Summary
The Gram‐negative bacterium X anthomonas euvesicatoria (X cv) is the causal agent of bacterial spot disease in pepper and tomato. X cv pathogenicity depends on a type III secretion (T3S) system that delivers effector proteins into host cells to suppress plant immunity and promote disease. The pool of known X cv effectors includes approximately 30 proteins, most identified in the 85‐10 strain by various experimental and computational techniques. To identify additional X cv 85‐10 effectors, we applied a genome‐wide machine‐learning approach, in which all open reading frames (ORFs) were scored according to their propensity to encode effectors. Scoring was based on a large set of features, including genomic organization, taxonomic dispersion, hypersensitive response and pathogenicity (hrp)‐dependent expression, 5′ regulatory sequences, amino acid composition bias and GC content. Thirty‐six predicted effectors were tested for translocation into plant cells using the hypersensitive response (HR)‐inducing domain of AvrBs2 as a reporter. Seven proteins (XopAU, XopAV, XopAW, XopAP, XopAX, XopAK and XopAD) harboured a functional translocation signal and their translocation relied on the HrpF translocon, indicating that they are bona fide T3S effectors. Remarkably, four belong to novel effector families. Inactivation of the xop AP gene reduced the severity of disease symptoms in infected plants. A decrease in cell death and chlorophyll content was observed in pepper leaves inoculated with the xop AP mutant when compared with the wild‐type strain. However, populations of the xop AP mutant in infected leaves were similar in size to those of wild‐type bacteria, suggesting that the reduction in virulence was not caused by impaired bacterial growth.
Keywords: effector, machine learning, pathogenomics, pepper, type III secretion system, Xanthomonas euvesicatoria
Introduction
Many Gram‐negative plant‐pathogenic bacteria utilize a type III secretion (T3S) system to deliver effector proteins into eukaryotic host cells (Galan et al., 2014). T3S effectors, which are selectively delivered by the secretion apparatus, modulate host cellular processes and contribute to bacterial virulence. They can subvert host signalling pathways, suppress immune responses, modify cytoskeleton structure and interfere with cellular trafficking (Dou and Zhou, 2012; Lindeberg et al., 2014).
Most known plant T3S effectors belong to conserved protein families that are represented in evolutionary distant bacteria encoding the T3S system (McCann and Guttman, 2008). However, a given bacterial pathogen species or pathovar may encode unique T3S effectors, which emerged during the evolution of its pathogenicity. The transcriptional activation of T3S effectors is co‐regulated with that of structural components of the T3S system by plant inducible cis‐acting regulatory elements present in their promoters (Fenselau and Bonas, 1995; Xiao and Hutcheson, 1994). At the protein level, the molecular architecture of most T3S effectors is modular and includes an N‐terminal translocation signal that conveys the effector to the T3S apparatus and then into the host cell, and one or more functional domains (Dean, 2011). N‐terminal translocation signals of T3S effectors are very diverse in their primary sequence, but they share common features, such as the frequency of amino acids and residues with certain physicochemical properties (Arnold et al., 2009; Petnicki‐Ocwieja et al., 2002). The translocation of T3S effectors is also facilitated by the binding of accessory chaperone proteins which are involved in the stabilization of the effectors and prevention of their premature interaction with other proteins (Lohou et al., 2013). T3S effectors often display eukaryotic features, such as post‐translational modifications, intracellular localization signals and enzymatic activities, that enable them to function within host cells (Dean, 2011).
Xanthomonas is a large genus of Gammaproteobacteria that cause disease in hundreds of plant hosts (Ryan et al., 2011). Xanthomonas species and pathovars display high specificity to their hosts and cause significant damage in economically important crops, such as rice, citrus, banana, cabbage, tomato, pepper and bean. With few exceptions (e.g. Xanthomonas albilineans, Xanthomonas sacchari and Xanthomonas cannabis), all Xanthomonas pathogenic strains require a functional T3S system to multiply in their host and cause disease. So far, sequence mining of Xanthomonas genomes has identified 52 different families of T3S effectors and a core set of nine effector genes (i.e. xopR, avrBs2, xopK, xopL, xopN, xopP, xopQ, xopX and xopZ) (Ryan et al., 2011; White et al., 2009). Nevertheless, each strain or pathovar encodes a different combination of effectors, which dictates the nature of the interaction with the host and its specificity.
Xanthomonas euvesicatoria (Xcv) is the causal agent of bacterial spot in tomato and pepper plants (Jones et al., 1998). This disease is characterized by the appearance of necrotic lesions on fruits and leaves, and causes significant economic loss in field‐grown crops in warm, temperate and tropical areas of the world (Stall et al., 2009). The repertoire of known Xcv T3S effectors includes approximately 30 proteins, most originally identified in strain 85‐10 (Thieme et al., 2005). Cellular processes affected by several Xcv 85‐10 effectors have been identified. For example, XopD has been implicated in transcriptional regulation (Kim et al., 2013); XopX, XopN, XopB and XopQ have been shown to suppress host immune responses (Kim et al., 2009; Metz et al., 2005; Sonnewald et al., 2012; Teper et al., 2014); and XopJ has been found to interfere with protein secretion and protein degradation (Bartetzko et al., 2009; Ustun et al., 2013). The identification of the entire pool of Xcv 85‐10 T3S effectors and an understanding of their mode of action will shed light on the virulence mechanisms of the bacterium and on host defence signalling.
Various approaches have been used previously for the identification of effectors from different important plant pathogens. Initially, the identification was limited to experimental screens based on specific functional characteristics of T3S effectors, such as translocation into the host cell or the ability to stimulate strong immune responses that are often associated with a rapid and localized programmed cell death (the hypersensitive response, HR) on resistant host or non‐host plants (Greenberg and Vinatzer, 2003). More recently, the availability of genome sequences for many bacterial phytopathogens has allowed us to uncover large sets of T3S effectors based on homologies to known effectors from other pathogens or the presence of eukaryotic characteristics indicative of a function within the host cell (e.g. da Silva et al., 2002; Salanoubat et al., 2002). Effectors have also been predicted by searching for conserved regulatory sequence motifs, such as hypersensitive response and pathogenicity (hrp) or plant‐inducible promoter (PIP) boxes, in the promoter regions, or characteristics of the putative N‐terminal translocation signal (Jiang et al., 2009; Petnicki‐Ocwieja et al., 2002). These approaches have all been successful to some extent, but are limited by the fact that they are based on a single prediction criterion.
In this study, we utilized a genome‐scale computational machine‐learning approach to predict novel T3S effectors of Xcv 85‐10. To this end, we analysed a large set of features that potentially differentiate effectors from non‐effectors. By this analysis, we identified seven novel effectors and experimentally demonstrated their translocation through the T3S system into pepper cells. Follow‐up characterization of the identified effectors revealed that XopAP contributes to the development of disease symptoms caused by Xcv on pepper plants. Our study is an important step in the effort to identify the entire pool of Xcv 85‐10 T3S effectors and to understand the virulence mechanisms of this bacterium.
Results
Prediction of T3S effectors of X cv strain 85‐10 by a machine‐learning approach
To identify novel T3S effectors of Xcv strain 85‐10 (Thieme et al., 2005), we used a machine‐learning approach similar to that employed previously for the identification of type III secreted proteins from Pseudomonas aeruginosa (Burstein et al., 2015) and type IV effectors from Legionella pneumophila and Coxiella burnetii (Burstein et al., 2009; Lifshitz et al., 2014). The machine‐learning algorithm is based on 79 features that potentially differentiate effectors from non‐effectors (Table S1A, see Supporting Information). These features cover a wide range of evolutionary, genomics and biochemical characteristics, including, for example, genomic proximity to other effectors, GC content, differential conservation among phytopathogens that do or do not encode a T3S system, amino acid composition at the N‐terminus and in the entire protein, T3S‐dependent regulation, homology to known T3S effectors of animal‐ and plant‐pathogenic bacteria and similarity to host proteins.
Our algorithm was first trained on known effectors (Table S1B). Specifically, the training step aims to find the features and their relative weights (i.e. the contribution of each feature to the accuracy of the classification) that best differentiate the set of effectors from the rest of the proteins encoded in the genome. Once trained, the algorithm uses the features and their weights to examine all the open reading frames (ORFs) in the genome and provides, as output, a score reflecting the prediction confidence that a certain ORF encodes an effector protein.
The features with the highest weights are listed in Table 1. The first two features in importance analysed the homology of the ORF to known effectors in other phytopathogens. Clearly, effectors are most easily predicted if they are homologous to previously validated effectors of other bacteria. The third best feature considered the similarity of the ORF to the average amino acid composition of Xcv 85‐10 T3S effectors. Notably, some highly informative features were negative. For example, the fourth feature in importance examined whether the ORF has a homologue in the genomes of phytopathogenic bacteria lacking the T3S system. This is an informative negative feature because such an ORF is highly unlikely to encode an effector. The fifth feature with high performance score examined whether the expression of the ORF is dependent on HrpG and HrpX, which are known master regulators of the T3S apparatus and its effectors in Xanthomonas spp. (Wengelnik and Bonas, 1996; Wengelnik et al., 1996). To predict HrpG‐ and HrpX‐dependent expression, we utilized the data published by Guo et al. (2011) that compared the transcriptomes of Xanthomonas axonopodis pv. citri (Xac 306), which is closely related to Xcv 85‐10, and its mutants in either the hrpX or hrpG genes. The high performance score obtained by this feature suggests that the T3S‐dependent expression profiles of effector genes are conserved between related bacterial species and have a strong predictive power for effector identification.
