Summary
Chemotaxis enables bacteria to move towards an optimal environment in response to chemical signals. In the case of plant‐pathogenic bacteria, chemotaxis allows pathogens to explore the plant surface for potential entry sites with the ultimate aim to prosper inside plant tissues and to cause disease. Chemoreceptors, which constitute the sensory core of the chemotaxis system, are usually transmembrane proteins which change their conformation when sensing chemicals in the periplasm and transduce the signal through a kinase pathway to the flagellar motor. In the particular case of the soft‐rot pathogen Dickeya dadantii 3937, jasmonic acid released in a plant wound has been found to be a strong chemoattractant which drives pathogen entry into the plant apoplast. In order to identify candidate chemoreceptors sensing wound‐derived plant compounds, we carried out a bioinformatics search of candidate chemoreceptors in the genome of Dickeya dadantii 3937. The study of the chemotactic response to several compounds and the analysis of the entry process to Arabidopsis leaves of 10 selected mutants in chemoreceptors allowed us to determine the implications of at least two of them (ABF‐0020167 and ABF‐0046680) in the chemotaxis‐driven entry process through plant wounds. Our data suggest that ABF‐0020167 and ABF‐0046680 may be candidate receptors of jasmonic acid and xylose, respectively.
Keywords: chemoreceptors, Dickeya dadantii 3937, entry, jasmonic acid, xylose
Introduction
Dickeya dadantii 3937 (Dd3937) is an enterobacterium included in the Top 10 List of phytopathogenic bacteria. This catalogue is based on the scientific/economic importance of the plant bacterial pathogens included (Mansfield et al., 2012). Dickeya dadantii causes soft rot in a wide range of plant species, including important crops, such as potato (Charkowski et al., 2012). Several mechanisms are needed for the adaptation and virulence of D. dadantii inside the plant. The most important virulence factor is the production of a large set of enzymes and isoenzymes (pectate lyases, polygalacturonases, pectin methylesterases, etc.) that disassemble the plant cell wall (Barras et al., 1994). Other processes involved in the adaptation of D. dadantii to the hostile plant conditions are as follows: (i) iron uptake, for which D. dadantii synthesizes two siderophores: chrysobactin and achromobactin (Munzinger et al., 2000; Persmark et al., 1989); (ii) mechanisms to counteract the presence of plant toxic substances, such as multidrug resistance (MDR) efflux pumps (Barabote et al., 2003; Maggiorani Valecillos et al., 2006); (iii) other mechanisms to overcome the presence of antimicrobial compounds (Costechareyre et al., 2013; Llama‐Palacios et al., 2003, 2005; López‐Solanilla et al., 1998; 2001; Rio‐Alvarez et al., 2012). Moreover, the hrp‐encoded type III secretion system (T3SS) is required for the full virulence of D. dadantii (Bauer et al., 1994; Yang et al., 2002; Yap et al., 2005).
All the above‐cited mechanisms are needed for the adaptation and virulence of D. dadantii inside the plant. Nevertheless, bacteria spend the majority of their life outside the plant and bacterial entry into the plant is critical. The life cycle of D. dadantii during the infection of potatoes requires adhesion to the plant surface and penetration into the plant tissues, either via wound sites or through natural openings, such as stomata (Reverchon and Nasser, 2013). For this, an active bacterial movement towards entry sites is required. Chemotaxis, which enables bacterial cells to move towards certain stimuli and away from others (Porter et al., 2011), is crucial for the colonization of the host and for the establishment of a successful infection (Moens and Vanderleyden, 1996).
The bacterial chemotaxis system is the best‐studied biological gradient sensor and has been analysed in detail in Escherichia coli (Sourjik and Wingreen, 2012). The system is composed of bacterial chemoreceptors or methyl‐accepting chemotaxis proteins (MCPs). Bacterial chemoreceptors are hexagonally packed trimers of receptor dimers (Briegel et al., 2012) that recognize specific chemicals (even redox state) at the periplasm and regulate the flagellar function, assisted by a signal transduction pathway composed of the so‐called Che proteins (Hazelbauer and Lai, 2010). The structure of a typical bacterial chemoreceptor is as follows: (i) a sensor domain or ligand‐binding region (LBR), which can be either periplasmic or cytoplasmic, and which detects chemical signals and induces a conformational change; (ii) a HAMP‐type adaptation domain (cytosolic), which transmits the molecular stimulus created by ligand binding at the LBR to the: (iii) MCP domain or signalling domain (cytosolic), which undergoes reversible methylation at multiple sites and transduces the signal to the downstream signalling cascade of Che proteins (Falke and Hazelbauer, 2001). Despite the fact that the chemotaxis system is conserved among bacteria, chemoreceptors differ largely in their protein topology. A large‐scale comparative genomic analysis of the MCP signalling and adaptation domain family (from 312 prokaryotic genomes) allowed the identification of seven major chemoreceptor classes and three distinct structural regions within the cytoplasmic domain: signalling, methylation and flexible bundle subdomains (Alexander and Zhulin, 2007). Moreover, although most LBRs are periplasmic, others are cytoplasmic, and sometimes the LBR is lacking in some chemoreceptors (Lacal et al., 2010a). Much of the knowledge about the ligands for bacterial chemoreceptors comes from studies in E. coli, which has four chemoreceptors (Tar, Tsr, Trg and Tap) and an aeroreceptor (Aer). The ligand profile of each of the four chemoreceptors has been determined: the Tar receptor is primarily for aspartate and maltose, the Tsr receptor for serine and leucine, the Trg receptor for ribose and galactose, and the Tap receptor for dipeptides and pyrimidines (Lacal et al., 2010a). The Aer receptor, which lacks the periplasmic domain, mediates aerotaxis in response to oxygen‐related cellular redox changes (Bibikov et al., 2004). Although there is little information on the ligands for other bacterial chemoreceptors, a considerable number of studies have focused on this issue, not only in animal‐pathogenic bacteria (Nishiyama et al., 2012; Rahman et al., 2014; Sweeney et al., 2013), but also in environmental bacteria (Lacal et al., 2010b; Nichols et al., 2012; Oku et al., 2012).
In the case of plant‐associated bacteria, active chemotaxis may represent an opportunity for the establishment of an interaction with the plant, either symbiotic or pathogenic. Nevertheless, fewer studies exist on chemotaxis and chemoreceptors in plant‐pathogenic bacteria. Chemotaxis is needed for virulence, biofilm formation and competitive fitness in Ralstonia solanacearum (Yao and Allen, 2006, 2007) and for pathogenicity in Agrobacterium tumefaciens (Hawes and Smith, 1989). The chemotactic behaviour towards several compounds has been described in Pseudomonas syringae (Cuppels, 1988) and Xanthomonas campestris (Kamoun and Kado, 1990), although the role of chemotaxis in the interaction with the host has not been elucidated to date. However, a particular case is Dd3937. The contribution of motility and chemotaxis to the pathogenicity of Dd3937 has been studied (Antúnez‐Lamas et al., 2009). Genes involved in the chemotaxis transduction system (cheW, B, Y and Z) and in the structure of the flagellar motor (motA) are required for swimming and entry into Arabidopsis leaves. Its capacity to mediate chemoattraction and chemorepellance in response to compounds such as sugars, amino acids and plant hormones, such as jasmonic acid (JA) (Antunez‐Lamas et al., 2009), has been assessed. JA, which participates in wound signalling in plants (León et al., 2001), is a strong chemoattractant for Dd3937. Furthermore, this perception seems to drive the ingress of this bacterium inside plant tissues through wounds. These results suggest that Dd3937 may have at least one chemoreceptor responsible for the perception of JA. Moreover, jasmonate‐dependent modifications of the pectin matrix of the plant cell wall during potato development function as a defence mechanism targeted by Dd3937 virulence factors (Taurino et al., 2014). This finding links the plant hormone, JA, with the plant cell wall modifications. In dicot secondary cell walls, the major hemicellulose is a polymer of β‐(1,4)‐linked xylose units, called xylan (Rennie and Scheller, 2014), and xylose is released when the plant cell wall is altered. Therefore, it can also be hypothesized that Dd3937 may possess chemoreceptors to detect plant cell wall‐derived compounds.
In this study, we carried out a bioinformatics analysis followed by an experimental approach to identify putative chemoreceptors involved in the perception of compounds released in plant wounds. Moreover, we analysed the role of these chemoreceptors in the entry to the plant apoplast and the colonization ability in Arabidopsis plants.
Results
Dd3937 possesses an unusually high number of chemoreceptors among Enterobacteria
We screened the Dd3937 genome (http://asap.ahabs.wisc.edu/asap/home.php) for the presence of proteins containing the highly conserved MCP signalling domain. The Dd3937 genome encodes 47 proteins containing the MCP domain. Despite the fact that Dd3937 belongs to Enterobacteria, this result contrasts with the average number of MCP genes found in other Enterobacteria: 29 MCPs per genome (Lacal et al., 2010a). The amino acid sequences of these 47 chemoreceptors were searched for transmembrane regions (TMs) using the DAS server (Table 1). According to the presence of TMs, Dd3937 chemoreceptors were classified into six different topologies (Fig. 1A), as described previously (Lacal et al., 2010a). In the case of Dd3937, all topologies, including class Ia, show similar results to those reported by Lacal et al. (2010a). Furthermore, in the most abundant topology Ia, we grouped the LBRs according to their length (Fig. 1B). Dd3937 LBRs (Ia) can be clustered into two groups as described previously by Lacal et al. (2010a). Most LBRs (60%) are characterized by sizes corresponding to cluster I, the majority of the LBRs being those with sizes between 150 and 159 amino acids (Fig. 1B).
