Summary
‘Gene‐for‐gene’ theory predicts that gain of virulence by an avirulent pathogen on plants expressing resistance (R) genes is associated with fitness loss in susceptible hosts. However, the validity of this prediction has been studied in only a few plant viral pathosystems. In this study, the Soybean mosaic virus (SMV)–Rsv4 pathosystem was exploited to test this prediction. In Rsv4‐genotype soybeans, P3 of avirulent SMV strains provokes an as yet uncharacterized resistance mechanism that restricts the invading virus to the inoculated leaves. A single amino acid substitution in P3 functionally converts an avirulent to a virulent strain, suggesting that the genetic composition of P3 plays a crucial role in virulence on Rsv4‐genotype soybeans. In this study, we examined the impact of gain of virulence mutation(s) on the fitness of virulent variants derived from three avirulent SMV strains in a soybean genotype lacking the Rsv4 gene. Our data demonstrate that gain of virulence mutation(s) by all avirulent viruses on Rsv4‐genotype soybean is associated with a relative fitness loss in a susceptible host. The implications of this finding on the durable deployment of the Rsv4 gene in soybean are discussed.
Keywords: avirulence, fitness penalty, Glycine max, P3, potyvirus, resistance durability
Plants have evolved resistance (R) genes to counter attacks by pathogens, including viruses (Flor, 1971; Hull, 2014; Jones and Dangl, 2006). Structurally and mechanistically known dominant R genes against plant viruses fall into two broad classes. One class contains classical nucleotide‐binding site leucine‐rich repeat (NBS‐LRR) genes with hypersensitive response (HR) or extreme resistance (ER) as the resistance mechanism(s) (Maule et al., 2007). The other class contains genes that differ structurally from NBS‐LRR genes and confer resistance by restricting avirulent viruses to inoculated leaves without the induction of ER or HR (Cosson et al., 2010; Decroocq et al., 2009; Gunduz et al., 2004; Ingvardsen et al., 2010; Khatabi et al., 2012; Saghai Maroof et al., 2010; Wang et al., 2015). Regardless of the class of R gene, or the mechanism of resistance, it has been demonstrated empirically that mutation(s) in a single virus‐encoded protein functionally converts avirulence to virulence on R‐genotype plants (Decroocq et al., 2009; Hajimorad et al., 2011; Khatabi et al., 2012; Wang et al., 2015; Wen et al., 2011, 2013).
‘Gene‐for‐gene’ theory predicts that the modification of a pathogen avirulence factor to gain virulence on an R‐genotype plant results in a fitness loss on susceptible hosts (Flor, 1971). This is based on the theory that plant immune systems have evolved to target crucial pathogen‐encoded proteins and that avirulence proteins are often pathogenicity factors (Dodds and Rathjen, 2010). For viruses, based on theoretical considerations, Sacristan and Garcia‐Arenal (2008) have predicted an even higher fitness cost relative to other cellular pathogens for gain of virulence on R‐genotype plants. This is presumably because of the multifunctional roles played by viral‐encoded proteins in the life cycle of viruses (Hull, 2014). However, for plant–virus pathosystems, in particular potyviral pathosystems, limited direct experimental evidence in support of these assumptions has been published (Fraile et al., 2011; Goulden et al., 1993; Janzac et al., 2010; Jenner et al., 2002; Khatabi et al., 2013; Kobayashi and Hohn, 2004; Rolland et al., 2009). In this study, we utilized the Soybean mosaic virus (SMV)–Rsv4 pathosystem to test whether gain of virulence by three avirulent SMV strains is associated with fitness loss in a susceptible soybean genotype lacking the Rsv4 gene.
