Summary
The Dothideomycetes represents a large and diverse array of fungi in which prominent plant pathogens are over‐represented. Species within the Cochliobolus, Alternaria, Pyrenophora and Mycosphaerella (amongst others) all cause diseases that threaten food security in many parts of the world. Significant progress has been made over the past decade in understanding how some of these pathogens cause disease at a molecular level. It is reasonable to suggest that much of this progress can be attributed to the increased availability of genome sequences. However, together with revealing mechanisms of pathogenicity, these genome sequences have also highlighted the capacity of the Dothideomycetes to produce an extensive array of secondary metabolites, far greater than originally thought. Indeed, it is now clear that we appear to have only scratched the surface to date in terms of the identification of secondary metabolites produced by these fungi. In the first half of this review, we examine the current status of secondary metabolite research in the Dothideomycetes and highlight the diversity of the molecules discovered thus far, in terms of both structure and biological activity. In the second part of this review, we survey the emerging techniques and technologies that will be required to shed light on the vast array of secondary metabolite potential that is encoded within these genomes. Experimental design, analytical chemistry and synthetic biology are all discussed in the context of how they will contribute to this field.
Keywords: Dothideomycetes, plant pathogens, secondary metabolites
Introduction
Plants and fungi have shared a long evolutionary history. There is evidence that plants first colonized the surface of the Earth by establishing symbiotic associations with fungi (Heckman et al., 2001; Humphreys et al., 2010). Over time, evolution has shaped and diversified the fungal–plant interaction into different forms: mycorrhizic, endophytic, saprophytic and pathogenic. Renowned by their plant‐associated members, the Dothideomycetes is the largest class of fungi within the biggest phylum, the Ascomycota (Kirk et al., 2008). The Dothideomycetes is a very heterogeneous class, occupying the most varied habitats and niches: from the Arctic to the tropics, over rocks facing the inclemency of weather (Ruibal et al., 2009), colonizing dung (Kruys et al., 2006), as lichenized symbionts (Nelsen et al., 2011) or living under water as either marine or freshwater organisms (Shearer et al., 2009; Suetrong et al., 2009). One notable feature of the Dothideomycete class is the over‐representation of devastating plant pathogens (Arnold, 2007; Schoch et al., 2009).
Members of the Dothideomycetes cause disease in every major crop (Ohm et al., 2012). Species from Alternaria, Botryosphaeria, Cochliobolus, Mycosphaerella, Phaeosphaeria and Pyrenophora affect the production of cereal crops, the base of human diet. Didymella is responsible for many major legume diseases, and Leptosphaeria is devastating in Brassicas. Consequently, members of the Dothidiomycetes represent a primary food biosecurity risk, through either crop yield losses or product quality issues caused by mycotoxin contaminations (Solomon, 2011).
Fungi infect plants using different strategies: biotrophs exploit plants' resources, whilst keeping the host alive; necrotrophs kill the host in order to thrive off dead or dying tissue; and hemibiotrophs have an asymptomatic phase followed by a necrotrophic stage (Horbach et al., 2011). Despite their lifestyle, a common mechanism for many plant‐infecting fungi is the production of effectors. Typically, these effectors interact with a specific host target, triggering a response that may lead to either the suppression or development of disease (Wolpert et al., 2002). For example, ToxA, a small secreted protein from Parastagonospora nodorum and Pyrenophora tritici‐repentis, is a classic example of an effector involved in an inverse gene‐for‐gene interaction (Ciuffetti et al., 2010; Friesen et al., 2006). When both ToxA and its host susceptibility gene Tsn1 are dominant, disease ensues. However, if either of these genes is missing, no susceptibility is observed (Oliver and Solomon, 2010). Although, in the above example, the term ‘effector’ refers to small secreted proteins, some definitions of plant–microbe effectors have been expanded. These expanded definitions consider all substances secreted into the plant by microorganisms (e.g. proteins, small molecules, small RNAs) which cause an effect on the host cell (Hogenhout et al., 2009). In consequence, this would therefore include some of the microbial secondary metabolites (SMs) (Collemare and Lebrun, 2011).
SMs, also known as natural products, have fascinated researchers for many decades. They are often small molecules that do not play an obvious role in the basic cellular functions, but have bio‐ecological roles in helping the producing organism adapt to its environmental niche (O'Brien and Wright, 2011; Williams et al., 1989). Studies of fungal SMs have traditionally been focused on two main areas: pharmaceutical and agronomical (Hanson, 2008). From ancient times, fungi have been recognized for their medicinal and toxicological properties. When Fleming discovered penicillin, the interest in fungal SMs as a source of drugs increased significantly (Aly et al., 2011). Partly driven by the invigorated pharmaceutical interests, agricultural researchers began looking for SMs involved in pathogenicity and mycotoxicosis, as well as new sources of agrochemicals. These SM studies have not only demonstrated that fungal SMs possess remarkable biological activities which can be harnessed for the benefit of humankind, they have also driven important developments in analytical, synthetic and medicinal chemistry (Nakanishi, 1999). However, there remains much to be learnt, especially with regard to the biological and ecological roles of SMs in fungal adaptation and survival.
We are only beginning to understand how fungal SMs work in plant pathosystems. For example, some phytotoxic compounds produced by plant‐pathogenic fungi can act as plant‐damaging photosensitizers, target host proteins to disrupt their function, affect the membrane or trigger apoptosis in plant cells (Möbius and Hertweck, 2009). Some of these are host‐specific toxins (HSTs), whereas others have activities against a broader range of organisms. Other roles of SMs from plant pathogens include mineral uptake (e.g. siderophores), protection against biotic and abiotic factors, mediation of interspecific interactions and even manipulation of the genetic regulation of the hosts (Scharf et al., 2014). However, regardless of the identified in vivo roles, SMs can exert physiological effects in different organisms. For example, SMs with a negative effect on humans and other higher animals, independent of their role in fungal biology, are called mycotoxins (Bennett and Klich, 2003; Reverberi et al., 2010).
Fungal SMs exhibit considerable structural diversity, but can be broadly grouped into four main categories based on their biosynthetic origin: polyketides, non‐ribosomal peptides (NRPs), terpenoids and tryptophan derivatives. These are made by the core enzymes polyketide synthases (PKSs), non‐ribosomal peptide synthases (NRPSs), prenyltransferases (PTs)/terpene synthases (TSs) and dimethylallyl tryptophan synthases (DMATSs) (Wiemann and Keller, 2014). The conserved sequence features, within the respective family of core enzymes, can be exploited to allow the rapid identification of genes encoding PKSs, NRPSs, PTs, TSs and DMATSs within fungal genomes. The fact that genes involved in fungal SM biosynthesis are often clustered together on the chromosomes, either to simplify regulation and horizontal gene transfer or to prevent the accumulation of toxic metabolic intermediaries, further facilitates the identification of these enzymes (Keller et al., 2005; McGary et al., 2013). From the number of fungal genomes that have been sequenced so far, it has been observed that the Dothideomycete fungi encode a remarkable number of SM gene clusters, suggesting their immense potential at producing vast chemical diversity. However, although the genetic template for this diversity is identifiable, the number of SM compounds known in each of the sequenced fungi remains limited. It is clear that what has been discovered to date is just a small portion of the fungal SM diversity.
The modern genomic era is transforming natural product research into an interdisciplinary area in which vastly different approaches are now being used to tackle the many facets of SMs from their evolutionary origins, genetic regulations and biosynthesis to their natural bio‐ecological functions. As Stadler and Keller (2008) wrote: there are new paradigms in fungal SMs. This review is an opportunity to survey the path we have walked and to consider where we are heading. The first half of this article presents an overview of what is known about Dothideomycete SMs in terms of compound diversity, biosynthesis and bioactivities. The second part reviews some of the new technologies for the study of SMs, with a focus on their application in the characterization and understanding of SMs and their roles from an ecological perspective (e.g. their role in plant–microbe or microbe–microbe interactions)
Pathogenic Dothideomycetes and Their SMs
The Dothideomycetes is a class so vast that it is not the aim of this review to describe each genera or to list all the known SMs. In this section, we summarize some particular examples within the Dothideomycetes with the aim of not only presenting what we currently understand, but, more importantly, highlighting what we do not.
Cochliobolus
Cochliobolus (teleomorph of Bipolaris and Curvularia) is a genus that contains a large number of devastating necrotrophic plant pathogens which are reliant on SMs to cause disease. Figure 1 shows examples of these SMs. Some of these compounds have been identified as HSTs and their biosynthetic genes and mode of action have been investigated and reviewed (Panaccione, 1993; Stergiopoulos et al., 2013). The three iconic HSTs from Cochliobolus are T‐toxin, HC‐toxin and victorin.
Figure 1.
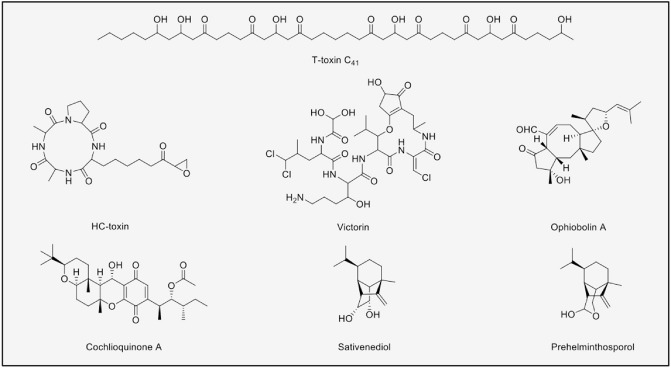
Secondary metabolites isolated from Cochliobolus.
The polyketide T‐toxin is a virulence determinant from the maize pathogen Cochliobolus heterostrophus race T and is encoded by a total of nine genes (Inderbitzin et al., 2010). Two of these are PKS genes located on two unlinked loci. T‐toxin triggers apoptosis when it interacts with a receptor, URF13, which is located on the inner mitochondrial membrane, resulting in small‐molecule leakage and necrotizing tissue (Inderbitzin et al., 2010). HC‐toxin is a cyclic NRP from the maize pathogen, Cochliobolus carbonarum race 1. This toxin is produced by the core NRPS, HST1, and other enzymes encoded in the TOX2 locus. HC‐toxin is an inhibitor of histone deacetylase that causes disease in maize lines which lack the carbonyl reductase enzymes, HM1/HM2, required for the detoxification of this compound (Walton, 2006). Victorin is an NRP from Cochliobolus victoriae, the causal agent of Victoria blight in oats. Although the genome of C. victoriae has been sequenced, the NRPS gene that is responsible for the production of victorin remains to be identified (Condon et al., 2013). Victorin elicits plant cell death in oat lines containing the resistance gene Pc‐2 against Puccinia coronata, allowing C. victoriae (a necrotroph) to proliferate (Lorang et al., 2007). Victorin also affects Arabidopsis thaliana, but the susceptibility of this host is caused by a different ‘resistance’ gene, LOV1, which encodes a protein containing a nucleotide‐binding domain and leucine‐rich repeat domain (NB‐LRR). This NB‐LRR protein is activated when victorin binds to a thioredoxin, TRX‐h5. Using Arabidopsis and virus‐induced gene silencing (VIGS), it was shown that the susceptibility response to this chemical toxin from C. victoriae is equivalent to the classical resistance response against proteinaceous effectors from biotrophs (Gilbert and Wolpert, 2013).
In addition, Cochliobolus produces non‐HSTs, such as ophiobolins, a large family of more than 20 phytotoxic sesterterpenoids with a characteristic structure of a 5‐8‐5 ring. Originally isolated from Cochliobolus miyabeanus (Tsuda et al., 1967), ophiobolins have also been reported in other Cochliobolus species (Strobel et al., 1988), and also in Aspergillus and Cephalosporium (Cutler et al., 1984; Itai et al., 1967). Ophiobolins are active against plants, microorganisms and animals, affecting different cellular processes (Au et al., 2000). Ophiobolin A targets the calcium receptor, calmodulin, causing ion leakage in corn roots (Leung et al., 1984, 1985), and also inhibits β‐1,3‐glucan synthase, affecting oomycete cell wall formation (Fukushima et al., 1993). Recently, the gene involved in ophiobolin F biosynthesis in Aspergillus clavatus has been reported, ACLA_076850; it is the first discovered sesterterpene synthase (Chiba et al., 2013). The identification of this ophiobolin gene will facilitate the discovery of homologues in Cochliobolus and other organisms. In addition, this could influence not only basic research, but also drug discovery, as the ophiobolin structure has been proposed as a promising anticancer template (Bury et al., 2013).
