Summary
Genome editing in plants has been boosted tremendously by the development of CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) technology. This powerful tool allows substantial improvement in plant traits in addition to those provided by classical breeding. Here, we demonstrate the development of virus resistance in cucumber (Cucumis sativus L.) using Cas9/subgenomic RNA (sgRNA) technology to disrupt the function of the recessive eIF4E (eukaryotic translation initiation factor 4E) gene. Cas9/sgRNA constructs were targeted to the N′ and C′ termini of the eIF4E gene. Small deletions and single nucleotide polymorphisms (SNPs) were observed in the eIF4E gene targeted sites of transformed T1 generation cucumber plants, but not in putative off‐target sites. Non‐transgenic heterozygous eif4e mutant plants were selected for the production of non‐transgenic homozygous T3 generation plants. Homozygous T3 progeny following Cas9/sgRNA that had been targeted to both eif4e sites exhibited immunity to Cucumber vein yellowing virus (Ipomovirus) infection and resistance to the potyviruses Zucchini yellow mosaic virus and Papaya ring spot mosaic virus‐W. In contrast, heterozygous mutant and non‐mutant plants were highly susceptible to these viruses. For the first time, virus resistance has been developed in cucumber, non‐transgenically, not visibly affecting plant development and without long‐term backcrossing, via a new technology that can be expected to be applicable to a wide range of crop plants.
Keywords: CRISPR/Cas9, cucumber, eIF4E, genome editing, Potyviridae, virus resistance
Introduction
Plant viruses cause extensive reductions in crop yields worldwide. There are several paths to the development of virus resistance in crop plants. One path is classical plant breeding by the introgression of genes for virus resistance from crop plant relatives. Many genes conferring virus resistance have been identified (Kang et al., 2005; Maule et al., 2007). Another path is genome editing, which permits the introduction of alleles conferring resistance directly into crop plants, without the many backcrosses required by classical breeding. As ‘gene‐edited crops’ do not necessarily include transgenic segments (Xu et al., 2015), they probably would not need extensive regulation (Jones, 2015), thereby opening up a new publicly acceptable method for the breeding of virus‐resistant crops.
RNA‐guided genome editing using Streptococcus pyogenes CRISPR‐Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) has renewed the concept of genome editing in plants (Belhaj et al., 2013, 2015; Bortesi and Fischer, 2015; Feng et al., 2013; Liu and Fan, 2014). Cas9‐induced double‐strand breaks in the plant genome are mainly repaired by non‐homologous end joining (Li et al., 2013; Nekrasov et al., 2013). The 20‐nucleotide DNA target sequence is followed by a protospacer adjacent motif (PAM) recognized by Cas9‐subgenomic RNA (sgRNA) that leads to DNA strand separation and breaking (Sternberg et al., 2014). During non‐homologous end joining repair, insertions or deletions (indels) may occur, which may alter a protein open reading frame or introduce a premature stop codon (Belhaj et al., 2013). Cas9/sgRNA editing has been demonstrated in various plant families, e.g. Cruciferae, Solanaceae, Poaceae and Fabaceae (Belhaj et al., 2013; Bortesi and Fischer, 2015; Jacobs et al., 2015; Xie et al., 2014), since it was first reported in 2013 (Li et al., 2013; Nekrasov et al., 2013). Recently, it has been shown that Cas9/sgRNA can target plant DNA viruses to develop virus resistance (Ali et al., 2015; Baltes et al., 2015).
Plant RNA viruses require host factors to maintain their life cycle. Many genes conferring resistance to viruses are recessive (Kang et al., 2005; Truniger and Aranda, 2009), including the eukaryotic translation initiation factors eIF4E and eIF(iso)4E (Lellis et al., 2002; Nicaise et al., 2003; Ruffel et al., 2006). The eIF4F complex [eIF4E and eIF4G (or their isoforms) and eIF4A] and other host factors, such as the polyA‐binding protein (PABP), bind to the potyviral 5′ m7G cap structure and 3′ polyA tail of mRNA for translation. In the eIF4F complex, the eIF4E and eIF(iso)4E proteins link to the 5′ of mRNA or viral RNA and to the scaffold protein of each. Both the eIF4E and eIF(iso)4E proteins occur in plant cytoplasm and have redundant functions (Jackson et al., 2010; Sanfaçon, 2015; Wang and Krishnaswamy, 2012). Viruses, especially potyviruses, can associate with one or both of these proteins through the viral‐encoded protein VPg (Duprat et al., 2002; Hwang et al., 2009; Ling et al., 2009; Ruffel et al., 2006; Sato et al., 2005). The copy numbers of the eIF4E and eIF(iso)4E genes differ among plant species (Le Gall et al., 2011). In Cucumis spp. (cucumber and melon), one gene each of eIF4E and eIF(iso)4E have been identified (A. Gal‐On et al., unpublished data; Rodríguez‐Hernández et al., 2012). Both eIF4E and eIF(iso)4E are recessive when mutated, and are essential for the translation of uncapped viruses having the VPg protein covalently linked to the viral RNA 5′ (Wittmann et al., 1997). eIF4E and eIF(iso)4E interact with VPg in different hosts (Jiang and Laliberté, 2011; Léonard et al., 2000; Sanfaçon, 2015) and disruption of this link by mutagenesis or silencing prevents virus infectivity (Duprat et al., 2002; Lellis et al., 2002; Rodríguez‐Hernández et al., 2012; Sato et al., 2005). The association of natural mutations in the eIF4E and eIF(iso)4E genes with potyvirus resistance has been observed in various crops and applied to breeding (Gómez et al., 2009). Broad RNA virus resistance has been demonstrated by silencing of the eIF4E gene in tomato and melon (Mazier et al., 2011; Rodríguez‐Hernández et al., 2012).
Members of the Potyviridae cause significant losses in a wide range of crops. The viruses in this family have an RNA genome of approximately 10 kb (with a 3′ polyA tail) that encodes a polyprotein which is cleaved by three viral proteases, resulting in 9–11 putative mature proteins (Revers and García, 2015). VPg is the amino part of the NIa protease, which is covalently attached to the genomic RNA 5′ end as an mRNA cap analogue. VPg plays a role in polyprotein translation and other functions in the virus life cycle. Mutations in the VPg gene have been shown to be associated with the breaking of natural resistance by a number of viruses (Ayme et al., 2006; Hébrard et al., 2006; Moury et al., 2004). Other potyvirus genes, such as cylindrical inclusion and P1, are known to be involved in the breaking of eIF4E‐mediated resistance (Abdul‐Razzak et al., 2009; Nakahara et al., 2010).
Here, we report for the first time targeted gene knockout in cucumber by the CRISPR/Cas9 system. We designed two sgRNAs (sgRNA1 and sgRNA2) to target the cucumber eIF4E gene at two different sites. Following their transgenic expression together with Cas9, a range of deletion and insertion indels were sequenced in both eIF4E targeted sites. Non‐transgenic T3 generation homozygotic plants harbouring 20 and four base deletions in the eIF4E gene were challenged with viruses from the Potyviridae family [Cucumber vein yellowing virus (CVYV), Zucchini yellow mosaic virus (ZYMV) and Papaya ring spot mosaic virus‐W (PRSV‐W)]. Progenies of T3 homozygotic mutant plants exhibited broad virus resistance compared with the susceptible heterozygous T3 plants.
Results
Efficacy of CRISPR/Cas9 in the T0 generation
To test the efficacy of CRISPR/Cas9 in cucumber and to develop a new strategy to generate virus resistance, we chose to disrupt eIF4E. eIF4E is a plant cellular translation factor essential for the Potyviridae life cycle, and natural point mutations in this gene can confer resistance to potyviruses (for a review, see Diaz‐Pendon et al., 2004; Kang et al., 2005; Le Gall et al., 2011; Sanfaçon, 2015). In cucumber, two eIF4E genes have been identified, eIF4E (accession no. XM_004147349) (236 amino acids) and eIF(iso)4E (accession no. XM_004147116.2) (204 amino acids), which share 56% nucleotide and 60% amino acid homology. We targeted two regions in the cucumber eIF4E gene by Cas9/sgRNA, which have no homology in the eIF(iso)4E gene. The Cas9/sgRNA1 construct was designed to target the sequence in the first exon of eIF4E (positions 65–86 in the coding region) (Fig. 1A). The Cas9/sgRNA2 construct was designed to target the third exon (positions 517–540) in the coding region to allow translation of approximately two‐thirds of the protein, perhaps without disrupting all of its functions (Fig. 1A).
