Summary
The ATP‐dependent Lon protease is involved in many physiological processes. In bacteria, Lon regulates pathogenesis and, in yeast, Lon protects mitochondia from oxidative damage. However, little is known about Lon in fungal phytopathogens. MAP1, a homologue of Lon in Magnaporthe oryzae, was recently identified to be important for stress resistance and pathogenesis. Here, we focus on a novel pathogenic pathway mediated by MAP1. Based on an interaction system between rice and a tandem affinity purification (TAP)‐tagged MAP1 complementation strain, we identified 23 novel fungal proteins from infected leaves using a TAP approach with mass spectrometry, and confirmed that 14 of these proteins physically interact with MAP1 in vivo. Among these 14 proteins, 11 candidates, presumably localized to the mitochondria, were biochemically determined to be substrates of MAP1 hydrolysis. Deletion mutants were created and functionally analysed to further confirm the involvement of these proteins in pathogenesis. The results indicated that all mutants showed reduced conidiation and sensitivity to hydrogen peroxide. Appressorial formations were not affected, although conidia from certain mutants were morphologically altered. In addition, virulence was reduced in four mutants, enhanced (with lesions forming earlier) in two mutants and remained unchanged in one mutant. Together with the known virulence‐related proteins alternative oxidase and enoyl‐CoA hydratase, we propose that most of the Lon‐interacting proteins are involved in the pathogenic regulation pathway mediated by MAP1 in M. oryzae. Perturbation of this pathway may represent an effective approach for the inhibition of rice blast disease.
Keywords: interacting proteins, Magnaporthe oryzae, mitochondrial ATP‐dependent Lon protease
Introduction
The ATP‐dependent Lon protease is the most highly conserved member of the energy‐dependent proteases in a myriad of organisms. A molecular mechanism of Lon common to both prokaryotes and eukaryotes is the proteolytic activity on substrates of misfolded, dysfunctional or aggregated proteins in cells (Venkatesh et al., 2012). However, the biological roles of Lon vary across different systems and circumstances. In prokaryotes, archaea, bacteria and Actinomyces, for instance, Lon proteases are mainly involved in basic physiological processes, such as the cell cycle, differentiation, sporulation, motility and survival during stress (Coleman et al., 2009; Lan et al., 2007; Su et al., 2006; Takeuchi et al., 2014; Tsilibaris et al., 2006). In eukaryotic cells, Lon is associated with fungal life span, ascospore germination, sexual reproduction and high temperature tolerance (Adam et al., 2012), as well as with plant post‐germinative growth and development (Rigas et al., 2009). Eukaryotic Lons are probably associated more with cell viability under stress conditions than with proteostasis maintenance in the mitochondrial matrix under normal conditions (Bota and Davies, 2001, 2002; Fukuda et al., 2007; Hori et al., 2002).
The early understanding of mitochondrial Lon protease was principally based on studies of bacteria, but eukaryotic systems employing fungi, animals and humans have now been established (Chen et al., 2008; Lu et al., 2007; Matsushima et al., 2010). Pim1, an extensively studied mitochondrial Lon in yeast, plays an essential role in maintaining oxidized protein substrates under normal metabolic conditions and in preventing the in vivo formation of accumulated enzymes or aggregates during environmental and cellular stresses. To characterize Lon substrates under non‐repressing conditions, comparative analyses of mitochondrial‐oxidized proteomes were performed using deletion mutants of pim1, and 14 different proteins were identified (Bayot et al., 2010). Except for two stress proteins, heat shock protein 60 and mitochondrial superoxide dismutase, most of the proteins were involved in mitochondrial energy pathways and basic metabolic adaptation. Temperature is one of the major environmental factors influencing growth and development in fungi (Kato and Kozaka, 1974). When mitochondria from Saccharomyces cerevisiae were subjected to a heat treatment of 42 °C, specific aggregation proteins were detected, with a majority of these determined to be metabolic enzymes, such as acetolactate synthase, α‐isopropylmalate synthase, glycerol‐3‐phosphate dehydrogenase, the β subunit of succinyl‐CoA ligase and two components of the α‐ketoglutarate dehydrogenase complex. Heat‐damaged proteins tend to be aggregated and cross‐linked (Bender et al., 2011). Once these proteins become irreversible aggregates, negative effects on cellular viability occur; therefore, a proteolytic degradation system is necessary to remove these aggregates. Indeed, Pim1 serves this function in yeast (Bayot et al., 2010; Bender et al., 2011), and similar mechanisms exist in Drosophila, rats and humans (Chen et al., 2008; Lu et al., 2007; Matsushima et al., 2010).
