Summary
Adhesion to host tissue is one of the key steps of the bacterial pathogenic process. Xanthomonas citri ssp. citri possesses a non‐fimbrial adhesin protein, XacFhaB, required for bacterial attachment, which we have previously demonstrated to be an important virulence factor for the development of citrus canker. XacFhaB is a 4753‐residue‐long protein with a predicted β‐helical fold structure, involved in bacterial aggregation, biofilm formation and adhesion to the host. In this work, to further characterize this protein and considering its large size, XacFhaB was dissected into three regions based on bioinformatic and structural analyses for functional studies. First, the capacity of these protein regions to aggregate bacterial cells was analysed. Two of these regions were able to form bacterial aggregates, with the most amino‐terminal region being dispensable for this activity. Moreover, XacFhaB shows features resembling pathogen‐associated molecular patterns (PAMPs), which are recognized by plants. As PAMPs activate plant basal immune responses, the role of the three XacFhaB regions as elicitors of these responses was investigated. All adhesin regions were able to induce basal immune responses in host and non‐host plants, with a stronger activation by the carboxyl‐terminal region. Furthermore, pre‐infiltration of citrus leaves with XacFhaB regions impaired X. citri ssp. citri growth, confirming the induction of defence responses and restraint of citrus canker. This work reveals that adhesins from plant pathogens trigger plant defence responses, opening up new pathways for the development of protective strategies for disease control.
Keywords: citrus canker, non‐fimbrial adhesin, Xanthomonas
Introduction
Xanthomonas citri ssp. citri (Xcc) is a Gram‐negative plant‐pathogenic bacterium responsible for citrus canker. This disease is distributed worldwide and, in severe cases, causes economic losses as a result of plant defoliation, twig dieback, premature fruit drop and general debilitation of the tree (Graham et al., 2004). Bacterial proteins related to attachment are important pathogenicity factors as adhesion to host tissues is a key step for plant colonization and disease development. We have determined that a non‐fimbrial adhesin of Xcc, named XacFhaB, is an important virulence factor for the development of citrus canker. The expression of this adhesin was found to be induced in planta during canker development. This adhesin is necessary for attachment to plant surfaces, epiphytic fitness and colonization of citrus leaves, and is involved in cell‐to‐cell attachment and biofilm formation (Gottig et al., 2009).
XacFhaB is a 4753‐amino‐acid protein encoded by XAC1815 which has high amino acid homology to FhaB from Bordetella pertussis (Gottig et al., 2009), which is secreted by the two‐partner secretion (TPS) pathway. FhaB has a signal peptide followed by the conserved ‘TPS domain’ which directs its secretion through its TPS partner, named FhaC. Beyond the TPS domain, FhaB has a predicted structure of an elongated β‐helix in which the β‐helical fold continues as a rod‐like structure ∼500 Å in length (Clantin et al., 2004; Kajava et al., 2001). The accepted model of FhaB secretion has established that the N‐terminus of FhaB recognizes the N‐terminal domain of FhaC and remains associated with it, whilst the rest of the protein is translocated through the channel. During secretion, the carboxyl‐terminus (C‐terminus) of FhaB is processed at the amino acid 3528 generating an ∼230‐kDa mature protein. The mature form of FhaB is anchored to the outer membrane through the interaction with FhaC, exposing the C‐terminal domain (Mazar and Cotter, 2006; Noel et al., 2012). It has been demonstrated that this region mediates adherence to epithelial and macrophage‐like cells in animals, and is required for colonization of host tissues and modulation of the inflammatory response (Julio et al., 2009). To our knowledge, no studies have been performed to infer plant pathogen non‐fimbrial adhesin domains involved in adherence.
In human bacterial pathogens, there is an emerging idea that adhesins modulate the host immune response. In Porphyromonas gingivalis, a bacterial pathogen associated with several forms of chronic marginal periodontitis, long fimbrial adhesins interact with host cell receptors and initiate intracellular signalling cascades leading to innate defence responses (Amano, 2010). Specifically, in the case of FhaB of B. pertussis, it has been observed that this adhesin activates the host interferon type I response (Dieterich and Relman, 2011). In addition, it is widely known that B. pertussis FhaB is able to trigger an immune defence response in its hosts, and is in fact used in vaccination as an immunogenic molecule (Pines et al., 1999; Sato and Sato, 1999). However, the role of this family of adhesins in plant immune responses has been studied only recently for a small filamentous haemagglutinin‐like protein (Fha1) of Xanthomonas campestris pv. vesicatoria (Xcv) (Choi et al., 2013). This protein is only 445 amino acids in length and lacks the signal peptide and TPS domain. Fha1 interacts with Capsicum annuum hypersensitive‐induced reaction protein (CaHIR1), a pepper plasma membrane‐localized protein that has been proposed to induce immunity‐associated cell death (Choi et al., 2013). Pathogen‐associated molecular patterns (PAMPs) are generally highly conserved molecules within a class of microbes that have an essential function in microbial fitness or survival. Plants have evolved the capacity to recognize these PAMPs through specific receptor triggering of a first line of defence, known as pattern‐triggered immunity (PTI). PTI restricts pathogen growth and thus hampers tissue colonization (Chisholm et al., 2006). The non‐fimbrial adhesins, such as XacFhaB, are important pathogenicity factors conserved in several plant and mammalian pathogens, suggesting that they may act as PAMPs (Mhedbi‐Hajri et al., 2011).
To gain an insight into the role of this family of adhesins, and taking into account that XacFhaB is a large protein, in this work, we expressed and purified different regions of this protein, and tested their functionalities, in order to unveil the regions responsible for bacterial adhesion and to characterize whether plants can recognize them as molecules capable of inducing defence responses.