Figure 1.
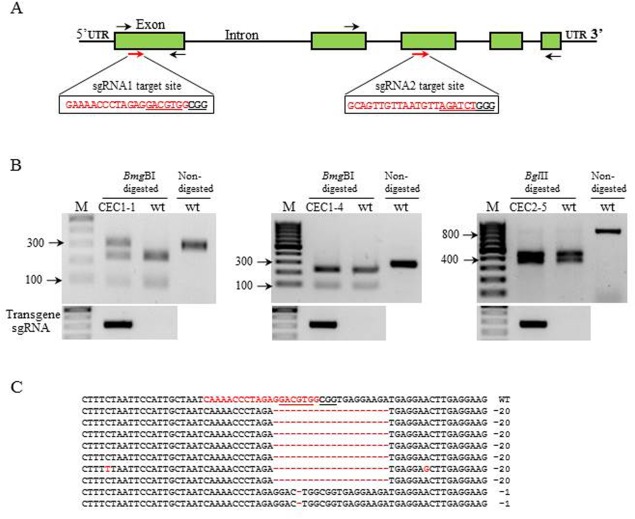
Gene editing of eIF4E mediated by CRISPR/Cas9 in transgenic cucumber plants. (A) Schematic representation of the cucumber eIF4E genomic map and the sgRNA1 and sgRNA2 target sites (red arrows). The target sequence is shown in red letters together with the restriction site (underlined), and the protospacer adjacent motif (PAM) is marked in bold underlined letters. The black arrows indicate the primers flanking the target sites used to detect the mutations. (B) Restriction analysis of T0 polymerase chain reaction (PCR) fragments of CEC‐1, CEC1‐4 and CEC2‐5. (C) Alignment of nine colony sequences from the undigested fragment of line 1 with the wild‐type (wt) genome sequence. DNA deletions are shown by red dashes and deletion sizes (nucleotides) are marked on the right side of the sequence.
Five independent T0 transgenic lines were generated by Agrobacterium‐mediated transformation. The presence of the transgene (Cas9/sgRNA) was confirmed by kanamycin resistance and polymerase chain reaction (PCR) using sgRNA‐specific primers (Fig 1B and Table S1, see Supporting Information). Three lines (1, 3 and 4) were identified as transgenic with Cas9/sgRNA1. To evaluate the types of mutations generated in sgRNA1 transgenic plants, PCR was performed in T0 plants using primers flanking the sgRNA1 target and subsequently digested with BmgBI (a site that would disappear if CAS9 and NHEJ were active in this location). In line 1, a distinct undigested fragment was observed following BmgBI restriction (Fig. 1B). The partial digestion observed indicated a heterozygous genome with both wild‐type and mutant eIF4E alleles. Cloning and sequencing of the uncut BmgBI fragment showed two types of mutation: a 20‐nucleotide deletion around the PAM sequence in seven colonies and a one‐nucleotide deletion 3 bp upstream of the PAM sequence in two colonies (Figs 1C, S2, see Supporting Information). In lines 4 (Fig. 1B) and 3 (data not shown), the amplified PCR was completely digested by BmgBI.
Two additional Cas9/sgRNA2 transgenic plants (2 and 5) did not show genome editing in T0 as determined by PCR and restriction analysis with BglII (Fig. 1B and data not shown). We continued our study with two sgRNA1 lines (1 and 4), designated CEC1‐1 and CEC1‐4, respectively (Cas/sgRNA1‐e IF4E‐Cucumber), and one sgRNA2 line (line 5), designated CEC2‐5.
Genotypes and segregation of T1 mutants of CEC1‐1
For propagation by seeds, the Cucumis sativus CEC1‐1 T0 mutant plant (derived from ‘Ilan’, a multi‐pistillate, parthenocarpic glasshouse cucumber) was cross‐pollinated with ‘Bet Alfa’, a monoecious, non‐parthenocarpic, open field cucumber. Indel polymorphisms were genotyped by PCR restriction analysis with BmgBI of the eIF4E gene in representative CEC1‐1 T1 plants (Fig. 2). The T1 progeny segregated into three groups (Fig. 2): (i) heterozygous plants that contained about equal amounts of undigested and digested DNA (plants 5, 8, 12, 16); (ii) plants with undigested DNA with an intensity stronger than that of digested DNA (plants 2, 9, 7, 20); (iii) non‐mutants (wild‐type), with most of the DNA digested (plants 3, 10). The intense undigested band in group (ii) and the faint undigested band of plant 10 might be caused by the continuing activity of cas9‐sgRNA1 in transgenic plants (Fig. 2A). The segregation of transgenic to non‐transgenic in the T1 population was approximately 1 : 1, as expected for a single transgene locus. The independent segregation of the transgene Cas9/sgRNA1 and the eIF4E mutation indicated that they were present on different chromosomes (Fig. 2A, bottom panel), allowing selection for non‐transgenic mutants in later generations. To evaluate the types of mutation generated in CEC1‐1 T1 plants, four representative plants [i.e. plants 1, 4, 7 (non‐transgenic, lacking the Cas9 transgene) and 5 (transgenic)] were chosen, and the undigested DNA was cloned and sequenced. Plant 1 had a 20‐nucleotide deletion and plants 4 and 5 had one‐nucleotide deletions (Figs 2B, S2). Plant 7 had both the 20‐ and one‐nucleotide deletions as observed in T0 (Figs 1C, S2). Hence, CRISPR/Cas9‐induced mutations in cucumber can be stably transmitted through the germ line. PCR genotyping of the T1 generation of CEC1‐1 (see Experimental Procedures) indicated that 20 plants had lengthy deletions (20 nucleotides) and 13 plants had a one‐nucleotide deletion (Fig. S2 and data not shown).
Figure 2.
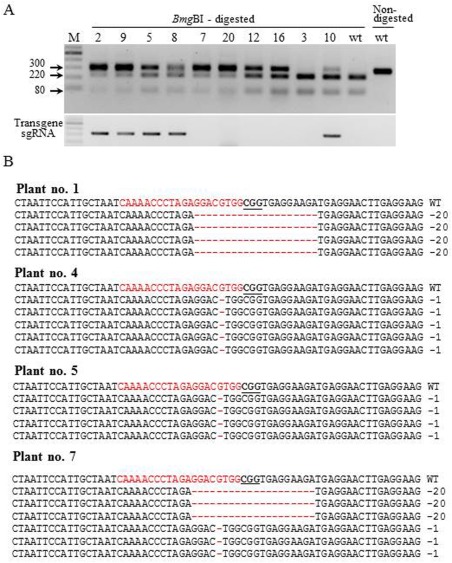
Genotyping of eif4e mutants in representative T1 progeny plants of the CEC1‐1 line. (A) Polymerase chain reaction (PCR) restriction analysis of Cas9/sgRNA1‐mediated mutations (top panel) and transgene insertion (bottom panel) in 10 representative T1 cucumber plants and non‐mutant wild‐type (wt). (B) Alignment of four representative eif4e mutant plants with the wild‐type sequence. The sequences of each plant represented clones from undigested fragments. The target sequence is shown in red letters and the protospacer adjacent motif (PAM) is marked by bold underlined letters. DNA deletions are marked with red dashes and deletion sizes (nucleotides) are indicated on the right side of the sequence.
The non‐transgenic CEC1 T1 plant 7 (CEC1‐1‐7) was grown to produce seeds for the production of homozygous eif4e mutant alleles. The all‐pistillate CEC1‐1‐7 plant was cross‐pollinated once again with the monoecious ‘Bet Alfa’. The resulting T2 progeny were genotyped and plants hemizygous for a 20‐nucleotide deletion (plant 1, Fig. 2B) (CEC1‐1‐7‐1) and one‐nucleotide deletion (plant 4; Fig. 2B) (CEC1‐1‐7‐4) were self‐pollinated to obtain a T3 generation. In the T3 generation, the 20‐nucleotide deletion segregated in a Mendelian manner 1 : 2 : 1 (homozygous : heterozygous : wild‐type without mutation). The homozygous, non‐transgenic T3 plant designated CEC1‐1‐7‐1, heterozygous and non‐mutant (wild‐type) plants were tested for virus resistance. Mutations in the T3 generation were confirmed by restriction analysis and sequencing.
