Summary
Five avirulence genes from Leptosphaeria maculans, the causal agent of blackleg of canola (Brassica napus), have been identified previously through map‐based cloning. In this study, a comparative genomic approach was used to clone the previously mapped AvrLm2. Given the lack of a presence–absence gene polymorphism coincident with the AvrLm2 phenotype, 36 L. maculans isolates were resequenced and analysed for single‐nucleotide polymorphisms (SNPs) in predicted small secreted protein‐encoding genes present within the map interval. Three SNPs coincident with the AvrLm2 phenotype were identified within LmCys1, previously identified as a putative effector‐coding gene. Complementation of a virulent isolate with LmCys1, as the candidate AvrLm2 allele, restored the avirulent phenotype on Rlm2‐containing B. napus lines. AvrLm2 encodes a small cysteine‐rich protein with low similarity to other proteins in the public databases. Unlike other avirulence genes, AvrLm2 resides in a small GC island within an AT‐rich isochore of the genome, and was never found to be deleted completely in virulent isolates.
Keywords: avirulence gene, Brassica napus, comparative genomics, Leptosphaeria maculans
Introduction
Blackleg is a major disease of the oilseed crop Brassica napus (canola/oilseed rape) and other Brassica crops worldwide. The disease is caused by the ascomycete pathogen Leptosphaeria maculans (Fitt et al., 2006). Infection starts on the cotyledons and leaves of plants and progresses systemically into the stem. Infection of the stem eventually leads to the formation of lesions at the base of the stem (stem canker) of adult plants, resulting in lodging of the crop and yield loss. Two types of genetic resistance to L. maculans have been described in Brassica species: qualitative resistance (race‐specific) and quantitative resistance (effective at the adult plant stage) (Ansan‐Melayah et al., 1998; Dion et al., 1995; Ferreira et al., 1995; Pilet et al., 1998; Rimmer, 2006). Qualitative resistance to L. maculans mostly follows the gene‐for‐gene model for plant–pathogen interactions described by Flor (1971) (Ansan‐Melayah et al., 1998), although some redundancy is also present within the system (Larkan et al., 2013; Parlange et al., 2009). Major resistance (R) genes against blackleg reported to date include Rlm1, Rlm2 and Rlm4 (Ansan‐Melayah et al., 1998), Rlm3, Rlm5, Rlm6 and Rlm8 (Balesdent et al., 2002), Rlm7 and Rlm9 (Delourme et al., 2004), LepR1, LepR2 and LepR3 (Larkan et al., 2013; Yu et al., 2005), and BLMR1 and BLMR2 (Long et al., 2011), although some redundancy amongst the reported R genes is likely (Larkan et al., 2013; Raman et al., 2013).
Leptosphaeria maculans avirulence genes, capable of triggering their cognate R genes to induce resistance, have been named ‘AvrLm’ or ‘AvrLepR’ to reflect their interaction with the corresponding R genes in Brassica. Genetic studies in L. maculans have identified the genomic location of AvrLm1, 2, 3, 4, 5, 6, 7, 9, 11 and AvrLepR1. Some of these genes are located within two genetic clusters, the AvrLm1‐2‐6 cluster (Balesdent et al., 2002) and the AvrLm3‐4‐7‐9‐AvrLepR1 cluster (Balesdent et al., 2005; Ghanbarnia et al., 2012), reflecting contrasting genomic situations. AvrLm1 and AvrLm6 are located in a region within the genome in which recombination is suppressed, and are separated by hundreds of kilobases (Fudal et al., 2007), whereas the AvrLm4 and AvrLm7 specificities are a result of two different alleles of the same gene, renamed AvrLm4‐7 (Parlange et al., 2009). Five avirulence genes, AvrLm1 (Gout et al., 2006), AvrLm4‐7 (Parlange et al., 2009), AvrLm6 (Fudal et al., 2007), AvrLm11 (Balesdent et al., 2013) and AvrLmJ1 (Van de Wouw et al., 2014), have been cloned. All encode small secreted proteins (SSPs) and, with the exception of AvrLm1, are cysteine rich. AvrLm11 is located on the smallest chromosome in L. maculans, which is a conditionally dispensable chromosome (CDC) extremely enriched in transposable elements (TEs) (Balesdent et al., 2013).
All five cloned avirulence genes identified to date from L. maculans reside in heterochromatin‐like regions with low GC, comprising mosaics of TEs intermingled, truncated and degenerated by repeat‐induced point (RIP) mutations (Balesdent et al., 2013; Fudal et al., 2007; Gout et al., 2006; Parlange et al., 2009; Van de Wouw et al., 2014). The TEs are a site of epigenetic control in which chromatin regulation allows the concerted expression of avirulence effectors at the onset of plant infection (Soyer et al., 2014). The transition of L. maculans races from avirulence to virulence is a result of multiple molecular events, with the most common being a complete deletion of the gene (Balesdent et al., 2013; Daverdin et al., 2012; Fudal et al., 2009; Gout et al., 2007). Other molecular events, such as truncation, RIP mutation or non‐RIP mutation, have been reported, resulting in either premature truncations or altered forms of the Avr protein, neither of which are capable of triggering a defence response in the host plant (Daverdin et al., 2012; Fudal et al., 2009; Parlange et al., 2009; Van de Wouw et al., 2009). One particular case is that of AvrLm4‐7 in which loss of recognition by the cognate Rlm4 is a result of a single non‐synonymous base mutation that maintains the integrity of the protein and the recognition by Rlm7 (Parlange et al., 2009).
Multiple approaches, such as linkage mapping (Fudal et al., 2007; Gout et al., 2006; Linning et al., 2004; Orbach et al., 2000; Parlange et al., 2009), reverse genetics (Rep et al., 2004; Rivas and Thomas, 2005), cDNA screening (Catanzariti et al., 2006; van Kan et al., 1991) or the combination of map‐based cloning and cDNA screening (Böhnert et al., 2004), have been applied to characterize Avr genes from plant‐pathogenic fungi. Map‐based cloning was the major approach utilized to clone five L. maculans avirulence genes (Balesdent et al., 2013; Fudal et al., 2007; Gout et al., 2006; Parlange et al., 2009; Van de Wouw et al., 2014). However, this approach is time consuming and has limitations, such as incompatibility of desired parental isolates for crossing. With the advent of next‐generation sequencing and the availability of whole genome sequences of pathogenic fungi (Dean et al., 2005; Rouxel et al., 2011), a combination of genetic mapping, high‐throughput phenotyping and intraspecies comparative genomics can facilitate the identification of avirulence genes. Here, we present the use of parallel genome resequencing as an alternative or complementary approach to map‐based cloning of effectors in L. maculans and, in particular, the cloning and characterization of the L. maculans AvrLm2 gene.
Results
Pathotyping of L. maculans isolates
Initially, 36 isolates were phenotyped for the occurrence of AvrLm1, AvrLm6 and AvrLm2 on differential lines harbouring the corresponding resistance genes at Agriculture and Agri‐Food Canada (AAFC) Saskatoon (Table 1, isolates 3R11 to 03‐02). We also added phenotypic data for another 120 isolates from the INRA‐Bioger collection (Table 1, isolates IBCN 18 to NzT4; Table S1, see Supporting Information), with 81 of these isolates being evaluated during the course of this study. The overall data were in accordance with what is currently known about the occurrence of AvrLm2 and other avirulence genes that are linked to AvrLm2. Most of the isolates were avirulent towards Rlm6 (or showed the presence of the unaltered sequence of AvrLm6). Only 18% of the isolates were avirulent towards Rlm2 genotypes, and they mostly originated from Canada.