Other noteworthy compounds from this genus are cochlioquinones from C. miyabeanus, Bipolaris cynodontis, Bipolaris bicolor, Bipolaris brizae and an endophytic Cochliobolus sp. These phytotoxins are hybrid compounds incorporating a p‐benzoquinone ring and an isoprenoid moiety (Carruthers et al., 1971). Cochlioquinones have been shown to inhibit the root growth of finger millet, rice and Italian rye grass, and also to affect complex I in electron transport in mitochondria by interfering with NADH‐ubiquinone reductase (Lim et al., 1996, 1998; Miyagawa et al., 1994). In addition, these SMs exhibit activities with medical relevance against cancer (Machida et al., 1995), human immunodeficiency virus (HIV) (Yoganathan et al., 2004) and Leishmania (Campos et al., 2008). Other important SMs are the sativenediol‐ and helminthosporol‐type compounds from the pathogenic Cochliobolus sativus (Nukina et al., 1975; Tamura et al., 1963). Unlike the cochlioquinones, these SMs promote seedling elongation in rice and their activity has been compared with the gibberellins (Kato et al., 1964). Structurally, the sativenediols, helminthosporins and gibberellins share a bicyclo(3.2.1)octane moiety with the same stereochemistry. Hence, it can be hypothesized that the growth‐promoting activity of these compounds is conferred by the bicyclo group (Briggs, 1966; Coombe et al., 1974).
The Cochliobolus genus has one of the highest representations amongst the Dothideomycete pathogens that have publicly available genomic sequences, including C. carbonarum, C. heterostrophus, Cochliobolus lunatus, C. miyabeanus, C. sativus, C. victoriae and Curvularia protuberata. This therefore provides an excellent opportunity for comparative genomic studies. A recent Cochliobolus comparative genomic study has shown that around 50% of the NRPS genes and 30% of the PKS genes are highly conserved among the first six species aforementioned (Condon et al., 2013). However, some of these enzymes are unique at a species level, making them good candidates for specific virulence factors. As it stands, most of the products of these genes are yet to be characterized (Condon et al., 2013).
Alternaria
Alternaria is a wide group of anamorphic fungi, mainly from the Lewia genus. Many of these are described as saprobes, but plant pathogens are also well represented in Alternaria (Thomma, 2003). Alternaria species are known for their prolific production of chemical structures (Fig. 2). The production of mycotoxins by this genus has long been studied. For instance, alternariol (AOH) was isolated and characterized in 1953 (Raistrick et al., 1953) and its biosynthetic origin was determined to be derived from a heptaketide chain by feeding 14C‐labelled acetate to Alternaria alternata (Thomas, 1961). Recently, an Al. alternata genome‐wide PKS expression analysis has shown that two PKS genes, pksH and pksJ, are expressed at the same time as AOH and alternariol methylether (AME) production. Down‐regulation of pksJ reduced the amount of AOH and AME produced, suggesting that pksJ is responsible for the biosynthesis of these polyketides (Saha et al., 2012). However, as pksJ encodes a highly reducing PKS that usually produces non‐aromatic compounds, it is surprising that it plays a role in AOH synthesis.
Figure 2.
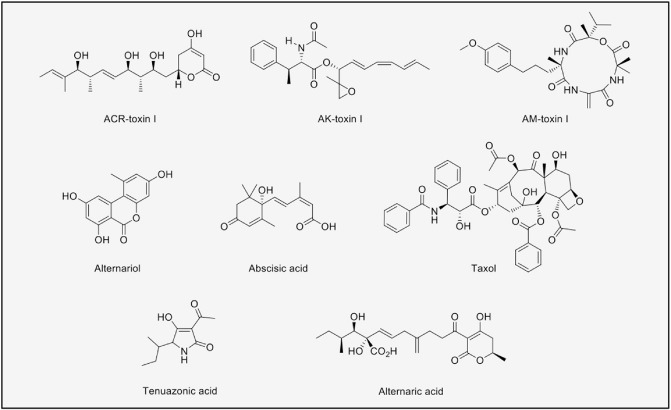
Secondary metabolites isolated from Alternaria.
As demonstrated for Cochliobolus, HSTs form part of the SM repertoire of Alternaria species. For example, Al. alternata pathotypes have specific toxins depending on their host, such as ACR toxins for citrus, AK toxins for Japanese pear, AM toxins for apples and pears and AT toxin for tobacco. For most of these toxins, the structures, effect, biological targets and biosynthetic genes have been described in two recent reviews (Akimitsu et al., 2013; Tsuge et al., 2013). However, the structure of some compounds, such as AT toxin, have not been elucidated.
Recently, Lou et al. (2013) have published a review with 268 SMs reported from Alternaria species (summarized in Table 1). Bioassays have identified phytotoxic and antimicrobial activities for some of these compounds. In addition, cytotoxic/antitumour effects have also been reported frequently, and specific bioactivities have arisen from targeted SM screenings (Lou et al., 2013). Included on this list are abscisic acid (a plant hormone involved in developmental processes), Taxol or Paclitaxel (the anticancer drug discovered in the bark of Taxus brevifolia) and tenuazonic acid (a mycotoxin with wide‐ranging activities, including antibiotic, phytotoxic, antitumour and antiviral properties). However, despite the many reports on Alternaria SMs, the functions for most of these compounds remain unknown.
Table 1.
Type and number of secondary metabolites (SMs) from Alternaria species reviewed by Lou et al. (2013)
| Alternaria SMs by chemical groups | |
|---|---|
| Alkaloids | 61 |
| Cyclopeptides | 22 |
| Steroids | 7 |
| Terpenoids | 49 |
| Pyranones | 56 |
| Quinones | 45 |
| Phenolics | 38 |
| Miscellaneous | 12 |
| Total | 268 |
Mycosphaerella
Mycosphaerella contains several anamorphic genera known to be economically important plant pathogens, including Septoria and the well‐known producer of SMs Cercospora (Crous, 2009). Some Mycosphaerella SMs are shown in Fig. 3. Probably the most renowned SM of this genus is the perylenequinone compound cercosporin, a non‐specific light‐activated phytotoxin first reported from Cercospora kikuchii (Kuyama and Tamura, 1957). A role for cercosporin in plant pathogenesis has been shown by using cercosporin‐deficient mutants (Upchurch et al., 1991). However, non‐pathogenic fungi also produce these perylenequinones which are likely to have other roles, including non‐plant‐associated interactions (e.g. microbe–microbe)(Daub and Chung, 2009; Daub and Ehrenshaft, 2000).
Figure 3.
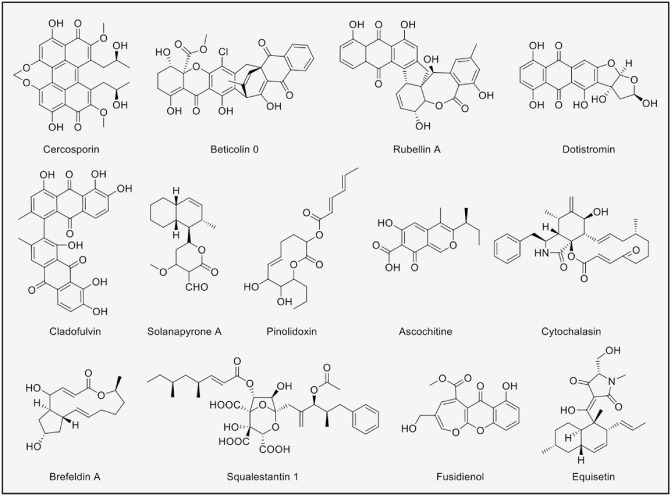
Secondary metabolites isolated from Mycosphaerella and Didymella.
Another interesting class of SMs in the Mycosphaerella are the beticolins (cebetins). These are a family of 20 polyketide‐non‐specific toxins produced by Cercospora beticola, a pathogen of sugar beet. These compounds are chlorinated heterodimers of anthraquinone and xanthone derivatives (Milat et al., 1992). Beticolins have shown activity against plants and bacteria, they complex different metals, interfere with H+ and K+ transport, affect ATPases and depolarize membranes (Jalal et al., 1992). These activities have been linked to the ability of beticolins to form pores in the plasma membrane (Goudet et al., 2000).
Similar to the Cercospora perylenequinones, the rubellins from the barley pathogen Ramularia collo‐cygni are also light‐activated toxins (Miethbauer et al., 2006). Rubellins are anthraquinone derivatives of polyketide origin which have demonstrated cytotoxicity in tumorous cell lines and also have light‐dependent antibiotic activity (Miethbauer et al., 2009). Although rubellins are not a determinant for the infection of the host (they are not host specific), their ability to induce necrosis in planta suggests that they are pathogenic factors, as R. collo‐cygni, a necrotroph, thrives in necrotic tissue (Heiser et al., 2004; Walters et al., 2008).
Another example of SMs from the Mycosphaerella genus is dothistromin, isolated from the pathogen Dothistroma septosporum (anamorphic M. pini), the causal agent of Dothistroma needle blight of Pinus ssp. (Barnes et al., 2004). Dothistromin is an aflatoxin‐related compound with a wide spectrum of toxicity; it exhibits activity against microorganisms and phytotoxicity against the host. Furthermore, it causes the characteristic red coloration of infected needles (Shain and Franich, 1981). However, it has been shown that dothistromin knockouts of M. pini maintain the ability to produce the disease, demonstrating that dothistromin is not needed for pathogenicity, but a possible role of this compound could be to compete against other Pinus‐infecting microorganisms (Schwelm et al., 2009). However, it is interesting to note that the genes for dothistromin synthesis are not clustered as typically observed for SM synthetic genes, but appear to be scattered throughout the genome (Zhang et al., 2007). This has been proposed to be the product of a gene cluster fragmentation which could be a mechanism for retooling SM biosynthetic machinery and, consequently, for niche adaptation (Bradshaw et al., 2013).
Cladosporium fulvum is a biotrophic pathogen of tomato and, presumably, an anamorphic Mycosphaerella (Thomma et al., 2005). A genomic analysis has shown that it has 21 SM core enzymes, including those coding for the perylenequinone and dothistromin structures (Collemare et al., 2014). Using comparative genomics and transcriptomics data, Collemare et al. showed that, in contrast with other biothrophs, which have a reduced number of SM genes, Cl. fulvum has a comparable number of SM biosynthetic genes to that found in hemibiotrophs and necrotrophs. However, their expression during the infection phase is down‐regulated, representing an alternative mechanism of biotrophic lifestyle (Collemare et al., 2014). The only major compound that can be detected and identified in vitro is a bianthraquinone, cladofulvin. Cladosporium fulvum pks6, whose expression correlates with the production of cladofulvin, has been proposed to be involved in the biosynthesis of this molecule. However, cladofulvin does not appear to play a role in causing necrosis of the plant host.
As a result of the polyphyly of Mycosphaerella, a new genus, Zymoseptoria, has been proposed for some of the species within this genus, most notably Zymoseptoria tritici (formerly known as M. graminicola) (Quaedvlieg et al., 2011). Interestingly, despite being one of the major wheat pathogens in Europe and considered by some as a model organism (Dean et al., 2012), there have been no reports describing toxins or SMs.
Didymella
A large number of Ascochyta and Phoma pathogens are Didymella anamorphs and have been reported to produce SMs; some of these compounds are presented in Fig. 3. Species within the Aschochyta represent the causal agents of many of the main diseases of legumes: Ascochyta pisi and Ascochyta pinodes infect peas, Ascochyta lathyri infects sweet peas, and chickpeas and beans are infected by Ascochyta rabiei and Ascochyta fabae, respectively.
The virulence of A. rabiei isolates has been correlated with their ability to produce the phytotoxic solanapyrones A, B and C (Kaur, 1995). However, when applying these compounds on chickpea shoots directly, they cannot be detected after symptoms develop, suggesting that these substances may be metabolized in planta. It has been suggested that solanapyrones react with glutathione‐S‐transferase, becoming covalently bonded to glutathione, and thereby reducing the toxicity (Hamid and Strange, 2000; Riaz et al., 2013). Pinodiloxines, five nonenolides from A. pinodes and ascosalitoxin, a precursor of ascochitine produced by A. pisi and A. fabae, have also been shown to exhibit phytotoxicity. These SMs produce similar symptoms to those observed during fungal infection. In addition, their effect on other microorganisms was tested and no inhibitory activity was found, suggesting that these compounds are plant‐specific toxins (Evidente et al., 1993a, b). In contrast, compounds from the cytochalasin family identified in A. lathyri (Vurro et al., 1992), A. rabiei (Latif et al., 1993) and Phoma exigua (Capasso et al., 1987, 1988; Evidente et al., 1992, 1996) displayed antimicrobial activities, but no visible effects on plants, when tested as pure compounds (Capasso et al., 1991). However, cytochalasins, which inhibit actin polymerization, have been shown to allow fungi to penetrate into non‐host plant cells (Kobayashi et al., 1997a, b) by suppression of the plant defence response, involving cytoskeleton reorganization (Hardham et al., 2007).
Phoma species from Didymella are also known to produce diverse SMs. For example, Phoma medicaginis has been shown to produce brefeldin A, originally a protein transport inhibitor isolated from Eupenicillium brefeldianum (Härri et al., 1963). Phoma medicaginis only produces brefeldin A in dead pre‐colonized plant tissue and in vitro, but not in living plants (Weber et al., 2004). As brefeldin A has been shown to suppress spore germination and the growth of other fungi, it has been suggested that it may play a role in substrate defence at the saprophytic stage (Weber et al., 2004).