Genotypes and segregation of T1 mutants of CEC1‐4
In addition to the derivatives of line 1 (CEC1‐1), we analysed Cas9/sgRNA1‐mediated mutations in the derivatives of line 4 (CEC1‐4) (Figs 1B and 3). In this CEC1‐4 T0 transgenic line, an indel mutation was not observed. To further confirm transgene inheritance, the plant CEC1‐4 was self‐pollinated and, in the T1 generation, only three of eight plants (Fig. 3, plants 4, 5 and 6) had a faint undigested band on digestion with BmgBI (Fig. 3A). Cloning and sequencing of the undigested faint band showed multiple mutations within the target gene in the same plant (Fig. 3B). All three plants appeared to be chimeric for the mutant allele with more than one type of mutation in the same plant. To verify whether CRISPR/Cas9 functions in the germ cell line, the CEC1‐4 plants 4, 5 and 6 were grown and self‐pollinated to produce a T2 generation. In the T2 generation 74 progeny plants were grown; PCR genotyping analysis showed that most of the plants had a faint undigested band, as observed in the T1 generation (CEC1‐4). Cloning and sequencing of two representative plants from the T2 generation showed that multiple mutations were present in the same plant (data not shown). Further analysis with CEC1‐4‐4, CEC1‐4‐5 and CEC1‐4‐6 plants was not performed, because it appeared that, in this CEC1‐4 line, Cas9/sgRNA1‐mediated cleavage occurred in somatic cells and not in the germ line.
Figure 3.
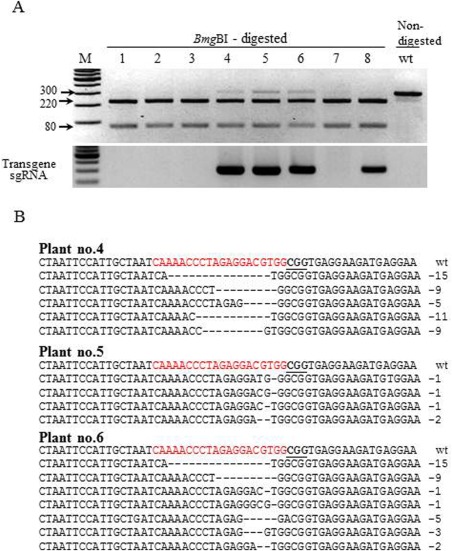
Genotyping of eif4e mutants in representative T1 progeny plants of the CEC1‐4 line. (A) Polymerase chain reaction (PCR) restriction analysis of Cas9/sgRNA1‐mediated mutations (top panel) and transgene insertion (bottom panel) in eight T1 cucumber plants and non‐mutant wild‐type (wt). (B) Alignment of three eif4e transgenic mutant plants 4, 5 and 6 with the wild‐type sequence. Sequences of each plant represent clones from undigested fragments. The target sequence is shown in red letters and the protospacer adjacent motif (PAM) is marked in bold underlined letters. DNA deletions are marked by red dashes and deletion sizes (nucleotides) are indicated on the right side of the sequence.
Genotypes and segregation of T1 mutants of CEC2‐5
To evaluate the types of mutation mediated by Cas9/sgRNA2 in line CEC2‐5, the flanking region was PCR amplified and the presence of indel mutations was tested by BglII restriction (Fig. 1B). The PCR fragment of CEC2‐5 was as completely digested as the wild‐type (Fig. 1B). CEC2‐5 was cross‐pollinated with ‘Bet Alfa’ and the progeny (T1) segregated approximately to 1 : 1 transgenic : non‐transgenic, suggesting a single transgenic locus. Mutations were observed only in transgenic progeny (Fig. 4A). Genotyping of CEC2‐5 T1 generation plants revealed three groups: (i) homozygous plants with completely undigested DNA (nine of 15 transgenic seedlings) (Fig. 4A, plants 6 and 14); (ii) heterozygous plants having similar intensities of undigested and digested DNA (Fig. 4A, plants 9 and 26); (iii) plants in which faintly digested DNA can be seen, which may reflect continuing activity of Cas9/sgRNA2 in heterozygous plants (Fig. 4A, plants 1 and 7). The DNA of non‐transgenic segregant plants lacking Cas9 was completely digested, as was the wild‐type non‐transgenic control (Fig. 4A, plants 2 and 3).
Figure 4.
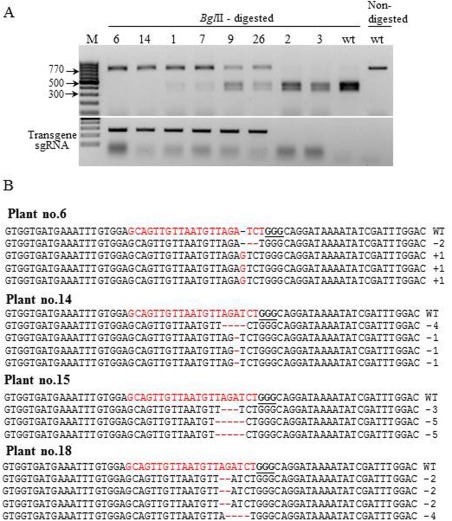
Genotyping of the Cas9/sgRNA2‐mediated mutation in T1 progeny plants of the CEC2‐5 line. (A) Polymerase chain reaction (PCR) restriction analysis of Cas9/sgRNA2‐mediated mutations (top panel) and the presence of the Cas9/sgRNA2 transgene (bottom panel) in eight representative T1 cucumber plants. (B) Alignment of four representative eif4e mutant plants with the wild‐type sequence. The target sequence is shown in red letters and the protospacer adjacent motif (PAM) is marked in bold underlined letters. DNA deletions or insertions are marked by red dashes and letters, and the sizes of the deletions or insertions (nucleotides) are indicated on the right side of the sequence.
To genotype the mutations in CEC2 T1 progeny plants, the complete uncut PCR product was cloned and sequenced from four representative plants (Fig. 4B, plants 6, 14, 15, 18). The results showed that each plant had two different types of mutation (Fig. 4B). Interestingly, in plant 6, we found an insertion of one nucleotide together with a two‐nucleotide deletion. All of the plants had bi‐allelic mutations, but the mutations differed from plant to plant. Mutations were found only in the T1 transgenic plants, but not in T0 plants. This suggests that editing occurred in the germ cell line in the T0 generation.
Bi‐allelic heterozygous mutant and mono‐allelic mutant T1 plants of CEC2‐5‐1 were cross‐pollinated with non‐transgenic ‘Bet Alfa’ plants and the resulting T2 progeny seeds were pooled [designated as Mix (M) CEC2‐5‐M] and germinated. The mutant plants were screened by PCR/BglII restriction analysis. To obtain homozygous mutant plants of CEC2‐5‐M, T2 homozygous CEC2‐5‐M‐9, CEC2‐5‐M‐16 and heterozygous CEC2‐5‐M‐8, CEC2‐5‐M‐21 seedlings were cross‐pollinated with T2 plants. Sequencing of T3 progeny of plants 9, 16, 8 and 21 showed four‐nucleotide deletions. The T3 progenies of plants 9, 16, 8 and 21 were used for virus resistance analysis.
Off‐target analysis
Cas9/sgRNA1 off‐targets were evaluated by the CRISPR‐P program (Lei et al., 2014) using the sgRNA1 sequence against the cucumber genome. Five candidate potential off‐targets were determined (Table 1). PCR and sequencing of these candidate targets revealed no changes in the genome of non‐transgenic T3 generation CEC1‐1‐7‐1.
Table 1.
Mutations in the putative eIF4E CRISPR/Cas9 sgRNA1 off‐target sites.
| Putative off‐target site | Putative off‐target locus | Sequence of the putativeoff‐target site* | No. of mismatching bases | Presence of mutations |
|---|---|---|---|---|
| Cucsa.300080 | scaffold02925:+101443 | CAAAACCGGAGAAGACGTGACGG | 4 | 0 |
| Intergenic | scaffold00919:1885568 | CATATGCCTAGAGTACGTGGGGG | 4 | 0 |
| Intergenic | scaffold00995:4267 | CAAAACCCTAGAGGGTTTGGGGG | 3 | 0 |
| Intergenic | scaffold02925:+41731 | CAAAACGCTAGATGTCTTGGAGG | 4 | 0 |
| Intergenic | scaffold03356:+3916928 | CAAAATACTAGAGGACGGTGTGG | 4 | 0 |
*The protospacer adjacent motif (PAM) is marked in italics.
