Summary
The definition of the precise molecular composition of membranous replication compartments is a key to understanding the mechanisms of virus multiplication. Here, we set out to investigate the protein composition of the potyviral replication complexes. We purified the potyviral 6K2 protein‐induced membranous structures from Potato virus A (PVA)‐infected Nicotiana benthamiana plants. For this purpose, the 6K2 protein, which is the main inducer of potyviral membrane rearrangements, was expressed in fusion with an N‐terminal Twin‐Strep‐tag and Cerulean fluorescent protein (SC6K) from the infectious PVA cDNA. A non‐tagged Cerulean‐6K2 (C6K) virus and the SC6K protein alone in the absence of infection were used as controls. A purification scheme exploiting discontinuous sucrose gradient centrifugation followed by Strep‐tag‐based affinity chromatography was developed. Both (+)‐ and (–)‐strand PVA RNA and viral protein VPg were co‐purified specifically with the affinity tagged PVA‐SC6K. The purified samples, which contained individual vesicles and membrane clusters, were subjected to mass spectrometry analysis. Data analysis revealed that many of the detected viral and host proteins were either significantly enriched or fully specifically present in PVA‐SC6K samples when compared with the controls. Eight of eleven potyviral proteins were identified with high confidence from the purified membrane structures formed during PVA infection. Ribosomal proteins were identified from the 6K2‐induced membranes only in the presence of a replicating virus, reinforcing the tight coupling between replication and translation. A substantial number of proteins associating with chloroplasts and several host proteins previously linked with potyvirus replication complexes were co‐purified with PVA‐derived SC6K, supporting the conclusion that the host proteins identified in this study may have relevance in PVA replication.
Keywords: 6K2 protein, Potato virus A, potyvirus, proteome, viral replication complex
Introduction
Positive‐strand RNA [(+)RNA] viruses replicate in association with host membranes (Miller and Krijnse‐Locker, 2008). This association results in the rearrangement of cellular membranes to accommodate viral replication machineries, hide viral RNA (vRNA) and proteins from host defence systems and provide an environment to concentrate host and viral factors for vRNA synthesis (den Boon and Ahlquist, 2010; Verchot, 2011). Among the different groups of (+)RNA viruses, the virus‐induced membranous replication complexes vary greatly in their origin, size and shape (reviewed in Paul and Bartenschlager, 2013). The formation of viral replication complexes (VRCs) in (+)RNA viruses requires an orchestrated assembly of many host and viral proteins (Mine and Okuno, 2012). Host factors involved in viral multiplication represent potential targets for virus control, and therefore their identification and functional characterization are important.
The genus Potyvirus is economically one of the most devastating groups of plant viruses in the world, affecting the production of nearly all cultivated plant species. The genome of potyviruses is a positive‐sense, single‐stranded RNA of approximately 10 kb. Ten of eleven potyviral proteins are synthesized from a large open reading frame (ORF), whereas the 11th protein, P3N‐PIPO, is produced from a separate, partially overlapping, ORF (reviewed in Revers and García, 2015). P3N‐PIPO expression is enabled by transcriptional slippage, specific for viral replicase (Olspert et al., 2015; Rodamilans et al., 2015). The formation of potyviral VRCs is initiated at the endoplasmic reticulum (ER), wherefrom the individual VRC vesicles are transported to chloroplasts via the endosomal trafficking pathway aided by the actomyosin system (Wei and Wang, 2008; Wei et al., 2010). As the infection progresses, these vesicles fuse with chloroplasts in a process requiring plant SNARE protein Syp71 (Wei et al., 2013). Finally, late in the infection, the chloroplast‐associated VRCs aggregate into large perinuclear globular structures (Grangeon et al., 2012).
The hydrophobic membrane‐associated potyviral protein 6K2 is a multifunctional protein participating in VRC formation, long‐distance movement and symptom development (Rajamäki and Valkonen, 1999; Spetz and Valkonen, 2004). 6K2 is able to induce vesicle formation at ER membranes even in the absence of infection (Beauchemin et al., 2007; Schaad et al., 1997; Thivierge et al., 2008). Most of the potyviral proteins, including P3, CI, 6K2, viral genome‐linked protein (VPg), NIapro and NIb, have been shown to associate with 6K2‐induced VRCs (Beauchemin et al., 2007; Cotton et al., 2009; Dufresne et al., 2008). The RNA helicase activity of CI (Carrington et al., 1998; Fernández et al., 1997; Kekarainen et al., 2002), the putative role of VPg as a primer for RNA synthesis (Anindya et al., 2005; Puustinen and Mäkinen, 2004; Rantalainen et al., 2011) and the RNA synthesis activity of NIb (Hong and Hunt, 1996) are essential for the amplification of vRNA. Although P3 is also required for replication (Klein et al., 1994), the molecular mechanism of how it exerts its replication‐associated function is not clear. P3N‐PIPO and coat protein (CP) are dispensable for replication (Mahajan et al., 1996; Wen and Hajimorad, 2010).
Host factors have various roles in replication, including RNA recruitment, assembly and activation of VRCs, (–)‐ and (+)‐strand synthesis activity, asymmetry in (+)‐strand production and adjustment of lipid composition (e.g. Barajas et al., 2014; Huang et al., 2012; Wei et al., 2013; reviewed by Nagy and Pogany, 2012 and Wang, 2015). The reconstitution of various replication reactions in yeast extracts supplemented with viral replication proteins has been a powerful tool to study the specific roles of the host proteins in tombusvirus replication (Pogany and Nagy, 2008). Although such an experimental tool is not available to study the replication reactions of potyviruses, a lot of biochemistry and cell biology work has been performed to identify host proteins associated with 6K2‐induced vesicles. The importance of these structures for viral replication is emphasized by the presence of double‐stranded vRNA and active vRNA synthesis in Turnip mosaic virus (TuMV)‐induced vesicles (Cotton et al., 2009).
Paul et al. (2013) used a membrane pull‐down approach to study the molecular composition of membranous replication compartments of human Hepatitis C virus (HCV; family Flaviviridae). They isolated double‐membrane vesicles (DMVs) and demonstrated active HCV RNA synthesis within them. Further biochemical and morphological studies revealed the presence of many viral and host proteins in DMVs, as well as details of their lipid composition.
We chose a similar approach to study the protein composition of potyviral VRCs. 6K2 protein fused to an affinity tag and Cerulean fluorescent protein (CFP) was used to purify 6K2‐induced membrane structures from Potato virus A (PVA)‐infected Nicotiana benthamiana plants. Although 6K2 appears to be an optimal marker protein for potyviral VRCs, it does not readily tolerate modifications in its natural genomic context (Spetz and Valkonen, 2004). To overcome this, an additional copy of the 6K2 gene is usually inserted into a different location in the genome. Successful visualization of TuMV VRCs has been achieved using this approach (Cotton et al., 2009; Thivierge et al., 2008; Wei et al., 2010). We inserted the Strep‐tagged CFP‐fused 6K2 protein in between NIb and CP coding regions and purified the membrane structures from infected and non‐infected 6K2 expressing control N. benthamiana leaves.
Our main goal was to identify the protein composition of the PVA VRCs. To achieve this, we performed a proteomic analysis of the purified 6K2‐induced membrane structures. The data produced will facilitate functional studies of the host proteins involved in potyviral replication in future.
Results
Establishment of infectious PVA cDNA encoding Twin‐Strep‐tagged 6K2
We set out to purify the putative PVA VRCs via the membrane‐associated 6K2 protein. To allow the visualization of the 6K2‐induced structures, we expressed 6K2 with a fluorescent marker from PVA infectious cDNA (icDNA). The PVA‐C6K construct allowed the expression of CFP in fusion with the N‐terminus and the PVA‐6KY construct allowed the expression of yellow fluorescent protein (YFP) in fusion with the C‐terminus of the 6K2 protein (Fig. 1). NIa protease cleavage sites were engineered to flank the 6K2 fusion protein to aid polyprotein processing.
Figure 1.
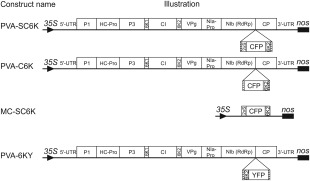
Schematic representation of the constructs. The PVA‐SC6K construct allows the expression of 2×Strep‐Cerulean fluorescent protein (CFP)‐6K2 (SC6K2) fusion protein in the context of Potato virus A (PVA) infection. The PVA‐C6K construct is similar, except that it lacks the 2×Strep‐tag. SC6K2 fusion is expressed from the MC‐SC6K construct (MC, membrane control) in a non‐infected background. The PVA‐6KY construct allows the expression of the 6K2‐yellow fluorescent protein (6KY) fusion in the context of PVA infection. Hatched rectangles flanking the fluorescent protein‐6K2 cassettes denote NIa protease cleavage sites.
Agrobacterium infiltration was used to introduce PVA‐C6K and PVA‐6KY icDNAs into N. benthamiana leaves. Both viruses spread approximately with a similar speed (Fig. S1, see Supporting Information). Next, we studied the infection of PVA‐C6K and PVA‐6KY by confocal microscopy (Fig. 2a). Scattered individual vesicles and hardly any aggregation with chloroplasts were observed with CFP‐6K2 (Fig. 2a, left panels), whereas 6K2‐YFP‐induced vesicles associated predominantly with chloroplasts (Fig. 2a, middle panels). The lack of CFP‐6K2 chloroplast labelling was a puzzling observation as VRC vesicle fusion with the outer chloroplast membrane is required for replication (Wei et al., 2010). Therefore, we studied the infection in systemic leaves by electron microscopy (EM) (Fig. 2b). The hallmarks of potyviral infection, cylindrical inclusions and virus particles, were observed in both PVA‐C6K‐ and PVA‐6KY‐infected cells. We concluded that both viruses, PVA‐C6K and PVA‐6KY, caused normal infection.
Figure 2.