Other Phoma SMs have been reviewed by Rai et al. (2009). Phoma SMs have shown activities that make them pharmaceutically attractive: squalestatins are cholesterol lowering (Dawson et al., 1992), fusidienol A has antitumour properties (Singh et al., 1997) and equisetin shows anti‐HIV properties (Hazuda et al., 1999). Unfortunately, many of the natural product screenings do not report on species identification and thus describe the fungi as Phoma sp., preventing their placement in the correct telemorphic group, as Phoma species could be either from the Didymella or Leptosphaeria genus.
Leptosphaeria
Leptosphaeria maculans, the causal agent of blackleg disease on canola, is the most prominent member of the Leptosphaeria. As such, several SMs from L. maculans have been identified with putative roles assigned (Howlett et al., 2001). Examples of these SMs can be found in Fig. 4. Sirodesmins are the hallmark compounds of L. maculans. The non‐HST, siderodesmin PL, is an antibacterial and antiviral compound of the epipolythiodioxopiperazine family. Sirodesmin biosynthesis is encoded by 18 genes in a cluster, including an NRPS responsible for the formation of the diketopiperazine core from a tyrosine and a serine (Ferezou et al., 1980; Gardiner et al., 2004). The disulphide bond bridging the diketopiperazine ring that is characteristic of epipolythiodioxopiperazine compounds is presumably responsible for the bioactivity, as it can interact with cysteine residues of proteins (Mullbacher et al., 1986). Another known compound from L. maculans is the antimicrobial polyketide phomenoic acid, which is found in stem and stubble. This long‐chain polyketide was isolated during a traditional natural product screen looking for compounds toxic to fungi (Devys et al., 1984). Using gene silencing, the PKS gene, together with the gene cluster responsible for its biosynthesis, has been found, PKS2 (Elliott et al., 2013). As phomenoic acid exhibits antifungal activity, but not phytotoxicity, it has been proposed that it plays a role in the suppression of competing fungi.
Figure 4.
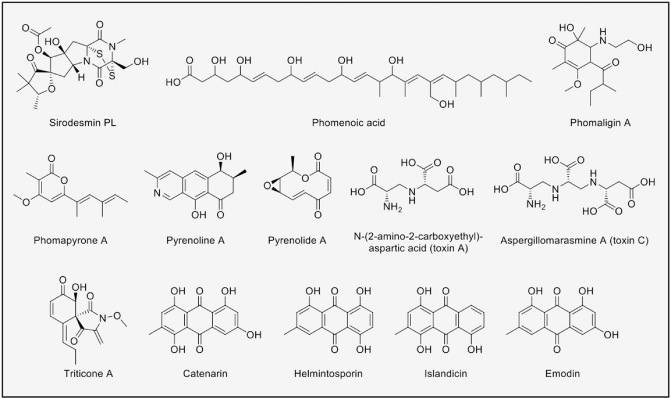
Secondary metabolites isolated from Leptosphaeria and Pyrenophora.
Initially considered to be the same species, Leptosphaeria biglobosa was separated from L. maculans (Shoemaker and Brun, 2001). Both fungi have distinct in vitro SM production (Pedras and Yu, 2009). Two groups of molecules, phomaligins and phomapyrones, have been isolated from L. biglobosa, but their biological activities and ecological roles have not been studied (Pedras et al., 1993; Soledade et al., 1994).
Pyrenophora
The Pyrenophora genus includes several significant cereal pathogens, such as the devastating Pyrenophora tritici‐repentis (wheat) and Pyrenophora teres (barley) (Ellwood et al., 2010; Manning et al., 2013). As a result of their potential role in facilitating disease, the SMs of these pathogens have received considerable attention. Some of these SMs are shown in Fig. 4. Pyrenophora teres produces the phytotoxic SMs isoquinoline pyrenolines (Coval et al., 1990), the antifungal macrolide pyrenolides (Nukina et al., 1980b, b) and five phytotoxic peptide alkaloids. Amongst the five peptide alkaloids, N‐(2‐amino‐2‐carboxythyl)‐aspartic acid (toxin A) and aspergillomarasmine A (toxin C) are the most phytotoxic, and their activity in planta correlates with the latter symptoms caused by the pathogen (Bach et al., 1979; Friis et al., 1991; Weiergang et al., 2002). Pyrenophora teres also produces proteinaceous toxins, and it has been reported that they work in a complementary way to the SMs of this pathogen. The symptoms observed at an early stage of P. teres infection correspond to those caused by the proteinaceous toxins, whereas the presence of small‐molecule toxins correlates with the latter stages (Sarpeleh et al., 2007).
Phytotoxic pirocyclic lactam triticone SMs have also been reported from P. tritici‐repentis (Hallock et al., 1993; Sugawara et al., 1988). The most active is triticone A, an intercellular toxin that causes necrosis, suppresses photosynthetic processes and has been reported to interfere with sulphydryl (SH)‐containing enzymes (Kenfield et al., 1988). In addition, seven anthraquinonic mycotoxins have been reported from P. tritici‐repentis: emodin, catenarin, helminthosporin, islandicin and the hydroxylated derivatives of the last three. These mycotoxins show antimicrobial activities, suggesting that they may play a role during the saprophytic stage of the interaction (Bouras and Strelkov, 2008; Kachlicki and Wakuliski, 2002).
Phaeosphaeria
The Phaeosphaeria genus contains pathogens of weeds and grasses; among them, Phaeosphaeria nodorum (anamorph Parastagonospora nodorum) is the causative agent of Stagonospora nodorum blotch (Shoemaker and Babcock, 1989). The genome sequence of P. nodorum reveals a large number of SM genes (Hane et al., 2007), some of which are expressed in planta during infection (Ipcho et al., 2012). To date, 11 SMs have been isolated and identified from P. nodorum (Fig. 5): mycophenolic acid, mellein and four of its derivatives (Devys et al., 1980, 1994), septorin and its two derivatives (Devys et al., 1982), AOH (Tan et al., 2009) and 4′‐methoxy‐2‐methylbutyrophenone (Yang et al., 2013). The mellein and septorin compound families have reported phytotoxic activities when tested as pure compounds. However, the necrotic effect of the proteinaceous toxins in Ph. nodorum (ToxA, Tox1 and Tox3) is so strong that the possible activity of the SMs may be masked in vivo.
Figure 5.
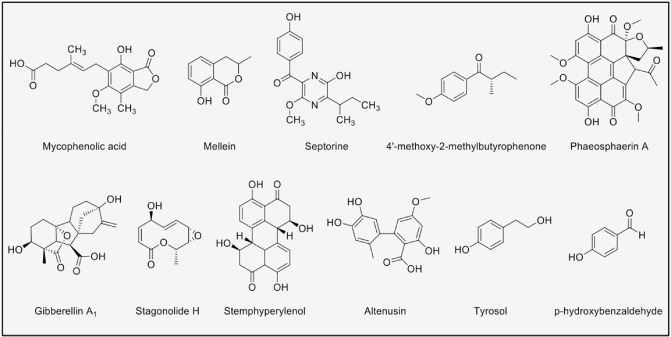
Secondary metabolites isolated from Phaeosphaeria and Botryosphaeria.
Other Phaeosphaeria species, including pathogens, endophytes, saprotrophs, endolichenic fungi as well as those found in aquatic environments, are the source of more than 75 reported natural products. These include the aforementioned perylenequinones (Li et al., 2012), the gibberellins (Sassa et al., 1989) and the phytotoxic stagonolides that could be involved in the bio‐control of weeds (Evidente et al., 2008) (Fig. 5). This indicates the potential of this genus to be a rich source of SMs with a wide diversity of activities and structures.
Botryosphaeria
Similar to Phaeosphaeria, most of the compounds reported from Botryosphaeria (Fig. 5) do not come from plant pathogens, even though many species from this genus are plant‐associated microorganisms. Botryosphaeria is mainly known for its forestry and agricultural pathogens. Amongst these, Botryosphaeria obtusa, a pathogen of apples and grapevines, has been studied chemically, leading to the commonly isolated mellein and its derivatives, as well as tyrosol and p‐hydroxybenzaldehyde. All these showed phytotoxicity in apple, but there was no correlation between the amount of compound produced by the strain and its pathogenicity (Djoukeng et al., 2009; Venkatasubbaiah and Chilton, 1990; Venkatasubbaiah et al., 1991). It is hypothesized that most, if not all, Botryosphaeria have an endophytic stage (Slippers and Wingfield, 2007); thus, it is not surprising that the majority of SMs reported from this genus come from endophytes. In a recent study in an endophytic Botryosphaeria dothidea from Melia azedarach, 18 SMs were isolated with a range of activities (Xiao et al., 2014). For instance, the perylenequinone derivative, stemphyperylenol, presented cytotoxic activity, and also showed, together with AOH, AME and other metabolites, toxicity against Alternaria solani; however, altenusin, with an opened lactone AME structure, showed an activity that its cyclic counterparts lacked: it scavenged free radicals (Xiao et al., 2014). This activity could be an important mechanism in plant‐associated microorganisms, as plant defence responses include the production of reactive oxygen species. It is interesting to consider, however, that many of the SMs reported from endophytic Botryosphaeria are phytotoxic. For example, Botryosphaeria mamane and Botryosphaeria parva, endophytes from Garcinia mangostana and Eugenia jambolana, respectively, produce mellein and other phytotoxic derivatives, raising the question as to what roles these compounds play during interaction (Araujo et al., 2012; Pongcharoen et al., 2007).
The Future of SM Research in the Dothideomycetes and the Role of Emerging Technologies
The above overview of Dothideomycete SMs provides a snapshot of their diversity, in terms of both their chemical structures and bioactivities. It suggests that members of the Dothideomycetes are a good source of phytotoxic compounds, which is not surprising given the numbers of plant‐associated fungi in this class. Interestingly, several of these SMs have been isolated from endophytes, such as the cochlioquinones and the mellein family, highlighting the fact that phytotoxicity may be a coarse measure to explain their biological role. It is plausible that these ‘phytotoxic’ substances may be involved in tuning the interaction between plant and microorganism at subtoxic concentrations, thus supporting the argument against equating laboratory‐observed biological activities to biological roles. In addition, there remains a lack of knowledge of their biosynthesis and the genes that encode them for many of the SMs presented above. From our perspective, one of the aspects of SMs in which our understanding is still lacking is the natural functions of SMs. What are their biological functions?
However, it would also seem clear that we have really only touched the surface in terms of the identification of what SMs are produced by Dothideomycete fungi. A large number of fungal SMs have been discovered by traditional application‐driven natural product screening programmes based on bioactivity‐guided isolations, e.g. many Dothideomycete phytotoxins have been found with the aim of obtaining mycoherbicides. Although these studies have enriched our understanding of SMs, they are also limited, as the fungus is typically grown in artificial conditions and the assays focus on a specific desired activity. Rarely do these studies consider the natural environment of the organism. During in vivo growth, SMs would be produced by fungi only when required and under specific conditions in order to economize resources. Furthermore, if we compare the number of SM gene clusters in the sequenced Dothideomycete genomes with the number of identified molecules, it is clear that our knowledge of secondary metabolism in this broad class of fungi is limited at best.
Indeed, the availability of fungal genome sequences has revolutionized SM research in many ways. It has allowed us to obtain a complete inventory of the SM biosynthesis genes of a fungus and to systematically determine the SM product(s) of each SM gene cluster (see below: Identification of cryptic SMs). Apart from facilitating the identification and characterization of the genes involved in the biosynthesis of SMs, comparative genomics and transcriptomics have allowed the inference of the evolutionary and functional role of SMs. For example, comparative genomics of 18 Dothideomycete fungi has revealed that the specialization of SMs and the diversification of their biosynthetic machinery play major roles in the adaptation of an organism to its lifestyle (Ohm et al., 2012). By comparing two closely related dothideomycete pathogens, Cl. fulvum and D. septosporum, with different hosts and lifestyles, it was shown that the evolution of SM biosynthesis genes, via gene gain, gene loss, changes in gene regulation and pseudogenization, plays an important role in the adaptation of the organisms to different environmental niches (de Wit et al., 2012). Nevertheless, to advance our understanding of the biological and ecological roles of SMs in fungi, it is important to: (i) study the SM molecules produced by the organisms in different natural environments and correlate with possible functions; (ii) identify these molecules; and (iii) elucidate or validate their biological functions.
In the next section, we focus on the current approaches and technologies available for the comparative study of the SM molecules produced by the organisms in different natural environments and the identification of these bio‐ecologically relevant molecules.