SMV is a species within the genus Potyvirus belonging to the Potyviridae family. Its genome is expressed as a single large polypeptide, which is cleaved post‐translationally by three viral‐encoded proteinases to yield a number of multifunctional proteins, including P3 (Urcuqui‐Inchima et al., 2001). In addition, a small open reading frame (ORF) embedded in the P3 cistron (i.e. pipo) encodes a protein in the +2 frame in relation to the polyprotein ORF that plays a role in virus movement (Chung et al., 2008; Vijayapalani et al., 2012; Wei et al., 2010; Wen and Hajimorad, 2010). Rsv4, a single atypical R gene in soybean, belonging to a previously uncharacterized type or class of R gene, mediates the restriction of the systemic movement of avirulent SMV strains to the inoculated leaves of soybean lines V94‐5152 and PI88788 without the expression of ER or HR (Buss et al., 1997; Gunduz et al., 2004; Khatabi et al., 2012; Saghai Maroof et al., 2010; Wang et al., 2015). V94‐5152 is an Essex isoline containing the Rsv4 allele from PI486355 (Buss et al., 1997), and PI88788 contains an allelic R gene to SMV at the Rsv4 locus in V94‐5152 (Gunduz et al., 2004). Essex (rsv4) is a soybean cultivar that is universally susceptible to SMV (Buss et al., 1997). For unknown reasons, the Rsv4 allele in V94‐5152 confers effective resistance against a larger number of SMV strains relative to the corresponding allele in PI88788 (Gunduz et al., 2004; Khatabi et al., 2012; Wang et al., 2015). Regardless, the virulence/avirulence determinants of SMV on Rsv4‐genotype soybeans reside solely on P3; however, the position of crucial virulence site(s) varies among strains (Ahangaran et al., 2013; Chowda‐Reddy et al., 2011; Khatabi et al., 2012; Wang et al., 2015). It has been suggested previously that the three‐dimensional context within which the virulence residue(s) resides is possibly crucial for gain of function (Wang et al., 2015). However, this possibility cannot be explored because the three‐dimensional structure of P3 has not been resolved for any potyvirus.
In this article, ‘fitness’ is defined as the ability of SMV to replicate and propagate in susceptible soybean cv. Essex (rsv4) measured on the basis of disease phenotype and accumulation level of virions (Holland et al. 1991). ‘Virulence’ is defined as the ability to overcome systemic movement restriction in PI88788 (Rsv4) or V94‐5152 (Rsv4), whereas pathogenicity is defined as the capability to induce uniformly distributed mosaic symptoms in soybean cv. Essex (rsv4) coupled with a high level of virion accumulation (Shaner et al., 1992).
Three molecularly cloned SMV strains [SMV‐N (GenBank accession number D00507), SMV‐G7 (AY216010) and SMV‐G7d (AY216987)], as well as the P3 derivative mutants used in this study, have all been described previously (Hajimorad et al., 2003, 2005; Khatabi et al., 2012; Wang et al., 2006, 2015).
SMV‐N is avirulent on V94‐5152 (Rsv4); however, it is virulent on PI88788 (Rsv4) (Fig. 1A; Khatabi et al., 2012). Nevertheless, a mutation at polypeptide position 1033 [glutamine (Gln) to lysine (Lys)] or 1054 [glycine (Gly) to arginine (Arg)], both within P3, confers virulent to avirulent SMV‐N on V94‐5152 (Rsv4) with no impact on its virulence on PI88788 (Rsv4) (Fig. 1A; Khatabi et al., 2012). However, infection of V94‐5152 (Rsv4) with SMV‐NG1054R is associated with a sequence polymorphism at polypeptide position 1045 with the presence of both valine and alanine (Khatabi et al., 2012). As expected, the simultaneous introduction of Q1033K and G1054R into P3 of SMV‐N also results in virulence on V94‐5152 (Rsv4) (Fig. 1A; Khatabi et al., 2012). Ahangaran et al. (2013) have reported that a mutation within P3 at polypeptide position 1053 [serine (Ser) to asparagine (Asn)] also confers virulence on Rsv4‐genotype soybeans to avirulent SMV strains from Iran, although SMV‐NS1053N is avirulent on V94‐5152 (Rsv4) (Fig. 1A; Wang et al., 2015).
Figure 1.
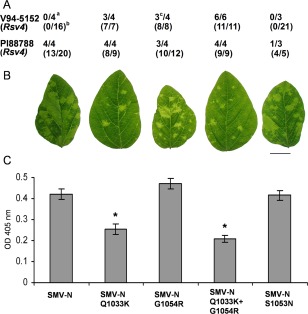
(A) Virulence of SMV‐N and its derivative P3 mutants on Rsv4‐genotype soybeans inoculated biolistically with an infectious cDNA clonea or mechanically with infectious sap derived from biolistically inoculated Essex (rsv4)b; the number of plants systemically infected/number of plants inoculated is shown (Khatabi et al., 2012; Wang et al., 2015). cInfection of V94‐5152 (Rsv4) with SMV‐NG1054R was associated with a sequence polymorphism in P3 of progeny viruses at position 3265, in which both GTA and GCA codons, encoding valine and alanine, respectively, were present at polyprotein position 1045 (Khatabi et al., 2012). (B) Systemic symptoms on central leaflets from trifoliate leaves of soybean cv. Essex (rsv4) inoculated mechanically on unifoliate leaves with progeny viruses derived from cDNA clones. Plants were maintained in a growth chamber at 22°C until being photographed at 21 days post‐inoculation (dpi). Scale bar, 2 cm. (C) Comparison of the accumulation level of viruses relative to SMV‐N in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) by enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated and maintained as in (B) until central leaflets from four fully developed trifoliate leaves of the inoculated plants were harvested at 21 dpi and analysed. Each bar represents the mean value of virion accumulation from two independent experiments, each with four replicate plants, with the standard errors indicated. Significant differences between the mean values for SMV‐N and each of its derivative P3 mutants were determined using Kruskal–Wallis test [non‐parametric statistical method from Data Processing System software (Tang and Zhang, 2013)] and are indicated by asterisks (P < 0.05). OD, optical density; SMV, Soybean mosaic virus.
