Summary
Research has established that mutations in highly conserved amino acids of the succinate dehydrogenase (SDH) complex in various fungi confer SDH inhibitor (SDHI) resistance. For Sclerotinia sclerotiorum (Lib.) de Bary, a necrotrophic fungus with a broad host range and a worldwide distribution, boscalid resistance has been attributed to the mutation H132R in the highly conserved SdhD subunit protein of the SDH complex. In our previous study, however, only one point mutation, A11V in SdhB (GCA to GTA change in SdhB), was detected in S. sclerotiorum boscalid‐resistant (BR) mutants. In the current study, replacement of the SdhB gene in a boscalid‐sensitive (BS) S. sclerotiorum strain with the mutant SdhB gene conferred resistance. Compared with wild‐type strains, BR and GSM (SdhB gene in the wild‐type strain replaced by the mutant SdhB gene) mutants were more sensitive to osmotic stress, lacked the ability to produce sclerotia and exhibited lower expression of the pac1 gene. Importantly, the point mutation was not located in the highly conserved sequence of the iron–sulfur subunit of SDH. These results suggest that resistance based on non‐conserved vs. conserved protein domains differs in mechanism. In addition to increasing our understanding of boscalid resistance in S. sclerotiorum, the new information will be useful for the development of alternative antifungal drugs.
Keywords: boscalid, fungicide resistance, Sclerotinia sclerotiorum, SDHI fungicides
Introduction
The plant‐pathogenic fungus Sclerotinia sclerotiorum (Lib.) de Bary is widely distributed and can infect more than 400 species of cultivated plant (Wang et al., 2009). Sclerotinia stem rot caused by S. sclerotiorum seriously reduces the yield and quality of rapeseed. For many years, the control of Sclerotinia stem rot in China has been mainly dependent on the benzimidazole fungicide carbendazim and the dicarboximide fungicide dimethachlon (Kuang et al., 2011). Because of the extensive and repeated application of these fungicides, however, resistance has been found in populations of S. sclerotiorum in China (Kuang et al., 2011; Ma et al., 2009; Shi et al., 2000). The rapid development of fungicide resistance by S. sclerotiorum has also been documented in other countries (Jo et al., 2006; Mueller et al., 2002; Smith et al., 1995).
Boscalid, a small molecule, belongs to a novel fungicide class of succinate dehydrogenase inhibitors (SDHIs) (Stammler et al., 2007). It inhibits fungal respiration by binding to the ubiquinone‐binding site (Q‐site) of the mitochondrial succinate dehydrogenase (SDH) complex II in the electron transport chain, which is a functional part of the tricarboxylic acid cycle (Keon et al., 1991). The mitochondrial SDH complex consists of four subunits: flavoprotein (Fp) (SdhA), iron–sulfur protein (Ip) (SdhB) and two membrane‐anchored proteins (SdhC and SdhD) (Hagerhall, 1997; Ito et al., 2004). Previous studies have demonstrated that the SDH binding site is highly conserved in bacteria and eukaryotes (Horsefield et al., 2006; Huang et al., 2006; Sun et al., 2005).
Because of their single site mode of action, SDH inhibitors (SDHIs) have selected for resistance in various fungi and bacteria. Numerous studies have indicated that mutations in highly conserved amino acid residues located in the SdhB, SdhC and SdhD subunits of SDH are responsible for SDHI resistance (Avenot and Michailides, 2010; Avenot et al., 2008; Matsson and Hederstedt, 2001; McGrath and Miazzi, 2008; Skinner et al., 1998). In Ustilago maydis, replacement of a highly conserved histidine residue [by either tyrosine or leucine (H257Y/L)] located at position 257 in the third cysteine‐rich cluster of the mitochondrial iron–sulfur subunit SdhB is correlated with carboxin resistance (Broomfield and Hargreaves, 1992; Keon et al., 1991). In Mycosphaerella graminicola, carboxin resistance is conferred by the alteration of the equivalent codon at position 267 to either tyrosine or leucine (H267Y; H267L) (Skinner et al., 1998). Boscalid resistance has also been detected in Alternaria alternata, Botrytis cinerea and Podosphaera xanthii (Avenot and Michailides, 2007; Miazzi and McGrath, 2008; Stammler et al., 2007).
For S. sclerotiorum, Glättli et al. (2009) and Stammler et al. (2011) have reported that boscalid field resistance is conferred by the point mutation H132R in SdhD. In our previous study, five boscalid‐resistant (BR) mutants, JK19R1, JK19R2, NT16R1, NT16R2 and CS51R, were obtained by fungicide induction. Interestingly, DNA sequence analysis showed that all BR mutants had only one point mutation A11V (GCA to GTA) in the iron–sulfur protein subunit (SdhB) (Wang et al., 2014). Therefore, the objectives of this study were to confirm whether the mutation confers resistance of S. sclerotiorum to boscalid, and to determine how the mutation affects the biological characteristics of S. sclerotiorum. We also analysed the phylogeny of the SdhB gene and determined whether the A11V mutation is located in a conserved or non‐conserved protein domain.
Results
Validation of the gene replacement mutants
To confirm whether the mutation in the SdhB gene confers resistance to boscalid, we generated targeted gene replacement mutants by transformation of the replacement cassette Trpc‐Neo‐SdhB (Fig. 1A). Gene replacement strains were subjected to polymerase chain reaction (PCR) amplification in order to detect the integration of the left and right portions of the replacement cassette. The primer pairs P9 + P10 and P11 + P12 amplified 1942‐bp and 2587‐bp fragments, respectively, from the gene replacement mutant, but did not amplify any fragments from the parental strain (Fig. 1B,C). When probed with the partial SdhB gene region, the genomic DNA of mutants digested with XhoI had a single 3.1‐kb hybridized DNA fragment instead of the 2.1‐kb fragment found in the parental strain (Fig. 1D). These sizes are consistent with accurate replacement of the SdhB regions. Two mutants, GRM1 and GRM2, were obtained by replacing the SdhB gene in an S. sclerotiorum boscalid‐sensitive (BS) strain with the mutant SdhB gene, and one mutant, GSM, was obtained by replacing the mutant SdhB gene with the SdhB gene from the BS S. sclerotiorum strain.
Figure 1.
