Summary
Members of the F usarium solani species complex (FSSC) are capable of causing disease in many agriculturally important crops. The genomes of some of these fungi include supernumerary chromosomes that are dispensable and encode host‐specific virulence factors. In addition to genomics, this review summarizes the known molecular mechanisms utilized by members of the FSSC in establishing disease.
Taxonomy
Kingdom Fungi; Phylum Ascomycota; Class Sordariomycetes; Order Hypocreales; Family Nectriaceae; Genus F usarium.
Host range
Members of the FSSC collectively have a very broad host range, and have been subdivided previously into formae speciales. Recent phylogenetic analysis has revealed that formae speciales correspond to biologically and phylogenetically distinct species.
Disease symptoms
Typically, FSSC causes foot and/or root rot of the infected host plant, and the degree of necrosis correlates with the severity of the disease. Symptoms on above‐ground portions of the plant can vary greatly depending on the specific FSSC pathogen and host plant, and the disease may manifest as wilting, stunting and chlorosis or lesions on the stem and/or leaves.
Control
Implementation of agricultural management practices, such as crop rotation and timing of planting, can reduce the risk of crop loss caused by FSSC. If available, the use of resistant varieties is another means to control disease in the field.
Useful websites
Keywords: cell wall‐degrading enzyme, conditionally dispensable, Fusarium solani, horizontal gene/chromosome transfer, phytoalexin tolerance, sudden death syndrome, supernumerary chromosome
Introduction
The genus Fusarium constitutes a large monophyletic group of several hundred species that, with the exception of a few, produce characteristic fusoid multiseptate macroconidia. The genus includes agriculturally important plant pathogens, endophytes and saprophytes capable of metabolizing diverse substrates, and emerging pathogens of clinical importance. Molecular phylogenetic analysis of this genus has revealed that it comprises at least 20 clades or ‘species complexes’ and that it originated in the middle Cretaceous period approximately 91.3 million years ago (O'Donnell et al., 2013).
The Fusarium solani species complex (FSSC) is a group currently estimated to contain at least 60 phylogenetically distinct species (Nalim et al., 2011; O'Donnell, 2000; O'Donnell et al., 2008; Zhang et al., 2006). As defined by Snyder and Hansen (1941), F. solani was the only species in what is now recognized as the FSSC. As members of the FSSC have been most extensively studied as plant pathogens, these fungi have been further subdivided into formae speciales (f. sp.) based on host specificity. Members of the FSSC represent a diverse set of self‐fertile (homothallic) and self‐sterile (heterothallic) species, although sexual cycles are only known for approximately one‐third of the species (O'Donnell, 2000; O'Donnell et al., 2008). Seven of the heterothallic species were originally classified as mating populations (MPs I–VII) of Nectria haematococca because of their ability to mate with other members of the same biological species (Matuo and Snyder, 1973); however, each mating population has been shown to represent a phylogenetically distinct species (O'Donnell, 2000). On the basis of molecular phylogenetic relationships among isolates, the FSSC comprises three major clades, where Clade 3 encompasses all of the isolates formerly characterized as N. haematococca (O'Donnell, 2000; Zhang et al., 2006). In addition to N. haematococca (Booth, 1971), several other teleomorphic names, including ‘Hypomyces solani’ (Reichle et al., 1964) and ‘Haematonectria haematococca’ (Rossman et al., 1999), have been applied to these seven mating populations. Recently, ‘Neocosmospora haematococca’ has been proposed for the teleomorph of these fungi, as it predates (from 1899) the traditionally used names (Lombard et al., 2015). Although previous nomenclatural rules dictated that the teleomorphic name of a fungus took preference over the anamorphic name, as of 2013 this stipulation no longer applies (Geiser et al., 2013). The abandonment of the teleomorphic names, the use of the name F. solani for only one species within the FSSC and the discovery of many phylogenetically distinct species have created a situation in which most species are unnamed. In lieu of Latin binomial names, a three‐locus haplotype nomenclatural system was developed for the members of Clade 3 of the FSSC based on polymorphisms in DNA sequences of translation elongation factor 1α (EF‐1α), the second largest subunit of RNA polymerase II (RPB2), and the nuclear ribosomal internal transcribed spacer region and domains D1 and D2 in the nuclear large subunit (Chang et al., 2006; O'Donnell et al., 2008). For the purpose of this review, the FSSC species will be referred to by the multilocus haplotype number and any traditionally used name when first introduced (Table 1).
Table 1.
Taxonomy of the F usarium solani species complex (FSSC) species of relevance to this review
| Phylogenetic species | Current scientific name | Traditional name(s) | Reference(s) |
|---|---|---|---|
| FSSC 1 | F. petroliphilum | Nectria haematococca MP V; F. solani f. sp. cucurbitae race 2 | Short et al. (2013) |
| FSSC 2 | F. keratoplasticum | Short et al. (2013) | |
| FSSC 3+4 | F. falcilforme | Cylindrocarpon lichenicola; Acremonium falciforme | Summerbell and Schroers (2002) |
| FSSC 8 | F. neocosmosporiellum | Neocosmospora vasinfectum | O'Donnell et al. (2013) |
| FSSC 10 | Unnamed | N. haematococca MP I; F. solani f. sp. cucurbitae race 1 | |
| FSSC 11 | Unnamed | N. haematococca MP VI; F. solani f. sp. pisi | |
| FSSC 13 | Unnamed | N. haematococca MP VII; F. solani f. sp. robiniae | |
| FSSC 17 | Unnamed | N. haematococca MP III; F. solani f. sp. mori | |
| FSSC 22 | Unnamed | N. haematococca MP IV; F. solani f. sp. xanthoxyli | |
| FSSC 23 | Unnamed | N. haematococca MP II; F. solani f. sp. batatas | |
| na | F. phaseoli | F. solani f. sp. phaseoli | |
| na |
F. virguliforme
F. tucumaniae F. brasiliense F. cuneirostrum |
F. solani f. sp. glycines | Aoki et al. (2003) |
MP, mating population. Phylogenetically distinct species within Clade 3 of the FSSC are distinguished by Arabic numerals (see O'Donnell et al., 2008).
na, not applicable.
Members of the FSSC are important pathogens of a number of agriculturally important crops. The diverse host range exists not only throughout the species complex, but also within a single species. For example, isolates of FSSC 11 (N. haematococca MP VI; F. solani f. sp. pisi) are best known for causing root rot of garden pea (Pisum sativum L.); however, they have a diverse habitat range and have been confirmed to be pathogenic on at least 10 other host plants (VanEtten, 1978). The four pathogens causing soybean sudden death syndrome (SDS), F. virguliforme, F. tucumaniae, F. brasiliense and F. cuneirostrum, are members of the FSSC (Aoki et al., 2003, 2005). Although traditionally thought to be only a pathogen on soybean, the host range of F. virguliforme is more diverse as it is able to cause root necrosis on alfalfa, pinto and navy bean, white and red clover, pea and Canadian milk vetch, and is asymptomatic on a number of other plants (Kolander et al., 2012). Investigation of the host range of F. solani f. sp. eumartii uncovered additional host plants besides potato, including other plants in the Solanaceae, such as tomato and pepper (Romberg and Davis, 2007). In addition to being plant pathogens, members of the FSSC are responsible for the majority of systemic fusarial infections in immunocompromised humans and animals (Muhammed et al., 2013; O'Donnell et al., 2008; Zhang et al., 2006). In immunocompetent individuals, diseases caused by FSSC isolates may manifest as localized, less invasive infections, including abscesses, onychomycosis and/or keratitis (Muhammed et al., 2013); for example, the 2005–2006 outbreaks of keratitis associated with a contact lens solution in North America and Asia were caused predominantly by members of the FSSC (Chang et al., 2006; Khor et al., 2006; O'Donnell et al., 2007).
Despite the agricultural importance and broad host range of members of the FSSC, few studies have focused on the identification of virulence factors in this species complex. The interaction between garden pea and isolates of FSSC 11 has been extensively utilized as a model system to study fungal virulence factors and plant defence responses at a molecular level (Hadwiger, 2008). This review focuses on the current understanding of the infection process and virulence factors for this fungus and other closely related members of the FSSC (Table 2).
Table 2.
Gene deletions and disruptions in members of the F usarium solani species complex (FSSC) and their phenotypic effects
| FSSC species | Gene | Product/function | Effect of gene inactivation/deletion | Reference |
|---|---|---|---|---|
| FSSC 11 | cut1 | Secreted cutinase | WT pathogenicity on pea; reduced virulence on pea | Stahl and Schäfer (1992); Rogers et al. (1994) |
| FSSC 1 | cutA | Cutinase | WT pathogenicity on cucurbits | Crowhurst et al. (1997) |
| FSSC 11 | dhc1 | Cytoplasmic dynein | Reduced growth | Inoue et al. (1998a) |
| F. virguliforme | fvtox1 | Secreted acidic protein | Reduced virulence on soybean | Pudake et al. (2013) |
| FSSC 11 | MAK1 | FAD mono‐oxygenase | Reduced virulence on chickpea; increased sensitivity to maackiain | Enkerli et al. (1998) |
| FSSC 11 | NhABC1 | ABC transporter | Reduced virulence on pea; increased sensitivity to pisatin and rishitin | Coleman et al. (2011) |
| FSSC 11 | Nhkin1 | Kinesin | Reduced growth | Wu et al. (1998) |
| FSSC 11 | PDA1 | Cytochrome P450 | Reduced virulence on pea; increased sensitivity to pisatin | Wasmann and VanEtten (1996) |
| FSSC 11 | pelA | Pectate lyase | Reduced virulence on pea | Rogers et al. (2000) |
| FSSC 11 | pelD | Pectate lyase | Reduced virulence on pea | Rogers et al. (2000) |
FAD, flavin adenine dinucleotide; WT, wild‐type.
Genome Organization and Supernumerary Chromosomes
Genome organization in members of the FSSC has been shown to be very dynamic. Pulse field gel electrophoretic profiles of members of the FSSC have revealed a high degree of variability in the size and number of small chromosomes between species and within a single species (Mahmoud and Taga, 2012; Miao et al., 1991a, 1991b; Suga et al., 2002). These small chromosomes have been most extensively studied in FSSC 11, where many of them have been shown to be dispensable supernumerary chromosomes (Covert, 1998; Covert et al., 1996; Enkerli et al., 1998; Funnell and VanEtten, 2002; Miao et al., 1991a; VanEtten et al., 1998; Wasmann and VanEtten, 1996).