To examine the impact of the virulence mutation(s) on the fitness of SMV‐N‐derived P3 mutants in the absence of the Rsv4 gene, progenies derived from molecularly cloned viruses were inoculated mechanically onto unifoliate leaves of soybean cv. Essex (rsv4) in parallel with the parental SMV‐N (the details of inoculum source, inoculation procedure and plant maintenance are presented in Materials and Methods S1, see Supporting Information). As expected, all mutants were replication competent in soybean cv. Essex (rsv4) and moved systemically (Fig. 1B). Interestingly, SMV‐NQ1033K and SMV‐NQ1033k+G1054R, both virulent on V94‐5152 (Rsv4), were less pathogenic in soybean cv. Essex (rsv4), evident by the failure to induce uniformly distributed mosaic symptoms on systemically infected leaves in comparison with avirulent SMV‐N or SMV‐NS1053N (Fig. 1B). The level of accumulation of all mutants was compared by enzyme‐linked immunosorbent assay (ELISA) relative to that of the parental SMV‐N in systemically infected leaves of soybean cv. Essex (rsv4) (Fig. 1C) (the details of the ELISA procedures are presented in Materials and Methods S1). SMV‐NQ1033K and SMV‐NQ1033k+G1054R accumulated to a significantly lesser extent than the other viruses. There was no significant difference in the accumulation level of virions from SMV‐NG1054R compared with that of avirulent SMV‐N, despite being virulent on V94‐5152 (Rsv4) (Fig. 1A,C). However, it should be noted that SMV‐NG1054R, unlike SMV‐NQ1033K and SMV‐NQ1033k+G1054R, is not stable in biolistically inoculated V94‐5152 (Rsv4) (Fig. 1A; Khatabi et al., 2012). We also evaluated the stability of all the mutant viruses in soybean cv. Essex (rsv4) after five successive passages by reverse transcription‐polymerase chain reaction (RT‐PCR) amplification of the full‐length P3 cistron and sequencing (the details of RNA extraction, RT‐PCR, sequencing and analysis are presented in Materials and Methods S1). Analysis showed that all mutant viruses, except SMV‐NQ1033k, in which Lys is substituted with Ser, were stable in the absence of the Rsv4 gene (data not shown). To evaluate the stability of SMV‐NQ1033k during the second to fourth passages in Essex (rsv4), P3 of progeny viruses was also RT‐PCR amplified and analysed. The analysis showed the absence of any newly emerged mutation during the second and third passages. However, a sequence polymorphism at polypeptide position 1043 was observed in P3 of progeny viruses derived from the fourth passage, resulting in the presence of both Asn and Ser at this position (data not shown).
To rule out the possibility that the fitness loss of SMV‐N‐derived virulent variants in the absence of the Rsv4 gene was not strain specific, we used two additional avirulent SMV strains. SMV‐G7 and SMV‐G7d are biologically and genetically distinct from SMV‐N, as well as from each other, and are both avirulent on the two Rsv4‐genotype soybeans (Hajimorad et al., 2003, 2005; Khatabi et al., 2012; Wang et al., 2006, 2015). SMV‐G7 and SMV‐G7d encode Gln at polypeptide position 1034, which corresponds to position 1033 on the SMV‐N polypeptide; however, at polypeptide position 1055, corresponding to position 1054 of SMV‐N, both SMV‐G7 and SMV‐G7d encode Ser (Wang et al., 2015). Among SMV‐G7‐derived mutants with a single mutation in P3, only SMV‐G7Q1034K is virulent on both PI88788 (Rsv4) and V94‐5152 (Rsv4), whereas SMV‐G7S1055R and SMV‐G7H1054N are avirulent (Fig. 2A; Wang et al., 2015). SMV‐G7Q1034K+S1055R, harbouring two mutations simultaneously, is also virulent on both Rsv4‐genotype soybeans (Fig. 2A; Wang et al., 2015). When the pathogenicity of SMV‐G7 was compared with that of its derivative P3 mutants in soybean cv. Essex (rsv4), only SMV‐G7Q1034K and SMV‐G7Q1034K+S1055R exhibited infection phenotypes that differed from those induced by the other SMV‐G7‐derived avirulent viruses (Fig. 2B). There was also a direct correlation between gain of virulence mutation on Rsv4‐genotype soybeans and significant reduction of virion accumulation in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) (Fig. 2A,C). Nevertheless, SMV‐G7Q1034K, SMV‐G7S1055R and SMV‐G7Q1034K+S1055R were all stable following five successive passages in soybean cv. Essex (rsv4) without the emergence of any compensatory mutation in their respective P3 cistrons.
