Summary
The bacterial agent of citrus canker disease (Xanthomonas citri ssp. citri, Xcc) has caused tremendous economic losses to the citrus industry around the world. Pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) is important to plant immunity. In this study, we compared the defence responses of citrus canker‐resistant and citrus canker‐susceptible genotypes to the Xcc‐derived PAMP flg22 (Xflg22) by analysing the expression of 20 citrus defence‐associated genes. We showed that, in the most resistant genotype, ‘Nagami’ kumquat, there was significant induction of several defence genes (EDS1, NDR1, PBS1, RAR1, SGT1, PAL1, NPR2 and NPR3) as early as 6 h and up to 72 h after Xflg22 treatment. At the other end of the spectrum, highly susceptible ‘Duncan’ grapefruit showed no induction of the same defence genes, even 120 h after treatment. Citrus genotypes with partial levels of resistance showed intermediate levels of transcriptional reprogramming that correlated with their resistance level. Xflg22 also triggered a rapid oxidative burst in all genotypes which was higher and accompanied by the induction of PTI marker genes (WRKY22 and GST1) only in the more resistant genotypes. Pretreatment with Xflg22 prior to Xcc inoculation inhibited bacterial growth in kumquat, but not in grapefruit. A flagellin‐deficient Xcc strain (XccΔfliC) showed greater growth increase relative to wild‐type Xcc in kumquat than in grapefruit. Taken together, our results indicate that Xflg22 initiates strong PTI in canker‐resistant genotypes, but not in susceptible ones, and that a robust induction of PTI is an important component of citrus resistance to canker.
Keywords: canker, citrus, flg22, PAMP, PTI, resistance, Xanthomonas citri ssp. citri
Introduction
Citrus is one of the most economically important crops for both the fresh and processed fruit markets. The bacterial disease citrus canker, caused by Xanthomonas citri ssp. citri (Xcc), has been damaging to the industry and has become endemic in many parts of the world. In certain areas (e.g. Florida), this has caused losses as a result of the eradication of millions of trees in an attempt to control this disease. In addition, restrictions have been imposed in the interstate and international transportation of citrus products because of quarantines and other regulatory measures (Bronson and Gaskalla, 2007), and has increased the costs of management of the groves. Most commercially grown citrus types, including sweet orange (C. sinensis Osb.), grapefruit (C. paradisi Macf.) and lemon [C. limon (L.) Burm. f.], are susceptible to canker (Goto, 1992; Gottwald et al., 1993, 2002), and disease control methods are limited in effectiveness (Dewdney and Graham, 2011; Graham and Leite, 2004; Granado et al., 1995; Leite and Mohan, 1990). Genetic resistance has the potential to become a solution for the management of this disease. Genotypes with disease‐resistant traits have been identified and serve as valuable resources for breeding. Kumquat [Fortunella margarita (L.) Swing.], a citrus relative that is highly resistant to canker, is one such example (Goto, 1992; Reddy, 1997). Moreover, the recent characterization of the kumquat–Xcc interaction offers insights into the canker resistance response at the cellular and molecular levels (Khalaf et al., 2007).
Plants have two levels of immunity to defend themselves against microbial invasions. At one level, plants respond to pathogen‐associated molecular patterns (PAMPs). Plant recognition of PAMPs is mediated by cell membrane‐localized pattern recognition receptors (PRRs), which trigger complex signalling events and a defence response resulting in the limitation of the microbe's multiplication and disease development (PAMP‐triggered immunity, PTI) (Asai et al., 2002; Jones and Dangl, 2006; Zipfel and Felix, 2005; Zipfel et al., 2004). At another level, adapted pathogens are able to secrete effector proteins into host cells and modulate plant defences. In turn, plants have evolved resistance (R) proteins to recognize such effectors and initiate a strong defence response that often leads to pathogen resistance (effector‐triggered immunity, ETI) (Dangl and Jones, 2001; Jones and Dangl, 2006; Qutob et al., 2006).
PTI is a broad‐spectrum immune response that can protect plants from a wide range of microbes by sensing PAMPs (Ebel and Cosio, 1994). Flagellin, one of the best studied PAMPs, is a protein component of the filament of the bacterial flagellum. Within the N‐terminus of flagellin, a conserved 22‐amino‐acid domain (flg22) is recognized by plants and is capable of initiating PTI (Felix et al., 1999). In Arabidopsis, flg22 perception results in the induction of defence‐associated genes (Navarro et al., 2004), reactive oxygen species (ROS) production (Felix et al., 1999), salicylic acid (SA) accumulation (Tsuda et al., 2008), and local and systemic acquired resistance (SAR) to pathogen infection (Mishina and Zeier, 2007b; Zipfel et al., 2004). To investigate PTI in citrus and its role in canker immunity, we used the flg22 derived from Xcc (Xflg22) to challenge and compare the expression responses of defence‐associated genes in different citrus genotypes, ranging from highly resistant to highly susceptible to canker. The genes selected represent key functional nodes of plant defence (Table 1): PTI and ETI perception and signalling [EDR1 (enhanced disease resistance 1), EDS1 (enhanced disease susceptibility 1), NDR1 (non‐specific disease resistance 1), PBS1 (avrPphB susceptible 1), RAR1 (required for Mla12 resistance 1) and SGT1 (suppressor of G2 allele of skp1)] (Azevedo et al., 2002; Century et al., 1995; Frye and Innes, 1998; Parker et al., 1996; Torp and Jorgensen, 1986; Warren et al., 1999), including established PTI markers [WRKY22 transcription factor and GST1 (glutathione S‐transferase 1)] (Asai et al., 2002); SA metabolism [EDS5, ICS1 (isochorismate synthase 1), PAL1 (phenylalanine ammonia‐lyase 1) and AZI1 (azelaic acid‐induced 1)] (Greenberg et al., 2009; Mauch‐Mani and Slusarenko, 1996; Nawrath and Métraux, 1999; Wildermuth et al., 2001); transcriptional regulators [NPR1 (nonexpressor of pathogenesis‐related gene 1), NPR2 and NPR3] (Cao et al., 1994; Hepworth et al., 2005; Norberg et al., 2005; Zhang et al., 2006); pathogenesis‐related proteins [PR1 (pathogenesis‐related 1), RdRp1 (RNA‐dependent RNA polymerase 1) and CHI (chitinase)] (Alexander et al., 1993; Samac et al., 1990; Xie et al., 2001) and jasmonic acid (JA) signalling [JAR1 (jasmonic acid‐resistant 1) and COI1 (coronatine insensitive 1)] (Staswick and Tiryaki, 2004; Yan et al., 2009). In addition, we compared the effect of Xflg22 on ROS accumulation and Xcc bacterial population growth in different citrus genotypes.
Table 1.
