Summary
Oomycetes form a deep lineage of eukaryotic organisms that includes a large number of plant pathogens which threaten natural and managed ecosystems. We undertook a survey to query the community for their ranking of plant‐pathogenic oomycete species based on scientific and economic importance. In total, we received 263 votes from 62 scientists in 15 countries for a total of 33 species. The Top 10 species and their ranking are: (1) Phytophthora infestans; (2, tied) Hyaloperonospora arabidopsidis; (2, tied) Phytophthora ramorum; (4) Phytophthora sojae; (5) Phytophthora capsici; (6) Plasmopara viticola; (7) Phytophthora cinnamomi; (8, tied) Phytophthora parasitica; (8, tied) Pythium ultimum; and (10) Albugo candida. This article provides an introduction to these 10 taxa and a snapshot of current research. We hope that the list will serve as a benchmark for future trends in oomycete research.
Keywords: oomycetes plant pathology, microbiology, diversity, genomics
Introduction
Oomycetes are eukaryotic organisms that superficially resemble filamentous fungi, but are phylogenetically related to diatoms and brown algae in the stramenopiles (Gunderson et al., 1987; Jiang and Tyler, 2012; Lamour and Kamoun, 2009; Thines, 2014; Thines and Kamoun, 2010). Fossil evidence indicates that a number of oomycetes emerged as endophytes of land plants at least by the Carboniferous period, approximately 300–350 million years ago (Krings et al., 2011). One species, Combresomyces williamsonii, described from ∼320‐million‐year‐old petrified stem cortex and rootlets of a seed fern, may have even been parasitic (Strullu‐Derrien et al., 2011). Phylogenetic analyses of modern taxa have revealed that plant parasitism has evolved independently in three lineages of oomycetes (Thines and Kamoun, 2010). Well‐known plant pathogens, namely downy mildews, Phytophthora and Pythium, appear to have radiated from a common plant‐parasitic ancestor (Thines and Kamoun, 2010). The impact of oomycetes on humankind is well documented as both a persistent threat to subsistence and commercial farming and as destructive pathogens of native plants (Agrios, 2005; Erwin and Ribeiro, 1996; Lamour and Kamoun, 2009). As a result, news related to plant diseases caused by oomycetes tends to capture the interest of the general public and is frequently featured in the media.
In the last two decades, increased awareness of the distinctive phylogeny and biology of oomycetes has driven the emergence of a specialist research community that is currently organized under the umbrella of the ‘Oomycete Molecular Genetics Network’. This community has moved the field beyond the gloomy view of the 1980s that oomycetes are a ‘fungal geneticist's nightmare’ (Shaw, 1983; discussed in Schornack et al., 2009). It has produced novel paradigms in understanding host–microbe interactions, effector biology and genome evolution (Bozkurt et al., 2012; Govers and Gijzen, 2006; Jiang and Tyler, 2012; Schornack et al., 2009; Vleeshouwers et al., 2011). The oomycete community was one of the first in plant pathology to initiate coordinated transcriptome and genome sequencing projects, and subsequently to exploit the resulting resources to drive conceptual advances (Govers and Gijzen, 2006; Pais et al., 2013; Schornack et al., 2009). These days, with the genomes serving as unique resources for basic and applied research, oomycetes are best portrayed as a ‘genomicist's dream’ (Govers and Gijzen, 2006; Jiang and Tyler, 2012; Pais et al., 2013; Schornack et al., 2009).
It is therefore particularly fitting that the oomycetes are at last covered by the Top 10 review series of Molecular Plant Pathology. The process to generate the list and the aim of this article are similar to those of previous contributions on plant‐pathogenic viruses (Scholthof et al., 2011), fungi (Dean et al., 2012), bacteria (Mansfield et al., 2012) and nematodes (Jones et al., 2013). We undertook a survey to query the community for their ranking of plant‐pathogenic oomycete taxa based on scientific and economic importance. In total, we received 263 votes from 62 scientists in 15 countries that yielded the Top 10 species (Table 1). To some degree, the results reflect the sizes of the subcommunities. Six of the ten species belong to the genus Phytophthora, which is more commonly studied than any other oomycete genus. Obligate parasites are also well represented with three species (Hyaloperonospora arabidopsidis, Plasmopara viticola and Albugo candida, Table 1). Another 23 species received votes but ranked outside the Top 10 (Table 2). The fish parasite Saprolegnia parasitica received enough votes to place it in the Top 10, but was removed, given that the list focuses on plant pathogens (Table 2).
Table 1.
Top 10 oomycetes in molecular plant pathology
| Rank | Species | Common disease name(s) | Number of papers (2005–2014) | Number of votes |
|---|---|---|---|---|
| 1 | Phytophthora infestans | Late blight | 1230 | 51 |
| = 2 | Hyaloperonospora arabidopsidis | Downy mildew | 137 | 25 |
| = 2 | Phytophthora ramorum | Sudden oak death; Ramorum disease | 378 | 25 |
| 4 | Phytophthora sojae | Stem and root rot | 276 | 22 |
| 5 | Phytophthora capsici | Blight; stem and fruit rot; various others | 541 | 17 |
| 6 | Plasmopara viticola | Downy mildew | 326 | 15 |
| 7 | Phytophthora cinnamomi | Root rot; dieback | 315 | 13 |
| = 8 | Phytophthora parasitica | Root and stem rot; various others | 142 | 10 |
| = 8 | Pythium ultimum | Damping off; root rot | 319 | 10 |
| 10 | Albugo candida | White rust | 65 | 9 |
The ‘=’ sign before the ranking indicates that the species tied for that position. The number of papers published in 2005–2014 is based on searches of the Scopus database (http://www.scopus.com) using the species names as a query. For H. arabidopsidis, a search for the alternative name ‘Peronospora parasitica’ was also performed and the combined number is shown. Searches with the terms ‘oomycete*’ and ‘Phytophthora’ yielded 2068 and 4059 articles, respectively.
Table 2.
Other oomycete species that received votes
| Rank | Species |
|---|---|
| 11 | Aphanomyces euteiches |
| 12 | Albugo laibachii |
| 13 | Bremia lactucae |
| 14 | Phytophthora palmivora |
| 15 | Pseudoperonospora cubensis |
| 16 | Plasmopara halstedii |
| 17 | Peronophythora litchi |
| 18 | Peronosclerospora sorghi |
| 19 | Peronospora belbahrii |
| 20 | Phytophthora alni |
| 21 | Phytophthora brassicae |
| 22 | Phytophthora cactorum |
| 23 | Phytophthora meadii |
| 24 | Phytophthora phaseoli |
| 25 | Phytophthora plurivora (formerly P. citricola) |
| 26 | Plasmopara obducens |
| 27 | Pythium aphanidermatum |
| 28 | Pythium oligandrum |
| 29 | Sclerophthora rayssiae |
| 30 | Hyaloperonospora brassicae |
| NR | Saprolegnia parasitica (fish parasite) |
| NR | Lagenidium giganteum (mosquito parasite) |
| NR | Pythium insidiosum (mammalian parasite) |
NR, not ranked because the species is not associated with plants.
This article is based on contributions from the voters to provide an introduction to the 10 taxa and a snapshot of current research. Each section starts with a brief overview of the selected species, e.g. its importance, pathology, host range and life cycle. This is followed by a review of current research themes, particularly the unique findings that have emerged from the study of particular species, and an outlook on future research. We hope that this article will serve as a reference and resource for novice and experienced readers alike, as well as provide a benchmark for future trends in oomycete research.
1. Phytophthora infestans
Phytophthora infestans causes potato late blight (Fig. 1), a disease with major historical impact. Providing twice the calories of rye and wheat per hectare, potato was central to European agriculture between 1750 and 1850. In 1844, the arrival of P. infestans changed this situation. In addition to the well‐documented Irish famine (Fig. 2), crop failures in 1845 and 1846 contributed to an estimated 750 000 hunger‐associated deaths in continental Europe (Zadoks, 2008). The discovery that late blight was caused by a microbial pathogen, 15 years before Pasteur's formal confirmation of Germ Theory, places the 19th century migration of P. infestans as a significant milestone in the foundation of plant pathology as a scientific discipline. Today, late blight remains a major constraint to the production of potato, the world's third largest staple crop, and is thus a constant threat to food security (Fisher et al., 2012; Haverkort et al., 2008).
Figure 1.

Potato plants with typical late blight lesions. Infection starts when a spore lands on the leaf and germinates. The germ tube forms an appressorium and an emerging penetration peg pushes into an epidermal cell. Then the inner cell layers are colonized. During the biotrophic phase, hyphae grow in the intercellular space, whereas haustoria enter plant cell cavities and invaginate host cell plasma membrane. Later, Phytophthora infestans switches to necrotrophic growth, resulting in the death of plant cells and the appearance of necrotic lesions on the infected tissues. In this phase, hyphae escape through the stomata and produce numerous asexual spores, named sporangia, that easily detach and disperse by wind or water. A sporangium that finds a new host can either germinate directly and initiate a new cycle or, at lower temperatures, undergo cleavage resulting in a zoosporangium from which six to eight flagellated spores are released. These zoospores can swim for several hours but, once they touch a solid surface, they encyst and germinate to initiate new infections. Under favourable conditions, the pathogen can complete the cycle from infection to sporulation in 4 days. In the field, this cycle is repeated multiple times during one growing season, resulting in billions of spores and a continuous increase in disease pressure. In addition to leaves, stems and tubers are also infected and P. infestans can continue to flourish on the decaying plant material. If not managed properly, infected seed potatoes or waste on refuse piles are often the sources of inoculum for new infections in the spring. An alternative route for surviving the winter is via oospores, sexual spores that can survive in the soil for many years. Phytophthora infestans is heterothallic; isolates are either A1 or A2 mating type, and sex organs only develop when isolates of opposite mating type sense the sex hormone produced by the mate.
Figure 2.

One of the many Great Famine memorials around the world. These sculptures on Customs House Quays in Dublin, by artist Rowan Gillespie, stand as if walking towards the emigration ships on the Dublin Quayside (courtesy of Michael Seidl).
In the 1840s, trade in potatoes probably facilitated the long‐distance migration of the pathogen. Whole‐genome sequencing of European herbarium samples has revealed that populations belonging to the HERB‐1 genotype were different from US‐1, the genotype dominating the population globally in the mid‐ to late 20th century (Goodwin et al., 1994; Martin et al., 2013, 2014; Yoshida et al., 2013, 2014). Waves of migration and genotype displacements in the 20th century have been well documented. The introduction of the A2 mating type to Europe in the 1970s (Drenth et al., 1994) and, more recently, the emergence of new, more aggressive lineages, such as 13_A2 (Blue 13) in Europe (Cooke et al., 2012), US‐22 in the eastern USA (Fry et al., 2013; Hu et al., 2012) and US‐23/US‐24 in Canada (Peters et al., 2014), impose constant challenges to disease resistance breeding.
In the 1990s, P. infestans research led to the development of molecular approaches to study oomycete pathology and biology. The first DNA transformation system (Judelson et al., 1991) and the first transcriptional profiling during infection (Pieterse et al., 1991) paved the way to detailed molecular investigations of sexual and asexual development and pathogenicity (reviewed in Judelson, 1997). DNA fingerprinting revolutionized population studies (Goodwin et al., 1992) and was a prelude to the first genome‐wide genetic map of a Phytophthora species (van der Lee et al., 1997). Observations that gene silencing occurred in P. infestans transformants (e.g. Judelson and Whittaker, 1995) sparked important studies of the mechanisms underlying transcriptional silencing (van West et al., 1999). The first large‐scale transcript sequencing studies (Kamoun et al., 1999) ushered in the genomics era of Phytophthora research.
In the last 15 years, advanced genomic and functional approaches have placed P. infestans at the forefront of research to understand oomycete development (Judelson and Blanco, 2005) and pathogenicity (Kamoun, 2006). An elegant combination of bioinformatics, proteomics and functional genomics has revealed secreted proteins as potential factors influencing host–P. infestans interactions (Torto et al., 2003). Apoplastic effectors that inhibit host secreted proteases were first described in P. infestans (Tian et al., 2004), and the apoplastic phytotoxin‐like SCR74 (Liu et al., 2005) provided support for the use of diversifying selection as a criterion for the identification of candidate effectors. The apoplastic effectors EPIC1 and EPIC2B were found to converge on the host protease RCR3, which is also targeted by an independently evolved effector from a fungal plant pathogen (Song et al., 2009). Recently, diversifying selection of the EPIC1 protease inhibitor was implicated in functional adaptation to the equivalent protease target after ‘jumps’ to a new host (Dong et al., 2014).
The first reported P. infestans avirulence effector was AVR3a, the counterpart of resistance protein R3a (Armstrong et al., 2005). Comparison with avirulence effectors from Phytophthora sojae and H. arabidopsidis identified conserved amino acid motifs RXLR and EER (Rehmany et al., 2005), which act as signals for translocation into host cells (Whisson et al., 2007). The genome sequence of P. infestans revealed hundreds of genes predicted to encode RXLR effectors, plus a second class of candidate effectors called Crinklers (CRNs) (Haas et al., 2009), which may also be delivered inside plant cells (Schornack et al., 2010). These effector genes are rapidly evolving and are typically variable between different P. infestans isolates (Cooke et al., 2012). RXLR and CRN reside in gene‐sparse, repeat‐rich regions, potentially subject to rearrangement and rapid mutation, raising the concept of a ‘two‐speed’ genome in evolutionary terms (Haas et al., 2009; Raffaele et al., 2010). Recent years have witnessed intense efforts to determine the biochemical function of RXLR effectors, notably by determining their protein targets in the host. The first target identified was ubiquitin E3 ligase CMPG1, which AVR3a stabilizes to prevent programmed cell death in response to elicitors such as INF1 (Bos et al., 2010).
Breeding for late blight resistance was initiated by James Torbitt of Belfast in the 1870s, inspired by the theories of Charles Darwin, with whom he shared considerable correspondence (DeArce, 2008). More than 130 years later, breeding efforts have yet to provide a durable solution. Occasionally, local successes are reported; for example, variety C88, widely cultivated in China for over a decade, retains its resistance (Li et al., 2011). Many resistance (R) genes effective against races of P. infestans have been identified (Rodewald and Trognitz, 2013). Only with the recent identification of avirulence genes, all of which encode RXLR effectors, can breeders incorporate knowledge on how P. infestans evades detection into their breeding strategies (Vleeshouwers et al., 2011). Such information is critical for the prediction of the durability of R genes. The use of effectors as tools to rapidly identify R genes is a powerful step towards rational selection and combination of lasting resistance (Vleeshouwers et al., 2008).
We still know relatively little about how P. infestans effectors manipulate host plants to establish disease. How do they translocate into host cells? Do they define host range? How do they work in concert to modulate complex networks of regulatory processes occurring in the host? Only recently have researchers started to adopt structural biology to fully investigate functional relationships between interacting pathogen and plant proteins (Wirthmueller et al., 2013). Could such research catalyse the modification of molecular interactions in favour of the plant immune system through synthetic biology approaches?
The basic knowledge gained on P. infestans in the last decade is starting to have an impact on the management of late blight disease. For the control of late blight, farmers largely rely on agrochemicals, many with unknown modes of action. The emergence of insensitive strains is possibly a result of high mutation rates in the pathogen (Randall et al., 2014). The phenylamide metalaxyl was one of the first chemicals to exhibit specificity to oomycetes. Soon after its introduction and widespread use, fully insensitive strains emerged, but only 35 years later was the molecular basis of this insensitivity uncovered (Randall et al., 2014). Mining of the P. infestans genome has revealed many novel proteins with domain combinations that are unique to oomycetes (Seidl et al., 2011). Several probably function in signalling and may be promising fungicide targets. Mode‐of‐action studies are more accessible thanks to the available genomics resources, transformation tools and marker strains with tags to visualize the cytoskeleton and various subcellular compartments (Ah Fong and Judelson, 2003; Meijer et al., 2014). In resistance breeding, the efficacy of stacking R genes into favoured cultivars, by either introgression or trans‐ or cis‐genesis, is being investigated (Tan et al., 2010). However, for success, the R genes to be combined should be selected carefully on the basis of our knowledge on the variation in the pathogen population, and the attendant vulnerability to resistance being overcome (Vleeshouwers et al., 2011). Future efforts should consider the introduction of multiple barriers, perhaps combining R proteins with, for instance, membrane‐localized pattern recognition receptors or components governing non‐host resistance to P. infestans. Even then, it is unlikely that plant resistance can fully control late blight. Agrochemicals should not be abandoned. On the contrary, for more rational fungicide design, novel targets should be identified, an endeavour that will be enhanced by an even more profound insight into the biology of P. infestans.
2. Hyaloperonospora arabidopsidis
Hyaloperonospora arabidopsidis (formerly H. parasitica and Peronospora parasitica) is one of 700 downy mildew species within the Peronosporaceae (Thines, 2014). Downy mildew pathogens cause harmful diseases on many important crops, notably Peronospora and Hyaloperonospora spp. on brassica crops, Plasmopara viticola on grape, Peronosclerospora spp. on maize and sorghum, Pseudoperonospora cubensis on cucurbits and Bremia lactucae on lettuce (Lucas et al., 1995).
