Summary
Several plant lipid transfer proteins (LTPs) act positively in plant disease resistance. Here, we show that LTP3 (At5g59320), a pathogen and abscisic acid (ABA)‐induced gene, negatively regulates plant immunity in Arabidopsis. The overexpression of LTP3 (LTP3‐OX) led to an enhanced susceptibility to virulent bacteria and compromised resistance to avirulent bacteria. On infection of LTP3‐OX plants with P seudomonas syringae pv. tomato, genes involved in ABA biosynthesis, NCED3 and AAO3, were highly induced, whereas salicylic acid (SA)‐related genes, ICS1 and PR1, were down‐regulated. Accordingly, in LTP3‐OX plants, we observed increased ABA levels and decreased SA levels relative to the wild‐type. We also showed that the LTP3 overexpression‐mediated enhanced susceptibility was partially dependent on AAO3. Interestingly, loss of function of LTP3 (ltp3‐1) did not affect ABA pathways, but resulted in PR1 gene induction and elevated SA levels, suggesting that LTP3 can negatively regulate SA in an ABA‐independent manner. However, a double mutant consisting of ltp3‐1 and silent LTP4 (ltp3/ltp4) showed reduced susceptibility to P seudomonas and down‐regulation of ABA biosynthesis genes, suggesting that LTP3 acts in a redundant manner with its closest homologue LTP4 by modulating the ABA pathway. Taken together, our data show that LTP3 is a novel negative regulator of plant immunity which acts through the manipulation of the ABA–SA balance.
Keywords: AAO3, ABA biosynthesis, Arabidopsis, disease development, lipid transfer protein 3 (LTP3)
Introduction
Despite constant exposure to microorganisms, plants are resistant to most potential pathogens and disease occurs rarely. Plants have developed multiple layers of defence (Chisholm et al., 2006; Jones and Dangl, 2006; Nishimura and Dangl, 2010). The recognition of pathogen‐associated molecular patterns (PAMPs) by cell surface receptors leads to the activation of basal defences effective against most microbes (PAMP‐triggered immunity, PTI). During evolution, pathogens have evolved effectors that play crucial roles in virulence by suppressing plant basal defence (da Cunha et al., 2007). In response, plants have evolved resistance (R) proteins that can specifically recognize such effectors [historically termed avirulence (AVR) factors] and activate a so‐called effector‐triggered immunity (ETI). ETI restricts pathogen proliferation (thus the host–pathogen interaction is incompatible), which is often, although not always, associated with a hypersensitive response (HR), a localized programmed cell death at the site of infection. In a compatible interaction, microbes can overcome plant defences, colonize the host plant and cause disease. Interestingly, plant signalling pathways leading to compatible and incompatible interactions largely overlap, but differ in timing, intensity and localization (Robert‐Seilaniantz et al., 2011; Tao et al., 2003).
In response to pathogen attack, plant hormones play crucial roles in the outcome of plant–microbe interactions (Grant and Jones, 2009). The fine‐tuning of plant immune responses is dependent on a complex interplay between various hormones (Grant and Jones, 2009; Robert‐Seilaniantz et al., 2011). Among them, abscisic acid (ABA), whose role in abiotic stress is well documented, has recently emerged as one of the key signalling molecules in plant–microbe interactions. ABA acts as a negative regulator of disease resistance, but also promotes plant defences via a complex network of synergistic and antagonistic interactions according to pathogen lifestyle and overall infection biology (Cao et al., 2011; Fan et al., 2009; Ton et al., 2009). In the Arabidopsis–Pseudomonas syringae pv. tomato (Pst) pathosystem, ABA positively regulates plant defence through the regulation of pre‐invasive stomata‐based responses (Melotto et al., 2006). However, at later stages of infection, some bacterial effectors target different steps of ABA biosynthesis or signalling to overcome plant basal defences (de Torres‐Zabala et al., 2007, 2009). ABA interacts antagonistically with salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signalling (Anderson et al., 2004; Audenaert et al., 2002; Grant and Jones, 2009; Mauch‐Mani and Mauch, 2005; Mohr and Cahill, 2007; Robert‐Seilaniantz et al., 2007; de Torres‐Zabala et al., 2009; Yasuda et al., 2008).
Plant lipid transfer proteins (LTPs) belong to a family of small (6.5–10.5 kDa), basic (isoelectric point usually between 8.5 and 12) and stable (with four conserved disulfide bonds) proteins and are characterized by an eight‐cysteine motif (C…C…CC…CXC…C…C) (Wang et al., 2012). LTPs possess the ability to exchange lipids between membranes in vitro (Kader, 1975, 1996). Because of the great variety of their lipid substrates, plant LTPs are also termed ‘non‐specific LTPs (nsLTPs), and vary greatly in sequence and expression patterns (Arondel et al., 2000; Chae et al., 2010). Several biological functions have been assigned to plant LTPs, including function in cuticle synthesis (Cameron et al., 2006; DeBono et al., 2009; Hollenbach et al., 1997), cell wall disruption and/or extension (Nieuwland et al., 2005) and modulators of plant growth and development (Chae et al., 2007, 2009; Kim et al., 2006; Lord, 2000). Their antimicrobial activities in vitro against fungi and bacteria are well documented (Cammue et al., 1995; Molina and Garcia‐Olmedo, 1993; Segura et al., 1993; Wang et al., 2004) and may result from the permeabilization of cell membranes (Kader, 1996). The overexpression of some LTPs derived from different plants leads to enhanced tolerance to pathogen infection in transgenic Arabidopsis, tobacco and rice (Jung et al., 2005; Molina and Garcia‐Olmedo, 1997; Patkar and Chattoo, 2006; Sarowar et al., 2009). Tobacco LTP1 is capable of binding JA; the LTP1–JA complex can bind some receptors of elicitins secreted by phytopathogenic microorganisms, therefore promoting resistance (Buhot et al., 2004).
In Arabidopsis, more than 100 proteins are annotated as LTPs, or putative LTPs, and their functions remain poorly documented (Chae et al., 2009, 2010). DIR1, an LTP‐like protein, is required for the transmission of a signal(s) from infected tissues via the vasculature to induce systemic acquired resistance (SAR) (Maldonado et al., 2002). Another LTP, AZI1, modulates the production and/or translocation of a mobile signal(s) during SAR (Jung et al., 2009). The Arabidopsis LTP3 is induced by abiotic stress and ABA (Seo et al., 2011; Wang et al., 2011) and, as a target of MYB96, is involved in plant tolerance to freezing and drought stress (Guo et al., 2013). LTP3 and its closest paralogue, LTP4, are highly induced during wilt disease development caused by the soil‐borne pathogen Ralstonia solanacearum (Hu et al., 2008).
In this report, we show that the overexpression of LTP3 results in increased endogenous ABA levels, enhanced susceptibility to the virulent P. syringae strain DC3000 and compromised HR‐associated resistance to the avirulent strain expressing avrRpm1. The enhanced susceptibility is reversed in aao3, an ABA biosynthesis mutant, indicating that the negative modulation of plant defence by LTP3 is AAO3 dependent. The double mutant ltp3/ltp4 displays reduced susceptibility to DC3000 and ABA biosynthesis gene expression, suggesting some level of redundancy between these two LTPs.
Results
LTP3 is induced by bacterial pathogens
In a previous study, LTP3 (At5g59320) was found to be strongly induced in Arabidopsis plants during wilt disease development caused by R. solanacearum (Hu et al., 2008). Two disease situations were considered: susceptible Col‐5 plants inoculated with the virulent GMI1000 strain, and Nd‐1 plants containing R genes (RRS1/RPS4) challenged with the same bacterial strain but with deletion of the corresponding avr PopP2 gene (GMI1000 ΔpopP2) (Deslandes et al., 2003; Heidrich et al., 2013; Narusaka et al., 2009). This induction was validated by real‐time polymerase chain reaction (PCR), as shown in Fig. 1A. LTP3 transcripts largely accumulated to high levels (up to 5000‐fold) during two cases of disease development in comparison with an incompatible interaction between Nd‐1 plants and the GMI1000 strain.
Figure 1.
