Summary
Dollar spot, caused by Sclerotinia homoeocarpa, is a prevalent turfgrass disease, and the fungus exhibits widespread fungicide resistance in North America. In a previous study, an ABC‐G transporter, ShatrD, was associated with practical field resistance to demethylation inhibitor (DMI) fungicides. Mining of ABC‐G transporters, also known as pleiotropic drug resistance (PDR) transporters, from RNA‐Seq data gave an assortment of transcripts, several with high sequence similarity to functionally characterized transporters from Botrytis cinerea, and others with closest blastx hits from Aspergillus and Monilinia. In addition to ShatrD, another PDR transporter showed significant over‐expression in replicated RNA‐Seq data, and in a collection of field‐resistant isolates, as measured by quantitative polymerase chain reaction. These isolates also showed reduced sensitivity to unrelated fungicide classes. Using a yeast complementation system, we sought to test the hypothesis that this PDR transporter effluxes DMI as well as chemically unrelated fungicides. The transporter (ShPDR1) was cloned into the Gal1 expression vector and transformed into a yeast PDR transporter deletion mutant, AD12345678. Complementation assays indicated that ShPDR1 complemented the mutant in the presence of propiconazole (DMI), iprodione (dicarboximide) and boscalid (SDHI, succinate dehydrogenase inhibitor). Our results indicate that the over‐expression of ShPDR1 is correlated with practical field resistance to DMI fungicides and reduced sensitivity to dicarboximide and SDHI fungicides. These findings highlight the potential for the eventual development of a multidrug resistance phenotype in this pathogen. In addition, this study presents a pipeline for the discovery and validation of fungicide resistance genes using de novo next‐generation sequencing and molecular biology techniques in an unsequenced plant pathogenic fungus.
Keywords: dicarboximide, DMI, multidrug resistance, PDR transporter, RNA‐Seq, SDHI
Introduction
Sclerotinia homoeocarpa (F.T. Bennett) causes dollar spot of turfgrass, which is the most prevalent and economically significant turfgrass disease (Vargas et al., 1992). Multiple fungicide applications are required to control dollar spot throughout the growing season (Smiley et al., 2005); however, the repeated use of fungicides has led to the development of fungicide resistance. Resistance to the benzimidazole and dicarboximide fungicide classes has been confirmed in field isolates of S. homoeocarpa (Cole et al., 1968; Detweiler et al., 1983). Recent reports have also indicated that S. homoeocarpa has developed location‐specific resistance to demethylation inhibitor (DMI) fungicides in the USA (Jo et al., 2006; Miller et al., 2002; Popko et al., 2012; Putman et al., 2010).
In the filamentous fungi, three genetic mechanisms of fungicide resistance have been widely reported. For site‐specific fungicides, in general, reducing affinity of the fungicide to the target site by non‐synonymous polymorphism in the target gene is known to confer resistance (Ma and Michailides, 2005). In addition, over‐expression of the target gene can be linked to resistance (Hamamoto et al., 2000). Another commonly reported mechanism is efflux of fungicide molecules by the activity of ATP‐binding cassette (ABC) efflux transporters, which are found in all eukaryotes, and can translocate compounds across cell membranes using the hydrolysis of ATP as an energy source (Del Sorbo et al., 2000; Driessen et al., 2000). The ABC transporters are thought to evolve through gene duplication events and are grouped into diverse subfamilies on the basis of protein domain organization (Kovalchuk and Driessen, 2010). In fungi, full size transporters in the ‘G’ subfamily (ABC‐G transporters) contain a nucleotide‐binding domain and transmembrane domain in duplicate, which is denoted by [NBD‐TMD]2 (Lamping et al., 2010). These transporters of toxic molecules, also known as pleiotropic drug resistance (PDR) transporters, are known to have differential specificity for fungicides, and some have been reported to mediate what has been termed a multidrug resistance (MDR) phenotype in Botrytis cinerea (Kretschmer et al., 2009). Although not necessarily analogous to MDR reported for human pathogenic bacteria and yeasts, this phenotype involves reduced in vitro sensitivity to multiple unrelated fungicide classes and has been proposed to negatively impact the efficacy of multiple fungicides for controlling plant pathogen populations (Leroux et al., 2013; Leroux and Walker, 2011).
Previously, Hulvey et al. (2012) utilized transcriptomic and molecular tools to investigate determinants of practical field resistance (PFR) to DMI fungicides in S. homoeocarpa. This work demonstrated that the over‐expression of the PDR transporter ShatrD was strongly associated with PFR to a DMI fungicide, whereas over‐expression of the gene target of DMIs, ShCYP51B, was a minor factor for PFR. Recently, we mined RNA‐Seq data for the complement of PDR transporter genes, and identified another with statistically significant over‐expression. This gene shares significant sequence similarity with the ABC transporters PMR1 from Penicillium digitatum and atrE from Aspergillus nidulans, both suspected determinants of DMI fungicide resistance (Hayashi et al., 2002; Nakaune et al., 1998).
We present evidence to support an MDR phenotype in field isolates of S. homoeocarpa from New England, similar to that presented by Kretschmer et al. (2009) for B. cinerea. We also demonstrate that decreased sensitivity to multiple fungicides is partially attributable to the newly described PDR transporter gene, ShPDR1, mined together with other PDR transporters from RNA‐Seq data. In order to confirm the involvement of ShPDR1 in reduced fungicide sensitivity, we utilized a yeast complementation assay to test the hypothesis that the cloned full‐length cDNA of ShPDR1 will complement a yeast mutant deficient for ABC efflux transporters. In summary, we show the utility of combining RNA‐Seq and molecular biology techniques for the discovery of novel fungicide resistance determinants, and discuss the implications of our findings.
Results
Sensitivity of S. homoeocarpa isolates to propiconazole, iprodione and boscalid
In order to investigate the MDR phenotype of S. homoeocarpa field isolates, the in vitro sensitivity of isolates to unrelated fungicide classes was tested. The in vitro sensitivity values determined using an initial panel of eight isolates for minimal medium (MM) amended with propiconazole, iprodione and boscalid were in the ranges 0.02–0.75, 0.40–0.82 and 3182.4–7432.7 μg/mL, respectively (Table 1). The propiconazole and iprodione EC50 and boscalid EC95 values of all insensitive initial isolates were higher than those of sensitive isolates, and the values of SMI27 were the lowest among the insensitive isolates. The mean EC50 and EC95 values of propiconazole, iprodione and boscalid for the group of four PFR isolates (HRI1–4) from Hickory Ridge Country Club (HRCC) were significantly higher than those for the group of four sensitive isolates (HRS1–4) (P < 0.0001). The group of four PFR isolates (HFI1–4) from Hartford Golf Club (HGC) showed significantly higher mean propiconazole and iprodione EC50 and boscalid EC95 values than those for the group of four sensitive isolates (HFS1–4) (P < 0.0001) (Table 1).
Table 1.