In 2009, genome analysis of FSSC 11 isolate 77‐13‐4 (FGSC 9596; NRRL 45880) was completed, the first genome for a member of the FSSC (Coleman et al., 2009). An optical map identified 17 chromosomes that ranged in size from 530 kb to 6.52 Mb (Fig. 1). The 54.43‐Mb genome encoding 15 707 predicted genes was significantly larger than the 36.45‐Mb genome of Fusarium graminearum (Cuomo et al., 2007), the only other Fusarium sequenced at the time, and 92.3% of the genome sequence was mapped to one of the 17 chromosomes. Comparative genomics revealed that enlargement of the genome of FSSC 11 was a result of genes not known to be present in F. graminearum and the presence of multiple copies of a gene that exists as a single copy in other fungi. The origin of these genes appears to have occurred through gene duplication events or may have been acquired by horizontal gene transfer. Gene families that showed a significant increase in number in the FSSC 11 genome included carbohydrate‐active enzymes, oxidoreductases, monooxygenases, dioxygenases and transporters; the increase in these gene families may be indicative of the increased niche colonization ability and adaptation of FSSC 11 when compared with F. graminearum.
Figure 1.
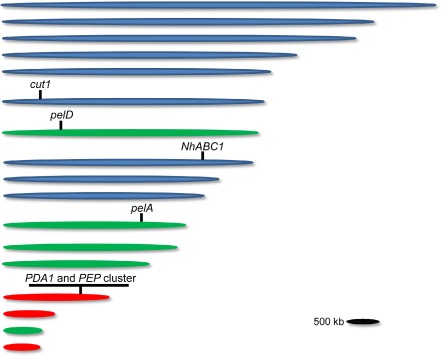
Profile and characteristics of chromosomes of the F usarium solani species complex (FSSC) 11 77‐13‐4 isolate genome. Chromosomes that contain a large number of ‘unique’ genes and in which >10% of the total number of genes were identified as potential pseudoparalogues are coloured green and red; red chromosomes have been confirmed experimentally to be dispensable. Approximate locations of genes involved in virulence are indicated.
Based on their loss in otherwise isogenic isolates, three chromosomes (14, 15 and 17) of the sequenced isolate were determined to be supernumerary. Chromosome 14 was previously known to be a supernumerary chromosome that was conditionally dispensable (CD). A CD chromosome is a chromosome that can be lost with no alteration in growth under laboratory conditions; however, it is able to broaden the habitat of the fungus when present (Covert, 1998). Chromosome 14 carries the PDA1 gene (discussed further below) and is referred to as the PDA1‐CD chromosome. Genomic analyses of these three supernumerary chromosomes showed that they were enriched for unique, duplicated genes, repeated sequences and had a decreased G + C content when compared with the other FSSC 11 chromosomes. Phylogenetic analysis of genes on the smaller chromosomes showed that they were more closely related to genes from the aspergilli than to genes within F. graminearum. In addition to unique genes on the supernumerary chromosomes, the population of repeated elements was enriched. A number of repeated elements have been characterized in FSSC 11 isolates that are associated with CD chromosomes: Nht1, Nht2 and Nrs1 (Enkerli et al., 1997; Kim et al., 1995; Shiflett et al., 2002). Analysis of the pogo‐like transposable elements (Fot2, Fot3, Fot4, Fot5 and Fot6) found 84 of these elements in the genome, with Fot5 the predominant pogo‐like transposable element in the FSSC 11 genome (n = 60; Dufresne et al., 2011). The chromosomal locations of these pogo‐like elements were enriched on the supernumerary chromosomes (36%) and unmapped scaffolds (37%). Similar characteristics (enrichment of specific gene families and repeated elements) of the supernumerary chromosomes in FSSC 11 were also found to occur for the lineage‐specific supernumerary chromosomes of Fusarium oxysporum f. sp. lycopersici (Ma et al., 2010).
The mitochondrial (mt) genome of FSSC 11 is ∼63 kb and encodes 15 genes expected to be in fungal mt genomes based on other analyses (Al‐Reedy et al., 2012); however, the FSSC 11 mt genome has two unidentified open reading frames (uORFs) and several more group I and II introns when compared with other sequenced fusarial mt genomes (Al‐Reedy et al., 2012). One of the unidentified ORFs was common to several Fusarium mt genomes and was exceptionally large (2013 amino acids in length in the FSSC 11 isolate). This ORF, termed LV‐uORF, is speculated to have been acquired by horizontal gene transfer based on a higher G + C content and a bias in codon usage (Al‐Reedy et al., 2012). Two linear plasmid‐like DNAs were identified that exist in the mitochondria of FSSC 10 (formerly N. haematococca MP I; F. solani f. sp. cucurbitiae ‘race 1’) and were termed pFSC1 (9.2 kb) and pFSC2 (8.3 kb) (Samac and Leong, 1988). When the isolate was cured of these plasmids, there was no significant difference in growth rate or conidiation between the wild‐type and plasmid‐cured isolates; however, the wild‐type isolate was significantly more virulent than the plasmid‐cured strains when assayed on a cucurbit using a low concentration of inoculum (Samac and Leong, 1988).
The genome of F. virguliforme, a member of the FSSC and one of the species responsible for SDS, has been completed recently (Srivastava et al., 2014). The Mont‐1 strain genome was 50.99 Mb in size, encoding for 14 845 predicted genes. Comparison between the F. virguliforme and FSSC 11 genomes revealed 13 068 putative orthologous genes between these two FSSC species (88% and 83% of their genes, respectively); many of these orthologous regions were in syntenic regions between the two. When these regions were anchored to the chromosomal karyotype of the FSSC 11 isolate, most were placed on the larger chromosomes, but there were small syntenic regions that aligned with the dispensable chromosomes of FSSC 11 strain 77‐13‐4 (chromosomes 14, 15 and 17), suggesting that there could be genetic elements in the F. virguliforme genome that could be supernumerary. As with the supernumerary chromosomes of the FSSC 11 isolate, an estimated 18% of the genome sequence of F. virguliforme was composed of repeated elements, a characteristic of supernumerary chromosomes. In addition, genomic comparison with the genomes of F. graminearum PH‐1, F. oxysporum f. sp. lycopersici 4287, F. verticillioides and FSSC 11 77‐13‐4 identified 1332 genes that were unique to F. virguliforme and not present in the other four genomes.
Adhesion and Penetration
The infection process of hypocotyls for several phylogenetic species within the FSSC has been described on their respective host plants (Bywater, 1959; Christou and Snyder, 1962; Samac and Leong, 1989; Stahl et al., 1994). Isolates of FSSC 10, FSSC 11 and F. phaseoli were able to penetrate into the hypocotyl, either directly through the epidermis (FSSC 10) or primarily through stomates (FSSC 11 and F. phaseoli). Microscopic examination and quantification of the fungal sterol ergosterol in planta allowed the spread of a FSSC 11 infection to be followed. After invasion of the hypocotyl, hyphae of FSSC 11 spread to the upper portion of the tap root and the epicotyl, eventually colonizing the xylem and causing wilting to the above‐ground portions of the host plant (Stahl et al., 1994).
Members of the FSSC are known to cause root rot on several plants, and root exudates can influence the germination and growth of fungi within the rhizosphere. Chlamydospores of FSSC 11 will germinate within 24 h when placed 7 mm away from a germinating seed (Short and Lacy, 1974); presumably, the carbohydrates and amino acids present in the root exudates are partially responsible for the stimulation of spore germination. One of the major root exudates from pea roots is homoserine (Van Egeraat, 1975), and the PDA1‐CD chromosome in FSSC 11 harbours a five‐gene locus that confers the ability for homoserine utilization (HUT) as a sole carbon and nitrogen source (Rodriguez‐Carres et al., 2008; White, 2008). Mutation in the HUT cluster renders FSSC 11 less competitive in the pea rhizosphere when compared with the isogenic isolate carrying the HUT cluster, demonstrating that the HUT cluster is a rhizosphere competence trait (Rodriguez‐Carres et al., 2008; White, 2008). In addition to carbohydrates and amino acids, flavonoids are capable of inducing spore germination in FSSC 11 and F. phaseoli (Ruan et al., 1995). These root exudates include flavones, isoflavones and pterocarpans that are synthesized as part of a defence mechanism. Stimulation of spore germination by these compounds involves the cyclic adenosine monophosphate (cAMP) pathway and is dependent on protein kinase A and regulated through cAMP phosphodiesterase (Bagga and Straney, 2000; Ruan et al., 1995).
The attachment of plant‐pathogenic fungal spores to a susceptible host is a common initial step for the establishment of disease. A number of mechanisms have been identified that are involved in this process in different phytopathogenic fungi (Tucker and Talbot, 2001); the molecular process of conidial adhesion has best been described for FSSC 10. Attachment of macroconidia to the unwounded surface of a susceptible host is a virulence factor for this FSSC species (Jones and Epstein, 1990), and is an active process that requires respiration and protein synthesis (Jones and Epstein, 1989). The production of spore tip mucilage is induced by treatment of the spores with zucchini fruit extract (a plant host of FSSC 10), conferring adherence at either or both apical tips of the macroconidium within 10 min after treatment (Kwon and Epstein, 1993). A 90‐kDa glycoprotein was associated with adhesion of the spore tip mucilage and, when bound by concanavalin A or an antibody against the 90‐kDa glycoprotein, adhesion of the macroconidium was abrogated (Kwon and Epstein, 1993, 1997). In addition, adherence was reduced in the presence of snowdrop lectin, a compound that specifically binds α‐mannose, and two transglutaminase inhibitors, suggesting that the 90‐kDa glycoprotein is cross‐linked to the spore tip mucilage (Kwon and Epstein, 1997). The production of spore tip mucilage has been implicated in adhesion in another member of the FSSC, F. phaseoli, and could be a common attachment mechanism in fungi (Schuerger and Mitchell, 1993). In addition to the glycoprotein, random mutagenesis and classical genetics have revealed that FSSC 10 utilizes another attachment mechanism encoded at a genetic locus termed Att1 (Epstein et al., 1994). Studies with an Att1– isolate indicated that macroconidium attachment was dependent on the concentration of zucchini extract applied to the conidia and the temperature (Epstein et al., 1994).
After spore attachment, the easiest route to invade the host plant is via the stomates. The role of direct penetration of plant tissue in disease development is uncertain. Passive barriers, such as cutin, a waxy polymer component of the cuticle, must be overcome to infect the plant. Variability in the role of cutinase in pathogenesis has been reported for plant‐pathogenic fungi. These enzymes have been studied extensively in FSSC 11, where isolates may have several cutinase genes (Rogers et al., 1994; Stahl and Schäfer, 1992). Expression of cutinase is induced in the presence of cutin monomers (Podila et al., 1988; Woloshuk and Kolattukudy, 1986), and the enzyme can be detected at infection sites on pea (Shaykh et al., 1977). A number of regulatory elements in the promoter of cutinases control expression, in particular a palindromic sequence which binds cutinase transcription factor 1α (CTF1α), which positively regulates expression in the presence of cutin (Kämper et al., 1994; Li and Kolattukudy, 1997; Li et al., 2002). A second transcription factor, termed CTF1β, is involved in the constitutive expression of some cutinase genes in FSSC 11 (Li et al., 2002). The expression of a FSSC 11 cutinase gene in Mycosphaerella spp., a fungus unable to infect unwounded papaya fruit, conferred to the transgenic fungus the ability to infect intact tissue (Dickman et al., 1989). An extracellular esterase in FSSC 11 is responsible for cutinase and esterase activity (Murphy et al., 1996; Rogers et al., 1994; Stahl and Schäfer, 1992). Disruption of the gene encoding this cutinase, cut1, demonstrated that the enzyme is not essential for virulence on pea (Stahl and Schäfer, 1992; Stahl et al., 1994), or results in a significant decrease in virulence (Rogers et al., 1994). The orthologue of cut1 has been studied in the closely related fungus FSSC 1 (F. petroliphilum; formerly N. haematococca MP V, F. solani f. sp. cucurbitiae ‘race 2’). As with FSSC 11, cutA is induced in the presence of cutin from various cucurbits (Hawthorne et al., 2001); however, disruption of the cutA gene in FSSC 1 did not cause a significant reduction in virulence on intact fruit of several varieties of Cucurbita maxima and C. moschata (Crowhurst et al., 1997). The induction of cutinase activity in FSSC 1 is dissimilar to that observed in FSSC 11, and therefore it appears that the regulation of cutA in FSSC 1 is different from that of cut1 in FSSC 11 (Hawthorne et al., 2001). Another cucurbit pathogen in the FSSC (FSSC 10) lacked cutinase activity, which may explain the difference in tissue specificity between these phylogenetically distinct cucurbit pathogens (Hawthorne et al., 2001).