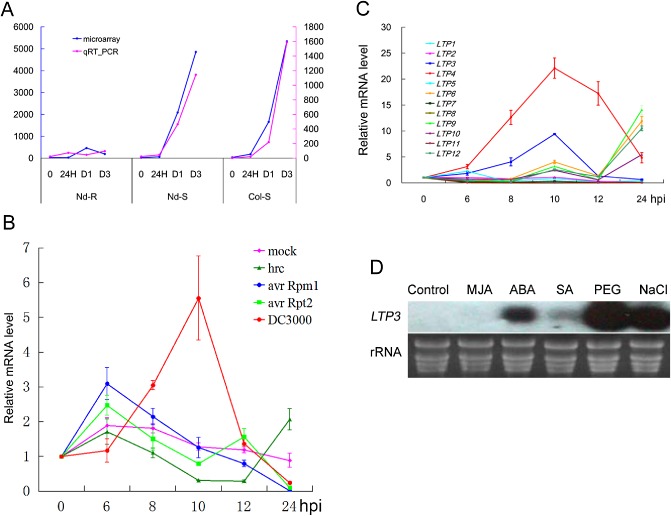
Expression analysis of the LTP3 gene. (A) Expression of the LTP3 gene identified by microarray analysis (blue lines) and validation by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) (pink lines) in both incompatible interaction (Nd‐R: Nd‐1/GMI1000) and compatible interactions (Col‐S: Col‐0/GMI1000; Nd‐S: Nd‐1/GMI1000 ΔpopP2) at different times after root inoculation by the R alstonia solanacearum GMI1000 strain. 24H, 24 h, no visible symptoms; D1, disease index 1, 25% of wilted leaves; D3, disease index 3, 75% of leaves completely wilted. The left and right scales correspond to the normalized Affymetrix dataset and the relative quantity of RNA measured by quantitative RT‐PCR, respectively. (B) Quantitative RT‐PCR analysis of LTP3 in Col‐0 plants on challenge with P seudomonas syringae pv. tomato (P st) DC3000 strain, avirulent P st avr R pm1 and avr R pt2 strains and type III secretion system (TTSS)‐deficient hrc C strain [1 × 108 colony‐forming units (CFU)/mL]. Mock, 10 mm MgCl2 as a control treatment. hpi, hours post‐inoculation. The data represent the means of three replicates ± standard deviation (SD). (C) Quantitative RT‐PCR analysis of lipid transfer protein (LTP) group I gene expression in Col‐0 plants on challenge with 1 × 108 CFU/mL of DC3000. The data represent the means of three replicates ± SD. (D) RNA gel blot analysis of LTP3 gene under methyl jasmonate (MJA), abscisic acid (ABA), salicylic acid (SA), polyethylene glycol (PEG) and NaCl treatment. Total RNA was extracted from 4‐week‐old plants treated with 100 μm ABA or 50 μm SA or MJA for 8 h; 10‐day‐old seedlings were treated with 20% PEG or 300 mm NaCl for 12 h.
The preferential expression of LTP3 during compatible interactions was further validated using the Pst–Arabidopsis pathosystem (Fig. 1B). In response to the virulent Pst DC3000 strain, LTP3 transcripts accumulated and reached maximum levels at 10 h post‐inoculation (hpi). In contrast, the induction of LTP3 was weaker following inoculation of the avirulent Pst avrRpm1 or avrRpt2 strains, which triggered visible HR around 18 hpi. The type III secretion system (TTSS)‐defective DC3000 mutant hrcC (Yuan and He, 1996) showed a limited effect on LTP3 induction. We confirmed this LTP3 expression pattern using pLTP3‐GUS plants (Fig. S1, see Supporting Information). Approximately 100 LTPs or putative LTPs identified in Arabidopsis have been classified into four groups according to phylogenetic analysis; LTP3 belongs to group I (Chae et al., 2009). The expression patterns of group I genes during disease development were studied (Fig. 1C). LTP4, the closest paralogue of LTP3, shares 85% nucleotide sequence and 90.2% amino acid sequence similarity with LTP3 and was up‐regulated by more than 20‐fold in response to Pst DC3000 at 10 hpi. LTP6, LTP9 and LTP12 transcripts were strongly induced at later stages, whereas the expression of the other LTPs tested was not altered significantly during infection.
Little is known about the effects of various hormones on LTP3 expression in Arabidopsis, except for its well‐characterized induction by ABA (Seo et al., 2011). We tested LTP3 expression in response to different hormones and abiotic stresses. Northern blot analysis showed that polyethylene glycol (PEG), NaCl and ABA treatment led to LTP3 transcript accumulation, SA had a weak effect, whereas methyl jasmonate (MJA) had no apparent effect (Fig. 1D). Taken together, our data indicate that LTP3 is involved in the plant response to biotic and some abiotic stresses.
Overexpression of LTP3 confers enhanced disease susceptibility and elevated ABA
In order to evaluate the functional importance of LTP3 in disease development, a homozygous T‐DNA line (SALK‐095248), named ltp3‐1, was characterized. T‐DNA was mapped in the first exon and no transcript was detected by reverse transcription‐polymerase chain reaction (RT‐PCR) (Fig. 2A), and so we considered it to be a knock‐out mutant. Transgenic LTP3‐overexpressing lines (LTP3‐OX) were generated and two independent transgenic lines (LTP3‐OX69 and LTP3‐OX88) exhibiting the highest LTP3 expression levels were chosen for further analyses (Fig. 2B). Under normal growth conditions, LTP3‐OX plants were similar to the wild‐type, except that the rosette leaves were slightly wider than those of wild‐type plants.
Figure 2.
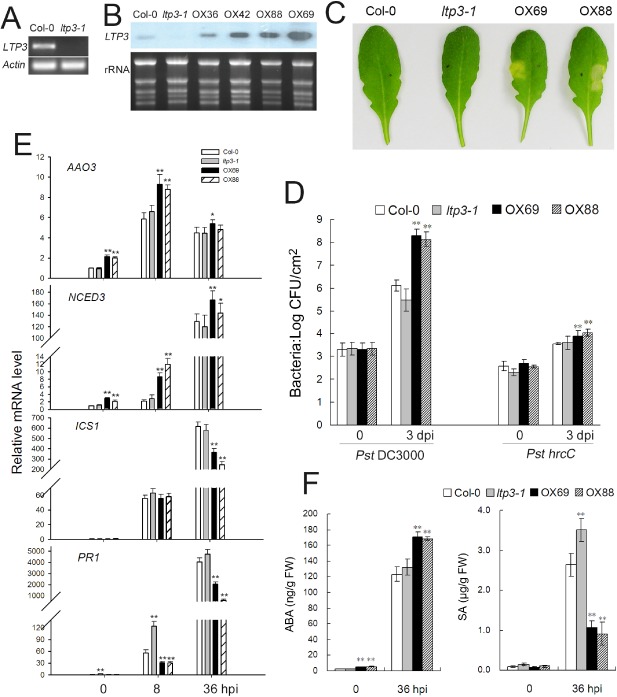
Overexpression of LTP3 enhances plant susceptibility and abscisic acid (ABA) biosynthesis. (A) Identification of lipid transfer protein 3 (LTP3) knock‐out mutant (ltp3‐1) by reverse transcription‐polymerase chain reaction (RT‐PCR). (B) Expression analysis of LTP3 gene in four transgenic lines (OX36, OX42, OX88, OX69) by Northern blot analysis. (C) Phenotypic responses of LTP3 overexpressors (OX69 and OX88), ltp3‐1 and wild‐type (Col‐0) were estimated 4 days after infiltration with P seudomonas syringae pv. tomato (P st) DC3000 strain [1 × 106 colony‐forming units (CFU)/mL]. (D) Bacterial growth in leaves inoculated with P st DC3000 and hrc C mutant strain (2 × 105 CFU/mL). The data represent the means of three independent experiments ± standard deviation (SD). Significant differences from the wild‐type are denoted by two asterisks, corresponding to P < 0.01, determined by Student's tests. dpi, days post‐inoculation. (E) Quantitative RT‐PCR analysis of ABA biosynthesis genes AAO3, NCED3, ICS1 and PR1 during disease development. Plants were infiltrated with DC3000 (5 × 106 CFU/mL). hpi, hours post‐inoculation. The data represent the means of three replicates ± SD. Significant differences from the wild‐type are denoted by one or two asterisks, corresponding to P < 0.05 and P < 0.01, respectively, by Student's test. (F) Quantification of ABA and salicylic acid (SA) levels in inoculated leaves. Data are the means of three biological replicates ± SD. Significant differences from the wild‐type are denoted by two asterisks, corresponding to P < 0.01, by Student's test. The experiments were repeated twice with similar results.
We next examined the responses of LTP3‐OX and ltp3‐1 lines to the virulent Pst DC3000 strain. Visible chlorosis in LTP3‐OX plants was observed 4 days after infiltration with DC3000, whereas wild‐type and ltp3‐1 plants did not show any symptoms at the same time point (Fig. 2C). At a high bacterial inoculum level [1 × 108 colony‐forming units (CFU)/mL], leaves of overexpressors exhibited severe wilting symptoms at 2 days post‐inoculation (dpi), whereas wild‐type and ltp3‐1 plants displayed limited local chlorosis (Fig. S2, see Supporting Information). Internal growth curves of Pst DC3000 were carried out and showed that the enhanced disease symptoms observed in the overexpressing lines correlated with an increased bacterial multiplication in planta (Fig. 2D). Pst DC3000 growth was 100‐fold greater in infected OX leaves than in wild‐type plants. No significant difference was observed between ltp3‐1 and wild‐type plants; however, LTP4 was drastically up‐regulated in ltp3‐1 relative to the wild‐type (Fig. S3, see Supporting Information), implying a functional redundancy of LTP4 in disease development. To further elucidate the role of LTP3 in PTI, hrcC was used to challenge plants of different genotypes (Fig. 2D); this non‐pathogenic strain multiplied to higher levels in LTP3‐OX plants than in wild‐type plants, but to a much lower extent relative to that observed in response to the virulent DC3000 strain.
