Summary
Pigeonpea sterility mosaic virus (PPSMV), a species of the genus Emaravirus, is the causal agent of sterility mosaic disease (SMD) of pigeonpea [Cajanus cajan (L.) Millsp]. This disease, dubbed the ‘green plague’, as the infected plants remain in the vegetative state without flower production, has been reported from India and a few other South‐East Asian countries. SMD is estimated to result in an annual yield loss of over US$300 million in India alone. The aetiology of SMD, which remained a mystery for over 70 years, was resolved with the discovery of PPSMV in 2000 and its complete genome sequence in 2014.
Aetiology and virus transmission
SMD is characterized by stunted and bushy plants, leaves of reduced size with chlorotic rings or mosaic symptoms, and partial or complete cessation of flower production (i.e. sterility). The causal agent of the disease is PPSMV, a virus with a segmented, negative‐sense, single‐stranded RNA genome, transmitted in a semi‐persistent manner by an eriophyid mite Aceria cajani Channabassavanna (Acari: Arthropoda). Both the virus and vector are highly specific to pigeonpea and a few of its wild relatives, such as C. scarabaeoides and C. cajanifolius. Under experimental conditions, PPSMV was transmitted to Nicotiana benthamiana by sap inoculation using fresh extract of SMD‐infected leaves (but not to pigeonpea); however, purified nucleoprotein preparations are not infectious. The virus was also transmitted to French bean (Phaseolus vulgaris L.) using viruliferous eriophyid mites. PPSMV is not seed transmitted in pigeonpea or other hosts known to be infected by this virus. On the basis of the differential host reactions in different geographical locations, the occurrence of diverse PPSMV strains was suspected.
Host range and epidemiology
PPSMV can infect several genotypes of cultivated and wild relatives of pigeonpea. Experimental hosts include N. benthamiana, N. clevelandii, P. vulgaris and Chrozophora rottleri. However, pigeonpea alone and a few wild species of Cajanus were found to support the vector A. cajani. SMD is endemic in most of the pigeonpea‐growing regions of India, but the incidence varies widely between regions and years. In nature, A. cajani populations were almost exclusively observed on SMD‐infected pigeonpea, but not on healthy plants, indicating a strong communalistic relationship between the virus‐infected plants and the vector. The epidemiology of SMD involves the virus, mite vector, cultivar and environmental conditions. Infected perennial and volunteer plants serve as a source for both the virus and its vector mites, and play an important role in the disease cycle.
Genome organization, gene function and taxonomy
The PPSMV genome contains five segments of single‐stranded RNA that are predicted to encode proteins in negative sense. The ribonucleoprotein complex is encased in quasi‐spherical, membrane‐bound virus particles of 100–150 nm. The largest segment, RNA‐1, is 7022 nucleotides in length and codes for RNA‐dependent RNA polymerase (2295 amino acids); RNA‐2, with a sequence length of 2223 nucleotides, codes for glycoproteins (649 amino acids); RNA‐3, with a sequence length of 1442 nucleotides, codes for nucleocapsid protein (309 amino acids); RNA‐4, with a sequence length of 1563 nucleotides, codes for a putative movement protein p4 (362 amino acids); and RNA‐5, with a sequence length of 1689 nucleotides, codes for p5 (474 amino acids), a protein with unknown function. PPSMV was recently classified as a species in the genus Emaravirus, a genus whose members show features resembling those of members of the genera Tospovirus (Family: Bunyaviridae) and Tenuivirus, both of which comprise single‐stranded RNA viruses that encode proteins by an ambisense strategy.
SMD control
The disease is mainly controlled using SMD‐resistant cultivars. However, the occurrence of distinct strains/isolates of PPSMV in different locations makes it difficult to incorporate broad‐spectrum resistance. Studies on the inheritance of SMD resistance in different cultivars against different isolates of PPSMV indicate that the resistance is mostly governed by recessive genes, although there are contrasting interpretations of the data. Genetic engineering through RNA‐interference (RNAi) and resistant gene‐based strategies are some of the potential approaches for the transgenic control of SMD. Seed treatment or soil and foliar application of a number of organophosphorus‐based insecticides or acaricides, which are recommended for the management of the vector mites, are seldom practised because of prohibitive costs and also their risks to human health and the environment.
Keywords: Emaravirus, eriophyid mite, Pigeonpea sterility mosaic virus, sterility mosaic disease
Introduction
Pigeonpea [Cajanus cajan (L.) Millsp.], also referred to as red gram or arhar, is a perennial shrub, with its centre of origin located in India. It is an important grain legume crop predominantly grown in the Indian subcontinent as an important source of dietary protein. Worldwide, pigeonpea is cultivated on approximately 6.22 million hectares, and 75% of this land area is presently in India (FAOSTAT, 2014). It is also cultivated in other parts of the world, including sub‐Saharan Africa, Latin America, the Caribbean and South‐East Asia. Its cultivation is increasing in semi‐arid areas because of the crop's ability to thrive under prolonged drought and in degraded lands (Upadhyaya et al., 2012). The crop is mainly grown as an annual using cultivars with different durations to maturity: 90 days (short), 120 days (medium) and 180 days (long). It is also cultivated as a perennial crop in several regions of India. Fresh pods are consumed as a vegetable; dried mature grains are used for cooking or in processing.
About 15 viruses are reported to naturally infect pigeonpea (Kumar et al., 2008). Sterility mosaic disease (SMD), caused by Pigeonpea sterility mosaic virus (PPSMV), is the economically most important viral disease in India, causing an estimated annual loss of more than US$300 million (Reddy et al., 1998). SMD was first reported in 1931 from Pusa (Bihar) (Mitra, 1931) and is mostly endemic to India, Nepal, Bangladesh and Myanmar. The disease has also been reported from Thailand (Nene et al., 1996) and Sri Lanka (Newton and Peiris, 1953). Symptoms similar to those of SMD were observed in pigeonpea grown in China (G. V. R. Rao, personal communication, ICRISAT, Patancheru, India), but there are no reports of virus confirmation. This disease is not known to occur outside Asia. SMD aetiology remained a mystery for about seven decades despite intensive efforts, making it one of the most challenging diseases of the 20th century in the Indian subcontinent (Nene, 1995). The first breakthrough was achieved in 1999, which eventually led to the complete characterization of the virus responsible for SMD (Kumar et al., 2000).
Aetiology: From Invisible Foe to a Member of the Genus Emaravirus
Complete to partial cessation of flower production (sterility), excessive vegetative growth, stunting, chlorotic rings or mosaic symptoms on the leaves and a reduction in their size are the characteristic features of SMD‐affected plants (Fig. 1A–C). However, the nature of the symptoms produced depends on the genotype and the time of virus infection. The fresh growth from ratooned pigeonpea, a method of harvesting plants by leaving the roots and the lower parts uncut to allow fresh growth from the stubble, shows the most severe symptoms. Ratooning is a common practice of farmers growing pigeonpea as a perennial or backyard crop. This disease is often referred to as the ‘green plague’ as the infected plants are pale green with excessive vegetative growth and no/poor flowering (Fig. 1B). Under favourable conditions it spreads across the fields in an epidemic form. In addition, infection predisposes the plants to subsequent infection by fungal diseases and colonization by spider mites. The yield losses caused by SMD vary and depend on the genotype and stage of infection; infection before flowering (<45 days after planting) can lead to a yield loss from 95% to 100%; late infections (>45 days after planting) can lead to a yield loss of between 26% and 97% (Kannaiyan et al., 1984).
Figure 1.

(A) Sterility mosaic disease (SMD)‐affected pigeonpea; symptomatic leaves are shown with arrows. (B) SMD‐affected plant (circled) is bushy and green without any flowers, whereas the uninfected plants (arrows) matured with pods. (C) Close‐up of SMD leaf symptoms. (D) Scanning electron micrograph of the Pigeonpea sterility mosaic virus (PPSMV) vector Aceria cajani on pigeonpea leaf.
The etiology of SMD remained a mystery for well over 70 years despite extensive efforts which yielded no clear answer, but suggested the likely involvement of a virus‐like agent based on the symptoms and transmission properties (Nene, 1995). The invariable association of the eriophyid mite species Aceria cajani (Fig. 1D) with SMD led to speculations that the disease could be caused by the toxic effect of eriophyid mite feeding. However, elaborate transmission studies with eriophyid mites reared on healthy plants demonstrated that the mosaic symptoms were not caused by mite feeding, strongly indicating the association of a causal agent (Ghanekar et al., 1992). The first evidence of the consistent association of a virus was provided in 1999, and it was named PPSMV (Kumar et al., 2000, 2002a, 2003).
