Summary
The accurate quantification of disease severity is important for the assessment of host–pathogen interactions in laboratory or field settings. The interaction between Arabidopsis thaliana and its naturally occurring downy mildew pathogen, Hyaloperonospora arabidopsidis (Hpa), is a widely used reference pathosystem for plant–oomycete interactions. Current methods for the assessment of disease severity in the Arabidopsis–Hpa interaction rely on measurements at the terminal stage of pathogen development; namely, visual counts of spore‐producing structures or the quantification of spore production with a haemocytometer. These assays are useful, but do not offer sensitivity for the robust quantification of small changes in virulence or the accurate quantification of pathogen growth prior to the reproductive stage. Here, we describe a quantitative real‐time polymerase chain reaction (qPCR) assay for the monitoring of Hpa growth in planta. The protocol is rapid, inexpensive and can robustly distinguish small changes in virulence. We used this assay to investigate the dynamics of early Hpa mycelial growth and to demonstrate the proof of concept that this assay could be used in screens for novel oomycete growth inhibitors.
Keywords: Arabidopsis, Hyaloperonospora arabidopsidis, oomycete, real‐time PCR
Introduction
Oomycete pathogens cause disease on many important crops and have a high evolutionary potential to circumvent host resistance and chemical control (Kamoun et al., 2015). Hyaloperonospora arabidopsidis (Hpa) causes downy mildew disease of Arabidopsis thaliana, which is characterized by the formation of aerial sporangiophores (asexual fruiting bodies) (Holub, 2008; Koch and Slusarenko, 1990). Like other downy mildew pathogens, Hpa is an obligate biotroph that extracts nutrients from living Arabidopsis cells and cannot be cultured apart from its host. Hpa is one of a few eukaryotic microbes that are specifically adapted to Arabidopsis (Holub, 2008; Koch and Slusarenko, 1990). The Arabidopsis–Hpa interaction occurs frequently in nature, exhibiting a high level of polymorphism for resistance/susceptibility, lending itself as a useful pathosystem to exploit Arabidopsis for a better understanding of the molecular basis of plant–pathogen interactions and the evolution of obligate biotrophy (Baxter et al., 2010; McDowell, 2014).
Many Arabidopsis accessions have been documented as resistant or susceptible to isolates of Hpa collected from field populations of Arabidopsis (Crute et al., 1993; Dangl et al., 1992; Holub and Beynon, 1996). Resistance is conditioned by genes encoding nucleotide‐binding, leucine‐rich repeat (NB‐LRR) immune surveillance proteins. Moreover, many induced mutants with altered responses to Hpa have been described (Coates and Beynon, 2010; Slusarenko and Schlaich, 2003). For example, loss‐of‐function mutations in several immune signalling genes produce an ‘enhanced disease susceptibility’ (eds) phenotype, in which the growth of virulent isolates is enhanced compared with that in the wild‐type host (Parker et al., 1996).
Typically, Hpa growth is quantified by the counting of sporangiophores (e.g. McDowell et al., 2011; Tome et al., 2014) or by the quantification of asexual spore production (e.g. Feys et al., 2001). Both techniques offer limited precision when determining small changes in host susceptibility. Furthermore, pathogen growth cannot be monitored with these techniques at the early stages of the host–pathogen interaction.
Quantitative real‐time polymerase chain reaction (qPCR) has been successfully utilized for plant pathogen genotyping and diagnostics, and for the quantification of pathogen growth in planta. Previous publications have described PCR assays for oomycetes (Alonso et al., 2010; Kokko et al., 2006; Landa et al., 2011), including Hpa (Brouwer et al., 2003; Lapin et al., 2012). Here, we describe a protocol utilizing qPCR of a single‐copy Hpa gene as a proxy for pathogen biomass. The assay employs standard qPCR primers and rapid genomic DNA (gDNA) preparations, with low cost and high throughput. The procedure also includes a qPCR assay for a single‐copy Arabidopsis gene, enabling normalization to host biomass and accurate resolution of small differences in susceptibility (e.g. Alonso et al., 2010; Kokko et al., 2006). We use this assay to investigate the dynamics of Hpa mycelial growth in an eds mutant, and demonstrate a practical application of this method by monitoring Hpa growth in a defence‐compromised Arabidopsis plant treated with the systemic benzenoid fungicide metalaxyl (Kerkenaar and Sijpesteijn, 1981).
