Summary
On absorption by plants, silicon (Si) offers protection against many fungal pathogens, including powdery mildews. The mechanisms by which Si exerts its prophylactic role remain enigmatic, although a prevailing hypothesis suggests that Si positively influences priming. Attempts to decipher Si properties have been limited to plants able to absorb Si, which excludes the model plant Arabidopsis because it lacks Si influx transporters. In this work, we were able to engineer Arabidopsis plants with an Si transporter from wheat (TaLsi1) and to exploit mutants (pad4 and sid2) deficient in salicylic acid (SA)‐dependent defence responses to study their phenotypic response and changes in defence expression against Golovinomyces cichoracearum (Gc) following Si treatment. Our results showed that TaLsi1 plants contained significantly more Si and were significantly more resistant to Gc infection than control plants when treated with Si, the first such demonstration in a plant transformed with a heterologous Si transporter. The resistant plants accumulated higher levels of SA and expressed higher levels of transcripts encoding defence genes, thus suggesting a role for Si in the process. However, TaLsi1 pad4 and TaLsi1 sid2 plants were also more resistant to Gc than were pad4 and sid2 plants following Si treatment. Analysis of the resistant phenotypes revealed a significantly reduced production of SA and expression of defence genes comparable with susceptible controls. These results indicate that Si contributes to Arabidopsis defence priming following pathogen infection, but highlight that Si will confer protection even when priming is altered. We conclude that Si‐mediated protection involves mechanisms other than SA‐dependent defence responses.
Keywords: Arabidopsis, defence genes, induced resistance, powdery mildew, priming, silicon, salicylic acid
Introduction
Silicon (Si) is considered to be a ‘quasi‐essential’ element (Epstein, 1999) for plant growth with multiple reports showing its many benefits (Bélanger et al., 1995; Liang, 1999; Liang et al., 2008; Richmond and Sussman, 2003; Zhang et al., 2008). The prophylactic effects of Si against fungal diseases, such as powdery mildews and rice blast (Fauteux et al., 2005), are arguably the most commonly described (Bélanger et al., 2003; Diogo and Wydra, 2007; Fawe et al., 1998; Ghanmi et al., 2004; Rodrigues et al., 2001).
A given plant species must be able to take up and accumulate Si in order to benefit from Si amendments, a disposition that appears to vary greatly among plants (Deshmukh et al., 2013). As expected, high accumulators, including most monocots, display the best results under Si feeding, whereas non‐ or low accumulators, including many dicots, such as Arabidopsis, benefit minimally (Ghanmi et al., 2004; Hodson et al., 2005; Montpetit et al., 2012). This ability to absorb Si is linked to an efficient uptake system mediated by an influx transporter, termed Lsi1, and an efflux transporter, termed Lsi2 (Ma, 2010). Since the discovery of these transporters, several reports have identified homologues in a number of primitive and higher plants known for their ability to absorb Si (Deshmukh et al., 2013; Grégoire et al., 2012; Ma and Yamaji, 2006; Mitani et al., 2011; Montpetit et al., 2012).
The mechanisms by which Si protects plants against diseases have been a source of debate for many years. Originally described as having a strict passive mechanical role, Si is now believed to be associated with the priming of the plant and induced defence strategies (Fauteux et al., 2005; Fawe et al., 1998; Ye et al., 2013). Indeed, many studies have shown that Si‐accumulating plants, such as cucumber and some cereals (rice and wheat), treated with Si, will resist pathogen attacks by the activation of defence responses, including the production of defence proteins, phenolic compounds and phytoalexins (Chérif et al., 1994; Fawe et al., 1998; Ghanmi et al., 2004; Rodrigues et al., 2004). More recently, priming of antiherbivore defence responses has been linked with Si and insect resistance (Reynolds et al., 2009; Ye et al., 2013).
Induced resistance (IR) is a mechanism allowing plants to synthesize new defence compounds in response to the presence of a pathogen (Glazebrook, 2005; Walters et al., 2013). During pathogen attack, plants initiate active defences by the perception of an elicitor signal, followed by transduction of the signal to the nucleus by a network of mitogen‐activated protein kinase (MAPK) cascades and the production of defence proteins (Benhamou, 2009; Jones and Dangl, 2006). In the case of biotrophic pathogens, most plants establish a type of IR called ‘systemic acquired resistance’ (SAR) that requires the activation of the salicylic acid (SA) signalling pathway (Delaney et al., 1994; Glazebrook, 2005; Zhou et al., 1998) and the presence of the defence regulatory protein Nonexpressor of Pathogenesis‐Related Protein1 (NPR1; Durrant and Dong, 2004). In addition, the IR mechanism is often associated with the augmented capacity to mobilize cellular defence responses only after contact with pathogens (Conrath et al., 2002). This phenomenon, often called ‘priming’, allows the plant to respond more quickly and effectively to an attack, with minimal metabolic cost (Katz et al., 1998; Van Hulten et al., 2006). In this context, several studies have now shown that pretreatment with Si will prime plants to better respond to pathogen infections (Chain et al., 2009; Fauteux et al., 2005; Van Bockhaven et al., 2013).
In an effort to better understand the molecular mechanisms of priming, Arabidopsis thaliana and a variety of useful mutants have been widely exploited (Glazebrook, 2005). For instance, Arabidopsis plants infected with powdery mildew have been shown to display SAR through the SA pathway and expression of defence genes, such as Pathogenesis‐related1 (PR1), PR5, β‐1,3‐Glucanase 2 (BGL2) and Glutathione S‐transferase (GST) (Reuber et al., 1998). SA [and analogues such as benzothiadiazole (BTH)] is an essential component of resistance against biotrophic pathogens, such as powdery mildews, in contrast with the jasmonic acid/ethylene‐dependent pathways that are involved in resistance against necrotrophs (Glazebrook, 2005; Vogel and Somerville, 2000). Indeed, mutation of the SA induction‐deficient2‐1 (SID2‐1) gene yields mutants that fail to accumulate SA and show an increased susceptibility to the powdery mildew pathogen (Dewdney et al., 2000). In addition, disease incidence is also enhanced by mutation of Enhanced Disease Susceptibility 1 (EDS1) and Phytoalexin Deficient 4 (PAD4), two lipase‐like proteins that interact upstream in the SA signalling pathway (Feys et al., 2001).