Electron microscopy studies on the purified virus particles indicated that these were thread‐like flexuous particles with an undetermined length, 3–10 nm in diameter, similar in appearance to tenuiviruses (Fig. 2A, C) (Kumar et al., 2000, 2003). These particles were shown to be composed of at least five to seven single‐stranded RNA segments and a protein with an estimated size of 32 kDa (Fig. 2B) (Kumar et al., 2003). Ultracytopathology studies of thin sections of SMD‐infected leaf tissues found amorphous electron‐dense material and large numbers of membrane‐bound bodies (MBBs) of 100–150 nm in diameter in all cell types of the leaf, except the phloem and bundle sheath parenchyma (Kumar et al., 2002a). In immunoelectron microscopy using gold‐labelled antibodies produced against the purified PPSMV preparations, these were bound specifically to MBBs within the cells, indicating the likely relationship between the thin, flexuous particles and MBBs (Fig. 2C) (Kumar et al., 2002a). The occurrence of MBBs was demonstrated in two pigeonpea genotypes that showed contrasting symptom response to PPSMV infection: ICP8863, in which PPSMV induced systemic severe mosaic symptoms, and ICP2376, in which the virus induced chlorotic ringspots as a result of incomplete systemic infection (Kumar et al., 2002a). In addition, MBB occurrence was also demonstrated in N. benthamiana experimentally infected by mechanical sap inoculation (Kumar et al., 2002a). Often, the MBBs containing ribonucleoproteins were found to be located near the endoplasmic reticulum (ER) and Golgi cisterns, indicating that particle morphogenesis might take place at these intracellular host membranes, as described for tospoviruses (Kikkert et al., 1999; Kumar et al., 2002a). For several years, only a small sequence length of 764 nucleotides (GenBank Acc. No. AJ439561) from RNA‐5 (renumbered RNA‐3 in Elbeaino et al., 2014) of the PPSMV genome was known (Kumar et al., 2003). This segment, and several other cDNA clones generated from PPSMV RNA preparations (P. L. Kumar, unpublished data), found no significant matches with any other sequence available at that time in the National Center for Biotechnology Information (NCBI) GenBank. As purified preparations of PPSMV were not infectious, Koch's postulates were not fulfilled. However, consistent association of PPSMV in SMD‐affected plants was demonstrated by virus detection in naturally infected plants in the fields, and also in plants experimentally infected with laboratory‐reared colonies of A. cajani (Kulkarni et al., 2002; Kumar et al., 2003).
Figure 2.
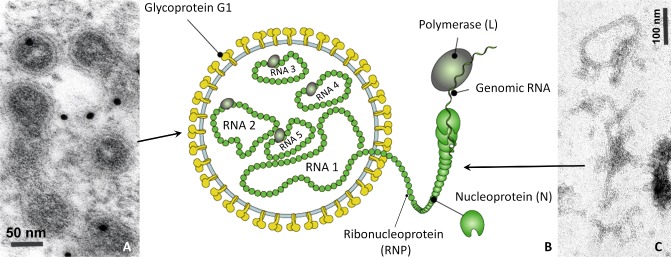
Diagrammatic representation of Pigeonpea sterility mosaic virus (PPSMV) particles. (A) Transmission electron micrograph (TEM) of membrane‐bound bodies of PPSMV in sterility mosaic disease (SMD)‐affected pigeonpea leaf. Particles were immunogold labelled with anti‐PPSMV polyclonal antiserum [source: James Hutton Institute (JHI), UK]. (B) Conceptual diagram of PPSMV particle structure (source: ViralZone, Swiss Institute of Bioinformatics). (C) TEM of ribonucleoprotein particles observed in purified preparations from SMD‐affected pigeonpea leaf samples (source: JHI).
The closest similarities to PPSMV were with the causal agent of High Plains disease of maize and wheat, and rose rosette disease, which shared similar cytopathology and vector transmission (Ahn et al., 1996). Based on this evidence, Kumar et al. (2003) postulated that PPSMV was a novel virus with properties similar to tospoviruses and tenuiviruses, and suggested the grouping of viral agents of similar properties into a separate genus. This was made possible following the complete genomic characterization of European mountain ash ringspot‐associated virus (EMARaV) from European mountain ash or rowan tree (Sorbus aucuparia L.) (Mielke‐Ehret and Muehlbach, 2007). Owing to the unique properties of EMARaV, this led to the creation of a new genus, Emaravirus (Mielke‐Ehret and Mühlbach, 2012). Based on the similarities in nucleotide sequence of RNA‐3, PPSMV was placed in this genus as a tentative species. Recently, the genome of a PPSMV isolate from pigeonpea cv. ICP8863 from International Crops Research Institute for the Semi‐Arid Tropics (ICRISAT)‐Patancheru (India) was completely sequenced by deep sequencing, and was shown to contain five RNA segments (Fig. 3); this provided evidence for its classification in the genus Emaravirus (Elbeaino et al., 2014). This sequence of PPSMV had a very high homology with EMARaV, Fig mosaic virus (FMV) and Rose rosette virus (RRV), as well as other members of Emaravirus (Mielke‐Ehret and Mühlbach, 2012) (Table 1).
Figure 3.
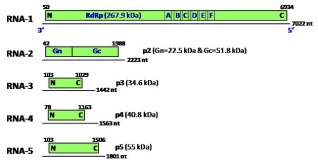
Schematic representation of the organization of the five Pigeonpea sterility mosaic virus (PPSMV) genomic RNA segments (linear lines); their open reading frames (ORFs) (boxes), expression products of each genomic RNA (p1, p2, p3, p4 and p5) and their estimated molecular weights (kDa) are indicated. Letters A–F represent the conserved motifs on the RNA‐dependent RNA polymerase (RdRp) gene on RNA‐1; Gn and Gc indicate the N‐ and C‐termini of the glycoprotein on RNA‐2; N and C are the N‐terminal and C‐terminal ends of the proteins, respectively; nt, nucleotides. (Modified from Elbeaino et al., 2014.)
Table 1.
Structural and biological features of the viruses belonging to the Emaravirus genusa
| Emaravirus species | Morphology of virus particle | Vector | Host species | Geographical distribution |
|---|---|---|---|---|
| EMARaV (European mountain ash ringspot‐associated virus) | dDMBs 80–120 nm | Phytoptus pyri b | Sorbus aucuparia | Europe |
| FMV (Fig mosaic virus) | DMBs 90–200 nm | Aceria ficus | Ficus carica (fig) | USA, Italy, Turkey, Serbia, Japan |
| RRV (Rose rosette virus) | DMBs 120–150 nm | Phyllocoptes fructiphilus | Rosa multiflora (hybrid roses) | USA, Canada |
| RYRSV (Redbud yellow ringspot virus) | DMBs | Aceria sp. | Cercis canadensis (Eastern redbud) | USA |
| RLBV (Raspberry leaf blotch virus) | Indistinct filamentous bodies | Phyllocoptes gracilis | Rubus spp. | UK |
| WMoV (Wheat mosaic virus) or MRSV/HPV (Maize red stripe virus or High plains virus) | Filamentous structures and enveloped particles 80–200 nm | Aceria tosichella | Zea mays, Triticum aestivum | North America, South America, Australia |
| PPSMV (Pigeonpea sterility mosaic virus) | Filamentous structures and DMBs 100–150 nm | Aceria cajani | Cajanus cajan | India, Nepal, Bangladesh, Myanmar, Sri Lanka, Thailandc |
| Coffee emaravirus diseasee | nif | nif | Coffea spp. | Hawaii (USA) |
Modified from Mielke‐Ehret and Mühlbach (2012).
Putative vector.
PPSMV confirmed only in India. It is likely that the same or similar virus would be involved in sterility mosaic disease (SMD) aetiology in other countries.
DMB, double membrane‐bound bodies.
New disease reported in 2014 (source: http://dailycoffeenews.com/2014/03/21/scientists‐find‐troubling‐new‐emaravirus‐coffee‐disease‐on‐hawaii‐farm/ 24Jun2014).
ni, no information.
The Emaravirus genus is not yet assigned to any family, and includes segmented, negative‐sense, single‐stranded RNA viruses with enveloped particles (double membrane‐bound bodies, DMBs; Fig. 2A), with a diameter of 80–200 nm (Mühlbach and Mielke‐Ehret, 2011). EMARaV is the type species of this genus. The other four accepted species are FMV, RRV, Redbud yellow ringspot virus (RYRV) and Raspberry leaf blotch virus (RLBV). The aetiological roles of all the emaraviruses identified to date were based on consistent detection in diseased plants; Koch's postulates were not completed (Mielke‐Ehret and Mühlbach, 2012). All the accepted or putative emaraviruses have segmented genomes consisting of four or more negative‐sense RNAs, and most are transmitted by eriophyid mites (Tables 1 and 2) (Mielke‐Ehret and Muehlbach, 2007). Other tentative emaraviruses have varying numbers of RNA segments ranging from four to eight, and the presence of subgenomic length mRNAs has been reported for Wheat mosaic virus (WMoV) (Tatineni et al., 2014) (Table 2). The presence of a diverse number of genomic segments among member species indicates that emaraviruses may evolve by acquiring additional genomic RNA segments so as to facilitate precise virus–host or virus–vector interactions. Each segment of the viral genome generally contains a single open reading frame (ORF) that codes for RNA‐dependent RNA polymerase (RdRp), glycoprotein (GP), nucleocapsid (NC) protein, movement protein (MP) and other proteins with unknown functions. RdRp, GP and NC protein of member species of the emaraviruses show low to moderate identities of 33%–67%, 24%–50% and 19%–60%, respectively (Tatineni et al., 2014). Members of the genera Emaravirus, Tospovirus and Tenuivirus share several features with respect to their genomic organization, and particle and NC protein morphology (Mielke‐Ehret and Mühlbach, 2012). Both tospoviruses and tenuiviruses are negative‐strand RNA viruses using an ambisense strategy for protein expression; Tospovirus is a genus of plant‐infecting viruses in the family Bunyaviridae, whereas the genus Tenuivirus is not assigned to any family (Kormelink et al., 2011).
Table 2.