In addition to cutinases, enzymes digesting pectin in the plant cell wall have been implicated in the penetration of plant barriers. A number of pectin lyases have been characterized in FSSC 11, including two that are involved in virulence on pea. The enzyme encoded by pelA is induced in the presence of pectin, whereas the enzyme encoded by pelD is only expressed in planta, where it is induced by asparagine and homoserine, two amino acids found at high levels in pea seedlings (Rogers et al., 2000; Yang et al., 2005). Deletion of either pelA or pelD individually did not cause a significant reduction in virulence; however, a ΔpelA/ΔpelD mutant was severely impaired in virulence, causing only a few mild lesions (Rogers et al., 2000).
Tolerance to Plant Defence Mechanisms
As a part of the defence reaction to pathogens, plants release small molecular weight antimicrobial compounds. These compounds can be either preformed and compartmentalized in an inactive state inside the plant cell, termed phytoanticipins, or synthesized de novo in response to the pathogen, termed phytoalexins (VanEtten et al., 1994). Regardless of the time of synthesis, a fungal pathogen must be able to tolerate these antimicrobial compounds in order to establish an infection on the host plant and, eventually, to develop disease. Pathogens primarily possess three mechanisms by which they accomplish this feat: (i) enzymatic degradation or modification of the compound to a less toxic molecule; (ii) efflux of the antimicrobial prohibiting an intracellular concentration that has a physiological effect; and (iii) modification of the target site of the compound (Coleman and Mylonakis, 2009; Morrissey and Osbourn, 1998; VanEtten et al., 2001). Of these, degradation/modification and efflux have been described in members of the FSSC.
Tolerance mechanisms of FSSC 11 against the phytoalexin (+)pisatin produced by garden pea have been studied extensively and represent one of the best examples in plant–pathogen interactions. On challenge with FSSC 11, pea plants begin to synthesize pisatin, which inhibits the pathogen. FSSC 11 isolates that are highly pathogenic on pea carry a gene encoding a cytochrome P450, termed pisatin demethylase (PDA; VanEtten et al., 1980). PDA catalyses the demethylation of pisatin at the C3 position, forming the much less toxic (+)‐6a‐hydroxymaackiain. Currently, nine alleles of PDA have been identified from FSSC 11 isolates by classical genetics (Funnell et al., 2002; Kistler and VanEtten, 1984; Mackintosh et al., 1989; Miao et al., 1991a, 1991b); however, only two of these, PDA1 and PDA4, are linked to high virulence on pea (Maloney and VanEtten, 1994; VanEtten et al., 1980). Deletion of PDA1 in FSSC 11 strains yielded transformants that were reduced in virulence on garden pea and more sensitive to pisatin (Wasmann and VanEtten, 1996). Alternatively, expression of PDA1 in an FSSC 11 isolate that was previously PDA – increased virulence on pea (Ciuffetti and VanEtten, 1996). Expression of PDA1 is induced in planta and on treatment with pisatin (Liu et al., 2003; Ruan et al., 1995; Straney and VanEtten, 1994). A pisatin‐responsive activating factor is hypothesized to bind to a 35‐bp region −514 to −480 bp relative to the start site of PDA1 activating transcription (Straney and VanEtten, 1994). Expression of PDA1 is repressed in vitro under high nutrient conditions, such as glucose and amino acids (Khan and Straney, 1999; Straney and VanEtten, 1994), and is independent of cAMP signalling (Ruan et al., 1995). This nutritional repression is mediated by a region in the PDA1 promoter that resides −287 to −429 bp relative to the start codon (Khan and Straney, 1999).
PDA resides on a small supernumerary chromosome that is not required for the fungus to grow under laboratory conditions, and has been called the PDA1‐CD chromosome (Miao et al., 1991a). The FSSC 11 77‐13‐4 isolate that has lost the 1.6‐Mb PDA1‐CD chromosome is non‐pathogenic on pea. However, when the isolate carries a disrupted PDA1 gene, it retains some pathogenicity, and therefore other virulence factors for pea must reside on the same CD chromosome as PDA1 (Wasmann and VanEtten, 1996). Subsequent studies have found that PDA1 indeed resides within an ∼25‐kb cluster of genes that are involved in pea pathogenicity (PEP; Fig. 2). Three genes (PEP1, PEP2 and PEP5) have been shown to contribute to virulence on pea when expressed in a non‐pathogenic isolate of FSSC 11 (Han et al., 2001). PEP1 may encode two proteins, 330 and 320 amino acids in length, that are in different reading frames, and PEP2 encodes a protein of 233 amino acids in length. PEP5 is the only PEP gene with a postulated function based on amino acid sequence, as PEP5 encodes an efflux pump of the major facilitator superfamily, although the substrate of PEP5 is unknown. As with PDA1, PEP1, PEP2 and PEP5 were expressed in planta and after treatment with pisatin (Liu et al., 2003). A correlation between the degree of virulence on pea and the PEP genes has been observed, where FSSC 11 isolates that are highly virulent carry all the PEP genes and, in many instances, multiple copies (Temporini and VanEtten, 2002).
Figure 2.
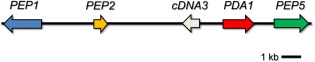
The PDA1 and PEP clusters in F usarium solani species complex (FSSC) 11. The PEP cluster is ∼20 kb in size and resides on chromosome 14, a conditionally dispensable chromosome. PDA1 encodes a cytochrome P450 and PEP5 encodes a putative major facilitator superfamily (MFS) transporter. The functions of PEP1, PEP2 and cDNA3 are unknown. PDA, pisatin demethylase; PEP, pea pathogenicity.
In addition to isolates of FSSC 11, functional PDA genes have been identified in isolates that are currently placed within FSSC 8 (Fusarium neocosmosporiellum; formerly Neocosmospora vasinfecta; O'Donnell et al., 2013) and other closely related fusaria (i.e. Neocosmospora bonniensis and Neocosmospora tenuicristata; Temporini and VanEtten, 2004; Milani et al., 2012). In addition to PDA, homologues of PEP1, PEP2 and PEP5 are present in Ne. bonniensis in a cluster as in FSSC 11 (Temporini and VanEtten, 2004). Although the Fusarium sp. originally described as Ne. bonniensis was not known to be a pea pathogen, when inoculated on pea roots, the fungus was able to cause necrosis to the root system and stunting (Temporini and VanEtten, 2004). The discontinuous phylogenetic distribution of PDA and PEP genes within the FSSC supports the potential evolutionary origin of these genes via horizontal gene transfer, as PDA genes are not found in other FSSC species that are closely related to FSSC 11 (Temporini and VanEtten, 2004). As observed with the dispensable chromosomes in the sequenced FSSC 11 77‐13‐4 isolate (Coleman et al., 2009), a G + C and codon bias exists between the PDA and PEP genes when compared with genes that have a conserved ‘housekeeping’ function (Liu et al., 2003; Milani et al., 2012).
In addition to PDA, members of FSSC 11 have a second tolerance mechanism to pisatin. In the absence of enzymatic detoxification, the fungus is able to overcome the inhibitory effects through a ‘non‐degradative’ mechanism that is induced by pisatin and other similarly structured compounds (Denny and VanEtten, 1983a, 1983b; Denny et al., 1987). The ABC transporter NhABC1, which is responsible for the efflux of pisatin, belongs to the pleotropic drug resistance (PDR or ABCG) family of efflux pumps (Coleman et al., 2011). ΔNhABC1 mutants were slightly reduced in sensitivity to pisatin; however, there was a significant reduction in virulence on pea. Double mutants of NhABC1 and PDA1 were more sensitive to pisatin than either single mutant. Virulence assays with the ΔNhABC1/ΔPDA1 mutants showed that they were significantly reduced in virulence when compared with either single mutant. Although the FSSC 11 isolate used in this study was not a pathogen of potato, NhABC1 also conferred tolerance to the potato phytoalexin rishitin. Phylogenetic analysis of NhABC1 revealed that it is a member of a Fusarium‐specific clade of ABC transporters that include other transporters involved in virulence on plants (Fleissner et al., 2002; Gardiner et al., 2013; Skov et al., 2004).
The phytoalexins (–)maackiain and (–)medicarpin are produced by chickpea (Cicer arietinum L.) in response to pathogens. Some FSSC 11 isolates are highly virulent on chickpea and are tolerant to phytoalexins; conversely, isolates more sensitive to maackiain are less virulent on this host plant (Lucy et al., 1988). Isolates of FSSC 11 were able to perform three metabolic modifications on maackiain and medicarpin via monooxygenases and were able to detoxify the phytoalexins to the less inhibitory (–)‐6a‐hydroxypterocarpan, the 1a‐hydroxydienone and the isoflavanone derivatives (Denny and VanEtten, 1981, 1982; Lucy et al., 1988). Three maackiain detoxification genes (MAK1–MAK3) have been identified by genetic studies (Covert et al., 1996; Miao and VanEtten, 1992). All FSSC 11 isolates that are capable of metabolizing maackiain are also able to modify medicarpin to analogous byproducts, suggesting that the enzymes encoded by the MAK genes are responsible for this modification (Denny and VanEtten, 1982). When MAK1 was hybridized to genomic DNA of isolates carrying the other MAK genes, no cross‐hybridization was observed, supporting divergence among the MAK genes in FSSC 11 (Covert et al., 1996). MAK1 encodes a flavin adenine dinucleotide (FAD)‐containing monooxygenase of 460 amino acids in length (Covert et al., 1996), which is responsible for the detoxification of maackiain to the less toxic compound 1a‐hydroxymaackiain (Covert et al., 1996; Miao and VanEtten, 1992). Despite the divergence between the MAK1 and MAK2 genes, MAK2 is also responsible for converting maackiain to 1a‐hydroxymaackiain, whereas MAK3 converts the phytoalexin to 6a‐hydroxymaackiain (Miao and VanEtten, 1992). Although all three MAK genes encode enzymes capable of metabolizing maackiain, only MAK1 and MAK2 correlate with high virulence on chickpea (Miao and VanEtten, 1992). When MAK1 was disrupted in a highly virulent isolate on chickpea, virulence was reduced. Conversely, when the gene was placed into a mak – isolate, there was an increase in virulence on chickpea (Enkerli et al., 1998). When MAK1‐disrupted mutants were placed on medium containing maackiain, growth was reduced by ∼20% (Enkerli et al., 1998). Genetic studies indicated that MAK1 was linked with PDA6‐1 and resided on the same dispensable chromosome (Covert et al., 1996; Miao and VanEtten, 1992). An isogenic isolate that had lost the 1.6‐Mb chromosome was reduced in virulence to a similar degree as the disrupted mak – mutant, demonstrating that there were no other virulence factors for chickpea on the MAK1/PDA6‐1 dispensable chromosome (Enkerli et al., 1998).