Thus, Lon is important for the maintenance of basic metabolism and stress tolerance; interestingly, Lon plays additional roles beyond these functions. For example, in bacterial pathogens, Lon proteases are associated with the infection process (Ingmer and Brøndsted, 2009). The type III secretion system involves a set of protein components that determine the virulence of pathogens (Cornelis, 2006; Van Melderen and Aertsen, 2009). During bacteria–animal interactions, Lon regulates the type III secretion system and promotes pathogenesis in Gram‐negative bacterial species, such as Pseudomonas aeruginosa and Salmonella enterica (Takaya et al., 2003, 2008). Moreover, the type III secretion regulation mechanism, in which the proteolysis of regulatory proteins is specifically controlled by Lon in response to host resistance, appears to be relatively conserved, because a similar mechanism exists in the Pseudomonas–Arabidopsis system (Bretz et al., 2002; Lan et al., 2007; Losada and Hutcheson, 2005). With regard to fungal pathogens, little is known about Lon‐mediated regulation, although fungi are the most prevalent pathogens of plants, and fungal diseases pose marked threats to grain production and food safety.
The fungus Magnaporthe oryzae is one of the most significant fungal pathogens in crops, causing severe blast disease in rice (Kadotani et al., 2003; Pennisi, 2010). When M. oryzae interacts with rice, it encounters a variety of harsh obstacles from environmental factors and plant‐derived passive or active resistance, such as the accumulation of reactive oxygen species, antimicrobial compounds, pathogenesis‐related proteins and resistance gene products (Egan et al., 2007; Hammond‐Kosack and Parker, 2003; Nürnberger et al., 2004; Osuna‐Canizalez et al., 1991). Thus, to colonize a host and complete the disease cycle, M. oryzae must survive these unfavourable conditions. In the past two decades, various genes essential for plant infection processes have been identified in M. oryzae (Fernandez and Wilson, 2014; Li et al., 2012), with many genes strangely ‘moonlighting’ in response to selected stressors (Ding et al., 2010; Huang et al., 2011; Sun et al., 2006; Zhang et al., 2014a). In our previous work, the mitochondrial ATP‐dependent Lon protease in M. oryzae (termed MAP1; Accession number: MGG_11141) was screened and analysed biologically and pathogenically (Li et al., 2015). To date, information regarding fungal Lon comes mainly from studies employing S. cerevisiae and Podospora anserina.
MAP1, a Lon‐like protease of fungal phytopathogens, shares common responses to environmental stresses with the fungal pathogens discussed above. However, the involvement of MAP1 in pathogenesis may be unique because S. cerevisiae and P. anserina are non‐pathogenic, and fungal pathogens generally lack a type III secretion system. We hypothesize that the involvement of MAP1 in pathogenesis results from the regulation of specific interacting proteins. In this study, we focused on 11 MAP1‐interacting proteins, which were degraded by MAP1 in the presence of ATP. Genes of these proteins were targeted for replacement with hygromycin resistance genes in the wild‐type. The pathogenic assay utilizing these mutants demonstrated that the majority of the proteins interacting with MAP1, together with the previously reported virulence‐related proteins alternative oxidase (AOX) (Avila‐Adame and Köller, 2002) and enoyl‐CoA hydratase (ECH1) (Patkar et al., 2012), were involved in the development of rice blast disease.
Results
The tandem affinity purification (TAP)‐tagged MAP1 strain possesses similar phenotype and infection ability to the wild‐type
In general, a protein that is TAP‐tagged and its endogenous form are co‐expressed in a transformed strain; thus, both protein forms compete for incorporation into multiple protein complexes, leading to specific biological or biochemical effects. To avoid this, it is necessary to have a TAP‐tagged strain in the absence of the corresponding endogenous protein. Based on a null mutant of MAP1 (ΔMAP1) that was generated with targeted gene replacement (Fig. S1, see Supporting Information) (Li et al., 2015), the pCBTAP‐MAP1 vector (Fig. S2A, see Supporting Information) was genetically transformed into ΔMAP1, and several TAP‐tagged MAP1 complementation strains were obtained through sulfonylurea resistance screening. The phenotype and pathogenicity of the resultant strains were compared with those of the wild‐type, and only unchanged strains were selected for further research. Among the transformed candidates, the strain termed TAP‐MAP1, which contained a single copy of the TAP‐tagged MAP1 fragment (Fig. S2B), was obtained because it was consistent with the wild‐type in colony growth rate, conidial phenotype (Fig. 1A), MAP1 protein levels (Fig. 1B), infection ability (Fig. 1C) and hydrogen peroxide (H2O2) tolerance (Fig. S2C).
Figure 1.