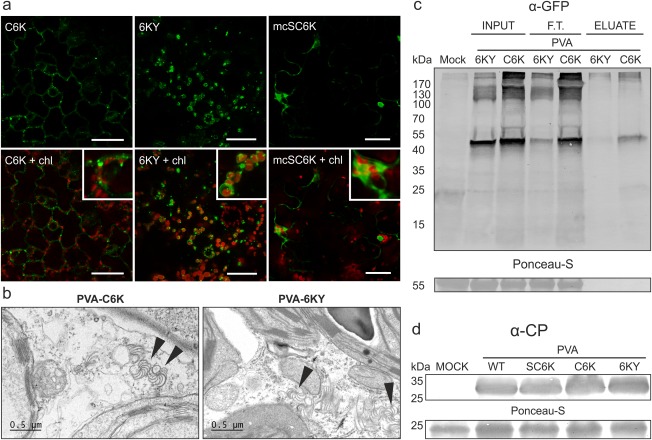
PVA‐C6K and PVA‐6KY are both infectious and the N‐terminal 6K2 tag is accessible for affinity purification. (a) Comparison of fluorescence derived from C6K (green) and 6KY (green) during Potato virus A (PVA) infection by confocal microscopy. The C6K protein localized mostly to scattered vesicles, whereas the 6KY signal was detected mostly in association with chloroplasts (chl., red). Magnified sections show 6K2 vesicle association with chloroplasts. (b) Electron microscopic images of the infected tissues. Both PVA‐C6K and PVA‐6KY produced cytoplasmic cylindrical inclusions which are indicated with arrowheads. (c) Affinity chromatography purification of the 6K2‐fusion protein from the infection context, revealing N‐terminally fused Cerulean fluorescent protein (CFP) to be better accessible for the green fluorescent protein (GFP)‐trap matrix compared with the C‐terminally fused yellow fluorescent protein (YFP). (d) Western blot analysis verified the presence of PVA coat protein (CP) in the upper leaves at 10 days post‐infiltration (DPI), indicating that PVA‐SC6K, PVA‐C6K and PVA‐6KY are all able to cause systemic infection.
To establish which virus, PVA‐C6K or PVA‐6KY, should be used for purification, we used green fluorescent protein (GFP)‐trap purification, which allows the isolation of fluorescent fusion proteins. The N‐terminally fused CFP gave better yields (Fig. 2c), suggesting that the tag is better exposed in this orientation, which led us to choose PVA‐C6K for the purification of VRCs.
Twin‐Strep‐tag (hereafter 2×Strep‐tag) consists of two copies of an eight‐amino‐acid (WSHPQFEK) long peptide, and allows efficient purification of proteins under native conditions (Schmidt and Skerra, 2007). We fused the 2×Strep‐tag encoding sequence to the CFP‐6K2 gene in PVA‐C6K, thus creating PVA‐SC6K icDNA. Systemic infection by PVA‐SC6K was confirmed by an immunoblot analysis (Fig. 2d). PVA‐C6K was used as a control to verify the tag‐specific purification of the proteins. In addition, we cloned a membrane control construct (MC‐SC6K; Fig. 1) to express monocistronic SC6K from a plant expression vector (mcSC6K). The purpose of the MC‐SC6K control was to reveal the host proteins associated with 6K2‐induced membranous structures in the absence of a replicating virus. The exclusion of these proteins from the final list should therefore reveal the proteins present in the membranous replication structures during infection.
Purification and characterization of membranous 6K2‐induced structures from PVA‐infected cells
To obtain PVA 6K2‐induced membranous structures, N. benthamiana plants were Agrobacterium infiltrated to initiate PVA‐SC6K and PVA‐C6K infections and MC‐SC6K expression. The systemically infected leaves were collected at 10 days post‐infiltration (DPI) and leaves transiently expressing mcSC6K were collected at 4 DPI. The purification scheme is depicted in Fig. 3a. The membranous SC6K‐ and C6K‐associated structures were enriched by sucrose gradient centrifugation prior to affinity purification. The presence of C6K or SC6K in the collected fractions was confirmed by Western blotting with an anti‐GFP antibody. The Western blot analysis revealed that most SC6K was concentrated to fraction 5 in the virus‐infected samples (Fig. 3b, left panel). In the non‐infected mcSC6K‐expressing sample, the signal concentrated to fractions 5–7 (Fig. 3b, right panel). This analysis also revealed the presence of an approximately 27‐kDa product, representing free CFP, which explains the cytoplasmic background fluorescence observed in confocal microscopy (see Fig. 2b). Free CFP in the top fractions showed that the SC6K‐containing fractions were well separated from those containing soluble cytosolic proteins. The affinity purification of SC6K‐containing membranes was carried out on a Strep‐Tactin matrix from fractions 5 of each sample. A clear enrichment of SC6K was observed in the PVA‐SC6K sample eluate, whereas no C6K was detected in the PVA‐C6K sample eluate (Fig. 3c), indicating efficient tag‐specific purification. A clear difference in the total protein content between the purified PVA‐SC6K proteins and the controls was detected in the silver‐stained gels (Fig. 3d). Protein quantification from the purified membranes showed that the PVA‐SC6K and MC‐SC6K samples contained a higher concentration of proteins than the PVA‐C6K samples (Fig. S2, see Supporting Information).
Figure 3.
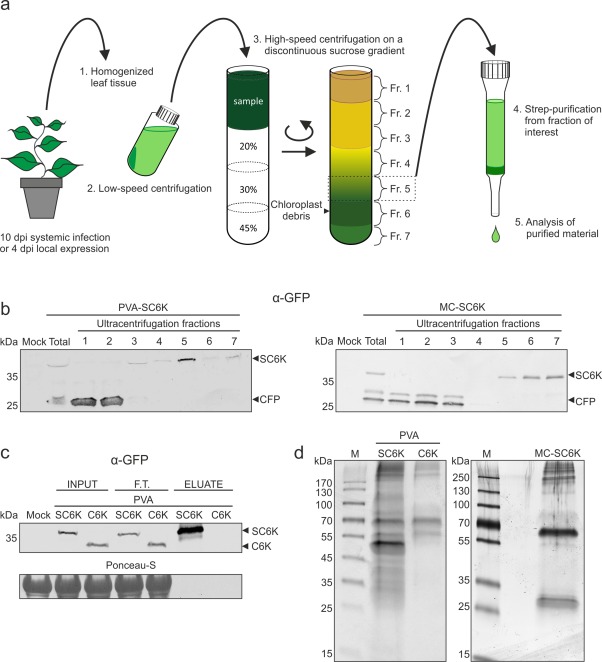
Purification of 6K2‐associated membranes from Potato virus A (PVA) infection. (a) A schematic representation of the purification protocol. PVA‐SC6K‐ and PVA‐C6K‐infected and SC6K‐expressing leaf tissues were homogenized and the cleared lysates were subjected to sucrose gradient centrifugation. Fraction 5 collected from the gradient was subjected to affinity purification via the 2×Strep‐tag. (b) The sucrose gradient fractions were analysed by Western blot analysis. SC6K concentrated to fraction 5 in the infection context and to fractions 5–7 when SC6K was expressed alone. (c) SC6K protein and its binding partners were subjected to 2×Strep‐tag affinity purification. SC6K protein was significantly enriched in the eluate. The 2× Strep‐tag‐specific purification was controlled with tag‐less C6K protein (F.T., flow through). (d) The outcome of the purification procedure was assessed by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), followed by silver staining, which revealed clear differences in the protein content between the purified PVA‐SC6K sample in the left panel and the controls: PVA‐C6K in the left panel and MC‐SC6K in the right panel.
Real‐time reverse transcriptase‐polymerase chain reaction (qRT‐PCR) showed that the PVA RNA copy number was greater in PVA‐C6K than in PVA‐SC6K input prior to purification, and vice versa after purification (Fig. S3, see Supporting Information), indicating that the yield of PVA RNA was significantly higher from the PVA‐SC6K sample than from the PVA‐C6K control. Approximately 5.2% of PVA RNA present in the input was recovered in the PVA‐SC6K sample, compared with approximately 0.6% in the PVA‐C6K control, the fold of enrichment being 8.7 (Fig. 4a). Reverse transcriptase‐PCR (RT‐PCR) with both (+)‐ and (–)‐strand PVA RNA‐specific primers revealed a strong (+)‐strand‐ and a weak (–)‐strand‐specific signal in the PVA‐SC6K sample, showing that it contained PVA RNA of both polarities (Fig. 4b). A weak (+)‐strand‐specific signal in the PVA‐C6K control sample suggested that some non‐specific binding of C6K‐containing membranes and/or PVA particles to the Strep‐Tactin matrix took place. Western blot analysis with VPg antibody revealed tag‐specific purification of the essential replication protein VPg in the PVA‐SC6K sample (Fig. 4c). The origin of the high‐molecular‐weight signal in the VPg blot is not clear. It may represent polyprotein intermediates, protein complexes that were not fully dissociated during sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) or VPg–RNA complexes.
Figure 4.
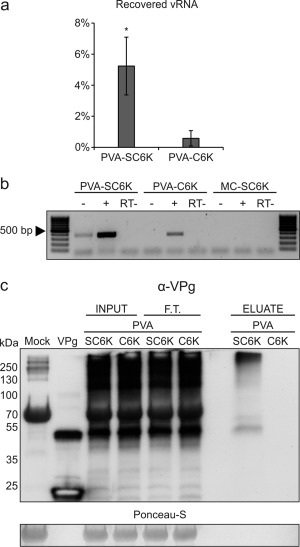
Potato virus A (PVA) RNA and viral genome‐linked protein (VPg) content in the purified 6K2‐associated membrane samples. (a) The percentages of recovered viral RNA (vRNA) indicate significant enrichment of PVA RNA in the PVA‐SC6K sample when compared with the PVA‐C6K sample. Mean values from three independent biological replicates are given. The error bars indicate the standard deviation. *P < 0.05. (b) The presence of viral (+) and (−)RNA in the column eluates of PVA‐SC6K, PVA‐C6K and MC‐SC6K samples was analysed by reverse transcriptase‐polymerase chain reaction (RT‐PCR). RNA samples from 2×Strep‐tag purifications were incubated prior to PCR with (+) or without (−) reverse transcriptase in the presence of either a (−)‐strand‐ or (+)‐strand‐specific primer. (c) Replication protein VPg was detected by Western blotting with anti‐VPg antibody in PVA‐SC6K samples, but not in the control PVA‐C6K.
We compared negatively stained 6K2 membranes from PVA‐SC6K infection and from the controls under EM (Fig. 5). The analysis showed that 2×Strep‐tagged samples, from PVA‐SC6K infection and mcSC6K expression, contained abundant vesicles and vesicle clusters (Fig. 5, top and bottom panels), whereas only very few vesicles were present in the control PVA‐C6K sample (Fig. 5, middle panels). The sizes of the small vesicles purified in all of the samples varied between 40 and 90 nm, the mean being 56 nm (Fig. 5). EM images from all samples also showed elongated membranous structures. Virions were present in both virus samples with no notable difference in their quantity between PVA‐SC6K and PVA‐C6K materials. This suggests that virions interact non‐specifically with the Strep‐Tactin matrix to some extent (Fig. 5, right panels).
Figure 5.
