454 grown in CLP production‐inducing conditions (bar graph). Results are reported as the fold difference relative to data obtained for the non‐producing corpeptin mutant strain GLRFIA. Values are means calculated from duplicate samples from three RNA extractions ± standard error (SE). The CFBP 5454 growth curve is displayed in the line graph (secondary y axis). (B, C) The cell‐free culture filtrates at each time point were tested by bioassays for N‐acyl‐homoserine lactone (AHL) production using Chromobacterium violaceum CV026 as biosensor to detect AHLs (B) and antimicrobial activity against the yeast Rhodotorula pilimanae and the Gram‐positive bacterium Bacillus megaterium (C).
Discussion
In this study, we identified part of the biosynthetic cluster responsible for corpeptin production, including genes transcriptionally joined coding for an NRPS and an ABC efflux system, designated crpCDE. We demonstrated that these genes, and hence corpeptins, greatly contribute to P. corrugata virulence and plant interaction. The introduction of a mutation in crpC yielded a P. corrugata strain, PCONRPS, which failed to produce corpeptins, thus demonstrating that crpC is part of the corpeptin biosynthesis locus via a thiotemplate mechanism (Gross and Loper, 2009). It is estimated that approximately 3 kb of DNA are required to code for each amino acid activation module (Gross and Loper, 2009); thus, it is possible to predict that the corpeptin NRPS system encompasses approximately 75 kb of DNA to code the 22‐amino‐acid activation modules in the same way as with the P. syringae pv. syringae syringopeptin biosynthetic cluster, which is the largest linear NRPS system described for prokaryotes (Scholz‐Schroeder et al., 2003).
Recently, we have reported the whole‐genome shotgun sequencing of P. corrugata CFBP 5454, and an initial analysis revealed that approximately 217 kb coded for putative NRPS biosynthetic clusters (Licciardello et al., 2014). Genes that code for these large multimodular enzymes with repetitive domain structures in next‐generation sequencing approaches are difficult to assemble and typically split between several contigs. Thus, a great deal of further work is required in order to obtain the entire CLP biosynthetic clusters.
Further analysis of the region downstream of crpC resulted in the identification of two other genes, which putatively code for an ABC transporter system. Gene disruption of crpD also affected the presence of corpeptins in the culture filtrates of P. corrugata CFBP 5454, supporting the assumption that crpDE is the unique transport system involved in corpeptin export. Phylogenetic analysis showed that CrpE groups with ABC transporters of bacteria that produce long‐chain peptides. These transporters may have evolved differently from those involved in the secretion of CLP with shorter peptide chains or with linear peptides.
In terms of the biological activity of corpeptins, we found that the PCONRPS and PCOMFP mutant strains still release substances into the culture medium whose antimicrobial activity can be attributed to the production of cormycin. Pathogenicity tests on tomato demonstrated that the mutant strains producing only cormycin were also clearly less virulent than the parent strain CFBP 5454, thus demonstrating the importance of corpeptins in the development of disease symptoms. Similarly, in the P. syringae pv. syringae strain B01D, a sypA mutant that produces syringomycin (but not syringopeptins) was still able to inhibit the growth of both B. megaterium and Geotrichum candidum and was less virulent in sweet cherry fruits (Scholz‐Schroeder et al., 2001). In P. syringae pv. syringae, three transporter systems, the SyrD (Quigley et al., 1993), PseABC (Kang and Gross, 2005) and PseEF (Cho and Kang, 2012) efflux systems, are mainly responsible for the secretion of the syringomycin and syringopeptins produced by strain B301D. Other natural functions of CLPs, mainly investigated in biocontrol strains, are their role in antagonism towards other (micro)organisms, motility and attachment to surfaces (Raaijmakers et al., 2010). Mutational analysis in CFBP 5454 mutants affected in CLP production raised interesting aspects with regard to their role in P. corrugata antimicrobial activity and motility that deserve further study.
The results of leaf inoculations of N. benthamiana deserve separate consideration. When strain CFBP 5454 is infiltrated into N. benthamiana mesophyll, it induces the collapse and necrosis of the leaf tissue and, as in an HR, its population declines rapidly. However, we observed that mutant strains that did not produce corpeptins only caused chlorosis, and the population titre of the crpCDE mutant (PCONRPS strains) was invariable over a 4‐day monitoring period. This result, and the fact that no T3SS was found in the P. corrugata CFBP 5454 genome, suggests that corpeptins play a role in the elicitation of HR in N. benthamiana. This is in accordance with recent studies, which showed that CLP may induce systemic resistance and which, taken together, suggest that CLPs constitute a novel class of microbial‐associated molecular patterns (MAMPs) (reviewed in Raaijmakers et al., 2010).