Unveiling the roles of fungal SMs
An alternative to activity‐based screenings could be ecological role‐based screenings. Ecological role‐based screenings look for SMs using the whole producer organism in different environmental conditions, rather than focusing on testing extracts, fractions and pure compounds. Broadly, these screenings compare SM profiles between a control state and a challenged state; SMs that are induced during the challenged state may have a role in organism adaptation to the challenge. A plant pathogen may be challenged with the presence of different hosts or tissues, by introducing an antagonistic microorganism, by changing the substrate in which it grows or by subjecting it to various abiotic stresses, such as water stress, heat stress, mineral deprivation, etc. This is followed by the analysis of the SM profiles of the organisms at different states. Similarly, instead of comparing SM production from challenged and unchallenged states, SM profiles from different developmental or life cycle stages can be compared.
New analytical techniques using mass spectrometry (MS) and nuclear magnetic resonance (NMR)
MS systems hyphenated with chromatographic devices are powerful tools for microbial SM profiling (Kersten et al., 2010). MS essentially gives the molecular mass of the compounds analysed. When high‐resolution MS (HRMS) is employed, the molecular formula can be deduced. MS can provide some structural data if fragmentation occurs; for this purpose tandem MS (MSn) is employed, which produces a characteristic fragmentation spectrum (Niessen, 2006). Extracts from the control and challenged samples are analysed, obtaining the retention time, exact mass and, if possible, fragmentation spectra of the components in these extracts. The collected data for each treatment are SM profiles that can be compared to highlight the differences and to select the compounds to be identified.
Nonetheless, it is seldom that either HRMS or MSn provides sufficient structural data to unequivocally identify a substance (Seger et al., 2013). Commonly, a purification process follows the screening and the purified compound is identified using NMR, the main technique to elucidate the structure of natural products (Halabalaki et al., 2014). Despite the advantages of MS (less expensive, greater sensitivity, shorter analysis times and easier data to interpret), NMR provides detailed structural information in a non‐destructive way that allows the de novo determination of a compound's structure (Sarker and Nahar, 2012; Wolfender, 2009). The full capability of NMR, however, can be limited by the time required (relative to MS) to generate sufficient data at low analyte concentrations. Because of this, NMR is being replaced by stop flow or online solid phase extraction coupled with liquid chromatography (LC‐SPE‐NMR) (Seger et al., 2013). The field has recently further advanced with the multiplexing of a combination of MS, NMR and other analytical techniques, providing platforms which are now being used for de‐replication, novel compound discovery and de novo identification (Sturm and Seger, 2012). Such analyses to date have been predominantly restricted to the plant natural product field and, to our knowledge, there are no reports in which these advanced analytical techniques have been applied to identify fungal SMs. However, these approaches offer immense potential for the deciphering of fungal SMs and their roles, and there is little doubt that, as they become more accessible, these techniques will improve the throughput of fungal SM discovery. For further information on these techniques, interested readers are directed to Li et al. (2013).
Imaging MS (IMS) is emerging as a powerful technique for the study of the ecological function of SMs in microorganisms by allowing the targeted or untargeted analysis of the spatial and temporal production of SMs in response to specific environmental stimuli. IMS overlaps spectrometric data of locally desorbed and ionized samples over the image of these samples, mapping the distribution of compounds over a two‐dimensional render (So et al., 2013; Watrous and Dorrestein, 2011). For example, IMS has been used to analyse the growth of Streptomyces coelicolor when co‐cultured with other actinomycetes. Several SMs were found to be selectively induced by the interaction, and some SMs were shown to be antagonist specific (Hopwood, 2013; Traxler et al., 2013). When such analytical systems are not available, targeted gas chromatography (GC‐MS) or liquid chromatography (LC‐MS) may be used to analyse extracts from different sections of a sample, allowing the localization of SM production. This approach identified O‐methylmellein in the interaction zones from plates containing Botryosphaeria obtusa and Eutypa lata using LC‐MS and NMR (Glauser et al., 2009). Both fungi are common elements in the mix of microorganisms causing esca disease in grapevines. Although extracts obtained from pure cultures did not show any activity, those present in the interaction zones were phytotoxic and fungitoxic. Consistent with this observation, O‐methylmellein showed germination inhibition of both plant and fungi. Researchers propose that the SMs produced by esca fungi in order to interact with each other could also drive the disease of the host (Glauser et al., 2009). It is clear that the emergence of these advanced analytical techniques is unveiling not just new SMs, but also novel biological and ecological interactions, highlighting their importance in the future of fungal SM research.
Identification of cryptic SMs
Even with the latest analytical technology described above for compound resolution and identification, at some point, all SM investigations require the isolation of the pure compound for structural elucidation and/or for the determination of the mechanism of action. However, these isolations may prove difficult if SMs are minor components in the extract or if they are not expressed in laboratory culture conditions. In the case of plant‐associated fungi, some SMs may only express in planta at levels that are difficult to isolate from the complex plant metabolic background. However, techniques are emerging to facilitate the biosynthesis and purification of these SMs for subsequent study (Lim et al., 2012; Scherlach and Hertweck, 2009; Takahashi et al., 2013).
Modification of culture conditions
As mentioned above, the in vitro culture of Dothideomycete fungi is likely to limit the expression of SMs. As some SMs are known to be antimicrobial and to inhibit the growth of competitors, perhaps growth in a heterogeneous environment is necessary to trigger the production of SMs not normally observed in homogeneous cultures (O'Brien and Wright, 2011). For example, it has been shown that Streptomyces rapamycinus triggers chromatin remodelling through histone modification in Aspergillus nidulans, leading to the expression of silent SM genes in the fungus (Nützmann et al., 2011). Several investigations to date have proven the success of co‐culturing studies, demonstrating their worth not just for scientific research, but also for SM production at industrial levels (Bader et al., 2010; Nützmann et al., 2012). Similarly, based on the fact that differences in the culture conditions can lead to different SM profiles, Bode et al. (2002) isolated more than 100 compounds from only six microbial strains, proposing ‘One Strain MAny Compounds’ (OSMAC), which refers to a systematic modification of the medium composition and culture conditions of the microbial fermentations.
Epigenetic regulation
Although the use of culture conditions to activate or promote cryptic and minor SMs has proven to be effective, perhaps most potential lies in the use of genetic‐based approaches to decrypt Dothideomycete secondary metabolism. For example, several global SM regulators have been identified and exploited for the activation of silent SM gene clusters in fungi (Brakhage and Schroeckh, 2011). It has been demonstrated that the overexpression of a global SM regulator, laeA, increases penicillin and lovastatin, but not sterigmatocystin, production in As. nidulans (Bok and Keller, 2004). Overexpression of laeA also leads to the production of terrequinone A, a cryptic metabolite not previously described from As. nidulans. LaeA works at a transcriptional level, but has no homology to any transcription factor; it has a methyltransferase domain and is involved in histone methylation, regulating SM production by restructuring the chromatin (Keller et al., 2005). LaeA, together with VeA and VelB, form the velvet complex, which is involved in sexual development and in the regulation of secondary metabolism at the global level (Bayram et al., 2008). Other chromatin‐regulating genes, a histone deacetylase hdaA and a histone methylase cclA from As. nidulans, appear to have contrasting action to laeA in that they suppress the expression of several SMs (Bok et al., 2009; Shwab et al., 2007). In addition, a mechanism of chromatin regulation was found in Fusarium graminearum involving silencing by a trimethylated histone H3 lysine 27 (H3K27me3); transcriptomic analysis of mutants without H3K27 methyltransferase showed that 14% more genes were expressed (Connolly et al., 2013). Epichloe festucae also presents this type of mechanism, which is linked to its characteristic endophytic lifestyle (Chujo and Scott, 2014). However, H3K27 methylation has not been observed in all organisms; Aspergillus, a model for the study of SM regulation, lacks this mechanism, showing how there is much still to be learned about the different layers of gene regulation (Connolly et al., 2013). Chromatin regulation of SM gene clusters has also been manipulated through the use of epigenetic modifiers (Williams et al., 2008). For example, supplementation of potato dextrose with histone deacetylase chemical inhibitors led to the discovery of 4′‐methoxy‐(2S)‐methylbutyrophenone in Ph. nodorum (Yang et al., 2013).
Molecular biology of SM pathway activation
The above‐mentioned methods are not SM specific and approaches targeting particular SM biosynthetic genes may be needed. Traditionally, the linkages between gene clusters and SMs are identified by genetic knockouts and mutant complementation, but this will not be applicable if the gene cluster of interest is silent. Recent advances in our understanding of SM biosynthetic genes and the development of bioinformatics tools, such as SMURF (Khaldi et al., 2010) and AntiSMASH 2.0 (Medema et al., 2011), have accelerated the identification of SM gene clusters in sequenced fungal genomes. These resources expedite the identification of SM gene clusters within individual fungal genomes and allow a targeted approach to acquire SM products encoded by specific gene clusters. One direct approach is by exchanging the promoters of individual genes of an SM gene cluster in the native host; however, this could be cumbersome if the gene cluster of interest contains many genes. Two main strategies can be exploited to characterize specific SMs or gene clusters of interest: (i) pathway‐specific regulatory gene manipulation; and (ii) heterologous expression of SM clusters. These two strategies not only allow the isolation of cryptic or minor SMs, but also simultaneously link the genes or clusters to the SM products.
The basis of manipulating the regulation of specific pathways lies in the fact that some clusters contain genes encoding for transcription factors that activate the biosynthesis of the SMs coded within that cluster (Yin and Keller, 2011). Pioneered by Bergmann et al. (2007), it was found that a single ectopic insertion of apdR, a putative Zn2Cys6‐type activator gene in a silent gene cluster, encodes for a hybrid PKS–NRPS pathway. The activation of apdR promotes the expression of the novel aspyridones and the cryptic products within the cluster. This strategy has been employed with success in several other laboratories, especially amongst the Aspergillus species (Chiang et al., 2009; Zabala et al., 2012). However, not all SM gene clusters are regulated by a pathway‐specific regulator and other complementing strategies are required.
Heterologous expression
As an alternative to overcome the SM pathway activation difficulties, an emerging approach is to transfer the SM genes of interest into a well‐characterized heterologous host, such as Saccharomyces cerevisiae or Escherichia coli. Successful attempts have been made in expressing SM genes in S. cerevisiae by incorporating a phosphopantetheinyl transferase gene, npgA, for post‐translational modification of acyl carrier protein domain‐containing enzymes, which include the PKSs and NRPSs (Ma et al., 2009; Tsunematsu et al., 2013). However the expression of fungal SMs in standard laboratory organisms is technically challenging because of the lack of appropriate intron splicing and post‐translational modification machinery in these models (Schümann and Hertweck, 2006). Thus, there is an advantage of using filamentous fungi, such as the well‐characterized Aspergillus spp., for the expression of SM gene clusters, as there is a high possibility that the fungal host has compatible intron splicing and post‐translational machinery. However, heterologous expression of SM clusters in a filamentous fungal host is often complicated by the metabolic complexity of the host and the possibility that the host biosynthetic enzymes cross‐talk with the heterologous pathway. For this reason, a strain of As. nidulans that lacks many of its SM clusters is being developed as a ‘clean host’ for the heterologous expression of fungal SMs (Chiang et al., 2013).
The Future of Dothideomycete SM Research Is Promising
A clear theme that can be garnered from this review is that, although we have made significant progress towards the understanding of secondary metabolism in Dothideomycete fungi, there remains much to learn. For example, cryptic SMs and orphan genes are a rule and not the exception, the biological role of individual SMs is unknown and their biosynthetic regulation is often poorly understood. However, the pieces of the puzzle are starting to come together. The next‐generation sequencing revolution means that access to genome sequences is no longer limited to models, and the bioinformatics tools are now available to readily identify SM clusters within these genomes. The analytical chemistry tools needed to identify and characterize SMs are rapidly advancing in resolution and, importantly, are becoming more easily available to researchers in the field. The emergence of synthetic biology tools to enable the expression and analysis of SM clusters that typically appear silent will start to shed light on SMs we previously could not study. It is also apparent that the field is now beginning to recognize that, in order to understand the roles of these SMs, we must look for and characterize them in their natural environment, and not necessarily in a laboratory fermentation. The future of this field is very promising. Given the convergence of resources and understanding, we feel that the study of Dothideomycete secondary metabolism is about to come of age.
Acknowledgements
PSS is an Australian Research Council Future Fellow (FT110100698). MJM‐G is a recipient of an Australian Government Endeavour Award and a Mexican CONACYT scholarship. Y‐HC is an Australian Research Council DECRA Fellow (DE130101350).