Pst can modulate plant ABA biosynthesis to suppress inducible defence responses by down‐regulating SA biosynthesis and the SA pathway (de Torres‐Zabala et al., 2009). LTP3 has been reported as a target of MYB96 (Guo et al., 2013) and the MYB96 transcription factor is involved in ABA signals in the stress response (Seo et al., 2009, 2010). In order to investigate the mechanism underlying the enhanced susceptibility of LTP3‐OX, we evaluated the expression of ABA‐ and SA‐related genes by quantitative RT‐PCR. Among all the ABA‐related genes tested including biosynthesis, signalling and ABA‐responsive genes, NCED3 (9‐cis‐epoxycarotenoid dioxygenase 3) and AAO3 (ABA adehyde oxidase 3), which catalyse the rate‐limiting and last step of ABA biosynthesis, respectively, were found to be up‐regulated two‐fold more strongly in un‐inoculated OX plants than in wild‐type plants (Fig. 2E). In response to pathogen infection, both NCED3 and AAO3 were transcriptionally activated in all plants; however, their levels remained higher in OX lines relative to wild‐type plants, indicating that the overexpression of LTP3 somehow promotes ABA biosynthetic gene expression. In contrast, the expression pattern of these two genes was unaffected in ltp3‐1 lines.
We next examined SA‐related genes. Transcript accumulation of the isochorismate synthase 1 (ICS1) gene, encoding an enzyme of SA biosynthesis, was significantly lower in OX lines than in wild‐type and ltp3‐1 plants at 36 hpi, and pathogenesis‐related 1 (PR1) transcript levels were lower than those in control plants at 8 and 36 hpi (Fig. 2E). Notably, in the ltp3‐1 mutant, basal PR1 levels were approximately two‐fold higher than in the wild‐type, and the induction of PR1 was enhanced in response to Pst DC3000, suggesting a negative role of LTP3 on PR1 induction.
In order to investigate the effect of LTP3 expression on hormones, we evaluated the ABA and SA levels in leaves (Fig. 2F). It is noteworthy that, in healthy LTP3‐overexpressing plants, ABA basal levels were higher than those found in Col‐0 plants. In response to DC3000, ABA levels increased in both wild‐type and LTP3‐OX plants, but to much higher levels in the latter at 36 hpi. In contrast, no significant difference was detected in ltp3‐1 relative to the wild‐type. This observation was consistent with the expression pattern of NCED3 and AAO3, suggesting that the ABA increases are the result of de novo synthesis. SA accumulated in all DC3000‐infected plants, but the levels were reduced by 60% in LTP3‐OX plants relative to the wild‐type at 36 hpi, which correlated with the enhanced susceptibility of the overexpressors.
Overexpression of LTP3 compromises Arabidopsis R gene‐mediated disease resistance
In order to check whether LTP3 modulation affects R gene‐mediated resistance, we studied the responses of LTP3‐OX lines to the avirulent strain Pst avrRpm1 which triggers a resistance response in Col‐0 plants possessing the cognate RPM1 gene (Fig. 3A). At a low bacterial inoculum level (3.5 × 105 CFU/mL), LTP3‐OX plants displayed chlorosis in the infected zone 4 days after infection, whereas no visible symptoms could be detected in both wild‐type and ltp3‐1 plants. Pst avrRpm1 growth was at least 10‐fold greater in LTP3‐OX than in control plants (Fig. 3B). Quantitative RT‐PCR analysis performed in OX plants following infection with Pst avrRpm1 revealed that PR1 expression was lower, whereas NCED3 and AAO3 gene expression was significantly up‐regulated, compared with the wild‐type (Fig. 3C).
Figure 3.
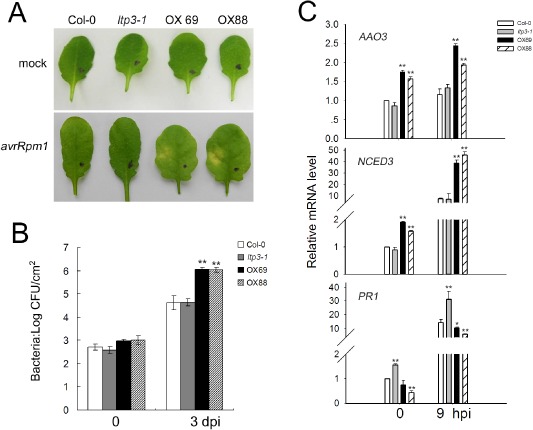
Overexpression of LTP3 compromises plant disease resistance to avirulent P seudomonas syringae pv. tomato (P st) strain infection. (A) Phenotypic responses were recorded 4 days after infiltration with P st avrRpm1 [3.5 × 105 colony‐forming units (CFU)/mL]. (B) Bacterial growth was determined at 3 days post‐inoculation (dpi). Plants were challenged with P st avr R pm1 (5 × 105 CFU/mL). Data shown are means of three biological replicates ± standard deviation (SD). Significant differences from the wild‐type are denoted by two asterisks, corresponding to P < 0.01, by Student's test. At least three independent experiments were performed with similar results. (C) The expression levels of AAO3, NCED3 and PR1 were determined by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) at 9 h post‐inoculation (hpi) on challenge with Pst avrRpm1 (7 × 107 CFU/mL). The data represent the means of three replicates ± SD. Significant differences from the wild‐type are denoted by one or two asterisks, corresponding to P < 0.05 and P < 0.01, respectively, by Student's test.
At a high bacterial inoculum level (7 × 107 CFU/mL), infected OX leaves rapidly exhibited necrotic symptoms which were indistinguishable from HR lesions in wild‐type plants (Fig. S4A, see Supporting Information). To check whether the necrotic symptoms observed in OX plants were HR related, the expression of an HR‐associated marker gene, HYPERSENSITIVITY‐RELATED 3 (HSR3) (Lacomme and Roby, 1999), was studied. At 3 hpi, HSR3 transcript levels increased dramatically in wild‐type plants, whereas no significant change was detected in OX plants (Fig. S4B). In addition, the cell death rate was monitored using the Evans blue method, which showed a decrease in Evans blue uptake in OX cells located at the inoculation site (Fig. S4C). Taken together, our data suggest that overexpression of LTP3 results in decreased R gene‐mediated defence and its associated HR.
LTP3 ‐ OX enhanced susceptibility is AAO3 dependent
To determine the importance of elevated ABA levels in the enhanced susceptibility observed in overexpressing lines, plants were treated with fluridone, an ABA biosynthesis inhibitor, before inoculation with Pst DC3000. We included lines overexpressing NCED3 (NCED3‐OX) and NCED5 (cds2, a gain‐of‐function mutant) that displayed an enhanced susceptibility to Pst DC3000 associated with elevated ABA levels (Fan et al., 2009). Fluridone treatment led to a significant decrease in bacterial multiplication in LTP3‐OX lines compared with Col‐0 plants, whereas this inhibitor showed less effect on the NCED3‐ and NCED5‐overexpressing lines (Fig. 4A).
Figure 4.
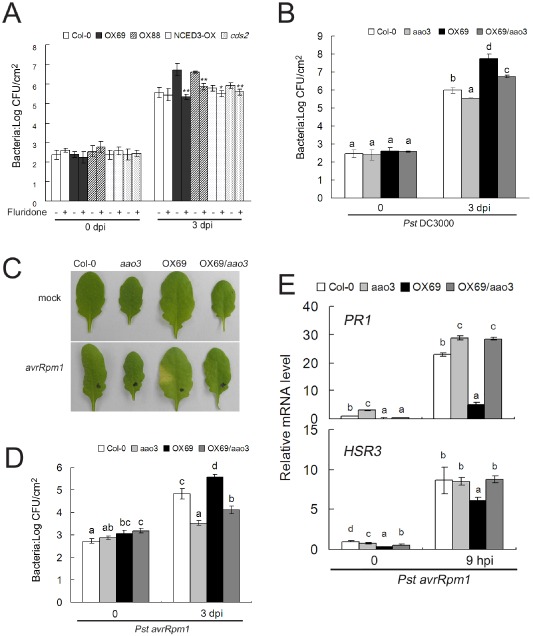
The function of LTP3 in the defence response is dependent on AAO3. (A) Pretreatment with fluridone suppresses in planta bacterial multiplication of P seudomonas syringae pv. tomato (P st) DC3000 in LTP3‐OX plants. Plants were pretreated with 100 μm fluridone (+) or buffer (−) 16 h prior to inoculation of P st DC3000 [2 × 105 colony‐forming units (CFU)/mL]. Bacterial growth was determined at 3 days post‐inoculation (dpi). Data are means ± standard deviation (SD) from three biological replicates. Significant differences from non‐fluridone treatment are denoted by two asterisks, corresponding to P < 0.01, by Student's test. (B) The enhanced susceptibility of LTP3‐OX69 plants is partially suppressed by AAO3. Plants were challenged with P st DC3000 (2 × 105 CFU/mL). Bacterial growth was determined at 3 dpi. Data shown are the means of three biological replicates ± SD. Different letters above SD indicate significant differences, as determined by Fisher's protected least significant difference (LSD) test (P < 0.05). At least three independent experiments were performed with similar results. (C–E) Compromised resistance of LTP3‐OX69 plants is suppressed by AAO3. (C) Phenotypic responses were recorded 4 days after plants had been challenged with P st avr R pm1 (3.5 × 105 CFU/mL). (D) Bacterial growth was determined at 3 dpi of P st avr R pm1 (5 × 105 CFU/mL). Different letters above SD indicate significant differences, as determined by Fisher's protected LSD test (P < 0.05). At least three independent experiments were performed with similar results. (E) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of PR1 and HSR3 transcripts. Plants were challenged with P st avr R pm1 (7 × 107 CFU/mL). The data represent the means of three replicates ± SD. Different letters above SD indicate significant differences, as determined by Fisher's protected LSD test (P < 0.05).