As useful as Arabidopsis is for the study of plant–pathogen interactions, its limited absorption of Si, owing to its lack of Lsi1 transporters, hinders its potential as a model plant to understand the prophylactic role of Si (Chain et al., 2009). However, Montpetit et al. (2012) were able to overcome this deficiency by expressing Lsi1 transporters from wheat into Arabidopsis and thus create plants able to accumulate Si. In this study, we sought to exploit these Si‐accumulating Arabidopsis transformants to investigate both the molecular and physiological aspects of Si amendment in the protection of Arabidopsis against Golovinomyces cichoracearum (DC.) V.P. Heluta (Gc). Our results show that Si+ Arabidopsis plants have a higher resistance to powdery mildew when treated with Si, and that the treatment leads to priming via the activation of SA and associated markers of resistance. At the same time, we observed that Si treatment of Si+ pad4 and sid2 mutants yielded similar resistant phenotypes, thus indicating that mechanisms other than SA‐dependent defence responses are involved in Si‐mediated resistance.
Results
Heterologous expression of TaLsi1 increases Si absorption in Arabidopsis
In order to determine whether the transformation of Arabidopsis plants with the Si influx gene from wheat (TaLsi1) increased Si accumulation, all six Arabidopsis lines used in this study were analysed for their Si levels when grown under Si fertilization (1.7 mm). Col‐0 and both mutant lines (pad4 and sid2) absorbed limited amounts of Si, all in the same proportions (Fig. 1). When the same lines were transformed with TaLsi1, they absorbed between three to four times more Si over the experimental period (Fig. 1).
Figure 1.
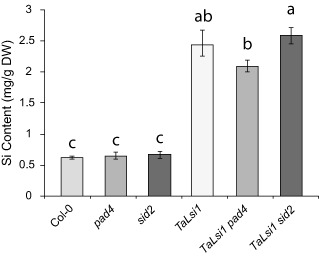
Silicon (Si) content in shoots of six Arabidopsis lines grown in a medium amended with an Si solution (1.7 mm). Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters are statistically significantly different by Tukey's test based on a significant analysis of variance (ANOVA) (P < 0.0001).
Si makes TaLsi1 plants more resistant to Gc
Col‐0 and TaLsi1 Arabidopsis plants, treated or not with Si, were inoculated with Gc to assess the protective effect of Si. In order to quantitatively assess powdery mildew infection over time, the standardized area under the disease progress curve (AUDPC) was calculated (see Experimental procedures). Seven days after inoculation, leaves of control (Si–) Col‐0 plants were heavily infested with colonies of the pathogen; similar results were obtained in Col‐0 plants regardless of Si treatment, as there was no significant difference in AUDPC between Si− and Si+ plants over the observation period (Fig. 2A). In the same manner, Si− TaLsi1 plants showed early signs of disease development that reached the levels found in Col‐0 plants after 7 days, which translated into a similar AUDPC over time (Fig. 2B). By contrast, TaLsi1 plants treated with Si prior to inoculation were less infected than Col‐0 plants or Si− TaLsi1 plants, and had a significantly lower AUDPC (Fig. 2B).
Figure 2.
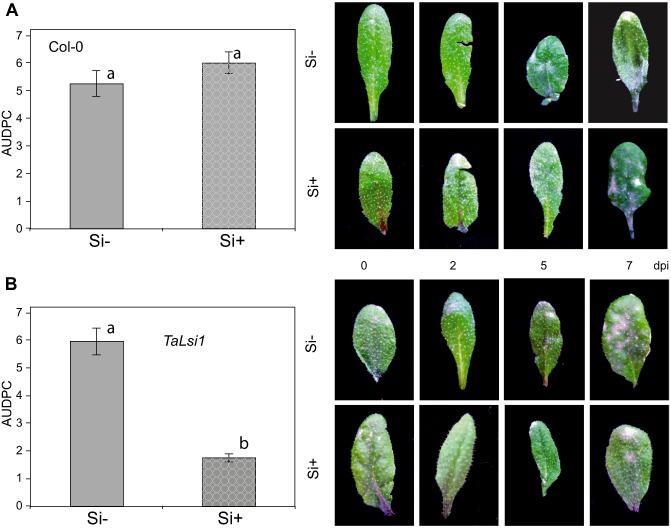
Powdery mildew (Golovinomyces cichoracearum) development and severity expressed as the area under the disease progress curve (AUDPC) over 7 days post‐inoculation (dpi) on Arabidopsis plants in response to silicon (Si) treatment. AUDPC on Col‐0 (A) and TaLsi1 (B) plants grown with (Si+) or without (Si–) Si and typical phenotypes over 7 days. Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters are statistically significantly different based on a t‐test (α = 0.05).
Si treatment primes SA biosynthesis and increases defence gene expression
In order to determine whether Si treatment acted as a primer of the SA pathway, we measured the accumulation of SA and the expression of NPR1 and three SA‐induced PR defence genes, PR1, PR2 and PR5, on all treated plants (Fig. 3). No significant differences in total SA accumulation were noted between Col‐0 and TaLsi1 plants, whether or not they were treated with Si before (Fig. S2, see Supporting Information) or over the first 2 days following inoculation (Fig. 3). A general increase in SA accumulation was observed in plants at 5 days post‐inoculation (dpi) compared with 2 dpi, with the largest increase in TaLsi1 plants treated with Si. At 7 dpi, Si+ TaLsi1 plants had accumulated significantly more SA than plants from all other treatments.
Figure 3.
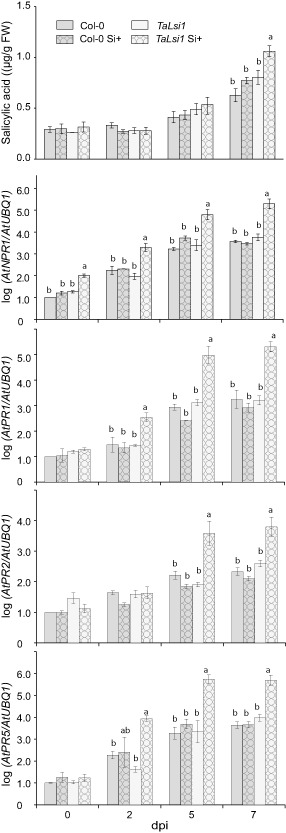
Effect of silicon (Si) amendments on the accumulation of salicylic acid (SA) and the expression of SA‐induced defence transcripts, NPR1, PR1, PR2 and PR5, in Arabidopsis plants Col‐0 and TaLsi1 infected with Golovinomyces cichoracearum at 0, 2, 5 and 7 days post‐inoculation (dpi). Levels of SA were quantified by high‐performance liquid chromatography (HPLC). Expression levels of AtNPR1, AtPR1, AtPR2 and AtPR5 transcripts were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to the transcript level of the internal control gene AtUBQ1. Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters within each time are statistically significantly different by Tukey's test based on a significant analysis of variance (ANOVA) (P < 0.05).