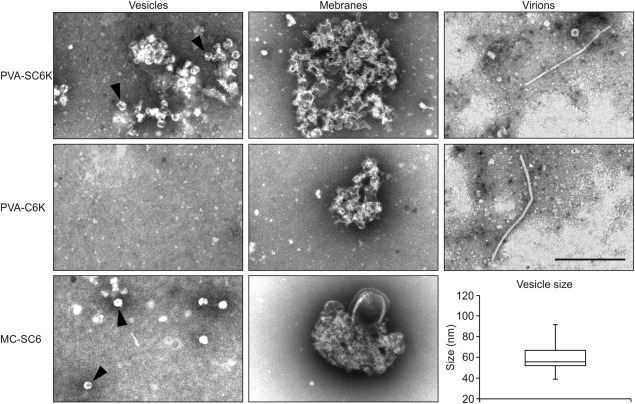
Morphological characterization of the purified 6K2‐associated membranes. Affinity‐purified PVA‐SC6K, PVA‐C6K and MC‐SC6K samples were negatively stained with uranyl acetate and examined by electron microscopy (EM). Two types of membrane structure were observed: individual vesicles (left panels; shown by arrowheads) and membrane clusters (middle panels). Strep‐Tactin matrix captured PVA particles non‐specifically from infected samples (right panels). The sizes of the purified individual vesicles vary between 50 and 100 nm, with the median size being 56 nm (n = 40). Scale bar, 500 nm.
Liquid chromatography‐tandem mass spectrometry (LC‐MS/MS) analysis of the affinity‐purified membranous 6K2‐induced structures
The affinity‐purified PVA‐SC6K samples were subjected to LC‐MS/MS analysis in three biological replicates, together with the corresponding PVA‐C6K and MC‐SC6K control samples. The MS analysis identified 729 proteins in the PVA‐SC6K samples, and 102 in PVA‐C6K and 49 in MC‐SC6K control samples. All unique proteins identified in these samples are given in Table S1 (see Supporting Information).
The number of peptide spectrum matches (PSMs) in the purified PVA‐SC6K vesicle sample was highest for the bait protein SC6K. Of the 40 identified SC6K peptides, seven corresponded to 6K2, four to 2×Strep and 29 to CFP. The second highest PSM scores were those of the viral proteins CI and HCpro with almost identical PSM scores of 137 and 136, respectively (see Table 1). Both CI and HCpro were also identified from the PVA‐C6K control sample, but had significantly lower PSM scores of 2 and 3, respectively. Therefore, it is clear that both CI and HCpro were purified in a tag‐specific manner and are abundantly present in SC6K‐induced vesicles during PVA infection. The viral proteins P3, 6K1, VPg, NIa‐pro and NIb were detected with high confidence and can be thought to associate genuinely with the 6K2‐induced vesicles during infection. The presence of CP is at least partially non‐specific. The CP PSM score in the PVA‐SC6K sample was 3.6 times higher than that in the PVA‐C6K control sample (18 vs. 5, respectively). Undoubtedly, virions contributed to the presence of CP in the MS data, as EM analysis revealed PVA particles in both PVA‐SC6K and PVA‐C6K samples (see Fig. 5). One viral P1 protein‐specific peptide was found in only one of the three biological replicates. Two N‐terminal peptides from P3 could be derived from P3N‐PIPO, but no peptides matching the PIPO part were identified.
Table 1.
Viral proteins identified by mass spectrometry analysis from PVA‐SC6K purification samples.
| Viral protein | Σ Coverage | Σ Unique peptides | Σ Peptides | Σ PSMs | PVA‐SC6K/PVA‐C6K* |
|---|---|---|---|---|---|
| SC6K2 | 79.49 | 27 | 40 | 613 | n.f. |
| CI | 54.02 | 27 | 27 | 137 | 68.5 |
| HCpro | 59.52 | 22 | 22 | 136 | 45.3 |
| P3 (P3N‐PIPO)† | 41.79 | 12 (2) | 12 | 66 | n.f. |
| VPg | 71.96 | 12 | 12 | 57 | n.f. |
| NIb | 43.99 | 14 | 14 | 48 | n.f. |
| NIa | 54.94 | 8 | 8 | 27 | n.f. |
| CP | 31.65 | 5 | 5 | 18 | 3.6 |
| 6K1 | 46.15 | 3 | 3 | 9 | n.f. |
| P1 | 7.72 | 1 | 1 | 1 | n.f. |
| PIPO‡ | 0.00 | 0 | 0 | 0 | n.f. |
*Enrichment of viral proteins in PVA‐SC6K over PVA‐C6K sample.
†Two peptides match to the N‐terminal part of P3 that can account for both P3 and P3N‐PIPO.
‡No peptides from the +2 frame of PIPO were identified.
n.f., not found in PVA‐C6K sample.
Next, the host proteins identified by LC‐MS/MS were sorted into a list presenting the host proteins which were identified with the highest confidence and fold of enrichment compared to the controls. The final list, presented in Table 2, contains proteins for which peptides were found in at least two biological replicates, amounting to at least four peptides, two of them unique. The threshold value for the fold of enrichment, calculated from PSM values, was set to at least 10 times higher than that in either of the controls. Altogether, 94 cellular proteins met these criteria.
Table 2.
Proteome of affinity‐purified PVA 6K2‐induced vesicles, location‐ and function‐based classification.
| Category | TAIR ID | N. benthamiana ID*(solgenomics.net) | Name† | Σ Unique peptides | Σ Peptides | Σ PSM | Σ Coverage |
|---|---|---|---|---|---|---|---|
| Chaperone | |||||||
| AT5G02500 | NbS00025223g0018.1 | HSC70‐1 | 6 | 18 | 95 | 42.47 | |
| AT5G02500 | NbS00016136g0003.1 | HSC70‐1 | 3 | 17 | 92 | 39.59 | |
| AT3G12580 | NbS00009983g0008.1 | HSP70 | 2 | 15 | 89 | 51.82 | |
| AT5G50920 | NbS00017400g0004.1 | ATP‐dependent Clp protease ATP‐binding subunit clpA homologue CD4B, chloroplastic | 2 | 26 | 55 | 38.15 | |
| AT5G28540 | NbS00040865g0006.1 | Luminal‐binding protein 5 | 6 | 8 | 26 | 14.81 | |
| AT5G56030 | NbS00025260g0001.1 | HSP81‐2 | 3 | 10 | 22 | 23.7 | |
| AT5G56030 | NbS00021897g0010.1 | HSP81‐2 | 4 | 11 | 21 | 18.99 | |
| AT4G24280 | NbS00052944g0006.1 | Chloroplast heat shock protein 70‐1 | 10 | 10 | 18 | 17.47 | |
| AT5G22060 | NbS00016695g0014.1 | DNAJ homologue 2 | 3 | 8 | 18 | 28.27 | |
| AT3G48870 | NbC25340200g0001.1 | ATP‐dependent Clp protease ATP‐binding subunit clpA homologue CD4B, chloroplastic | 2 | 7 | 17 | 64.43 | |
| AT5G22060 | NbS00004100g0004.1 | DNAJ homologue 2 | 2 | 7 | 15 | 22.15 | |
| AT3G13470 | NbS00034791g0001.1 | Chaperonin 60 subunit beta 2, chloroplastic | 4 | 4 | 8 | 9.05 | |
| AT4G24190 | NbS00013845g0026.1 | Heat shock protein 90 | 4 | 4 | 5 | 5.65 | |
| Chloroplast | |||||||
| AT2G39730 | NbS00047700g0013.1 | Ribulose bisphosphate carboxylase/oxygenase activase 2, chloroplastic | 2 | 15 | 62 | 43.53 | |
| AT1G06950 | NbS00009678g0004.1 | Protein TIC110, chloroplastic | 9 | 23 | 59 | 32.34 | |
| AT2G39730 | NbS00009714g0011.1 | Ribulose bisphosphate carboxylase/oxygenase activase 2, chloroplastic | 2 | 14 | 56 | 47.8 | |
| AT3G01500 | NbS00016159g0006.1 | Carbonic anhydrase, chloroplastic | 3 | 10 | 53 | 43.71 | |
| AT3G46740 | NbS00002677g0011.1 | Protein TOC75‐3 | 14 | 14 | 50 | 30.02 | |
| AT1G06950 | NbS00021174g0005.1 | Protein TIC110, chloroplastic | 3 | 17 | 38 | 23.96 | |
| AT4G02510 | NbS00024250g0002.1 | Translocase of chloroplast 159, chloroplastic | 13 | 13 | 30 | 16.47 | |
| AT3G47520 | NbS00008675g0002.1 | Malate dehydrogenase, chloroplastic | 5 | 8 | 18 | 29.25 | |
| AT5G38410 | NbS00022486g0001.1 | Ribulose bisphosphate carboxylase small chain 8B, chloroplastic | 5 | 6 | 18 | 44.75 | |
| AT2G20260 | NbS00003075g0011.1 | Photosystem I reaction centre subunit IV B, chloroplastic | 2 | 6 | 18 | 53.15 | |
| AT2G24820 | NbS00029739g0004.1 | Translocon at the inner envelope membrane of chloroplasts 55‐II | 2 | 5 | 16 | 18.2 | |
| AT3G56940 | NbS00020307g0015.1 | Magnesium‐protoporphyrin IX monomethyl ester [oxidative] cyclase, chloroplastic protein, putative (Crd1) | 7 | 7 | 15 | 19.55 | |
| AT5G16620 | NbS00017610g0108.1 | Translocon at the inner envelope membrane of chloroplasts 40 | 3 | 5 | 15 | 15.41 | |
| AT1G61520 | NbS00021892g0003.1 | Chlorophyll a/b‐binding protein 8 | 3 | 4 | 13 | 30.56 | |
| AT1G74470 | NbS00056940g0001.1 | Geranylgeranyl diphosphate reductase, chloroplastic | 5 | 5 | 12 | 14.22 | |
| AT5G05000 | NbS00005332g0008.1 | Translocon at the outer envelope membrane of chloroplasts 34 | 4 | 4 | 12 | 25.58 | |
| AT4G23430 | NbS00010186g0028.1 | Short‐chain dehydrogenase TIC 32 | 4 | 4 | 11 | 25.08 | |
| AT2G16640 | NbS00010608g0012.1 | Translocase of chloroplast 132, chloroplastic | 3 | 7 | 10 | 5.77 | |
| AT2G28000 | NbS00033391g0006.1 | RuBisCO large subunit‐binding protein subunit alpha, chloroplastic | 4 | 4 | 10 | 11.29 | |
| AT2G16640 | NbS00004829g0016.1 | Translocase of chloroplast 132, chloroplastic | 2 | 6 | 9 | 5.17 | |
| AT2G38040 | NbS00026142g0005.1 | Acetyl‐coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic | 4 | 5 | 8 | 12.61 | |
| AT1G77590 | NbS00027106g0001.1 | LACS9, long‐chain acyl‐CoA synthetase 9, chloroplastic | 4 | 4 | 8 | 9.95 | |