Virus resistance analysis
To test whether CRISPR/Cas9‐mediated mutations in eIF4E confer virus resistance, T3 progenies of CEC1 and CEC2 seedlings were inoculated with the ipomovirus CVYV, two potyviruses ZYMV and PRSV‐W, Cucumber mosaic cucumovirus (CMV) and Cucumber green mottle mosaic tobamovirus (CGMMV). T3 non‐transgenic progenies of CEC1‐1‐7‐1 showed a Mendelian segregation ratio of 1 : 2 : 1 for the homozygous mutant allele (eif4e), heterozygous mutant allele and homozygous non‐mutant. In the case of CEC2‐5‐M‐9, CEC2‐5‐M‐16, CEC2‐5‐M‐8 and CEC2‐5‐M‐21 (mixture of four lines: 4n) (designated CEC2‐5‐M‐4n), the T3 progenies of plants 9 and 16 segregated 1 : 1 (homozygous : heterozygous) and plants 8 and 21 segregated 1 : 2 : 1 (homozygous : heterozygous : non‐mutant).
CVYV resistance analysis
Whitefly inoculation (natural vector) of the T3 generation with CVYV showed that both CEC1‐1‐7‐1 and CEC2‐5‐M‐4n homozygous mutant plants were immune to CVYV infection (0/20 and 0/32, respectively), whereas, in the heterozygous mutant and wild‐type plants, severe symptoms were observed at 7–10 days post‐infection (dpi) (Table 2 and Fig. 5A). The mutant homozygous plants remained healthy through 45 dpi. The experiment was repeated four times with consistent results (Table 2). Reverse transcription‐polymerase chain reaction (RT‐PCR) analyses revealed no viral RNA accumulation in the homozygous mutant plants, whereas, in the heterozygous plants, viral RNA accumulation was observed in similar quantities as to the wild‐type (Fig. 5B).
Table 2.
Response of T3 generation plants of non‐transgenic CEC‐1‐7‐1 and CEC2‐5‐M‐4n lines to Cucumber vein yellowing virus (CVYV), Zucchini yellow mosaic virus (ZYMV), Papaya ring spot mosaic virus‐W (PRSV‐W), Cucumber mosaic cucumovirus (CMV) and Cucumber green mottle mosaic tobamovirus (CGMMV) infection at different days post‐infection (dpi).
| CEC1‐1‐7‐1 | ||||
|---|---|---|---|---|
| Virus | Homozygous* | Non‐homozygous† | ||
| 14 dpi | 25–45 dpi‡ | 14 dpi | ||
| CVYV | 0/20 | 0/12 | 60/60 | |
| ZYMV | 0/28 | 10/21 (0.48 ± 0.04)* | 97/97 | |
| PRSV | 1/14 | n.t. | 40/41 | |
| CMV | 11/11 | – | 9/9 | |
| CGMMV | 10/10 | – | 10/10 | |
| CEC2‐5‐M‐4n | ||||
| Homozygous | Non‐homozygous | |||
| CVYV | 0/32 | 0/10 | 42/43 | |
| ZYMV | 0/64 | 16/63 (0.26 ± 0.05)* | 67/69 | |
| ZYMV§ | 0/8 | 0/8 | 6/7 | |
| PRSV | 1/55 | 7/18 | 33/37 | |
n.t., not tested.
*Infectivity rates were scored as the number of symptomatic plants vs. the total number of plants inoculated; mean ± standard deviation of three biological repeats.
†Non‐homozygous plants include heterozygous and non‐mutant plants.
‡Some of the resistant plants were kept in a net house for further observation.
§Plants were inoculated by five to seven aphids per plant.
Figure 5.
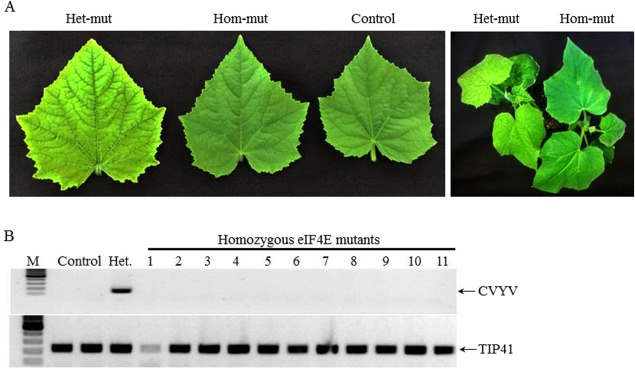
Homozygous eif4e mutant plants exhibited immunity to Cucumber vein yellowing virus (CVYV) infection. (A) Disease symptoms (leaves and plants) of heterozygous (Het‐mut), homozygous (Hom‐mut) and non‐inoculated (Control) plants of the CEC1‐1‐7‐1 T3 generation at 10 days post‐infection (dpi). (B) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of CVYV RNA accumulation at 14 dpi in homozygous eif4e mutant plants (plants 1–11), heterozygous eif4e mutant plant (Het.) and non‐inoculated plant (Control). TIP41 (tonoplast intrinsic protein) was used as a reference gene for RT‐PCR amplification. A molecular marker 100‐bp ladder is shown (M).
ZYMV resistance analysis
Following ZYMV inoculation (mechanical and by aphids) of CEC1‐1‐7‐1 and CEC2‐5‐M‐4n T3 lines, seedlings showed mosaic symptom development at 7–10 dpi that exacerbated to severe symptoms of leaf deformation and stunting at 20 dpi in heterozygous and non‐mutant plants (Fig. 6A); in contrast, the eif4e mutant plants did not display disease symptoms at 20 dpi (Fig. 6A). Accordingly, resistance to ZYMV systemic infection was observed (zero of 28 plants were infected) in four separate biological repeat experiments (Table 2). However, at 25–45 dpi, mild symptoms could be observed in 48% (10 of 21 plants) of the T3 homozygous plants in three biological experiments with CEC1‐1‐7‐1 and in 25% of CEC2‐5‐M‐4n plants (16 of 63 plants; Table 2). However, the mild symptoms appeared only in patches (Fig. 6A) and the plants developed normally, similar to non‐infected plants, compared with the stunted growth with deformed fruit of the infected heterozygous and wild‐type plants. ZYMV RNA was not detected in the immune homozygous resistant plants (Fig. 6B). The late appearance of mild symptoms was accompanied by the accumulation of ZYMV RNA in the upper leaves (plants 4 and 10, Fig. 6B). The level of ZYMV RNA accumulation in mild symptomatic plants was lower in CEC1‐1‐7‐1 and CEC2‐5‐M‐21 homozygous plants than in the wild‐type (Fig. 6C). Interestingly, resistance breaking was not observed when inoculation was made by the natural vector of ZYMV, Aphis gossypii (Table 2).
Figure 6.
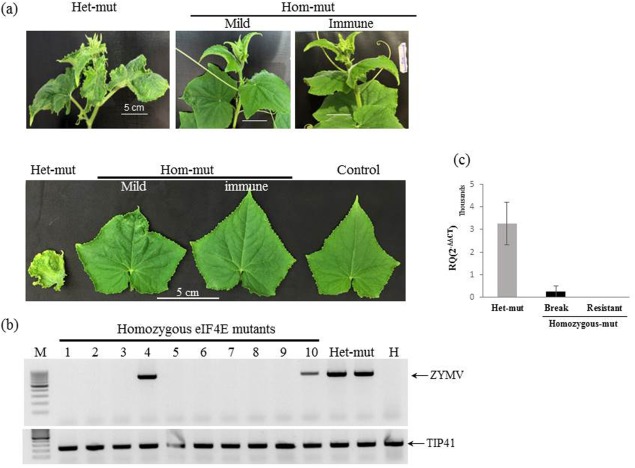
Homozygous eif4e mutant plants exhibited resistance to Zucchini yellow mosaic virus (ZYMV) infection. (A) Disease symptoms of heterozygous (Het‐mut), homozygous (Hom‐mut) and non‐inoculated (Control) plants of the CEC1‐1‐7‐1 T3 generation at 25 days post‐infection (dpi). (B) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of ZYMV RNA accumulation in homozygous eif4e mutant plants (1–10), heterozygous plants (Het‐mut) and non‐inoculated plant (H) at 14 dpi. Tip41 was used as a reference gene for RT‐PCR amplification. A molecular marker 100‐bp ladder is shown (M). (C) Relative (real‐time quantitative RT‐PCR) ZYMV RNA accumulation in CEC1‐1‐7‐1 heterozygous (Het‐mut) and two classes of homozygous mutant: resistant (Resistant) and breaking (Break). RNA was extracted from three plants (third top leaf) and the ZYMV level was calculated using the ΔΔCT method normalized to the F‐box gene expression level.