Genome organization of emaraviruses, the size of their genomic segments and the putative proteins and their sizes encoded by thema
| Emaravirus species | RNA‐1 RNA‐dependent RNA polymerase (RdRp) (p1) | RNA‐2 glycoprotein precursor (p2) | RNA‐3 nucleocapsid protein (p3 or NC) | RNA‐4 movement protein (p4) | RNA‐5 unknown (p5) | RNA‐6 unknown (p6) |
|---|---|---|---|---|---|---|
| PPSMV | 7022 nt | 2223 nt | 1442 nt | 1563 nt | 1801 nt | — |
| 268 kDa | 74.3 kDa | 34.6 kDa | 40.8 kDa | 55 kDa | ||
| FMV | 7093 nt | 2252 nt | 1490 nt | 1472 nt | 1752 nt | 1212 nt |
| 264 kDa | 73 kDa | 35 kDa | 40.5 kDa | 59 kDa | 22 kDa | |
| RRV | 7026 nt | 2220 nt | 1544 nt | 1541 nt | —b | — |
| 265 kDa | 74 kDa | 36 kDa | 41 kDa | |||
| EMARaV | 7040 nt | 2335 nt | 1559 nt | 1348 nt | — | — |
| 266 kDa | 75 kDa | 35 kDa | 27 kDa | |||
| RLBV | 7062 nt | 2135 nt | 1365 nt | 1675 nt | 1718 nt | — |
| 269 kDa | 75 kDa | 32 kDa | 42 kDa | 56 kDa | ||
| WMoVc | 6981 nt | 2211 nt | 1439/1441 nt | 1682 nt | 1715 nt | 1752 nt |
| 266 kDa | 77 kDa | 33 kDa | 42 kDa | 56 kDa | 58 kDa |
EMARaV, European mountain ash ringspot‐associated virus; FMV, Fig mosaic virus; PPSMV, Pigeonpea sterility mosaic virus; RLBV, Raspberry leaf blotch virus; RRV, Rose rosette virus; WMoV, Wheat mosaic virus. nt, nucleotides.
Modified from Mielke‐Ehret and Mühlbach (2012).
Not detected.
The WMoV genome has eight RNA segments; RNA‐7 (1434 nt) and RNA‐8 (1339 nt) are predicted to code for 36‐ and 21‐kDa proteins. WMoV is also known to have two different sized RNA‐3 forms (1439 and 1441 nt).
PPSMV Genome Organization, Gene Function and Taxonomy
The genome of a PPSMV isolate from the SMD‐affected genotype ICP8863 from ICRISAT‐Patancheru (India) was completely sequenced by deep sequencing of double‐stranded RNA preparations, and was shown to contain five RNA segments (Elbeaino et al., 2014) (Figs 2B and 3). The largest segment, RNA‐1, is 7022 nucleotides in length and codes for RdRp (p1, 267.9 kDa). RNA‐2, with a sequence length of 2223 nucleotides, codes for GPs (p2, 74.3 kDa); RNA‐3, with a length of 1442 nucleotides, codes for NC protein (p3, 34.6 kDa); RNA‐4, with a length of 1563 nucleotides, codes for a putative MP (p4, 40.8 kDa); and RNA‐5, with a length of 1689 nucleotides, codes for p5 (55 kDa), with unknown function (Fig. 3) (Elbeaino et al., 2014). Although the length of RNA‐3 of PPSMV is shorter than those of RNA‐4 and RNA‐5, by analogy with other emaraviruses, it is referred to as RNA‐3, based on the similarities of the functional domains of the ORFs encoded by respective RNA segments (Elbeaino et al., 2014). All of the five RNA segments of PPSMV show the highest identity with orthologues of FMV and RRV.
The PPSMV RdRp (2294 amino acids), encoded by RNA, contains all the conserved motifs common to members of the Bunyaviridae family (Fig. 3). The RdRp of PPSMV has amino acid sequence similarities ranging from 37% to 54% with the RdRp of other emaraviruses, and contains the core polymerase module with five conserved motifs (motifs A–E) of the RdRp active site (Fig. 3). The RdRp motifs A (DASKWS, 1125–1130) and C (SDD, 1183–1185) are part of the palm domain of the replicase protein involved in divalent metal cation binding, and motif B (QGNLNHLSS, 1210–1218) is involved in RNA binding (Bruenn, 2003; Elbeaino et al., 2014). Motif D (KK, 1276–1277) is thought to have catalytic activity because of its tertiary structure, and motif E (EFLST, 1312–1316) is thought to be involved in cap‐snatching activity, characteristic of the Bunyaviridae family (Duijsings et al., 2001). Cap‐snatching is a strategy of certain RNA viruses to express their genes, wherein the RdRp, with its inherent endonuclease property, cleaves the capped‐RNA leader sequences of host mRNA and subsequently uses them to prime the transcription of the viral genome (Kormelink et al., 2011).
RNA‐2 encodes for a GP precursor of 648 amino acids and this protein shares 31%–45% identity with the GP precursors of FMV, RRV, RYRV, RLBV and EMARaV (Elbeaino et al., 2014). The GPs are responsible for the formation of spike projections on the envelope membrane of tospoviruses and other Bunyaviridae members (Kormelink et al., 2011). Amino acid analysis indicated the presence of three putative transmembrane helices, four putative glycosylation sites and a signal peptide sequence with one predicted cleavage site, in which the GP precursor may be cleaved into two single GPs, Gn (22.5 kDa) and Gc (51.8 kDa), in the Golgi complex (Elbeaino et al., 2014; Kormelink et al., 2011). RNA‐3 codes for NC protein (nucleoprotein, N), with amino acid identities comparable with the N proteins of FMV (44%), RRV (43%), RYRSV (37%), EMARaV (35%), RLBV (27%) and WMoV (20%) (Fig. 3). The PPSMV NC protein contains three stretches of amino acids, NVLSFNK (134–140), NRLA (183–186) and GYEF (204–207), most probably involved in RNA binding, which are also conserved in other emaraviruses (Elbeaino et al., 2014). The NC protein functions to encapsidate the viral genomic RNAs and is also a component of the viroplasms and polymerase complex (Kormelink et al., 2011). Recent studies on the macromolecular trafficking of the virus particles of FMV, employing live imaging and ultrastructural analysis, have revealed that the agglomerates of the N protein are passively dragged by the actomyosin‐mediated streaming of the ER, resulting in their rapid movement in plant cells (Ishikawa et al., 2015). In virus‐infected cells, the N protein agglomerates are surrounded by ER membranes, which may indicate that they form the basis of enveloped virus particles (Ishikawa et al., 2015). The PPSMV N protein may perform similar functions in viral protein trafficking through the ER. Although studies are yet to be performed, ultracytopathology using immunogold‐labelled antibodies raised against the N protein of PPSMV heavily labelled the ER, indicating the localization of the N protein on the ER and MBBs (Kumar et al., 2002a; Fig. 2A). The size of the NC proteins varied among the PPSMV isolates; those from Patancheru (P) in Andhra Pradesh and Bengaluru (B; formerly Bangalore) contained a 32‐kDa N protein, whereas another from Coimbatore (C) in Tamil Nadu was characterized by a larger protein of size c. 35 kDa (Jones et al., 2004; Kumar et al., 2004).
RNA‐4 encodes for a protein (p4) with a molecular mass of 40.8 kDa, with homologies with the p4 proteins encoded by RRV (41%), FMV (40%), RYRSV (30%) and RLBV (24%). However, the amino acid analysis predicted the expression product of PPSMV RNA‐4 to be a possible membrane‐located protein lacking transmembrane helices, which might be involved in the cell‐to‐cell movement of the virus, similar to the MPs encoded by RNA‐4 of RLBV and RRV (McGavin et al., 2012; Yu et al., 2013). The plant virus MPs modify plasmodesmata to allow the passage of viral genomes from one cell to another. In addition, the p4 protein could act as a gene silencing suppressor. The PPSMV RNA‐5 encodes a polypeptide (p5) of 473 amino acids with a molecular mass of 55 kDa, sharing a sequence identity of 33% with FMV RNA‐5 (Elbeaino et al., 2014). The p5 protein has not been described for EMARaV, RRV and RYRSV, and no amino acid sequence motifs were identified that might indicate its function. However, p5 has been reported in RLBV. When the reporter gene green fluorescent protein (GFP) was fused to p5, the fusion protein was localized in the cytoplasm as aggregated structures, but its role in the life cycle of RLBV remains unknown (McGavin et al., 2012).
The phylogenetic analysis of amino acid sequences of RdRp, GPs and NC proteins encoded by RNA‐1, RNA‐2 and RNA‐3 of emaraviruses and selected members of Bunyaviridae indicated that emaraviruses formed a separate cluster and that, within them, there were three clades: the first contained WMoV and RLBV, the second RYRV and EMARaV, and the third PPSMV, RRV and FMV (Fig. 4). Proteins encoded by PPSMV showed close resemblance to FMV and RRV proteins. The 5′ and 3′ termini of all RNA segments of PPSMV possess untranslated regions (UTRs) that extend from 42 to 103 nucleotides at the 5′ end and from 88 to 413 nucleotides at the 3′ end. Sequences of the first 13 nucleotides at both the 5′ and 3′ termini of each RNA segment are similar and exhibit sequence complementarities that are unbroken in all the RNAs, except for two nucleotides at positions 8 and 9 (U8–U9) (Elbeaino et al., 2014). These stretches of nucleotides are conserved in all 5′ and 3′ genomic RNA ends of all the emaraviruses (Kormelink et al., 2011). Within each genomic RNA segment, the nucleotide complementarity extends to ∼65 nucleotides, which aids in the formation of a panhandle structure, giving a pseudo‐circular appearance to the ribonucleoproteins. The terminal sequences are conserved among members of the same genus, but differ among genera of the Bunyaviridae family (Elliott and Blakqori, 2011).
Figure 4.