Figure 2.
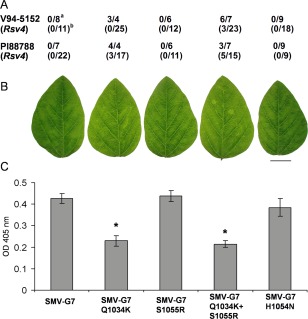
(A) Virulence of SMV‐G7 and its derivative P3 mutants on Rsv4‐genotype soybeans inoculated biolistically with infectious cDNA clonea or mechanically with infectious sap derived from biolistically inoculated Essex (Rsv4)b; the number of plants systemically infected/number of plants inoculated is shown (Wang et al., 2015). (B) Systemic symptoms on central leaflets from trifoliate leaves of soybean cv. Essex (rsv4) inoculated mechanically on unifoliate leaves with progeny viruses derived from cDNA clones. Plants were maintained in a growth chamber at 22°C until being photographed at 21 days post‐inoculation (dpi). Scale bar, 2 cm. (C) Comparison of the accumulation level of viruses relative to SMV‐G7 in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) by enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated and maintained as in (B) until central leaflets from four fully developed trifoliate leaves of the inoculated plants were harvested at 21 dpi and analysed. Each bar represents the mean value of virion accumulation from two independent experiments, each with four replicate plants, with the standard errors indicated. Significant differences between the mean values for SMV‐G7 and each of its derivative P3 mutants were determined using Kruskal–Wallis test [non‐parametric statistical method from Data Processing System software (Tang and Zhang, 2013)] and are indicated by asterisks (P < 0.05). OD, optical density; SMV, Soybean mosaic virus.
Unlike SMV‐G7, in which Gln to Lys substitution at polypeptide position 1034 results in a gain of virulence, SMV‐G7dQ1034K and SMV‐G7dS1055R are both avirulent on PI88788 (Rsv4) and V94‐5152 (Rsv4) (Fig. 3A; Wang et al., 2015). However, SMV‐G7dQ1034K+S1055R, which harbours two simultaneous mutations in P3, is virulent on both Rsv4‐genotype soybeans, albeit to a limited extent (Fig. 3A; Wang et al., 2015). When inoculated onto soybean cv. Essex (rsv4), SMV‐G7dQ1034K and SMV‐G7dQ1034K+S1055R differed phenotypically from the parental SMV‐G7d (Fig. 3B). Furthermore, both mutants accumulated to a significantly lesser extent relative to the parental SMV‐G7d (Fig. 3C). However, all the SMV‐G7d‐derived P3 mutants were stable on five successive passages in soybean cv. Essex (rsv4), except SMV‐G7dQ1034K, in which Lys was replaced with Asn. This substitution was accompanied by an additional compensatory mutation at polyprotein position 768 residing on P3 (E768K). This mutation (E768K) is located outside of pipo. It should be noted that the reversion of Q1034K to Q1034N in P3 of SMV‐G7dQ1034K has also been reported previously in biolistically inoculated Essex (rsv4) (Wang et al., 2015). Analyses of P3 from SMV‐G7dQ1034K‐derived progeny viruses in the second to fourth passages in Essex (rsv4) also showed only the presence of Q1034N.
Figure 3.
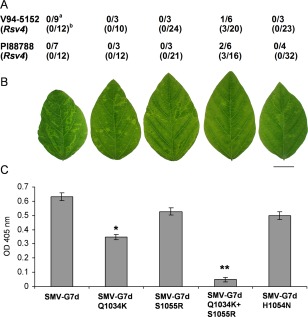
(A) Virulence of SMV‐G7d and its derivative P3 mutants on Rsv4‐genotype soybeans inoculated biolistically with infectious cDNA clonea or mechanically with infectious sap derived from biolistically inoculated Essex (rsv4)b; the number of plants systemically infected/number of plants inoculated is shown (Wang et al., 2015). (B) Systemic symptoms on central leaflets from trifoliate leaves of soybean cv. Essex (rsv4) inoculated mechanically on unifoliate leaves with progeny viruses derived from cDNA clones. Plants were maintained in a growth chamber at 22°C until being photographed at 21 dpi. Scale bar, 2 cm. (C) Comparison of accumulation level of viruses relative to SMV‐G7d in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) by enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated and maintained as in (B) until central leaflets from four fully developed trifoliate leaves of the inoculated plants were harvested at 21 days post‐inoculation (dpi) and analysed. Each bar represents the mean value of virion accumulation from two independent experiments, each with four replicate plants, with the standard errors indicated. Significant differences between the mean values for SMV‐G7d and each of its derivative P3 mutants were determined using Kruskal–Wallis test [non‐parametric statistical method from Data Processing System software (Tang and Zhang, 2013)] and are indicated by single (P < 0.05) or double (P < 0.01) asterisks. OD, optical density; SMV, Soybean mosaic virus.