Prior to inoculation by the fungus, Si treatment did not affect the expression of any of the SA‐induced genes in leaves of Col‐0 and transgenic plants relative to Si− control plants (Fig. S2). Following Gc infection, NPR1 expression was significantly higher in Si+ TaLsi1 plants than in Si− plants or Si+ Col‐0 plants as early as 2 dpi (Fig. 3). Similarly, transcripts of PR genes PR1, PR2 and PR5, markers of SA‐induced defence responses in the Gc–Arabidopsis interaction, were significantly more abundant as early as 2 dpi in Si+ TaLsi1 plants compared with all other treated plants (Fig. 3).
Camalexin concentrations were analysed in all plants at all time points, and were found to be less than 0.5 μg/g fresh weight (FW) and never statistically significantly different between Col‐0 and TaLsi1 plants at any sampled time point regardless of Si treatment.
Si treatment primes expression of PAD4 and EDS1
As PAD4 and EDS1 are involved in SA synthesis, we monitored the transcript levels of PAD4 and EDS1 genes to determine whether Si influenced their activity (Fig. 4). In the absence of Si, Col‐0 and TaLsi1 plants showed no significant differences in transcript levels of PAD4 for each time point following Gc infection, except for a reduced level in TaLsi1 at 0 dpi (Fig. 4). However, when plants were treated with Si, Si+ TaLsi1 plants showed transcript levels approximately 15 times higher than all other treated plants at 5 and 7 dpi (Fig. 4). The same pattern was observed for EDS1 where transcript levels were 15 times higher at 7 dpi in Si+ TaLsi1 plants (Fig. 4).
Figure 4.
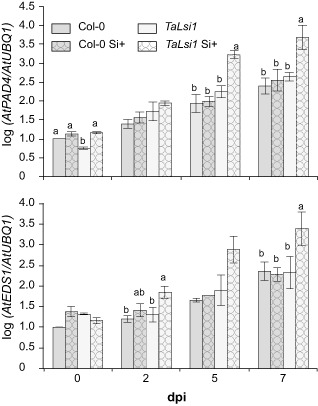
Effect of silicon (Si) amendments on the expression of PAD4 and EDS1 in Arabidopsis plants Col‐0 and TaLsi1 infected with Golovinomyces cichoracearum at 0, 2, 5 and 7 days post‐inoculation (dpi). Expression levels of AtPAD4 and AtEDS1 transcripts were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to the transcript level of the internal control gene AtUBQ1. Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters within each time are statistically significantly different by Tukey's test based on a significant analysis of variance (ANOVA) (P < 0.05).
Si‐enhanced resistance to Gc is maintained in pad4 and sid2 mutants engineered to better absorb Si
In Arabidopsis, the mutation of genes whose proteins participate in SA production will impair SA biosynthesis and enhance disease susceptibility to powdery mildew (Guo et al., 2013; Massoud et al., 2012). In order to test how Si‐enhanced resistance was influenced by the priming of the SA pathway, we crossed mutants pad4 and sid2 with the line TaLsi1, and homozygous T3 transformants were obtained for each. Seven days after inoculation with Gc, powdery mildew severity reached near‐maximum levels on sid2 (Fig. 5A) and pad4 (Fig. 5B) plants whether or not they were treated with Si. Interestingly, these levels were nearly 50% higher than those observed on Col‐0 plants (see Fig. 2), thus confirming the higher susceptibility of the mutants and indicating that impaired SA production in Arabidopsis leads to higher disease incidence. However, Si treatment led to a significant reduction in AUDPC in both TaLsi1 sid2 (Fig. 5A) and TaLsi1 pad4 (Fig. 5B), a result all the more surprising considering the high susceptibility of the mutant plants.
Figure 5.
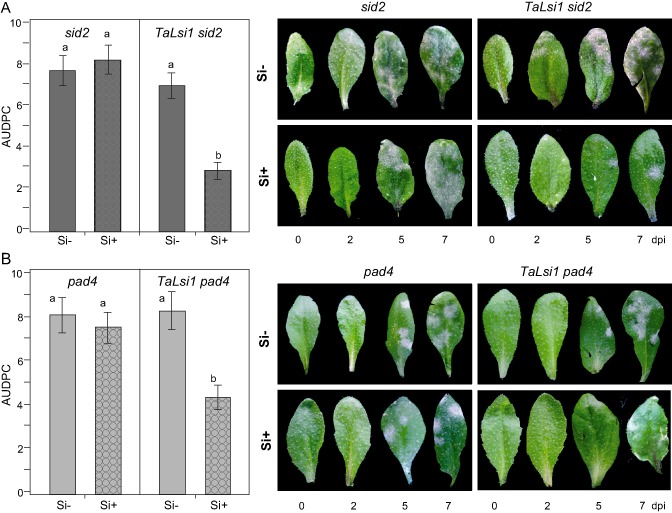
Powdery mildew (Golovinomyces cichoracearum) development and severity expressed as the area under the disease progress curve (AUDPC) over 7 days post‐inoculation (dpi) on Arabidopsis plants in response to silicon (Si) treatment. AUDPC on sid2 and TaLsi1 sid2 plants (A) and pad4 and TaLsi1 pad4 plants (B) grown with (Si+) or without (Si–) Si and typical phenotypes over 7 days. Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters are statistically significantly different based on a t‐test (α = 0.05).
Si treatment does not influence SA accumulation and SA‐induced gene expression in pad4 and sid2 mutants
SA accumulation and SA‐induced gene expression were monitored over time in mutant plants treated or not with Si during infection to determine whether they were altered by Si (Fig. 6). No difference in SA accumulation was observed in TaLsi1 sid2 plants as a result of Si treatment (Fig. 6). The same pattern was observed with TaLsi1 pad4 plants (Fig. 6). By contrast, TaLsi1 plants accumulated significantly more SA than TaLsi1 pad4 and TaLsi1 sid2 plants at 5 and 7 dpi regardless of Si treatment. In the same manner, Si treatment did not influence the expression of SA‐induced NPR1 in the mutant plants, and all values were significantly lower than those observed in TaLsi1 plants as early as 2 dpi (Fig. 6). The same pattern emerged with PR1, PR2 and PR5 expression, where levels did not appear to be influenced by Si and were consistently lower in mutant plants than in TaLsi1 plants, treated or not with Si (Fig. 6).
Figure 6.