| AT5G16620 | NbS00033277g0017.1 | Protein TIC 40, chloroplastic | 2 | 4 | 8 | 20.29 | |
| AT2G05100 | NbS00019403g0016.1 | Chlorophyll a/b‐binding protein 37, chloroplastic | 3 | 4 | 7 | 22.26 | |
| Cytoplasm | |||||||
| AT1G62020 | NbS00031319g0001.1 | Coatomer subunit alpha‐1 | 2 | 4 | 8 | 4.75 | |
| Cytoskeleton | |||||||
| AT1G75780 | NbS00056603g0002.1 | Tubulin beta‐1 chain | 2 | 12 | 49 | 39.39 | |
| AT5G23860 | NbS00001148g0004.1 | Tubulin beta‐8 chain | 1 | 9 | 44 | 30.72 | |
| AT4G14960 | NbS00001594g0015.1 | Tubulin alpha‐6 chain | 9 | 9 | 27 | 32.55 | |
| AT5G12250 | NbS00023211g0007.1 | Tubulin beta‐6 chain | 2 | 8 | 19 | 30.15 | |
| Metabolic enzyme | |||||||
| AT1G12900 | NbS00001169g0153.1 | Glyceraldehyde 3‐phosphate dehydrogenase A subunit 2 | 10 | 12 | 59 | 38.42 | |
| AT4G35000 | NbS00062043g0007.1 | l‐Ascorbate peroxidase 3, peroxisomal | 4 | 10 | 31 | 37.09 | |
| AT3G27820 | NbS00023324g0005.1 | Probable monodehydroascorbate reductase, cytoplasmic isoform | 10 | 10 | 30 | 23.81 | |
| AT4G35000 | NbS00035202g0004.1 | l‐Ascorbate peroxidase 3, peroxisomal | 2 | 7 | 26 | 37.77 | |
| AT3G16950 | NbS00014159g0003.1 | Dihydrolipoyl dehydrogenase | 4 | 8 | 21 | 19.07 | |
| AT2G34590 | NbS00007894g0004.1 | Pyruvate dehydrogenase E1 component subunit beta | 6 | 6 | 17 | 22.74 | |
| AT1G01090 | NbS00017334g0012.1 | Pyruvate dehydrogenase E1 component subunit alpha | 4 | 6 | 15 | 20.29 | |
| AT1G01090 | NbS00034265g0006.1 | Pyruvate dehydrogenase E1 component subunit alpha | 4 | 6 | 12 | 17.45 | |
| AT2G22780 | NbS00002298g0008.1 | Malate dehydrogenase, glyoxysomal | 5 | 5 | 11 | 16.87 | |
| AT3G14420 | NbS00005125g0015.1 | Peroxisomal (S)−2‐hydroxy‐acid oxidase | 6 | 6 | 8 | 24.44 | |
| AT4G35090 | NbS00006116g0019.1 | Catalase isozyme 1 | 5 | 5 | 8 | 17.97 | |
| AT1G44170 | NbS00047628g0009.1 | Aldehyde dehydrogenase | 4 | 4 | 7 | 10.48 | |
| Nucleotide binding | |||||||
| AT4G17170 | NbS00004361g0010.1 | Ras‐related protein RABB1c | 6 | 6 | 19 | 37.44 | |
| AT5G27540 | NbS00036282g0010.1 | MIRO‐related GTP‐ase 1 | 4 | 11 | 17 | 28.59 | |
| AT2G27600 | NbS00006021g0010.1 | AAA‐type ATPase family protein | | 8 | 8 | 17 | 31.34 | |
| AT1G78900 | NbS00033958g0004.1 | V‐type proton ATPase catalytic subunit A | 6 | 6 | 13 | 14.36 | |
| Ribosome/protein biosynthesis | |||||||
| AT4G10450 | NbS00002134g0112.1 | Ribosomal protein L6 family | 2 | 11 | 34 | 61.47 | |
| AT2G27710 | NbS00029619g0005.1 | 60S acidic ribosomal protein P2 | 3 | 5 | 13 | 42.95 | |
| AT2G19730 | NbS00054743g0008.1 | Ribosomal L28e protein family | 5 | 6 | 12 | 27.68 | |
| AT4G09800 | NbS00056355g0003.1 | S18 ribosomal protein | 2 | 4 | 12 | 32.24 | |
| AT1G61580 | NbS00018918g0016.1 | R‐protein L3 B | 3 | 7 | 11 | 18.77 | |
| AT3G13920 | NbS00044851g0011.1 | Eukaryotic translation initiation factor 4A1 | 5 | 5 | 10 | 15.98 | |
| AT1G48830 | NbS00039588g0004.1 | Ribosomal protein S7e family protein | 4 | 4 | 10 | 30.96 | |
| AT1G61580 | NbS00045996g0004.1 | 60S ribosomal protein L3 | 3 | 7 | 9 | 18.77 | |
| AT3G49910 | NbS00036380g0001.1 | 60S ribosomal protein L26‐1 | 2 | 5 | 9 | 32.19 | |
| AT1G57720 | NbS00006811g0211.1 | Probable elongation factor 1‐gamma 2 | 4 | 4 | 9 | 9.38 | |
| AT3G11250 | NbS00018504g0008.1 | 60S acidic ribosomal protein P0 | 5 | 5 | 8 | 30.87 | |
| Other | |||||||
| AT1G04690 | NbS00043430g0001.1 | Probable voltage‐gated potassium channel subunit beta; KAB1, KV‐BETA1 | 13 | 13 | 71 | 64.75 | |
| AT1G34430 | NbC26077604g0003.1 | Dihydrolipoyllysine‐residue acetyltransferase component of pyruvate dehydrogenase complex | 2 | 8 | 26 | 30.49 | |
| AT5G08540 | NbS00000164g0018.1 | Unknown protein | 9 | 9 | 21 | 34.58 | |
| AT3G13930 | NbS00000479g0101.1 | Dihydrolipoyllysine‐residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial | 8 | 8 | 19 | 19.78 | |
| AT1G11910 | NbS00028367g0004.1 | Aspartic proteinase A1 | 2 | 6 | 19 | 17.98 | |
| AT5G24650 | NbS00016191g0009.1 | Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein | 6 | 6 | 18 | 26.92 | |
| AT1G11910 | NbS00031482g0006.1 | Aspartic proteinase A1 | 2 | 6 | 17 | 17.39 | |
| AT3G01280 | NbS00027615g0006.1 | Voltage‐dependent anion channel 1 | 2 | 4 | 17 | 23.91 | |
| AT5G12470 | NbS00005727g0008.1 | Unknown | 6 | 7 | 15 | 26.77 | |
| AT1G17860 | NbC24872723g0001.1 | Kunitz family trypsin and protease inhibitor protein | 5 | 5 | 15 | 35.85 | |
| AT1G05270 | NbS00011400g0014.1 | TraB family protein | 3 | 7 | 14 | 25.92 | |
| AT2G38040 | NbS00048878g0013.1 | CAC3, acetyl Co‐enzyme A carboxylase carboxyltransferase alpha subunit | 9 | 10 | 13 | 16.33 | |
| AT5G22640 | NbS00044837g0004.1 | emb1211, MORN (Membrane Occupation and Recognition Nexus) repeat‐containing protein | 7 | 7 | 13 | 14.47 | |
| AT1G33970 | NbS00011570g0012.1 | P‐loop‐containing nucleoside triphosphate hydrolases superfamily protein | 5 | 5 | 13 | 13.86 | |
| AT2G28900 | NbS00045823g0014.1 | Outer plastid envelope protein 16‐1 | 4 | 4 | 13 | 26.75 | |
| AT5G62810 | NbS00031622g0006.1 | Peroxisomal membrane protein PEX14 | 2 | 5 | 11 | 36.63 | |
| AT1G10510 | NbS00019582g0003.1 | emb2004, RNI‐like superfamily protein | 6 | 6 | 10 | 19.64 | |
| AT2G20580 | NbS00013228g0016.1 | 26S proteasome regulatory subunit S2 1A | 2 | 5 | 8 | 9.7 | |
| AT1G11910 | NbS00050653g0003.1 | Aspartic proteinase A1 | 4 | 4 | 8 | 11.42 | |
| AT2G17840 | NbS00004850g0017.1 | ERD7, senescence/dehydration‐associated protein‐related | 5 | 5 | 7 | 16.82 | |
| AT3G57090 | NbS00059906g0004.1 | BIGYIN, FIS1A; tetratricopeptide repeat (TPR)‐like superfamily protein | 4 | 4 | 7 | 27.97 | |
| AT5G62810 | NbS00005874g0019.1 | Peroxisomal membrane protein PEX14 | 3 | 4 | 7 | 18.94 | |
| AT2G44640 | NbS00028734g0022.1 | Uncharacterized protein | 4 | 4 | 6 | 13.8 | |
| AT4G36720 | NbS00015416g0013.1 | HVA22‐like protein K | 4 | 4 | 5 | 30.48 | |
| AT2G32730 | NbS00029577g0007.1 | 26S proteasome regulatory complex, non‐ATPase subcomplex, Rpn2/Psmd1 subunit | 4 | 4 | 5 | 8.39 | |
*Derived from the Nicotiana benthamiana annotated transcriptome list v0.4.4 (solgenomics.net).
†Derived from the N. benthamiana annotated transcriptome list v0.4.4 (solgenomics.net) or from the closest WU‐BLAST match in the TAIR web‐page.
The list of host proteins identified in VRCs was sorted on the basis of both their cellular localization and molecular function (see Table 2). According to the functional annotation program DAVID (Huang et al., 2009a, 2009b), less than 1% of the host proteins were ER‐associated and approximately 14% were chloroplast‐associated in our proteomic data. DAVID often places the same protein into several categories, e.g. based on both its cellular location and molecular function, which hampers the calculations. When all the proteins having a localization or molecular function associated with chloroplasts were combined manually, the amount of these proteins among all of the identified proteins was increased to 25%. Among the most abundant ER proteins in the sample were Luminal binding protein 5 (BiP5) and calreticulin (Table S1). However, calreticulin, which is an HCpro binding partner (Shen et al., 2010), did not meet all the criteria to enter the final list presented in Table 2. Of the chloroplast‐associated proteins, chloroplastic glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), carbonic anhydrase, a peripheral protein of the translocon at the chloroplast inner envelope, TIC110, and the outer envelope translocation channel, TOC75‐3, were identified with high confidence (Table 2). Although the relevance of some identified proteins, such as HSP70, HSP90, HSP40, eEF1A and GAPDH, is already known in (+)RNA virus multiplication (Castorena et al., 2007; Davis et al., 2007; Huang and Nagy, 2011; Li et al., 2010; Okamoto et al., 2006; Pogany et al., 2008; Tomita et al., 2003; Wang and Nagy, 2008; Wang et al., 2009; Weeks et al., 2010; Yi et al., 2011), for most of the proteins, the exact molecular function needs to be worked out in order to understand potyviral replication.
