Summary
Natural and synthetic elicitors have contributed significantly to the study of plant immunity. Pathogen‐derived proteins and carbohydrates that bind to immune receptors, allow the fine dissection of certain defence pathways. Lipids of a different nature that act as defence elicitors, have also been studied, but their specific effects have been less well characterized, and their receptors have not been identified. In animal cells, nanoliposomes of the synthetic cationic lipid 3‐tetradecylamino‐tert‐butyl‐N‐tetradecylpropionamidine (diC14) activate the TLR4‐dependent immune cascade. Here, we have investigated whether this lipid induces Arabidopsis defence responses. At the local level, diC14 activated early and late defence gene markers (FRK1, WRKY29, ICS1 and PR1), acting in a dose‐dependent manner. This lipid induced the salicylic acid (SA)‐dependent, but not jasmonic acid (JA)‐dependent, pathway and protected plants against Pseudomonas syringae pv. tomato (Pst), but not Botrytis cinerea. diC14 was not toxic to plant or pathogen, and potentiated pathogen‐induced callose deposition. At the systemic level, diC14 induced PR1 expression and conferred resistance against Pst. diC14‐induced defence responses required the signalling protein EDS1, but not NDR1. Curiously, the lipid‐induced defence gene expression was lower in the fls2/efr/cerk1 triple mutant, but still unchanged in the single mutants. The amidine headgroup and chain length were important for its activity. Given the robustness of the responses triggered by diC14, its specific action on a defence pathway and the requirement for well‐known defence components, this synthetic lipid is emerging as a useful tool to investigate the initial events involved in plant innate immunity.
Keywords: Arabidopsis, lipid elicitors, local and systemic defences, PAMP/DAMP, SA‐ and JA‐dependent pathways
Introduction
Plants detect potential pathogenic microbes at different cellular levels. At the cell surface, pattern recognition receptors (PRRs) perceive pathogen/microbe‐associated molecular patterns (PAMPs) or compounds released during infection (damage‐associated molecular patterns, DAMP) (Albert et al., 2010) to activate pattern‐triggered immunity (PTI) (Macho and Zipfel, 2014; Zhang and Thomma, 2013). This warning system confers broad‐spectrum defences against non‐adapted invaders, but fails to counteract successful pathogens that deliver effectors to suppress PTI (Block and Alfano, 2011). Inside plant cells, effectors can be recognized by host resistance (R) proteins to therefore induce effector‐triggered immunity (ETI), a second layer of defence that provides race‐specific resistance (McDowell and Simon, 2008).
PRRs are single‐pass transmembrane proteins carrying a ligand‐binding ectodomain. At the intracellular level, these proteins contain either a kinase domain (receptor‐like kinases, RLKs) or a short tail lacking kinase function (receptor‐like proteins, RLPs). Most PRRs act in concert with other receptors forming active multi‐component complexes that signal defences on ligand recognition (Macho and Zipfel, 2014; Zhang and Thomma, 2013). PRRs that bind known ligands have been identified in various plant species. In Arabidopsis, flagellin and the flagellin‐derived peptide flg22 bind to the FLS2 (flagellin sensing 2) receptor (Chinchilla et al., 2006), elongation factor Tu (EF‐Tu) binds to EFR (EF‐Tu receptor) (Zipfel et al., 2006), chitin binds to CERK1 (chitin elicitor receptor kinase 1) (Miya et al., 2007) and peptidoglycans (PGNs) bind to LYM1 and LYM3 (lysin‐motif proteins 1 and 3) (Willmann et al., 2011). In turn, R proteins contain a central nucleotide‐binding site (NBS) and a C‐terminal leucine‐rich repeat (LRR) and, based on their N‐terminus, are classified into two subfamilies. One subfamily includes homologues of Drosophila Toll/mammalian interleukin receptors (TIR), and the second contains receptors with a coiled‐coil (CC) domain. TIR‐NBS‐LRR and CC‐NBS‐LRR proteins function through distinct signalling cascades, with the first group requiring the lipase‐like protein EDS1 (enhanced disease susceptibility 1) and the second the NDR1 (non‐race‐specific disease resistance 1) protein (Aarts et al., 1998).
ETI and PTI trigger certain common responses, such as the increase in reactive oxygen species, nitric oxide and salicylic acid (SA), the activation of mitogen‐activated protein kinase cascades, the alteration of the cell wall and the induction of pathogen‐responsive genes (Asai et al., 2002; Block and Alfano, 2011; Dempsey et al., 2011; Tsuda et al., 2013). In most cases, ETI is faster and stronger than PTI, and generates cell death associated with the hypersensitive response (HR) (Mur et al., 2008). Both defence programmes induce systemic acquired resistance (SAR), which protects the entire plant against further microbial infections (Dempsey and Klessig, 2012; Shah and Zeier, 2013). Interestingly, some endogenous signalling components, such as the EDS1 protein, participate in ETI and PTI (Rietz et al., 2011), reinforcing the notion of some convergence between them (Tsuda et al., 2013).
The use of elicitors has allowed the discovery of many key features of the plant immune programmes. Bacteria‐derived elicitors, including peptides from flagellin (flg22) and EF‐Tu (elf18), and the sugar moiety of cell wall‐derived PGNs, have assisted the identification and characterization of their corresponding receptors. The Xanthomonas outer membrane protein Ax21 is another well‐studied natural PAMP, previously suspected to bind the XA21 receptor (see Macho and Zipfel, 2014). Elicitors derived from fungi and oomycetes include the EIX (ethylene‐inducing xylanase) protein, the 13‐amino‐acid peptide from cell wall transglutaminase (Pep13) and the cellulose‐binding domains of cell wall proteins, β‐glycans and chitin. PRRs that bind these elicitors have also been identified, including the tomato EIX receptor (EIX2) and the Arabidopsis and rice chitin receptors (CERK1 and CEBip, respectively) (Bar and Avni, 2009; Kaku et al., 2006; Miya et al., 2007). In contrast, the effects of lipid elicitors present in bacteria, fungi or oomycetes have been less well characterized, and their receptors have not been described so far. Rhamnolipids and surfactin stimulate the SA and jasmonic acid (JA) pathways, protecting plants against Botrytis cinerea, Hyaloperonospora arabidopsidis and Pseudomonas syringae (Bais et al., 2004; Ongena et al., 2007; Sanchez et al., 2012). Massetolide A triggers induced systemic resistance, but not SA signalling, and enhances defences to Phytophthora infestans (Tran et al., 2007). Arachidonic acid activates JA‐mediated responses and represses the SA pathway, affecting resistance to aphids and necrotrophic as well as biotrophic pathogens (Savchenko et al., 2010). Ergosterol induces alkalinization of the cell growth medium and may activate a host receptor, as its application desensitizes host cells to a second treatment (Granado et al., 1995). At present, the best‐characterized lipid elicitor is lipopolysaccharide (LPS), which induces defences in monocots and dicots (Newman et al., 2002; Sun et al., 2012; Zeidler et al., 2004). LPS potentiates the oxidative burst, nitric oxide generation, callose deposition, defence gene expression, and local and/or systemic pathogen resistance, although not all of these responses are observed in all systems (Newman et al., 2002, 2007; Sun et al., 2012). The lipid A moiety is conserved in many plant‐associated bacteria and itself acts as a PAMP, whereas the oligosaccharide core and O‐specific chains also trigger defences. Moreover, in Arabidopsis, lipid A and oligosaccharide domains display agonistic effects on defence gene expression (Madala et al., 2012). The mechanisms involved in LPS perception by plants are unknown, and receptors for this elicitor have not been described to date. In animals, LPS is detected by intracellular nucleotide‐binding oligomerization domain (NOD)‐like receptors and the surface Toll‐like receptor 4 (TLR4), which requires accessory proteins (Chen et al., 2009).