Results
XacFhaB triggers plant defence responses
In order to analyse whether XacFhaB could trigger plant defence responses, the transcript levels of several genes related to the plant defence response in citrus canker (Garavaglia et al., 2010) were analysed in citrus plants infiltrated with Xcc wild‐type and the ΔfhaB mutant at 107 colony‐forming units (CFU)/mL. Infiltrations at this bacterial concentration do not yield differences in either virulence or bacterial growth between the two strains (Gottig et al., 2009), and therefore differences in defence gene expression level may be related to the lack of XacFhaB. However, and considering that the disease process is complex, the possibility that other factors, such as modified bacterial growth or impaired fitness in the mutant strain, may contribute to the differences in defence gene expression cannot be ruled out. Plant RNA was extracted from citrus leaves at 24 h post‐infiltration (hpi) and real‐time quantitative reverse‐transcriptase polymerase chain reactions (RT‐qPCRs) were performed. Several genes were analysed: oxidative stress marker genes, such as PEROXIREDOXIN (PrxA), NADPH OXIDASE (RbohB), GLUTATHIONE‐S‐TRANSFERASE (GST) and SUPEROXIDE DISMUTASE (SOD), and defence response‐related genes, including MITOGEN‐ACTIVATED PROTEIN KINASE 3 (MAPK3), MAP KINASE KINASE 4 (MKK4), WRKY30 transcription factor, LIPOXYGENASE 2 (Lox2), PATHOGENESIS‐RELATED 1 (PR1), PR3 and PR4. RT‐qPCR analysis showed that all transcripts, except Lox2, were significantly (P < 0.05) less abundant at 24 hpi in plants infected with the ΔfhaB mutant than in plants infected with the Xcc wild‐type (Fig. 1). The lower expression levels in defence genes in the strain lacking XacFhaB indicated that this adhesin may have a role in the elicitation of the plant defence response.
Figure 1.
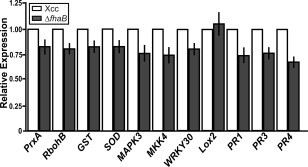
Analysis of the expression levels of genes related to the defence response in citrus leaves infected with Xanthomonas citri ssp. citri (Xcc) and the ΔfhaB mutant by real‐time quantitative reverse‐transcriptase polymerase chain reaction (RT‐qPCR). RNA extracted from citrus leaves at 1 day post‐infiltration (dpi) with 107 colony‐forming units (CFU)/mL of Xcc and ΔfhaB mutant was subjected to RT‐qPCR to identify citrus genes related to the defence response. Grey bars indicate the expression levels of the genes in the ΔfhaB mutant relative to the expression levels of the wild‐type‐infiltrated leaves (white bars). Values are the means of three biological replicates with three technical replicates each. Error bars indicate standard deviations. Results were analysed by Student's t‐test (P < 0.05) and one‐way analysis of variance (ANOVA) (P < 0.05).
Analysis and structural modelling of the XacFhaB sequence and functional domains
Structural modelling of XacFhaB with I‐TASSER (Roy et al., 2010; Yang et al., 2015; Zhang, 2008) suggests that this 4753‐amino‐acid protein adopts a long β‐helix structure that basically extends a TPS β‐helix fold from its C‐terminus (Fig. 2). Based on this and on PFAM annotations, XacFhaB would be an extended, flexible β‐helix with several type‐1 haemagglutinin features up to residue 3400–3500, and a disordered structure with type‐2 haemagglutinin features after that. PFAM further shows that the segments between haemagglutinin domains are rich in low‐complexity regions, often involved in the creation of flexible hinges and in non‐specific binding to other molecules (Coletta et al., 2010; Rado‐Trilla and Alba, 2012). Assuming a continuous β‐helix structure for the full folded segment until residue 3500, this would make up a filament‐like structure around 35–45 Å thick and around 600 Å (0.06 μm) in length when extended, a size similar to that proposed for B. pertussis FhaB from modelling and electron microscopy data (40 Å × 500 Å; Kajava et al., 2001). Interestingly, the sequence segment PLFETRIKFID in XacFhaB, which is cleaved in B. pertussis FhaB to achieve the final mature form (Noel et al., 2012), is conserved in XacFhaB between positions 3542 and 3550, i.e. immediately C‐terminal to the last structured region. Such a maturation point for XacFhaB would imply: (i) the full exposure of the last folded segment, including all the type‐1 haemagglutinin domains and the Fha‐1‐like region that is 97% similar and 95% identical to the already studied Fha1 from Xcv (Fig. S1, see Supporting Information); and (ii) the release of a disordered polypeptide of ∼1200 residues with type‐2 haemagglutinin signatures. These domains have not been characterized in detail, but they compose a family of secreted bacterial exotoxins (InterPro025157). [For a further description of XacFhaB structure, see Text S1 (Supporting Information)].
Figure 2.
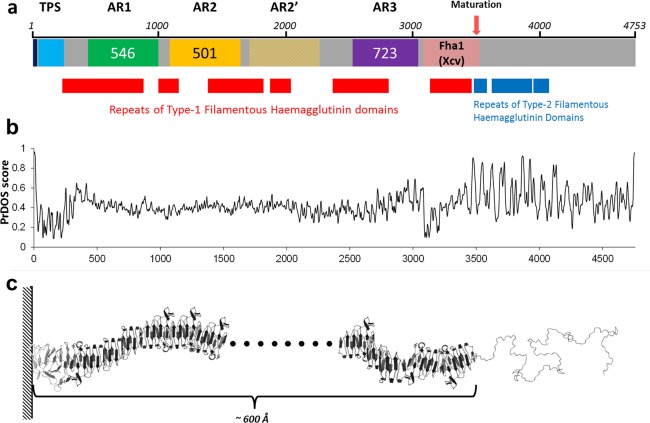
Schematic representation and modelling of FhaB. (a) Schematic representation of Xanthomonas citri ssp. citri (Xcc) FhaB. The dark blue box indicates the signal peptide; the light blue box is the two‐partner secretion (TPS) domain. Adhesin Regions (ARs) AR1, AR2 and AR3 are indicated as green, yellow and purple boxes, respectively, with their lengths indicated inside. The light pink box indicates a region highly similar to Xanthomonas campestris pv. vesicatoria (Xcv) Fha1. The pink arrow indicates the putative protease‐processing site. Red and blue boxes below the representation of the full protein show type‐1 and type‐2 filamentous haemagglutinin signatures, respectively, as detected by PFAM. (b) Disorder predictions by the PrDOS server for XacFhaB. This plot is aligned to the diagram shown in (a). (c) Schematic representation of the structure of XacFhaB based on the disorder predictions and on partial modelling of 600‐residue‐long segments with the I‐TASSER server. The first ∼3500 residues are predicted to form a large flexible β‐fiber, whereas the whole segment C‐terminal to the processing site is expected to be highly disordered.
