Summary
Hemibiotrophic plant pathogens, such as the oomycete Phytophthora infestans, employ a biphasic infection strategy, initially behaving as biotrophs, where minimal symptoms are exhibited by the plant, and subsequently as necrotrophs, feeding on dead plant tissue. The regulation of this transition and the breadth of molecular mechanisms that modulate plant defences are not well understood, although effector proteins secreted by the pathogen are thought to play a key role. We examined the transcriptional dynamics of P. infestans in a compatible interaction with its host tomato (Solanum lycopersicum) at three infection stages: biotrophy; the transition from biotrophy to necrotrophy; and necrotrophy. The expression data suggest a tight temporal regulation of many pathways associated with the suppression of plant defence mechanisms and pathogenicity, including the induction of putative cytoplasmic and apoplastic effectors. Twelve of these were experimentally evaluated to determine their ability to suppress necrosis caused by the P. infestans necrosis‐inducing protein PiNPP1.1 in Nicotiana benthamiana. Four effectors suppressed necrosis, suggesting that they might prolong the biotrophic phase. This study suggests that a complex regulation of effector expression modulates the outcome of the interaction.
Keywords: hemibiotroph, in planta effectors, Phytophthora infestans, transcriptome
Introduction
Plant pathogens can be classified by their infection strategy as biotrophs, hemibiotrophs or necrotrophs. Biotrophic pathogens proliferate within and feed on living tissues, necrotrophs kill host tissue and feed on dead cells (Glazebrook, 2005) and hemibiotrophic pathogens, such as the oomycete Phytophthora infestans, show an initial asymptomatic biotrophic phase of infection followed by a necrotrophic phase. During the biotrophic stage, P. infestans sequentially forms appressoria, primary and secondary hyphae and, finally, specialized structures called haustoria, through which proteins and small molecules, called effectors, are delivered into the apoplast or adjacent plant cells (Dou et al., 2008; Grenville‐Briggs et al., 2005). These effectors are thought to enable pathogens to manipulate host metabolism and suppress host defences (Abramovitch and Martin, 2004; Axtell and Staskawicz, 2003; Hahn and Mendgen, 2001; Mackey et al., 2003; Restrepo et al., 2005; Tian et al., 2004). The subsequent necrotrophic phase is characterized by hyphal ramification and water soaking, followed by necrosis of the tissue (Grenville‐Briggs et al., 2005). However, marked differences in the duration of both biotrophic and necrotrophic growth, depending on the host–P. infestans isolate interaction, have been reported previously (Berg, 1926; Cai et al., 2013; Smart et al., 2003; Vega‐Sanchez et al., 2000). There is an extended period of biotrophy between tomato and certain tomato‐specialized isolates of P. infestans, (Berg, 1926; Cai et al., 2013; Smart et al., 2003; Vega‐Sanchez et al., 2000).
The diversity of processes that regulate each pathogenicity stage and the molecular mechanisms that trigger the transition from biotrophy to necrotrophy (Lee and Rose, 2010) are not well understood. The first detailed global insights have come from recent studies of the hemibiotrophic oomycete P. capsici–tomato interaction (Jupe et al., 2013), suggesting defined shifts in gene expression during the different stages of hemibiotrophy and a dynamic transcriptional regulation of effector‐coding genes. In addition, studies of the hemibiotrophic fungus Colletotrichum (Gan et al., 2013; O'Connell et al., 2012) have resulted in the identification of suites of genes that are associated with stage transitions, with effectors and secondary metabolism‐related genes predominating during biotrophy, and lytic hydrolases and transporters featured more during necrotrophy. However, it is not known whether such features are common to all hemibiotrophic infections. The goal of this study was to determine whether similar transcriptional signatures are evident in P. infestans over the time course of its interaction with its tomato host.
The characterization of some effectors/proteins suggests that Phytophthora actively induces or suppresses cell death in plants (van Damme et al., 2012; Kelley et al., 2010; Lee et al., 2014; Lee and Rose, 2010; Wang et al., 2011). Moreover, the analysis of apoplastic effectors overexpressed in planta indicates that INF1 causes a hypersensitive response (HR) and cell death in Nicotiana benthamiana, and is highly expressed at later stages of the interaction of P. infestans with potato (Kamoun et al., 1997). Likewise, the P. infestans Nep‐1‐like protein PiNPP1.1 (Kanneganti et al., 2006), which acts synergistically with INF1, induces necrosis late in the infection of tomato or N. benthamiana (Kanneganti et al., 2006), and the RXLR effector PexRD2 causes cell death in N. benthamiana (Oh et al., 2009) and suppresses both mitogen‐activated protein kinase kinase kinase (MPKKK)‐triggered cell death and cell death elicited by effector/resistant protein interactions that are MPKKK dependent (King et al., 2014). Recently, the CRN8 effector, which shows similarity to a serine/threonine kinase, has been found to cause cell death when transiently expressed in planta (van Damme et al., 2012). In contrast, the AVR3aKI (Bos et al., 2006), PexED8 and PexRD36 (Oh et al., 2009) cytoplasmic effectors from P. infestans suppress the INF1‐induced HR in N. benthamiana, and the RXLX effector SNE1 of P. infestans suppresses necrosis caused by either PiNPP1.1 or PsojNIP (Kelley et al., 2010). SNE1 is secreted during the biotrophic phase of the tomato–P. infestans interaction and has been hypothesized to promote biotrophy by suppressing necrosis (Kelley et al., 2010; Lee and Rose, 2010). Another example of the modulation of host responses by P. infestans comes from the interaction between the P. infestans effector protein IpiO‐1 and the Solanum bulbocastanum resistance gene protein RB. After recognition of IpiO‐1 by RB, HR is elicited in a typical gene‐for‐gene interaction, but this effector‐triggered immunity is suppressed by pathogen genotypes expressing the competing effector, IpiO‐4, which abolishes the HR and leads to susceptibility (Halterman et al., 2010). Other components of this intricate regulatory system include the Crinkler (CRN) effector proteins, which were initially identified by their ability to cause necrosis in planta (Torto et al., 2003). It has been shown recently that, although one of these (PsCRN63) caused necrosis in N. benthamiana, the other (PsCRN115) suppressed necrosis caused by PsojNIP or PsCRN63 (Liu et al., 2011).
To gain a more comprehensive insight into the processes that mediate the hemibiotrophic infection, we used the infection of tomato (Solanum lycopersicum) by P. infestans as a model pathosystem. The P. infestans–tomato interaction can be highly compatible, partially compatible or incompatible, and these differences can be a consequence of both the plant genotype and whether or not the P. infestans strain is tomato specialized (Smart et al., 2003). Importantly, it has been demonstrated previously that the P. infestans–tomato interaction is characterized by a prolonged biotrophic phase and a distinct necrotrophic phase at later stages of infection with tomato‐specialized isolates (Berg, 1926; Cai et al., 2013; Smart et al., 2003; Vega‐Sanchez et al., 2000). We analysed the transcriptomes of the tomato‐specialized P. infestans isolate (US‐11) and its host tomato (M82) in a highly compatible interaction using RNA sequencing (RNA‐Seq), in order to identify suites of co‐expressed genes that could be associated with each stage of the interaction and to infer regulated pathways from both the pathogen and host plant. Here, we compare the changes that occur in gene expression as the pathogen progresses from biotrophic to necrotrophic growth in planta and present the transcriptional dynamics of P. infestans effectors and other pathogenicity genes that are expressed during different stages of infection. We also describe the evaluation of candidate secreted effectors that were expressed at different infection stages for their ability to suppress or induce necrosis.
Results
Dynamics of P. infestans infection of tomato plants
We first defined the time points at which the various stages of hemibiotrophic growth were exhibited by P. infestans (US‐11 clonal lineage) during infection of a compatible tomato cultivar (Solanum lycopersicum, cv. M82). To do this, we harvested samples from the inoculation site (7‐mm‐diameter discs of tomato leaves every 12 h over a 7‐day period (Fig. S1, see Supporting Information). Three methods were used to evaluate the time points: macroscopic observation of symptoms in leaves; microscopic evaluation of the pathogen developmental stage; and molecular assessment based on biotrophic and necrotrophic stage‐specific genes of P. infestans. Using these methods (as described below), we identified 48, 96 and 144 h after inoculation (hai) as being representative of biotrophy, transition to necrotrophy and necrotrophy for this particular interaction. Once the time points had been selected, we performed a large‐scale experiment with four biological replicates with 25 plants per time point in each replicate. Plant samples from each time point were then pooled (see Experimental procedures).
In the macroscopic assessment in tomato leaflets, we observed no symptoms at the inoculation site up to 60 hai (Fig. 1A shows a macroscopic assessment at 48 hai; Fig. S1 shows various time points). Water soaking was visible at the inoculation site starting at 96 hai (Figs 1B, S1), indicating the transition from biotrophy to necrotrophy. Necrosis was seen at the 7‐mm‐diameter inoculation site at the centre of the lesion at 144 hai (Figs 1C, S1), indicating a well‐established necrotrophic phase. To determine the developmental stage of the pathogen in planta, we observed microscopically the inoculation site that had been stained using trypan blue (Chung et al., 2010; Knox‐Davies, 1974). In the biotrophic phase (48 hai), germinating sporangia and invading hyphae were seen penetrating plant tissue (Fig. 1D), whereas, at 96 hai, hyphal growth and some mycelial branching were apparent (Fig. 1E). Finally, in the necrotrophic phase (144 hai), there was abundant mycelial growth and ramification (Fig. 1F).
Figure 1.