Table 1.
Amino acid coordinates of methyl‐accepting chemotaxis protein (MCP) domains and transmembrane regions (TMs) determine the topology of the Dd3937 chemoreceptor
| ID | TM | MCP | Topology classc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ASAPa | UNIPROTb | i1 | j1 | i2 | j2 | i3 | j3 | i | j | |
| ABF‐0014536 | E0SHS4 | 10 | 32 | 181 | 203 | 334 | 501 | Ia | ||
| ABF‐0014618 | E0SM30 | 36 | 58 | 214 | 232 | 367 | 531 | Ia | ||
| ABF‐0014722 | E0SF47 | 7 | 29 | 190 | 209 | 344 | 511 | Ia | ||
| ABF‐0014824 | E0SDP4 | 10 | 32 | 183 | 205 | 340 | 507 | Ia | ||
| ABF‐0015168 | E0SH41 | 5 | 24 | 177 | 199 | 335 | 501 | Ia | ||
| ABF‐0015513 | E0SBF1 | 20 | 42 | 200 | 222 | 360 | 523 | Ia | ||
| ABF‐0015600 | E0SE76 | 12 | 34 | 278 | 295 | 431 | 598 | Ia | ||
| ABF‐0015603 | E0SE77 | 38 | 60 | 300 | 317 | 451 | 618 | Ia | ||
| ABF‐0016115 | E0SN86 | 13 | 35 | 269 | 285 | 421 | 588 | Ia | ||
| ABF‐0016585 | E0SIA2 | 10 | 32 | 189 | 211 | 343 | 510 | Ia | ||
| ABF‐0016979 | E0SEW8 | 10 | 32 | 183 | 205 | 343 | 510 | Ia | ||
| ABF‐0017097 | E0SCJ7 | 17 | 39 | 272 | 290 | 427 | 593 | Ia | ||
| ABF‐0017662 | E0SFJ0 | 5 | 27 | 187 | 204 | 342 | 508 | Ia | ||
| ABF‐0017665 | E0SFJ2 | 36 | 58 | 213 | 235 | 372 | 538 | Ia | ||
| ABF‐0017668 | E0SFJ3 | 10 | 32 | 184 | 206 | 344 | 511 | Ia | ||
| ABF‐0017672 | E0SFJ4 | 10 | 32 | 184 | 206 | 347 | 514 | Ia | ||
| ABF‐0017674 | E0SFJ5 | 20 | 39 | 201 | 223 | 359 | 526 | Ia | ||
| ABF‐0017863 | E0SI11 | 10 | 32 | 189 | 211 | 346 | 510 | Ia | ||
| ABF‐0017896 | E0SHY9 | 11 | 33 | 178 | 200 | 337 | 503 | Ia | ||
| ABF‐0018502 | E0SEF0 | 10 | 32 | 187 | 209 | 346 | 512 | Ia | ||
| ABF‐0018541 | E0SFF6 | 10 | 32 | 182 | 204 | 336 | 505 | Ia | ||
| ABF‐0018585 | E0SJX6 | 20 | 42 | 199 | 221 | 357 | 524 | Ia | ||
| ABF‐0018754 | E0SIM4 | 10 | 32 | 179 | 201 | 336 | 503 | Ia | ||
| ABF‐0018765 | E0SIN3 | 20 | 42 | 204 | 226 | 356 | 518 | Ia | ||
| ABF‐0018892 | E0SJ51 | 20 | 42 | 194 | 216 | 355 | 522 | Ia | ||
| ABF‐0019050 | E0SAP6 | 15 | 37 | 270 | 292 | 427 | 591 | Ia | ||
| ABF‐0019306 | E0SF78 | 13 | 35 | 282 | 299 | 435 | 601 | Ia | ||
| ABF‐0019718 | E0SHT9 | 7 | 29 | 180 | 202 | 336 | 502 | Ia | ||
| ABF‐0019790 | E0SF98 | 10 | 32 | 187 | 209 | 344 | 510 | Ia | ||
| ABF‐0019851 | E0SM37 | 7 | 29 | 179 | 194 | 333 | 499 | Ia | ||
| ABF‐0019852 | E0SM36 | 20 | 42 | 195 | 214 | 348 | 515 | Ia | ||
| ABF‐0019855 | E0SM35 | 13 | 35 | 185 | 204 | 340 | 507 | Ia | ||
| ABF‐0019858 | E0SM34 | 31 | 53 | 202 | 222 | 357 | 522 | Ia | ||
| ABF‐0020167 | E0SDB0 | 10 | 29 | 178 | 200 | 340 | 507 | Ia | ||
| ABF‐0020252 | E0SF13 | 25 | 47 | 288 | 314 | 439 | 604 | Ia | ||
| ABF‐0020431 | E0SDX8 | 5 | 27 | 168 | 190 | 327 | 489 | Ia | ||
| ABF‐0046680 | E0SMA1 | 58 | 77 | 348 | 370 | 504 | 670 | Ia | ||
| ABF‐0017537 | E0SD21 | 10 | 32 | 142 | 147 | 421 | 586 | Ia | ||
| ABF‐0017419 | E0SJN1 | 138 | 160 | 295 | 462 | Ib | ||||
| ABF‐0014726 | E0SF49 | 159 | 178 | 188 | 200 | 335 | 497 | II | ||
| ABF‐0019309 | E0SF75 | 164 | 181 | 186 | 205 | 341 | 506 | II | ||
| ABF‐0014843 | E0SDM9 | 147 | 151 | 165 | 180 | 185 | 196 | 337 | 499 | III |
| ABF‐0016380 | E0SEN9 | 30 | 52 | 183 | 205 | 336 | 350 | 490 | 656 | III |
| ABF‐0018511 | E0SEE2 | 13 | 30 | 196 | 203 | 315 | 330 | 468 | 633 | III |
| ABF‐0016436 | E0SMM7 | 297 | 417 | IVa | ||||||
| ABF‐0017090 | E0SCK2 | 474 | 615 | IVa | ||||||
| ABF‐0017824 | E0SI40 | 17 | 131 | IVa | ||||||
Nomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php).
Nomenclature from the UNIPROT database (http://www.uniprot.org/).
Topology class according to Lacal et al. (2010a).
Figure 1.
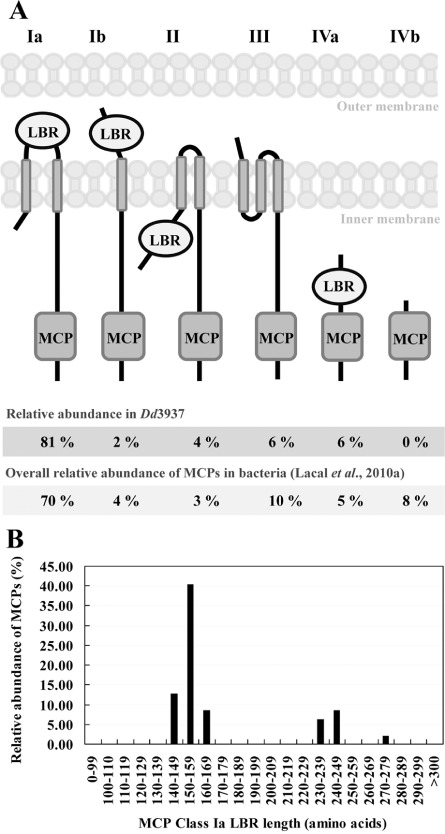
(A) Classification of Dd3937 chemoreceptors into six different topologies (roman numerals); 47 MCP sequences from Dd3937 were analysed. Classification is based on clustering annotated by Lacal et al. (2010a). The prediction of transmembrane regions (TMs) was performed using the DAS server (Cserzo et al., 1997). Arabic numbers indicate the relative abundance of receptors with a given topology in Dd3937 and in bacteria according to Lacal et al. (2010a). MCP, methyl‐accepting chemotaxis protein; LBR, ligand‐binding region. (B) Relative abundance of Dd3937 MCP sequences (belonging to class Ia) according to the size of the LBR (amino acids).
LBR sequences were analysed by Clustal W2 (Larkin et al., 2007), searching for diversity among the sequences (Table S1, see Supporting Information). The results showed that the level of sequence conservation was generally low and no duplications were observed. Moreover, LBRs of each of the 47 MCP proteins in Dd3937 were searched for Pfam components (Finn et al., 2014), using a customized BioPerl (Stajich et al., 2002) script. Only hits with an E‐value of less than or equal to 0.005 were retained (Table S2, see Supporting Information). The results showed that 23 of the LBRs fall into the annotated domain 4HB_MCP_1 (four helix bundle sensory module for signal transduction) (Ulrich and Zhulin, 2005), which is a ubiquitous sensory module in prokaryotic signal transduction. Five are homologues to the annotated domain TarH, a member of a family of transmembrane receptors that mediate the chemotactic response in certain enteric bacteria, such as Salmonella typhimurium and E. coli (Kim et al., 1996). Homology with other annotated domains involved in sensory processes can be found.