It has been suggested that potyviral VRCs contain ribosomes translating PVA RNA (Grangeon et al., 2010). To compare the content of ribosomal proteins, we looked at each proteome individually with more relaxed parameters. Ribosomal proteins for which peptides were found in at least two biological replicates amounting to at least three peptides, one of them unique, were selected. LC‐MS/MS data showed that 16 of the 32 small and 38 of the 47 large ribosomal subunit proteins fulfilled these criteria in the PVA‐SC6K samples (Table 3). Importantly, the MC‐SC6K control membranes pulled down none of the small and only one of the 47 large ribosomal subunit proteins (L7a‐1). The PVA‐C6K control also contained ribosomes as many ribosomal proteins were identified (17/47, 3/32), but most were less abundantly present than in the PVA‐SC6K samples. These data suggest that a small portion of VRCs also bind non‐specifically to the Strep‐Tactin matrix. These data indicate that the ribosomal proteins were very specifically associated with membranes active in virus replication.
Table 3.
Ribosomal proteins found in the PVA‐SC6K sample.
| TAIR ID | Name | Σ PSM | Σ Coverage | PVA‐SC6K/PVA‐C6* | PVA‐SC6K/MC‐SC6K† |
|---|---|---|---|---|---|
| AT1G58684 | 40S ribosomal protein S2‐2 | 5 | 15.36 | 1.25 | |
| AT2G31610 | 40S ribosomal protein S3‐1 | 8 | 23.21 | ||
| AT4G34670 | 40S ribosomal protein S3a‐2 | 6 | 17.77 | 2.00 | |
| AT3G11940 | 40S ribosomal protein S5‐2 | 10 | 22.97 | ||
| AT4G31700 | 40S ribosomal protein S6‐1 | 3 | 12.5 | ||
| AT1G48830 | 40S ribosomal protein S7‐1 | 10 | 30.96 | ||
| AT5G59240 | 40S ribosomal protein S8‐2 | 10 | 19.13 | ||
| AT4G00100 | 40S ribosomal protein S13‐2 | 11 | 26.32 | ||
| AT3G11510 | 40S ribosomal protein S14‐2 | 12 | 38.67 | 2.40 | |
| AT2G09990 | 40S ribosomal protein S16‐1 | 4 | 17.01 | ||
| AT5G04800 | 40S ribosomal protein S17‐4 | 7 | 16.67 | ||
| AT4G09800 | 40S ribosomal protein S18 | 10 | 26.32 | ||
| AT3G09680 | 40S ribosomal protein S23‐1 | 4 | 20.18 | ||
| AT4G39200 | 40S ribosomal protein S25‐4 | 10 | 33.02 | ||
| AT2G40590 | 40S ribosomal protein S26‐1 | 4 | 25 | ||
| AT5G56670 | 40S ribosomal protein S30 | 3 | 16.13 | ||
| AT3G11250 | 60S acidic ribosomal protein P0‐3 | 8 | 30.87 | 2.67 | |
| AT5G47700 | 60S acidic ribosomal protein P1‐3 | 5 | 18.18 | ||
| AT2G27710 | 60S acidic ribosomal protein P2‐2 | 7 | 30.09 | 1.40 | |
| AT1G61580 | 60S ribosomal protein L3‐2 | 9 | 18.77 | ||
| AT3G09630 | 60S ribosomal protein L4‐1 | 17 | 20.51 | 1.70 | |
| AT1G74060 | 60S ribosomal protein L6‐2 | 12 | 28.63 | ||
| AT1G74050 | 60S ribosomal protein L6‐3 | 5 | 12.31 | ||
| AT2G01250 | 60S ribosomal protein L7‐2 | 12 | 29.17 | ||
| AT3G13580 | 60S ribosomal protein L7‐4 | 12 | 26.03 | ||
| AT2G47610 | 60S ribosomal protein L7a‐1 | 7 | 22.09 | 1.75 | |
| AT3G62870 | 60S ribosomal protein L7a‐2 | 7 | 22.09 | ||
| AT4G36130 | 60S ribosomal protein L8‐3 | 13 | 28.85 | 2.60 | |
| AT1G33140 | 60S ribosomal protein L9‐1 | 28 | 55.15 | ||
| AT4G10450 | 60S ribosomal protein L9‐2 | 34 | 61.47 | 11.33 | |
| AT1G26910 | 60S ribosomal protein L10‐2 | 9 | 17.81 | ||
| AT1G08360 | 60S ribosomal protein L10a‐1 | 11 | 20.83 | 1.22 | |
| AT5G45775 | 60S ribosomal protein L11‐2 | 4 | 8.81 | ||
| AT2G37190 | 60S ribosomal protein L12‐1 | 11 | 36.63 | 2.75 | |
| AT3G49010 | 60S ribosomal protein L13‐1 | 3 | 26.67 | 1.13 | |
| AT3G07110 | 60S ribosomal protein L13a‐1 | 3 | 15.48 | 0.75 | |
| AT2G20450 | 60S ribosomal protein L14‐1 | 4 | 15.69 | ||
| AT4G16720 | 60S ribosomal protein L15‐1 | 8 | 19.61 | 2.67 | |
| AT1G27400 | 60S ribosomal protein L17‐1 | 3 | 14.6 | ||
| AT3G05590 | 60S ribosomal protein L18‐2 | 6 | 21.61 | 1.13 | |
| AT3G16780 | 60S ribosomal protein L19‐2 | 5 | 19.07 | 1.67 | |
| AT1G09690 | 60S ribosomal protein L21‐1 | 6 | 32.32 | ||
| AT3G05560 | 60S ribosomal protein L22‐2 | 6 | 32.26 | ||
| AT3G04400 | 60S ribosomal protein L23 | 11 | 46.43 | ||
| AT3G55280 | 60S ribosomal protein L23a‐2 | 10 | 37.14 | 3.33 | |
| AT2G36620 | 60S ribosomal protein L24‐1 | 3 | 16.44 | 0.90 | |
| AT3G49910 | 60S ribosomal protein L26‐1 | 9 | 32.19 | ||
| AT4G15000 | 60S ribosomal protein L27‐3 | 4 | 19.23 | ||
| AT2G19730 | 60S ribosomal protein L28‐1 | 12 | 27.68 | ||
| AT4G18100 | 60S ribosomal protein L32‐1 | 5 | 50.62 | ||
| AT1G26880 | 60S ribosomal protein L34‐1 | 5 | 27.5 | 2.50 | |
| AT1G74270 | 60S ribosomal protein L35a‐3 | 4 | 28.57 | ||
| AT4G14320 | 60S ribosomal protein L36a | 5 | 20.88 | ||
| AT3G10950 | Putative60S ribosomal protein L37a‐1 | 5 | 27.17 | ||
| AT3G52590 | NEDD8‐like protein RUB1;60S ribosomal protein L40;40S ribosomal protein S27a‐1;40S ribosomal protein S27a‐3; Ubiquitin | 3 | 14.93 |
*Enrichment of viral proteins in PVA‐SC6K over PVA‐C6K sample.
†Enrichment of viral proteins in PVA‐SC6K over MC‐SC6K sample.
Discussion
The focus of this work was to reveal the protein composition of the potyviral VRCs. To enable the determination of the full viral and host protein content of the 6K2‐induced membranous vesicles, we developed a purification strategy based on an affinity tag fused to 6K2 and exposed on the VRCs during PVA infection. Interesting questions relate to the orientation of 6K2 in the ER membranes. According to Lerich et al. (2011), Tobacco etch virus (TEV) 6K2 consists of an N‐terminal cytoplasmic domain of 23 amino acids, a transmembrane (TM) domain of 19 amino acids and a C‐terminal putatively luminal domain of 11 amino acids. Our data from SC6K carrying the N‐terminal 2×Strep‐CFP fusion supports this orientation (see Fig. 3). Achieving this orientation would require the insertion of SC6K with its C‐terminus first into the ER lipid bilayer. The TM domain of potyviral 6K2 protein is located near the C‐terminus, which is typical for the tail‐anchored (TA) ER membrane proteins (reviewed in Johnson et al., 2013). These proteins have a single C‐terminal TM domain required to target and anchor them to the ER membrane. TA proteins form a topological group of membrane proteins and, like potyviral 6K2 protein, many are involved in vesicular transport (Ungar and Hughson, 2003). The N‐terminus of TEV 6K2 contains a D(X)E motif which is essential for ER exit of the 6K2‐induced vesicles (Aniento et al., 2006; Hanton et al., 2005; Lerich et al., 2011). However, the orientation question becomes more complicated when 6K2 is expressed from its natural context in the polyprotein. It is likely that the 6K2‐VPg‐Pro precursor serves as a scaffolding protein leading to the formation of the vesicle structures capable of supporting viral replication (Beauchemin et al., 2007; Schaad et al., 1997; Thivierge et al., 2008). In this context, the 6K2 TM domain would not be C‐terminal any longer. Therefore, the question of 6K2‐VPg‐Pro orientation, which is probably a key factor determining the site of replication either inside the vesicles or on their surface, remains open for further studies.
In our analysis, we aimed to identify proteins from vesicles that contained PVA replication complexes. Despite many attempts, we could not demonstrate RNA synthesis activity within the purified PVA‐SC6K vesicles. Nevertheless, an approximately nine‐fold enrichment of PVA (+)‐strand RNA was observed in PVA‐SC6K compared with the PVA‐C6K eluates. This demonstrates that a major part of PVA RNA detected in PVA‐SC6K was there as a result of tag‐specific purification. As demonstrated by EM analysis of the eluates (see Fig. 5), some PVA particles were purified as a result of non‐specific binding of particles to the Strep‐Tactin matrix, which probably explains the presence of PVA RNA in the PVA‐C6K eluate. The statistically significant difference in the fold of PVA RNA enrichment, together with the presence of PVA (–)‐strand RNA in PVA‐SC6K eluates, suggests that the PVA‐SC6K sample probably contains membranes active in replication.