The AAO3 gene was significantly up‐regulated in LTP3‐OX plants. The ABA‐deficient aao3 mutant (SALK_072361), affected in the final step of ABA biosynthesis, exhibited reduced ABA accumulation and decreased susceptibility to Pst DC3000 (Seo et al., 2004; de Torres‐Zabala et al., 2009). In order to further address the role of LTP3 in ABA biosynthesis, we constructed the LTP3‐OX69/aao3 double mutant. On infection with Pst DC3000, the in planta bacterial growth of LTP3‐OX69/aao3 was intermediate between that of LTP3‐OX69 and aao3 plants (Fig. 4B), indicating that the LTP3‐mediated negative regulation of defence is exerted through its impact on de novo ABA biosynthesis, at least partially via AAO3.
Studies on the role of ABA in R gene‐based resistance remain limited, as do investigations of the response of aao3 to avirulent bacterial strains. On inoculation with Pst avrRpm1, visible lesions were only observed in LTP3‐OX69‐infected leaves (Fig. 4C). Internal growth curves of Pst avrRpm1 were obtained (Fig. 4D). The data showed that bacterial multiplication in aao3 was at least 10‐fold less than in wild‐type plants, indicating a negative role of ABA in RPM/AvrRpm1‐mediated resistance. On challenge with Pst avrRpm1, bacterial multiplication was significantly restricted in LTP3‐OX69/aao3 plants, which resembled the enhanced resistance of aao3 in comparison with the wild‐type (Fig. 4D). Consistent with these data, the induction of PR1 and HSR3 in LTP3‐OX69/aao3 plants was also comparable with aao3 (Fig. 4E).
LTP3 positively mediates ABA signalling via AAO3 in seed germination
The higher ABA basal levels in LTP3‐OX relative to wild‐type plants led us to examine whether these plants exhibited altered responses to ABA treatment and more generally to abiotic stress. Seeds of LTP3‐OX, ltp3‐1 and wild‐type plants were sown on medium supplemented with various concentrations of exogenous ABA (Fig. 5A,B). On normal Murashige and Skoog (MS) medium without ABA, there was no visible difference between wild‐type and OX lines for either germination rate or timing. However, on ABA‐containing medium, seed germination of three OX lines was significantly impaired relative to that in Col‐0 plants, and this increased sensitivity to ABA was ABA concentration independent, suggesting that the elevated basal level of ABA in LTP3‐OX plants renders them more sensitive to additional exogenous ABA relative to the wild‐type. This ABA inhibition of seed germination conferred by LTP3 overexpression was considerably attenuated in LTP3‐OX/aao3 relative to LTP3‐OX (Fig. 5C,D), indicating that LTP3 modulation of ABA signalling in seed germination is also AAO3 dependent.
Figure 5.
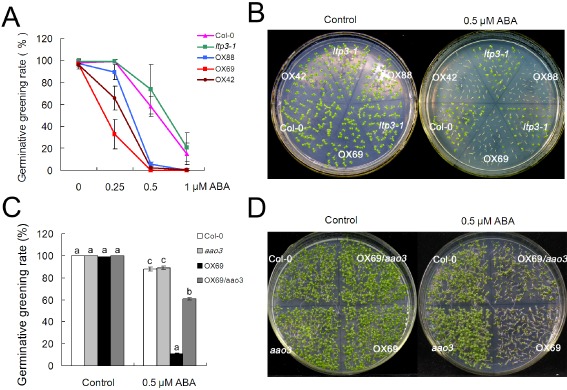
LTP3 mediates abscisic acid (ABA) signalling via AAO3 in seed germination. (A) ABA dose–response analysis of germination in Col‐0, ltp3‐1 and LTP3‐OX lines. Seeds were plated on Murashige and Skoog (MS) medium containing various amounts of ABA, and the germinative greening rate was scored 5 days after stratification by counting the seedlings with greening cotyledons. Data are means of three independent experiments ± standard deviation (SD) (each with at least 100 seeds for each line). (B) LTP3‐OX hypersensitivity to ABA on MS medium containing 0.5 μm ABA. Photographs were taken 7 days after stratification. (C) aao3 reduced LTP3‐OX69 hypersensitivity to ABA. Seeds were germinated and grown on MS medium containing 0.5 μm ABA for 7 days. Data are means ± SD from three biological replicates. Different letters above SD indicate significant differences, as determined by Fisher's protected least significant difference (LSD) test (P < 0.05). (D) Photographs were taken 9 days after stratification.
Previous studies have shown that LTP3‐overexpressing lines are more tolerant to drought (Guo et al., 2013). In order to check whether this phenotype is associated with an alteration of stomatal closure, we analysed stomatal responses to ABA. Compared with wild‐type plants, LTP3‐OX stomatal closure was more sensitive to ABA treatment at the different concentrations tested (Fig. S5A, see Supporting Information). Stomatal closure in OX plants was indeed detectable at an ABA concentration of 1 μm, whereas, in Col‐0 plants, the same degree of stomatal closure was only observed on plant treatment with 10 μm ABA (Fig. S5B). These data suggest that elevated endogenous ABA levels in OX plants elicit stomatal closure, leading to enhanced drought tolerance (Fig. S5C). Taken together, our data confirm that LTP3 is a positive regulator of the ABA pathway and is involved in the modulation of biotic and abiotic stress responses.
Double mutant ltp3/ltp4 is more resistant than the wild‐type to P st DC3000
The redundancy between LTP3 and its closest homologue LTP4 may explain the absence of phenotypes of null LTP3 lines in response to the pathogen. To test this hypothesis, double ltp3/ltp4 mutants were generated using an RNA interference (RNAi) approach to silence LTP4 in the ltp3‐1 background because LTP3 (At5g59320) and LTP4 (At5g59310) are physically adjacent. The ltp3/ltp4‐5 and ltp3/ltp4‐7 lines were chosen for further characterization (Fig. 6A). Under normal conditions, no developmental difference was observed between these lines and ltp3‐1 plants. On challenge with Pst DC3000, bacterial multiplication in ltp3/ltp4‐5 and ltp3/ltp4‐7 lines was significantly restricted compared with that found in wild‐type and ltp3‐1 plants (Fig. 6B). Consistent with this decreased susceptibility, a significant reduction in AAO3, NCED3 and NCED5, coupled with enhanced ICS1 and PR1 induction, was detected in ltp3/ltp4 (Fig. 6C). Furthermore, the insensitivity to ABA of seed germination observed in ltp3/ltp4 (Fig. 6D,E) strengthens the evidence for the redundancy of LTP3 and LTP4 in ABA signalling.
Figure 6.