Most downy mildews have narrow host ranges and are completely dependent on their host for growth and reproduction. They can survive in the soil as quiescent oospores that initiate infection through roots. Spread of the pathogen mostly occurs through airborne sporangiospores that are formed on the lower leaf surface, giving downy patches (Fig. 3). These spores germinate on the plant surface and penetrate by forming appressoria (Koch and Slusarenko, 1990). Once past the epidermis, hyphae grow intercellularly and, similar to Phytophthora spp. and other downy mildews, develop haustoria, specialized structures that may function in feeding and the suppression of host defence by targeted secretion of effectors (Fig. 4, Whisson et al., 2007).
Figure 3.

Hyaloperonospora arabidopsidis disease symptoms on a 2‐week‐old Arabidopsis seedling. Mature sporangiophores are visible as white structures on the right side of the leaf.
Figure 4.
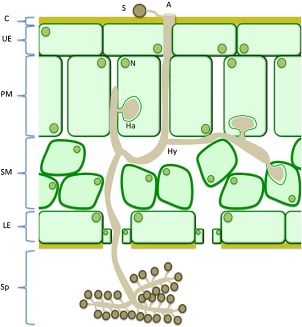
Diagram depicting a compatible interaction between Hyaloperonospora arabidopsidis and Arabidopsis initiated by a sporangiospore landing on the leaf surface. A, appressorium; C, cuticle; Ha, haustorium; Hy, hyphae; LE, lower epidermis; N, nucleus; PM, palisade mesophyll cells; S, sporangiospore; SM, spongy mesophyll cells; Sp, mature sporangiophore; UE, upper epidermis. Note: sporangiophores are not drawn to scale.
Hyaloperonospora arabidopsidis is a prominent pathogen in natural populations of Arabidopsis thaliana (Coates and Beynon, 2010; Holub, 2008). As such, it was adopted in the 1980s as one of two pathogens of Arabidopsis, together with the bacterium Pseudomonas syringae (Koch and Slusarenko, 1990). The Top 10 ranking of H. arabidopsidis reflects the subsequent success of the Arabidopsis–H. arabidopsidis pathosystem. H. arabidopsidis was initially utilized as a ‘physiological probe’ of the Arabidopsis immune system (Holub et al., 1994). This research led to the cloning of the first plant disease R genes against an oomycete, better understanding of the evolutionary dynamics of R genes, the definition of broadly important immune system regulators, the identification of downy mildew‐resistant mutants and genetic definition of the complexity of the plant immune signalling network (reviewed in Coates and Beynon, 2010; Lapin and Van den Ackerveken, 2013; Slusarenko and Schlaich, 2003). On the pathogen side, research is hampered by the lack of protocols for culture and genetic transformation, established techniques with other oomycetes such as P. infestans. However, work in the early 2000s led to the development of genetic maps and DNA libraries that enabled the discovery of the first avirulence effector (Allen et al., 2004), and later to the RXLR effector family (Rehmany et al., 2005).
Genome sequencing of H. arabidopsidis isolate Emoy2, completed in 2010, unveiled 134 predicted RXLR effectors and other components of the H. arabidopsidis secretome (Baxter et al., 2010). Notably, this report also revealed important genomic signatures of obligate biotrophy that have evolved convergently in other obligate oomycete and fungal lineages (reviewed in McDowell, 2011). Protein interaction assays have shown that H. arabidopsidis effectors target a highly interconnected host machinery, helping to define a representative plant–pathogen interaction network (Mukhtar et al., 2011). In addition, several high‐throughput functional studies have investigated effector subcellular localizations, suppression of immune responses, molecular targets and cognate immune receptors (Cabral et al., 2011, 2012; Caillaud et al., 2011; Fabro et al., 2011).
Future studies with the H. arabidopsidis experimental system will include: (i) direct or Agrobacterium‐mediated transformation for genetic manipulation required for the molecular analysis of downy mildew pathogenicity; (ii) the establishment of the temporal hierarchy of effectors during penetration, colonization and sporulation, which may serve as a blueprint for a better understanding of the molecular basis of biotrophy; (iii) the role of genetic recombination and epigenetics on the emergence of new effectors; (iv) the development of tools to understand how plant‐originated molecules regulate pathogen response; and (v) the relevance of interspecies transfer of small RNAs. These investigations on H. arabidopsidis will continue to provide new insights into the molecular mechanisms of downy mildew pathogenicity, and contribute to comparative and functional analysis of (obligate) biotrophic oomycete and fungal pathogens.
3. Phytophthora ramorum
Phytophthora ramorum is the most destructive disease of oaks worldwide and the cause of sudden oak death, sudden larch death and ramorum blight (Brasier and Webber, 2010; Grünwald et al., 2008; Rizzo et al., 2005; Werres et al., 2001) (Fig. 5). The host range of P. ramorum is one of the widest of any Phytophthora spp., and includes many species of hardwood trees and ornamentals. To date, four distinct clonal lineages have been recognized by various molecular markers and two mating types have been found (Grünwald et al., 2009; Van Poucke et al., 2012). These lineages appear to be anciently diverged, yet have emerged repeatedly in both North America and Europe (Goss et al., 2009a; Grünwald et al., 2012) (Fig. 6). Lineages NA1, NA2 and EU1 are currently found in Canada and the USA, whereas EU1 and EU2 exist in Europe (Goss et al., 2009b, 2011; Grünwald et al., 2012; Van Poucke et al., 2012). These lineages differ in aggressiveness in controlled assays, but field experiments have not been conducted given quarantine restrictions (Elliott et al., 2011; Grünwald et al., 2008; Hüberli et al., 2012; Kasuga et al., 2012; Van Poucke et al., 2012). Unique ecological attributes, such as a wide host range and survival in dry, hot Mediterranean summers, combined with the ability to reproduce from chlamydospores, are thought to provide the basis for sudden oak death in California (Garbelotto and Hayden, 2012).
Figure 5.

Impact of sudden oak death in California. Tanoak mortality evidenced by defoliated or wilted canopies on the Bolinas Ridge at Mt. Tamalpais, Marin County, CA, USA. Photograph courtesy of Janet Klein (Marin Municipal Open Space District).
Figure 6.
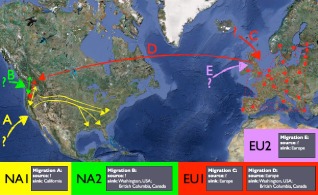
Inferred pattern of migration of the four clonal lineages of Phytophthora ramorum. Modified from Grünwald et al. (2012).
The availability of the genome sequence just a few years after the identification of the pathogen provided rapid and novel insights into the biology of this pathogen (Tyler et al., 2006). Like other Phytophthora spp., P. ramorum has a large number of candidate effectors interacting with the plant hosts, including RXLR, CRNs and the necrosis and ethylene‐inducing peptide 1 (Nep1)‐like protein (NLP) gene families (Goss et al., 2013; Tyler et al., 2006). RXLR effectors diversify rapidly, despite the clonality of this organism, using mechanisms such as loss or gain of repeated domains, recombination or gene conversion among paralogues and selection on point mutations (Goss et al., 2013). Recent work has focused on the discovery of endogenous small RNAs and description of the silencing machinery (Fahlgren et al., 2013). Phytophthora ramorum and two other Phytophthora spp. examined produce two primary, distinct 21‐ and 25‐nucleotide small RNA classes, including a novel microRNA family. Two argonaute classes and two dicer‐like proteins appear to be involved in each pathway (Fig. 7), but this remains to be formally tested. Epigenetic mechanisms have been implicated recently in phenotypic diversification, namely colony morphology, colony senescence and virulence on coast live oak and California bay laurel (Kasuga et al., 2012).
Figure 7.
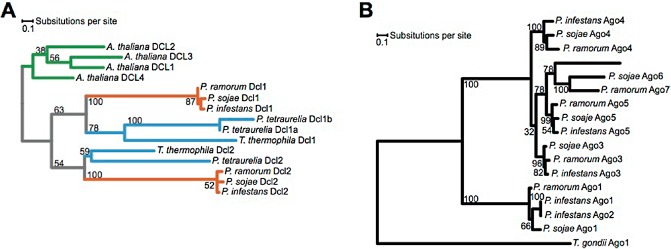
Silencing machinery in Phytophthora. Phylogenetic placement of dicer‐like (DCL) (A) and argonaute (Ago) (B) proteins in the genus Phytophthora. For more details, see Fahlgren et al. (2013). Species correspond to: Arabidopsis thaliana, Paramecium tetraurelia, Phytophthora infestans, Phytophthora ramorum, Phytophthora sojae, Tetrahymena thermophila and Toxoplasma gondii.
Phytophthora ramorum provides a unique opportunity for the study of the evolution of a genome that has two mating types, yet appears to lack sexual reproduction in the known field populations. The epigenome, transcriptome and proteome of P. ramorum remain a mystery. The apparent documented phenotypic variation driven by host association might be epigenetic in nature and invites further study. Several basic questions remain unanswered. Why does P. ramorum have such a large number of RXLR effectors, given that the genome is adapted to a wide host range? What are the genes conferring a wide host range compared with P. infestans and P. sojae? Is there a centre of origin? This centre might be located in Asia where the closest relative, P. lateralis, has been found in an old growth Chamaecyparis forest (Brasier et al., 2010). The discovery of a centre of origin might also reveal a host with which P. ramorum might have co‐evolved, providing a mechanism of R gene discovery, a critical tool needed for the management of sudden oak death. Comparative genomics of related species, such as P. lateralis, P. syringae and P. hibernalis, amongst others, will provide novel insights into core effectors and genes under purifying selection in the clade, but diverged among clades.
4. Phytophthora sojae
Root rot disease of soybean caused by P. sojae was first recognized in North America in the 1950s (Hildebrand, 1959; Kaufmann and Gerdemann, 1958). The alternative names P. megasperma var. sojae and P. megasperma f.sp. glycinea were commonly used for this species in the past (Erwin and Ribeiro, 1996). Infection of soybean by P. sojae, which is a hemibiotroph, typically initiates below ground and eventually produces spreading cankers that destroy root tissues and travel up the stem (Fig. 8). The pathogen thrives in wet conditions and in compacted or heavy clay soils. Motile, water‐borne zoospores are released from sporangia and are attracted to soybean root exudates (Morris and Ward, 1992). Phytophthora sojae is homothallic and creates abundant, thick‐walled sexual oospores that are long lived and provide a soil‐borne inoculum. Phytophthora sojae has a narrow host range and its economic damage is limited to soybean. It has been suggested that P. sojae originated in North America as a pathogen of lupins, as some 26 species of the genus Lupinus are susceptible to infection (Erwin and Ribeiro, 1996). In laboratory tests, P. sojae can also be made to infect lima bean (Phaseolus lunatus), string bean (Phaseolus vulgaris) and cranesbill (Geranium carolinianum) (Erwin and Ribeiro, 1996; Hildebrand, 1959). Phytophthora sojae is one of the most damaging disease problems confronting soybean growers (Wrather and Koenning, 2006). The management of P. sojae in soybean has primarily relied on breeding for resistance.
Figure 8.
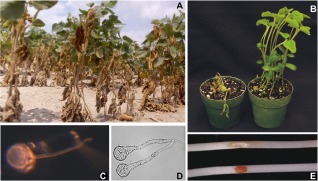
Phytophthora sojae. (A) Diseased soybean plants in the field, infected with P. sojae. Plant height is 20–30 cm. (B) Susceptible (left) and resistant (right) soybean plants inoculated in the stem with P. sojae, 7 days after infection, illustrating R‐gene‐mediated resistance. Pots are 10 cm in diameter. (C) Germinating oospore of P. sojae growing on water agar. Oospore is 35 μm in diameter. (D) Germinating P. sojae cysts growing on water agar. Cysts are 15 μm in diameter. (E) Etiolated soybean hypocotyls inoculated with a 10‐μL water droplet containing 103 zoospores from a virulent (top) and avirulent (bottom) strain of P. sojae, 48 h after infection, illustrating strain‐specific variation in avirulence effectors and the hypersensitive response. Soybean hypocotyls are 5 mm in diameter.
Research on P. sojae has focused on the mechanisms of pathogen virulence, host resistance and the molecular basis of recognition between P. sojae and soybean (Dorrance and Grünwald, 2009; Gijzen and Qutob, 2009; Tyler, 2007). Phytophthora sojae, together with P. ramorum, was the first oomycete to have its genome sequenced (Tyler et al., 2006). Genomic information, including gene expression data and multiple genome sequences, have driven rapid progress (Qutob et al., 2000; Torto‐Alalibo et al., 2007; Wang et al., 2011). The genome of P. sojae, and later other oomycetes (Haas et al., 2009), was found to be partitioned into stable regions rich in housekeeping genes displaying extensive synteny with other oomycete genomes, interspersed with dynamic, transposon‐rich regions that contained rapidly evolving genes implicated in virulence (Jiang & Tyler, 2012; Tyler et al., 2006) (Fig. 9). Multiple large, rapidly diversifying families of genes with functions in virulence were discovered (Jiang and Tyler, 2012; Tyler et al., 2006), including hydrolases, hydrolase inhibitor proteins, toxin‐like proteins, such as the NLP family (Qutob et al., 2006), and two huge, diverse classes of effector proteins (RXLR and CRN effectors) (Haas et al., 2009; Jiang et al., 2008; Tyler et al., 2006) that can cross into the cytoplasm of host plant cells (Dou et al., 2008; Kale et al., 2010) (Fig. 9). All 11 avirulence genes cloned from P. sojae proved to encode RXLR effectors detected by host R proteins (Jiang and Tyler, 2012) (Fig. 9). Genetic inheritance studies in P. sojae using molecular markers led to discoveries of high‐frequency gene conversion (also referred to as loss of heterozygosity) (Chamnanpunt et al., 2001) and epigenetic silencing (Qutob et al., 2013) as important mechanisms underlying pathogen variation.
Figure 9.
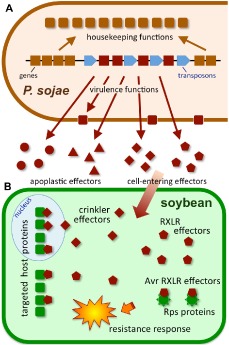
Large diverse families of virulence proteins encoded by the Phytophthora sojae genome. (A) The Phytophthora sojae genome contains clusters of conserved housekeeping genes (brown) that have conserved orders among Phytophthora species, separated by dynamic transposon‐rich regions that contain genes (red) encoding virulence proteins, many of which are secreted. (B) Secreted virulence proteins (effectors) may act in the apoplast, or be transported inside the cell. Cell‐entering effectors may have targets in the nucleus or cytoplasm, and may be detected by resistance proteins (Rps; resistance against P. sojae).
Many areas of future research into P. sojae will be shared with those of other oomycete and fungal pathogens. One area will be to define in detail how apoplastic and host cell‐entering effectors of P. sojae manipulate soybean physiology to enable infection, including the soybean molecules targeted by the effectors (Fig. 9) and the mechanisms by which RXLR and CRN effectors gain entry (Fig. 9). The identification of the P. sojae effectors most important for infection (e.g. using new tools for genetic manipulation, such as tailored nucleases) should lead to new targets for disease control. The cell biology of P. sojae infection (Enkerli et al., 1997) must be defined in much more detail, incorporating the spatial distributions of pathogen and host transcripts, proteins and metabolites. Understanding fully the role of epigenetic mechanisms in the generation of variation in pathogen populations will also be important for the design of effective strategies for disease control. Sequencing the genomes and transcriptomes of numerous isolates of P. sojae and its nearest sister species will shed light on the genetic adaptability of the pathogen. In practical terms, a large translational research effort will be required to convert the rapidly accumulating knowledge about the basic biology and pathology of P. sojae and other oomycetes into effective disease control solutions.
5. Phytophthora capsici
Phytophthora capsici is a highly destructive invasive pathogen that attacks solanaceous (pepper, tomato), legume (lima and snap beans) and most cucurbit hosts (Hausbeck and Lamour, 2004; Leonian, 1922). Disease is favoured by warm (25–28 °C) and wet conditions, and the asexual epidemiology is often explosive (Granke et al., 2009). Initial infection is biotrophic, followed by transitions within 24–48 h to necrotrophy and the production of deciduous sporangia on the surface of infected tissues (Fig. 10). The importance of the sexual stage differs by region. In Peru and Argentina and across much of China, long‐lived and widely dispersed clonal lineages dominate (Gobena et al., 2012; Hu et al., 2013; Hurtado‐Gonzales et al., 2008; Sun et al., 2008). This is in contrast with the USA, South Africa and the northern provinces of China, where populations have high levels of genotypic diversity and outcrossing is frequent (Dunn et al., 2010; Gobena et al., 2012; Meitz et al., 2010). Once introduced to a field site, P. capsici is difficult to control and often impossible to eradicate. Phytophthora capsici grows rapidly and sporulates heavily on simple media, and isolates from sexual field populations are often fecund (Gobena et al., 2012; Lamour and Hausbeck, 2000). Phytophthora capsici is one of the most genetically diverse eukaryotic organisms yet described, and there is significant genetic variation in the form of single nucleotide polymorphisms within individual genomes and across world populations (Lamour et al., 2012).
