Summary
Cucumber mosaic virus (CMV) 2b suppresses RNA silencing primarily through the binding of double‐stranded RNA (dsRNA) of varying sizes. However, the biologically active form of 2b remains elusive. Here, we demonstrate that the single and double alanine substitution mutants in the N‐terminal 15th leucine and 18th methionine of CMV 2b exhibit drastically attenuated virulence in wild‐type plants, but are efficiently rescued in mutant plants defective in RNA‐dependent RNA polymerase 6 (RDR6) and Dicer‐like 4 (DCL4). Moreover, the transgenic plants of 2b, but not 2blm (L15A/M18A), rescue the high infectivity of CMV‐Δ2b through the suppression of antiviral silencing. L15A, M18A or both weaken 2b suppressor activity on local and systemic transgene silencing. In contrast with the high affinity of 2b to short and long dsRNAs, 2blm is significantly compromised in 21‐bp duplex small interfering RNA (siRNA) binding ability, but maintains a strong affinity for long dsRNAs. In cross‐linking assays, 2b can form dimers, tetramers and oligomers after treatment with glutaraldehyde, whereas 2blm only forms dimers, rather than tetramers and oligomers, in vitro. Together, these findings suggest that L15 and M18 of CMV 2b are required for high affinity to ds‐siRNAs and oligomerization activity, which are essential for the suppression activity of 2b on antiviral silencing.
Keywords: 2b, Cucumber mosaic virus, oligomerization, RNA binding, RNA silencing, suppressor
Introduction
RNA silencing is a conserved and sequence‐specific pathway that triggers RNA degradation, protein translation inhibition, DNA methylation and heterochromatin formation in eukaryotic genomes (Baulcombe, 2004, 2005; Kanazawa, 2008; Malone and Hannon, 2009; Meins et al., 2005; Voinnet, 2005). In addition to regulating eukaryotic development, RNA silencing also contributes to antiviral immunity in plants and invertebrates, as well as in mammals (Ding and Voinnet, 2007; Li et al., 2013; Maillard et al., 2013; Pumplin and Voinnet, 2013). In antiviral silencing pathways, 21–30‐nucleotide‐long viral small interfering RNAs (siRNAs) from viral‐specific double‐stranded RNAs (dsRNAs) are produced by RNaseIII enzymes called Dicers, and these siRNAs are subsequently assembled with Argonaute (AGO) proteins into RNA‐induced silencing complexes (RISCs) that guide the specific cleavage of genomic RNA of invading viruses (Baulcombe, 2005; Ding and Voinnet, 2007; Malone and Hannon, 2009). In addition to Dicer and AGO proteins, cellular RNA‐dependent RNA polymerases (RDRs) are also involved in reinforcing the host silencing responses through the amplification of secondary siRNAs (Andika et al., 2013; Dalmay et al., 2000; Díaz‐Pendón and Ding, 2008; Garcia‐Ruiz et al., 2010; Li et al., 2014; Qu, 2010; Qu et al., 2005; Schwach et al., 2005; Vaistij and Jones, 2009; Wang et al., 2010, 2011; Xie et al., 2001; Ying et al., 2010). To counter RNA silencing and infect hosts effectively, viruses have evolved more than 35 families of viral suppressors of RNA silencing (VSRs) to interfere with various steps of antiviral silencing (Díaz‐Pendón and Ding, 2008; Li and Ding, 2006; Pumplin and Voinnet, 2013). Although VSRs have strikingly diverse sequences and specific functions, they share some similar suppression strategies and biochemical properties (Díaz‐Pendón and Ding, 2008; Ding and Voinnet, 2007; Li and Ding, 2006). For instance, interactions with long dsRNAs and duplex siRNAs are common features of a large number of VSRs (Duan et al., 2012; González et al., 2012; Lakatos et al., 2006). In addition, protein components of silencing processes are also targeted by some VSRs to inhibit RNA silencing (Burgyán and Havelda, 2011; Wu et al., 2010).
Cucumber mosaic virus (CMV) is an economically important pathogen with a broad host range of more than 1200 plant species (Palukaitis and Garcia‐Arenal, 2003). The CMV‐encoded 2b protein was one of the first identified VSRs (Brigneti et al., 1998), and is also an essential pathogenesis factor for viral infection that manifests its activities by a markedly reduced virulence of 2b‐deficient mutant viruses (Díaz‐Pendón and Ding, 2008; Ding et al., 1994, 1995; González et al., 2010; Wang et al., 2010, 2011). The 2b protein is a unique suppressor that exhibits multiple features, including direct interaction with AGO proteins and the binding of short and long dsRNAs, as well as targeting of the nucleus and nucleolus activities (Duan et al., 2012; Goto et al., 2007; Hamera et al., 2012; Zhang et al., 2006). The N‐terminus of the 2b protein contains one or two nuclear localization signals (NLSs) that are required for RNA binding activities (Lucy et al., 2000; Wang et al., 2004). Recently, Duan et al. (2012) demonstrated the detailed characterization of the 2b protein of the CMV SD strain and determined specific domains required for multiple features. In spite of the multiple functions of 2b, only dsRNA binding, but not nuclear targeting or interaction with AGOs, is required for suppression of post‐transcriptional gene silencing (Duan et al., 2012; González et al., 2010, 2012). In the context of viral pathogenicity, however, 2b nuclear localization is indispensable for virulence and inhibition of RNA‐directed DNA methylation (RdDM) (Du et al., 2014; Duan et al., 2012).
To date, the crystal structures of several classical suppressors have been characterized. For instance, tombusvirus p19 forms head‐to‐tail homodimers, such as an ‘siRNA caliper’ that specifically sequesters 21‐bp dsRNAs (Vargason et al., 2003; Ye et al., 2003). The B2 dimeric structure of Flock house virus (FHV) shows affinity to one face of dsRNAs, but not the duplex ends, indicating that B2 binds to dsRNAs independent of their length (Chao et al., 2005). Similar to p19, the NS3 protein of Rice hoja blanca tenuivirus (RHBV) binds to duplex siRNA in head‐to‐tail homodimers (Yang et al., 2011). However, the crystal structure reveals that Tomato aspermy virus (TAV) 2b first forms a dimer and binds to dsRNAs, and then two dimers and dsRNA complexes form tetramers through hydrogen bonds between the conserved leucine‐rich motifs at the 2b N‐terminus (Chen et al., 2008; Hoffmann et al., 2008).
Compared with the L8/I11/L15/M18 signature required for TAV 2b tetramer formation (Chen et al., 2008), only L15 and M18 are well conserved in 2b of CMV subgroup I. In this work, we generated a point mutant in the 2b gene of the CMV Fny strain, in which the conserved L15 and M18 residues were substituted with two alanines (2blm), and examined the active form associated with suppressor activity and viral pathogenicity. We demonstrated that the L15 and M18 residues of 2b are essential for 21‐bp duplex siRNA binding and oligomerization activity, which play pivotal roles in interfering with RDR6‐ and Dicer‐like 4 (DCL4)‐dependent antiviral silencing. Our results also suggest that other amino acids in addition to those of the R‐rich domain of 2b are also required for siRNA binding activity. The possible function of 2b oligomerization in the suppression of RNA silencing is discussed.