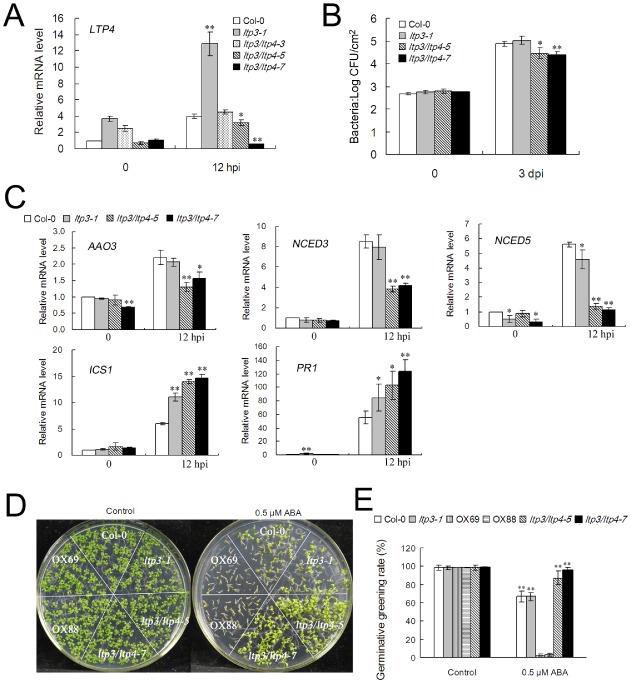
Double mutant ltp3/ltp4 is more resistant to P seudomonas syringae pv. tomato (P st) DC3000 and insensitive to abscisic acid (ABA) (A) LTP4 transcript levels in different double mutants ltp3/ltp4 were determined by quantitative reverse transcription‐polymerase chain reaction (RT‐PCR) Plants were challenged with P st DC3000 [5 × 106 colony‐forming units (CFU)/mL]. The data represent the means of three replicates ± standard deviation (SD). Significant differences from the wild‐type are denoted by one or two asterisks, corresponding to P < 0.05 and P < 0.01, respectively, by Student's test. hpi, hours post‐inoculation. (B) ltp3/ltp4 was more resistant to P st DC3000 than the wild‐type. In planta bacterial growth was determined at 3 days post‐inoculation (dpi) at a cell density of 2 × 105 CFU/mL. Data shown are the means of three biological replicates ± SD. Significant differences from the wild‐type are denoted by one or two asterisks, corresponding to P < 0.05 and P < 0.01, respectively, by Student's test. At least three independent experiments were performed with similar results. (C) The expression levels of AAO3, NCED3, NCED5, ICS1 and PR1 were determined by quantitative RT‐PCR at 12 hpi with P st DC3000 (5 × 106 CFU/mL). The data represent the means of three replicates ± SD. Significant differences from the wild‐type are denoted by one or two asterisks, corresponding to P < 0.05 and P < 0.01, respectively, by Student's test. (D, E) Insensitivity of ltp3/ltp4 to ABA in seed germination. (D) Photographs were taken 9 days after stratification. (E) Germination rate was scored 7 days after stratification. Data are means of three independent experiments ± SD (each with at least 100 seeds for each line). Significant differences from the wild‐type are denoted by two asterisks, corresponding to P < 0.01, by Student's test.
Discussion
Several plant LTPs function in plant immunity as positive factors and, as a result of their inducible expression patterns on pathogen challenge, have been included in the PR14 family of pathogenesis‐related proteins (Sels et al., 2008; Van Loon and Van Strien, 1999). LTP‐like proteins, DIR1 and AZI1, also play crucial roles in systemic plant immunity (Jung et al., 2009; Maldonado et al., 2002). However, our findings demonstrate a negative role for LTP3 in plant immunity by manipulation of the ABA–SA balance.
LTP3 exerts a negative role in disease resistance by positively regulating ABA biosynthesis
ABA regulates many aspects of plant growth and development, e.g. the inhibition of germination and induction of stomatal closure (Finkelstein et al., 2002), and has emerged as a multifaceted and pivotal role player in plant–microbe interactions (Cao et al., 2011). Our results showed that LTP3 overexpression resulted in the up‐regulation of AAO3 and NCED3, genes encoding two key enzymes in ABA biosynthesis, ultimately leading to elevated ABA levels, which corroborates previous findings indicating that NCED3 and AAO3 activation increases ABA levels (Schwartz et al., 2003; Seo et al., 2004). Compared with the wild‐type, higher endogenous ABA contents in OX indeed confer OX lines with increased sensitivity to exogenous ABA in terms of seed germination and stomatal closure. During plant–pathogen interactions, ABA biosynthesis fulfils an important function in the regulation of plant defence. Disruption of ABA biosynthesis enhances the plant response to necrotrophic pathogens, such as the fungi Botrytis cinerea and Plectrosphaerella cucumerina, or the bacterium Dickeia dadantii. The enhanced resistance of some ABA biosynthesis mutants is associated with an increased cuticle permeability and a faster reactive oxygen species (ROS) accumulation (Asselbergh et al., 2007, 2008; Audenaert et al., 2002; Curvers et al., 2010; L'Haridon et al., 2011; Sánchez‐Vallet et al., 2012). During disease development, ABA biosynthesis is targeted by P. syringae effectors to promote virulence (Fan et al., 2009; de Torres‐Zabala et al., 2007, 2009). In our study, the significantly enhanced growth of Pst DC3000 in LTP3 overexpressors was accompanied by the activation of ABA biosynthesis and suppression of SA biosynthesis. The dynamic changes in ABA and SA levels conferred by LTP3 overexpression support the ABA–SA antagonism described previously in the literature (de Torres‐Zabala et al., 2009). Pretreatment with an ABA biosynthesis inhibitor abolished the enhanced bacterial growth in LTP3‐OX, suggesting that activated ABA biosynthesis contributes to the OX phenotype. This hypothesis was supported by the observation that the generation of LTP3‐OX/aao3 double mutants indeed partially rescued the resistance to DC3000. All of these findings strongly suggest that LTP3 positively regulates AAO3 to fulfil its negative role in plant defence. By contrast, the ABA pathway was not affected in ltp3‐1, which is thought to be a result of the overexpression of its paralogue LTP4 (Fig. S3). Consistently, when LTP4 was silenced in the ltp3‐1 background, the suppression of ABA biosynthesis coincided with decreased bacterial multiplication, indicating that LTP3 and LTP4 cooperate to modulate ABA biosynthesis during disease development. However, despite the unaffected ABA in ltp3‐1, enhanced PR1 gene expression and elevated SA levels suggest that LTP3 somehow negatively modulates SA biosynthesis, at least in part in an ABA‐independent manner.
Recently, ABA has been shown to negatively modulate R gene‐mediated resistance by interfering with R protein localization (Mang et al., 2012). In this study, ABA accumulation promoted bacterial growth, even in the context of the AvrRpm1/Rpm1‐mediated resistance response. Thus, reduced ABA levels in the aao3 mutant impair the growth of Pst avrRpm1 to levels even lower than those observed in wild‐type Col‐0. Moreover, the rescued resistance of OX69 in the aao3 background in the context of Rpm1/AvrRpm1 interaction highlights the important role of ABA in plant immunity. Collectively, LTP3 may act on the plant general defence response to pathogens via the attenuation of SA‐related defences. LTP3‐mediated ABA induction seems to be a strategy used by virulent bacterial strains to promote virulence and by avirulent strains to interfere with avrRpm1‐induced resistance and even HR.
Given the strong induction of LTP3 by ABA and the elevated ABA levels in LTP3 overexpressors, it appears that the accumulation of this hormone prevents the establishment of SA‐related defences and, meanwhile, induces LTP3 expression through a positive feedback (regulatory circuit) to create a favourable environment for the invading pathogens. On the basis of these findings, we present a model of LTP3 modulation in plant defence (Fig. 7).
Figure 7.
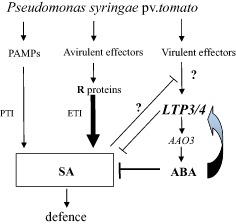
Proposed model of lipid transfer protein 3 (LTP3) modulation of abscisic acid (ABA) in the Arabidopsis–P seudomonas syringae pv. tomato interaction. Pathogen‐associated molecular patterns (PAMPs) induce PAMP‐triggered immunity (PTI) in the plant; specific avirulent effectors are recognized by immune receptors (R proteins) to induce effector‐triggered immunity (ETI). Salicylic acid (SA) plays an important role in the activation of plant general defence; pathogen‐modulated ABA signalling antagonizes SA‐mediated defences. LTP3 might be targeted by virulent effectors to promote ABA biosynthesis through AAO3, and thus down‐regulate the SA pathway. A positive regulatory loop exists between ABA and LTP3. LTP3 also negatively modulates SA through an ABA‐independent way.
LTP3, a disease‐related marker gene?
Sustained and strong LTP3 induction has been observed during disease development in different pathosystems (http://www.genevestigator.com). In our study, LTP3 was activated at high levels during compatible interactions between Arabidopsis and pathogens such as R. solanacearum and P. syringae, which suggests that LTP3 constitutes a marker for disease development. More evidence from infected ltp3‐1 plants indicated that PR1 expression was largely increased compared with the wild‐type in response to both virulent and avirulent strains. Consistently, our data clearly show that the overexpression of LTP3 is positively correlated with increased bacterial multiplication in response to both virulent and avirulent bacteria, reinforcing the evidence that LTP3 is pathogenesis associated.
ABA negatively regulates basal resistance and the activation of callose deposition. Interestingly, it has been reported previously that the pathogen effector, AvrPto, can target NCED3 to regulate de novo ABA biosynthesis (Clay et al., 2009; Fan et al., 2009; de Torres‐Zabala et al., 2007, 2009). In this study, we showed that hrcC, a TTSS‐defective DC3000 mutant, was less effective on LTP3 induction relative to DC3000 (Fig. 1B), implying that the induction of LTP3 is at least partially effector dependent. In the compatible interaction, the significant induction of LTP3 and the remarkable increased susceptibility of LTP3‐OX indicate that a strong LTP3 induction by the effector(s) is required to provoke ABA accumulation to exert its negative role on basal defence. Hence, we speculate that LTP3, a host component, is targeted by Pseudomonas effector(s) in order to disturb the ABA pathway (Fig. 7). In the case of an incompatible interaction, it is likely that R/Avr recognition preceding LTP3 manipulation by a virulent effector(s) triggers a strong SA signal and defence response, which is able to inhibit virulent effectors, so that LTP3 is not up‐regulated and ABA remains at a normal level; thereby, an appropriate ABA–SA balance is constructed for the resistance outcome. However, when LTP3 is overexpressed in transgenic plants, de novo ABA biosynthesis can be triggered, which, in turn, interferes with SA‐mediated defence and HR, ultimately leading to decreased resistance. Taken together, our data demonstrate that the shared general defence across PTI and ETI could be manipulated by an effector(s) via LTP3, and that LTP3 accumulation is probably crucial to its impact on the modulation of ABA biosynthesis and final disease outcome.