Figure 10.
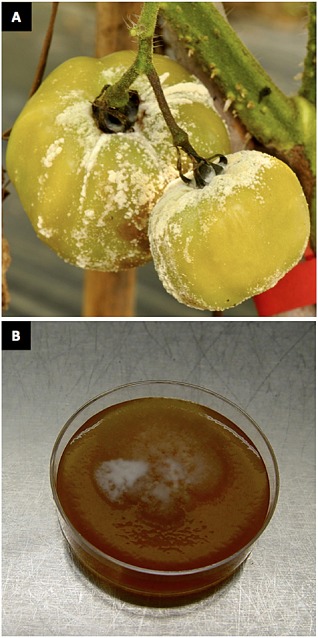
Heavy sporulation and spontaneous morphological variation in the vegetable pathogen Phytophthora capsici. (A) Naturally infected tomato fruit with sporangium production on the surface of the fruit. (B) A single zoospore‐derived field isolate of P. capsici sectoring on V8 agar medium following long‐term storage.
Current research includes the investigation of resistance (host and nonhost) in commercial vegetable and experimental plants, the discovery and characterization of genes and proteins driving pathogenicity and virulence, and studies to measure the evolution of natural and laboratory populations. Under controlled conditions, P. capsici infects at least 26 plant families (Granke et al., 2012). Natural resistance to P. capsici in pepper and cucurbits appears to be rare (Mallard et al., 2013) but, in tomato, it may be extensive (Quesada‐Ocampo and Hausbeck, 2010). Recent molecular studies of the CRN class of effectors have indicated that they are often localized to the host nucleus and play an important role in the infection process (Chen et al., 2013; Stam et al., 2013a, 2013b). Transcriptome studies using RNAseq and microarrays have indicated dramatic changes from pre‐infective spores through the early biotrophic and later necrotrophic stages of infection (Chen et al., 2013; Jupe et al., 2013). Studies of individual effectors have suggested that some manipulate the host to allow infection, whereas others trigger plant cell death (Chen et al., 2013; Feng and Li, 2013; Feng et al., 2010; Sun et al., 2009).
A high‐quality reference genome and a high‐density single nucleotide polymorphism‐based genetic linkage map have been completed recently (Lamour et al., 2012). These important resources illuminated an important source of asexual genetic variation, known as loss of heterozygosity (Lamour et al., 2012). Loss of heterozygosity occurs when variable length tracts (300 bp to >1 Mbp) of the diploid genome switch to one of the two possible haplotypes. Loss of heterozygosity has been described in laboratory‐produced sexual progeny, field isolates maintained on agar medium and field isolates genotyped directly from naturally infected tissue (Gobena et al., 2010; Hu et al., 2013; Hulvey et al., 2010). Loss of heterozygosity is associated with spontaneous switches from the A2 to the A1 mating type, loss of pathogenicity and reduced virulence (Hu et al., 2013; Lamour et al., 2012).
The diversity and plasticity of P. capsici present challenges and unique opportunities for research (Fig. 10). Future research questions include the following. What is the significance of a highly diverse effector arsenal? Are these effectors important for host‐specific interactions and the broad host range? How important is loss of heterozygosity in laboratory and field populations? The genomic plasticity of P. capsici and the ease of laboratory manipulation provide a unique opportunity to measure genome stability and adaptive evolution at a fine scale, and may provide useful insights into this and other oomycete pathogens (Lamour and Hu, 2013).
6. Plasmopara viticola
The obligate biotrophic oomycete Pl. viticola, the causal agent of grape downy mildew, is native to North America and was inadvertently introduced into Europe at the end of the 19th century (Millardet, 1881; Viennot‐Bourgin, 1949), causing severe damage to Vitis vinifera, which had evolved in the absence of this pathogen (Galet, 1977; Gessler et al., 2011). Plasmopara viticola belongs to the family Peronosporaceae; its life cycle includes an asexual multiplication phase occurring during the plant vegetative period and a sexual phase that ensures pathogen overwinter survival (Wong et al., 2001). The pathogen overwinters as oospores in dead leaf lesions or as mycelium in infected twigs. In spring, and in particular during rainy periods, oospores germinate to produce sporangia, which are transported by wind or water to the wet leaves near the ground, which they infect through stomata on the lower surface. The mycelium then spreads into the intercellular spaces of the leaf from which sporangiophores arise and emerge through the stoma, ready to start new infection cycles (Gobbin et al., 2005). Downy mildew affects leaves, fruits and shoots, with young tissues being particularly susceptible to infection (Fig. 11) (Kennelly et al., 2007). A disease cycle may take from 5 to 18 days, depending on the temperature, humidity and varietal susceptibility (Agrios, 2005; Gessler et al., 2011).
Figure 11.

Downy mildew symptoms with well‐evident sporangiophores on the lower side of a grape leaf (A) and a young cluster (B).
Downy mildew is still most destructive in Europe and in the eastern half of the USA, where it may cause severe epidemics year after year (Agrios, 2005; Madden et al., 1995). When the weather is favourable and no protection against the disease is provided, downy mildew can easily destroy up to 75% of production in a single season (Madden et al., 2000; Rossi and Caffi, 2012). Occasionally, Pl. viticola can be destructive in other humid parts of the world in which V. vinifera is cultivated.
In the last 50 years, research on grapevine downy mildew has focused on the pathogen life cycle and epidemiology (reviewed in Gessler et al., 2011), and on the genetic identification of host resistance loci for the establishment of molecular markers (revised in Töpfer et al., 2011). In the last decade, studies on Pl. viticola have highlighted its enormous potential in the development of fungicide resistance (Blum et al., 2010; Chen et al., 2007) and in breaking down plant resistance of interspecific hybrids, such as ‘Bianca’ (Casagrande et al., 2011; Peressotti et al., 2010) and ‘Regent’ (Delmotte et al., 2013). Population genetic studies have been carried out (Fontaine et al., 2013 and references therein) to assess pathogen dynamics and diversity in Europe. Moreover, it has been proposed that grapevine downy mildew is not caused by a single species, but instead by a complex of cryptic species that have diverged on Vitaceae (Rouxel et al., 2013). Trade‐offs between the size and number of sporangia produced have been reported, leading to ecological advantages for this pathogen (Delmotte et al., 2013). For these reasons, the Vitis–Pl. viticola pathosystem was elected as a prime candidate for the study of host specialization of biotrophic plant pathogens (Rouxel et al., 2013) and pathogen adaptation to partial host resistance (Delmotte et al., 2013).
Control methods against Pl. viticola are essentially based on the use of chemical fungicides and the application of disease models that serve as decision support systems (Lalancette et al., 1987; Rossi et al., 2009, 2013; Vercesi et al., 2010). As a result of the growing public concern on the use of chemical pesticides, and the need to adhere to the European Directive 2009/128/EC encompassing a framework ‘to achieve a sustainable use of pesticides’ by promoting Integrated Pest Management (IPM), the search for alternative methods for the control of Pl. viticola on grapevine appears to be urgent. Several alternative approaches have been proposed in the last 20 years, but none has been transferred to practical use (Gessler et al., 2011). Moreover, the lack of precise characterization of Pl. viticola isolates is dramatically limiting the reliable control of the pathogen. Although molecular studies have previously confirmed a high diversity in the pathogen population, there is a surprising lack of phenotypic characterization of pathotype strains or races which could be used to study the mechanisms of interaction with host genotypes with different levels of resistance (Gómez‐Zeledón et al., 2013). In conclusion, interdisciplinary studies and consistent resources should be invested for the study of the Pl. viticola–V. vinifera pathosystem in order to find new, effective alternatives for the IPM of this highly adaptive and destructive pathogen.
7. Phytophthora cinnamomi
Some call it the ‘biological bulldozer’ for its capacity to destroy natural plant communities across the globe, and disease caused by P. cinnamomi has broad and economically important impacts in forestry and horticulture, and in the nursery industry. Like other Phytophthora spp., P. cinnamomi has a number of strategies for survival, propagation and dissemination. The motile zoospore is recognized as the main infective propagule following encystment and attachment to roots and stems (Fig. 12). Recent work has suggested that, in addition to chlamydospores, P. cinnamomi is also associated with lignituber formation which enables survival under harsh conditions (Jung et al., 2013). Both A1 and A2 mating types are pathogenic, but wide differences in the distribution and frequency of occurrence suggest that the A2 mating type is more invasive and is generally recognized as being the more aggressive of the two mating types. Where introduced, this pathogen has had enormous impacts on natural systems, including those of Australia, southern Europe and the USA. In Australia, for example, it has been estimated that >3000 largely endemic plant species from numerous plant families are under threat, and a new National Threat Abatement Plan has been urgently implemented (Australian Government Department of Environment, 2014) (Fig. 13). The control of disease, especially across the large areas of native vegetation affected, is still a great problem, but some success has been achieved by the use of phosphite in both natural systems and in agriculture (Akinsanmi and Drenth, 2013; Crane and Shearer, 2014), by the use of calcium amendments to soil (Serrano et al., 2013) and by containment and eradication of spot infections (Dunstan et al., 2010). Climate change is predicted to have a significant influence on the intensity and distribution of disease (Thompson et al., 2014).
Figure 12.
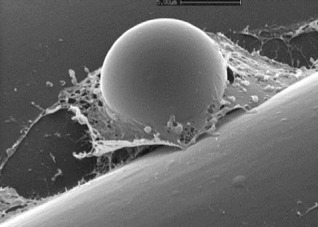
Cryo‐scanning electron micrograph of a Phytophthora cinnamomi cyst between two epidermal cells on a root of tobacco. Note the adhesive material that surrounds the cyst that has been expelled by zoospore peripheral vesicles. Photograph courtesy of Adrienne Hardham, Australian National University.
Figure 13.

(A) Individual plants of Xanthorrheoa australis (austral grass tree), a highly suscpetible native Australian species, infected by Phytophthora cinnamomi within a dry sclerophyll eucalypt forest at Anglesea, Victoria, Australia. Note the dead and dying plants that have brown, collapsed leaves compared with the healthy green and erect leaves of plants which are yet to be killed. These individual plants range in age from approximately 20 years (the smallest in the centre) to around 70 years (green individual on the right of the image). (B) Advancing disease front caused by invasion by P. cinnamomi in Xanthorrhoea australis‐dominated understorey in eucalyptus open forest at Wilsons Promontory, Victoria, Australia. The disease has moved from the foregound of the picture, where all susceptible vegetation including X. australis has been killed, and its progress can be seen as a line of dead and dying X. australis (brown collapsed plants) at the disease margin. Healthy green plants behind them will soon be killed. Loss of the major understorey components, as in the forground, results in complete structural change and loss of all susceptible species.
Phytophthora cinnamomi is a challenging organism to work with. Sequencing of its genome has recently been completed (JGI; http://genome.jgi‐psf.org/) and has opened up a variety of opportunities to investigate the critical features that enable this pathogen to be so wide‐ranging in its hosts. A real change in emphasis in research on P. cinnamomi has occurred in recent times, with an increasing number of groups using genomic, transcriptomic and proteomic approaches to understand the pathogen and/or its interaction with the host (for example, Reeksting et al., 2014). Experimental plants, including Arabidopsis, Zea mays and Eucalyptus, for which comprehensive genomic information is available, are increasingly being used as host–pathogen platforms (for example, Allardyce et al., 2013; Dempsey et al., 2012; Rookes et al., 2008). Fundamental studies on P. cinnamomi plant cell wall‐degrading enzymes and the genes that encode them are being undertaken (Hee et al., 2013; Adrienne Hardham, Australian National University, Canberra, ACT, Australia, personal communication), as well as examination, at the molecular level, of defence pathways and their control (Eshraghi et al., 2014; Gunning et al., 2013).
High‐throughput, genome sequencing is emerging as an exciting way to examine the pathogen, its hosts and the soil environment, and is now being used to examine pathogen ecology and diversity within soils, and for the analysis of the response of host plants (Treena Burgess, Murdoch University, Perth, WA, Australia, personal communication). Much of our progress in understanding host–pathogen interactions will come from such approaches. We still lack knowledge on why plants are resistant to this pathogen and what is the basis for induced resistance, for example, following phosphite treatment. The search for host resistance to P. cinnamomi is an active area in which signalling pathways and their control are being elucidated. Mapping the disease over the large invaded areas is still not straightforward, although advances in remote sensing, high‐resolution digital photography, hyperspectral imaging (David Guest, Sydney University, Sydney, NSW, Australia, personal communication) and ‘drone’ technology will probably see our ability in this area being greatly enhanced in the near future. The era of nanotechnology offers new opportunities for the delivery of molecules that can influence disease outcome, yet this area has not been explored to date, although preliminary research with various nanoparticle systems (Hussain et al., 2013; Nadiminti et al., 2013) has indicated that these may be useful in delivering molecules, including pathogen effectors, DNA and miRNA, which may modify host processes and their response to pathogen infection.
Future research will focus on the following questions. (i) What are the virulence factors that enable P. cinnamomi to infect and colonize susceptible species and therefore should be targeted for control? (ii) What constitutes host resistance and how can it be manipulated? Can we look to nanotechnology for some answers? (iii) Is phosphite the only chemical that we can use to control P. cinnamomi, and how does phosphite alter the host response?
8. Phytophthora parasitica
Phytophthora parasitica (= P. nicotianae) is a worldwide distributed pathogen (Erwin and Ribeiro, 1996). Primarily known to cause tobacco black shank and citrus root rot and gummosis (Fig. 14), it is also responsible for severe foliar and fruit diseases, as well as root and crown rots on herbaceous and perennial plant species in more than 250 genera, including solanaceous crops (Fig. 15), and horticultural and fruit trees (Cline et al., 2008). Phytophthora parasitica produces both asexual zoospores and thick‐walled sexual oospores. Oospores constitute a potential source of genetic variation and, with resting chlamydospores, contribute to survival in unfavourable conditions in soil or within infected plant tissues.
Figure 14.
Impact of Phytophthora parasitica infection on citrus plants. (A, B) Five‐year‐old citrus plants not infected and infected, respectively, with Phytophthora parasitica. (C, D) Symptoms of P. parasitica on stems: (C) not infected; (D) infected plant displaying gummosis symptoms. (E, F) Leaves and fruits of infected (left) and healthy (right) plants. (G, H) Scanning electron microscope images of citrus fine roots infected with P. parasitica. Yellow arrows show encysted zoospores and germ tube of P. parasitica. Bar represents 20 μm. (A–F) Photographs courtesy of R. J. D. Dalio. (G, H) Photographs courtesy of M. E. Escanferla.
Figure 15.

Severe infection of brinjal fruit with Phytophthora parasitica. Typical symptoms are brown, soft, water‐soaked patches which rapidly cover the whole fruit. Brinjal is also known as aubergine or eggplant (Solanum melongena). Photograph courtesy of S. Guha Roy.
Contrasting with the broad host range observed at the species level, most individual P. parasitica isolates display host preference (Erwin and Ribeiro, 1996). Frequent cases of differential virulence on a range of hosts have been revealed (Colas et al., 1998; Matheron and Matejka, 1990), pointing out the need to decipher the genetic structure of P. parasitica on a global scale. In agreement with pathogenicity tests, recent single nucleotide polymorphism analyses conducted with mitochondrial and nuclear genes have revealed a specific association between the host of origin and genetic grouping which was particularly evident for tobacco and citrus isolates (Mammella et al., 2011, 2013). In contrast, no clear genetic structure was revealed for isolates from other hosts, especially potted ornamentals in nurseries. A significant geographical structuring was revealed for tobacco, but not for citrus, isolates (Bonnet et al., 1994; Colas et al., 2001; Mammella et al., 2013). Further studies relying on whole‐genome sequencing programmes (see below) are necessary to determine whether these molecular groups represent evidence of physiological races, pathotypes or even subspecies within P. parasitica.
As some isolates may also infect many hosts, including Arabidopsis thaliana (Attard et al., 2008, 2010), P. parasitica has emerged as an ideal pathogen to develop specific studies with the aim to advance our knowledge on the mechanisms underlying general pathogenicity and those governing host specificity. Furthermore, P. parasitica is phylogenetically related to P. infestans in Phytophthora clade 1, and overlaps its host range, which includes potato (Taylor et al., 2008). Comparative analyses on these two species varying in genome size (83 and 240 Mb for P. parasitica and P. infestans, respectively) will facilitate the understanding of the evolution behind pathogenicity and host range among Phytophthora spp. Through the sequencing of the genome of isolates of diverse host range and geographical origins, the international ‘Phytophthora parasitica genome initiative’ project will enable the characterization of genes that determine host range. In‐depth genomic and transcriptomic analyses of 14 sequenced genomes are currently being performed. Special efforts are being devoted to characterize the repertoire of effector proteins. As a broad host range pathogen, P. parasitica provides a unique opportunity for intra‐ and interspecific comparative analyses, looking for the presence of various effector families, their organization, their role in plant recognition and infection, and their evolution among strains and species that display broad or restricted host ranges. The identification of conserved effector groups, as well as of other pathogenicity genes, will allow the evaluation of the evolutionary pressures of exposure to different host defence responses to the diversification of effectors and their role in adaptation to host plants.