Global blast of LBRs from Dd3937 chemoreceptors suggests putative receptors for plant compounds
In order to identify Dd3937 chemoreceptors involved in the sensing of plant‐specific compounds, LBRs were further analysed by bioinformatics methods. LBR sequences were extracted from the protein sequence as described in Experimental procedures. LBR sequences were subjected to local blast against a bacterial genome database (this database contains complete bacterial genomes available as of 14 February 2014 from GeneBank Ref Seq) using an ad hoc PERL script (for more details, see Experimental procedures). Table S3 (see Supporting Information) shows similar LBR sequences (E‐value ≤ 10−30) to those found in other bacteria. Figure 2A shows the distribution of Dd3937 LBRs among the following groups of bacteria: (i) other Dickeya strains; (ii) plant‐pathogenic bacteria; (iii) animal‐pathogenic bacteria (considering the occurrence of a high number of similar LBR sequences in animal pathogens); and (iv) non‐pathogenic bacteria (including bacteria present in plants, soil and water). Among the 47 LBRs of Dd3937, nine were widely distributed among bacteria regardless of their niche (animals, plants, soils and water). Another seven LBRs, apart from being present in plant‐pathogenic bacteria, were also found in environmental (non‐pathogenic) bacteria. Moreover, another four LBRs, apart from being present in Dickeya strains, were also found in environmental (non‐pathogenic) bacteria. Only for one LBR were similar sequences found exclusively in animal and environmental bacteria. The largest group, 13 LBRs, was found in other plant‐pathogenic bacteria besides the Dickeya genus, and another large group containing 12 members included LBRs exclusively present in the Dickeya genus. This means that 56% of Dd3937 LBRs show similarity only with other LBRs from phytopathogenic bacteria(Fig. 2B). Interestingly, one Dd3937 LBR was not found in other Dickeya strains, but was found in other plant‐pathogenic bacteria.
Figure 2.
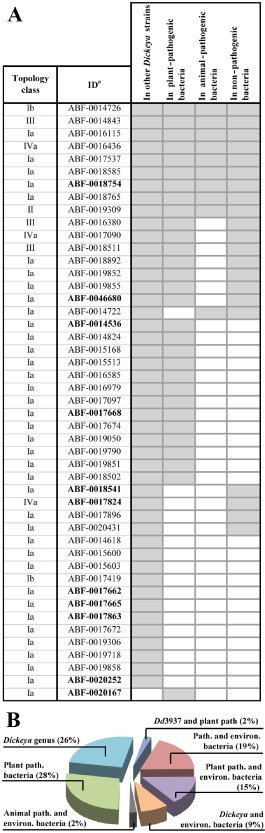
(A) Distribution of Dd3937 ligand‐binding regions (LBRs) among bacteria. aNomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php). In bold, genes selected for further analyses. Grey boxes indicate that similar sequences to a given Dd3937 LBR were found in the global blast analysis. (B) Schematic representation of the distribution of Dd3937 LBRs among bacteria according to their ecological niche. path., pathogenic; environ., environmental.
Mutants in individual chemoreceptors of Dd3937 are affected in the perception of JA and xylose
In order to unveil the role of the perception of plant molecules during the interaction of Dd3937 with plants, chemoreceptors from the different groups shown in Fig. 1 were selected. Thus, ABF‐0018754 was selected as a control for chemotaxis assays as it is the candidate for serine perception in Dd3937. ABF‐0046680 was chosen from the group of chemoreceptors whose LBRs showed similarity with those of plant‐pathogenic bacteria, but also environmental bacteria. ABF‐0014536 and ABF‐0017668 were selected from the group of chemoreceptors whose LBRs showed similarity only with those of plant‐pathogenic bacteria; ABF‐0018541 and ABF‐0017824 were selected from the group of chemoreceptors whose LBRs showed similarity with those of Dickeya strains (not with other plant‐pathogenic bacteria) and environmental bacteria. ABF‐0017662, ABF‐0017665, ABF‐0017863 and ABF‐0020252 were selected from the group of chemoreceptors whose LBRs showed similarity only with those of Dickeya strains. Finally, ABF‐0020167 was chosen because of its particularity of not showing similarity with other Dickeya strains, but with other plant‐pathogenic bacteria. These chemoreceptors were mutagenized by different approaches (for more details, see Experimental procedures). ABF‐0018754, chosen as a control for chemotaxis assays, restored serine perception when heterologously expressed in the non‐chemotactic E. coli UU1250 strain (Bibikov et al., 2004) (Fig. S1, see Supporting Information).
In order to elucidate how the mutations in the chemoreceptors cited above affected the movement of Dd3937 towards putative entry sites, such as wounds, the chemotactic response of wild‐type (WT) and mutant strains towards JA and xylose was analysed. As mentioned previously, JA secreted in plant wounds is a strong chemoattractant for Dd3937 (Antunez‐Lamas et al., 2009) and xylose is released when the plant cell wall is altered.
The analysis of the chemotaxis towards these plant compounds was performed using the classical ‘in‐plug’ assay (Tso and Adler, 1974), which is based on the ability of bacteria to swim in soft agar and move towards/away from a given chemical. The results were recorded from soft agar–bacteria plates based on either the observation of attraction haloes or of the absence of a chemotactic response (Fig. 3). Independent of the intensity of the haloes, these were considered as a positive chemoattraction when an apparent ring of accumulated cells was found at a certain distance from the plug.
Figure 3.
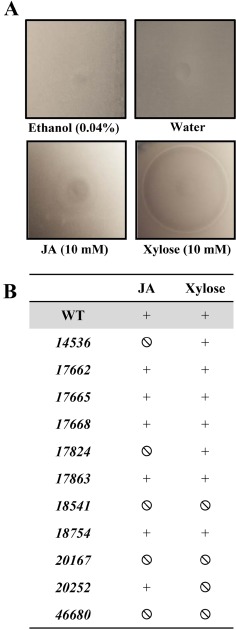
(A) Typical attraction haloes of Dd3937 wild‐type (WT) towards jasmonic acid (JA) and xylose in the ‘in‐plug’ assay. A drop containing a given chemical, either JA (10 mm) or xylose (10 mm), or their corresponding controls, ethanol (0.04% v/v) and water, was deposited on the (0.25%) soft‐agar bacteria, and haloes were recorded after 20 min. (B) Chemotactic response of Dd3937 strains towards JA and xylose. Chemotactic response of Dd3937 WT and mutant strains was evaluated in the presence of a concentration gradient of JA (10 mm) and xylose (10 mm). The chemotactic response was considered as attraction (+) or non‐chemotactic response (Ø) in comparison with that of the WT strain for each chemical. Similar results were obtained in three independent experiments.
From the results of the ‘in‐plug’ assays, we selected mutants 14536, 17824, 18541, 20167 and 46680 for quantitative chemotaxis assays towards JA, and mutants 18541, 20167, 20252 and 46680 for quantitative chemotaxis assays towards xylose. For this purpose, a Palleroni‐like assay was performed using 5‐μL tips instead of glass capillaries, as handling allows a high‐throughput screening in comparison with the classic Palleroni assay (Palleroni, 1976). This assay allows the calculation of the chemotaxis index (CI). As bacterial cells can move actively towards/away from a given chemical contained in the capillary, values of CI greater than 1.0 are considered as chemoattraction and values less than 1.0 as chemorepulsion (Fig. 4). The results in Fig. 4 indicate that, in the case of JA, only 18541 and 20167 mutants showed a reduced chemoattraction towards this plant hormone. In the case of xylose, 20252 and 46680 mutants were affected in the perception of this sugar. As a control for the quantitative (capillary) assays, chemotaxis towards serine of the WT and mutant 18754 was assayed in each experiment (data not shown). Strains overexpressing the WT chemoreceptors restored the ability to develop a chemoattraction response in the presence of the respective compound (Fig. 4B, D).
Figure 4.
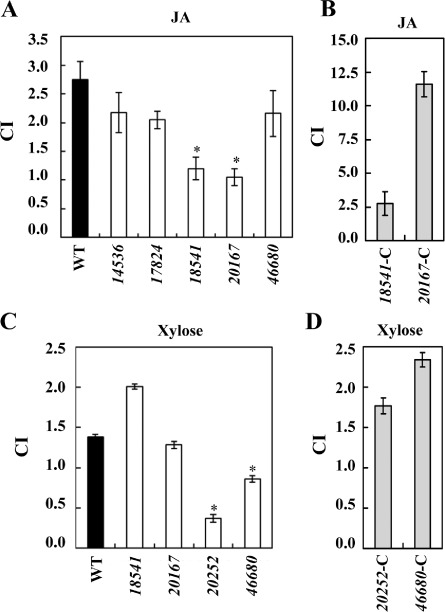
Quantitative chemotaxis assays of Dd3937 strains towards jasmonic acid (JA) and xylose. Bars represent the chemotaxis index (CI) for wild‐type (WT) and mutant strains towards 1 mm JA (A) and 1 mm xylose (C). Those mutants showing a different chemotaxis towards JA or xylose were complemented and the chemotaxis towards 1 mm JA (B) or xylose (D) was tested using the same procedure. CI is defined as the number of cells that accumulate after 20 min within a tip containing a given compound divided by the number of cells accumulated within a tip containing the solvent. The means and standard errors are shown. Error bars represent the standard error of the mean (SEM). Asterisk indicates significant differences between WT and mutant CI determined according to Student's t‐test (P < 0.05). Similar results were obtained in three independent experiments.