References
- Akimitsu, K. , Tsuge, T. , Kodama, M. , Yamamoto, M. and Otani, H. (2013) Alternaria host‐selective toxins: determinant factors of plant disease. J. Gen. Plant Pathol. 80, 1–14. [Google Scholar]
- Aly, A. , Debbab, A. and Proksch, P. (2011) Fifty years of drug discovery from fungi. Fungal Divers. 50, 3–19. [Google Scholar]
- Araujo, A.R. , Monfardini, J.D. , Chapla, V.M. , Lopes, M.N. , Silva, D.H.S. , Cavalheiro, A.J. and da S Bolzani, V. (2012) Dihydroisocoumarins produced by Botryosphaeria parva an endophytic fungus from Eugenia jambolana . Planta Med. 78, PI87. [Google Scholar]
- Arnold, A.E. (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol. Rev. 21, 51–66. [Google Scholar]
- Au, T.K. , Chick, W.S.H. and Leung, P.C. (2000) The biology of ophiobolins. Life Sci. 67, 733–742. [DOI] [PubMed] [Google Scholar]
- Bach, E. , Christensen, S. , Dalgaard, L. , Larsen, P.O. , Olsen, C.E. and Smedegård‐Petersen, V. (1979) Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres . Physiol. Plant Pathol. 14, 41–46. [Google Scholar]
- Bader, J. , Mast‐Gerlach, E. , Popović, M.K. , Bajpai, R. and Stahl, U. (2010) Relevance of microbial coculture fermentations in biotechnology. J. Appl. Microbiol. 109, 371–387. [DOI] [PubMed] [Google Scholar]
- Barnes, I. , Crous, P.W. , Wingfield, B.D. and Wingfield, M.J. (2004) Multigene phylogenies reveal that red band needle blight of Pinus is caused by two distinct species of Dothistroma, D. septosporum and D. pini . Stud. Mycol. 50, 551–565. [Google Scholar]
- Bayram, Ö. , Krappmann, S. , Ni, M. , Bok, J.W. , Helmstaedt, K. , Valerius, O. , Braus‐Stromeyer, S. , Kwon, N.‐J. , Keller, N.P. , Yu, J.‐H. and Braus, G.H. (2008) VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science, 320, 1504–1506. [DOI] [PubMed] [Google Scholar]
- Bennett, J.W. and Klich, M. (2003) Mycotoxins. Clin. Microbiol. Rev. 16, 497–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, S. , Schumann, J. , Scherlach, K. , Lange, C. , Brakhage, A.A. and Hertweck, C. (2007) Genomics‐driven discovery of PKS‐NRPS hybrid metabolites from Aspergillus nidulans . Nat. Chem. Biol. 3, 213–217. [DOI] [PubMed] [Google Scholar]
- Bode, H.B. , Bethe, B. , Höfs, R. and Zeeck, A. (2002) Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem, 3, 619–627. [DOI] [PubMed] [Google Scholar]
- Bok, J.W. and Keller, N.P. (2004) LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell, 3, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok, J.W. , Chiang, Y.‐M. , Szewczyk, E. , Reyes‐Dominguez, Y. , Davidson, A.D. , Sanchez, J.F. , Lo, H.‐C. , Watanabe, K. , Strauss, J. , Oakley, B.R. , Wang, C.C.C. and Keller, N.P. (2009) Chromatin‐level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5, 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras, N. and Strelkov, S.E. (2008) The anthraquinone catenarin is phytotoxic and produced in leaves and kernels of wheat infected by Pyrenophora tritici‐repentis . Physiol. Mol. Plant Pathol. 72, 87–95. [Google Scholar]
- Bradshaw, R.E. , Slot, J.C. , Moore, G.G. , Chettri, P. , de Wit, P.J.G.M. , Ehrlich, K.C. , Ganley, A.R.D. , Olson, M.A. , Rokas, A. , Carbone, I. and Cox, M.P. (2013) Fragmentation of an aflatoxin‐like gene cluster in a forest pathogen. New Phytol. 198, 525–535. [DOI] [PubMed] [Google Scholar]
- Brakhage, A.A. and Schroeckh, V. (2011) Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. [DOI] [PubMed] [Google Scholar]
- Briggs, D.E. (1966) Gibberellin‐like activity of helminthosporol and helminthosporic acid. Nature, 210, 418–419.5963237 [Google Scholar]
- Bury, M. , Novo‐Uzal, E. , Andolfi, A. , Cimini, S. , Wauthoz, N. , Heffeter, P. , Lallemand, B. , Avolio, F. , Delporte, C. , Cimmino, A. , Dubois, J. , Van Antwerpen, P. , Zonno, M.C. , Vurro, M. , Poumay, Y. , Berger, W. , Evidente, A. , De Gara, L. , Kiss, R. and Locato, V. (2013) Ophiobolin A, a sesterterpenoid fungal phytotoxin, displays higher in vitro growth‐inhibitory effects in mammalian than in plant cells and displays in vivo antitumor activity. Int. J. Oncol. 43, 575–585. [DOI] [PubMed] [Google Scholar]
- Campos, F.F. , Rosa, L.H. , Cota, B.B. , Caligiorne, R.B. , Teles Rabello, A.L. , Alves, T.M.A. , Rosa, C.A. and Zani, C.L. (2008) Leishmanicidal metabolites from Cochliobolus sp., an endophytic fungus isolated from Piptadenia adiantoides (Fabaceae). Plos Negl. Trop. Dis. 2, e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso, R. , Evidente, A. , Randazzo, G. , Ritieni, A. , Bottalico, A. , Vurro, M. and Logrieco, A. (1987) Isolation of cytochalasins A and B from Ascochyta heteromorpha . J. Nat. Prod. 50, 989–990. [DOI] [PubMed] [Google Scholar]
- Capasso, R. , Evidente, A. , Ritieni, A. , Randazzo, G. , Vurro, M. and Bottalico, A. (1988) Ascochalasin, a new cytochalasin from Ascochyta heteromorpha . J. Nat. Prod. 51, 567–571. [DOI] [PubMed] [Google Scholar]
- Capasso, R. , Evidente, A. and Vurro, M. (1991) Cytochalasins from Phoma exigua var. heteromorpha . Phytochemistry, 30, 3945–3950. [DOI] [PubMed] [Google Scholar]
- Carruthers, J.R. , Cerrini, S. , Fedeli, W. , Casinovi, C.G. , Galeffi, C. , Vaccaro, A.M.T. and Scala, A. (1971) Structures of cochlioquinones A and B, new metabolites of Cochliobolus miyabeanus: chemical and X‐ray crystallographic determination. J. Chem. Soc. D, 3, 164–166. [Google Scholar]
- Chiang, Y.‐M. , Szewczyk, E. , Davidson, A.D. , Keller, N. , Oakley, B.R. and Wang, C.C.C. (2009) A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans . J. Am. Chem. Soc. 131, 2965–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, Y.‐M. , Oakley, C.E. , Ahuja, M. , Entwistle, R. , Schultz, A. , Chang, S.‐L. , Sung, C.T. , Wang, C.C.C. and Oakley, B.R. (2013) An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans . J. Am. Chem. Soc. 135, 7720–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, R. , Minami, A. , Gomi, K. and Oikawa, H. (2013) Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus . Org. Lett. 15, 594–597. [DOI] [PubMed] [Google Scholar]
- Chujo, T. and Scott, B. (2014) Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte–plant symbiosis. Mol. Microbiol. 92, 413–434. [DOI] [PubMed] [Google Scholar]
- Ciuffetti, L.M. , Manning, V.A. , Pandelova, I. , Betts, M.F. and Martinez, J.P. (2010) Host‐selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici‐repentis–wheat interaction. New Phytol. 187, 911–919. [DOI] [PubMed] [Google Scholar]
- Collemare, J. and Lebrun, M.‐H. (2011) Fungal secondary metabolites: ancient toxins and novel effectors in plant–microbe interactions In: Effectors in Plant–Microbe Interactions (Martin F. and Kamoun S., eds.), pp. 377–400. Oxford, UK: Wiley‐Blackwell. [Google Scholar]
- Collemare, J. , Griffiths, S. , Iida, Y. , Karimi Jashni, M. , Battaglia, E. , Cox, R.J. and de Wit, P.J.G.M. (2014) Secondary metabolism and biotrophic lifestyle in the tomato pathogen Cladosporium fulvum . PLoS ONE, 9, e85877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon, B.J. , Leng, Y. , Wu, D. , Bushley, K.E. , Ohm, R.A. , Otillar, R. , Martin, J. , Schackwitz, W. , Grimwood, J. , MohdZainudin, N. , Xue, C. , Wang, R. , Manning, V.A. , Dhillon, B. , Tu, Z.J. , Steffenson, B.J. , Salamov, A. , Sun, H. , Lowry, S. , LaButti, K. , Han, J. , Copeland, A. , Lindquist, E. , Barry, K. , Schmutz, J. , Baker, S.E. , Ciuffetti, L.M. , Grigoriev, I.V. , Zhong, S. and Turgeon, B. (2013) Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. Plos Genet. 9, e1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, L.R. , Smith, K.M. and Freitag, M. (2013) The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. Plos Genet. 9, e1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe, B. , Mander, L. , Paleg, L. and Turner, J. (1974) Gibberellin‐like activity of helminthosporic acid analogues. Funct. Plant Biol. 1, 473–481. [Google Scholar]
- Coval, S.J. , Hradil, C.M. , Lu, H.S.M. , Clardy, J. , Satouri, S. and Strobel, G.A. (1990) Pyrenoline‐A and ‐B, two new phytotoxins from Pyrenophora teres . Tetrahedron Lett. 31, 2117–2120. [Google Scholar]
- Crous, P.W. (2009) Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Divers. 38, 1–24. [Google Scholar]
- Cutler, H.G. , Crumley, F.G. , Cox, R.H. , Springer, J.P. , Arrendale, R.F. , Cole, R.J. and Cole, P.D. (1984) Ophiobolins G and H: new fungal metabolites from a novel source, Aspergillus ustus . J. Agric. Food Chem. 32, 778–782. [Google Scholar]
- Daub, M. and Chung, K.‐R. (2009) Photoactivated perylenequinone toxins in plant pathogenesis In: Plant Relationships (Deising H., ed.), pp. 201–219. Berlin, Heidelberg: Springer. [Google Scholar]
- Daub, M.E. and Ehrenshaft, M. (2000) The photoactivated Cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 38, 461–490. [DOI] [PubMed] [Google Scholar]
- Dawson, M.J. , Farthing, J.E. , Marshall, P.S. , Middleton, R.F. , O'Neill, M.J. , Shuttleworth, A. , Stylli, C. , Tait, R.M. , Taylor, P.M. , Wildman, H.G. , Buss, A.D. , Langley, D. and Hayes, M.V. (1992) The squalestatins, novel inhibitors of squalene synthase produced by a species of Phoma. I. Taxonomy, fermentation, isolation, physico‐chemical properties and biological activity. J. Antibiot. 45, 639–647. [DOI] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys, M. , Bousquet, J.F. , Kollmann, A. and Barbier, M. (1980) Dihydroisocoumarines et acide mycophenolique du milieu de culture du champignon phytopathogene Septoria nodorum . Phytochemistry, 19, 2221–2222. [Google Scholar]
- Devys, M. , Barbier, M. , Kollmann, A. and Bousquet, J.F. (1982) Septorine and N‐methoxy septorine, substituted pyrazines from the fungus Septoria nodorum Berk. Tetrahedron Lett. 23, 5409–5412. [Google Scholar]
- Devys, M. , Ferezou, J.‐P. , Topgi, R.S. , Barbier, M. , Bousquet, J.‐F. and Kollmann, A. (1984) Structure and biosynthesis of phomenoic acid, an antifungal compound isolated from Phoma lingam Tode. J. Chem. Soc., Perkin Trans. 1, 2133–2137. [Google Scholar]
- Devys, M. , Barbier, M. , Bousquet, J.F. and Kollmann, A. (1994) Isolation of the (–)‐(3R)‐5‐hydroxymellein from the fungus Septoria nodorum . Phytochemistry, 35, 825–826. [Google Scholar]
- Djoukeng, J. , Polli, S. , Larignon, P. and Abou‐Mansour, E. (2009) Identification of phytotoxins from Botryosphaeria obtusa, a pathogen of black dead arm disease of grapevine. Eur. J. Plant Pathol. 124, 303–308. [Google Scholar]
- Elliott, C.E. , Callahan, D.L. , Schwenk, D. , Nett, M. , Hoffmeister, D. and Howlett, B.J. (2013) A gene cluster responsible for biosynthesis of phomenoic acid in the plant pathogenic fungus, Leptosphaeria maculans . Fungal Genet. Biol. 53, 50–58. [DOI] [PubMed] [Google Scholar]
- Ellwood, S. , Liu, Z. , Syme, R. , Lai, Z. , Hane, J. , Keiper, F. , Moffat, C. , Oliver, R. and Friesen, T. (2010) A first genome assembly of the barley fungal pathogen Pyrenophora teres f. teres . Genome Biol. 11, R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evidente, A. , Lanzetta, R. , Capasso, R. , Vurro, M. and Bottalico, A. (1992) Cytochalasins U and V, two new cytochalasans, from Phoma exigua var. heteromorpha . Tetrahedron, 48, 6317–6324. [Google Scholar]