Breaking down XacFhaB into three main regions for functional studies
Taking into account the analysis of the XacFhaB sequence presented above (Fig. 2) and considering that it is a large protein difficult to express completely in Escherichia coli as a recombinant protein, we divided the protein into three polypeptides, named AR (Adhesin Region) 1–3: AR1 from amino acid 454 to 999 (53 kDa); AR2 from 1113 to 1613 (48 kDa); and AR3 from 2324 to 3046 (72 kDa). AR1 comprises the region immediately C‐terminal to the TPS domain and covers most of the first large series of type‐1 haemagglutinin domains. Immediately after AR1, partially overlapping with the second large series of type‐1 haemagglutinin domains, there is a long region made up of two large repeats that are 46% identical (62% similar) to each other (Fig. S1). Our AR2 region corresponds to the first of these two repeats. Finally, AR3 begins after the second repeat‐like region of AR2 and finishes before the more ordered region highly homologous to the already studied Fha1 from Xcv (Choi et al., 2013). Together, AR3 and the subsequent Fha1‐like segment comprise a region which, in B. pertussis FhaB, was proposed to be its most exposed part (Noel et al., 2012).
AR2 and AR3 regions of XacFhaB promote bacterial aggregation
The three XacFhaB regions were expressed as recombinant proteins in E. coli and purified to homogeneity fused to a 6×His‐Tag. First, the ability of each AR to aggregate bacterial cells was analysed. This was evaluated using a green fluorescent protein (GFP)‐expressing Xcc strain cultured in XVM2 in the presence of AR1, AR2 and AR3. The different proteins were incubated at 5 μm with the bacterial suspensions for 3 h and visualized by confocal microscopy. As a control, the bacterial suspensions were incubated for 3 h with 5 μm 6×His‐Trx which was purified in the same conditions as AR1, AR2 and AR3. The incubation with AR2 and AR3 caused a bacterial association similar to macrocolonies, an effect not observed in either the incubation with AR1 or the control (Fig. 3). The ability of AR1, AR2 and AR3 to promote bacterial aggregation was also analysed with the ΔfhaB mutant strain (Gottig et al., 2009), and results similar to those for the Xcc wild‐type were obtained. Nevertheless, these XacFhaB regions could not mediate any aggregation in E. coli (Fig. S2, see Supporting Information). When the proteins were boiled for 10 min in order to denature them, no agglutination was observed in any case, suggesting that a structured fold is required for this activity. The proteins were checked by sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) after denaturation to ascertain that the heat treatment did not result in their degradation (data not shown).
Figure 3.
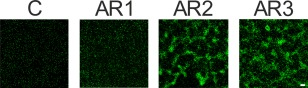
Analysis of the ability of Adhesin Regions (ARs) AR1, AR2 and AR3 to aggregate Xanthomonas citri ssp. citri (Xcc) cells. Representative photographs of confocal laser scanning microscopy of green fluorescent protein (GFP)‐expressing Xcc wild‐type in the presence of 5 µm AR1, AR2 and AR3. As control (C), 6×His‐Trx was used. Bar, 10 µm.
XacFhaB regions elicit plant defence responses when infiltrated
The role of XacFhaB regions as elicitors of plant defence responses was also analysed. AR1, AR2 and AR3 at 5 μm were infiltrated into citrus host plants as well as non‐host plants, such as tomato and pepper. In pepper and tomato tissues infiltrated with AR1 and AR2, marked chlorotic lesions were observed, and AR3‐infiltrated tissues displayed necrotic lesions that were larger in tomato than in pepper leaves. In citrus infiltrated leaves, AR1 caused no visible reaction, whereas AR2 and AR3 both caused mild chlorosis (Fig. 4a). Once again, when the proteins were boiled (without protein degradation) for 10 min before infiltration in tomato leaves, the observed lesions almost disappeared (Fig. S3, see Supporting Information), suggesting that they need to be folded to exert their function. Moreover, callose deposition, a known marker for PAMP‐triggered immunity (Nguyen et al., 2010), was evaluated in the presence of the three regions at 6 hpi in tomato and pepper and at 16 hpi in citrus. The three regions induced significant callose deposition which was not displayed in the 6×His‐Trx‐infiltrated control. A major response was observed in all AR3 infiltrated leaves with a larger response for tomato than for pepper and citrus (Fig. 4b,c). Another marker of plant defence responses analysed was the production of apoplastic reactive oxygen species (ROS). Infiltrations of the three XacFhaB regions in leaves elicited the production of hydrogen peroxide in citrus (6 hpi), pepper and tomato (1 hpi) as visualized by 3,3′‐diaminobenzidine (DAB) staining (Fig. 4d). Infiltrations with the control 6×His‐Trx did not show any DAB staining. Altogether, these results show that the three identified regions individually have varying potential to initiate the response, and therefore suggest a role for full XacFhaB in the induction of plant defence responses.
Figure 4.