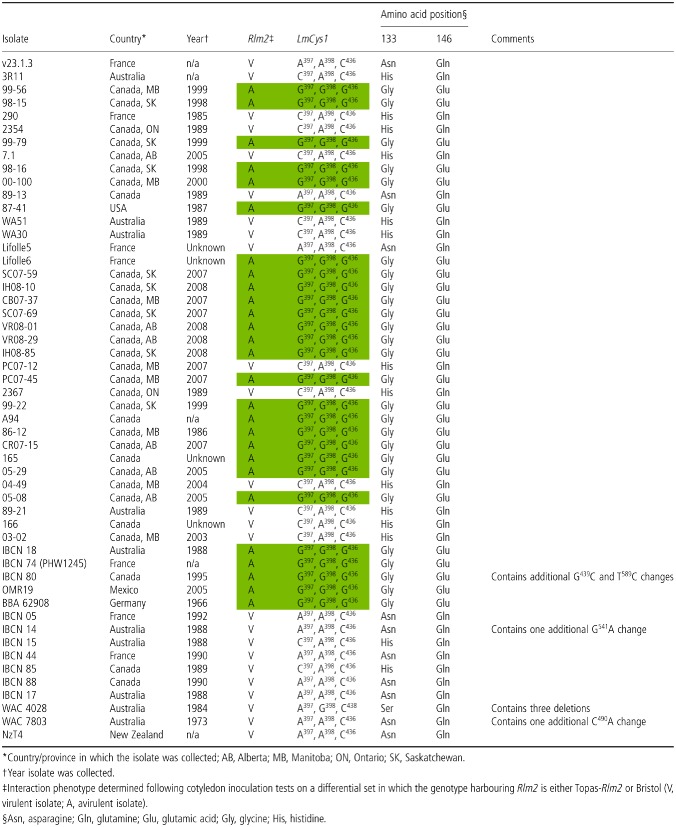
Table 1.
Allelic variation at the LmCys1 locus in a collection of 51 isolates and v23.1.3 (reference isolate), virulent or avirulent towards R lm2 and the polymorphic site in its protein
Identification of AvrLm2
A genetic map of the AvrLm1‐AvrLm6‐AvrLm2 region was built previously using an F1 population consisting of 249 progeny derived from a cross between isolates v23.1.3 and v29.3.1 (Fudal et al., 2007; Gout et al., 2006; Table S1). These studies placed the AvrLm2 locus 0.8 cM distant from AvrLm6 in a genomic region which mostly corresponded to a large AT‐rich isochore in which very few genes were present. In this study, the genomes of 36 L. maculans isolates from the AAFC collection (14 avrLm2 and 22 AvrLm2 isolates) were resequenced, and the resulting sequence reads were mapped to the reference genome v23.1.3. Our analysis focused on the AvrLm2 genomic interval, corresponding to a 270‐kb region containing AvrLm6, together with two predicted effector genes LmCys1 and LmCys2, and three other genes (LmTrans, LmGT and LmMFS) (Fig. 1A). As L. maculans virulence is often linked to the deletion of the SSP‐encoding avirulence gene, we first evaluated the presence–absence polymorphism for two possible candidates, LmCys1 and LmCys2, and examined the correlation with the AvrLm2 phenotype. LmCys1 was found to be present in all isolates, whereas LmCys2 was ruled out as AvrLm2 based on its absence in all 22 avirulent isolates. After determining that the presence–absence polymorphism in the candidate genes could not account for the observed variation in the AvrLm2 phenotype, we examined single nucleotide polymorphism (SNP) events within the target genomic region. We found that several SNPs within the LmCys1 locus perfectly correlated with the phenotypic variation of AvrLm2 within the first 36 sequenced isolates. Three SNPs were identified that differentiated the AvrLm2 alleles from the virulent (avrLm2) isolates, including the reference isolate v23.1.3. These mutations were SNP397 (G in avirulent isolates versus A or C in virulent isolates), SNP398 (G in avirulent isolates versus A in virulent isolates) and SNP436 (G in avirulent isolates versus C in virulent isolates) (Fig. 1C). We also investigated these SNP polymorphisms in 15 additional isolates by sequencing the polymerase chain reaction (PCR)‐amplified allele of LmCys1. Nine haplotypes were found in the total of 51 isolates analysed (Table 1). All avirulent isolates contained an invariant LmCys1 allele specific to avirulent isolates, except for isolate IBCN80, which also contained additional mutations. Further examination of the sequence variation in virulent isolates showed that A398 and C436 were invariant in all virulent isolates, except for isolate WAC4028 (see below) (Table 1). The G397 polymorphism was more variable and resulted in either an A or C at that position (Table 1). Other mutations were found in the LmCys1 coding sequence of virulent isolates, but were restricted to one isolate each. All of these mutations led to predicted changes in amino acids (Table 1). The non‐synonymous point mutations at bases A397/C397 and A398 corresponded to either amino acid changes Gly133 → Asn133 or Gly133 → His133, whereas C436 corresponded to amino acid changes Glu146 → Gln146 (Fig. 1C; Table 1). Two cases diverging from this simple scheme were found among the sequenced isolates: the AvrLm2 isolate IBCN80 had two additional point mutations, one of which introduced a stop codon at amino acid 197, producing a truncated predicted protein missing the last 35 amino acids (Table 1). The avrLm2 isolate WAC 4028 had point mutations leading to a Gly133 → Ser133 change. However, the LmCys1 sequence in this isolate showed the deletion of a dinucleotide at bases 399–400, followed by a single base mutation at base 454. This introduces multiple mutations between amino acids 133 and 151, including the loss of a cysteine residue.
Figure 1.
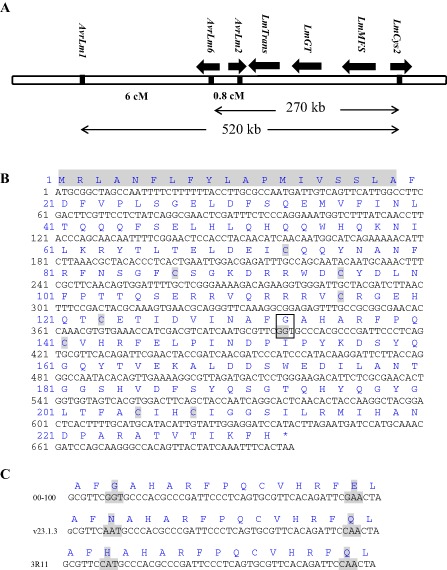
(A) Schematic representation of genetic and physical distance bordered by AvrLm1‐LmCys2 and putative candidate genes for AvrLm2. It should be noted that the gene size and intervals are not to scale. It should also be noted that large AT‐rich regions separating AvrLm1 from AvrLm2 (LmCys1)‐LmTrans‐LmGT, the latter from LmMSF and the latter from LmCys2 are not represented. (B) Nucleotide sequence of the 699 nucleotide region encoding AvrLm2 and its predicted amino acid sequence. The predicted signal peptide (19 amino acids) and eight cysteine residues are illustrated by grey shading. The mutation of codons G 397 G 398 → A/C 397 A 398, which results in a G 133 → N 133/H 133 change in the amino acids in the protein, leading to the loss of Rlm2‐mediated recognition specificity, is indicated in a box. (C) Comparison of different haplotypes at the AvrLm2 locus. Non‐synonymous base substitutions and their corresponding amino acid changes are indicated by grey shading.
Functional complementation assay
A genomic LmCys1 amplicon, including the native promoter region (1109 bp upstream of the ATG start codon) and 42 bp downstream of the predicted open reading frame (ORF) (total length, 1850 bp), from the AvrLm2 isolate ‘00‐100’ was transferred into the fungal transformation vector pNL11. After transforming the virulent isolate v23.1.3 with the LmCys1 construct, the restoration of the avirulence phenotype was evaluated by the inoculation of transgenic isolates on Topas‐Rlm2 and other Rlm2 cultivars (Glacier DH24287, Bristol, Tapidor and Samourai, Table 2). Twelve transformant selections were tested on the B. napus differential lines. Eight of the 12 transformants showed avirulence on cotyledons of Rlm2 plants, but remained virulent on the susceptible Topas DH16516 and Westar control lines (Fig. 2). Positive transformants showed a wild‐type interaction phenotype with the differential lines harbouring other resistance genes (Table 3). This confirmed the identity of LmCys1 as AvrLm2.
Table 2.