Sclerotinia homoeocarpa field isolates used in this study
| Isolatesb | Propiconazole | Propiconazole | Iprodione | Boscalid |
|---|---|---|---|---|
| Sensitivityc | EC50 (μg/mL) | EC50 (μg/mL) | EC95 (μg/mL) | |
| HRI11 | I | 0.75 | 0.76 | 7432.7 |
| HFI40 | I | 0.60 | 0.76 | 6599.5 |
| WBI7 | I | 0.54 | 0.82 | 7069.1 |
| SMI27 | I | 0.20 | 0.55 | 4895.7 |
| HRS10 | S | 0.02 | 0.53 | 3499.6 |
| HFS35 | S | 0.03 | 0.54 | 3182.4 |
| JTS30 | S | 0.02 | 0.52 | 3695.3 |
| SMS23 | S | 0.07 | 0.40 | 3564.6 |
| HRS1‐4 | S | 0.05 | 0.41 | 3819.1 |
| HRI1‐4 | I (PFR) | 0.74 | 0.83 | 8891.8 |
| P valued | a | a | a | |
| HFS1‐4 | S | 0.03 | 0.50 | 3516.5 |
| HFI1‐4 | I (PFR) | 0.74 | 0.80 | 6629.9 |
| P valuee | a | a | a |
Significant at P < 0.0001.
Isolates are named according to Hulvey et al. (2012). Location abbreviations: HF, Hartford Golf Club; HR, Hickory Ridge Country Club; JT, Joseph Troll Turf Research Center at University of Massachusetts, Amherst, MA, USA; SM, Shuttle Meadow Country Club; WB, Wintonbury Hills Golf Club.
Propiconazole sensitivity previously determined by Hulvey et al. (2012): I, insensitive; PFR, practical field resistance; S, sensitive.
Statistical differences between mean EC50 and EC95 values of sensitive and insensitive groups from Hickory Ridge Country Club.
Statistical differences between mean EC50 and EC95 values of sensitive and insensitive groups from Hartford Golf Club.
RNA‐Seq data and ABC‐G transporters from the transcriptomic data of S. homoeocarpa
The RNA‐Seq method allows for the profiling of the total complement of expressed transcripts in an organism and, in this study, was used to define the PDR transportome of S. homoeocarpa and which PDR transcripts showed differential expression following exposure to a DMI fungicide. From the assembled RNA‐Seq data, 12 494 contigs over 500 bp were generated. For each library of RNA‐Seq data, over 30 million reads were mapped back in pairs for both biological replicates of treated and untreated samples of HRS10 and HRI11. Of the assembled contigs, seven were found to represent PDR transporters (Table 2). Six of the seven contigs had a member of the Sclerotiniaceae as a top blastx hit, whereas ShPDR1 (contig 6887) had a top blast hit from Aspergillus kawachii (Trichocomaceae). Of the expression data, only two contigs, one representing ShatrD (contig 4492) and the other ShPDR1, were found to show significant (P < 0.05) over‐expression in HRI11 as determined by two‐tailed P values derived from Baggerly's test (Table 2). The ShatrD and ShPDR1 genes showed high over‐expression in both untreated and treated samples in the DMI‐insensitive isolate HRI11. Conversely, only one of the identified PDR transporters (contig 2174) showed significant, although numerically low, over‐expression (less than three‐fold) in the DMI‐sensitive isolate HRS10 (Table 2).
Table 2.
Summary of transcriptome contigs encoding pleiotropic drug resistance (PDR) transporters from S clerotinia homoeocarpa
| Contig # | Contig length (bp) | Top blastx hit in Botrytis cinerea † (e value, % query coverage, % identity, # of amino acid matches/total amino acids aligned) | Top blastx hit (e value, % query coverage, % identity, # of amino acid matches/total amino acids aligned) | Baggerly's test, weighted proportions fold difference‡ | |
|---|---|---|---|---|---|
| Untreated | Treated | ||||
| 2174 | 4063 | BcatrA, CCD52170.1 (0, 99, 83, 1124/1358) | Botrytis cinerea, CCD52170.1 (0, 99, 83, 1124/1358) | −2.3*** | −2.3NS |
| 4492 | 7129 | BcatrD, CAC41639.1 (0, 87, 84, 1242/1482) | Botrytis cinerea, CAC41639.1 (0, 87, 84, 1242/1482) | 8.7* | 7.3* |
| 4985 | 5241 | BMR1, BAA93677.1 (0, 85, 85, 1257/1485) | Sclerotinia borealis, ESZ99265.1 (0, 87, 86, 1262/1476) | −1.1NS | 1.3NS |
| 6230 | 2984 | BcatrB, CCD54655.1 (0, 99, 83, 847/1020) | Monilinia fructicola, AAL80009.1 (0, 99, 84, 860/1020) | −1.6NS | −1.4NS |
| 6887 | 5223 | BcatrD, CAC41639.1 (0, 83, 64, 937/1471) | Aspergillus kawachii, GAA89082.1 (0, 87, 77, 1143/1485) | 11.0*** | 22.5NS |
| 7104 | 2804 | Unnamed, BAC67160.1 (0, 89, 87, 728/837) | Sclerotinia borealis, ESZ91198.1 (0, 91, 85, 733/858) | 1.0NS | 2.0NS |
| 8448 | 2401 | Unnamed, EMR87975.1 (0, 89, 80, 580/723) | Sclerotinia borealis, ESZ91198.1 (0, 88, 82, 586/716) | 1.6NS | 3.4NS |
*, *** and NS refer to significance at P ≤ 0.05, P ≤ 0.001 and not significant, respectively.
†Top blast hits are denoted by either gene name in B. cinerea as in the case of functionally characterized genes, or GenBank accession number as in the case of uncharacterized genes.
‡Fold differences between HRS10 and HRI11 isolates were calculated with HRS10 as the calibrator; therefore, positive values mean greater than HRS10, and negative values mean lower than HRS10.
Sequence and phylogenetic analyses
In order to characterize the PDR transporters identified from the assembled transcripts, we utilized blast searches, domain characterization of the encoded amino acids and phylogenetic analysis with other characterized PDR transporters. blastx search results of ShPDR1 and other PDR transporters mined from the transcriptome data are presented in Table 2. Conserved domain searches confirmed that ShPDR1, ShatrD and three other PDR transporters encoded two NBDs, two ABC2 domains and a PDR domain, as expected (Lamping et al., 2010). The other two were partial sequences, but both included ABC2 domains, indicative of PDR transporters, and top blast hits that are PDR transporters. Phylogenetic analysis indicated that ShPDR1, AnatrE, PMR1, PMR4 and an unnamed PDR transporter from A. kawachii cluster together in a group supported by a bootstrap value of 100% (Fig. 1).
Figure 1.

Neighbour‐joining phylogenetic tree of amino acid sequences of pleiotropic drug resistance (PDR) transporters from the dataset of Hulvey et al. (2012), including five additional full‐length PDR transporters from Sclerotinia homoeocarpa (ShPDR1, ShPDR1 deletion, contigs 2174, 4985 and 6230), Penicillium digitatum PMR3 and PMR4, and Aspergillus kawachii PDR transporter (Lamping et al., 2010). The scale bar equals the proportion of amino acid substitutions.
Comparison of sequences obtained from polymerase chain reaction (PCR) and sequencing of genomic DNA and from RNA‐Seq data revealed that the 4572 bp of ShPDR1 is interrupted with one intron (78 bp). Single‐nucleotide polymorphisms (SNPs) from comparison with the JTS30 sequence were detected in the coding region of ShPDR1 at the following base pair positions: +487, +490, +1335, +1638 and +3825 for WBI7 and HFI40; +4181 for SMS23 and SMI27. Three non‐synonymous SNPs (+487, +490 and +4181) were identified (T163A, V163I and S1394T, respectively). The sequences of the coding region in the sensitive isolate HRS10 revealed an 81‐bp deletion from +1069 to +1149. In the upstream sequence of ShPDR1, an SNP was detected in one position +325 for WBI7 and HFI40; however, no promoters were detected in the upstream region. The ShPDR1 coding sequences from HRS10 and HRI11 and upstream region of ShPDR1 from JTS30 were deposited in GenBank with accession numbers KJ128076–8.