PRSV‐W resistance analysis
Resistance to PRSV‐W (Israeli isolate) was assessed following mechanical inoculation of T3 generation seedlings of CEC1‐1‐7‐1 and CEC2‐5‐M‐4n. PRSV‐W symptoms in wild‐type cucumber were less aggressive than those of ZYMV, and severe symptoms appeared at 14 dpi. Resistance to PRSV‐W can be seen in CEC1‐1‐7‐1 (Fig. 7A) and CEC2‐5‐M‐4n at 14 dpi (data not shown). In about 40% (seven of 18 in one experiment) of the resistant eif4e plants, mild symptoms appeared at 25 dpi (Table 2), although such resistance breaking did not affect plant development. In most of the asymptomatic resistant plants, PRSV‐W RNA accumulation was detected (Fig. 7B); however, its RNA levels were significantly lower than those in infected heterozygous plants (Fig. 7C). A recovery phenomenon was observed in four of seven resistance‐breaking plants, together with a significant decrease in viral RNA level at 35 dpi.
Figure 7.
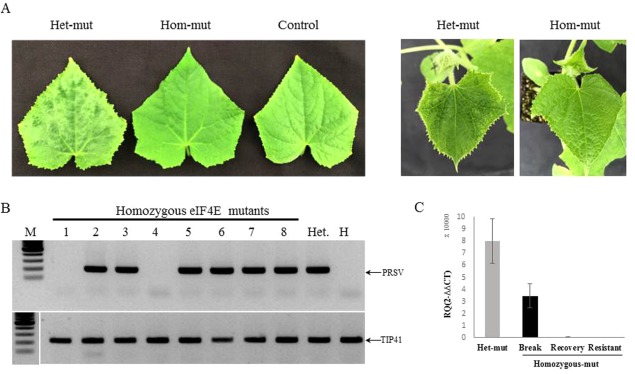
Homozygous eif4e mutants exhibited resistance to Papaya ring spot mosaic virus‐W (PRSV‐W) infection. (A) Disease symptoms of heterozygous (Het‐mut), homozygous (Hom‐mut) and non‐inoculated (Control) plants of CEC1‐1‐7‐1 T3 generation at 21 days post‐infection (dpi). (B) Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis of PRSV‐W RNA accumulation in homozygous plants (1–8), heterozygous plant (Het.) and non‐inoculated plant (H) at 14 dpi. Tip41 was used as a reference gene for RT‐PCR amplification. A molecular marker 100‐bp ladder is shown (M). (C) Relative (real‐time quantitative RT‐PCR) accumulation of PRSV‐W RNA in CEC2‐5‐M‐9 heterozygous (Het‐mut) and three classes of homozygous mutant: resistant (Resistant), breaking (Break) and recovering (Recovery). RNA was extracted from the second top leaf of three plants and the PRSV‐W RNA level was calculated using the ΔΔCT method normalized to the F‐box gene expression level.
CMV and CGMMV resistance analysis
CEC1‐1‐7‐1 T3 progenies were tested for resistance to CMV (Cucumovirus) and CGMMV (Tobamovirus) as these viruses have 5′ capped RNA. Virus symptoms were observed in all plants (homozygous mutants, heterozygous mutants and wild‐type) (Table 2) without significant differences in symptom appearance. The levels of CMV and CGMMV RNAs of these mutants were not tested.
Discussion
A major goal of plant biotechnology is to improve crop yield and quality in a sustainable manner, more rapidly than classical breeding, which may require many generations. Studies of plant–virus interactions have produced a list of host genes associated with virus resistance (Diaz‐Pendon et al., 2004; Gómez et al., 2009; Kang et al., 2005). Moreover, the transgenic approach to the development of resistance in different crops has been very successfully demonstrated against many viruses in the last two decades (see Cillo and Palukaitis, 2014). However, difficulties in meeting regulatory requirements and the public opposition to transgenic plants have limited the implementation of these biotechnological methods. For this purpose, the development of efficient plant genome editing by CRISPR/Cas9 opens up many opportunities for crop improvement. Here, for the first time using CRISPR/Cas9 editing technology, we have developed eif4e non‐transgenic cucumber mutants that exhibit resistance to three economically important viruses.
We designed two Cas9/sgRNA constructs to target the cucumber eIF4E gene: one (sgRNA1) was expected to disrupt the intact eIF4E protein, and the other (sgRNA2) to permit translation of two‐thirds of the protein product.
In Agrobacterium‐transformed T0 lines, we found deletions in the eIF4E target gene in one line (CEC1‐1) out of five (Fig. 1 and data not shown). In the CEC1‐1 line, the same mutations were observed in the T1 generation, which implies a heterozygous mono‐allellic CEC1‐1 T0 plant, as observed in tomato (Brooks et al., 2014) and rice (Zhang et al., 2014). In the transgenic line CEC1‐4, all of the progeny from the T1 and T2 generations showed partial cleavage activity of Cas9 (Fig. 4), which implies that cleavage occurred only in somatic cells and not in the germ cell line. Such phenomena may be a result of the transgene insertion site of Cas9 or spatial specificity of the 35S promoter in the plant genome that causes a low level of expression and activity.
In the transgenic T0 generation of the CEC2‐5 line, a mutation was not detected by PCR or restriction analysis (Fig. 1B), although, in the T1 generation (Fig. 4), homozygous, heterozygous and non‐mutant plants were observed; mutations in homozygous plants were bi‐allelic, with two mutations in the same plant. It is possible that Cas9 activity in T0 was undetectable, although active, in the germ cell line. Alternatively, the T0 plant was chimeric, in which expression of Cas9/sgRNA2 occurred in the germ cell line, as observed in rice (Zhang et al., 2014). The differences in Cas9 targeting between the three T0 lines (CEC1‐1, CEC1‐2 and CEC2‐5) may be a result of differential activities of Cas9 in different transgenic lines, depending on the transgene insertion site.
Off‐target cleavage of Cas9/sgRNA occurs in mammalian and plant cells, and is more frequent if the first 12 nucleotides linked to the NGG sequence (PAM) are identical to the target sequence (Fu et al., 2013; Zhang et al., 2014). In the T3 generation of CEC1‐1‐7‐1 plants, off‐target mutations were not found (Table 1), perhaps attributable to the sequence changes close to PAM in all five putative off‐target sites (Table 1). This suggests that, in our non‐transgenic homozygous mutant plants, the only mutation occurred in the eIF4E gene, and the remainder of the genome was unchanged.
Cucumber has a diploid genome and a single eIF4E gene and, to knock out eIF4E gene expression, we propagated homozygous mutant plants. We transformed a parthenocarpic, gynoecious hybrid, ‘Ilan’ (Gal‐On et al., 2005), and therefore the production of non‐transgenic homozygous mutations required breeding for three generations (T3), using the monoecious ‘Bet Alfa’ as a pollen source.
The T3 progeny plants of the non‐transgenic CEC1‐1‐7‐1 and CEC2‐5‐M‐4n lines segregated for eif4e mutants into homozygous, heterozygous and non‐mutant. This allowed an evaluation of virus resistance with internal susceptible controls (heterozygous and non‐mutant).
Progeny from the two independent lines, CEC1‐1‐7‐1 and CEC2‐5‐M‐4n, showed immunity to CVYV (Ipomovirus) infection by the natural whitefly vector. As CVYV‐RNA was not detected in inoculated and systemic leaves at different times post‐inoculation, we assume that initial viral translation probably could not be established in the cells containing virus particles. Similar resistance to CVYV has been shown in transgenic melon in which the eIF4E gene was silenced (Rodríguez‐Hernández et al., 2012). This demonstrates that eIF4E has a crucial function in the CVYV life cycle in Cucumis spp. (cucumber and melon); notably, the eIF4E protein of cucumber shares 99% similarity with that of melon.