Phytophthora parasitica has not been as extensively studied as its economic importance should warrant. However, it is likely to gain importance in the foreseeable future for the following reasons. First, it is prominent in nurseries of potted ornamentals and fruit tree species, the trade of which seems to represent one of the most efficient dissemination pathways of Phytophthora (Moralejo et al., 2009; Olson and Benson, 2011). This makes P. parasitica an ideal species to study diffusion pathways of Phytophthora and other soil‐borne pathogens on a global scale. For aesthetic reasons, the ornamental industry requires the extensive use of anti‐oomycete chemicals (Olson et al., 2013). The global nursery trade thus increases the risk of the rapid spread of resistant P. parasitica strains to new areas following the inappropriate use of fungicides, and constitutes a growing threat to local agriculture and natural ecosystems (Brasier, 2008). In addition, P. parasitica will probably benefit from the warming climate. Its host range generally includes those of other species of prime economic importance (P. infestans, P. capsici, P. citrophthora), but generally requires warmer conditions than these potential competitors (Erwin and Ribeiro, 1996). Consequently, the foreseen global warming is likely to provide a catalyst for the geographical expansion of this species, as has already been proposed in Mediterranean climates (Andres et al., 2003; Saadoun and Allagui, 2008) and in eastern India (Guha Roy et al., 2009) .
9. Pythium ultimum
Pythium resides in the peronosporalean lineage of oomycetes, with Phytophthora and downy mildews (Dick, 2001). One of its most significant members is Py. ultimum, which causes damping off and root rot on >300 diverse hosts, including corn, soybean, wheat, Douglas fir and ornamentals (Farr and Rossman, 2014). Pythium ultimum is a common inhabitant of fields, ponds and streams, and of decomposing vegetation worldwide. Contributing to the ubiquity of the species is its ability to grow saprotrophically in soil and plant residue, a trait shared by most Pythium spp., but not other peronosporaleans, which must colonize living hosts. Pythium ultimum is also homothallic, producing oospores capable of long‐term survival (Martin and Loper, 1999). Mycelia and oospores in soil can thus initiate infections of seeds or roots, leading to wilting, reduced yield and mortality (Fig. 16A). Disease management is difficult, but focuses on sanitation, fungicides and biological control, particularly in glasshouses. Quantitative resistance has been reported in several hosts, but has a limited effect (Lucas and Griffiths, 2004; Wang and Davis, 1997).
Figure 16.

Pythium ultimum var. ultimum. (A) Pre‐emergence damping‐off in okra, resulting in the death of most plants at the front of the row. At the back, disease was controlled using metalaxyl. (B) Terminal hyphal swellings; bar, 10 μm. (C) Oospore within oogonium; bar, 10 μm. Images adapted from Kida et al. (2006) with permission.
The role of asexual spores depends on the strain. Pythium ultimum is a species complex that includes Py. ultimum var. ultimum and var. sporangiiferum, which have indistinguishable internal transcribed spacer (ITS) sequences (Schroeder et al., 2013). Sporangia and zoospores are produced rarely by the former, but abundantly by the latter. Pythium ultimum var. ultimum forms sporangia‐like hyphal swellings which germinate to yield infective hyphae (Fig. 16B; Stanghellini and Hancock, 1971). Similar to other oomycetes, it also produces sexual oospores (Fig. 16C).
Genome sequencing of several Pythium spp., including Py. ultimum var. ultimum and var. sporangiiferum, has provided insight into their biology (Adhikari et al., 2013; Levesque et al., 2010). Annotated in the c. 43‐Mb genomes of two subspecies are 15 290 and 14 086 genes, respectively, which is less than the smallest Phytophthora, but more than downy mildews. Whether this difference in gene number is significant requires more analysis, as the two assemblies vary in quality and only single isolates were sequenced. Large size variation between isolates was reported on the basis of electrophoretic karyotyping (Martin, 1995).
Nevertheless, it is evident from genome analysis that there are many differences between Pythium and Phytophthora, including a 25% reduction in Pythium of the fraction of proteins that are secreted. In part, this reflects the absence of RXLR effectors, which may not be needed for necrotrophic lifestyles. Pythium also lacks haustoria, which appear to be used by biotrophic and hemibiotrophic oomycetes to deliver RXLRs to host cells (Whisson et al., 2007). Another likely sign of the necrotrophy of Pythium is its greater abundance of lipases and proteases. However, Pythium lacks cutinases and pectin esterases, but this is probably because it infects primarily nonsuberized tissue, whereas Phytophthora (which encodes these enzymes) can penetrate plant cuticles. Pythium encodes sufficient enzymes for the maceration of host cell walls, but cannot degrade some ubiquitous wall components, such as xylans, which suggests that soluble sugars may be a more important carbon source (Zerillo et al., 2013).
Markers derived from Py. ultimum genomes should help reveal what distinguishes the subspecies and provide more resolution to population studies, which can help to identify the sources of outbreaks. Studies of Py. ultimum var. ultimum have indicated that populations are not clonal and some outcrossing may occur (Francis and St. Clair, 1997). A greater evolutionary question concerns the organization of the Pythium genus as a whole. Division of the >120 Pythium species over five new genera has been proposed on the basis of the sequence analysis of two loci (Uzuhashi et al., 2010). If accepted by the community, Py. ultimum would be renamed Globisporangium ultimum.
Of much interest is the interaction between Py. ultimum and biocontrol agents, such as Trichoderma, Streptomyces, Pythium oligandrum (a mycoparasitic member of the genus) and others (Gracia‐Garza et al., 2003; Martin and Loper, 1999). The extent to which these organisms function by altering rhizosphere microflora or attacking Py. ultimum directly is an open question (Naseby et al., 2000; Vallance et al., 2012); Pythium ultimum competes poorly with organisms having higher saprotrophic capabilities.
Pythium ultimum is just one of many destructive members of the genus, with Py. aphanidermatum and Py. irregulare also topping lists of important pathogens (Martin and Loper, 1999). Opportunities for understanding their biology, ecology and evolution have been enhanced by genome sequencing. DNA‐mediated transformation has been achieved for several species, but few studies of gene function have been reported (Grenville‐Briggs et al., 2013; Weiland, 2003). As core promoter structure appears to be conserved within the Peronosporales, vectors and technologies applied to species, such as Phytophthora, should be transferable to Pythium (Roy et al., 2013). Pythium spp. are easily grown and have the potential to develop into valuable experimental systems for necrotrophic and saprotrophic oomycete lifestyles. Their growth on byproducts of the agro‐food industry has also attracted interest as sources of polyunsaturated fatty acids for human consumption (Stredansky et al., 2000).
10. Albugo candida
White blister rust is a disease caused in many dicotyledonous plant species by obligate biotrophic parasites. For example, A. candida infection of Brassica juncea (Indian mustard) has resulted in significant crop losses in India (Awasthi et al., 2012), Canada (Rimmer et al., 2000) and Australia (Kaur et al., 2008). The white rusts, order Albuginales, are phylogenetically distant from the Peronosporales, and probably represent an independent acquisition of biotrophy (Thines and Kamoun, 2010; Thines and Spring, 2005). All Albugo species infecting the Brassicaceae were thought to be races of A. candida, but molecular studies of isolates from various hosts and locations have led to the description of specialists, for example Albugo laibachii on Arabidopsis thaliana (Thines, 2014; Thines et al., 2009). Specific A. candida races can grow on diverse plant hosts, including Brassicaceae, Cleomaceae and Capparaceae (Thines, 2014). Albugo spp. provide an interesting experimental system for the study of plant immune suppression, disease resistance and host–pathogen co‐evolution.
Albugo spp. reproduce asexually via zoosporangia, which release flagellated motile zoospores on incubation in water. On the surface of a plant leaf, zoospores settle in stomata, and each extends a germ tube into the substomatal chamber (Holub et al., 1994). Coenocytic hyphae then grow intercellularly through the plant. Small globose haustoria penetrate into plant cells (Soylu et al., 2003). When an Albugo infection is mature, zoosporangia rupture the plant epidermis with force and enzymatic digestion (Heller and Thines, 2009). This results in characteristic ‘white blister’ pustules. Albugo also has a sexual cycle, producing tough oospores that can survive difficult environmental conditions (Petrie, 1975). During the systemic infection of Brassicaceae hosts, the inflorescences become misshapen, forming so‐called ‘stagheads’ (Fig. 17A).
Figure 17.

Disease symptoms of Albugo infections. (A) Disease resulting from the infection of Brassica juncea with an unknown Albugo species. The misshapen inflorescence phenotype is known as a ‘staghead’. (B) An example of immunity suppression by Albugo laibachii: A. thaliana Col‐0 is resistant to Hyaloperonospora arabidopsidis Emoy2 via RPP1 but, when pre‐infected with A. laibachii, can support the growth of both pathogens.
Albugo infection has long been associated with ‘green islands’, where infected tissue appears to be healthy and senescence is delayed. Infection by Albugo also greatly enhances susceptibility to co‐infections with downy mildews (Bains and Jhooty, 1985; Crute et al., 1994). Cooper et al. (2008) investigated the ability of A. laibachii and A. candida to suppress host immunity. They showed that A. laibachii can suppress the ‘runaway cell death’ of Arabidopsis lsd1 mutants after inoculation with avirulent H. arabidopsidis. Furthermore, when pre‐infected with virulent A. laibachii, resistant Arabidopsis accessions were no longer resistant to avirulent H. arabidopsidis isolates (Fig. 17B), lettuce downy mildew or powdery mildew. Suppression was also observed on B. juncea with A. candida and Brassica downy mildew (Cooper et al., 2008). These results suggest that Albugo is effective at broad suppression of plant immunity, including effector‐triggered immunity.
The first step in understanding how Albugo spp. impose such susceptibility is to examine their genomes. Links et al. (2011) and Kemen et al. (2011) sequenced A. candida and A. laibachii genomes, respectively. The genomes are around 40 Mb and compact; about 50% of the assemblies consist of coding sequences (see Fig. 18). Both genomes show adaptations to obligate biotrophy; they are missing sulfite oxidases, nitrate and nitrite reductases and, in the case of A. laibachii, the whole molybdopterin biosynthesis pathway. The A. candida secretome consists of 929 proteins (without transmembrane domains), compared with 672 in A. laibachii, perhaps reflecting its wider host range. Within the secretomes, there is no enrichment of putative RXLR effectors. Kemen et al. (2011) discovered the CHXC (cysteine, histidine, any amino acid, cysteine) motif at the N‐terminus of a class of candidate effectors. The CHXC‐containing N‐terminus is sufficient to translocate the C‐terminus of P. infestans AVR3a (an RXLR effector) into host cells (Kemen et al., 2011).
Figure 18.
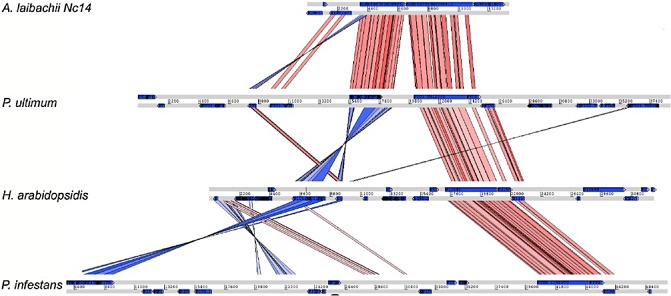
Albugo spp. have compact genomes. Synteny between Albugo laibachii, Pythium ultimum, Hyaloperonospora arabidopsidis and Phytophthora infestans. The region shown is an example of the relatively dense clustering of genes in Albugo species. With increasing genome size, the distance between both genes increases and re‐organizations occur (red, synteny without inversion; blue, inverted regions). Reproduced from Kemen et al. (2011).
Several A. candida races can infect some, but not all, A. thaliana accessions and, from crosses between resistant and susceptible accessions, an R gene against four A. candida races, WRR4 [encoding a toll interleukin 1 receptor‐nucleotide‐binding‐leucine‐rich repeat (TIR‐NB‐LRR) R protein], was identified (Borhan et al., 2008). WRR4 can also provide resistance to A. candida when transformed into susceptible cultivars of B. napus and B. juncea (Borhan et al., 2010). In A. thaliana, RAC1 (also encoding a TIR‐NB‐LRR R protein) confers resistance to A. laibachii (Borhan et al., 2004). The inheritance of avirulence of a B. juncea isolate (Ac2V) was studied through a cross between two A. candida isolates; this work predicted a single avirulence gene for the incompatibility between Ac2V and B. rapa (Adhikari et al., 2003).
There are open questions about Albugo from both fundamental and translational perspectives. Thines (2014) speculated that the broad host range of the A. candida meta‐population is maintained through frequent genetic exchange where the host range of individual isolates overlap. Comparing the genomes of multiple isolates from different hosts would test this hypothesis and build up a clear picture of population variation. This would also aid the discovery of new effector candidates through the identification of secreted proteins under strong selective pressure. A more extensive analysis of Albugo effectors should be made. The presence of the CHXC effectors inside host cells needs to be confirmed and the translocation mechanism elucidated. It is unclear which Albugo effector proteins are recognized by the few known R proteins. Arabidopsis thaliana cannot be colonized by most A. candida isolates. The molecular basis for this resistance could be exploited to introduce durable resistance to Brassica crops.
Lastly, and perhaps most interestingly, is the question: how does the remarkable defence suppression by Albugo work? We need to define the extent of this suppression. What other pests or pathogens can grow on plants infected with Albugo? What changes occur in the microbiome of Albugo‐infected leaves in the field? Recent data have suggested that Albugo could be widespread as an asymptomatic endophyte (Ploch and Thines, 2011). The implications of these infections as a reservoir for (i) Albugo and (ii) further Albugo suppression‐enabled infections by other species remain to be discovered.
Conclusion
Although the study of oomycete plant pathogens has always been an important topic in plant pathology, it has recently taken an even more central role with the advent of genomics and the discovery of large effector repertoires in oomycete genomes. This article provides a benchmark for future trends. Many topics remain to be investigated in more depth. For example, the large number of pathogenic species and the wide diversity of host species and host ranges should provide a rich basis for the investigation of the genetic and physiological mechanisms of host adaptation and specialization. It will also be interesting to track how the Top 10 list will develop in the coming years. Many pathogenic oomycetes among the 33 listed (Table 2) have evolved unique adaptations in their parasitic lifestyle that will undoubtedly reveal fascinating processes and mechanisms. Thus, future research efforts should take into account a diverse spectrum of taxa beyond the widely studied species.
Author Contributions
All authors voted and contributed to the writing of the 10 sections. SK oversaw the voting and writing. Other authors are listed in reverse order based on the species ranking. The species coordinators are listed last in their section and are preceded by other contributors listed in alphabetical order. The species coordinators are: Albugo candida (OF and JDGJ), Pythium ultimum (HSJ), Phytophthora parasitica (FP), Phytophthora cinnamomi (DC), Plasmopara viticola (MR and AF), Phytophthora capsici (KL), Phytophthora sojae (MG and BMT), Phytophthora ramorum (NJG), Hyaloperonospora arabidopsidis (GvdA and JMcD) and Phytophthora infestans (PRJB and FG).
Acknowledgements
The authors would like to thank Dr Diane Hird for assistance with several aspects of the project.