Quantitative PCR analysis of ShPDR1 for the initial panel of eight isolates and PFR isolates
ShPDR1 expression of the initial panel of eight isolates and PFR isolates was assayed to investigate the possible involvement of the over‐expression of ShPDR1 in reduced sensitivity to propiconazole, iprodione and boscalid. Significant differences (P < 0.0001) in constitutive relative expression (RE) values of ShPDR1 were found between the group of four insensitive isolates from the initial panel of eight isolates [mean ± standard error (SE) = 22.8 ± 1.2] and the group of four sensitive isolates (mean ± SE = 3.1 ± 1.2). Three insensitive isolates (HRI11, HFI40 and WBI27) significantly increased RE of ShPDR1 after exposure to propiconazole, iprodione and boscalid, but one insensitive isolate (SMI27) and four sensitive isolates did not increase RE of ShPDR1 in the presence of the three fungicides, except for SMS23 in response to propiconazole (Fig. 2).
Figure 2.
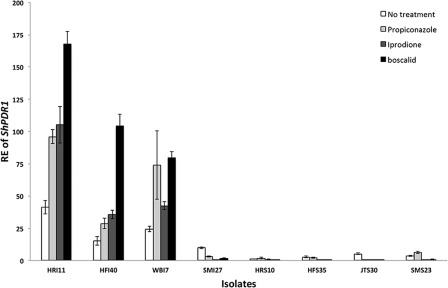
Relative expression (RE) of ShPDR1 in the initial panel of eight isolates from five sites in New England in the absence and presence of propiconazole (0.1 μg/mL), iprodione (1 μg/mL) and boscalid (10 μg/mL) for 1 h. All bars (white, constitutive RE; light grey, propiconazole‐induced RE; dark grey, iprodione‐induced RE; black, boscalid‐induced RE) represent mean RE values. Error bars indicate one standard error of the mean. Isolates with an ‘I’ indicate propiconazole insensitive and with an ‘S’ represent propiconazole sensitive.
The differences in the constitutive RE values of ShPDR1 were highly significant (P < 0.0001) between the group of PFR isolates (mean ± SE = 29.8 ± 1.7) and the group of sensitive isolates (mean ± SE = 3.4 ± 1.7) from HRCC. The differences in the constitutive RE values of ShPDR1 were also highly significant (P < 0.0001) between the group of PFR isolates (mean ± SE = 17.0 ± 0.9) and the group of sensitive isolates (mean ± SE = 5.3 ± 0.9) from HGC (Fig. 3).
Figure 3.
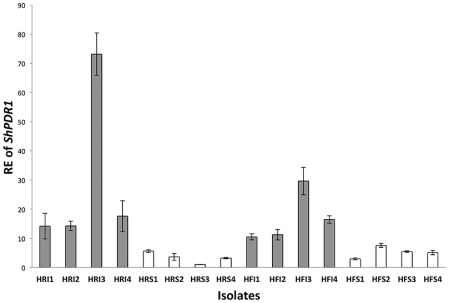
Relative expression (RE) values for ShPDR1 in practical field‐resistant (PFR) and sensitive Sclerotinia homoeocarpa isolates from Hickory Ridge Country Club and Hartford Golf Club. All bars (grey, constitutive RE of PFR isolates from propiconazole applied plots; white, constitutive RE of sensitive isolates from control plots) represent mean RE values. Error bars indicate one standard error of the mean.
Quantitative PCR analysis of ShPDR1 and ShatrD at different concentrations of propiconazole
The gene expression of the two ABC transporters in two DMI‐insensitive isolates at different doses of propiconazole was assessed to help gain a better understanding of the role of the two ABC transporters in DMI resistance. The induced RE values of ShPDR1 for both isolates HRI11 and HFI40 (HRI11, mean ± SE = 1321.4 ± 78.4; HFI40, mean ± SE = 679.0 ± 48.7) were significantly higher than the RE values of ShatrD for the isolates (HRI11, mean ± SE = 265.2 ± 78.4; HFI40, mean ± SE = 84.0 ± 48.7) in the presence of propiconazole at 5 μg/mL (P < 0.0001). Significant differences (P < 0.0001) were found between the induced RE values of ShPDR1 and ShatrD after treatment of propiconazole at 0.1 μg/mL for HRI11 and 1 μg/mL for HFI40. No differences were found for HRI11 between the RE values of ShPDR1 and ShatrD before and after treatment of propiconazole at 1 μg/mL, and for HFI40 between the RE values of the two genes in the absence and presence of propiconazole at 0.1 μg/mL (Fig. 4).
Figure 4.
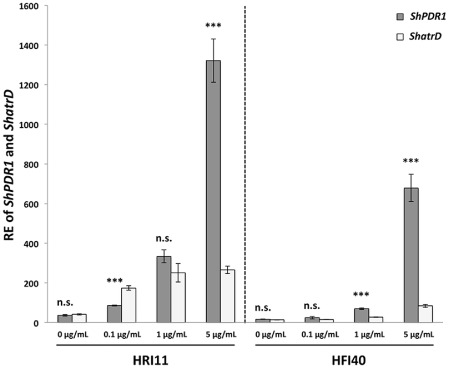
Relative expression (RE) of ShPDR1 and ShatrD in two insensitive Sclerotinia homoeocarpa isolates (HRI11 and HFI40) in the absence and presence of propiconazole (0.1, 1 and 5 μg/mL) for 1 h. All bars (grey, RE of ShPDR1; white, RE of ShatrD) represent mean RE values. Error bars indicate one standard error of the mean. Significant differences between RE values of ShPDR1 and ShatrD are indicated: n.s., not significant; ***, significant at P < 0.0001.
Linear regression of ShPDR1 expression and EC50 values for propiconazole and iprodione and EC95 values for boscalid
The linear regression analysis was conducted to examine whether the constitutive expression of ShPDR1 is correlated with the sensitivity to the three fungicides. The linear regression of the log10‐transformed mean RE values for the constitutive expression of ShPDR1 and EC50 values for propiconazole and iprodione showed a significant relationship (P < 0.0001) (Fig. 5A, B). In addition, the linear relationship between log10 RE of ShPDR1 and EC95 values of boscalid was highly significant (P < 0.0001) (Fig. 5C).
Figure 5.
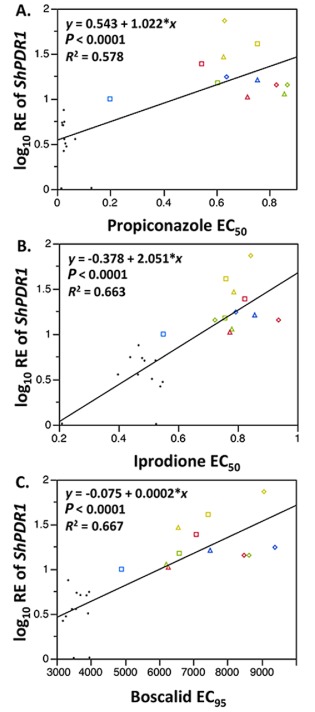
Relationship between log10 relative expression (RE) values of ShPDR1 and in vitro sensitivities to propiconazole (A), iprodione (B) or boscalid (C) (as measured by EC50 for propiconazole and iprodione and EC95 for boscalid) for 24 isolates from New England. The notation of isolates was according to Hulvey et al. (2012) [triangles, practical field‐resistant (PFR) isolates from Hartford Golf Club; diamonds, PFR isolates from Hickory Ridge Country Club; squares, insensitive isolates from four sites; black dots, sensitive isolates from five sites]. Each coloured shape indicates the same isolates for (A), (B) and (C).