In mammals, lipid‐based nanoparticles stimulate the immune system (Landesman‐Milo and Peer, 2012). In mouse and human bone marrow‐derived dendritic cells, the cationic lipid diC14 (3‐tetradecylamino‐tert‐butyl‐N‐tetradecylpropionamidine) activates the TLR4‐dependent pathways, leading to the secretion of the cytokines tumour necrosis factor‐α (TNF‐α), interleukin‐6 (IL‐6), interferon‐β (IFN‐β), IFN‐γ‐induced protein 10 (IP‐10) and IL‐12p40, and the induction of CD80/CD86 expression of co‐stimulatory factors (Jacquet et al., 2005; Tanaka et al., 2008; Wilmar et al., 2012). This cytokine secretion pattern is reminiscent of the TLR4‐dependent cytokine pattern induced by bacterial LPS, the natural ligand of TLR4 (Tanaka et al., 2008). To date, the effect of synthetic cationic lipid‐based nanoparticles on plant immunity has not been analysed. Here, we describe the ability of diC14 nanoparticles to stimulate the Arabidopsis defence signalling cascades. A single application of the lipid induces long‐term defences and pathogen resistance at local and systemic levels. The activation of these responses requires endogenous defence components, such as EDS1.
Results
diC14 induces SA‐dependent gene expression
We tested whether a single application of diC14 nanoliposomes was sufficient to elicit Arabidopsis defence responses. Leaves infiltrated with different lipid concentrations [5, 40 and 100 μg diC14/mL in 0.2 mm N‐2‐hydroxyethylpiperazine‐N’‐2‐ethanesulfonic acid (HEPES)] were collected at 8 and 24 h post‐treatment and used to monitor defence gene expression by semi‐quantitative reverse transcription‐polymerase chain reaction (sqRT‐PCR). WRKY29 (transcription factor) and FRK1 (flg22‐induced receptor‐like kinase 1) were used as PTI markers, ICS1 (isochorismate synthase 1) and PR1 (pathogen‐related 1) as SA‐sensitive genes, and PDF1.2 (plant defensin 1.2), VSP2 (vegetative storage protein 2) and LOX2 (lipoxygenase 2) as JA‐responsive genes (Asai et al., 2002; Dempsey et al., 2011). In parallel, mock (0.2 mm HEPES)‐treated leaves were used to control the effects of mechanical stress associated with the inoculation process.
diC14 nanoliposomes induced the expression of all PTI/SA gene markers (Fig. 1a). In different experiments, WRKY29 was consistently activated at 8 h post‐infiltration (hpi), even with the lowest lipid dose. FRK1, ICS1 and PR1 were induced at 8 and 24 hpi in a dose‐dependent manner, and PR1 showed the strongest overall response. In turn, VSP2 was induced at 8 hpi by low lipid concentrations (5 and 40 μg/mL), and PDF1.2 and LOX2 responded to both diC14 and mock treatment in a similar manner (Fig. 1b). These results indicated that diC14 produced a strong and sustained activation of the SA pathway (at least 24 h post‐treatment), whereas it induced the JA pathway in a weak and transient manner.
Figure 1.
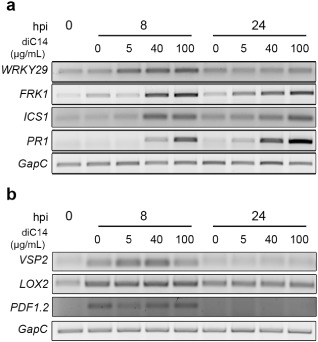
Expression of salicylic acid (SA)‐ and jasmonic acid (JA)‐sensitive genes in diC14‐treated leaves analysed by semi‐quantitative reverse transcription‐polymerase chain reaction (sqRT‐PCR). The pattern triggered immunity (PTI) and SA gene markers WRKY29, FRK1, ICS1 and PR1 (a) and the JA‐sensitive genes VSP2, LOX2 and PDF1.2 (b) were studied. GapC was used as internal control. Untreated [0 h post‐infiltration (hpi)] and diC14‐treated (5, 40 or 100 μg/mL HEPES, 0.2 mm] samples were analysed at 8 and 24 hpi, using HEPES (0.2 mm) as control (0 μg/mL diC14; mock inoculation). One representative from three independent biological experiments is shown.
diC14 enhances resistance to Pseudomonas syringae pv. tomato (Pst) DC3000
To examine the physiological relevance of the previous responses, we tested whether diC14 (20 μg/mL) conferred resistance to hemibiotrophic pathogens. In this assay, leaves were first treated with the lipid; 24 h later they were challenged with Pst DC3000 [105 colony‐forming units (cfu)/mL] and, at 3 days post‐inoculation (dpi), they were excised to quantify the bacterial content. As a control, leaves pre‐treated with mock solution were used to determine pathogen growth. As shown in Fig. 2a, diC14 restricted Pst proliferation in planta. At 3 dpi, mock‐ and diC14‐treated leaves contained 1.4 × 105 and 9.2 × 103 cfu/cm2, respectively. Interestingly, the lipid did not affect bacterial growth in vitro (Fig. S1, see Supporting Information), suggesting that, in the plant, it reduced pathogen proliferation by activating defence responses.
Figure 2.
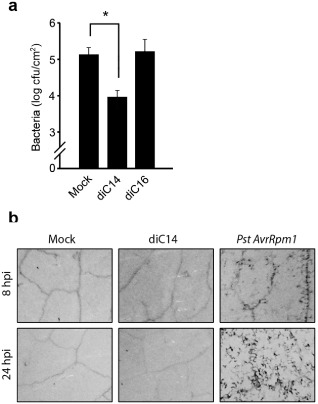
diC14 induces local resistance to Pseudomonas syringae pv. tomato (Pst). (a) Leaves pre‐treated with HEPES (0.2 mm) (mock), diC14 or diC16 (20 μg/mL each) were inoculated 24 h later with Pst [105 colony‐forming units (cfu)/mL] to quantify the bacterial content at 3 days post‐infection. Values represent mean ± standard deviation (SD) of two technical replicates. Similar results were obtained in two independent infection experiments. *Significant differences between mock and diC14 treatments (P < 0.05 by t‐test). (b) Cell death was evaluated by trypan blue staining on leaves sampled at 8 or 24 h post‐infiltration (hpi). The effects of mock solution (HEPES, 0.2 mm), diC14 (20 μg/mL) and Pst AvrRpm1 (107 cfu/mL) are compared. Similar results were obtained in three independent experiments.
Tissues treated with diC14 (20 μg/mL), which were able to activate SA‐responsive genes from 8 hpi (Fig. 1a), showed no cell death or damage at 8 or 24 hpi (Fig. 2b). In contrast, tissues challenged with a classical inducer of SA defences and cell death responses, the avirulent bacterium Pst AvrRpm1, showed cell death from 8 hpi (Fig. 2b).
diC14 primes callose deposition
Reinforcement of the cell wall through callose deposition is a classical PTI marker, usually accompanied by the activation of the Callose Synthase 12 gene (CalS12) (Dong et al., 2008). Callose deposits reach high levels in tissues treated with the Pst hrpC – mutant lacking the capacity to inject effectors though the type III secretion system (TTSS) (Hauck et al., 2003), and lower levels in tissues treated with Pst whose effectors may inhibit their generation (DebRoy et al., 2004).