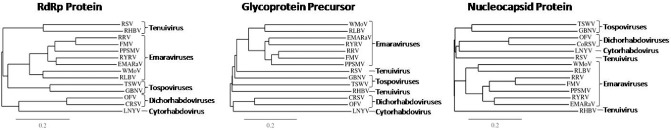
Phylogenetic trees drawn with predicted amino acid sequences of RNA‐1 (RNA‐dependent RNA polymerase), RNA‐2 (glycoprotein precursor) and RNA‐3 (nucleocapsid) of Pigeonpea sterility mosaic virus (PPSMV), together with orthologues of members of the genera Emaravirus, Tenuivirus, Cytorhabdovirus, Dichorrhabdovirus and Tospovirus. All the sequences of the polypeptides were retrieved from the National Center for Biotechnology Information (NCBI) database and the multiple sequence alignments were performed using the MUSCLE (MUltiple Sequence Comparison by Log‐Expectation) program; the phylograms were drawn using the TreeDyn program. TSWV (Tomato spotted wilt virus, Acc. No. L20953) and GBNV (Groundnut bud necrosis virus, Acc. No. NC003619) are tospoviruses; RSV (Rice stripe virus, Acc. No. JQ927420) and RHBV (Rice hoja blanca virus, Acc. No. AF004658) are tenuiviruses; CoRSV (Coffee ringspot virus, Acc. No. KF812525) and OFV (Orchid fleck virus, Acc. No. AB244417) are dichorhabdoviruses (unassigned negative‐sense, single‐stranded RNA viruses that were previously proposed to be included in the family Rhabdoviridae); LNYV (Lettuce necrotic yellow virus, Acc. No. L30103) is used as an outgroup and is a member of the genus Cytorhabdovirus (family Rhabdoviridae). EMARaV, European mountain ash ringspot‐associated virus; FMV, Fig mosaic virus; RLBV, Raspberry leaf blotch virus; RRV, Rose rosette virus; RYRV, Redbud yellow ringspot virus; WMoV, Wheat mosaic virus.
Evidence from the multilocational evaluation of a set of differential pigeonpea genotypes across different locations in India suggests the probable occurrence of several variants of PPSMV (Kumar et al., 2004; Reddy et al., 1993). For instance, genotype ICP2376 showed ringspot symptoms in Patancheru, but severe mosaic in Bengaluru and Coimbatore (Kumar et al., 2004; Reddy et al., 1993). The occurrence of different isolates (strains or species) of the causal agent was hypothesized for this phenomenon. Unpublished studies on PPSMV isolates from SMD‐affected plants in Bengaluru (Karnataka state) and Coimbatore (Tamil Nadu state) showed differences in the cytopathology and size of the ribonucleoprotein (P. L. Kumar, T. K. S. Latha, A. T. Jones and D. V. R. Reddy, unpublished data; Kumar et al., 2004). Recently, four genomic segments of a PPSMV variant, named PPSMV‐II (Isolate: Chevela), have been released in the NCBI GenBank (Acc. Nos. LM652701, LM652702, LM652703 and LM652704) (S. Kumar, B. L. Subbarao, A. A. Zaidi and V. Hallan, unpublished data). These sequences corresponding to RNA‐3 and RNA‐4 share less than 60% homology at the nucleotide level with PPSMV, confirming the occurrence of diverse strains or even different species of the virus involved in SMD in India. Evidence from this and previous studies underscores the need for nucleotide sequencing of the various geographical isolates in order to understand the diversity of viruses associated with SMD.
Virus Diagnostics
SMD symptoms in pigeonpea are unique and form a key diagnostic feature for disease identification. However, virus characterization has led to the development of cost‐effective and sensitive diagnostic tools. Polyclonal antibodies to PPSMV particle preparations are effective in detecting PPSMV in plant tissues by the double antibody sandwich enzyme‐linked immunosorbent assay (DAS‐ELISA) using enzyme‐labelled (alkaline phosphatase) immunogammaglobulin (Kumar et al., 2003, 2007). Serological diagnostic methods, such as DAS‐ELISA and dot‐immunobinding assay (DIBA), have been developed to detect the presence of PPSMV from the viruliferous vector using whole mite extract as an antigen (Latha and Doraiswamy, 2008). Both DAS‐ELISA and DIBA are sensitive for virus detection in vector mites; however, detection is possible only in groups of 10 or more mites. Reverse transcription‐polymerase chain reaction (RT‐PCR) assay has been developed based on the amplification of a 230‐nucleotide region of the RNA‐3 segment (Kumar et al., 2003). PCR‐based methods involving specific degenerate PCR primers for the amplification of partial RdRp sequences of emaraviruses have also been developed, which have been shown to detect a wide range of emaraviruses (Elbeaino et al., 2013). Diagnostic methods, such as lateral flow devices (LFDs), isothermal amplification methods, such as loop‐mediated isothermal amplification (LAMP), and the recombinase amplification reaction (Boonham et al., 2014) can offer unique capabilities for future diagnostics of SMD viruses. Last but not least, because of the significant reduction in their cost and advancement in their performance, next‐generation sequencing (NGS) strategies have accelerated the speed at which new viruses are being discovered. This technology has already been employed to unravel the first genome sequence of PPSMV from Patancheru, and has great potential to characterize geographical variants.
Virus Transmission
PPSMV is transmitted by the eriophyid mite A. cajani (Channabasavanna) (Arthropoda: Acari: Eriophyidae) (Fig. 1D) in a semi‐persistent manner (Channabasavanna, 1981; Kulkarni et al., 2002). Eriophyid mites have been shown to vector several emaraviruses and also members of the genus Tritimovirus (family Potyviridae), such as Wheat streak mosaic virus (WSMV). These microscopic eriophyid mites are one of the smallest arthropods, white to yellowish in colour, and their primary method of population spread is by the wind. They are obligate plant pests during active stages of their life cycle and affect a wide range of crops, causing substantial economic damage (Oldfield and Proeseler, 1996). Aceria cajani mites inhabit the lower surface of leaves and are predominantly found on the symptomatic leaves of PPSMV‐infected plants; their feeding causes no mechanical damage to the host plant.
Information available on the transmission mechanism of emaraviruses by eriophyid mites is limited to PPSMV and WMoV. Some of the eriophyid mites are known to transmit together both emaraviruses and potyviruses, leading to mixed infections. The wheat curl mite (Aceria tosichella Keifer) is the vector for both WMoV and WSMV (Ahn et al., 1998; Mahmood et al., 1998). Transmission studies of PPSMV by eriophyid mites have been reported by Kulkarni et al. (2002), and it has been shown that the transmission efficiency of a single A. cajani mite is up to 53%. However, when more than five mites per plant were used, it was 100%. The A. cajani mites acquired PPSMV after a minimum acquisition access period (AAP) of 15 min, and transmitted the virus after a minimum inoculation access period (IAP) of 90 min. There was no latent period (Kulkarni et al., 2002). However, there is no evidence of PPSMV replication within the mite, which stays infectious for about 6–13 h, and no transovarial transmission was observed (Kulkarni et al., 2002). PPSMV infection of pigeonpea significantly increased the proliferation of A. cajani compared with their numbers on healthy plants (Reddy and Nene, 1980). Similar observations were made for Blackcurrant reversion virus (genus Nepovirus) and its mite vector Cecidophyopsis ribis (Thresh, 1964). Thus, in such instances, there could be a beneficial relationship between the vector mite and the virus it transmits. As a result of their short stylet, eriophyid mites predominantly reach the epidermal cells or adjacent layers of the mesophyll tissue on the undersurface of plant leaves. However, for effective transmission of the virus, the cell type to be infected is critical. The stringent host specificity of eriophyid mites makes it difficult to study them on experimental host plants from the genus/families of Chenopodium, Solanaceae and Cucurbitaceae. The mites die quickly without feeding on their host and A. cajani has been shown to survive for only 13 h (Kulkarni et al., 2002). However, once established on PPSMV‐susceptible pigeonpea genotypes, the mites can multiply to high densities in only a few weeks. The dispersal of these mites is passive, mainly assisted by wind currents. Of the different abiotic factors, such as temperature, relative humidity and rainfall, the relative humidity has a significant effect on the mite population (Kaushik et al., 2013; Singh and Rathi, 1997). A mean temperature of about 20–30 °C was found to be favourable for the proliferation of the mites. However, high temperatures and heavy rainfall were unfavourable for their growth.
The accurate identification of eriophyid mites from their morphological features is very difficult because of their minute size (∼200 mm) and their morphological uniformity. PCR amplification of the primary transcription unit of the ribosomal (r)RNA‐encoding genes (3’ end of the 18S gene, ITS1, 5.8S gene, ITS2 and 5’ end of the 28S gene; collectively known as rDNA) is widely used in the diagnostics of microorganisms. rDNA is a very well‐studied gene family with a highly conserved sequence and structure of the coding regions across different species. The two internal transcribed spacer (ITS) regions between the coding regions diverge rapidly between species, but are highly conserved within eukaryotic species, and this has also been confirmed in acarids (Fenton et al., 1997; Hillis and Dixon, 1991; Navajas et al., 1994, 2001) (Fig. 5). Other important eriophyid mites prevalent in India are A. guerreronis (Keifer) in coconut, A. litchii (Keifer) in lychee, A. lycopersici (Wolff.) in tomato and aubergine, A. mangiferae (Sayed) in mango, and Eriophyes cernuus (Massee) in ber (Singh and Raghuraman, 2011). Several A. cajani populations were collected from various SMD endemic locations in India, Nepal and Myanmar, and were subjected to PCR‐restriction fragment length polymorphism (RFLP) and sequencing for the ITS regions of rDNA (Kumar et al., 2001; Latha and Doraiswamy, 2008). No significant variation was found in either the ITS regions of rDNA or the morphological features of the eriophyid mites. These results strongly suggest that A. cajani on pigeonpea constitutes only one species across the Indian subcontinent, and that no other Aceria species, and probably no A. cajani biotypes differing in their vectoring ability, are involved in the transmission of PPSMV.
Figure 5.