To check that these results were not the product of an altered chemotactic ability, Dd3937 mutants were tested for chemotaxis towards serine. All mutants, except 18754 (candidate serine receptor of Dd3937), showed the same chemotaxis towards serine as did the WT strain (Fig. S2, see Supporting Information). Moreover, to demonstrate that these mutants do not have a general defect in motility, we carried out a swimming assay in soft agar‐rich medium. This assay, which reflects the sum of taxis towards compounds present in the medium, showed that the spreading of these strains was not altered with respect to that of the WT strain (Fig. S3, see Supporting Information).
Dd3937 chemoreceptors contribute in a different manner to entry into Arabidopsis leaves
As stated above, how pathogens enter plant tissues is critical for pathogenesis. To ascertain the particular relevance of the selected Dd3937 chemoreceptors in this process, and how this could affect the subsequent development of symptoms, WT and mutant strains were challenged to entry and colonization assays in Arabidopsis leaves. Arabidopsis thaliana has been described and characterized previously as a susceptible host of Dd3937 (formerly Erwinia chrysanthemi 3937) (Dellagi et al., 2005; Fagard et al., 2007).
To compare the ability to detect plant compounds released in a wound, we first performed entry assays in A. thaliana Col‐0 leaves, as described previously (Antunez‐Lamas et al., 2009). For each particular strain, the cells which entered the plant apoplast were measured relative to the initial inoculum of cells (cells recovered from the leaf/cells put on the wound; expressed as a percentage), and these ratios were used for comparison with the WT strain ratio. Figure 5A shows the differences in entry ability of the 10 MCP mutants tested in comparison with the WT strain. Two mutants, 20167 and 46680, were significantly affected in their ability to enter the plant apoplast in contrast with the WT strain. Entry assays in A. thaliana leaves were performed as described previously. 20167‐C and 46680‐C complemented strains showed entry percentages close to those of the WT levels (Fig. 5B).
Figure 5.
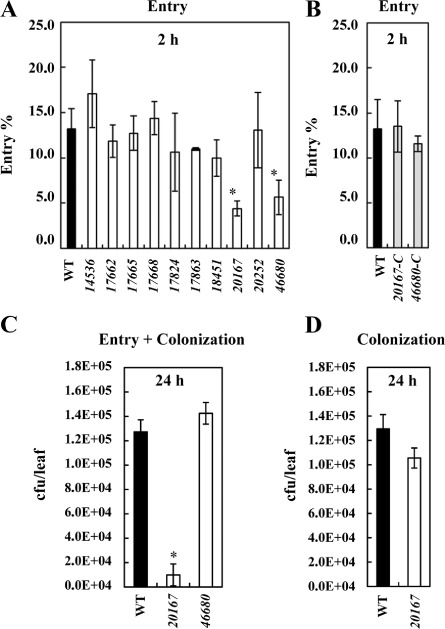
(A) Entry of Dd3937 wild‐type (WT) and mutant strains in Arabidopsis thaliana leaves. For each strain, 105 cells were placed on a wound. This process was repeated in 15 leaves from at least five different plants. Bacterial populations inside the leaf were estimated after 2 h. (B) Entry of Dd3937 WT and complemented strains in A. thaliana leaves. For each strain, 105 cells were placed on a wound. This process was repeated in 15 leaves from at least five different plants. Bacterial populations inside the leaf were estimated after 2 h. (C) Entry‐dependent colonization assay in A. thaliana leaves. 105 cells of WT or mutant (20167 and 46680) strains were placed on a wound. This process was repeated in 15 leaves from at least five different plants. Bacterial populations inside the leaf were estimated after 24 h. (D) Non‐entry‐dependent pathogenicity assay in A. thaliana leaves. 2 × 106 cells of WT or 20167 strains were infiltrated into the abaxial side of leaves. This process was repeated in 15 leaves from at least five different plants. The bacterial population inside the leaf was estimated after 24 h. cfu, colony‐forming unit. For (A–D), means and standard errors are shown. Error bars represent standard error of the mean (SEM). Asterisk indicates significant differences between WT and mutant population (estimated either as percentage entry or total population inside the leaves) determined according to Student's t‐test (P < 0.05). Similar results were obtained in three independent experiments.
To ascertain whether the delay in entry into the plant apoplast exhibited by 20167 and 46680 mutants could affect the subsequent establishment of infection, the same assay described above was performed, changing the incubation time from 2 to 24 h (Fig. 5C). Figure 5C shows that the delay in entry of 20167 observed at 2 h affected the population inside the leaves at 24 h, whereas 46680 reached WT population levels at 24 h. To check that the decrease in the 20167 population inside the leaves at 24 h was not a result of impairment in colonization ability, but a direct consequence of reduced entry, a non‐entry‐dependent colonization assay was performed. For this, WT and 20167 cell suspensions were infiltrated into the abaxial side of A. thaliana leaves and bacterial populations were estimated at 24 h. Figure 5D shows that, when infiltrated, the population of the 20167 mutant is at the same level as that of the WT strain.
Discussion
The E. coli chemotactic pathway has provided a paradigm for sensory systems in general. However, the increasing number of sequenced bacterial genomes shows that, although the central sensory mechanism seems to be common to all bacteria, there is added complexity in a wide range of species (Krell et al., 2011; Wadhams and Armitage, 2004). Indeed, the analysis of bacterial genomes reveals that motile bacteria differ enormously in the number of chemoreceptors. Chemoreceptor abundance in a bacterial genome ranges from 64 in Magnetospirillum magnetotacticum (Alexandre et al., 2004) to a single chemoreceptor in other bacteria (Lacal et al., 2010a). Forty‐seven chemoreceptors have been identified in Dd3937 through bioinformatics analyses. In comparison with its close Enterobacteria relatives (Shigella, Salmonella, Yersinia, Escherichia spp.), which show a small number of chemoreceptors, for instance five in E. coli (Lacal et al., 2010a; Wadhams and Armitage, 2004), Dd3937 possesses a large number of chemoreceptors. This is congruent with the hypothesis that microorganisms living in complex and variable environments must be able to detect a greater variability of changing conditions. Indeed, genome sequencing has revealed that many microorganisms from aquatic and soil environments possess large numbers of chemoreceptors (Alexandre et al., 2004). Moreover, to assess the relationship between the abundance of chemoreceptors and lifestyle, regardless of taxonomic classification, Lacal et al. (2010a) analysed 264 bacterial genomes for the presence of chemoreceptors, and similar results were obtained, showing that the number of chemoreceptors per genome depends mostly on bacterial lifestyle and metabolic diversity, but weakly on genome size. Among the 264 bacterial genomes analysed, only three phytopathogenic bacteria, Xylella fastidiosa, Xanthomonas oryzae pv. oryzae and Agrobacterium tumefaciens C58, were included. These authors also analysed the topology of the chemoreceptors defining several classes. In the case of Dd3937, the most abundant topology is that of class Ia, as it mainly occurs in bacteria (Lacal et al., 2010a). For the rest of the classes, similar results to those shown for bacteria were obtained in Dd3937. Moreover, the analysis of the LBR length in the most abundant topology class (Ia) also showed the same clustering into two defined groups described by Lacal et al. (2010a).
The detailed analysis of the distribution of Dd3937 LBRs among known bacterial genomes allowed us to determine that more than 50% of the LBRs show high similarity only with sequences from other plant‐pathogenic bacteria. These in silico‐based data suggest that these chemoreceptors would be implicated in the perception of plant‐derived compounds. The large number of putative chemoreceptors involved in this process demonstrates the relevance of the perception of plant compounds during the interaction. Indeed, the identification of host–microbe interaction factors in the genome of Dd3937 with supervised machine learning shows that methyl‐accepting chemotaxis genes are highly enriched among the predicted host–microbe interaction factors in this bacterium (Ma et al., 2014).
The results from the ‘in‐plug’ chemotaxis assays indicated that more than one mutant strain showed an altered chemotactic response towards JA or xylose with respect to the WT strain. A possible explanation of these findings would be that a given compound can be detected by more than one chemoreceptor. Indeed, there are several examples in the literature showing this (Kato et al., 2008). The best studied example is the recognition of amino acids by the paralogous receptors PctA, PctB and PctC of Pseudomonas aeruginosa (Rico‐Jiménez et al., 2013). However, the opposite phenomenon has also been described: one chemoreceptor is able to sense more than one compound. For example, McpS, a Pseudomonas putida KT2440 chemoreceptor for tricarboxylic acid (TCA) cycle intermediates, is involved in the chemotactic response towards different ligands, such as malate, succinate and fumarate (Lacal et al., 2010b). Other examples of multi‐ligand chemoreceptors are as follows: Tcp receptor in S. typhimurium, which mediates positive chemotaxis to citrate and negative chemotaxis to phenol (Yamamoto and Imae, 1993), PA2652 of P. aeruginosa, which mediates chemotaxis to malate, but not to any of the remaining TCA cycle intermediates (Alvarez‐Ortega and Harwood, 2007), and McpX of Vibrio cholera, which functions as a chemoreceptor for multiple amino acids (Nishiyama et al., 2012). Another hypothesis which cannot be ruled out is that a mutation in a given chemoreceptor alters the chemotactic response of the remaining receptors, through gene expression reprogramming. There is much information missing about the gene regulation of chemoreceptors. For example, it is unknown whether all receptors are constitutively expressed, whether they are inducible under certain stimuli and whether there is fine regulation depending on the metabolic needs. It has been shown that the responses of E. coli to two opposing chemoattractant gradients are dependent on the chemoreceptor ratio (Kalinin et al., 2010). Therefore, if Dd3937 chemoreceptor ratios are being altered through individual mutations, one can expect several changing chemotactic behaviours, as found in this study.