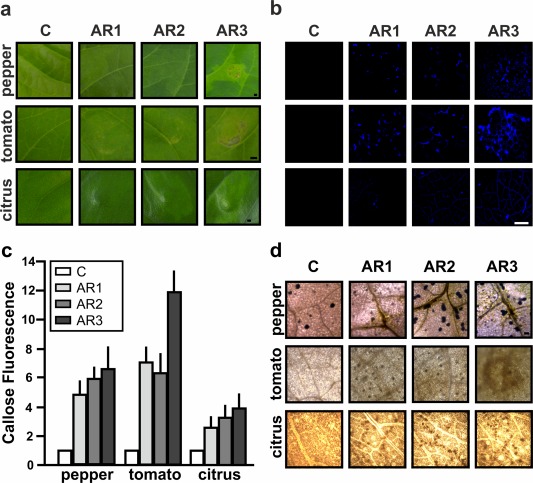
Analysis of the response in pepper, tomato and citrus leaves infiltrated with Adhesin Regions (ARs) AR1, AR2 and AR3. (a) Representative photographs of the responses at 1 day post‐infiltration (dpi) with 5 µm AR1, AR2 and AR3. Bar indicates 1 mm. (b) Representative fluorescence microscopy photographs of aniline blue staining of callose deposition in pepper and tomato leaves at 6 h post‐infiltration (hpi) and citrus at 16 hpi. Bar indicates 20 µm. (c) Relative callose fluorescence intensities were quantified as described in Experimental procedures. Values represent means standardized to the mean callose intensity in control‐treated leaves. Error bars indicate standard deviations. The results are representative of three independent experiments. Results were analysed by one‐way analysis of variance (ANOVA) (P < 0.05). (d) Representative optical microscopy photographs of 3,3′‐diaminobenzidine (DAB) staining in pepper, tomato and citrus leaves infiltrated with the three XacFhaB regions (1 hpi in pepper and tomato and 6 hpi for citrus leaves). Bars indicates 1 mm. In all cases, 6×His‐Trx was used as a control (C).
XacFhaB regions increase the expression of genes for the basal immune response
The expression of genes involved in the basal immune response was analysed in citrus and tomato, the non‐host plant that showed the most significant phenotypes (Sgro et al., 2012). For this purpose, leaves were infiltrated with 5 μm AR1, AR2, AR3 and buffer as a control. Plant RNA from infiltrated citrus and tomato tissues was extracted, and RT‐qPCR was performed. In citrus leaves, the induction of several genes was observed in the presence of the three AR proteins (Fig. 5a). The induction of MAPK3 and WRKY30 was two‐ to three‐fold, whereas MKK4 was induced by the three regions more than five‐fold (P < 0.05) (Fig. 5a). Consistent with the oxidative burst observed (Fig. 4d), RbohB, GST, SOD and PrxA were also induced (P < 0.05). Finally, the defence‐related proteins Lox2 and PR1 also showed increased expression, the latter only with AR2 and AR3 (P < 0.05), whereas PR3 and PR4 showed no significant changes (Fig. 5a). In tomato leaves, MAPK3 and MKK4 showed a two‐fold induction with the three AR proteins at 6 hpi (P < 0.05). In addition, the expression of two genes previously developed as markers for PTI in tomato, Gras2 and Pti5 (Nguyen et al., 2010), was assayed and showed more than a three‐fold induction with the three ARs (P < 0.05) (Fig. 5b).
Figure 5.
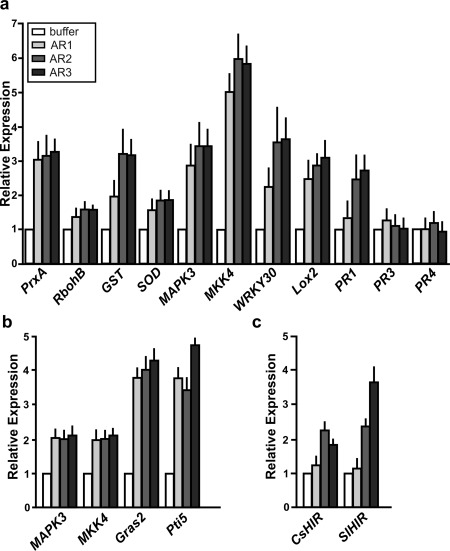
Analysis of the expression levels of genes related to the defence response in citrus and tomato leaves infiltrated with Adhesin Regions (ARs) AR1, AR2 and AR3 by real‐time quantitative reverse‐transcriptase polymerase chain reaction (RT‐qPCR). RNA was extracted from citrus leaves at 16 h post‐infiltration (hpi) (a) or tomato at 6 hpi (b). (c) Relative expression levels of CsHIR and SlHIR in citrus and tomato leaves infiltrated with AR1, AR2 and AR3. Bars indicate the expression levels of the genes relative to the expression levels of the buffer‐infiltrated control. Values are the means of four biological replicates with three technical replicates each. Error bars indicate standard deviations. Results were analysed by one‐way analysis of variance (ANOVA) (P < 0.05).
Next, the ability of AR1, AR2 and AR3 to induce the expression of the recently identified CaHIR1 homologues in citrus and tomato leaves, as occurs with Fha1 from Xcv (Choi et al., 2013), was analysed. With specific primers, transcript levels of CsHIR and SlHIR were quantified by RT‐qPCR on RNA samples obtained from AR‐infiltrated citrus and tomato leaves, respectively. The results showed that AR2 and AR3 induced HIR1 expression in both plants (P < 0.05), whereas AR1 did not (Fig. 5c).
Pre‐infiltration of citrus leaves with XacFhaB regions impairs Xcc infection
Our results indicate that XacFhaB regions can promote defence responses in citrus leaves. Therefore, the potential of these regions to enhance canker disease resistance was investigated. Citrus leaves were pre‐infiltrated with 1 µm of AR1, AR2, AR3 and 6×His‐Trx as a control. The pre‐infiltrated tissues were then infiltrated with Xcc at 106 CFU/mL and bacterial growth was monitored up to 5 dpi. We observed that, at both 3 and 5 dpi, XacFhaB regions were able to induce a defence response, reducing significantly (P < 0.05) the population of bacteria in the three cases (Fig. 6).
Figure 6.
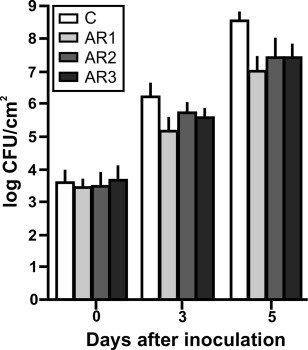
Analysis of Xanthomonas citri ssp. citri (Xcc) growth in citrus leaves pre‐infiltrated with Adhesin Regions (ARs) AR1, AR2 and AR3. Quantification of Xcc growth in citrus leaves pre‐infiltrated with 1 µm of AR1, AR2 and AR3, or with 6×His‐Trx as control. Values are the means obtained from 10 infiltrated citrus leaves at different days post‐infiltration (dpi). Error bars show the standard deviation. Results were analysed by one‐way analysis of variance (ANOVA) (P < 0.05). CFU, colony‐forming units.