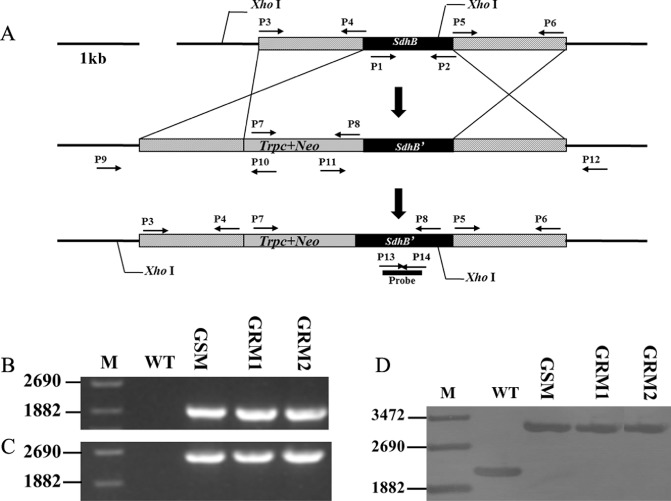
Generation and identification of Sclerotinia sclerotiorum SdhB gene replacement mutants. (A) Schematic representation of the gene replacement strategy. The top part represents the genomic locus target of the replacement construct. Two grey fragments represent the left and right homologous arms, respectively, of the SdhB gene. The black fragment represents the target SdhB gene. The middle part represents the gene replacement cassette Trpc + Neo + SdhB′ connecting the left and right homologous arms of SdhB, and the prime represents the selected SdhB mutation. The cassette Trpc + Neo + SdhB contains the neomycin resistance gene. Primer binding sites are indicated by arrows (see Table 1 for primer sequences). The bottom part of Fig. 1A represents the digested position with Xho1 in the genomic DNA. (B) Polymerase chain reaction (PCR) performed with primer pair P9/P10; a 1942‐bp amplified fragment indicates integration of the replacement cassette Trpc + Neo + SdhB at the left junction. (C) PCR performed with primer pair P11/P12; a 2587‐bp amplified fragment indicates integration of the replacement cassette Trpc + Neo + SdhB at the right junction. (D) Southern blot hybridization analysis of JK19 (WT, wild‐type), GSM, GRM1 and GRM2 using a 400‐bp fragment from the SdhB gene as probe; genomic DNA was digested with Xho1.
Fungicide sensitivity
The EC50 values for boscalid differed significantly (P > 0.05) among the strains (Table 2). As expected, the five BR mutants, GRM1 and GRM2 were resistant to boscalid, with RF (resistance factor: ratio of the EC50 value of the mutant to that of the wild‐type) values between 86.5 and 127.6. The sensitivity to boscalid did not differ significantly among GSM and BS strains. We also assessed the susceptibility to other SDHI fungicides, carboxin and fluopyram. Pearson correlation analysis indicated that boscalid EC50 values were correlated with carboxin and fluopyram EC50 values.
Table 2.
Mycelial growth and sensitivity of Sclerotinia sclerotiorum strains or mutants exposed to succinate dehydrogenase inhibitor (SDHI) fungicides
| Strain or mutant | EC50 (μg/mL) | |||
|---|---|---|---|---|
| Boscalid | Carboxin | Fluopyram | Colony diameter (cm) | |
| JK19 | 0.134f* | 0.883f | 0.113f | 8.09ab |
| NT16 | 0.126f | 0.819f | 0.124f | 7.98b |
| CS51 | 0.158f | 0.932f | 0.098f | 8.15a |
| JK19R1 | 12.7d | 3.52e | 3.65c | 7.32dc |
| JK19R2 | 13.5cd | 4.34bc | 3.42cd | 7.07d |
| NT16R1 | 11.2e | 3.96d | 2.99e | 7.23cd |
| NT16R2 | 10.9e | 4.00d | 3.17de | 7.25cd |
| CS51R | 14.3c | 4.17cd | 3.28d | 7.18cd |
| GSM | 0.153f | 0.915f | 0.155f | 7.34c |
| GRM1 | 15.6b | 4.52b | 4.07b | 8.12a |
| GRM2 | 17.1a | 5.01a | 4.75a | 8.05ab |
*Values in a column followed by the same letter are not significantly different (P > 0.05) according to Fisher's least‐significant difference.
Hyphal growth and sclerotial production
Hyphal growth was slower for the BR and GSM mutants than for the BS strains, but did not differ among GRM1, GRM2 and BS strains (Table 2). In our previous study, BR mutants lacked the ability to produce sclerotia. In the current study, however, the ability of GRM1 and GRM2 to produce sclerotia was similar to that of the BS strains (Fig. 2). Interestingly, GSM lacked the ability to produce sclerotia. These results indicate that the mutation A11V (GCA to GTA) in the SdhB gene does not affect hyphal growth or sclerotial development directly.
Figure 2.
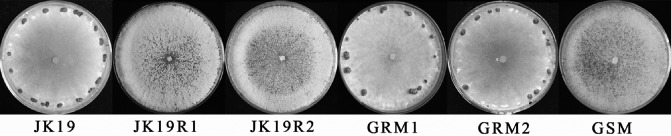
Sclerotial production by Sclerotinia sclerotiorum strains and mutants.
Sensitivity to temperature, osmotic stress and oxidative stress
The BR strain and GSM mutants were more sensitive than the BS strain JK19 to osmotic stress. The sensitivity to osmotic stress, however, did not differ significantly among GRM1, GRM2 and BS strains (Fig. 3). Previous studies have demonstrated that paraquat can induce oxidative stress in organisms by generating superoxide ions in the cytoplasm or mitochondria (Betarbet et al., 2002; Yanase et al., 2002). In the current study, the strains did not differ in their sensitivity to oxidative stress induced by paraquat (data not shown). The strains also did not differ in their sensitivity to temperature (data not shown).
Figure 3.
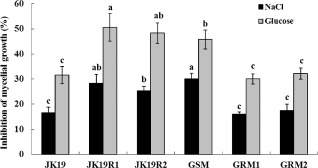
Sensitivity of the wild‐type strain JK19, boscalid‐resistant (BR) mutants (JK19R1 and JK19R2) and gene replacement mutants (GSM, GRM1 and GRM2) of Sclerotinia sclerotiorum to osmotic stress (generated by 0.4 m NaCl or 1 m glucose). Values on the bars followed by the same letter are not significantly different (P > 0.05) according to Fisher's least significant difference.