We tested whether diC14 nanoliposomes (20 μg/mL) activated callose deposition at the infiltration site. Lipid‐treated leaves were sampled at 0, 12 and 24 hpi, and stained with aniline blue to detect callose deposits (Cecchini et al., 2011). diC14 itself was unable to induce this response (Fig. 3a), but slightly activated CalS12 gene expression (Fig. 3b). Interestingly, when diC14 was combined with Pst, the lipid potentiated the effect of pathogen in both responses. At 12 and 24 hpi, callose deposits increased to 100% and 25%, respectively, in tissues simultaneously inoculated with lipid and bacteria compared with those treated with pathogen only (Fig. 3a).
Figure 3.
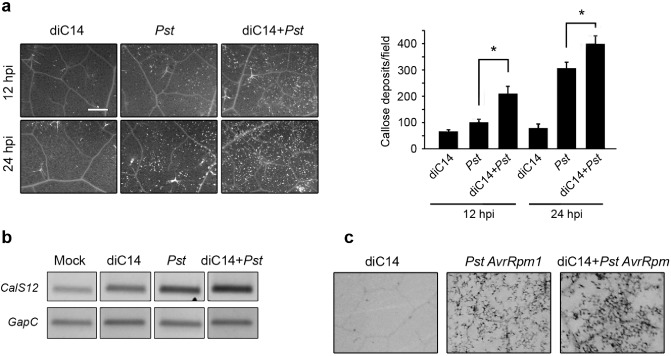
diC14 potentiates callose deposition. (a) Left: callose deposits detected by aniline blue staining at 12 and 24 h post‐infiltration (hpi) with diC14 (20 μg/mL), Pseudomonas syringae pv. tomato (Pst) [107 colony‐forming units (cfu)/mL] or diC14 + Pst. Scale bar, 0.5 mm. Right: number of callose deposits per field (4.4 mm2) determined with ImageJ software; values indicate mean ± standard error (SE) (12 photographs from six leaves). *Significant differences between Pst and diC14 + Pst treatments (P < 0.05 by t‐test). (b) Semi‐quantitative reverse transcription‐polymerase chain reaction (sqRT‐PCR) was used to determine the CalS12 transcript levels at 24 hpi in the samples described in (a). Mock: HEPES, 0.2 mm. (c) Trypan blue staining of tissues treated with diC14 (20 μg/mL), Pst AvrRpm1 (107 cfu/mL) or diC14 + Pst AvrRpm1 analysed at 24 hpi.
We wondered whether the potentiation effect of the lipid was caused by the inhibition of effector secretion. To test this possibility, we performed two experiments. First, we quantified callose deposits in tissues simultaneously treated with diC14 and Pst hrpC –. The lipid maintained a potentiation effect under this condition (130% increase in callose deposits at 24 hpi in diC14 + Pst hrpC – relative to Pst hrpC –; Fig. S2, see Supporting Information). Next, we evaluated whether diC14 reduced the capacity of Pst AvrRpm1 to trigger ETI in Col‐0 plants, where intracellular recognition of the TTSS effector AvrRpm1 by the host R protein RPM1 leads to cell death. As shown in Fig. 3c, cell death was not reduced by the co‐inoculation of diC14 with the avirulent bacteria. These results indicate that diC14 does not impair the TTSS of Pst, suggesting that the lipid sensitizes the tissues to rapidly trigger callose deposition after PAMP sensing.
diC14 requires EDS1 to activate defences
To learn more about the action of diC14, we tested whether lipid‐induced FRK1 and PR1 activation required endogenous defence signalling components. First, we used the triple mutant fls2/efr/cerk1 lacking the FLS2, EFR and CERK1 receptors (Gimenez‐Ibanez et al., 2009). At 24 hpi, the fls2/efr/cerk1 mutant showed lower capacity than wild‐type plants to induce FRK1 and PR1 by diC14 [Figs 4a and S3a (top panels, see Supporting Information)], accumulating lower levels of both transcripts in lipid‐treated tissues (Fig. S3a, bottom panels). However, the mutant retained the ability to activate both genes during stimulation of ETI by Pst AvrRpm1 (Fig. S3b). Next, we evaluated the effect of diC14 on the fls2, efr and cerk1 single mutants, and observed that all of these plants activated FRK1 and PR1 similarly to control plants (Fig. 4b).
Figure 4.
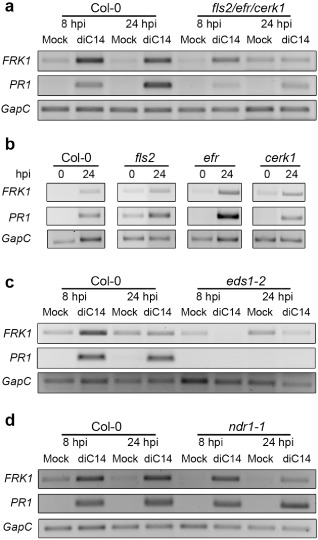
Plant signalling components required to activate defences by diC14. FRK1 and PR1 expression was evaluated as described in Fig. 1. Leaves treated with mock solution or diC14 (20 μg/mL) were sampled at 0, 8 or 24 h post‐infiltration (hpi). Responses of wild‐type plants were compared with those of fls2/efr/cerk1 (a), fls2, efr, cerk1 (b), eds1‐2 (c) and ndr1‐1 (d) mutants. One representative experiment from three independent assays is shown. Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) assays, confirming the reduced capacity of the fls2/efr/cerk1 mutant to induce FRK1 and PR1 by diC14, are shown in Fig. S3a (see Supporting Information).
Finally, we assessed how the absence of EDS1 and NDR1 affected the induction of FRK1 and PR1 by diC14. EDS1 participates in both PTI and ETI, whereas NDR1 is only involved in ETI (Aarts et al., 1998; Wiermer et al., 2005). Interestingly, the eds1‐2 mutant abolished PR1 induction by diC14. In addition, eds1‐2 plants reduced diC14‐mediated FRK1 activation (Fig. 4c). In contrast, ndr1‐1 plants maintained the capacity to activate both genes in response to diC14 (Fig. 4d).
These results indicate that the function of EDS1, but not NDR1, is required for full activation of FRK1 and PR1 by diC14 nanoliposomes.
diC14 induces systemic resistance
To test the effect of diC14 on systemic defences, we treated two leaves per plant with diC14 or mock solution and, 8 and 24 h later, sampled systemic untreated leaves to analyse PR1 and FRK1 expression. Both gene markers were clearly induced at 24 hpi, demonstrating the effect of diC14 on systemic tissues (Fig. 5a), which could derive from the generation of a systemic signal or mobility of the lipid in the plant (see Discussion). To assess the susceptibility of these systemic tissues, we treated plants with diC14 as before. One day later, we inoculated distal leaves with Pst (105 cfu/mL) to determine the bacterial content at 3 days post‐infection. A single application of diC14 was sufficient to induce SAR, as indicated by a 10‐fold reduction in pathogen content in diC14‐ relative to mock‐pre‐inoculated plants (Fig. 5b). DIR1 is an Arabidopsis lipid transport protein necessary to induce systemic PR1 expression after local inoculation of avirulent bacteria (Maldonado et al., 2002). To test whether diC14 requires DIR1 to signal PR1 and FRK1 activation, we quantified these transcripts in distal tissues of the null mutant plant dir1‐1. The mutant behaved as the control plant (Fig. 5c), indicating that DIR1 is dispensable for these effects.