- Evidente, A. , Capasso, R. , Abouzeid, M.A. , Lanzetta, R. , Vurro, M. and Bottalico, A. (1993a) Three new toxic pinolidoxins from Ascochyta pinodes . J. Nat. Prod. 56, 1937–1943. [Google Scholar]
- Evidente, A. , Capasso, R. , Vurro, M. and Bottalico, A. (1993b) Ascosalitoxin, a phytotoxic trisubstituted salicylic aldehyde from Ascochyta pisi . Phytochemistry, 34, 995–998. [Google Scholar]
- Evidente, A. , Capasso, R. , Vurro, M. and Bottalico, A. (1996) Cytochalasin W, a new 24‐oxa[14]cytochalasan from Phoma exigua var. heteromorpha . Nat. Toxins, 4, 53–57. [DOI] [PubMed] [Google Scholar]
- Evidente, A. , Cimmino, A. , Berestetskiy, A. , Andolfi, A. and Motta, A. (2008) Stagonolides G−I and modiolide A, nonenolides produced by Stagonospora cirsii, a potential mycoherbicide for Cirsium arvense . J. Nat. Prod. 71, 1897–1901. [DOI] [PubMed] [Google Scholar]
- Ferezou, J.‐P. , Quesneau‐Thierry, A. , Servy, C. , Zissmann, E. and Barbier, M. (1980) Sirodesmin PL biosynthesis in Phoma lingam tode. J. Chem. Soc., Perkin Trans. 1, 1739–1746. [Google Scholar]
- Friesen, T.L. , Stukenbrock, E.H. , Liu, Z. , Meinhardt, S. , Ling, H. , Faris, J.D. , Rasmussen, J.B. , Solomon, P.S. , McDonald, B.A. and Oliver, R.P. (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. [DOI] [PubMed] [Google Scholar]
- Friis, P. , Olsen, C.E. and Moller, B.L. (1991) Toxin production in Pyrenophora teres, the ascomycete causing the net‐spot blotch disease of barley (Hordeum vulgare L.). J. Biol. Chem. 266, 13 329–13 335. [PubMed] [Google Scholar]
- Fukushima, Y. , Sakagami, Y. and Marumo, S. (1993) β‐Glucan biosynthesis inhibitors isolated from fungi as hyphal malformation inducer. Bioorg. Med. Chem. Lett. 3, 1219–1222. [Google Scholar]
- Gardiner, D.M. , Cozijnsen, A.J. , Wilson, L.M. , Pedras, M.S.C. and Howlett, B.J. (2004) The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans . Mol. Microbiol. 53, 1307–1318. [DOI] [PubMed] [Google Scholar]
- Gilbert, B.M. and Wolpert, T.J. (2013) Characterization of the LOV1‐mediated, victorin‐induced, cell‐death response with virus‐induced gene silencing. Mol. Plant–Microbe Interact. 26, 903–917. [DOI] [PubMed] [Google Scholar]
- Glauser, G. , Gindro, K. , Fringeli, J. , De Joffrey, J.‐P. , Rudaz, S. and Wolfender, J.‐L. (2009) Differential analysis of mycoalexins in confrontation zones of grapevine fungal pathogens by ultrahigh pressure liquid chromatography/time‐of‐flight mass spectrometry and capillary nuclear magnetic resonance. J. Agric. Food Chem. 57, 1127–1134. [DOI] [PubMed] [Google Scholar]
- Goudet, C. , Milat, M.‐L. , Sentenac, H. and Thibaud, J.‐B. (2000) Beticolins, nonpeptidic, polycyclic molecules produced by the phytopathogenic fungus Cercospora beticola, as a new family of ion channel‐forming toxins. Mol. Plant–Microbe Interact. 13, 203–209. [DOI] [PubMed] [Google Scholar]
- Halabalaki, M. , Vougogiannopoulou, K. , Mikros, E. and Skaltsounis, A.L. (2014) Recent advances and new strategies in the NMR‐based identification of natural products. Curr. Opin. Biotechnol. 25, 1–7. [DOI] [PubMed] [Google Scholar]
- Hallock, Y.F. , Lu, H.S.M. , Clardy, J. , Strobel, G.A. , Sugawara, F. , Samsoedin, R. and Yoshida, S. (1993) Triticones, spirocyclic lactams from the fungal plant pathogen Drechslera tritici‐repentis . J. Nat. Prod. 56, 747–754. [Google Scholar]
- Hamid, K. and Strange, R.N. (2000) Phytotoxicity of solanapyrones A and B produced by the chickpea pathogen Ascochyta rabiei (Pass.) Labr. and the apparent metabolism of solanapyrone A by chickpea tissues. Physiol. Mol. Plant Pathol. 56, 235–244. [Google Scholar]
- Hane, J.K. , Lowe, R.G.T. , Solomon, P.S. , Tan, K.‐C. , Schoch, C.L. , Spatafora, J.W. , Crous, P.W. , Kodira, C. , Birren, B.W. , Galagan, J.E. , Torriani, S.F.F. , McDonald, B.A. and Oliver, R.P. (2007) Dothideomycete–plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum . Plant Cell, 19, 3347–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, J.R. (2008) The Chemistry of Fungi. Cambridge: The Royal Society of Chemistry. [Google Scholar]
- Hardham, A.R. , Jones, D.A. and Takemoto, D. (2007) Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10, 342–348. [DOI] [PubMed] [Google Scholar]
- Härri, E. , Loeffler, W. , Sigg, H.P. , Stähelin, H. and Tamm, C. (1963) Über die Isolierung neuer stoffwechselprodukte aus Penicillium brefeldianum DODGE. Helv. Chim. Acta, 46, 1235–1243. [Google Scholar]
- Hazuda, D. , Blau, C.U. , Felock, P. , Hastings, J. , Pramanik, B. , Wolfe, A. , Bushman, F. , Farnet, C. , Goetz, M. , Williams, M. , Silverman, K. , Lingham, R. and Singh, S. (1999) Isolation and characterization of novel human immunodeficiency virus integrase inhibitors from fungal metabolites. Antivir. Chem. Chemother. 10, 63–70. [DOI] [PubMed] [Google Scholar]
- Heckman, D.S. , Geiser, D.M. , Eidell, B.R. , Stauffer, R.L. , Kardos, N.L. and Hedges, S.B. (2001) Molecular evidence for the early colonization of land by fungi and plants. Science, 293, 1129–1133. [DOI] [PubMed] [Google Scholar]
- Heiser, I. , Heß, M. , Schmidtke, K.‐U. , Vogler, U. , Miethbauer, S. and Liebermann, B. (2004) Fatty acid peroxidation by rubellin B, C and D, phytotoxins produced by Ramularia collo‐cygni (Sutton et Waller). Physiol. Mol. Plant Pathol. 64, 135–143. [Google Scholar]
- Hogenhout, S.A. , Van der Hoorn, R.A.L. , Terauchi, R. and Kamoun, S. (2009) Emerging concepts in effector biology of plant‐associated organisms. Mol. Plant–Microbe Interact. 22, 115–122. [DOI] [PubMed] [Google Scholar]
- Hopwood, D.A. (2013) Imaging mass spectrometry reveals highly specific interactions between actinomycetes to activate specialized metabolic gene clusters. MBio, 4, e00612‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horbach, R. , Navarro‐Quesada, A.R. , Knogge, W. and Deising, H.B. (2011) When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J. Plant Physiol. 168, 51–62. [DOI] [PubMed] [Google Scholar]
- Howlett, B.J. , Idnurm, A. and Pedras, M.S.C. (2001) Leptosphaeria maculans, the causal agent of blackleg disease of brassicas. Fungal Genet. Biol. 33, 1–14. [DOI] [PubMed] [Google Scholar]
- Humphreys, C.P. , Franks, P.J. , Rees, M. , Bidartondo, M.I. , Leake, J.R. and Beerling, D.J. (2010) Mutualistic mycorrhiza‐like symbiosis in the most ancient group of land plants. Nat. Commun. 1, 103. [DOI] [PubMed] [Google Scholar]
- Inderbitzin, P. , Asvarak, T. and Turgeon, B.G. (2010) Six new genes required for production of T‐toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant–Microbe Interact. 23, 458–472. [DOI] [PubMed] [Google Scholar]
- Ipcho, S.V.S. , Hane, J.K. , Antoni, E.A. , Ahren, D.A.G. , Henrissat, B. , Friesen, T.L. , Solomon, P.S. and Oliver, R.P. (2012) Transcriptome analysis of Stagonospora nodorum: gene models, effectors, metabolism and pantothenate dispensability. Mol. Plant Pathol. 13, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itai, A. , Nozoe, S. , Tsuda, K. , Okuda, S. , Iitaka, Y. and Nakayama, Y. (1967) The structure of cephalonic acid, a pentaprenyl terpenoid. Tetrahedron Lett. 8, 4111–4112. [DOI] [PubMed] [Google Scholar]
- Jalal, M.A.F. , Bilayet Hossain, M. , Robeson, D.J. and Van der Helm, D. (1992) Cercospora beticola phytotoxins: cebetins that are photoactive, Mg2+‐binding, chlorinated anthraquinone–xanthone conjugates. J. Am. Chem. Soc. 114, 5967–5971. [Google Scholar]
- Kachlicki, P. and Wakuliński, W. (2002) Analysis of anthraquinone pigments of Pyrenophora tritici‐repentis . Acta Agrobot. 53, 97–105. [Google Scholar]
- Kato, J. , Shiotani, Y. , Tamura, S. and Sakurai, A. (1964) A new plant growth‐promoting substance: helminthosporol. Naturwissenschaften, 51, 341. [Google Scholar]
- Kaur, S. (1995) Phytotoxicity of solanapyrones produced by the fungus Ascochyta rabiei and their possible role in blight of chickpea (Cicer arietinum). Plant Sci. 109, 23–29. [Google Scholar]
- Keller, N.P. , Turner, G. and Bennett, J.W. (2005) Fungal secondary metabolism: from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. [DOI] [PubMed] [Google Scholar]
- Kenfield, D. , Strobel, S. , Sugawara, F. , Berglund, D. and Strobel, G. (1988) Triticone A: a novel bioactive lactam with potential as a molecular probe. Biochem. Biophys. Res. Commun. 157, 174–182. [DOI] [PubMed] [Google Scholar]
- Kersten, R.D. , Meehan, M.J. and Dorrestein, P.C. (2010) Applications of modern mass spectrometry techniques in natural products chemistry In: Comprehensive Natural Products II (Lew M. and Hung‐Wen L., eds), pp. 389–456. Oxford: Elsevier. [Google Scholar]
- Khaldi, N. , Seifuddin, F.T. , Turner, G. , Haft, D. , Nierman, W.C. , Wolfe, K.H. and Fedorova, N.D. (2010) SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, P.M. , Ainsworth, G.G.C. and Bisby, G.R. (2008) Dictionary of the Fungi. UK: CAB International. [Google Scholar]
- Kobayashi, Y. , Kobayashi, I. , Funaki, Y. , Fujimoto, S. , Takemoto, T. and Kunoh, H. (1997a) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non‐host resistance in barley coleoptile cells. Plant J. 11, 525–537. [Google Scholar]
- Kobayashi, Y. , Yamada, M. , Kobayashi, I. and Kunoh, H. (1997b) Actin microfilaments are required for the expression of nonhost resistance in higher plants. Plant Cell Physiol. 38, 725–733. [Google Scholar]
- Kruys, Å. , Eriksson, O.E. and Wedin, M. (2006) Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycol. Res. 110, 527–536. [DOI] [PubMed] [Google Scholar]
- Kuyama, S. and Tamura, T. (1957) Cercosporin. A pigment of Cercosporina kikuchii Matsumoto et Tomoyasu. I. Cultivation of fungus, isolation and purification of pigment. J. Am. Chem. Soc. 79, 5725–5726. [Google Scholar]
- Latif, Z. , Strange, R.N. , Bilton, J. and Riazuddin, S. (1993) Production of the phytotoxins, solanapyrones A and C and cytochalasin D among nine isolates of Ascochyta rabiei . Plant Pathol. 42, 172–180. [Google Scholar]
- Leung, P.C. , Taylor, W.A. , Wang, J.H. and Tipton, C.L. (1984) Ophiobolin A. A natural product inhibitor of calmodulin. J. Biol. Chem. 259, 2742–2747. [PubMed] [Google Scholar]
- Leung, P.C. , Taylor, W.A. , Wang, J.H. and Tipton, C.L. (1985) Role of calmodulin inhibition in the mode of action of ophiobolin A. Plant Physiol. 77, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Wang, H. , Zhu, R. , Sun, L. , Wang, L. , Li, M. , Li, Y. , Liu, Y. , Zhao, Z. and Lou, H. (2012) Phaeosphaerins A–F, cytotoxic perylenequinones from an endolichenic fungus, Phaeosphaeria sp. J. Nat. Prod. 75, 142–147. [DOI] [PubMed] [Google Scholar]
- Li, P. , Zhang, Z. , Hu, X. and Zhang, Q. (2013) Advanced hyphenated chromatographic‐mass spectrometry in mycotoxin determination: current status and prospects. Mass Spectrom. Rev. 32, 420–452. [DOI] [PubMed] [Google Scholar]