Table 1.
Performance scores of the top 25 features
| Feature | Feature performance* |
|---|---|
| Degree of homology to a known effector of phytopathogenic bacteria | 0.100 |
| Number of homologues among known effectors of phytopathogenic bacteria | 0.051 |
| Similarity to the average amino acid profile of effectors from Xcv 85‐10 | 0.041 |
| Degree of homology to the closest homologue in the proteomes of phytopathogenic bacteria not encoding a type III secretion (T3S) system | 0.037 |
| HrpG/HrpX‐dependent regulation | 0.029 |
| Degree of homology to a known effector of Xcv 85‐10 | 0.029 |
| Number of homologues among known effectors of Xcv 85‐10 | 0.025 |
| Number of homologues in the proteomes of phytopathogenic bacteria not encoding a T3S system | 0.024 |
| Genomic distance from the closest effector | 0.014 |
| Number of effectors in the proximity of 10 open reading frames (ORFs) | 0.011 |
| Relative abundance of valine in the full protein | 0.011 |
| Degree of homology to the closest homologue in bacteria | 0.010 |
| Number of effectors in the proximity of five ORFs | 0.008 |
| Length | 0.008 |
| Hydrophobicity score in the N‐terminus | 0.007 |
| Degree of homology to a known effector of Xanthomonas spp. | 0.006 |
| Relative abundance of serine in the full protein | 0.005 |
| Relative abundance of leucine in the N‐terminus | 0.005 |
| Number of effectors in the proximity of 15 ORFs | 0.004 |
| GC content | 0.004 |
| Number of homologues among known effectors from pathogenic bacteria of mammals | 0.003 |
| Relative abundance of proline in the N‐terminus | 0.002 |
| Number of homologues among known effectors of Xanthomonas spp. | 0.002 |
| Number of effectors in the proximity of 20 ORFs | 0.002 |
| Relative abundance of histidine in the full protein | 0.002 |
*Values obtained from the second training cycle using randomForest quantify the contribution of the feature to the learning accuracy (compared with a learning in which the feature is artificially made non‐informative).
The overall accuracy of the algorithm performance is assessed by a cross‐validation procedure. In this procedure, the algorithm is repeatedly trained on different parts of the known data (effectors and non‐effectors), and its performance is measured by comparing the resulting prediction with the data that were omitted from the training. Our algorithm achieved high accuracy: the area under the precision recall curve (AUPRC; Davis and Goadrich, 2006) obtained was over 0.82.
To predict novel effectors, the analysis included learning (Table S1C) and validation phases. The ORFs with the highest scores that were not previously reported as effectors were tested for translocation into plant cells. Two of the ORFs predicted as effectors by our machine‐learning approach, XCV0324 (XopS) and XCV0442 (XopM), were reported as translocated T3S effectors in parallel to our study (Schulze et al., 2012). After effector validation, we conducted an additional learning analysis (Table S1D) that included the newly identified effectors. The list of high scoring predictions represents a pool of putative additional effectors.
Seven predicted X cv T3S effector proteins are translocated into plant cells
To examine the translocation of predicted effectors, we utilized a reporter system based on the delivery into plant cells of a truncated form of the Xcv T3S effector AvrBs2 (amino acids 62–574). AvrBs262–574 lacks a translocation signal, but is sufficient to elicit HR in plants expressing the Bs2 resistance gene (Roden et al., 2004). A DNA fragment containing 24 base pairs upstream to the start codon and the complete ORFs of each of 36 predicted effector genes (Table S2, see Supporting Information) were cloned upstream to an AvrBs262–574::haemagglutinin (HA) tag fusion. Plasmids were mobilized into the Xcv hrpG* ΔavrBs2 strain, which constitutively expresses the T3S apparatus and contains a mutation in the avrBs2 gene (Roden et al., 2004). Constitutive expression of the T3S apparatus results in an earlier and stronger HR (Wengelnik et al., 1999), enhancing the sensitivity of the translocation assay. The bacterial strains obtained were tested for elicitation of the HR in the pepper line ECW20R, which encodes a functional Bs2 resistance gene. As expected, Xcv hrpG* ΔavrBs2 expressing the AvrBs262–574::HA fusion did not elicit the HR in Bs2‐expressing leaves (Fig. 1A, top panel). However, Xcv hrpG*ΔavrBs2 expressing XCV1196 (XopAU), XCV1197 (XopAV), XCV3093 (XopAW), XCV3138 (XopAP), XCVd0086 (XopAX), XCV3786 (XopAK) or XCV4315 (XopAD) fused to AvrBs2::HA induced the HR at 36 h after infection, similar to AvrBs21–61 fused to AvrBs262–574::HA (Fig. 1A, top panel). No HR was observed in leaf areas inoculated with Xcv strains expressing the other tested constructs (data not shown).
Figure 1.
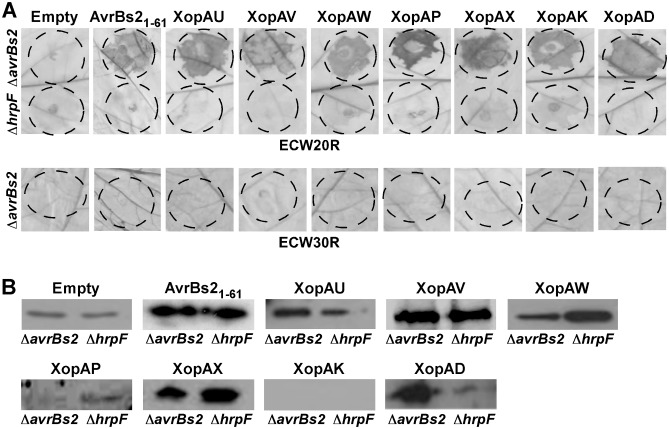
Translocation assay for predicted Xanthomonas euvesicatoria (X cv) type III effectors. (A) Suspensions [5 × 107 colony‐forming units (CFU)/mL] of X cv 85‐10 hrp G *ΔavrBs2 (Δavr B s2) or X cv 85‐10 hrp G * ΔhrpF (Δhrp F) strains containing AvrBs262–574::HA (Empty), the indicated candidate effectors fused to AvrBs262–574::HA or AvrBs21–61 fused to AvrBs262–574::HA were infiltrated into ECW20R (carrying the B s2 resistance gene) or ECW30R (lacking the B s2 resistance gene) pepper leaves. Infected leaves were monitored for the appearance of cell death, washed in bleaching solution and photographed at 36 h after infection. (B) Total protein was extracted from overnight cultures of X cv 85‐10 hrp G * Δavr B s2 (Δavr B s2) or X cv 85‐10 hrp G * Δhrp F (Δhrp F) strains containing AvrBs262–574::HA (Empty), the indicated effectors fused to AvrBs262–574::HA or AvrBs21–61 fused to AvrBs262–574::HA, and analysed by Western blot analysis using anti:HA‐specific antibodies. HA, haemagglutinin.
To confirm that the HR observed in ECW20R pepper leaves resulted from the specific recognition of the translocated AvrBs262–574 fusion protein, bacterial strains were infiltrated into the pepper line ECW30R, which lacks a functional Bs2 gene. As expected, in this background, the HR was not detected for any of the tested genes (Fig. 1A, bottom panel). To validate that translocation of the identified effectors occurred through the T3S system, the plasmids harbouring the various effector–AvrBs262–574::HA fusions were mobilized into the Xcv hrpG*ΔhrpF strain, which carries a deletion in the hrpF translocon gene, and therefore cannot translocate effector proteins into host cells (Buttner et al., 2002). The Xcv strains obtained were tested for the elicitation of HR in pepper ECW20R leaves. None of the fusion proteins elicited the HR when expressed in the Xcv hrpG* ΔhrpF mutant (Fig. 1A, top panel), indicating that the identified effectors require a functional T3S system for translocation. We assessed the expression of the fusion proteins in Xcv ΔavrBs2 and Xcv ΔhrpF strains by Western blot analysis. As shown in Fig. 1B, all the fusion proteins were expressed in both bacterial strains, with the exception of XopAK, whose expression was not detected despite the fact that the HR was observed in areas infiltrated with Xcv hrpG* ΔavrBs2 carrying the XopAK–AvrBs262–574::HA fusion. Together, these results indicate that seven of the 36 tested ORFs encode bona fide T3S effectors.