Figure 5.
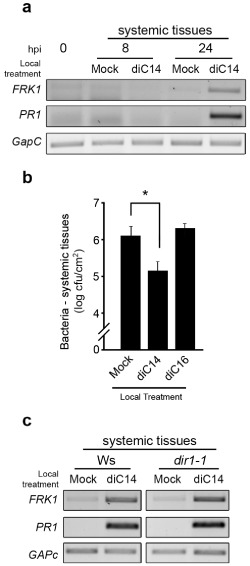
diC14 induces systemic resistance. Two leaves per plant were infiltrated with mock solution, diC14 or diC16 (20 μg/mL each). One day later, three untreated secondary leaves were used to evaluate either gene expression (a, c) or pathogen resistance (b). Expression of PR1 and FRK1 genes was studied as described in Fig. 1. (b) Leaves infiltrated with Pst (105 cfu/mL) were analysed at 3 days post‐infection, as described in Fig. 2. One representative experiment from two biological replicates is shown. *Significant differences between mock and diC14 treatments (P < 0.05 by t test). (c) Responses of wild‐type (Ws) and dir1‐1 mutant plants are compared.
Features of the diC14 molecule affecting defence induction
Finally, we evaluated which features of the diC14 molecule (charge, hydrophilic moiety or chain length) affected its capability to activate plant defences. Nanoliposomes of diC14 and four other synthetic related lipids (Table S1, see Supporting Information) were infiltrated in different sets of plants (20 μg/mL of each lipid) to test their effect on FRK1, ICS1 and PR1 expression. diC16 (3‐hexadecyl‐amino‐tert‐butyl‐N‐hexadecyl‐propionamidine), a longer tailed derivative that conserves the cationic amidine group, but contains 16 instead of 14 carbon residues in each lipid chain, also had an effect, albeit much smaller than that of diC14, as it activated FRK1 at 8 but not 24 hpi, and weakly induced ICS1 and PR1 at these time points (Fig. 6a). However, diC16 did not enhance resistance against Pst, as pre‐treatment with this lipid did not reduce pathogen growth (Fig. 2a). Neither was diC16 able to enhance systemic resistance to Pst (Fig. 5b). These results indicate that structural differences in the amidine molecule can alter the lipid capacity of defence activation.
Figure 6.
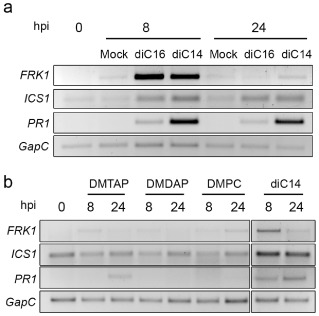
Effect of other synthetic lipids on FRK1, ICS1 and PR1 activation. Gene expression was monitored as in Fig. 1. All lipids were used at 20 μg/mL. The structures of diC16 (a), DMTAP, DMDAP and DMPC (b) are described in Table S1 (see Supporting Information).
To assess the role of the hydrophilic lipid moiety, we compared FRK1, ICS1 and PR1 expression in samples treated with diC14, the cationic lipids DMTAP and DMDAP (1,2‐dimyristoyl‐3‐trimethyl‐ammonium‐propane and 1,2‐dimyristoyl‐3‐dimethyl‐ammonium‐propane, respectively), and neutral lipid DMPC (1,2‐dimyristoyl‐sn‐glycero‐3‐phosphocholine), all sharing 14‐C acyl chains (Table S1). None of these lipids, except diC14, activated the gene markers in a strong or sustained manner (Fig. 6b), suggesting that the amidine chemical function is required for gene induction. Moreover, pre‐inoculation of DMTAP, DMDAP or DMPC on leaf tissues did not confer resistance against Pst (Fig. S4, see Supporting Information). In addition, diC16, which does possess an amidine chemical function, did not produce robust PR1 expression, suggesting that lipid chain length is also important for this effect.
Discussion
This study examines how diC14 impacts on the Arabidopsis immune pathways. The lipid proved to be a good inducer of local and systemic defences. A single application of diC14 produced rapid (8 hpi) and durable (24 hpi) activation of FRK1, WRKY29, ICS1 and PR1 genes in treated tissues. Gene induction was dose sensitive, increasing with lipid concentration. The extent of PR1 activation was similar to that produced by Pst AvrRpm1 infection (Fig. S5, see Supporting Information). Thus, diC14 appears to activate signalling cascades that normally function in disease resistance, using amplification events to mount a range of lasting defences. Supporting this possibility, several defence marker genes were induced in parallel and remained up‐regulated for at least 24 h. In addition, diC14 boosted pathogen‐induced defences by potentiating Pst‐mediated callose deposition. Similarly, LPS and rhamnolipids activate SA‐sensitive genes for at least 24 h (Sanchez et al., 2012; Zeidler et al., 2004), and LPS primes both the synthesis of antimicrobial compounds and the expression of defence genes induced by bacterial infection (Newman et al., 2002).
Lipid elicitors characterized to date have different effects on defence. Rhamnolipids and surfactin, mostly used as mixtures of compounds, activate SA‐ and JA‐dependent pathways, enhancing resistance to biotrophic and necrotrophic pathogens (Bais et al., 2004; Ongena et al., 2007; Sanchez et al., 2012; Varnier et al., 2009). Riboflavin (Dong and Beer, 2000) and ultrashort cationic lipopeptides (Brotman et al., 2009), used as pure compounds, also activate both pathways. Extracts of LPS induce the SA cascade, protecting plants against hemi/biotrophic pathogens (Sun et al., 2012; Zeidler et al., 2004). Under particular conditions, massetolide A (Tran et al., 2007), surfactin (Bais et al., 2004; Ongena et al., 2007), ultrashort cationic lipopeptides (Makovitzki et al., 2006) and rhamnolipids (Varnier et al., 2009) display antimicrobial activity, whereas none of these elicitors cause death or toxicity in plant cells at the lowest concentration that induces defence (Dong and Beer, 2000; Makovitzki et al., 2007; Newman et al., 2007; Ongena et al., 2007; Tran et al., 2007; Varnier et al., 2009). Meanwhile, diC14 induced SA‐sensitive defences signalled by EDS1, but not NDR1, and protected plants against Pst, having a mild and transient effect on the JA pathway and no consequences on resistance to B. cinerea (Fig. S6, see Supporting Information). diC14 was not toxic for bacteria or plant cells. Comparing the effects of diC14 with those of other lipid elicitors, LPS shows the greatest similarity. However, both compounds have differential actions (only LPS activates callose deposition). Hence, diC14 appears to be a nontoxic lipid elicitor that can be used as a pure compound to study early signalling events that activate plant immunity.