Heterologous expression of ShPDR1 in Saccharomyces cerevisiae
The hypersensitive mutant strain AD12345678 (AD1‐8) of S. cerevisiae (Decottignies et al., 1998) was transformed with full‐length ShPDR1 cDNA cloned in the yeast expression vector pYES2 in order to test whether ShPDR1 is involved in decreased sensitivity to multiple fungicides in yeast. A clear qualitative difference in fungicide sensitivity between two ShPDR1 transformants (AD1‐8:PDR1‐1 and AD1‐8:PDR1‐2) and the control transformant (AD1‐8‐pYES2) was observed with propiconazole, iprodione and boscalid (Fig. 6).
Figure 6.
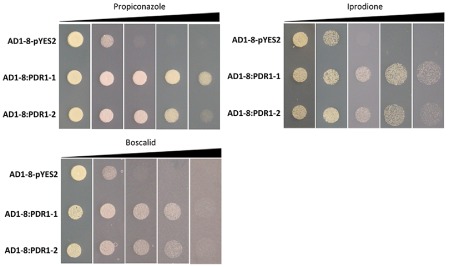
Complementation of Saccharomyces cerevisiae strain AD1‐8 with ShPDR1 from Sclerotinia homoeocarpa. Sensitivity assays of the transformants were conducted on bacto‐yeast nitrogen base (YNB) agar medium lacking uracil, containing 2% galactose and amended with increasing concentrations of propiconazole, iprodione and boscalid fungicides.
Discussion
This study provides key pieces of evidence confirming, for the first time in S. homoeocarpa, that the over‐expression of a PDR transporter is correlated with reduced sensitivity to multiple fungicide classes. This evidence includes constitutive and induced over‐expression of ShPDR1 in a diverse panel of field‐resistant isolates (Figs 2 and 3), high correlation of fungicide sensitivity with RE of ShPDR1 (Fig. 5), and ShPDR1 complementation of the yeast transporter mutant AD1‐8 for growth in the presence of different fungicide classes (Fig. 6). Previously, over‐expression of ShCYP51B and ShatrD has been reported for S. homoeocarpa isolates exhibiting PFR (Hulvey et al., 2012). Taken together, the observations of Hulvey et al. (2012) and of the current study illustrate that insensitivity to DMI fungicides in S. homoeocarpa is governed by multiple genes (multigenic), as opposed to monogenic resistance, which would entail only polymorphism and/or over‐expression of the gene target of DMI fungicides, ShCYP51B. Recent RNA‐Seq studies on pesticide resistance in bedbug (Cimex lectularius) and monoterpene resistance in the fungus Grossmania have suggested that there are certainly additional genes from multiple gene families that are driving this phenotype in S. homoeocarpa (Mamidala et al., 2012; Wang et al., 2013). Further studies involving RNA‐Seq data and the methods presented here will seek to verify additional novel genes and gene families underpinning both qualitaitve and quatitative resistance to multiple fungicides.
In the human pathogenic fungus, Candida albicans, an MDR phenotype is routinely reported, when prolonged treatments of the DMI fungicide fluconazole can select for strains showing constitutive over‐expression of the PDR transporters CDR1 and CDR2 (Morschhäuser et al., 2007). The activity of these transporters also renders the pathogen resistant to additional fungicide classes. A similar trend has also been observed in plant pathogenic fungi, whereby DMI‐resistant strains exhibiting over‐expression of PDR transporters show decreased sensitivity to multiple fungicide classes (Nakaune et al., 1998). Indeed, the isolates tested in this study were from the populations repeatedly exposed to DMIs and, interestingly, the PFR isolates were collected before application of boscalid at HRCC and HGC. A similar phenomenon has been reported for DMI resistance and boscalid insensitivity in a plant pathogen Monilinia fructicola by Chen et al. (2013). Chen et al. (2013) suggested that over‐expression of a multidrug transporter is linked to insensitivity to both propiconazole and boscalid, as boscalid resistance was not accompanied by target site mutations. In the current study, we have shown that PFR to DMI fungicides is also associated with reduced sensitivity to multiple unrelated fungicides, which is reminiscent of the MDR phenotypes reported by Kretschmer et al. (2009) for B. cinerea. However, in B. cinerea, MDR and fungicide efflux has been shown to have a tangible negative impact on the efficacy of fungicides in the field, although this is the only case in the published literature. Although iprodione EC50 and boscalid EC95 values are statistically greater for PFR isolates, we cannot confirm that these isolates are capable of exhibiting field resistance or reduced efficacy to the respective fungicides. Field studies to demonstrate the selection of isolates with the MDR phenotype by non‐DMI fungicide applications are ongoing and will provide more insights into how these isolates cause disease in a practical field setting.
Two ABC transporters, ShatrD and ShPDR1, are constitutively over‐expressed in the S. homoeocarpa isolates with an MDR phenotype, and the exact role of ShatrD in this phenotype is currently unknown. There is currently limited evidence on the role of ShatrD in the reduced sensitivity to individual fungicides, although its expression is increased following exposure to propiconazole, but not the two other fungicides used in this study (Fig. S1, see Supporting Information). In addition, the induced expression of these two PDR transporters was incrementally increased in response to an increasing dose of propiconazole, with ShPDR1 showing significantly higher over‐expression than ShatrD with the highest concentration (5 μg/mL) of propiconazole exposure. Care must be taken in interpreting these results, which could imply that ShatrD expression patterns indicate a lesser role in the detoxification of iprodione, boscalid and higher propiconazole concentrations. In B. cinerea, over‐expression of BcatrD is induced in the presence of iprodione, although BcatrD was experimentally shown to be unable to efflux this fungicide (Hayashi et al., 2001, 2002). Future research should include ShatrD yeast complementation studies to better understand substrate specificity. In addition, more detailed studies, such as fungicide accumulation assays with 14C‐labelled fungicides and the fungal transformation system for the generation of gene knock‐outs, will be essential for further functional characterization of PDR transporters in S. homoeocarpa.
The analysis of the coding sequences of ShPDR1 detected non‐synonymous polymorphisms in two DMI‐insensitive isolates (WBI7 and HFI40); however, there are no previous reports of polymorphisms in a coding region of a PDR transporter relating to differential fungicide sensitivity in plant pathogenic fungi. A similar phenomenon was found for the P. digitatum PMR1 sequence diversity among a panel of isolates, providing further evidence that PDRs can exhibit non‐synonymous polymorphism within a species that is not indicative of a phenotypic difference in fungicide sensitivity (Paloma and Juan, 2011). Of greatest interest is a deletion of 27 amino acids spanning the N‐terminus of a partial ABC signature motif, complete Walker B motif and partial D‐loop in HRS10. At first, we surmised this result to be an anomaly, as it was only observed in one isolate of the eight that were sequenced, but, following blastx of both variants of the gene, we found PDR transporter sequence variants in other distantly related fungal species with similar deletions in the Walker B motif in A. kawachii IFO 4308 (GenBank# GAA90510.1) and Ajellomyces capsulatus (GenBank# EER44595.1). Rai et al. (2005, 2006) found that conserved Trp326 and Asp327 residues of the Walker B motif in the ABC transporter CDR1 from C. albicans are important for nucleotide binding and ATP hydrolysis, respectively. Thus, we hypothesize that the deleted Walker B motif in ShPDR1 of HRS10 may affect the function of nucleotide binding and ATP hydrolysis, although the evolutionary consequences of this deletion are not clear. Further yeast complementation experiments will be necessary to test these hypotheses.