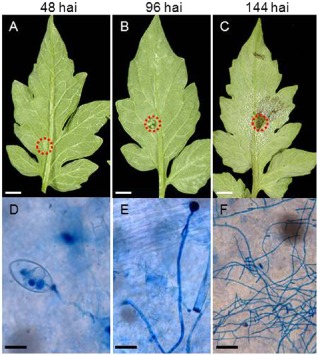
Macroscopic assessment of symptoms of Phytophthora infestans development on tomato leaflets. (A–C) Macroscopic symptoms of P. infestans development on tomato leaflets. (D–F) Pathogen developmental stages (trypan blue staining). Detached M82 tomato leaves were inoculated with P. infestans (US‐11). Symptoms were monitored at 48 h after inoculation (hai; A, D), 96 hai (B, E) and 144 hai (C, F). The red circles denote examples of the 7‐mm‐diameter sample areas from which RNA was subsequently extracted. Scale bars: (A‐C) 7 mm; (D, E) 20 μm; (F) 100 μm.
The expression of three P. infestans genes that represent markers for the stages of infection was assessed using reverse transcription‐polymerase chain reaction (RT‐PCR). Two P. infestans RXLR/RXLX effector genes, IpiO (also known as Avr‐blb1; van West et al., 1998) and SNE1 (Kelley et al., 2010), have been shown previously to be expressed predominantly during the biotrophic phase of the interaction. The PiNPP1.1 gene was used as the necrotrophic marker as its expression is restricted to the necrotrophic phase (Kanneganti et al., 2006). The expression of both biotrophic markers was detected during the early stages of the interaction (48 hai) and transcript accumulation increased during the biotrophic and transition phase, before decreasing at the necrotrophic phase [Figs 2, S2 (see Supporting Information)]. In contrast, PiNPP1.1 was first detected at 96 hai and was predominantly expressed at later stages of the interaction (Figs 2, S2). Expression of the P. infestans Actin‐A gene was used to show that the apparent reduction in SNE1 and IpiO expression during the time course did not reflect less pathogen biomass. Overall, we conclude that, for this specific pathogen–host interaction, 48, 96 and 144 hai represent biotrophic growth, transition to necrotrophic growth and mainly necrotrophic growth, respectively. These time points were therefore used for the more intensive analysis described below.
Figure 2.
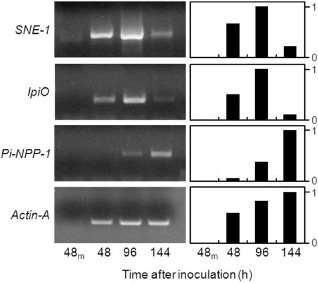
Expression profile of the molecular markers used to characterize Phytophthora infestans developmental stage. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) evaluation of the expression of P. infestans biotrophic (IpiO and SNE1) and necrotrophic (PiNPP‐1) stage‐specific marker genes in infected tomato leaves at 48, 96 and 144 h after inoculation and in water control tomato mock‐inoculated leaves after 48 h (48m). The left panels show gels of the reverse transcription products and the right panels show the corresponding quantification of relative band intensity, with the band showing the maximal intensity set at a value of 1.0. Phytophthora infestans Actin‐A was used as control to assess pathogen abundance.
Analysis of 454‐reads and mapping to P. infestans unigenes
We generated cDNA libraries from 7‐mm discs of tomato leaves inoculated with P. infestans at 48, 96 and 144 hai. These were then sequenced using the 454‐pyrosequencing technology to generate transcriptome profiles of biotrophic, transition and necrotrophic growth.
A summary of the number of 454‐reads before and after quality filtering, as well as the average length in nucleotides (nt) for the three samples, is shown in Table 1. Based on these criteria, 98%–99% of the sequences from each sample were determined to be of high quality. A total of 86 319 cDNA sequences were aligned to the genomes of P. infestans, P. sojae and P. ramorum, 85 952 (99.57%) of which mapped to the P. infestans T30‐4 reference genome, 21 967 (25.45%) to the genome of P. sojae and 18 699 (21.66%) to the genome of P. ramorum, with at least 90% sequence identity and 50% length coverage (Fig. S3, see Supporting Information). After a blastx database analysis, 737, 8472 and 51 732 predicted Phytophthora spp. sequences were identified from the biotrophic, transition and necrotrophic stages, respectively (Table 1). As expected, the number of P. infestans sequences detected increased as infection progressed, presumably reflecting the increase in pathogen biomass. Sequences from P. infestans represented 0.3%, 6% and 28% of the total number of sequences identified at these three time points. The remaining sequences corresponded to those from the host tomato plant.
Table 1.
Summary of the number of 454‐sequence reads and blastx hits to the Phytophthora infestans genome (Broad Institute, Cambridge, MA, USA)
| Sample | Total number of reads | High‐quality reads | Length average (nt) |
P. infestans
blast hits to reference genomes (e‐value ≤ 9e‐7) |
Match to existing P. infestans unigenes (%) |
|---|---|---|---|---|---|
| 48 hai | 248 172 | 245 631 | 225 | 737 | 0.3 |
| 96 hai | 154 842 | 151 863 | 209 | 8472 | 6 |
| 144 hai | 187 459 | 184 620 | 201 | 51 732 | 28 |
hai, hours after inoculation; nt, nucleotides.
Inferring transcription levels using transcript abundance
The estimation of relative transcript levels by 454‐pyrosequencing has been reported previously (Fernandez et al., 2012; Vega‐Arreguín et al., 2009). In order to confirm the accuracy of transcript abundance from the RNA‐Seq data, we analysed the expression of the four P. infestans genes that were initially used as markers to characterize the stage of infection: SNE1, IpiO, PiNPP1.1 and Actin‐A (Fig. 2). A similar pattern of expression for these genes was seen using either RT‐PCR or 454‐reads (Fig. S4, see Supporting Information), although it appeared that RT‐PCR was more sensitive. SNE1 was detected at 48 hai using RT‐PCR, but was not identified until 96 hai in the 454‐sequence profiles. A peak of expression of SNE1 at 96 hai was observed using both techniques and its expression then declined at 144 hai. Similarly, IpiO was first detected at 48 hai using RT‐PCR and at 96 hai using the RNA‐Seq data. With both approaches, the peak of expression for this marker was at the transition phase, whereas the necrotrophic marker PiNPP1.1 was maximally expressed at 144 hai. Finally, when we analysed the P. infestans Actin‐A expression profile, we observed, as expected, an increase in expression over the infection time course, reflecting the increasing relative amount of pathogen biomass. Thus, RNA‐Seq‐based transcript abundance analysis was considered to be an accurate measure of the steady state of transcript levels.
Transcriptome of P. infestans during infection of tomato leaves
Of the 18 178 predicted genes in the genome of P. infestans (Haas et al., 2009), we detected the expression of 9109 over the time course of the interaction with tomato. In subsequent analyses, only unigenes that had at least five reads were considered, in order to focus on genes that were identified with the highest confidence, reducing the number studied to 3495 (filtered file: File S1, see Supporting Information; all the sequence data described in this article can be found in the GenBank Sequence Read Archive (SRA) under accession number SRP041501; Fasta files are also available at: http://solgenomics.net/download/data/secretom/Secretome_Phytophthora_tomato_interactions/P.infestans_unigenes.fasta). Approximately 38% of these correspond to hypothetical proteins, 10% of which are predicted to be secreted based on the presence of the N‐terminal signal peptide (SP), as determined using SignalP software (Bendtsen et al., 2004). A total of 818 genes (23%) listed in the filtered file had not been previously annotated in the T30‐4 P. infestans reference genome sequence. The two genotypes (US‐11 and T30‐4) are unlikely to be closely related because US‐11 was collected in the USA (see below) and T30‐4 is a recombinant between parents collected in Europe. Some new genes were detected at each time point (32% of the genes at 48 hai, 23% of the genes at 96 hai and 23% of the genes at 144 hai). Many (11% of the 818 genes) are predicted ribosomal proteins, some are putative effectors (six RXLR, six CRN and 12 correspond to elicitins) and one is annotated as carbonic anhydrase (CA). The majority (43% of the 818) correspond to hypothetical or unknown proteins, whereas 29% have no homology to known genes using blastx (File S2, see Supporting Information).
In order to determine which pathogen genes were differentially expressed throughout the interaction with tomato, we compared all the genes expressed at 48 hai with the genes expressed at 96 hai. Similarly, we compared the genes expressed at 96 hai with those expressed at 144 hai. The relative transcript abundance of a particular gene was assessed by comparing its absolute abundance with the absolute abundance of all genes (see Experimental procedures). Ten per cent of the P. infestans genes were differentially expressed (defined as two‐fold or greater change in transcript abundance) using a false discovery rate (FDR) of 0.05 (File S3, see Supporting Information). Of these, 34% are classified as hypothetical proteins, 6% are putative effectors (14 RXLR, five CRN, three elicitins and an NPP‐1‐like protein), 3% are categorized as detoxification genes (including ATP‐binding cassette and a cytochrome P‐450) and 10% correspond to ribosomal proteins. Of the differentially expressed genes, 8% show no homology to known genes in public databases. The remaining genes (39%) are associated with primary metabolism.