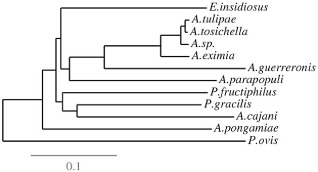
Phylogenetic tree for the internal transcribed spacer (ITS) sequences of the eriophyid mites. The ITS sequences of ∼300–400 nucleotides, corresponding to the pigeonpea mite Aceria cajani (Acc. No. AJ251693), were used to draw the phylogenetic analysis, and the sequences were retrieved from the National Center for Biotechnology Information (NCBI) database. The ITS sequences used are A. cajani (AJ251693), A. eximia (JF920113), A. guerreronis (KJ461967; host plant, coconut), A. parapopuli (JF792237; host plant, poplar), A. pongamiae (AJ251696; host plant, pongamia), Aceria sp. (JQ512784; host plants, sweetpotato and Eastern redbud), A. tosichella (JF960158; host plant, wheat), A. tulipae (AJ251695; host plant, tulip), Eriophyes insidiosus (AJ251694; host plant, peach), Phyllocoptes fructiphilus (AJ251692; host plant, rose), P. gracilis (AJ251697; host plant, raspberry) and Psoroptes ovis (AF270823; hosts: sheep, cattle, goats, horses, rabbits and camelids). Psoroptes ovis, an animal mite (Arthropod; family: Psoroptidae) which causes the contagious sheep scab, was used as an outgroup. Multiple sequence alignments were performed using the MUSCLE (MUltiple Sequence Comparison by Log‐Expectation) program, and the phylograms were drawn using the TreeDyn program.
Mechanical Transmission and Host Range
The purified preparations of PPSMV were not infectious to plants (Kumar et al., 2003). However, it was possible to transmit (10%–30% infection) PPSMV to N. benthamiana and N. clevelandii by sap inoculation using fresh extract of SMD‐infected pigeonpea leaves, but not to pigeonpea or other herbaceous hosts (Kumar et al., 2002b). PPSMV was also transmitted experimentally by viruliferous A. cajani to French bean (Kulkarni et al., 2003a) and by grafting to pigeonpea (Ghanekar et al., 1992; Kumar et al., 2002a). None of the emaraviruses, including PPSMV, have been shown to be transmitted by seed or pollen (Divya et al., 2005; Mielke‐Ehret and Mühlbach, 2012). In pigeonpea seed itself, PPSMV was detectable only in the seed coat, but not in the cotyledons. Three commonly used methods for experimental transmission of PPSMV from infected to healthy pigeonpea plants are: (i) leaf stapling (Nene and Reddy, 1976a); (ii) infector hedge (Nene and Reddy, 1976a); and (iii) spreader row inoculation (Nene and Reddy, 1976b). The leaf stapling technique can be used for evaluation in pots and in the field, whereas infector hedge and spreader row techniques are used in field screening. The leaf stapling technique requires more inoculum and labour, and can also cause damage to the leaf.
Host range studies on SMD indicate that PPSMV can infect several accessions of cultivated and wild pigeonpea, N. benthamiana, N. clevelandii, Phaseolus vulgaris and Chrozophora rottleri (Kulkarni et al., 2003b; Kumar et al., 2004). In field conditions, pigeonpea and its wild relatives are infected with PPSMV, whereas only a few wild species of Cajanus support the vector A. cajani (Kumar et al., 2007). Although Hibiscus panduriformis (Malvaceae) was infested with A. cajani, it could not support PPSMV. These studies show that PPSMV can infect plants outside the genus Cajanus. However, because of the host specificity of the mites, only the accessions of the Cajanus genus were found to support the proliferation of PPSMV under natural conditions (Kumar et al., 2007). Therefore, only the cultivated and wild accessions of pigeonpea serve as potential sources of PPSMV under field conditions, and the weed species, such as H. panduriformis, act as a refuge for mite survival and thus aid the spread of SMD (Kumar et al., 2007).
Epidemiology
Studies on SMD epidemiology are limited. In India, SMD is an endemic disease in most of the pigeonpea‐producing regions, but its incidence varies widely from season to season and from one region to another (Kumar et al., 2008). In India, SMD has been reported from the states of Andhra Pradesh, Bihar, Chhattisgarh, Gujarat, Karnataka, Maharashtra, Punjab, Tamil Nadu, Telangana, Uttar Pradesh and West Bengal (Kannaiyan et al., 1984; Narayana et al., 2000; Singh and Raghuraman, 2011; Zote et al., 1991). PPSMV is not seed borne; however, the disease is introduced into the newly sown fields by viruliferous mites spreading from perennial pigeonpea or volunteer plants. Therefore, the occurrence and incidence of SMD depends entirely on the proximity of new fields to inoculum sources, weather conditions favouring eriophyid mite multiplication and genotype susceptibility. Although there are contrasting opinions about the influence of climatic conditions on the epidemiology of SMD, plants cultivated under irrigation are more vulnerable to early infection by SMD (Dharmaraj et al., 2004).
The vast diversity in cropping seasons and patterns makes it difficult to identify the primary sources of the inoculum of SMD. Infected plants left in the field after harvest, and the occurrence of perennial pigeonpea and its wild relatives, such as C. scarabaeoides, in the field are considered to be potential sources of primary inoculum (Kumar et al., 2008; Narayana et al., 2000). In rain‐fed agriculture, the left‐over stubble of pigeonpea in the field after harvest, and plants thriving on the banks of canals, near wells or any such water bodies, still have a reasonable amount of green foliage to harbour and support the proliferation of viruliferous mites (Dharmaraj et al., 2004). Immediately after the early or summer rains, the left‐over stubble, previously infected by SMD, sprouts back with sufficient foliage to act as a primary source of SMD inoculum, thus providing an opportunity for repeated cycles of infection. The spread of SMD within fields mainly depends on the proximity to sources of inoculum, plant age, genotype, weather conditions and the population of vector mites.
Management of SMD
Insecticides and acaricides, commonly used for the chemical management of vectors transmitting viral diseases (Hoy, 2011; Marcic, 2012; Van Leeuwen et al., 2010), are seldom used for the control of eriophyid mites to manage SMD. Biological control through the use of entomopathogenic fungi is also an option, as the eriophyid mites are soft‐bodied organisms with no cuticular barrier and the fungi generally invade through their cuticle (McCoy, 1996). However, the most effective and realistic approach to reduce losses caused by disease is the use of host plant resistance or the deployment of less susceptible cultivars. Recent advances in SMD research have facilitated the selection of high‐yielding varieties with durable resistance to SMD. Sources of resistance have been identified in the germplasm collection at ICRISAT, Patancheru, India (Kumar et al., 2008; Sharma et al., 2012). However, distinct PPSMV isolates occur in different geographical regions of India, and broad‐based resistance to all of these isolates is scarce in cultivated genotypes. Broad‐based resistance in the wild species C. scarabaeoides has been reported for mild and severe strains of PPSMV. However, it is still not clear whether the observed resistance is to the mites or to PPSMV (Kulkarni et al., 2003a). Studies have been performed on the inheritance of resistance in different cultivars against different isolates, and there are major differences in the interpretations made on the nature of resistance (Ganapathy et al., 2009; Gnanesh et al., 2011a, 2011b; Kumar et al., 2005; Srinivas et al., 1997). Studies by Kumar et al. (2005) have shown that some wild Cajanus species are resistant to three isolates of PPSMV: Patancheru, Bengaluru and Coimbatore. Recently, the nature of inheritance of SMD was studied in the segregating populations of two crosses: Gullyal white (susceptible) × BSMR 736 (resistant) and BSMR 736 (resistant) × ICP 8863 (susceptible) (Bhairappanavar et al., 2014; Daspute et al., 2014). The above studies showed that the resistant trait was governed by two independent non‐allelic genes, designated SV1 and SV2, with inhibitory gene interaction (Bhairappanavar et al., 2014; Daspute et al., 2014). Fifteen accessions resistant to SMD were identified by screening 115 accessions from six Cajanus species (C. albicans, C. platycarpus, C. cajanifolius, C. lineatus, C. scarabaeoides and C. sericeus) against the three PPSMV isolates prevailing in peninsular India, through mite‐mediated virus inoculation under glasshouse conditions (Kulkarni et al., 2003a; Kumar et al., 2005). Most of the wild species did not support the multiplication of mites. However, the majority of the accessions resistant to PPSMV, when challenged with viruliferous mites, were susceptible by graft inoculation. This suggests that vector resistance confers resistance to infection with PPSMV (Kumar et al., 2008). Some of the wild species which are resistant to infestation by mites have a thicker leaf cuticle and epidermal cell wall that prevent the stylet from reaching epidermal cells (Reddy et al., 1995). Cajanus scarabaeoides (ICPW 94), which is resistant to all isolates of PPSMV, is used in the crossing programme and the progeny are screened for resistance. Lines derived from crosses with C. acutifolius and C. platycarpus have shown resistance to the Patancheru isolate of PPSMV under field conditions (Mallikarjuna et al., 2011). Recently, new sources of resistance to Fusarium wilt and SMD have been identified in a mini‐core collection of pigeonpea germplasm at ICRISAT (Sharma et al., 2012). A high level of resistance was found in 24 accessions. Combined resistance to Fusarium wilt and SMD was found in five accessions (ICPs 6739, 8860, 11015, 13304 and 14819), and these diverse accessions could be useful for the resistance breeding programme (Sharma et al., 2012).
Amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR)‐based molecular markers have been developed for SMD resistance, and mapping populations are also being developed (Ganapathy et al., 2009; Gnanesh et al., 2011a; Naik et al., 2012). Using composite interval mapping, different quantitative trait loci (QTLs) have been identified for resistance to Patancheru and Bengaluru isolates, and this is an indication of the involvement of different genes conferring resistance to these isolates (Gnanesh et al., 2011b). Recently, the draft genome sequence for the inbred line ICPL 87119, popularly known as Asha, has been published, and this is a widely cultivated Indian variety tolerant to SMD (Singh et al., 2011; Varshney et al., 2012). A number of genetic and genomic resources are available for this genotype and the genome sequence information should help in the development of novel and reliable molecular markers for resistance, and thus expedite breeding for resistance. Of the available transgenic approaches to control plant viral diseases, RNA‐interference (RNAi) is a very promising strategy that has been successfully employed to control numerous plant viral diseases (Sudarshana et al., 2007). RNAi can also be employed in the development of transgenic pigeonpea resistant to PPSMV. However, this will require significant progress in tissue culture and transformation technologies to develop SMD‐resistant transgenics (Krishna et al., 2010, 2011).
Future Prospects
SMD of pigeonpea has been reported from South‐East Asia alone, and it is interesting that there are no reports of any other Emaravirus‐associated diseases from this region, either because of a lack of exhaustive surveys or because of the absence of a similar Emaravirus. This makes it difficult to develop a hypothesis on the origin and evolution of this virus in the area. It is most important to study the biodiversity of PPSMV by sequencing the full‐length genomes of isolates across the Indian subcontinent. Studies suggest the possible occurrence of several strains, if not species, in SMD aetiology. The identification of alternative hosts of PPSMV and retrieval of the viral sequences from symptomatic alternative host plants or weed plants will help in our understanding of the evolution of PPSMV and its isolates. In addition, in the entire world, PPSMV is the only Emaravirus reported to infect legumes. Thus, there is an urgent need to search for emaraviruses in other hosts of eriophyid mites, such as pongamia, jasmine and sugarcane.
Detailed studies on the characterization of the proteins translated by PPSMV will be a priority for the understanding of the biology of this novel Emaravirus, including the involvement of any other genomic or subgenomic RNAs encoding additional ORFs, as Kumar et al. (2003) have shown the association of five to seven single‐stranded RNA segments with purified preparations from PPSMV‐infected pigeonpea. Studies need to be carried out on the replication of the viral genome, transcription and translation of their genes, movement of the viral genome from cell to cell and across the plant, and virus encapsidation. More importantly, the identification and characterization of the PPSMV proteins involved in the suppression of gene silencing will be critical for strategies to be developed for the genetic engineering of SMD resistance. Further studies on virus–host and virus–insect interactions are also essential for a long‐term solution to this menace. Eriophyid mites, as vectors of PPSMV, require further detailed studies, particularly at the molecular level, as little molecular detail of virus–vector interaction is known for these mites.
Tissue culture and transformation require significant progress to realize transgenic pigeonpea which is stable across several generations. More studies are required on the genetics and inheritance of SMD resistance, as past studies are conflicting and it is difficult to draw any concrete conclusions. Efforts are required to map SMD resistance loci by employing molecular marker technology in cultivated plants and/or wild‐types. The identification and characterization of SMD‐resistant gene(s) should help in the development of virus‐resistant pigeonpea. RNAi is another successful technology that is being used widely to achieve transgenic virus resistance (Patil et al., 2011); this has been successfully used for the control of tospoviruses, which are also negative‐strand RNA viruses (Peng et al., 2014). However, until now, there have been no reports on the transgenic control of any Emaravirus. RNAi‐based resistance is highly sequence specific and the identification of an appropriate target sequence will be critical for its success. The latest techniques, such as NGS of the PPSMV transcriptome or the use of NGS for profiling of its small RNA, should help significantly in the identification of the correct target sequence. Studies on host–pathogen interactions will also be important and, in particular, it will be interesting to understand the molecular basis of the two characteristic features of SMD symptoms in pigeonpea, i.e. sterility and increased vegetative growth.
The discovery of the causal agent of SMD and the unravelling of the PPSMV genome sequence are two important milestones, which are just at the start of their development. There is a long way to go for this novel Emaravirus to be thoroughly understood in terms of its diversity in the Indian subcontinent, its vectors and, ultimately, the delivery of SMD‐resistant pigeonpea to the poor and marginal farmers of South‐East Asia.
Acknowledgements
P. L. Kumar worked at ICRISAT, Patancheru, India, and the John Hutton Institute (JHI, formerly Scottish Crop Research Institute), Dundee, UK, from 1996 to 2007 and gratefully acknowledges D. V. R. Reddy, A. T. Jones, B. Fenton, P. Sreenivasulu, N. K. Kulkarni and other colleagues and collaborators for years of interaction and contributions to research on SMD of pigeonpea. We are grateful to ViralZone (http://www.ncbi.nlm.nih.gov/pubmed?term=20947564) of the Swiss Institute of Bioinformatics for according permission to use the Emaravirus line diagram with modifications (Fig. 2B), and also to JHI for permission to use electron micrographs (Figs 1D, 2A and 2C).
References
- Ahn, K.K. , Kim, K.S. , Gergerich, R.C. , Jensen, S.G. and Anderson, E.J. (1996) Comparative ultrastructure of double membrane‐bound particles and inclusions associated with eriophyid mite‐borne plant diseases of unknown etiology: a potentially new group of plant viruses. J. Submicrosc. Cytol. Pathol. 28, 345–355. [Google Scholar]
- Ahn, K.K. , Kim, K.S. , Gergerich, R.C. and Jensen, S.G. (1998) High Plains disease of corn and wheat: ultrastructural and serological aspects. J. Submicrosc. Cytol. Pathol. 30, 563–571. [PubMed] [Google Scholar]
- Bhairappanavar, S.B. , Fakrudin, B. and Sambasiva Rao, K.R.S. (2014) Inheritance studies of sterility mosaic disease (SMD) resistance cross Gullyal White×BSMR736 in pigeonpea (Cajanus cajan (L.) Millsp.). Plant Gene Trait, 5, 27–32. [Google Scholar]
- Boonham, N. , Kreuze, J. , Winter, S. , van der Vlugt, R. , Bergervoet, J. , Tomlinson, J. and Mumford, R. (2014) Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 186, 20–31. [DOI] [PubMed] [Google Scholar]
- Bruenn, J.A. (2003) A structural and primary sequence comparison of the viral RNA‐dependent RNA polymerases. Nucleic Acids Res. 31, 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channabasavanna, G.P. (1981) Contributions to acarology in India In: Proceedings of the 1st All India Symposium in Acarology (ChannaBasavanna G.P., Mallik B. and Ghopardé K.D., eds), p. 256 Bangalore: Acarological Society of India. [Google Scholar]
- Daspute, A. , Fakrudin, B. , Bhairappanavar, S.B. , Kavil, S.P. , Narayana, Y.D. , Muniswamy, Kumar, A. , Krishnaraj, P.U. , Yerimani, A. and Khadi, B.M. (2014) Inheritance of pigeonpea sterility mosaic disease resistance in pigeonpea. Plant Pathol. J. 30, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaraj, P.S. , Narayana, Y.D. , Kumar, P.L. , Waliyar, F. and Jones, A.T. (2004) Pigeonpea sterility mosaic disease: an emerging problem in northern Karnataka. Int. Chickpea Pigeonpea Newsl. 11, 47–49. [Google Scholar]
- Divya, P. , Kumar, L.L. , Rangaswamy, K.T. and Muniyappa, V. (2005) Detection of pigeonpea sterility mosaic virus in floral parts and seeds. Indian J. Virol. 16, 36. [Google Scholar]
- Duijsings, D. , Kormelink, R. and Goldbach, R. (2001) In vivo analysis of the TSWV cap‐snatching mechanism: single base complementarity and primer length requirements. EMBO J. 20, 2535–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeaino, T. , Whitfield, A. , Sharma, M. and Digiaro, M. (2013) Emaravirus‐specific degenerate PCR primers allowed the identification of partial RNA‐dependent RNA polymerase sequences of Maize red stripe virus and Pigeonpea sterility mosaic virus. J. Virol. Methods, 188, 37–40. [DOI] [PubMed] [Google Scholar]
- Elbeaino, T. , Digiaro, M. , Uppala, M. and Sudini, H. (2014) Deep sequencing of pigeonpea sterility mosaic virus discloses five RNA segments related to emaraviruses. Virus Res. 188, 27–31. [DOI] [PubMed] [Google Scholar]
- Elliott, R.M. and Blakqori, G. (2011) Molecular biology of orthobunyaviruses In: Bunyaviridae: Molecular and Cellular Biology (Plyusnin A. and Elliot R.M., eds), pp. 1–39. Norfolk, UK: Caister Academic Press. [Google Scholar]
- FAOSTAT . (2014) Food and Agriculture Organisation Statistics Database. http://faostat3.fao.org/faostat‐gateway/go/to/home/E. Accessed May 09, 2014.