RNA‐Seq data of S. homoeocarpa revealed that seven identified PDR transporters have high amino acid sequence similarities of greater than 83% with functionally characterized PDR transporters from B. cinerea (Table 2). Surprisingly, ShPDR1 is not shown to have a putative homologue in B. cinerea, but shares high amino acid similarity to a PDR transporter from A. kawachii, and clusters with high bootstrap support with PDR transporters from P. digitatum and A. nidulans (Fig. 1). In previous phylogenetic analyses, S. homoeocarpa is shown to fall within a group sister to the clade containing B. cinerea and S. sclerotiorum (Amselem et al., 2011), which are phylogenetically distinct from Penicillium and Aspergillus species (Schoch et al., 2009). The number of PDR transporter genes in the genomes of related fungal species can be highly variable, and a plausible explanation is gene loss and differential gene duplication events among fungal lineages (Lamping et al., 2010).
In the light of this and previous studies, there is no doubt that research on the molecular basis of fungicide resistance stands to benefit tremendously with the application of next‐generation sequencing tools, such as RNA‐Seq (Cools and Hammond‐Kosack, 2012). However, there are few published papers that have utilized RNA‐Seq for such purposes, and those that are available are centered on laboratory‐generated mutants in model fungal species (Sun et al., 2013). From this study and the previous work of Hulvey et al. (2012), it is clear that de novo RNA‐Seq offers great advantages when applied to unsequenced plant pathogenic fungi, especially when considering the labor and resources required for genome sequencing and assembly. Most importantly, de novo RNA‐Seq offers the advantage of the identification of mRNAs representing novel genes and for the simultaneous measurement of the expression of transcripts following exposure to fungicides. When combined with additional molecular biology tools, such as yeast complementation and quantitative PCR, researchers can begin to quickly make significant progress towards the confirmation and discovery of the molecular mechanisms of fungicide resistance by a functional genomics approach.
Experimental Procedures
Isolates and in vitro sensitivity assay
Two sets of field isolates were utilized in this study for in vitro fungicide sensitivity assays and quantitative PCR measurements. One panel of eight isolates was recovered from an initial sampling of five sites, and the other panel of isolates consists of two sets of eight isolates that were sampled from two golf courses in New England during the study of Popko et al. (2012) (Hulvey et al., 2012). The panel of isolates sampled initially was collected from four golf course sites with previous exposure to fungicides (HRCC, HGC, Wintonbury Hills Golf Club and Shuttle Meadow Country Club) and from one baseline site with no previous exposure (Joseph Troll Turf Research Center at the University of Massachusetts, Amherst, MA, USA). Two additional sets (from HRCC and HGC, respectively) of isolates were included, and each set consisted of four isolates that were observed to exhibit PFR and four sensitive isolates as per Popko et al. (2012). Isolates were sampled from dollar spot infection centers during the control period of propiconazole (7 days after treatment) and were therefore considered to cause PFR (Popko et al., 2012). The initial panel of isolates and two sets of PFR and sensitive isolates had been characterized previously for EC50 values of propiconazole on potato dextrose agar (PDA) (Difco Laboratories, Detroit, MI, USA) (Hulvey et al., 2012).
In this study, we used MM for fungicide sensitivity assays in order to minimize the effect of additional nutrients in the medium on mycelial growth. MM has been shown to be preferential for the assessment of succinate dehydrogenase inhibitor (SDHI) fungicide sensitivity in other filamentous plant pathogens, such as M. fructicola (Hu et al., 2011). In vitro sensitivity assays of a total of 24 isolates were conducted with DMI (propiconazole), dicarboximide (iprodione) and SDHI (boscalid) fungicides on MM (Hu et al., 2011). Commercial formulations of propiconazole (Banner MAXX 1.3ME, Syngenta Crop Protection, Greensboro, NC, USA) (0.001, 0.01, 0.1, 1 and 10 μg/mL), iprodione (Chipco 26GT 2SC, Bayer, Research Triangle Park, NC, USA) (0.01, 0.1, 1, 10 and 100 μg/mL) and boscalid (Emerald 70WG, BASF, Research Triangle Park, NC, USA) (1, 1000, 3000, 5000 and 10 000 μg/mL) were added to autoclaved MM that had been cooled to 55–60 °C, and fungicide‐amended and non‐amended media were poured into plates. Isolates from long‐term storage were grown on PDA for 2 days, and agar plugs (5 mm in diameter) were transferred from the margin of actively growing colonies to the edge of fungicide‐amended MM and non‐amended MM Petri plates. Transferred isolates were replicated in quadruplicate and two separate experiments were performed for a total of eight plates per isolate. Eight days after transfer, one diameter from the agar plug to the actively growing colony margin was measured with 16EX digital callipers (Mahr, Göttingen, Germany). Propiconazole and iprodione EC50 values (the mean effective concentration for the reduction of mycelial growth by 50%) and EC95 values for boscalid were calculated according to Jo et al. (2006).
DNA extraction, PCR amplification and DNA sequencing
The extraction of genomic DNA was conducted according to the methods described previously by Hulvey et al. (2012). For PCR amplification and DNA sequencing, primers were designed using primer3 (Rozen and Skaletsky, 2000). The five sets of primers, including internal primers for the coding region of ShPDR1 (4492 bp) and two sets of primers for the partial upstream region (459 bp) of ShPDR1, were designed from transcriptomic and partial genome data (Hulvey et al., 2012). Total volumes of PCR were 20 μL, with a final concentration of 1 × PCR buffer, 0.2 μm of each primer, 0.2 mm deoxynucleoside triphosphate (dNTP), 2.5 nm MgCl2, 1 unit Taq DNA polymerase (New England BioLabs, Ipswich, MA, USA) and ∼250 ng of genomic DNA. The PCR cycling conditions were used as described by Hulvey et al. (2012). Amplicons purified using ExoSAP‐IT PCR cleanup reagent (Affymetrix, Santa Clara, CA, USA) were sequenced at the Genomic Resource Laboratory (University of Massachusetts, Amherst).
Mycelial preparation, total RNA isolation and cDNA synthesis
The extraction of RNA was performed by modifying the methods described previously by Hulvey et al. (2012). In brief, isolates were inoculated in 25 mL of half‐strength potato dextrose broth (PDB) and grown for 3 days at 23 °C. Commercial formulations of propiconazole (0.1 μg/mL), iprodione (1 μg/mL) and boscalid (10 μg/mL) were added to treated samples, and the same volume of water was placed in untreated samples. Tubes were lightly shaken on a benchtop shaker for 1 h. Approximately 100 mg of mycelial mat were placed in a 2.0‐mL screw cap tube with zirconia/silica beads (BioSpec, Bartlesville, OK, USA) and immediately dropped into liquid nitrogen. Frozen samples were stored in a −80 °C freezer.
For RNA extraction, 1 mL of TRIzol (Invitrogen, Carlsbad, CA, USA) reagent was added to the tubes with frozen mycelia from the freezer, and the tubes were inserted into a Mini‐beadbeater (BioSpec, Bartlesville, OK, USA) to homogenize the samples for 40 s. After leaving the samples at room temperature for 3 min, 200 μL of chloroform were added to the tubes, which were shaken vigorously by hand for 15 s. The tubes were incubated at room temperature for 5 min and centrifuged at 10 000 × g for 10 min at 4 °C. The upper aqueous phase was removed into a new tube and 1 vol of 1.2 m NaCl–0.8 m Na3C6H5O7 solution and isopropanol were added to the aqueous phase to precipitate total nucleic acid. After inverting the tubes several times, the tubes were incubated at 4 °C for 10 min, and the nucleic acids were pelleted by centrifugation at 9200 × g at 4 °C for 10 min. Pellets were air dried at room temperature for 10 min and suspended in 50 μL of RNase‐free water at 62 °C for 10 min. cDNA was synthesized from each sample of RNA using a QuantiTect reverse transcription kit (Qiagen Inc., Valencia, CA, USA).