It is now well established that virulence determinant(s) of SMV on Rsv4‐genotype soybean reside solely on P3, with the genetic composition of P3 playing a crucial role (Ahangaran et al., 2013; Chowda‐Reddy et al., 2011; Khatabi et al., 2012; Wang et al., 2015). An SMV‐G7d‐derived recombinant having the precise P3 cistron from SMV‐G7, SMV‐G7d/G7P3, similar to the parental viruses, is avirulent on both Rsv4‐genotype soybeans (Fig. 4A; Wang et al., 2015). However, SMV‐G7d/G7P3Q1034K and SMV‐G7d/G7P3Q1034K+S1055R are both virulent (Fig. 4A; Wang et al., 2015). Interestingly, the pathogenicity of these two virulent mutants on soybean cv. Essex (rsv4) also differed from those of avirulent SMV‐G7d and SMV‐G7d/G7P3, and both accumulated at significantly lower levels (Fig. 4B,C).
Figure 4.
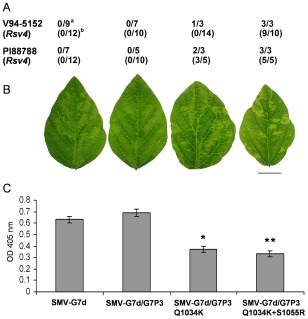
(A) Virulence of SMV‐G7d, recombinant SMV‐G7d/G7P3 and its derivative P3 mutants on Rsv4‐genotype soybeans inoculated biolistically with infectious cDNA clonea or mechanically with infectious sap derived from biolistically inoculated Essex (rsv4)b; number of plants systemically infected/number of plants inoculated is shown (Wang et al., 2015). (B) Systemic infection on central leaflets from trifoliate leaves of soybean cv. Essex (rsv4) inoculated mechanically on unifoliate leaves with progeny viruses derived from cDNA clones. Plants were maintained in a growth chamber at 22°C until being photographed at 21 days post‐inoculation (dpi). Scale bar, 2 cm. (C) Comparison of the accumulation level of viruses relative to SMV‐G7d in systemically infected trifoliate leaves of Essex (rsv4) by enzyme‐linked immunosorbent assay (ELISA). Plants were inoculated and maintained as in (B) until central leaflets from four fully developed trifoliate leaves of the inoculated plants were harvested at 21 dpi and analysed. Each bar represents the mean value of virion accumulation from two independent experiments, each with four replicate plants, with the standard errors indicated. Significant differences between the mean values for SMV‐G7d and each of its derivative P3 mutants were determined using Kruskal–Wallis test [non‐parametric statistical method from Data Processing System software (Tang and Zhang, 2013)] and are indicated by single (P < 0.05) or double (P < 0.01) asterisks. OD, optical density; SMV, Soybean mosaic virus.
Our data demonstrate that gain of virulence on Rsv4‐genotype soybean by avirulent SMV strains is associated with a loss of fitness in a susceptible host. In addition, our data provide experimental evidence that mutation at polypeptide position 1033 is not beneficial for the pathogenicity of SMV, regardless of its direct (SMV‐N and SMV‐G7) or indirect (SMV‐G7d) impact on virulence on Rsv4‐genotype soybeans (Figs 1, 2, 3, 4). This conclusion is further supported by the absence of Lys at polypeptide position 1033 among field isolates of SMV originating from different geographical regions (Ahangaran et al., 2013; Khatabi et al., 2012). It should be noted that, although SMV‐G7dQ1034K was avirulent on Rsv4‐genotype soybean, SMV‐G7d/G7P3Q1034K was virulent (compare Fig. 3 with Fig. 4).