Results
Alignment of 2b sequences and construction of CMV 2b point mutants
Although previous studies have revealed many features of CMV 2b in RNA silencing suppression activity, the biologically active form of CMV 2b in the complex is largely unknown. The crystal structure of TAV 2b has revealed that the tetrameric structure consists of four molecules of 2b and two dsRNAs in a complex mediating high suppressor activity (Chen et al., 2008). Although TAV 2b shares a homologous sequence with CMV 2b, the two suppressors possess certain different features in RNA silencing suppression activity and symptom induction. A leucine‐zipper‐like motif (residues L8, I11, L15 and M18) in the N‐terminus of TAV 2b contributes to tetramer formation (Chen et al., 2008). Sequence alignments of the 2b protein between TAV 2b and CMV subgroups I and II show that L15 is conserved in 2b sequences and M18 is only present in the 2b sequences of TAV and CMV subgroup I (Fig. 1A). In previous studies, none of these residues has been shown to be involved in dsRNA binding activity in crystallographic data of TAV 2b (Chen et al., 2008) or in the nucleus and nucleolus localization (Duan et al., 2012).
Figure 1.
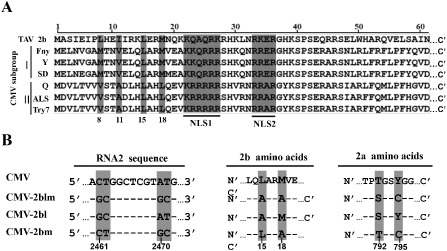
Alignment of 2b protein sequences and schematics showing Cucumber mosaic virus (CMV) 2b point mutants. (A) Alignment of 2b protein sequences from Tomato aspermy virus (TAV) and different CMV subgroup I and II isolates. All of the sequence alignments are from the N‐terminal 61 amino acids of the diverse 2b derivatives. The two nuclear localization signal (NLS) sequences are highlighted in grey and underlined. The N‐terminal eighth, 11th, 15th and 18th amino acids are indicated in grey. (B) The nucleotide substitutions (left panel) in the CMV‐2blm, CMV‐2bl and CMV‐2bm virus mutants and the resulting amino acid point mutations in the 2b (middle panel) and 2a (right panel) regions. The point mutations are highlighted with a grey background and their positions in the genome sequences are indicated at the bottom.
To determine whether the residues are required for viral infectivity, we generated the alanine substitutions of L15 and M18 in the 2b gene of CMV Fny strain (Fig. 1B). As the overlapping sequences of 2b and 2a are in alternative reading frames, the 2b point mutant (L15A/M18A) also results in substitutions of T792 and Y795 with S792 and C795 in the C‐terminus of the 2a protein (Fig. 1B). The single and double point mutant viruses are referred to as CMV‐2bl (L15A), CMV‐2bm (M18A) and CMV‐2blm (L15A/M18A), respectively (Fig. 1B).
CMV‐2blm mutant virus is suppressed by RDR6‐dependent antiviral silencing
To investigate the functions of L15 and M18 in viral infection, we first compared the responses of wild‐type Arabidopsis with single, double and triple null mutants of RDRs (RDR1, RDR2 and RDR6) inoculated with wild‐type CMV and CMV‐2blm, respectively, at 21 days post‐infiltration (dpi) (Fig. 2A). In agreement with previous studies (Wang et al., 2010), wild‐type plants and all the rdr mutant plants developed obvious susceptible symptoms after infection with wild‐type CMV (Fig. 2A). By sharp contrast, CMV‐2blm only induced severe disease symptoms in plants harbouring RDR6‐deficient mutations, including rdr6, rdr1/6, rdr2/6 and rdr1/2/6, but not in wild‐type, rdr1, rdr2 and rdr1/2 plants (Fig. 2A). These results clearly demonstrate that RDR6 has a dominant role in defence against CMV‐2blm. Moreover, similar responses of the rdr6 single mutant with rdr1/6 and the rdr2/6 double mutant to CMV‐2blm challenge suggest that RDR1 and RDR2 do not confer resistance to CMV‐2blm (Fig. 2A).
Figure 2.
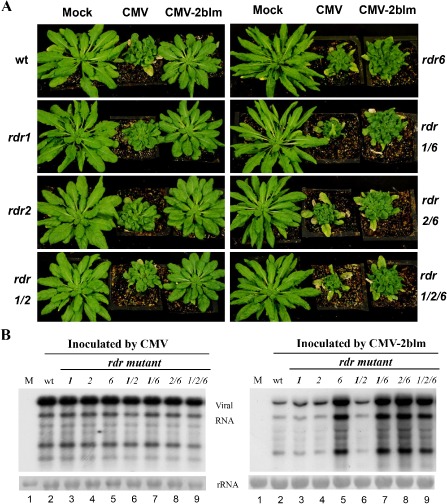
Symptom development and viral accumulation in plants infected with Cucumber mosaic virus (CMV) and CMV‐2blm. (A) CMV‐2blm only induced severe disease symptoms in plants harbouring RDR6‐deficient mutations, including rdr6, rdr1/6, rdr2/6 and rdr1/2/6, but not in wild‐type, rdr1, rdr2 and rdr1/2 plants. Seedlings were photographed at 21 days after inoculation with purified CMV or CMV‐2blm viruses at a concentration of 20 μg/mL. (B) Accumulation of viral genomic RNA in systemically infected leaves of wild‐type and mutant plants infected with CMV or CMV‐2blm. Methylene blue‐stained rRNA was used as a loading control. It should be noted that 5 μg and 2 μg of total RNAs were used for the Northern blot analyses of CMV‐2blm and wild‐type CMV viral RNAs. wt, wild‐type.
We further carried out RNA gel blot hybridizations to compare the accumulation of viral RNAs in wild‐type and mutant plants infected with CMV or CMV‐2blm at 21 dpi. The viral genomic RNA and sub‐genomic RNA (sgRNA) of the wild‐type virus accumulated to similar levels in both wild‐type and various rdr mutant plants (Fig. 2B, left panel), consistent with the symptom observations. However, the presence of rdr6 in either a single mutant or in combination with rdr1 and rdr2 was reproducibly associated with a markedly increased accumulation of CMV‐2blm in the systemically infected leaves (Fig. 2B, right panel, compare lanes 5, 7, 8 and 9 with lanes 2, 3, 4 and 6).
Collectively, these results clearly show that CMV‐2blm is deprived of the ability to suppress RDR6‐dependent antiviral immunity, but still retains the ability to overcome RDR1‐ and RDR2‐dependent secondary silencing or host RDR‐independent primary antiviral silencing. However, the wild‐type CMV can induce severe symptoms and accumulate to high levels regardless of whether RDR6 is functional or inactivated, indicating that CMV 2b can inhibit the RDR6‐mediated antiviral pathway.
DCL4‐mediated secondary siRNA production is required for potent antiviral silencing against CMV‐2blm, CMV‐2bl and CMV‐2bm mutant viruses
During RNA silencing against plus single‐stranded RNA viruses, DCL4 is the dominant antiviral Dicer for the production of 21‐nucleotide viral siRNAs (viRNAs) in wild‐type plants. When DCL4 is genetically inactivated or suppressed, DCL2 rescues antiviral silencing by generating a large amount of 22‐nucleotide viRNAs (Bouche et al., 2006; Deleris et al., 2006; Diaz‐Pendon et al., 2007). Accordingly, the inactivation of both DCL4 and DCL2 is necessary to decrease viRNA production and rescue viral infection (Bouche et al., 2006; Deleris et al., 2006; Diaz‐Pendon et al., 2007).
Here, we inoculated the dcl2, dcl4 and dcl2/4 mutants with CMV‐2blm, and carried out Northern blotting to compare the accumulation levels of viral RNA. CMV‐2blm accumulated to higher levels of viral RNA in rdr6, rdr1/6 and dcl4 than in wild‐type, rdr1 and dcl2 mutant plants (Fig. 3, top panel), suggesting that the RDR6‐ and DCL4‐dependent pathway is required for potent antiviral silencing against CMV‐2blm. However, the dcl2/4 double mutant plants allowed the highest accumulation of CMV‐2blm RNA among all of the other infected mutants. This suggests that DCL2 is functional in primary silencing and/or RDR1‐dependent silencing in DCL4‐inactivated mutant plants (Fig. 3, top panel).