The analysis of the chemotactic response to xylose and JA using a quantitative methodology based on capillary assays showed that, of the five mutants found to be altered in chemotaxis towards JA in the in‐plug assay, only two were affected in the capillary assays. In the case of xylose, of the four mutants altered, only two consistently showed a reduced chemotaxis in the capillary assays. This discrepancy could be a result of the nature of the medium employed, i.e. semi‐solid agar in the ‘in‐plug’ assay and liquid medium in the capillary assay. Both the availability of the chemoattractant to be perceived and the effects of the conditions during the assay on the motility ability could be responsible for the different results obtained. An important element to take into account is that chemotaxis is a flagellum‐dependent phenomenon and the number of flagella is intimately linked to the characteristics of the medium (Jarrell and McBride, 2008).
Although other chemoreceptors could be involved in JA perception, we focused our studies on the role of ABF‐0018541 and ABF‐0020167 in the active entry process through wounds as the corresponding mutant strains consistently showed a decreased chemotaxis towards this hormone in the two assays performed. Our experiments showed that only the 20167 mutant displayed impaired entry ability to the plant apoplast, which suggested a pivotal role for this receptor during this process. In the case of xylose, although 20252 and 46680 mutants showed reduced chemoattraction towards this sugar in both ‘in‐plug’ and quantitative assays, only 46680 was impaired in the entry process, which also suggested a key role for this receptor during the entry to the plant apoplast. However, when analysing the colonization ability, although 20167 and 46680 mutants displayed impaired entry ability, only the 20167 strain was altered in the establishment of a sufficiently large population to prosper in the hostile plant conditions. When 20167 was infiltrated, the bacterial population reached the same level as that of the WT strain. Therefore, the altered perception of compounds produced in wounds, such as JA, is the main contributor to the impaired colonization ability of this strain, which does not seem to be related to the colonization ability or fitness inside the plant tissue. In the case of 46680, although under laboratory conditions the mutant is not affected in its colonization ability, a role in this process under natural conditions cannot be ruled out.
It is noteworthy that, in our in silico analysis, similar sequences to the LBR from ABF‐0020167 were only found in plant‐pathogenic bacterial genomes. It is also noteworthy that no matches with other Dickeya strains were found. Moreover global blast analyses did not show any match of this LBR with Pectobacterium carotovorum, a phylogenetically related bacterium which does not sense JA (Antunez‐Lamas et al., 2009). Therefore, ABF‐0020167 could be considered as a potential receptor for JA, although further analyses are required to demonstrate the interaction of its LBR with this plant hormone. It is noteworthy that ABF‐0018541 has also been identified as a putative JA chemoreceptor in our capillary assays. The sequence similarity between the LBR of these two chemoreceptors is very low, but it cannot be ruled out that a similar tertiary structure exists, allowing the binding of the same type of molecules. Indeed, both LBR sequences show homology with the annotated Pfam domain 4HB_MCP_1 (four helix bundle sensory module for signal transduction) (Ulrich and Zhulin, 2005), which is a ubiquitous sensory module in prokaryotic signal transduction. A detailed structural analysis of these regions will shed light on their implications in JA binding.
With regard to the ABF‐0046680 chemoreceptor, the 46680 mutant was impaired in the ability to sense xylose, but further analyses are required to demonstrate whether: (i) the LBR of ABF‐0046680 binds directly to xylose; and (ii) xylose is being released at wound sites.
Moreover, it is expected that the contribution of these chemoreceptors to the efficiency of infection by Dd3937 will depend on the host. As demonstrated previously, the chemotaxis process depends on the presence of a concentration gradient of the compound being sensed (Kalinin et al., 2010), and the gradient created in a wound for a given compound may be different for different hosts.
In summary, the study of Dd3937 chemoreceptors by bioinformatics analyses and experimental approaches has allowed us to identify the ABF‐0020167 and ABF‐0046680 chemoreceptors as putative receptors for biomolecules of great importance in plant defence: JA and xylose, respectively. Moreover, entry assays through wounds performed in A. thaliana leaves suggest that the perception of plant compounds released from a wound by the chemoreceptors ABF‐0020167 and ABF‐0046680 is pivotal to the entry into the plant apoplast and to the achievement of a population density sufficient to prosper in the plant apoplast. Further research is needed to demonstrate the interaction between the LBRs and these molecules.
Experimental Procedures
Identification of MCP domains
For sequence analyses, the Dd3937 genome sequence was downloaded from the ASAP database (http://asap.ahabs.wisc.edu/asap/home.php). The Dd3937 genome was searched for genes encoding chemoreceptors. The search for the MCP domain was performed using an internal pipeline. The MCP domain was downloaded from the Pfam website of the Sanger Centre (http://pfam.sanger.ac.uk/) using the Pfam identifier PF00015. This alignment was used to build hidden Markov models (HMMs) by means of HMMER v2.3.2 (Eddy, 1998). Finally, a customized BioPerl (Stajich et al., 2002) script combined with HMMER searched for HMM hits using an E‐value of 0.005 to search for MCP‐containing proteins.
Identification of TMs and determination of LBR sequence
TM domains were identified one by one using the DAS algorithm at the DAS server (Cserzo et al., 1997, 2002). The complete sequences of chemoreceptors were searched for TMs considering a cut‐off value of –2.5. Then, the LBR was extracted from the protein sequence as: (i) the sequence in between two TM domains when present; (ii) the sequence in between the amino terminus and the TM when only one TM was present; (iii) the sequence in between the amino terminus and the MCP domain when no TM was present; or (iv) the sequence in between the carboxyl terminus and the MCP domain when no TM was present. When three TMs were found, only the periplasmic LBR was considered. LBRs were identified in all MCPs and aligned with Clustal W2, which renders the percentage identity matrix. The sizes of LBRs for the chemoreceptors belonging to topology Ia were clustered into groups of 10‐amino‐acid intervals.
Identification of Pfam domains in the LBR sequence
HMMs based on protein families were downloaded from the Pfam (Finn et al., 2014) website (ftp://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/). We focused on the Pfam component, so‐called Pfam‐A, corresponding to a number of high‐quality, manually curated families. LBRs of each of the 47 MCP‐associated proteins in Dd3937 were searched for the above Pfam domains using a customized BioPerl (Stajich et al., 2002) script. Only hits with an E‐value of less than or equal to 0.005 were retained.
Global blast analysis of LBR sequences
Amino acid sequences of LBRs were searched against a database of bacterial genome sequences. For this, the complete bacterial sequences of 2997 chromosomes and 2164 plasmids, available as of 14 February 2014, were downloaded from GenBank Refseq. Local blast (E‐value ≤ 10−30) was performed using an ad hoc PERL script. The output file was processed manually.
Microbiological methods
The bacterial strains and plasmids used in this study are listed in Table 2. Unless indicated, strains of E. coli were grown at 37 °C in Luria–Bertani (LB) medium (Sambrook et al., 1989). Unless indicated, strains of Dd3937 were cultivated at 28 °C in nutrient broth (NB) (per litre: yeast extract, 1 g; beef extract, 2 g; NaCl, 5 g; bactopeptone, 5 g) or King's B (KB) medium (King et al., 1954). When required, antibiotics were used at the following final concentrations (μg/mL): ampicillin, 100; chloramphenicol, 30; gentamycin, 20; kanamycin, 20; spectinomycin, 50; streptomycin, 20. Bacterial growth was monitored at an optical density at 600 nm (OD600) using a Jenway 6300 spectrophotometer Jenway (Stone/Staffordshire/United Kingdom). For Dd3937 strains used in this study, OD600 = 1.0 corresponds approximately to 5 × 108 colony‐forming units (cfu).