Results
qPCR requires robust primers that amplify the target sequence with high efficiency. Our target genes were actin in Hpa (Hpa gene_ID_807716) and in Arabidopsis (AtActin2, At1g49240) (Table S1, see Supporting Information). To determine the efficiency of these primer sets, a dilution series of gDNA from infected tissue was used as a template for qPCR (Fig. S1, see Supporting Information). The Hpa and Arabidopsis primer sets exhibited efficient amplification over a 200‐fold range of template input (5 ng to 1 μg) (Fig. S1a). A standard curve of C t values plotted against the logarithmic value of gDNA yielded slope values (M) of −2.322 and −2.895 for AtActin and HpaActin, respectively (Fig. S1b). Both primer sets yielded linear amplification over the range of template concentration (correlation coefficient R 2 > 0.99) with similar slopes, indicating matching efficiencies for both primer sets (Fig. S1b) (Livak and Schmittgen, 2001).
To confirm primer specificity, a dissociation curve analysis was performed on products amplified from infected and uninfected tissue. HpaActin primers did not amplify any products from uninfected Arabidopsis tissue, as expected. Conversely, HpaActin primers produced a single peak (T m = 82.93 °C) from Arabidopsis tissue infected with Hpa (Fig. S2, see Supporting Information). The AtActin primer set produced a single peak, distinct from the HpaActin peak (T m = 79.77 °C) from infected and uninfected templates. These results demonstrate that both primers sets are specific for their respective target genes.
Seedlings of Arabidopsis genotypes Col‐0, Oy‐1 and the mutant Ws eds1‐1 were inoculated with the Hpa isolate Emoy2. These genotypes are resistant, moderately susceptible and highly susceptible, respectively, to Hpa Emoy2 (Aarts et al., 1998; van der Biezen et al., 2002; Holub et al., 1994). We developed a protocol to homogenize tissue with a bead beater, to increase sample throughput and reduce variability in the efficiency of tissue disruption. We used DNA from this protocol and HpaActin and AtActin primer sets to monitor Hpa growth over the course of infection up to 6 days post‐inoculation, just prior to the onset of sporulation (Fig. 1). In addition, we stained plants from the same time course with trypan blue, which highlights pathogen hyphae and host cell death (McDowell et al., 2011). These samples were used to visually compare hyphal growth at each time point with the values from the qPCR assay.
Figure 1.
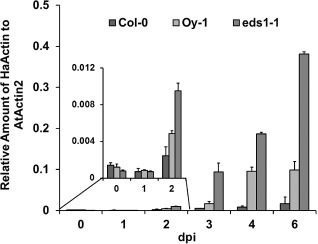
Time course of Hyaloperonospora arabidopsidis (Hpa) Emoy2 growth during infection and colonization of resistant (Col‐0), moderately susceptible (Oy‐1) and highly susceptible (Ws eds1‐1) Arabidopsis. Hpa growth was monitored over a 6‐day time course on three different genotypes using real‐time polymerase chain reaction (PCR). Growth is plotted as the relative quantity of HpaActin to AtActin [2‐ΔΔCT]. Black, Col‐0; pale grey, Oy‐1; dark grey, eds1‐1. Error bars represent standard deviation of three technical replicates. This experiment was replicated three times.
Col‐0 resistance to Hpa Emoy2 is caused by the RPP4 gene, which encodes a Toll/interleukin‐1 receptor, nucleotide‐binding, leucine‐rich repeat (TIR‐NB‐LRR) protein that recognizes the Atr4 effector from Emoy2 (van der Biezen et al., 2002). RPP4‐mediated hypersensitive response (HR) was macroscopically visible at 3 dpi in cotyledons stained with trypan blue (Fig. 2), became more pronounced by 4 dpi and was profuse at 7 dpi. Correspondingly, the accumulation of Hpa Emoy2, as measured by qPCR, in Col‐0 plateaued by 2–3 dpi (Fig. 1). qPCR indicated that Hpa Emoy2 growth was insignificant by 7 dpi.