- Fenton, B. , Malloch, G. and Moxey, E. (1997) Analysis of eriophyid mite rDNA internal transcribed spacer sequences reveals variable simple sequence repeats. Insect Molecular Biology. 6, 23–32. [DOI] [PubMed] [Google Scholar]
- Ganapathy, K.N. , Byregowda, M. , Venkatesha, S.C. , Rama Chandra, R. , Gnanesh, B.N. and Girish, G. (2009) Identification of AFLP markers linked to sterility mosaic disease in pigeonpea Cajanus cajan (L.) Millsp. Int. J. Integr. Biol. 7, 145–149. [Google Scholar]
- Ghanekar, A.M. , Sheila, V.K. , Beniwal, S.P.S. , Reddy, M.V. and Nene, Y.L. (1992) Sterility mosaic of pigeonpea In: Plant Diseases of International Importance, Vol. 1, Diseases of Cereals and Pulses (Singh U.S., Mukhopadhyay A.N., Kumar J. and Chaube H.S., eds), pp. 415–428. Englewood Cliffs, NJ, USA: Prentice Hall, Inc. [Google Scholar]
- Gnanesh, B.N. , Abhishek, B. , Sarma, M. , Wesley, V. , Byre Gowda, M. , Pande, S. , Saxena, R. , Saxena, K.B. and Varshney, R.K. (2011a) Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan (L) Millsp.]. Field Crop Res. 123, 56–61. [Google Scholar]
- Gnanesh, B.N. , Ganapathy, K.N. , Ajay, B.C. and Byre Gowda, M. (2011b) Inheritance of sterility mosaic disease resistance to Bangalore and Patancheru isolates in pigeonpea (Cajanus cajan (L.) Millsp.). Electron. J. Plant Breeding, 2, 218–223. [Google Scholar]
- Hillis, D.M. and Dixon, M.T. (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol. 66, 411–453. [DOI] [PubMed] [Google Scholar]
- Hoy, M.A. (2011) Agricultural Acarology: Introduction to Integrated Mite Management. Boca Raton, FL: CRC Press, Taylor & Francis Group. [Google Scholar]
- Ishikawa, K. , Miura, C. , Maejima, K. , Komatsu, K. , Hashimoto, M. , Tomomitsu, T. , Fukuoka, M. , Yusa, A. , Yamaji, Y. and Namba, S. (2015) Nucleocapsid protein from Fig mosaic virus forms cytoplasmic agglomerates that are hauled by endoplasmic reticulum streaming. J. Virol. 89, 480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.T. , Kumar, P.L. , Saxena, K.B. , Kulkarni, N.K. , Muniyappa, V. and Waliyar, F. (2004) Sterility mosaic disease—the ‘green plague’ of pigeonpea: advances in understanding the etiology, transmission and control of a major virus disease. Plant Dis. 88, 436–445. [DOI] [PubMed] [Google Scholar]
- Kannaiyan, J. , Nene, Y.L. , Reddy, M.V. , Ryan, J.G. and Raju, T.N. (1984) Prevalence of pigeonpea diseases and associated crop losses in Asia, Africa and the Americas. Trop. Pest Manag. 30, 62–71. [Google Scholar]
- Kaushik, D. , Srivastava, S. , Nath, B.C. , Chauhan, V.B. and Singh, R. (2013) Correlation between mite population (Aceria cajani) and environmental factors causing sterility mosaic disease of Pigeon pea. Int. J. Life Sci. 1, 228–232. [Google Scholar]
- Kikkert, M. , van Lent, J. , Storms, M. , Bodegom, P. , Kormelink, R. and Goldbach, R. (1999) Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73, 2288–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormelink, R. , Garcia, M.L. , Goodin, M. , Sasaya, T. and Haenni, A.L. (2011) Negative‐strand RNA viruses: the plant‐infecting counterparts. Virus Res. 162, 184–202. [DOI] [PubMed] [Google Scholar]
- Krishna, G. , Reddy, P.S. , Ramteke, P.W. and Bhattacharya, P.S. (2010) Progress of tissue culture and genetic transformation research in pigeon pea [Cajanus cajan (L.) Millsp.]. Plant Cell Rep. 29, 1079–1095. [DOI] [PubMed] [Google Scholar]
- Krishna, G. , Reddy, P.S. , Ramteke, P.W. , Sohrab, S.S. , Rana, D. and Bhattacharya, P.S. (2011) In vitro regeneration through organogenesis and somatic embryogenesis in pigeon pea [Cajanus cajan (L.) Millsp.] cv. JKR105. Physiol. Mol. Biol. Plants, 17, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, N.K. , Kumar, P.L. , Muniyappa, V. , Jones, A.T. and Reddy, D.V.R. (2002) Transmission of Pigeonpea sterility mosaic virus by the eriophyid mite, Aceria cajani (Acari: Arthropoda). Plant Dis. 86, 1297–1302. [DOI] [PubMed] [Google Scholar]
- Kulkarni, N.K. , Kumar, P.L. , Muniyappa, V. , Jones, A.T. and Reddy, D.V.R. (2003a) Broad‐based resistance to pigeonpea sterility mosaic disease in the accessions of Cajanus scarabaeoides . Indian J. Plant Protect. 31, 6–11. [Google Scholar]
- Kulkarni, N.K. , Kumar, P.L. , Muniyappa, V. , Jones, A.T. and Reddy, D.V.R. (2003b) Studies on host range of Pigeonpea sterility mosaic virus. J. Mycol. Plant Pathol. 33, 141–145. [Google Scholar]
- Kumar, P.L. , Fenton, B. , Duncan, G. , Jones, A.T. , Sreenivasulu, P. and Reddy, D.V.R. (2001) Assessment of variation in Aceria cajani (Acari: Eriophyidae) using analysis of nuclear rDNA ITS regions and scanning electron microscopy: implications for the variability observed in host plant resistance to pigeonpea sterility mosaic disease. Ann. Appl. Biol. 139, 61–73. [Google Scholar]
- Kumar, P.L. , Duncan, G.H. , Roberts, I.M. , Jones, A.T. and Reddy, D.V.R. (2002a) Cytopathology of Pigeonpea sterility mosaic virus in pigeonpea and Nicotiana benthamiana: similarities with those of eriophyid mite‐borne agents of undefined aetiology. Ann. Appl. Biol. 140, 87–96. [Google Scholar]
- Kumar, P.L. , Jones, A.T. and Reddy, D.V.R. (2002b) Mechanical transmission of Pigeonpea sterility mosaic virus. J. Mycol. Plant Pathol. 32, 88–89. [Google Scholar]
- Kumar, P.L. , Jones, A.T. and Reddy, D.V.R. (2003) A novel mite‐transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology, 93, 71–81. [DOI] [PubMed] [Google Scholar]
- Kumar, P.L. , Jones, A.T. , Sreenivasulu, P. and Reddy, D.V.R. (2000) Breakthrough in the identification of the causal agent of pigeonpea sterility mosaic disease. J. Mycol. Plant Pathol. 30, 249. [Google Scholar]
- Kumar, P.L. , Jones, A.T. and Waliyar, F. (2004) Biology, etiology and management of pigeonpea sterility mosaic disease. Annu. Rev. Plant Pathol. 3, 77–100. [Google Scholar]
- Kumar, P.L. , Latha, T.K.S. , Kulkarni, N.K. , Raghavendra, N. , Saxena, K.B. , Waliyar, F. , Rangaswamy, K.T. , Muniyappa, V. , Doriswamy, S. and Jones, A.T. (2005) Broad based resistance to pigeonpea sterility mosaic disease in wild relatives of pigeonpea (Cajanus: Phaseoleae). Ann. Appl. Biol. 146, 371–379. [Google Scholar]
- Kumar, P.L. , Jones, A.T. , Waliyar, F. , Sreenivasulu, P. , Muniyappa, V. , Latha, T.K.S. and Saxena, K.B. (2007) Pigeonpea sterility mosaic disease In: Diagnosis and Detection of Viruses Infecting ICRISAT Mandate Crops (Kumar P.L. and Waliyar F., eds), pp. 15–21. Patancheru, India: ICRISAT. ICRISAT's Methods Manual. [Google Scholar]
- Kumar, P.L. , Jones, A.T. and Waliyar, F. (2008) Virus diseases of pigeonpea In: Characterization, Diagnosis and Management of Plant Viruses. Vol. 3: Vegetable and Pulse Crops (Rao G.P., Kumar P.L. and Holguin‐Pena R.J., eds), pp. 235–258. Texas, USA: Studium Press. [Google Scholar]
- Latha, T.K.S. and Doraiswamy, S. (2008) Detection of pigeonpea sterility mosaic virus, the causal agent of sterility mosaic disease of pigeonpea in viruliferous mite vector by DAS‐ELISA and DIBA. Arch. Phytopathol. Plant Protect. 41, 537–541. [Google Scholar]
- Mahmood, T. , Hein, G.L. and Jensen, S.G. (1998) Mixed infection of hard red winter wheat with High Plains virus and Wheat streak mosaic virus from wheat curl mites in Nebraska. Plant Dis. 82, 311–315. [DOI] [PubMed] [Google Scholar]
- Mallikarjuna, N. , Saxena, K.B. and Jadhav, D.R. (2011) Cajanus In: Wild Crop Relatives: Genomic and Breeding Resources, Legume Crops and Forages (Kole C., ed.), pp. 21–33. Berlin, Heidelberg: Springer‐Verlag. [Google Scholar]
- Marcic, D. (2012) Acaricides in modern management of plant‐feeding mites. J. Pest. Sci. 85, 395–408. [Google Scholar]
- McCoy, C.W. (1996) Pathogens of eriophyoid mites In: Eriophyoid Mites—Their Biology, Natural Enemies and Control (Lindquist E.E., Sabelis M.W. and Bruin J., eds), pp. 481–490. Amsterdam: Elsevier. [Google Scholar]
- McGavin, W.J. , Mitchell, C. , Cock, P.J.A. , Wright, K.M. and MacFarlane, S.A. (2012) Raspberry leaf blotch virus, a putative new member of the genus Emaravirus, encodes a novel genomic RNA. J. Gen. Virol. 93, 430–437. [DOI] [PubMed] [Google Scholar]