Phylogenetic analysis and determination of the position of the mutation
The SdhB gene from S. sclerotiorum was compared with the SdhB genes from other selected plant pathogens based on amino acid sequence. A phylogenetic tree was generated by alignment and cluster analysis. This analysis grouped SdhB of S. sclerotiorum in a branch together with SdhB from B. cinerea (AAW52509.1) (Fig. 4). The identities between the S. sclerotiorum SdhB amino acid sequence and that of other plant pathogens in the National Center for Biotechnology Information (NCBI) GenBank database were different, as shown in Fig. 4.
Figure 4.
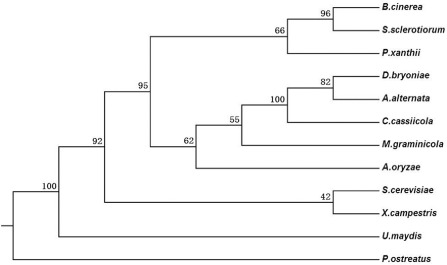
Phylogenetic relationship of the SdhB gene among Sclerotinia sclerotiorum and other plant pathogens based on amino acid sequence. The identities between the S. sclerotiorum SdhB amino acid sequence and that of other plant pathogens are as follows: Botrytis cinerea, 94% identity; Podosphaera xanthii (AB547417), 90% identity; Didymella bryoniae (HQ156462.1), 80% identity; Mycosphaerella graminicola (Mg, AAB97419.1), 77% identity; Pleurotus ostreatus (AB007363.1), 75% identity; Alternaria alternata (B2BZ64), 74% identity; Saccharomyces cerevisiae (Sc, B3LTD3), 73% identity; Corynespora cassiicola (AB548738), 73% identity; Aspergillus oryzae (Ao, Q2TWM0), 71% identity; Xanthomonas campestris SdhB (WP_017113179.1), 69% identity; Ustilago maydis SdhB (GenBank: CAA44612.1), 65% identity.
To determine whether the mutation is located in the highly conserved amino acid sequences, we analysed the conserved domain of the SdhB and SdhD gene in S. sclerotiorum. As shown in Table 3, the conserved amino acids of SdhB in S. sclerotiorum ranged from position 44 to 300, and the conserved amino acids of SdhD in S. sclerotiorum ranged from position 88 to 191 (Table 3). This indicates that the new point mutation A11V is not located in the highly conserved domain of the SdhB subunit, whereas the mutation H132R is located in the highly conserved domain of the SdhD subunit.
Table 3.
Analysis of the conserved domains of the SdhB and SdhD genes in S clerotinia sclerotiorum
| List of domain hitsa | |||||
|---|---|---|---|---|---|
| Subunit | Name | Accession | Descriptionb | Interval | E‐value |
| SdhB | Fer2‐3 | pfam 13085 | 2Fe–2S iron–sulfur cluster binding domain | 71–177 | 2.52e‐39 |
| Fer4‐17 | pfam 13534 | 4Fe–4S dicluster domain | 214–287 | 8.16e‐07 | |
| PLN00129 | PLN00129 | Succinate dehydrogenase [ubiquinone] iron–sulfur subunit | 44–300 | 5.52e‐160 | |
| SdhD | SQR‐TypeC‐Cybs | cd03496 | SQR catalyses the oxidation of succinate to fumarate coupled to the reduction of quinone to quinol | 88–191 | 1.78e‐49 |
Amino acid sequences of the SdhB and SdhD genes in S. sclerotiorum were submitted and analysed on the website http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
More detailed information of the description can be found on the above website.
Gene expression level analysis
In S. sclerotiorum, the pac1 gene (AY005467) is required for sclerotial development; transcription of the smk1 gene (AY351633) and mitogen‐activated protein kinase (MAPK) enzyme activity are dramatically induced during sclerotiogenesis and are required for sclerotial development; and the adenylate cyclase sac1 gene (DQ526020) affects multiple developmental pathways and pathogenicity (Chen et al., 2004; Erental et al., 2007; Jurick and Rollins, 2007; Rollins, 2003). Quantitative reverse transcriptase‐polymerase chain reactiuon (qRT‐PCR) was used to determine whether the expression of pac1, smk1 and sac1 differs among the BR and BS strains and mutants. qRT‐PCR was also used to assess the effect of the point mutation on the expression of the SdhB gene in these strains and mutants. The expression of pac1 was lower in JK19R1, JK19R2 and GSM than in JK19, GRM1 or GRM2 (Fig. 5); the expression of smk1 and sac1 did not differ among these strains and mutants (data not shown). The expression of the SdhB gene was higher in the other five mutants than in JK19 (Fig. 5).
Figure 5.
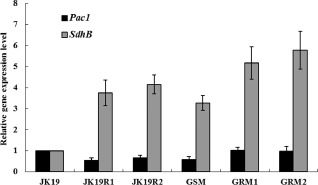
Relative expression levels of the pac1 and SdhB genes in JK19, JK19R1, JK19R2, GSM, GRM1 and GRM2 of Sclerotinia sclerotiorum. Values are means and standard errors.
Discussion
Boscalid resistance has been reported for A. alternata on pistachio, B. cinerea on grapevine and strawberry, P. xanthii on cucurbits, and for other fungi on various crops in several countries. Mutation in SdhB is the most frequent cause of boscalid resistance in B. cinerea (Avenot and Michailides, 2007; Avenot et al., 2008; McGrath, 2008; McGrath and Miazzi, 2008; Miazzi and McGrath, 2008; Stammler, 2008). In S. sclerotiorum, however, boscalid resistance in one field strain resulted from the mutation H132R in the SdhD gene. In our previous study, a new point mutation A11V (GCA to GTA) in the SdhB subunit protein was detected in S. sclerotiorum BR mutants (Wang et al., 2014). Because the SdhB gene in S. sclerotiorum has over 90% identity with the SdhB gene in B. cinerea, this new point mutation in SdhB was not unexpected. To confirm whether the mutation in SdhB confers resistance to boscalid, we replaced the SdhB gene in the sensitive strain with that from the BR mutant in the current study. The gene replacement mutants exhibited significant resistance to boscalid, as well as to carboxin and fluopyram, which was consistent with previous findings of positive cross‐resistance among boscalid, carboxin and fluopyram (Wang et al., 2014). Similarly, replacement of the mutant SdhB gene with the SdhB gene from an S. sclerotiorum BS strain conferred sensitivity. These results indicate that the point mutation A11V in SdhB (as a result of a GCA to GTA change in the SdhB gene) confers resistance not only to boscalid, but also to carboxin and fluopyram, which could be explained by the similar chemical structures of these fungicides. In this research, we also attempted to knock out the SdhB gene from the BR mutant. Unfortunately, no gene knockout mutant was obtained (data not shown). We therefore infer that the SdhB gene is essential for S. sclerotiorum survival and that the A11V mutation leads to a functional SdhB protein.