The deployment of R genes is one of the most effective, environmentally friendly and economically sound approaches to control plant diseases caused by viruses. However, the main concern is the durability of R genes, in particular those against plant RNA viruses, because of their potential for a high mutation rate and rapid adaptation (Domingo and Holland, 1997). Nevertheless, the role and significance of avirulence determinants in the life cycle of viruses, the number of mutations required to gain virulence on R‐genotype plants and the high fitness cost in susceptible hosts associated with gain of virulence mutation(s), individually or in combination, contribute to the durability of R genes (Bornemann and Varrelmann, 2013; Harrison, 2002; Janzac et al., 2009, 2010; Jenner et al., 2002; Khatabi et al., 2013; Leach et al., 2001). Durability could also be enhanced by careful selection and deployment of R genes targeting different functional proteins of a virus, as avirulence factors, via a gene stacking approach. In soybean, the complex Rsv1 locus confers functional immunity against the majority of known SMV strains by recognizing two SMV‐encoded proteins, HC‐Pro and P3 (Ahangaran et al., 2013; Eggenberger et al., 2008; Hajimorad et al., 2006, 2011, Khatabi et al., 2012; Seo et al., 2009; Viel et al., 2009; Wen et al., 2013). For gain of virulence of SMV on Rsv1‐genotype soybeans, multiple mutations in HC‐Pro and P3 are required (Eggenberger et al., 2008; Hajimorad et al., 2011; Wen et al., 2013). In addition, a fitness penalty in susceptible hosts is also associated with gain of virulence by SMV on Rsv1‐genotype soybean that is a consequence of mutation(s) in HC‐Pro, but not P3 (Khatabi et al., 2013). In this study, we also demonstrated a fitness penalty in a susceptible soybean genotype associated with gain of virulence by SMV on Rsv4‐genotype soybeans as a consequence of mutation in P3. HC‐Pro and P3 are both multifunctional proteins playing a number of essential roles in the life cycle of potyviruses (Revers and Garcia, 2015). Assuming that the fitness costs of overcoming Rsv1 and Rsv4 by SMV are soybean genotype independent, it is very likely that the stacking of Rsv1 and Rsv4 genes together will result in durable resistance against the virus.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Materials and Methods S1 Details of viruses, soybean genotype, inoculation, maintenance, evaluation of virion accumulation by enzyme‐linked immunosorbent assay (ELISA), RNA extraction, reverse transcription‐polymerase chain reaction (RT‐PCR), sequencing and analysis.
Fig. S1 Comparison of the accumulation level of mutant viruses relative to parental viruses in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) in two independent experiments by enzyme‐linked immunosorbent assay (ELISA).
Acknowledgements
Dr Y. Wang wishes to express his gratitude to Jilin Province Science Foundation (awards 20130101089JC and 20150414044GH) for partial financial support. This project was supported in part by The University of Tennessee Agricultural Experimental Station, Knoxville, TN, USA. The authors are grateful to Dr M. Mazarei of the Plant Sciences Department for assistance with statistical analysis and Dr P. Lambdin of the Entomology and Plant Pathology Department for comments on the manuscript.
References
- Ahangaran, A. , Habibi, M.K. , Mosahebi Mohammadi, G.‐H. , Winter, S. and Garcia‐Arenal, F. (2013) Analysis of Soybean mosaic virus genetic diversity in Iran allows the characterization of a new mutation resulting in overcoming Rsv4‐resistance. J. Gen. Virol. 94, 2557–2568. [DOI] [PubMed] [Google Scholar]
- Bornemann, K. and Varrelmann, M. (2013) Effect of sugar beet genotype on the Beet necrotic yellow vein virus P25 pathogenicity factor and evidence for a fitness penalty in resistance‐breaking strains. Mol. Plant Pathol. 14, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss, G.R. , Ma, G. , Chen, P. and Tolin, S.A. (1997) Registration of V94‐5152 soybean germplasm resistant to soybean mosaic potyvirus. Crop Sci. 37, 1987–1988. [Google Scholar]
- Chowda‐Reddy, R.V. , Sun, H. , Chen H., Poysa, V. , Ling, H. , Gijzen, M. and Wang, A. (2011) Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4‐genotype soybean. Mol. Plant–Microbe Interact. 24, 37–43. [DOI] [PubMed] [Google Scholar]