The mechanisms underlying diC14‐mediated plant defence activation are unknown. At the local level, the lipid targets particular signalling cascades. Based on our data and other published studies, we can envisage different effects. Surfactin binds to the plasma membrane (PM) of tobacco cells, showing high affinity for phospholipids. Its insertion into the membrane may either disturb lipid compartmentalization, or generate curvature constraints, thus affecting mechanosensitive channels or proteins involved in defence signalling (Henry et al., 2011). Cryptogein alters the PM of tobacco cells by modifying lateral compartmentalization and biophysical properties (fluidity), suggesting the generation of signalling platforms at the cell surface (Gerbeau‐Pissot et al., 2014). In addition, cryptogein (Stanislas et al., 2009), chitin (Fujiwara et al., 2009) and flg22 (Keinath et al., 2010) modify the protein composition of detergent‐resistant PM (DRM) fractions. In the latter case, 64 proteins (including RLKs‐like, FLS2, H+‐ATPAses and others) are enriched in this fraction 15 min after elicitation. Therefore, the interaction of diC14 with plant PM may alter the organization, compartmentalization or composition of this membrane to somehow boost the activity of the defence components targeted by this lipid.
Eventually, as suggested for animal cells (Tanaka et al., 2008), diC14 may bind a plant receptor. Cationic lipids sharing structural similarities with diC14 (charge, acyl chain length or headgroup), such as DMTAP, DMDAP, DMPC and diC16, were unable to stimulate significantly defence genes or confer resistance against Pst, as observed for diC14, arguing against broad‐range effects of cationic lipids on plant immunity. Low‐affinity receptors that perceive lipid PAMPs/DAMPs might be present in plants. Lipid elicitors require higher concentrations than peptide elicitors to activate JA/SA/PTI markers [rhamnolipids, 200 μg/mL (approximately 300 μm); diC14, 5–40 μg/mL (approximately 10–75 μm); peptide elicitors, subnanomolar concentration] (Boller and Felix, 2009; Sanchez et al., 2012). The dose of LPS that triggers plant PTI (5–100 μg/mL) is higher than that stimulating the TLR4 pathway in animal cells (pg/mL to ng/mL) (Zeidler et al., 2004). Competition experiments analysing the internalization of labelled molecules suggested the existence of a low‐affinity LPS receptor in tobacco cells (Gross et al., 2005). However, this possibility was questioned by other studies (Zeidler et al., 2004). Interestingly, lipid receptors operate in plant–insect interactions. The linolenic acid derivative volicitin from beet armyworm caterpillar elicits defences in maize, displaying high‐affinity interaction (K d = 1.3 nm) with a PM protein. Such interaction is reversible and saturable, and involves close to 3000 binding sites per cell (Truitt et al., 2004). Therefore, it is feasible that plants use receptors to detect pathogen‐derived lipids, as they are essential components of fungal (ergosterol) or bacterial (LPS) PMs that can function as PAMPs/DAMPs. Moreover, diC14 may mimic the effect of such PAMPs/DAMPs.
diC14‐mediated PR1 and FRK1 activation was lower in the fls2/efr/cerk1 mutant, but not in the fls2, efr or cerk1 single mutants, indicating that these proteins do not function by themselves as the lipid receptor. This is consistent with the notion that PRRs with LRRs at the ectodomain (FLS2, EFR) bind proteins or peptides, whereas those with lysine motifs (CERK1) bind chitin or PGN (Macho and Zipfel, 2014). Interestingly, the fls2/efr/cerk1 mutant was able to induce PR1 and FRK1 during Pst AvrRpm1‐mediated ETI activation, suggesting that the low response of these genes to diC14 is not a result of a general effect of the plant on the activation of these genes. As mentioned previously, diC14 may stimulate defences at different levels. We do not know why diC14‐induced defence gene expression is weaker in the fls2/efr/cerk1 mutant. There is much evidence suggesting that PRR association is necessary for elicitor‐triggered defences. In rice, hetero‐oligomeric complexes formed by dimers of the binding receptor OsCEBiP (chitin‐elicitor binding protein) and the non‐ligand‐binding receptor OsCERK1 signal chitin perception (Macho and Zipfel, 2014). In Arabidopsis, perception of flg22 requires heterodimerization of FLS2 with the co‐receptor BAK1 (Sun et al., 2013), whereas recognition of PGN requires the ligand‐binding receptors LYM1/LYM3, and CERK1 which does not bind itself to the elicitor (Willmann et al., 2011). However, to date, there is no direct link between the PRRs FLS2, CERK1 and EFR related to the activation of defence responses.
At the systemic level, diC14 induces defences against pathogens, as do LPS, flagellin, surfactin, fengycin and massetolide A (Mishina and Zeier, 2007; Ongena et al., 2007; Tran et al., 2007). This may result from the generation of plant signals acting at the systemic level in response to diC14, as well as from lipid movement to systemic tissues. Even if diC14 could be transported to other leaves, the second possibility seems unlikely as its capacity to activate defence genes is reduced at low dose (5 vs. 40 μg/mL) and may be negligible once the inoculum (40 μg/mL) is diluted into the plant. Concerning the involvement of plant systemic signals, diC14 may use any reported mobile SAR signal, such as methyl salicylate (MeSA), glycerol‐3‐phosphate (G3P), dehydroabietinal (DA), azelaic acid (AzA) and pipecolic acid (Pip) (Dempsey and Klessig, 2012; Shah and Zeier, 2013). However, diC14 does not require DIR1, at least exclusively, for systemic PR1 activation. This lipid transport protein participates in MeSA, G3P, DA and AzA mobilization (Shah and Zeier, 2013), suggesting a priori that these signals would not be involved. Assuming that DIR is capable of transporting Pip, it would be interesting to determine whether diC14 uses Pip to induce SAR, as suspected for LPS and flagellin, which cause Pip accumulation (Navarova et al., 2012).
Experimental Procedures
Plant material
Arabidopsis thaliana (Col‐0 and Ws) wild‐type, and dir1‐1, fls2 (Zipfel et al., 2004), efr (efr1; SALK_044334) (Zipfel et al., 2006), cerk1 (cerk1‐2; GABI_096F09) and fls2/efr/cerk1 (Gimenez‐Ibanez et al., 2009), ndr1‐1 (Shapiro and Zhang, 2001) and eds1‐2 (Bartsch et al., 2006) mutants were grown in soil under cycles of 8 h light and 16 h dark at 23 °C for 8 weeks in incubators with strict hygiene.
Pathogen growth and inoculation
Pseudomonas syringae pv. tomato DC3000 strains (virulent, hrpC – mutant; avirulent, AvrRpm1) were grown on King's B medium supplemented with kanamycin (50 μg/mL) and rifampicin (100 μg/mL). Bacterial pathogens were inoculated into leaf tissues (Pavet et al., 2005) at 105 cfu/mL for bacterial growth curves, and 107 cfu/mL for all other studies. Bacterial growth curves were obtained as reported previously (Pavet et al., 2005). Botrytis cinerea B05.10 was obtained from Dr F. Pieckenstain (INTECH‐CONICET, Buenos Aires, Argentina) and used for plant inoculation as described previously (Rossi et al., 2011). Fungal proliferation was analysed by trypan blue staining (Pavet et al., 2005).
Liposome preparation
diC14 and diC16 nanoliposomes were synthesized as described previously (Ruysschaert et al., 1994). The same protocol was used to prepare liposomes of diC14, DMTAP, DMDAP and DMPC (Avanti Polar Lipids, Alabaster, AL, USA). Lipid films (formed after resuspension of lipids into chloroform and evaporation of solvent under nitrogen flux) and HEPES (10 mm, pH 7.2) were independently incubated at 55–60 °C for 10 min. The buffer was then added to the lipids without mixing at a final concentration of 1 mg/mL and incubated for an additional 20 min at 55–60 °C. A similar protocol was used to prepare diC16 liposomes, but both incubation steps were made at 60–65 °C. After incubation, all resuspended lipids were vortexed for 1 min to obtain liposomes. Next, all lipids were diluted in water at 20 μg/mL and infiltrated in leaves by needleless syringe. In the experiments in Fig. 1, diC14 was also used at 5, 40 and 100 μg/mL. Mock treatment included infiltration of HEPES (0.2 mm) used under identical conditions.