In this work, we also showed that the transcriptional regulators PcoR and RfiA play a pivotal role in the expression of crpC and crpD genes. LuxR‐type transcriptional regulators positioned up‐ and downstream of the CLP biosynthetic cluster also play a pivotal role in the production of syringomycin and syringopeptins (Lu et al., 2002; Wang et al., 2006), viscosin and massetolide (de Bruijn and Raaijmakers, 2009), syringafactins (Berti et al., 2007) and putisolvins (Dubern et al., 2008). Pseudomonas corrugata RfiA is highly homologous to a number of these CLP‐associated Pseudomonas LuxR regulators, including P. syringae pv. syringae SalA, which is located in the syr‐syp genomic island (Lu et al., 2002). SalA positively regulates all the genes of the syr‐syp cluster associated with the biosynthesis, secretion and regulation of syringomycin and syringopeptins, and is crucial in the development of disease symptoms (Lu et al., 2002, 2005). The RfiA gene is regulated directly by P. corrugata QS by co‐transcription with pcoI, which codes for the LuxI homologue AHL synthase (Licciardello et al., 2009). Similarly, in Pseudomonas sp. DF41, the RfiA mutant strain, in which rfiA is also co‐transcribed with the LuxI homologue gene, was depleted of antifungal activity and showed markedly reduced CLP sclerosin production (Berry et al., 2014).
QS is probably involved in the production of corpeptins, given the evidence that, in P. corrugata strain CFBP 5454, crpC transcription greatly increases with a high population density and following the trend in AHL signal molecule production. It presumably reaches large amounts of transcripts as a consequence of the QS positive‐feedback regulatory loop. To date, the involvement of QS in CLP production has only been demonstrated in a few Pseudomonas strains, namely in the plant pathogen P. fluorescens strain 5064 and saprophytic strain P. putida PCL1445, where AHL QS was shown to be involved in viscosin and putisolvin biosynthesis, respectively (Cui et al., 2005; Dubern et al., 2006).
The mutagenesis of P. corrugata CFBP 5454 genes in this study and the bioinformatic analysis revealed that the P. corrugata corpeptin biosynthetic locus is highly similar to that of other CLP‐producing Pseudomonas. Functional analysis revealed that corpeptins, long‐chain CLPs, as with P. syringae syringopeptins, are important for virulence. As the syr‐syp cluster in P. syringae represents approximately 2% of its genome, it is also important to study the biosynthesis and regulation in P. corrugata which, taxonomically, is more strongly related to the fluorescent oxidase‐positive Pseudomonas biocontrol strains than to P. syringae pv. syringae. However, as, unlike P. syringae pv. syringae, QS is involved in corpeptin production via PcoR and Rfia, and crpCDE also seems to be involved in HR, P. corrugata represents an interesting and intriguing study model.
Experimental Procedures
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas corrugata strains and Chromobacterium violaceum strain CV026 (AHL bacterial biosensor) were routinely grown at 28 °C in nutrient agar (Oxoid, Milan, Italy) plus 1% dextrose (NDA) or in Luria–Bertani (LB) agar. Escherichia coli strain DH5α and CC118λpir were used as hosts for the plasmids and for insertional mutagenesis. Escherichia coli strains were grown at 37 °C on LB plates or in LB broth. Antibiotics were added as required at the following final concentrations: ampicillin, 100 μg/mL; kanamycin, 50 μg/mL (C. violaceum) or 100 μg/mL; tetracycline, 15 μg/mL (E. coli) or 40 μg/mL (Pseudomonas spp.).
DNA recombinant techniques
DNA manipulations, including digestions with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 DNA ligase, DNA hybridization, radioactive labelling by random priming and E. coli transformation, were performed as described by Sambrook et al. (1989). Southern hybridizations were performed using Amersham Hybond N+ membranes (GE Healthcare Bio‐Sciences, Pittsburgh, USA). Total DNA from Pseudomonas spp. was isolated using the Gentra Puregene Cell Kit (Qiagen Inc., Valencia, CA, USA). Triparental matings from E. coli to P. corrugata were carried out with the helper strain E. coli DH5α (pRK2013) (Figurski and Helinski, 1979).