List of primer and probe sequences used to analyse the expression of defence‐associated genes in citrus. The assay names are based on homology to genes characterized in Arabidopsis. Loci of the respective genes are from the C itrus sinensis genomic database available at the National Center for Biotechnology Information (NCBI). Forward primer (f), reverse primer (r) and probe (p) sequences were generated using Primer Express Software 3.0 (Applied Biosystems, Foster City, CA, USA)
| Assay | Primer/probe | Citrus sinensis locus |
|---|---|---|
| 5.8S rRNA | f: CGACTCTCGGCAACGGATA | JQ990165 |
| r: CGCATTTCGCTACGTTCTTCA | ||
| p: CTCGGCTCTCGCATC | ||
| AZI1 | f: CCATCAAAGCGAACATTTTGG | LOC102617861 |
| r: CGTTCAAAAGAAGGCTGAGTGA | ||
| p: ATCAACCTTAATATCCC | ||
| CHI | f: GCCGGCTTCCGGGATAC | Z70032 |
| r: CTTGGCCACATTCAATTCCA | ||
| p: CTAACCACAAATATAATCAACG | ||
| COI1 | f: GGGAATGGAGGATGAAGAAGGT | LOC102620384 |
| r: GCCCTGAGCCAAAGCAATTA | ||
| p: TTGTCTCGCAAAGAGGA | ||
| EDR1 | f: TCCAGGAGTGCTTTGAGTGGTA | LOC102618775 |
| r: GCCCATTTAACTGACTTGTGCTAGA | ||
| p: TGGCCCATCATTGG | ||
| EDS1 | f: GGCTCGAGTATGCCCTGAAG | LOC102618041 |
| r: CTTGCCCAGAAACATGATTCC | ||
| p: ATCGGCAGGATCCAG | ||
| EDS5 | f: ATCGAAGTTTGGTAAAGGCAAGA | LOC102615350 |
| r: AGCGTGGATCCAATGAGAAGA | ||
| p: TGCTGCTGAAGTCAC | ||
| GST1 | f: GCCCGTTTGTCTCAGTCCAA | LOC102614737 |
| r: TGCAAATCGACCAAGGTGAA | ||
| p: ACTTGGCGTGCGACAG | ||
| ICS1 | f: CAGCGCTGGCCTTGGA | LOC102630235 |
| r: GGAGGTGGGTTGGATTTCAA | ||
| p: AAACTTCACTCTGCCATTT | ||
| JAR1 | f: AAGGCGATGCAGTCACAATG | LOC102611440 |
| r: TGGTGGAAATCAGGACCAAAG | ||
| p: AGCCCTGATGAAGTAA | ||
| NDR1 | f: GCGCCGACCGATCAGA | LOC102630232 |
| r: CGCCGACCCATTTGCA | ||
| p: TTTCCCGGGCGGTTT | ||
| NPR1 | f: CGTGGCATATTGTGATGCAAA | LOC102617188 |
| r: GTTGACATCAGCAAGTCCAAGATC | ||
| p: ACCACAACTGAGCTTC | ||
| NPR2 | f: ACCTTAGACGAAGCCAATGCA | LOC102621158 |
| r: CAGACAACACCTTGGGATCACA | ||
| p: TCCATTATGCTGCAGCGTA | ||
| NPR3 | f: TTTTATACTGGCCTTTCAGCATCA | LOC102624339 |
| r: CGTCTCAACTGTTTTAAGCAAAGC | ||
| p: TGCAAGCCAAGAGAC | ||
| PAL1 | f: CTCGGCCCTCAGATCGAA |
LOC102620173 LOC102620464 |
| r: CCGAGTTGATCTCCCGTTCA | ||
| p: TGATTCGGTTTGCAACCA | ||
| PBS1 | f: TCCAAAAGAACCAACTGCACAT | LOC102630702 |
| r: AGCAGCAAGCTCCCGAAAT | ||
| p: CCGCCCAAACGTTTA | ||
| PR1 | f: AAGGAAAGCGGATTGCAAACT | LOC102622841 |
| r: CTCGCCAAGCTTGAAATTGTC | ||
| p: CAGCATTCGTTCCCG | ||
| RAR1 | f: GTCAGAACGACGACGCTTTG | LOC102626257 |
| r: GCGTTGCAGCCAATTCG | ||
| p: AGCGTCTTCGATGCC | ||
| RdRp1 | f: CGGCAGCGCTTTATTGTTC | LOC102608479 |
| r: ACATCACCTGGATGCAAACAAG | ||
| p: ACTGGTGGTTGTTGCAA | ||
| SGT1 | f: GCTGATGCAGATGAGGACACA | LOC102623505 |
| r: CCCGTTTGACTCGACGAAAG | ||
| p: ACGAGCCATGAAAAA | ||
| WRKY22 | f: GCGGATTGTCTCGCATGTG | LOC102612567 |
| r: TTATGGGTTTCTGCCCGTATTT | ||
| p: AAGTGGGCTTGGCG |
Our results show that Xflg22 induces transcriptional reprogramming in resistant but not susceptible genotypes. The differences in response between genotypes were mostly independent of the Xflg22 concentration used; however, the intensity of the reprogramming correlated with the level of resistance of each citrus genotype. Higher levels of ROS production induced by Xflg22 also correlated with canker resistance. Furthermore, we showed that the genotype with the most intense transcriptional reprogramming (kumquat) exhibited enhanced immunity when pretreated with Xflg22, and was less resistant to a flagellin mutant Xcc strain. The role of PTI in resistance against Xcc is discussed.
Results
Xflg22 triggers transcriptional reprogramming of defence‐associated genes in ‘Nagami’ kumquat, but not in ‘Duncan’ grapefruit
To determine the role of Xflg22 in the immune response of citrus against Xcc, highly susceptible ‘Duncan’ grapefruit and highly resistant ‘Nagami’ kumquat were challenged with the peptide. Subsequently, challenged leaf tissue was collected in a time course from 0 to 120 h post‐Xflg22 treatment and subjected to gene expression analysis by real‐time reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Collection time points were selected on the basis of a previous study (Khalaf et al., 2007) and our own empirical data, and aimed to study the extent and long‐term defence response. Two concentrations of Xflg22 (10 and 100 μm) were used to rule out the possibility that any lack of response was caused by insufficient peptide. The genes chosen for analysis were categorized as: established markers of PTI (WRKY22 and GST1), PTI/ETI perception and signalling genes (EDR1, EDS1, NDR1, PBS1, RAR1 and SGT1), SA metabolism (EDS5, ICS1, PAL1 and AZI1), transcriptional regulators (NPR1, NPR2 and NPR3), pathogenesis‐related protein genes (PR1, RdRp1 and CHI) and JA signalling genes (JAR1 and COI1). Gene expression levels were considered to be significantly different at a particular time point when their relative quantification (RQ) values were statistically significantly different from pre‐inoculation levels (0 h) and the water control at the same time point.
WRKY22 and GST1 are inducible by flg22 treatment and have been used as early markers of flg22‐triggered innate immunity in Arabidopsis (Asai et al., 2002). In this study, we observed a higher level of WRKY22 only with 100 μm Xflg22 at 24 h after treatment and no significant induction of GST1 in susceptible ‘Duncan’ grapefruit (Fig. 1a,b). However, in ‘Nagami’ kumquat, WRKY22 expression was significantly induced at 24 h by 10 μm Xflg22 and at 24 and 72 h by 100 μm Xflg22. In addition, the levels of induction were several orders of magnitude higher than in ‘Duncan’ grapefruit (Fig. 1c). The other marker gene, GST1, was also significantly up‐regulated at 24 and 72 h by both Xflg22 treatments (Fig. 1d).
Figure 1.
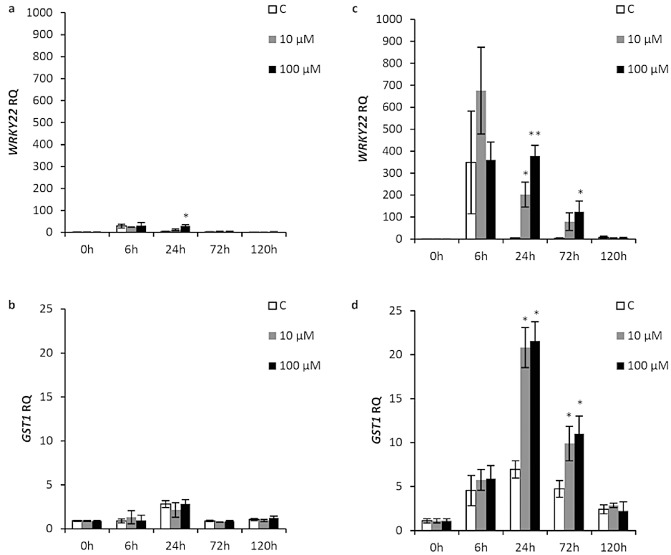
The effect of Xflg22 on the expression of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) marker genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. A single asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. A double asterisk indicates that RQ of the 100 μm Xflg22 treatment is significantly higher (P < 0.05) than that of the control and 10 μm Xflg22 at the indicated time point. Bars are means ± standard error (n = 3).
In susceptible ‘Duncan’ grapefruit, no significant induction of PTI/ETI perception and signalling genes by Xflg22 at either concentration was observed (Figs 2a–e and S1a, see Supporting Information). In contrast, Xflg22 in ‘Nagami’ kumquat significantly induced the expression of these genes as early as 6 h (EDS1, NDR1, RAR1), at 24 h (EDS1, NDR1, PBS1, RAR1, SGT1) and up to 72 h (RAR1, SGT1) after treatment (Fig. 2f–j). The only gene that did not show a significant change in expression was EDR1 (Fig. S1b). Essentially no difference in the levels of gene expression was observed between the 10 and 100 μm treatments in ‘Nagami’ kumquat (Fig. 2f–j).