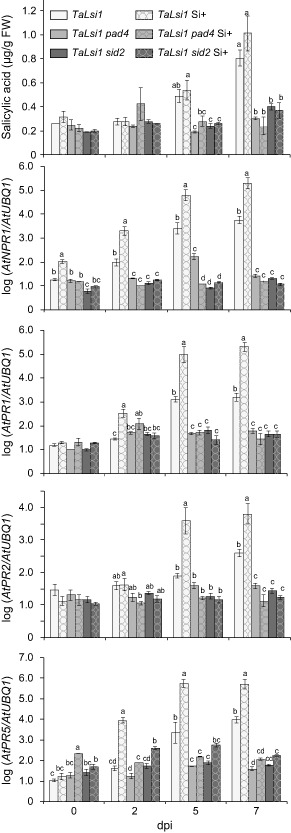
Effect of silicon (Si) amendments on the accumulation of salicylic acid (SA) and the expression of SA‐induced defence transcripts, NPR1, PR1, PR2 and PR5, in Arabidopsis plants TaLsi1, TaLsi1 pad4 and TaLsi1 sid2 infected with Golovinomyces cichoracearum at 0, 2, 5 and 7 days post‐inoculation (dpi). Levels of SA were quantified by high‐performance liquid chromatography (HPLC). Expression levels of AtNPR1, AtPR1, AtPR2 and AtPR5 transcripts were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to the transcript level of the internal control gene AtUBQ1. Values represent the means ± standard error (SE) of nine biologically independent experimental units. Means with different letters within each time are statistically significantly different by Tukey's test based on a significant analysis of variance (ANOVA) (P < 0.05).
Discussion
This work yields new and unexpected results regarding the elusive and debated prophylactic role of Si against plant diseases. Although it provides further support for the concept of priming being associated with Si, it highlights an additional phenomenon that could very well become pivotal in describing a unifying hypothesis for the mode of action of Si.
As convenient and versatile as Arabidopsis is as a model plant, its usefulness for Si studies is compromised by its limited absorption of the element owing to the absence of influx transporters (Deshmukh et al., 2013). To circumvent this problem, we relied on the methodology proposed by Montpetit et al. (2012), whereby it is possible to increase Si absorption in Arabidopsis by the insertion of heterologous influx transporters, such as that from wheat in this specific situation.
The concept that it is possible to transform a non‐accumulator plant species into an accumulating one opens up a wide array of possibilities to exploit the beneficial properties of Si. However, this presupposes that such transformed plants will display the expected phenotype in the presence of Si. Our results present the first demonstration of this outcome, whereby transformed plants were much more resistant to powdery mildew when fed with Si compared with control plants or transformed plants deprived of Si. This suggests that the beneficial effects of Si are universal among plant species as long as a plant can absorb the element through the presence of influx transporters.
Considering the impressive body of information available on the Arabidopsis–powdery mildew interaction, we took advantage of our resistant phenotypes to investigate the probable mechanisms behind the protective role of Si. As stated earlier, Si was initially described as providing a mechanical barrier impeding fungal penetration (Fauteux et al., 2005) and, for a long period, this mode of action stood uncontested. However, this hypothesis was first challenged by Menzies et al. (1991) and Chérif et al. (1992a, b, 1994), who associated the protective role of Si with the elicitation of defence mechanisms. Fawe et al. (1998) further confirmed the link between Si feeding and IR, and the priming role of Si has since been shown in numerous plant–pathogen interactions (Van Bockhaven et al., 2013) and even in plant–insect interactions (Reynolds et al., 2009; Ye et al., 2013). For these reasons, it was relevant to analyse the specific and well‐described markers of resistance in the Arabidopsis–powdery mildew interaction and to determine whether Si was involved in their expression. Our results clearly showed an increase in expression of genes encoding enzymes involved in the SA pathway directly associated with Si feeding and resistant phenotypes. At the same time, SA concentrations were also augmented, thus strengthening the hypothesis that priming occurred through this pathway. Interestingly, the production of camalexin remained unchanged, a result consistent with reports that camalexin production is useful against necrotrophs but not involved with biotrophs (Rogers et al., 1996). These results thus suggest that the response is aligned with the specific pathogen under study and that Si somehow facilitates this response, but does not elicit directly the priming machinery.
Other factors militate in support of this indirect role of Si associated with the manifestation of priming. Silicic acid is an uncharged molecule for which no evidence of biochemical activity has ever been obtained. It has been argued that soluble Si could somehow be directly involved in the elicitation of defence responses, namely as a secondary messenger, much in the same manner as SA (Fauteux et al., 2005; Fawe et al., 2001; Van Bockhaven et al., 2013). However, the presence of silicic acid in the symplastic environment and subsequent interactions with key defence molecules have simply not been corroborated by scientific data and remain speculative. Furthermore, the fact that the expression of NPR1 and other defence‐related genes was unchanged in pad4 and sid2 mutants under Si treatment would indicate that silicic acid does not act as a surrogate for SA as previously suggested (Fawe et al., 2001; Van Bockhaven et al., 2013).
The large array of available Arabidopsis mutants offered the unique opportunity to validate the hypothesis that the priming of defence reactions explains how Si protects plants against diseases. Indeed, by using mutants able to absorb larger quantities of Si, but deficient in the activation of the SA pathway, we observed that plants transformed for high Si absorption and fed with Si displayed resistant phenotypes in spite of having lost the ability to produce defence reactions through the SA pathway. These results strongly suggest that the defence reactions observed in TaLsi1 plants following inoculation with Gc and feeding with Si are not directly involved in reducing powdery mildew incidence, and that other undefined factors are at play. However, this does not preclude that priming may happen as a result of Si feeding in Si‐accumulating plants and that the observed defence reactions are involved in protection against subsequent infections, emanating from secondary infections for example.
Based on our observations, it might be tempting to conclude that the resistance conferred by Si on SA‐deficient mutants supports the concept of a physical barrier. Although there is no direct evidence that amorphous Si deposition in the apoplast can actually physically interfere with fungal penetration, the hypothesis that Si played a mechanical role originally prevailed until data supporting a priming role supplanted it (Fauteux et al., 2005). However, compelling evidence argued against the mechanical barrier hypothesis as early as 1960, when leaf puncture measurements led to the conclusion that Si deposits in the apoplast were not sufficiently resistant to impede fungal progress. In addition, a mechanical barrier physically stopping a germinating spore would not lead to the elicitation of defence mechanisms as observed here and in previous scientific reports (Bi et al., 2006; Kanto et al., 2007; Qin and Tian, 2005). Nevertheless, our results provide a unique perspective as they show that, if we can alter the priming state associated with Si feeding, we can still obtain resistant phenotypes.