Cell death and callose analysis
Trypan blue and aniline blue staining were used to quantify plant cell death and callose deposition, respectively (Cecchini et al., 2009).
Gene expression
Reverse transcription was performed using 2 μg of total RNA treated with RQ1 DNAsa (Promega, USA), random hexamer primers and MMLV reverse transcriptase (Promega) to synthesize cDNA. sqPCR was performed with Taq polymerase (Promega, USA) as follows: 3 min at 95 °C, n cycles of 35 s at 95 °C, 35 s at 60 °C and 45 s at 72 °C. GapC (GADPH C subunit; At3g04120) was used as reference gene. The primers, number of cycles and annealing conditions are listed in Table S2 (see Supporting Information), where cycle numbers correspond to the exponential amplification phase for each gene. qPCR was performed with Promega Master Mix as follows: 10 min at 95 °C; 40 cycles of 35 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. Reaction efficiency was in the range 97%–105% for all analysed genes, including UBQ5 (Ubiquitin 5; At3g62250) used as internal control. The relative expression of FRK1 and PR1 in each sample was calculated by the 2–ΔCt method using UBQ5 as reference gene (ΔCt = Ct target − Ct reference). The 2–ΔΔCt method was used to evaluate FRK1 and PR1 expression relative to UBQ5 in diC14‐treated samples by normalizing to mock samples [ΔΔCt = (Ct target − Ct reference)elicited − (Ct target − Ct reference)mock].
Supporting information
Fig. S1 diC14 does not reduce Pseudomonas syringae pv. tomato (Pst) proliferation in vitro. Pst was inoculated at 5 × 106 colony‐forming units (cfu)/mL in liquid KB medium supplemented with diC14 (20 μg/mL) or HEPES (0.2 mm) solution (mock). Bacterial content was determined by plating aliquots of culture at 0, 2, 4, 8, 10, 12 and 24 h post‐infiltration (hpi).
Fig. S2 diC14 potentiates callose deposition induced by Pseudomonas syringae pv. tomato (Pst) hrpC –. Leaves were infiltrated with diC14 (20 μg/mL), Pst hrpC – (107 cfu/mL) or diC14 + Pst hrpC –. (a) Callose deposits were detected by aniline blue staining at 12 and 24 h post‐infiltration (hpi). Scale bar, 0.5 mm. (b) Amount of deposits per field (4.4 mm2) determined with ImageJ software as indicated in Fig. 3. Values indicate mean ± standard error (12 photographs from six leaves). *Significant differences between Pst and diC14 + Pst treatments (P < 0.05 by t‐test).
Fig. S3 FRK1 and PR1 expression in wild‐type and fls2/efr/cerk1 plants treated with diC14 (a) or Pseudomonas syringae pv. tomato (Pst) AvrRpm1 (b). (a) Top panels: reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) comparing diC14‐mediated gene activation in each plant, relative to mock treatment (ΔΔCt method), at 7 and 24 h post‐infiltration (hpi). Different letters indicate significant differences among samples [P < 0.05; two‐way analysis of variance (ANOVA) followed by Tukey test]. Insets show the indicated samples in different scales. Bottom panels: RT‐qPCR comparing the FRK1 and PR1 mRNA levels in both plants, relative to UBQ5 mRNA content (ΔCt method), at 24 hpi. *Significant differences relative to Col‐0 samples (P < 0.05 by t‐test). (b) RT‐sqPCR (left) and RT‐qPCR (right) assays used to compare the response of plants to mock or Pst AvrRpm1 (10−7 cfu/mL) (Avr) treatments at the indicated time points. One representative experiment from two independent assays is shown for each study.
Fig. S4 Leaves pre‐treated with HEPES (0.2 mm) (Mock), diC14, DMTAP, DMDAP or DMPC (20 μg/mL each) were inoculated 24 h later with Pseudomonas syringae pv. tomato (Pst) [105 colony‐forming units (cfu)/mL] to quantify bacterial content at 3 days post‐infection. Values represent mean ± standard deviation of two technical replicates. Similar results were obtained in two independent infection experiments. *Significant differences between mock and diC14 treatments (P < 0.05 by t‐test).
Fig. S5 PR1 induction by diC14 and Pseudomonas syringae pv. tomato (Pst) AvrRpm1. PR1 expression was analysed as described in Fig. 1 in leaves infiltrated with diC14 (20 μg/mL) or Pst AvrRpm1 [2 × 107 colony‐forming units (cfu)/mL]. Two independent experiments produced similar results.
Fig. S6 diC14 does not protect against Botrytis cinerea infection. Leaves were infiltrated with diC14 (20 μg/mL) or mock solution (HEPES, 0.2 mm) and inoculated 24 h later with Botrytis cinerea (104 and 103 conidia/mL; three sites for each concentration). Trypan blue staining was used to monitor fungal proliferation at 3 days post‐inoculation (dpi).
Table S1 Chemical structure of the lipids used in this work.
Table S2 Primers and conditions used in reverse transcription‐polymerase chain reaction (RT‐PCR) experiments.