DNA sequencing and sequence analysis
Two contigs were obtained sequencing the cosmid pLC3.34 DNA insert on both strands (Macrogen, Inc., Seoul, South Korea). A set of primers, Primerseq‐fw/rev, overlapping the ends of the two sequences, was used to merge the gap between the two contigs, yielding a 2284‐bp amplicon (Table 1). The resulting amplicon was sequenced on both strands by BMRCRIBI (University of Padua, Italy). Homology searches of nucleotide and protein sequences were performed using the blast searching program in the National Center for Biotechnology Information (NCBI) database (Altschul et al., 1990). For phylogenetic analysis, alignments were made with ClustalW incorporated into the mega 5.01 software package (Tamura et al., 2011). The previous sequence, Accession number EF189721, was thus substituted with the complete insert with the Accession number KF192265.
Cell‐free culture filtrate and RNA sample preparation
Bacterial strains were grown in Improved Minimal Medium (IMM) (Surico et al., 1998) at 28 °C in still culture. Time‐course analysis of P. corrugata CFBP 5454 was determined by sampling aliquots from still cultures incubated in IMM at 28 °C from triplicate flasks at 17, 24, 48, 72 and 96 hpi. To obtain cell‐free culture filtrates after centrifugation (9000 × g, 20 min), the supernatant was passed through a 0.22‐μm Millipore filter (Millipore, Billerica, MA, USA). Aliquots of all samples were dried and resuspended in sterile water to obtain a 10 × culture filtrate.
Total RNAs were prepared from P. corrugata CFBP 5454 and mutant strain cell aliquots, using the RNeasy Mini kit (Qiagen Inc.) according to the manufacturer's directions.
Construction of P . corrugata crpD and crpC knock‐out mutants
The central parts of crpD and crpC genes were amplified by PCR as 486‐bp and 1234‐bp fragments, respectively, using the primers MFPkn‐fw/rev and NRPSkn‐fw/rev (Table 1). These fragments were first cloned into pCR2.1 vector (Invitrogen, Milan, Italy) according to the manufacturer's instructions, and then subcloned by BamHI/XbaI digestions into pKnock‐Km suicide vector (Alexeyev, 1999), generating PKMMFP and PKMNRPS, respectively. These latter plasmids were transferred into P. corrugata CFBP 5454 by triparental mating, generating PCOMFP and PCONRPS, respectively. Transformants were selected on LB agar plates supplemented with kanamycin (100 μg/mL) and confirmed by Southern blot analysis. PCOMFP mutant strain was complemented by introducing pBBR1MCS‐5 containing the full‐length P. corrugata crpDE genes (3224 bp). The sequence was first amplified by PCR using oligonucleotides ABC‐fw/rev (Table 1), cloned in pGEM‐T easy vector (Promega, Madison, WI, USA) and removed as a HindIII/BamHI fragment. The insert was then cloned in the corresponding sites in pBBR1MCS‐5 (Kovach et al., 1995), yielding pBBR‐CrpDE.
Reverse transcription‐polymerase chain reaction (RT‐PCR)
RT‐PCR analysis was performed using two sets of specific primers, PCR1‐fw/rev (523 bp) and PCR2‐fw/rev (512 bp) (Table 1), to identify the putative transcripts overlapping the crpC‐crpD and crpD‐crpE regions, respectively. Following a DNAse purification step by DNAse I (Invitrogen), 1 μg of RNA was used for cDNA synthesis with Superscript III (Invitrogen) according to the manufacturer's protocol. Genomic DNA was used to test the fidelity of the primer pairs, whereas samples in which reverse transcriptase was not added were used as negative controls. PCRs were performed using a Gene Amp PCR system 9700 (PE Applied Biosystem, Milan, Italy) under the following conditions: an initial 94 °C for 2 min; followed by 35 cycles of 94 °C for 45 s, 56 °C for 45 s and 72 °C for 45 s; and a final extension of 72 °C for 5 min. The RT‐PCR products were subjected to electrophoresis with a 1.5% agarose gel.