If our results appear contradictory at first, they provide an opportunity to consider an alternative hypothesis that would unify the modes of action behind the observed phenomena. It is well known that the prophylactic role of Si has been more extensively documented and is more efficient against pathogens with a biotrophic phase (e.g. powdery mildews, oomycetes, rice blast). In the last few years, with the advent of high‐throughput sequencing, the annotation of plant pathogen genomes has highlighted the presence and importance of effector proteins, most notably in the case of biotrophs and hemibiotrophs. Effectors modify host cell structure, metabolism and function, and interfere with signal pathways required for host invasion or for the triggering of host resistance (Giraldo and Valent, 2013). Recent developments have located effectors in the apoplast, the extrahaustorial matrix or the host cytoplasm after translocation across the plant membrane. Interestingly, amorphous Si deposition in plants is located mostly in the apoplast and, more precisely, at the interface of the plasma membrane and the cell wall (Bauer et al., 2011; Zhang et al., 2013). This area is the site of intense interactions of many effectors with plant targets and sites of attempted penetration by biotrophic fungi (Bozkurt et al., 2012). Indeed, the appressorium and haustorium of powdery mildew fungi are structures of active release of effectors (Giraldo and Valent, 2013); the appressorium releases effectors in the apoplastic compartment to prevent the action of plant proteases, and the haustorium releases them into the cytoplasm through the extrahaustorial matrix (EHMx) to alter plant defences. Given that the apoplast and EHMx are within the confines of Si deposition (Ghanmi et al., 2004), and based on our observations, it thus seems not only plausible, but also logical, that Si would interfere with effectors reaching their targets. This would thus prevent the invading fungus from inhibiting plant defence, resulting in the expression of the complete array of defence mechanisms as observed in this work and elsewhere. In addition, the intercellular space is a hostile environment for a fungal pathogen, and the latter will rely on apoplastic effectors to inhibit the release of a wide array of proteases and other plant molecules that adversely affect its development (Giraldo and Valent, 2013; Win et al., 2012). From our results, it appears that this initial barrier is indeed quite efficient and significantly delays fungal infection. Considering the superior prophylactic role of Si against biotrophs, the heavy reliance of biotrophs on effectors to maintain their virulence and the site of Si deposition coinciding with effector release, our results support a link between Si and effectors, and certainly future efforts in testing this hypothesis.
In conclusion, our work confirms the association between Si and priming in plant–pathogen interactions, but has also uncovered a new phenomenon suggesting that mechanisms other than SA‐dependent plant defence priming are involved. These unforeseen results may be helpful in defining a unifying theory explaining the elusive and debated mode of action of Si in plant–pathogen interactions.
Experimental Procedures
Plant material
Six different Arabidopsis genotypes [Colombia (Col‐0; Ohio State University, Columbus, OH, USA), TaLsi1 lines (Montpetit et al., 2012), mutants pad4 (pad4‐1) (provided by Dr Roger Innes, Indiana University, Bloomington, IN, USA) and sid2 (sid2‐1) (obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH, USA), and TaLsi1 pad4 and TaLsi1 sid2 lines] were used in this work. For all experiments, Col‐0 and T3 transgenic seeds were stored at 4 °C for 4 days to break dormancy and placed in pasteurized Connaisseur® Tropical Plant Potting Soil (Fafard et frères, St‐Bonaventure, QC, Canada) in a growth chamber under long‐day conditions (16 h of light at 22 °C, 8 h of dark at 20 °C, 60–70% humidity and a light intensity of 150 μmol/m2/s) and covered with plastic sheets for 1 week. At 7 days, seedlings of uniform size were transferred to pots containing Connaisseur® Soil at a density of four plants per pot. Plants were treated in one of two different treatments: (i) nutrient solution (as per Tocquin et al., 2003) without soluble Si (control); or (ii) nutrient solution containing 1.7 mm potassium silicate (K2SiO3). For Si‐negative treatments, potassium chloride was used to replenish potassium. The plants were maintained in a growth chamber with the conditions as described above. Arabidopsis plants of different genotypes were used for experiments 10 days after transplanting.
Generation of transgenic plants
To determine whether Si‐mediated resistance to Gc was linked to the priming of defence responses, pad4 and sid2 mutants were transformed in order to allow Si uptake by introducing TaLsi1 (a wheat Si transporter gene) under the control of a root‐specific AtNIP5;1 promoter. The TaLsi1 pad4 and TaLsi1 sid2 lines were generated as described by Montpetit et al. (2012) for pNIP5‐TaLsi1 Arabidopsis plants.
Agrobacterium tumefaciens (strain GV3101/pMP90) harbouring pNIP5;1::TaLsi1 binary vectors (Montpetit et al., 2012) was used for the transformation of Arabidopsis according to the floral dip method (Clough and Bent, 1998). Transformants (T1) were selected on Murashige and Skoog Basal Medium with Gamborg's Vitamins (MS) (Sigma‐Aldrich, St. Louis, MO, USA) containing hygromycin (15 mg/L), and the presence of the TaLsi1 transgene was verified by polymerase chain reaction (PCR) using TaLsi1‐Fow and TaLsi1‐Rev primers (Montpetit et al., 2012). T2 seeds were harvested and sown on MS medium containing hygromycin (15 mg/L) to select single locus lines. For all experiments, one T3 homozygous TaLsi1 pad4 and TaLsi1 sid2 line was used.
Determination of Si concentration in leaves
The mutants pad4 and sid2, transgenic lines TaLsi1, TaLsi1 pad4 and TaLsi1 sid2 and Col‐0 plants treated or not with Si were analysed in this study. The Si content in experimental plants was measured by inductively coupled plasma‐optical emission spectrometry (ICP‐OES, Jobin‐Yvon Horiba JY2000‐2, Longjumeau, France). Aerial parts of the plants from each treatment (three replicates of 10 plants each) were collected and freeze–dried 1 week following the beginning of Si amendment. Samples were ground to a powder and total Si analysis was carried out at 251.611 nm by ICP‐OES as described by Côté‐Beaulieu et al. (2009).
SA and camalexin analyses
All lines treated or not with Si amendment before or after infection by Gc were analysed for SA and camalexin content. One leaf from Gc‐treated plants and one leaf from untreated plants were harvested 2, 1 and 0 days before or 0, 2, 5 and 7 days after infection. Samples were collected, immersed immediately in liquid nitrogen and stored at −80 °C before analysis. Three plants for each treatment at each time point were sampled and the experiment was performed three times. Total SA and camalexin were extracted and measured by high‐performance liquid chromatography (HPLC) as described by Massoud et al. (2012). Standards of SA were from Sigma‐Aldrich, whereas authentic camalexin was a gift from Dr A. J. Buchala (University of Fribourg, Fribourg, Switzerland).
Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis in Arabidopsis
The differential expression of selected genes in all six different Arabidopsis genotypes (Col‐0, TaLsi1, pad4, sid2, TaLsi1 pad4 and TaLsi1 sid2) in response to Si and Gc infection was determined by qRT‐PCR. Total RNA from the leaves of three independent plants for each condition was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and stored at −80 °C until use. First‐strand cDNAs were prepared from 1 μg of total RNA treated with RQ1RNase‐free DNase (Promega Corp., Madison, WI, USA), and then reverse transcribed with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligodT‐(18) primers. qRT‐PCR experiments were performed with 2 μL of a 1:5 dilution of cDNA using a Mastercycler EP Realplex 2S (Eppendorf) and the Quantitect SYBR green PCR kit (Qiagen), according to the manufacturer's instructions. PCR was performed as follows: 15 min at 95 °C plus 35–45 cycles of 20 s at 95 °C, 20 s at 58 °C and 20 s at 72 °C. Fluorescence data were collected during the cycle at 72 °C. Amplicon specificity was verified by melting curve analysis, and relative transcript levels were analysed twice by normalizing the PCR threshold cycle number of each gene with that of the Ubiquitin (Ubq1) reference gene. The efficiencies of all qRT‐PCR primer pairs used in this work (Table 1) were determined and the quantification of the relative changes in gene expression was performed using the Pfaffl method (Pfaffl et al., 2002). Three independent biological replicates for each treatment were used for qRT‐PCR analyses.
Table 1.
List of primers used in this study
| Name | Sequence 5′–3′ |
|---|---|
| AtUBQ1 fow | GGCCGTACTTTGGCTGACTA |
| AtUBQ1 rev | ACAGCTCTTGGGTGAAGACG |
| AtNPR1 fow | CTTGCGGAGAAGACGACACT |
| AtNPR1 rev | CACCGACGACGATGAGAGAG |
| AtPR1 fow | ACGGGGAAAACTTAGCCTGG |
| AtPR1 rev | TTGGCACATCCGAGTCTCAC |
| AtPR2 fow | ATCGGACGTTGTGGCTCTTT |
| AtPR2 rev | AACCGCGTTCTCGATGTTCT |
| AtPR5 fow | TGTGTCTCTGACCTCAACGC |
| AtPR5 rev | TCCGGTACAAGTGAAGGTGC |
| AtPAD4 fow | TATGGTCGACGCTGCCATAC |
| AtPAD4 rev | CACGTGGCAGAAGTTGTGTG |
| AtEDS1 fow | TGAGCACAAGAGGCAGACAG |
| AtEDS1 rev | GGGCTTGACACTTTGGCTTG |
| AtEDR1 fow | TGCTGCTTTAGCCGAGTTCA |
| AtEDR1 rev | GGGCCGATGAAGGATTCGAT |
| AtPDF1.2 fow | CGAGAAGCCAAGTGGGACAT |
| AtPDF1.2 rev | ACTTGTGTGCTGGGAAGACA |
Fungal material and maintenance on plants
Golovinomyces cichoracearum (Gc; formerly Erysiphe cichoracearum) UCSC1 was obtained from Dr Roger Innes (Indiana University) and maintained for inoculation on pad4 mutants. Mature fungal conidia on infected plants were removed by blowing them with air 3 days before inoculation to stimulate the regeneration of fresh spores.
Gc inoculation and evaluation of disease severity
The mutants pad4 and sid2, transgenic lines TaLsi1, TaLsi1 pad4 and TaLsi1 sid2 and Col‐0 plants, treated or not with Si as described above, were inoculated with Gc. Ten days after Si treatment, three leaves on each plant were individually inoculated according to the method of Adam and Somerville (1996). Low‐density inoculations (5–10 conidia/mm2) were performed by the transfer of conidia from one heavily infected pad4 mutant onto three healthy leaves from three separate plants. Inoculated plants were incubated for 1 day at 100% relative humidity at 21 °C to promote the germination of conidia, after which the plants were returned to the normal growth conditions described above.
Inoculated leaves from three plants of all plant material and for each treatment were monitored for disease development. Disease severity was quantified 0, 2, 5 and 7 days after inoculation on a continuous disease scale from 0 to 4 based on the percentage of leaf coverage (Fig. S1, see Supporting Information). The experiment was repeated three times. To compare disease severity between the plant material and the Si treatment, AUDPC was calculated according to Shanner and Finney (1977).
Statistical analyses
Descriptive statistics, including the mean and standard error, together with the Tukey range test for multiple comparison procedure, were used when the analysis of variance (ANOVA) was significant (P < 0.05).
Supporting information
Fig. S1 Continuous disease scale used to score Golovinomyces cichoracearum infection on Arabidopsis plants. Disease severity was measured with a personal scale from 0 to 4 based on the percentage of leaf coverage.
Fig. S2 Effect of silicon (Si) amendments on the accumulation of salicylic acid (SA) and the expression of SA‐induced defence transcripts, NPR1, PR1, PR2 and PR5, in Arabidopsis plants Col‐0 and TaLsi1 at 0, 5 and 10 days following Si treatment. Levels of SA were quantified by high‐performance liquid chromatography (HPLC). Expression levels of AtNPR1, AtPR1, AtPR2 and AtPR5 transcripts were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to the transcript level of the internal control gene AtUBQ1. Values are means ± standard error (SE) (n = 9) from three independent experiments.
Acknowledgements
We thank Dr Patrick Saindrenan and Dr Sejir Chaouch (CNRS‐Université Paris‐Sud 11, Paris, France) for total SA and camalexin analyses, and Dr A. J. Buchala (University of Fribourg, Fribourg, Switzerland) for the gift of camalexin. We are grateful to Dr Roger Innes (Indiana University, Bloomington, IN, USA) for pad4 and the inoculum of Golovinomyces cichoracearum. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Research Chairs Program to RRB.