All PVA proteins, except the PIPO part of P3N‐PIPO, were detected in PVA‐SC6K eluates. Therefore, the two peptides from the P3N region, which are common to both P3 and P3N‐PIPO, are derived from P3 protein with high probability. As a result of the very small amount of P1 peptides in LC‐MS/MS data, we do not consider P1 to be specifically present. Because PVA particles were co‐purified, CP was present partially non‐specifically. CP was, however, enriched 3.6‐fold compared with the PVA‐C6K control, and therefore it is a borderline case whether it should be regarded as present in VRCs. P1, CP and P3N‐PIPO are the three proteins that are not needed for potyviral replication (Mahajan et al., 1996; Verchot and Carrington, 1995; Wen and Hajimorad, 2010), which is supported by our findings. Peptides derived from HCpro and CI were abundantly found in the PVA‐SC6K eluates and were enriched 68‐ and 45‐fold, respectively, in comparison with the PVA‐C6K control. The presence of CI and HCpro in the PVA‐C6K control may even result partially from the virions being non‐specifically co‐purified, as both CI and HCpro have been located at one of the extremities of PVA particles (Gabrenaite‐Verkhovskaya et al., 2008; Torrance et al., 2006). The presence of NIb, VPg and NIa‐Pro is a prerequisite to state that VRCs have been purified. All these were found unquestionably from the PVA‐SC6K eluates. As P3 of TEV has previously been shown to form punctate inclusions that co‐localize with the 6K2 vesicles (Cui et al., 2010), the specific detection of P3 supports our hypothesis. Taken together, HCpro, P3, 6K1, CI, 6K2, VPg‐NIpro and NIb are present in the purified 6K2‐induced vesicles during PVA infection. However, further studies are required to investigate their relative abundances within the VRC.
It is proposed in the model for potyviral TuMV replication vesicle biogenesis, presented by Grangeon et al. (2010), that viral translation and replication are tightly coupled within the virus‐induced vesicles. This suggestion is based on the finding that vesicles derived from a single viral genome contain the proteins synthesized from that same vRNA (Cotton et al., 2009). This model states that ribosomes associated with viral factors and RNA on the ER membranes become trapped within the replication vesicles, where viral protein synthesis then continues. Our LC‐MS/MS data revealed the presence of 38 60S and 16 40S ribosomal proteins in the PVA‐SC6K eluates. By application of the same conditions to the MC‐SC6K control proteome list, there was only one ribosomal protein present, namely the 60S ribosomal protein L7a‐1. Therefore, it can be suggested that ribosomes associate with VRCs in infected cells, but not with 6K2‐induced vesicles in the absence of infection. However, whether the ribosomes are internalized or exist on the outer surface of VRCs cannot be concluded from our data. Nevertheless, tight association between replication and translation, as suggested in Cotton et al. (2009) and Hafrén et al. (2010), is supported in the light of these data. Some of the ribosomal proteins found from the proteomic data may also have more specific functions in potyviral multiplication. Yang et al. (2009) have shown that the replication of TuMV is inhibited in plants in which RPL19, RPL13, RPL7, RPS2 and RPS6 are silenced. In addition, acidic ribosomal protein P0 was present in the VRC proteome. In our earlier work, we have identified P0 from the PVA RNP complex associated with replication membranes, and have shown it to be essential for PVA RNA and virion accumulation in infection (Hafrén et al., 2013).
Despite the fact that the PVA‐SC6K‐induced vesicles were not found to associate with chloroplasts in the same way as the PVA‐6KY vesicles when imaged by confocal microscopy, 25% of the host proteins in the final proteome were categorized as chloroplast‐associated proteins. In the light of the proteomic data, it is therefore conceivable that PVA replication within the SC6K‐containing vesicles occurs in association with chloroplasts, and there is no contradiction with the previous literature stating that disruption of the VRC–chloroplast fusion is detrimental for potyviral replication (Wei et al., 2013). Several host factors that are linked with potyviral replication and localize to potyviral VRCs (reviewed in Revers and García, 2015) were identified. From these host factors, HSP70 was the host protein most abundantly found in the samples. Moreover, eEF1A and initiation factor 4A (IF4A) were both found from the VRC proteome, confirming the previous findings. Many other cellular proteins with a confirmed role in (+)RNA virus replication were identified. GAPDH, which is essential for the determination of the (+)/(–)RNA synthesis ratio during tombusvirus infection (Huang and Nagy, 2011), is an example of such a protein. In summary, we conclude that the PVA‐SC6K sample consists mostly of purified VRCs, and the associated host proteins may have relevance in PVA replication. The next essential step will be to screen for those host proteins which have a role in PVA multiplication, and to further analyse the detailed molecular function of each host protein.
Experimental Procedures
Plants, growth conditions, agroinfiltration
Nicotiana benthamiana plants were kept under glasshouse conditions with 22 °C daytime and 18 °C night‐time temperatures. Plants were infiltrated with Agrobacterium tumefaciens in infiltration buffer [10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), 10 mm MgCl2, 150 μm acetosyringone] at the four‐ to six‐leaf stage as described in Eskelin et al. (2010). Systemically infected plant leaves were harvested at 10 DPI; locally expressed constructs were harvested 4 days after agroinfiltration.
Plasmid construction
Viral and protein expression constructs used in this study were based on the full‐length icDNA copies of PVA strain B11 (GenBank accession number AJ296311).
To prepare PVA‐C6K, the CFP‐6K2 fusion sequence was amplified from the pSITEII‐2C1 vector using primer XbaKpnCer, which adds XbaI and KpnI restriction sites in front of the cerulean sequence, and primer 6KR_Mlu, which adds the MluI restriction site at the 3′‐end of the 6K2 sequence (Table S2, see Supporting Information). The PCR product was cloned to the pGEM‐T Easy vector, producing the pGEM‐T Easy::Cerulean‐6K2 vector. SacII and MluI sites were used to transfer CFP‐6K2 from pGEM‐T Easy to pUC18::PVAWT between the NIb and CP sequences, resulting in the pUC18::PVA‐Cerulean‐6K2 (pUC18::PVA‐C6) vector. PVA‐Cerulean‐6K2 was cloned to the pRD400::PVAWT agro vector using SalI and AgeI restriction sites, resulting in the pRD400::PVA‐CFP‐6K2 (PVA‐C6K) vector.
To prepare PVA‐SC6K, the Twin‐Strep‐tag II (2×Strep) sequence was amplified with PCR from pGEM‐T Easy::2×Strep using SP6 and T7 primers. The PCR fragment was cut with XbaI and KpnI and inserted into the same sites in pGEM‐T Easy::CFP‐6K2, resulting in the pGEM‐T Easy::2×Strep‐CFP‐6K2 vector. SacII and MluI sites were used to transfer 2×Strep‐CFP‐6K2 from pGEM‐T Easy to pUC18::PVAWT between the NIb and CP sequences, resulting in the pUC18::PVA‐2×Strep‐CFP‐6K2 (pUC18::PVA‐SC6) vector. PVA‐2×Strep‐CFP‐6K2 was cloned into the pRD400::PVAWT agro vector using SalI and AgeI restriction sites, resulting in the pRD400::PVA‐2×Strep‐CFP‐6K2 (PVA‐SC6K) vector.
To prepare PVA‐6KY, the potyviral 6K2 protein was PCR amplified from the PVA genome and inserted into pGWB41. The 6K2‐YFP fusion was then amplified from the pGWB41‐6K2 plasmid with primers Afl_6K and YFP_R (Table S2), which introduced AflII and MluI restriction sites in front and at the end of 6K2‐YFP. These restriction sites were used to clone the PCR fragment to the icDNA of PVA in the pUC18 vector. From there, the complete PVA icDNA with the CaMV 35S promoter and nos terminator was cloned to the pRD400 binary vector using KpnI and SalI restriction sites, yielding the PVA‐6KY construct. NIa protease cleavage sites flanked all insertions in between NIb and CP genes.
To prepare MC‐SC6K, the 2×Strep‐CFP‐6K2 sequence was amplified from the pGEM‐T Easy::2×Strep‐CFP‐6K2 vector using primers 2×Strep_ATG_Xho_Fw and 6K2_stop_Bam_Rev (Table S2), resulting in a fragment that contained the XhoI restriction site and ATG translation initiation codon in front of the 2×Strep sequence and a double TAA translation termination codon and BamHI restriction site at the 3′‐end of 6K2. This fragment was cloned into the pANU vector that contained a 35S promoter and nos transcription terminator, using XhoI and BamHI sites. The resulting 35S‐2×Strep‐CFP‐6K2‐term cassette was inserted into the pRD400 agro‐vector using the HindIII site. The correctness of all constructs was confirmed by sequencing.
Sample preparation
For LC‐MS/MS analysis, the vesicles were prepared as follows: 10 g of N. benthamiana leaves were homogenized in sampling buffer [13% sucrose, 50 mm Tris‐HCl, 10 mm KCl, 3 mm MgCl2, 1 mm ethylenediaminetetraacetic acid (EDTA), 1 mm dithiothreitol (DTT), 0.3% dextran, 0.1% bovine serum albumin (BSA), pH 8] in the ratio of 1 g of leaves to 3 mL of buffer, yielding 30 mL of sample. Initial centrifugation of the homogenized material was carried out at 3000 g for 10 min at 4 °C. The resulting supernatant, 13 mL per centrifugation tube, was loaded on top of a discontinuous sucrose gradient (from the bottom: 8 mL 45%; 9 mL 30%; 9 mL 20%). Ultracentrifugation of samples on a sucrose gradient was carried out in a Beckman‐Coulter (Brea, California, USA) SW‐28 rotor at 83 000 g for 5 h at 4 °C. The fraction enriched in the CFP signal (fraction 5) was used in the affinity purification step. Fraction 5 from two ultracentrifugation tubes, 5 mL each, was combined. All samples were performed in three biological replicates.
Affinity purification
Avidin (100 µg/mL) was added to the fraction of interest from ultracentrifugation and the sample was incubated on ice for 10 min. Affinity chromatography was carried out at room temperature by letting the sample flow through 0.5 mL (initially 1 mL of 50% slurry) of Strep‐Tactin® Sepharose (IBA) resin by gravity flow. The column was washed with 5 mL (10 × column bed volume) of ice‐cold washing buffer (50 mm Tris‐HCl, 10 mm KCl, 3 mm MgCl2, pH 8). Samples were eluted with 2 mL of ice‐cold washing buffer containing 1 mm biotin (Thermo Scientific, Waltham, MA, USA). Samples were concentrated using Amicon® Ultra‐4 Centrifugal Filter Units (Merck Millipore, Darmstadt, Germany) with a 10‐kDa cut‐off limit.