- Chung, B.Y.W. , Miller, W.A. , Atkins, J.F. and Firth, A.E. (2008) An overlapping essential gene in the potyviridae. Proc. Natl. Acad. Sci. USA, 105, 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, P. , Sofer, L. , Le, Q.H. , Leger, V. , Schurdi‐Levraud, V. , Whitham, S.A. , Yamamoto, M.L. , Gopalan, S. , Gall. O.L. , Candresse, T. , Carrington, J.C. and Reverse, F. (2010) RTM3, which controls long‐distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain‐containing protein. Plant Physiol. 154, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq, V. , Salvador, B. , Sicard, O. , Glasa, M. , Cosson, P. , Svanella‐Dumas, L. , Reverse, F. , Garcia, J.A. and Candresse, T. (2009) The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N‐terminal region of the coat protein. Mol. Plant–Microbe Interact. 22, 1302–1311. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Domingo, E. and Holland, J.J. (1997) RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51, 151–178. [DOI] [PubMed] [Google Scholar]
- Eggenberger, A.L. , Hajimorad, M.R. and Hill, J.H. (2008) Gain of virulence on Rsv1‐genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC‐Pro. Mol. Plant–Microbe Interact. 21, 931–936. [DOI] [PubMed] [Google Scholar]
- Flor, H.H. (1971) Current status of the gene‐for‐gene concept. Annu. Rev. Phytopathol. 9, 275–296. [Google Scholar]
- Fraile, A. , Pagan, I. , Anastasio, G. , Saez, E. and Garcia‐Arenal, F. (2011) Rapid genetic diversification and high fitness penalties associated with pathogenicity evolution in a plant virus. Mol. Biol. Evol. 28, 1425–1437. [DOI] [PubMed] [Google Scholar]
- Goulden, M.G. , Köhm, B.A. , Santa Cruz, S. , Kavanagh, T.A. and Baulcombe, D.C. (1993) A feature of the coat protein of potato virus X affects both induced virus resistance in potato and viral fitness. Virology, 197, 293–302. [DOI] [PubMed] [Google Scholar]
- Gunduz, I. , Buss, G.R. , Chen, P. and Tolin, S.A. (2004) Genetic and phenotypic analysis of Soybean mosaic virus resistance in PI88788 soybean. Phytopathology, 94, 687–692. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2003) Evolution of Soybean mosaic virus‐G7 molecularly cloned genome in Rsv1‐genotype soybean results in emergence of a mutant capable of evading Rsv1‐mediated recognition. Virology, 314, 497–509. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2005) Loss and gain of elicitor function of Soybean mosaic virus G7 provoking Rsv1‐mediated lethal systemic hypersensitive response maps to P3. J. Virol. 79, 1215–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Eggenberger, A.L. and Hill, J.H. (2006) Strain‐specific P3 of Soybean mosaic virus elicits Rsv1‐mediated extreme resistance, but absence of P3 elicitor function alone is insufficient for virulence on Rsv1‐genotype soybean. Virology, 345, 156–166. [DOI] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Wen, R.‐H. , Eggenberger, A.L. , Hill, J.H. and Saghai Maroof, M.A. (2011) Experimental adaptation of an RNA virus mimics natural evolution. J. Virol. 85, 2557–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B.D. (2002) Virus variation in relation to resistance‐breaking in plants. Euphytica, 124, 181–192. [Google Scholar]
- Holland, J.J. , Torre, J.C. , Clarke, D.K. and Duarte, E. (1991) Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65, 2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2014) Plant Virology. New York: Academic Press. [Google Scholar]
- Ingvardsen, C.R. , Xing, Y. , Frei, U.K. and Lubberstedt, T. (2010) Genetic and physical fine mapping of Scmv2, a potyvirus resistance gene in maize. Theor. Appl. Genet. 120, 1621–1634. [DOI] [PubMed] [Google Scholar]
- Janzac, B. , Fabre, F. , Palloix, A. and Moury, B. (2009) Constraints on evolution of virus avirulence factors predict the durability of corresponding plant resistances. Mol. Plant Pathol. 10, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzac, B. , Montarry, J. , Palloix, A. , Navaud, O. and Moury, B. (2010) A point mutation in the polymerase of Potato virus Y confers virulence toward the Pvr4 resistance of pepper and a high competitiveness cost in susceptible cultivar. Mol. Plant–Microbe Interact. 23, 823–830. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E. , Wang, X. , Ponz, F. and Walsh, J.A. (2002) A fitness cost for Turnip mosaic virus to overcome host resistance. Virus Res. 86, 1–6. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Khatabi, B. , Fajolu, O.L. , Wen, R.‐H. and Hajimorad, M.R. (2012) Evaluation of North American isolates of Soybean mosaic virus for gain of virulence on Rsv‐genotype soybeans with special emphasis on resistance‐breaking determinants on Rsv4 . Mol. Plant Pathol. 13, 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatabi, B. , Wen, R.‐H. and Hajimorad, M.R. (2013) Fitness penalty in susceptible host is associated with virulence of Soybean mosaic virus on Rsv1‐genotype soybean: a consequence of perturbation of HC‐Pro and not P3. Mol. Plant Pathol. 14, 885–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, K. and Hohn, T. (2004) The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol. Plant–Microbe Interact. 17, 475–483. [DOI] [PubMed] [Google Scholar]