Transcriptome sequencing, assembly and annotation
For the RNA‐Seq experiment, two biological replicates of sequencing‐by‐synthesis (SBS) reads were generated by the method of Hulvey et al. (2012). In brief, RNA was extracted from treated and untreated mycelia of HRS10 and HRI11 and sent to Macrogen Inc. (Seoul, South Korea) for mRNA isolation and cDNA library preparation using the TruSeq mRNA kit, and sequencing with the Illumina v7 HiSeq2000 platform (Life Technologies, Carlsbad, CA, USA ). For Illumina sequencing, 100 cycle reactions were performed yielding over 50 million 2 × 100‐bp paired‐end (PE) SBS reads per cDNA library. For each biological replicate, one lane of sequencing was performed for four cDNA libraries (HRS10 untreated, HRI11 untreated, HRS10 treated and HRI11 treated), as described previously (Hulvey et al., 2012). The PE SBS reads from both replicates were combined with 454 GS FLX Titanium data from Hulvey et al. (2012) to perform a de novo transcriptome assembly using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark). The alignment contigs were blastx searched against the National Center for Biotechnology Information (NCBI) non‐redundant protein database (August 2013) using blast2go software with the default settings, except for an e‐value threshold of less than e−30. In addition, gene ontology and Pfam domain searches were performed on the assembled contigs in blast2go.
The PDR transporters analysed in this study were parsed from the dataset based on several criteria: gene ontology (GO) terms, e‐value threshold of e−60 for blastx result, nucleotide length greater than 2 kb and presence of ABC‐G protein domains in the translated amino acid sequence. Such stringent criteria ensure that mined sequences represent full‐size ABC‐G transporters suitable for phylogenetic analysis and high confidence in RNA‐Seq analysis.
RNA‐Seq transcriptome profiling
RNA‐Seq analysis was performed by mapping read pairs from each biological replicate of SBS PE data back to the hybrid assembly in CLC Genomics Workbench with the following criteria: Mismatch cost = 2, Insertion cost = 3, Deletion cost = 3, Length fraction = 0.5 and Similarity fraction = 0.95. In order to analyse differential expression between samples, RNA‐Seq experiments were set up in CLC Genomics Workbench to look for differences between HRS10 and HRI11 in untreated and treated samples separately. As we analysed reads per kilobase per million mapped reads (RPKM), and thus count‐based data, values were first normalized by read count totals (Bolstad et al., 2003). Baggerly's test, a proportions‐based test, was used to detect significant differences in the expression of mined ABC‐G transporters for each experiment (Baggerly et al., 2003). Normalized RPKM values were analysed by Baggerly's test to look for significant differences in expression data of untreated and treated samples. The mean fold change was calculated from the mean normalized RPKM values, and indicates how many times larger expression values were for HRI11 than for HRS10.
Upstream and coding sequence analyses of ShPDR1 and phylogenetic analysis of ABC‐G transporters
Sequence analysis of the upstream and coding regions of ShPDR1 followed the method of Hulvey et al. (2012). The Neural Network Promoter Prediction (NNPP) software at the Berkeley Drosophila Genome Project website was employed to search a 500‐bp upstream region of ShPDR1 for promoters.
A phylogenetic analysis was performed for the ABC‐G transporter amino acid sequences obtained from Hulvey et al. (2012) and GenBank, with the addition of PDR transporter contigs from RNA‐Seq data. The MAFFT server was used to align amino acid sequences with automated method selection and the Blosum 62 amino acid scoring matrix (Katoh and Toh, 2008). Neighbour joining phylogenetic analysis with a Poisson model, Gamma distributed rates among sites, complete deletion of missing data and 1000 bootstrap replicates was conducted in mega v5 (Tamura et al., 2011).
Quantitative real‐time PCR analysis
The RE of ShPDR1 was assayed with quantitative real‐time PCR before and after treatment with propiconazole (0.1 μg/mL), iprodione (1 μg/mL) and boscalid (10 μg/mL) for 1 h in the initial panel of eight isolates. In addition, constitutive expression of ShPDR1 was assayed for PFR and sensitive isolates from two sites. Moreover, the effect of propiconazole dose on the expression of ShPDR1 and ShatrD genes was assessed in two insensitive isolates, HRI11 and HFI40, in the absence and presence of propiconazole at 0.1, 1 and 5 μg/mL. The Actin (Shact) gene in S. homoeocarpa was selected as a housekeeping gene. Primers for quantitative PCR were designed using the Integrate DNA Technologies (Coralville, IA, USA) qPCR primer design web tool server to amplify 113‐bp amplicons from the ShPDR1 gene, and primers for ShatrD and Shact gene amplification were used according to Hulvey et al. (2012). Quantitative reverse transcription‐PCR was conducted in 25‐μL reactions containing 12.5 μL of Absolute Blue SYBR qPCR MasterMix (Thermo Fisher Scientific, Waltham, MA, USA), 1.75 μL of each primer at 1 μm, 8 μL of microbiological grade water (Thermo Fisher Scientific, Waltham, MA, USA) and 1 μL of cDNA at 250 ng/μL. Mastercycler ep realplex thermocycler (Eppendorf, Hamburg, Germany) was used for PCRs. PCR conditions were as follows: one cycle of 15 min at 95 °C, and 40 cycles of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. The comparative C T method was used to calculate relative gene expression (Livak and Schmittgen, 2001). Two biological replicates and three technical replicates per biological replicate were performed for each isolate and treatment.
Heterologous expression of ShPDR1 in yeast
The full‐length cDNA sequences of ShPDR1 were amplified using primers ShPDR1_cloning_F and ShPDR1_cloning_R (Table S1, see Supporting Information), which introduce KpnI and SphI sites at the 5′ and 3′ ends of the amplified product, respectively. PCR was performed using Phusion high‐fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) in a PCR with an initial denaturation step at 98 °C for 30 s, followed by 53 cycles of denaturation (at 98 °C for 15 s) and annealing (at 73 °C for 30 s, decreasing at increments of 1.0 °C per cycle for the first eight cycles, and at 65 °C for the remaining 45 cycles) and extension (at 72 °C for 2 min), with a final extension step at 72 °C for 5 min. The purified 4514‐bp PCR product and the plasmid pYES2 (Invitrogen, Carlsbad, CA, USA) were digested with KpnI and SphI, and the two digested products were gel purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA) and ligated. Escherichia coli DH5a was used for the propagation of vector pYES2 containing ShPDR1.
Saccharomyces cerevisiae strain AD1‐8 (MATα, pdr1‐3, his1, ura3, Δpdr5::hisG, Δyor1::hisG, Δsnq2::hisG, Δycf1::hisG, Δpdr10::hisG, Δpdr11::hisG, Δpdr15::hisG, Δpdr3::hisG) (Decottignies et al., 1998) was transformed with the empty expression vector pYES2 (AD1‐8‐pYES2) and the vector containing ShPDR1 (AD1‐8:PDR1‐1, AD1‐8:PDR1‐2) by a high‐efficiency polyethylene glycol/lithium acetate (PEG/LiAc)‐based method using the Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA, USA). Yeast transformants were selected on solid synthetic MM containing bacto‐yeast nitrogen base (YNB) without amino acids (6.7 g/L), drop‐out mix (2 g/L containing amino acids minus uracil), glucose (20 g/L) and agar (20 g/L). Transformants with the empty vector pYES2 and the vector pYES2 with ShPDR1 were grown at 30 °C for 3 days in liquid MM lacking uracil and containing 2% galactose.