Electron microscopy
Thin sections were prepared as follows. An infected leaf sample was taken under 0.1 m phosphate buffer (pH 7.4). Leaf discs were degassed in 0.1 m phosphate buffer containing 2.5% glutaraldehyde and left for fixation at room temperature for 2 h. After washing with 0.1 m phosphate buffer, the samples were osmicated in the same buffer containing 1% OsO4 for 1 h at room temperature. The samples were then dehydrated with ethanol and acetone, and embedded in Epon 812 (Taab Laboratories, Reading, Berkshire, UK); 90‐nm sections were post‐stained with 2% uranyl acetate and Reynold's lead citrate. For negative staining of purified vesicles, the sample was prepared as for LC‐MS/MS, with the exception that elution was carried out with washing buffer containing 2.5 mm desthiobiotin. Purified vesicle samples were viewed on a Jeol (JEM‐1400, Tokyo, Japan) 1400 transmission electron microscope; 1% phosphotungstic acid was used as the negative staining agent for purified 6K2 membrane samples.
Confocal microscopy
Confocal laser scanning microscopy was performed on a Leica (Wetzlar, Germany) TCS SP5II confocal microscope. Systemically infected N. benthamiana leaves were selected for confocal microscopy analysis at 10 DPI for PVA icDNA constructs and locally infiltrated N. benthamiana leaves were selected for confocal microscopy analysis at 4 DPI for the MC‐SC6K construct. Small discs were cut from the N. benthamiana leaves, mounted between cover and objective glass, immersed in water and viewed with a 20× objective. YFP excitation was carried out using an argon laser at 514 nm with emission recorded at 525–555 nm (DD 458/514 beam splitter). CFP excitation was carried out using an argon laser at 458 nm with emission recorded at 470–500 nm. Images represent 5‐µm Z‐stacks taken at 1‐µm intervals. All images were deconvoluted using Autoquant X3 software and the presented images were obtained using Imaris software.
RT‐PCR
Total RNA was extracted from 200 µL of sample using Trizol (Thermo Scientific, Waltham, MA, USA) and taken up in 20 µL of nuclease‐free water. The purified RNA was DNase treated for 10 min at room temperature using 1 µL of DNaseI (1 U/µL) and RDD buffer (Qiagen, Venlo, Netherlands). cDNA was synthesized from 4 µL of RNA using Superscript III reverse transcriptase (Life Technologies, Waltham, MA, USA). Primer RT_CPminus_F was used to detect PVA genomic RNA of negative polarity and primer RT_CPplus_R was used to detect PVA genomic RNA of positive polarity (Table S2). Phusion polymerase (Finnzymes, Thermo Fisher Scientific; Waltham, MA, USA), with the same RT primers, was used to amplify the coding region of the PVA CP gene.
qRT‐PCR
RNA was extracted using Trizol (Thermo Scientific) from 100 µL of input and affinity‐purified sample in three biological replicates for each construct and taken up in 20 µL of nuclease‐free water. The purified RNA was DNase treated for 10 min at room temperature using 1 µL of DNaseI (1 U/µL) and RDD buffer (Qiagen). cDNA was synthesized from 5 µL of purified RNA using a RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) and random hexamers. Quantitative real‐time PCR was performed using a Maxima SYBR Green qPCR kit (Thermo Scientific) and primers specific to the PVA CP sequence, namely qPCR_CP_F and qPCR_CP_R (Table S2).
Protein identification by LC‐MS/MS and proteome analysis
Disulfide bridges in proteins were reduced with 50 mm TCEP [tris(2‐carboxyethol)phosphine hydrochloride salt, Sigma‐Aldrich, St. Louis, Missouri, United States] for 20 min at 37 °C. To block cysteine residues, iodoacetamide (Fluka, St. Louis, Missouri, United States (owned by Sigma‐Aldrich)) was added to a final concentration of 150 mm and the samples were incubated at room temperature in the dark for 30 min. A total of 0.75 μg of trypsin (Promega, Fitchburg, Wisconsin, United States) was added, and the samples were incubated overnight at 37 °C. Tryptic digests were quenched with 10% (v/v) trifluoroacetic acid (TFA) and purified using C18 microspin columns (Harvard Apparatus, Holliston, Massachusetts, USA). Columns were eluted with 0.1% (v/v) TFA in 50% (v/v) acetonitrile (ACN), and the volume of the eluted samples was reduced to approximately 2 μL in a vacuum centrifuge. The peptides were reconstituted to a final volume of 30 μL with 0.1% (v/v) TFA, 1% (v/v) ACN and vortexed thoroughly. LC‐MS/MS analysis was carried out using an EASY‐nLC nano‐HPLC system (Thermo Fisher Scientific) connected to a Velos Pro‐Orbitrap Elite hybrid mass spectrometer (Thermo Fisher Scientific) with a nano‐electrospray ion source (Thermo Fisher Scientific). A two‐column set‐up was used, consisting of a 2‐cm C18‐A1 trap column (Thermo Fisher Scientific), followed by a 10‐cm C18‐A2 analytical column (Thermo Fisher Scientific). The linear separation gradient was 5% (v/v) buffer B (0.1% TFA in 98% ACN) in 5 min, 35% (v/v) buffer B in 60 min, 80% (v/v) buffer B in 5 min and 100% buffer B in 10 min at a flow rate of 0.3 µL/min; 5 µL of sample were injected for SC6K and C6K and 3 µL for MC‐SC6K LC‐MS/MS runs. A full MS scan was acquired with a resolution of 60 000 over a normal mass range of the Orbitrap analyser; the method was set to fragment the 20 most intense precursor ions with CID (energy 35). Data were acquired using LTQ Tune software. The acquired MS2 scans were searched against the N. benthamiana annotated protein database derived from solgenomics.net using Sequest search algorithms in Proteome Discoverer software (Thermo Fisher Scientific). The allowed mass error was 15 ppm for precursor ions and 0.8 Da for fragment ions. Carbamidomethylation (+57.021 Da) of cysteine was set as a static modification, and oxidation of methionine (+15.995 Da) was set as a dynamic modification. Database searches were limited to fully tryptic peptides with a maximum of one missed cleavage. The web‐based bioinformatics database DAVID (Huang et al., 2009a, 2009b) was used as a guide for the classification of the proteins. For ribosomal protein lists, the TAIR IDs of the discovered proteins were submitted to the DAVID functional annotation tool and the proteins classified as ‘ribosomal protein’ were selected for the ribosomal protein lists.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot of Potato virus A (PVA) coat protein (CP) in PVA‐C6K‐ and PVA‐6KY‐infected plants.
Fig. S2 Protein concentrations in the affinity‐purified samples.
Fig. S3 Potato virus A (PVA) RNA amount in the affinity purification input and pull‐down of PVA‐SC6K and PVA‐C6K samples.
Table S1 List of the unique proteins, with summation of the peptide spectrum matches (PSMs) and peptides and unique peptides.
Table S2 List of primers used in this study.
Acknowledgements
We thank Anders Hafrén for valuable discussions throughout this study, Minna Pöllänen for taking care of the plants and Sini Miettinen for assistance with the LC‐MS/MS analysis. Dr Eija Jokitalo and Ms Mervi Lindman at the EM unit of the Institute of Biotechnology, University of Helsinki, are acknowledged for their help in EM imaging. Financial support given by the Academy of Finland (grant 1138329 to K.M. and grant 1258978 to M.V.) and the Jenny and Antti Wihuri Foundation is gratefully acknowledged. A.L. was supported by the Integrative Life Science Doctoral Program and Research Foundation of the University of Helsinki. We declare that we have no conflicts of interest.
References
- Aniento, F. , Matsuoka, K. and Robinson, D.G. (2006) ER‐to‐Golgi transport: the COPII‐pathway In: The Plant Endoplasmic Reticulum (Robinson D.G., ed.), pp. 99–124. Plant Cell Monographs Berlin, Heidelberg: Springer. [Google Scholar]
- Anindya, R. , Chittori, S. and Savithri, H.S. (2005) Tyrosine 66 of Pepper vein banding virus genome‐linked protein is uridylylated by RNA‐dependent RNA polymerase. Virology, 336, 154–162. [DOI] [PubMed] [Google Scholar]
- Barajas, D. , Martín, I.F. de C., Pogany, J. , Risco, C. and Nagy, P.D. (2014) Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of tomato bushy stunt virus replicase. PLoS Pathog. 10, e1004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin, C. , Boutet, N. and Laliberté, J.‐F. (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon, J.A. and Ahlquist, P. (2010) Organelle‐like membrane compartmentalization of positive‐strand RNA virus replication factories. Annu. Rev. Microbiol. 64, 241–256. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. , Jensen, P.E. and Schaad, M.C. (1998) Genetic evidence for an essential role for potyvirus CI protein in cell‐to‐cell movement. Plant J. 14, 393–400. [DOI] [PubMed] [Google Scholar]
- Castorena, K.M. , Weeks, S.A. , Stapleford, K.A. , Cadwallader, A.M. and Miller, D.J. (2007) A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J. Virol. 81, 8412–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton, S. , Grangeon, R. , Thivierge, K. , Mathieu, I. , Ide, C. , Wei, T. , Wang, A. and Laliberté, J.‐F. (2009) Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments, and are each derived from a single viral genome. J. Virol. 83, 10 460–10 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X. , Wei, T. , Chowda‐Reddy, R.V. , Sun, G. and Wang, A. (2010) The tobacco etch virus P3 protein forms mobile inclusions via the early secretory pathway and traffics along actin microfilaments. Virology, 397, 56–63. [DOI] [PubMed] [Google Scholar]
- Davis, W.G. , Blackwell, J.L. , Shi, P.‐Y. and Brinton, M.A. (2007) Interaction between the cellular protein eEF1A and the 3′‐terminal stem‐loop of West Nile virus genomic RNA facilitates viral minus‐strand RNA synthesis. J. Virol. 81, 10 172–10 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne, P.J. , Thivierge, K. , Cotton, S. , Beauchemin, C. , Ide, C. , Ubalijoro, E. , Laliberté, J.‐F. and Fortin, M.G. (2008) Heat shock 70 protein interaction with Turnip mosaic virus RNA‐dependent RNA polymerase within virus‐induced membrane vesicles. Virology, 374, 217–227. [DOI] [PubMed] [Google Scholar]
- Eskelin, K. , Suntio, T. , Hyvärinen, S. , Hafren, A. and Mäkinen, K. (2010) Renilla luciferase‐based quantitation of Potato virus A infection initiated with Agrobacterium infiltration of N. benthamiana leaves. J. Virol. Methods, 164, 101–110. [DOI] [PubMed] [Google Scholar]
- Fernández, A. , Guo, H.S. , Sáenz, P. , Simón‐Buela, L. , Gómez de Cedrón, M. and García, J.A. (1997) The motif V of plum pox potyvirus CI RNA helicase is involved in NTP hydrolysis and is essential for virus RNA replication. Nucleic Acids Res. 25, 4474–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrenaite‐Verkhovskaya, R. , Andreev, I.A. , Kalinina, N.O. , Torrance, L. , Taliansky, M.E. and Mäkinen, K. (2008) Cylindrical inclusion protein of potato virus A is associated with a subpopulation of particles isolated from infected plants. J. Gen. Virol. 89, 829–838. [DOI] [PubMed] [Google Scholar]