Resistance to ZYMV and PRSV‐W was detected in the two eif4e homozygous lines CEC1‐1‐7‐1 and CEC2‐5‐M‐4n (Table 2). In the case of ZYMV, resistance breaking was observed late in infection (25 dpi) in some T3 progeny, which was associated with low virus titre, mild symptoms and plant development similar to uninfected cucumber. However, silencing of eIF4E in melon showed resistance to ZYMV without resistance breaking (Rodríguez‐Hernández et al., 2012). The differences in the resistance observed here and in melon could be explained by the changes in eIF4E biology between cucumber and melon or by the differences in the ZYMV strains. ZYMV‐NAT‐Is (Gal‐On et al., 1992) was used in the current study, and ZYMV‐C71 was used in melon (Rodríguez‐Hernández et al., 2012). In addition, low ZYMV accumulation was described in some watermelon plants which have a natural single nucleotide polymorphism (SNP) in the eIF4E gene (Ling et al., 2009). Similarly, others have observed instances of resistance breaking as a result of eIF4E silencing in tomato infected with strains of Potato virus Y (PVY‐LYE84) and Pepper severe mosaic virus (Mazier et al., 2011).
Two explanations may account for the breaking of resistance in eif4e homozygous mutant plants. First, ZYMV may use eIF(iso)4E with less efficiency than eIF4E for replication and systemic movement, and therefore is able to cause mild symptoms. It has been shown that several potyviruses on different hosts can use eIF4E and eIF(iso)4E proteins (Duprat et al., 2002; Hwang et al., 2009; Lellis et al., 2002; Nicaise et al., 2003; Piron et al., 2010; Ruffel et al., 2002; Sato et al., 2005). In some cases, the potyvirus requires both genes, e.g. for the infection of Pepper veinal mottle virus in pepper with natural mutants of pvr2 and pvr6 (Ruffel et al., 2006). Second, a mutation may have occurred in the ZYMV genome which allowed interaction of ZYMV with eIF(iso)4E. A mutation breaking resistance was demonstrated in Lettuce mosaic virus potyvirus in lettuce (Abdul‐Razzak et al., 2009).
Interestingly, resistance breaking by ZYMV was not observed by inoculation with the natural aphid vector. This may imply that the few virus particles (0.5–3.2 per cell) (Moury et al., 2007) transmitted by aphids may not be sufficient to overcome the eif4e mutation, whereas several hundred virus particles are transmitted per cell by mechanical inoculation (Sacristán et al., 2003).
In the case of PRSV‐W, the resistance of T3 progenies of CEC1 and CEC2 was weaker than the resistance to ZYMV, and a low level of viral RNA could be detected in most plants (Fig. 7B). However, similar to ZYMV, in most eif4e mutant plants, a lower virus titre was associated with delayed mild symptom appearance and plant development was similar to that of uninfected cucumber. The accumulation of PRSV‐W in most of the homozygous eif4e mutants indicated that PRSV‐W can use both eIF4E and eIF(iso)4E for its life cycle, as demonstrated in pepper for Chilli veinal mottle virus (Hwang et al., 2009). In melon, it has been shown that eIF4E is not essential for PRSV‐W infection (Rodríguez‐Hernández et al., 2012), which implies functional differences in this gene between cucumber and melon with regard to PRSV‐W infection.
Homozygous eif4e mutants (CEC1 and CEC2) were susceptible to CMV, as observed by symptom development, similar to the report by Rodríguez‐Hernández et al. (2012) in melon silenced for eIF4E. The susceptibility of homozygous eif4e mutants (CEC1 and CEC2) to CGMMV was expected, as no tobamovirus resistance has been found so far in eIF4E and eIF(iso)4E mutants.
Conclusions
Here, we show for the first time that CRISPR/Cas9 is an efficient tool for genome editing in cucumber. Disruption of the eIF4E gene in cucumber by CRISPR/Cas9 sgRNA led to the development of virus‐resistant plants without otherwise affecting the plant genome. Three generations of backcrossing produced virus‐resistant plants free of genetic modification, and thus would be considered safe for human consumption and for release into the environment. We believe that this novel technology has the potential to expedite the development of pest resistance in many crops without the need for extensive backcrossing and genetic manipulation with wild sources of resistance.
Experimental Procedures
CRISPR/Cas9 binary construct design
We used the pRCS binary vector (Dafny‐Yelin and Tzfira, 2007), which contained 35S:Cas9‐AtU6:sgRNA‐PDS, where the Cas9 gene was optimized for plant codon usage (Li et al., 2013; Nekrasov et al., 2013). The nptII (kanamycin) selection marker gene, under the control of the 35S promoter and nos terminator, was cloned into the AscI site (Fig. S1, see Supporting Information).
eIF4E sgRNA design and cloning
The eIF4E gene (GenBank accession no. XM_004147349) of cucumber (Cucumis sativus) was selected as the target gene. Each target sequence of 20 nucleotides was designed according to the criteria described previously (Mali et al., 2013) upstream of the NGG trinucleotide, known as the protospacer adjacent motif (PAM). Two different sgRNA forward primers were designed for the eIF4E target gene (Table S1). Each primer contained a SalI site as part of the U6 Arabidopsis promoter (Fig. S1). The eIF4E target sequence, together with the sgRNA scaffold, was amplified using sgRNA1 or sgRNA2 as forward primer (Table S1) and a reverse primer of the PolIII‐terminator sequence that contained a HindIII site and pRCS‐35S:Cas9‐AtU6:sgRNA‐PDS as a template. The amplified DNAs (∼130 bp) were cloned into SalI and HindIII sites of the pRCS‐35S:Cas9‐AtU6:sgRNA‐nptII binary plasmid (Fig. S1). The clones obtained were confirmed by sequencing.
Agrobacterium‐mediated transformation
Agrobacterium tumefaciens‐mediated transformation of cucumber ‘Ilan’ (Zera‘im Gedera, Israel) was performed according to Gal‐On et al. (2005). Cut cotyledons without embryo were pre‐cultured for 1 day, followed by inoculation with A. tumefaciens EHA105 containing the CRISPR/cas9 constructs (pRCS‐35S:Cas9‐AtU6:CECsgRNA1 or CECsgRNA2). The cotyledon segments were transferred to a selective regeneration medium that contained 100 mg/L kanamycin. Shoots regenerated from explants were transferred to an elongation medium, followed by a rooting medium with 100 mg/L kanamycin. Well‐rooted plants were transferred to moist Jiffy 7 peat pellets and covered with transparent plastic boxes for hardening in a growth chamber under continuous white fluorescent light at 25 °C.
Transgenic plant growth conditions and propagation
Transgenic lines were transferred to coir medium (Pelemix Ltd. Ashkelon, Israel) 2–3 weeks post‐hardening and grown in glasshouse conditions under natural daylight at 26 ºC. Water and fertilizer (120 ppm of 5 : 3 : 8 NPK) were supplied twice daily by drip irrigation according to the size of the plants. T0 transgenic cucumber lines were hand pollinated with male flowers of the monoecious ‘Bet Alfa’, because the transformed ‘Ilan’ is gynoecious.
Genotyping and mutant verification
Genomic DNA was isolated from T0 transgenic and non‐transgenic cucumber plants by a Gen Elute™ Plant Genomic DNA Miniprep kit (Sigma Aldrich, St. Louis, MO, USA) and by the method of Dellaporta et al. (1983). The presence of the Cas9/sgRNA1/sgRNA2 transgene in T0 lines was confirmed by PCR using specific primers (Table S1). The transgenic lines were genotyped for indel polymorphisms using primers flanking sgRNA1 or sgRNA2 of the eIF4E target regions (Table S1). PCR products were digested with restriction enzymes BmgBI or BglII for sgRNA1 and sgRNA2, respectively. The digested products were separated on 1.5% agarose gel and the undigested PCR products were excised, purified and cloned into pJET1.2/blunt (Thermo Fisher Scientific, Waltham, MA, USA). Five colonies were sequenced to discriminate indel polymorphisms and the sequences were aligned to the intact eIF4E using the ClustalW BioEdit software program (Copyright©1997–2013, Tom Hall Ibis Biosciences, Carlsbad, CA, USA).