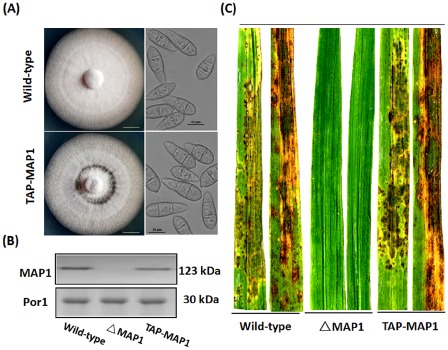
Tandem affinity purification (TAP)‐tagged MAP1 strain possesses characteristics similar to those of the wild‐type. (A) Morphological comparison of the wild‐type and TAP‐tagged MAP1 complemented strains. Left panel: both strains were cultured in complete agar medium (CM) under standard incubation conditions (25 °C) for 7 days. Representative colonies are shown photographed at 7 days post‐inoculation. Scale bar, 10 mm. Right panel: conidia were harvested from 14‐day‐old oatmeal agar cultures, and conidia from both strains were observed under a light microscope. Scale bar, 10 μm. (B) MAP1 protein levels. Western blot analysis with total proteins extracted from the wild‐type strain, MAP1 deletion mutant strain (ΔMAP1) and TAP‐tagged MAP1 complemented strain. MAP1 was detected with an anti‐MAP1 antibody, and the presence of Porin (Por1) was detected as an internal control using an anti‐Por1 antibody. (C) Pathogenicity assay with the wild‐type strain, MAP1 deletion mutant strain (ΔMAP1) and TAP‐tagged MAP1 complemented strain (TAP‐MAP1). Rice disease symptoms were assessed after 7 days.
Identification of MAP1‐interacting proteins
To characterize MAP1‐binding proteins during M. oryzae–rice interactions, a biochemical approach was adopted. The TAP‐MAP1 strain‐infected rice leaves were collected as test samples 5 days after inoculation, and the wild‐type‐infected leaves were used as controls. A set of extracted proteins was mixed with immunoglobulin G (IgG)‐agarose beads to generate a TAP‐MAP1‐coupled column. The control was processed in parallel. Protein complexes associated with TAP‐tagged MAP1 were then specifically released from the resin using Tobacco etch virus (TEV) protease cleavage. MAP1 and binding proteins were purified through calcium chelation.
The pulled‐down proteins were separated with sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) followed by Coomassie brilliant blue staining. The TAP‐tagged MAP1 column bound many prominent proteins, whereas the control column bound almost no proteins (Fig. 2A). Twenty‐eight detectable protein bands (absent in the control column) were excised and analysed using matrix‐assisted laser desorption ionization‐tandem time of flight‐mass spectrometry (MALDI‐TOF/TOF‐MS/MS). The results indicated that a majority of identified proteins were derived from the rice blast fungus, except for five proteins (data not shown) which originated from rice. The 23 fungal proteins were further characterized using bioinformatics. Their functional annotation, molecular weights and suborganellar locations are listed in Table 1.
Figure 2.
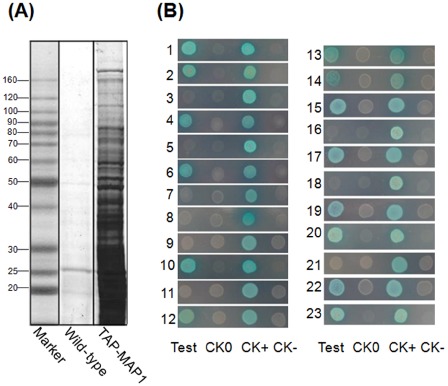
Identification of MAP1‐interacting proteins. (A) Tandem affinity purification (TAP) of MAP1‐interacting proteins. Elution fractions were analysed using sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) with Coomassie blue staining. (B) Yeast two‐hybrid assay. Test, interaction between MAP1 and each interacting candidate; CK0, self‐activation control; CK+, positive control; CK–, negative control. Each interacting protein is represented by a number (1–23) and is listed in Table 1.
Table 1.
Characterization of the MAP1‐interacting proteins
| No. | Protein | Predicted ORFa | Function or contained domainb | MWc (kDa) | Organelle locationd |
|---|---|---|---|---|---|
| 1 | ECH1 | MGG_12868 | Enoyl‐CoA hydratase | 31.4 | M |
| 2 | CGT | MGG_10668 | Ceramide glucosyltransferase | 55.1 | Cyto/M |
| 3 | LRS | MGG_04042 | Leucyl‐tRNA synthetase | 99.2 | Extracellular |
| 4 | C3HC | MGG_04317 | C3HC‐zinc finger domain‐containing protein | 58.9 | N/M |
| 5 | hisHF | MGG_00253 | Imidazole glycerol phosphate synthase hisHF | 59.2 | Cyto/N/M |
| 6 | C2H2 | MGG_15393 | C2H2‐zinc finger domain‐containing protein | 43.9 | N/M |
| 7 | MFT | MGG_07808 | Major facilitator superfamily | 59.0 | PM/ER |
| 8 | ACT | MGG_05587 | Mitochondrial actin | 53.1 | Cyto/N/M |
| 9 | Myo | MGG_03060 | Myosin N‐terminal SH3‐like domain | 272.8 | N/M |
| 10 | MMT | MGG_06247 | Mitochondrial metal transporter | 44.1 | Cyto/N |
| 11 | VRS | MGG_04396 | Valyl‐tRNA synthetase | 118.0 | Cyto/N |
| 12 | SHM | MGG_13781 | Serine hydroxymethyltransferase | 56.8 | M |
| 13 | Bud6p | MGG_11773 | Bud site selection or actin‐interacting protein | 109.5 | N/Cyto/M |
| 14 | PrePL | MGG_07599 | Predicted protein/leucine‐rich repeat | 14.6 | N/M |
| 15 | CHP1 | MGG_10140 | Conserved hypothetical protein/RP S21 | 28.4 | M/N |
| 16 | RPL10 | MGG_03136 | 60S ribosomal protein L10‐B | 25.0 | M/Cyto |
| 17 | TauD | MGG_07619 | Taurine catabolism dioxygenase TauD | 44.9 | Cyto/N |
| 18 | CHP2 | MGG_02919 | Conserved hypothetical protein | 45.0 | Golgi/M |
| 19 | AnnA7 | MGG_06847 | Annexin A7 | 48.9 | Cyto/PM/M |
| 20 | RanBP | MGG_00448 | RanGTP‐binding protein | 70.3 | N/Cyto/M |
| 21 | CysP1 | MGG_02695 | Cysteine proteinase1, mitochondrial precursor | 57.1 | Cyto/N |
| 22 | AOX | MGG_12936 | Alternative oxidase | 42.7 | M/Cyto |
| 23 | CHS6 | MGG_13013 | Chitin synthase 8 (CHS6) | 208.2 | Cyto/PM |
Open reading frame (ORF): Broad Institute Magnaporthe grisea database (http://www.broad.mit.edu/annotation/fungi/magnaporthe/).