Figure 3.
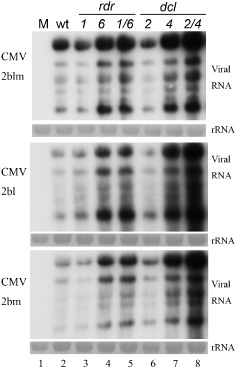
Accumulation of viral genomic RNA in systemic infected leaves of wild‐type and rdr and dcl mutant plants at 21 days after inoculation with 20 μg/mL of CMV‐2bl, CMV‐2bm or CMV‐2blm. Total RNA (5 μg) was used for the detection of viral genomic RNAs by Northern blot analyses. Methylene blue‐stained rRNA was used as a loading control. CMV, Cucumber mosaic virus; wt, wild‐type.
To detect the individual effects of L15 or M18 on viral infection, we also inoculated rdr and dcl mutant plants with CMV‐2bl or CMV‐2bm. In line with the CMV‐2blm results, CMV‐2bl and CMV‐2bm were also inhibited specifically by potent antiviral silencing that is dependent on RDR6 and DCL4, but not RDR1 and DCL2 (Fig. 3, two bottom panels). Consistently, CMV‐2bl and CMV‐2bm induced severe disease symptoms in rdr6 and rdr1/6, rather than in wild‐type and rdr1 plants (Fig. S1, see Supporting Information). These results indicate that both L15 and M18 are necessary for 2b to suppress RDR6‐ and DCL4‐mediated antiviral immunity.
CMV 2b, but not CMV‐2blm, causes developmental defects and rescues the accumulation of CMV‐Δ2b in transgenic Arabidopsis
In addition to the 2blm mutant changes, the substitutions also affect the overlapping portions of the CMV 2a polymerase gene by introducing two amino acid substitutions (Fig. 1B). Although the C‐terminus of 2a is not absolutely required for viral RNA replication, secondary effects of the 2a mutant cannot be ruled out completely (Wang et al., 2010, 2011). To elucidate the distinct features of the 2b and 2blm mutant infection phenotypes, transgenic Arabidopsis plants were generated by transformation with binary vectors in which 2b and 2blm were under the control of the double 35S promoter. In agreement with a previous study (Zhang et al., 2006), the 2b transgenic plants exhibited abnormal development (Fig. 4A). By contrast, the 2blm transgenic plants did not exhibit visible morphological changes and appeared to have the same growth phenotypes as the wild‐type plants and the empty vector transgenic plants (Fig. 4A).
Figure 4.
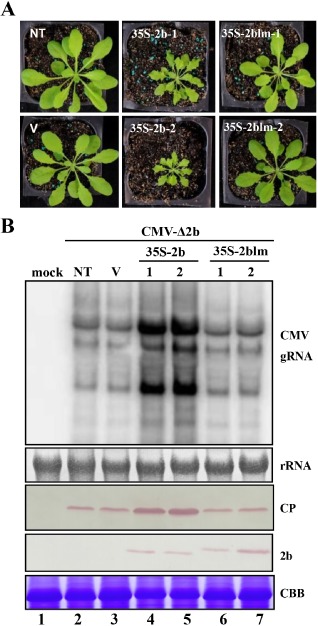
Transgenic expression of 2b, but not 2blm, affects plant development (A) and rescues the high levels of infectivity of CMV‐Δ2b (B). Two independent transgenic 2b and 2blm lines are shown and were mechanically inoculated with CMV‐Δ2b at a concentration of 20 μg/mL. Total RNAs were extracted at 3 weeks after inoculation and used for Northern blotting. The extracted proteins were used in Western blots to assess the accumulation of CMV CP and transgenic 2b proteins. Methylene blue‐stained rRNA and Coomassie brilliant blue (CBB)‐stained protein were used as loading controls for RNA and protein, respectively. NT and V indicate non‐transgenic plants and empty vector transgenic plants, respectively. CMV, Cucumber mosaic virus; CP, coat protein.
To investigate the suppressor activity of the constitutively expressed proteins, two independent transgenic lines of 2b and 2blm were inoculated with CMV‐Δ2b, a virus mutant specifically inhibited by RDR6 in wild‐type plants (Wang et al., 2011). RNA gel hybridization and Western blotting were performed to compare viral RNA accumulation and viral coat protein levels. The results revealed that transgenic 2b rather than transgenic 2blm supported higher accumulation levels of viral RNA and coat protein after infection with CMV‐Δ2b (Fig. 4B, compare lanes 4 and 5 with lanes 6 and 7). These results further demonstrate that the L15 and M18 residues play essential roles in the induction of the symptom phenotype and rescue of CMV‐Δ2b infectivity by interfering with RDR6‐dependent antiviral immunity.
The 2b L15 and M18 residues are critical for the inhibition of transgene‐induced local and systemic RNA silencing
In the absence of virus infection, the silencing suppressor activity of virus‐encoded proteins was identified through the Agrobacterium co‐infiltration assay in Nicotiana benthamiana (Li and Ding, 2006). As RDR1 is naturally mutated in N. benthamiana, RNA silencing induced by the sense green fluorescent protein (GFP) transgene is dominantly dependent on RDR6 (Yang et al., 2004; Ying et al., 2010). Therefore, we examined the suppression of GFP‐induced silencing through co‐infiltration with 2b, 2blm and single mutants 2bl and 2bm. The green fluorescence from the transient expression of GFP co‐infiltrated with the empty vector disappeared quickly, indicating the potent silencing induced by GFP mRNA at 3 dpi (Fig. 5A). In contrast, the higher intensity of GFP fluorescence in plants agroinfiltrated with p19 and 2b at 3 and 6 dpi indicated that GFP silencing is inhibited efficiently by p19 and 2b (Fig. 5A). In the case of the 2b mutants, the GFP fluorescence signals after transient co‐expression with 2bl, 2bm and 2blm decreased substantially from 3 to 6 dpi, and exhibited markedly lower levels than in leaves agroinfiltrated with 2b, but were slightly more intense than the empty vector infiltrations. These results demonstrate that 2bl, 2bm and 2blm are compromised remarkably in suppressor activity compared with 2b, but still retain partial suppressor activity in contrast with the empty vector (Fig. 5A).
Figure 5.
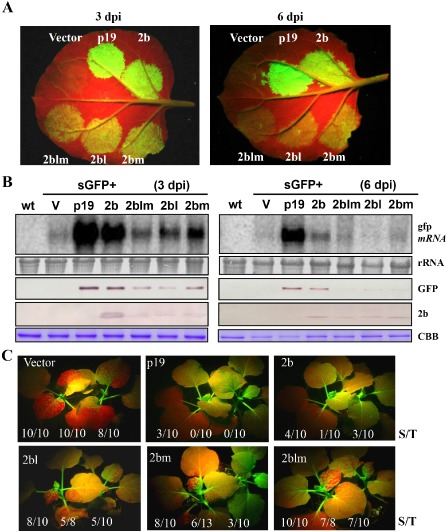
Local and systemic green fluorescent protein (GFP) silencing suppression activity of 2b and 2b mutants in the transient expression systems. (A) Fluorescence in agroinfiltrated leaves of Nicotiana benthamiana for co‐expression of GFP from 2b and 2b mutants at 3 and 6 days post‐infiltration (dpi). The empty vector and p19 were used as negative and positive controls, respectively. (B) Northern and Western blot analysis with samples extracted from co‐infiltrated leaves at 3 and 6 dpi. A GFP DNA probe labelled with [α‐32 P]dCTP was used for the detection of GFP mRNA. Anti‐GFP and anti‐flag antibodies were used to detect the accumulation of GFP and 2b, respectively, in the infiltrated leaves. Methylene blue‐stained rRNA and Coomassie brilliant blue (CBB)‐stained protein were used as loading controls for RNA and protein, respectively. V, vector; wt, wild‐type. (C) Systemic silencing of GFP transgene in 16c plants co‐infiltrated with GFP and 2b or various 2b mutants. The empty vector and p19 were used as negative and positive controls, respectively. The number ratios (S/T) of plants exhibiting systemic silencing (S) among the total number of infiltrated plants (T) in three repeats are shown at the bottom of each panel.