- Leach, J.E. , Cruz, C.M.V. , Bai, J. and Leung, H. (2001) Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. [DOI] [PubMed] [Google Scholar]
- Maule, A.J. , Caranta, C. and Boulton, M.I. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol. Plant Pathol. 8, 223–231. [DOI] [PubMed] [Google Scholar]
- Revers, F. and Garcia, J.A. (2015) Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. [DOI] [PubMed] [Google Scholar]
- Rolland, M. , Kerlan, C. and Jacquot, E. (2009) The acquisition of molecular determinants involved in potato virus Y necrosis capacity leads to fitness reduction in tobacco plants. J. Gen. Virol. 90, 244–252. [DOI] [PubMed] [Google Scholar]
- Sacristan, S. and Garcia‐Arenal, F. (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai Maroof, M.A. , Tucker, D.M. , Skoneczka, J.A. , Bowman, B.C. , Tripathy, S. and Tolin, S.A. (2010) Fine mapping and candidate gene discovery of the Soybean mosaic virus resistance gene, Rsv4 . Plant Genome, 3, 14–22. [Google Scholar]
- Seo, J.‐K. , Ohshima, K. , Lee, H.‐G. , Son, M. , Choi, H.‐S. , Lee, S.‐H. , Sohn, S.‐H. and Kim, K.‐H. (2009) Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology, 393, 91–103. [DOI] [PubMed] [Google Scholar]
- Shaner, G. , Stromberg, E.L. , Lacy, G.H. , Barker, K.R. and Pirone, T.P. (1992) Nomenclature and concepts of pathogenicity and virulence. Annu. Rev. Phytopathol. 30, 47–66. [DOI] [PubMed] [Google Scholar]
- Tang, Q.Y. and Zhang, C.X. (2013) Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 20, 254–260. [DOI] [PubMed] [Google Scholar]
- Urcuqui‐Inchima, S. , Haenni, A.L. and Bernardi, F. (2001) Potyvirus proteins: a wealth of functions. Virus Res. 74, 157–175. [DOI] [PubMed] [Google Scholar]
- Viel, C. , Ide, C. , Cui, X. , Wang, A. , Farsi, M. , Michelutti, R. and Stromvik, M. (2009) Isolation, partial sequencing, and phylogenetic analyses of Soybean mosaic virus (SMV) in Ontario and Quebec. Can. J. Plant Pathol. 31, 108–113. [Google Scholar]
- Vijayapalani, P. , Maeshima, M. , Nagasaki‐Takekuchi, N. and Miller, W.A. (2012) Interaction of the trans‐frame potyvirus protein P3N‐PIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8, e1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Eggenberger, A. , Hill, J. and Bogdanove, A.J. (2006) Pseudomonas syringae effector avrB confers soybean cultivar‐specific avirulence on Soybean mosaic virus adapted for transgene expression but effector avrPto does not. Mol. Plant–Microbe Interact. 19, 304–312. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Khatabi, B. and Hajimorad, M.R. (2015) Amino acid substitution in P3 of Soybean mosaic virus to convert avirulence to virulence on Rsv4‐genotype soybeans is influenced by the genetic composition of P3. Mol. Plant Pathol. 16, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Zhang, C. , Hong, J. , Xiong, R. , Kasschau, K.D. , Zhou, X. , Carrington, J.C. and Wang, A. (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N‐PIPO. PLoS Pathog. 6, e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R.‐H. and Hajimorad, M.R. (2010) Mutational analysis of the putative pipo of soybean mosaic virus suggests disruption of PIPO protein impedes movement. Virology, 400, 1–7. [DOI] [PubMed] [Google Scholar]
- Wen, R.‐H. , Saghai Maroof, M.A. and Hajimorad, M.R. (2011) Amino acid changes in P3, and not the overlapping pipo‐encoded protein, determine virulence of Soybean mosaic virus on functionally immune Rsv1‐genotype soybean. Mol. Plant Pathol. 12, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, R.‐H. , Khatabi, B. , Ashfield, T. , Saghai Maroof, M.A. and Hajimorad, M.R. (2013) The HC‐Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different genes at the complex Rsv1 locus. Mol. Plant–Microbe Interact. 26, 203–215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Materials and Methods S1 Details of viruses, soybean genotype, inoculation, maintenance, evaluation of virion accumulation by enzyme‐linked immunosorbent assay (ELISA), RNA extraction, reverse transcription‐polymerase chain reaction (RT‐PCR), sequencing and analysis.
Fig. S1 Comparison of the accumulation level of mutant viruses relative to parental viruses in systemically infected trifoliate leaves of soybean cv. Essex (rsv4) in two independent experiments by enzyme‐linked immunosorbent assay (ELISA).