Figure 2.
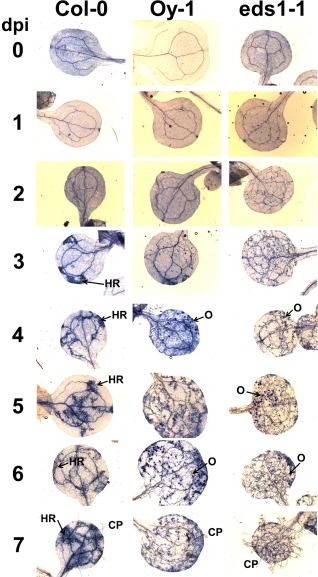
Hyaloperonospora arabidopsidis (Hpa) Emoy2 growth correlates with real‐time results. Hpa‐infected tissue was stained with trypan blue and visualized with a light microscope during a time course of 7 days. CP, conidiophore; dpi, days post‐inoculation; HR, hypersensitive response; O, oospore. This experiment was replicated three times.
Oy‐1 is a naturally occurring ecotype of Arabidopsis that is susceptible to Hpa Emoy2 (Holub et al., 1994). Growth was steady over the 7‐day period (Fig. 1) and little to no cell death was visible in samples stained with trypan blue. Oospore (sexual spores) production was apparent by 4 dpi. As expected, Hpa biomass was much higher in Oy‐1 than in the resistant ecotype Col‐0.
The third genotype tested was the immunocompromised mutant Ws eds1‐1. EDS1 is an important regulator of effector‐triggered immunity and of basal resistance that restricts the growth of virulent Hpa strains (Wiermer et al., 2005). Loss‐of‐function mutations in this gene cause a phenotype of enhanced susceptibility, compared with the wild‐type and susceptible Arabidopsis genotypes (Parker et al., 1996). We included Ws eds1‐1 to test whether the qPCR assay could accurately resolve small increases in Hpa virulence in heavily colonized tissue. The Ws eds1‐1 mutant supported higher levels of Hpa Emoy2, and the difference in growth compared with Oy‐1 was clear by 2 dpi. At 6 dpi, the overall levels of Hpa Emoy2 were approximately three‐fold higher in Ws eds1‐1 than in Oy‐1. Examination of leaves stained with trypan blue revealed abundant hyphal growth at 2 dpi that was clearly greater in Ws eds1‐1 than in Oy‐1 and Col‐0 (Figs 1 and 2). These results demonstrate that the qPCR assay resolved small changes in virulence at early time points. This is not possible with assays that are based on pathogen reproduction at the terminal stage of the interaction.
To further validate the qPCR assay, we compared the measurements acquired by qPCR with estimates of growth based on traditional sporangiophore counts. We counted sporangiophores per cotyledon at 7 dpi from the same experiments as used for the qPCR assays. The qPCR assay of Hpa growth at 6 dpi correlated with previous time points, with little biomass in Col‐0, moderate biomass in Oy‐1 and enhanced growth in Ws eds1‐1 (Fig. 3b). As expected, sporulation was low on resistant Col‐0, moderate on Oy‐0 and high on Ws eds1‐1 (Fig. 3a). The measurements derived from qPCR and sporangiophore counts correlated well (correlation coefficient of 0.99, Fig. 3c).
Figure 3.
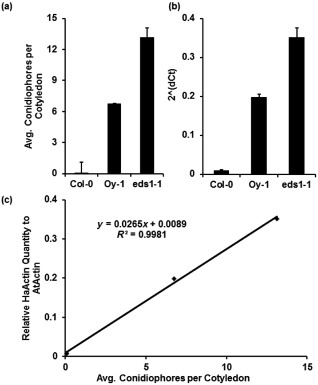
Hyaloperonospora arabidopsidis (Hpa) sporulation correlates with polymerase chain reaction (PCR)‐based quantification of hyphal growth in infected tissue. (a) Sporangiophores counted at 7 days post‐inoculation (dpi). (b) Relative abundance of HpaActin to AtActin of infected plants at 6 dpi. (c) Results of real‐time PCR quantification depicted in (b) plotted against the spore counts depicted in (a). R 2, correlation coefficient. Error bars represent standard error in (a) and standard deviation in (b). This experiment was replicated three times.