Phytophthora infestans gene expression during stages of hemibiotrophy
Transcriptome of P. infestans at the biotrophic stage: 48 hai
Phytophthora infestans acts as a biotrophic pathogen during the early stages of the interaction and as a necrotroph at later stages. We hypothesized that P. infestans effectors modulate the outcome of the interaction by either blocking host defence mechanisms or avoiding recognition by the plant, or a combination of both. Therefore, we examined a temporal profile of the transcript accumulation of some of the genes that are known or that have been hypothesized previously to be involved in the P. infestans–tomato or P. infestans–potato interaction, using the scheme adopted by Torto‐Alalibo et al. (2007). We examined the abundance of putative pathogenicity genes from eight functional categories in each of the three stages of the interaction (Fig. 3 depicts the abundance trend for each category; for individual gene ID and number of reads for each gene, see File S4 in Supporting Information). Putative effectors, elicitins and elicitin‐like (INFs), CRNs, necrosis‐inducing protein (NPP) and RXLRs were analysed separately, as shown in Fig. 4 (for individual gene ID and number of reads for each gene, see File S5 in Supporting Information).
Figure 3.
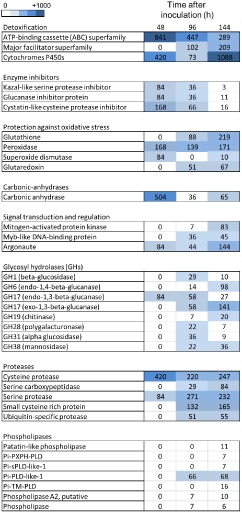
Relative expression of classes of Phytophthora infestans putative pathogenicity genes based on RNA‐Seq data during a time course of infection of tomato. The units are the number of normalized number of reads for each category, representing all the detected homologues for each gene. The individual datasets for each homologue are provided in File S4.
Figure 4.
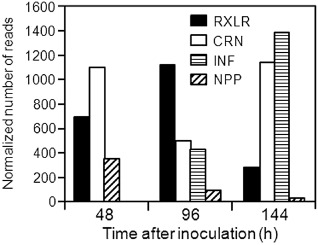
Temporal profile of the expression of Phytophthora infestans putative effectors. Expression of P. infestans elicitins and elicitin‐like (INF), Crinkler (CRN), necrosis inducing‐like (NPP1.1) genes and RXLR effectors based on the RNA‐Seq data during a time course of infection of tomato leaves (hours after inoculation). Units are normalized to the number of reads representing all the detected homologues for each gene category. The individual datasets for each homologue are provided in File S5.
After penetration of the host tissue, the pathogen must overcome preformed or induced host defences. Accordingly, the P. infestans RNA‐Seq data revealed high levels of transcript accumulation during the biotrophic phase of genes associated with detoxification, such as cytochrome P450 and ATP‐binding cassette transporters (ABC‐transporters; Coleman et al., 2011; Matthews and VanEtten, 1983), and protection against oxidative stress (peroxidase and superoxide dismutase) (Fig. 3; File S4). Another abundant class of genes encodes enzyme inhibitors, including a Kazal‐like serine protease inhibitor, which inhibits a subtilisin‐like serine protease from tomato (Tian et al., 2004), glucanase inhibitor proteins that bind and inhibit host endo‐β‐1,3‐glucanases of glycosyl hydrolase (GH) family 17 (Damasceno et al., 2008; Rose et al., 2002), and a cystatin‐like cysteine protease inhibitor that targets a tomato papain‐like apoplastic protease (Tian et al., 2007; Fig. 3; File S4). A cysteine protease, a homologue of which has been suggested to inhibit reactive oxygen species by interfering with photosynthesis and to suppress the plant innate immune responses in Pseudomonas syringae (Rodríguez‐Herva et al., 2012), also showed the highest accumulation at 48 hai (Fig. 3).
Hemibiotrophic pathogens, such as P. infestans, avoid causing host cell death at early stages of the interaction. Consistent with this suggestion, only a few examples of cell wall hydrolase transcripts were detected at this stage. One example is predicted to encode the GH17 protein endo‐1,3‐β‐glucanase (Fig. 3; File S4), which is suggested to be involved in remodelling the P. infestans cell wall for hyphal tip growth and branching in the initial colonization steps (McLeod et al., 2003). Serine protease homologues were also abundant, reaching a peak at 96 hai, as were members of the family of cysteine proteases (cathepsin‐B and papain‐like) (Fig. 3; File S4). Genes that are involved in pathogen cell wall formation also accumulated to high levels at this stage. Similarly, elicitin‐like (INF) genes, which are sterol carriers (Mikes et al., 1998), and are also known to cause HR‐like cell death in N. benthamiana (Kamoun et al., 1997) (Fig. 4), as well as five RXLR genes and nine CRN cytoplasmic putative effectors, were highly abundant. One of the P. infestans genes with the highest transcript abundance at 48 hai is a predicted CA (Fig. 3), an enzyme that has been reported to be involved in the pathogen sensing of CO2 concentrations (Schlicker et al., 2009), and was described as a candidate virulence factor (Raffaele et al., 2010).
Transcriptome of P. infestans at 96 hai
At 96 hai, we observed a shift in the categories of expressed P. infestans genes (Fig. 3; File S4). There was an increase in the number and diversity of families of GHs that probably facilitate plant cell wall breakdown, including members of GH‐1 (β‐glucosidase), GH‐6 (endo‐1,4‐β‐glucanase), GH‐16 [endo‐1,3(4)‐β‐glucanase], GH‐28 (polygalacturonase), GH‐31 (α‐glucosidase), GH‐38 (α‐mannosidase) and pectin esterases, or pathogen wall modification, such as GH‐17 (exo‐ and endo‐1,3‐β‐glucosidase) and GH‐19 (chitinase). Other putative degradative enzymes, such as lipases and serine proteases, were also first detected at this stage (Fig. 3; File S4), as were major facilitator superfamily (MFS) transporters, which may be involved in pathogen protection (Hayashi et al., 2002; Torto‐Alalibo et al., 2007; Zwiers et al., 2003). In addition, members of four phospholipase D subfamilies, including the putatively secreted Pi‐sPLD‐like‐1, were first detected at 96 hai, and their transcript abundance increased at 144 hai (Fig. 3). Recent characterization of some members of the P. infestans phospholipase D family has suggested that some members are secreted and may be involved in pathogenicity (Meijer et al., 2011).
The relative transcript abundance of RXLR cytoplasmic effectors, which are hypothesized to modulate the host defences, was highest at 96 hai (Fig. 4). Among these RXLR effectors were the previously characterized IpiO (also known as Avr‐blb1) (Vleeshouwers et al., 2008), SNE1 (Kelley et al., 2010), Avr1, Avr2 and two members of the Avr‐blb2 superfamily (Oh et al., 2009). In contrast, the CRN effectors showed the lowest relative transcript abundance at this time point (Fig. 4). Other genes that had a reduction in transcript abundance relative to the previous time point were genes related to protection against oxidative stress (Fig. 3; File S4).
Transcriptome of P. infestans at the necrotrophic stage: 144 hai
The 7‐mm tomato tissue discs were entirely necrotic at 144 hai and, at this time point, there was an increase in the number and expression of genes that have previously been associated with necrosis. Among these were small cysteine‐rich proteins, NPP‐like family proteins, NAPDH oxidases and GHs. Similarly, the transcript abundance of genes involved in signal transduction, protection against oxidative stress, detoxification and the cytoplasmic effectors CRNs and INFs was highest at this stage, whereas that of the RXLRs was lowest (Figs 3, 4; for individual gene ID and number of reads for each gene, see File S5). Among the RXLRs detected was PexRD2, which has been shown to induce plant cell death (King et al., 2014; Oh et al., 2009).
Cluster analysis
Hierarchical cluster analysis was performed in order to identify genes and pathways with similar expression profiles. Regardless of the level of transcript accumulation, profiles were characterized as the number of reads for each gene at each time point compared with the mean of that gene across all time points (Fig. S5, see Supporting Information). Homologues of a particular gene may show differences in their transcription profiles, and so not all homologues are in the same cluster. Using all the genes of the filtered file, five major clusters were identified (Fig. S5). Clusters P2 (76 genes) and P5 (362 genes) include genes whose transcript accumulation was predominant at the biotrophic stage (Fig. S5; File S6, see Supporting Information). Among the genes that were abundant in this category were many (24%) predicted to code for unknown or hypothetical proteins. Others are predicted to encode proteins involved in transcription, such as helicases, translation (ribosomal proteins and elongation factors) and the initiation of protein synthesis, including eukaryotic initiation factor 4A‐III, DEAD/DEAH box RNA helicase and eukaryotic translation initiation factor 3 subunit C. Clusters P1 (947 genes) and P4 (695 genes) include genes whose transcript abundance peaked at the transition stage. Finally, cluster P3 (1415 genes) includes genes that are most abundant at the necrotrophic stage (Fig. S5). Approximately 38% of the genes with higher transcript abundance in the necrotrophic stage correspond to unknown or hypothetical proteins.
Functional characterization of putative effectors: RXLR, CRN and hypothetical proteins
To test the hypothesis that P. infestans effectors differentially modulate the outcome of the interaction with tomato, depending on the infection phase in which they are expressed, a subset of putative effectors identified in this study was evaluated by transient expression in N. benthamiana leaves using agro‐infiltrations. As several reports have indicated that induction or suppression of cell death mediated by RXLR effectors occurs regardless of the presence or absence of an N‐terminal secretory SP, the full‐length sequence was used for each candidate gene (Bos et al., 2006; Dong et al., 2009). Each selected gene was expressed in a region overlapping an area of a leaf in which the necrosis‐inducing PiNPP1.1 effector was also transiently expressed (Fig. 5A,B). Three possible outcomes were anticipated: (i) necrosis in both agro‐infiltrated areas; (ii) necrosis only in the area infiltrated with PiNPP1.1; or (iii) no necrosis in the overlapping area if the putative effector suppresses necrosis caused by PiNPP1.1.