- Lim, C.‐H. , Ueno, H. , Miyoshi, H. , Miyagawa, H. , Iwamura, H. and Ueno, T. (1996) Phytotoxic compounds cochlioquinones are inhibitors of mitochondrial NADH‐ubiquinone reductase. J. Pestic. Sci. 21, 213–215. [Google Scholar]
- Lim, C.‐H. , Miyagawa, H. , Akamatsu, M. , Nakagawa, Y. and Ueno, T. (1998) Structures and biological activities of phytotoxins produced by the plant pathogenic fungus Bipolaris cynodontis cynA. J. Pestic. Sci. 23, 281–288. [Google Scholar]
- Lim, F.Y. , Sanchez, J.F. , Wang, C.C.C. and Keller, N.P. (2012) Toward awakening cryptic secondary metabolite gene clusters in filamentous fungi In: Methods Enzymol (David A.H., ed.), pp. 303–324. USA: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorang, J.M. , Sweat, T.A. and Wolpert, T.J. (2007) Plant disease susceptibility conferred by a ‘resistance’ gene. Proc. Natl. Acad. Sci. USA, 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, J. , Fu, L. , Peng, Y. and Zhou, L. (2013) Metabolites from Alternaria fungi and their bioactivities. Molecules, 18, 5891–5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, S.M. , Li, J.W.‐H. , Choi, J.W. , Zhou, H. , Lee, K.K.M. , Moorthie, V.A. , Xie, X. , Kealey, J.T. , Da Silva, N.A. , Vederas, J.C. and Tang, Y. (2009) Complete reconstitution of a highly reducing iterative polyketide synthase. Science, 326, 589–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida, T. , Higashi, K. and Ogawara, H. (1995) Cochlioquinone A, an inhibitor of diacylglycerol kinase. J. Antibiot. 48, 1076–1080. [DOI] [PubMed] [Google Scholar]
- Manning, V.A. , Pandelova, I. , Dhillon, B. , Wilhelm, L.J. , Goodwin, S.B. , Berlin, A.M. , Figueroa, M. , Freitag, M. , Hane, J.K. , Henrissat, B. , Holman, W.H. , Kodira, C.D. , Martin, J. , Oliver, R.P. , Robbertse, B. , Schackwitz, W. , Schwartz, D.C. , Spatafora, J.W. , Turgeon, B.G. , Yandava, C. , Young, S. , Zhou, S. , Zeng, Q. , Grigoriev, I.V. , Ma, L.‐J. and Ciuffetti, L.M. (2013) Comparative genomics of a plant‐pathogenic fungus, Pyrenophora tritici‐repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3 (Bethesda), 3, 41–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary, K.L. , Slot, J.C. and Rokas, A. (2013) Physical linkage of metabolic genes in fungi is an adaptation against the accumulation of toxic intermediate compounds. Proc. Natl. Acad. Sci. USA, 110, 11 481–11 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema, M.H. , Blin, K. , Cimermancic, P. , de Jager, V. , Zakrzewski, P. , Fischbach, M.A. , Weber, T. , Takano, E. and Breitling, R. (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethbauer, S. , Haase, S. , Schmidtke, K.‐U. , Günther, W. , Heiser, I. and Liebermann, B. (2006) Biosynthesis of photodynamically active rubellins and structure elucidation of new anthraquinone derivatives produced by Ramularia collo‐cygni . Phytochemistry, 67, 1206–1213. [DOI] [PubMed] [Google Scholar]
- Miethbauer, S. , Gaube, F. , Möllmann, U. , Dahse, H.‐M. , Schmidtke, M. , Gareis, M. , Pickhardt, M. and Liebermann, B. (2009) Antimicrobial, antiproliferative, cytotoxic, and Tau inhibitory activity of rubellins and caeruleoramularin produced by the phytopathogenic fungus Ramularia collo‐cygni . Planta Med. 75, 1523–1525. [DOI] [PubMed] [Google Scholar]
- Milat, M.L. , Prange, T. , Ducrot, P.H. , Tabet, J.C. , Einhorn, J. , Blein, J.P. and Lallemand, J.Y. (1992) Structures of the beticolins, the yellow toxins produced by Cercospora beticola . J. Am. Chem. Soc. 114, 1478–1479. [Google Scholar]
- Miyagawa, H. , Nagai, S. , Tsurushima, T. , Sato, M. , Ueno, T. and Fukami, H. (1994) Phytotoxins produced by the plant pathogenic fungus Bipolaris bicolor El‐1. Biosci. Biotechnol. Biochem. 58, 1143–1145. [Google Scholar]
- Möbius, N. and Hertweck, C. (2009) Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 12, 390–398. [DOI] [PubMed] [Google Scholar]
- Mullbacher, A. , Waring, P. , Tiwari‐Palni, U. and Eichner, R.D. (1986) Structural relationship of epipolythiodioxopiperazines and their immunomodulating activity. Mol. Immunol. 23, 231–235. [DOI] [PubMed] [Google Scholar]
- Nakanishi, K. (1999) A brief history of natural products chemistry In: Comprehensive Natural Products Chemistry (Otto M.‐C., Sir Derek B. and NakanishiA2 K., eds), pp. 1–31. Oxford: Pergamon. [Google Scholar]
- Nelsen, M.P. , Lücking, R. , Mbatchou, J.S. , Andrew, C.J. , Spielmann, A.A. and Lumbsch, H.T. (2011) New insights into relationships of lichen‐forming Dothideomycetes. Fungal Divers. 51, 155–162. [Google Scholar]
- Niessen, W.M.A. (2006) Liquid Chromatography‐Mass Spectrometry. USA: Taylor & Francis. [Google Scholar]
- Nukina, M. , Hattori, H. and Marumo, S. (1975) cis‐Sativenediol, a plant growth promotor, produced by fungi. J. Am. Chem. Soc. 97, 2542–2543. [Google Scholar]
- Nukina, M. , Ikeda, M. and Sassa, T. (1980a) Two new pyrenolides, fungal morphogenic substances produced by Pyrenophora teres (Diedicke) Drechsler. Agric. Biol. Chem. 44, 2761–2762. [Google Scholar]
- Nukina, M. , Sassa, T. and Ikeda, M. (1980b) A new fungal morphogenic substance, pyrenolide A from Pyrenophora teres . Tetrahedron Lett. 21, 301–302. [Google Scholar]
- Nützmann, H.‐W. , Reyes‐Dominguez, Y. , Scherlach, K. , Schroeckh, V. , Horn, F. , Gacek, A. , Schümann, J. , Hertweck, C. , Strauss, J. and Brakhage, A.A. (2011) Bacteria‐induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada‐mediated histone acetylation. Proc. Natl. Acad. Sci. USA, 108, 14 282–14 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann, H.‐W. , Schroeckh, V. and Brakhage, A.A. (2012) Regulatory cross talk and microbial induction of fungal secondary metabolite gene clusters In: Methods Enzymol (David A.H., ed.), pp. 325–341. USA: Academic Press. [DOI] [PubMed] [Google Scholar]
- O'Brien, J. and Wright, G.D. (2011) An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 22, 552–558. [DOI] [PubMed] [Google Scholar]
- Ohm, R.A. , Feau, N. , Henrissat, B. , Schoch, C.L. , Horwitz, B.A. , Barry, K.W. , Condon, B.J. , Copeland, A.C. , Dhillon, B. , Glaser, F. , Hesse, C.N. , Kosti, I. , LaButti, K. , Lindquist, E.A. , Lucas, S. , Salamov, A.A. , Bradshaw, R.E. , Ciuffetti, L. , Hamelin, R.C. , Kema, G.H.J. , Lawrence, C. , Scott, J.A. , Spatafora, J.W. , Turgeon, B.G. , de Wit, P.J.G.M. , Zhong, S. , Goodwin, S.B. and Grigoriev, I.V. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. Plos Pathog. 8, e1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, R.P. and Solomon, P.S. (2010) New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 13, 415–419. [PubMed] [Google Scholar]
- Panaccione, D.G. (1993) The fungal genus Cochliobolus and toxin‐mediated plant disease. Trends Microbiol. 1, 14–20. [DOI] [PubMed] [Google Scholar]
- Pedras, M.S.C. and Yu, Y. (2009) Phytotoxins, elicitors and other secondary metabolites from phytopathogenic ‘blackleg’ fungi: structure, phytotoxicity and biosynthesis. Nat. Prod. Commun. 4, 1291–1304. [PubMed] [Google Scholar]
- Pedras, M.S.C. , Morales, V.M. and Taylor, J.L. (1993) Phomaligols and phomaligadiones: new metabolites from the blackleg fungus. Tetrahedron, 49, 8317–8322. [Google Scholar]
- Pongcharoen, W. , Rukachaisirikul, V. , Phongpaichit, S. and Sakayaroj, J. (2007) A new dihydrobenzofuran derivative from the endophytic fungus Botryosphaeria mamane PSU‐M76. Chem. Pharm. Bull. 55, 1404–1405. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg, W. , Kema, G.H.J. , Groenewald, J.Z. , Verkley, G.J.M. , Seifbarghi, S. , Razavi, M. , Gohari, A.M. , Mehrabi, R. and Crous, P.W. (2011) Zymoseptoria gen. nov.: a new genus to accommodate Septoria‐like species occurring on graminicolous hosts. Persoonia, 26, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, M. , Deshmukh, P. , Gade, A. , Ingle, A. , Kövics, G.J. and Irinyi, L. (2009) Phoma Saccardo: distribution, secondary metabolite production and biotechnological applications. Crit. Rev. Microbiol. 35, 182–196. [DOI] [PubMed] [Google Scholar]
- Raistrick, H. , Stickings, C.E. and Thomas, R. (1953) Studies in the biochemistry of micro‐organisms. 90. Alternariol and alternariol monomethyl ether, metabolic products of Alternaria tenuis . Biochem. J. 55, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverberi, M. , Ricelli, A. , Zjalic, S. , Fabbri, A. and Fanelli, C. (2010) Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 87, 899–911. [DOI] [PubMed] [Google Scholar]
- Riaz, A. , Nicklin, J. , Haque, I. , Rauf, C.A. , Qadir, G. and Naz, F. (2013) Toxicity induced by solanapyrone A in chickpea shoots and its metabolism through glutathione/glutathione‐S‐transferase system. Pak. J. Bot. 45, 135–139. [Google Scholar]
- Ruibal, C. , Gueidan, C. , Selbmann, L. , Gorbushina, A.A. , Crous, P.W. , Groenewald, J.Z. , Muggia, L. , Grube, M. , Isola, D. , Schoch, C.L. , Staley, J.T. , Lutzoni, F. and de Hoog, G.S. (2009) Phylogeny of rock‐inhabiting fungi related to Dothideomycetes. Stud. Mycol. 64, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, D. , Fetzner, R. , Burkhardt, B. , Podlech, J. , Metzler, M. , Dang, H. , Lawrence, C. and Fischer, R. (2012) Identification of a polyketide synthase required for alternariol (AOH) and alternariol‐9‐methyl ether (AME) formation in Alternaria alternata . PLoS ONE, 7, e40564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker, S. and Nahar, L. (2012) Hyphenated techniques and their applications in natural products analysis In: Natural Products Isolation (Sarker S. D. and Nahar L., eds.), pp. 301–340. USA: Humana Press. [DOI] [PubMed] [Google Scholar]
- Sarpeleh, A. , Wallwork, H. , Catcheside, D.E.A. , Tate, M.E. and Able, A.J. (2007) Proteinaceous metabolites from Pyrenophora teres contribute to symptom development of barley net blotch. Phytopathology, 97, 907–915. [DOI] [PubMed] [Google Scholar]
- Sassa, T. , Suzuki, K. and Haruki, E. (1989) Isolation and identification of gibberellins A4 and A9 from a fungus Phaeosphaeria sp. Agric. Biol. Chem. 53, 303–304. [Google Scholar]
- Scharf, D.H. , Heinekamp, T. and Brakhage, A.A. (2014) Human and plant fungal pathogens: the role of secondary metabolites. Plos Pathog. 10, e1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherlach, K. and Hertweck, C. (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760. [DOI] [PubMed] [Google Scholar]
- Schoch, C.L. , Crous, P.W. , Groenewald, J.Z. , Boehm, E.W.A. , Burgess, T.I. , de Gruyter, J. , de Hoog, G.S. , Dixon, L.J. , Grube, M. , Gueidan, C. , Harada, Y. , Hatakeyama, S. , Hirayama, K. , Hosoya, T. , Huhndorf, S.M. , Hyde, K.D. , Jones, E.B.G. , Kohlmeyer, J. , Kruys, Å. , Li, Y.M. , Lücking, R. , Lumbsch, H.T. , Marvanová, L. , Mbatchou, J.S. , McVay, A.H. , Miller, A.N. , Mugambi, G.K. , Muggia, L. , Nelsen, M.P. , Nelson, P. , Owensby, C.A. , Phillips, A.J.L. , Phongpaichit, S. , Pointing, S.B. , Pujade‐Renaud, V. , Raja, H.A. , Plata, E.R. , Robbertse, B. , Ruibal, C. , Sakayaroj, J. , Sano, T. , Selbmann, L. , Shearer, C.A. , Shirouzu, T. , Slippers, B. , Suetrong, S. , Tanaka, K. , Volkmann‐Kohlmeyer, B. , Wingfield, M.J. , Wood, A.R. , Woudenberg, J.H.C. , Yonezawa, H. , Zhang, Y. and Spatafora, J.W. (2009) A class‐wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 64, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schümann, J. and Hertweck, C. (2006) Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J. Biotechnol. 124, 690–703. [DOI] [PubMed] [Google Scholar]