A correlation between tolerance to the bean (Phaseolus vulgaris) phytoalexin kievitone and virulence has been observed for F. phaseoli (formerly F. solani f. sp. phaseoli), in which isolates that produce extracellular kievitone hydratase are highly virulent on bean and, conversely, are less virulent when kievitone hydratase activity is abolished (Smith et al., 1982, 1984). Kievitone hydratase is an acidic glycoprotein that catalyses a hydration reaction on the dimethylallyl moiety, forming the less toxic compound kievitone hydrate (Cleveland and Smith, 1983; Zhang and Smith, 1983). The gene encoding kievitone hydratase, khs, generates a preprotein of 350 amino acids in length, in which the N‐terminal 19 amino acids are hydrolysed during maturation and secretion (Li et al., 1995). Pulse field gel electrophoresis has revealed that khs resides on chromosomes usually ranging in size from approximately 2 to 3.5 Mb in F. phaseoli and F. cuneirostrum (reported as F. phaseoli), whereas it is absent in almost all of the other FSSC isolates included in the analysis (Suga et al., 2002), suggesting that the chromosomes in F. phaseoli and F. cuneirostrum that harbour khs may be supernumerary. It should be noted that, in addition to kievitone, P. vulgaris produces other phytoalexins, including phaseollidin, phaseollin and phaseollin isoflavan. Phaseollidin is hydroxylated in a similar manner to kievitone by an extracellular enzyme, termed phaseollidin hydratase, forming phaseollidin hydrate (Turbek et al., 1992). Biochemical analysis of F. phaseoli indicates that phaseollin is detoxified to the less toxic compound 1a‐hydroxyphaseollone by a monooxygenase (Kistler and VanEtten, 1981).
Effectors, Secreted Proteins and Host‐Specific Virulence Factors
Although the tolerance mechanisms to phytoalexins represent the best‐studied host‐specific virulence factors in members of the FSSC, only a few effectors have been identified in these fungi. An extracellular DNase has been identified in F. phaseoli and FSSC 11 which is capable of eliciting a defence response in pea (Hadwiger, 2008; Hadwiger et al., 1995). Produced by the macroconidia, the single‐strand nicking enzyme, termed Fsph DNase, enters the pea cell and eventually the nucleus (Gerhold et al., 1993), where it induces the expression of several pathogenesis‐related (PR) proteins, including defensin DRR230 and the phytoalexin pisatin (Hadwiger, 2008; Hadwiger et al., 1995).
There were 746 small secreted proteins identified by SignalP in the sequenced genome of FSSC 11, which represents 4.75% of the entire annotated proteome (Coleman et al., 2009). FSSC 11 isolates are virulent on carrot and tomato in addition to pea. Genes conferring pathogenicity on carrot and ripe tomato are on chromosomes other than the PDA1‐CD chromosome (Funnell and VanEtten, 2002). A number of effectors have been identified in F. oxysporum f. sp. lycopersici that are involved in virulence on tomato (van der Does et al., 2008; Rep et al., 2004), and multiple homologues of these proteins exist in the FSSC 11 genome.
Analysis of the F. virguliforme genome using SignalP identified 1155 putative secreted proteins (Srivastava et al., 2014). A host‐selective toxin has been identified in F. virguliforme, a causative agent of SDS. This small secreted protein, termed FvTox1, is responsible for causing the foliar symptoms observed in plants infected with the fungus, i.e. chlorosis and necrosis, ultimately leading to the loss of leaves and pods of the infected plant (Brar et al., 2011). A single gene, fvtox1, is expressed in infected root tissue that encodes a pro‐peptide. On secretion into culture medium, the N‐terminal 32 amino acids are cleaved and the protein is predicted to contain a sequence for translocation through the endoplasmic reticulum and a putative chloroplast transit peptide sequence (Brar et al., 2011). Targeted mutation of fvtox1 resulted in strains that caused foliar disease symptoms and chlorophyll losses that were over two‐fold less than in soybean plants infected with the wild‐type isolate (Pudake et al., 2013). A homologue of fvtox1 is present in the genome of the sequenced FSSC 11 isolate (Nh Protein ID 122357 located on chromosome 2), encoding a protein that is 91% identical to FvTox1 (Brar et al., 2011). Proteomic studies of xylem sap from F. virguliforme‐infected soybean plants uncovered five fungal proteins that were present in the xylem (Abeysekara and Bhattacharyya, 2014). All five proteins possessed an N‐terminal secretion signal amino acid sequence and were all less than 30 kDa in size. The most frequently identified and abundant F. virguliforme protein found in the soybean xylem sap encoded a protein with a high degree of similarity to cerato‐platanin, a known phytotoxin that was originally identified in Ceratocystis fimbriata f. sp. platani. Although FvTox1 was not identified in the xylem sap, it appears that multiple protein toxins are secreted into the xylem during F. virguliforme infection of soybean (Abeysekara and Bhattacharyya, 2014).
Secondary Metabolites
As with all fusaria, members of the FSSC are capable of producing an abundant array of secondary metabolites. Unlike species belonging to the Fusarium sambucinum and fujikuroi species complexes, members of the FSSC are not known to produce trichothecene and fumonisin mycotoxins. Therefore, the biosynthetic pathways for these secondary metabolites are postulated to have evolved after the divergence of the FSSC from other fusaria (O'Donnell et al., 2013).
There were 12 polyketide synthase (PKS), 12 non‐ribosomal peptide synthase (NRPS) and one PKS–NRPS hybrid encoding genes in the genome of FSSC 11 (Coleman et al., 2009). Homologues of eight NRPS genes are present in three other Fusarium genomes (F. graminearum PH‐1, F. verticillioides 7600 and F. oxysporum 4287). Based on similarity, it can be predicted that three of these NRPS‐containing biosynthetic clusters are responsible for the production of the siderophores ferricrocin, fusarinine and malonichrome or other similarly structured secondary metabolites (Hansen et al., 2012; O'Donnell et al., 2013). Of the PKS and NRPS genes in the FSSC 11 genome, seven PKS and four NRPS genes were unique to FSSC isolate 77‐13‐4 (Hansen et al., 2012). Comparison of the F. virguliforme Mont‐1 genome with the four Fusarium genomes identified a single PKS (Fv14626) that was unique to F. virguliforme (Srivastava et al., 2014).
A homologue of the PKS–NRPS hybrid in FSSC 11 has been characterized in F. verticillioides (FUS1, FVEG_11086) and is responsible for the production of fusarin (Brown et al., 2012). FUS1 and the homologue in the FSSC 11 isolate, Nh70660, are 64% identical in amino acid sequence, and homologues of seven of the eight other genes in the FUS biosynthetic cluster are present in the FSSC 11 genome as a 27‐kb cluster, the exception being FUS5 (Brown et al., 2012; Hansen et al., 2012). Although genes within the FUS gene cluster are homologous, the arrangement of genes within the cluster is different, as well as the genes immediately flanking the clusters, indicating a difference in genomic position (Brown et al., 2012; Hansen et al., 2012). Although fusarin production has not been reported by members of the FSSC, the similarity between the FUS biosynthetic gene clusters of these fungi suggests that members of the FSSC may be able to produce fusarin or a similarly structured compound.
A plethora of naphthoquinones are produced by members of the FSSC. These pigmented compounds have a wide range of structural variation and biological action, including antimicrobial and phytotoxic activity (reviewed in Medentsev and Akimenko, 1998). Some of the naphthoquinones produced by the FSSC include fusarubin, javanicin, norjavanicin, solaniol, bostrycoidin, fusarubinoic acid, novarubin, marticin, nectriafurone, nectriachrysone and derivatives of these metabolites (Medentsev and Akimenko, 1998; Short et al., 2013). The role of naphthoquinones in the pathogenicity of FSSC on host plants is unknown. Although some of these compounds are toxic to plants and can be found in diseased tissue (Kern and Naef‐Roth, 1965), genetic studies have shown that pathogenicity on pea does not correlate with naphthoquinone production (Holenstein and Défago, 1983). The purple pigment associated with the more derived clades of Fusarium is produced by a biosynthetic cluster containing the PKS PGL1 (Proctor et al., 2007), and data suggest that this pigmented compound is either fusarubin or synthesized from the compound (Brown et al., 2012). Although FSSC isolates are associated with a red pigment (discussed further below), a homologue of PGL1 is present in the FSSC 11 genome. Only three of the six genes in the PGL cluster were present in FSSC 11 [PGL1, NhOMT1 (PGL2) and NhFDM1 (PGL3]; however, they were present in a syntenic block and there was a high degree of sequence similarity (>70%) between the FSSC 11 homologues and those in F. graminearum and F. verticillioides (Proctor et al., 2007). Although members of the FSSC do not produce the purple pigment, they are known to produce fusarubin and multiple structural derivatives (Medentsev and Akimenko, 1998; Short et al., 2013).
One of the PKS genes that might be unique to the FSSC is pksN (Graziani et al., 2004b). pksN, Nh33672 in the sequenced genome, was found to be one of two genetic loci that are involved in the production of the red perithecial wall pigment that is characteristic of FSSC species with known sexual reproduction. Mutation of pksN did not cause any phenotypic alteration in mycelial growth, conidiation or ascospore production in the FSSC CBS 225.53 isolate (Graziani et al., 2004a, 2004b). Although the chemical structure of the final product of pksN is unknown, the similarity of pksN to PGL1, and the known diverse array of pigmented naphthoquinones produced by members of the FSSC, allows speculation that pksN is responsible for the production of a red naphthoquinone.