The identification of novel compounds that can inhibit pathogen growth is valuable for disease control. qPCR is potentially useful for the accurate quantification of the effects of growth inhibitors. As a proof of concept, we infiltrated 4‐week‐old Ws eds1‐1 plants with the systemic benzenoid fungicide metalaxyl and subsequently inoculated these plants with Hpa Emoy2. Leaf punches were collected at 6 dpi and gDNA was extracted and used as the template for qPCR. The assay demonstrated that metalaxyl inhibited Hpa growth on Ws eds1‐1 plants compared with control Ws eds1‐1 plants infiltrated with water (Fig. 4). This demonstrates that the qPCR assay is potentially useful for the identification of novel growth inhibitors.
Figure 4.

Polymerase chain reaction (PCR)‐based quantification of the extent to which metalaxyl inhibits Hyaloperonospora arabidopsidis (Hpa) growth. Arabidopsis Ws eds1‐1 plants were infiltrated with water or the fungicide metalaxyl and subsequently inoculated with Hpa Emoy2. Growth was quantified at 5 days post‐inoculation (dpi) with quantitative real‐time PCR. Error bars represent standard deviation. This experiment was replicated three times.
Discussion
The Hpa–Arabidopsis interaction is a valuable reference pathosystem, and its utility has been enhanced by recent sequencing of the Hpa genome, the identification of effector gene candidates and screens for effector targets (Baxter et al., 2010; Cabral et al., 2011; Caillaud et al., 2011; Fabro et al., 2011; Mukhtar et al., 2011; McDowell, 2014). These and other advances have created a critical need for new assays that accurately measure small differences in Hpa growth in planta. Quantification of sporangiophores can be used to accurately distinguish differences in pathogen growth when sporangiophore density is relatively low, such as when testing mutants that incrementally disable resistance to an incompatible isolate. However, sporangiophores are difficult to count accurately when they grow densely from the surface of heavily infected leaves. Alternatively, spores can be collected, counted with a haemocytometer and normalized to fresh tissue weight. In our hands, this method has potential for high variability introduced during spore collection, as Hpa conidia are easily dispersed under ambient conditions. Thus, this method is less robust for distinguishing small changes in virulence. Recent protocols to quantify pathogen growth with qPCR have shown sensitivity, precision and accuracy. qPCR procedures offer the added advantage of quantifying pathogen growth at early time points in the interaction.
For these reasons, we developed an inexpensive, high‐throughput procedure for qPCR quantification of Hpa growth. This procedure complements and extends a previously published qPCR method for Hpa quantification in several respects (Brouwer et al., 2003): First, the qPCR protocol includes an assay for a single‐copy Arabidopsis gene, which enables accurate normalization of pathogen biomass to plant biomass, and eliminates the need for normalization based on tissue weight or other measures. In addition, we used a simple method for gDNA extraction that employs common, low‐cost reagents. The gDNA extraction uses a leaf punch to collect tissue and a bead beater for tissue disruption, allowing 32 samples to be processed in parallel (96 tubes in the bead beater/three replicate tubes per sample). Thus, this procedure enables large numbers of samples to be processed simultaneously with minimal expense.
Our optimization experiments demonstrate that the AtActin and HpaActin primers are specific for their targets and provide linear amplification through a large concentration range of gDNA template. Importantly, the slopes generated by the standard curve of each primer set are similar, allowing a relative quantification of Hpa biomass (Livak and Schmittgen, 2001). The estimates of Hpa biomass from the qPCR method correlated well with hyphal growth observed with trypan blue and with sporangiophore production. These correlations held true in resistant, moderately susceptible and highly susceptible interactions. This indicates that amplification from the single‐copy HpaActin target serves as an unbiased and faithful proxy for Hpa growth in planta. The qPCR method is also sensitive, as demonstrated during the early time points of the Hpa–Arabidopsis interaction. RPP4 resistance to Hpa Emoy2 is manifested through HR and is visible with trypan blue staining by 3 dpi. However, differences in Hpa growth rate were clear as early as 2 dpi with qPCR, suggesting that RPP4 resistance is established earlier than can be visually detected.