Transcriptional analysis by Q‐PCR
To determine whether crpC and crpD expression is regulated by the transcriptional regulators PcoR and RfiA, P. corrugata CFBP 5454 and derivative mutants GL2 and GLRFIA were analysed by Q‐PCR. The same technique was also used to determine crpC and crpD transcript levels in PCOMFP and PCONRPS strains. The primers for Q‐PCR are listed in Table 1 (Abc1 fw/rev for crpD and Nrps fw/rev for crpC). Reactions were conducted with the Bio‐Rad (Bio‐Rad, Hercules, CA,USA) iQ5 Cycler and the SYBR GreenER qPCR Super Mix iCycler (Invitrogen), according to the manufacturers' protocols. The P. corrugata 16S rRNA (Conte et al., 2006) gene was used as housekeeping gene. The relative expression was determined using the comparative CT (cycle threshold) method, also known as the ΔΔCT method (Livak and Schmittgen, 2001). Q‐PCR analysis was performed in duplicate on three independent RNA isolations.
I n vitro bioassay for AHL and CLP production
Pseudomonas corrugata CFBP 5454 and respective mutant culture filtrates were used to asses AHL and CLP production. Culture filtrates of P. corrugata CFBP 5454 were added to the AHL CV026 biosensor to determine AHL production by visual detection of violacein formation (Martinelli et al., 2004). Antimicrobial activity was assessed by well‐diffusion assay in plates containing a double layer of solidified potato dextrose agar (PDA) (Oxoid) containing the two CLP‐sensitive bioindicator strains R. pilimanae ATTC26432 and B. megaterium ITM100 (Licciardello et al., 2009). The region around the well in which growth was prevented was measured. All tests were carried out at least twice in triplicate each time. The presence of CLP in P. corrugata CFBP 5454 and derivative mutant cell‐free culture filtrates was detected by MALDI‐TOF‐MS analysis according to Licciardello et al. (2012).
Plant inoculations
Pseudomonas corrugata CFBP 5454 and derivative mutants were tested for pathogenicity on tomato cv. Marmande plants grown in nursery flats, 1 month after germination. During the trials, plants were maintained in a growth chamber with a 16 h/8 h photoperiod and a temperature of 26 °C. Tomato plants were pin‐pricked on the stem at the axil of the first true leaf with bacterial cells from 48‐h culture on NDA (Licciardello et al., 2007). At various time points (3 and 7 dpi), 1 cm of stem was cut either at the inoculation site or at different distances above and below the inoculation site and homogenized in 1 mL of distilled water; serial 10‐fold dilutions were plated onto NDA plates with appropriate antibiotics for the selection of the strains using a spiral plater (Eddy Jet, IUL,S.A. Barcelona, Spain). At each sampling time, six plants per strain were analysed. The length of the stem discoloration/necrosis was assessed at 15 dpi. An HR test was performed by infiltration of N. benthamiana leaf mesophyll with a bacterial suspension of 108 cfu/mL using a blunt syringe. Twenty leaf panels were inoculated per strain. After inoculation, plants were placed at 25 °C in a growth chamber and the collapse/necrosis of the mesophyll was recorded daily. Bacterial populations were estimated daily for 4 days in a single leaf disc cut with a boring tool (inner diameter, 0.7 cm) from three individual lesions from three different leaves (Licciardello et al., 2007).
Statistical analysis
Data were analysed by one‐ or three‐way ANOVA with CosTat software (CoHort Software, Berkeley, CA, USA). Mean values were compared using Student–Newman–Keuls test.
Supporting information
Table S1 Characteristics of CrpE homologous ABC transporters in plant‐associated Pseudomonas.
Acknowledgements
This work was funded in part by Ministero dell′Istruzione, dell′Università e della Ricerca, Programma Operativo Nazionale ‘Ricerca e Competitività’ 2007–2013 (MIUR, PON R&C2007‐2013), project ‘PolyBioPlast—Technologies and Processes for the Production of Diversely Functionalized Sheets based on Microbial Biopolymers and Biosurfactants (PON01_1377)’.