Conservation of the novel T3S effectors among plant‐associated bacteria
Four novel effectors (XopAV, XopAU, XopAX and XopAW) do not have any homologues that have been reported previously as T3S effectors in other bacteria, and therefore define novel effector families. Homologues of three novel effectors have been reported to be translocated in other bacterial species. XopAD and XopAP share homology to the Ralstonia solanacearum effectors RipS and RipAL (Peeters et al., 2013), respectively. In addition, XopAK is homologous to the Pseudomonas syringae effector HopK1 (Li et al., 2014). A search for homologues within the Xanthomonas genus and in other bacterial species revealed different conservation patterns for the newly identified effectors (Fig. 2 and Table 2). Both XopAV and XopAD have homologues in the genomes of most Xanthomonas spp. Unlike XopAD, which is conserved in several other phytopathogenic bacteria and in Rhizobium, XopAV is unique to Xanthomonas spp. XopAP and XopAK show a similar conservation pattern within the Xanthomonas genus and are found in all the examined strains of the axonopodis clade and in a few other Xanthomonas spp. These effectors also appear in other bacteria with different conservation patterns. XopAU has homologues in all the strains of the axonopodis and oryzae clades. In addition, it is conserved in the genomes of X. gardneri and X. vesicatoria strains which are causal agents of spot disease in tomato and pepper plants, similar to Xcv. Among other bacterial strains, XopAU was found only in Acidovorax spp. XopAX is present in Xcv, but not in other Xanthomonas spp., and only in a few other bacterial species. In this regard, it is important to note that XopAX is encoded in the pXCV183 plasmid of Xcv 85‐10 (Thieme et al., 2005), suggesting that this effector was recently acquired by the bacterium via horizontal gene transfer. Finally, XopAW is present in a few Xanthomonas spp. of the axonopodis clade and in several other plant‐pathogenic bacteria and Rhizobium. The presence of these novel effector families in several bacterial species encoding the T3S system suggests that they may play important roles in the interaction with host plants and in the establishment of disease.
Figure 2.
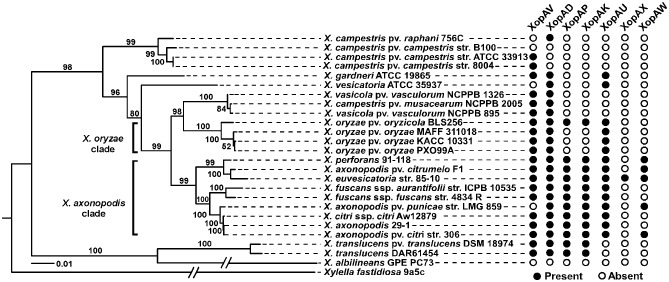
Phyletic pattern of the identified effectors across the X anthomonas genus tree. The presence or absence of each newly validated effector is presented on a tree of X anthomonas strains with X ylella fastidiosa as an outgroup.
Table 2.
Conservation of novel effectors in plant‐associated bacteria
| Effector | Synonym | Putative function | Rs | Ps | Aa | Rz |
|---|---|---|---|---|---|---|
| XopAV | XCV1197 | Hypothetical protein | − * | − | − | − |
| XopAD | XCV4315 | SKW repeat protein | + | + | − | + |
| XopAP | XCV3138 | Class III lipase | + | − | + | − |
| XopAK | XCV3786 | Deamidase | + | + | − | − |
| XopAU | XCV1196 | Serine/threonine kinase | − | − | + | − |
| XopAX | XCVd0086 | Hypothetical protein | − | + | − | + |
| XopAW | XCV3093 | Calcium‐binding protein | + | + | + | + |
*+ and − represent the presence or absence of homologues in the bacterial spp. Ralstonia solanacearum (Rs), Pseudomonas syringae (Ps), Acidovorax avena (Aa) and Rhizobium (Rz).
The genes encoding XopAV (XCV1197) and XopAD (XCV4315) in Xcv 85‐10 share high similarity to the N‐terminus of homologues from other Xanthomonas strains. Interestingly, the C‐terminus of these homologues is highly similar to the adjacent ORFs of XopAV and XopAD (XCV1198 for XopAV; XCV4314 and XCV4313 for XopAD). Therefore, we hypothesize that the ORFs annotated as XCV1197 (XopAV) and XCV1198, and XCV4315 (XopAD), XCV4314 and XCV4313, were originally two complete ORFs that were later truncated by the introduction of early stop codons.
The analysis of conserved domains revealed that four of the novel T3S effectors contain a putative catalytic domain (Fig. S1, see Supporting Information; Table 2): XopAP contains a class III lipase domain, XopAU has a serine/threonine kinase domain, XopAW contains an EF‐hand calcium‐binding motif and XopAK has a deamidase domain.
New insights into shared characteristics of X cv T3S effectors
The analysis of features that can distinguish between effectors and non‐effectors identified several characteristics that are shared by Xcv T3S effectors, but not by the other ORFs in the genome. The analysis of the amino acid composition revealed that several amino acids are differentially represented in the 25 N‐terminal residues and in the whole effector protein. The N‐terminus of effectors was enriched in serine, but displayed a lower content of leucine as compared with the other ORFs (Fig. 3A). When inspecting the entire protein, 10 amino acids were differentially represented in Xcv effectors as compared with the other ORFs (Fig. 3B). Notably, depletion of valine and enrichment of serine were found as key distinguishing features. The enrichment of serine residues was maintained even when excluding from the analysis the 25 N‐terminal amino acids. Thus, the amino acid composition, in both the 25 N‐terminal residues and the entire protein, is different between effectors and non‐effectors, and contributes to the accurate identification of effectors (Table 1). Additional distinguishing characteristics are the GC content and the distribution of effectors in the genome. The average GC content of effector genes (60.3%) was significantly lower than that of other genes (64.6%) (P < 8 × 10−5, two‐tailed t‐test) (Fig. 4A). Moreover, the distribution of effector along the genome is not random (P < 1.6 × 10−27, Wald–Wolfowitz test) (Fig. 4B), but rather characterized by genomic islands enriched in effectors (Fig. 4C). Notably, seven effector genes (hpaG, hpaF, xopF1, hpaA, xopD, xopA and xopM) are clustered in the hrp gene cluster (positions 459 555 to 495 209) and several others reside in small genomic islands.
Figure 3.
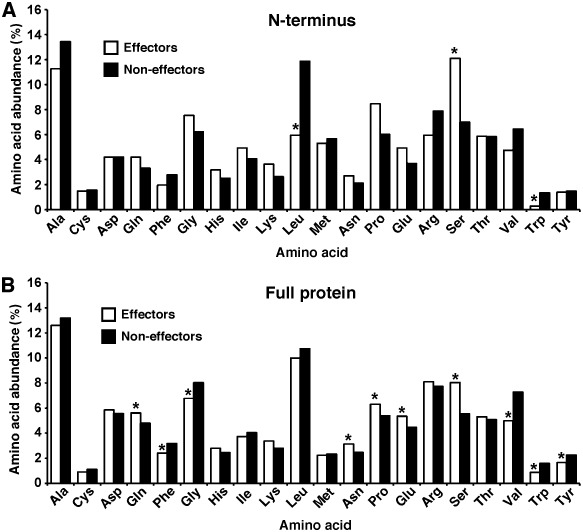
Amino acid composition of Xanthomonas euvesicatoria (X cv) type III secretion (T3S) effectors. The abundance of each amino acid was quantified for effector and non‐effector open reading frames (ORFs). The results represent the quantification of amino acid abundance in the first 25 amino acids at the N‐terminus (A) or in the whole protein (B). Asterisks indicate a significant difference (P < 0.01) between effectors and non‐effectors in both Student's t‐test and Wilcoxon signed‐rank test.
Figure 4.
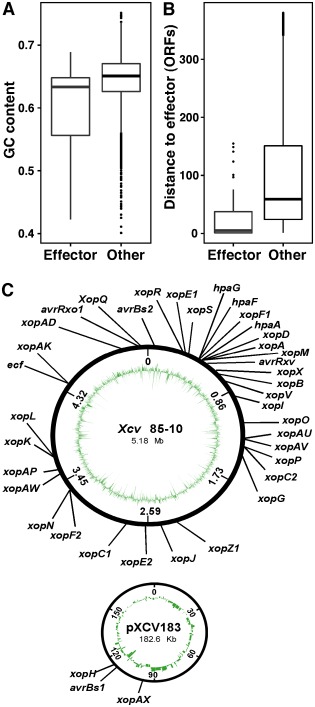
Comparison of GC content and genome organization between effectors and non‐effectors. (A) GC content distribution of effectors vs. non‐effectors. The box represents the 25th and 75th percentiles and the bold line within the box is the median. (B) Distribution of the distance in open reading frames (ORFs) to the closest effector on the genome. (C) Localization of the effectors in the X cv 85‐10 chromosome and plasmid. The inside circles represent the GC content.