Complementation assays with AvrLm2 candidate gene. Phenotypic interaction of wild‐type and AvrLm2‐transgenic isolates on different B rassica lines harbouring Rlm2
| Isolates/transformantsb | B. napus lines/cultivarsa | ||||||
|---|---|---|---|---|---|---|---|
| Westar | Topas | T‐Rlm2 | Glacier | Bristol | Samourai | Tapidor | |
| Control | Control | Rlm2 | Rlm2,Rlm3 | Rlm2,Rlm9 | RLm2,Rlm9 | Rlm2 | |
| v23.1.3 (A1a2a3a9) | V | V | V | V | V | V | V |
| 00‐100 (a1A2A3A9) | V | V | A | A | A | A | A |
| v23.1.3: LmCys1(AW1) | V | V | A | A | A | A | A |
| v23.1.3: LmCys1(AW2) | V | V | A | A | A | A | A |
| v23.1.3: LmCys1(SNP1)c | V | V | V | V | V | V | V |
| v23.1.3: LmCys1(SNP2) | V | V | V | V | V | V | V |
| v23.1.3: LmCys1(SNP3) | V | V | V | V | V | V | V |
| v23.1.3: LmCys1(SNP4) | V | V | A | A | A | A | A |
| v23.1.3: LmCys1(Control) | V | V | A | A | A | A | A |
Pathogenicity test on differential lines/cultivar carrying different resistance genes. Each Leptosphaeria maculans isolate was tested on 12 seedlings of the differential lines and 12 seedlings of ‘Westar’ and Topas as highly susceptible controls. The disease reactions were scored 14 days after inoculation and rated using the 0–9 scale described by Williams (1985). T‐Rlm2 stands for Topas‐Rlm2.
Isolate v23.1.3 (A1a2a3a9) is avirulent on lines harbouring Rlm1, but virulent on lines harbouring Rlm2, Rlm3 and Rlm9, and isolate 00‐100 (a1A2A3A9) is virulent on the line harbouring Rlm1, but avirulent on lines harbouring Rlm2, Rlm3 and Rlm9. v23.1.3: LmCys1(AW1) is a transformant with the pNL11‐LmCys1 construct (LmCys1 avirulent allele was amplified from isolate 00‐100). V23.1.3: LmCys1(AW2) is a transformant with the pLM4‐LmCys1 construct (LmCys1 avirulent allele coding region was amplified from isolate 00‐100). All constructs were introduced into isolate v23.1.3 by Agrobacterium‐mediated transformation.
The four various alleles of AvrLm2 (single nucleotide polymorphisms, SNPs) were synthesized and cloned in a Gateway vector (pLM4) downstream of the promoter of the AvrLm1 effector. These SNPs are: SNP1, G397 → A397 (amino acid change Gly133 → Ser133); SNP2, G397 → C397 (amino acid change Gly133 → Arg133); SNP3, G398 → A398 (amino acid change Gly133 → Asp133); SNP4, G436 → C436 (amino acid change Glu146 → Gln146). The AvrLm2 open reading frame (ORF) from isolate 00‐100 was cloned in the same vector, as a positive control.
Figure 2.
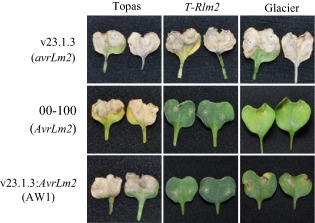
Phenotypic interaction of wild‐type and complemented Leptosphaeria maculans isolates on the cotyledons of control (Topas), Topas‐Rlm2 (T‐Rlm2) and Glacier DH24287 harbouring Rlm2 and Rlm3. Photographs of the infected cotyledons were taken at 14 days post‐inoculation. v23.1.3: AvrLm2 (AW1) contains an avirulent allele from isolate 00‐100.
Table 3.
Pathogenicity test on wild‐type isolates and positive transformants on B rassica napus lines/cultivars carrying diverse resistance genes
| Isolates/transformantsb | B. napus or B. juncea lines/cultivarsa | |||||||
|---|---|---|---|---|---|---|---|---|
| Westar | Topas | T‐Rlm2 | Quantum | JetNeuf | Vulcan‐1S | Roxet | Goeland | |
| Control | Control | Rlm2 | Rlm3 | Rlm4 | Rlm6 | Rlm7 | Rlm9 | |
| v23.1.3 | V | V | V | V | A | A | A | V |
| 00‐100 | V | V | A | A | V | A | A | A |
| v23.1.3: LmCys1(AW1) | V | V | A | V | A | A | A | V |
| v23.1.3: LmCys1(AW2) | V | V | A | V | A | A | A | V |
Pathogenicity test on differential lines/cultivar carrying different resistance genes. Each Leptosphaeria maculans isolate was tested on 12 seedlings of the differential lines and ‘Westar’ and Topas as susceptible controls. The disease reactions were scored 14 days after inoculation and rated using the 0–9 scale described by Williams (1985). T‐Rlm2 stands for Topas‐Rlm2.
v23.1.3: LmCys1(AW1) is a transformant with the pNL11‐LmCys1 construct (LmCys1 avirulent allele was amplified from isolate 00‐100). v23.1.3: LmCys1(AW2) is a transformant with the pLM4‐LmCys1 construct (LmCys1 avirulent allele coding region was amplified from isolate 00‐100, driven by the AvrLm1 promoter). All constructs were introduced into isolate v23.1.3 by Agrobacterium‐mediated transformation.
Gene annotation and sequence analysis
The AvrLm2 (accession: KM073975) ORF is 699 bp with 46% GC content and comprises a single exon. AvrLm2 encodes a predicted 232‐amino‐acid protein of 27.046 kDa and contains a predicted N‐terminal signal peptide of 19 amino acids. Eight cysteine residues were present in AvrLm2 and three to four disulfide bonds were predicted, depending on the software used: SCRATCH (Cheng et al., 2005), amino acid positions 87–96, 123–141 and 73–116; DISULFIND (Ceroni et al., 2006), amino acid positions 73–116, 87–96, 123–208 and 141–205; DIANA (Ferre and Clote, 2005), amino acid positions 73–96, 87–116, 123–205 and 141–208. A search of the National Center for Biotechnology Information (NCBI) non‐redundant protein database with AvrLm2 identified distantly related hypothetical proteins in genomes of several Fusarium oxysporum forma speciales GenBank accessions (EXK23710.1, EXL90101.1, EXA28588.1 and EXA28676.1). E‐values ranged between 3 × 10−4 and 1 × 10−6 with 28%–30% identities. However, in all cases, the number and spacing of cysteine residues were identical between all of the proteins. These F. oxysporum genes encode for potential secreted proteins based on the presence of the N‐terminal signal peptide, lack of a transmembrane domain and subcellular targeting signals. Blastp of the NCBI database was performed on the L. maculans isolate v23.1.3 genome sequence. (E‐value, 8 × 10−12; locus tag, LEMA_P09120.1; accession, XP_003845604). LEMA_P09120.1 encodes for a predicted secreted protein with 29% amino acid identity to AvrLm2, and the same number and position of cysteine residues. AvrLm2 was absent from all other species of the L. maculans–L. biglobosa species complex, except L. biglobosa ‘thlaspii’ (Grandaubert et al., 2014b). Compared with this latter species, however, the homologue of LmCys1 has been translocated to another genome location, as observed previously for other avirulence genes of L. maculans (Grandaubert et al., 2014a, b).
Functional validation of SNPs associated with AvrLm2 specificity
Comparison of AvrLm2 alleles among 51 reference isolates identified SNPs in three locations. These SNPs gave rise to four potential combinations: SNP1, G397 → A397 (amino acid change Gly133 → Ser133); SNP2, G397 → C397 (amino acid change Gly133 → Arg133); SNP3, G398 → A398 (amino acid change Gly133 → Asp133); SNP4, G436 → C436 (amino acid change Glu146 → Gln146). These potential allelic variants of AvrLm2 were synthesized and transferred to the fungal transformation vector pLM4 under the control of the AvrLm1 promoter. The AvrLm2 ORF from isolate 00‐100 was used as a positive control. Transgenic v23.1.3 isolates harbouring individual SNP constructs or the AvrLm2 ORF were tested for the function of the transgene on the B. napus differential lines described above. The results showed that only the non‐synonymous changes of G397 → A/C397 (SNP1 and SNP2) or G398 → A398 (SNP3), both of which lead to a change in amino acid at Gly133, were responsible for the loss of Rlm2‐mediated recognition specificity (Table 2).