Figure 5.
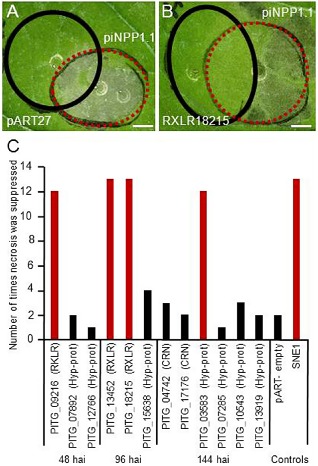
Necrosis induction or suppression assays in Nicotiana benthamiana. (A, B) Areas of an N. benthamiana leaf infiltrated with Agrobacterium tumefaciens to transiently express RXLR18215, a suppressor of necrosis (B, black circle), or a pART27 empty vector control which does not suppress necrosis (A, black circle), and 24 h later infiltrated to express a necrosis‐inducing protein (PiNPP1.1; red circles in A and B), such that an overlapping region of infiltration is generated. (C) Number of times a candidate gene (RXLRs, CRNs and hypothetical proteins) suppressed cell death caused by PiNPP1.1 (results of at least 15 infiltrations per gene) in the transient expression assays, with those showing suppression activity 80% of the time or more highlighted in red. hai, hours after inoculation. Positive control SNE1 without a signal peptide (SP).
Based on their high transcript abundance at a particular stage, three putative RXLR effectors, PITG_09216 (48 hai), PITG_13452 and PITG_18215 (96 hai), as well as two CRNs, PITG_04742 and PITG_17176 (144 hai), were selected for functional analysis. We hypothesized that effectors with high transcript abundance during the biotrophic and transition phase might suppress plant cell death and delay the onset of necrosis. In agreement with our model, the three candidates expressed during the biotrophic and transitional phase suppressed necrosis caused by the PiNPP1.1 effector (Fig. 5C). As several CRN families have been shown to cause necrosis in planta (Haas et al., 2009), we hypothesized that the two CRN effectors expressed during the necrotrophic phase might cause necrosis; however, the expression of these two candidate genes neither suppressed nor induced necrosis in the N. benthamiana infiltration assays (Fig. 5C).
As P. infestans cytoplasmic effectors have an SP for secretion from the pathogen cell and additional peptide signals that direct translocation into the host cytoplasm (e.g. RXLR/RXLX, LQLFLAK) (Haas et al., 2009), some of the hypothetical proteins with an SP predicted by SignalP software (Bendtsen et al., 2004), but lacking the RXLR/RXLX motif, were selected to investigate whether they played a role as effectors. In addition, hypothetical proteins lacking an SP, but showing a high number of reads at the different stages, were selected to evaluate whether they induced or suppressed necrosis. The selected proteins comprised: two hypothetical proteins with the highest transcript abundance at 48 hai; PITG_07892 lacking an SP; the secreted PITG_12766; a 60s ribosomal protein lacking an SP which was highly expressed at the transitional stage (PITG_15638) and four that were highly expressed at the necrotrophic phase (PITG_03583 with SP, PITG_07285 lacking SP, PITG_10543 without an SP and PITG_13919 without an SP). The candidates were agro‐infiltrated into N. benthamiana leaves and any induction of necrosis, or suppression of PiNPP1.1‐mediated necrosis, was noted. None of these genes induced necrosis and only one putative effector (PITG_03583, encoding a hypothetical secreted protein, which was expressed at the necrotrophic stage) suppressed necrosis in the majority of trials (13/15) (Fig. 5C).
Discussion
The primary goal of this study was to test the hypothesis that the evaluation of patterns of gene expression in P. infestans during the various stages of hemibiotrophy would provide insights into the underlying mechanisms of the transition from biotrophy to necrotrophy.
One of the unexpected results was the identification of 818 unigenes that had not previously been annotated as genes in the P. infestans sequenced genome. There are several possible explanations for this: (i) these ‘orphan’ genes are located in areas of the P. infestans genome that were not sequenced or, if sequenced, might have been incorrectly assembled and so incorrectly annotated; (ii) they correspond to non‐coding RNAs; (iii) they represent untranslated region (UTR) sequences that were not included in the predicted genes; (iv) they correspond to novel genes of this particular strain; (v) they are expressed pseudogenes; and/or (vi) they are the products of alternative splicing. Indeed, approximately one‐third of the P. infestans genes have introns (Win et al., 2006). These results are consistent with previously reported and well‐annotated genome sequences, where 454‐sequencing‐derived reads could not be mapped to previously predicted genes. This occurred for 36% of the reads in humans (Mane et al., 2009), 13% of the reads in Arabidopsis thaliana (Weber et al., 2007) and 28% of the reads in cucumber (Guo et al., 2010), and confirms the value of RNA‐Seq transcriptome sequencing as a gene discovery platform.
To date, many of the analyses of P. infestans effectors have been based on the selection of candidates in silico and functional analysis by expression in a heterologous system (Haas et al., 2009; Oh et al., 2009). This has been a successful strategy for both the discovery of expressed effectors and the assignment of function (Haas et al., 2009; Oh et al., 2009; Whisson et al., 2007). The RNA‐Seq analysis presented here provides additional identification of predicted effectors. For example, we found 31 RXLRs expressed during the tomato–P. infestans interaction, 14 of which had not been identified previously among the 79 RXLRs that were differentially expressed in a potato–P. infestans interaction study (Haas et al., 2009), and only three of which were found in common with those identified through a screen using a P. infestans–Solanum bulbocastanum pathosystem (Oh et al., 2009). Similarly, although a P. infestans–potato analysis identified 10 CRN proteins as being induced during infection (Haas et al., 2009), our study identified 51 and, indeed, these were the most abundant family of effectors expressed in the tomato–P. infestans interaction. Remarkably, none of the 51 CRN effectors identified here was present in the expression profile of P. infestans genes expressed during infection of potato (Haas et al., 2009). This highlights the importance of using complementary approaches when studying plant–pathogen interactions to obtain a more comprehensive coverage of putative in planta expressed effectors, the expression of which may be strongly influenced by the nature of the host.
The expression patterns of predicted pathogenicity effectors observed in this study suggest that there might be a coordinated regulation of these effectors over time. During the biotrophic phase, the pathogen must overcome potential plant defences. Accordingly, we observed a large induction of genes involved in detoxification, enzyme inhibitors and protection against oxidative stress during this stage. In addition, the pathogen modulates the plant response by the secretion of predicted cytoplasmic effectors. Similarly, a dynamic expression pattern of effectors throughout a time course has also been described for the P. capsici–tomato interaction (Jupe et al., 2013), P. sojae–soybean interaction (Wang et al., 2011), Melampsora larici‐populina–poplar interaction (Duplessis et al., 2011) and Hemileia vastatrix–Coffea arabica pathosystem (Fernandez et al., 2012) in the transition from biotrophy to sporulation.
Two families of putative cytoplasmic effectors (RXLR and CRN proteins) have been the subject of many studies because of their presumed role in pathogenicity (e.g. Haas et al., 2009; Kelley et al., 2010; Oh et al., 2009; Rietman et al., 2012; van West et al., 1998; Whisson et al., 2007). We found that many of these genes were expressed during tomato infection, with a higher proportion of annotated CRN genes (25%) than of RXLR genes (5%), a phenomenon that has also been observed in the infection of Helianthus annuus by Plasmopara halstedii (As‐sadi et al., 2011).
The timing of expression of previously characterized effectors coincided in general with their predicted function. The RXLR gene products that are either associated with the biotrophic phase, or that have been shown to suppress necrosis (such as SNE1, Kelley et al., 2010; IpiO, van West et al., 1998) were detected early in the interaction (48 and 96 hai, respectively) in this study, whereas transcript abundance for genes whose products are thought to be involved in necrosis (INF‐like elicitins, Kamoun et al., 1997; PiNPP‐like, Kanneganti et al., 2006; and PexRD2, Oh et al., 2009) and GHs was higher at later stages (144 hai). We showed that three RXLRs expressed at 48 and 96 hai suppressed necrosis caused by the PiNPP1.1 effector, supporting the hypothesis that effectors secreted at the early stages of the interaction may allow the pathogen to extend its biotrophic phase. However, contrary to our expectation that proteins expressed at 144 hai would contribute to necrosis, the hypothetical protein (PITG_03583), expressed at 144 hai, suppressed rather than induced necrosis. None of the effectors evaluated caused necrosis in N. benthamiana.
The use of agro‐infiltration proved to be an effective tool to assess the putative function of several candidate effector proteins, although there are potential limitations. For example, it is possible that the putative effectors interact with other effectors or with plant‐specific genes, in which case the use of heterologous systems would not be appropriate. A next step in the characterization of these candidate effectors is to find their targets in the affected plant cell and to elucidate how they function in the context of other virulence genes.
Our expression data provide support to other investigations of pathogenesis by P. infestans. For instance, Meijer et al. (2011) suggested that members of the P. infestans phospholipase D (PLD)‐like family may have a role in pathogenicity, and our data show that the expression of phospholipases is induced at 96 hai in the interaction with the host. As another example, one of the P. infestans transcripts that was most abundantly expressed during the biotrophic phase corresponds to a putative CA, which was found to be one of the most abundant proteins identified in in silico analyses (Raffaele et al., 2010). This class of enzyme has been widely studied in mammalian pathogenic microbes, such as the yeasts Cryptococcus neoformans and Candida albicans and the malaria pathogen Plasmodium (Schlicker et al., 2009). Silencing of CA in N. benthamiana resulted in a faster growth of P. infestans, suggesting a role for this gene in resistance (Restrepo et al., 2005). Thus, CA may be important to both host and pathogen, although the underlying mechanisms remain mysterious.