XopAP contributes to the development of disease symptoms caused by X cv in pepper leaves
To evaluate the contribution of the novel effectors to Xcv 85‐10 virulence, we inactivated each of the corresponding genes, with the exception of xopAD (for technical reasons), by insertion mutagenesis. The mutant strains obtained were used to infect susceptible pepper plants which were then monitored for the development of disease symptoms and bacterial growth. Disease symptoms were estimated visually and quantified by measuring ion leakage and chlorophyll content, as parameters of cell death and leaf bleaching, respectively. Inactivation of xopAP, but not that of other genes, reduced the severity of the disease symptoms (Figs 5A and S2A, see Supporting Information), whereas bacterial growth was not affected in any of the mutants (Fig. S3, see Supporting Information). The appearance of reduced symptoms in leaves inoculated with the Xcv xopAP:GnR mutant was reflected by lower ion leakage and higher chlorophyll content when compared with plants infected with the wild‐type Xcv (Fig. 5B, C). Conversely, ion leakage and chlorophyll content in leaves inoculated with the other mutants were similar to that in leaves infected with the wild‐type Xcv (Fig. S2B, C). To confirm that the effect on Xcv virulence resulted from inactivation of the xopAP gene, the Xcv xopAP:GnR mutant was complemented with a plasmid carrying xopAP driven by the lacZ promoter. As shown in Fig. 5A–C, leaf areas inoculated with the complemented strain displayed similar disease symptoms, ion leakage and chlorophyll content as the wild‐type Xcv. Together, these results suggest that XopAP acts as a virulence factor that contributes to the development of disease symptoms, but not to bacterial growth, in plants infected with Xcv.
Figure 5.
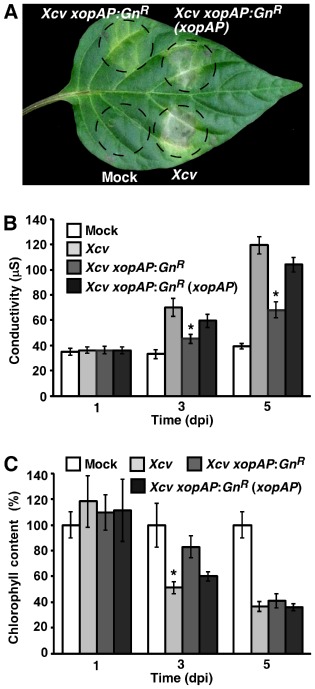
Development of disease symptoms in pepper plants inoculated with the X cv 85‐10 xop AP:GnR mutant. Pepper ECW30R plants were syringe infiltrated with a mock solution or with suspensions [5 × 106 colony‐forming units (CFU)/mL] of X cv 85–10, wild‐type, mutated in xop AP (X cv xopAP:GnR) or mutated in xop AP and complemented with a plasmid carrying the xop AP gene. Inoculated areas were monitored for the appearance of disease symptoms and photographed at 6 days post‐inoculation (dpi) (A). Cell death (B) and leaf bleaching (C) in inoculated areas were quantified at different dpi by measuring electrolyte leakage and chlorophyll content, respectively. Values are the means ± standard error (SE) of five independent leaf areas. Asterisks indicate a significant difference (Student's t‐test, P < 0.05) relative to the X cv 85‐10 wild‐type treatment. The experiment was repeated three times with similar results.
We then examined whether XopAP can cause a detectable phenotype when overexpressed in the non‐host plant Nicotiana benthamiana. The benefit of this system is that the effect of the overexpressed effector can be evaluated in the absence of other Xcv bacterial proteins. Expression of xopAP in N. benthamiana leaves by an Agrobacterium‐mediated transient expression assay caused a bleaching phenotype (Fig. 6A), which was apparent 3 days after Agrobacterium infiltration and was reflected by reduced chlorophyll content compared with a green fluorescent protein (GFP) control (Fig. 6B). However, this phenotype was not accompanied by a significant increase in ion leakage in the inoculated areas (Fig. 6C). Together, these results suggest that XopAP functions as a virulence determinant in Xcv bacteria, and contributes to the development of disease symptoms.
Figure 6.
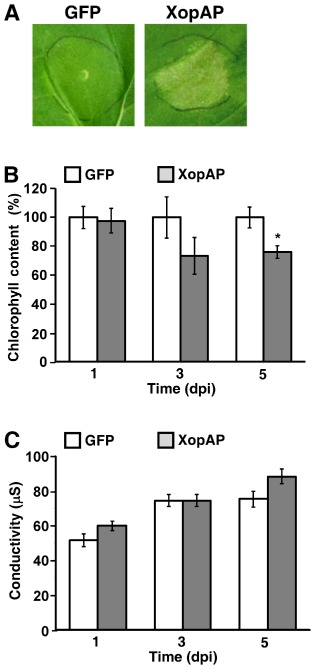
Transient expression of XopAP in N icotiana benthamiana leaves. (A) N icotiana benthamiana leaves were infiltrated with Agrobacterium expressing XopAP or a green fluorescent protein (GFP) control driven by the CaMV 35S promoter, and photographed at 5 days post‐inoculation (dpi). (B) Quantification of leaf bleaching by measurement of the chlorophyll content at 1, 3 and 5 dpi. (C) Cell death was quantified by measurement of electrolyte leakage at 1, 3 and 5 dpi. Values are the means ± standard error (SE) of five independent plants. Asterisks indicate a significant difference (Student's t‐test, P < 0.05) relative to the GFP control treatment. The experiment was repeated three times with similar results.
Discussion
We utilized a machine‐learning approach combined with in planta translocation assays to predict and validate novel T3S effectors of the bacterial pathogen Xcv strain 85‐10. This analysis led to the identification of seven novel Xcv T3S effectors, four of which belong to previously unknown classes of effector families. Functional analysis of the novel effectors revealed that XopAP contributes to the development of disease symptoms.
Previous studies aimed at the identification of T3S effector proteins have been based on the analysis of a single molecular characteristic to differentiate between effectors and non‐effectors. These include, for example, homology to known effectors (Buell et al., 2003; Thieme et al., 2005), the amino acid composition of the protein at the N‐terminus (Petnicki‐Ocwieja et al., 2002) and the presence of cis‐acting elements in the promoter sequence, such as the PIP box or the hrp box in ORFs of Xanthomonas spp. and Pseudomonas spp., respectively (Jiang et al., 2009; Koebnik et al., 2006; Vencato et al., 2006; Xiao and Hutcheson, 1994). However, none of these characteristics is universal to all T3S effectors. For example, we found a perfect or imperfect (containing one mismatch in the TTCGC motif) PIP box only in 26% of Xcv 85‐10 effector genes. In line with this finding, only one of the newly identified effectors, XopAU, contained a PIP box. Moreover, a PIP box was also present in 3% of non‐effector genes and thus, if used as a single differentiating feature, it would have pointed to false‐positive candidate effectors. The strength of our learning approach is the combination of a large set of genomic, regulatory and homology‐based attributes leading to the identification of T3S effectors that were not discovered by other methods.
The selection of features that can distinguish between effectors and non‐effectors sheds new light into the characteristics of Xcv effectors. The analysis of the amino acid composition of Xcv effectors revealed that several amino acids are differentially represented in the 25 N‐terminal amino acids. This is in agreement with observations related to the composition of the translocation signal of T3S effectors of various pathogens (Arnold et al., 2009). Interestingly, several amino acids were also differentially represented in the entire Xcv effector proteins when compared with the other ORFs. This phenomenon may reflect physical characteristics that are required for the recognition and translocation of the effectors by the T3S apparatus. Alternatively, it may reflect selective constraints related to the stability or function of the effector within the host cell. An additional notable distinguishing feature identified by machine learning is the presence of homologues in phytopathogens that encode a T3S system. Phytopathogenic bacteria, which are evolutionarily related to Xcv but lack a T3S system, such as Xylella fastidiosa and Xanthomonas albilineans, encode homologues of Xcv ORFs involved in housekeeping or niche adaptation, but not effector‐encoding genes. Conversely, effector‐encoding genes are more likely to have homologues in more evolutionarily distant bacteria that rely on the T3S system.
Several lines of evidence indicate that at least some of the newly discovered effectors have undergone horizontal gene transfer. For example, XopAU and XopAV are located in the same genomic island; XopAX is located on a plasmid and is present exclusively in Xcv 85‐10 and not in other Xanthomonas strains. In addition, the irregular phyletic pattern of other newly identified effectors (Fig. 2) provides a strong indication that these effectors are readily transferred among different Xanthomonas strains or lost by certain strains during evolution. Finally, the average GC content of the Xcv effectors (60.3%) is significantly (t‐test P = 5.56 × 10−5) lower than the average of non‐effectors (64.6%). On the basis of these findings, it is expected that less‐studied strains of Xanthomonas may encode novel effectors that are yet to be discovered.
Functional analysis of the novel effectors by insertion mutagenesis revealed that XopAP is a virulence factor in Xcv 85‐10 that contributes to the development of symptoms in infected pepper plants, but not to bacterial growth. Interestingly, XopAP encodes a putative class III lipase and shares homology to the plant protein DAD1 (Fig. S4, see Supporting Information), which is involved in the biosynthesis of the plant hormone jasmonic acid (JA) (Ishiguro et al., 2001). JA regulates signalling pathways that mediate resistance against necrotrophic pathogens and increases susceptibility to biotrophic pathogens through its antagonistic effect on salicylic acid (SA) defence pathways. It is possible that XopAP manipulates host lipid biosynthesis to enhance JA production, and thereby activates JA‐induced defences whilst suppressing SA‐induced defences. This hypothesis is supported by recent findings showing that JA signalling components are targeted by several bacterial virulence determinants, including toxins and T3S effectors (Gimenez‐Ibanez et al., 2014; Jiang et al., 2013; Uppalapati et al., 2007). The effectors HopX1 and HopZ1, and the coronatine toxin from different P. syringae pathovars, directly or indirectly degrade JAZ transcriptional repressors leading to the transcription of JA‐dependent genes and the repression of SA‐dependent defence responses against bacteria.