For fungicide sensitivity assays, cell suspensions were diluted to an optical density at 600 nm (OD600) of 0.5 in liquid MM using the VERSAmax™ microplate reader (Molecular Devices Corporation, Sunnyvale, CA, USA), and 5 and 10 μL of each yeast transformant were plated as spots on YNB agar medium lacking uracil, containing 2% galactose and amended with different concentrations of fungicides. The sensitivity of yeast transformants to fungicides was qualitatively assessed after incubation at 30 °C for 3 days. The fungicide concentrations (μg/mL) used in this study were as follows: propiconazole (0, 0.0005, 0.001, 0.002, 0.003), iprodione (0, 10, 100, 250, 500) and boscalid (0, 10, 100, 250, 500). Two biological replicates and four technical replicates per biological replicate were conducted for each transformant and treatment.
Statistical analyses of in vitro sensitivity assay and quantitative PCR data
EC50 values for propiconazole and iprodione and EC95 values for boscalid were generated for the two sets of isolates. The sets consisted of four PFR and four sensitive isolates from HRCC and HGC, respectively. Analysis of variance was conducted on the two sets to examine differences between the mean EC50 and EC95 values of sensitive and PFR groups of isolates. Analysis of variance was also conducted between biological replications of EC50 and EC95 values for all sets.
For the statistical analysis of RE data for ShPDR1 and ShatrD, four independent sets were generated. The first set contained the initial panel of isolates before and after treatment of propiconazole, boscalid and iprodione. The second and third sets contained PFR and sensitive isolates from HRCC and HGC, respectively. The last set included two insensitive isolates from the initial panel of isolates in the absence and presence of different concentration of propiconazole. Analysis of variance was conducted on RE levels for ShPDR1 between the sensitive and insensitive groups of isolates and between constitutive and induced expression for the first set. For the second and third sets, analysis of variance was conducted on RE values for ShPDR1 between the sensitive and insensitive groups of isolates. Analysis of variance was conducted to examine differences in RE values between ShPDR1 and ShatrD in the last set. Analysis of variance was conducted between biological replications of RE data for all aforementioned sets of isolates.
Linear regression analysis was conducted with propiconazole and iprodione EC50 and boscalid EC95 values and log10‐transformed mean RE values of ShPDR1 constitutive expression for all 24 isolates. All statistical analyses were conducted using JMP software package, version 9.0 (SAS Institute Inc., Cary, NC, USA).
Supporting information
Fig. S1 Relative expression (RE) of ShatrD in the initial panel of eight isolates from five sites in New England before and after iprodione (1 μg/mL) and boscalid (10 μg/mL) incubation for 1 h. All bars (white, constitutive RE; dark grey, iprodione‐induced RE; black, boscalid‐induced RE) represent mean RE values. Error bars indicate one standard error of the mean. Isolates with an ‘I’ indicate propiconazole insensitive and with an ‘S’ represent propiconazole sensitive.
Table S1 Primers used in this study.
Acknowledgements
We gratefully acknowledge Dr Mohan Gupta (Department of Molecular Genetics and Cell Biology, University of Chicago, Chicago, IL, USA) for providing yeast strains. Funding for this work comes from the National Institute of Food and Agriculture, US Department of Agriculture, Massachusetts Agricultural Experiment Station and Stockbridge School of Agriculture under project number MAS00436.
References
- Amselem, J. , Cuomo, C. , van Kan, J. , Viaud, M. , Benito, E. , Couloux, A. , Coutinho, P. , de Vries, R. , Dyer, P. , Fillinger, S. , Fournier, E. , Gout, L. , Hahn, M. , Kohn, L. , Lapalu, N. , Plummer, K. , Pradier, J.‐M. , Quévillon, E. , Sharon, A. , Simon, A. and Dickman, M. (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . PLoS Genet. 7, e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggerly, K. , Morris, J. , Wang, J. , Gold, D. , Xiao, L.‐C. and Coombes, K. (2003) A comprehensive approach to the analysis of matrix‐assisted laser desorption/ionization‐time of flight proteomics spectra from serum samples. Proteomics, 3, 1667–1672. [DOI] [PubMed] [Google Scholar]
- Bolstad, B. , Irizarry, R. , Astrand, M. and Speed, T. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Chen, F. , Liu, X. , Chen, S. , Schnabel, E. and Schnabel, G. (2013) Characterization of Monilinia fructicola strains resistant to both propiconazole and boscalid. Plant Dis. 97, 645–651. [DOI] [PubMed] [Google Scholar]
- Cole, H.B. , Taylor, B. and Duich, J. (1968) Evidence of differing tolerances to fungicides among isolates of Sclerotinia homoeocarpa . Phytopathology, 58, 683–686. [Google Scholar]
- Cools, H. and Hammond‐Kosack, K. (2012) Exploitation of genomics in fungicide research: current status and future perspectives. Mol. Plant Pathol. 14, 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies, A. , Grant, A. , Nichols, J. , de Wet, H. , McIntosh, D. and Goffeau, A. (1998) ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273, 12 612–12 622. [DOI] [PubMed] [Google Scholar]
- Del Sorbo, G. , Schoonbeek, H. and De Waard, M. (2000) Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol. 30, 1–15. [DOI] [PubMed] [Google Scholar]
- Detweiler, A.R. , Vargas, J.M.J. and Danneberger, T.K. (1983) Resistance of Sclerotinia homoeocarpa to iprodione and benomyl. Plant Dis. 67, 627–630. [Google Scholar]
- Driessen, A. , Rosen, B. and Konings, W. (2000) Diversity of transport mechanisms: common structural principles. Trends Biochem. Sci. 25, 397–401. [DOI] [PubMed] [Google Scholar]
- Hamamoto, H. , Hasegawa, K. , Nakaune, R. , Lee, Y.J. , Makizumi, Y. , Akutsu, K. and Hibi, T. (2000) Tandem repeat of a transcriptional enhancer upstream of the sterol 14alpha ‐demethylase gene (CYP51) in Penicillium digitatum . Appl. Environ. Microbiol. 66, 3421–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, K. , Schoonbeek, H.‐J. , Sugiura, H. and De Waard, M.A. (2001) Multidrug resistance in Botrytis cinerea associated with decreased accumulation of the azole fungicide oxpoconazole and increased transcription of the ABC transporter gene BcatrD . Pestic. Biochem. Physiol. 70, 168–179. [Google Scholar]
- Hayashi, K. , Schoonbeek, H. and De Waard, M. (2002) Expression of the ABC transporter BcatrD from Botrytis cinerea reduces sensitivity to sterol demethylation inhibitor fungicides. Pestic. Biochem. Physiol. 73, 110–121. [Google Scholar]
- Hu, M. , Luo, C. , Grabke, A. and Schnabel, G. (2011) Selection of a suitable medium to determine sensitivity of Monilinia fructicola mycelium to SDHI fungicides. J. Phytopathol. 159, 616–620. [Google Scholar]