Table 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi‐1 relA1 | Hanahan (1983) |
| CC118 λpir | Δ (ara, leu) araD ΔlacX74 galE galK phoA20 thi‐1 rps rpoB argE(Am) recA1 lysogenized with λpir phage; Spr | Herrero et al. (1990) |
| S17‐1λpir | λpir lysogen of S17‐1 | de Lorenzo, CIB, Madrid, Spain |
| UU1250 | Δaer‐1 Δ(tar‐tap)5201 Δtsr‐7028 Δtrg‐100 ygjG::Gm zbd::Tn5 | Bibikov et al. (2004) |
| K12 | Wild‐type strain | CBGP, Madrid Spain |
| D. dadantii 3937 | ||
| 3937 | Wild‐type strain | Kotoujansky et al. (1985) |
| 14536 | 14536::Tn5 Spr derivative of 3937 | This work |
| 17662 | 17662::Tn5 Spr derivative of 3937 | This work |
| 17665 | 17665::Tn7 Kmr derivative of 3937 | This work |
| 17668 | 17668::Tn7 Kmr derivative of 3937 | This work |
| 17824 | 17824::miniTn5 Spr derivative of 3937 | This work |
| 17863 | 17863::miniTn5 Spr derivative of 3937 | This work |
| 18541 | 18541::miniTn5 Spr derivative of 3937 | This work |
| 18754 | 18754::miniTn5 Spr derivative of 3937 | This work |
| 20167 | 20167::pKNG101 Smr derivative of 3937 | This work |
| 20252 | 20252::Tn5 Spr derivative of 3937 | This work |
| 46680 | 46680::pKNG101 Smr derivative of 3937 | This work |
| Plasmids and BACs | ||
| pKNG101 | R6K origin sacB marker exchange vector; mob+, Smr | Kaniga et al. (1991) |
| pGEM‐T easy® | Apr | Invitrogen |
| pGEM‐T easy‐18754 | Apr | This work |
| pGEM‐T easy‐4387 | Apr | This work |
| TrueBlue‐BAC2® | Cmr | Genomics One |
| TrueBlue‐BAC2‐18541 | Cmr | This work |
| TrueBlue‐BAC2‐20167 | Cmr | This work |
| TrueBlue‐BAC2‐20252 | Cmr | This work |
| TrueBlue‐BAC2‐46680 | Cmr | This work |
DNA manipulation
The plasmids used in this study are described in Table 2. To analyse the functions of the genes of interest in Dd3937, mutants in these genes were tracked by genomic polymerase chain reaction (PCR) from a mutant grid available in our laboratory using two gene‐specific primers and a transposon‐specific primer. Those mutants not found in the grid were made by two different approaches: In short, the 17662 and 17668 mutants were constructed by Tn7 in vitro mutagenesis using a Genome Priming System (GPS‐1) kit (New England Biolabs, Ipswich/Massachusetts/USA), and marker exchange was performed as described previously by Roeder and Collmer (1985). Construction of the 20167 and 46680 mutants was carried out by amplifying with Pfu DNA polymerase (Biotools, Madrid/Spain) an internal sequence between 700 and 800 bp from the Dd3937 genome and cloning into pKNG101 (Kaniga et al., 1991). The resulting plasmid was transferred into Dd3937 via triparental mating (de Lorenzo and Timmis, 1994). As pKNG101 cannot replicate in Dd3937, single crossover integrants were selected by resistance to streptomycin. In all cases, the mutations were verified by DNA sequencing.
For gain‐of‐function assays, ABF‐0018754 and AIH‐0004387 genes were amplified using a Pfu DNA polymerase from Dd3937 and E. coli K12 genomic DNA, respectively. The primers (5′→3′) used were as follows: 18754F, GCCCATGTCATTTGATAC; 18754R, AGCGATTAAAAGGTTTCCC; 04387F, ATTCACTCGCGCTAATCTCC; 04387R, CTTATCGGGCATTTTCATGGC. PCR products were cloned into pGEM‐Teasy® vector and the resulting plasmids were transferred into E. coli UU1250 via electroporation (Sambrook et al., 1989).
For complementation of the mutants, ABF‐0020167 and ABF‐0046680 genes were amplified using a Pfu DNA polymerase from Dd3937 genomic DNA employing the following primers (5′→3′): 18541F_PstI, CTGCAGTAGCAATTGTCTCCAATTG; 18541R_XmaI, CCCGGGTTGTAACGCCATCATC; 20167F_PstI, CTGCAGTTTGCGTTCAAGAGCCTC; 20167R_XmaI, CCCGGGTATCAAGGTGCAGTGC; 20252F_PstI, CTGCAGTACATGGAATGACGTTCACA; 20252R_XmaI, CCCGGGCCAAACAGGTGTGATGGTTA; 46680F_BamHI, GGATCCATCATTAATTCTCATGTTG; 46680R_PstI, CTGCAGTGTATCAGCGTTGACCGC; the restriction sites used for cloning into TrueBlueBAC2© (Genomics One) are shown in italic type. The cloned genes were sequenced to confirm that no PCR errors had occurred, and TrueBlueBAC2‐20167 or TrueBlueBAC2‐46680 was introduced into the corresponding mutant by electroporation (Sambrook et al., 1989).
Gain‐of‐function assays
Starter cultures of E. coli cells were grown overnight in liquid LB medium to the late exponential phase and then 20‐fold diluted into liquid LB medium and incubated at 37 °C to an OD600 of 0.6. Cultures were centrifuged at 6500 g and washed twice with 10 mm MgCl2. Sediments were used for inoculation. Tryptone broth (TB) (Parkinson, 1976) 0.35% agar plates, supplemented or not with 1 mm serine (final concentration), were inoculated with bacterial sediments using a toothpick. Plates were incubated for 48 h at 37 °C. Five plates were inoculated for each experiment.
Swimming assays
Dd3937 cells were scraped from NB plates incubated overnight and suspended in 10 mm MgCl2 to an OD600 of 1.0. A 5‐μL drop from each bacterial suspension was inoculated at the centre of each of five KB 0.25% agar plates. Plates were incubated for 24 h at 28 °C with high humidity. After that, bacterial haloes were recorded. Significant differences between the CIs of WT and mutant strains were determined and statistically compared according to Student's t‐test (P < 0.05).
The chemical ‘in‐plug’ bacterial chemotaxis assay
Dd3937 cells were scraped from NB plates incubated overnight and suspended in 10 mm MgCl2 at the desired final concentration. Then the bacterial suspension was mixed with Minimal Medium A (MMA) agar medium (Antúnez‐Lamas et al., 2009) to a final concentration of 0.25% agar and 2.5 × 108 cfu/mL. Once polymerized, a 5‐μL drop containing a given chemical was placed on the agar surface (Tso and Adler, 1974). Xylose, serine and JA were used at 10 mm (final concentration). Plates were incubated for 20 min at room temperature. After that, bacterial haloes were recorded. Five replicates were analysed for each experiment. Water was used as a solvent for xylose and serine, and JA was dissolved in ethanol.
Capillary assays
The chemotaxis of Dd3937 towards several compounds was measured using a modified Palleroni assay (Palleroni, 1976). CI was defined as the number of cells that accumulated after 20 min within a 5‐μL tip containing 2 μL of a given compound divided by the number of cells accumulated within a 5‐μL tip containing 2 μL of the solvent in which the compound was dissolved. Values of CI significantly greater than 1.0 were identified as a positive response of the cells to the chemical compound. Values of CI significantly less than 1.0 were identified as a negative response of the cells to the chemical compound. Three replicates (capillaries) were analysed in each experiment. Xylose, serine and JA were tested at 1 mm. Significant differences between the CIs of WT and mutant strains were determined and statistically compared according to Student's t‐test (P < 0.05).
Plant bioassays
Entry and entry‐dependent colonization assays of Dd3937 strains in A. thaliana leaves were carried out as described previously by Antunez‐Lamas et al. (2009) using 4‐week‐old Col‐0 ecotype plants. A 10‐μL drop containing 105 cfu was placed on a hole made with a needle on the adaxial side of leaves. Ten leaves were inoculated with each strain. Three leaves were mock infiltrated with 10 mm MgCl2. After 2 or 24 h at 28 °C, leaves were cut and washed with sterile water, and viable cell counts in leaves were determined by serial dilution and plating.
For non‐entry‐dependent colonization assays, 10 leaves were infiltrated into the abaxial side with a suspension containing 2 × 106 cfu/mL by syringe infiltration. Three leaves were mock infiltrated with 10 mm MgCl2. After 24 h at 28 °C, leaves were processed as described above.
In both kinds of experiment, significant differences between WT and mutant cells inside the leaves were determined and statistically compared according to Student's t‐test (P < 0.05).
Supporting information
Fig. S1 Complementation of non‐chemotactic strain Escherichia coli UU1250 with putative serine chemoreceptors. Tryptone broth (TB) 0.35% agar plates (A) and TB 0.35% agar plates supplemented with 1 mm serine (B) were inoculated with wild‐type (WT), UU1250 or transformed E. coli UU1250 strains expressing the E. coli serine chemoreceptor (AIH‐0004387) or the putative serine chemoreceptor of Dd3937 (ABF‐0018754). Results were recorded at 48 h. Similar results were obtained in three independent experiments.
Fig. S2 ‘In‐plug’ chemotactic response of Dd3937 strains towards serine. A drop containing water or serine (10 mm) was deposited on the (0.25%) soft‐agar bacteria. All the Dd3937 strains tested [wild‐type (WT) and mutants] showed no chemotactic response towards water; a representative image is shown. Haloes were recorded after 20 min. Similar results were obtained in three independent experiments.
Fig. S3 Swimming ability of Dd3937 strains in rich medium. King's B (KB) 0.25% agar plates were inoculated with Dd3937 wild‐type (WT) and mutant strains at the exponential phase. Diameters of haloes were recorded at 24 h. The means and standard errors are shown. Error bars represent standard error of the mean (SEM). Similar results were obtained in three independent experiments.
Table S1 Percentage identity matrix of the Dd3937 ligand‐binding region (LBR).
Table S2 Pfam matches for Dd3937 ligand‐binding regions (LBRs). (a) Nomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php). In grey, methyl‐accepting chemotaxis proteins (MCPs) under study in this work.
Table S3 Global blast of Dd3937 ligand‐binding regions (LBRs). Amino acid sequences of LBRs were searched against a database of bacterial genome sequences using blast (E‐value ≤ 10−30). * indicates that the periplasmic LBR was selected when two LBRs were present. (a) Nomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php).