Further dissection of AR3, the region with the major eliciting activity
Considering the higher response observed for AR3 in the three plant species analysed, we dissected this region taking into account the information obtained by Pfam analysis. Three subregions were expressed and purified as recombinant polypeptides in E. coli: AR3‐1 (2273–2448), which encompasses only three type‐1 haemagglutinin domains, to analyse the minimal filamentous region; AR3‐2 (2324−2576), which bears four type‐1 haemagglutinin domains and is predicted to adopt a larger filamentous structure than AR3‐1; and AR3‐3 (2709–3046), which mainly comprises disordered predicted regions (Fig. 7a). The analysis of the responses of pepper, tomato and citrus leaves to these three AR3 subregions revealed that the three were able to elicit a response in pepper and tomato (Fig. 7b), whereas no response was observed in citrus leaves (data not shown), consistent with the mild chlorosis observed in these leaves when the complete AR3 was used (Fig. 4a). The production of hydrogen peroxide was analysed by DAB staining in the three plant species (Fig. 7c). The three subregions caused hydrogen peroxide production in tomato and pepper. In citrus, AR3‐2 and AR3‐3 showed stronger DAB staining than AR3‐1. Finally, the expression of characteristic basal immune response genes in tomato was analysed by RT‐qPCR (Fig. 7d). The results showed that MAPK3 and WRKY28 were induced with the three subregions (P < 0.05), whereas Pti5 expression was induced only with AR3‐2 and AR3‐3 (P < 0.05) and Gras2 showed no changes. None was able to increase SlHIR expression (Fig. 7d).
Figure 7.
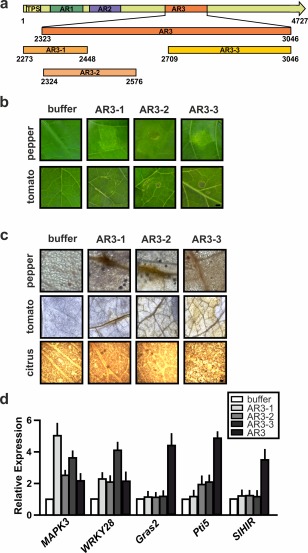
Analysis of the responses in tomato leaves infiltrated with Adhesin Regions (ARs) AR3‐1, AR3‐2 and AR3‐3. (a) Schematic representation of XacFhaB AR3 subregions. Numbers indicate amino acids in the full‐length protein. (b) Representative photographs of the responses at 1 day post‐infiltration (dpi) of 5 µm AR3‐1, AR3‐2 and AR3‐3 in tomato and pepper. (c) 3,3′‐Diaminobenzidine (DAB) detection of H2O2 accumulation in tomato leaves infiltrated with AR3‐1, AR3‐2 and AR3‐3 at 1 dpi. Representative optical microscopy photographs of DAB staining of leaves. Bars indicate 1 mm. (d) Real‐time quantitative reverse‐transcriptase polymerase chain reaction (RT‐qPCR) of tomato genes related to the defence response. Bars indicate the expression levels relative to control of the genes from RNA extracted from leaves infiltrated with AR3 and AR3 subregions at 6 h post‐infiltration (hpi). Values are the means of four biological replicates with three technical replicates each. Error bars indicate standard deviations. Results were analysed by one‐way analysis of variance (ANOVA) (P < 0.05).
Discussion
Many bacterial pathogens have haemagglutinin proteins at their surfaces that allow them to adhere to host tissue. The most studied among these proteins is FhaB from B. pertussis, which constitutes the pathogen's major adhesion factor for lung colonization. Indeed, FhaB is one of the components of highly protective vaccines against whooping cough (Pines et al., 1999; Sato and Sato, 1999). FhaB is able to interact with complement receptor 3 (CR3) integrins of macrophages, crucial for pathogen phagocytosis (Mobberley‐Schuman and Weiss, 2005), as well as to activate the host interferon type I response (Dieterich and Relman, 2011). The fact that FhaB is an important adhesion factor and immune response activator led us to hypothesize that XacFhaB may have a similar function in plant infections. To perform the analyses, and as full XacFhaB is a large protein difficult to express in E. coli as a recombinant protein, we dissected it into three regions.
Our results indicate that AR2 and AR3 are the main regions responsible for bacterial agglutination and, moreover, that the folded structures of AR2 and AR3 are required for agglutination function. As AR2 and AR3 can mediate Xcc bacterial agglutination when added extracellularly, these results suggest that XacFhaB may be secreted in a similar manner to FhaB of B. pertussis or adhere to the cell surface (Mazar and Cotter, 2006). The fact that AR2 and AR3 also promote bacterial aggregation in the ΔfhaB mutant strain, but not in E. coli, suggests that other Xcc molecules are involved in mediating XacFhaB agglutination. Considering that mature XacFhaB is a long β‐helical fiber with several flexible hinges, as predicted and consistent with previous studies in B. pertussis FhaB, and is anchored through its N‐terminal TPS domain, it is reasonable that the more exposed regions AR2 and AR3 are those involved in adhesion, as observed here. Moreover, AR3 and its continuing Fha1‐like region are predicted to be the most exposed areas in XacFhaB, resembling the most exposed region of B. pertussis FhaB and having the highest bacterial agglutination and defence elicitor activities (Mazar and Cotter, 2006). It should be noted that the lengths estimated for the mature, folded parts of these proteins are close to 0.05–0.06 μm, i.e. only one to two orders of magnitude smaller than bacterial sizes. XacFhaB could therefore form thin layers of filaments, thick as 2‐3% the cell diameter (assuming a cell length of 2 μm) providing soft, flexible and sticky surfaces to facilitate unspecific adhesion.