Members of the FSSC are capable of producing several other secondary metabolites. The diversity of secondary metabolites produced by members of the FSSC varies not only between species, but also within a single phylogenetic species (Short et al., 2013). The non‐polar cyclic peptides cyclosporin A and cyclosporin C are produced by members of the FSSC, in particular isolates of FSSC 1 (F. petroliphilum), FSSC 2 (F. keratoplasticum), FSSC 3+4 (F. falciforme) and FSSC 5 (Short et al., 2013), and perhaps others (Sugiura et al., 1999). A unique NRPS in the genome of FSSC 11 showed 25.1% amino acid identity to SimA of the cyclosporin‐producing fungus Tolypocladium inflatum (J. J. Coleman, unpublished data). Fusaric acid (5‐butylpicolinic acid) production has been reported in the FSSC (Bacon et al., 1996), although a homologue of the PKS in the fusaric acid biosynthetic gene cluster in F. verticillioides, FUB1, was not present in the FSSC 11 genome. The secondary metabolite bikaverin is produced by several members of the FSSC (Chelkowski et al., 1992; Short et al., 2013), although a homologue of the PKS responsible for bikaverin synthesis (BIK) in F. fujikuroi was not found in the FSSC 11 genome (Hansen et al., 2012).
Members of the FSSC as a Model System for Cell Biology
Although the supernumerary chromosomes encoding host‐specific virulence factors of the FSSC have been a focal point of research, members of the FSSC have been used as a system for cell biology. FSSC 11 was developed as a model system to study mitosis in fungi, in particular the formation and function of the spindle pole body and asters in anaphase (Aist, 2002). FSSC 11 was the first fungus to show that spindle pushing and aster pulling forces were both involved in mitotic spindle elongation (Aist and Bayles, 1991). Further studies have indicated that the microtubule‐associated motor protein dynein is involved in the formation of mitotic asters, as mutants of cytoplasmic dynein show impaired hyphal growth, reduced astral microtubules and limited post‐mitotic nuclear migration (Inoue et al., 1998a, 1998b). Mutants of the other major microtubule motor protein, kinesin, resulted in morphological changes in hyphae, as well as a reduced growth rate, and subcellular abnormalities in location and size of mitochondria and the Spitzenkörper (Wu et al., 1998). A kinesin‐related motor protein in FSSC 11, termed NhKRP1, is responsible for producing an inwardly directed force in the mitotic spindle. Mutation of NhKRP1 significantly increased the rate of spindle elongation during anaphase A and B, as the lack of the inward force from NhKRP1 allows the astral pulling forces to elongate the spindle more rapidly (Aist, 2002; Sandrock et al., 1999).
Epigenetic factors have been shown to be responsible for two different morphological alterations in FSSC 11 that appear either as a typical sector (‘Secteur’) in the culture or as a ring surrounding the culture (‘Anneau’). The factors responsible for these changes can appear spontaneously, and the frequency is dependent on cultural conditions, in particular temperature (Silar and Daboussi, 1999). Once established, transmission can occur to an undifferentiated culture at the growing edge of the mycelium through anastomoses, thereby transmitting the infectious cytoplasmic determinants, termed α and σ, for the ‘Anneau’ and ‘Secteur’ morphologies, respectively. Both determinants are under the control of two unlinked nuclear loci. Genetic studies revealed that ‘Anneau’ is controlled by locus A composed of a single gene that exists in three allelic forms: wild‐type, a and 58 (Daboussi‐Bareyre, 1980). The locus controlling the ‘Secteur’ morphology, S or Ses, is controlled by two genes, termed SesA and SesB (Graziani et al., 2004a). The function of the protein encoded by SesA (210 amino acids in length) is unknown, although homologues are only present in other filamentous fungi, whereas the SesB encoded protein (386 amino acids in length) has a domain that may confer esterase/lipase/thioesterase activity. The C‐terminal regions of both SesA and SesB share a homology sequence that is also present in the N‐terminal region of an adjacent gene, designated het‐eN (Daskalov et al., 2012; Graziani et al., 2004a). These three proteins have characteristics of prion forming domains, and it has been proposed that the HET‐eN protein induces a conformational change in sesA which then modifies sesB by interacting through the shared domain.
Conclusions
The FSSC is a diverse group of fungi capable of causing disease on a wide array of plants. As several specific virulence factors have been identified in these fungi that reside on CD supernumerary chromosomes, genomics and other next‐generation technologies should greatly advance the current understanding of the interaction between members of the FSSC and their respective host plants. Further knowledge of the molecular mechanisms responsible for conferring virulence to members of the FSSC could facilitate the development of alternative disease management strategies.
Acknowledgements
The author would like to thank Hans VanEtten for comments and suggestions on the manuscript. There are no conflicts of interest to declare.
References
- Abeysekara, N.S. and Bhattacharyya, M.K. (2014) Analyses of the xylem sap proteomes identified candidate Fusarium virguliforme proteinacious toxins. PLoS ONE, 9, e93667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist, J.R. (2002) Mitosis and motor proteins in the filamentous ascomycete, Nectria haematococca, and some related fungi. Int. Rev. Cytol. 212, 239–263. [DOI] [PubMed] [Google Scholar]
- Aist, J.R. and Bayles, C.J. (1991) Detection of spindle pushing forces in vivo during anaphase B in the fungus Nectria haematococca . Cell Motil. Cytoskeleton, 19, 18–24. [Google Scholar]
- Al‐Reedy, R.M. , Malireddy, R. , Dillman, C.B. and Kennell, J.C. (2012) Comparative analysis of Fusarium mitochondrial genomes reveals a highly variable region that encodes an exceptionally large open reading frame. Fungal Genet. Biol. 49, 2–14. [DOI] [PubMed] [Google Scholar]
- Aoki, T. , O'Donnell, K. , Homma, Y. and Lattanzi, A.R. (2003) Sudden‐death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex—F. virguliforme in North America and F. tucumaniae in South America. Mycologia, 95, 660–684. [PubMed] [Google Scholar]
- Aoki, T. , O'Donnell, K. and Scandiani, M.M. (2005) Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. nov., F. cuneirostrum sp. nov., F. tucumaniae, and F. virguliforme . Mycoscience, 46, 162–183. [Google Scholar]
- Bacon, C.W. , Porter, J.K. , Norred, W.P. and Leslie, J.F. (1996) Production of fusaric acid by Fusarium species. Appl. Environ. Microbiol. 62, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S. and Straney, D. (2000) Modulation of cAMP and phosphodiesterase activity by flavonoids which induce spore germination of Nectria haematococca MP VI (Fusarium solani). Physiol. Mol. Plant Pathol. 56, 51–61. [Google Scholar]
- Booth, C. (1971) The Genus Fusarium. Kew: Commonwealth Mycological Institute. [Google Scholar]
- Brar, H.K. , Swaminathan, S. and Bhattacharyya, M. (2011) The Fusarium virguliforme toxin FvTox1 causes foliar sudden death syndrome‐like symptoms in soybean. Mol. Plant–Microbe Interact. 24, 1179–1188. [DOI] [PubMed] [Google Scholar]
- Brown, D.W. , Butchko, R.A.E. , Busman, M. and Proctor, R.H. (2012) Identification of gene clusters associated with fusaric acid, fusarin, and perithecial pigment production in Fusarium verticillioides . Fungal Genet. Biol. 49, 521–532. [DOI] [PubMed] [Google Scholar]
- Bywater, J. (1959) Infection of peas by Fusarium solani var. martii forma 2 and the spread of the pathogen. Trans. Br. Mycol. Soc. 42, 201–212. [Google Scholar]
- Chang, D.C. , Grant, G.B. , O'Donnell, K. , Wannemuehler, K.A. , Nobel‐Wang, J. , Rao, C.Y. , Jacobson, L.M. , Crowell, C.S. , Sneed, R.S. , Lewis, F.M.T. , Schaffzin, J.K. , Kainer, M.A. , Genese, C.A. , Alfonso, E.C. , Jones, D.B. , Srinivasan, A. , Fridkin, S.K. and Park, B.J. (2006) A multistate outbreak of Fusarium keratitis associated with use of a new contact lens solution. J. Am. Med. Assoc. 296, 953–963. [DOI] [PubMed] [Google Scholar]
- Chelkowski, J. , Zajkowski, P. and Visconti, A. (1992) Bikaverin production by Fusarium species. Mycotoxin Res. 8, 73–76. [DOI] [PubMed] [Google Scholar]
- Christou, T. and Snyder, W.C. (1962) Penetration and host–parasite relationships of Fusarium solani f. phaseoli in the bean plant. Phytopathology, 52, 219–226. [Google Scholar]
- Ciuffetti, L.M. and VanEtten, H.D. (1996) Virulence of a pisatin demethylase‐deficient Nectria haematococca MPVI isolate is increased by transformation with a pisatin demethylase gene. Mol. Plant–Microbe Interact. 9, 787–792. [Google Scholar]
- Cleveland, T.E. and Smith, D.A. (1983) Partial purification, and further characterization, of kievitone hydratase from cell‐free culture filtrates of Fusarium solani f. sp. phaseoli . Physiol. Plant Pathol. 22, 129–142. [Google Scholar]
- Coleman, J.J. and Mylonakis, E. (2009) Efflux in fungi: la pièce de résistance. PLoS Pathog. 5, e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.J. , Rounsley, S.D. , Rodriguez‐Carres, M. , Kuo, A. , Wasmann, C.C. , Grimwood, J. , Schmutz, J. , Taga, M. , White, G.J. , Zhou, S. , Schwartz, D.C. , Freitag, M. , Ma, L.‐J. , Danchin, E.G.J. , Henrissat, B. , Coutinho, P.M. , Nelson, D.R. , Straney, D. , Napoli, C.A. , Barker, B.M. , Gribskov, M. , Rep, M. , Kroken, S. , Molnár, I. , Rensing, C. , Kennell, J.C. , Zamora, J. , Farman, M.L. , Selker, E.U. , Salamov, A. , Shapiro, H. , Pangilinan, J. , Lindquist, E. , Lamers, C. , Grigoriev, I.V. , Geiser, D.M. , Covert, S.F. , Temporini, E. and VanEtten, H.D. (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5, e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.J. , White, G.J. , Rodriguez‐Carres, M. and VanEtten, H.D. (2011) An ABC transporter and a cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant–Microbe Interact. 24, 368–376. [DOI] [PubMed] [Google Scholar]