blastx search of protein databases using a translated nucleotide query according to database (http://blast.be‐md.ncbi.nlm.nih.gov/).
Molecular weight (MW) was predicted according to the database http://web.expasy.org/compute_pi/. The actual size may vary as a result of cleavage of the mitochondrial targeting signal after import.
Organelle location was predicted according to the PSORT prediction database (psort.ims.u‐tokyo.ac.jp/form2.html). Cyto, cytoplasm; ER, endoplasmic reticulum; Golgi, Golgi apparatus; M, mitochondrion; N, nucleus; PM, plasma membrane.
To further confirm interactions between MAP1 and each of the pulled‐down proteins in vivo, a yeast two‐hybrid system was applied using MAP1 in‐frame fused bait and candidate interacting proteins in‐frame fused prey. The results demonstrated that 14 of the 23 candidates bound directly to MAP1 in yeast, including AnnA7, SHM, RanBP, C3HC, CGT, C2H2, AOX, ECH1, Bud6p, PrePL, CHP1, TauD, MMT and CHS6 (Fig. 2B).
Proteins interacting with MAP1 are substrates for degradation
Based on their biochemistry, Lon and its homologues are enzymes that hydrolyse proteins. To analyse whether these binding proteins can be degraded by MAP1, we established a series of proteolytic reaction systems to detect proteolytic activity in vitro. The candidate proteins were selected based primarily on their location in mitochondria as predicted with PSORT II. Among the 14 interacting proteins confirmed using the yeast two‐hybrid assay, 11 proteins were probably localized to the mitochondria (Table 1). Recombinant MAP1 and the histidine (His)‐tagged binding proteins were expressed in Escherichia coli and purified to electrophoretic homogeneity using affinity chromatography. Representative SDS‐PAGE results of recombinant MAP1 and its interacting proteins are shown in Fig. S3 (see Supporting Information).
A reaction system consisting of MAP1 and one binding protein in the presence or absence of 2.5 mm ATP was prepared and incubated at 25 °C for 1 h. From the SDS‐PAGE results (Fig. 3A), we found that all the binding proteins were degraded to varying degrees in the presence of ATP, indicating that MAP1, as an ATP‐dependent protease, indeed processes the interacting proteins as substrates for hydrolysis. We also examined whether MAP1 utilized denatured proteins as substrates for hydrolysis. We found that, when the interacting proteins were treated at 45 °C prior to incubation, they were completely degraded in the presence of ATP within 1 h (Fig. 3A), suggesting the highly efficient degradation of the denatured proteins by MAP1. Importantly, under normal hydrolytic conditions, three of the proteins (C2H2, C3HC and PrePL) were probably truncated at the carboxyl‐terminus, the amino‐terminus or both termini, according to the appearance of a lower band (Fig. 3A), implying a specific regulatory function of MAP1.
Figure 3.
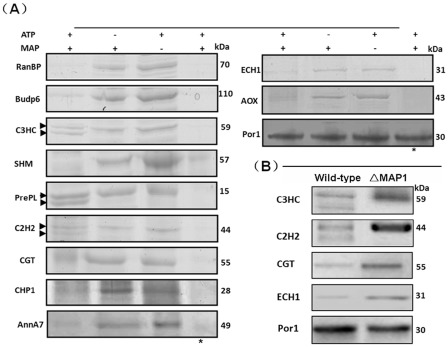
Interacting proteins are substrates for MAP1 hydrolysis. (A) Coomassie brilliant blue‐stained sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis. The symbol ‘+’ indicates that ATP or MAP1 was added to the reaction system, and ‘–’ indicates that ATP or MAP1 was not added. The symbol ‘*’ indicates that individual protein substrates (MAP1‐interacting proteins) were denatured before being added to the reaction system. Two protein bands in the reaction system of C3HC, C2H2 and PrePL are marked with arrows. (B) Western blot analysis. Equal amounts of protein were loaded in each lane on one gel. Porin1 (Por1) was used as a loading control. The experiment was performed twice, and a representative blot is presented.