Four novel effectors contain conserved eukaryotic domains which may participate in their molecular function. XopAU contains a putative serine/threonine kinase domain at the C‐terminus. This is the first identified T3S effector of phytopathogenic bacteria that displays a domain of this kind. Effectors that are protein kinases have been reported previously in bacterial pathogens of mammals. For example, the Yersinia effector YopO has been found to interfere with host G‐protein signalling by direct phosphorylation of the Gq α subunit (Navarro et al., 2007). XopAW contains a canonical EF‐hand calcium‐binding motif and can potentially interfere with host calcium signalling. A homologous EF‐hand containing protein, CasA, from Rhizobium etli has been reported to be required for bacteroid development and symbiotic nitrogen fixation in bean (Xi et al., 2000). XopAD appears in Xcv 85‐10 as a truncated protein when compared with other Xanthomonas homologues. In several Xanthomonas spp. the effector size spans from 2200 to 2950 amino acids and contains two conserved domains. The N‐terminal domain, which shares homology with Xcv XopAD, consists of multiple SKW repeats of semi‐conserved 42 amino acids. The C‐terminal domain, which is absent in Xcv 85‐10 XopAD, encodes a putative RelA‐like nucleotidyltransferase domain, which has been found to be involved in the metabolism of the ppGpp nucleotide in Escherichia coli bacteria (Cashel et al., 1996). It is yet to be determined whether XopAD is still functional in Xcv 85‐10 or whether the loss of the RelA‐like domain confers a selective benefit to the bacterium. XopAK is predicted to contain a deaminase catalytic domain at the C‐terminus. HopK1, the P. syringae homologue of XopAK, has been reported recently to suppress host immune responses and to localize to the chloroplast (Li et al., 2014). However, the involvement of deaminase activity in the function of HopK1 and XopAK remains to be investigated. It is interesting to note that the toxin PMT/ToxA of the bacterium Pasteurella multocida activates G‐protein signalling in the host by deamidation of the Gαi2 protein (Orth et al., 2009).
In summary, by employing an efficient machine‐learning approach based on multiple features, we extended the repertoire of known T3S Xcv effectors and paved the way to explore further the molecular strategies used by this pathogen to cause disease in host plants.
Experimental Procedures
Bacterial strains and plant material
The bacterial strains and plasmids used in this study are listed in Table S3 (see Supporting Information). Escherichia coli and Agrobacterium tumefaciens were grown in Luria–Bertani (LB) medium at 37 and 28 °C, respectively. Xcv was grown in nutrient yeast glycerol medium (Daniels et al., 1984) at 28 °C. The plant cultivars used were as follows: pepper (Capsicum annuum) ECW20R (Kearney and Staskawicz, 1990) and ECW30R (Minsavage et al., 1990), and Nicotiana benthamiana (Goodin et al., 2008). Plants were grown in the glasshouse at 25 °C and kept in long‐day conditions (16 h light, 8 h dark).
Machine‐learning scheme
A machine‐learning approach was employed as described previously (Burstein et al., 2009; Lifshitz et al., 2014). The following classification algorithms were used: naïve Bayes (Langley et al., 1992; Morrison, 1990), Bayesian networks (Heckerman et al., 1995), support vector machine (SVM) (Burges, 1998; Vapnik, 1999) and random forest (Breiman, 2001). The ORFs in the genome of Xcv 85‐10 were divided into effector ORFs (Table S1B—newly identified effectors were added to the list in the second learning cycle) and non‐effector ORFs (after filtering highly similar ORFs). This training set was used to train the four classification algorithms for a predetermined set of features (Table S1A). The ‘Wrapper’ procedure for feature selection (Kohavi and John, 1997) was carried out for each classifier, excluding random forest, which performs feature selection internally. During the training procedure, the performance of each classifier was estimated using the mean AUPRC over 10‐fold cross‐validation, as described in Lifshitz et al. (2014). The final classification score of an ORF was a weighted mean of its scores from the four classifiers trained on the full training set. The performance estimate of each classifier was used as the classifier's weight. For random forest, we used the randomForest R package (Liaw and Wiener, 2002), based on the original Fortran implementation described by Breiman (2001). For the other classifiers, the WEKA Java library was used with default parameters. Feature importance was estimated by random forest. The AUPRC values were calculated using AUCCalculator 0.2 (Davis and Goadrich, 2006).
Features and feature selection
For each ORF in the Xcv 85‐10 genome, 79 features measuring different characteristics were analysed (Table S1A). The features were divided into five groups.
Homology features. The protein sequence of each ORF was compared with tailored datasets of protein sequences (Tables S1E–G) and the complete proteome of selected bacteria (Table S1F) using blast‐p (Altschul et al., 1997). For sequence similarity, two measures were used: (i) the blast bit score of the most similar blast hit; (ii) the number of blast hits with E < 0.01.
Physical features. Each ORF was scored for protein length, GC content, amino acid composition in the full protein sequence or in the first 25 amino acids at the N‐terminus. The presence of a secretion signal peptide was predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). The hydrophobic/hydrophilic/amphipathic scores were calculated according to the AAindex of the first 25 amino acids at the N‐terminus of the protein (Kawashima et al., 2008).
Genome organization features. The genomic distance between each ORF and the nearest known effector was quantified by the number of ORFs present between them. In addition, the number of effector ORFs in the proximity of 5, 10, 15, 20, 25 and 30 ORFs upstream and downstream to the selected ORF was measured.
Taxonomic grouping information was obtained using BLink (http://www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blink.html).
Regulatory features. The 300‐bp region upstream of the annotated start codon of each ORF was analysed for the presence of the −10 consensus motif (TATAAT), the perfect PIP box motif (TTCGC‐N15‐TTCGC) and a PIP box motif containing one nucleotide mismatch. In addition, we utilized the list of differentially regulated genes in the hrpG or hrpX deletion mutants, obtained from transcriptomic data of X. axonopodis pv. citri 306 (Xac) (Guo et al., 2011). Specifically, for each ORF, we examined whether its Xac homologue was differentially regulated.
Phylogenetic reconstruction
The phylogenetic tree of Xanthomonas was reconstructed on the basis of a concatenated alignment of three genes: atpD, gyrB and rpoB. The nucleic acid sequences of these genes were globally aligned using Mafft with the ‘ginsi’ scheme (Katoh and Standley, 2013). The maximum likelihood tree and 100 bootstrap resampling were reconstructed using RAxML (Stamatakis, 2014) under the GTRGAMMA evolutionary model.
Translocation assay
For construction of the pAvrBs2‐HR plasmid, the HR domain of avrBs2 (amino acids 62–574), fused to an haemagglutinin (HA) tag, was polymerase chain reaction (PCR) amplified from the pMDD1(avrBs262–574::HA) plasmid (Roden et al., 2004) and cloned into the pBBR1MCS2 plasmid (Kovach et al., 1995). Candidate effector genes and avrBs21–61 were amplified from Xcv 85‐10 using gene‐specific primers (Table S4, see Supporting Information) and cloned into pAvrBs2‐HR. The plasmids obtained were mobilized into the Xcv hrpG* ΔavrBs2 (Roden et al., 2004) and Xcv hrpG* ΔhrpF (Casper‐Lindley et al., 2002) strains by triparental mating (Rott et al., 1996).
For translocation assays, overnight bacterial cultures were suspended in 10 mm MgCl2 at an optical density at 600 nm (OD600) of 0.1 and infiltrated into the leaves of 7‐week‐old ECW20R (carrying the Bs2 gene) and ECW30R (not carrying the Bs2 gene) pepper using a needleless syringe. The elicitation of HR was monitored at 36 h post‐inoculation. For the visualization of cell death, leaves were harvested and soaked for 24 h in a bleaching solution (40% ethanol, 40% chloroform, 10% acetic acid), and then transferred to a recovery solution (40% glycerol, 10% ethanol). For each translocation assay, three leaves of at least three pepper plants were infiltrated. Experiments were repeated three times with similar results.
Mutant construction and complementation
For the generation of insertion mutants by double crossover in XCV1196 (xopAU), XCV1197 (xopAV), XCV3093 (xopAW), XCV3138 (xopAP) and XCVd0086 (xopAX), the gene of interest and flanking regions were PCR amplified and subcloned into pBluescript KSII. The gentamycin‐3‐acetyltransferase gene (GnR) was then inserted into the gene ORF and the construct obtained was cloned into the pVIK165 suicide vector (Table S3) (Kalogeraki and Winans, 1997). For construction of the insertion mutant in XCV3786 (xopAK) by single crossover, a 368‐bp fragment of the xopAK ORF was inserted into the pVIK165 plasmid. Plasmids were mobilized into Xcv 85‐10 bacteria which were plated on solid NYGB medium (0.5% peptone, 0.3% yeast extract, 2% glycerol) with the appropriate selectable markers (i.e. gentamycin for xopAU, xopAV, xopAW, xopAP and xopAX; kanamycin for xopAK). Gene disruption was verified by PCR using primers corresponding to regions flanking the insertions (Table S4).