Sophien Kamoun, Oliver Furzer and Jonathan D. G. Jones received funding from the European Research Council (ERC), the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the Gatsby Charitable Foundation. Ronaldo Dalio thanks the National Council for Scientific and Technological Development (CNPq/CsF Brazil—313139/2013‐0) for financial support. Michelina Ruocco receives support from Conoscenze Integrate per Sostenibilità ed Innovazione del Made in Italy Agroalimentare (CISIA–MIUR). Leonardo Schena was supported by grant FIRB 2010—RBFR10PZ4N from the Italian Ministry of Education, University and Research (MIUR). Andreia Figueiredo receives support from the Portuguese Foundation for Science and Technology (Grant n° SFRH/BPD/63641/2009). Xiao‐Ren Chen was financed by the National Natural Science Foundation of China (Grant no. 31101395) and Jiangsu Province Basic Research Program (Natural Science Foundation) of China (Grant no. BK2011443). Howard S. Judelson and Brett M. Tyler were supported by grants 2011‐68004‐30154 and 2011‐68004‐30104, respectively, from the Agriculture and Food Research Institute of the National Institute of Food and Agriculture of the US Department of Agriculture. Mark Gijzen was supported by the Agriculture and Agri‐Food Canada GRDI program. Niklaus J. Grünwald was supported by grant 2011‐68004‐30154 from the US Department of Agriculture National Institute of Food and Agriculture, the National Research Initiative Competitive Grants Program grant 2008‐35600‐18780, the US Department of Agriculture Agricultural Research Service CRIS 5358‐22000‐039‐00D, the Northwest Center for Nursery Crop Research, the US Forest Service, the US Department of Agriculture Agricultural Research Service Floriculture Nursery Initiative and Oregon Department of Agriculture/Oregon Association of Nurseries. Daniel F. A. Tomé is funded by the UK Biotechnology and Biological Sciences Research Council grant BB/G015066/1. John McDowell is supported by the US Department of Agriculture–Agriculture and Food Research Initiative (2009‐03008 and 2011‐68004), the National Science Foundation (ABI‐1146819) and the Virginia Tech Institute for Critical Technology and Applied Sciences. Fouad Daayf received funding from acronyms are: The Natural Sciences and Engineering Research Council of Canada (NSERC), Manitoba Agri‐Food Research and Development Initiative (MB‐ARDI), McCain Foods, Keystone Potato Producers Association (KPPA) and Peak of the Market. Harold J. G. Meijer is funded by The Dutch Technology Foundation STW‐NWO (VIDI grant 10281). Benjamin Petre is supported by an INRA Contrat Jeune Scientifique and has received the support of the European Union, in the framework of the Marie‐Curie FP7 COFUND People Programme, through the award of an AgreenSkills fellowship (under grant agreement n°267196). Francine Govers receives support from the Food for Thought Program, Wageningen University Fund.
References
- Adhikari, B.N. , Hamilton, J.P. , Zerillo, M.M. , Tisserat, N. , Levesque, C.A. and Buell, C.R. (2013) Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS ONE, 8, e75072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari, T.B. , Liu, J.Q. , Mathur, S. , Wu, C.R.X. and Rimmer, S.R. (2003) Genetic and molecular analyses in crosses of race 2 and race 7 of Albugo candida . Phytopathology, 93, 959–965. [DOI] [PubMed] [Google Scholar]
- Agrios, G.N. (2005) Plant Pathology. Burlington, Massachusetts: Elsevier Academic Press Publications. [Google Scholar]
- Ah Fong, A.M.V. and Judelson, H.S. (2003) Cell cycle regulator Cdc14 is expressed during sporulation but not hyphal growth in the fungus‐like oomycete Phytophthora infestans . Mol. Microbiol. 50, 487–494. [DOI] [PubMed] [Google Scholar]
- Akinsanmi, O.A. and Drenth, A. (2013) Phosphite and metalaxyl rejuvenate macadamia trees in decline caused by Phytophthora cinnamomi . Crop Protect. 53, 29–36. [Google Scholar]
- Allardyce, J.A. , Rookes, J.E. , Hussain, H.I. and Cahill, D.M. (2013) Transcriptional profiling of Zea mays roots reveals roles for jasmonic acid and terpenoids in resistance against Phytophthora cinnamomi . Funct. Integr. Genomics, 13, 217–228. [DOI] [PubMed] [Google Scholar]
- Allen, R.L. , Bittner‐Eddy, P.D. , Grenvitte‐Briggs, L.J. , Meitz, J.C. , Rehmany, A.P. , Rose, L.E. and Beynon, J.L. (2004) Host–parasite coevolutionary conflict between Arabidopsis and downy mildew. Science, 306, 1957–1960. [DOI] [PubMed] [Google Scholar]
- Andres, J.L. , Rivera, A. and Fernandez, J.A. (2003) Phytophthora nicotianae pathogenic to pepper in northwest spain. J. Plant Pathol. 85, 91–98. [Google Scholar]
- Armstrong, M.R. , Whisson, S.C. , Pritchard, L. , Bos, J.I. , Venter, E. , Avrova, A.O. , Rehmany, A.P. , Bohme, U. , Brooks, K. , Cherevach, I. , Hamlin, N. , White, B. , Fraser, A. , Lord, A. , Quail, M.A. , Churcher, C. , Hall, N. , Berriman, M. , Huang, S. , Kamoun, S. , Beynon, J.L. and Birch, P.R. (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA, 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard, A. , Gourgues, M. , Galiana, E. , Panabieres, F. , Ponchet, M. and Keller, H. (2008) Strategies of attack and defense in plant–oomycete interactions, accentuated for Phytophthora parasitica Dastur (syn P. nicotianae Breda de Haan). J. Plant Physiol. 165, 83–94. [DOI] [PubMed] [Google Scholar]
- Attard, A. , Gourgues, M. , Callemeyn‐Torre, N. and Keller, H. (2010) The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 187, 449–460. [DOI] [PubMed] [Google Scholar]
- Australian Government Department of Environment (2014) Threat Abatement Plan for Disease in Natural Ecosystems Caused by Phytophthora cinnamomi. Canberra: Commonwealth of Australia. [Google Scholar]
- Awasthi, R.P. , Nashaat, N.I. , Kolte, S.J. , Tewari, A.K. , Meena, P.D. and Renu, B. (2012) Screening of putative resistant sources against Indian and exotic isolates of Albugo candida inciting white rust in rapeseed‐mustard. J. Oilseed Brassica, 3, 27–37. [Google Scholar]
- Bains, S.S. and Jhooty, J.S. (1985) Association of Peronospora‐parasitica with Albugo‐candida on Brassica‐juncea leaves. J. Phytopathol. 112, 28–31. [Google Scholar]
- Baxter, L. , Tripathy, S. , Ishaque, N. , Boot, N. , Cabral, A. , Kemen, E. , Thines, M. , Ah‐Fong, A. , Anderson, R. , Badejoko, W. , Bittner‐Eddy, P. , Boore, J.L. , Chibucos, M.C. , Coates, M. , Dehal, P. , Delehaunty, K. , Dong, S. , Downton, P. , Dumas, B. , Fabro, G. , Fronick, C. , Fuerstenberg, S.I. , Fulton, L. , Gaulin, E. , Govers, F. , Hughes, L. , Humphray, S. , Jiang, R.H.Y. , Judelson, H. , Kamoun, S. , Kyung, K. , Meijer, H. , Minx, P. , Morris, P. , Nelson, J. , Phuntumart, V. , Qutob, D. , Rehmany, A. , Rougon‐Cardoso, A. , Ryden, P. , Torto‐Alalibo, T. , Studholme, D. , Wang, Y. , Win, J. , Wood, J. , Clifton, S.W. , Rogers, J. , Van den Ackerveken, G. , Jones, J.D.G. , McDowell, J.M. , Beynon, J. and Tyler, B.M. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science, 330, 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, M. , Waldner, M. and Gisi, U. (2010) A single point mutation in the novel PvCesA3 gene confers resistance to the carboxylic acid amide fungicide mandipropamid in Plasmopara viticola . Fungal Genet. Biol. 47, 499–510. [DOI] [PubMed] [Google Scholar]
- Bonnet, P. , Lacourt, I. , Venard, P. and Ricci, P. (1994) Diversity in pathogenicity to tobacco and in elicitin production among isolates of Phytophthora parasitica . J. Phytopathol. 141, 25–37. [Google Scholar]
- Borhan, M.H. , Holub, E.B. , Beynon, J.L. , Rozwadowski, K. and Rimmer, S.R. (2004) The Arabidopsis TIR‐NB‐LRR gene RAC1 confers resistance to Albugo candida (white rust) and is dependent on EDS1 but not PAD4. Mol. Plant–Microbe Interact. 17, 711–719. [DOI] [PubMed] [Google Scholar]
- Borhan, M.H. , Gunn, N. , Cooper, A. , Gulden, S. , Tör, M. , Rimmer, S.R. and Holub, E.B. (2008) WRR4 encodes a TIR‐NB‐LRR protein that confers broad‐spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida . Mol. Plant–Microbe Interact. 21, 757–768. [DOI] [PubMed] [Google Scholar]
- Borhan, M.H. , Holub, E.B. , Kindrachuk, C. , Omidi, M. , Bozorgmanesh‐Frad, G. and Rimmer, S.R. (2010) WRR4, a broad‐spectrum TIR‐NB‐LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed brassica crops. Mol. Plant Pathol. 11, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, J.I. , Armstrong, M.R. , Gilroy, E.M. , Boevink, P.C. , Hein, I. , Taylor, R.M. , Zhendong, T. , Engelhardt, S. , Vetukuri, R.R. , Harrower, B. , Dixelius, C. , Bryan, G. , Sadanandom, A. , Whisson, S.C. , Kamoun, S. and Birch, P.R. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA, 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt, T.O. , Schornack, S. , Banfield, M.J. and Kamoun, S. (2012) Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. [DOI] [PubMed] [Google Scholar]
- Brasier, C. and Webber, J. (2010) Plant pathology: sudden larch death. Nature, 466, 824–825. [DOI] [PubMed] [Google Scholar]
- Brasier, C.M. (2008) The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57, 792–808. [Google Scholar]
- Brasier, C.M. , Vettraino, A.M. , Chang, T.T. and Vannini, A. (2010) Phytophthora lateralis discovered in an old growth Chamaecyparis forest in Taiwan. Plant Pathol. 59, 595–603. [Google Scholar]
- Cabral, A. , Stassen, J.H. , Seidl, M.F. , Bautor, J. , Parker, J.E. and Van den Ackerveken, G. (2011) Identification of Hyaloperonospora arabidopsidis transcript sequences expressed during infection reveals isolate‐specific effectors. PLoS ONE, 6, e19328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral, A. , Oome, S. , Sander, N. , Kufner, I. , Nurnberger, T. and Van den Ackerveken, G. (2012) Nontoxic Nep1‐like proteins of the downy mildew pathogen Hyaloperonospora arabidopsidis: repression of necrosis‐inducing activity by a surface‐exposed region. Mol. Plant–Microbe Interact. 25, 697–708. [DOI] [PubMed] [Google Scholar]
- Caillaud, M.‐C. , Piquerez, S.J.M. , Fabro, G. , Steinbrenner, J. , Ishaque, N. , Beynon, J. and Jones, J.D. (2011) Subcellular localization of the H. arabidopsidis RxLR effector repertoire identifies the extrahaustorial membrane‐localized HaRxL17 that confers enhanced plant susceptibility. Plant J. 69, 252–265. [DOI] [PubMed] [Google Scholar]
- Casagrande, K. , Falginella, L. , Castellarin, S. , Testolin, R. and Di Gaspero, G. (2011) Defence responses in Rpv3‐dependent resistance to grapevine downy mildew. Planta, 234, 1097–1109. [DOI] [PubMed] [Google Scholar]
- Chamnanpunt, J. , Shan, W.X. and Tyler, B.M. (2001) High frequency mitotic gene conversion in genetic hybrids of the oomycete Phytophthora sojae . Proc. Natl. Acad. Sci. USA, 98, 14 530–14 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Delmotte, F. , Richard‐Cervera, S. , Douence, L. , Greif, C. and Corio‐Costet, M. (2007) At least two origins of fungicide resistance in grapevine downy mildew populations. Appl. Environ. Microbiol. 73, 5162–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X.R. , Xing, Y.P. , Li, Y.P. , Tong, Y.H. and Xu, J.Y. (2013) RNA‐Seq reveals infection‐related gene expression changes in Phytophthora capsici . PLoS ONE, 8, e74588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, E.T. , Farr, D.F. and Rossman, A.Y. (2008) Synopsis of Phytophthora with accurate scientific names, host range, and geographic distribution. Plant Health Progress, doi: 10.1094/PHP-2008-0318-01-RS. [DOI] [Google Scholar]
- Coates, M.E. and Beynon, J.L. (2010) Hyaloperonospora arabidopsidis as a Pathogen Model. Annu. Rev. Phytopathol. 48, 329–345. [DOI] [PubMed] [Google Scholar]
- Colas, V. , Lacourt, I. , Ricci, P. , Vanlerberghe‐Masutti, F. , Poupet, A. and Panabieres, F. (1998) Diversity of virulence in Phytophthora parasitica on tobacco, as reflected by nuclear RFLPs. Phytopathology, 88, 205–212. [DOI] [PubMed] [Google Scholar]
- Colas, V. , Conrod, S. , Venard, P. , Keller, H. , Ricci, P. and Panabieres, F. (2001) Elicitin genes expressed in vitro by certain tobacco isolates of Phytophthora parasitica are down regulated during compatible interactions. Mol. Plant–Microbe Interact. 14, 326–335. [DOI] [PubMed] [Google Scholar]
- Cooke, D.E.L. , Cano, L.M. , Raffaele, S. , Bain, R.A. , Cooke, L.R. , Etherington, G.J. , Deahl, K.L. , Farrer, R.A. , Gilroy, E.M. , Goss, E.M. , Grünwald, N.J. , Hein, I. , Maclean, D.J. , McNicol, J.W. , Randall, E. , Oliva, R.F. , Pel, M.A. , Shaw, D.S. , Squires, J.N. , Taylor, M.C. , Vleeshouwers, V.G.A.A. , Birch, P.R.J. , Lees, A.K. and Kamoun, S. (2012) Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog. 8, e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, A.J. , Latunde‐Dada, A.O. , Woods‐Tör, A. , Lynn, J. , Lucas, J.A. , Crute, I.R. and Holub, E.B. (2008) Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad‐spectrum suppression of innate immunity. Mol. Plant–Microbe Interact. 21, 745–756. [DOI] [PubMed] [Google Scholar]
- Crane, C.E. and Shearer, B.L. (2014) Comparison of phosphite application methods for control of Phytophthora cinnamomi in threatened communities. Australas. Plant Pathol. 43, 143–149. [Google Scholar]
- Crute, I. , Beynon, J. , Dangl, J. , Holub, E. , Mauch‐Mani, B. , Slusarenko, A. , Staskawicz, B. and Ausubel, F. (1994) Microbial pathogenesis of Arabidopsis In: Arabidopsis (Meyerowitz E.M. and Somerville C.R., eds), pp. 705–747. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Dean, R. , Van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeArce, M. (2008) Correspondence of Charles Darwin on James Torbitt's project to breed blight‐resistant potatoes. Arch. Nat. Hist. 35, 208–222. [DOI] [PubMed] [Google Scholar]
- Delmotte, F. , Mestre, P. , Schneider, C. , Kassemeyer, H.‐H. , Kozma, P. , Richart‐Cervera, S. , Rouxela, M. and Delière, L. (2013) Rapid and multiregional adaptation to host partial resistance in a plant pathogenic oomycete: evidence from European populations of Plasmopara viticola, the causal agent of grapevine downy mildew. Infect. Genet. Evol. 27, 500–508. [DOI] [PubMed] [Google Scholar]
- Dempsey, R.W. , Merchant, A. and Tausz, M. (2012) Differences in ascorbate and glutathione levels as indicators of resistance and susceptibility in Eucalyptus trees infected with Phytophthora cinnamomi . Tree Physiol. 32, 1148–1160. [DOI] [PubMed] [Google Scholar]
- Dick, M.W. (2001) Straminipilous Fungi. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Dong, S. , Stam, R. , Cano, L.M. , Song, J. , Sklenar, J. , Yoshida, K. , Bozkurt, T.O. , Oliva, R. , Liu, Z. , Tian, M. , Win, J. , Banfield, M.J. , Jones, A.M.E. , van der Hoorn, R.A.L. and Kamoun, S. (2014) Effector specialization in a lineage of the Irish Potato Famine Pathogen. Science, 343, 552–555. [DOI] [PubMed] [Google Scholar]
- Dorrance, A. and Grünwald, N.J. (2009) Phytophthora sojae: diversity among and within populations In: Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools (Lamour K. and Kamoun S., eds), pp. 197–212. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- Dou, D. , Kale, S.D. , Wang, X. , Jiang, R.H.Y. , Bruce, N.A. , Arredondo, F.D. , Zhang, X. and Tyler, B.M. (2008) RXLR‐mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen‐encoded machinery. Plant Cell, 20, 1930–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenth, A. , Tas, I.C.Q. and Govers, F. (1994) DNA fingerprinting uncovers a new sexually reproducing population of Phytophthora infestans in the Netherlands. Eur. J. Plant Pathol. 100, 97–107. [Google Scholar]
- Dunn, A. , Milgroom, M. , Meitz, J. , McLeod, A. , Fry, W. , McGrath, M. , Dillard, H. and Smart, C. (2010) Population structure and resistance to mefenoxam of Phytophthora capsici in New York State. Plant Dis. 94, 1461–1468. [DOI] [PubMed] [Google Scholar]
- Dunstan, W.A. , Rudman, T. , Shearer, B.L. , Moore, N.A. , Paap, T. , Calver, M.C. , Dell, B. and Hardy, G.E.S.J. (2010) Containment and spot eradication of a highly destructive, invasive plant pathogen (P. cinnamomi) in natural ecosystems. Biol. Invasions, 12, 913–925. [Google Scholar]
- Elliott, M. , Sumampong, G. , Varga, A. , Shamoun, S.F. , James, D. , Masri, S. and Gruenwald, N.J. (2011) Phenotypic differences among three clonal lineages of Phytophthora ramorum . For. Pathol. 41, 7–14. [Google Scholar]
- Enkerli, K. , Hahn, M.G. and Mims, C.W. (1997) Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae . Can. J. Bot. 75, 1494–1508. [Google Scholar]
- Erwin, D.C. and Ribeiro, O.K. (1996) Phytophthora Diseases Worldwide. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Eshraghi, L. , Anderson, J.P. , Aryamanesh, N. , McComb, J.A. , Shearer, B.S. and Hardy, G.St.J.E. (2014) Suppression of the auxin response pathway enhances susceptibility to Phytophthora cinnamomi while phosphite‐mediated resistance stimulates the auxin signalling pathway. BMC Plant Biol. 14, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabro, G. , Steinbrenner, J. , Coates, M. , Ishaque, N. , Baxter, L. , Studholme, D.J. , Körner, E. , Allen, R.L. , Piquerez, S.J.M. , Rougon‐Cardoso, A. , Greenshields, D. , Lei, R. , Badel, J.L. , Caillaud, M.‐C. , Sohn, K.‐H. , Van den Ackerveken, G. , Parker, J.E. , Beynon, J. and Jones, J.D.G. (2011) Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog. 7, e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren, N. , Bollmann, S.R. , Kasschau, K.D. , Cuperus, J.T. , Press, C.M. , Sullivan, C.M. , Chapman, E.J. , Hoyer, J.S. , Gilbert, K.B. , Grünwald, N.J. and Carrington, J.C. (2013) Phytophthora have distinct endogenous small RNA populations that include short interfering and microRNAs. PLoS ONE, 8, e77181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, D.F. and Rossman, A.Y. (2014) Fungal databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. Available at: http://nt.ars‐grin.gov/fungaldatabases/ [accessed on Oct 6, 2014].