Figure 2.
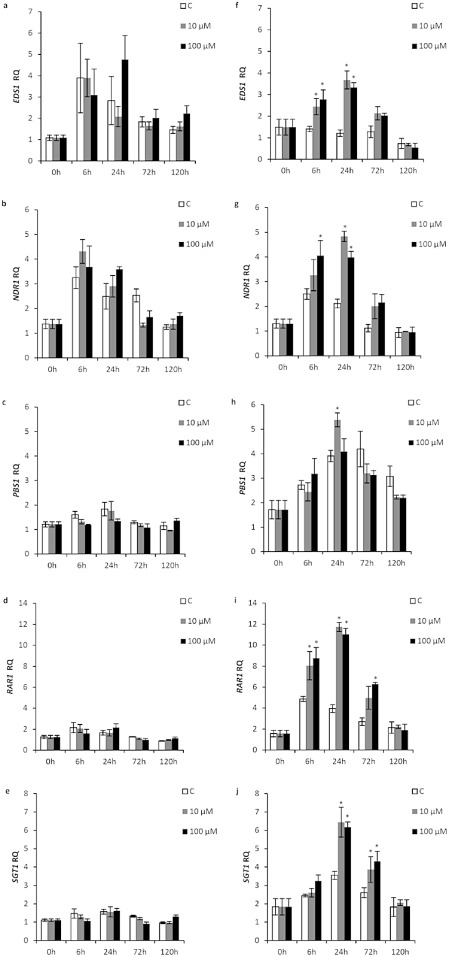
The effect of Xflg22 on the expression of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI)/effector‐triggered immunity (ETI) perception and signalling genes in ‘Duncan’ grapefruit (a–e) and ‘Nagami’ kumquat (f–j). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
The expression of SA metabolism genes in ‘Duncan’ grapefruit was no different from that of water controls at any time point and any Xflg22 concentration, except for a significantly lower level of EDS5 with 10 μm Xflg22 and ICS1 with 100 μm Xflg22 at 72 h (Fig. 3a‐c). In ‘Nagami’ kumquat, EDS5 remained unchanged (Fig. 3d). ICS1 levels were lower than those of water controls at 24, 72 and 120 h, but not significantly lower than the pre‐inoculation levels (Fig. 3e). In contrast, PAL1 expression was significantly induced at 24 h (10 and 100 μm treatments) and 72 h (100 μm treatment) after inoculation (Fig. 3f). Expression of AZI1 was not significantly induced compared with the controls by any of the treatments in any of the two genotypes (Fig. S2, see Supporting Information). In a separate, replicate experiment, however, significant induction of ICS1 at 24 and 72 h, and of AZI1 at 72 h, by Xflg22 was observed in ‘Duncan’ grapefruit (Fig. S6, j1 and l1, see Supporting Information).
Figure 3.
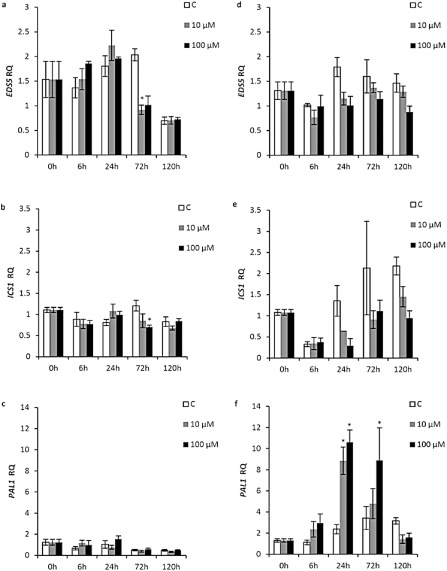
The effect of Xflg22 on the expression of salicylic acid (SA) biosynthesis and signalling genes in ‘Duncan’ grapefruit (a–c) and ‘Nagami’ kumquat (d–f). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
The inoculation of ‘Duncan’ grapefruit with either 10 or 100 μm of Xflg22 did not change the expression levels of the transcriptional regulators NPR1, NPR2 and NPR3 significantly (Figs 4a,b and S3a, see Supporting Information). In ‘Nagami’ kumquat, no significant changes were observed in the expression of NPR1 by any of the treatments (Fig. S3b). However, NPR2 and NPR3 were significantly induced at 6 and 24 h after inoculation. The induction at 6 h of NPR2 was significantly higher with 100 μm of Xflg22 relative to that with 10 μm of Xflg22 (Fig. 4c,d).
Figure 4.
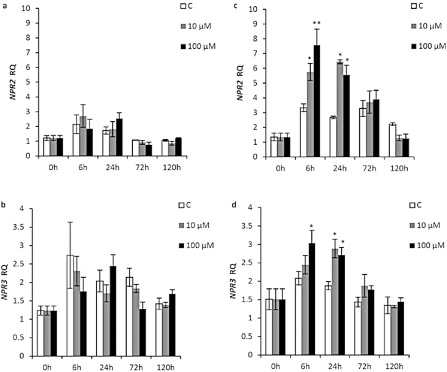
The effect of Xflg22 on the expression of transcription regulation genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. A single asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. The double asterisk indicates that the RQ of the 100 μm Xflg22 treatment is significantly higher (P < 0.05) than that of the control and 10 μm Xflg22 at the indicated time point. Bars are means ± standard error (n = 3).
In the case of the PR gene responses, we observed higher expression levels of PR1 in ‘Duncan’ grapefruit after Xflg22 treatment compared with the water controls; however, the induction of this gene was not significant relative to the levels at 0 h (Fig. 5a), although, in a separate experiment, Xflg22 significantly induced PR1 at 24 and 72 h (Fig. S6, p1). In ‘Nagami’ kumquat, Xflg22 induced significantly lower expression of PR1 at 24 h (Fig. 5b), but did not significantly affect the expression of RdRp1 (Fig. S4d, see Supporting information). However, RdRp1 was significantly up‐regulated by Xflg22 at 6, 24 and 72 h in a separate experiment (Fig. S6, r2). The expression of CHI and RdRp1 was not affected in ‘Duncan’ grapefruit (Fig. S4a,b), and CHI expression was unaffected in ‘Nagami’ kumquat, by Xflg22 treatment (Fig. S4c).
Figure 5.
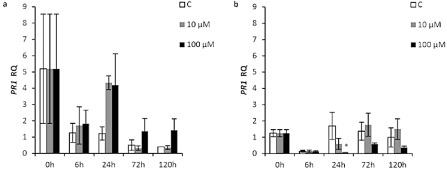
The effect of Xflg22 on the expression of PR1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
In ‘Duncan’ grapefruit, neither the expression of JAR1 nor COI1 was affected by Xflg22 treatment; similar results were also observed in ‘Nagami’ kumquat, except for the induction of JAR1 at 24 h by 100 μm of Xflg22 (Fig. S5, see Supporting Information).
In a separate, replicate experiment, Xflg22 treatment (10 μm) consistently induced defence‐associated genes WRKY22, GST1, EDS1, NDR1, PBS1, RAR1, SGT1, PAL1, NPR2 and NPR3 in ‘Nagami’ kumquat, but did not affect the expression of these genes in ‘Duncan’ grapefruit. However, the expression of WRKY22, EDS5, ICS1, AZI1 and PR1 in ‘Duncan’ grapefruit and RdRp1 and JAR1 in ‘Nagami’ kumquat by Xflg22 (Fig. S6) showed inconsistencies with the results described previously (Figs 1, 2, 3, 4, 5).