Numerous studies have shown that the ubiquinone‐binding site in the mitochondrial matrix is highly conserved between fungi and bacteria, and is formed by residues from subunits B, C and D (Horsefield et al., 2006; Sun et al., 2005; Yankovskaya et al., 2003). Previous studies have also demonstrated that mutations in highly conserved amino acids located in the SdhB, SdhC or SdhD subunit of the SDH complex are responsible for SDHI resistance (Avenot and Michailides, 2010; Avenot et al., 2008; Matsson and Hederstedt, 2001; McGrath and Miazzi, 2008; Skinner et al., 1998). Unexpectedly, the new point mutation, A11V in SdhB, is not located in the highly conserved sequence of the iron–sulfur subunit of the SDH complex. A docking model, which has been used to predict the binding pockets formed by the SdhB, SdhC and SdhD subunits in other studies, has also demonstrated that the new mutation is not located in these binding pockets (Fraaije et al., 2012; Scalliet et al., 2012; Shima et al., 2009). In addition, the expression levels of the SdhB gene were higher in BR and GRM mutants than in the parental strain, which may be consistent with three possible hypotheses. First, over‐expression of the SdhB gene may prevent boscalid from reaching the binding site. Second, the amino acid substitution by this mutation may alter the conformation of the binding site of the SDH complex, which, in turn, may reduce the binding affinity to boscalid (Shima et al., 2009). Third, the A11V mutation may reduce the efficiency of the SdhB protein instead of inactivating it, and therefore the cells may be trying to overcompensate for this inferior protein by making more of it. These results suggest that different mechanisms operate for non‐conserved and conserved protein domains to gain resistance.
Sclerotia are hard, asexual, resting structures that facilitate the survival and spread of S. sclerotiorum. Sclerotia of S. sclerotiorum can germinate carpogenically to form apothecia from which ascospores are liberated, or myceliogenically to produce hyphae (Abawi and Grogan, 1979; Steadman, 1979). We have reported previously that the BR mutants induced in our laboratory lacked the ability to produce sclerotia, and thus we suspected that resistance was associated with this inability. The gene replacement mutants GRM1 and GRM2 obtained in this research, however, exhibited no loss in the ability to produce sclerotia, even though they were quite resistant to boscalid. Interestingly, GSM still lacked the ability to produce sclerotia, even though it was quite sensitive to boscalid. As noted earlier, the pac1 gene is required for sclerotial development in S. sclerotiorum, and qRT‐PCR has shown that pac1 expression is lower in BR strains and the GSM mutant than in the wild‐type strain JK19 and GRM mutants. These differences in pac1 expression may account for our observations. Further work concerning the molecular relationship between sclerotium production and the mutation A11V in SdhB is underway in our laboratory.
In previous studies, benomyl resistance has been positively correlated with cold sensitivity in Fusarium moniliforme and Monilinia fructicola (Ma, 2003; Yan and Dickman, 1996). In addition, BR mutants of A. alternata or Saccharomyces cerevisiae were more sensitive to oxidative stress than BS A. alternata or Sa. cerevisiae strains (Avenot et al., 2009; Szeto et al., 2007). In the present study, in contrast, BS and BR strains of S. sclerotiorum did not differ in their sensitivity to temperature or oxidative stress. Compared with BS strains and the GRM mutants, however, BR and GSM mutants grew more slowly, could not produce sclerotia and were more sensitive to osmotic stress. Previous research has also shown that BR mutants exhibited lower virulence than their parental strains (Wang et al., 2014). At least three possible explanations could account for our observations. First, the new point mutation located in the non‐conserved domain of SdhB may reduce S. sclerotiorum fitness. Second, the non‐conserved domain may regulate indirectly a variety of signal transduction pathways in S. sclerotiorum and may have a major impact on biodiversity in biological evolution. Third, altered phenotypes may also be a result of multiple mutations induced in the fungicide‐treated mutants described in our previous study, and further research is still needed. We therefore infer that the conserved domains explain the commonalities among species, whereas the non‐conserved domains explain the differences among species, and that there is a balance between conserved and non‐conserved domains (Dickman and Figueiredo, 2011).
In conclusion, our results indicate that the new point mutation A11V in the SdhB subunit protein confers resistance to boscalid in S. sclerotiorum. Importantly, the point mutation is not located in the highly conserved sequence of the SdhB subunit of the SDH complex. These results suggest that mechanisms differ for resistance arising from non‐conserved vs. conserved protein domains. This information increases our understanding of boscalid resistance in plant‐pathogenic fungi and should be useful for the development of new fungicides.
Experimental Procedures
Fungal strains and media
The strains of S. sclerotiorum used in this study were collected from oilseed rape fields in Jiangsu Province, China during 2010 and 2013. Five BR mutants (JK19R1, JK19R2, NT16R1, NT16R2 and CS51R) were induced from their parental strains (JK19, NT16 and CS51) in our previous study (Wang et al., 2014).
Regeneration medium (RM) was prepared with 0.5 g of yeast extract, 0.5 g of casein hydrolysate, 0.7 m sucrose and 16 g of agar per litre of distilled water. Selective regeneration medium (SRM) was prepared with 0.5 g of yeast extract, 0.5 g of casein hydrolysate, 1 m sucrose and 10 g of agarose per litre of distilled water. Potato dextrose agar (PDA) was prepared with 200 g of potato, 20 g of agar and 20 g of dextrose per litre of distilled water (Duan et al., 2013).