Expression of AvrLm2
AvrLm2 expression was investigated using quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) during infection and in vitro growth with the isolate 00‐100. The gene expression level was measured relative to that of actin (Fig. 3). AvrLm2 expression was very low in germinating conidia compared with other stages. It was expressed at a higher level in infected plants at 3 days post‐inoculation (dpi) and peaked at 5 dpi. It then decreased after 5 dpi, but remained at a higher level than during in vitro mycelial growth (Fig. 3). The level of expression was higher in mycelium grown axenically than in germinated pycnidiospores, and was comparable with the expression exhibited at 12 dpi (Fig. 3). The virulent allele, avrLm2, showed a similar expression pattern (Fig. S1, see Supporting Information).
Figure 3.
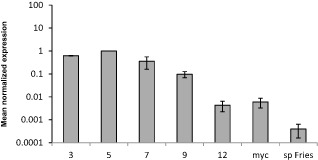
Expression of representative AvrLm2 (avirulent) alleles during in vitro growth of Leptosphaeria maculans and oilseed rape infection. Expression of the AvrLm2 alleles was analysed in the avirulent isolate 00‐100 by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR). 3–12, RT‐PCR products obtained from RNA isolated from oilseed rape cotyledons (Westar) at 3–12 days post‐infection; myc, RT‐PCR product obtained from RNA isolated from mycelial culture; sp Fries, RT‐PCR product obtained from RNA isolated from conidia germinating in rich medium (Fries). RNA extracted from uninfected cotyledons and water were used as negative controls. Gene expression levels are relative to actin. Each data point is the average of three biological repeats (extractions from different biological material). The standard error of the mean normalized expression level is indicated by error bars.
Genome environment and heterochromatin‐based regulation of expression of AvrLm2
Recently, Soyer et al. (2014) related the heterochromatin‐like genome environment of the avirulence gene with an efficient repression of expression during axenic growth, and showed that the repression was relieved during in planta infection or in mutants silenced in the expression of two key players in heterochromatin assembly and maintenance, LmHP1 and LmDIM‐5. Reassessment of the data including other avirulence genes, such as the newly cloned AvrLmJ1, confirmed that the silencing of LmHP1 or LmDIM‐5 results in increased expression of most of the avirulence genes located in AT isochores, but has little or no effect on AvrLm6 (Table 4). Surprisingly, a reverse effect was observed for AvrLm2 with an increased repression of expression in s.LmHP1 or s.LmDIM‐5 backgrounds (Table 4).
Table 4.
Influence of the silencing of LmHP1 and LmDIM5 on the expression of selected avirulence genes in axenic culture
| Avirulence gene | Location† | Fold change (P value) in:‡ | |
|---|---|---|---|
| Silenced mHP1 | Silenced LmDIM5 | ||
| AvrLm2 | GC island | −5.7* | −9.6* |
| AvrLm1 a | AT‐HB | 5.6* | 16.9* |
| AvrLm4‐7 a | AT‐HB | 8.8* | 16.8* |
| AvrLmJ1 | AT‐HB | 4.2* | 4.2 |
| AvrLm6 | AT‐HB | 1.1 | 1.15 |
| AvrLm11 | AT‐HB | 4.1* | 2.9 |
†AT‐HB refers to AT isochores; GC islands refer to regions of more than 1 kb within AT isochores with a GC content of more than 50%.
‡Genes with fold change of less than −1.5 or more than 1.5 in transcript level and an associated P value of less than 0.05 (indicated by *) were considered to be significantly down‐ or up‐regulated in the silenced LmHP1 or silenced LmDIM5 transformants compared with the wild‐type v23.1.3 isolate in axenic culture.
AvrLm1 and AvrLm4‐7 data are from Soyer et al. (2014).
Discussion
In this article, we report the identification of the L. maculans AvrLm2 gene from the AvrLm1‐AvrLm2‐AvrLm6 genetic cluster (Balesdent et al., 2002; Fudal et al., 2007; Gout et al., 2006). AvrLm2, like the majority of effector proteins identified from plant‐pathogenic fungi, is a small cysteine‐rich secreted protein with limited homology to other proteins in the public protein databases.
Taking an intraspecific comparative genomics approach, as an alternative or complementary approach to map‐based cloning, we were able to rapidly identify AvrLm2 by focusing on a gene that did not show the typical presence–absence polymorphism found for AvrLm1, AvrLm6, AvrLm7 and AvrLm11. Thus, we could associate SNPs within the predicted effector LmCys1 with the phenotypic responses of L. maculans isolates on B. napus lines containing the Rlm2 resistance gene. The cloning of AvrLm2 described here provides an example of the rapid cloning of effector genes through comparative genomics as an alternative or complementary approach to map‐based cloning. Map‐based cloning has been used successfully to clone fungal Avr genes; however, it is a time‐consuming approach, limited by the compatibility of parental isolates for crossing and, in the case of fungal and oomycete plant pathogens, has been applied mainly to species that can be grown axenically. More importantly, the majority of the effectors of L. maculans and many other fungal and oomycete plant pathogens sequenced to date are located within a repeat‐rich part of the genome that may impede fine mapping or even correct positioning of the target gene. Although a knowledge of the map position of AvrLm2 facilitated our genome comparison approach to target AvrLm2, at the same time the identification of LmCys1 as AvrLm2 pointed us to the error in the previously published mapping data (Fudal et al., 2007), which excluded LmCys1 as a candidate.
In the case of L. maculans, genome‐assisted map‐based cloning has been facilitated recently by the availability of a complete repertoire of putative effector‐encoding genes (Rouxel et al., 2011), which helped in the cloning process of AvrLm11 and AvrLmJ1 (Balesdent et al., 2013; Van de Wouw et al., 2014).
The identification of effectors by comparative genomics requires a high‐quality reference genome that has been correctly annotated and contains the gene of interest. In the case in which a reference isolate is virulent because of total deletion of the Avr gene, we would be unable to use this method. The production of an alternative reference genome with a complementary Avr profile to the current reference (v23.1.3) and the mapping of the unknown target genes can rectify these shortcomings. Rapid advances in DNA/RNA sequencing technologies, the expansion of genome databases and the improvement of bioinformatics software promise significant improvement and more common use of such tools in the near future.
Another advantage of the intraspecific comparison approach described here is that it provides detailed information on the allelic variation of all candidate effector genes unearthed in the reference genome, eliminating the need for the time‐consuming and repetitive amplification and sequencing of individual candidate genes. Here, we were able to rapidly assign sequence polymorphism to the virulence/avirulence phenotype and showed that only two adjacent nucleotide changes were sufficient for the loss of recognition by Rlm2.
Other rare allelic variants of characterized avirulence genes, such as the C‐terminal truncation predicted in the AvrLm2 isolate IBCN80 described above, could help to define functional effector domains. The allelic information collected could be applied to the design of SNP markers for the rapid genotyping of new L. maculans isolates with practical application for the management of blackleg disease. For example, Carpezat et al. (2014) have recently used a very limited nucleotide polymorphism observed for avirulent versus virulent alleles of the AvrLm7 gene to develop a diagnostic method based on high‐resolution melting (HRM) analysis.