Acknowledgements
This research was supported by the Spanish Plan Nacional I+D+I grants AGL‐2009‐12757 and AGL2012‐32516. I. Río‐Álvarez was supported by the FPI program (MICINN‐Spain). C. Muñoz‐Gómez was supported by the FPI program (MINECO‐Spain). P. M. Martínez‐García was supported by the Campus de Excelencia Internacional Andalucía Tech and AGL2011‐30343‐C02‐01. We thank Dr Tino Krell for critical reading of the manuscript and helpful comments, and for providing the E. coli UU1250 strain. We also thank Dr Miguel Angel Torres for helpful review of the manuscript.
References
- Alexander, R.P. and Zhulin, I.B. (2007) Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA, 104, 2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre, G. , Greer‐Phillips, S. and Zhulin, I.B. (2004) Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 28, 113–126. [DOI] [PubMed] [Google Scholar]
- Alvarez‐Ortega, C. and Harwood, C.S. (2007) Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis‐deficient mutant. Appl. Environ. Microbiol. 73, 7793–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunez‐Lamas, M. , Cabrera, E. , Lopez‐Solanilla, E. , Solano, R. , Gonzalez‐Melendi, P. , Chico, J.M. , Toth, I. , Birch, P. , Pritchard, L. , Liu, H. and Rodriguez‐Palenzuela, P. (2009) Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol. Microbiol. 74, 662–671. [DOI] [PubMed] [Google Scholar]
- Antúnez‐Lamas, M. , Cabrera‐Ordonez, E. , Lopez‐Solanilla, E. , Raposo, R. , Trelles‐Salazar, O. , Rodriguez‐Moreno, A. and Rodriguez‐Palenzuela, P. (2009) Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology, 155, 434–442. [DOI] [PubMed] [Google Scholar]
- Barabote, R.D. , Johnson, O.L. , Zetina, E. , San Francisco, S.K. , Fralick, J.A. and San Francisco, M.J.D. (2003) Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J. Bacteriol. 185, 5772–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras, F. , van Gijsegem, F. and Chatterjee, A.K. (1994) Extracellular enzymes and pathogenesis of soft‐rot Erwinia . Annu. Rev. Phytopathol. 32, 201–234. [Google Scholar]
- Bauer, D.W. , Bogdanove, A.J. , Beer, S.V. and Collmer, A. (1994) Erwinia chrysanthemi hrp genes and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant–Microbe Interact. 7, 573–581. [DOI] [PubMed] [Google Scholar]
- Bibikov, S.I. , Miller, A.C. , Gosink, K.K. and Parkinson, J.S. (2004) Methylation‐independent aerotaxis mediated by the Escherichia coli Aer protein. J. Bacteriol. 186, 3730–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel, A. , Li, X. , Bilwes, A.M. , Hughes, K.T. , Jensen, G.J. and Crane, B.R. (2012) Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA, 109, 3766–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charkowski, A. , Blanco, C. , Condemine, G. , Expert, D. , Franza, T. , Hayes, C. , Hugouvieux‐Cotte‐Pattat, N. , López‐Solanilla, E. , Low, D. , Moleleki, L. , Pirhonen, M. , Pitman, A. , Perna, N. , Reverchon, S. , Rodríguez‐Palenzuela, P. , San Francisco, M. , Toth, I. , Tsuyumu, S. , van der Waals, J. , van der Wolf, J. , Van Gijsegem, F. , Yang, C.H. and Yedidia, I. (2012) The role of secretion systems and small molecules in soft‐rot Enterobacteriaceae pathogenicity. Annu. Rev. Phytopathol. 50, 425–449. [DOI] [PubMed] [Google Scholar]
- Costechareyre, D. , Chich, J.F. , Strub, J.M. , Rahbé, Y. and Condemine, G. (2013) Transcriptome of Dickeya dadantii infecting Acyrthosiphon pisum reveals a strong defense against antimicrobial peptides. PLoS ONE, 8, e54118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserzo, M. , Wallin, E. , Simon, I. , von Heijne, G. and Elofsson, A. (1997) Prediction of TM alpha‐helices in prokaryotic membrane proteins: the Dense Alignment Surface method. Protein Eng. 10, 673–676. [DOI] [PubMed] [Google Scholar]
- Cserzo, M. , Eisenhaber, F. , Eisenhaber, B. and Simon, I. (2002) On filtering false positive transmembrane protein predictions. Protein Eng. 15, 745–752. [DOI] [PubMed] [Google Scholar]
- Cuppels, D.A. (1988) Chemotaxis by Pseudomonas syringae pv tomato. Appl. Environ. Microbiol. 54, 629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo, V. and Timmis, K.N. (1994) Analysis and construction of stable phenotypes in gram‐negative bacteria with Tn5‐ and Tn10‐derived minitransposons. Methods Enzymol. 235, 386–405. [DOI] [PubMed] [Google Scholar]
- Dellagi, A. , Rigault, M. , Segond, D. , Roux, C. , Kraepiel, Y. , Cellier, F. , Briat, J.F. , Gaymard, F. and Expert, D. (2005) Siderophore‐mediated upregulation of A. thaliana ferritin expression in response to Erwinia chrysanthemi infection. Plant J. 43, 262–272. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- Fagard, M. , Dellagi, A. , Roux, C. , Perino, C. , Rigault, M. , Boucher, V. , Shevchik, V.E. and Expert, D. (2007) Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi . Mol. Plant–Microbe Interact. 20, 794–805. [DOI] [PubMed] [Google Scholar]
- Falke, J.J. and Hazelbauer, G.L. (2001) Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D. , Bateman, A. , Clements, J. , Coggill, P. , Eberhardt, R.Y. , Eddy, S.R. , Heger, A. , Hetherington, K. , Holm, L. , Mistry, J. , Sonnhammer, E.L. , Tate, J. and Punta, M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hawes, M.C. and Smith, L.Y. (1989) Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil‐grown pea plants. J. Bacteriol. 171, 5668–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer, G.L. and Lai, W.C. (2010) Bacterial chemoreceptors, providing enhanced features to two‐component signaling. Curr. Opin. Microbiol. 13, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, M. , de Lorenzo, V. and Timmis, K.N. (1990) Transposon vectors containing non‐antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram‐negative bacteria. J. Bacteriol. 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell, K.F. and McBride, M.J. (2008) The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6, 466–476. [DOI] [PubMed] [Google Scholar]
- Kalinin, Y. , Neumann, S. , Sourjik, V. and Wu, M. (2010) Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J. Bacteriol. 192, 1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S. and Kado, C.I. (1990) Phenotypic switching affecting chemotaxis, xanthan production, and virulence in Xanthomonas campestris . Appl. Environ. Microbiol. 56, 3855–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga, K. , Delor, I. and Cornelis, G.R. (1991) A wide‐host‐range suicide vector for improving reverse genetics in gram‐negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica . Gene, 109, 137–141. [DOI] [PubMed] [Google Scholar]
- Kato, J. , Kim, H.E. , Takiguchi, N. , Kuroda, A. and Ohtake, H. (2008) Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J. Biosci. Bioeng. 106, 1–7. [DOI] [PubMed] [Google Scholar]
- Kim, S.H. , Prive, G.G. , Pandit, J. , Koshland, D.E. , Yeh, J.I. and Biemann, H.P. (1996) High‐resolution structures of the ligand binding domain of the wild‐type bacterial aspartate receptor. J. Mol. Biol. 262, 186–201. [DOI] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kotoujansky, A. , Diolez, A. , Boccara, M. , Bertheau, Y. , Andro, T. and Coleno, A. (1985) Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 4, 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell, T. , Lacal, J. , Muñoz‐Martínez, F. , Reyes‐Darias, J.A. , Cadirci, B.H. , García‐Fontana, C. and Ramos, J. (2011) Diversity at its best: bacterial taxis. Environ. Microbiol. 13, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Lacal, J. , García‐Fontana, C. , Muñoz‐Martínez, F. , Ramos, J.L. and Krell, T. (2010a) Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 12, 2873–2884. [DOI] [PubMed] [Google Scholar]
- Lacal, J. , Alfonso, C. , Liu, X. , Parales, R.E. , Morel, B. , Conejero‐Lara, F. , Rivas, G. , Duque, E. , Ramos, J.L. and Krell, T. (2010b) Identification of a chemoreceptor for TCA cycle intermediates: ligand induced dimerization of chemoreceptor sensor domain determines magnitude of chemotaxis. J. Biol. Chem. 285, 23 126–23 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. , Chenna, R. , McGettigan, P.A. , McWilliam, H. , Valentin, F. , Wallace, I.M. , Wilm, A. , Lopez, R. , Thompson, J.D. , Gibson, T.J. and Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- León, J. , Rojo, E. and Sánchez‐Serrano, J.J. (2001) Wound signalling in plants. J. Exp. Bot. 52, 1–9. [DOI] [PubMed] [Google Scholar]
- Llama‐Palacios, A. , López‐Solanilla, E. , Poza‐Carrión, C. , García‐Olmedo, F. and Rodríguez‐Palenzuela, P. (2003) The Erwinia chrysanthemi phoP‐phoQ operon plays an important role in growth at low pH, virulence and bacterial survival in plant tissue. Mol. Microbiol. 49, 347–357. [DOI] [PubMed] [Google Scholar]