References
- Alexeyev, M.F. (1999) The pKNOCK series of broad‐host‐range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram‐negative bacteria. Biotechniques, 26, 824–828. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ballio, A. , Barra, D. , Bossa, F. , Collina, A. , Grgurina, I. , Marino, G. , Moneti, G. , Paci, M. , Pucci, P. , Segre, A. and Simmaco, M. (1991) Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae . FEBS Lett. 291, 109–112. [DOI] [PubMed] [Google Scholar]
- Ballio, A. , Bossa, F. , Camoni, L. , Di Giorgio, D. , Flamand, M.C. , Maraite, H. , Nitti, G. , Pucci, P. and Scaloni, A. (1996) Structure of fuscopeptins, phytotoxic metabolites of Pseudomonas fuscovaginae . FEBS Lett. 381, 213–216. [DOI] [PubMed] [Google Scholar]
- Baltrus, D.A. , Nishimura, M.T. , Romanchuk, A. , Chang, J.H. , Mukhtar, M.S. Cherkis, K. , Roach, J. , Grant, S.R. , Jones, C.D. and Dangl, J.L. (2011) Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7, e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, C.L. , Alarcon‐Chaidez, F. and Gross, D.C. (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry, C.L. , Nandi, M. , Manuel, J. , Brassinga, A.K.C. , Fernando, W.G.D. , Loewena, P.C. and de Kievit Teresa, R. (2014) Characterization of the Pseudomonas sp. DF41 quorum sensing locus and its role in fungal antagonism. Biological Control, 69, 82–89. [Google Scholar]
- Berti, A.D. , Greve, N.J. , Christensen, Q.H. and Thomas, M.G. (2007) Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 189, 6312–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn, I. and Raaijmakers, J.M. (2009) Diversity and functional analysis of LuxR‐type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens . Appl. Environ. Microb. 75, 4753–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn, I. , de Kock, M.J.D. , de Waard, P. , van Beek, T.A. and Raaijmakers, J.M. (2008) Massetolide A biosynthesis in Pseudomonas fluorescens . J. Bacteriol. 190, 2777–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catara, V. (2007) Pseudomonas corrugata: plant pathogen and/or biological resource? Mol. Plant Pathol. 8, 233–244. [DOI] [PubMed] [Google Scholar]
- Catara, V. , Gardan, L. and Lopez, M.M. (1997) Phenotypic heterogeneity of Pseudomonas corrugata strains from southern Italy. J. Appl. Microbiol. 83, 576–586. [Google Scholar]
- Catara, V. , Sutra, L. , Morineeau, A. , Achouak, W. , Christan, R. and Gardan, L. (2002) Phenotypic and genomic evidence for the revision of Pseudomonas corrugata and proposal of Pseudomonas mediterranea species sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Cho, H. and Kang, H. (2012) The PseEF efflux system is a virulence factor of Pseudomonas syringae pv. syringae . J. Microbiol. 50, 79–90. [DOI] [PubMed] [Google Scholar]
- Conte, E. , Catara, V. , Greco, S. , Russo, M. , Alicata, R. , Strano, L. , Lombardo, A. , Di Silvestro, S. and Catara, A. (2006) Regulation of polyhydroxyalkanoate synthases (phaC1 and phaC2) gene expression in Pseudomonas corrugata . Appl. Microbiol. Biotechnol. 72, 1054–1062. [DOI] [PubMed] [Google Scholar]
- Coraiola, M. , Lo Cantore, P. , Lazzaroni, S. , Evidente, A. , Iacobellis, N.S. and Dalla Serra, M. (2006) WLIP and tolaasin I, lipodepsipeptides from Pseudomonas reactans and Pseudomonas tolaasii, permeabilise model membranes. Biochim. Biophys. Acta, 1758, 1713–1722. [DOI] [PubMed] [Google Scholar]
- Cui, X. , Harling, R. , Mutch, P. and Darling, D. (2005) Identification of N‐3‐hydroxyoctanoyl‐homoserine lactone production in Pseudomonas fluorescens 5064, pathogenic to broccoli, and controlling biosurfactant production by quorum sensing. Eur. J. Plant Pathol. 111, 297–308. [Google Scholar]
- Dubern, J.F. , Lugtenberg, B.J.J. and Bloemberg, G.V. (2006) The ppuI‐rsaL‐ppuR quorum‐sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J. Bacteriol. 188, 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubern, J.F. , Coppoolse, E.R. , Stiekema, W.J. and Bloemberg, G.V. (2008) Genetic and functional characterization of the gene cluster directing the biosynthesis of putisolvin I and II in Pseudomonas putida strain PCL1445. Microbiology, 154, 2070–2083. [DOI] [PubMed] [Google Scholar]