- Hulvey, J. , Popko, J.T. , Sang, H. , Berg, A. and Jung, G. (2012) Overexpression of ShCYP51B and ShatrD in Sclerotinia homoeocarpa isolates exhibiting practical field resistance to a demethylation inhibitor fungicide. Appl. Environ. Microbiol. 78, 6674–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, Y. , Niver, A. , Rimelspach, J. and Boehm, M.J. (2006) Fungicide sensitivity of Sclerotinia homoeocarpa from golf courses in Ohio. Plant Dis. 90, 807–813. [DOI] [PubMed] [Google Scholar]
- Katoh, K. and Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, A. and Driessen, A. (2010) Phylogenetic analysis of fungal ABC transporters. BMC Genomics, 11, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer, M. , Leroch, M. , Mosbach, A. , Walker, A.‐S. , Fillinger, S. , Mernke, D. , Schoonbeek, H.‐J. , Pradier, J.‐M. , Leroux, P. , De Waard, M. and Hahn, M. (2009) Fungicide‐driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea . PLoS Pathog. 5, e1000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping, E. , Baret, P.V. , Holmes, A.R. , Monk, B.C. , Goffeau, A. and Cannon, R.D. (2010) Fungal PDR transporters: phylogeny, topology, motifs and function. Fungal Genet. Biol. 47, 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux, P. and Walker, A.‐S. (2011) Multiple mechanisms account for resistance to sterol 14α‐demethylation inhibitors in field isolates of Mycosphaerella graminicola . Pest Manag. Sci. 67, 44–59. [DOI] [PubMed] [Google Scholar]
- Leroux, P. , Gredt, M. , Remuson, F. , Micoud, A. and Walker, A.‐S. (2013) Fungicide resistance status in French populations of the wheat eyespot fungi Oculimacula acuformis and Oculimacula yallundae . Pest Manag. Sci. 69, 15–26. [DOI] [PubMed] [Google Scholar]
- Livak, K. and Schmittgen, T. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(–Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, Z. and Michailides, T.J. (2005) Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot. 24, 853–863. [Google Scholar]
- Mamidala, P. , Wijeratne, A. , Wijeratne, S. , Kornacker, K. , Sudhamalla, B. , Rivera‐Vega, L. , Hoelmer, A. , Meulia, T. , Jones, S. and Mittapalli, O. (2012) RNA‐Seq and molecular docking reveal multi‐level pesticide resistance in the bed bug. BMC Genomics, 13, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G.L. , Stevenson, K.L. and Burpee, L.L. (2002) Sensitivity of Sclerotinia homoeocarpa isolates to propiconazole and impact on control of dollar spot. Plant Dis. 86, 1240–1246. [DOI] [PubMed] [Google Scholar]
- Morschhäuser, J. , Barker, K. , Liu, T. , BlaB‐Warmuth, J. , Homayouni, R. and Rogers, P. (2007) The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans . PLoS Pathog. 3, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaune, R. , Adachi, K. , Nawata, O. , Tomiyama, M. , Akutsu, K. and Hibi, T. (1998) A novel ATP‐binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum . Appl. Environ. Microbiol. 64, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloma, S.‐T. and Juan, J.T. (2011) Molecular insights into fungicide resistance in sensitive and resistant Penicillium digitatum strains infecting citrus. Postharvest Biol. Technol. 59, 159–165. [Google Scholar]
- Popko, J.T. , Ok, C. , Campbell‐Nelson, K. and Jung, G. (2012) The association between in vitro propiconazole sensitivity and field efficacy of five New England Sclerotinia homoeocarpa populations. Plant Dis. 96, 552–561. [DOI] [PubMed] [Google Scholar]
- Putman, A. , Jung, G. and Kaminski, J. (2010) Geographic distribution of fungicide‐insensitive Sclerotinia homoeocarpa isolates from golf courses in the northeastern United States. Plant Dis. 94, 186–195. [DOI] [PubMed] [Google Scholar]
- Rai, V. , Shukla, S. , Jha, S. , Komath, S. and Prasad, R. (2005) Functional characterization of N‐terminal nucleotide binding domain (NBD‐1) of a major ABC drug transporter Cdr1p of Candida albicans: uncommon but conserved Trp326 of Walker B is important for ATP binding. Biochemistry, 44, 6650–6661. [DOI] [PubMed] [Google Scholar]
- Rai, V. , Gaur, M. , Shukla, S. , Shukla, S. , Ambudkar, S. , Komath, S. and Prasad, R. (2006) Conserved Asp327 of walker B motif in the N‐terminal nucleotide binding domain (NBD‐1) of Cdr1p of Candida albicans has acquired a new role in ATP hydrolysis. Biochemistry, 45, 14 726–14 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Schoch, C. , Sung, G.‐H. , López‐Giráldez, F. , Townsend, J. , Miadlikowska, J. , Hofstetter, V. , Robbertse, B. , Matheny, P. , Kauff, F. , Wang, Z. , Gueidan, C. , Andrie, R. , Trippe, K. , Ciufetti, L. , Wynns, A. , Fraker, E. , Hodkinson, B. , Bonito, G. , Groenewald, J. , Arzanlou, M. , de Hoog, G. , Crous, P. , Hewitt, D. , Pfister, D. , Peterson, K. , Gryzenhout, M. , Wingfield, M. , Aptroot, A. , Suh, S.‐O. , Blackwell, M. , Hillis, D. , Griffith, G. , Castlebury, L. , Rossman, A. , Lumbsch, H. , Lücking, R. , Büdel, B. , Rauhut, A. , Diederich, P. , Ertz, D. , Geiser, D. , Hosaka, K. , Inderbitzin, P. , Kohlmeyer, J. , Volkmann‐Kohlmeyer, B. , Mostert, L. , O'Donnell, K. , Sipman, H. , Rogers, J. , Shoemaker, R. , Sugiyama, J. , Summerbell, R. , Untereiner, W. , Johnston, P. , Stenroos, S. , Zuccaro, A. , Dyer, P. , Crittenden, P. , Cole, M. , Hansen, K. , Trappe, J. , Yahr, R. , Lutzoni, F. and Spatafora, J. (2009) The Ascomycota tree of life: a phylum‐wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 58, 224–239. [DOI] [PubMed] [Google Scholar]
- Smiley, R.W. , Dernoeden, P.P. and Clarke, B.B. (2005) Compendium of Turfgrass Diseases. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Sun, N. , Fonzi, W. , Chen, H. , She, X. , Zhang, L. , Zhang, L. and Calderone, R. (2013) Azole susceptibility and transcriptome profiling in Candida albicans mitochondrial electron transport chain complex I mutants. Antimicrob. Agents Chemother. 57, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, J.M.J. , Golembiewski, R. and Detweiler, A.R. (1992) Dollar spot resistance to DMI fungicides. Golf Course Manage. 60, 50–54. [Google Scholar]
- Wang, Y. , Lim, L. , DiGuistini, S. , Robertson, G. , Bohlmann, J. and Breuil, C. (2013) A specialized ABC efflux transporter GcABC‐G1 confers monoterpene resistance to Grosmannia clavigera, a bark beetle‐associated fungal pathogen of pine trees. New Phytol. 197, 886–898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Relative expression (RE) of ShatrD in the initial panel of eight isolates from five sites in New England before and after iprodione (1 μg/mL) and boscalid (10 μg/mL) incubation for 1 h. All bars (white, constitutive RE; dark grey, iprodione‐induced RE; black, boscalid‐induced RE) represent mean RE values. Error bars indicate one standard error of the mean. Isolates with an ‘I’ indicate propiconazole insensitive and with an ‘S’ represent propiconazole sensitive.
Table S1 Primers used in this study.