For complementation of the Xcv xopAP:GnR mutant, the ORF of xopAP was PCR amplified, cloned into the pBBR1MCS2 plasmid and mobilized into Xcv xopAP:GnR.
Plant inoculation and measurement of bacterial growth, relative chlorophyll content and ion leakage
For virulence assays, 7‐week‐old pepper plants were infiltrated with bacterial cultures [105 colony‐forming units (CFU)/mL when monitoring bacterial growth; 107 CFU/mL when measuring ion leakage and relative chlorophyll content] suspended in 10 mm MgCl2 using a needleless syringe.
For measurement of bacterial growth, three 1‐cm2 leaf discs were sampled from at least three plants and ground in 1 mL of 10 mm MgCl2. Bacterial numbers were determined by plating 10 μL from 10‐fold serial dilutions and counting the resulting colonies.
For the quantification of leaf bleaching, the relative chlorophyll ratio was measured using a CCM‐200 plus chlorophyll meter (Opti‐Sciences Inc., Hudson, NH, USA) in three leaves of at least five plants. Chlorophyll readings were standardized to mock treatment.
For the quantification of ion leakage, three 1.5‐cm‐diameter leaf discs were sampled from infiltrated areas of at least five plants, and floated in 10‐mL tubes containing 5 mL of double‐distilled water for 4 h at 25 °C with shaking. Conductivity was measured using a DDS‐12DW conductivity meter (BANTE Instruments, Shanghai, China).
Agrobacterium‐mediated transient expression
The xopAP gene was cloned with a C‐terminal myc epitope into the pBTEX binary vector (Frederick et al., 1998) and transformed into Agrobacterium GV2260. For transient expression, an overnight culture of Agrobacterium was pelleted, resuspended in induction medium (10 mm MgCl2, 10 mm 2‐(N‐morpholino)ethanesulfonic acid (MES), pH 5.6, 200 mm acetosyringone) and incubated at 25 °C with shaking for 4 h. Bacterial cultures were diluted to OD600 = 0.05 and infiltrated into leaves of N. benthamiana plants using a needleless syringe.
Supporting information
Fig. S1 Conserved domains of newly identified effectors. (A) Schematic representation of the XopAP, XopAU, XopAW, XopAD and XopAK effectors. Boxes represent conserved catalytic domains. A blue box represents an SKW repeat domain. aa, amino acid. (B) Amino acid conservation in the six SKW repeats (each consisting of 42 amino acids) extending from amino acid 360 to 612 of XopAD and represented by a sequence logo.
Fig. S2 Development of disease symptoms in pepper plants inoculated with Xanthomonas euvesicatoria (Xcv) strains carrying mutations in novel effector genes. Pepper ECW30R plants, which are susceptible to Xcv 85‐10, were syringe infiltrated with a mock solution or with suspensions [5 × 106 colony‐forming units (CFU)/mL] of Xcv 85‐10 strains, either wild‐type or mutated in the indicated effector genes by insertion of a gentamycin cassette (GnR). Inoculated areas were monitored for the appearance of disease symptoms and photographed at 6 days post‐inoculation (dpi) (A). Cell death (B) and leaf bleaching (C) in inoculated areas were quantified at different dpi by measuring electrolyte leakage and relative chlorophyll content, respectively. Values are means ± standard error (SE) of five independent leaf areas. The experiment was repeated three times with similar results.
Fig. S3 Contribution of the newly identified effectors to Xanthomonas euvesicatoria (Xcv) virulence. Pepper ECW30R plants were syringe infiltrated with suspensions [5 × 105 colony‐forming units (CFU)/mL] of wild‐type 85‐10 and the following mutant strains obtained by insertion of a gentamycin cassette (GnR): Xcv xopAU:GnR and Xcv xopAW:GnR (A); Xcv xopAP:GnR and Xcv xopAX:GnR (B); Xcv xopAV:GnR and Xcv xopAK:GnR (C). Bacterial populations were determined at the indicated days post‐inoculation (dpi). Values are means ± standard error (SE) of five independent plants. The experiment was repeated three times with similar results.
Fig. S4 Amino acid sequence alignment of Xcv XopAP (XCV3138) and Arabidopsis thaliana DAD1 (Q948R1.1). Protein sequences were aligned with the ClustalX1.83 program using default parameters. Conserved amino acids are shaded in black. Amino acids shaded in grey share similar biochemical properties.
Table S1 Datasets used in the machine‐learning scheme. (A) Features used in the machine‐learning analysis. (B) Type III secretion (T3S) effectors of Xanthomonas euvesicatoria (Xcv) 85‐10 used as the training dataset. (C) Prediction scores of Xcv 85‐10 open reading frames (ORFs) in the first learning phase. (D) Prediction scores of Xcv 85‐10 ORFs in the second learning phase. (E) T3S effectors of phytopathogenic bacteria. (F) T3S effectors of pathogenic bacteria of mammals. (G) T3S effectors of Xanthomonas spp. (H) Proteomes used for homology analysis.
Table S2 Xanthomonas euvesicatoria (Xcv) 85‐10 candidate effectors tested for translocation into pepper cells.
Table S3 Bacterial strains and plasmids used in this study.
Table S4 Primers used in this study.
Acknowledgements
This research was supported by the Israel Science Foundation (ISF; grant no. 326/10 to G.S.) and by the Chief Scientist Office of the Israeli Ministry of Science and Technology (grant no. 3‐8178 to G.S. and T.P.). D.B. was a fellow of the Converging Technologies Program of the Israeli Council for Higher Education. M.G., D.B. and T.P. were supported by the Edmond J. Safra Center for Bioinformatics at Tel Aviv University. This work benefited from interactions promoted by COST Action FA1208 (https://www.cost‐sustain.org).
References
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, R. , Brandmaier, S. , Kleine, F. , Tischler, P. , Heinz, E. , Behrens, S. , Niinikoski, A. , Mewes, H.W. , Horn, M. and Rattei, T. (2009) Sequence‐based prediction of type III secreted proteins. PLoS Pathog. 5, e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartetzko, V. , Sonnewald, S. , Vogel, F. , Hartner, K. , Stadler, R. , Hammes, U.Z. and Bornke, F. (2009) The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: evidence for interference with cell wall‐associated defense responses. Mol. Plant–Microbe Interact. 22, 655–664. [DOI] [PubMed] [Google Scholar]
- Breiman, L. (2001) Random forest. Mach. Learn. 45, 5–32. [Google Scholar]
- Buell, C.R. , Joardar, V. , Lindeberg, M. , Selengut, J. , Paulsen, I.T. , Gwinn, M.L. , Dodson, R.J. , Deboy, R.T. , Durkin, A.S. , Kolonay, J.F. , Madupu, R. , Daugherty, S. , Brinkac, L. , Beanan, M.J. , Haft, D.H. , Nelson, W.C. , Davidsen, T. , Zafar, N. , Zhou, L. , Liu, J. , Yuan, Q. , Khouri, H. , Fedorova, N. , Tran, B. , Russell, D. , Berry, K. , Utterback, T. , Van Aken, S.E. , Feldblyum, T.V. , D'Ascenzo, M. , Deng, W.L. , Ramos, A.R. , Alfano, J.R. , Cartinhour, S. , Chatterjee, A.K. , Delaney, T.P. , Lazarowitz, S.G. , Martin, G.B. , Schneider, D.J. , Tang, X. , Bender, C.L. , White, O. , Fraser, C.M. and Collmer, A. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 100, 10 181–10 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burges, C.J.C. (1998) A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2, 121–167. [Google Scholar]
- Burstein, D. , Zusman, T. , Degtyar, E. , Viner, R. , Segal, G. and Pupko, T. (2009) Genome‐scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 5, e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein, D. , Satanower, S. , Simovitch, M. , Belnik, Y. , Zehavi, M. , Yerushalmi, G. , Ben‐Aroya, S. , Pupko, T. and Banin, E. (2015) Novel type III effectors in Pseudomonas aeruginosa . MBio, 6, e00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner, D. , Nennstiel, D. , Klusener, B. and Bonas, U. (2002) Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 184, 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel, M. , Gentry, D.R. , Hernandez, V.J. and Vinella, D. (1996) The stringent response In: Escherichia coli and Salmonella: Cellular and Molecular Biology (Neidhart F.C., Curtiss R., III, Ingraham J.L., Lin E.C.C., Low K.B., Magasanik B., Reznikoff W.S., Riley M., Schaechter M. and Umbarger H.E., eds), pp. 1458–1496, Washington DC: AMS Press. [Google Scholar]