The observation was further verified by Northern blots of GFP mRNA accumulation in the infiltrated patches, as well as the corresponding GFP protein levels. CMV 2b showed increased levels of GFP mRNAs and proteins relative to 2bl, 2bm and 2blm at 3 and 6 dpi (Fig. 5B). However, 2bl, 2bm and 2blm showed slightly increased GFP levels relative to the empty vector (Fig. 5B). The protein levels of the 2b point mutants were also lower than those of 2b, implying that 2b could inhibit the silencing targeted to itself (Fig. 5B). Nonetheless, it cannot be ruled out that the 2b point mutants are less stable than 2b in vivo, resulting in lower accumulation and less efficient suppression activity of RNA silencing.
We further evaluated the effect of 2b and mutants on systemic silencing. The fluorescence derived from transgenic 16c GFP was reproducibly silenced by infiltrated GFP and empty vector in the upper non‐infiltrated leaves, which was suppressed efficiently by co‐expressed p19 at 14 dpi (Fig. 5C). The three independent assays to detect the extent of systemic silencing revealed that 2b inhibits systemic silencing more efficiently than 2blm, 2bl and 2bm (Fig. 5C). In conclusion, 2blm, 2bl and 2bm show substantially compromised suppressor activity of transgene local and systemic silencing.
2blm is compromised in the binding of small duplex RNAs, but maintains high affinity to long dsRNAs
Given that dsRNA binding is important for suppressor activity, we subsequently investigated dsRNA binding by CMV 2b and CVM‐2blm in vitro. Unfortunately, we could not purify the full‐length CMV 2b because of the low expression of 2b in Escherichia coli. Given that previous studies have shown that the N‐terminal 61 amino acids of SD CMV 2b retain complete suppressor activity and dsRNA binding ability (Duan et al., 2012), truncated 2bN61 and 2blmN61 with N‐terminal glutathione‐S‐transferase (GST) fusions were expressed in E. coli and purified for binding assays. GST tag and GST‐p19 were also purified as negative and positive controls for electrophoretic mobility shift assays (EMSAs) (Fig. 6A). GST‐2bN61 exhibited high binding affinity to 21‐bp dsRNAs after the addition of 0.025 μg to the reactions. In the case of GST‐2blmN61, dsRNA binding was only detected in the presence of more than 0.4 μg of GST‐2blmN61 (Fig. 6B), indicating that the bound 21‐bp binding capacity of GST‐2blmN61 was strongly decreased compared with that of GST‐2bN61. The negative GST control did not show detectable dsRNA binding activity in vitro, whereas p19 exhibited high binding activity to the 21‐bp duplex siRNAs (Fig. 6B). Similarly, GST‐2blm, GST‐2bl and GST‐2bm also exhibited a much lower binding to the 21‐bp duplex siRNAs than did GST‐2b (Fig. S2, see Supporting Information). To examine the long dsRNA binding ability of 2bN61 and 2blmN61, 55‐bp dsRNA probes were synthesized and used in the EMSAs. GST‐2bN61 and GST‐2blmN61 exhibited no notable differences in the high affinities to the 55‐bp dsRNAs (Fig. 6C).
Figure 6.
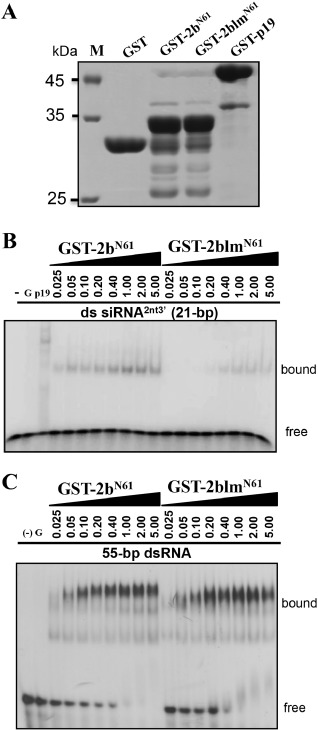
Detection of duplex RNA binding affinity of 2b and 2blm by electrophoretic mobility shift assay (EMSA). (A) Purified glutathione‐S‐transferase (GST)‐tagged proteins used in EMSAs were separated and stained by Coomassie brilliant blue. M, marker. (B) Comparative EMSAs carried out with increasing amounts (0.025–5.00 μg) of GST‐2b or GST‐2blm and a constant amount (1 nm) of [γ‐32P]‐labelled, 21‐bp, double‐stranded (ds), small interfering RNAs (siRNAs) with 2‐nucleotide 3′ overhangs. (C) EMSAs to compare the binding activities of 2b and 2blm with 55‐bp dsRNAs. Bound and free probes are indicated. GST (G) and GST‐p19 were used as negative and positive controls, respectively.
Collectively, our results demonstrate that L15 and M18 are critical for the binding affinity of the 21‐bp duplex siRNAs, and for suppressor activity and symptom induction. Although 2blm exhibits affinities for long dsRNAs, the binding affinity for 21‐bp ds‐siRNAs is much lower. The 2blm mutant is also deficient in suppression of RDR6‐amplified silencing induced by viral infection and the GFP transgene. Therefore, the long dsRNA binding activity of 2b is not critical for suppressor activity and symptom induction.
L15/M18‐dependent oligomerization of CMV 2b in vitro
Chemical cross‐linking is widely used to characterize protein–protein interactions (Sinz, 2006). To gain an insight into the oligomerization activity of 2b and 2blm in vitro, the truncated 2bN61‐His and 2blmN61‐His were expressed and purified for cross‐linking assays. Purified 2bN61‐His and 2blmN61‐His were incubated with 0%, 0.05%, 0.10% and 0.20% of glutaraldehyde solution, and analysed on sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels (Fig. 7A), and confirmed by Western blots with an anti‐His‐tag antibody (Fig. 7B). Most of the untreated 2bN61‐His and 2blmN61‐His were monomers (Fig. 7A,B, lanes 1 and 5). Following treatment with glutaraldehyde solution, 2bN61‐His formed dimer‐ and tetramer‐sized bands, and oligomers were observed in Coomassie brilliant blue‐stained gels and Western blots with the anti‐His antibody (Fig. 7A,B, lanes 2–4). In contrast, the 2blmN61‐His protein primarily formed dimers, rather than tetramers and oligomers, after treatment with glutaraldehyde (Fig. 7A,B, lanes 6–8). Collectively, wild‐type 2b is prone to the formation of dimers, tetramers and oligomers in vitro, whereas 2blm only forms dimers. Therefore, L15 and M18 are required for the oligomerization activity of CMV 2b in vitro.
Figure 7.