- Feng, B.Z. and Li, P.Q. (2013) Molecular characterization and functional analysis of the Nep1‐like protein‐encoding gene from Phytophthora capsici . Genet. Mol. Res. 12, 1468–1478. [DOI] [PubMed] [Google Scholar]
- Feng, B.Z. , Li, P.Q. , Wang, H.M. and Zhang, X.G. (2010) Functional analysis of Pcpme6 from oomycete plant pathogen Phytophthora capsici . Microb. Pathog. 49, 23–31. [DOI] [PubMed] [Google Scholar]
- Fisher, M.C. , Henk, D.A. , Briggs, C.J. , Brownstein, J.S. , Madoff, L.C. , McCraw, S.L. and Gurr S.J. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature, 484, 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine, M. , Austerlitz, F. , Giraud, T. , Labbe, F. , Papura, D. , Richard‐Cervera, S. and Delmotte, F. (2013) Genetic signature of a range expansion and leap‐frog event after the recent invasion of Europe by the grapevine downy mildew pathogen Plasmopara viticola . Mol. Ecol. 22, 2771–2786. [DOI] [PubMed] [Google Scholar]
- Francis, D.M. and St. Clair, D.A. (1997) Population genetics of Pythium ultimum . Phytopathology, 87, 454–461. [DOI] [PubMed] [Google Scholar]
- Fry, W.E. , McGrath, M.T. , Seaman, A. , Zitter, T.A. , McLeod, A. , Danies, G. , Small, I.M. , Myers, K. , Everts, K. , Gevens, A.J. , Gugino, B.K. , Johnson, S.B. , Judelson, H. , Ristaino, J. , Roberts, P. , Secor, G. , Seebold, K. , Snover‐Clift, K. , Wyenandt, A. , Grünwald, N.J. and Smart, C.D. (2013) The 2009 late blight pandemic in the eastern United States—causes and results. Plant Dis. 97, 296–306. [DOI] [PubMed] [Google Scholar]
- Galet, P. (1977) Mildiou. Paris: Lavoisier. [Google Scholar]
- Garbelotto, M. and Hayden, K.J. (2012) Sudden oak death: interactions of the exotic oomycete Phytophthora ramorum with naïve North American hosts. Eukaryot. Cell, 11, 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler, C. , Pertot, I. and Perazzolli, M. (2011) Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50, 3–44. [Google Scholar]
- Gijzen, M. and Qutob, D. (2009) Phytophthora sojae and Soybean In: Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools (Lamour K. and Kamoun S., eds), pp. 303–329. Hoboken, New Jersey: John Wiley & Sons. [Google Scholar]
- Gobbin, D. , Jermini, M. , Loskill, B. , Pertot, I. , Raynal, M. and Gessler, C. (2005) Importance of secondary inoculum of Plasmopara viticola to epidemics of grapevine downy mildew. Plant Pathol. 54, 522–534. [Google Scholar]
- Gobena, D. , McGrath, M.T. and Lamour, K.H. (2012) Survival and spread of Phytophthora capsici on Long Island, New York. Mycol. Prog. 11, 761–768. [Google Scholar]
- Gobena, D.J. , Roig, J. , Hulvey, J. and Lamour, K. (2010) Genetic diversity of the vegetable pathogen Phytophthora capsici in Argentina. Phytopathology, 100, S41. [Google Scholar]
- Gómez‐Zeledón, J. , Zipper, R. and Spring, O. (2013) Assessment of phenotypic diversity of Plasmopara viticola on Vitis genotypes with different resistance. Crop Protection, 54, 221–228. [Google Scholar]
- Goodwin, S.B. , Drenth, A. and Fry, W.E. (1992) Cloning and genetic analyses of two highly polymorphic, moderately repetitive nuclear DNAs from Phytophthora infestans . Curr. Genet. 22, 107–115. [DOI] [PubMed] [Google Scholar]
- Goodwin, S.B. , Cohen, B.A. and Fry, W.E. (1994) Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc. Natl. Acad. Sci. USA, 91, 11 591–11 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss, E.M. , Carbone, I. and Grünwald, N.J. (2009a) Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum . Mol. Ecol. 18, 1161–1174. [DOI] [PubMed] [Google Scholar]
- Goss, E.M. , Larsen, M. , Chastagner, G.A. , Givens, D.R. and Grünwald, N.J. (2009b) Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathog. 5, e1000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss, E.M. , Larsen, M. , Vercauteren, A. , Werres, S. , Heungens, K. and Grünwald, N.J. (2011) Phytophthora ramorum in Canada: evidence for migration within North America and from Europe. Phytopathology, 101, 166–171. [DOI] [PubMed] [Google Scholar]
- Goss, E.M. , Press, C.M. and Grünwald, N.J. (2013) Evolution of RXLR‐class effectors in the oomycete plant pathogen Phytophthora ramorum . PLoS ONE, 8, e79347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers, F. and Gijzen, M. (2006) Phytophthora genomics: the plant destroyers' genome decoded. Mol. Plant–Microbe Interact. 19, 1295–1301. [DOI] [PubMed] [Google Scholar]
- Gracia‐Garza, J.A. , Little, M. , Brown, W. , Blom, T.J. , Schneider, K. , Allen, W. and Potter, J. (2003) Efficacy of various biological control agents and biorationals against Pythium root rot in poinsettia. Horttechnology, 13, 149–153. [Google Scholar]
- Granke, L. , Windstam, S. , Hoch, H. , Smart, C. and Hausbeck, M. (2009) Dispersal and movement mechanisms of Phytophthora capsici sporangia. Phytopathology, 99, 1258–1264. [DOI] [PubMed] [Google Scholar]
- Granke, L. , Quesada‐Ocampo, L. , Lamour, K. and Hausbeck, M.K. (2012) Advances in research on Phytophthora capsici on vegetable crops in the United States. Plant Dis. 95, 1588–1600. [DOI] [PubMed] [Google Scholar]
- Grenville‐Briggs, L.J. , Horner, N.R. , Phillips, A.J. , Beakes, G.W. and Van West, P. (2013) A family of small tyrosine rich proteins is essential for oogonial and oospore cell wall development of the mycoparasitic oomycete Pythium oligandrum . Fungal Biol. 117, 163–172. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. , Goss, E.M. and Press, C.M. (2008) Phytophthora ramorum: a pathogen with a remarkably wide host‐range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 9, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald, N.J. , Goss, E.M. , Ivors, K. , Garbelotto, M. , Martin, F.N. , Prospero, S. , Hansen, E.M. , Bonants, P.J.M. , Hamelin, R.C. , Chastagner, G. , Werres, S. , Rizzo, D.M. , Abad, G. , Beales, P. , Bilodeau, G.J. , Blomquist, C.L. , Brasier, C. , Brière, S.C. , Chandelier, A. , Davidson, J.M. , Denman, S. , Elliott, M. , Frankel, S.J. , Goheen, E.M. , de Gruyter, H. , Heungens, K. , James, D. , Kanaskie, A. , McWilliams, M.G. , Veld, W.M. , Moralejo, E. , Osterbauer, N.K. , Palm, M.E. , Parke, J.L. , Sierra, A.M.P. , Shamoun, S.F. , Shishkoff, N. , Tooley, P.W. , Vettraino, A.M. , Webber, J. and Widmer, T.L. (2009) Standardizing the nomenclature for clonal lineages of the sudden oak death pathogen, Phytophthora ramorum . Phytopathology, 99, 792–795. [DOI] [PubMed] [Google Scholar]
- Grünwald, N.J. , Garbelotto, M. , Goss, E.M. , Heungens, K. and Prospero, S. (2012) Emergence of the sudden oak death pathogen Phytophthora ramorum . Trends Microbiol. 20, 131–138. [DOI] [PubMed] [Google Scholar]
- Guha Roy, S. , Mukherjee, S.K. , Bhattacharyya, S. and Khatua, D.C. (2009) Molecular identification of Phytophthora sp. affecting some economicaly important crops in eastern India through ITS‐RFLP and sequencing of the ITS region. J. Phytopathol. 157, 666–674. [Google Scholar]
- Gunderson, J.H. , Elwood, H. , Ingold, H. , Kindle, A. and Sogin, M.L. (1987) Phylogenetic relationships between chlororphytes, chrysophytes, and oomycetes. Proc. Natl. Acad. Sci. USA, 84, 5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning, K. , Conlan, X.A. , Parker, R.M. , Dyson, G.A. , Adams, M.J. , Barnett, N.W. and Cahill, D.M. (2013) Profiling of secondary metabolites in blue lupin inoculated with Phytophthora cinnamomi following phosphite treatment. Funct. Plant Biol. 40, 1089–1097. [DOI] [PubMed] [Google Scholar]
- Haas, B.J. , Kamoun, S. , Zody, M.C. , Jiang, R.H.Y. , Handsaker, R.E. , Cano, L.M. , Grabherr, M. , Kodira, C.D. , Raffaele, S. , Torto‐Alalibo, T. , Bozkurt, T.O. , Ah‐Fong, A.M.V. , Alvarado, L. , Anderson, V.L. , Armstrong, M.R. , Avrova, A.O. , Baxter, L. , Beynon, J.L. , Boevink, P.C. , Bollmann, S.R. , Bos, J.I.B. , Bulone, V. , Cai, G. , Cakir, C. , Carrington, J.C. , Chawner, M. , Conti, L. , Costanzo, S. , Ewan, R. , Fahlgren, N. , Fischbach, M.l.A. , Fugelstad, J. , Gilroy, E.M. , Gnerre, S. , Green, P.J. , Grenville‐Briggs, L.J. , Griffith, J.M. , Grünwald, N.J. , Horn, K. , Horner, N.R. , Hu, C.H. , Huitema, E. , Jeong, D.H. , Jones, A.M.E. , Jones, J.D.G. , Jones, R.W. , Karlsson, E.K. , Kunjeti, S.G. , Lamour, K. , Liu, Z. , Ma, L.J. , Maclean, D.J. , Chibucos, M.C. , McDonald, H. , McWalters, J. , Meijer, H.J.G. , Morgan, W. , Morris, P.F. , Munro, C.A. , O'Neill, K. , Ospina‐Giraldo, M.D. , Pinzon, A. , Pritchard, L. , Ramsahoye, B. , Ren, Q. , Restrepo, S. , Roy, S. , Sadanandom, A. , Savidor, A. , Schornack, S. , Schwartz, D.C. , Schumann, U.D. , Schwessinger, B. , Seyer, L. , Sharpe, T. , Silvar, C. , Song, J. , Studholme, D.J. , Sykes, S. , Thines, M. , van de Vondervoort, P.J.I. , Phuntumart, V. , Wawra, S. , Weide, R. , Win, J. , Young, C. , Zhou, S. , Fry, W.E. , Meyers, B.C. , van West, P. , Ristaino, J.B. , Govers, F. , Birch, P.R.J. , Whisson, S.C. , Judelson, H.S. and Nusbaum, C. (2009) Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature, 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hausbeck, M.K. and Lamour, K.H. (2004) Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 88, 1292–1303. [DOI] [PubMed] [Google Scholar]
- Haverkort, A. , Boonekamp, P. , Hutten, R. , Jacobsen, E. , Lotz, L. , Kessel, G. , Visser, R. and van der Vossen, E. (2008) Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 51, 47–57. [Google Scholar]
- Hee, W.Y. , Torreña, P.S. , Blackman, L.M. and Hardham, A.R. (2013) Phytophthora cinnamomi in Australia In: Phytophthora: A Global Perspective (Lamour K., ed.), CABI Plant Protection Series, No. 2, pp. 124–134. Wallingford, UK: CABI. [Google Scholar]
- Heller, A. and Thines, M. (2009) Evidence for the importance of enzymatic digestion of epidermal walls during subepidermal sporulation and pustule opening in white blister rusts (Albuginaceae). Mycol. Res. 113 (Pt 6–7), 657–667. [DOI] [PubMed] [Google Scholar]
- Hildebrand, A.A. (1959) A root and stalk rot of soybeans caused by Phytophthora megasperma Drechsler var. sojae var. nov. Can. J. Bot. 37, 927–957. [Google Scholar]
- Holub, E. , Brose, E. , Tör, M. , Clay, C. , Crute, I.R. and Beynon, J.L. (1994) Phenotypic and genotypic variation in the interaction between Arabidopsis thaliana and Albugo candida . Mol. Plant–Microbe Interact. 8, 916–928. [DOI] [PubMed] [Google Scholar]
- Holub, E.B. (2008) Natural history of Arabidopsis thaliana and oomycete symbioses. Eur. J. Plant Pathol. 122, 91–109. [Google Scholar]
- Hu, C.H. , Perez, F.G. , Donahoo, R. , McLeod, A. , Myers, K. , Ivors, K. , Secor, G. , Roberts, P.D. , Fry, W.E. , Deahl, K.L. and Ristaino, J.B. (2012) Recent genotypes of Phytophthora infestans in eastern USA reveal clonal populations and reappearance of mefenoxam sensitivity. Plant Dis. 96, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Diao, Y. , Zhou, Y. , Lin, D. , Bi, Y. , Pang, Z. , Trout Fryxell, R. , Liu, X. and Lamour, K. (2013) Loss of heterozygosity drives clonal diversity of Phytophthora capsici in China. PLoS ONE, 8, e82691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüberli, D. , Hayden, K.J. , Calver, M. and Garbelotto, M. (2012) Intraspecific variation in host susceptibility and climatic factors mediate epidemics of sudden oak death in western US forests. Plant Pathol. 61, 579–592. [Google Scholar]
- Hulvey, J. , Young, J. , Finley, L. and Lamour, K. (2010) Loss of heterozygosity in Phytophthora capsici after N‐ethyl‐nitrosourea mutagenesis. Mycologia, 102, 27–32. [DOI] [PubMed] [Google Scholar]
- Hurtado‐Gonzales, O. , Aragon‐Caballero, L. , Apaza‐Tapia, W. , Donahoo, R. and Lamour, K. (2008) Survival and spread of Phytophthora capsici in coastal Peru. Phytopathology, 98, 688–694. [DOI] [PubMed] [Google Scholar]
- Hussain, H.I. , Yi, Z. , Rookes, J.E. , Kong, L.X. and Cahill, D.M. (2013) Mesoporous silica nanoparticles as a biomolecule delivery vehicle in plants. J. Nanopart. Res. 15, 1–15. [Google Scholar]
- Jiang, R.H.Y. and Tyler, B.M. (2012) Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318. [DOI] [PubMed] [Google Scholar]
- Jiang, R.H.Y. , Tripathy, S. , Govers, F. and Tyler, B.M. (2008) RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving super‐family with more than 700 members. Proc. Natl. Acad. Sci. USA, 105, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.T. , Haegeman, A. , Danchin, E.G. , Gaur, H.S. , Helder, J. , Jones, M.G. , Kikuchi, T. , Manzanilla‐López, R. , Palomares‐Rius, J.E. , Wesemael, W.M. and Perry, R.N. (2013) Top 10 plant‐parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson, H.S. (1997) The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fungal Genet. Biol. 22, 65–76. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. and Blanco, F.A. (2005) The spores of Phytophthora: weapons of the plant destroyer. Nat. Rev. Microbiol. 3, 47–58. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. and Whittaker, S.L. (1995) Inactivation of transgenes in Phytophthora infestans is not associated with their deletion, methylation, or mutation. Curr. Genet. 28, 571–579. [DOI] [PubMed] [Google Scholar]
- Judelson, H.S. , Tyler, B.M. and Michelmore, R.W. (1991) Transformation of the oomycete pathogen, Phytophthora infestans . Mol. Plant–Microbe Interact. 4, 602–607. [DOI] [PubMed] [Google Scholar]
- Jung, T. , Colquhoun, I.J. and Hardy, G.E.S.J. (2013) New insights into the survival strategy of the invasive soilborne pathogen Phytophthora cinnamomi in different natural ecosystems in Western Australia. Forest Pathol. 43, 266–288. [Google Scholar]
- Jupe, J. , Stam, R. , Howden, A.J. , Morris, J.A. , Zhang, R. , Hedley, P.E. and Huitema, E. (2013) Phytophthora capsici–tomato interaction features dramatic shifts in gene expression associated with a hemi‐biotrophic lifestyle. Genome Biol. 14, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale, S.D. , Gu, B. , Capelluto, D.G.S. , Dou, D.‐L. , Feldman, E. , Rumore, A. , Arredondo, F.D. , Hanlon, R. , Fudal, I. , Rouxel, T. , Lawrence, C.B. , Shan, W. and Tyler, B.M. (2010) External lipid PI‐3‐P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell, 142, 284–295. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. , Hraber, P. , Sobral, B. , Nuss, D. and Govers, F. (1999) Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet. Biol. 28, 94–106. [DOI] [PubMed] [Google Scholar]