- Casper‐Lindley, C. , Dahlbeck, D. , Clark, E.T. and Staskawicz, B.J. (2002) Direct biochemical evidence for type III secretion‐dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA, 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, M.J. , Barber, C.E. , Turner, P.C. , Sawczyc, M.K. , Byrde, R.J. and Fielding, A.H. (1984) Cloning of genes involved in pathogenicity of Xanthomonas campestris pv. campestris using the broad host range cosmid pLAFR1. EMBO J. 3, 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J. and Goadrich, M. (2006) The relationship between Precision‐Recall and ROC curves In: Proceedings of the 23rd International Conference on Machine Learning, pp. 233–240, New York, NY, USA: Association for Computing Machinery. [Google Scholar]
- Dean, P. (2011) Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35, 1100–1125. [DOI] [PubMed] [Google Scholar]
- Dou, D. and Zhou, J.M. (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe, 12, 484–495. [DOI] [PubMed] [Google Scholar]
- Fenselau, S. and Bonas, U. (1995) Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant–Microbe Interact. 8, 845–854. [DOI] [PubMed] [Google Scholar]
- Frederick, R.D. , Thilmony, R.L. , Sessa, G. and Martin, G.B. (1998) Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr‐204 in the activation loop of the tomato Pto kinase. Mol. Cell, 2, 241–245. [DOI] [PubMed] [Google Scholar]
- Galan, J.E. , Lara‐Tejero, M. , Marlovits, T.C. and Wagner, S. (2014) Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernandez‐Barbero, G. , Chini, A. , Rathjen, J.P. and Solano, R. (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M. , Zaitlin, D. , Naidu, R.A. and Lommel, S.A. (2008) Nicotiana benthamiana: its history and future as a model for plant–pathogen interactions. Mol. Plant–Microbe Interact. 21, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. and Vinatzer, B.A. (2003) Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6, 20–28. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Figueiredo, F. , Jones, J. and Wang, N. (2011) HrpG and HrpX play global roles in coordinating different virulence traits of Xanthomonas axonopodis pv. citri . Mol. Plant–Microbe Interact. 24, 649–661. [DOI] [PubMed] [Google Scholar]
- Heckerman, D. , Geiger, D. and Chickering, D. (1995) Learning Bayesian networks: the combination of knowledge and statistical data. Mach. Learn. 20, 197–243. [Google Scholar]
- Ishiguro, S. , Kawai‐Oda, A. , Ueda, J. , Nishida, I. and Okada, K. (2001) The DEFECTIVE IN ANTHER DEHISCENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell, 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S. , Yao, J. , Ma, K.W. , Zhou, H. , Song, J. , He, S.Y. and Ma, W. (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9, e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Jiang, B.L. , Xu, R.Q. , Huang, J.D. , Wei, H.Y. , Jiang, G.F. , Cen, W.J. , Liu, J. , Ge, Y.Y. , Li, G.H. , Su, L.L. , Hang, X.H. , Tang, D.J. , Lu, G.T. , Feng, J.X. , He, Y.Q. and Tang, J.L. (2009) Identification of six type III effector genes with the PIP box in Xanthomonas campestris pv. campestris and five of them contribute individually to full pathogenicity. Mol. Plant–Microbe Interact. 22, 1401–1411. [DOI] [PubMed] [Google Scholar]
- Jones, J.B. , Stall, R.E. and Bouzar, H. (1998) Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36, 41–58. [DOI] [PubMed] [Google Scholar]
- Kalogeraki, V.S. and Winans, S.C. (1997) Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene, 188, 69–75. [DOI] [PubMed] [Google Scholar]
- Katoh, K. and Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, S. , Pokarowski, P. , Pokarowska, M. , Kolinski, A. , Katayama, T. and Kanehisa, M. (2008) AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 36, D202–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, B. and Staskawicz, B.J. (1990) Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2 . Nature, 346, 385–386. [DOI] [PubMed] [Google Scholar]
- Kim, J.G. , Li, X. , Roden, J.A. , Taylor, K.W. , Aakre, C.D. , Su, B. , Lalonde, S. , Kirik, A. , Chen, Y. , Baranage, G. , McLane, H. , Martin, G.B. and Mudgett, M.B. (2009) Xanthomonas T3S effector XopN suppresses PAMP‐triggered immunity and interacts with a tomato atypical receptor‐like kinase and TFT1. Plant Cell, 21, 1305–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.G. , Stork, W. and Mudgett, M.B. (2013) Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe, 13, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik, R. , Kruger, A. , Thieme, F. , Urban, A. and Bonas, U. (2006) Specific binding of the Xanthomonas campestris pv. vesicatoria AraC‐type transcriptional activator HrpX to plant‐inducible promoter boxes. J. Bacteriol. 188, 7652–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi, R. and John, G.H. (1997) Wrappers for feature subset selection. Artif. Intell. 97, 273–324. [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. 2nd and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Langley, P. , Iba, W. and Thompson, K. (1992) An analysis of Bayesian classifiers. In: Aaai‐92 Proceedings: Tenth National Conference on Artificial Intelligence, pp. 223–228, Palo Alto, CA, USA: Association for the Advancement of Artificial Intelligence. [Google Scholar]
- Li, G. , Froehlich, J.E. , Elowsky, C. , Msanne, J. , Ostosh, A.C. , Zhang, C. , Awada, T. and Alfano, J.R. (2014) Distinct Pseudomonas type‐III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77, 310–321. [DOI] [PubMed] [Google Scholar]
- Liaw, A. and Wiener, M. (2002) Classification and regression by randomForest. R News, 2, 18–22. [Google Scholar]
- Lifshitz, Z. , Burstein, D. , Schwartz, K. , Shuman, H.A. , Pupko, T. and Segal, G. (2014) Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen‐activated protein kinase pathway. Infect. Immun. 82, 3740–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg, M. , Cunnac, S. and Collmer, A. (2014) Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. [DOI] [PubMed] [Google Scholar]
- Lohou, D. , Lonjon, F. , Genin, S. and Vailleau, F. (2013) Type III chaperones & Co in bacterial plant pathogens: a set of specialized bodyguards mediating effector delivery. Front. Plant Sci. 4, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, H.C. and Guttman, D.S. (2008) Evolution of the type III secretion system and its effectors in plant–microbe interactions. New Phytol. 177, 33–47. [DOI] [PubMed] [Google Scholar]
- Metz, M. , Dahlbeck, D. , Morales, C.Q. , Al Sady, B. , Clark, E.T. and Staskawicz, B.J. (2005) The conserved Xanthomonas campestris pv. vesicatoria effector protein XopX is a virulence factor and suppresses host defense in Nicotiana benthamiana . Plant J. 41, 801–814. [DOI] [PubMed] [Google Scholar]
- Minsavage, G.V. , Dahlbeck, D. , Morales, C.Q. , Whalen, M.C. , Kearny, B. , Bonas, U. , Staskawicz, B.J. and Stall, R.E. (1990) Gene‐for‐gene reletionships specifying disease resistance in Xanthomonas campestris pv. vesicatoria–pepper interactions. Mol. Plant–Microbe Interact. 3, 41–47. [Google Scholar]
- Morrison, D.F. (1990) Multivariate Statistical Methods, New York, NY: McGraw‐Hill. [Google Scholar]
- Navarro, L. , Koller, A. , Nordfelth, R. , Wolf‐Watz, H. , Taylor, S. and Dixon, J.E. (2007) Identification of a molecular target for the Yersinia protein kinase A. Mol. Cell, 26, 465–477. [DOI] [PubMed] [Google Scholar]
- Orth, J.H. , Preuss, I. , Fester, I. , Schlosser, A. , Wilson, B.A. and Aktories, K. (2009) Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc. Natl. Acad. Sci. USA, 106, 7179–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N. , Carrere, S. , Anisimova, M. , Plener, L. , Cazale, A.C. and Genin, S. (2013) Repertoire, unified nomenclature and evolution of the type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics, 14, 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petnicki‐Ocwieja, T. , Schneider, D.J. , Tam, V.C. , Chancey, S.T. , Shan, L. , Jamir, Y. , Schechter, L.M. , Janes, M.D. , Buell, C.R. , Tang, X. , Collmer, A. and Alfano, J.R. (2002) Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA, 99, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden, J.A. , Belt, B. , Ross, J.B. , Tachibana, T. , Vargas, J. and Mudgett, M.B. (2004) A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA, 101, 16 624–16 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott, P.C. , Costet, L. , Davis, M.J. , Frutos, R. and Gabriel, D.W. (1996) At least two separate gene clusters are involved in albicidin production by Xanthomonas albilineans . J. Bacteriol. 178, 4590–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorholter, F.J. , Potnis, N. , Jones, J.B. , Van Sluys, M.A. , Bogdanove, A.J. and Dow, J.M. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat. Rev. Microbiol. 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Salanoubat, M. , Genin, S. , Artiguenave, F. , Gouzy, J. , Mangenot, S. , Arlat, M. , Billault, A. , Brottier, P. , Camus, J.C. , Cattolico, L. , Chandler, M. , Choisne, N. , Claudel‐Renard, C. , Cunnac, S. , Demange, N. , Gaspin, C. , Lavie, M. , Moisan, A. , Robert, C. , Saurin, W. , Schiex, T. , Siguier, P. , Thebault, P. , Whalen, M. , Wincker, P. , Levy, M. , Weissenbach, J. and Boucher, C.A. (2002) Genome sequence of the plant pathogen Ralstonia solanacearum . Nature, 415, 497–502. [DOI] [PubMed] [Google Scholar]