LTP and LTP‐like proteins belong to a large and ubiquitous family. It appears that individual isoforms are likely to play specific or multiple biological roles. Given that all other characterized LTPs are positive regulators of plant defence, an intriguing possibility is that the negative role of LTP3 in general defence means that it could be hijacked by pathogens to promote virulence by up‐regulation of the ABA pathway. Further characterization of LTP3 and identification of downstream components interacting directly with the ABA pathway will need to be performed in the future to explain the exact role of LTP3 in ABA biosynthesis. Moreover, gaining a further insight into how LTP3 interplays with the SA signalling network will also be of great interest in understanding plant disease development.
Experimental Procedures
Plant material and treatments
Arabidopsis thaliana ecotypes Col‐0 and Nd‐1 were used in these experiments. Seeds of ltp3 plants (SALK_095248) were obtained from The Arabidopsis Information Resource (TAIR). aao3, cds2 and NCED3‐OX plants were kindly provided by Dr Murray Grant (University of Exeter, Exeter, Devon, UK), Fan Jun (China Agricultural University, Beijing, China) and Gary Creissen (John Innes Centre, Norwich, Norfolk, UK), respectively. All plants were grown at 22 °C under short‐day conditions (9 h light/15 h dark), and 4‐ or 5‐week‐old plants were used for bacterial inoculations.
For seed germination assays, at least 100 seeds from each plant genotype were sterilized and sown in triplicate on MS plates supplemented with 30 g/L sucrose, 6 g/L agar and different concentrations of ABA. After a 2‐day stratification at 4 °C, the plates were cultured in a growth chamber at 22 °C under long‐day conditions (16 h light/8 h dark) at 80 μmol/m2/s light. The percentage of germination was scored at 5 days, with expansion and greening of cotyledons used as a criterion for germination, as described previously (Nambara et al., 2002).
Hormone treatments were performed by spraying 4‐week‐old plants with ABA (100 μm), SA or MJA (50 μm) solutions. Ten‐day‐old seedlings were treated with 20% PEG and 300 mm NaCl (Lee et al., 2001). For fluridone treatment, 4–5‐week‐old plants were sprayed with 100 μm fluridone containing 0.1% ethanol and 0.005% (v/v) Tween‐20, 16 h before inoculation with Pst DC3000 (Zhou et al., 2008).
Generation of transgenic plants overexpressing LTP3 and of LTP3‐OX69/aao3 double mutants
The full‐length LTP3 was amplified from cDNAs synthesized from 1 μg of total RNA from 4‐week‐old Col‐0 plants using oligo(dT) primer and SuperScript reverse transcriptase II (Invitrogen, Carlsbad, CA, USA). The sense primer (AttB1‐LTP3) used in the amplification was 5′‐CAAAAAAGCAGGCTTAATGGCTTTCGCTTTGAGGTTCTTC‐3′, and the antisense primer (AttB2‐LTP3) was 5′‐CAAGAAAGCTGGGTCCTTGATGTTGTTGCAGTTAGTGCTCAT‐3′. The AttB1‐LTP3‐AttB2 PCR product was recombined into the pDONR207 vector (Invitrogen) via a BP reaction to produce the pENTR‐LTP3 construct. This construct was recombined with the pAM‐PAT‐35SS‐GW‐StrepII destination vector to generate pAM‐35SS‐LTP3‐StrepII* binary plasmid, which was then introduced into the Agrobacterium tumefaciens GV3101 strain. Plant transformation was performed by floral dipping. Independent transgenic lines were selected by growing T1 seeds on MS plates containing 5 μg/mL of phosphinothricin (PPT, Duchefa, Haarlem, Netherlands). Homozygous T3 transgenic seeds selected by segregation analyses were used for analyses.
Double mutants LTP3‐OX69/aao3 were generated by standard genetic crosses. The presence of the aao3 mutation was confirmed by detection of the T‐DNA insert using the flanking primers (5′‐TTCTATTGGAAATGCATTGCC‐3′ and 5′‐CCATGTCTGCATGTTTCTGTG‐3′) and the T‐DNA internal primer (5′‐CCCTTTAGGGTTCCGATTTAGTGCT‐3′ reference). The presence of the LTP3 overexpressor was detected by the following two primers: 5′‐ATGACGCACAATCCCACTATCCTTCGCA‐3′ and 5′‐CTTATCTGGGAACTACTCACACAT‐3′.
Construction of double mutant ltp3/ltp4
The double mutant ltp3/ltp4 was generated by knocking down LTP4 in the ltp3‐1 mutant. The pRNAi‐LIC vector for the generation of the LTP4 silencing construct was kindly provided by Dr Liu Yule (Tisnghua University, Beijing, China). The sense (LIC1‐LTP4) and antisense (LIC2‐LTP4) primers used for PCR Product 1 amplification were 5′‐CGACGACAAGACCCTATGTGGCACAGTGGCAAGT‐3′ and 5′‐GAGGAGAAGAGCCCTCTCTTCAGGCAAATGATGTC‐3′, respectively. PCR Product 2 was amplified using purified PCR Product 1 as template and the universal oligo pair LIC3‐TT‐LIC2 and LIC4‐TT‐LIC1, as described previously (Xu et al., 2010). The PCR Products 1 and 2 were recombined into the pRNAi‐LIC vector to generate the pRNAi‐LTP4 binary plasmids. The constructs were then introduced into the Agrobacterium tumefaciens GV3101 strain and plants were transformed by floral dipping. Independent transgenic lines were selected by growing T1 seeds on MS plates containing 50 mg/L kanamycin. Homozygous T3 seeds were selected by segregation analyses, and the LTP4 transcript level was determined by quantitative RT‐PCR.
Bacterial strains and inoculation
The R. solanacearum strain GMI1000 is a wild‐type strain originally isolated from tomato. The ΔPopP2 strain has already been described (Deslandes et al., 2002, 2003). Disease resistance phenotypes were determined by root inoculation of 4‐week‐old plants with R. solanacearum strains. Disease symptoms were scored according to the percentage of wilted leaves, using the following 0–4 scale (0, no wilting; D1, 0–25%; D2, 25–50%; D3, 50–75%; D4, 75–100%).
The Pseudomonas syringae stains used in this study were Pst DC3000, Pst avrRpm1, Pst avrRpt2 and Pst DC3000 HrcC– (hrcC). All strains were grown at 28 °C on King's B plates containing the appropriate antibiotics for selection.
Four‐ or five‐week‐old plants were used for bacterial inoculation. For this objective, they were kept at high humidity 12 h before infection. Plants were injected with a bacterial suspension of 2 × 105–1 × 106 CFU/mL for the phenotype test, and 2 × 105 CFU/mL (DC3000) or 5 × 105 CFU/mL (Pst avrRpm1) for determination of in planta bacterial growth, using a blunt syringe on the abaxial side of the leaves (Lorrain et al., 2004).
pLTP3‐GUS activity analysis
A promoter fragment of 1669 bp of the LTP3 gene was amplified from Col‐0 genomic DNA by PCR using the primer pair 5′‐CTGGAAGAGGAAAAACAAGTAAATGCAAGT‐3′ and 5′‐TGTGTTTGAACTTCTTTTTGGGTGATAACAAA‐3′. The amplified fragment was cloned into the pCAMBIA 1391 (Cambia, Canberra, Australia) vector, resulting in a transcriptional fusion of the LTP3 promoter with the β‐glucuronidase (GUS) coding region. The LTP3 promoter–GUS fusion construct was introduced into Agrobacterium tumefaciens and transferred into plants. Independent T2 transgenic lines were grown on MS medium containing hygromycin (30 μg/mL) and subjected to GUS activity assays as described by Jefferson et al. (1987).
Gene expression studies
Total RNA was extracted from Arabidopsis leaf samples with Trizol reagent (Invitrogen). Transcript levels of the LTP3 gene were determined by RNA gel blot analyses. Total RNA was separated on a 1.2% formaldehyde–3‐(N‐morpholino) propanesulfonic acid (MOPS) agarose gel and blotted onto nylon membranes, and the blot was probed, washed and exposed to X‐ray film for autoradiography.
For quantitative RT‐PCR analyses, 2.5 μg of total RNA was used for first‐strand cDNA synthesis employing M‐MLV reverse transcriptase (Invitrogen) and gene‐specific primers listed in Table S1. Quantitative PCR analyses were performed using SYBR Green PCR Master Mix (Takara, Dalian, China), SYBR Premix Ex Taq (Takara) and an Applied Biosystems 7500 real‐time PCR system (Life Technologies, USA). Relative transcript abundance was calculated using the comparative CT method (Miura et al., 2007), and the value for the wild‐type without pathogen infection (0 h) was normalized to unity. At least three biological replicates were used.