Thus, our results indicate that Xflg22 induced transcriptional reprogramming of defence‐associated genes in ‘Nagami’ kumquat, but not in ‘Duncan’ grapefruit. In general, a larger number of genes showed expression levels significantly higher in ‘Nagami’ kumquat, particularly at 24 h, than those of pre‐inoculation and water control levels, compared with ‘Duncan’ grapefruit. ‘Nagami’ kumquat responded more rapidly (as early as 6 h post‐treatment) than ‘Duncan’ grapefruit to Xflg22 treatment. Most genes had similar expression levels between the two Xflg22 concentrations tested (10 and 100 μm) in both citrus genotypes. A few exceptions were WRKY22 at 24 h and NPR2 at 6 h in ‘Nagami’ kumquat, with significantly higher levels in the 100 μm treatment. In addition, 100 μm of Xflg22 resulted in longer lasting induction of WRKY22, RAR1 and PAL1 than 10 μm of Xflg22 in ‘Nagami’ kumquat.
The level of Xflg22‐induced transcriptional reprogramming is related to the level of Xcc resistance/susceptibility in citrus
In citrus, resistance to canker can be divided into five levels: highly resistant, resistant, less susceptible, susceptible and highly susceptible (Gottwald et al., 2002). ‘Nagami’ kumquat and ‘Duncan’ grapefruit represent the highly resistant and highly susceptible levels, respectively (Goto, 1992; Gottwald et al., 1993, 2002). In order to determine whether the level of resistance/susceptibility to canker correlated with PTI induced by Xflg22, two further citrus genotypes, susceptible ‘Navel’ sweet orange and resistant ‘Sun Chu Sha’ mandarin, were studied. For simplicity, we chose only genes which, in the experiments described above, were differentially expressed after Xflg22 treatment, and we only used the 10 μm Xflg22 concentration.
In susceptible ‘Navel’ sweet orange, the expression of most genes was not significantly different from pre‐inoculation levels and water controls. However, the transcript levels of GST1, EDS1, NDR1 and RAR1 were significantly higher and ICS1 was significantly lower (Fig. 6a). In contrast, resistant ‘Sun Chu Sha’ mandarin showed significantly higher levels of GST1, EDS1, NDR1, RAR1, SGT1, PAL1 and NPR3 transcripts, and much higher induction of WRKY22, than that of ‘Navel’ sweet orange (Fig. 6b) This pattern was very similar (with the sole exception of NPR2) to the observations with the highly resistant ‘Nagami’ kumquat.
Figure 6.
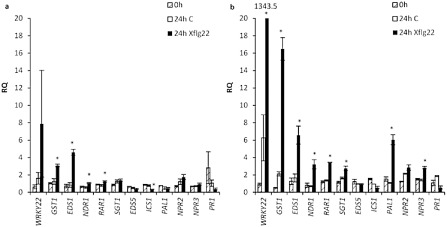
The effect of 10 μm Xflg22 on the expression of defence‐associated genes in (a) ‘Navel’ sweet orange and (b) ‘Sun Chu Sha’ mandarin, 24 h after treatment. RQ values 24 h after Xflg22 treatment (black) are shown in comparison with pre‐inoculation (shaded) and water control (white) levels. The genes studied are shown on the x axis. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation (0 h) and control levels. Bars are means ± standard error (n = 3).
Xflg22 induces a higher oxidative burst in canker‐resistant genotypes
flg22 elicits a rapid oxidative burst in plants, one of the earliest observable events in plant immunity (Felix et al., 1999). In order to study the early Xflg22 response in citrus and its association with canker resistance, we compared Xflg22‐induced ROS accumulation between canker‐resistant (‘Nagami’ kumquat and ‘Sun Chu Sha’ mandarin) and canker‐susceptible (‘Duncan’ grapefruit and ‘Navel’ sweet orange) genotypes during the first 60 min of Xflg22 exposure. Xflg22 treatment caused the transient production of ROS in all four genotypes relative to water controls (Fig. 7a‐d). In most genotypes, ROS production peaked at around 15 min, quickly declining to pretreatment levels. Interestingly, this was not the case for ‘Sun Chu Sha’ mandarin (Fig. 7c), in which the decline after the 15‐min peak was slower and more variable. Overall, ROS reached higher values in the resistant genotypes (‘Nagami’ kumquat and ‘Sun Chu Sha’ mandarin, Fig. 7a,c) compared with the susceptible genotypes (‘Duncan’ grapefruit and ‘Navel’ sweet orange, Fig. 7b,d).
Figure 7.
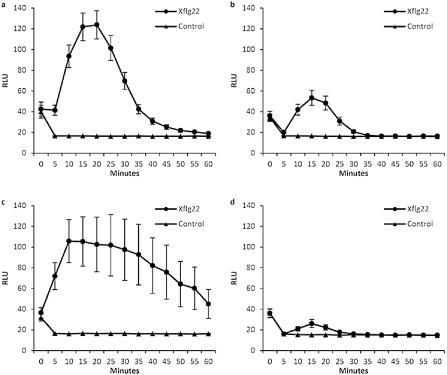
Xflg22‐triggered oxidative burst in (a) ‘Nagami’ kumquat, (b) ‘Duncan’ grapefruit, (c) ‘Sun Chu Sha’ mandarin and (d) ‘Navel’ sweet orange. Reaction solutions containing 100 nm Xflg22 were added to citrus leaf discs at 0 min. No Xflg22 was added to the control reactions. Relative light unit (RLU) was measured every 5 min for 60 min after treatment. Values are means ± standard error (n = 40).
Xflg22 pretreatment increases immunity to citrus canker in ‘Nagami’ kumquat, but not in ‘Duncan’ grapefruit
Our gene expression analysis indicated that ‘Nagami’ kumquat was able to induce a series of defence‐associated genes in response to Xflg22 treatment. This response was more robust than that observed in ‘Duncan’ grapefruit. Moreover, we found a correlation between the extent of gene induction by Xflg22 and the level of canker resistance in the four citrus genotypes studied. In order to determine whether the observed Xflg22‐triggered defence (gene induction) had an effect on citrus canker resistance, we inoculated ‘Duncan’ grapefruit and ‘Nagami’ kumquat leaves with 5 × 105 colony‐forming units (CFU)/mL of Xcc 24 h after infiltration with Xflg22. The bacterial population growth in each genotype was compared (Fig. 8a,b). In ‘Duncan’ grapefruit, Xcc grew to higher levels than those in ‘Nagami’ kumquat, and pretreatment with Xflg22 had little effect on bacterial growth (Fig. 8a). In contrast, ‘Nagami’ kumquat showed significantly reduced Xcc bacterial growth at 2 and 4 days post‐inoculation (DPI) (by up to 27‐fold at 2 DPI) in leaves pretreated with Xflg22 compared with those pretreated with water. Subsequently, this difference in bacterial populations was reduced (Fig. 8b), but remained higher than that observed in ‘Duncan’ grapefruit.
Figure 8.
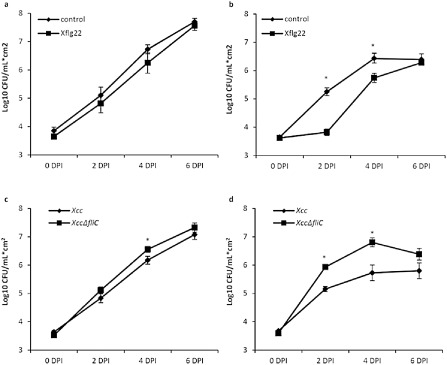
Role of flagellin in the citrus defence response. Top: effect of Xflg22 pretreatment on Xanthomonas citri ssp. citri (Xcc) population growth in (a) ‘Duncan’ grapefruit and (b) ‘Nagami’ kumquat. Xcc at a concentration of 5 × 105 CFU/mL was inoculated into leaves that had been pre‐treated 24 h before with Xflg22 (10 μm). Leaves pre‐treated with water were used as controls. Bottom: comparison of population growth between Xcc wild‐type and fliC mutant (XccΔfliC) in (c) ‘Duncan’ grapefruit and (d) ‘Nagami’ kumquat. Xcc strains at a concentration of 5 × 105 CFU/mL were inoculated into leaves. DPI is days post‐Xcc inoculation. Values shown are means ± standard error (n = 9). An asterisk indicates a significant difference (P < 0.05) at the particular time point.