Our time course experiment comparing Hpa growth in Arabidopsis lines with moderate and high susceptibility provides a previously unattainable quantification of Hpa growth from the beginning to the end of the infection cycle. The amount of growth in Ws eds1‐1 and Oy‐1 was similar during the first day after inoculation. During this interval, the Hpa spores germinate and hyphae emerge to penetrate between epidermal cells into the mesophyll layer. The first haustoria are formed during this time interval. At 2 dpi, hyphae have extended further into the mesophyll. This was the time point at which differences in Hpa biomass became apparent between Oy‐1 and Ws eds1‐1, suggesting that EDS1‐dependent basal defences are activated after the initial penetration steps. It should be noted that Ws eds1‐1 is in a different genetic background and is therefore not isogenic with Oy‐1. Thus, relative rates of Hpa Emoy2 growth in the two host backgrounds could be affected by other genetic differences in addition to the eds1‐1 mutation. However, this experiment still provides proof of concept that the qPCR assay can enable accurate measurements of small differences in growth in planta.
This assay allows the possibility to infiltrate candidate anti‐oomycete compounds and to observe the effect on Hpa growth. As a proof of concept, we infiltrated adult leaves of Arabidopsis with metalaxyl, a systemic benzenoid fungicide that inhibits nucleic acid synthesis. A large reduction in growth was observed, even in immunocompromised Ws eds1‐1, demonstrating that metalaxyl's inhibitory effects are caused entirely by its inherent toxicity rather than the activation of plant immunity. This general approach could be extended to recombinant proteins or extracts that cannot be applied with a foliar spray.
Experimental Procedures
Plant growth and maintenance of Hpa
Arabidopsis plants for pathogen assays were grown under 8 h of light at 22 °C, 16 h of dark at 20 °C in Sunshine Mix #1. The Hpa isolate Emoy2 was maintained on Oy‐1 and eds1‐1 Arabidopsis plants as described by McDowell et al. (2011). Conidial suspensions of 5 × 104 spores/mL were prepared from sporulating plants and applied with a Preval™ spray unit (Preval, Inc., Coal City, IL) to 10–12‐day‐old seedlings. Inoculated seedlings were covered for 24 h, and then uncovered and kept under short‐day conditions. At 6 dpi, infected seedlings were covered and the relative humidity was raised to 100% to trigger sporulation.
Sample collection and gDNA extraction
Five seedlings from each genotype were pooled in a 1.2‐mL library tube (VWR 83009‐678). This constituted one sample. Three samples were collected per treatment, constituting one biological replicate. Three 2‐mm glass balls were added to each library tube with 50 μL of extraction buffer [200 mm tris(hydroxymethyl)aminomethane (Tris), pH 7.5, 25 mm ethylenediaminetetraacetic acid (EDTA), pH 7.5, 250 mm NaCl, 0.5% sodium dodecylsulfate (SDS)]. Samples were placed into the corresponding library tube rack and homogenized for 2 min in a Mini Beadbeater 96+ (BioSpec, Bartlesville, OK, USA). Samples were subjected to a second round of homogenization if needed, based on visual inspection. Samples were then spun at 21,000 X g for 1 min. Library caps were removed and discarded. Extraction buffer (400 μL) was added to each sample, new library caps were added and the samples were briefly mixed. Samples were then spun at 21,000 X g for 3 min to pellet the cell debris. Supernatant (200 μL) was added to equal parts of 100% isopropanol in a 1.5‐mL plastic tube, mixed gently and incubated at room temperature for 2 min. Samples were spun at 21,000 X g for 5 min and the supernatant was discarded. Pellets were air dried for 5 min and resuspended overnight at 4 °C in 30 μL of sterile water. gDNA samples were quantified with a NanoDrop spectrophotometer (Thermo Scientific Waltham, MA USA), diluted to 10 ng/μL and stored at −20 °C.