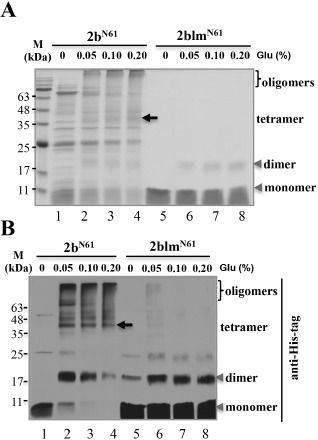
Multimerization of CMV 2b and CMV‐2blm in vitro. The purified 2b and 2blm N‐terminal 61 amino acids were treated with different concentrations of glutaraldehyde (Glu) and separated in sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gels. Samples were detected by Coomassie brilliant blue staining (A) and Western blotting (B) with His antibody to evaluate the oligomeric forms of 2b and 2blm. The monomer and dimer forms are indicated by arrow heads. The tetramer forms are labelled by arrows. The oligomer forms are labelled by brackets. CMV, Cucumber mosaic virus.
Discussion
According to previous studies, the NLS motifs of 2b are immersed in an α‐helix that is also directly involved in dsRNA binding (Chen et al., 2008; Duan et al., 2012). Therefore, it is nearly impossible to uncouple the functions of RNA binding and nuclear localization in viral infection through alteration of the NLS motifs (González et al., 2012). In addition to the NLS regions, conserved P41 and L55 residues of CMV 2b are also required for binding activity and VSR activity (Chen et al., 2008; González et al., 2012; Xu et al., 2013). In the current study, we found that the N‐terminal 15th leucine and 18th methionine of FNY CMV 2b were essential for 21‐bp duplex RNA binding activity, but not for long dsRNA binding activity (Fig. 6). Therefore, our results demonstrate that ds‐siRNA binding, but not long dsRNA binding, has a pivotal role in suppressor activity. Moreover, our results also indicate that the basic amino acid regions at the NLS motifs are not sufficient for effective RNA binding activity of 2b. As 2blm also targets to the nucleus and nucleolus (Fig. S3, see Supporting Information), our results convincingly demonstrate that 21‐bp duplex RNA binding activity, rather than nuclear localization, is required for suppressor activity.
Previous studies have revealed that the self‐interaction of CMV 2b is required for VSR activity and viral symptom induction (González et al., 2010; Xu et al., 2013). Moreover, the B2, p19 and NS3 suppressors have been reported to form dimers that actively contribute to dsRNA duplex binding via different strategies (Chao et al., 2005; Vargason et al., 2003; Yang et al., 2011; Ye et al., 2003). Here, we show that 2blm has weak silencing suppressor activity even though it can form dimers, and some diminished RNA binding activity. Therefore, the dimeric structure is required, but not sufficient, for VSR activity of 2b (Fig. 7). Interestingly, 2blm lacks the ability to form tetramers and oligomers compared with 2b oligomerization. Our results cannot establish the direct effect of tetramers and/or oligomers on 2b suppressor activity. It is unclear at present whether oligomerization of 2b improves the stability of 2b and the dsRNA complex. In previous studies, the tobacco calmodulin‐like protein has been shown to bind to the dsRNA binding domains of 2b and other VSRs, which are then degraded by autophagy‐related systems (Nakahara et al., 2012). Accordingly, it is possible that the tetrameric and/or oligomeric structures facilitate the hiding of the binding domain to escape from degradation systems.
In this study, we also showed that the single substitution mutants, 2bl and 2bm, exhibit similar performance to double point mutants in both the GFP‐associated patch assays and virus infection (Fig. 5). The 2b L15 residue is well conserved in most CMV and TAV strains (Fig. 1A). However, the M18 residue is only present in the 2b region of CMV subgroup I, and is replaced by the L18 amino acid in CMV subgroup II (Fig. 1A). It remains to be investigated whether 2b L18 of CMV subgroup II influences viral pathogenesis. Previous studies have shown that 2b of CMV Q strain, a typical strain of subgroup II, has little effect on the function of miRNA in transgenic plants, probably as a result of its instability or deficiency in the necessary domains for miRNA function (Lewsey et al., 2007; Zhang et al., 2006). In contrast, 2b of CMV Fny strain of subgroup I is very stable and disrupts miRNA function (Lewsey et al., 2007; Zhang et al., 2006). Here, our results imply that 2b of CMV Q strain, similar to 2blm, may form oligomers with low efficiency, resulting in degradation by host‐specific proteases.
Most previous studies of VSRs have been performed to assess the inhibition of silencing induced by exogenous dsRNAs or overexpressed sense transgenes (Li and Ding, 2006). The mechanistic effects of suppressor activity on viral pathogenicity have been largely ignored. However, recent studies are beginning to establish specific genetic roles for VSRs in the silencing of interference pathways through the rescue of VSR‐deficient viruses in specific mutant hosts that are defective in RNA silencing (Ding and Voinnet, 2007). We have shown that either RDR6 or RDR1 mediates secondary silencing sufficiently to confer resistance to CMV‐2aTΔ2b, a mutant deleted in the 2b region and an overlapping region of the 2a gene (Wang et al., 2010). In contrast, the CMV‐Δ2b point mutant with an intact 2a gene is suppressed by RDR6‐dependent antiviral silencing (Wang et al., 2011). These studies have demonstrated that RDR‐mediated silencing exhibits antiviral functions in the absence of 2b expression. In the current study, comparative analyses of CMV and CMV‐2blm directly demonstrated that 2b exerts VSRs through high affinity to ds‐siRNAs amplified by RDR6 and DCL4 (Figs 2 and 3). However, the diminished ds‐siRNA binding ability of 2blm is not able to sequester the secondary siRNAs away from RISC. Given that the basic amino acid motifs close to NLS regions confer the main binding ability of 2b, the low oligomerization of 2blm may directly weaken ds‐siRNA binding activity or independently affect the stability of the 2b protein. Nonetheless, 2blm still maintains compromised dsRNA binding and partial VSR activities through its dimeric structure. Indeed, CMV‐2blm could overcome primary silencing and RDR1‐dependent secondary silencing.
In summary, our data indicate that wild‐type CMV 2b has high ds‐siRNA affinity and favours tetrameric and oligomeric structures. Both features require the presence of L15 and M18 amino acids and exert essential roles in 2b suppressor activities and viral virulence, cooperatively or independently. An alternative explanation is that the secondary viral siRNAs amplified by RDR6 and DCL4 are sequestered by 2b oligomers more efficiently than by dimeric 2blm. Accordingly, the CMV‐2blm strain is only rescued in the presence of inactivated mutants of RDR6 and DCL4. This feature of CMV‐2blm will facilitate future genetic studies to screen novel genes required for RDR6‐ and DCL4‐dependent immunity.
Experimental Procedures
Plant materials
Arabidopsis thaliana mutant lines rdr1‐1, rdr2‐1, rdr6‐15, dcl2‐1 and dcl4‐2, and their double and triple mutants, are all in the Columbia (Col) ecotype, as described previously (Wang et al., 2011). CMV Fny 2b and 2blm transgenic plants were transformed with a binary vector harbouring 2b or 2blm fused with the N‐terminus of a 6 × Myc tag under the control of the 35S promoter. A line was also transformed with the empty vector as a negative control. Arabidopsis thaliana wild‐type, mutants and transgenic lines were vernalized in the dark at 4 °C, and then transferred into a growth room at 22–23 °C with a 10 h/14 h light/dark cycle. The N. benthamiana transgenic line 16c constitutively expresses mGFP as described previously (Baulcombe, 2004). Wild‐type N. benthamiana and 16c plants were grown in a growth room at 25 °C under a 16 h/8 h light/dark cycle.