- Grangeon, R. , Cotton, S. and Laliberté, J.‐F. (2010) A model for the biogenesis of turnip mosaic virus replication factories. Commun. Integr. Biol. 3, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon, R. , Agbeci, M. , Chen, J. , Grondin, G. , Zheng, H. and Laliberté, J.‐F. (2012) Impact on the endoplasmic reticulum and Golgi apparatus of turnip mosaic virus infection. J. Virol. 86, 9255–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén, A. , Hofius, D. , Rönnholm, G. , Sonnewald, U. and Mäkinen, K. (2010) HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell, 22, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén, A. , Eskelin, K. and Mäkinen, K. (2013) Ribosomal protein P0 promotes potato virus A infection and functions in viral translation together with VPg and eIF(iso)4E. J. Virol. 87, 4302–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton, S.L. , Renna, L. , Bortolotti, L.E. , Chatre, L. , Stefano, G. and Brandizzi, F. (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell, 17, 3081–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y. and Hunt, A.G. (1996) RNA polymerase activity catalyzed by a potyvirus‐encoded RNA‐dependent RNA polymerase. Virology, 226, 146–151. [DOI] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. and Lempicki, R.A. (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W. , Sherman, B.T. and Lempicki, R.A. (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang, T.‐S. and Nagy, P.D. (2011) Direct inhibition of tombusvirus plus‐strand RNA synthesis by a dominant negative mutant of a host metabolic enzyme, glyceraldehyde‐3‐phosphate dehydrogenase, in yeast and plants. J. Virol. 85, 9090–9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.W. , Hu, C.C. , Liou, M.R. , Chang, B.Y. , Tsai, C.H. , Meng, M. , Lin, N.S. and Hsu, Y.H. (2012) Hsp90 interacts specifically with viral RNA and differentially regulates replication initiation of Bamboo mosaic virus and associated satellite RNA. PLoS Pathog. 8, e1002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, N. , Powis, K. and High, S. (2013) Post‐translational translocation into the endoplasmic reticulum. Biochim. Biophys. Acta BBA ‐ Mol. Cell Res. 1833, 2403–2409. [DOI] [PubMed] [Google Scholar]
- Kekarainen, T. , Savilahti, H. and Valkonen, J.P.T. (2002) Functional genomics on potato virus A: virus genome‐wide map of sites essential for virus propagation. Genome Res. 12, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P.G. , Klein, R.R. , Rodriguez‐Cerezo, E. , Hunt, A.G. and Shaw, J.G. (1994) Mutational analysis of the tobacco vein mottling virus genome. Virology, 204, 759–769. [DOI] [PubMed] [Google Scholar]
- Lerich, A. , Langhans, M. , Sturm, S. and Robinson, D.G. (2011) Is the 6 kDa tobacco etch viral protein a bona fide ERES marker? J. Exp. Bot. 62, 5013–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Pogany, J. , Tupman, S. , Esposito, A.M. , Kinzy, T.G. and Nagy, P.D. (2010) Translation elongation factor 1A facilitates the assembly of the tombusvirus replicase and stimulates minus‐strand synthesis. PLoS Pathog. 6, e1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, S. , Dolja, V.V. and Carrington, J.C. (1996) Roles of the sequence encoding tobacco etch virus capsid protein in genome amplification: requirements for the translation process and a cis‐active element. J. Virol. 70, 4370–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. and Krijnse‐Locker, J. (2008) Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine, A. and Okuno, T. (2012) Composition of plant virus RNA replicase complexes. Curr. Opin. Virol. 2, 669–675. [DOI] [PubMed] [Google Scholar]
- Nagy, P.D. and Pogany, J. (2012) The dependence of viral RNA replication on co‐opted host factors. Nat. Rev. Microbiol. 10, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T. , Nishimura, Y. , Ichimura, T. , Suzuki, K. , Miyamura, T. , Suzuki, T. , Moriishi, K. and Matsuura, Y. (2006) Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25, 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olspert, A. , Chung, B.Y.‐W. , Atkins, J.F. , Carr, J.P. and Firth, A.E. (2015) Transcriptional slippage in the positive‐sense RNA virus family Potyviridae. EMBO Rep. 16, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, D. and Bartenschlager, R. (2013) Architecture and biogenesis of plus‐strand RNA virus replication factories. World J. Virol. 2, 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, D. , Hoppe, S. , Saher, G. , Krijnse‐Locker, J. and Bartenschlager, R. (2013) Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87, 10 612–10 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany, J. and Nagy, P.D. (2008) Authentic replication and recombination of tomato bushy stunt virus RNA in a cell‐free extract from yeast. J. Virol. 82, 5967–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany, J. , Stork, J. , Li, Z. and Nagy, P.D. (2008) In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc . Natl. Acad. Sci. USA, 105, 19 956–19 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen, P. and Mäkinen, K. (2004) Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 279, 38 103–38 110. [DOI] [PubMed] [Google Scholar]
- Rajamäki, M.‐L. and Valkonen, J.P.T. (1999) The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides . Mol. Plant–Microbe Interact. 12, 1074–1081. [DOI] [PubMed] [Google Scholar]
- Rantalainen, K.I. , Eskelin, K. , Tompa, P. and Mäkinen, K. (2011) Structural flexibility allows the functional diversity of potyvirus genome‐linked protein VPg. J. Virol. 85, 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revers, F. and García, J.A. (2015) Molecular biology of potyviruses In: Advances in Virus Research (Maramorosch K. and Thomas C. M. eds), pp. 101–199. Academic Press, Cambridge, Massachusetts, USA. [DOI] [PubMed] [Google Scholar]
- Rodamilans, B. , Valli, A. , Mingot, A. , San León, D. , Baulcombe, D. , López‐Moya, J.J. and García, J.A. (2015) RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the potyviridae family. J. Virol. 89, 6965–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C. , Jensen, P.E. and Carrington, J.C. (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum‐targeted viral protein. EMBO J. 16, 4049–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T.G. and Skerra, A. (2007) The Strep‐tag system for one‐step purification and high‐affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535. [DOI] [PubMed] [Google Scholar]
- Shen, W. , Yan, P. , Gao, L. , Pan, X. , Wu, J. and Zhou, P. (2010) Helper component‐proteinase (HC‐Pro) protein of papaya ringspot virus interacts with papaya calreticulin. Mol. Plant Pathol. 11, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetz, C. and Valkonen, J.P.T. (2004) Potyviral 6K2 protein long‐distance movement and symptom‐induction functions are independent and host‐specific. Mol. Plant–Microbe Interact. 17, 502–510. [DOI] [PubMed] [Google Scholar]
- Thivierge, K. , Cotton, S. , Dufresne, P.J. , Mathieu, I. , Beauchemin, C. , Ide, C. , Fortin, M.G. and Laliberté, J.‐F. (2008) Eukaryotic elongation factor 1A interacts with turnip mosaic virus RNA‐dependent RNA polymerase and VPg‐Pro in virus‐induced vesicles. Virology, 377, 216–225. [DOI] [PubMed] [Google Scholar]
- Tomita, Y. , Mizuno, T. , Díez, J. , Naito, S. , Ahlquist, P. and Ishikawa, M. (2003) Mutation of host DnaJ homolog inhibits brome mosaic virus negative‐strand RNA synthesis. J. Virol. 77, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance, L. , Andreev, I.A. , Gabrenaite‐Verhovskaya, R. , Cowan, G. , Mäkinen, K. and Taliansky, M.E. (2006) An unusual structure at one end of potato potyvirus particles. J. Mol. Biol. 357, 1–8. [DOI] [PubMed] [Google Scholar]
- Ungar, D. and Hughson, F.M. (2003) SNARE protein structure and function. Annu. Rev. Cell Dev. Biol. 19, 493–517. [DOI] [PubMed] [Google Scholar]
- Verchot, J. (2011) Wrapping membranes around plant virus infection. Curr. Opin. Virol. 1, 388–395. [DOI] [PubMed] [Google Scholar]
- Verchot, J. and Carrington, J.C. (1995) Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69, 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. (2015) Dissecting the molecular network of virus–plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66. [DOI] [PubMed] [Google Scholar]
- Wang, R.Y.‐L. and Nagy, P.D. (2008) Tomato bushy stunt virus co‐opts the RNA‐binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe, 3, 178–187. [DOI] [PubMed] [Google Scholar]
- Wang, R.Y.‐L. , Stork, J. and Nagy, P.D. (2009) A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, S.A. , Shield, W.P. , Sahi, C. , Craig, E.A. , Rospert, S. and Miller, D.J. (2010) A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J. Virol. 84, 330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. and Wang, A. (2008) Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI‐ and COPII‐dependent manner. J. Virol. 82, 12 252–12 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Huang, T.‐S. , McNeil, J. , Laliberté, J.‐F. , Hong, J. , Nelson, R.S. and Wang, A. (2010) Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hou, X. , Sanfaçon, H. and Wang, A. (2013) The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus‐induced vesicles with chloroplasts. PLoS Pathog. 9, e1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R.‐H. and Hajimorad, M.R. (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology, 400, 1–7. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Zhang, C. , Dittman, J.D. and Whitham, S.A. (2009) Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology, 390, 163–173. [DOI] [PubMed] [Google Scholar]
- Yi, Z. , Sperzel, L. , Nürnberger, C. , Sperzel, L. , Nürnberger, C. , Bredenbeek, P.J. , Lubick, K.J. , Best, S.M. , Stoyanov, C.T. , Law, L.M. , Yuan, Z. , Rice, C.M. and MacDonald, M.R. (2011) Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS Pathog. 7, e1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1 Western blot of Potato virus A (PVA) coat protein (CP) in PVA‐C6K‐ and PVA‐6KY‐infected plants.
Fig. S2 Protein concentrations in the affinity‐purified samples.
Fig. S3 Potato virus A (PVA) RNA amount in the affinity purification input and pull‐down of PVA‐SC6K and PVA‐C6K samples.
Table S1 List of the unique proteins, with summation of the peptide spectrum matches (PSMs) and peptides and unique peptides.
Table S2 List of primers used in this study.