A flagellin‐deficient Xcc mutant strain shows increased growth compared with the wild type in ‘Nagami’ kumquat, but not in ‘Duncan’ grapefruit
The previous experiment showed that Xflg22 treatment triggered defence‐associated gene induction and Xcc growth inhibition in highly resistant ‘Nagami’ kumquat, but not in highly susceptible ‘Duncan’ grapefruit, suggesting that Xflg22 perception plays an important role in the immune response against Xcc. In order to corroborate the role of Xflg22 and, further, flagellin in the initiation of PTI as a component of canker resistance, we used an Xcc flagellin mutant (XccΔfliC) which contains a deletion in the flagellin‐encoding fliC gene, including a truncation of the Xflg22 region. We inoculated this mutant in citrus leaves and compared bacterial growth with that in wild‐type Xcc. In ‘Duncan’ grapefruit, XccΔfliC and Xcc populations grew similarly up to 6 DPI (the same time frame studied during the gene expression experiments), with the XccΔfliC population significantly higher than that of the wild‐type strain (by two‐fold) at 4 DPI (Fig. 8c). In ‘Nagami’ kumquat, XccΔfliC growth levels were significantly higher than those of Xcc starting at 2 DPI (by six‐fold), and the difference in populations between the two bacterial strains reached 12‐fold at 4 DPI (Fig. 8d).
Discussion
Previous work comparing the response of ‘Nagami’ kumquat and ‘Duncan’ grapefruit has shown that ‘Nagami’ kumquat exhibits an active defence response against Xcc, rendering it highly resistant. This response was found to be associated with physiological (H2O2 production), structural (thickening of the cell wall) and molecular (gene expression reprogramming) changes (Khalaf et al., 2007). In the present study, we set out to dissect this immune response to better understand which components play a role in citrus defence against canker. One of these components is PTI induced by flg22, a well‐characterized epitope in flagellin, the main protein constituent of the bacterial flagellar filament. Is PTI elicited during the Xcc–citrus interaction? This is considered an early response. Is it similar in citrus genotypes with different levels of resistance, hinting at adaptation by the pathogen in the form of effectors that would target the plant immune response at later stages of pathogenicity? Or is the level of PTI distinct, indicating genotypic differences in the perception of this important PAMP and suggestive of a more basic adaptation by the pathogen?
In the first part of this study, we compared the expression of 20 genes directly associated with plant defence between highly resistant ‘Nagami’ kumquat and highly susceptible ‘Duncan’ grapefruit in the presence and absence of Xflg22. Xflg22 significantly and highly induced the expression of the early PTI marker genes WRKY22 and GST1 in ‘Nagami’ kumquat. In contrast, only WRKY22 was significantly induced by 100 μm Xflg22 in ‘Duncan’ grapefruit, and to a fraction of the level observed in ‘Nagami’ kumquat (Fig. 1). This is suggestive of a robust initiation of PTI by Xflg22 in resistant ‘Nagami’ kumquat, but not in susceptible ‘Duncan’ grapefruit, within the studied time course.
In ‘Nagami’ kumquat, Xflg22 also significantly increased the expression of EDS1, NDR1, PBS1, RAR1 and SGT1, whereas none of these genes were induced in ‘Duncan’ grapefruit, at the time points analysed (Fig. 2). EDS1, NDR1, RAR1 and SGT1 have been shown to be involved in the early events of pathogen recognition in plants and are important to both R gene‐mediated resistance (ETI) (Aarts et al., 1998; Azevedo et al., 2002) and PTI‐associated basal or non‐host resistance (Fu et al., 2009; Peart et al., 2002; Yun et al., 2003; Zhang et al., 2010). Moreover, microarray analysis of Arabidopsis seedlings treated with flg22 has shown transcriptional changes in the abundance of genes, including NDR1 and EDS1, as early as 30 min after treatment (Navarro et al., 2004; Zipfel et al., 2004). In our study, we observed a similar induction of these genes at 6 and 24 h after treatment during PTI in the resistant citrus genotypes.
Following pathogen recognition, plants accumulate SA, which has been shown to be important in the establishment of the immune response and defence signalling (Malamy et al., 1990; Metraux et al., 1990). Two separate pathways are involved in SA biosynthesis, each initiated by different catalytic enzymes: ICS1 and PAL1 (Mauch‐Mani and Slusarenko, 1996; Wildermuth et al., 2001). In model plant systems, it has been proposed that the ICS1 pathway is that which is predominantly involved in SA biosynthesis (in the chloroplast) during plant immunity (Strawn et al., 2007; Wildermuth et al., 2001). EDS5 is also critical, as it has been shown recently to function as the transporter of chloroplast‐accumulated SA into the cytoplasm (Serrano et al., 2013). However, AZI1 is involved in the priming of SAR by SA (Greenberg et al., 2009). In ‘Nagami’ kumquat, the expression of EDS5, ICS1 and AZI1 was mostly unaffected, whereas the expression level of PAL1 was increased, by Xflg22 treatment (Figs 3d–f and S2b). PAL1 catalyses the production of trans‐cinnamic acid, a precursor of SA. However, trans‐cinnamic acid is also a precursor in the biosynthesis of lignin and several antimicrobial compounds (flavonoids and anthocyanins) which are part of the defence response in plants, although they do not play a direct role in signalling and the establishment of immunity (Bate et al., 1994; Mauch‐Mani and Slusarenko, 1996; Mishina and Zeier, 2007a). Whether our results indicate that the alternative PAL1‐mediated SA biosynthetic pathway is preferred in ‘Nagami’ kumquat or whether the increase in PAL1 transcripts is merely part of the secondary metabolite defence response, or both, will need to be confirmed in further experiments, but is certainly a compelling idea.
Downstream of SA accumulation, NPR1 regulates plant defence by transcriptional modulation of the expression of PR1 in an SA‐dependent manner (Cao et al., 1994), a process negatively controlled by the SA receptors NPR3 and NPR4 (homologous to citrus NPR2 and NPR3, see Experimental procedures) (Fu et al., 2012; Zhang et al., 2006). In addition to PR1, SA can also induce RdRp1 and CHI, two proteins known for their antimicrobial activities against viral and fungal pathogens, respectively (Samac et al., 1990; Xie et al., 2001). In ‘Nagami’ kumquat, Xflg22 did not affect the expression of NPR1 and CHI, up‐regulated NPR2 and NPR3 and down‐regulated PR1. However, none of these genes was affected by Xflg22 in ‘Duncan’ grapefruit (Figs 4, 5, S3 and S4), although PR1 expression increased in another replicate experiment (Fig. S6, p1). In our study, Xflg22 treatment did not change the expression of NPR1 in either of the genotypes, even when PR1 was induced, suggesting that, in citrus, this gene may be regulated at the protein level rather than at the mRNA level. In Arabidopsis, conversion of the NPR1 protein from an oligomer to a monomer and subsequent translocation from the cytoplasm to the nucleus are required in order to activate PR1 expression (Kinkema et al., 2000; Tada et al., 2008). It has been shown recently that Arabidopsis NPR3 and NPR4 are important in the regulation of SAR by binding SA and mediating the degradation of NPR1 coordinately (Fu et al., 2012). The induction of the citrus orthologues (NPR2 and NPR3) suggests involvement of these two genes during PTI and, perhaps, the activation of SAR. The induction of PR1 in systemic tissues is frequently used as the marker for SAR. In our study, only local expression of PR1 was analysed and its induction was not observed in ‘Nagami’ kumquat or consistently observed in ‘Duncan’ grapefruit. However, it would be interesting to compare Xflg22‐triggered SAR between canker‐resistant and canker‐susceptible genotypes by measuring PR1 expression systemically, and to determine whether citrus NPR1, NPR2 and NPR3 play any role in the induction of SAR in resistant genotypes.
We also studied the expression of JAR1, an important enzyme necessary for the activation of JA signalling (Staswick and Tiryaki, 2004), and COI1, a receptor of jasmonate (Yan et al., 2009). Neither gene was consistently induced in ‘Nagami’ kumquat or ‘Duncan’ grapefruit (Figs S5, S6, s1, t1, s2, t2), suggesting no involvement of JA signalling during Xflg22‐initiated PTI in citrus.