- Emanuele, M.C. , Scaloni, A. , Lavermicocca, P. , Jacobellis, N.S. , Camoni, L. , Di Giorgio, D. , Pucci, P. , Paci, M. , Segre, A. and Ballio, A. (1998) Corpeptins, new bioactive lipodepsipeptides from cultures of Pseudomonas corrugata . FEBS Lett. 433, 317–320. [DOI] [PubMed] [Google Scholar]
- Figurski, D.H. and Helinski, D.R. (1979) Replication of an origin‐containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA, 76, 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finking, R. and Marahiel, M.A. (2004) Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58, 453–488. [DOI] [PubMed] [Google Scholar]
- Gross, H. and Loper, J.E. (2009) Genomics of secondary metabolite production by Pseudomonas spp. Natl. Prod. Rep. 26, 1408–1446. [DOI] [PubMed] [Google Scholar]
- Gross, H. , Stockwell, V.O. , Henkels, M.D. , Nowak‐Thompson, B. , Loper, J.E. and Gerwick, W.H. (2007) The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters. Chem. Biol. 14, 53–63. [DOI] [PubMed] [Google Scholar]
- Gustine, D.L. , Sherwood, R.T. , Lukezic, F.L. and Moyer, B.G. (1990) Metabolites from Pseudomonas corrugata elicit phytoalexin biosynthesis in white clover. Phytopathology, 80, 1427–1432. [Google Scholar]
- Gustine, D.L. , Sherwood, R.T. , Lukezic, F.L. , Moyer, B.G. and Devlin, W.S. (1994) Metabolites of Pseudomonas corrugata elicit plant defense reactions In: Bioregulators and Natural Products. ACS Symposium Series (Hedin P.A., ed.), pp. 169–181. Washington DC: American Chemical Society. [Google Scholar]
- Gustine, D.L. , Sherwood, R.T. and Moyer, B.G. (1995) Evidence for a new class of peptide elicitor of the hypersensitive reaction from the tomato pathogen Pseudomonas corrugata . Phytopathology, 85, 848–853. [Google Scholar]
- Herrero, M. , de Lorenzo, V. and Timmis, K.N. (1990) Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertions of foreign genes in gram‐negative bacteria. J. Bacteriol. 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. and Gross, D.C. (2005) Characterization of a resistance‐nodulation‐cell division transporter system associated with the syr‐syp genomic island of Pseudomonas syringae pv. syringae . Appl. Environ. Microbiol. 71, 5056–5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M., II and Peterson, K.M. (1995) Four new derivatives of the broad‐host range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Licciardello, G. , Bertani, I. , Steindler, L. , Bella, P. , Venturi, V. and Catara, V. (2007) Pseudomonas corrugata contains a conserved N‐acyl homoserine lactone quorum sensing system; its role in tomato pathogenicity and tobacco hypersensitivity response. FEMS Microbiol. Ecol. 61, 222–234. [DOI] [PubMed] [Google Scholar]
- Licciardello, G. , Bertani, I. , Steindler, L. , Bella, P. , Venturi, V. and Catara, V. (2009) The transcriptional activator rfiA is quorum sensing regulated by cotranscription with the luxI homolog pcoI and is essential for plant virulence in Pseudomonas corrugata . Mol. Plant–Microbe Interact. 22, 1514–1522. [DOI] [PubMed] [Google Scholar]
- Licciardello, G. , Strano, C.P. , Bertani, I. , Bella, P. , Fiore, A. , Fogliano, V. , Venturi, V. and Catara, V. (2012) N‐Acyl‐homoserine lactones quorum sensing in tomato phytopathogenic Pseudomonas spp. is involved in lipodepsipeptide production. J. Biotechnol. 159, 274–282. [DOI] [PubMed] [Google Scholar]
- Licciardello, G. , Jackson, R.W. , Bella, P. , Strano, C.P. , Catara, A.F. , Arnold, D.L. , Venturi, V. , Silby, M.W. and Catara, V. (2014) Draft genome sequence of Pseudomonas corrugata, a phytopathogenic bacterium with potential industrial applications. J. Biotech. 175, 65–66. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, S.E. , Scholz‐Schroeder, B.K. and Gross, D.C. (2002) Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol. Plant–Microbe Interact. 15, 43–53. [DOI] [PubMed] [Google Scholar]