To conclude, these results confirm previous suggestions that effectors modulate the outcome of the interaction with tomato, and we have identified a large number of such effectors that show stage‐predominant expression patterns. The identification of an effector that apparently suppresses necrosis late in the interaction suggests that the interaction is more complex than originally assumed by our initial hypothesis. This is in agreement with Dong et al. (2012), who found some NLP toxin proteins of P. sojae to be expressed at early stages (cyst germination) and only 50% of all the NLPs studied caused necrosis in N. benthamiana (Dong et al., 2012). However, our proposal that necrosis‐inducing factors operate late in the interaction was confirmed by detection late in the interaction of the abundant expression of apoplastic effectors, such as GHs, PiNPP and others. The dynamics of the plant host transcriptome that accompany the expression of the pathogen effectors are described in an accompanying paper (Zuluaga et al., in press), and may help guide the identification of molecular targets and identify the consequences of effector action.
Experimental Procedures
Plant material
Four‐week‐old tomato (Solanum lycopersicum cv. M82) glasshouse‐grown plants were used. Natural light was supplemented with 400‐W high‐pressure sodium lamps for 12 h and temperatures were maintained between 24 and 29 °C. Plants were grown in a soil‐less mix (Cornell mix) consisting of a 1 : 1 (v/v) peat–vermiculite mix supplemented with nitrogen, phosphorus and potassium (0.4 kg each per cubic metre of mix).
Inoculum preparation and P. infestans isolate
The tomato‐specialized P. infestans US‐11 (US050007) was used in this study, a genotype that most likely originated from a cross between US‐6 and US‐7 in the western USA (Gavino et al., 2000) and that has been sporadically problematic in the USA since the 1990s (Gavino et al., 2000). Sporangia used for inoculations were obtained from lesions on detached tomato leaflets in order to ensure that the inocula were robust and that P. infestans was virulent, as axenically grown inocula loses virulence throughout time (Mizubuti et al., 2000). Sporangia were harvested in distilled water and the concentration was adjusted to 4000 sporangia per millilitre using a haemacytometer. Subsequently, the sporangia were incubated at 4 °C for 1 h to release zoospores. After 1 h at 4 °C, a mixture of sporangia and zoospores is typically generated, as most of the sporangia germinate indirectly (forming zoospores), which is typically the case with field infections. Inoculation involved the application of a 20‐μL drop of this mixture of sporangia and zoospores to the abaxial side of the leaflet, and the inoculated leaflet was placed in a Petri dish containing water agar as a humid chamber. The drop was left on the leaflet until the tissue was harvested to ensure that the inoculated site was sampled, especially at early time points when symptoms of infection were not apparent.
Assessment of the biotrophic, transition to necrotrophic and necrotrophic phases
The interaction between P. infestans and tomato was studied over a time course spanning 7 days, in order to define the biotrophic, transition to necrotrophic and necrotrophic phases. To define the three pathogenicity stages, samples were collected every 12 h and analysed using three methods: (i) macroscopic observation of the tomato symptoms after P. infestans inoculation; (ii) microscopic observation of P. infestans development using trypan blue; and (iii) the use of P. infestans molecular markers that are specifically expressed at the biotrophic and necrotrophic stages of infection.
Macroscopic observation
The biotrophic period was defined as the time of inoculation until just before the tissue was first observed to be water soaked. The period from just prior to the appearance of water soaking to just before the appearance of necrosis was designated as the transition phase, and the necrotic phase occurred when the entire inoculation site (7 mm in diameter) was necrotic (Fig. S1).
Microscopic observation using trypan blue staining
Trypan blue staining was based on an established technique (Knox‐Davies, 1974), modified by Chung et al. (2010). Briefly, leaflets were submerged in a clearing solution A (acetic acid–ethanol, 1 : 3 v/v) overnight. After 16 h, the clearing solution A was discarded and replaced by clearing solution B (acetic acid–ethanol–glycerol, 1 : 5 : 1 v/v/v) for 3 h. Clearing solution B was replaced by staining solution (0.01% trypan blue in lactophenol) overnight. The staining solution was removed and the leaves were rinsed with sterile 60% glycerol. After rinsing, the glycerol was removed and new 60% glycerol was added to the leaflets for 2 h prior to microscopic observation.
RT‐PCR for stage‐specific P. infestans genes
For the RT‐PCR analyses, total RNA was extracted using the protocol of Perry and Francki (1992), as modified by Gu et al. (2000). DNaseI‐treated RNA (1 μg) was employed for cDNA synthesis, using the ImProm‐IITM Reverse Transcription System (Promega Madison, WI, USA), following the manufacturer's instructions. PCR was carried out with 2 μL of the cDNA synthesis reaction in a 30‐μL volume containing 0.2 mm of each deoxynucleoside triphosphate (dNTP), 2 μm of each of the primers (Table S1, see Supporting Information) and 0.5 U Taq polymerase (Invitrogen Grand Island, NY, USA). PCR conditions consisted of one cycle of 95 °C for 5 min, followed by 35 cycles of a three‐step procedure (1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C) and a final step of 5 min at 72 °C. As a control, RT‐PCR of the P. infestans Actin‐A gene was performed. The PCR conditions for Actin‐A were as described above. The relative intensities of the PCR amplified bands were assessed using ImageJ software (Abramoff et al., 2004).
Tissue collection and 454‐sequencing
Tissue was collected from P. infestans‐inoculated leaves at 48, 96 and 144 hai. Leaf discs from the centre of the inoculation sites were harvested using a paper hole puncher (7 mm in diameter; Fig. S6, see Supporting Information) and immediately frozen in liquid nitrogen. Twenty‐five tomato plants per time point were used for each experimental trial and the experiment was repeated three times. The leaf discs from the four trials were then pooled (100 plants per time point) and total RNA was extracted as above.
mRNA was isolated from 250 ng of total RNA and amplified using a TargetAmp™ One‐Round aRNA Amplification Kit 103 (Epicentre Biotechnologies Madison, WI, USA). First, poly‐A RNA was transcribed from total RNA to generate first‐strand cDNAs. The reaction was primed with a synthetic oligo (dT) primer containing a phage T7 RNA polymerase promoter sequence at its 5′ end. The first‐strand cDNA synthesis was catalysed by SuperScript III reverse transcriptase (Invitrogen) generating a cDNA:RNA hybrid. The RNA component of the cDNA:RNA hybrid was then digested using the RNase H enzyme and the RNA fragments primed the second‐strand cDNA synthesis. The resulting product was a double‐stranded cDNA containing T7 transcriptional promoter in an orientation that generated anti‐sense RNA. High yields of anti‐sense RNA were produced in a rapid in vitro transcription reaction (amplified RNA) using the double‐stranded cDNAs (Epicentre Biotechnologies).
cDNA was synthesized from three reactions of 5 μg of amplified RNA, yielding a total of 15 μg of amplified RNA per sample, using 100 ng of random hexamers in each reaction. The SuperScript choice system was used for cDNA synthesis (Invitrogen) following the manufacturer's instructions. Following second‐strand synthesis, the cDNA samples were purified using a PureLink™ PCR purification kit (Invitrogen Grand Island, NY, USA) following the manufacturer's instructions and quantified using a Nanodrop 2000 apparatus (Thermos Scientific), Madison, WI, USA., with a minimum of 9 μg of cDNA per sample. The construction of cDNA libraries for 454‐sequencing took place at the Cornell University Life Sciences Core Laboratories Center. The raw 454‐sequencing reads have been deposited at the National Center for Biotechnology Information (NCBI) SRA under accession SRA027389.
cDNA sequence processing and assembly
The raw 454‐sequence files in SFF format were base called using the Pyrobayes base caller (Quinlan et al., 2008). The sequences were then processed to remove low‐quality regions and adaptor sequences using LUCY (Chou and Holmes, 2001) and SeqClean. The resulting high‐quality sequences were then screened against the NCBI UniVec database, the Escherichia coli genome sequences and Phytophthora ribosomal RNA, to remove contaminants. Sequences shorter than 30 bp were discarded. To identify P. infestans transcript sequences, the cDNA sequences were aligned with the P. infestans, P. sojae and P. ramorum genomes (Broad Institute, Cambridge, MA, USA), using SPALN (Gotoh, 2008) for those longer than 100 bp and BLAT (Kent, 2002) for those shorter than 100 bp. Sequences that could be aligned with any of the three Phytophthora genomes with at least 90% sequence identity and 50% length coverage were regarded as derived from P. infestans, whereas the remaining sequences were designated as derived from tomato. Phytophthora infestans cDNA sequences, together with P. infestans transcripts predicted from the genome sequences (Haas et al., 2009), were assembled into unigenes using the iAssembler program (Zheng et al., 2011).
Unigene annotation and pathway prediction
Phytophthora infestans unigenes were employed to interrogate the GenBank non‐redundant protein and UniProt databases using blastx with a cut‐off e‐value of 1e‐5. The unigene sequences were also translated into proteins using ESTScan (Iseli et al., 1999) and the translated protein sequences were then compared with InterPro and pfam domain databases. The gene ontology (GO) terms were assigned to each unigene based on the GO terms annotated to the corresponding homologues in the UniProt database (Camon et al., 2004), as well as to the InterPro and pfam domains (cut‐off value of 1e‐5), using interpro2go and pfam2go mapping files, respectively, provided by the GO website. The CAZy database was used to define the members of each GH family.