Acknowledgements
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2008‐1542; PICT 2012‐2117) and Secretaría de Ciencia y Tecnología, Universidad Nacional de Córdoba to MEA. DAC is a Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) fellow. MEA is a Senior Career Investigator of CONICET. CL is an Intra‐European Fellowships (IEF) Marie Curie Actions Research Fellow and would like to thank the Wiener‐Anspach Foundation for financial support.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert, M. , Jehle, A.K. , Lipschis, M. , Mueller, K. , Zeng, Y. and Felix, G. (2010) Regulation of cell behaviour by plant receptor kinases: pattern recognition receptors as prototypical models. Eur. J. Cell Biol. 89, 200–207. [DOI] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Bais, H.P. , Fall, R. and Vivanco, J.M. (2004) Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 134, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar, M. and Avni, A. (2009) EHD2 inhibits ligand‐induced endocytosis and signaling of the leucine‐rich repeat receptor‐like protein LeEix2. Plant J. 59, 600–611. [DOI] [PubMed] [Google Scholar]
- Bartsch, M. , Gobbato, E. , Bednarek, P. , Debey, S. , Schultze, J.L. , Bautor, J. and Parker, J.E. (2006) Salicylic acid‐independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell, 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block, A. and Alfano, J.R. (2011) Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Brotman, Y. , Makovitzki, A. , Shai, Y. , Chet, I. and Viterbo, A. (2009) Synthetic ultrashort cationic lipopeptides induce systemic plant defense responses against bacterial and fungal pathogens. Appl. Environ. Microbiol. 75, 5373–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, N.M. , Monteoliva, M.I. , Blanco, F. , Holuigue, L. and Alvarez, M.E. (2009) Features of basal and race‐specific defences in photosynthetic Arabidopsis thaliana suspension cultured cells. Mol. Plant Pathol. 10, 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, N.M. , Monteoliva, M.I. and Alvarez, M.E. (2011) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol. 155, 1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , Shaw, M.H. , Kim, Y.G. and Nuñez, G. (2009) NOD‐like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol. 4, 365–398. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D. , Bauer, Z. , Regenass, M. , Boller, T. and Felix, G. (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell, 18, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy, S. , Thilmony, R. , Kwack, Y.B. , Nomura, K. and He, S.Y. (2004) A family of conserved bacterial effectors inhibits salicylic acid‐mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA, 101, 9927–9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D.A. and Klessig, D.F. (2012) SOS—too many signals for systemic acquired resistance? Trends Plant Sci. 17, 538–545. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A. , Vlot, A.C. , Wildermuth, M.C. and Klessig, D.F. (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book, 9, e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. and Beer, S.V. (2000) Riboflavin induces disease resistance in plants by activating a novel signal transduction pathway. Phytopathology, 90, 801–811. [DOI] [PubMed] [Google Scholar]
- Dong, X. , Hong, Z. , Chatterjee, J. , Kim, S. and Verma, D.P. (2008) Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta, 229, 87–98. [DOI] [PubMed] [Google Scholar]
- Fujiwara, M. , Hamada, S. , Hiratsuka, M. , Fukao, Y. , Kawasaki, T. and Shimamoto, K. (2009) Proteome analysis of detergent‐resistant membranes (DRMs) associated with OsRac1‐mediated innate immunity in rice. Plant Cell Physiol. 50, 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau‐Pissot, P. , Der, C. , Thomas, D. , Anca, I.A. , Grosjean, K. , Roche, Y. , Perrier‐Cornet, J.M. , Mongrand, S. and Simon‐Plas, F. (2014) Modification of plasma membrane organization in tobacco cells elicited by cryptogein. Plant Physiol. 164, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Ntoukakis, V. and Rathjen, J.P. (2009) The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 4, 539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granado, J. , Felix, G. and Boller, T. (1995) Perception of fungal sterols in plants (subnanomolar concentrations of ergosterol elicit extracellular alkalinization in tomato cells). Plant Physiol. 107, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, A. , Kapp, D. , Nielsen, T. and Niehaus, K. (2005) Endocytosis of Xanthomonas campestris pathovar campestris lipopolysaccharides in non‐host plant cells of Nicotiana tabacum . New Phytol. 165, 215–226. [DOI] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA, 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, G. , Deleu, M. , Jourdan, E. , Thonart, P. and Ongena, M. (2011) The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune‐related defence responses. Cell. Microbiol. 13, 1824–1837. [DOI] [PubMed] [Google Scholar]
- Jacquet, A. , Vanderschrick, J.F. , Vandenbranden, M. , Elouahabi, A. , Magi, M. , Garcia, L. and Ruysschaert, J.M. (2005) Vaccination with the recombinant allergen ProDer p 1 complexed with the cationic lipid diC14‐amidine prevents allergic responses to house dust mite. Mol. Ther. 11, 960–968. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , kimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. and Shibuya, N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA, 103, 11 086–11 091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath, N.F. , Kierszniowska, S. , Lorek, J. , Bourdais, G. , Kessler, S.A. , Shimosato‐Asano, H. , Grossniklaus, U. , Schulze, W.X. , Robatzek, S. and Panstruga, R. (2010) PAMP (pathogen‐associated molecular pattern)‐induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39 140–39 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landesman‐Milo, D. and Peer, D. (2012) Altering the immune response with lipid‐based nanoparticles. J. Control. Release, 161, 600–608. [DOI] [PubMed] [Google Scholar]
- Macho, A.P. and Zipfel, C. (2014) Plant PRRs and the activation of innate immune signaling. Mol. Cell, 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Madala, N.E. , Molinaro, A. and Dubery, I.A. (2012) Distinct carbohydrate and lipid‐based molecular patterns within lipopolysaccharides from Burkholderia cepacia contribute to defense‐associated differential gene expression in Arabidopsis thaliana . Innate. Immun. 18, 140–154. [DOI] [PubMed] [Google Scholar]
- Makovitzki, A. , Avrahami, D. and Shai, Y. (2006) Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA, 103, 15 997–16 002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovitzki, A. , Viterbo, A. , Brotman, Y. , Chet, I. and Shai, Y. (2007) Inhibition of fungal and bacterial plant pathogens in vitro and in planta with ultrashort cationic lipopeptides. Appl. Environ. Microbiol. 73, 6629–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado, A.M. , Doerner, P. , Dixon, R.A. , Lamb, C.J. and Cameron, R.K. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature, 419, 399–403. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. and Simon, S.A. (2008) Molecular diversity at the plant–pathogen interface. Dev. Comp. Immunol. 32, 736–744. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. , Kawakami, N. , Kaku, H. and Shibuya, N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA, 104, 19 613–19 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur, L.A. , Kenton, P. , Lloyd, A.J. , Ougham, H. and Prats, E. (2008) The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. [DOI] [PubMed] [Google Scholar]
- Navarova, H. , Bernsdorff, F. , Doring, A.C. and Zeier, J. (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell, 24, 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M.A. , von Roepenack‐Lahaye, E. , Parr, A. , Daniels, M.J. and Dow, J.M. (2002) Prior exposure to lipopolysaccharide potentiates expression of plant defenses in response to bacteria. Plant J. 29, 487–495. [DOI] [PubMed] [Google Scholar]
- Newman, M.A. , Dow, J.M. , Molinaro, A. and Parrilli, M. (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13, 69–84. [DOI] [PubMed] [Google Scholar]