- Lu, S.E. , Wang, N. , Wang, J.L. , Chen, Z.J. and Gross, D.C. (2005) Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae . Mol. Plant–Microbe Interact. 18, 324–333. [DOI] [PubMed] [Google Scholar]
- Martinelli, D. , Grossmann, G. , Séquin, U. , Brandl, H. and Bachofen, R. (2004) Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum . BMC Microbiol. 4, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean, K.H. , Winson, M.K. , Fish, L. , Taylor, A. , Chhabra, S.R. , Camara, M. , Daykin, M. , Lamb, J.H. , Swift, S. , Bycroft, B.W. , Stewart, G.S.A. and Williams, P. (1997) Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N‐acylhomoserine lactones. Microbiology, 143, 3703–3711. [DOI] [PubMed] [Google Scholar]
- Quigley, N.B. , Mo, Y.Y. and Gross, D.C. (1993) ) SyrD is required for syringomycin production by Pseudomonas syringae pathovar syringae and is related to a family of ATP‐binding secretion proteins. Mol. Microbiol. 9, 787–801. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , de Bruijn, I. and De Kock, M.J.D. (2006) Cyclic lipopeptide production by plant associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Mol. Plant–Microbe Interact. 19, 699–710. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , de Bruijn, I. , Nybroe, O. and Ongena, M. (2010) Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 34, 1037–1062. [DOI] [PubMed] [Google Scholar]
- Roongsawang, N. , Hase, K. , Haruki, M. , Imanaka, T. , Morikawa, M. and Kanaya, S. (2003) Cloning and characterization of the gene cluster encoding arthrofactin synthetase from Pseudomonas sp. MIS38. Chem. Biol. 10, 869–880. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scaloni, A. , Dalla Serra, M. , Amodeo, P. , Mannina, L. , Vitale, R.M. , Segre, A.L. , Cruciani, O. , Lodovichetti, F. , Greco, M.L. , Fiore, A. , Gallo, M. , D'Ambrosio, C. , Coraiola, M. , Menestrina, G. , Granit, A. and Fogliano, V. (2004) Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: Cormycin A. Biochem. J. 384, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett, C.M. , Fletcher, J.T. , Roberts, P. and Lelliott, R.A. (1978) Tomato pith necrosis caused by Pseudomonas corrugata. n. sp. Ann. Appl. Biol. 88, 105–114. [Google Scholar]
- Scholz‐Schroeder, B.K. , Hutchison, M.L. , Grgurina, I. and Gross, D.C. (2001) The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant–Microbe Interact. 14, 336–348. [DOI] [PubMed] [Google Scholar]
- Scholz‐Schroeder, B.K. , Soule, J.D. and Gross, D.C. (2003) The sypA, sypB, and sypC synthetase genes encode twenty‐two modules involved in the nonribosomal peptide synthesis of syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant–Microbe Interact. 16, 271–280. [DOI] [PubMed] [Google Scholar]
- Siverio, F. , Cambra, M. , Gorris, M.T. , Corzo, J. and Lopez, M.M. (1993) Lipopolysaccharides as determinant of serological variability in Pseudomonas corrugata . Appl. Environ. Microbiol. 59, 1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surico, G. , Lavermicocca, P. and Iacobellis, N.S. (1998) Produzione di siringomicina e siringotossina in colture di Pseudomonas syringae pv. syringae . Phytopathol. Mediterr. 27, 163–168. [Google Scholar]
- Sutra, L. , Siverio, F. , Lopez, M.M. , Hanaul, T.G. , Bollet, C. and Gardan, L. (1997) Taxonomy of Pseudomonas strains isolated from tomato pith necrosis: emended description of Pseudomonas corrugata and proposal of three unnamed fluorescent Pseudomonas genomo‐species. Int. J. Syst. Bacteriol. 47, 1020–1033. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet‐Gely, I. , Novikov, A. , Augusto, L. , Liehl, P. , Bolbach, G. , Pe′chy‐Tarr, M. , Cosson, P. , Keel, C. , Caroff, M. and Lemaitre, B. (2010) Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl. Environ. Microbiol. 73, 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Lu, S.E. , Records, A.R. and Gross, D.C. (2006) Characterization of the transcriptional activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae . J. Bacteriol. 188, 3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Characteristics of CrpE homologous ABC transporters in plant‐associated Pseudomonas.