Identification of differentially expressed genes
The 454‐reads were normalized with the calculation: number of reads of a unigene from the specific sample × 100 000 (that is, the number of reads if 100 000 reads are collected)/total number of reads collected from that specific sample. The significance of differential gene expression was determined using the R statistic described in Stekel et al. (2000), and the resulting raw P values were adjusted for multiple testing using FDR (Benjamin and Hochberg, 1995). Genes with a fold change of >2.0 from one infection time point to another and FDR < 0.05 were considered to be differentially expressed genes. GO terms enriched in the set of differentially expressed genes were identified using GO::TermFinder (Boyle et al., 2004), requiring P values adjusted for multiple testing to be less than 0.05.
A hierarchical clustering was performed to identify groups of genes with similar expression patterns, using the expression values from the three pathogenicity stages of P. infestans. The analysis was conducted using MATLAB version 7.8 (R2009a).
Cloning of P. infestans putative effector genes
Putative P. infestans effector genes were cloned using total RNA from the three pathogenicity stages. A SuperScript™III One‐step RT‐PCR system with Platinum® Taq High Fidelity (Invitrogen) was used to synthesize the full‐length cDNA of each candidate gene with gene‐specific primers (Table S2, see Supporting Information). The PCR amplification conditions were as described above for the stage‐specific primers. Amplified PCR fragments were purified using a QIAquick® PCR purification kit (Qiagen Grand Island, NY, USA) following the manufacturer's instructions. Candidates were cloned into pGEMT®‐easy vector with 2 × rapid ligation buffer (Promega). Plasmids were sequenced using T7 universal primers in the Genomics Facility at Cornell University. Once the sequence was confirmed, the candidate genes were excised from the plasmid using the respective restriction enzymes (Table S2) and purified in a 1.2% agarose gel using the E.Z.N.A™ Gel extraction kit (Omega bio‐Tek Norcross, GA, USA). Candidate genes were ligated into the pART plasmid (Kelley et al., 2010) at 15 °C overnight. After ligation, the plasmids were transformed into E. coli and the inserts were confirmed by sequencing using the 35S promoter primer 5′‐CTATCCTTCGCAAGACCCTTC‐3′. After sequence confirmation, the plasmid was transformed into Agrobacterium tumefaciens strain GV3101.
Transient expression assays in Nicotiana benthamiana
Transient expression of recombinant proteins in N. benthamiana was performed as described previously (Bos et al., 2006). Agrobacterium tumefaciens strains were grown at 29 °C for 24 h in induction medium (Sessa et al., 2000) and then centrifuged for 5 min at 10 000 g; the pellet was resuspended in 5 mL of infiltration medium [10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.5, and 200 μm acetosyringone] and centrifuged again as above. Cells were resuspended in new infiltration medium and the optical density at 600 nm (OD600) was adjusted to 0.3. For cell death assays, A. tumefaciens GV3101 carrying the gene of interest was infiltrated into N. benthamiana leaves with a 1‐mL needle‐less syringe. After 24 h, the necrosis inducer PiNPP1‐1 (kindly provided by Dr S. Kamoun, Sainsbury Laboratory, John Innes Centre, Norwich, UK) was infiltrated at the same OD. The plants were scored for necrosis symptoms every 24 h for 4 days.
Supporting information
File S1 Filtered file with normalized number of reads from the Phytophthora infestans RNA‐Seq dataset.
File S2 Orphan Phytophthora infestans genes.
File S3 Differentially expressed Phytophthora infestans genes.
File S4 Putative Phytophthora infestans pathogenicity genes.
File S5 Normalized number of reads for each effector homologue.
File S6 Hierarchical cluster analysis of Phytophthora infestans genes.
Fig. S1 Macroscopic observation of the time course of the Phytophthora infestans–tomato interaction. Time course of the P. infestans–tomato interaction showing macroscopic evaluation at 48, 72, 96, 120, 144 and 168 h after inoculation (hai). Tissues were analysed every 12 hai, and the time points shown here illustrate a summary of disease development. The pathogen biotrophic growth can be seen up to 72 hai (no symptoms on the leaflets) and water‐soaking lesions are evident at 96 hai. Sporulation surrounding the necrotic lesion is seen at 120 hai and a well‐established necrotrophic stage can be seen at 144 hai.
Fig. S2 Expression profile of the molecular markers used to characterize the Phytophthora infestans developmental stage. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) evaluation of the expression of P. infestans biotrophic (IpiO and SNE1) and necrotrophic (PiNPP1.1) stage‐specific marker genes in infected tomato leaves at 12, 24, 36, 48, 96, 108, 120 and 132 h after inoculation.
Fig. S3 Percentage of genes mapping to the Phytophthora infestans, P. sojae and P. ramorum genomes. Of the 86 319 454 transcripts identified as Phytophthora, 99.57% were mapped to the P. infestans reference genome T30‐4, 25.45% to the P. sojae reference genome and 21.66% to the P. ramorum reference genome, with at least 90% sequence identity and 50% length coverage.
Fig. S4 RNA‐Seq representation of Phytophthora infestans stage‐specific gene expression. Normalized 454‐reads for the three stage‐specific genes IpiO, SNE1, PiNPP1.1 and P. infestans Actin‐A. Units are normalized to the number of reads.
Fig. S5 Hierarchical cluster analysis of Phytophthora infestans gene expression using the total number of genes from the filtered file. Based on the transcript profile, five clusters were generated identified as yellow circles (P1–P5). Red represents values above the mean, black represents the mean, and green represents values below the mean of a row (gene) across all columns (time points, shown as hours after inoculation).
Fig. S6 Sample collection method using a paper hole puncher. Leaf discs from the centre of the inoculation sites were harvested using a paper hole puncher (7 mm in diameter) and immediately frozen in liquid nitrogen. The red dotted line corresponds to 7 mm.
Table S1 Primers used to determine the pathogenicity stage of Phytophthora infestans.
Table S2 Primers used for cloning candidate effectors for heterologous expression in Nicotiana benthamiana.
Acknowledgements
We thank Drs Jose C. Huguet, Rafael O. Tinoco‐López and Gregory Martin for valuable advice and Kent Loeffler for photographic assistance. This work was supported by a grant from the National Science Foundation (Plant Genome Program; DBI‐0606595) and by support from the College of Agriculture and Life Sciences at Cornell University. The authors declare no conflict of interest.
References
- Abramoff, M.D. , Magalhaes, P.J. and Ram, S.J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42. [Google Scholar]
- Abramovitch, R.B. and Martin, G.B. (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7, 356–364. [DOI] [PubMed] [Google Scholar]
- As‐sadi, F. , Carrere, S. , Gascuel, Q. , Hourlier, T. , Rengel, D. , Le Paslier, M.C. , Bordat, A. , Boniface, M.C. , Brunel, D. , Gouzy, J. , Godiard, L. and Vincourt, P. (2011) Transcriptomic analysis of the interaction between Helianthus annuus and its obligate parasite Plasmopara halstedii shows single nucleotide polymorphisms in CRN sequences. BMC Genomics, 12, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J. and Staskawicz, B.J. (2003) Initiation of RPS2‐specified disease resistance in Arabidopsis is coupled to the AvrRpt2‐directed elimination of RIN4. Cell, 112, 369–377. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D. , von Heijne, H.N.G. and Brunak, S.J. (2004) Improved prediction of signal peptides: SignalP 3.0. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B, 57, 289–300. [Google Scholar]
- Berg, A. (1926) Tomato late blight and its relation to late blight of potato. Experimental Station Bulletin 205, pp. 1–31. Charleston, WV: West Virginia Agriculture Department. [Google Scholar]
- Bos, J.I. , Kanneganti, T.D. , Young, C. , Cakir, C. , Huitema, E. , Win, J. , Armstrong, M.R. , Birch, P.R. and Kamoun, S. (2006) The C‐terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a‐mediated hypersensitivity and suppress INF1‐induced cell death in Nicotiana benthamiana . Plant J. 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Boyle, E.I. , Weng, S. , Gollub, J. , Jin, H. , Botstein, D. , Cherry, J.M. and Sherlock, G. (2004) GO:TermFinder: open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics, 20, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, G. , Restrepo, S. , Myers, K. , Zuluaga, A.P. , Danies, G. , Smart, C. and Fry, W.E. (2013) Gene profiling in partially resistant and susceptible near‐isogenic tomatoes in response to late blight in the field. Mol. Plant Pathol. 14, 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon, E. , Magrane, M. , Barrell, D. , Lee, V. , Dimmer, E. , Maslen, J. , Binns, D. , Harte, N. , Lopez, R. and Apweiler, R. (2004) The Gene Ontology Annotation (GOA) Database: sharing knowledge in uniprot with gene ontology. Nucleic Acids Res. 32, D262–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, H.H. and Holmes, M.H. (2001) DNA sequence quality trimming and vector removal. Bioinformatics, 17, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Chung, C.L. , Longfellow, J.M. , Walsh, E.K. , Kerdieh, Z. , Van Esbroeck, G. , Balint‐Kurti, P. and Nelson, R.J. (2010) Resistance loci affecting distinct stages of fungal pathogenesis: use of introgression lines for QTL mapping and characterization in the maize–Setosphaeria turcica pathosystem. BMC Plant Biol. 10, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.J. , White, G.J. , Rodriguez‐Carres, M. and VanEtten, H.D. (2011) An ABC transporter and a Cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant–Microbe Interact. 24, 368–376. [DOI] [PubMed] [Google Scholar]