The evaluation of indel mutations in T1, T2 and T3 progeny seedlings was performed as described above for T0 by PCR with specific primers, restriction analysis and sequencing. Screening for mutations of 20‐nucleotide deletions was performed with specific primers (Table S1) flanking the 20‐nucleotide deletion. This primer can only bind to DNA having a 20‐nucleotide deletion and not a one‐nucleotide deletion.
Inoculation of plants with viruses
The following viruses were used for resistance analysis: ZYMV (accession no. EF062582) (Gal‐On, 2000); PRSV‐W (accession no. JF737858); CVYV (accession no. AY290865) (Martínez‐García et al., 2004); CMV Fny‐strain (accession no. D10538) (Rizzo and Palukaitis, 1990); and CGMMV (accession no. KF155232) (Reingold et al., 2015). Squash (Cucurbita pepo L. ‘Ma‘yan) plants were used as a source of inoculum of ZYMV, PRSV and CMV. Cucumber ‘Bet Alfa’ was used as a source of inoculum for CVYV and CGMMV. Cucumber seedlings at the cotyledon stage with small true leaves (about 3–5 days post‐emergence) were dusted with carborundum (320 mesh grit powder, Fisher Scientific, Springfield, NJ, USA) prior to mechanical inoculation with virus‐bearing sap (c. 1 : 10 ratio of g tissue : H2O) of ZYMV, PRSV‐W, CMV and CGMMV. CVYV inoculation was performed with whiteflies (Bemisia tabaci) exposed for 24 h to CVYV‐infected cucumber leaves, followed by 24‐h inoculation of cucumber seedlings with one true leaf (more than 10 whiteflies per seedling). Aphid inoculation of cucumber with ZYMV was performed with Aphis gossypii according to Gal‐On et al. (1992) with five to seven aphids per plant.
Evaluation of virus resistance
The response of the tested plants to virus infection was determined by visual monitoring of symptoms from 28 to 45 dpi following RT‐PCR for the presence of viral RNA.
Virus accumulation was determined by RT‐PCR and real‐time quantitative RT‐PCR. RNA samples were collected from the second and third leaves of cucumber (two leaf discs per plant). Total RNAs were extracted by a TRI‐REAGENT kit (Molecular Research Center, Inc., Cincinnati, OH, USA) and adjusted to the same concentration as prior to RT‐PCR, measured by a NanoDrop ND1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). First‐strand cDNA was synthesized from 2 µg of total RNA using a Verso™ cDNA Kit (Thermo Fisher Scientific, Epsom, UK) with oligo(dT) primer (100 pmol) for ZYMV‐, PRSV‐W‐ and CVYV‐inoculated plants and specific virus reverse primers for CMV and CGMMV analysis (Table S1). PCR conditions were 2 min at 94 °C, followed by 30 cycles of 30 s each at 94, 58 and 72 °C, and a final elongation step of 5 min at 72 °C. Quantitative PCRs were performed in a volume of 15 µL with 4 µL of diluted cDNA (1 : 4), 3 pmol of each primer and 7.5 µL of Absolute QPCR SYBR Green Mix (Thermo Scientific). Quantitative analysis was performed using Rotor‐Gene 3000 (Qiagen, Gaithersburg, MD, USA) with PCR conditions of 20 min at 95 °C (‘hot start’), followed by 40 cycles of 15 s at 96 °C, 15 s at 60 °C and 15 s at 72 °C. The relative expression level of gene accumulation was calculated using the ΔΔCT method normalized to the reference genes using Rotor Gene Series 3000 software version 1.7.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Map of the binary vector with Cas9‐sgRNA.
Fig. S2 Chromatogram of eIF4E mutants in representative T1 progeny plants of CEC1‐1 (A) and CEC2‐5 (B).
Table S1 List of primers and their applications.
Acknowledgements
The authors declare that they have no conflicts of interest with respect to this work. The authors are grateful to Dr Moshe Lapidot for performing the CVYV inoculations by whitefly, and Drs Victor Gaba and Harry S. Paris for critical reading of the manuscript. This research was supported by the Chief Scientist of the Israeli Ministry of Agriculture and Rural Development (Project 20‐10‐0039) to A.G.‐O., T.A. and A.S. Contribution from the Agricultural Research Organization, The Volcani Center, Bet‐Dagan, Israel, No. 552/15.
References
- Abdul‐Razzak, A. , Guiraud, T. , Peypelut, M. , Walter, J. , Houvenaghel, M.‐C. , Candresse, T. , Le Gall, O. and German‐Retana, S. (2009) Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E‐mediated resistance against Lettuce mosaic potyvirus . Mol. Plant Pathol. 10, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, Z. , Abulfaraj, A. , Idris, A. , Ali, S. , Tashkandi, M. and Mahfouz, M.M. (2015) CRISPR/Cas9‐mediated viral interference in plants. Genome Biol. 16, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadœuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of potato virus Y confer virulence on the pvr23 resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Baltes, N.J. , Hummel, A.W. , Konecna, E. , Cegan, R. , Bruns, A.N. , Bisaro, D.M. and Voytas, D.F. (2015) Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants, 1, doi: 10.1038/nplants.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj, K. , Chaparro‐Garcia, A. , Kamoun, S. and Nekrasov, V. (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods, 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhaj, K. , Chaparro‐Garcia, A. , Kamoun, S. , Patron, N.J. and Nekrasov, V. (2015) Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. [DOI] [PubMed] [Google Scholar]
- Bortesi, L. and Fischer, R. (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. [DOI] [PubMed] [Google Scholar]
- Brooks, C. , Nekrasov, V. , Lippman, Z.B. and Eck, J.V. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR‐associated9 system. Plant Physiol. 166, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo, F. and Palukaitis, P. (2014) Transgenic resistance. Adv. Virus Res. 90, 35–146. [DOI] [PubMed] [Google Scholar]
- Dafny‐Yelin, M. and Tzfira, T. (2007) Delivery of multiple transgenes to plant cells. Plant Physiol. 145, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L. , Wood, J. and Hicks, J.B. (1983) A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- Diaz‐Pendon, J.A. , Truniger, V. , Nieto, C. , Garcia‐Mas, J. , Bendahmane, A. and Aranda, M.A. (2004) Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 5, 223–233. [DOI] [PubMed] [Google Scholar]
- Duprat, A. , Caranta, C. , Revers, F. , Menand, B. , Browning, K.S. and Robaglia, C. (2002) The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. [DOI] [PubMed] [Google Scholar]
- Feng, Z. , Zhang, B. , Ding, W. , Liu, X. , Yang, D.‐L. , Wei, P. , Cao, F. , Zhu, S. , Zhang, F. , Mao, Y. and Zhu, J.‐K. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Foden, J.A. , Khayter, C. , Maeder, M.L. , Reyon, D. , Joung, J.K. and Sander, J.D. (2013) High‐frequency off‐target mutagenesis induced by CRISPR‐Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal‐On, A. (2000) A point mutation in the FRNK motif of the Potyvirus helper component‐protease gene alters symptom expression in cucurbits and elicits protection against the severe homologous virus. Phytopathology, 90, 467–473. [DOI] [PubMed] [Google Scholar]
- Gal‐On, A. , Antignus, Y. , Rosner, A. and Raccah, B. (1992) A zucchini yellow mosaic virus coat protein gene mutation restores aphid transmissibility but has no effect on multiplication. J. Genet. Virol. 73, 2183–2187. [DOI] [PubMed] [Google Scholar]
- Gal‐On, A. , Wolf, D. , Antignus, Y. , Patlis, L. , Ryu, K.H. , Min, B.E. , Pearlsman, M. , Lachman, O. , Gaba, V. , Wang, Y. , Shiboleth, Y.M. , Yang, J. and Zelcer, A. (2005) Transgenic cucumbers harboring the 54‐kDa putative gene of Cucumber fruit mottle mosaic tobamovirus are highly resistant to viral infection and protect nontransgenic scions from soil infection. Transgenic Res. 14, 81–93. [DOI] [PubMed] [Google Scholar]
- Gómez, P. , Rodríguez‐Hernández, A.M. , Moury, B. and Aranda, M.A. (2009) Genetic resistance for the sustainable control of plant virus diseases: breeding, mechanisms and durability. Eur. J. Plant Pathol. 125, 1–22. [Google Scholar]