- Ongena, M. , Jourdan, E. , Adam, A. , Paquot, M. , Brans, A. , Joris, B. , Arpigny, J.L. and Thonart, P. (2007) Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Pavet, V. , Olmos, E. , Kiddle, G. , Mowla, S. , Kumar, S. , Antoniw, J. , Alvarez, M.E. and Foyer, C.H. (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 139, 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz, S. , Stamm, A. , Malonek, S. , Wagner, S. , Becker, D. , Medina‐Escobar, N. , Vlot, A.C. , Feys, B.J. , Niefind, K. and Parker, J.E. (2011) Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191, 107–119. [DOI] [PubMed] [Google Scholar]
- Rossi, F.R. , Garriz, A. , Marina, M. , Romero, F.M. , Gonzalez, M.E. , Collado, I.G. and Pieckenstain, F.L. (2011) The sesquiterpene botrydial produced by Botrytis cinerea induces the hypersensitive response on plant tissues and its action is modulated by salicylic acid and jasmonic acid signaling. Mol. Plant–Microbe Interact. 24, 888–896. [DOI] [PubMed] [Google Scholar]
- Ruysschaert, J.M. , el, O.A. , Willeaume, V. , Huez, G. , Fuks, R. , Vandenbranden, M. and Di, S.P. (1994) A novel cationic amphiphile for transfection of mammalian cells. Biochem. Biophys. Res. Commun. 203, 1622–1628. [DOI] [PubMed] [Google Scholar]
- Sanchez, L. , Courteaux, B. , Hubert, J. , Kauffmann, S. , Renault, J.H. , Clement, C. , Baillieul, F. and Dorey, S. (2012) Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiol. 160, 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko, T. , Walley, J.W. , Chehab, E.W. , Xiao, Y. , Kaspi, R. , Pye, M.F. , Mohamed, M.E. , Lazarus, C.M. , Bostock, R.M. and Dehesh, K. (2010) Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell, 22, 3193–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J. and Zeier, J. (2013) Long‐distance communication and signal amplification in systemic acquired resistance. Front. Plant. Sci. 4, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, A.D. and Zhang, C. (2001) The role of NDR1 in avirulence gene‐directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 127, 1089–1101. [PMC free article] [PubMed] [Google Scholar]
- Stanislas, T. , Bouyssie, D. , Rossignol, M. , Vesa, S. , Fromentin, J. , Morel, J. , Pichereaux, C. , Monsarrat, B. and Simon‐Plas, F. (2009) Quantitative proteomics reveals a dynamic association of proteins to detergent‐resistant membranes upon elicitor signaling in tobacco. Mol. Cell. Proteomics, 8, 2186–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, A. , Nie, S. and Xing, D. (2012) Nitric oxide‐mediated maintenance of redox homeostasis contributes to NPR1‐dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiol. 160, 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Macho, A.P. , Han, Z. , Hu, Z. , Zipfel, C. , Zhou, J.M. and Chai, J. (2013) Structural basis for flg22‐induced activation of the Arabidopsis FLS2‐BAK1 immune complex. Science, 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Tanaka, T. , Legat, A. , Adam, E. , Steuve, J. , Gatot, J.S. , Vandenbranden, M. , Ulianov, L. , Lonez, C. , Ruysschaert, J.M. , Muraille, E. , Tuynder, M. , Goldman, M. and Jacquet, A. (2008) diC14‐amidine cationic liposomes stimulate myeloid dendritic cells through Toll‐like receptor 4. Eur. J. Immunol. 38, 1351–1357. [DOI] [PubMed] [Google Scholar]
- Tran, H. , Ficke, A. , Asiimwe, T. , Hofte, M. and Raaijmakers, J.M. (2007) Role of the cyclic lipopeptide massetolide A in biological control of Phytophthora infestans and in colonization of tomato plants by Pseudomonas fluorescens . New Phytol. 175, 731–742. [DOI] [PubMed] [Google Scholar]
- Truitt, C.L. , Wei, H.X. and Pare, P.W. (2004) A plasma membrane protein from Zea mays binds with the herbivore elicitor volicitin. Plant Cell, 16, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. , Mine, A. , Bethke, G. , Igarashi, D. , Botanga, C.J. , Tsuda, Y. , Glazebrook, J. , Sato, M. and Katagiri, F. (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana . Plos Genet. 9, e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnier, A.L. , Sanchez, L. , Vatsa, P. , Boudesocque, L. , Garcia‐Brugger, A. , Rabenoelina, F. , Sorokin, A. , Renault, J.H. , Kauffmann, S. , Pugin, A. , Clement, C. , Baillieul, F. and Dorey, S. (2009) Bacterial rhamnolipids are novel MAMPs conferring resistance to Botrytis cinerea in grapevine. Plant Cell Environ. 32, 178–193. [DOI] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Willmann, R. , Lajunen, H.M. , Erbs, G. , Newman, M.A. , Kolb, D. , Tsuda, K. , Katagiri, F. , Fliegmann, J. , Bono, J.J. , Cullimore, J.V. , Jehle, A.K. , Gotz, F. , Kulik, A. , Molinaro, A. , Lipka, V. , Gust, A.A. and Nurnberger, T. (2011) Arabidopsis lysin‐motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA, 108, 19 824–19 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmar, A. , Lonez, C. , Vermeersch, M. , Andrianne, M. , Perez‐Morga, D. , Ruysschaert, J.M. , Vandenbranden, M. , Leo, O. and Temmerman, S.T. (2012) The cationic lipid, diC14 amidine, extends the adjuvant properties of aluminum salts through a TLR‐4‐ and caspase‐1‐independent mechanism. Vaccine, 30, 414–424. [DOI] [PubMed] [Google Scholar]
- Zeidler, D. , Zahringer, U. , Gerber, I. , Dubery, I. , Hartung, T. , Bors, W. , Hutzler, P. and Durner, J. (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA, 101, 15 811–15 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. and Thomma, B.P. (2013) Structure–function aspects of extracellular leucine‐rich repeat‐containing cell surface receptors in plants. J. Integr. Plant Biol. 55, 1212–1223. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 diC14 does not reduce Pseudomonas syringae pv. tomato (Pst) proliferation in vitro. Pst was inoculated at 5 × 106 colony‐forming units (cfu)/mL in liquid KB medium supplemented with diC14 (20 μg/mL) or HEPES (0.2 mm) solution (mock). Bacterial content was determined by plating aliquots of culture at 0, 2, 4, 8, 10, 12 and 24 h post‐infiltration (hpi).
Fig. S2 diC14 potentiates callose deposition induced by Pseudomonas syringae pv. tomato (Pst) hrpC –. Leaves were infiltrated with diC14 (20 μg/mL), Pst hrpC – (107 cfu/mL) or diC14 + Pst hrpC –. (a) Callose deposits were detected by aniline blue staining at 12 and 24 h post‐infiltration (hpi). Scale bar, 0.5 mm. (b) Amount of deposits per field (4.4 mm2) determined with ImageJ software as indicated in Fig. 3. Values indicate mean ± standard error (12 photographs from six leaves). *Significant differences between Pst and diC14 + Pst treatments (P < 0.05 by t‐test).
Fig. S3 FRK1 and PR1 expression in wild‐type and fls2/efr/cerk1 plants treated with diC14 (a) or Pseudomonas syringae pv. tomato (Pst) AvrRpm1 (b). (a) Top panels: reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) comparing diC14‐mediated gene activation in each plant, relative to mock treatment (ΔΔCt method), at 7 and 24 h post‐infiltration (hpi). Different letters indicate significant differences among samples [P < 0.05; two‐way analysis of variance (ANOVA) followed by Tukey test]. Insets show the indicated samples in different scales. Bottom panels: RT‐qPCR comparing the FRK1 and PR1 mRNA levels in both plants, relative to UBQ5 mRNA content (ΔCt method), at 24 hpi. *Significant differences relative to Col‐0 samples (P < 0.05 by t‐test). (b) RT‐sqPCR (left) and RT‐qPCR (right) assays used to compare the response of plants to mock or Pst AvrRpm1 (10−7 cfu/mL) (Avr) treatments at the indicated time points. One representative experiment from two independent assays is shown for each study.
Fig. S4 Leaves pre‐treated with HEPES (0.2 mm) (Mock), diC14, DMTAP, DMDAP or DMPC (20 μg/mL each) were inoculated 24 h later with Pseudomonas syringae pv. tomato (Pst) [105 colony‐forming units (cfu)/mL] to quantify bacterial content at 3 days post‐infection. Values represent mean ± standard deviation of two technical replicates. Similar results were obtained in two independent infection experiments. *Significant differences between mock and diC14 treatments (P < 0.05 by t‐test).
Fig. S5 PR1 induction by diC14 and Pseudomonas syringae pv. tomato (Pst) AvrRpm1. PR1 expression was analysed as described in Fig. 1 in leaves infiltrated with diC14 (20 μg/mL) or Pst AvrRpm1 [2 × 107 colony‐forming units (cfu)/mL]. Two independent experiments produced similar results.
Fig. S6 diC14 does not protect against Botrytis cinerea infection. Leaves were infiltrated with diC14 (20 μg/mL) or mock solution (HEPES, 0.2 mm) and inoculated 24 h later with Botrytis cinerea (104 and 103 conidia/mL; three sites for each concentration). Trypan blue staining was used to monitor fungal proliferation at 3 days post‐inoculation (dpi).
Table S1 Chemical structure of the lipids used in this work.
Table S2 Primers and conditions used in reverse transcription‐polymerase chain reaction (RT‐PCR) experiments.