We compared changes in gene expression with pre‐inoculation (0 h) levels after treatment with Xflg22 and water controls. In certain cases (EDS1, NPR1 and NPR3 in ‘Duncan’ grapefruit and RdRp1 in ‘Nagami’ kumquat), we observed higher levels for both Xflg22‐treated and water controls (Figs 2a, S3a, 4b and S4d) compared with 0 h, suggesting that the inoculation procedure or other factors, such as circadian modulation, may have been the cause of the induction. We also compared the effect of two Xflg22 concentrations (10 and 100 μm). In ‘Nagami’ kumquat, the treatment with the lower concentration of 10 μm of Xflg22 induced transcriptional reprogramming. For most genes and time points, there was no significant difference in gene expression with a Xflg22 concentration of 100 μm. Only NPR2 and NPR3 were induced earlier (6 h) (Fig. 4c,d) and WRKY22, RAR1 and PAL1 remained induced for longer (72 h) (Figs 1c, 2i and 3f) with the higher Xflg22 concentration in ‘Nagami’ kumquat. PR1 was down‐regulated at 24 h only by the higher concentration treatment (Fig. 5b). In ‘Duncan’ grapefruit, we did not observe transcriptional reprogramming even with the higher concentration (100 μm) of Xflg22 (Figs 1, 2, 3, 4, 5). These results suggest that the observed absence of a response in ‘Duncan’ grapefruit is not a result of the application of insufficient Xflg22, but rather of a lack of sensitivity to this PAMP.
We tested two further citrus genotypes with intermediate levels of canker resistance/susceptibility using the Xflg22‐responsive genes (WRKY2, GST1, EDS1, NDR1, RAR1, SGT1, EDS5, ICS1, PAL1, NPR2, NPR3 and PR1) identified from the first experiment. At 24 h after treatment, the response observed in resistant ‘Sun Chu Sha’ mandarin was similar to that of the highly resistant ‘Nagami’ kumquat, whereas susceptible ‘Navel’ sweet orange responded similarly to highly susceptible ‘Duncan’ grapefruit (Fig. 6). In addition, we studied the oxidative burst induced by Xflg22 among the four citrus genotypes within the first hour of treatment, and found that ROS reached higher levels and lasted longer in the two resistant genotypes (‘Nagami’ kumquat and ‘Sun Chu Sha’ mandarin) than in the susceptible genotypes (‘Duncan’ grapefruit and ‘Navel’ sweet orange) (Fig. 7). These results, together with the previous ones, indicate a correlation between the levels of resistance in citrus genotypes and the intensity of the flagellin‐triggered defence gene reprogramming. Moreover, resistant genotypes, but not susceptible ones, showed a strong early flagellin‐induced oxidative burst, an indicator of the establishment of PTI. Although our experiments do not rule out the role of other Xcc‐derived PAMPs, which were not part of this study, in citrus canker resistance, they underscore the importance of flagellin perception. Reduced gene induction and weak ROS production in susceptible citrus genotypes may indicate their lower sensitivity to flagellin, which renders Xcc less ‘visible’ to the plant defence system. In Arabidopsis, perception of flagellin is mediated by the receptor FLS2 (Gomez‐Gomez and Boller, 2000), a protein that affects the susceptibility of this plant to adapted and non‐adapted Pseudomonas syringae strains (Forsyth et al., 2010; Hann and Rathjen, 2007; Zipfel et al., 2004). In the case of the citrus–Xcc pathosystem, characterization of the citrus FLS2 orthologue, such as differences in the protein primary structure and expression levels between genotypes, and how these differences affect canker resistance are interesting future areas of study.
Xcc bacterial growth was inhibited in ‘Nagami’ kumquat when plants were pretreated with Xflg22 24 h prior to inoculation (Fig. 8a,b). This indicates that the changes in defence gene expression induced by Xflg22 were accompanied by an enhanced immune response against Xcc in this genotype. The bacterial growth retardation diminished after 2 DPI. This transient effect suggests that Xflg22‐triggered PTI has a role in canker resistance as an early signal that is subsequently amplified into stronger immunity during ETI. The Xcc with a mutated flagellin fliC gene (XccΔfliC) showed markedly higher bacterial growth than wild‐type Xcc in ‘Nagami’ kumquat at all time points, but not in ‘Duncan’ grapefruit (Fig. 8c,d), again indicating that perception of Xflg22 is important to the resistant genotype. This confirms our previous results that Xflg22‐induced PTI contributes to canker resistance. It is worth noting that, because natural infection requires flagellum‐based motility for bacterial entry, for which XccΔfliC is most likely to be compromised, by direct infiltration into the intercellular spaces we can bypass this limitation. The XccΔfliC strain contains a 305‐amino‐acid deletion in fliC (a 399‐amino‐acid protein), including 16 amino acids at the C‐terminus of Xflg22. In ‘Duncan’ grapefruit, negligible gene induction, weak oxidative burst, low bacterial growth inhibition by Xflg22 and little difference in response between Xcc and XccΔfliC suggest that this genotype has a very low response to Xflg22, which may contribute to its susceptibility. It is well known that virulent pathogens evolve effectors capable of suppressing plant defence, including PTI, rendering plants susceptible (Jones and Dangl, 2006; Nomura et al., 2005). However, our study shows evidence that the induction of PTI in citrus plays an important role in defence against Xcc, and contributes towards the final outcome in this plant–pathogen interaction.
In conclusion, the Xcc PAMP Xflg22 induces PTI in the citrus canker‐resistant genotypes but not in susceptible ones, as indicated by the Xflg22‐initiated defence gene expression reprogramming and enhanced immune response against Xcc bacterial growth. Among the citrus defence‐associated genes studied, GST1, EDS1, NDR1, PBS1, RAR1, SGT1, PAL1, NPR2 and NPR3 were significantly induced by Xflg22 in ‘Nagami’ kumquat, but not in susceptible ‘Duncan’ grapefruit. Further, the intensity of defence gene reprogramming (number of genes induced and levels of induction) correlated with the levels of citrus canker resistance observed in the genotypes studied. It will be important to determine the role of these genes in the response to Xcc infection, and any promising gene(s) could potentially be used to engineer citrus‐susceptible genotypes by genetic transformation to increase disease tolerance or even to achieve resistance. Alternatively, chemical induction of these genes may also be a way to increase resistance in susceptible genotypes.
The flagellin‐triggered defence response is widespread among higher plants and is most probably an ancient adaptation (Boller and Felix, 2009). On the basis of our results, there are two possible reasons why Xflg22 does not trigger PTI in susceptible genotypes. One is that there is a deficiency in the detection of flagellin as a result of missing, defective or not fully adapted FLS2‐mediated perception. Another possibility is a deficiency in the initiation of PTI caused by mutations in any of the downstream genes, which would render the plant generally insensitive to PAMPs. One way to differentiate between these two possibilities is to study how canker‐resistant and canker‐susceptible genotypes respond to other PAMPs from Xcc. The fact that we observed genotypes with intermediate levels of Xflg22 sensitivity is more in line with different levels of FLS2‐mediated adaptation, rather than a more global defect in PTI. Our efforts to further characterize PTI and PAMP perception in citrus will continue.
Experimental Procedures
Plant material
The citrus genotypes used in this study were ‘Duncan’ grapefruit (C. paradisi Macf.), ‘Nagami’ kumquat [F. margarita (L.) Swing.], ‘Navel’ sweet orange (C. sinensis Osb.) and ‘Sun Chu Sha’ mandarin (C. reticulalta Blanco). All citrus plants were grown in pots under glasshouse conditions. Before each experiment, the plants were pruned and fertilized weekly until new flushes were produced (4–6 weeks).
Xflg22 peptide treatment
The Xcc flagellin conserved domain (Xflg22: QRLSSGLRINSAKDDAAGLAIS), based on GenBank accession number 21242719, was synthesized by GenScript USA Inc., Piscataway, NJ, USA. The Xflg22 solution was prepared by dissolving the lyophilized peptide in sterile distilled water to a final concentration of either 10 or 100 μm (Zipfel et al., 2004). Young, fully expanded leaves were used for the experiments. The Xflg22 solution was infiltrated into the abaxial surface of leaves using a 1‐cm3 insulin syringe with a needle until half of the leaf was saturated. Infiltration with distilled water was used as a control. Leaf tissue was collected at 0 (before infiltration), 6, 24, 72 and 120 h after infiltration, and RNA was subsequently extracted from infiltrated areas. Three different plants of each genotype were used as biological replicates. The experiments were repeated twice with similar results.