Construction of the SdhB gene replacement vector
The gene replacement vector for the targeted gene was generated as described previously (Laleve et al., 2014; Zheng et al., 2014). First, the primer pair P1 + P2 was used to amplify the entire SdhB gene (Gene ID: SS1G_04384) from the genomic DNA of the S. sclerotiorum BR strain JK19R1. Similarly, primer pairs P3 + P4 and P5 + P6 were used to amplify the sequence 1.0 kb upstream (1.0‐up) and 1.0 kb downstream (1.0‐down) of the SdhB gene, respectively (all primers are listed in Table 1). In addition, primer pair P7 + P8 was used to amplify a 1.2‐kb Neo cassette containing a Trpc promoter (resistance to neomycin) according to a previous study (Duan et al., 2013). The first two fragments (1.2‐kb Trpc + Neo, SdhB) were connected by double‐joint PCR, and then the three fragments (1.0‐up, Trpc + Neo + SdhB and 1.0‐down) were mixed in a 1:3:1 molar ratio and employed as a template for the fusion round, which was performed using La Taq Polymerase (TaKaRa, Dalian, China) without primers (Yu et al., 2004). A 1‐μL volume of product from the second PCR round by the three fragments was used as DNA template to amplify a 4.1‐kb DNA fragment employing the primers P3 + P6. The PCR products were purified with the AxyPreTP DNA Gel Extraction Kit (Axygen, USA), and the entire gene replacement vector was confirmed by sequencing. In addition, we also replaced the mutant SdhB gene with the SdhB gene from the BS S. sclerotiorum strain as described above.
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′) | Use |
|---|---|---|
| P1 | CCTTCTTGACGAGTTCTTCTGAATGGCATCTCTCCGCACAAG | Amplify the full DNA sequence of the SdhB gene (SS1G_04384.3) |
| P2 | TGCTTTCCCATCATATCCC | |
| P3 | AGGTCGCCAGTGCACTGCC | Amplification of the upstream SdhB fragment (1.0 kb) |
| P4 | TTCAATATCATCTTCTGTCGACAGTTGTGGAGAAGGCGAAA | |
| P5 | GGGATATGATGGGAAAGCAGAAGCCTTTAGCGAGATG | Amplification of the downstream SdhB fragment (1.0 kb) |
| P6 | TCCATTCCCACAACAGAAGG | |
| P7 | GTCGACAGAAGATGATATTG | Amplify the Neo cassette containing a Trpc promoter (1165 bp) |
| P8 | TCAGAAGAACTCGTCAAGAAGGCG | |
| P9 | CATCGAAGCGGCCATTCTG | Identification of Trpc‐Neo‐SdhB* cassette integrated at left junction (1942 bp) |
| P10 | CGATGCCTGCTTGCCGAATA | |
| P11 | ACACGGCGGCATCAGAGCAG | Identification of Trpc‐Neo‐SdhB* cassette integrated at right junction (2587 bp) |
| P12 | TGGAGGGCAATTAGATAGTC | |
| P13 | TACCATTTCACCATCTCGTCCTG | Amplify a probe for Southern blotting (400 bp) |
| P14 | GGTCTCGTGCTTGGCGTCT | |
| P15 | CCCCAGCGTTCTACGTCT | Amplify the actin gene (SS1G_08733) for quantitative real‐time polymerase chain reaction |
| P16 | CATGTCAACACGAGCAATG | |
| P17 | TGAACATCGACGGAGTAAA | Amplify the SdhB gene for quantitative real‐time polymerase chain reaction |
| P18 | ATCTTGGTCTCGTGCTTGG | |
| P19 | GCAGCATACCCTAATCTTCC | Amplify the Pac1 gene (AY005467) for quantitative real‐time polymerase chain reaction |
| P20 | GTCCACCAGCACTTCTTTG | |
| P21 | GGGAAGGGAAGACAACAGT | Amplify the Sac1 gene (DQ526020) for quantitative real‐time polymerase chain reaction |
| P22 | TAGTGCAATCGGAATAGTGA | |
| P23 | ATCCGATGATCATTGCCAG | Amplify the Smk1 gene (AY351633) for quantitative real‐time polymerase chain reaction |
| P24 | ATCTAGCAAGACCAAAATCGCA |
Fungal transformation and confirmation of the gene replacement mutants
Transformation was carried out as described previously (Duan et al., 2013). Mycelial plugs taken from the edge of a 2‐day‐old colony of the BS strain JK19 were placed in 250‐mL flasks (five plugs per flask) containing 100 mL of liquid PDB (PDA without agar). After the flasks had been shaken at 175 rpm and 25 °C for 36 h, the mycelia were collected and washed twice with distilled water. Then, 0.1 g of fresh mycelia was treated with 20 mL of protoplast solution (10 mg/mL of lysing enzyme, Sigma, USA) and digested at 85 rpm and 30 °C for 2 h. The enzyme solution was then filtered to eliminate mycelial residues. The protoplasts in the filtrate were washed twice with STC (0.8 m sorbitol, 0.05 m Tris, pH 8.0, 50 mm CaCl2) and resuspended in SPTC (STC with 40% w/v PEG 6000) buffer (STC : SPTC = 4:1). Protoplasts were transformed as described previously (Zheng et al., 2014). For transformation, 107 protoplasts in 200 μL of SPTC buffer and 30 μg gene replacement vector DNA in 10 μL of heparin sodium were mixed and incubated on ice for 30 min; 1 mL SPTC was mixed with the suspension and incubated at room temperature for 20 min. Protoplasts were mixed into 200 mL RM at 43 °C, poured into 9‐cm‐diameter Petri plates (20 mL per plate) and incubated at 25 °C. After 12 h, RM plates were overlaid with 10 mL of SRM containing 100 μg/mL neomycin. Four days post‐transformation, the transformants were transferred to fresh PDA containing 100 μg/mL neomycin.
The total DNA of the transformants was extracted by the cetyltrimethylammonium bromide (CTAB) method (Nicholson and Parry, 1996). Primer pairs P9 + P10 and P11 + P12 (Table 1) were used to amplify the fragment of the connecting area to confirm that the gene replacement vector had been inserted into the site in which SdhB is normally located in the genome, and the PCR products amplified by primer pairs P9 + P12 from transformants genome were sequenced. The transformants were further confirmed by Southern analysis of genomic DNA digested with Xho1 and hybridized with a labelled probe obtained by PCR amplification with the primers P13/P14 (Table 1, Fig. 1A). Three gene replacement mutants, GSM, GRM1 and GRM2, were obtained (see Results).