Finally, we observed that processing of the precursor XacFhaB protein would not only lead to the full exposure of the last folded part, which includes all the type‐1 haemagglutinin domains and the Fha‐1‐like region, to mediate binding, but also the release of a disordered polypeptide of ∼1200 residues with type‐2 haemagglutinin signatures. These domains have not been characterized in detail, but they compose a family of secreted bacterial exotoxins and, as such, could contribute to additional infective mechanisms which need to be explored.
The non‐fimbrial plant pathogen adhesins share several characteristics with PAMP molecules characterized previously (Boller and Felix, 2009). For example, they are widely conserved in animal and plant pathogens, are localized at the outer membrane and are required for bacterial pathogenicity. In particular, the idea that XacFhaB, as occurs with FhaB in animal hosts, may behave as an immune elicitor in plants is also supported by the recent evidence that Fha1 from Xcv (which is 95% identical to amino acids 3067–3508 of XacFhaB) interacts with the positive regulator of pathogen‐induced cell death CaHIR1 (Choi et al., 2013). In this context, we analysed markers of the PTI response in tissues infiltrated with the different regions of XacFhaB. Phenotypic observation of cell death, mainly in the non‐host plants tomato and pepper, callose deposition, DAB staining of ROS produced by the oxidative burst during the basal defence response and induction of the expression of genes previously observed to be involved in the basal defence response confirm that XacFhaB acts as a PAMP in the bacterial interaction with host and non‐host tissue. We also analysed the capacity of the different XacFhaB regions to induce HIR1 expression, such as Fha1 of Xcv (Choi et al., 2013). In the XacFhaB sequence, the homologous region to Fha1 begins 20 amino acids after the AR3 C‐terminal end. Even so, AR2 and AR3 induced the expression of HIR1, suggesting a role for HIR1 in the defence responses triggered by AR2 and AR3.
A further confirmation of the role of the different XacFhaB regions in triggering the plant defence response is the observation that the pre‐infiltration of citrus leaves with these regions impairs Xcc growth, inducing citrus canker disease resistance. These results indicate that XacFhaB regions may be used the future as molecules to prevent canker development.
AR3 is the most reactive region of XacFhaB. This is consistent with the proposed folding model for FhaB of B. pertussis (Noel et al., 2012), which indicates that AR3 may be more exposed to plant cell membranes. The aim of the dissection of AR3 into different subregions was to further study this region. Like full AR3, the three subregions elicited a response. The main difference between the subregions and AR3 was the lack of expression induction of gene markers for PTI. However, AR3 subregions induced the expression of MAPK3 and WRKY28, proteins involved in signal transduction and transcriptional processes related to a wide range of stress responses, including biotic and abiotic stresses (Atkinson and Urwin, 2012; Taj et al., 2010). Thus, these results suggest that these subregions induce a stress response, but do not elicit a PAMP‐like reaction as does AR3.
In summary, Xcc, like other animal and plant bacterial pathogens, requires proteins, such as FhaB, to allow it to adhere to host tissues and, in the case of Xcc, to form a biofilm to complete the disease cycle and to be able to colonize new niches (Gottig et al., 2009). However, our results demonstrate that plants have evolved strategies to recognize this virulence factor and thus hamper the disease process by mounting the basal PTI response. In particular, in the case of non‐host plants, this response is sufficient to hinder disease, whereas, in host plants, as a result of Xcc pathogenic mechanisms, the bacterium can colonize the host and cause disease. Further studies of plant molecules that may interact with XacFhaB regions will clarify how plants can recognize this important Xcc virulence factor.
Experimental Procedures
Strains, culture conditions and media
Escherichia coli JM109 used for DNA subcloning and S‐17 expressing GFP (Gottig et al., 2009) were cultivated at 37 °C in Luria–Bertani (LB) medium. Xcc (Xcc99‐1330), the derivative fhaB mutant (ΔfhaB) and ΔfhaB expressing GFP were grown at 28 °C in Silva Buddenhagen (SB) medium (5 g/l sucrose, 5 g/l yeast extract, 5 g/l peptone, and 1 g/l glutamic acid, pH 7.0) or XVM2 (20 mM NaCl, 10 mM (NH4)2SO4, 1 mM CaCl2, 10 mM FeSO4, 5 mM MgSO4, 0.16 mM KH2PO4, 0.32 mM K2HPO4, 10 mM fructose, 10 mM sucrose and 0.03% (w/v) casein acid hydrolysate (casaminoacid), pH 6.7) (Gottig et al., 2009). Antibiotics were used at the following final concentrations: ampicillin (Ap), 100 µg/mL for E. coli and 25 µg/mL for Xcc; kanamycin (Km), 40 µg/mL for both strains; gentamycin (Gm), 20 µg/mL for both strains; chloramphenicol (Cm), 30 µg/mL for E. coli. Xcc expressing GFP was constructed previously (Gottig et al., 2009).
Expression and purification of recombinant FhaB regions
Regions AR1, AR2, AR3, AR3‐1, AR3‐2 and AR3‐3 were amplified by PCR from Xcc genomic DNA using the oligonucleotides AR1LB and AR1RH, AR2LB and AR2RH, AR3LB and AR3RH, AR3‐1LB and AR3‐1RH, AR3LB and AR3‐2RH, and AR3‐3LB and AR3RH, respectively (Table S1, see Supporting Information), and cloned into the pET28a vector (Novagen, Merck KGaA, Darmstadt, Germany) previously digested with the restriction enzymes BamHI and HindIII. After transformation into E. coli strain BL21 (pLysS), the synthesis of recombinant polypeptides and also of 6×His‐Trx (thioredoxin) was induced by Isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) (0.1 mm) for 16 h at 18 °C. The proteins were purified by affinity chromatography from the soluble fraction of the bacterial lysates using Ni2+‐nitrilotriacetate (Ni‐NTA) agarose (Qiagen, Hilden, Germany). The purity of recombinant proteins was checked by SDS‐PAGE.
Plant material and plant infiltrations
Citrus sinensis cv. Valencia was grown in a glasshouse at 26 ± 2 °C and tomato (Solanum lycopersicum cv. Victoria) and pepper (Capsicum annuum cv. Grossum) at 24 ± 2 °C, all with a photoperiod of 16 h. Proteins were infiltrated with needleless syringes at 5 μm. Bacteria were grown in SB broth to an optical density of unity at 600 nm, harvested by centrifugation and resuspended in 10 mm MgCl2 at the required density. Infiltrations into leaves were performed with needleless syringes.