AvrLm2 genotypes are: (i) currently absent in Europe and found only for two isolates obtained before 1975 (Balesdent et al., 2006; Stachowiak et al., 2006; this study); (ii) rare in Australia with only one isolate of 65 showing the AvrLm2 phenotype (Dilmaghani et al., 2009; this study); (iii) common in current and older populations from western and eastern Canada, but not in Ontario (Dilmaghani et al., 2009; Kutcher et al., 2007, 2010), and becoming increasingly less common through southern Manitoba and North Dakota (Chen and Fernando, 2006; Nepal et al., 2014); and (iv) prevalent in Mexican populations (Dilmaghani et al., 2012) (Table S1). Adaptive mechanisms used by fungi to escape R gene recognition have been analysed in detail for those Avr genes which have been submitted to cognate R gene selection in either agronomic practice or experimental fields, namely AvrLm1 (Gout et al., 2007), AvrLm2 (this study), alleles AvrLm4 (Parlange et al., 2009) and AvrLm7 (Daverdin et al., 2012) of AvrLm4‐7 and AvrLm6 (Fudal et al., 2009). At present, such data are not available for the recently cloned AvrLm11 (Balesdent et al., 2013) and AvrLmJ1 (Van de Wouw et al., 2014), whose cognate R genes are not used commercially. In Europe, Rlm2 is likely to have been the oldest resistance gene to be used against L. maculans at the time of the re‐introduction of winter oilseed rape in the 1960s and 1970s. Thus, cv. Ramses that harboured Rlm2 was described as being resistant to French populations in 1968–1970 (Rouxel et al., 2003b). The selection pressure exerted on populations of L. maculans at this time is consistent with the fact that only the two oldest French isolates in collections harbour AvrLm2 and that current European populations are 100% virulent towards Rlm2 (Balesdent et al., 2006). A similar situation was found later for Rlm4 with the release of the market‐leader cultivar Major in 1971, followed by cv. Jet Neuf in 1977, which was grown on more than 80% of the French acreages (Clement, 1981; Rouxel et al., 2003a). Again, this led to a strong selection and to a drastic impoverishment of isolates avirulent towards AvrLm4 in France, and more generally in Europe (Balesdent et al., 2006; Stachowiak et al., 2006). This is paralleled by the observation that both AvrLm2 and the AvrLm4 allele of AvrLm4‐7 escape from recognition because of point mutations (Parlange et al., 2009; this study). Although the intrinsic effector function of AvrLm4‐7 is still unknown, the importance of the effector protein in fungal fitness has been established previously (Huang et al., 2010). Here, we show that escape from recognition by Rlm2 also depends on a few point mutations that all contribute to change the single Gly133 residue, and that AvrLm2 deletion is never observed. The importance of non‐synonymous point mutations in these two genes (AvrLm2 and AvrLm4‐7) contrasts with that observed by examining the evolution of populations submitted to a new selection exerted by Rlm1, Rlm6 or Rlm7. In these cases, the immediate response to the novel pressure was multiple inactivating RIP mutations or complete deletion, with only very few cases of non‐synonymous mutations (Daverdin et al., 2012; Fudal et al., 2007; Gout et al., 2006). This suggests the following sequence of events when an avirulence gene is submitted to a novel selection: (i) immediate gene inactivation as a result of multiple RIP mutations of the gene, permitted by the obligate sexual cycle of the fungus and linked with the genome location of Avr genes; (ii) simultaneous or secondary deletion of the inactivated gene and its surrounding genomic region comprising only the inactivated gene and its inactivated TE surroundings; and (iii) at a later stage, surge and dissemination of a less detrimental allelic version of the gene, maintaining the effector function whilst preventing recognition by the cognate resistance gene (Daverdin et al., 2012). Such a compensatory mechanism would only be set up for genes of importance for fungal pathogenicity, such as AvrLm2 and AvrLm4‐7, in contrast with genes, such as AvrLm1, whose loss has only a limited effect on fungal fitness (Huang et al., 2010).
In L. maculans, all currently known avirulence genes occur isolated as single genes within large (typically hundreds of kilobases) AT‐rich isochores, genomic regions made up of mosaics of RIP‐degenerated and truncated TEs (Fudal et al., 2007; Gout et al., 2006). This has strong consequences on their mode of evolution under selection, mechanisms of allelic diversification and concerted overexpression at the onset of plant infection (Rouxel et al., 2011; Soyer et al., 2014). Compared with this common scheme, AvrLm2 has the unique feature of being accompanied by a second head‐to‐head gene in the AT isochore, making up a small GC isochore. This single‐copy gene, LmTrans, has been described to contain a DDE superfamily endonuclease domain predicted to be involved in efficient DNA transposition, and its best match is a putative transposase from Stagonospora nodorum (Van de Wouw et al., 2010). However, this gene also has strong similarity with the Fot5 transposase of F. oxysporum f. sp. lycopersici (accession, CAE55867.1; E‐value, 8e−67; identity, 60%). Interestingly, in the genome of F. oxysporum f. sp. lycopersici, the transposase is located two genes away from the gene FoSIX1 (accession CAE55870.1) encoding an effector protein which shows limited identity to AvrLm2 (E‐value, 6e−05; identity, 25%), but a good conservation of the cysteine residues. It is unknown whether this may indicate an ancient horizontal gene transfer event.
The fact that AvrLm2 is not a typical ‘lost in the middle of nowhere’ Avr gene seems to have consequences on its regulation of expression. Repression of L. maculans effector expression in axenic culture has been shown to be a result of the AT‐rich isochore environment behaving like heterochromatin (Soyer et al., 2014). In addition, the heterochromatin regulation of expression was relieved during plant infection and favoured a concerted and very high overexpression of the avirulence gene in the first stages of plant infection (Balesdent et al., 2013; Fudal et al., 2007; Gout et al., 2006; Parlange et al., 2009; Soyer et al., 2014; Van de Wouw et al., 2014). Here, we show that AvrLm2 expression is not subject to heterochromatin‐based epigenetic control, being adversely affected in a silenced LmHP1 and LmDIM5 genetic background compared with most other avirulence genes. Finally, as discussed previously, specific adaptive features (lack of RIP mutations, lack of deletions) may also be linked to the occurrence of AvrLm2 within a GC island in the middle of an AT isochore. Additional investigations are now needed to confirm that the characteristics of AvrLm2 (regulation and adaptive features) are linked to this peculiar genome environment, and/or are strongly interlinked and interdependent on the importance of the gene for fungal fitness.
Experimental Procedure
Fungal material and inoculum preparation
Leptospaeria maculans isolates in this study were sourced from the Rimmer Collection, AAFC Saskatoon, representing isolates collected over 30 years from different geographical regions in the world and two progenies of in vitro crosses (Table 1; isolates 3R11 to 03‐02). Some of the Canadian isolates in the collection were kindly provided by R. Kutcher, AAFC Melfort, Canada. Isolate ‘3R11’ was kindly provided by Drs A. Van de Wouw and B. Howlett, University of Melbourne, Australia. All isolates were purified as single‐spore cultures. Pycnidiospores were harvested from the single‐spore cultures after 7–10 days of incubation at 22 °C on V8 juice agar containing 100 μg/mL streptomycin sulfate, as described by Chen and Fernando (2006). All other isolates were from the INRA‐Bioger collection and included a series of reference isolates, such as isolates from the International Blackleg of Crucifers Network (IBCN) collection (described in Balesdent et al., 2005), one isolate obtained from cabbage in Mexico (OMR19; Dilmaghani et al., 2012), a collection of isolates from Western Australia [WTxx isolates; described in Vincenot et al. (2008) and Dilmaghani et al. (2009)] and a series of collections obtained from samplings performed in France between 1994 (SAM xxxxx collection) and 2003 (V03‐Mx‐xxx from Versailles and CB‐X‐x.xx from four locations in France) (Table 1; isolates IBCN 18 to NzT4; Table S1). The fungal culture conditions for growth and sporulation were as reported by Ansan‐Melayah et al. (1995). For long‐term conservation, isolates were stored at 4 °C in 1% malt‐agar slant tubes.