- Llama‐Palacios, A. , Lopez‐Solanilla, E. and Rodriguez‐Palenzuela, P. (2005) Role of the PhoP‐PhoQ system in the virulence of Erwinia chrysanthemi strain 3937: involvement in sensitivity to plant antimicrobial peptides, survival at acid pH, and regulation of pectolytic enzymes. J. Bacteriol. 187, 2157–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Solanilla, E. , García‐Olmedo, F. and Rodríguez‐Palenzuela, P. (1998) Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell, 10, 917–924. [PMC free article] [PubMed] [Google Scholar]
- López‐Solanilla, E. , Llama‐Palacios, A. , Collmer, A. , García‐Olmedo, F. and Rodríguez‐Palenzuela, P. (2001) Relative effects on virulence of mutations in the sap, pel, and hrp loci of Erwinia chrysanthemi . Mol. Plant–Microbe Interact. 14, 386–393. [DOI] [PubMed] [Google Scholar]
- Ma, B. , Charkowski, A.O. , Glasner, J.D. and Perna, N.T. (2014) Identification of host–microbe interaction factors in the genomes of soft rot‐associated pathogens Dickeya dadantii 3937 and Pectobacterium carotovorum WPP14 with supervised machine learning. BMC Genomics, 15, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiorani Valecillos, A. , Rodriguez Palenzuela, P. and Lopez‐Solanilla, E. (2006) The role of several multidrug resistance systems in Erwinia chrysanthemi pathogenesis. Mol. Plant–Microbe Interact. 19, 607–613. [DOI] [PubMed] [Google Scholar]
- Mansfield, J. , Genin, S. , Magori, S. , Citovsky, V. , Sriariyanum, M. , Ronald, P. , Dow, M. , Verdier, V. , Beer, S.V. , Machado, M.A. , Toth, I. , Salmond, G. and Foster, G.D. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, S. and Vanderleyden, J. (1996) Functions of bacterial flagella. Crit. Rev. Microbiol. 22, 67–100. [DOI] [PubMed] [Google Scholar]
- Munzinger, M. , Budzikiewicz, H. , Expert, D. , Enard, C. and Meyer, J.M. (2000) Achromobactin, a new citrate siderophore of Erwinia chrysanthemi . Z. Naturforsch. 55, 328–332. [DOI] [PubMed] [Google Scholar]
- Nichols, N.N. , Lunde, T.A. , Graden, K.C. , Hallock, K.A. , Kowalchyk, C.K. , Southern, R.M. , Soskin, E.J. and Ditty, J.L. (2012) Chemotaxis to furan compounds by furan‐degrading Pseudomonas strains. Appl. Environ. Microbiol. 78, 6365–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, S. , Suzuki, D. , Itoh, Y. , Suzuki, K. , Tajima, H. , Hyakutake, A. , Homma, M. , Butler‐Wu, S.M. , Camilli, A. and Kawagishi, I. (2012) Mlp24 (McpX) of Vibrio cholera implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 80, 3170–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku, S. , Komatsu, A. , Tajima, T. , Nakashimada, Y. and Kato, J. (2012) Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0‐1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 27, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleroni, N.J. (1976) Chamber for bacterial chemotaxis experiments. Appl. Environ. Microbiol. 32, 729–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, J. (1976) cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persmark, M. , Expert, D. and Neilands, J.B. (1989) Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi . J. Biol. Chem. 264, 3187–3193. [PubMed] [Google Scholar]
- Porter, S.L. , Wadhams, G.H. and Armitage, J.P. (2011) Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9, 153–165. [DOI] [PubMed] [Google Scholar]
- Rahman, H. , King, R.M. , Shewell, L.K. , Semchenko, E.A. , Hartley‐Tassell, L.E. , Wilson, J.C. , Day, C.J. and Korolik, V. (2014) Characterization of a multi‐ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni . PLoS Pathog. 10, e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie, E.A. and Scheller, H.V. (2014) Xylan biosynthesis. Curr. Opin. Biotechnol. 26C, 100–107. [DOI] [PubMed] [Google Scholar]
- Reverchon, S. and Nasser, W. (2013) Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 5, 622–636. [DOI] [PubMed] [Google Scholar]
- Rico‐Jiménez, M. , Muñoz‐Martínez, F. , García‐Fontana, C. , Fernandez, M. , Morel, B. , Ortega, A. , Ramos, J.L. and Krell, T. (2013) Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non‐protein amino acid gamma‐aminobutyrate (GABA). Mol. Microbiol. 88, 1230–1243. [DOI] [PubMed] [Google Scholar]
- Rio‐Alvarez, I. , Rodríguez‐Herva, J.J. , Cuartas‐Lanza, R. , Toth, I. , Pritchard, L. , Rodriquez‐Palenzuela, P. and López‐Solanilla, E. (2012) Genome‐wide analysis of the response of Dickeya dadantii 3937 to plant antimicrobial peptides. Mol. Plant Microbe Interact. 25, 523–533. [DOI] [PubMed] [Google Scholar]
- Roeder, D.L. and Collmer, A. (1985) Marker‐exchange mutagenesis of a pectate lyase isozyme gene in Erwinia chrysanthemi . J. Bacteriol. 164, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Frits, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sourjik, V. and Wingreen, N.S. (2012) Responding to chemical gradients: bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J.E. , Block, D. , Boulez, K. , Brenner, S.E. , Chervitz, S.A. , Dagdigian, C. , Fuellen, G. , Gilbert, J.G. , Korf, I. , Lapp, H. , Lehväslaiho, H. , Matsalla, C. , Mungall, C.J. , Osborne, B.I. , Pocock, M.R. , Schattner, P. , Senger, M. , Stein, L.D. , Stupka, E. , Wilkinson, M.D. and Birney, E. (2002) The Bioperl toolkit: perl modules for the life sciences. Genome Res. 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, E.G. , Henderson, J.N. , Goers, J. , Wreden, C. , Hicks, K.G. , Foster, J.K. , Parthasarathy, R. , Remington, S.J. and Guillemin, K. (2013) Structure and proposed mechanism for the pH sensing Helicobacter pylori chemoreceptor TlpB. Structure, 20, 1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino, M. , Abelenda, J.A. , Río‐Alvarez, I. , Navarro, C. , Vicedo, B. , Farmaki, T. , Jiménez, P. , García‐Agustín, P. , López‐Solanilla, E. , Prat, S. , Rojo, E. , Sanchez‐Serrano, J. and Sanmartin, M. (2014) Jasmonate‐dependent modifications of the pectin matrix during potato development function as a defense mechanism targeted by Dickeya dadantii virulence factors. Plant J. 77, 418–429. [DOI] [PubMed] [Google Scholar]
- Tso, W.W. and Adler, J. (1974) Negative chemotaxis in Escherichia coli . J. Bacteriol. 118, 560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, L.E. and Zhulin, I.B. (2005) Four‐helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics, 21 (Suppl 3), iii45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams, G.H. and Armitage, J.P. (2004) Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K. and Imae, Y. (1993) Cloning and characterization of the Salmonella typhimurium‐specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc. Natl. Acad. Sci. USA, 90, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.H. , Gavilanes‐Ruiz, M. , Okinaka, Y. , Vedel, R. , Berthuy, I. , Boccara, M. , Wei‐Ta Chen, J. , Perna, N.T. and Keen, N.T. (2002) hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant–Microbe Interact. 5, 472–480. [DOI] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2006) Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum . J. Bacteriol. 188, 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2007) The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap, M.N. , Yang, C.H. , Barak, J.D. , Jahn, C.E. and Charkowski, A.O. (2005) The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Complementation of non‐chemotactic strain Escherichia coli UU1250 with putative serine chemoreceptors. Tryptone broth (TB) 0.35% agar plates (A) and TB 0.35% agar plates supplemented with 1 mm serine (B) were inoculated with wild‐type (WT), UU1250 or transformed E. coli UU1250 strains expressing the E. coli serine chemoreceptor (AIH‐0004387) or the putative serine chemoreceptor of Dd3937 (ABF‐0018754). Results were recorded at 48 h. Similar results were obtained in three independent experiments.
Fig. S2 ‘In‐plug’ chemotactic response of Dd3937 strains towards serine. A drop containing water or serine (10 mm) was deposited on the (0.25%) soft‐agar bacteria. All the Dd3937 strains tested [wild‐type (WT) and mutants] showed no chemotactic response towards water; a representative image is shown. Haloes were recorded after 20 min. Similar results were obtained in three independent experiments.
Fig. S3 Swimming ability of Dd3937 strains in rich medium. King's B (KB) 0.25% agar plates were inoculated with Dd3937 wild‐type (WT) and mutant strains at the exponential phase. Diameters of haloes were recorded at 24 h. The means and standard errors are shown. Error bars represent standard error of the mean (SEM). Similar results were obtained in three independent experiments.
Table S1 Percentage identity matrix of the Dd3937 ligand‐binding region (LBR).
Table S2 Pfam matches for Dd3937 ligand‐binding regions (LBRs). (a) Nomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php). In grey, methyl‐accepting chemotaxis proteins (MCPs) under study in this work.
Table S3 Global blast of Dd3937 ligand‐binding regions (LBRs). Amino acid sequences of LBRs were searched against a database of bacterial genome sequences using blast (E‐value ≤ 10−30). * indicates that the periplasmic LBR was selected when two LBRs were present. (a) Nomenclature from the ASAP database (https://asap.ahabs.wisc.edu/asap/logon/php).