- Damasceno, C.M. , Bishop, J.G. , Ripoll, D.R. , Win, J. , Kamoun, S. and Rose, J.K.C. (2008) Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo‐beta‐1,3‐glucanases. Mol. Plant–Microbe Interact. 21, 820–830. [DOI] [PubMed] [Google Scholar]
- van Damme, M. , Bozkurt, T.O. , Cakir, C. , Schornack, S. , Sklenar, J. , Jones, A.M.E. and Kamoun, S. (2012) The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. Plos Pathog. 8, e1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Qutob, D. , Tedman‐Jones, J. , Kuflu, K. , Wang, Y. , Tyler, B.M. and Gijzen, M. (2009) The Phytophthora sojae avirulence locus Avr3c encodes a multi‐copy RXLR effector with sequence polymorphisms among pathogen strains. Plos ONE, 4, e5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S. , Kong, G. , Qutob, D. , Yu, X. , Tang, J. , Kang, J. , Dai, T. , Wang, H. , Gijzen, M. and Wang, Y. (2012) The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis‐inducing activity. Mol. Plant–Microbe Interact. 25, 896–909. [DOI] [PubMed] [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis, S. , Hacquard, S. , Delaruelle, C. , Tisserant, E. , Frey, P. , Martin, F. and Kohler, A. (2011) Melampsora larici‐populina transcript profiling during germination and time‐course infection of poplar leaves reveals dynamic expression patterns associated with virulence and biotrophy. Mol. Plant–Microbe Interact. 24, 808–818. [DOI] [PubMed] [Google Scholar]
- Fernandez, D. , Tisserant, E. , Talhinhas, P. , Azinheira, H. , Vieira, A. , Petitot, A.S. , Loureiro, A. , Poulain, J. , Da Silva, C. , Silva, M.C. and Duplessis, S. (2012) 454‐pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta‐expressed pathogen‐secreted proteins and plant functions in a late compatible plant–rust interaction. Mol. Plant Pathol. 13, 17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, P. , Ikeda, K. , Irieda, H. , Narusaka, M. , O'Connell, R.J. , Narusaka, Y. , Takano, Y. , Kubo, Y. and Shirasu, K. (2013) Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197, 1236–1249. [DOI] [PubMed] [Google Scholar]
- Gavino, P.D.C.D. , Smart, C.D. , Sandrock, R.W. , Miller, J.S. , Hamm, P.B. , Lee, T.Y. , Davis, R.M. and Fry, W.E. (2000) Implications of sexual reproduction for Phytophthora infestans in the United States: generation of an aggressive lineage. Plant Disease, 84, 731–735. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gotoh, O. (2008) A space‐efficient and accurate method for mapping and aligning cDNA sequences onto genomic sequence. Nucleic Acids Res. 36, 2630–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Avrova, A.O. , Bruce, C.R. , Williams, A. , Whisson, S.C. , Birch, P.R. and van West, P. (2005) Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet. Biol. 42, 244–256. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q. , Yang, C. , Thara, V.K. , Zhou, J. and Martin, G.B. (2000) The Pti4 gene is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell, 12, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. , Zheng, Y. , Joung, J.G. , Liu, S. , Zhang, Z. , Crasta, O.R. , Sobral, B.W. , Xu, Y. , Huang, S. and Fei, Z. (2010) Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics, 11, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A. , Baxter, L. , Beynon, J. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J. , Grunwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M. , Jones, J.D. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L. , Maclean, D. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J. , Morgan, W. , Morris, P.F. , Munro, C.A. , O'Neill, K. , Ospina‐Giraldo, M. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W. , Meyers, B.C. , van West, P. , Ristaino, J. , Govers, F. , Birch, P.R. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hahn, M. and Mendgen, K. (2001) Signal and nutrient exchange at biotrophic plant–fungus interfaces. Curr. Opin. Plant Biol. 4, 322–327. [DOI] [PubMed] [Google Scholar]
- Halterman, D.A. , Chen, Y. , Sopee, J. , Berduo‐Sandoval, J. and Sánchez‐Pérez, A. (2010) Competition between Phytophthora infestans effectors leads to increased aggressiveness on plants containing broad‐spectrum late blight resistance. Plos ONE, 5, e10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K. , Schoonbeek, H. and De Waard, M.A. (2002) Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 68, 4996–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli, C. , Jongeneel, C.V. and Bucher, P. (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 138–148. [PubMed] [Google Scholar]
- Jupe, J. , Stam, R. , Howden, A.J.M. , Morris, J.A. , Zhang, R. , Hedley, P.E. and Huitema, E. (2013) Phytophthora capsici–tomato interaction features dramatic shifts in gene expression associated with a hemi‐biotrophic lifestyle. Genome Biol. 14, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. , Lindqvist, H. and Govers, F. (1997) A novel class of elicitin‐like genes from Phytophthora infestans . Mol. Plant–Microbe Interact. 10, 1028–1030. [DOI] [PubMed] [Google Scholar]
- Kanneganti, T.D. , Huitema, E. , Cakir, C. and Kamoun, S. (2006) Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl‐like protein PiNPP1.1 and INF1 elicitin. Mol. Plant–Microbe Interact. 19, 854–863. [DOI] [PubMed] [Google Scholar]
- Kelley, B.S. , Lee, S.J. , Damasceno, C.M. , Chakravarthy, S. , Kim, B.D. , Martin, G.B. and Rose, J.K.C. (2010) A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J. 62, 357–366. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. (2002) BLAT—the BLAST‐like alignment tool. Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.R.F. , McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Bukharova, T. , Sukarta, O. , Win, J. , Kamoun, S. , Birch, P.R.J. and Banfield, M.J. (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell, 26, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox‐Davies, P.S. (1974) Penetration of maize leaves by Helminthosporium turicum . Phytopathology, 64, 1468–1470. [Google Scholar]
- Lee, H.A. , Kim, S.Y. , Oh, S.K. , Yeom, S.I. , Kim, S.B. , Kim, M.S. , Kamoun, S. and Choi, D. (2014) Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans . New Phytol. 203, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.J. and Rose, J.K.C. (2010) Mediation of the transition from biotrophy to necrotrophy in hemibiotrophic plant pathogens by secreted effector proteins. Plant Signal Behav. 5, 769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Ye, W. , Ru, Y. , Yang, X. , Gu, B. , Tao, K. , Lu, S. , Dong, S. , Zheng, X. , Shan, W. , Wang, Y. and Dou, D. (2011) Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant Physiol. 155, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D. , Belkhadir, Y. , Alfonso, J.M. , Ecker, J.R. and Dangl, J.L. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2‐mediated resistance. Cell, 112, 379–389. [DOI] [PubMed] [Google Scholar]
- Mane, S.P. , Evans, C. , Cooper, K.L. , Crasta, O.R. , Folkerts, O. , Hutchison, S.K. , Harkins, T.T. , Thierry‐Mieg, D. , Thierry‐Mieg, J. and Jensen, R.V. (2009) Transcriptome sequencing of the Microarray Quality Control (MAQC) RNA reference samples using next generation sequencing. BMC Genomics, 10, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.E. and VanEtten, H.D. (1983) Detoxification of the phytoalexin pisatin by a fungal cytochrome P‐450. Arch. Biochem. Biophys. 224, 494–505. [DOI] [PubMed] [Google Scholar]
- McLeod, A. , Smart, C.D. and Fry, W.E. (2003) Characterization of 1,3‐β‐glucanase and 1,3;1,4‐β‐glucanase genes from Phytophthora infestans . Fungal Genet. Biol. 38, 250–263. [DOI] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Hassen, H.H. and Govers, F. (2011) Phytophthora infestans has a plethora of phospholipase D enzymes including a subclass that has extracellular activity. Plos ONE, 6, e1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikes, V. , Milat, M.L. , Ponchet, M. , Panabieres, F. , Ricci, P. and Blein, J.P. (1998) Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem. Biophys. Res. Commun. 245, 133–139. [DOI] [PubMed] [Google Scholar]
- Mizubuti, E.S.G. , Aylor, D.E. and Fry, W.E. (2000) Survival of Phytophthora infestans sporangia exposed to solar radiation. Phytopathology, 90, 78–84. [DOI] [PubMed] [Google Scholar]
- O'Connell, R.J. , Thon, M.R. , Hacquard, S. , Amyotte, S.G. , Kleemann, J. , Torres, M.F. , Damm, U. , Buiate, E.A. , Epstein, L. , Alkan, N. , Altmüller, J. , Alvarado‐Balderrama, L. , Bauser, C.A. , Becker, C. , Birren, B.W. , Chen, Z. , Choi, J. , Crouch, J.A. , Duvick, J.P. , Farman, M.A. , Gan, P. , Heiman, D. , Henrissat, B. , Howard, R.J. , Kabbage, M. , Koch, C. , Kracher, B. , Kubo, Y. , Law, A.D. , Lebrun, M.H. , Lee, Y.H. , Miyara, I. , Moore, N. , Neumann, U. , Nordström, K. , Panaccione, D.G. , Panstruga, R. , Place, M. , Proctor, R.H. , Prusky, D. , Rech, G. , Reinhardt, R. , Rollins, J.A. , Rounsley, S. , Schardl, C.L. , Schwartz, D.C. , Shenoy, N. , Shirasu, K. , Sikhakolli, U.R. , Stüber, K. , Sukno, S.A. , Sweigard, J.A. , Takano, Y. , Takahara, H. , Trail, F. , van der Does, H.C. , Voll, L.M. , Will, I. , Young, S. , Zeng, Q. , Zhang, J. , Zhou, S. , Dickman, M.B. , Schulze‐Lefert, P. , Ver Loren van Themaat, E. , Ma, L.J. and Vaillancourt, L.J. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S.K. , Young, C. , Lee, M. , Oliva, R. , Bozkurt, T.O. , Cano, L.M. , Win, J. , Bos, J.I. , Liu, H.Y. , van Damme, M. , Morgan, W. , Choi, D. , Van der Vossen, E.A. , Vleeshouwers, V.G. and Kamoun, S. (2009) In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi‐blb2. Plant Cell, 21, 2928–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, K.L. and Francki, R.I.B. (1992) Insect‐mediated transmission of mixed and reassorted cucumovirus genomic RNAs. J. Gen. Virol. 73, 2105–2114. [DOI] [PubMed] [Google Scholar]