To verify the proteolytic effect in vivo, four proteins of interest, C2H2, C3HC, CGT and ECH1, were selected and analysed using Western blot analysis. We generated rabbit polyclonal anti‐serum against each of the proteins and identified the corresponding protein bands on a Western blot by comparing the total proteins from the MAP1 deletion mutant (ΔMAP1) with those from the wild‐type strain (Fig. 3B). When blots were probed using the anti‐C3HC antibody, two bands at approximately 55 and 60 kDa were present in the wild‐type, whereas only the upper band (60 kDa) was present in the mutant. Similarly, with the anti‐C2H2 antibody‐probed blots, two bands appeared in the wild‐type at 45 and 38 kDa, but only the upper band persisted in the mutant. Based on the molecular weights of C3HC and C2H2, the upper bands in each blot were probably the intact proteins or precursors and the lower bands were probably mature or active forms derived from the proteins in the upper bands. To rule out the possibility of the antibodies binding nonspecifically to the lower bands, we excised the bands and applied MS analysis. The results showed that the peptide fragments were indeed from intact proteins. In contrast with the results for C2H2 and C3HC, when we probed blots with antibodies specific for CGT or ECH1, single bands appeared in the blots with the mutants that corresponded to those on the blots of the wild‐type (Fig. 3A,B). These results indicate that the regulation of the identified proteins is specifically controlled by MAP1.
Deletion mutants of individual MAP1‐interacting proteins and their biological characteristics
MAP1 plays multiple roles during stress and infection processes (Li et al., 2015). To gain an insight into the functional relationships between MAP1 and its interacting proteins, a series of mutants was generated with targeted gene deletions for each of the interacting proteins. Among the 11 interacting proteins which were specifically controlled by MAP1, information regarding AOX and ECH has been reported previously (Avila‐Adame and Köller, 2002; Patkar et al., 2012). Thus, this study focused on the nine remaining proteins. Their corresponding gene replacement vectors were constructed and transferred into protoplasts of M. oryzae (Fig. S4, see Supporting Information). We successfully obtained seven deletion mutants targeting the C3HC, C2H2, SHM, RanBP, CGT, AnnA7 and Bud6p genes (Figs S5 and S6, see Supporting Information), but failed to obtain PrePL and CHP1 mutants.
We then compared biological phenotypes among the deletion and wild‐type strains. When cultured on complete agar medium at 25 °C, except for the Bud6p and RanBP deletion mutants with severely reduced growth rates, all the other strains grew at a rate similar to that of the wild‐type (Table S2, see Supporting Information). However, the colony morphology for the AnnA7 and C3HC mutants appeared to be slightly different from that of the wild‐type (Fig. 4A). When all strains were inoculated on complete agar medium (CM) plates supplemented with 5 mm H2O2, the size of the colony for all mutants was smaller than that of the wild‐type (Fig. 4B), implying that the targeted genes were associated with oxidation resistance. This characteristic is consistent with that of a MAP1 deletion mutant. Conidiation was affected to different degrees in the mutants (Fig. 4C). In comparison with those of the wild‐type, the numbers of conidia from the Bud6p, C2H2 and CGT deletion mutants were reduced by 34%–48% and by more than 70% for the other mutants (Fig. 4C). The C3HC and RanBP mutants produced so few conidia that 30 times more plates were inoculated with these mutants than with the wild‐type to obtain similar amounts of conidia. When we analysed the conidial germination and appressorium formation after 24 h of incubation on an inductive surface, however, we found that the germinated conidia of all the mutants produced germ tubes and formed appressoria free of defects (Fig. 4D; Table S2).
Figure 4.
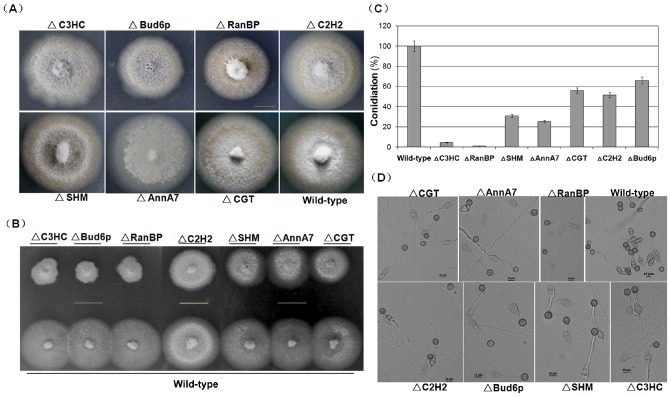
Biological analysis of seven MAP1‐interacting protein deletion mutants. (A) Comparison of colony morphology. The growth of both Bud6p and RanBP deletion mutants (ΔBud6p and ΔRanBP) was inhibited, whereas all other strains grew at a rate similar to that of the wild‐type. The experiment was performed thrice, and a representative blot is presented. Bar, 20 mm. (B) Sensitivity of the mutants to oxidative stress. The experiment was performed thrice, and a representative blot is presented. Bar, 20 mm. (C) Statistical analysis of the conidial productivity for all mutants. (D) Conidial germination and appressorium formation analysis. Conidia for all deletion mutants germinate and form appressoria after 24 h of incubation on inductive surfaces of glass coverslips. Bar, 10 μm.