Fungicide susceptibility test
Sensitivity of the strains listed in Table 2 to the SDHIs boscalid, carboxin, and fluopyram was assessed as previously described (Leroux et al., 1999, 2010; Wang et al., 2009). Fungicide concentrations used for the BR or the gene replacement mutants were 0, 1.56, 3.13, 6.25, 12.5, 25, and 50 μg/mL. Fungicide concentrations used for the BS strains were 0, 0.0625, 0.125, 0.25, 0.5, 1, and 2 μg/mL. After the cultures were incubated at 25 °C for 2 days, the colony diameters were measured, and the EC50 (50% of mycelia growth inhibition) values were calculated by regressing the percentage of growth inhibition against the log of fungicide concentration (Kuang et al., 2011).
Hyphal growth and sclerotial production
Mycelial plugs (5 mm in diameter) taken from the margin of a 2‐day‐old colony of the strains used for the sensitivity test described in the previous section were placed on PDA plates (one plug per plate). Colony diameters were measured perpendicularly after incubation at 25 °C for 2 days. Sclerotial production was assessed as follows: the colonies of the wild‐type strain JK19, BR mutants (JK19R1 and JK19R2) and gene replacement mutants (GSM, GRM1 and GRM2) were photographed 1 month post‐inoculation. Each strain or mutant was represented by three plates, and the experiment was performed three times.
Sensitivity to temperature, osmotic and oxidative stresses
The strains used to test sclerotial production were also used to test the sensitivity to temperature, osmotic and oxidative stresses. Mycelial plugs (5 mm in diameter) taken from the edge of a 2‐day‐old colony were transferred to PDA plates (one plug per 9‐cm‐diameter plate) and incubated at 5, 15 and 35 °C. Plates incubated at 25 °C were used as controls. Colony diameters were measured after 2 days of incubation.
Similar mycelial plugs were also transferred to plates containing PDA amended with 0.4 m NaCl, 1 m glucose or paraquat (1, 10 or 50 μg/mL); NaCl and glucose were used to generate osmotic stress and paraquat was used to generate oxidative stress (Duan et al., 2013; Wang et al., 2014). Plates without NaCl, glucose or paraquat served as controls. The plates were incubated at 25 °C in a growth chamber (12‐h photoperiod) for 2 days before colony diameters were measured. The percentage of inhibition of mycelial radial growth (PIMG) was calculated using the formula, PIMG = [(C – N)/(C – 5)] × 100, where C is the colony diameter of the untreated control and N is that of the NaCl, glucose or paraquat treatment. Each combination of strain and treatment was represented by three plates, and the experiment was performed three times.
Phylogenetic analysis and determination of the position of the mutation
Phylogenetic analysis was carried out as described previously (Chen et al., 2009). Amino acid sequences of the SdhB gene from S. sclerotiorum and other plant pathogens were obtained from the public database GenBank. For phylogenetic analysis, the deduced protein sequences were aligned with ClustalW using the Blosum matrix and standard default parameters. The phylogenetic tree was established using mega software with the neighbour‐joining method, excluding positions with gaps.
The positions of the mutations A11V in the SdhB gene and H132R in the SdhD gene, which conferred resistance to boscalid, were determined. Amino acid sequences of the SdhB and SdhD genes in S. sclerotiorum were submitted and analysed on the website http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. The conserved domains of the SdhB and SdhD genes are listed in Table 3.
qRT‐PCR
Total RNA was extracted with the RNeasy kit (Tiangen Biotech. Co., Beijing, China) from 2‐day‐old mycelia grown in potato sucrose broth. First‐strand cDNA was synthesized with the PrimeScript® RT reagent kit (TaKaRa) (Zheng et al., 2014). qRT‐PCRs were performed with an ABI 7500 real‐time detection system (Applied Biosystems, Foster City, CA, USA). The primers used for qRT‐PCR are listed in Table 1. All data were normalized to actin gene expression, and relative changes in gene expression levels were analysed by ABI 7500 SDS software (Applied Biosystems, Foster City, CA, USA), which automatically sets the baseline. Data from three biological replicates were used to calculate the means and standard deviations.
Acknowledgements
This work was supported by the Special Fund for Agro‐scientific Research in the Public Interest (201103016 and 201303023), the National Natural Science Foundation of China (31401764) and the Natural Science Foundation of Jiangsu Province, China (BK20140679).
References
- Abawi, G.S. and Grogan, R.G. (1979) Epidemiology of diseases caused by Sclerotinia species. Phytopathology, 69, 899–904. [Google Scholar]
- Avenot, H. , Morgan, D.P. and Michailides, T.J. (2008) Resistance to pyraclostrobin, boscalid and multiple resistance to Pristine (pyraclostrobin + boscalid) fungicide in Alternaria alternata causing Alternaria late blight of pistachios in California. Plant Pathol. 57, 135–140. [Google Scholar]
- Avenot, H. , Sellam, A. and Michailides, T.J. (2009) Characterization of mutations in the membrane‐anchored subunits AaSDHC and AaSDHD of succinate dehydrogenase from Alternaria alternata isolates conferring field resistance to the fungicide boscalid. Plant Pathol. 58, 1134–1143. [Google Scholar]
- Avenot, H.F. and Michailides, T.J. (2007) Resistance to boscalid fungicide in Alternaria alternata isolates from pistachio in California. Plant Dis. 91, 1345–1350. [DOI] [PubMed] [Google Scholar]
- Avenot, H.F. and Michailides, T.J. (2010) Progress in understanding molecular mechanisms and evolution of resistance to succinate inhibiting (SDHI) fungicides in phytopathogenic fungi. Crop Prot. 29, 643–651. [Google Scholar]
- Betarbet, R. , Sherer, T.B. and Greenamyre, J.T. (2002) Animal models of Parkinson's disease. Bioessays, 24, 308–318. [DOI] [PubMed] [Google Scholar]
- Broomfield, P.L.E. and Hargreaves, J.A. (1992) A single amino‐acid change in the iron sulphur protein subunit of succinate dehydrogenase confers resistance to carboxin in Ustilago maydis . Curr. Genet. 22, 117–121. [DOI] [PubMed] [Google Scholar]
- Chen, C.B. , Harel, A. , Gorovoits, R. , Yarden, O. and Dickman, M.B. (2004) MAPK regulation of sclerotial development in Sclerotinia sclerotiorum is linked with pH and cAMP sensing. Mol. Plant–Microbe Interact. 17, 404–413. [DOI] [PubMed] [Google Scholar]
- Chen, C.J. , Yu, J.J. , Bi, C.W. , Zhang, Y.N. , Xu, J.Q. , Wang, J.X. and Zhou, M.G. (2009) Mutations in a β‐tubulin confer resistance of Gibberella zeae to benzimidazole fungicides. Phytopathology, 99, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Dickman, M.B. and Figueiredo, P.D. (2011) Comparative pathobiology of fungal pathogens of plants and animals. Plos Pathog. 7, e1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y.B. , Ge, C.Y. , Liu, S.M. , Wang, J.X. and Zhou, M.G. (2013) A two‐component histidine kinase Shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum . Mol. Plant Pathol. 14, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erental, A. , Harel, A. and Yarden, O. (2007) Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum . Mol. Plant–Microbe Interact. 20, 944–954. [DOI] [PubMed] [Google Scholar]
- Fraaije, B.A. , Bayon, C. , Atkins, S. , Cools, H.J. , Lucas, J.A. and Fraaije, M.W. (2012) Risk assessment studies on succinate dehydrogenase inhibitors, the new weapons in the battle to control Septoria leaf blotch in wheat. Mol. Plant Pathol. 13, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glättli, A. , Stammler, G. and Schlehuber, S. (2009) Mutations in the target proteins of succinate‐dehydrogenase inhibitors (SDHI) and 14delta‐demethylase inhibitors (DMI) conferring changes in the sensitivity – structural insights from molecular modelling. In Association Française de Protection des Plantes, 9ème conférence international sur les maladies des plantes, Tours, France, 8 et 9 Décembre 2009. (pp. 670–681). Association Française de Protection des Plantes (AFPP).