qPCR
Samples (25 μL) were prepared by mixing 5 μL (50 ng) of gDNA with 12.5 μL of 2 × Sybr Green Mastermix (ABI, Carlsbad, CA, USA), with the appropriate primers added to a final concentration of 5 μm each and water. PCRs were performed in triplicate for each biological sample, using the default relative quantification program of the ABI 7300 device. Stage 1, 50.0 °C for 2 h, followed by 95.0 °C for 10 h in stage 2. Following the initial activation and denaturation steps, 40 cycles of stage 3 were performed at 95.0 °C for 15 min, followed by 53.0 °C for 1 h. C t values were determined using the included ABI software. Hpa primers were derived from Huibers et al. (2009). Relative abundance to AtActin was calculated as 2‐ΔΔCT (Livak and Schmittgen, 2001).
Trypan blue staining
Trypan blue staining was used to visualize hyphal growth and regions of cell death, as described by McDowell et al. (2011), and imaged with a Zeiss Axio Imager M1 (Carl Zeiss, Jena, Germany).
Metalaxyl treatment
Four‐week‐old eds1‐1 plants, grown under 8 h light/16 h dark, were infiltrated with water or metalaxyl (0.1 g/L) using needle‐less syringes, allowed to dry for 1 h and inoculated with Emoy2. Tissue was collected at 6 dpi using a hole punch. Two hole punches were collected per leaf from six leaves in total and pooled into three samples. gDNA was extracted for qPCR analysis as described above.
Supporting information
Fig. S1 Quantification of HpaActin and AtActin genes in genomic DNA extracted from Arabidopsis infected with Hyaloperonospora arabidopsidis (Hpa). (a) Quantitative real‐time PCR amplification profiles of normalized florescence (R n) plotted against cycle number with AtActin and HpaActin primers. Black, 500 ng; green, 250 ng; orange, 100 ng; red, 50 ng; purple, 25 ng; blue, 5 ng. (b) Standard curve assays to measure the efficiency of AtActin and HpaActin primers from dilution series of genomic DNA extracted from Arabidopsis infected with Hpa. M, slope; R 2, correlation coefficient. This experiment was replicated three times.
Fig. S2 Dissociation curve analysis of PCR amplicons generated by the AtActin and HpaActin primers demonstrate target specificity. Inverse fluorescence (derivative reporter) plotted against temperature. Light blue, uninfected Arabidopsis; dark blue, infected Arabidopsis; yellow, uninfected Arabidopsis; green, infected Arabidopsis. This experiment was replicated three times.
Table S1 Primers used in this study.
Acknowledgements
This research was supported by the National Science Foundation (IOS‐0744875) and the US Department of Agriculture–Agriculture and Food Research Initiative (2009‐03008 and 2011‐68004). We thank Janet Donahue and David Schmale for technical advice. The authors declare that they have no conflicts of interest.
References
- Aarts, N. , Metz, M. , Holub, E. , Staskawicz, B.J. , Daniels, M.J. and Parker, J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene‐mediated pathways in Arabidopsis . Proc. Natl. Acad. Sci. USA, 95, 10 306–10 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, G.D. , Llorente, B. , Bravo‐Almonacid, F. , Cvitanich, C. , Orlowska, E. , Torres, H.N. and Flawia, M.M. (2010) A quantitative real‐time PCR method for in planta monitoring of Phytophthora infestans growth. Lett. Appl. Microbiol. 51, 603–610. [DOI] [PubMed] [Google Scholar]