Conidium morphology was also analysed. Except for Bud6p and AnnA7 mutants, most mutants produced conidia with normal morphology. For the AnnA7 mutant, 55% of the conidia were dysmorphic (Fig. 5A), demonstrating more than 29 different shapes, for example the septum formation altered type, the basal cell deformed type, the columnar type and the line‐shaped type (Fig. 5B). Interestingly, for the Bud6p mutant, more than 85% of the conidia (Fig. 5A) showed greater spore length and width than those of the wild‐type, but these conidia still maintained pyriform shapes (Fig. 5C,D), suggesting that Bud6p is associated with the growth potential of spores.
Figure 5.
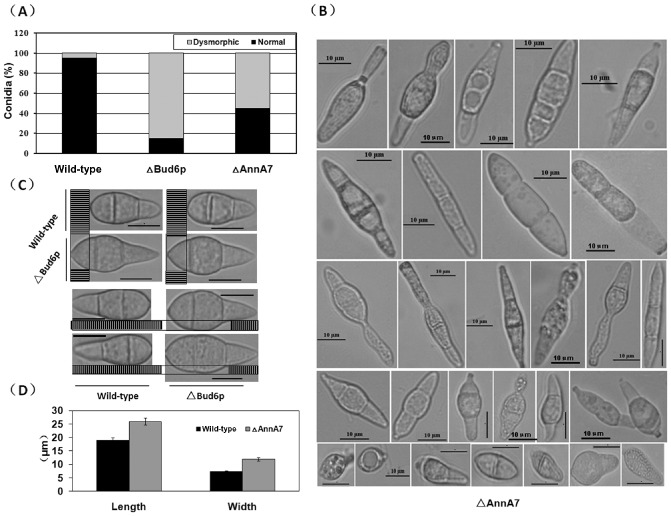
Dysmorphic conidia in the AnnA7 and Bud6p deletion mutants. (A) The percentage of normal or dysmorphic conidia in the wild‐type strain, AnnA7 deletion mutant (ΔAnnA7) and Bud6p deletion mutant (ΔBud6p). For the AnnA7 deletion mutant, 55% of the conidia are dysmorphic, demonstrating 29 different shapes. For the Bud6p deletion mutant, the spores in 85% of the conidia are longer and wider than those in the wild‐type. (B) The 29 types of conidial morphology demonstrated by the AnnA7 deletion mutant. Bar, 10 μm. (C) Conidial volume comparison between the wild‐type and Bud6p deletion mutant. Bar, 10 μm. (D) Statistical analysis of the spore length and width between the wild‐type and mutants.
MAP1‐interacting proteins are involved in pathogenesis
Our previous research has indicated that MAP1 plays essential roles in disease development (Li et al., 2015); thus, proteins controlled by MAP1 probably have biological functions associated with the development of rice blast. ECH1 and AOX have been biologically characterized to be relevant to virulence in M. oryzae (Avila‐Adame and Köller, 2002; Patkar et al., 2012). Both studies confirmed our hypothesis; therefore, we focused on the other MAP1‐interacting proteins. Mutants of C3HC, C2H2, SHM, RanBP, CGT, AnnA7 and Bud6p were successfully generated in this study (Figs S5 and S6). To identify their involvement in plant infection, conidia of each mutant strain were applied as a spray on rice seedlings. As predicted, four mutants, ΔSHM, ΔRanBP, ΔAnnA7 and ΔC3HC, were less virulent than the wild‐type strain. Furthermore, the ΔRanBP strain was no longer virulent because no typical symptoms of infection were observed 7 days after inoculation. The virulence of ΔBud6p remained unchanged throughout the entire interaction with rice (Fig. 6A). However, ΔCGT and ΔC2H2 mutants appeared to be more aggressive than the wild‐type because symptoms similar to those observed after inoculation with the wild‐type formed earlier, although after 7 days the severity of disease was the same as that in the wild‐type (Fig. 6A,B), implying that CGT and C2H2 may be suppressors in pathogenesis. We sought to reproduce this result in a more susceptible rice variety, Lijiangxintuanheigu (LTH) (Fig. 6B), and found that the lesions caused by ΔCGT and ΔC2H2 also appeared earlier than those induced by the wild‐type, and the other mutants showed reduced virulence (Fig. 6B). This infection assay revealed that, of the seven MAP1‐interacting proteins, at least four candidates are involved in the process of pathogenesis mediated by MAP1.