RNA extraction and cDNA synthesis
Total RNA was extracted using TriZol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, followed by DNase treatment and clean up with the RNeasy Plant Mini Kit (Qiagen, Gaithersburg, MD, USA). The RNA concentration and purity were determined using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The cDNA synthesis reaction was performed using 1 μg of RNA and M‐MLV reverse transcriptase (Invitrogen) with random decamers.
Gene expression analysis
Gene expression was measured by RT‐qPCR using a StepOnePlus instrument (Applied Biosystems, Foster City, CA, USA). The reactions were set to comparative C T (ΔΔC T) with the fast amplification (95 °C for 20 s and 40 cycles of 95 °C for 1 s and 60 °C for 20 s). TaqMan MGB probe (250 nm) labelled with 6‐carboxyfluorescein (FAM), primers (900 nm each) and fast universal PCR master mix (Applied Biosystems) were used for target sequence amplification from 5 ng of cDNA (Table 1). Amplification of 5.8S RNA (150 mm of 4,7,2′‐trichloro‐7′‐phenyl‐6‐carboxyfluorescein (VIC)‐labelled probe and 250 mm of each primer) was used as endogenous control. One 0‐h sample selected at random from the three biological replicates was used as the reference and to calculate the RQ values. The data obtained (RQ) were first subjected to a Q‐test (Rorabacher, 1991) for the evaluation of outliers, and subsequently analysed with JMP Genomics 5.0 (SAS Institute Inc., Cary, NC, USA) for model fitting of standard least‐square means (LS means) and Student's t‐test statistical significance analysis (P < 0.05).
ROS production assay
Young, fully expanded citrus leaves were used for the ROS production assay. Leaf discs with a diameter of 3.8 mm were obtained (n = 8 per treatment) from at least four plants and kept in 150 μL of sterile water overnight in a 96‐well plate at room temperature. The next day, the water was replaced with 100 μL of assay solution (100 μm of luminol, 10 μg/mL of horseradish peroxidase and 100 nm of Xflg22). Assay solution without Xflg22 was used as control. Light emission (relative light unit, RLU) was measured in 5‐min intervals for 60 min using a luminescence microplate reader (BioTek, Winooski, VT, USA). Means and standard errors were calculated on the basis of five independent experiments (n = 40).
Bacterial population dynamics
Xflg22‐pretreated leaves were inoculated with Xcc (strain 306) bacterial suspension adjusted to 5 × 105 CFU/mL. Inoculated leaves were collected from three plants (biological replicates) at 0, 2, 4 and 6 DPI. One leaf disc (0.554 cm2) was sampled from the inoculation area from each leaf and ground in 1 mL of sterile tap water. A total of 50 μL from three serial dilutions (up to 10−5) of suspension was spread on Petri dishes with solid nutrient agar medium and incubated at 28 °C for 48 h. CFU from each sample were counted and converted to log10 CFU/mL cm2. Means and standard errors were calculated on the basis of three independent experiments with similar results (n = 9). Student's t‐test was used to determine statistical significance (P < 0.05).
Xcc fliC mutant
A fliC deletion mutant (from nucleotide 106 to 1015) was produced by complementing XccΔfliCΔhrpG strain 306 with a wild‐type hrpG from strain Aw (with an hrpG identical to strain 306) to generate XccΔfliCΔhrpG::hrpG (herein referred to as XccΔfliC).
Supporting information
Fig. S1 The effect of Xflg22 on the expression of EDR1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S2 The effect of Xflg22 on the expression of AZI1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S3 The effect of Xflg22 on the expression of NPR1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S4 The effect of Xflg22 on the expression of pathogenesis‐related genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S5 The effect of Xflg22 on the expression of jasmonic acid (JA) signalling genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
Fig. S6 Replicate experiment for the effect of Xflg22 on the expression of defence‐associated genes in ‘Duncan’ grapefruit (a1–t1) and ‘Nagami’ kumquat (a2–t2). RQ is the relative quantification of gene expression levels after water control (white) or 10 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
Acknowledgements
We would like to thank Dr José Chaparro for providing the ‘Sun Chu Sha’ mandarin plants, Dr Karen Koch for the glasshouse space, Dr Christine Chase for the luminometer and Jerry Minsavage for the XccΔfliC strain. Our thanks also go to Kimberly Niblett for her technical assistance in the laboratory. This research was financially supported by the Citrus Research and Development Foundation, Inc. (CRDF), Lake Alfred, FL, USA. The authors declare that they have no conflicts of interest.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, D. , Goodman, R.M. , Gut‐Rella, M. , Glascock, C. , Weymann, K. , Friedrich, L. , Maddox, D. , Ahl‐Goy, P. , Luntz, T. , Ward, E. and Ryals, J. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis‐related protein 1a. Proc. Natl. Acad. Sci. USA, 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T. , Tena, G. , Plotnikova, J. , Willmann, M.R. , Chiu, W.L. , Gomez‐Gomez, L. , Boller, T. , Ausubel, F.M. and Sheen, J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature, 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Azevedo, C. , Sadanandom, A. , Kitagawa, K. , Freialdenhoven, A. , Shirasu, K. and Schulze‐Lefert, P. (2002) The RAR1 interactor SGT1, an essential component of R gene‐triggered disease resistance. Science, 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Bate, N.J. , Orr, J. , Ni, W.T. , Meromi, A. , Nadlerhassar, T. , Doerner, P.W. , Dixon, P.A. , Lamb, C.J. and Elkind, Y. (1994) Quantitative relationship between phenylalanine ammonia‐lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate‐determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA, 91, 7608–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Bronson, C.H. and Gaskalla, R. (2007) Comprehensive Report on Citrus Canker in Florida. Gainesville, FL: Division of Plant Industry, Florida Department of Agriculture and Consumer Services. [Google Scholar]
- Cao, H. , Bowling, S.A. , Gordon, A.S. and Dong, X. (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell, 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S. , Holub, E.B. and Staskawicz, B.J. (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. USA, 92, 6597–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dewdney, M.M. and Graham, J.H. (2011) 2011 Florida Citrus Pest Management Guide: Citrus Canker, p. 6 Gainesville, FL: Electronic Data Information Source, University of Florida Extension. [Google Scholar]
- Ebel, J. and Cosio, E.G. (1994) Elicitors of plant defense responses. Int. Rev. Cytol. 148, 1–36. [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Forsyth, A. , Mansfield, J.W. , Grabov, N. , de Torres, M. , Sinapidou, E. and Grant, M.R. (2010) Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Mol. Plant–Microbe Interact. 23, 1545–1552. [DOI] [PubMed] [Google Scholar]
- Frye, C.A. and Innes, R.W. (1998) An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell, 10, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, D.‐Q. , Ghabrial, S. and Kachroo, A. (2009) GmRAR1 and GmSGT1 are required for basal, R gene‐mediated and systemic acquired resistance in soybean. Mol. Plant–Microbe Interact. 22, 86–95. [DOI] [PubMed] [Google Scholar]
- Fu, Z.Q. , Yan, S. , Saleh, A. , Wang, W. , Ruble, J. , Oka, N. , Mohan, R. , Spoel, S.H. , Tada, Y. , Zheng, N. and Dong, X. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature, 486, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Goto, M. (1992) Citrus Canker. Englewood Cliffs, NJ: Prentice Hall. [Google Scholar]
- Gottwald, T.R. , Graham, J.H. , Civerolo, E.L. , Barrett, H.C. and Hearn, C.J. (1993) Differential host‐range reaction of citrus and citrus relatives to citrus canker and citrus bacterial spot determined by leaf mesophyll susceptibility. Plant Dis. 77, 1004–1009. [Google Scholar]
- Gottwald, T.R. , Graham, J.H. and Schubert, T.S. (2002) Citrus canker: the pathogen and its impact. Online. Plant Health Progress. doi: 10.1094/PHP-2002-0812-01-RV [DOI]
- Graham, J.H. and Leite, R.P. (2004) Lack of control of citrus canker by induced systemic resistance compounds. Plant Dis. 88, 745–750. [DOI] [PubMed] [Google Scholar]