- Kasuga, T. , Kozanitas, M. , Bui, M. , Hüberli, D. , Rizzo, D.M. and Garbelotto, M. (2012) Phenotypic diversification is associated with host‐induced transposon derepression in the sudden oak death pathogen Phytophthora ramorum . PLoS ONE, 7, e34728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, M.J. and Gerdemann, J.W. (1958) Root and stem rot of soybeans caused by Phytophthora sojae n. sp. Phytopathology, 48, 201–208. [Google Scholar]
- Kaur, P. , Sivasithamparam, K. and Barbetti, M.J. (2008) Pathogenic behaviour of strains of Albugo candida from Brassica juncea (Indian mustard) and Raphanus raphanistrum (wild radish) in Western Australia. Australas. Plant Pathol. 37, 353–356. [Google Scholar]
- Kemen, E. , Schultz‐Larsen, T. , Kemen, A.C. , Balmuth, A. , Robert‐Seilaniantz, A. , Bailey, K. , Holub, E. , Studholme, D. , MacLean, D. and Jones, J.D.G. (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis. PLoS Biol. 9, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennelly, M.M. , Gadoury, D.M. , Wilcox, W.F. , Magarey, P.A. and Seem, R.C. (2007) Primary infection, lesion productivity, and survival of sporangia in the grapevine downy mildew pathogen Plasmopara viticola . Phytopathology, 97, 512–522. [DOI] [PubMed] [Google Scholar]
- Kida, K. , Tojo, M. , Yano, K. and Kotani, S. (2006) First report of Pythium ultimum var. ultimum causing damping‐off on okra in Japan. Plant Pathol. 56, 1042. [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings, M. , Taylor, T.N. and Dotzler, N. (2011) The fossil record of the Peronosporomycetes (Oomycota). Mycologia, 103, 455–457. [DOI] [PubMed] [Google Scholar]
- Lalancette, N. , Madden, L.V. and Ellis, M.A. (1987) A model for predicting the sporulation of Plasmopara viticola based on temperature and duration of high relative‐humidity. Phytopathology, 77, 1699. [Google Scholar]
- Lamour, K.H. and Hausbeck, M.K. (2000) Mefenoxam insensitivity and the sexual stage of Phytophthora capsici in Michigan cucurbit fields. Phytopathology, 90, 396–400. [DOI] [PubMed] [Google Scholar]
- Lamour, K.H. and Hu, J. (2013) Diversity and Phytophthora: a threat to forests, crops and traditional laboratory research—mini review. CAB Rev. 8, Number 38. [Google Scholar]
- Lamour, K.H. and Kamoun, S. (2009) Oomycete Genetics and Genomics: Diversity, Interactions, and Research Tools. Hoboken, NJ: John Wiley &; Sons. [Google Scholar]
- Lamour, K.H. , Mudge, J. , Gobena, D. , Hurtado‐Gonzales, O.P. , Schmutz, J. , Kuo, A. , Miller, N.A. , Rice, B.J. , Raffaele, S. , Cano, L.M. , Bharti, A.K. , Donahoo, R.S. , Finley, S. , Huitema, E. , Hulvey, J. , Platt, D. , Salamov, A. , Savidor, A. , Sharma, R. , Stam, R. , Storey, D. , Thines, M. , Win, J. , Haas, B.J. , Dinwiddie, D.L. , Jenkins, J. , Knight, J.R. , Affourtit, J.P. , Han, C.S. , Chertkov, O. , Lindquist, E.A. , Detter, C. , Grigoriev, I.V. , Kamoun, S. and Kingsmore, S. (2012) Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici . Mol. Plant–Microbe Interact. 25, 1350–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin, D. and Van den Ackerveken, G. (2013) Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 10, 546–554. [DOI] [PubMed] [Google Scholar]
- Leonian, L.H. (1922) Stem and fruit blight of peppers caused by Phytophthora capsici sp. nov. Phytopathology, 12, 401–408. [Google Scholar]
- Levesque, C.A. , Brouwer, H. , Cano, L. , Hamilton, J.P. , Holt, C. , Huitema, E. , Robideau, G.P. , Thines, M. , Win, J. , Zerillo, M.M. , Beakes, G.W. , Boore, J.L. , Busam, D. , Dumas, B. , Ferriera, S. , Fuerstenberg, S.I. , Gachon, C.M. , Gaulin, E. , Govers, F. , Grenville‐Briggs, L. , Horner, N. , Hostetler, J. , Jiang, R.H. , Johnson, J. , Krajaejun, T. , Lin, H. , Meijer, H.J. , Moore, B. , Morris, P. , Phuntmart, V. , Puiu, D. , Shetty, J. , Stajich, J.E. , Tripathy, S. , Wawra, S. , van West, P. , Whitty, B.R. , Coutinho, P.M. , Henrissat, B. , Martin, F. , Thomas, P.D. , Tyler, B.M. , De Vries, R.P. , Kamoun, S. , Yandell, M. , Tisserat, N. and Buell, C.R. (2010) Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Wang, J. , Chien, D. , Chujoy, E. , Song, B. and VanderZaag, P. (2011) Cooperation‐88: a high yielding, multi‐purpose, late blight resistant cultivar growing in southwest China. Am. J. Pot. Res. 88, 190–194. [Google Scholar]
- Links, M.G. , Holub, E. , Jiang, R.H. , Sharpe, A.G. , Hegedus, D. , Beynon, E. , Sillito, D. , Clarke, W.E. , Uzuhashi, S. and Borhan, M.H. (2011) De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics, 12, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Bos, J.I. , Armstrong, M. , Whisson, S.C. , da Cunha, L. , Torto‐Alalibo, T. , Win, J. , Avrova, A.O. , Wright, F. , Birch, P.R. and Kamoun, S. (2005) Patterns of diversifying selection in the phytotoxin‐like scr74 gene family of Phytophthora infestans . Mol. Biol. Evol. 22, 659–672. [DOI] [PubMed] [Google Scholar]
- Lucas, B. and Griffiths, P.D. (2004) Evaluation of common bean accessions for resistance to Pythium ultimum . Hortscience, 39, 1193–1195. [Google Scholar]
- Lucas, J. , Hayter, J. and Crute, I. (1995) The downy mildews: host specificity and pathogenesis In: Pathogenesis and Host Specificity in Plant Diseases (Singh U. and Singh R., eds), pp. 217–234. Oxford, UK: Permagon. [Google Scholar]
- Madden, L.V. , Hughes, G. and Ellis, M.A. (1995) Spatial heterogeneity of the incidence of grape downy mildew. Phytopathology, 85, 269–275. [Google Scholar]
- Madden, L.V. , Ellis, M.A. , Lalancette, N. , Hughes, G. and Wilson, L.L. (2000) Evaluation of a disease warning system for downy mildew of grapes. Plant Dis. 84, 549–554. [DOI] [PubMed] [Google Scholar]
- Mallard, S. , Cantet, M. , Massire, A. , Bachellez, A. , Ewert, S. and Lefebvre, V. (2013) A key QTL cluster is conserved among accessions and exhibits broad‐spectrum resistance to Phytophthora capsici: a valuable locus for pepper breeding. Mol. Breed. 32, 349–364. [Google Scholar]
- Mammella, M.A. , Cacciola, S.O. , Martin, F. and Schena, L. (2011) Genetic characterization of Phytophthora nicotianae by the analysis of polymorphic regions of the mitochondrial DNA. Fungal Biol. 115, 432–442. [DOI] [PubMed] [Google Scholar]
- Mammella, M.A. , Martin, F.N. , Cacciola, S.O. , Coffey, M.D. , Faedda, R. and Schena, L. (2013) Analyses of the population structure in a global collection of Phytophthora nicotianae isolates inferred from mitochondrial and nuclear DNA sequences. Phytopathology, 103, 610–622. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I. , Salmond, G. and Foster, G.D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F.N. (1995) Electrophoretic karyotype polymorphisms in the genus Pythium . Mycologia, 87, 333–353. [Google Scholar]
- Martin, F.N. and Loper, J.E. (1999) Soilborne plant diseases caused by Pythium spp: ecology, epidemiology, and prospects for biological control. Crit. Rev. Plant Sci. 18, 111–181. [Google Scholar]
- Martin, M.D. , Cappellini, E. , Samaniego, J.A. , Zepeda, M.L. , Campos, P.F. , Seguin‐Orlando, A. , Wales, N. , Orlando, L. , Ho, S.Y.W. , Dietrich, F.S. , Mieczkowski, P.A. , Heitman, J. , Willerslev, E. , Krogh, A. , Ristaino, J.B. and Gilbert, M.T.P. (2013) Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 4, 2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.D. , Ho, S.Y. , Wales, N. , Ristaino, J.B. and Gilbert, M.T.P. (2014) Persistence of the mitochondrial lineage responsible for the Irish potato famine in extant new world Phytophthora infestans . Mol. Biol. Evol. 31, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Matheron, M.E. and Matejka, J.C. (1990) Differential virulence of Phytophthora parasitica recovered from Citrus and other plants to rough lemon and tomato. Plant Dis. 74, 138–140. [Google Scholar]
- McDowell, J.M. (2011) Genomes of obligate plant pathogens reveal adaptations for obligate parasitism. Proc. Natl. Acad. Sci. USA, 108, 8921–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, H.J.G. , Hua, C. , Kots, K. , Ketelaar, T. and Govers, F. (2014) Actin dynamics in Phytophthora infestans; rapidly reorganizing cables and immobile, long‐lived plaques. Cell. Microbiol. 16, 948–961. [DOI] [PubMed] [Google Scholar]
- Meitz, J.C. , Linde, C.C. , Thompson, A. , Langenhoven, S. and McLeod, A. (2010) Phytophthora capsici on vegetable hosts in South Africa: distribution, host range and genetic diversity. Australas. Plant Pathol. 39, 431–439. [Google Scholar]
- Millardet, A. (1881) Notes sur les vignes américaines et opuscules divers sur le même sujet. Bordeaux.
- Moralejo, E.P.‐S.A.M. , Alvarez, L.A. , Belbahri, L. , Lefort, F. and Descals, E. (2009) Multiple alien Phytophthora taxa discovered on diseased ornamental plants in Spain. Plant Pathol. 58, 100–110. [Google Scholar]
- Morris, P.F. and Ward, E.W.B. (1992) Chemoattraction of zoospores of the soybean pathogen Phytophthora‐sojae by isoflavones. Physiol. Molec. Plant Pathol. 40, 17–22. [Google Scholar]
- Mukhtar, M.S. , Carvunis, A.R. , Dreze, M. , Epple, P. , Steinbrenner, J. , Moore, J. , Tasan, M. , Galli, M. , Hao, T. , Nishimura, M.T. , Pevzner, S.J. , Donovan, S.E. , Ghamsari, L. , Santhanam, B. , Romero, V. , Poulin, M.M. , Gebreab, F. , Gutierrez, B.J. , Tam, S , Monachello, D. , Boxem, M. , Harbort, C.J. , McDonald, N. , Gai, L. , Chen, H. , He, Y. ; European Union Effectoromics Consortium , Vandenhaute, J. , Roth, F.P. , Hill, D.E. , Ecker, J.R. , Vidal, M. , Beynon, J. , Braun, P. and Dangl, J.L. (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science, 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminti, P. , Dong, Y. , Sayer, C. , Hay, P. , Rookes, J.E. , Boyd, B. and Cahill, D.M. (2013) Nanostructured liquid crystalline particles as an alternative delivery vehicle for plant agrochemicals. ACS Adv. Mater. Interfaces, 5, 1818–1826. [DOI] [PubMed] [Google Scholar]
- Naseby, D.C. , Pascual, J.A. and Lynch, J.M. (2000) Effect of biocontrol strains of Trichoderma on plant growth, Pythium ultimum populations, soil microbial communities and soil enzyme activities. J. Appl. Microbiol. 88, 161–169. [DOI] [PubMed] [Google Scholar]
- Olson, H.A. and Benson, D.M. (2011) Characterization of Phytophthora spp. on floriculture crops in North Carolina. Plant Dis. 95, 1013–1020. [DOI] [PubMed] [Google Scholar]
- Olson, H.A. , Jeffers, S.N. , Ivors, K.L. , Steddom, K.C. , Williams‐Woodward, J.L. , Mmbaga, M.T. , Benson, D.M. and Hong, C.X. (2013) Diversity and mefenoxam sensitivity of Phytophthora spp. associated with the ornamental horticulture industry in the southeastern United States. Plant Dis. 97, 86–92. [DOI] [PubMed] [Google Scholar]
- Pais, M. , Win, J. , Yoshida, K. , Etherington, G.J. , Cano, L.M. , Raffaele, S. , Banfield, M.J. , Jones, A. , Kamoun, S. and Saunders, D.G.O. (2013) From pathogen genomes to host plant processes: the power of plant parasitic oomycetes. Genome Biol. 14, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peressotti, E. , Wiedemann‐Merdinoglu, S. , Delmotte, F. , Bellin, D. , Di Gaspero, G. , Testolin, R. , Merdinoglu, D. and Mestre, P. (2010) Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 10, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, R.D. , Al‐Mughrabi, K.I. , Kalischuk, M.L. , Dobinson, K. , Conn, K. , Alkher, H.A. , Islam, M.R. , Daayf, F. , Lynn, J. , Bizimungu, B. , De Koeyer, D.L. , Lévesque, A. and Kawchuk, L.M. (2014) Migration and recombination increases Phytophthora infestans population diversity and independent segregation of genotype specific loci. Can. J. Plant Pathol. 36, 73–82. [Google Scholar]
- Petrie, G.A. (1975) Prevalence of oospores of Albugo cruciferarum in Brassica seed samples from western Canada 1967–1973. Can. Plant Dis. Surv. 55, 19–24. [Google Scholar]
- Pieterse, C.M.J. , Risseeuw, E.P. and Davidse, L.C. (1991) An in planta induced gene of Phytophthora infestans codes for ubiquitin. Plant Mol. Biol. 17, 799–811. [DOI] [PubMed] [Google Scholar]
- Ploch, S. and Thines, M. (2011) Obligate biotrophic pathogens of the genus Albugo are widespread as asymptomatic endophytes in natural populations of Brassicaceae. Mol. Ecol. 20, 3692–3699. [DOI] [PubMed] [Google Scholar]
- Quesada‐Ocampo, L.M. and Hausbeck, M.K. (2010) Resistance in tomato and wild relatives to crown and root rot caused by Phytophthora capsici . Phytopathology, 100, 619–627. [DOI] [PubMed] [Google Scholar]
- Qutob, D. , Hraber, P. , Sobral, B. and Gijzen, M. (2000) Comparative analysis of expressed sequences in Phytophthora sojae . Plant Phys. 123, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Kemmerling, B. , Brunner, F. , Kufner, I. , Engelhardt, S. , Gust, A.A. , Luberacki, B. , Seitz, H.U. , Stahl, D. , Rauhut, T. , Glawischnig, E. , Schween, G. , Lacombe, B. , Watanabe, N. , Lam, E. , Schlichting, R. , Scheel, D. , Nau, K. , Dodt, G. , Hubert, D. , Gijzen, M. and Nürnberger, T. (2006) Phytotoxicity and innate immune responses induced by Nep1‐like proteins. Plant Cell, 18, 3721–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qutob, D. , Patrick Chapman, B. and Gijzen, M. (2013) Transgenerational gene silencing causes gain of virulence in a plant pathogen. Nat. Commun. 4, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S. , Farrer, R.A. , Cano, L.M. , Studholme, D.J. , MacLean, D. , Thines, M. , Jiang, R.H.Y. , Zody, M.C. , Kunjeti, S.G. , Donofrio, N.M. , Meyers, B.C. , Nusbaum, C. and Kamoun, S. (2010) Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science, 330, 1540–1543. [DOI] [PubMed] [Google Scholar]
- Randall, E. , Young, V. , Sierotzki, H. , Scalliet, G. , Birch, P.R.J. , Cooke, D.E.L. , Csukai, M. and Whisson, S.C. (2014) Sequence diversity in the large subunit of RNA polymerase I contributes to Mefenoxam insensitivity in Phytophthora infestans . Mol. Plant Pathol. 15, 664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeksting, B.J. , Coetzer, N. , Mahomed, W. , Engelbrecht, J. and van den Berg, N. (2014) De novo sequencing, assembly, and analysis of the root transcriptome of Persea americana (Mill.) in response to Phytophthora cinnamomi and flooding. PLoS ONE, 9, e86399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmany, A.P. , Gordon, A. , Rose, L.E. , Allen, A.E. , Armstrong, M.R. , Whisson, S.C. , Kamoun, S. , Tyler, B.M. , Birch, P.R. and Beynon, J.L. (2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell, 17, 1839–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer, S.R. , Mathur, S. and Wu, C.R. (2000) Virulence of isolates of Albugo candida from western Canada to Brassica species. Can. J. Plant Pathol. 22, 235. [Google Scholar]