- Quinlan, A.R. , Stewart, D.A. , Strömberg, M.P. and Marth, G.T. (2008) Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat. Methods, 5, 179–181. [DOI] [PubMed] [Google Scholar]
- Raffaele, S. , Win, J. , Cano, L.M. and Kamoun, S. (2010) Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans . BMC Genomics, 11, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo, S. , Myers, K.L. , del Pozo, O. , Martin, G.B. , Hart, A.L. , Buell, C.R. , Fry, W.E. and Smart, C.D. (2005) Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol. Plant–Microbe Interact. 18, 913–922. [DOI] [PubMed] [Google Scholar]
- Rietman, H. , Bijsterbosch, G. , Cano, L.M. , Lee, H.R. , Vossen, J.H. , Jacobsen, E. , Visser, R.G.F. , Kamoun, S. and Vleeshouwers, V.G.A.A. (2012) Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol. Plant–Microbe Interact. 25, 910–919. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Herva, J.J. , González‐Melendi, P. , Cuartas‐Lanza, R. , Antúnez‐Lamas, M. , Rio‐Alvarez, I. , Li, Z. , López‐Torrejón, G. , Díaz, I. , del Pozo, J.C. , Chakravarthy, S. , Collmer, A. , Rodríguez‐Palenzuela, P. and López‐Solanilla, E. (2012) A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 14, 669–681. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C. , Ham, K.‐S. , Darvill, A.G. and Albersheim, P. (2002) Molecular cloning and characterization of glucanase inhibitor proteins (GIPs): co‐evolution of a counter‐defense mechanism by plant pathogens. Plant Cell, 14, 1329–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker, C. , Hall, R.A. , Vullo, D. , Middelhaufe, S. , Gertz, M. , Supuran, C.T. , Muhlschlegel, F.A. and Steegborn, C. (2009) Structure and inhibition of the CO2‐sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans . J. Mol. Biol. 385, 1207–1220. [DOI] [PubMed] [Google Scholar]
- Sessa, G. , D'Ascenzo, M. and Martin, G.B. (2000) Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto–Pto‐mediated hypersensitive response. EMBO J. 19, 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart, C.D. , Myers, K.L. , Restrepo, S. , Martin, G.B. and Fry, W.E. (2003) Partial resistance of tomato to Phytophthora infestans is not dependent upon ethylene, jasmonic acid or salicylic acid signaling pathways. Mol. Plant–Microbe Interact. 16, 141–148. [DOI] [PubMed] [Google Scholar]
- Stekel, D.J. , Git, Y. and Falciani, F. (2000) The comparison of gene expression from multiple cDNA libraries. Genome Res. 10, 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Huitema, E. , Da Cunha, L. , Torto‐Alalibo, T. and Kamoun, S. (2004) A Kazal‐like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis‐related protease P69B. J. Biol. Chem. 279, 26 370–26 377. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Win, J. , Song, J. , van der Hoorn, R. , van der Knaap, E. and Kamoun, S. (2007) A Phytophthora infestans cystatin‐like protein targets a novel tomato papain‐like apoplastic protease. Plant Physiol. 143, 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto, T. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A.R. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from Phytophthora . Genome Res. 13, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto‐Alalibo, T.A. , Tripathy, S. , Smith, B.M. , Arredondo, F.D. , Zhou, L. , Li, H. , Chibucos, M.C. , Qutob, D. , Gijzen, M. , Mao, C. , Sobral, B.W. , Waugh, M.E. , Mitchell, T.K. , Dean, R.A. and Tyler, B.M. (2007) Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol. Plant–Microbe Interact. 20, 781–793. [DOI] [PubMed] [Google Scholar]
- Vega‐Arreguín, J.C. , Ibarra‐Laclette, E. , Jimenez‐Moraila, B. , Martinez de la Vega, O. , Vielle‐Calzada, J.P. , Herrera‐Estrella, L. and Herrera‐Estrella, A. (2009) Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics, 10, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega‐Sanchez, M.E. , Erselius, L.J. , Rodriguez, A.M. , Bastidas, O. , Hohl, H.R. , Ojiambo, P.S. , Mukalazi, J. , Vermeulen, T. , Fry, W.E. and Forbes, G.A. (2000) Host adaptation to potato and tomato within the US‐1 clonal lineage of Phytophthora infestans in Uganda and Kenya. Plant Pathol. 49, 531–539. [Google Scholar]
- Vleeshouwers, V.G. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.K. , Wang, M. , Bouwmeester, K. , Vosman, B. , Visser, R.G. , Jacobsen, E. , Govers, F. , Kamoun, S. and Van der Vossen, E.A. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. Plos ONE, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. , Kale, S.D. , Gu, B. , Sheng, Y. , Sui, Y. , Wang, X. , Zhang, Z. , Cheng, B. , Dong, S. , Shan, W. , Zheng, X. , Dou, D. , Tyler, B.M. and Wang, Y. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.P. , Weber, K.L. , Carr, K. , Wilkerson, C. and Ohlrogge, J.B. (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 144, 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van West, P. , de Jong, A.J. , Judelson, H.S. , Emons, A.M. and Govers, F. (1998) The IpiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet. Biol. 23, 126–138. [DOI] [PubMed] [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Win, J. , Kanneganti, T.D. , Torto‐Alalibo, T. and Kamoun, S. (2006) Computational and comparative analyses of 150 full‐length cDNA sequences from the oomycete plant pathogen Phytophthora infestans . Fungal Genet. Biol. 43, 20–33. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Zhao, L. , Gao, J. and Fei, Z. (2011) iAssembler: a package for de novo assembly of Roche‐454/Sanger transcriptome sequences. BMC Bioinform. 12, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuluaga, A.P. , Vega‐Arreguín, J.C. , Fei, Z. , Matas, A.J. , Fry, W.E. and Rose, J.K.C. (in press) Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans . Mol. Plant Pathol. DOI: 10.1111/mpp.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.H. , Stergiopoulos, I. , Gielkens, M.M.C. , Goodall, S.D. and De Waard, M.A. (2003) ABC transporters of the wheat pathogen Mycosphaerella graminicola function as protectants against biotic and xenobiotic toxic compounds. Mol. Genet. Genomics, 269, 499–507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1 Filtered file with normalized number of reads from the Phytophthora infestans RNA‐Seq dataset.
File S2 Orphan Phytophthora infestans genes.
File S3 Differentially expressed Phytophthora infestans genes.
File S4 Putative Phytophthora infestans pathogenicity genes.
File S5 Normalized number of reads for each effector homologue.
File S6 Hierarchical cluster analysis of Phytophthora infestans genes.
Fig. S1 Macroscopic observation of the time course of the Phytophthora infestans–tomato interaction. Time course of the P. infestans–tomato interaction showing macroscopic evaluation at 48, 72, 96, 120, 144 and 168 h after inoculation (hai). Tissues were analysed every 12 hai, and the time points shown here illustrate a summary of disease development. The pathogen biotrophic growth can be seen up to 72 hai (no symptoms on the leaflets) and water‐soaking lesions are evident at 96 hai. Sporulation surrounding the necrotic lesion is seen at 120 hai and a well‐established necrotrophic stage can be seen at 144 hai.
Fig. S2 Expression profile of the molecular markers used to characterize the Phytophthora infestans developmental stage. Semi‐quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) evaluation of the expression of P. infestans biotrophic (IpiO and SNE1) and necrotrophic (PiNPP1.1) stage‐specific marker genes in infected tomato leaves at 12, 24, 36, 48, 96, 108, 120 and 132 h after inoculation.
Fig. S3 Percentage of genes mapping to the Phytophthora infestans, P. sojae and P. ramorum genomes. Of the 86 319 454 transcripts identified as Phytophthora, 99.57% were mapped to the P. infestans reference genome T30‐4, 25.45% to the P. sojae reference genome and 21.66% to the P. ramorum reference genome, with at least 90% sequence identity and 50% length coverage.
Fig. S4 RNA‐Seq representation of Phytophthora infestans stage‐specific gene expression. Normalized 454‐reads for the three stage‐specific genes IpiO, SNE1, PiNPP1.1 and P. infestans Actin‐A. Units are normalized to the number of reads.
Fig. S5 Hierarchical cluster analysis of Phytophthora infestans gene expression using the total number of genes from the filtered file. Based on the transcript profile, five clusters were generated identified as yellow circles (P1–P5). Red represents values above the mean, black represents the mean, and green represents values below the mean of a row (gene) across all columns (time points, shown as hours after inoculation).
Fig. S6 Sample collection method using a paper hole puncher. Leaf discs from the centre of the inoculation sites were harvested using a paper hole puncher (7 mm in diameter) and immediately frozen in liquid nitrogen. The red dotted line corresponds to 7 mm.
Table S1 Primers used to determine the pathogenicity stage of Phytophthora infestans.
Table S2 Primers used for cloning candidate effectors for heterologous expression in Nicotiana benthamiana.