Plant material and virulence phenotyping
AAFC isolates were first phenotyped using the B. napus differential line Topas‐Rlm2, harbouring the Rlm2 resistance gene from the B. napus line Glacier DH24287. Topas‐Rlm2 is one of several introgression lines containing individual blackleg R genes in the completely susceptible (no R gene against L. maculans) doubled haploid B. napus line Topas DH16516 (N. J. Larkan, unpublished data). This material allowed us to accurately determine the phenotypic response of the isolates to Rlm2 (Tables 1 and 2). Transgenic isolates were phenotyped on Topas‐Rlm2 (Rlm2), Glacier DH24287 (Rlm2, Rlm3) and Bristol (Rlm2, Rlm9) (Table 2). Positive transformants (showing restored phenotypic reaction on Topas‐Rlm2, Glacier and Bristol) were also tested on additional differential cultivars to confirm the interaction of AvrLm2 only with lines harbouring Rlm2 (Table 3). Westar and Topas DH16516 (no R genes) were used as positive controls for infection by L. maculans. The inoculation of B. napus cotyledons was performed as described previously (Chen and Fernando, 2006). Each L. maculans isolate was tested on 12 seedlings of the differential lines and 12 seedlings of ‘Westar’ and Topas as highly susceptible controls. The disease reactions were scored 14 days after inoculation and rated using the 0–9 scale described by Williams (1985). Isolates from the INRA‐Bioger collection were phenotyped for the presence of AvrLm1, AvrLm6 and AvrLm2 using a cotyledon inoculation test on the set of differential cultivars as described by Dilmaghani et al. (2009) (Table 1, isolates IBCN 18 to NzT4; Table S1).
DNA and RNA manipulation
For PCR and sequencing, DNA was extracted following the cetyltrimethylammonium bromide (CTAB) method of Rogers and Bendich (1985) as modified by O'Gorman et al. (1994). Primer sequences were designed using Vector NTI software (Table S2, see Supporting Information). Candidate DNA fragments were sent for synthesis (Eurofins, Huntsville, AL, USA) or amplified through PCR. PCR amplifications were performed using AmpliTaq Gold 360 Master Mix (Applied Biosystems, Foster City, CA, USA) or Q5 DNA Polymerase Master Mix (New England Biolabs, Ipswich, MA, USA) on a C1000 Touch Thermal Cycler (BIO‐RAD, Berkeley, CA, USA).
Total RNA was extracted from mycelium grown for 1 week in Fries liquid medium, from germinating conidia grown for 36 h in Fries or from infected leaf tissue, using a PureLink RNA Mini kit (Ambion, Carlsbad, CA, USA) or TRIzol reagent (Invitrogen, CergyPontoise, France) according to the manufacturer's protocol. Total RNA was treated with DNase I RNase‐Free (Invitrogen). The RNA concentration was adjusted to 4 μg, and single‐strand cDNA was generated using oligo‐dT‐primed reverse transcription with PowerScript Reverse Transcriptase (Clontech, Palo Alto, CA, USA) or a ThermoScript RT‐PCR system (Invitrogen) according to the manufacturer's protocol.
Identification, cloning and transformation of the candidate gene
Leptospaeria maculans isolates were sequenced under contract at the National Research Council of Canada (Saskatoon) by running a paired‐end multiplex of 36 isolates across two lanes of Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. The depth of sequencing among the 36 isolates provided between nine‐ and 59‐fold coverage of the L. maculans genome (Table S3, see Supporting Information). Sequence reads for each isolate were mapped to the reference genome v23.1.3 using Bowtie2 (Langmead and Salzberg, 2012) and visualized using GBrowse 2.0 (Stein, 2013). Candidate gene sequences were either synthesized (Eurofins) or amplified via PCR with Gateway attB‐tagged primers. These fragments were cloned using the TOPO TA cloning kit (Invitrogen), confirmed by sequencing and transferred to the Gateway entry vector pDONR‐Zeo (Invitrogen). Inserts, either including the native promoter or containing only the coding region of the gene, were transferred to the Gateway‐compatible fungal expression vectors pNL11 (Larkan et al., 2013) and pLM4 (a Gateway vector with the AvrLm1 promoter; Ma, L., AAFC, Saskatoon, unpublished data), respectively (Tables 2 and 3). Cloning of each insert was confirmed by sequencing. Confirmed pLM4‐AvrLm2 or pNL11‐AvrLm2 constructs were transferred to Agrobacterium tumefaciens strain ‘AGL1 pTiBo542’. Transformation of pycnidiospores from the L. maculans isolate v23.1.3 was performed as described by Utermark and Karlovsky (2008). Transformed colonies were selected after 5–10 days of incubation on Czapek Dox medium enriched with 200 μm cefotaxime and 150 μg/mL hygromycin B, and transferred to V8 medium supplemented with the above antibiotics. For the functional validation of SNP involvement in AvrLm2 specificity, different mutagenized allelic forms of LmCys1 were synthesized (Eurofins) and transferred to pLM4 or pNL11 transformation destination vectors (Table 2).
qRT‐PCR
qRT‐PCR was performed using a 7700 real‐time PCR machine (Applied Biosystems) and SsoFast EvaGreen Supermix (BIO‐RAD). RT‐PCR was performed for the AvrLm2 allele with three biological samples. The primers used for qRT‐PCR are described in Table S2. Ct values were analysed according to the 2−ΔΔCt method (Livak and Schmittgen, 2001). The L. maculans actin was targeted as a reference gene.
Supporting information
Fig. S1 Expression of representative AvrLm2 (virulent) alleles during the in vitro growth of Leptosphaeria maculans and oilseed rape infection.
Table S1 List and characteristics of the isolates used here for the identification of AvrLm2.
Table S2 List of polymerase chain reaction (PCR) primers used in this study.
Table S3 The genome coverage for each of the Leptosphaeria maculans isolates used in this study.
Acknowledgements
We thank Dr Angela Van de Wouw, Dr Barbara Howlett and Dr Randy Kutcher for kindly providing some of the blackleg isolates. Funding for this research at Agriculture and Agri‐Food Canada (AAFC) was provided by the Government of Saskatchewan Agricultural Development Fund (ADF) and Growing Forward 2 (SaskCanola and AAFC). The work at INRA was funded by the Marie Curie Training Site FunGene HPMT‐CT‐2001‐00395 to M. H. Balesdent. Bronislava Profotova was funded by the FunGene project. Thanks are due to Françoise Blaise, Martin Willisecker, Laurent Coudard (INRA‐Bioger) and Elena Beynon (AAFC) for technical assistance. The authors declare that they have no conflict of interest.