Extraction of SA and ABA
The evaluation of SA and ABA concentrations was performed in triplicate, with each replicate consisting of a minimum of five infected leaves from three plants. Samples were collected at appropriate times after inoculation and frozen immediately in liquid nitrogen. Free and total SA were extracted and measured as described previously (Li et al., 1999). ABA was extracted from 200 mg of fresh inoculated or healthy plant tissues and measured using ultrahigh‐performance liquid chromatography–triple quadrupole mass spectrometry (UPLC‐MS/MS), as described previously (Fu et al., 2012).
Cell death assay
Cell death was quantified by monitoring the uptake of Evans blue (0.25%) by leaf discs from inoculated or healthy plants, as described previously (Baker and Mock, 1994). The assay was performed with eight leaf discs (diameter, 5 mm) from the inoculated zone of five plants. Three replicates per point were performed.
Supporting information
Fig. S1 pLTP3:GUS is induced by Pseudomonas syringae pv. tomato (Pst) infection and abscisic acid (ABA) treatment. hpi, hours post‐inoculation.
Fig. S2 LTP3‐OX plants exhibit enhanced disease symptoms on challenge with Pseudomonas syringae pv. tomato (Pst) DC3000.
Fig. S3 LTP4 is up‐regulated in ltp3‐1 plants on challenge with Pseudomonas syringae pv. tomato (Pst) DC3000.
Fig. S4 Overexpression of LTP3 compromises hypersensitive response (HR) development. hpi, hours post‐inoculation.
Fig. S5 LTP3‐OX stomatal closure is more sensitive to abscisic acid (ABA) treatment. hpi, hours post‐inoculation.
Table S1 List of primers used for transcript analysis by real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐PCR).
Acknowledgements
This work was supported by the National Science Foundation of China (Grants 31071675 and 30671179) and the State Key Laboratory of Plant Physiology and Biochemistry (Grant SKLPPBKF1401). ABA dosage experiments were carried out at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China by Shuang Fang, Cunyu Yan and Jinfang Chu (National Centre for Plant Gene Research). The authors thank Dr Yves Marco (CNRS/INRA, Toulouse, France) and Maud Bernoux (CSIRO, Canberra, Australia) for critical reading, stimulating discussions and suggestions on the manuscript. We thank Dr Laurent Deslandes for some plasmid constructions.
References
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arondel, V. , Vergnolle, C. , Cantrel, C. and Kader, J.C. (2000) Lipid transfer proteins are encoded by a small multigene family in Arabidopsis thaliana . Plant Sci. 157, 1–12. [DOI] [PubMed] [Google Scholar]
- Asselbergh, B. , Curvers, K. , Franca, S.C. , Audenaert, K. , Vuylsteke, M. , Van Breusegem, F. and Höfte, M. (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid‐deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144, 1863–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh, B. , Achuo, A.E. , Höfte, M. and Van Gijsegem, F. (2008) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi . Mol. Plant Pathol. 9, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K. , De Meyer, G.B. and Hofte, M.M. (2002) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid‐dependent signaling mechanisms. Plant Physiol. 128, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.J. and Mock, N.M. (1994) An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 39, 7–12. [Google Scholar]
- Buhot, N. , Gomes, E. , Milat, M.L. , Ponchet, M. , Marion, D. , Lequeu, J. , Delrot, S. , Coutos‐Thevenot, P. and Blein, J.P. (2004) Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol. Biol. Cell, 15, 5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, K.D. , Teece, M.A. and Smart, L.B. (2006) Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140, 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue, B.P. , Thevissen, K. , Hendriks, M. , Eggermont, K. , Goderis, I.J. , Proost, P. , Van Damme, J. , Osborn, R.W. , Guerbette, F. and Kader, J.C. (1995) A potent antimicrobial protein from onion seeds showing sequence homology to lipid transfer proteins. Plant Physiol. 109, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, F.Y. , Yoshioka, K. and Desveaux, D. (2011) The roles of ABA in plant–pathogen interactions. J. Plant Res. 124, 489–499. [DOI] [PubMed] [Google Scholar]
- Chae, K. , Li, Z. , Li, K. , Morikis, D. , Kim, S.T. , Mollet, J.C. , de la Rosa, N. , Tan, K. and Lord, E.M. (2007) Two SCA (stigma/style cysteine‐rich adhesin) isoforms show structural differences that correlate with their levels of in vitro pollen tube adhesion activity. J. Biol. Chem. 282, 33 845–33 858. [DOI] [PubMed] [Google Scholar]
- Chae, K. , Kieslich, C. , Morikis, D. , Kim, S. and Lord, E.M. (2009) A gain‐of‐function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell, 21, 3902–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, K. , Gonong, B.J. , Kim, S.C. , Kieslich, C.A. , Morikis, D. , Balasubramanian, S. and Lord, E.M. (2010) A multifaceted study of stigma/style cysteine‐rich adhesion (SCA)‐like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 61, 4277–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Clay, N.K. , Adio, A.M. , Denoux, C. , Jander, G. and Ausubel, F.M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science, 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha, L. , Sreerekha, M.V. and Mackey, D. (2007) Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 10, 349–357. [DOI] [PubMed] [Google Scholar]
- Curvers, K. , Seifi, H. , Mouille, G. , de Rycke, R. , Asselbergh, B. , Van Hecke, A. , Vanderschaeghe, D. , Höfte, H. , Callewaert, N. , Van Breusegem, F. and Höfte, M. (2010) Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea . Plant Physiol. 154, 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono, A. , Yeats, T.H. , Rose, J.K.C. , Bird, D. , Jetter, R. , Kunst, L. and Samuelsa, L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol‐anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell, 21, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Theulieres, F. , Hirsch, J. , Feng, D.X. , Bittner‐Eddy, P. , Beynon, J. and Marco, Y. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1‐R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA, 99, 2404–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes, L. , Olivier, J. , Peeters, N. , Feng, D.X. , Khounlotham, M. , Boucher, C. , Somssich, I. , Genin, S. and Marco, Y. (2003) Physical interaction between RRS1‐R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA, 100, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Hill, L. , Crooks, C. , Doerner, P. and Lamb, C. (2009) Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. , Gampala, S.S. and Rock, C.D. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell, 14 (Suppl.), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Chu, J. , Sun, X. , Wang, J. and Yan, C. (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC‐MS/MS using single SPE purification and isotope dilution. Anal. Sci. 28, 1081–1087. [DOI] [PubMed] [Google Scholar]
- Grant, M.R. and Jones, J.D.G. (2009) Hormone (dis)harmony moulds plant health and disease. Science, 324, 750–752. [DOI] [PubMed] [Google Scholar]
- Guo, L. , Yan, H.B. , Zhang, X.Y. and Yang, S.H. (2013) Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis . J. Exp. Bot. 64, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich, K. , Tsuda, K. , Blanvillain‐Baufume, S. , Wirthmueller, L. , Bautor, J. and Parker, J.E. (2013) Arabidopsis TNL‐WRKY domain receptor RRS1 contributes to temperature‐conditioned RPS4 auto‐immunity. Front. Plant Sci. 4, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach, B. , Schreiber, L. , Hartung, W. and Dietz, K.J. (1997) Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: implications for the involvement of lipid transfer proteins in wax assembly. Planta, 203, 9–19. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Barlet, X. , Deslandes, L. , Hirsch, J. , Feng, D.X. , Somssich, I. and Marco, Y. (2008) Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil‐borne phytopathogenic bacterium, Ralstonia solanacearum . PLoS ONE, 3, e2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. , Kavanagh, T.A. and Bevan, M.W. (1987) GUS fusions: beta‐glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. , Kim, K.D. and Hwang, B.K. (2005) Identification of pathogen‐responsive regions in the promoter of a pepper lipid transfer protein gene (CALTP1) and the enhanced resistance of the CALTP1 transgenic Arabidopsis against pathogen and environment stresses. Planta, 221, 361–373. [DOI] [PubMed] [Google Scholar]
- Jung, H.W. , Tschaplinski, T.J. , Wang, L. , Glazebrook, J. and Greenberg, J.T. (2009) Priming in systemic plant immunity. Science, 324, 89–91. [DOI] [PubMed] [Google Scholar]
- Kader, J.C. (1975) Proteins and the intracellular exchange of lipids: stimulation of phospholipid exchange between mitochondria and microsomal fractions by proteins isolated from potato tuber. Biochim. Biophys. Acta, 380, 31–44. [PubMed] [Google Scholar]
- Kader, J.C. (1996) Lipid‐transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 627–654. [DOI] [PubMed] [Google Scholar]