References
- Adam, L. and Somerville, S.C. (1996) Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . Plant J. 9, 341–356. [DOI] [PubMed] [Google Scholar]
- Bauer, P. , Elbaum, R. and Weiss, I.M. (2011) Calcium and silicon mineralization in land plants: transport, structure and function. Plant Sci. 180, 746–756. [DOI] [PubMed] [Google Scholar]
- Bélanger, R.R. , Bowen, P.A. , Ehret, D.L. and Menzies, J.G. (1995) Soluble silicon: its role in crop and disease management of greenhouse crops. Plant Dis. 79, 329–336. [Google Scholar]
- Bélanger, R.R. , Benhamou, N. and Menzies, J.G. (2003) Cytological evidence of an active role of silicon in wheat resistance to powdery mildew (Blumeria graminis f. sp. tritici). Phytopathology, 93, 402–412. [DOI] [PubMed] [Google Scholar]
- Benhamou, N. (2009) La Résistance chez les Plantes. Principes de la Stratégie Défensive et Applications Agronomiques. Paris: Éditions TEC and DOC, Lavoisier. [Google Scholar]
- Bi, Y. , Tian, S.P. , Guo, Y.R. , Ge, Y.H. and Qin, G.Z. (2006) Sodium silicate reduces postharvest decay on Hami melons: induced resistance and fungistatic effects. Plant Dis. 90, 279–283. [DOI] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Banfield, M.J. and Kamoun, S. (2012) Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. [DOI] [PubMed] [Google Scholar]
- Chain, F. , Côté‐Beaulieu, C. , Belzile, F. , Menzies, J.G. and Bélanger, R.R. (2009) A comprehensive transcriptomic analysis of the effect of silicon on wheat plants under control and pathogen stress conditions. Mol. Plant–Microbe Interact. 22, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Chérif, M. , Benhamou, N. , Menzies, J.G. and Bélanger, R.R. (1992a) Silicon induced resistance in cucumber plants against Pythium ultimum . Physiol. Mol. Plant Pathol. 41, 411–425. [Google Scholar]
- Chérif, M. , Menzies, J.G. , Benhamou, N. and Bélanger, R.R. (1992b) Studies of silicon distribution in wounded and Pythium ultimum infected cucumber plants. Physiol. Mol. Plant Pathol. 41, 371–385. [Google Scholar]
- Chérif, M. , Asselin, A. and Bélanger, R.R. (1994) Defense responses induced by soluble silicon in cucumber roots infected by Pythium spp. Phytopathology, 84, 236–242. [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conrath, U. , Pieterse, C.M.J. and Mauch‐Mani, B. (2002) Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. [DOI] [PubMed] [Google Scholar]
- Côté‐Beaulieu, C. , Chain, F. , Menzies, J.G. , Kinrade, S.D. and Bélanger, R.R. (2009) Absorption of aqueous inorganic and organic silicon compounds by wheat and their effect on growth and powdery mildew control. Environ. Exp. Bot. 65, 155–161. [Google Scholar]
- Delaney, T.P. , Uknes, S. , Vernooij, B. , Friedrich, L. , Weymann, K. , Negrotto, D. , Gaffney, T. , Gut‐Rella, M. , Kessmann, H. , Ward, E. and Ryals, J. (1994) A central role of salicylic acid in plant disease resistance. Science, 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Deshmukh, R.K. , Vivancos, J. , Guérin, V. , Sonah, H. , Labbé, C. , Belzile, F. and Bélanger, R.R. (2013) Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 83, 303–315. [DOI] [PubMed] [Google Scholar]
- Dewdney, J. , Reuber, T.L. , Wildermuth, M.C. , Devoto, A. , Cui, J. , Stutius, L.M. , Drummond, E.P. and Ausubel, F.M. (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. [DOI] [PubMed] [Google Scholar]
- Diogo, R.V.C. and Wydra, K. (2007) Silicon‐induced basal resistance in tomato against Ralstonia solanacearum is related to modification of pectic cell wall polysaccharide structure. Physiol. Mol. Plant Pathol. 70, 120–129. [Google Scholar]
- Durrant, W.E. and Dong, X. (2004) Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Epstein, E. (1999) Silicon. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 641–664. [DOI] [PubMed] [Google Scholar]
- Fauteux, F. , Rémus‐Borel, W. , Menzies, J.G. and Bélanger, R.R. (2005) Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol. Lett. 249, 1–6. [DOI] [PubMed] [Google Scholar]
- Fawe, A. , Abou‐Zaid, M. , Menzies, J.G. and Bélanger, R.R. (1998) Silicon‐mediated accumulation of flavonoid phytoalexins in cucumber. Phytopathology, 88, 396–401. [DOI] [PubMed] [Google Scholar]
- Fawe, A. , Menzies, J.G. , Chérif, M. and Bélanger, R.R. (2001) Silicon and disease resistance in dicotyledons In: Silicon in Agriculture (Datnoff L.E., Snyder G.H. and Korndörfer G.H., eds), pp. 323–341. Amsterdam: Elsevier Science. [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.‐A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanmi, D. , McNally, D.J. , Benhamou, N. , Menzies, J.G. and Bélanger, R.R. (2004) Powdery mildew of Arabidopsis thaliana: a pathosystem for exploring the role of silicon in plant–microbe interactions. Physiol. Mol. Plant Pathol. 64, 189–199. [Google Scholar]
- Giraldo, M.C. and Valent, B. (2013) Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Grégoire, C. , Rémus‐Borel, W. , Vivancos, J. , Labbé, C. , Belzile, F. and Bélanger, R.R. (2012) Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense . Plant J. 72, 320–330. [DOI] [PubMed] [Google Scholar]
- Guo, C. , Wu, G. , Xing, J. , Li, W. , Tang, D. and Cui, B. (2013) A mutation in a coproporphyrinogen III oxidase gene confers growth inhibition, enhanced powdery mildew resistance and powdery mildew‐induced cell death in Arabidopsis. Plant Cell Rep. 32, 687–702. [DOI] [PubMed] [Google Scholar]
- Hodson, M.J. , White, P.J. , Mead, A. and Broadley, M.R. (2005) Phylogenetic variation in the silicon composition of plants. Ann. Bot. 96, 1027–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanto, T. , Maekawa, K. and Aino, M. (2007) Suppression of conidial germination and appressorial formation by silicate treatment in powdery mildew of strawberry. J. Gen. Plant Pathol. 73, 1–7. [Google Scholar]
- Katz, V.A. , Thulke, O.U. and Conrath, U. (1998) A benzothiadiazole primes parsley cells for augmented elicitation of defense responses. Plant Physiol. 117, 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. (1999) Effects of silicon on enzyme activity and sodium, potassium and calcium concentration in barley under salt stress. Plant Soil, 209, 217–224. [Google Scholar]
- Liang, Y. , Zhu, J. , Li, Z. , Chu, G. , Ding, Y. , Zhang, J. and Sun, W. (2008) Role of silicon in enhancing resistance to freezing stress in two contrasting winter wheat cultivars. Environ. Exp. Bot. 64, 286–294. [Google Scholar]