Bacterial aggregation assays
For bacterial aggregation assays, 20 μL of cultured bacterial strains expressing GFP were incubated with or without 5 µm of each of the XacFhaB regions for 3 h on glass slides in a humidity chamber. Then, bacteria were visualized by confocal laser scanning microscopy (Nikon Eclipse TE‐2000‐E2, Tokyo, Japan).
Callose staining and DAB staining
Callose staining was performed as described previously (Sgro et al., 2012) and examined by confocal laser scanning microscopy (Nikon Eclipse TE‐2000‐E2). Average callose measurements were based on at least 20 photographs from three independent experiments, and were analysed for statistical differences by one‐way analysis of variance (ANOVA) (P < 0.05). DAB staining was performed as described previously (Piazza et al., 2015). Cleared leaves were observed and photographed in an optical microscope.
RNA preparation and real‐time PCR
Total RNA was isolated from plant infiltrated leaves using TRIzol® reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer's instructions. At least 100 mg of frozen tissue were used for each total RNA extraction, and samples were stored at −80 ºC until use. The RT‐qPCRs were performed as described previously (Sgro et al., 2012). Oligonucleotide primers complementary to defence‐associated genes were used to amplify the tomato and citrus transcripts. As control, primers for the ribosomal protein L2 (Rpl2) were used in tomato and, in the case of citrus, for actin (Table S1). The primers used for tomato (Wei et al., 2015) and citrus (Piazza et al., 2015) are described in Table S1. Oligonucleotides for the amplification of HIR genes in both species were designed, looking for the homologues of pepper HIR (CaHIR). Gene‐specific cDNA amounts were calculated from threshold cycle (Ct) values, expressed relative to controls, and normalized with respect to actin cDNA, used as internal reference. Values were normalized by an internal reference (Ctr) according to the equation ΔCt = Ct – Ctr and quantified as 2–ΔCt. A second normalization by a control (Ctc), ΔΔCt = Ct – Ctc, produces a relative quantification: 2–ΔΔCt. The results were analysed with one‐way ANOVA and by Student's t‐test.
Analysis of Xcc growth in citrus leaves pre‐infiltrated with different XacFhaB polypeptide regions
Citrus leaves were pre‐infiltrated with needleless syringes with purified 6×His‐Trx and the different XacFhaB regions at 1 μm. After 16 h, these leaves were infiltrated with Xcc suspension at 106 CFU/mL. Growth assays were performed at 0, 3 and 5 dpi from 10 infiltrated leaves for each treatment at the indicated times by grinding 0.8‐cm‐diameter leaf discs in 1 mL of 10 mm MgCl2, followed by serial dilutions, and plating onto SB agar plates. Colonies were counted after 48 h of incubation at 28 ºC, and the results are presented as log CFU/cm2 of leaf tissue. In all cases, data were statistically analysed by one‐way ANOVA (P < 0.05).
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Text S1. Complete description of the analysis and structural modelling of the XacFhaB sequence and functional domains.
Fig. S1. XacFhaB sequence alignments. The sequences of XacFhaB and Fha1 from Xanthomonas campestris pv. vesicatoria (Xcv) (a) and AR2 and AR2′ (b) were aligned with ClustalW2.
Fig. S2. Ability of Adhesin Regions (ARs) AR1, AR2 and AR3 to aggregate ΔfhaB mutant and Escherichia coli cells. Representative photographs of confocal laser scanning microscopy of green fluorescent protein (GFP)‐expressing ΔfhaB mutant and E. coli S‐17 cells in the presence of 5 µm AR1, AR2 and AR3. As control (C), 6×His‐Trx was used. Bar, 10 µm.
Fig. S3. The response of tomato leaves infiltrated with Adhesin Regions (ARs) AR1, AR2 and AR3 requires the polypeptides to be folded. Representative photographs of the responses at 1 day post‐infiltration (dpi) of 5 µm AR1, AR2 and AR3 at room temperature and previously boiled for 10 min. Bar indicates 1 mm.
Table S1. Oligonucleotides used in this study.
Acknowledgements
We thank Rodrigo Vena for assistance with the confocal microscopy facility, Catalina Anderson (INTA Concordia, Argentina), Gastón Alanis and Rubén Díaz Vélez (Proyecto El Alambrado) for the citrus plants and Diego Aguirre for plant technical assistance. This work was supported by grants from the Argentine Federal Government (PICT2013‐0625 to JO and PICT2013‐0556 to NG). BSG, JO and NG are staff members of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
Contributor Information
Jorgelina Ottado, Email: ottado@ibr-conicet.gov.ar.
Natalia Gottig, Email: gottig@ibr-conicet.gov.ar.