- Hagerhall, C. (1997) Succinate: quinone oxidoreductases variations on a conserved theme. Biochim. Biophys. Acta, 1320, 107–141. [DOI] [PubMed] [Google Scholar]
- Horsefield, R. , Yankovskaya, V. , Sexton, G. , Whittingham, W. , Shiomi, K. , Omura, S. , Byrne, B. , Cecchini, G. and Iwata, S. (2006) Structural and computational analysis of the quinone‐binding site of complex II (succinate‐ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 281, 7309–7316. [DOI] [PubMed] [Google Scholar]
- Huang, L.S. , Sun, G. , Cobessi, D. , Wang, A.C. , Shen, J.T. , Tung, E.Y. , Anderson, V.E. and Berry, E.A. (2006) 3‐Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 281, 5965–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y. , Muraguchi, H. , Seshime, Y. , Oita, S. and Yanagi, S.O. (2004) Flutolanil and carboxin resistance in Coprinus cinereus conferred by a mutation in the cytochrome b560 subunit of succinate dehydrogenase complex (complex II). Mol. Genet. Genomics, 272, 328–335. [DOI] [PubMed] [Google Scholar]
- Jo, Y.K. , Niver, A.L. , Rimelspach, J.W. and Boehm, M.J. (2006) Fungicide sensitivity of Sclerotinia homoeocarpa from golf courses in Ohio. Plant Dis. 90, 807–813. [DOI] [PubMed] [Google Scholar]
- Jurick, W.M. and Rollins, J.A. (2007) Deletion of the adenylate cyclase (sac1) gene affects multiple developmental pathways and pathogenicity in Sclerotinia sclerotiorum . Fungal Genet. Biol. 44, 521–530. [DOI] [PubMed] [Google Scholar]
- Keon, J.P.R. , White, G.A. and Hargreaves, J.A. (1991) Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen, Ustilago maydis . Curr. Genet. 19, 475–481. [DOI] [PubMed] [Google Scholar]
- Kuang, J. , Hou, Y.P. , Wang, J.X. and Zhou, M.G. (2011) Sensitivity of Sclerotinia sclerotiorum to fludioxonil: in vitro determination of baseline sensitivity and resistance risk. Crop Prot. 30, 876–882. [Google Scholar]
- Laleve, A. , Gamet, S. , Walker, A.S. , Debieu, D. , Toquin, V. and Fillinger, S. (2014) Site‐directed mutagenesis of the P225, N230 and H272 residues of succinate dehydrogenase subunit B from Botrytis cinerea highlights different roles in enzyme activity and inhibitor binding. Environ. Microbiol. 16, 2253–2266. [DOI] [PubMed] [Google Scholar]
- Leroux, P. , Chapeland, F. , Desbrosses, D. and Gredt, M. (1999) Patterns of cross‐resistance to fungicides in Botryotinia fuckeliana (Botrytis cinerea) isolates from French vineyards. Crop Prot. 18, 687–697. [Google Scholar]
- Leroux, P. , Gredt, M. , Leroch, M. and Walker, A.S. (2010) Exploring mechanisms of resistance to respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl. Environ. Microbiol. 76, 6615–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H.X. , Feng, X.J. , Chen, Y. , Chen, C.J. and Zhou, M.G. (2009) Occurrence and characterization of dimethachlon insensitivity in Sclerotinia sclerotiorum in Jiangsu Province of China. Plant Dis. 93, 36–42. [DOI] [PubMed] [Google Scholar]
- Ma, Z.H. (2003) Identification and characterization of benzimidazole resistance in Monilinia fructicola from stone fruit orchards in California. Appl. Environ. Microb. 69, 7145–7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson, M. and Hederstedt, L. (2001) The carboxin‐binding site on Paracoccus denitrificans succinate:quinone reductase identified by mutations. J. Bioenerg. Biomembr. 33, 99–105. [DOI] [PubMed] [Google Scholar]
- McGrath, M.T. (2008) Fungicide sensitivity in Podosphaera xanthii and efficacy for cucurbit powdery mildew in NY, USA, in 2003–2006. J. Plant Pathol. 90 (Suppl. 2), 90. Abstract. [Google Scholar]
- McGrath, M.T. and Miazzi, M.M. (2008) Sensitivity of Podosphaera xanthii to registered fungicides at‐risk for resistance related to their efficacy for powdery mildew in pumpkin. Phytopathology, 98, S102 (abstract). [Google Scholar]
- Miazzi, M. and McGrath, M.T. (2008) Sensitivity of Podosphaera xanthii to registered fungicides and experimentals in GA and NY, USA, in 2007. J. Plant Pathol. 90 (Suppl. 2), 90. Abstract. [Google Scholar]
- Mueller, D.S. , Dorrance, A.E. , Derksen, R.C. , Ozkan, E. , Kurle, J.E. , Grau, C.R. , Gaska, J.M. , Hartman, G.L. , Bradley, C.A. and Pedersen, W.L. (2002) Efficacy of fungicides on Sclerotinia sclerotiorum and their potential for control of Sclerotinia stem rot on soybean. Plant Dis. 86, 26–31. [DOI] [PubMed] [Google Scholar]
- Nicholson, P. and Parry, D.W. (1996) Development and use of a PCR assay to detect Rhizoctonia cerealis, the cause of sharp eyespot in wheat. Plant Pathol. 45, 872–883. [Google Scholar]
- Rollins, J.A. (2003) The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant–Microbe Interact. 16, 785–795. [DOI] [PubMed] [Google Scholar]