Virus inoculation
CMV‐2blm was obtained by introducing four point mutations at nucleotides 2461, 2462, 2470 and 2471 in the cDNA clone of CMV RNA2, which resulted in an L15A/M18A mutation in the 2b coding region, and T792 Y795 to S792 C795 substitutions in the 2a C‐terminal region (Fig. 1). To obtain CMV‐2blm, Fny209 was used as a template to amplify a mutated fragment with primers 2blm‐F and 2blm‐R (Table 1), and then self‐ligated to obtain the Fny209‐2blm mutant. The in vitro transcripts of RNA1, RNA3 and RNA2‐2blm were inoculated on Nicotiana clevelandii for propagation of CMV‐2blm virions. The single mutant CMV‐2bl and CMV‐2bm mutants were constructed by point mutations with different primers (Table 1). For 2b detection, we fused a Flag tag to the C‐terminus of 2b and 2blm in the virus context. Purified virions were propagated in N. clevelandii and used for inocula at a concentration of 20 μg/mL (Wang et al., 2011).
Table 1.
Primers used in this study
| DNA or RNA Oligos | Sequences | Putative function |
|---|---|---|
| siGFP‐1 RNA | 5′GUCACUACUAUGGGUUAUGAG3′ | 21‐bp duplex siRNAs for EMSA |
| siGFP‐2 RNA | 5′CAUAACCCAUAGUAGUGACUG3′ | |
| 55‐nt sense RNA | 5′GAUGCAUUGUGCACUACGACUACUGCUCACUCGACGUAUAUAUCAGGCGACACAG3′ | Long dsRNA for EMSA |
| 55‐nt antisense RNA | 5′CUGUGUCGCCUGAUAUAUACGUCGAGUGAGCAGUAGUCGUAGUGCACAAUGCAUC3′ | |
| 2b‐F1 | 5′CAAGCTTATGGAATTGAACGTAGGTGC3′ | pGD‐2b/2blm/2bl/2bm |
| 2b‐R1 | 5′CGGATCCTCATTTGTCGTCATCGTCTTTG3′ | |
| 2b‐F2 | 5′TCCCCCGGGATGGAATTGAACGTAGGTGC3′ | 35S‐6myc‐Fny2blm |
| 2b‐R2 | 5′CGAGCTCTCAGGCATAGTCTGGGACGTC3′ | |
| 2bN61‐F1 | 5′CGCGGATCCATGGAATTGAACGTAGG3′ | pGEX‐KG‐Fny2b N61/2blmN61 |
| 2bN61‐R1 | 5′CCCAAGCTTTCAATCCACTTGATAGAACG3′ | |
| 2bN61‐F2 | 5′ CATGCCATGGATGGAATTGAACGTAGGTG 3′ | pET28a‐ Fny2b N61/2blmN61 |
| 2bN61‐R2 | 5′ CCGCTCGAGGAAAGCACCTTCCG 3′ | |
| GFP‐F | 5′TAATACGACTCACTATAGACGGAAACATCCTCGGCCAC3′ | GFP probe for Northern blot |
| GFP‐R | 5′TTATTTGTATAGTTCATCCATGCCATG3′ | |
| 2blm‐F | 5′CGTGCGGTGGAGGCGAAGAAGCAGAG3′ | Obtain CMV‐2blm |
| 2blm‐R | 5′AGCCGCTTGGAGTTCGACGTTTGTCATTG3′ | |
| 2bl‐F | 5′CGTATGGTGGAGGCGAAGAAGCAGAG3′ | With 2blm‐R to obtain CMV‐2bl |
| 2bm‐R | 5′AGCCAGTTGGAGTTCGACGTTTG3′ | With 2blm‐F to obtain CMV‐2bm |
CMV, Cucumber mosaic virus; dsRNA, double‐stranded RNA; EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; nt, nucleotide; siRNA, small interfering RNA.
Agroinfiltration and GFP imaging
For transient expression in N. benthamiana, CMV 2b, CMV‐2blm, CMV‐2bl and CMV‐2bm cDNAs fused to the N‐terminus of a Flag tag, as well as full‐length p19, were introduced into the pGD binary vector (Goodin et al., 2002). Leaves of 4–5‐week‐old wild‐type and 16c transgenic N. benthamiana were co‐infiltrated with equal volumes of two Agrobacterium cultures harbouring sense GFP plus p19‐, 2b‐, 2blm‐, 2bl‐ or 2bm‐expressing binary vectors or an empty vector control. GFP fluorescence in the agroinfiltrated plants was observed under a long‐wavelength UV lamp (UVP, Upland, CA, USA) and photographed using a 600D Canon digital camera (Canon, Tokyo, Japan) equipped with a yellow filter. For local silencing assays, GFP expression was monitored at 3 and 6 dpi. For systemic silencing assays, GFP was monitored and counted at 14 dpi. Local and systemic silencing assays were independently performed at least three times.
RNA blot analyses
Systemically infected leaves were collected from a pool of 15–20 plants for RNA extractions with Trizol reagent as instructed by the manufacturer (Invitrogen‐Life Technologies, Inc., Carlsbad, CA, USA). We used 5 and 2 μg of total RNA for RNA detection of mutant virus and wild‐type virus by Northern blot analyses, respectively. Northern blots were performed with [α‐32P]dCTP randomly labelled cDNA probes from 3′‐terminal 240 nucleotides of CMV RNA2 as described previously (Wang et al., 2011). Similarly, 5 μg of total RNA were extracted from infiltrated N. benthamiana and used for GFP detection with a probe corresponding to the P region of GFP cDNA.
Western blot analysis
Soluble protein samples extracted from viral‐inoculated or agroinfiltrated leaf tissue were suspended in SDS buffer [100 mm Tris (pH 6.8), 20% glycerol, 4% SDS, 0.2% bromophenol blue, 10% β‐mercaptoethanol]. Total proteins were separated in 12.5% SDS‐PAGE gels, transferred onto nitrocellulose membranes, and tested for GFP and 2b expression using anti‐GFP antibodies (1 : 1000) and an anti‐flag antibody (1 : 5000, Abmart, Shanghai, China), respectively. Goat anti‐rabbit immunoglobulin G (IgG) conjugated with alkaline phosphatase was used as secondary antibody, and the membranes were incubated in 5‐bromo‐4‐chloro‐3‐indolyl phosphate disodium salt (NBT‐BCIP) solution as instructed (Sigma‐Aldrich Corp, St. Louis, MO, USA).
Protein expression and EMSA
To generate GST‐2bN61 and GST‐2blmN61 proteins, coding sequences for the N‐terminal 61 amino acids of 2b and 2blm were individually amplified, engineered into the pGEX‐KG vector and transformed into E. coli strain BL21 (DE3 plyss strain). Recombinant proteins were expressed at 20 °C by induction with 0.4 mm isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) and purified with Glutathione Sepharose 4B by affinity columns, according to the manufacturer's instructions (GE Healthcare, Uppsala, Sweden). EMSA was carried out as described previously (Duan et al., 2012). RNA probes (Table 1) used for EMSAs were synthesized by the JiMa Company (JiMa, Shanghai, China). The protein–RNA complexes were resolved on 5% native polyacrylamide gels and exposed with a storage phosphor screen and X‐ray films.
Glutaraldehyde cross‐linking
The cDNAs of 2bN61 and 2blmN61 were cloned into pET28a and fused to the N‐terminus of a 6 × His tag, and then transformed into E. coli strain BL21 (DE3 strain). Recombinant 2bN61 and 2blmN61 were expressed in BL21 overnight at 20 °C induced by 0.4 mm IPTG. The cells were harvested by centrifugation and resuspended in lysis buffer [50 mm Tris‐HCl (pH 8.0), 150 mm NaCl, 0.2 mm phenylmethanesulfonylfluoride (PMSF), 1 mm dithiothreitol]. Following cell disruption by ultrasound and centrifugation, the supernatant was applied to Ni2+‐nitriloacetic acid agarose beads (Bio‐Rad, California, USA) by affinity columns. After washing with lysis buffer, the bound proteins were eluted with lysis buffer plus 50 mm imidazole. The purified proteins were incubated with 0%, 0.05%, 0.10% and 0.20% concentrations of freshly prepared glutaraldehyde solution for 1 h in the dark at 28 °C, and then boiled in SDS buffer. The samples were separated in 15% SDS‐PAGE gel, and detected by Coomassie brilliant blue staining and Western blots with His antibody.