Figure 6.

Infection assays with seedlings from rice cultivars Jijing88 and Lijiangxintuanheigu (LTH). Two‐week‐old seedlings from rice cultivars Jijing88 and LTH were spray inoculated with conidial suspensions of the wild‐type strain and one of seven deletion mutants (representative leaves were photographed at 7 days post‐inoculation) (A), or with conidial suspensions of the wild‐type strain and C2H2 and CGT deletion mutants (representative leaves were photographed 72 and 96 h after inoculation) (B).
Discussion
Lons are conserved and have been extensively studied in diverse organisms, but few studies have examined their involvement in fungal pathogenesis. To our knowledge, MAP1 is the first studied Lon in the fungal phytopathogens. This study delineates a pathway mediated by MAP1: under normal conditions, MAP1 exerts specific and controllable proteolytic activity to modulate substrates to a certain activity or maturation state; however, under stress conditions, MAP1 exerts a protective function to clean up damaged proteins and to inhibit the accumulation of aggregates. Indeed, in MAP1 deletion mutants, the C2H2 and C3HC proteins were not altered because MAP1 was missing, whereas, in the wild‐type strain, C2H2 and C3HC proteins were truncated (Fig. 3A,B).
To clarify the biological relationship between MAP1 and its interacting proteins, we created several mutants with gene deletions corresponding to these proteins. Consistent with the ΔMAP1 mutant, four mutants (ΔAnnA7, ΔSHM, ΔRanBP and ΔC3HC) showed reduced virulence, indicating their involvement in pathogenesis. However, the early occurrence of lesions, namely so‐called enhanced virulence, observed in both the CGT and C2H2 deletion mutants was unanticipated. CGT (ceramide glycosyltransferase), which transfers glucose from UDP‐glucose to ceramide, is widely distributed throughout animals (Nomura et al., 2011), but is also present in Cryptococcus neoformans and is required for host infection (Rittershaus et al., 2006). The Magnaporthe CGT gene (also named GCS, AF364402) has also been analysed in the glucosylceramide (GlcCer)‐deficient Pichia pastoris GCS null mutant strain system (Leipelt et al., 2001), but very little is known about the role and function of GCS/CGT. In our research, therefore, the deletion of CGT in M. oryzae is probably helpful to elucidate the function of CGT in the fungal kingdom. Considering that ceramides are major components of extracellular lipids that are derived from GlcCer on extrusion from lamellar granules into the extracellular space in the upper layers of the epidermis (Nomura et al., 2011; Sando et al., 1996), we hypothesized that CGT in the fungus that causes rice blast also participates in the regulation of ceramide metabolism, through which pathogenicity is determined. Less is known about the C2H2 protein than the CGT protein. C2H2 is homologous to the zinc C2H2 finger domain‐containing protein EPE08389 of Ophiostoma piceae, with 48% (115/241) identity and 63% (152/241) similarity (Haridas et al., 2013). C2H2 motifs are generally contained in transcription factors. However, it is currently uncertain whether C2H2 is involved in the regulation of a virulence or related gene. Thus, the elucidation of MAP1‐mediated pathogenesis will require additional study of the C2H2 protein.
AnnA7, SHM, RanBP and C3HC are four novel proteins in fungal phytopathogens, and no fungal pathogen systems for these proteins have been established. However, their known functions in other organisms inform their roles in M. oryzae. AnnA7 belongs to a member of the multifunctional calcium/phospholipid‐binding annexin family. It plays critical roles in the regulation of cellular exocytosis, interaction with the receptor for activated C kinase 1 (RACK1) and the development of lymphatic metastasis in cancer progression (Nir et al., 1987; Pollard et al., 1992). Thus, there is a link between AnnA7 expression and tumorigenesis in proliferation (Jin et al., 2013). AnnA7 probably plays an essential role in the promotion of conidial development and formation in M. oryzae because, in our research, conidial formation was significantly affected in this deletion mutant (Fig. 5B). The enzyme serine hydroxymethyltransferase (SHM) is of primary importance for the production of one‐carbon units. In plants, SHM is mainly involved in photorespiratory conversion (Jamai et al., 2009). Recently, Arabidopsis SHM1 and ubiquitin‐specific protease 16 (UBP16) were found to prevent cell death and reduce the accumulation of reactive oxygen species in the process of salt resistance. Interestingly, the involvement of SHM1 in salt resistance partially depends on the regulation of UBP16 (Zhou et al., 2012), suggesting that SMH hydrolysis is important for stress tolerance in the cells of organisms, with which our research is consistent. Ran is a Ras‐related G protein, which is associated with the transport of macromolecules in and out of the nucleus. Ran and Ran‐binding proteins (RanBP) regulate cell cycle progression in higher eukaryotes (Görlich et al., 1997). In yeast, RanBP participates in processes such as catabolite repression, calcium signalling, mating, cell proliferation and phosphate utilization (Alepuz et al., 1999). The RanBP of M. oryzae identified in this research is homologous to Yrb30p, which is a RanGTP‐binding protein identified in S. cerevisiae (Braunwarth et al., 2003). However, unlike Yrb30p, RanBP is related to conidial development, and the RanBP‐deleted strain is non‐pathogenic. The identified C3HC protein contains a zinc‐finger C3HC domain (Ouyang et al., 2003) as well as an Rsm1‐like domain (Yoon, 2004). Both domains are associated with gene transcription, like C2H2, but their actions appear to be diametrically opposed based on the difference in virulence.