- Rizzo, D.M. , Garbelotto, M. and Hansen, E.M. (2005) Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Annu. Rev. Phytopathol. 43, 309–335. [DOI] [PubMed] [Google Scholar]
- Rodewald, J. and Trognitz, B. (2013) Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol. Plant Pathol. 14, 740–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rookes, J.E. , Wright, M.L. and Cahill, D.M. (2008) Elucidation of defence responses and signalling pathways induced in Arabidopsis thaliana following challenge with Phytophthora cinnamomi . Physiol. Mol. Plant Pathol. 72, 151–161. [Google Scholar]
- Rossi, V. and Caffi, T. (2012) The role of rain in dispersal of the primary inoculum of Plasmopara viticola . Phytopathology, 102, 158–165. [DOI] [PubMed] [Google Scholar]
- Rossi, V. , Giosue, S. and Caffi, T. (2009) Modelling the dynamics of infections caused by sexual and asexual spores during Plasmopara viticola epidemics. J. Plant Pathol. 91, 615–627. [Google Scholar]
- Rossi, V. , Caffi, T. and Gobbin, D. (2013) Contribution of molecular studies to botanical epidemiology and disease modelling: grapevine downy mildew as a case‐study. Eur. J. Plant Pathol. 135, 641–654. [Google Scholar]
- Rouxel, M. , Mestre, P. , Comont, G. , Lehman, B. , Schilder, A. and Delmotte, F. (2013) Phylogenetic and experimental evidence for host‐specialized cryptic species in a biotrophic oomycete. New Phytol. 197, 251–263. [DOI] [PubMed] [Google Scholar]
- Roy, S. , Poidevin, L. , Jiang, T. and Judelson, H.S. (2013) Novel core promoter elements in the oomycete pathogen Phytophthora infestans and their influence on expression detected by genome‐wide analysis. BMC Genomics, 14, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadoun, M. and Allagui, M.B. (2008) Pathogenic variability of Phytophthora nicotianae on pepper in tunisia. J. Plant Pathol. 90, 351–355. [Google Scholar]
- Scholthof, K.B.G. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. , Hohn, B. , Saunders, K. , Candresse, T. , Ahlquist, P. , Hemenway, C. and Foster, G. (2011) Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , Huitema, E. , Cano, L.M. , Bozkurt, T.O. , Oliva, R. , van Damme, M. , Schwizer, S. , Raffaele, S. , Chaparro‐Garcia, A. , Farrer, R. , Segretin, M.E. , Bos, J. , Haas, B.J. , Zody, M.C. , Nusbaum, C. , Win, J. , Thines, M. and Kamoun, S. (2009) Ten things to know about oomycete effectors. Mol. Plant Pathol. 10, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack, S. , van Damme, M. , Bozkurt, T.O. , Cano, L.M. , Smoker, M. , Thines, M. , Gaulin, E. , Kamoun, S. and Huitema, E. (2010) Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA, 107, 17 421–17 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, K.L. , Martin, F.N. , de Cock, A.W.A.M. , Levesque, C.A. , Spies, C.F.J. , Okubara, P.A. and Paulitz, T.C. (2013) Molecular detection and quantification of Pythium species: evolving taxonomy, new tools, and challenges. Plant Dis. 97, 4–20. [DOI] [PubMed] [Google Scholar]
- Seidl, M.F. , Van den Ackerveken, G. , Govers, F. and Snel, B. (2011) A domain‐centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 155, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, M.S. , Fernández‐Rebollo, P. , De Vita, P. and Esperanza Sánchez, M. (2013) Calcium mineral nutrition increases the tolerance of Quercus ilex to Phytophthora root disease affecting oak rangeland ecosystems in Spain. Agroforestry Systems, 87, 173–179. [Google Scholar]
- Shaw, D.S. (1983) The perenosporales: a fungal geneticist's nightmare In: Zoosporic Plant Pathogens (Buczacki S.T., ed.), pp. 85–121. London: Academic Press. [Google Scholar]
- Slusarenko, A.J. and Schlaich, N.L. (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Patol. 4, 159–170. [DOI] [PubMed] [Google Scholar]
- Song, J. , Win, J. , Tian, M. , Schornack, S. , Kaschani, F. , Ilyas, M. , van der Hoorn, R.A.L. and Kamoun, S. (2009) Two effectors secreted by unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA, 106, 1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu, S. , Keshavarzi, M. , Brown, I. and Mansfield, J.W. (2003) Ultrastructural characterisation of interactions between Arabidopsis thaliana and Albugo candida . Physiol. Mol. Plant Pathol. 63, 201–211. [Google Scholar]
- Stam, R. , Howden, A.J. , Delgado‐Cerezo, M.T.M.M.M.A. , Motion, G.B. , Pham, J. and Huitema, E. (2013a) Characterization of cell death inducing Phytophthora capsici CRN effectors suggests diverse activities in the host nucleus. Front. Plant Sci. 4, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, R. , Jupe, J. , Howden, A.J. , Morris, J.A. , Boevink, P.C. , Hedley, P.E. and Huitema, E. (2013b) Identification and characterisation of CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE, 8, e59517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanghellini, M.E. and Hancock, J.G. (1971) Sporangium of Pythium ultimum as survival structure in soil. Phytopathology, 61, 157–164. [Google Scholar]
- Stredansky, M. , Conti, E. and Salaris, A. (2000) Production of polyunsaturated fatty acids by Pythium ultimum in solid‐state cultivation. Enzyme Microb. Technol. 26, 304–307. [DOI] [PubMed] [Google Scholar]
- Strullu‐Derrien, C. , Kenrick, P. , Rioult, J.P. and Strullu, D.G. (2011) Evidence of parasitic oomycetes (Peronosporomycetes) infecting the stem cortex of the Carboniferous seed fern Lyginopteris oldhamia . Proc. R. Soc. B, 278, 675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W. , Jia, Y. , O'Neill, N. , Feng, B. and Zhang, X. (2008) Genetic diversity in Phytophthora capsici from eastern China. Can. J. Plant Pathol. 30, 414–424. [Google Scholar]
- Sun, W.X. , Jia, Y.J. , Feng, B.Z. , O'Neill, N.R. , Zhu, X.P. , Xie, B.Y. and Zhang, X.G. (2009) Functional analysis of Pcipg2 from the straminopilous plant pathogen Phytophthora capsici . Genesis, 47, 535–544. [DOI] [PubMed] [Google Scholar]
- Tan, M.Y.A. , Hutten, R.C.B. , Visser, R.G.F. and Eck, H.J. (2010) The effect of pyramiding Phytophthora infestans resistance genes RPi‐mcd1 and RPi‐ber in potato. Theor. Appl. Genet. 121, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R.J. , Pasche, J.S. , Gallup, C.A. , Shew, H.D. and Gudmestad, N.C. (2008) A foliar blight and tuber rot of potato caused by Phytophthora nicotianae: new occurrences and characterization of isolates. Plant Dis. 92, 492–503. [DOI] [PubMed] [Google Scholar]
- Thines, M. (2014) Phylogeny and evolution of plant pathogenic oomycetes, a global overview. Eur. J. Plant Pathol. 138, 431–447. [Google Scholar]
- Thines, M. and Kamoun, S. (2010) Oomycete–plant coevolution: recent advances and future prospects. Curr. Opin. Plant Biol. 13, 427–433. [DOI] [PubMed] [Google Scholar]
- Thines, M. and Spring, O. (2005) A revision of Albugo (chromista, peronosporomycetes). Mycotaxon, 92, 443–458. [Google Scholar]
- Thines, M. , Choi, Y.J. , Kemen, E. , Ploch, S. , Holub, E.B. , Shin, H.D. and Jones, J.D. (2009) A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae). Persoonia, 22, 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, S.E. , Levin, S. and Rodriguez‐Iturbe, I. (2014) Rainfall and temperature changes have confounding impacts on Phytophthora cinnamomi occurrence risk in the southwestern USA under climate change scenarios. Global Change Biol. 20, 1299–1312. [DOI] [PubMed] [Google Scholar]
- Tian, M. , Huitema, E. , Da Cunha, L. , Torto‐Alalibo, T. and Kamoun, S. (2004) A Kazal‐like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis‐related protease P69B. J. Biol. Chem. 279, 26 370–26 377. [DOI] [PubMed] [Google Scholar]
- Töpfer, R. , Hausmann, L. and Eibach, R. (2011) Molecular breeding In: Genetics, Genomics and Breeding of Grapes (Adam‐Blondon A.F., Martinez‐Zapater J.M. and Kole C., eds), pp. 160–185. Enfield: Science Publishers. [Google Scholar]
- Torto, T.A. , Li, S. , Styer, A. , Huitema, E. , Testa, A. , Gow, N.A. , van West, P. and Kamoun, S. (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora . Genome Res. 13, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto‐Alalibo, T. , Tripathy, S. , Smith, B.M. , Arredondo, F. , Zhou, L. , Li, H. , Qutob, D. , Gijzen, M. , Mao, C. , Sobral, B.W.S. , Waugh, M.E. , Mitchell, T.K. , Dean, R.A. and Tyler, B.M. (2007) Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol. Plant–Microbe Interact. 20, 781–793. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. [DOI] [PubMed] [Google Scholar]
- Tyler, B.M. , Tripathy, S. , Zhang, X. , Dehal, P. , Jiang, R.H.Y. , Aerts, A. , Arredondo, F.D. , Baxter, L. , Bensasson, D. , Beynon, J.L. , Chapman, J. , Damasceno, C.M.B. , Dorrance, A.E. , Dou, D. , Dickerman, A.W. , Dubchak, I.L. , Garbelotto, M. , Gijzen, M. , Gordon, S.G. , Govers, F. , Grünwald, N.J. , Huang, W. , Ivors, K.L. , Jones, R.W. , Kamoun, S. , Krampis, K. , Lamour, K.H. , Lee, M.‐K. , McDonald, W.H. , Medina, M. , Meijer, H.J.G. , Nordberg, E.K. , Maclean, D.J. , Ospina‐Giraldo, M.D. , Morris, P.F. , Phuntumart, V. , Putnam, N.H. , Rash, S. , Rose, J.K.C. , Sakihama, Y. , Salamov, A.A. , Savidor, A. , Scheuring, C.F. , Smith, B.M. , Sobral, B.W.S. , Terry, A. , Torto‐Alalibo, T.A. , Win, J. , Xu, Z. , Zhang, H. , Grigoriev, I.V. , Rokhsar, D.S. and Boore, J.L. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science, 313, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Uzuhashi, S. , Tojo, M. and Kakishima, M. (2010) Phylogeny of the genus Pythium and description of new genera. Mycoscience, 51, 337–365. [Google Scholar]
- Vallance, J. , Deniel, F. , Barbier, G. , Guerin‐Dubrana, L. , Benhamou, N. and Rey, P. (2012) Influence of Pythium oligandrum on the bacterial communities that colonize the nutrient solutions and the rhizosphere of tomato plants. Can. J. Microbiol. 58, 1124–1134. [DOI] [PubMed] [Google Scholar]
- Van der Lee, T. , De Witte, I. , Drenth, A. , Alfonso, C. and Govers, F. (1997) AFLP linkage map of the oomycete Phytophthora infestans . Fungal Genet. Biol. 21, 278–291. [DOI] [PubMed] [Google Scholar]
- Van Poucke, K. , Franceschini, S. , Webber, J.F. , Vercauteren, A. , Turner, J.A. , McCracken, A.R. , Heungens, K. and Brasier, C.M. (2012) Discovery of a fourth evolutionary lineage of Phytophthora ramorum: EU2. Fungal Biol. 116, 1178–1191. [DOI] [PubMed] [Google Scholar]
- Van West, P. , Kamoun, S. , van 't Klooster, J.W. and Govers, F. (1999) Internuclear gene silencing in Phytophthora infestans . Mol. Cell, 3, 339–348. [DOI] [PubMed] [Google Scholar]
- Vercesi, A. , Toffolatti, S.L. , Zocchi, G. , Guglielmann, R. and Ironi, L. (2010) A new approach to modelling the dynamics of oospore germination in Plasmopara viticola . Eur. J. Plant Pathol. 128, 113–126. [Google Scholar]
- Viennot‐Bourgin, G. (1949) Les Champignons Parasites des Plantes Cultivées. Paris: Librairies de l'Académie de Médecine. [Google Scholar]
- Vleeshouwers, V.G.A. , Rietman, H. , Krenek, P. , Champouret, N. , Young, C. , Oh, S.‐K. , Wang, M.C. , Bouwmeester, K. , Vosman, B. , Visser, R.G.F. , Jacobsen, E. , Govers, F. , Kamoun, S. and Van der Vossen, E.A.G. (2008) Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE, 3, e2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers, V.G.A. , Raffaele, S. , Vossen, J. , Champouret, N. , Oliva, R. , Segretin, M.E. , Rietman, H. , Cano, L. , Lokossou, A.A. , Kessel, G. , Pel, M.A. and Kamoun, S. (2011) Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49, 507–531. [DOI] [PubMed] [Google Scholar]
- Wang, H.P. and Davis, R.M. (1997) Susceptibility of selected cotton cultivars to seedling disease pathogens and benefits of chemical seed treatments. Plant Dis. 81, 1085–1088. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. , Kale, S.D. , Gu, B. , Sheng, Y. , Sui, Y. , Wang, X. , Zhang, Z. , Cheng, B. , Dong, S. , Shan, W. , Zheng, X. , Dou, D. , Tyler, B.M. and Wang. Y. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland, J.J. (2003) Transformation of Pythium aphanidermatum to geneticin resistance. Curr. Genet. 42, 344–352. [DOI] [PubMed] [Google Scholar]
- Werres, S. , Marwitz, R. , Veld, W. , De Cock, A. , Bonants, P.J.M. , De Weerdt, M. , Themann, K. , Ilieva, E. and Baayen, R.P. (2001) Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum . Mycol. Res. 105, 1155–1165. [Google Scholar]
- Whisson, S.C. , Boevink, P.C. , Moleleki, L. , Avrova, A.O. , Morales, J.G. , Gilroy, E.M. , Armstrong, M.R. , Grouffaud, S. , van West, P. , Chapman, S. , Hein, I. , Toth, I.K. , Pritchard, L. and Birch, P.R.J. (2007) A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature, 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Wirthmueller, L. , Maqbool, A. and Banfield, M.J. (2013) On the front line: structural insights into plant–pathogen interactions. Nat. Rev. Microbiol. 11, 761–776. [DOI] [PubMed] [Google Scholar]
- Wong, F. , Burr, H. and Wilcox, W. (2001) Heterothallism in Plasmopara viticola . Plant Pathol. 50, 427–432. [Google Scholar]
- Wrather, J.A. and Koenning, S.R. (2006) Estimates of disease effects on soybean yields in the United States 2003 to 2005. J. Nematol. 38, 173–180. [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Schuenemann, V. , Cano, L. , Pais, M. , Mishra, B. , Sharma, R. , Kamoun, S. , Krause, J. , Thines, M. , Weigel, D. and Burbano, H. (2013) The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife, 2, e00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K. , Burbano, H.A. , Krause, J. , Thines, M. , Weigel, D. and Kamoun, S. (2014) Mining herbaria for plant pathogen genomes: back to the future. PLoS Pathog. 10, e1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks, J.C. (2008) The potato murrain on the European continent and the revolutions of 1848. Potato Res. 51, 5–45. [Google Scholar]
- Zerillo, M.M. , Adhikari, B.N. , Hamilton, J.P. , Buell, C.R. , Levesque, C.A. and Tisserat, N. (2013) Carbohydrate‐active enzymes in Pythium and their role in plant cell wall and storage polysaccharide degradation. PLoS ONE, 8, e72572. [DOI] [PMC free article] [PubMed] [Google Scholar]