Acknowledgements
We thank Shou‐Wei Ding (Department of Plant Pathology and Microbiology, University of California, Riverside, CA, USA), Andrew O. Jackson (Department of Plant and Microbial Biology, University of California at Berkeley, CA, USA), and Jialin Yu, Chenggui Han, Yongliang Zhang and other laboratory members for their helpful suggestions and constructive criticism. This work was supported by grants from the Natural Science Foundation of China (31322004, 31370176) and Chinese Universities Scientific Fund (2014RC014) to X‐BW. We declare that we have no conflict of interest.
Supporting information
Fig. S1. Symptom development in plants infected with Cucumber mosaic virus (CMV) (A, B), CMV‐2bl (A) or CMV‐2bm (B). The seedlings of wild‐type, rdrl, rdr6 and rdrl/6 plants were photographed at 21 days after inoculation with purified CMV, CMV‐2bl or CMV‐2blm viruses at a concentration of 20 μg/mL.
Fig. S2. Detection of duplex RNA binding affinity of 2bl and 2bm by electrophoretic mobility shift assay (EMSA). (A) Purified GST‐tagged proteins used in EMSAs were separated and stained by Coomassie brilliant blue. M, marker. (B) Comparative EMSAs carried out with increasing amounts (0.05–1.00 μg) of GST‐2b, GST‐2bl or GST‐2bm and a constant amount (1 nm) of [γ‐32P]‐labelled, 21‐bp, double‐stranded, small interfering RNAs (siRNAs) with 2‐nucleotide 3′ overhangs. Glutathione‐S‐transferase (GST) was used as negative control.
Fig. S3. Subcellular distribution of 2b‐YFP and 2blm‐YFP in the epidermal cells of agroinfiltrated Nicotiana benthamiana. The cDNA of 2b or 2blm was cloned into the gateway vector pEarleyGate 101, and transformed into EHA105. The transient expression of proteins was achieved by agroinfiltration of N. benthamiana leaves. Confocal micrographs were taken at 2 days post‐infiltration (dpi). YFP, yellow fluorescent protein. Bar, 10 μm.
References
- Andika, I.B. , Sun, L. , Xiang, R. , Li, J. and Chen, J. (2013) Root‐specific role for Nicotiana benthamiana RDR6 in the inhibition of Chinese wheat mosaic virus accumulation at higher temperatures. Mol. Plant–Microbe Interact. 26, 1165–1175. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2005) RNA silencing. Trends Biochem. Sci. 30, 290–293. [DOI] [PubMed] [Google Scholar]
- Bouche, N. , Lauressergues, D. , Gasciolli, V. and Vaucheret, H. (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti, G. , Voinnet, O. , Li, W.X. , Ji, L.H. , Ding, S.W. and Baulcombe, D.C. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burgyán, J. and Havelda, Z. (2011) Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272. [DOI] [PubMed] [Google Scholar]
- Chao, J.A. , Lee, J.H. , Chapados, B.R. , Debler, E.W. , Schneemann, A. and Williamson, J.R. (2005) Dual modes of RNA‐silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 12, 952–957. [DOI] [PubMed] [Google Scholar]
- Chen, H.‐Y. , Yang, J. , Lin, C. and Yuan, Y.A. (2008) Structural basis for RNA‐silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 9, 754–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T. , Hamilton, A. , Rudd, S. , Angell, S. and Baulcombe, D.C. (2000) An RNA‐dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Deleris, A. , Gallego‐Bartolome, J. , Bao, J. , Kasschau, K.D. , Carrington, J.C. and Voinnet, O. (2006) Hierarchical action and inhibition of plant Dicer‐like proteins in antiviral defense. Science, 313, 68–71. [DOI] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. , Li, F. , Li, W.‐X. and Ding, S.‐W. (2007) Suppression of antiviral silencing by Cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell, 19, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz‐Pendón, J.A. and Ding, S.‐W. (2008) Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326. [DOI] [PubMed] [Google Scholar]
- Ding, S.‐W. and Voinnet, O. (2007) Antiviral immunity directed by small RNAs. Cell, 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S.W. , Anderson, B.J. , Haase, H.R. and Symons, R.H. (1994) New overlapping gene encoded by the cucumber mosaic virus genome. Virology, 198, 593–601. [DOI] [PubMed] [Google Scholar]
- Ding, S.‐W. , Li, W.‐X. and Symons, R.H. (1995) A novel naturally occuring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14, 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z. , Chen, A. , Chen, W. , Liao, Q. , Zhang, H. , Bao, Y. , Roossinck, M.J. and Carr, J.P. (2014) Nuclear‐cytoplasmic partitioning of cucumber mosaic virus protein 2b determines the balance between its roles as a virulence determinant and an RNA‐silencing suppressor. J. Virol. 88, 5228–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, C.‐G. , Fang, Y.‐Y. , Zhou, B.‐J. , Zhao, J.‐H. , Hou, W.‐N. , Zhu, H. , Ding, S.‐W. and Guo, H.‐S. (2012) Suppression of Arabidopsis ARGONAUTE1‐mediated slicing, transgene‐induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell, 24, 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Ruiz, H. , Takeda, A. , Chapman, E.J. , Sullivan, C.M. , Fahlgren, N. , Brempelis, K.J. and Carrington, J.C. (2010) Arabidopsis RNA‐dependent RNA polymerases and Dicer‐like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell, 22, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, I. , Martínez, L. , Rakitina, D.V. , Lewsey, M.G. , Atencio, F.A. , Llave, C. , Kalinina, N.O. , Carr, J.P. , Palukaitis, P. and Canto, T. (2010) Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Mol. Plant–Microbe Interact. 23, 294–303. [DOI] [PubMed] [Google Scholar]
- González, I. , Rakitina, D. , Semashko, M. , Taliansky, M. , Praveen, S. , Palukaitis, P. , Carr, J.P. , Kalinina, N. and Canto, T. (2012) RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA, 18, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin, M.M. , Dietzgen, R.G. , Schichnes, D. , Ruzin, S. and Jackson, A.O. (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383. [DOI] [PubMed] [Google Scholar]
- Goto, K. , Kobori, T. , Kosaka, Y. , Natsuaki, T. and Masuta, C. (2007) Characterization of silencing suppressor 2b of Cucumber mosaic virus based on examination of its small RNA‐binding abilities. Plant Cell Physiol. 48, 1050–1060. [DOI] [PubMed] [Google Scholar]
- Hamera, S. , Song, X. , Su, L. , Chen, X. and Fang, R. (2012) Cucumber mosaic virus suppressor 2b binds to AGO4‐related small RNAs and impairs AGO4 activities. Plant J. 69, 104–115. [DOI] [PubMed] [Google Scholar]
- Hoffmann, J. , Brutschy, B. , Piehler, J. and Chen, J.C.H. (2008) Multiple targets for suppression of RNA interference by Tomato aspermy virus protein 2B. Biochemistry, 47, 12 655–12 657. [DOI] [PubMed] [Google Scholar]
- Kanazawa, A. (2008) RNA silencing manifested as visibly altered phenotypes in plants. Plant Biotechnol. 25, 423–435. [Google Scholar]
- Lakatos, L. , Csorba, T. , Pantaleo, V. , Chapman, E.J. , Carrington, J.C. , Liu, Y.P. , Dolja, V.V. , Calvino, L.F. , Lopez‐Moya, J.J. and Burgyan, J. (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewsey, M. , Robertson, F.C. , Canto, T. , Palukaitis, P. and Carr, J.P. (2007) Selective targeting of miRNA‐regulated plant development by a viral counter‐silencing protein. Plant J. 50, 240–252. [DOI] [PubMed] [Google Scholar]