- Covert, S.F. (1998) Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33, 311–319. [DOI] [PubMed] [Google Scholar]
- Covert, S.F. , Enkerli, J. , Miao, V.P.W. and VanEtten, H.D. (1996) A gene for maackiain detoxification from a dispensable chromosome of Nectria haematococca . Mol. Gen. Genet. 251, 397–406. [DOI] [PubMed] [Google Scholar]
- Crowhurst, R.N. , Binnie, S.J. , Bowen, J.K. , Hawthorne, B.T. , Plummer, K.M. , Rees‐George, J. , Rikkerink, E.H.A. and Templeton, M.D. (1997) Effect of disruption of a cutinase gene (cutA) on virulence and tissue specificity of Fusarium solani f. sp. cucurbitae race 2 toward Cucurbita maxima and C. moschata . Mol. Plant–Microbe Interact. 10, 355–368. [DOI] [PubMed] [Google Scholar]
- Cuomo, C.A. , Güldener, U. , Xu, J.‐R. , Trail, F. , Turgeon, B.G. , Di Pietro, A. , Walton, J.D. , Ma, L.‐J. , Baker, S.E. , Rep, M. , Adam, G. , Antoniw, J. , Baldwin, T. , Calvo, S. , Chang, Y.‐L. , DeCaprio, D. , Gale, L.R. , Gnerre, S. , Goswami, R.S. , Hammond‐Kosack, K. , Harris, L.J. , Hilburn, K. , Kennell, J.C. , Kroken, S. , Magnuson, J.K. , Mannhaupt, G. , Mauceli, E. , Mewes, H.‐W. , Mitterbauer, R. , Muehlbauer, G. , Münsterkötter, M. , Nelson, D. , O'Donnell, K. , Ouellet, T. , Qi, W. , Quesneville, H. , Roncero, M.I.G. , Seong, K.‐Y. , Tetko, I.V. , Urban, M. , Waalwijk, C. , Ward, T.J. , Yao, J. , Birren, B.W. and Kistler, H.C. (2007) The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science, 317, 1400–1402. [DOI] [PubMed] [Google Scholar]
- Daboussi‐Bareyre, M.‐J. (1980) Heterokaryosis in Nectria haematococca: complementation between mutants affecting the expression of two differentiated states. J. Gen. Microbiol. 116, 425–433. [Google Scholar]
- Daskalov, A. , Paoletti, M. , Ness, F. and Saupe, S.J. (2012) Genomic clustering and homology between HET‐S and the NWD2 STAND protein in various fungal genomes. PLoS ONE, 7, e34854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny, T.P. and VanEtten, H.D. (1981) Tolerance by Nectria haematococca MP VI of the chickpea (Cicer arietinium) phytoalexins medicarpin and maackiain. Physiol. Plant Pathol. 19, 419–437. [Google Scholar]
- Denny, T.P. and VanEtten, H.D. (1982) Metabolism of the phytoalexins medicarpin and maackiain by Fusarium solani . Phytochemistry, 21, 1023–1028. [Google Scholar]
- Denny, T.P. and VanEtten, H.D. (1983a) Tolerance of Nectria haematococca MP VI to the phytoalexin pisatin in the absence of detoxification. J. Gen. Microbiol. 129, 2893–2901. [Google Scholar]
- Denny, T.P. and VanEtten, H.D. (1983b) Characterization of an inducible, nondegradative tolerance of Nectria haematococca MP VI to phytoalexins. J. Gen. Microbiol. 129, 2903–2913. [Google Scholar]
- Denny, T.P. , Matthews, P.S. and VanEtten, H.D. (1987) A possible mechanism of nondegradative tolerance of pisatin in Nectria haematococca MP VI. Physiol. Mol. Plant Pathol. 30, 93–107. [Google Scholar]
- Dickman, M.B. , Podila, G.K. and Kolattukudy, P.E. (1989) Insertion of cutinase gene into a wound pathogen enables it to infect intact host. Nature, 342, 446–448. [Google Scholar]
- van der Does, H.C. , Lievens, B. , Claes, L. , Houterman, P.M. , Cornelissen, B.J.C. and Rep, M. (2008) The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 10, 1475–1485. [DOI] [PubMed] [Google Scholar]
- Dufresne, M. , Lespinet, O. , Daboussi, M.‐J. and Hua‐Van, A. (2011) Genome‐wide comparative analysis of pogo‐like transposable elements in different Fusarium species. J. Mol. Evol. 73, 230–243. [DOI] [PubMed] [Google Scholar]
- Enkerli, J. , Bhatt, G. and Covert, S.F. (1997) Nht1, a transposable element cloned from a dispensable chromosome in Nectria haematococca . Mol. Plant–Microbe Interact. 10, 742–749. [DOI] [PubMed] [Google Scholar]
- Enkerli, J. , Bhatt, G. and Covert, S.F. (1998) Maackiain detoxification contributes to the virulence of Nectria haematococca MP VI on chickpea. Mol. Plant–Microbe Interact. 11, 317–326. [Google Scholar]
- Epstein, L. , Kwon, Y.H. , Almond, D.E. , Schached, L.M. and Jones, M.J. (1994) Genetic and biochemical characterization of Nectria haematococca strains with adhesive and adhesion‐reduced macroconidia. Appl. Environ. Microbiol. 60, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner, A. , Sopalla, C. and Weltring, K.‐M. (2002) An ATP‐binding cassette multidrug‐resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol. Plant–Microbe Interact. 15, 102–108. [DOI] [PubMed] [Google Scholar]
- Funnell, D.L. and VanEtten, H.D. (2002) Pisatin demethylase genes are on dispensable chromosomes while genes for pathogenicity on carrot and ripe tomato are on other chromosomes in Nectria haematococca . Mol. Plant–Microbe Interact. 15, 840–846. [DOI] [PubMed] [Google Scholar]
- Funnell, D.L. , Matthews, P.S. and VanEtten, H.D. (2002) Identification of new pisatin demethylase genes (PDA5 and PDA7) in Nectria haematococca and non‐Mendelian segregation of pisatin demethylating ability and virulence on pea due to loss of chromosomal elements. Fungal Genet. Biol. 37, 121–133. [DOI] [PubMed] [Google Scholar]
- Gardiner, D.M. , Stephens, A.E. , Munn, A.L. and Manners, J.M. (2013) An ABC pleiotropic drug resistance transporter in Fusarium graminearum with a role in crown and root diseases in wheat. FEMS Microbiol. Lett. 348, 36–45. [DOI] [PubMed] [Google Scholar]
- Geiser, D.M. , Aoki, T. , Bacon, C.W. , Baker, S.E. , Bhattacharyya, M.K. , Brandt, M.E. , Brown, D.W. , Burgess, L.W. , Chulze, S. , Coleman, J.J. , Correll, J.C. , Covert, S.F. , Crous, P.W. , Cuomo, C.A. , De Hoog, G.S. , Di Pietro, A. , Elmer, W.H. , Epstein, L. , Frandsen, R.J.N. , Freeman, S. , Gagkaeva, T. , Glenn, A.E. , Gordon, T.R. , Gregory, N.F. , Hammond‐Kosack, K. , Hanson, L.E. , Jímenez‐Gasco, M.D.M. , Kang, S. , Kistler, H.C. , Kuldau, G.A. , Leslie, J.F. , Logrieco, A. , Lu, G. , Lysøe, E. , Ma, L.‐J. , McCormick, S.P. , Migheli, Q. , Moretti, A. , Munaut, F. , O'Donnell, K. , Pfenning, L. , Ploetz, R.C. , Proctor, R.H. , Rehner, S.A. , Robert, V.A.R.G. , Rooney, A.P. , Salleh, B.B. , Scandiani, M.M. , Scauflaire, J. , Short, D.P.G. , Steenkamp, E. , Suga, H. , Summerall, B.A. , Sutton, D.A. , Thrane, U. , Trail, F. , Van Diepeningen, A. , VanEtten, H.D. , Viljoen, A. , Waalwijk, C. , Ward, T.J. , Wingfield, M.J. , Xu, J.‐R. , Yang, X.‐B. , Yli‐Mattila, T. and Zhang, N. (2013) One name, one fungus: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology, 103, 400–408. [DOI] [PubMed] [Google Scholar]
- Gerhold, D.L. , Pettinger, A.J. and Hadwiger, L.A. (1993) Characterization of a plant‐stimulated nuclease from Fusarium solani . Physiol. Mol. Plant Pathol. 43, 33–46. [Google Scholar]
- Graziani, S. , Silar, S. and Daboussi, M.‐J. (2004a) Bistability and hysteresis of the ‘Secteur’ differentiation are controlled by a two‐gene locus in Nectria haematococca . BMC Biol. 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani, S. , Vasnier, C. and Daboussi, M.‐J. (2004b) Novel polyketide synthase from Nectria haematococca . Appl. Environ. Microbiol. 70, 2984–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger, L.A. (2008) Pea–Fusarium solani interactions: contributions of a system toward understanding disease resistance. Phytopathology, 98, 372–379. [DOI] [PubMed] [Google Scholar]
- Hadwiger, L.A. , Chang, M.‐M. and Parsons, M.A. (1995) Fusarium solani DNase is a signal for increasing expression of nonhost disease resistance response genes, hypersensitivity, and pisatin production. Mol. Plant–Microbe Interact. 8, 871–879. [DOI] [PubMed] [Google Scholar]
- Han, Y. , Liu, X. , Benny, U. , Kistler, H.C. and VanEtten, H.D. (2001) Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca . Plant J. 25, 305–314. [DOI] [PubMed] [Google Scholar]
- Hansen, F.T. , Sørensen, J.L. , Giese, H. , Sondergaard, T.E. and Frandsen, R.J.N. (2012) Quick guide to polyketide synthase and nonribosomal synthase genes in Fusarium . Int. J. Food Microbiol. 155, 128–136. [DOI] [PubMed] [Google Scholar]
- Hawthorne, B.T. , Rees‐George, J. and Crowhurst, R.N. (2001) Induction of cutinolytic esterase activity during saprophytic growth of cucurbit pathogens, Fusarium solani f. sp. cucurbitiae races one and two (Nectria haematococca MPI and MPV, respectively). FEMS Microbiol. Lett. 194, 135–141. [DOI] [PubMed] [Google Scholar]
- Holenstein, J. and Défago, G. (1983) Inheritance of napthazarin production and pathogenicity to pea in Nectria haematococca . J. Exp. Bot. 34, 927–935. [Google Scholar]
- Inoue, S. , Turgeon, B.G. , Yoder, O.C. and Aist, J.R. (1998a) Role of fungal dynein in hyphal growth, microtubule organization, spindle pole body motility and nuclear migration. J. Cell Sci. 111, 1555–1566. [DOI] [PubMed] [Google Scholar]
- Inoue, S. , Yoder, O.C. , Turgeon, B.G. and Aist, J.R. (1998b) A cytoplasmic dynein required for mitotic aster formation in vivo. J. Cell Sci. 111, 2607–2614. [DOI] [PubMed] [Google Scholar]
- Jones, M.J. and Epstein, L. (1989) Adhesion of Nectria haematococca macroconidia. Physiol. Mol. Plant Pathol. 35, 453–461. [Google Scholar]
- Jones, M.J. and Epstein, L. (1990) Adhesion of macroconidia to the plant surface and virulence of Nectria haematococca . Appl. Environ. Microbiol. 56, 3772–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper, J.T. , Kämper, U. , Rogers, L.M. and Kolattukudy, P.E. (1994) Identification of regulatory elements in the cutinase promoter from Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 269, 9195–9204. [PubMed] [Google Scholar]