- Schulze, S. , Kay, S. , Buttner, D. , Egler, M. , Eschen‐Lippold, L. , Hause, G. , Kruger, A. , Lee, J. , Muller, O. , Scheel, D. , Szczesny, R. , Thieme, F. and Bonas, U. (2012) Analysis of new type III effectors from Xanthomonas uncovers XopB and XopS as suppressors of plant immunity. New Phytol. 195, 894–911. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C. , Ferro, J.A. , Reinach, F.C. , Farah, C.S. , Furlan, L.R. , Quaggio, R.B. , Monteiro‐Vitorello, C.B. , Van Sluys, M.A. , Almeida, N.F. , Alves, L.M. , do Amaral, A.M. , Bertolini, M.C. , Camargo, L.E. , Camarotte, G. , Cannavan, F. , Cardozo, J. , Chambergo, F. , Ciapina, L.P. , Cicarelli, R.M. , Coutinho, L.L. , Cursino‐Santos, J.R. , El‐Dorry, H. , Faria, J.B. , Ferreira, A.J. , Ferreira, R.C. , Ferro, M.I. , Formighieri, E.F. , Franco, M.C. , Greggio, C.C. , Gruber, A. , Katsuyama, A.M. , Kishi, L.T. , Leite, R.P. , Lemos, E.G. , Lemos, M.V. , Locali, E.C. , Machado, M.A. , Madeira, A.M. , Martinez‐Rossi, N.M. , Martins, E.C. , Meidanis, J. , Menck, C.F. , Miyaki, C.Y. , Moon, D.H. , Moreira, L.M. , Novo, M.T. , Okura, V.K. , Oliveira, M.C. , Oliveira, V.R. , Pereira, H.A. , Rossi, A. , Sena, J.A. , Silva, C. , de Souza, R.F. , Spinola, L.A. , Takita, M.A. , Tamura, R.E. , Teixeira, E.C. , Tezza, R.I. , Trindade dos Santos, M. , Truffi, D. , Tsai, S.M. , White, F.F. , Setubal, J.C. and Kitajima, J.P. (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 417, 459–463. [DOI] [PubMed] [Google Scholar]
- Sonnewald, S. , Priller, J.P. , Schuster, J. , Glickmann, E. , Hajirezaei, M.R. , Siebig, S. , Mudgett, M.B. and Sonnewald, U. (2012) Regulation of cell wall‐bound invertase in pepper leaves by Xanthomonas campestris pv. vesicatoria type three effectors. PLoS ONE, 7, e51763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stall, R.E. , Jones, J.B. and Minsavage, G.V. (2009) Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 47, 265–284. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper, D. , Salomon, D. , Sunitha, S. , Kim, J.G. , Mudgett, M.B. and Sessa, G. (2014) Xanthomonas euvesicatoria type III effector XopQ interacts with tomato and pepper 14‐3‐3 isoforms to suppress effector‐triggered immunity. Plant J. 77, 297–309. [DOI] [PubMed] [Google Scholar]
- Thieme, F. , Koebnik, R. , Bekel, T. , Berger, C. , Boch, J. , Buttner, D. , Caldana, C. , Gaigalat, L. , Goesmann, A. , Kay, S. , Kirchner, O. , Lanz, C. , Linke, B. , McHardy, A.C. , Meyer, F. , Mittenhuber, G. , Nies, D.H. , Niesbach‐Klosgen, U. , Patschkowski, T. , Ruckert, C. , Rupp, O. , Schneiker, S. , Schuster, S.C. , Vorholter, F.J. , Weber, E. , Puhler, A. , Bonas, U. , Bartels, D. and Kaiser, O. (2005) Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187, 7254–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati, S.R. , Ishiga, Y. , Wangdi, T. , Kunkel, B.N. , Anand, A. , Mysore, K.S. and Bender, C.L. (2007) The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant–Microbe Interact. 20, 955–965. [DOI] [PubMed] [Google Scholar]
- Ustun, S. , Bartetzko, V. and Bornke, F. (2013) The Xanthomonas campestris type III effector XopJ targets the host cell proteasome to suppress salicylic‐acid mediated plant defence. PLoS Pathog. 9, e1003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik, V. (1999) The Nature of Statistical Learning Theory. New York, NY: Springer. [Google Scholar]
- Vencato, M. , Tian, F. , Alfano, J.R. , Buell, C.R. , Cartinhour, S. , DeClerck, G.A. , Guttman, D.S. , Stavrinides, J. , Joardar, V. , Lindeberg, M. , Bronstein, P.A. , Mansfield, J.W. , Myers, C.R. , Collmer, A. and Schneider, D.J. (2006) Bioinformatics‐enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant–Microbe Interact. 19, 1193–1206. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. and Bonas, U. (1996) HrpXv, an AraC‐type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria . J. Bacteriol. 178, 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik, K. , Van den Ackerveken, G. and Bonas, U. (1996) HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two‐component response regulators. Mol. Plant–Microbe Interact. 9, 704–712. [DOI] [PubMed] [Google Scholar]
- Wengelnik, K. , Rossier, O. and Bonas, U. (1999) Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181, 6828–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, F.F. , Potnis, N. , Jones, J.B. and Koebnik, R. (2009) The type III effectors of Xanthomonas . Mol. Plant Pathol. 10, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, C. , Schoeters, E. , Vanderleyden, J. and Michiels, J. (2000) Symbiosis‐specific expression of Rhizobium etli casA encoding a secreted calmodulin‐related protein. Proc. Natl. Acad. Sci. USA, 97, 11 114–11 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. and Hutcheson, S.W. (1994) A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae . J. Bacteriol. 176, 3089–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Conserved domains of newly identified effectors. (A) Schematic representation of the XopAP, XopAU, XopAW, XopAD and XopAK effectors. Boxes represent conserved catalytic domains. A blue box represents an SKW repeat domain. aa, amino acid. (B) Amino acid conservation in the six SKW repeats (each consisting of 42 amino acids) extending from amino acid 360 to 612 of XopAD and represented by a sequence logo.
Fig. S2 Development of disease symptoms in pepper plants inoculated with Xanthomonas euvesicatoria (Xcv) strains carrying mutations in novel effector genes. Pepper ECW30R plants, which are susceptible to Xcv 85‐10, were syringe infiltrated with a mock solution or with suspensions [5 × 106 colony‐forming units (CFU)/mL] of Xcv 85‐10 strains, either wild‐type or mutated in the indicated effector genes by insertion of a gentamycin cassette (GnR). Inoculated areas were monitored for the appearance of disease symptoms and photographed at 6 days post‐inoculation (dpi) (A). Cell death (B) and leaf bleaching (C) in inoculated areas were quantified at different dpi by measuring electrolyte leakage and relative chlorophyll content, respectively. Values are means ± standard error (SE) of five independent leaf areas. The experiment was repeated three times with similar results.
Fig. S3 Contribution of the newly identified effectors to Xanthomonas euvesicatoria (Xcv) virulence. Pepper ECW30R plants were syringe infiltrated with suspensions [5 × 105 colony‐forming units (CFU)/mL] of wild‐type 85‐10 and the following mutant strains obtained by insertion of a gentamycin cassette (GnR): Xcv xopAU:GnR and Xcv xopAW:GnR (A); Xcv xopAP:GnR and Xcv xopAX:GnR (B); Xcv xopAV:GnR and Xcv xopAK:GnR (C). Bacterial populations were determined at the indicated days post‐inoculation (dpi). Values are means ± standard error (SE) of five independent plants. The experiment was repeated three times with similar results.
Fig. S4 Amino acid sequence alignment of Xcv XopAP (XCV3138) and Arabidopsis thaliana DAD1 (Q948R1.1). Protein sequences were aligned with the ClustalX1.83 program using default parameters. Conserved amino acids are shaded in black. Amino acids shaded in grey share similar biochemical properties.
Table S1 Datasets used in the machine‐learning scheme. (A) Features used in the machine‐learning analysis. (B) Type III secretion (T3S) effectors of Xanthomonas euvesicatoria (Xcv) 85‐10 used as the training dataset. (C) Prediction scores of Xcv 85‐10 open reading frames (ORFs) in the first learning phase. (D) Prediction scores of Xcv 85‐10 ORFs in the second learning phase. (E) T3S effectors of phytopathogenic bacteria. (F) T3S effectors of pathogenic bacteria of mammals. (G) T3S effectors of Xanthomonas spp. (H) Proteomes used for homology analysis.
Table S2 Xanthomonas euvesicatoria (Xcv) 85‐10 candidate effectors tested for translocation into pepper cells.
Table S3 Bacterial strains and plasmids used in this study.
Table S4 Primers used in this study.