- Mielke‐Ehret, N. and Muehlbach, H.P. (2007) A novel, multipartite, negative‐strand RNA virus is associated with the ringspot disease of European mountain ash (Sorbus aucuparia L.). J. Gen. Virol. 88, 1337–1346. [DOI] [PubMed] [Google Scholar]
- Mielke‐Ehret, N. and Mühlbach, H.P. (2012) Emaravirus: a novel genus of multipartite, negative strand RNA plant viruses. Viruses, 4, 1515–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, M. (1931) Report of the Imperial Mycologist. Sci. Rep. Agric. Res. Inst. Pusa 19, 58–71. [Google Scholar]
- Mühlbach, H.P. and Mielke‐Ehret, N. (2011) Emaravirus In: Virus Taxonomy. Classification and Nomenclature of Viruses, Ninth Report of the International Committee on Taxonomy of Viruses (King A.M.Q., Adams M.J., Carstens E.B. and Lefkovitz E.J., eds), pp. 767–770. Oxford: Elsevier. [Google Scholar]
- Naik, S.S.J. , Byre Gowda, M. , Venkatesha, S.C. , Ramappa, H.K. , Pramila, C.K. , Mary Reena, G.A. and Ramesh, S. (2012) Molecular diversity among pigeonpea genotypes differing in response to PSMD. J. Food Legumes, 25, 194–199. [Google Scholar]
- Narayana, Y.D. , Mahalinga, D.M. , Jayalakshmi, S.K. and Benagi, V.I. (2000) Prevalence of SMD of pigeonpea in northern Karnataka. Karnataka J. Agric. Sci. 13, 470–472. [Google Scholar]
- Navajas, M. , Gutierrez, J. , Bonato, O. , Bolland, H.R. and Mapangou‐Divassa, S. (1994) Intraspecific diversity of the Cassava green mite Mononychellus progresivus (Acari: Tetranychidae) using comparisons of mitochondrial and nuclear ribosomal DNA sequences and cross‐breeding. Exp. Appl. Acarol. 18, 351360. [DOI] [PubMed] [Google Scholar]
- Navajas, M. , Gutierrez, J. , Williams, M. and Gotoh, T. (2001) Synonymy between two spider mite species, Tetranychus kanazawai and T. hydrangeae (Acari: Tetranychidae), shown by ribosomal ITS2 sequences and cross‐breeding experiments. Bull Entomol Res. 91, 117–123. [PubMed] [Google Scholar]
- Nene, Y.L. (1995) Sterility mosaic of pigeonpea: the challenge continues. Indian J. Mycol. Plant Pathol. 25, 1–11. [Google Scholar]
- Nene, Y.L. and Reddy, M.V. (1976a) Screening for resistance to sterility mosaic of pigeonpea. Plant Dis. Rep. 60, 1034–1036. [Google Scholar]
- Nene, Y.L. and Reddy, M.V. (1976b) A new technique to screen pigeonpea for resistance to sterility mosaic for pigeonpea. Trop. Grain Legume Bull. 5, 23–24. [Google Scholar]
- Nene, Y.L. , Sheila, V.K. and Sharma, S.B. (1996) A World List of Chickpea (Cicer arietinum) and Pigeonpea (Cajanus cajan). Pathogens, 5th edn ICRISAT, Patancheru, India: ICRISAT Publication. [Google Scholar]
- Newton, W. and Peiris, J.W.L. (1953) Virus diseases of plants in Ceylon. FAO Plant Protect. Bull. 2, 17–21. [Google Scholar]
- Oldfield, G. and Proeseler, G. (1996) Eriophiod mites as vectors of plant pathogens In: Eriophyoid Mites—Their Biology, Natural Enemies and Control (Lindquist E.E., Sabelis M.W. and Bruin J., eds), pp. 259–275. Amsterdam: Elsevier. [Google Scholar]
- Patil, B.L. , Ogwok, E. , Wagaba, H. , Mohammed, I.U. , Yadav, J.S. , Bagewadi, B. , Taylor, N.J. , Alicai, T. , Kreuze, J.F. , Gowda, M.N. and Fauquet, C.M. (2011) RNAi mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. Plant Pathol. 12, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.C. , Chen, T.C. , Raja, J.A. , Yang, C.F. , Chien, W.C. , Lin, C.H. , Liu, F.L. , Wu, H.W. and Yeh, S.D. (2014) Broad‐spectrum transgenic resistance against distinct tospovirus species at the genus level. PLoS ONE, 9, e96073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, M.V. and Nene, Y.L. (1980) Influence of sterility mosaic resistant pigeonpeas on multiplication of the mite vector. Indian Phytopathol. 33, 61–63. [Google Scholar]
- Reddy, M.V. , Raju, T.N. , Nene, Y.L. , Ghanekar, A.M. , Amin, K.S. , Arjunan, G. , Astaputre, J.V. , Sinha, B.K. , Reddy, S.V. , Gupta, R.P. and Gangadharan, K. (1993) Variability in sterility mosaic pathogen in pigeonpea in India. Indian Phytopathol. 46, 206–212. [Google Scholar]
- Reddy, M.V. , Sheila, V.K. , Murthy, A.K. and Padma, P. (1995) Mechanism of resistance to Aceria cajani in pigeonpea. Int. J. Trop. Plant Dis. 13, 51–57. [Google Scholar]
- Reddy, M.V. , Raju, T.N. and Lenne, J.M. (1998) Diseases of pigeonpea In: The Pathology of Food and Pasture Legumes (Allen D.J. and Lenne J.M., eds), pp. 517–558. Wallingford, Oxfordshire: CAB International. [Google Scholar]
- Sharma, M. , Rathore, A. , Mangala, U.N. , Ghosh, R. , Sharma, S. , Upadhyay, H. and Pande, S. (2012) New sources of resistance to Fusarium wilt and sterility mosaic disease in a mini‐core collection of pigeonpea germplasm. Eur. J. Plant Pathol. 133, 707–714. [Google Scholar]
- Singh, A.K. and Rathi, Y.P.S. (1997) Epidemiology of vector of pigeon pea sterility mosaic virus. Indian J. Virol. 13, 143–145. [Google Scholar]
- Singh, J. and Raghuraman, M. (2011) Emerging scenario of important mite pests in north India. Zoosymposia, 6, 170–179. [Google Scholar]
- Singh, N.K. , Gupta, D.K. , Jayaswal, P.K. , Mahato, A.K. , Dutta, S. , Singh, S. , Bhutani, S. , Dogra, V. , Singh, B.P. , Kumawat, G. , Pal, J.K. , Pandit, A. , Singh, A. , Rawal, H. , Kumar, A. , Rama Prashat, G. , Khare, A. , Yadav, R. , Raje, R.S. , Singh, M.N. , Datta, S. , Fakrudin, B. , Wanjari, K.B. , Kansal, R. , Dash, P.K. , Jain, P.K. , Bhattacharya, R. , Gaikwad, K. , Mohapatra, T. , Srinivasan, R. and Sharma, T.R. (2011) The first draft of the pigeonpea genome sequence. J. Plant Biochem. Biotech. 21, 98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas, T. , Reddy, M.V. , Jain, K.C. and Reddy, M.S.S. (1997) Studies on inheritance of resistance and allelic relationships for strain 2 of Pigeonpea sterility mosaic pathogen. Ann. Appl. Biol. 130, 105–110. [Google Scholar]
- Sudarshana, M.R. , Roy, G. and Falk, B.W. (2007) Methods for engineering resistance to plant viruses. Methods Mol. Biol. 354, 183–195. [DOI] [PubMed] [Google Scholar]
- Tatineni, S. , McMechan, A.J. , Wosula, E.N. , Wegulo, S.N. , Graybosch, R.A. , French, R. and Hein, G.L. (2014) An eriophyid mite‐transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J. Virol. 88, 11 834–11 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresh, J.M. (1964) Increased susceptibility to the mite vector (Phytoptus ribis Nal.) caused by infection with black‐currant reversion virus. Nat. Lond. 202, 1028. [DOI] [PubMed] [Google Scholar]
- Upadhyaya, H.D. , Kashiwagi, J. , Varshney, R.K. , Gaur, P.M. , Saxena, K.B. , Krishnamurthy, L. , Gowda, C.L.L. , Pundir, R.P.S. , Chaturvedi, S.K. , Basu, P.S. and Singh, I.P. (2012) Phenotyping chickpeas and pigeonpeas for adaptation to drought. Front Physiol. 3, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leeuwen, T. , Witters, J. , Nauen, R. , Duso, C. and Tirry, L. (2010) The control of eriophyoid mites: state of the art and future challenges. Exp. Appl. Acarol. 51, 205–224. [DOI] [PubMed] [Google Scholar]
- Varshney, R.K. , Chen, W. , Li, Y. , Bharti, A.K. , Saxena, R.K. , Schlueter, J.A. , Donoghue, M.T. , Azam, S. , Fan, G. , Whaley, A.M. , Farmer, A.D. , Sheridan, J. , Iwata, A. , Tuteja, R. , Penmetsa, R.V. , Wu, W. , Upadhyaya, H.D. , Yang, S.P. , Shah, T. , Saxena, K.B. , Michael, T. , McCombie, W.R. , Yang, B. , Zhang, G. , Yang, H. , Wang, J. , Spillane, C. , Cook, D.R. , May, G.D. , Xu, X. and Jackson, S. (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource‐poor farmers. Nat. Biotechnol. 30, 83–89. [DOI] [PubMed] [Google Scholar]
- Yu, C. , Karlin, D.G. , Lu, Y. , Wright, K. , Chen, J. and MacFarlane, S. (2013) Experimental and bioinformatic evidence that Raspberry leaf blotch emaravirus p4 is a movement protein of the 30K superfamily. J. Gen. Virol. 94, 2117–2128. [DOI] [PubMed] [Google Scholar]
- Zote, K.K. , Mali, V.R. , Mayee, C.D. , Kulkarni, S.V. and Mote, T.S. (1991) Outbreak of sterility mosaic of pigeonpea in Marathwada region Maharashtra, India. Int. Pigeonpea Newsl. 14, 19–21. [Google Scholar]