- Kern, H. and Naef‐Roth, S. (1965) Zur Bildung phytotoxischer Farbstoffe durch Fusarien der Gruppe Martiella. Phytopathol. Z. 53, 45–64. [Google Scholar]
- Khan, K. and Straney, D.C. (1999) Regulatory signals influencing expression of the PDA1 gene of Nectria haematococca MPVI in culture and during pathogenesis of pea. Mol. Plant–Microbe Interact. 12, 733–742. [Google Scholar]
- Khor, W.B. , Aung, T. , Saw, S.M. , Wong, T.Y. , Tambyah, P.A. , Tan, A.L. , Beuerman, R. , Lim, L. , Chan, W.K. , Heng, W.J. , Lim, J. , Loh, R.S.K. , Lee, S.B. and Tan, D.T.H. (2006) An outbreak of Fusarium keratitis associated with contact lens wear in Singapore. J. Am. Med. Assoc. 295, 2867–2873. [DOI] [PubMed] [Google Scholar]
- Kim, H.‐G. , Meinhardt, L.W. , Benny, U. and Kistler, H.C. (1995) NRS1, a middle repetitive sequence linked to pisatin demethylase genes in Nectria haematococca . Mol. Plant–Microbe Interact. 8, 524–531. [DOI] [PubMed] [Google Scholar]
- Kistler, H.C. and VanEtten, H.D. (1981) Phaseollin metabolism and tolerance in Fusarium solani f. sp. phaseoli . Physiol. Plant Pathol. 19, 257–271. [Google Scholar]
- Kistler, H.C. and VanEtten, H.D. (1984) Three non‐allelic genes for pisatin demethylation in the fungus Nectria haematococca . J. Gen. Microbiol. 130, 2595–2603. [Google Scholar]
- Kolander, T.M. , Bienapfl, J.C. , Kurle, J.E. and Malvick, D.K. (2012) Symptomatic and asymptomatic host range of Fusarium virguliforme, the causative agent of soybean sudden death syndrome. Plant Dis. 96, 1148–1153. [DOI] [PubMed] [Google Scholar]
- Kwon, Y.H. and Epstein, L. (1993) A 90‐kDa glycoprotein associated with adhesion of Nectria haematococca macroconidia to substrata. Mol. Plant–Microbe Interact. 6, 481–487. [Google Scholar]
- Kwon, Y.H. and Epstein, L. (1997) Involvement of the 90 kDa glycoprotein in adhesion of Nectria haematococca macroconidia. Physiol. Mol. Plant Pathol. 51, 287–303. [Google Scholar]
- Li, D. and Kolattukudy, P.E. (1997) Cloning of cutinase transcription factor 1, a transactivating protein containing Cys6Zn2 binuclear cluster DNA‐binding motif. J. Biol. Chem. 272, 12 462–12 467. [DOI] [PubMed] [Google Scholar]
- Li, D. , Chung, K.‐R. , Smith, D.A. and Schardl, C.L. (1995) The Fusarium solani gene encoding kievitone hydratase, a secreted enzyme that catalyzes detoxification of a bean phytoalexin. Mol. Plant–Microbe Interact. 8, 388–397. [DOI] [PubMed] [Google Scholar]
- Li, D. , Sirakova, T. , Rogers, L. , Ettinger, W.F. and Kolattukudy, P.E. (2002) Regulation of constitutively expressed and induced cutinase genes by different zinc finger transcription factors in Fusarium solani f. sp. pisi (Nectria haematococca). J. Biol. Chem. 277, 7905–7912. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Inlow, M. and VanEtten, H.D. (2003) Expression profiles of pea pathogenicity (PEP) genes in vivo and in vitro, characterization of the flanking regions of the PEP cluster and evidence that the PEP cluster region resulted from horizontal gene transfer in the fungal pathogen Nectria haematococca . Curr. Genet. 44, 95–103. [DOI] [PubMed] [Google Scholar]
- Lombard, L. , van der Merwe, N.A. , Groenewald, J.Z. and Crous, P.W. (2015) Generic concepts in Nectriaceae . Stud. Mycol. 80, 189–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy, M.C. , Matthews, P.S. and VanEtten, H.D. (1988) Metabolic detoxification of the phytoalexins maackiain and medicarpin by Nectria haematococca field isolates: relationship to virulence on chickpea. Physiol. Mol. Plant Pathol. 33, 187–199. [Google Scholar]
- Ma, L.‐J. , van der Does, H.C. , Borkovich, K.A. , Coleman, J.J. , Daboussi, M.‐J. , Di Pietro, A. , Dufresne, M. , Freitag, M. , Grabherr, M. , Henrissat, B. , Houterman, P.M. , Kang, S. , Shim, W.‐B. , Woloshuk, C. , Xie, X. , Xu, J.‐R. , Antoniw, J. , Baker, S.E. , Bluhm, B.H. , Breakspear, A. , Brown, D.W. , Butchko, R.A.E. , Chapman, S. , Coulson, R. , Coutinho, P.M. , Danchin, E.G.J. , Diener, A. , Gale, L.R. , Gardnier, D.M. , Goff, S. , Hammond‐Kosack, K.E. , Hilburn, K. , Hua‐Van, A. , Jonkers, W. , Kazan, K. , Kodira, C.D. , Koehrsen, M. , Kumar, L. , Lee, Y.‐H. , Li, L. , Manners, J.M. , Miranda‐Saavedra, D. , Mukherjee, M. , Park, G. , Park, J. , Park, S.‐Y. , Proctor, R.H. , Regev, A. , Ruiz‐Roldan, M.C. , Sain, D. , Sakthikumar, S. , Sykes, S. , Schwartz, D.C. , Turgeon, B.G. , Wapinski, I. , Yoder, O. , Young, S. , Zeng, Q. , Zhou, S. , Galagan, J. , Cuomo, C.A. , Kistler, H.C. and Rep, M. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature, 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh, S.F. , Matthews, D.E. and VanEtten, H.D. (1989) Two additional genes for pisatin demethylation and their relationship to the pathogenicity of Nectria haematococca on pea. Mol. Plant–Microbe Interact. 4, 341–349. [Google Scholar]
- Mahmoud, A.M. and Taga, M. (2012) Cytological karyotyping and characterization of a 410‐Kb minichromosome in Nectria haematococca MPI. Mycologia, 104, 845–856. [DOI] [PubMed] [Google Scholar]
- Maloney, A.P. and VanEtten, H.D. (1994) A gene from the fungal pathogen Nectria haematococca that encodes the phytoalexin‐detoxifying enzyme pisatin demethylase defines a new cytochrome P450 family. Mol. Gen. Genet. 243, 506–514. [DOI] [PubMed] [Google Scholar]
- Matuo, T. and Snyder, W.C. (1973) Use of morphology and mating populations in the identification of formae speciales in Fusarium solani . Phytopathology, 63, 562–565. [Google Scholar]
- Medentsev, A.G. and Akimenko, V.K. (1998) Naphthoquinone metabolites of the fungi. Phytochemistry, 47, 935–959. [DOI] [PubMed] [Google Scholar]
- Miao, V.P. , Covert, S.F. and VanEtten, H.D. (1991a) A fungal gene for antibiotic resistance on a dispensable (‘B’) chromosome. Science, 254, 1773–1776. [DOI] [PubMed] [Google Scholar]
- Miao, V.P.W. and VanEtten, H.D. (1992) Three genes for metabolism of the phytoalexin maackiain in the plant pathogen Nectria haematococca: meiotic instability and relationships to a new gene for pisatin demethylase. Appl. Environ. Microbiol. 58, 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, V.P.W. , Matthews, D.E. and VanEtten, H.D. (1991b) Identification and chromosomal locations of a family of cytochrome P‐450 genes for pisatin detoxification in the fungus Nectria haematococca . Mol. Gen. Genet. 226, 214–223. [DOI] [PubMed] [Google Scholar]
- Milani, N.A. , Lawrence, D.P. , Arnold, A.E. and VanEtten, H.D. (2012) Origin of pisatin demethylase (PDA) in the genus Fusarium . Fungal Genet. Biol. 49, 933–942. [DOI] [PubMed] [Google Scholar]
- Morrissey, J.P. and Osbourn, A.E. (1998) Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63, 708–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammed, M. , Anagnostou, T. , Desalermos, A. , Kourkoumpetis, T.K. , Carneiro, H.A. , Glavis‐Bloom, J. , Coleman, J.J. and Mylonakis, E. (2013) Fusarium infection: report of 26 cases and review of 97 cases from the literature. Medicine (Baltimore), 92, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C.A. , Cameron, J.A. , Huang, S.J. and Vinopal, R.T. (1996) Fusarium polycaprolactone depolymerase is cutinase. Appl. Environ. Microbiol. 62, 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalim, F.A. , Samuels, G.J. , Wijesundera, R.L. and Geiser, D.M. (2011) New species from the Fusarium solani species complex derived from perithecia and soil in the Old World tropics. Mycologia, 103, 1302–1330. [DOI] [PubMed] [Google Scholar]
- O'Donnell, K. (2000) Molecular phylogeny of the Nectria haematococca–Fusarium solani species complex. Mycologia, 92, 919–938. [Google Scholar]
- O'Donnell, K. , Sarver, B.A.J. , Brandt, M. , Chang, D.C. , Noble‐Wang, J. , Park, B.J. , Sutton, D.A. , Benjamin, L. , Lindsley, M. , Padhye, A. , Geiser, D.M. and Ward, T.J. (2007) Phylogenetic diversity and microsphere array‐based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens‐associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 45, 2235–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K. , Sutton, D.A. , Fothergill, A. , McCarthy, D. , Rinaldi, M.G. , Brandt, M.E. , Zhang, N. and Geiser, D.M. (2008) Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J. Clin. Microbiol. 46, 2477–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K. , Rooney, A.P. , Proctor, R.H. , Brown, D.W. , McCormick, S.P. , Ward, T.J. , Frandsen, R.J.N. , Lysøe, E. , Rehner, S.A. , Aoki, T. , Robert, V.A.R.G. , Crous, P.W. , Groenewald, J.Z. , Kang, S. and Geiser, D.M. (2013) Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet. Biol. 52, 20–31. [DOI] [PubMed] [Google Scholar]
- Podila, G.K. , Dickman, M.B. and Kolattukudy, P.E. (1988) Transcriptional activation of a cutinase gene in isolated fungal nuclei by plant cutin monomers. Science, 242, 922–925. [Google Scholar]
- Proctor, R.H. , Butchko, R.A. , Brown, D.W. and Moretti, A. (2007) Functional characterization, sequence comparisons and distribution of a polyketide synthase gene required for perithecial pigmentation in some Fusarium species. Food Addit. Contam. 24, 1076–1087. [DOI] [PubMed] [Google Scholar]
- Pudake, R.N. , Swaminathan, S. , Sahu, B.B. , Leandro, L.F. and Bhattacharyya, M.K. (2013) Investigation of the Fusarium virguliforme fvtox1 mutants revealed that the FvTox1 toxin is involved in foliar sudden death syndrome development in soybean. Curr. Genet. 59, 107–117. [DOI] [PubMed] [Google Scholar]
- Reichle, R.E. , Snyder, W.C. and Matuo, T. (1964) Hypomyces stage of Fusarium solani f. pisi . Nature, 203, 664–665. [Google Scholar]