References
- Ansan‐Melayah, D. , Balesdent, M.H. , Buèe, M. and Rouxel, T. (1995) Genetic characterization of AvrLm1, the first avirulence gene of Leptosphaeria maculans . Phytopathology, 85, 1525–1529. [Google Scholar]
- Ansan‐Melayah, D. , Balesdent, M.H. , Delorume, R. , Pilet, M.L. , Tanguy, X. , Renard, M. and Rouxel, T. (1998) Genes for race‐specific resistance against blackleg disease in Brassica napus L. Plant Breed. 117, 373–378. [Google Scholar]
- Balesdent, M.H. , Attard, A. , Kuhn, M.L. and Rouxel, T. (2002) New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans . Phytopathology, 92, 1122–1133. [DOI] [PubMed] [Google Scholar]
- Balesdent, M.H. , Barbetti, M.J. , Hua, L. , Sivasithamparam, K. , Gout, L. and Rouxel, T. (2005) Analysis of Leptosphaeria maculans race structure in a worldwide collection of isolates. Phytopathology, 95, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Balesdent, M.H. , Louvard, K. , Pinochet, X. and Rouxel, T. (2006) A large scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur. J. Plant Pathol. 114, 53–65. [Google Scholar]
- Balesdent, M.H. , Fudal, I. , Ollivier, B. , Bally, P. , Grandaubert, J. , Eber, F. , Chèvre, A.M. , Leflon, M. and Rouxel, T. (2013) The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa . New Phytol. 198, 887–898. [DOI] [PubMed] [Google Scholar]
- Böhnert, H.U. , Fudal, I. , Dioh, W. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2004) A putative polyketide synthase peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell, 16, 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpezat, J. , Bothorel, S. , Daverdin, G. , Balesdent, M.H. and Leflon, M. (2014) Use of high resolution melting analysis to genotype the avirulence AvrLm4‐7 gene of Leptosphaeria maculans, a fungal pathogen of Brassica napus . Ann. Appl. Biol. 3, 430–440. [Google Scholar]
- Catanzariti, A.M. , Dodds, P.N. , Lawrence, G.J. , Ayliffe, M.A. and Ellis, J.G. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell, 18, 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni, A. , Passerini, A. , Vullo, A. and Frasconi, P. (2006) DISULFIND, a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 34 (Suppl. 2), W177–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. and Fernando, W.G.D. (2006) Prevalence of pathogenicity groups of Leptosphaeria maculans in Western Canada and North Dakota, USA. Can. J. Plant Pathol. 28, 533–539. [Google Scholar]
- Cheng, J. , Randall, A.Z. , Sweredoski, M.J. and Baldi, P. (2005) SCRATCH, a protein structure and structural feature prediction server. Nucleic Acids Res. 33 (Suppl. 2), W72–W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, J.M. (1981) Larousse Agricole. Paris: Librairie Larousse. [Google Scholar]
- Daverdin, G. , Rouxel, T. , Gout, L. , Aubertot, J.N. , Fudal, I. , Meyer, M. , Parlange, F. , Carpezat, J. and Balesdent, M.H. (2012) Genome structure and reproductive behaviour influence the evolutionary potential of a fungal phytopathogen. PLoS Pathog. 8, e1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A. , Talbot, N.J. , Ebbole, D.J. , Farman, M.L. , Mitchell, T.K. , Orbach, M.J. , Thon, M. , Kulkarni, R. , Xu, J.R. , Pan, H. , Read, N.D. , Lee, Y.H. , Carbone, I. , Brown, D. , Oh, Y.Y. , Donofrio, N. , Jeong, J.S. , Soanes, D.M. , Djonovic, S. , Kolomiets, E. , Rehmeyer, C. , Li, W. , Harding, M. , Kim, S. , Lebrun, M.H. , Bohnert, H. , Coughlan, S. , Butler, J. , Calvo, S. , Ma, L.J. , Nicol, R. , Purcell, S. , Nusbaum, C. , Galagan, J.E. and Birren, B.W. (2005) The genome sequence of the rice blast fungus Magnaporthe grisea . Nature, 434, 980–986. [DOI] [PubMed] [Google Scholar]
- Delourme, R. , Pilet‐Nayel, M.L. , Archipiano, M. , Horvais, R. , Tanguy, X. , Rouxel, T. , Brun, H. , Renard, M. and Balesdent, M.H. (2004) A cluster of major specific resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Des.) Ces. et de Not.] in canola (Brassica napus L.). Theor. Appl. Genet. 91, 1190–1194. [Google Scholar]
- Dilmaghani, A. , Balesdent, M.H. , Didier, J.P. , Wu, C. , Davey, J. , Barbetti, M.J. , Li, H. , Moreno‐Rico, O. , Philips, D. , Despeghel, J.P. , Gout, L. and Rouxel, T. (2009) The Leptosphaeria maculans–Leptosphaeria biglobosa species complex in the American continent. Plant Pathol. 58, 1044–1058. [Google Scholar]
- Dilmaghani, A. , Gout, L. , Moreno‐Rico, O. , Dias, J.S. , Coudard, L. , Castillo‐Torres, N. , Balesdent, M.H. and Rouxel, T. (2012) Clonal populations of Leptosphaeria maculans contaminating cabbage in Mexico. Plant Pathol. 62, 520–532. [Google Scholar]
- Dion, Y. , Gugel, R.K. , Rakow, G.F.W. , Séguin‐Swartz, G. and Landry, B.S. (1995) RFLP mapping of resistance to blackleg disease [causal agent, Leptosphaeria maculans (Desm) Ces et de Not] in canola (Brassica napus L.). Theor. Appl. Genet. 91, 1190–1194. [DOI] [PubMed] [Google Scholar]
- Ferre, F. and Clote, P. (2005) DIANNA, a web server for disulfide connectivity prediction. Nucleic Acids Res. 33 (Suppl. 2), W230–W232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M. , Rimmer, S.R. , Williams, P. and Osborn, T. (1995) Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology, 85, 213–217. [Google Scholar]
- Fitt, B.D.L. , Brun, H. , Barbetti, M.J. and Rimmer, S.R. (2006) World‐wide importance of phoma stems canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 114, 3–15. [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fudal, I. , Ross, S. , Gout, L. , Blaise, F. , Kuhn, M.L. , Eckert, M.R. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2007) Heterochromatin‐like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome, map‐based cloning of AvrLm6 . Mol. Plant–Microbe Interact. 20, 459–470. [DOI] [PubMed] [Google Scholar]
- Fudal, I. , Ross, S. , Brun, H. , Besnard, A.L. , Ermel, M. , Kuhn, M.L. , Balesdent, M.H. and Rouxel, T. (2009) Repeat‐induced point mutation (RIP) as an alternative mechanism of evolution toward virulence in Leptosphaeria maculans . Mol. Plant–Microbe Interact. 22, 932–941. [DOI] [PubMed] [Google Scholar]
- Ghanbarnia, K. , Lydiate, J.L. , Rimmer, S.R. , Li, G. , Kutcher, H.R. , Larkan, N.J. , McVetty, P.B.E. and Fernando, W.G.D. (2012) Genetic mapping of the Leptosphaeria maculans avirulence gene corresponding to the LepR1 resistance gene of Brassica napus . Theor. Appl. Genet. 124, 505–513. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Fudal, I. , Kuhn, M.L. , Blaise, F. , Eckert, M. , Cattolico, L. , Balesdent, M.H. and Rouxel, T. (2006) Lost in the middle of nowhere, the AvrLm1 avirulence gene of the Dothideomycetes Leptosphaeria maculans . Mol. Microbiol. 60, 67–80. [DOI] [PubMed] [Google Scholar]
- Gout, L. , Kuhn, M.L. , Vincenot, L. , Bernard‐Samain, S. , Cattolico, L. , Barbetti, M. , Moreno‐Rico, O. , Balesdent, M.H. and Rouxel, T. (2007) Genome structure impacts molecular evolution at the AvrLm1 avirulence locus of the plant pathogen Leptosphaeria maculans . Environ. Microbiol. 9, 2978–2992. [DOI] [PubMed] [Google Scholar]
- Grandaubert, J. , Balesdent, M.H. and Rouxel, T. (2014a) Evolutionary and adaptive role of transposable elements in fungal genomes. Ann. Bot. Res. 70, 79–107. [Google Scholar]
- Grandaubert, J. , Lowe, R.G.T. , Soyer, J.L. , Schoch, C.L. , Van deWouw, A.P. , Fudal, I. , Robbertse, B. , Lapalu, N. , Links, M.G. , Ollivier, B. , Linglin, J. , Barbe, V. , Mangenot, S. , Cruaud, C. , Borhan, H. , Howlett, B.J. , Balesdent, M.H. and Rouxel, T. (2014b) Transposable element‐assisted evolution and adaptation to host plant within the Leptosphaeria maculans–Leptosphaeria biglobosa species complex of fungal pathogens. BMC Genomics, 15, 891–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y.J. , Balesdent, M.H. , Li, Z.Q. , Evans, N. , Rouxel, T. and Fitt, B.D.L. (2010) Fitness cost of virulence differs between the AvrLm1 and AvrLm4 loci in Leptosphaeria maculans (phoma stem canker of oilseed rape). Eur. J. Plant Pathol. 126, 279–291. [Google Scholar]