- Li, F. and Ding, S.‐W. (2006) Virus counterdefense: diverse strategies for evading the RNA‐silencing immunity. Annu. Rev. Microbiol. 60, 503–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. , Huang, C. , Li, Z. and Zhou, X. (2014) Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin‐like protein to repress RDR6 expression. Plos Pathog. 10, e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Lu, J.F. , Han, Y.H. , Fan, X.X. and Ding, S.W. (2013) RNA interference functions as an antiviral immunity mechanism in mammals. Science, 342, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy, A.P. , Guo, H.S. , Li, W.X. and Ding, S.W. (2000) Suppression of post‐transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard, P.V. , Ciaudo, C. , Marchais, A. , Li, Y. , Jay, F. , Ding, S.W. and Voinnet, O. (2013) Antiviral RNA interference in mammalian cells. Science, 342, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C.D. and Hannon, G.J. (2009) Small RNAs as Guardians of the Genome. Cell, 136, 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins, F., Jr , Si‐Ammour, A. and Blevins, T. (2005) RNA silencing systems and their relevance to plant development. Annu. Rev. Cell Dev. Biol. 21, 297–318. [DOI] [PubMed] [Google Scholar]
- Nakahara, K.S. , Masuta, C. , Yamada, S. , Shimura, H. , Kashihara, Y. , Wada, T.S. , Meguro, A. , Goto, K. , Tadamura, K. , Sueda, K. , Sekiguchi, T. , Shao, J. , Itchoda, N. , Matsumura, T. , Igarashi, M. , Ito, K. , Carthew, R.W. and Uyeda, I. (2012) Tobacco calmodulin‐like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc. Natl. Acad. Sci. USA, 109, 10 113–10 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palukaitis, P. and Garcia‐Arenal, F. (2003) Cucumoviruses. Adv. Virus Res. 62, 241–323. [DOI] [PubMed] [Google Scholar]
- Pumplin, N. and Voinnet, O. (2013) RNA silencing suppression by plant pathogens: defence, counter‐defence and counter‐counter‐defence. Nat. Rev. Microbiol. 11, 745–760. [DOI] [PubMed] [Google Scholar]
- Qu, F. (2010) Antiviral role of plant‐encoded RNA‐dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin. Mol. Plant–Microbe Interact. 23, 1248–1252. [DOI] [PubMed] [Google Scholar]
- Qu, F. , Ye, X. , Hou, G. , Sato, S. , Clemente, T.E. and Morris, T.J. (2005) RDR6 has a broad‐spectrum but temperature‐dependent antiviral defense role in Nicotiana benthamiana . J. Virol. 79, 15 209–15 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach, F. , Vaistij, F.E. , Jones, L. and Baulcombe, D.C. (2005) An RNA‐dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138, 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz, A. (2006) Chemical cross‐linking and mass spectrometry to map three‐dimensional protein structures and protein–protein interactions. Mass Spectrom. Rev. 25, 663–682. [DOI] [PubMed] [Google Scholar]
- Vaistij, F.E. and Jones, L. (2009) Compromised virus‐induced gene silencing in RDR6‐deficient plants. Plant Physiol. 149, 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M. , Szittya, G. , Burgyan, J. and Hall, T.M. (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell, 115, 799–811. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. (2005) Non‐cell autonomous RNA silencing. FEBS Lett. 579, 5858–5871. [DOI] [PubMed] [Google Scholar]
- Wang, X.‐B. , Wu, Q. , Ito, T. , Cillo, F. , Li, W.‐X. , Chen, X. and Ding, S.‐W. (2010) RNAi‐mediated viral immunity requires amplification of virus‐derived siRNAs in Arabidopsis thaliana . Proc. Natl. Acad. Sci. USA, 107, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.‐B. , Jovel, J. , Udomporn, P. , Wang, Y. , Wu, Q. , Li, W.‐X. , Gasciolli, V. , Vaucheret, H. and Ding, S.‐W. (2011) The 21‐nucleotide, but not 22‐nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana . Plant Cell, 23, 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Tzfira, T. , Gaba, V. , Citovsky, V. , Palukaitis, P. and Gal‐On, A. (2004) Functional analysis of the Cucumber mosaic virus 2b protein: pathogenicity and nuclear localization. J. Gen. Virol. 85, 3135–3147. [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Wang, X. and Ding, S.W. (2010) Viral suppressors of RNA‐based viral immunity: host targets. Cell Host Microbe, 8, 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z. , Fan, B. , Chen, C. and Chen, Z. (2001) An important role of an inducible RNA‐dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. USA, 98, 6516–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, A. , Zhao, Z. , Chen, W. , Zhang, H. , Liao, Q. , Chen, J. , Carr, J.P. and Du, Z. (2013) Self‐interaction of the cucumber mosaic virus 2b protein plays a vital role in the suppression of RNA silencing and the induction of viral symptoms. Mol. Plant Pathol. 14, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.J. , Carter, S.A. , Cole, A.B. , Cheng, N.H. and Nelson, R.S. (2004) A natural variant of a host RNA‐dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana . Proc. Natl. Acad. Sci. USA, 101, 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Tan, S.H. , Teh, Y.J. and Yuan, Y.A. (2011) Structural implications into dsRNA binding and RNA silencing suppression by NS3 protein of Rice Hoja Blanca Tenuivirus. RNA, 17, 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K. , Malinina, L. and Patel, D.J. (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature, 426, 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, X.‐B. , Dong, L. , Zhu, H. , Duan, C.‐G. , Du, Q.‐S. , Lv, D.‐Q. , Fang, Y.‐Y. , Garcia, J.A. , Fang, R.‐X. and Guo, H.‐S. (2010) RNA‐dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana . Plant Cell, 22, 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Yuan, Y.‐R. , Pei, Y. , Lin, S.‐S. , Tuschl, T. , Patel, D.J. and Chua, N.‐H. (2006) Cucumber mosaic virus‐encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 20, 3255–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Symptom development in plants infected with Cucumber mosaic virus (CMV) (A, B), CMV‐2bl (A) or CMV‐2bm (B). The seedlings of wild‐type, rdrl, rdr6 and rdrl/6 plants were photographed at 21 days after inoculation with purified CMV, CMV‐2bl or CMV‐2blm viruses at a concentration of 20 μg/mL.
Fig. S2. Detection of duplex RNA binding affinity of 2bl and 2bm by electrophoretic mobility shift assay (EMSA). (A) Purified GST‐tagged proteins used in EMSAs were separated and stained by Coomassie brilliant blue. M, marker. (B) Comparative EMSAs carried out with increasing amounts (0.05–1.00 μg) of GST‐2b, GST‐2bl or GST‐2bm and a constant amount (1 nm) of [γ‐32P]‐labelled, 21‐bp, double‐stranded, small interfering RNAs (siRNAs) with 2‐nucleotide 3′ overhangs. Glutathione‐S‐transferase (GST) was used as negative control.
Fig. S3. Subcellular distribution of 2b‐YFP and 2blm‐YFP in the epidermal cells of agroinfiltrated Nicotiana benthamiana. The cDNA of 2b or 2blm was cloned into the gateway vector pEarleyGate 101, and transformed into EHA105. The transient expression of proteins was achieved by agroinfiltration of N. benthamiana leaves. Confocal micrographs were taken at 2 days post‐infiltration (dpi). YFP, yellow fluorescent protein. Bar, 10 μm.