- Hébrard, E. , Pinel‐Galzi, A. , Bersoult, A. , Siré, C. and Fargette, D. (2006) Emergence of a resistance‐breaking isolate of Rice yellow mottle virus during serial inoculations is due to a single substitution in the genome‐linked viral protein VPg. J. Genet. Virol. 87, 1369–1373. [DOI] [PubMed] [Google Scholar]
- Hwang, J. , Li, J. , Liu, W.‐Y. , An, S.‐J. , Cho, H. , Her, N.H. , Yeam, I. , Kim, D. and Kang, B.‐C. (2009) Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells, 27, 329–336. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J. , Hellen, C.U.T. and Pestova, T.V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, T.B. , LaFayette, P.R. , Schmitz, R.J. and Parrott, W.A. (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J. and Laliberté, J.‐F. (2011) The genome‐linked protein VPg of plant viruses—a protein with many partners. Curr. Opin. Virol. 1, 347–354. [DOI] [PubMed] [Google Scholar]
- Jones, H.D. (2015) Regulatory uncertainty over genome editing. Nat. Plants, 1, DOI: 10.1038/NPLANTS.2014.11. [DOI] [PubMed] [Google Scholar]
- Kang, B.‐C. , Yeam, I. and Jahn, M.M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Le Gall, O. , Aranda, M.A. and Caranta, C. (2011) Plant resistance to viruses mediated by translation initiation factors. In: Caranta C., Aranda M.A., Tepfer M., Lopez‐Moya J., editors. Recent Advances in Plant Virology. Caister Academic Press; Wymondham, Norfolk, VA, USA: 2011. pp. 177–194. [Google Scholar]
- Lei, Y. , Lu, L. , Liu, H.‐Y. , Li, S. , Xing, F. and Chen, L.‐L. (2014) CRISPR‐P: a web tool for synthetic single‐guide RNA design of CRISPR‐system in plants. Mol. Plant, 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during Potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Léonard, S. , Plante, D. , Wittmann, S. , Daigneault, N. , Fortin, M.G. and Laliberté, J.‐F. (2000) Complex formation between Potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.‐F. , Norville, J.E. , Aach, J. , McCormack, M. , Zhang, D. , Bush, J. , Church, G.M. and Sheen, J. (2013) Multiplex and homologous recombination‐mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, K.‐S. , Harris, K.R. , Meyer, J.D.F. , Levi, A. , Guner, N. , Wehner, T.C. , Bendahmane, A. and Havey, M.J. (2009) Nonsynonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus. Theor. Appl. Genet. 120, 191–200. [DOI] [PubMed] [Google Scholar]
- Liu, L. and Fan, X.‐D. (2014) CRISPR–Cas system: a powerful tool for genome engineering. Plant Mol. Biol. 85, 209–218. [DOI] [PubMed] [Google Scholar]
- Mali, P. , Aach, J. , Stranges, P.B. , Esvelt, K.M. , Moosburner, M. , Kosuri, S. , Yang, L. and Church, G.M. (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez‐García, B. , Marco, C.F. , Goytia, E. , López‐Abella, D. , Serra, M.T. , Aranda, M.A. and López‐Moya, J.J. (2004) Development and use of detection methods specific for cucumber vein yellowing virus (CVYV). Eur. J. Plant Pathol. 110, 811–821. [Google Scholar]
- Maule, A.J. , Caranta, C. and Boulton, M.I. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol. Plant Pathol. 8, 223–231. [DOI] [PubMed] [Google Scholar]
- Mazier, M. , Flamain, F. , Nicolaï, M. , Sarnette, V. and Caranta, C. (2011) Knock‐down of both eIF4E1 and eIF4E2 genes confers broad‐spectrum resistance against potyviruses in tomato. PLoS One, 6, e29595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Fabre, F. and Senoussi, R. (2007) Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA, 104, 17 891–17 896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K.S. , Shimada, R. , Choi, S.‐H. , Yamamoto, H. , Shao, J. and Uyeda, I. (2010) Involvement of the P1 cistron in overcoming eIF4E‐mediated recessive resistance against clover yellow vein virus in pea. Mol. Plant–Microbe Interact. 23, 1460–1469. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Staskawicz, B. , Weigel, D. , Jones, J.D.G. and Kamoun, S. (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA‐guided endonuclease. Nat. Biotechnol. 31, 691–693. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , German‐Retana, S. , Sanjuán, R. , Dubrana, M.‐P. , Mazier, M. , Maisonneuve, B. , Candresse, T. , Caranta, C. and LeGall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus lettuce mosaic virus. Plant Physiol. 132, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron, F. , Nicolaï, M. , Minoïa, S. , Piednoir, E. , Moretti, A. , Salgues, A. , Zamir, D. , Caranta, C. and Bendahmane, A. (2010) An induced mutation in tomato eif4e leads to immunity to two Potyviruses. PLoS One, 5, e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reingold, V. , Lachman, O. , Blaosov, E. and Dombrovsky, A. (2015) Seed disinfection treatments do not sufficiently eliminate the infectivity of Cucumber green mottle mosaic virus (CGMMV) on cucurbit seeds. Plant Pathol. 64, 245–255. [Google Scholar]
- Revers, F. and García, J.A. (2015) Chapter three—Molecular biology of Potyviruses. In: Advances in Virus Research (Maramorosch K. and Mettenleiter T.C., eds), pp. 101–199. New York: Academic Press. [WorldCat] [DOI] [PubMed] [Google Scholar]
- Rizzo, T.M. and Palukaitis, P. (1990) Construction of full‐length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222, 249–256. [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Hernández, A.M. , Gosalvez, B. , Sempere, R.N. , Burgos, L. , Aranda, M.A. and Truniger, V. (2012) Melon RNA interference (RNAi) lines silenced for Cm‐eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel, S. , Dussault, M.‐H. , Palloix, A. , Moury, B. , Bendahmane, A. , Robaglia, C. and Caranta, C. (2002) A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Ruffel, S. , Gallois, J.‐L. , Moury, B. , Robaglia, C. , Palloix, A. and Caranta, C. (2006) Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Genet. Virol. 87, 2089–2098. [DOI] [PubMed] [Google Scholar]
- Sacristán, S. , Malpica, J.M. , Fraile, A. and García‐Arenal, F. (2003) Estimation of population bottlenecks during systemic movement of tobacco mosaic virus in tobacco plants. J. Virol. 77, 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon, H. (2015) Plant translation factors and virus resistance. Viruses, 7, 3392–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M. , Nakahara, K. , Yoshii, M. , Ishikawa, M. and Uyeda, I. (2005) Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 579, 1167–1171. [DOI] [PubMed] [Google Scholar]
- Sternberg, S.H. , Redding, S. , Jinek, M. , Greene, E.C. and Doudna, J.A. (2014) DNA interrogation by the CRISPR RNA‐guided endonuclease Cas9. Nature, 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger, V. and Aranda, M.A. (2009) Recessive resistance to plant viruses In: Advances in Virus Research (Loebenstein G. and Carr J.P., eds), pp. 119–231. Wymondham, Norfolk: Caister Academic Press. [DOI] [PubMed] [Google Scholar]
- Wang, A. and Krishnaswamy, S. (2012) Eukaryotic translation initiation factor 4E‐mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, S. , Chatel, H. , Fortin, M.G. and Laliberté, J.‐F. (1997) Interaction of the viral protein genome linked of turnip mosaic Potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two‐hybrid system. Virology, 234, 84–92. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Zhang, J. and Yang, Y. (2014) Genome‐wide prediction of highly specific guide RNA spacers for CRISPR–Cas9‐mediated genome editing in model plants and major crops. Mol. Plant, 7, 923–926. [DOI] [PubMed] [Google Scholar]
- Xu, R.‐F. , Li, H. , Qin, R.‐Y. , Li, J. , Qiu, C.‐H. , Yang, Y.‐C. , Ma, H. , Li, L. , Wei, P.‐C. and Yang, J.‐B. (2015) Generation of inheritable and “transgene clean” targeted genome‐modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5, doi: 10.1038/srep11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. , Yang, L. , Zhang, H. , Xu, N. and Zhu, J.‐K. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website.
Fig. S1 Map of the binary vector with Cas9‐sgRNA.
Fig. S2 Chromatogram of eIF4E mutants in representative T1 progeny plants of CEC1‐1 (A) and CEC2‐5 (B).
Table S1 List of primers and their applications.