- Ma, J.F. (2010) Silicon transporters in higher plants In: MIPS and their Role in the Exchange of Metalloids (Jahn P.T. and Bienert G., eds), pp. 99–109. New York: Springer. [Google Scholar]
- Ma, J.F. and Yamaji, N. (2006) Silicon uptake and accumulation in higher plants. Trends Plant Sci. 11, 392–397. [DOI] [PubMed] [Google Scholar]
- Massoud, K. , Barchietto, T. , Le Rudulier, T. , Pallandre, L. , Didierlaurent, L. , Garmier, M. , Ambard‐Bretteville, F. , Seng, J.‐M. and Saindrenan, P. (2012) Dissecting phosphite‐induced priming in Arabidopsis infected with Hyaloperonospora arabidopsidis . Plant Physiol. 159, 286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies, J.G. , Ehret, D.L. , Glass, A.D.M. and Samuels, A.L. (1991) The influence of silicon on cytological interactions between Sphaerotheca fuliginea and Cucumis sativus . Physiol. Mol. Plant Pathol. 39, 403–414. [Google Scholar]
- Mitani, N. , Yamaji, N. , Ago, Y. , Iwasaki, K. and Ma, J.F. (2011) Isolation and functional characterization of an influx silicon transporter in two pumpkin cultivars contrasting in silicon accumulation. Plant J. 66, 231–240. [DOI] [PubMed] [Google Scholar]
- Montpetit, J. , Vivancos, J. , Mitani‐Ueno, N. , Yamaji, N. , Rémus‐Borel, W. , Belzile, F. , Ma, J.F. and Bélanger, R.R. (2012) Cloning, functional characterization and heterologous expression of TaLsi1, a wheat silicon transporter gene. Plant Mol. Biol. 79, 35–46. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. , Horgan, G.W. and Dempfle, L. (2002) Relative expression software tool (REST©) for group‐wise comparison and statistical analysis of relative expression results in real‐time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, G.Z. and Tian, S.P. (2005) Enhancement of biocontrol activity of Cryptococcus laurentii by silicon and the possible mechanisms involved. Phytopathology, 95, 69–75. [DOI] [PubMed] [Google Scholar]
- Reuber, T.L. , Plotnikova, J.M. , Dewdney, J. , Rogers, E.E. , Wood, W. and Ausubel, F.M. (1998) Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Reynolds, O.L. , Keeping, M.G. and Meyer, J.H. (2009) Silicon‐augmented resistance of plants to herbivorous insects: a review. Ann. Appl. Biol. 155, 171–186. [Google Scholar]
- Richmond, K.E. and Sussman, M. (2003) Got silicon? The non‐essential beneficial plant nutrient. Curr. Opin. Plant Biol. 6, 268–272. [DOI] [PubMed] [Google Scholar]
- Rodrigues, F. , Datnoff, L.E. , Korndörfer, G.H. , Seebold, K.W. and Rush, M.C. (2001) Effect of silicon and host resistance on sheath blight development in rice. Plant Dis. 85, 827–832. [DOI] [PubMed] [Google Scholar]
- Rodrigues, F. , McNally, D.J. , Datnoff, L.E. , Jones, J.B. , Labbé, C. , Benhamou, N. , Menzies, J.G. and Bélanger, R.R. (2004) Silicon enhances the accumulation of diterpenoid phytoalexins in rice: a potential mechanism for blast resistance. Phytopathology, 94, 177–183. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E. , Glazebrook, J. and Ausubel, F.M. (1996) Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis–pathogen interactions. Mol. Plant–Microbe Interact. 9, 748–757. [DOI] [PubMed] [Google Scholar]
- Shanner, G. and Finney, R.E. (1977) The effect of nitrogen fertilization on the expression of slow‐mildewing resistance in Knox wheat. Phytopathology, 67, 1051–1056. [Google Scholar]
- Tocquin, P. , Corbesier, L. , Havelange, A. , Pieltain, A. , Kurtem, E. , Bernier, G. and Perilleux, C. (2003) A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana . BMC Plant Biol. 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockhaven, J. , De Vleesschauwer, D. and Höfte, M. (2013) Towards establishing broad‐spectrum disease resistance in plants: silicon leads the way. J. Exp. Bot. 64, 1281–1293. [DOI] [PubMed] [Google Scholar]
- Van Hulten, M. , Pelser, M. , van Loon, L.C. , Pieterse, C.M.J. and Ton, J. (2006) Costs and benefits of priming for defense in Arabidopsis . Proc. Natl. Acad. Sci. USA, 103, 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J. and Somerville, S. (2000) Isolation and characterization of powdery mildew‐resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA, 97, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, D.R. , Ratsep, J. and Havis, N.D. (2013) Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280. [DOI] [PubMed] [Google Scholar]
- Win, J. , Chaparro‐Garcia, A. , Belhaj, K. , Saunders, D.G.O. , Yoshida, K. , Dong, S. , Schornack, S. , Zipfel, C. , Robatzek, S. , Hogenhout, S.A. and Kamoun, S. (2012) Effector biology of plant‐associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247. [DOI] [PubMed] [Google Scholar]
- Ye, M. , Song, Y. , Long, J. , Wang, R. , Baerson, S.R. , Pan, Z. , Zhu‐Salzman, K. , Xie, J. , Cai, K. , Luo, S. and Zeng, R. (2013) Priming of jasmonate‐mediated antiherbivore defense responses in rice by silicon. Proc. Natl. Acad. Sci. USA, 110, E3631–E3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Wang, L. , Nie, Q. , Zhang, W. and Zhang, F. (2008) Long‐term effects of exogenous silicon on cadmium translocation and toxicity in rice (Oryza sativa L.). Environ. Exp. Bot. 62, 300–307. [Google Scholar]
- Zhang, C. , Wang, L. , Zhang, W. and Zhang, F. (2013) Do lignification and silicification of the cell wall precede silicon deposition in the silica cell of the rice (Oryza sativa L.) leaf epidermis? Plant Soil, 372, 137–149. [Google Scholar]
- Zhou, N. , Tootle, T.L. , Tsui, F. , Klessig, D.F. and Glazebrook, J. (1998) PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell, 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Continuous disease scale used to score Golovinomyces cichoracearum infection on Arabidopsis plants. Disease severity was measured with a personal scale from 0 to 4 based on the percentage of leaf coverage.
Fig. S2 Effect of silicon (Si) amendments on the accumulation of salicylic acid (SA) and the expression of SA‐induced defence transcripts, NPR1, PR1, PR2 and PR5, in Arabidopsis plants Col‐0 and TaLsi1 at 0, 5 and 10 days following Si treatment. Levels of SA were quantified by high‐performance liquid chromatography (HPLC). Expression levels of AtNPR1, AtPR1, AtPR2 and AtPR5 transcripts were quantified by quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) and normalized to the transcript level of the internal control gene AtUBQ1. Values are means ± standard error (SE) (n = 9) from three independent experiments.