- Granado, J. , Felix, G. and Boller, T. (1995) Perception of fungal sterols in plants—subnanomolar concentrations of ergosterol elicit extracellular alkalinization in tomato cells. Plant Physiol. 107, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T. , Jung, H.W. , Tschaplinski, T.J. , Wang, L. and Glazebrook, J. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Hann, D.R. and Rathjen, J.P. (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana . Plant J. 49, 607–618. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R. , Zhang, Y. , McKim, S. , Li, X. and Haughn, G.W. (2005) BLADE‐ON‐PETIOLE‐dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell, 17, 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Khalaf, A. , Moore, G.A. , Jones, J.B. and Gmitter, J.F.G. (2007) New insights into the resistance of Nagami kumquat to canker disease. Physiol. Mol. Plant Pathol. 71, 240–250. [Google Scholar]
- Kinkema, M. , Fan, W. and Dong, X. (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell, 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, R.P. and Mohan, S.K. (1990) Integrated management of the citrus bacterial canker disease caused by Xanthomonas campestris pv. citri in the State of Paraná, Brazil. Crop Prot. 9, 3–7. [Google Scholar]
- Malamy, J. , Carr, J.P. , Klessig, D.F. and Raskin, I. (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science, 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Mauch‐Mani, B. and Slusarenko, A.J. (1996) Production of salicylic acid precursors is a major function of phenylalanine ammonia‐lyase in the resistance of Arabidopsis to Peronospora parasitica . Plant Cell, 8, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metraux, J.P. , Signer, H. , Ryals, J. , Ward, E. , Wyssbenz, M. , Gaudin, J. , Raschdorf, K. , Echmid, E. , Blum, W. and Inverardi, B. (1990) Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science, 250, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007a) Bacterial non‐host resistance: interactions of Arabidopsis with non‐adapted Pseudomonas syringae strains. Physiol. Plant. 131, 448–461. [DOI] [PubMed] [Google Scholar]
- Mishina, T.E. and Zeier, J. (2007b) Pathogen‐associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50, 500–513. [DOI] [PubMed] [Google Scholar]
- Navarro, L. , Zipfel, C. , Rowland, O. , Keller, I. , Robatzek, S. , Boller, T. and Jones, J.D.G. (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene‐dependent defense responses and bacterial pathogenesis. Plant Physiol. 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C. and Métraux, J. (1999) Salicylic acid induction‐deficient mutants of Arabidopsis express PR‐2 and PR‐5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell, 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, K. , Melotto, M. and He, S.Y. (2005) Suppression of host defense in compatible plant–Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8, 361–368. [DOI] [PubMed] [Google Scholar]
- Norberg, M. , Holmlund, M. and Nilsson, O. (2005) The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development, 132, 2203–2213. [DOI] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R. , Lu, R. , Sadanandom, A. , Malcuit, I. , Moffett, P. , Brice, D.C. , Schauser, L. , Jaggard, D.A.W. , Xiao, S. , Coleman, M.J. , Dow, M. , Jones, J.D.G. , Shirasu, K. and Baulcombe, D.C. (2002) Ubiquitin ligase‐associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA, 99, 10 865–10 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Tedman‐Jones, J. and Gijzen, M. (2006) Effector‐triggered immunity by the plant pathogen Phytophthora . Trends Microbiol. 14, 470–473. [DOI] [PubMed] [Google Scholar]
- Reddy, M.R.S. (1997) Sources of resistance to bacterial canker in citrus. J. Mycol. Plant Pathol. 27, 80–81. [Google Scholar]
- Rorabacher, D.B. (1991) Statistical treatment for rejection of deviant values—critical values of Dixon Q parameter and related subrange ratios at the 95‐percent confidence level. Anal. Chem. 63, 139–146. [Google Scholar]
- Samac, D.A. , Hironaka, C.M. , Yallaly, P.E. and Shah, D.M. (1990) Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana . Plant Physiol. 93, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, M. , Wang, B.J. , Aryal, B. , Garcion, C. , Abou‐Mansour, E. , Heck, S. , Geisler, M. , Mauch, F. , Nawrath, C. and Métraux, J. (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion‐like transporter EDS5. Plant Physiol. 162, 1815–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. and Tiryaki, I. (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell, 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn, M.A. , Marr, S.K. , Inoue, K. , Inada, N. , Zubieta, C. and Wildermuth, M.C. (2007) Arabidopsis isochorismate synthase functional in pathogen‐induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. [DOI] [PubMed] [Google Scholar]
- Tada, Y. , Spoel, S.H. , Pajerowska‐Mukhtar, K. , Mou, Z. , Song, J. , Wang, C. , Zuo, J. and Dong, X. (2008) Plant immunity requires conformational changes of NPR1 via S‐nitrosylation and thioredoxins. Science, 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torp, J. and Jorgensen, J.H. (1986) Modification of barley powdery mildew resistance gene Ml‐a12 by induced mutation. Can. J. Genet. Cytol. 28, 725–731. [Google Scholar]
- Tsuda, K. , Sato, M. , Glazebrook, J. , Cohen, J.D. and Katagiri, F. (2008) Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J. 53, 763–775. [DOI] [PubMed] [Google Scholar]
- Warren, R.F. , Merritt, P.M. , Holub, E. and Innes, R.W. (1999) Identification of three putative signal transduction genes involved in R gene‐specified disease resistance in Arabidopsis. Genetics, 152, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Fan, B. , Chen, C. and Chen, Z. (2001) An important role of an inducible RNA‐dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA, 98, 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J.B. , Zhang, C. , Gu, M. , Bai, Z.Y. , Zhang, W.G. , Qi, T.C. , Cheng, Z.W. , Peng, W. , Luo, H.B. , Nan, F.J. , Wang, Z. and Xie, D.X. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell, 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, B.‐W. , Atkinson, H.A. , Gaborit, C. , Greenland, A. , Read, N.D. , Pallas, J.A. and Loake, G.J. (2003) Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non‐host resistance in Arabidopsis against wheat powdery mildew. Plant J. 34, 768–777. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Lu, H. , Li, X. , Li, Y. , Cui, H. , Wen, C.‐K. , Tang, X. , Su, Z. and Zhou, J.M. (2010) Effector‐triggered and pathogen‐associated molecular pattern‐triggered immunity differentially contribute to basal resistance to Pseudomonas syringae . Mol. Plant–Microbe Interact. 23, 940–948. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Cheng, Y.T. , Qu, N. , Zhao, Q. , Bi, D. and Li, X. (2006) Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. and Felix, G. (2005) Plants and animals: a different taste for microbes? Curr. Opin. Plant Biol. 8, 353–360. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 The effect of Xflg22 on the expression of EDR1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S2 The effect of Xflg22 on the expression of AZI1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S3 The effect of Xflg22 on the expression of NPR1 in ‘Duncan’ grapefruit (a) and ‘Nagami’ kumquat (b). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S4 The effect of Xflg22 on the expression of pathogenesis‐related genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. Bars are means ± standard error (n = 3).
Fig. S5 The effect of Xflg22 on the expression of jasmonic acid (JA) signalling genes in ‘Duncan’ grapefruit (a,b) and ‘Nagami’ kumquat (c,d). RQ is the relative quantification of gene expression levels after water control (white), 10 μm Xflg22 (grey) or 100 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).
Fig. S6 Replicate experiment for the effect of Xflg22 on the expression of defence‐associated genes in ‘Duncan’ grapefruit (a1–t1) and ‘Nagami’ kumquat (a2–t2). RQ is the relative quantification of gene expression levels after water control (white) or 10 μm Xflg22 (black) infiltration. An asterisk indicates RQ values significantly different (P < 0.05) from pre‐inoculation levels (0 h) and from the water controls at the particular time point. Bars are means ± standard error (n = 3).