- Rep, M. , van der Does, H.C. , Meijer, M. , van Wijk, R. , Houterman, P.M. , Dekker, H.L. , de Koster, C.G. and Cornelissen, B.J.C. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rogers, L.M. , Flaishman, M.A. and Kolattukudy, P.E. (1994) Cutinase gene disruption in Fusarium solani f.sp. pisi decreases its virulence on pea. Plant Cell, 6, 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, L.M. , Kim, Y.‐K. , Guo, W. , Gonzalez‐Candelas, L. , Li, D. and Kolattukudy, P.E. (2000) Requirement for either a host‐ or pectin‐induced pectate lyase for infection of Pisum sativum by Nectria haematococca . Proc. Natl. Acad. Sci. USA, 97, 9813–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Carres, M. , White, G. , Tsuchiya, D. , Taga, M. and VanEtten, H.D. (2008) The supernumerary chromosome of Nectria haematococca that carries pea‐pathogenicity‐related genes also carries a trait for pear rhizosphere competitiveness. Appl. Environ. Microbiol. 74, 3849–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg, M.K. and Davis, R.M. (2007) Host range and phylogeny of Fusarium solani f. sp. eumartii from potato and tomato in California. Plant Dis. 91, 585–592. [DOI] [PubMed] [Google Scholar]
- Rossman, A.Y. , Samuels, G.J. , Rogerson, C.T. and Lowen, R. (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud. Mycol. 42, 1–248. [Google Scholar]
- Ruan, Y. , Kotraiah, V. and Straney, D.C. (1995) Flavonoids stimulate spore germination in Fusarium solani pathogenic on legumes in a manner sensitive to inhibitors of cAMP‐dependent protein kinase. Mol. Plant–Microbe Interact. 8, 929–938. [Google Scholar]
- Samac, D.A. and Leong, S.A. (1988) Two linear plasmids in mitochondria of Fusarium solani f. sp. cucurbitae . Plasmid, 19, 57–67. [DOI] [PubMed] [Google Scholar]
- Samac, D.A. and Leong, S.A. (1989) Disease development in Cucurbita maxima (squash) infected with Fusarium solani f. sp. cucurbitae . Can. J. Bot. 67, 3486–3489. [Google Scholar]
- Sandrock, T.M. , Yoder, O.C. , Turgeon, B.G. and Aist, J.R. (1999) Subtle mitotic phenotypes of a Kar3/KlpA‐like mutant of Nectria haematococca . Mol. Biol. Cell, 10 (Suppl. ), 128a. [Google Scholar]
- Schuerger, A.C. and Mitchell, D.J. (1993) Influence of mucilage secreted by macroconidia of Fusarium solani f. sp. phaseoli on spore attachment to roots of Vigna radiata in hydroponic nutrient solution. Phytopathology, 83, 1162–1170. [Google Scholar]
- Shaykh, M. , Soliday, C.L. and Kolattukudy, P.E. (1977) Proof for the production of cutinase by Fusarium solani f.sp. pisi during penetration into its host, Pisum sativum . Plant Physiol. 60, 170–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett, A.M. , Enkerli, J. and Covert, S.F. (2002) Nht2, a copia LTR retrotransposon from a conditionally dispensable chromosome in Nectria haematococca . Curr. Genet. 41, 99–106. [DOI] [PubMed] [Google Scholar]
- Short, D.P.G. , O'Donnell, K. , Thrane, U. , Nielsen, K.F. , Zhang, N. , Juba, J.H. and Geiser, D.M. (2013) Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet. Biol. 53, 59–70. [DOI] [PubMed] [Google Scholar]
- Short, G.E. and Lacy, M.L. (1974) Germination of Fusarium solani f. sp. pisi chlamydospores in the spermosphere of pea. Phytopathology, 64, 558–562. [Google Scholar]
- Silar, P. and Daboussi, M.‐J. (1999) Non‐conventional infectious elements in filamentous fungi. Trends Genet. 15, 141–145. [DOI] [PubMed] [Google Scholar]
- Skov, J. , Lemmens, M. and Giese, H. (2004) Role of a Fusarium culmorum ABC transporter (FcABC1) during infection of wheat and barley. Physiol. Mol. Plant Pathol. 64, 245–254. [Google Scholar]
- Smith, D.A. , Harrer, J.M. and Cleveland, T.E. (1982) Relation of production of extracellular kievitone hydratase by isolates of Fusarium and their pathogenicity on Phaseolus vulgaris . Phytopathology, 72, 1319–1323. [Google Scholar]
- Smith, D.A. , Wheeler, H.E. , Banks, S.W. and Cleveland, T.E. (1984) Association between lowered kievitone hydratase activity and reduced virulence to bean in variants of Fusarium solani f. sp. phaseoli . Physiol. Plant Pathol. 25, 135–147. [Google Scholar]
- Snyder, W.C. and Hansen, H.N. (1941) The species concept in Fusarium with reference to section Martiella . Am. J. Bot. 28, 738–742. [Google Scholar]
- Srivastava, S.K. , Huang, X. , Brar, H.K. , Fakhoury, A.M. , Bluhm, B.H. and Bhattacharyya, M.K. (2014) The genome sequence of the fungal pathogen Fusarium virguliforme that causes sudden death syndrome in soybean. PLoS ONE, 9, e81832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, D.J. and Schäfer, W. (1992) Cutinase is not required for fungal pathogenicity on pea. Plant Cell, 4, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, D.J. , Theuerkauf, A. , Heitefuss, R. and Schäfer, W. (1994) Cutinase of Nectria haematococca (Fusarium solani f. sp. pisi) is not required for fungal virulence or organ specificity on pea. Mol. Plant–Microbe Interact. 7, 713–725. [Google Scholar]
- Straney, D.C. and VanEtten, H.D. (1994) Characterization of the PDA1 promoter of Nectria haematococca and the identification of a region that binds a pisatin‐responsive DNA binding factor. Mol. Plant–Microbe Interact. 7, 256–266. [DOI] [PubMed] [Google Scholar]
- Suga, H. , Ikeda, S. , Taga, M. , Kageyama, K. and Hyakumachi, M. (2002) Electrophoretic karyotyping and gene mapping of seven formae speciales in Fusarium solani . Curr. Genet. 41, 254–260. [DOI] [PubMed] [Google Scholar]
- Sugiura, Y. , Barr, J.R. , Barr, D.B. , Brock, J.W. , Elie, C.M. , Ueno, Y. , Patterson, D.G. Jr. , Potter, M.E. and Reiss, E. (1999) Physiological characteristics and mycotoxins of human clinical isolates of Fusarium species. Mycol. Res. 103, 1462–1468. [Google Scholar]
- Summerbell, R.C. and Schroers, H.‐J. (2002) Analysis of phylogenetic relationship of Cylindrocarpon lichenicola and Acremonium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic infections caused by these fungi. J. Clin. Microbiol., 40, 2866–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temporini, E.D. and VanEtten, H.D. (2002) Distribution of the pea pathogenicity (PEP) genes in the fungus Nectria haematococca mating population VI. Curr. Genet. 41, 107–114. [DOI] [PubMed] [Google Scholar]
- Temporini, E.D. and VanEtten, H.D. (2004) An analysis of the phylogenetic distribution of the pea pathogenicity genes of Nectria haematococca MPVI supports the hypothesis of their origin by horizontal gene transfer and uncovers a potentially new pathogen of garden pea: Neocosmospora boniensis . Curr. Genet. 46, 29–36. [DOI] [PubMed] [Google Scholar]
- Tucker, S.L. and Talbot, N.J. (2001) Surface attachment and pre‐penetration stage development in plant pathogenic fungi. Annu. Rev. Phytopathol. 39, 385–417. [DOI] [PubMed] [Google Scholar]
- Turbek, C.S. , Smith, D.A. and Schardl, C.L. (1992) An extracellular enzyme from Fusarium solani f. sp. phaseoli, which catalyzes hydration of the isoflavonoid phytoalexin, phaseollidin. FEMS Microbiol. Lett. 94, 187–190. [DOI] [PubMed] [Google Scholar]
- Van Egeraat, A.W.S.M. (1975) The possible role of homoserine in the development of Rhizobium leguminosarum in the rhizospheres of pea seedlings. Plant Soil, 42, 37–47. [Google Scholar]
- VanEtten, H. , Jorgensen, S. , Enkerli, J. and Covert, S.F. (1998) Inducing the loss of conditionally dispensable chromosomes in Nectria haematococca during vegetative growth. Curr. Genet. 33, 299–303. [DOI] [PubMed] [Google Scholar]
- VanEtten, H. , Temporini, E. and Wasmann, C. (2001) Phytoalexin (and phytoanticipin) tolerance as a virulence trait: why is it not required by all pathogens? Physiol. Mol. Plant Pathol. 59, 83–93. [Google Scholar]
- VanEtten, H.D. (1978) Identification of additional habitats of Nectria haematococca mating population VI. Phytopathology, 68, 1552–1556. [Google Scholar]
- VanEtten, H.D. , Matthews, P.S. , Tegtmeier, K.J. , Dietert, M.F. and Stein, J.F. (1980) The association of pisatin tolerance and demethylation with virulence on pea in Nectria haematococca . Physiol. Plant Pathol. 16, 257–269. [Google Scholar]
- VanEtten, H.D. , Mansfield, J.W. , Bailey, J.A. and Farmer, E.E. (1994) Two classes of plant antibiotics: phytoalexins versus ‘phytoanticipins’. Plant Cell, 6, 1191–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmann, C.C. and VanEtten, H.D. (1996) Transformation‐mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol. Plant–Microbe Interact. 9, 793–803. [Google Scholar]
- White, G.J. (2008) The effect of conditionally dispensable chromosomes of Nectria haematococca MPVI on rhizosphere colonization and the identification of a gene cluster for homoserine utilization. PhD Dissertation.
- Woloshuk, C.P. and Kolattukudy, P.E. (1986) Mechanism by which contact with plant cuticle triggers cutinase gene expression in the spores of Fusarium solani f. sp. pisi . Proc. Natl. Acad. Sci. USA, 83, 1704–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. , Sandrock, T.M. , Turgeon, B.G. , Yoder, O.C. , Wirsel, S.G. and Aist, J.R. (1998) A fungal kinesin required for organelle motility, hyphal growth, and morphogenesis. Mol. Biol. Cell, 9, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Rogers, L.M. , Song, Y. , Guo, W. and Kolattukudy, P.E. (2005) Homoserine and asparagine are host signals that trigger in planta expression of a pathogenesis gene in Nectria haematococca . Proc. Natl. Acad. Sci. USA, 102, 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. , O'Donnell, K. , Sutton, D.A. , Nalim, F.A. , Summerbell, R.C. , Padhye, A.A. and Geiser, D.M. (2006) Members of the Fusarium solani species complex that cause infections in humans and plants are common in the environment. J. Clin. Microbiol. 44, 2186–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. and Smith, D.A. (1983) Concurrent metabolism of the phytoalexins phaseollodin, kievitone and phaseollinisoflavan by Fusarium solani f. sp. phaseoli . Physiol. Plant Pathol. 23, 89–100. [Google Scholar]