- Baxter, L. , Tripathy, S. , Ishaque, N. , Boot, N. , Cabral, A. , Kemen, E. , Thines, M. , Ah‐Fong, A. , Anderson, R. , Badejoko, W. , Bittner‐Eddy, P. , Boore, J.L. , Chibucos, M.C. , Coates, M. , Dehal, P. , Delehaunty, K. , Dong, S. , Downton, P. , Dumas, B. , Fabro, G. , Fronick, C. , Fuerstenberg, S.I. , Fulton, L. , Gaulin, E. , Govers, F. , Hughes, L. , Humphray, S. , Jiang, R.H. , Judelson, H. , Kamoun, S. , Kyung, K. , Meijer, H. , Minx, P. , Morris, P. , Nelson, J. , Phuntumart, V. , Qutob, D. , Rehmany, A. , Rougon‐Cardoso, A. , Ryden, P. , Torto‐Alalibo, T. , Studholme, D. , Wang, Y. , Win, J. , Wood, J. , Clifton, S.W. , Rogers, J. , Van den Ackerveken, G. , Jones, J.D. , McDowell, J.M. , Beynon, J. and Tyler, B. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science, 330, 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A. , Freddie, C.T. , Kahn, K. , Parker, J.E. and Jones, J.D. (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR‐NB‐LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Brouwer, M. , Lievens, B. , Van Hemelrijck, W. , Van den Ackerveken, G. , Cammue, B.P. and Thomma, B.P. (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real‐time fluorescence PCR. FEMS Microbiol. Lett. 228, 241–248. [DOI] [PubMed] [Google Scholar]
- Cabral, A. , Stassen, J.H. , Seidl, M.F. , Bautor, J. , Parker, J.E. and Van den Ackerveken, G. (2011) Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate‐specific effectors. Plos ONE, 6, e19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud, M.‐C. , Piquerez, S.J.M. , Fabro, G. , Steinbrenner, J. , Ishaque, N. , Beynon, J. and Jones, J.D.G. (2011) Subcellular localization of the Hpa RxLR effector repertoire identifies the extrahaustorial membrane‐localized HaRxL17 that confers enhanced plant susceptibility. Plant J. 69, 252–265. [DOI] [PubMed] [Google Scholar]
- Coates, M.E. and Beynon, J.L. (2010) Hyaloperonospora arabidopsidis as a pathogen model. Annu. Rev. Phytopathol. 48, 329–345. [DOI] [PubMed] [Google Scholar]
- Crute, I.R. , Holub, E.B. , Tor, M. , Brose, E. and Beynon, J.L. (1993) The identification and mapping of loci in Arabidopsis thaliana for recognition of the fungal pathogens Peronospora parasitica (downy mildew) and Albugo candida (white blister) In: Advances in Molecular Genetics of Plant–Microbe Interactions (Nester E.W. and Verma D.P.S., eds), pp. 437–444. Dordrecht: Kluwer Academic. [Google Scholar]
- Dangl, J.L. , Holub, E.B. , Debener, T. , Lehnackers, H. , Ritter, C. and Crute, I.R. (1992) Genetic definition of loci involved in Arabidopsis–pathogen interactions In: Methods in Arabidopsis Research (Koncz C., Chua N.‐H. and Schell J., eds), pp. 393–418. London: World Scientific Publishing Co. [Google Scholar]
- Fabro, G. , Steinbrenner, J. , Coates, M. , Ishaque, N. , Baxter, L. , Studholme, D.J. , Korrner, E. , Allen, R.L. , Piquerez, S.J.M. , Rougon‐Cardoso, A. , Greenshields, D. , Lei, R. , Badel, J.L. , Caillaud, M.‐C. , Sohn, K.‐H. , Van den Ackerveken, G. , Parker, J.E. , Beynon, J. and Jones, J. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog. 7, e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J. , Moisan, L.J. , Newman, M.A. and Parker, J.E. (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B. (2008) Natural history of Arabidopsis thaliana and oomycete symbioses. Eur. J. Plant Pathol. 122, 91–109. [Google Scholar]
- Holub, E.B. and Beynon, J.L. (1996) Symbiology of mouse ear cress (Arabidopsis thaliana) and oomycetes. Adv. Bot. Res. 24, 228–273. [Google Scholar]
- Holub, E.B. , Beynon, J.L. and Crute, I.R. (1994) Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana . Mol. Plant–Microbe Interact. 7, 223–239. [Google Scholar]