References
- Amano, A. (2010) Bacterial adhesins to host components in periodontitis. Periodontol 2000, 52, 12–37. [DOI] [PubMed] [Google Scholar]
- Atkinson, N.J. and Urwin, P.E. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. [DOI] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Choi, H.W. , Kim, D.S. , Kim, N.H. , Jung, H.W. , Ham, J.H. and Hwang, B.K. (2013) Xanthomonas filamentous hemagglutinin‐like protein Fha1 interacts with pepper hypersensitive‐induced reaction protein CaHIR1 and functions as a virulence factor in host plants. Mol. Plant–Microbe. Interact. 26, 1441–1454. [DOI] [PubMed] [Google Scholar]
- Clantin, B. , Hodak, H. , Willery, E. , Locht, C. , Jacob‐Dubuisson, F. and Villeret, V. (2004) The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two‐partner secretion pathway. Proc. Natl. Acad. Sci. USA, 101, 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta, A. , Pinney, J.W. , Solis, D.Y. , Marsh, J. , Pettifer, S.R. and Attwood, T.K. (2010) Low‐complexity regions within protein sequences have position‐dependent roles. BMC Syst. Biol. 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich, C. and Relman, D.A. (2011) Modulation of the host interferon response and ISGylation pathway by B. pertussis filamentous hemagglutinin. PLoS One, 6, e27535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia, B.S. , Thomas, L. , Gottig, N. , Dunger, G. , Garofalo, C.G. , Daurelio, L.D. , Ndimba, B. , Orellano, E.G. , Gehring, C. and Ottado, J. (2010) A eukaryotic‐acquired gene by a biotrophic phytopathogen allows prolonged survival on the host by counteracting the shut‐down of plant photosynthesis. PLoS One, 5, e8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottig, N. , Garavaglia, B.S. , Garofalo, C.G. , Orellano, E.G. and Ottado, J. (2009) A filamentous hemagglutinin‐like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One, 4, e4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. and Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Julio, S.M. , Inatsuka, C.S. , Mazar, J. , Dieterich, C. , Relman, D.A. and Cotter, P.A. (2009) Natural‐host animal models indicate functional interchangeability between the filamentous haemagglutinins of Bordetella pertussis and Bordetella bronchiseptica and reveal a role for the mature C‐terminal domain, but not the RGD motif, during infection. Mol. Microbiol. 71, 1574–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava, A.V. , Cheng, N. , Cleaver, R. , Kessel, M. , Simon, M.N. , Willery, E. , Jacob‐Dubuisson, F. , Locht, C. and Steven, A.C. (2001) Beta‐helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42, 279–292. [DOI] [PubMed] [Google Scholar]
- Mazar, J. and Cotter, P.A. (2006) Topology and maturation of filamentous haemagglutinin suggest a new model for two‐partner secretion. Mol. Microbiol. 62, 641–654. [DOI] [PubMed] [Google Scholar]
- Mhedbi‐Hajri, N. , Jacques, M.A. and Koebnik, R. (2011) Adhesion mechanisms of plant‐pathogenic Xanthomonadaceae. Adv. Exp. Med. Biol. 715, 71–89. [DOI] [PubMed] [Google Scholar]
- Mobberley‐Schuman, P.S. and Weiss, A.A. (2005) Influence of CR3 (CD11b/CD18) expression on phagocytosis of Bordetella pertussis by human neutrophils. Infect. Immun. 73, 7317–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.P. , Chakravarthy, S. , Velasquez, A.C. , McLane, H.L. , Zeng, L. , Nakayashiki, H. , Park, D.H. , Collmer, A. and Martin, G.B. (2010) Methods to study PAMP‐triggered immunity using tomato and Nicotiana benthamiana . Mol. Plant–Microbe. Interact. 23, 991–999. [DOI] [PubMed] [Google Scholar]
- Noel, C.R. , Mazar, J. , Melvin, J.A. , Sexton, J.A. and Cotter, P.A. (2012) The prodomain of the Bordetella two‐partner secretion pathway protein FhaB remains intracellular yet affects the conformation of the mature C‐terminal domain. Mol. Microbiol. 86, 988–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza, A. , Zimaro, T. , Garavaglia, B.S. , Ficarra, F.A. , Thomas, L. , Marondedze, C. , Feil, R. , Lunn, J.E. , Gehring, C. , Ottado, J. and Gottig, N. (2015) The dual nature of trehalose in citrus canker disease: a virulence factor for Xanthomonas citri subsp. citri and a trigger for plant defence responses. J. Exp. Bot. 66, 2795–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, E. , Barrand, M. , Fabre, P. , Salomon, H. , Blondeau, C. , Wood, S.C. and Hoffenbach, A. (1999) New acellular pertussis‐containing paediatric combined vaccines. Vaccine, 17, 1650–1656. [DOI] [PubMed] [Google Scholar]
- Rado‐Trilla, N. and Alba, M. (2012) Dissecting the role of low‐complexity regions in the evolution of vertebrate proteins. BMC Evol. Biol. 12, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A. , Kucukural, A. and Zhang, Y. (2010) I‐TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y. and Sato, H. (1999) Development of acellular pertussis vaccines. Biologicals, 27, 61–69. [DOI] [PubMed] [Google Scholar]
- Sgro, G.G. , Ficarra, F.A. , Dunger, G. , Scarpeci, T.E. , Valle, E.M. , Cortadi, A. , Orellano, E.G. , Gottig, N. and Ottado, J. (2012) Contribution of a harpin protein from Xanthomonas axonopodis pv. citri to pathogen virulence. Mol. Plant Pathol. 13, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj, G. , Agarwal, P. , Grant, M. and Kumar, A. (2010) MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 5, 1370–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Wang, L. , Zhou, X. , Ren, X. , Dai, X. and Liu, H. (2015) PopW activates PAMP‐triggered immunity in controlling tomato bacterial spot disease. Biochem. Biophys. Res. Commun. 463, 746–750. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Yan, R. , Roy, A. , Xu, D. , Poisson, J. and Zhang, Y. (2015) The I‐TASSER Suite: protein structure and function prediction. Nat. Methods, 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. (2008) I‐TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Text S1. Complete description of the analysis and structural modelling of the XacFhaB sequence and functional domains.
Fig. S1. XacFhaB sequence alignments. The sequences of XacFhaB and Fha1 from Xanthomonas campestris pv. vesicatoria (Xcv) (a) and AR2 and AR2′ (b) were aligned with ClustalW2.
Fig. S2. Ability of Adhesin Regions (ARs) AR1, AR2 and AR3 to aggregate ΔfhaB mutant and Escherichia coli cells. Representative photographs of confocal laser scanning microscopy of green fluorescent protein (GFP)‐expressing ΔfhaB mutant and E. coli S‐17 cells in the presence of 5 µm AR1, AR2 and AR3. As control (C), 6×His‐Trx was used. Bar, 10 µm.
Fig. S3. The response of tomato leaves infiltrated with Adhesin Regions (ARs) AR1, AR2 and AR3 requires the polypeptides to be folded. Representative photographs of the responses at 1 day post‐infiltration (dpi) of 5 µm AR1, AR2 and AR3 at room temperature and previously boiled for 10 min. Bar indicates 1 mm.
Table S1. Oligonucleotides used in this study.