- Scalliet, G. , Bowler, J. , Luksch, T. , Kirchhofer‐Allan, L. , Steinhauer, D. , Ward, K. , Niklaus, M. , Verras, A. , Csukai, M. , Daina, A. and Fonne‐Pfister, R. (2012) Mutagenesis and functional studies with succinate dehydrogenase inhibitors in the wheat pathogen Mycosphaerella graminicola . Plos ONE, 7, e35429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Z.Q. , Zhou, M.G. and Ye, Z.Y. (2000) Resistance of Sclerotinia sclerotiorum to carbendazim and dimethachlon. Chin. J. Oil Crop Sci. 22, 54–57. [Google Scholar]
- Shima, Y. , Ito, Y. , Kaneko, S. , Hatabayashi, H. , Watanabe, Y. , Adachi, Y. and Yabe, K. (2009) Identification of three mutant loci conferring carboxin‐resistance and development of a novel transformation system in Aspergillus oryzae . Fungal Genet. Biol. 46, 67–76. [DOI] [PubMed] [Google Scholar]
- Skinner, W. , Bailey, A. , Renwick, A. , Keon, J. , Gurr, S. and Hargreaves, J. (1998) A single amino‐acid substitution in the iron–sulphur protein subunit of succinate dehydrogenase determines resistance to carboxin in Mycosphaerella graminicola . Curr. Genet. 34, 393–398. [DOI] [PubMed] [Google Scholar]
- Smith, F.D. , Phipps, P.M. , Stipes, R.J. and Brenneman, T.B. (1995) Significance of insensitivity of Sclerotinia minor to Iprodione in control of Sclerotinia blight of peanut. Plant Dis. 79, 517–523. [Google Scholar]
- Stammler, G. (2008) Mode of action, biological performance and latest monitoring results of boscalid sensitivity. In: Abstracts of the 18th Symposium of Research Committee on Fungicide Resistance. (Dehne H.W., Deising H.B., Gisi U., Kuck K.‐H., Russell P.E. and Lyr H., eds), pp. 30–43. Matsue: Phytopathological Society of Japan. [Google Scholar]
- Stammler, G. , Brix, H.D. , Glattli, A. , Semar, M. and Schoefl, U. (2007) Biological properties of the carboxamide boscalid including recent studies on its mode of action. In: Proceedings of the 16th International Plant Protection Congress, Glasgow. (Dehne H.W., Deising H.B., Gisi U., Kuck K.‐H., Russell P.E. and Lyr H., eds), pp. 16–21. Alton, Hampshire, UK: British Crop Protection Council Publications. [Google Scholar]
- Stammler, G. , Glättli, A. , Koch, A. and Schlehuber, S. (2011) Mutations in the target protein conferring resistance to SDHI fungicides. Modern fungicides and antifungal compounds VI. In: Proceedings of the 16th International Reinhardsbrunn Symposium. (Dehne H.W., Deising H.B., Gisi U., Kuck K.H., Russell P.E. and Lyr H., eds), pp. 195–198. Braunschweig, Germany: DPG Selbstverlag. [Google Scholar]
- Steadman, J.R. (1979) Control of plant disease caused by Sclerotinia species. Phytopathology, 69, 904–907. [Google Scholar]
- Sun, F. , Huo, X. , Zhai, Y. et al (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell, 121, 1043–1057. [DOI] [PubMed] [Google Scholar]
- Szeto, S.S.W. , Reinke, S.N. , Sykes, B.D. and Lemire, B.D. (2007) Ubiquinone‐binding site mutations in the Saccharomyces cerevisiae succinate dehydrogenase generate superoxide and lead to the accumulation of succinate. J. Biol. Chem. 37 (Suppl. 282), 27 518–27 526. [DOI] [PubMed] [Google Scholar]
- Wang, J.X. , Ma, H.X. , Chen, Y. , Zhu, X.F. , Yu, W.Y. , Tang, Z.H. , Chen, C.J. and Zhou, M.G. (2009) Sensitivity of Sclerotinia sclerotiorum to boscalid in Jiangsu Province of China. Crop Prot. 28, 882–886. [Google Scholar]
- Wang, Y. , Duan, Y.B. and Zhou, M.G. (2014) Molecular and biochemical characterization of boscalid resistance in laboratory mutants of Sclerotinia sclerotiorum . Plant Pathol. doi: 10.1111/ppa.12246. [DOI] [Google Scholar]
- Yan, K.Y. and Dickman, M. (1996) Isolation of a β‐tubulin gene from Fusarium moniliforme that confers cold‐sensitive benomyl resistance. Appl. Environ. Microb. 62, 3053–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanase, S. , Yasuda, K. and Ishii, N. (2002) Adaptive response to oxidative damage in three mutants of Caenorhabditis elegans (age‐1, mev‐1 and daf‐16) that affect life span. Mech. Ageing Dev. 123, 1579–1587. [DOI] [PubMed] [Google Scholar]
- Yankovskaya, V. , Horsefield, R. , Tornroth, S. et al (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science, 299, 700–704. [DOI] [PubMed] [Google Scholar]
- Yu, J.H. , Hamari, Z. , Han, K.H. , Seo, J.A. , Reyes‐Dominguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 11, 973–981. [DOI] [PubMed] [Google Scholar]
- Zheng, Z.T. , Gao, T. , Zhang, Y. , Hou, Y.P. , Wang, J.X. and Zhou, M.G. (2014) FgFim, a key protein regulating resistance to the fungicide JS399‐19, asexual and sexual development, stress responses and virulence in Fusarium graminearum . Mol. Plant Pathol. 15, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]