- Schwelm, A. , Barron, N.J. , Baker, J. , Dick, M. , Long, P.G. , Zhang, S. and Bradshaw, R.E. (2009) Dothistromin toxin is not required for dothistroma needle blight in Pinus radiata . Plant Pathol. 58, 293–304. [Google Scholar]
- Seger, C. , Sturm, S. and Stuppner, H. (2013) Mass spectrometry and NMR spectroscopy: modern high‐end detectors for high resolution separation techniques—state of the art in natural product HPLC‐MS, HPLC‐NMR, and CE‐MS hyphenations. Nat. Prod. Rep. 30, 970–987. [DOI] [PubMed] [Google Scholar]
- Shain, L. and Franich, R.A. (1981) Induction of Dothistroma blight symptoms with dothistromin. Physiol. Plant Pathol. 19, 49–55. [Google Scholar]
- Shearer, C.A. , Raja, H.A. , Miller, A.N. , Nelson, P. , Tanaka, K. , Hirayama, K. , Marvanová, L. , Hyde, K. D. and Zhang, Y. (2009) The molecular phylogeny of freshwater Dothideomycetes. Stud. Mycol. 64, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, R.A. and Babcock, C.E. (1989) Phaeosphaeria . Can. J. Bot. 67, 1500–1599. [Google Scholar]
- Shoemaker, R.A. and Brun, H. (2001) The teleomorph of the weakly aggressive segregate of Leptosphaeria maculans . Can. J. Bot. 79, 412–419. [Google Scholar]
- Shwab, E.K. , Bok, J.W. , Tribus, M. , Galehr, J. , Graessle, S. and Keller, N.P. (2007) Histone deacetylase activity regulates chemical diversity in Aspergillus . Eukaryot. Cell, 6, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S.B. , Ball, R.G. , Zink, D.L. , Monaghan, R.L. , Polishook, J.D. , Sanchez, M. , Pelaez, F. , Silverman, K.C. and Lingham, R.B. (1997) Fusidienol A: a novel Ras farnesyl‐protein transferase inhibitor from Phoma sp. J. Org. Chem. 62, 7485–7488. [DOI] [PubMed] [Google Scholar]
- Slippers, B. and Wingfield, M.J. (2007) Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol. Rev. 21, 90–106. [Google Scholar]
- So, P.‐K. , Hu, B. and Yao, Z. (2013) Mass spectrometry: towards in vivo analysis of biological systems. Mol. BioSyst. 9, 915–929. [DOI] [PubMed] [Google Scholar]
- Soledade, M. , Pedras, C. , Morales, V.M. and Taylor, J.L. (1994) Phomapyrones: three metabolites from the blackleg fungus. Phytochemistry, 36, 1315–1318. [Google Scholar]
- Solomon, P. (2011) Assessing the mycotoxigenic threat of necrotrophic pathogens of wheat. Mycotoxin Res. 27, 231–237. [DOI] [PubMed] [Google Scholar]
- Stadler, M. and Keller, N.P. (2008) Paradigm shifts in fungal secondary metabolite research. Mycol. Res. 112, 127–130. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos, I. , Collemare, J. , Mehrabi, R. and De Wit, P.J.G.M. (2013) Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 37, 67–93. [DOI] [PubMed] [Google Scholar]
- Strobel, G. , Kenfield, D. and Sugawara, F. (1988) The incredible fungal genus—Drechslera—and its phytotoxic ophiobolins. Phytoparasitica, 16, 145–152. [Google Scholar]
- Sturm, S. and Seger, C. (2012) Liquid chromatography–nuclear magnetic resonance coupling as alternative to liquid chromatography–mass spectrometry hyphenations: curious option or powerful and complementary routine tool? J. Chromatogr. A, 1259, 50–61. [DOI] [PubMed] [Google Scholar]
- Suetrong, S. , Schoch, C.L. , Spatafora, J.W. , Kohlmeyer, J. , Volkmann‐Kohlmeyer, B. , Sakayaroj, J. , Phongpaichit, S. , Tanaka, K. , Hirayama, K. and Jones, E.B.G. (2009) Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 64, 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, F. , Takahashi, N. , Strobel, G.A. , Strobel, S.A. , Lu, H.S.M. and Clardy, J. (1988) Triticones A and B, novel phytotoxins from the plant pathogenic fungus Drechslera tritici‐repentis . J. Am. Chem. Soc. 110, 4086–4087. [Google Scholar]
- Takahashi, J. , Teles, A. , Almeida Pinto Bracarense, A. and Gomes, D. (2013) Classical and epigenetic approaches to metabolite diversification in filamentous fungi. Phytochem. Rev. 12, 773–789. [Google Scholar]
- Tamura, S. , Sakurai, A. , Kainuma, K. and Takai, M. (1963) Isolation of helminthosporol as a natural plant growth regulator and its chemical structure. Agric. Biol. Chem. 27, 738–739. [Google Scholar]
- Tan, K.C. , Trengove, R.D. , Maker, G.L. , Oliver, R.P. and Solomon, P.S. (2009) Metabolite profiling identifies the mycotoxin alternariol in the pathogen Stagonospora nodorum . Metabolomics, 5, 330–335. [Google Scholar]
- Thomas, R. (1961) Studies in the biosynthesis of fungal metabolites. 2. The biosynthesis of alternariol and its relation to other fungal phenols. Biochem. J. 78, 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P. , VAN Esse, H.P. , Crous, P.W. and DE Wit, P.J. (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6, 379–393. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J. (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4, 225–236. [DOI] [PubMed] [Google Scholar]
- Traxler, M.F. , Watrous, J.D. , Alexandrov, T. , Dorrestein, P.C. and Kolter, R. (2013) Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio, 4, e00459‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. , Nozoe, S. , Morisaki, M. , Hirai, K. , Itai, A. , Okuda, S. , Canonica, L. , Fiecchi, A. , Kienle, M.G. and Scala, A. (1967) Nomenclature of ophiobolins. Tetrahedron Lett. 8, 3369–3370. [Google Scholar]
- Tsuge, T. , Harimoto, Y. , Akimitsu, K. , Ohtani, K. , Kodama, M. , Akagi, Y. , Egusa, M. , Yamamoto, M. and Otani, H. (2013) Host‐selective toxins produced by the plant pathogenic fungus Alternaria alternata . FEMS Microbiol. Rev. 37, 44–66. [DOI] [PubMed] [Google Scholar]
- Tsunematsu, Y. , Ishiuchi, K. , Hotta, K. and Watanabe, K. (2013) Yeast‐based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat. Prod. Rep. 30, 1139–1149. [DOI] [PubMed] [Google Scholar]
- Upchurch, R.G. , Walker, D.C. , Rollins, J.A. , Ehrenshaft, M. and Daub, M.E. (1991) Mutants of Cercospora kikuchii altered in cercosporin synthesis and pathogenicity. Appl. Environ. Microbiol. 57, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubbaiah, P. and Chilton, W.S. (1990) Phytotoxins of Botryosphaeria obtusa . J. Nat. Prod. 53, 1628–1630. [Google Scholar]
- Venkatasubbaiah, P. , Sutton, T.B. and Chilton, W.S. (1991) Effect of phytotoxins produced by Botryosphaeria obtusa, the cause of black rot of apple fruit and frogeye leaf spot. Phytopathology, 81, 243–247. [Google Scholar]
- Vurro, M. , Zonno, M.C. , Evidente, A. , Capasso, R. and Bottaiico, A. (1992) Isolation of cytochalasins A and B from Ascochyta lathyri . Mycotoxin Res. 8, 17–20. [DOI] [PubMed] [Google Scholar]
- Walters, D.R. , Havis, N.D. and Oxley, S.J.P. (2008) Ramularia collo‐cygni: the biology of an emerging pathogen of barley. FEMS Microbiol. Lett. 279, 1–7. [DOI] [PubMed] [Google Scholar]
- Walton, J.D. (2006) HC‐toxin. Phytochemistry, 67, 1406–1413. [DOI] [PubMed] [Google Scholar]
- Watrous, J.D. and Dorrestein, P.C. (2011) Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 9, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, R.W.S. , Stenger, E. , Meffert, A. and Hahn, M. (2004) Brefeldin A production by Phoma medicaginis in dead pre‐colonized plant tissue: a strategy for habitat conquest? Mycol. Res. 108, 662–671. [DOI] [PubMed] [Google Scholar]
- Weiergang, I. , Lyngs Jorgensen, H.J. , Moller, I.M. , Friis, P. and Smedegaard‐petersen, V. (2002) Correlation between sensitivity of barley to Pyrenophora teres toxins and susceptibility to the fungus. Physiol. Mol. Plant Pathol. 60, 121–129. [Google Scholar]
- Wiemann, P. and Keller, N.P. (2014) Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 41, 301–313. [DOI] [PubMed] [Google Scholar]
- Williams, D.H. , Stone, M.J. , Hauck, P.R. and Rahman, S.K. (1989) Why are secondary metabolites (natural products) biosynthesized? J. Nat. Prod. 52, 1189–1208. [DOI] [PubMed] [Google Scholar]
- Williams, R.B. , Henrikson, J.C. , Hoover, A.R. , Lee, A.E. and Cichewicz, R.H. (2008) Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 6, 1895–1897. [DOI] [PubMed] [Google Scholar]
- de Wit, P.J.G.M. , van der Burgt, A. , Ökmen, B. , Stergiopoulos, I. , Abd‐Elsalam, K.A. , Aerts, A.L. , Bahkali, A.H. , Beenen, H.G. , Chettri, P. , Cox, M. , Datema, E. , de Vries, R. , Dhillon, B. , Ganley, A.R. , Griffiths, S.A. , Guo, Y. , Hamelin, R.C. , Henrissat, B. , Kabir, M.S. , Jashni, M.K. , Kema, G. , Klaubauf, S. , Lapidus, A. , Levasseur, A. , Lindquist, E. , Mehrabi, R. , Ohm, R.A. , Owen, T.J. , Salamov, A. , Schwelm, A. , Schijlen, E. , Sun, H. , van den Burg, H.A. , van Ham, R.C.H.J. , Zhang, S. , Goodwin, S.B. , Grigoriev, I.V. , Collemare, J. and Bradshaw, R. (2012) The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. Plos Genet. 8, e1003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfender, J.‐L. (2009) HPLC in natural product analysis: the detection issue. Planta Med. 75, 719–734. [DOI] [PubMed] [Google Scholar]
- Wolpert, T.J. , Dunkle, L.D. and Ciuffetti, L.M. (2002) Host‐selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
- Xiao, J. , Zhang, Q. , Gao, Y.‐Q. , Tang, J.‐J. , Zhang, A.‐L. and Gao, J.‐M. (2014) Secondary metabolites from the endophytic Botryosphaeria dothidea of Melia azedarach and their antifungal, antibacterial, antioxidant, and cytotoxic activities. J. Agric. Food Chem. 62, 3584–3590. [DOI] [PubMed] [Google Scholar]
- Yang, X.‐L. , Awakawa, T. , Wakimoto, T. and Abe, I. (2013) Induced biosyntheses of a novel butyrophenone and two aromatic polyketides in the plant pathogen Stagonospora nodorum . Nat. Prod. Bioprospect. 3, 141–144. [Google Scholar]
- Yin, W. and Keller, N. (2011) Transcriptional regulatory elements in fungal secondary metabolism. Microbiology, 49, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoganathan, K. , Yang, L.‐K. , Rossant, C. , Huang, Y. , Ng, S. , Butler, M.S. and Buss, A.D. (2004) Cochlioquinones and Epi‐cochlioquinones. Antagonists of the human chemokine receptor CCR5 from Bipolaris brizae and Stachybotrys chartarum . J. Antibiot. 57, 59–63. [DOI] [PubMed] [Google Scholar]
- Zabala, A.O. , Xu, W. , Chooi, Y.‐H. and Tang, Y. (2012) Characterization of a silent azaphilone gene cluster from Aspergillus niger ATCC 1015 reveals a hydroxylation‐mediated pyran‐ring formation. Chem. Biol. 19, 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Schwelm, A. , Jin, H. , Collins, L.J. and Bradshaw, R.E. (2007) A fragmented aflatoxin‐like gene cluster in the forest pathogen Dothistroma septosporum . Fungal Genet. Biol. 44, 1342–1354. [DOI] [PubMed] [Google Scholar]