- Huibers, R.P. , de Jong, M. , Dekter, R.W. and Van den Ackerveken, G. (2009) Disease‐specific expression of host genes during downy mildew infection of Arabidopsis. Mol. Plant–Microbe Interact. 22, 1104–1115. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Furzer, O. , Jones, J.D.G. , Judelson, H.S. , Ali, G.S. , Dalio, R.J.D. , Roy, S.G. , Schena, L. , Zambounis, A. , Panabières, F. , Cahill, D. , Ruocco, M. , Figueiredo, A. , Chen, X.‐R. , Hulvey, J. , Stam, R. , Lamour, K. , Gijzen, M. , Tyler, B.M. , Grünwald, N.J. , Mukhtar, M.S. , Tomé, D.F.A. , Tör, M. , Van den Ackerveken, G. , McDowell, J. , Daayf, F. , Fry, W.E. , Lindqvist‐Kreuze, H. , Meijer, H.J.G. , Petre, B. , Ristaino, J. , Yoshida, K. , Birch, P.R.J. and Govers, F. (2015) The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkenaar, A. and Sijpesteijn, A.K. (1981) Antifungal activity of metalaxyl and furalaxyl. Pestic. Biochem. Physiol. 15, 71–78. [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokko, H. , Hukkanen, A. , Pietikainen, L. and Karenlampi, S. (2006) Quantification of downy mildew (Peronospora sparsa) in Rubus species using real‐time PCR. Eur. J. Plant Pathol. 116, 225–235. [Google Scholar]
- Landa, B.B. , Montes‐Borrego, M. , Munoz‐Ledesma, F.J. and Jimenez‐Diaz, R.M. (2011) Real‐time PCR quantification of Peronospora arborescens, the opium poppy downy mildew pathogen, in seed stocks and symptomless infected plants. Plant Dis. 95, 143–152. [DOI] [PubMed] [Google Scholar]
- Lapin, D. , Meyer, R. , Takhashi, H. , Bechtold, U. and Van den Ackerveken, G. (2012) Broad‐spectrum resistance of Arabidopsis C24 to downy mildew is mediated by different combinations of isolate‐specific loci. New Phytol. 196, 1171–1181. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M. (2014) Hyaloperonospora arabidopsidis: a model pathogen of Arabidopsis In: Genomics of Plant Associated Fungi and Oomcyetes: Dicot Pathogens (Dean R.A., Kole C. and Lichens‐Park A., eds), pp. 209–234. New York: Springer. [Google Scholar]
- McDowell, J.M. , Hoff, T. , Anderson, R.G. and Deegan, D. (2011) Propagation, storage, and assays with Hyaloperonospora arabidopsidis: a model oomycete pathogen of Arabidopsis. Methods Mol. Biol. 712, 137–151. [DOI] [PubMed] [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S. , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. , Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E. , Holub, E.B. , Frost, L.N. , Falk, A. , Gunn, N.D. and Daniels, M.J. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell, 8, 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarenko, A. and Schlaich, N. (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Pathol. 4, 159–170. [DOI] [PubMed] [Google Scholar]
- Tome, D.F. , Steinbrenner, J. and Beynon, J.L. (2014) A growth quantification assay for Hyaloperonospora arabidopsidis isolates in Arabidopsis thaliana . Methods Mol. Biol. 1127, 145–158. [DOI] [PubMed] [Google Scholar]
- Wiermer, M. , Feys, B.J. and Parker, J.E. (2005) Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Quantification of HpaActin and AtActin genes in genomic DNA extracted from Arabidopsis infected with Hyaloperonospora arabidopsidis (Hpa). (a) Quantitative real‐time PCR amplification profiles of normalized florescence (R n) plotted against cycle number with AtActin and HpaActin primers. Black, 500 ng; green, 250 ng; orange, 100 ng; red, 50 ng; purple, 25 ng; blue, 5 ng. (b) Standard curve assays to measure the efficiency of AtActin and HpaActin primers from dilution series of genomic DNA extracted from Arabidopsis infected with Hpa. M, slope; R 2, correlation coefficient. This experiment was replicated three times.
Fig. S2 Dissociation curve analysis of PCR amplicons generated by the AtActin and HpaActin primers demonstrate target specificity. Inverse fluorescence (derivative reporter) plotted against temperature. Light blue, uninfected Arabidopsis; dark blue, infected Arabidopsis; yellow, uninfected Arabidopsis; green, infected Arabidopsis. This experiment was replicated three times.
Table S1 Primers used in this study.