- van Kan, J.A.L. , van den Ackerveken, F.J.M. and de Wit, P.J.G.M. (1991) Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant–Microbe Interact. 4, 52–59. [DOI] [PubMed] [Google Scholar]
- Kutcher, H.R. , Keri, M. , McLaren, D.L. and Rimmer, S.R. (2007) Pathogenic variability of Leptosphaeria maculans in Western Canada. Can. J. Plant Pathol. 29, 388–393. [Google Scholar]
- Kutcher, H.R. , Balesdent, M.H. , Rimmer, S.R. , Rouxel, T. , Chèvre, A.M. , Delourme, R. and Brun, H. (2010) Frequency of avirulence genes in Leptosphaeria maculans in Western Canada. Can. J. Plant Pathol. 32, 77–85. [Google Scholar]
- Langmead, B. and Salzberg, S. (2012) Fast gapped‐read alignment with Bowtie2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkan, N.J. , Lydiate, D.J. , Parkin, I.A.P. , Nelson, M.N. , Epp, D.J. , Cowling, W.A. , Rimmer, S.R. and Borhan, M.H. (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor‐like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. [DOI] [PubMed] [Google Scholar]
- Linning, R. , Lin, D. , Lee, N. , Abdennadher, M. , Gaudet, D. , Thomas, P. , Mills, D. , Kronstad, J.W. and Bakkeren, G. (2004) Marker‐based cloning of the region containing the UhAvr1 avirulence gene from the basidiomycete barley pathogen Ustilago hordei . Genetics, 166, 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCt method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Long, Y. , Wang, Z. , Sun, Z. , Fernando, D.W.G. , McVetty, P.B.E. and Li, G. (2011) Identification of two blackleg resistance genes and fine mapping of one of these two genes in a Brassica napus canola cultivar ‘Surpass 400’. Theor. Appl. Genet. 122, 1223–1231. [DOI] [PubMed] [Google Scholar]
- Nepal, A. , Markell, S. , Knodel, J. , Bradley, C.A. and del Río Mendoza, L.E. (2014) Prevalence of blackleg and pathogenicity groups of Leptosphaeria maculans in North Dakota. Plant Dis. 98, 328–335. [DOI] [PubMed] [Google Scholar]
- O'Gorman, D. , Xue, B. , Hsiang, T. and Goodwin, P.H. (1994) Detection of Leptosphaeria korrae with the polymerase chain reaction and primers from ribosomal internal transcribed spacers. Can. J. Bot. 72, 342–346. [Google Scholar]
- Orbach, M.J. , Farrall, L. , Sweigard, J.A. , Chumley, F.G. and Valent, B. (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi‐ta . Plant Cell, 12, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlange, F. , Daverdin, G. , Fudal, I. , Kuhn, M.L. , Balesdent, M.H. , Blaise, F. , Grezes‐Besset, B. and Rouxel, T. (2009) Leptosphaeria maculans avirulence gene AvrLm4‐7 confers dual recognition specificity by the Rlm4 and Rlm7 resistance genes of oilseed rape, and circumvents Rlm4‐mediated recognition through a single amino acid change. Mol. Microbiol. 71, 851–863. [DOI] [PubMed] [Google Scholar]
- Pilet, M.L. , Delourme, R. , Foisset, N. and Renard, M. (1998) Identification of QTL involved in field resistance to light leaf spot (Pyrenopeziza brassicae) and blackleg resistance (Leptosphaeria maculans) in winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 97, 398–406. [Google Scholar]
- Raman, H. , Raman, R. and Larkan, N. (2013) Genetic dissection of blackleg resistance loci in rapeseed (Brassica napus L.) In: Plant Breeding from Laboratories to Fields (Andersen S.B., ed.), pp. 85–120. Rijeka, Croatia: InTech. [Google Scholar]
- Rep, M. , Van Der Does, H.C. , Meijer, M. , Van Wijk, R. , Houterman, P.M. , Dekker, H.L. , de Koster, C.G. and Cornelissen, B.J. (2004) A small, cysteine‐rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I‐3‐mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rimmer, S.R. (2006) Resistance genes to Leptosphaeria maculans in Brassica napus . Can. J. Plant Pathol. 28, 288–297. [Google Scholar]
- Rivas, S. and Thomas, C.M. (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum . Annu. Rev. Phytopathol. 43, 395–436. [DOI] [PubMed] [Google Scholar]
- Rogers, S.O. and Bendich, A.J. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissue. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Rouxel, T. , Penaud, A. , Pnochet, X. , Brun, H. , Gout, L. , Delourme, R. , Schmit, J. and Balesdent, M. (2003a) A 10‐year survey of population of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape. Eur. J. Plant Pathol. 109, 871–881. [Google Scholar]
- Rouxel, T. , Willner, E. , Coudard, L. and Balesdent, M.H. (2003b) Screening and identification of resistance to Leptosphaeria maculans in Brassica napus accessions. Euphytica, 133, 219–231. [Google Scholar]
- Rouxel, T. , Grandaubert, J. , Hane, J.K. , Hoede, C. , Van de Wouw, A. , Couloux, A. , Dominguez, V. , Anthouard, V. , Bally, P. , Bourras, S. , Cozijnsen, A.J. , Ciuffetti, L.M. , Degrave, A. , Dilmaghani, A. , Duret, L. , Fudal, I. , Goodwin, S.B. , Gout, L. , Glaser, N. , Liglin, J. , Kema, G.H. , Lapalu, N. , Lawrence, C.B. , May, K. , Meyer, M. , Ollivier, B. , Poulian, J. , Schoch, C.L. , Simon, A. , Spatafora, J.W. , Stachowiak, A. , Turgeon, B.G. , Tyler, B.M. , Vincent, D. , Weissenbach, J. , Amselem, J. , Quesneville, H. , Oliver, R.P. , Wincker, P. , Balesdent, M.H. and Howlett, B.J. (2011) Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat‐Induced Point mutations. Nat. Commun. 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyer, J.L. , El Ghalid, M. , Glaser, N. , Ollivier, B. , Linglin, J. , Grandaubert, J. , Balesdent, M.H. , Connolly, L.R. , Freitag, M. , Rouxel, T. and Fudal, I. (2014) Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans . PLoS Genet. 10, e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak, A. , Olechowicz, J. , Jedryczka, M. , Rouxel, T. , Balesdent, M.H. , Happstadius, I. , Gladders, P. , Latunde‐Dada, A. and Evans, N. (2006) Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur. J. Plant Pathol. 114, 67–75. [Google Scholar]
- Stein, L.D. (2013) Using GBrowse 2.0 to visualize and share next‐generation sequence data. Brief. Bioinform. 14, 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermark, J. and Karlovsky, P. (2008) Genetic transformation of filamentous fungi by Agrobacterium tumefaciens . Nat. Protoc. 10.1038/nprot.2008.83. [DOI] [Google Scholar]
- Van de Wouw, A.P. , Marcroft, S.J. , Barbetti, M.J. , Li, H. , Salisbury, P.A. , Gout, L. , Rouxel, T. , Howlett, B.J. and Balesdent, M.H. (2009) Dual control of avirulence effectors in Leptosphaeria maculans towards a Brassica napus cultivar with sylvestris‐derived resistance suggests involvement on two resistance genes. Plant Pathol. 58, 305–313. [Google Scholar]
- Van de Wouw, A.P. , Cozijnsen, A.J. , Hane, J.K. , Bruner, P.C. , McDonald, B.A. , Oliver, R.P. and Howlett, B.J. (2010) Evolution of linked avirulence effectors in Leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog. 6, e1001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wouw, A.P. , Lowe, R.G.T. , Elliott, C.E. , Dubois, D.J. and Howlett, B.J. (2014) An avirulence gene, AvrLmJ1, from the blackleg fungus, Leptosphaeria maculans, confers avirulence to Brassica juncea cultivars. Mol. Plant Pathol. 15, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenot, L. , Balesdent, M.H. , Li, H. , Barbetti, M.J. , Sivasithamparam, K. , Gout, L. and Rouxel, T. (2008) Occurrence of a new subclade of Leptosphaeria biglobosa in Western Australia. Phytopathology, 98, 321–329. [DOI] [PubMed] [Google Scholar]
- Williams, P.H. (1985) Crucifer genetics cooperative. Plant Mol. Boil. Rep. 3, 129–144. [Google Scholar]
- Yu, F. , Lydiate, D.J. and Rimmer, S.R. (2005) Identification of two novel genes for blackleg resistance in Brassica napus . Theor. Appl. Genet. 110, 969–979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression of representative AvrLm2 (virulent) alleles during the in vitro growth of Leptosphaeria maculans and oilseed rape infection.
Table S1 List and characteristics of the isolates used here for the identification of AvrLm2.
Table S2 List of polymerase chain reaction (PCR) primers used in this study.
Table S3 The genome coverage for each of the Leptosphaeria maculans isolates used in this study.