- Kim, S.T. , Zhang, K.L. , Dong, J. and Lord, E.M. (2006) Exogenous free ubiquitin enhances lily pollen tube adhesion to an in vitro stylar matrix and may facilitate endocytosis of SCA. Plant Physiol. 142, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme, C. and Roby, D. (1999) Identification of new early markers of the hypersensitive response in Arabidopsis thaliana . FEBS Lett. 459, 149–153. [DOI] [PubMed] [Google Scholar]
- Lee, H. , Xiong, L.M. , Gong, Z.Z. , Ishitani, M. , Stevenson, B. and Zhu, J.K. (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold‐regulated nucleo‐cytoplasmic partitioning. Genes Dev. 15, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Haridon, F. , Besson‐Bard, A. , Binda, M. , Serrano, M. , Abou‐Mansour, E. , Balet, F. , Schoonbeek, H.J. , Hess, S. , Mir, R. , Léon, J. , Lamotte, O. and Métraux, J.P. (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 7, e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhang, Y. , Clarke, J.D. , Li, Y. and Dong, X. (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1‐1. Cell, 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Lord, E. (2000) Adhesion and cell movement during pollination: cherchez la femme. Trends Plant Sci. 5, 368–373. [DOI] [PubMed] [Google Scholar]
- Lorrain, S. , Lin, B. , Auriac, M.C. , Kroj, T. , Saindrenan, P. , Nicole, M. , Balagué, C. and Roby, D. (2004) Vascular associated death1, a novel GRAM domain‐containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell, 16, 2217–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado, A.M. , Doerner, P. , Dixon, R.A. , Lamb, C.J. and Cameron, R.K. (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis . Nature, 419, 399–403. [DOI] [PubMed] [Google Scholar]
- Mang, H.G. , Qian, W. , Zhu, Y. , Qian, J. , Kang, H.G. , Klessig, D.F. and Hua, J. (2012) Abscisic acid deficiency antagonizes high‐temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell, 24, 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch‐Mani, B. and Mauch, F. (2005) The role of abscisic acid and plant–pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Miura, K. , Jin, J.B. , Lee, J. , Yoo, C.Y. , Stirm, V. , Miura, T. , Ashworth, E.N. , Bressan, R.A. , Yun, D.J. and Hasegawa, P.M. (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . Plant Cell, 19, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr, P.G. and Cahill, D.M. (2007) Suppression by ABA of salicylic acid and lignin accumulation and the expression of multiple genes, in Arabidopsis infected with Pseudomonas syringae pv. tomato . Funct. Integr. Genomics, 7, 181–191. [DOI] [PubMed] [Google Scholar]
- Molina, A. and Garcia‐Olmedo, F. (1993) Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 316, 119–122. [DOI] [PubMed] [Google Scholar]
- Molina, A. and Garcia‐Olmedo, F. (1997) Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12, 669–675. [DOI] [PubMed] [Google Scholar]
- Nambara, E. , Suzuki, M. , Abrams, S. , McCarty, D.R. , Kamiya, Y. and McCourt, P. (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana . Genetics, 161, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka, M. , Shirasu, K. , Noutoshi, Y. , Kubo, Y. , Shiraishi, T. and Iwabuchi, M. (2009) RRS1 and RPS4 provide a dual resistance‐gene system against fungal and bacterial pathogens. Plant J. 60 (2), 218–226. [DOI] [PubMed] [Google Scholar]
- Nieuwland, J. , Feron, R. , Huisman, B.A.H. , Fasolino, A. , Hilbers, C.W. , Derksen, J. and Mariani, C. (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell, 17, 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M.T. and Dangl, J.L. (2010) Arabidopsis and the plant immune system. Plant J. 61, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patkar, R.N. and Chattoo, B.B. (2006) Transgenic indica rice expressing nsLTP‐like protein shows enhanced resistance to both fungal and bacterial pathogens. Mol. Breed. 17, 159–171. [Google Scholar]
- Robert‐Seilaniantz, A. , Navarro, L. , Bari, R. and Jones, J.D. (2007) Pathological hormone imbalances. Curr. Opin. Plant Biol. 10, 372–379. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylated antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , López, G. , Ramos, B. , Delgado‐Cerezo, M. , Riviere, M.P. , Llorente, F. , Fernández, P.V. , Miedes, E. , Estevez, J.M. , Grant, M. and Molina, A. (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant Physiol. 160, 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarowar, S. , Kim, Y.J. , Kim, K.D. , Hwang, B.K. , Ok, S.H. and Shin, J.S. (2009) Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long‐distance systemic signaling in tobacco. Plant Cell Rep. 28, 419–427. [DOI] [PubMed] [Google Scholar]
- Schwartz, S.H. , Qin, X. and Zeevaart, J.A. (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura, A. , Moreno, M. and Garcia‐Olmedo, F. (1993) Purification and antipathogenic activity of lipid transfer proteins from the leaves of Arabidopsis and spinach. FEBS Lett. 332, 243–246. [DOI] [PubMed] [Google Scholar]
- Sels, J. , Mathys, J. , De Coninck, B.M. , Cammue, B.P. and De Bolle, M.F. (2008) Plant pathogenesis‐related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. [DOI] [PubMed] [Google Scholar]
- Seo, M. , Aoki, H. , Koiwai, H. , Kamiya, Y. , Nambara, E. and Koshiba, T. (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 45, 1694–1703. [DOI] [PubMed] [Google Scholar]
- Seo, P.J. , Lee, S.B. , Suh, M.C. , Park, M.J. , Go, Y.S. and Park, C.M. (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell, 23, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, P.J. and Park, C.M. (2010) MYB96‐mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis . New Phytol. 186, 471–483. [DOI] [PubMed] [Google Scholar]
- Seo, P.J. , Xiang, F. , Qiao, M. , Park, J.Y. , Lee, Y.N. , Kim, S.G. , Lee, Y.H. , Park, W.J. and Park, C.M. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis . Plant Physiol. 151, 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y. , Xie, Z. , Chen, W. , Glazebrook, J. , Chang, H.S. , Han, B. , Zhu, T. , Zou, G. and Katagiri, F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae . Plant Cell, 15, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- de Torres‐Zabala, M. , Truman, W. , Bennett, M.H. , Lafforguel, G. , Mansfield, J.W. , Egea, P.R. , Bogre, L. and Grant, M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signaling pathway to cause disease. EMBO J. 26, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres‐Zabala, M. , Bennett, M.H. , Truman, W. and Grant, M. (2009) Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. and Van Strien, E.A. (1999) The family of pathogenesis‐related proteins, their activities, and comparative analysis of PR1‐type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Wang, N.J. , Lee, C.C. , Cheng, C.S. , Lo, W.C. , Yang, Y.F. , Chen, M.N. and Lyu, P.C. (2012) Construction and analysis of a plant non‐specific lipid transfer protein database (nsLTPDB). BMC Genomics, 13 (Suppl. 1), S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.Y. , Wu, J.H. , Ng, T.B. , Ye, X.Y. and Rao, P.F. (2004) A non‐specific lipid transfer protein with antifungal and antibacterial activities from the mung bean. Peptides, 25, 1235–1242. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y. , Xiong, L. , Li, W. , Zhu, J.K. and Zhu, J.H. (2011) The plant cuticle is required for osmotic stress regulation of abscisic acid biosynthesis and osmotic stress tolerance in Arabidopsis. Plant Cell, 23, 1971–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Sui, N. , Tang, Y. , Xie, K. , Lai, Y. and Liu, Y. (2010) One‐step, zero‐background ligation‐independent cloning intron‐containing hairpin RNA constructs for RNAi in plants. New Phytol. 187, 240–250. [DOI] [PubMed] [Google Scholar]
- Yasuda, M. , Ishikawa, A. , Jikumaru, Y. , Umezawa, T. , Asami, T. , Maruyama‐Nakashita, A. , Kudo, T. , Shinozaki, K. , Yoshida, S. and Nakashita, H. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid‐mediated abiotic stress responses in Arabidopsis. Plant Cell, 20, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J. and He, S.Y. (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J. Bacteriol. 78, 6399–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Zhang, H. , Yang, Y. , Zhang, Z. , Zhang, H. , Hu, X. , Chen, J. , Wang, X.C. and Huang, R. (2008) Abscisic acid regulates TSRF1‐mediated resistance to Ralstonia solanacearum by modifying the expression of GCC box‐containing genes in tobacco. J. Exp. Bot. 59, 645–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 pLTP3:GUS is induced by Pseudomonas syringae pv. tomato (Pst) infection and abscisic acid (ABA) treatment. hpi, hours post‐inoculation.
Fig. S2 LTP3‐OX plants exhibit enhanced disease symptoms on challenge with Pseudomonas syringae pv. tomato (Pst) DC3000.
Fig. S3 LTP4 is up‐regulated in ltp3‐1 plants on challenge with Pseudomonas syringae pv. tomato (Pst) DC3000.
Fig. S4 Overexpression of LTP3 compromises hypersensitive response (HR) development. hpi, hours post‐inoculation.
Fig. S5 LTP3‐OX stomatal closure is more sensitive to abscisic acid (ABA) treatment. hpi, hours post‐inoculation.
Table S1 List of primers used for transcript analysis by real‐time quantitative reverse transcription‐polymerase chain reaction (RT‐PCR).