Among the seven mutants created in this study, the Bud6p‐deleted strain showed no changes in virulence, although more than 85% of the conidia produced by this strain increased in volume (Fig. 5A,C,D). Bud6p is a regulator of cell and cytoskeletal polarity in yeast. It was previously identified as an actin‐interacting protein called Aip3p (Amberg et al., 1997). Bud6p/Aip3p localizes at the cell cortex, where cytoskeleton assembly must be achieved to execute polarized cell growth, and AIP3 deletion causes gross defects in cell and cytoskeletal polarity (Glynn et al., 2001). We did not identify cytoskeletal polarity in the Magnaporthe Bud6p mutant, but Bud6p is probably associated with this defect because of the morphological alterations observed in the conidia. However, the conidial size may not be essential for pathogenesis, because Magnaporthe microconidia are also infectious (Zhang et al., 2014b).
In addition to the seven mutants examined in this research, AOX (Avila‐Adame and Köller, 2002) and ECH1 (Patkar et al., 2012) are also relevant to virulence in M. oryzae. ECH1 is a key enzyme in mitochondrial β‐oxidation, but, in M. oryzae, ECH1 is required for conidial germination and is related to the viability of older hyphae. In addition, ECH1‐deleted mutants failed to infect susceptible rice plants (Patkar et al., 2012). The AOX gene was also examined in M. oryzae (Avila‐Adame and Köller, 2002). In terms of pathogenicity, AOX appeared to be less important than ECH1 because the AOX deletion strain produced symptoms on barley leaves similar to those produced by the wild‐type; however, when barley leaves were pretreated with the fungicide azoxystrobin, the AOX mutant caused fewer lesions than the wild‐type, indicating the involvement of AOX in the process of disease development.
Unfortunately, we were unable to examine the effects on virulence following gene deletions of two proteins (PrePL and CHP1) which are known to interact with MAP1, because we were unsuccessful at generating their deletion mutants. The gene deletion may be lethal. Knockdown transformant strains will be required in future studies.
In summary, we have discovered a novel pathogenic pathway mediated by MAP1 in M. oryzae. In this pathway, the proteins associated with pathogenesis are under the control of MAP1. However, many specific questions regarding this pathway, for example, the subcellular location of the binding proteins, the functions of the binding proteins not examined in this study and the mechanisms the interacting proteins use to contribute to pathogenicity, await answers in future studies.
Experimental Procedures
Strains and growth conditions
The wild‐type strain M. oryzae JL0910 has been isolated and purified previously from Oryza sativa cultivar Jijing88, which is a widely planted variety in Jilin Province, China. All M. oryzae strains, including the mutants created in this study, were stored on filter paper at −20 °C in our laboratory. An oatmeal–tomato agar medium (OTA; 4% w/v oatmeal, 150 mL tomato juice per litre and 2.0% w/v agar) was used for conidia harvesting. The strains were cultured for 7 days on CM for the assessment of the growth rate. Each test was repeated at least three times. The mycelia used for nucleic acid extraction were prepared by growing the relevant strains in 100 mL of liquid CM for 3 days at 25 °C at 150 rpm under bright light.
Escherichia coli strains XL1 Blue and DH5α were used as hosts for plasmid DNA amplification and protein expression. Saccharomyces cerevisiae strain AH109 (Clontech, Palo Alto, CA, USA) was used for yeast two‐hybrid assay. Recombinant yeasts were selected by prototrophy to leucine, tryptophan or uracil. Yeast experiments and plasmid isolation were conducted using standard protocols [Clontech Yeast Protocol Handbook, PT4084‐1 (PR033493)].
Nucleic acid manipulation
Gene cloning, nucleic acid restriction, polymerase chain reaction (PCR), propagation of recombinant plasmids and Southern blotting analysis were conducted using standard protocols, as described previously (Sambrook and Russell, 2001). The mycelial samples were collected by filtration and washed three times with deionized water for DNA and RNA extraction. For DNA extraction, a cetyltrimethylammonium bromide procedure was used. The fragments that included the hygromycin B phosphotransferase gene (HPH) or MAP1 cDNA gene were marked as hybridization probes, and the probe fragment was amplified using PCR. Genomic DNA of the wild‐type and transformant strains was digested overnight using corresponding restriction enzymes. Total RNA was extracted using an RNA extraction kit, and residual genomic DNA was digested with RNase‐free DNase I (Sangon, Shanghai, China). First‐strand cDNA was synthesized from 2.0 mg of total RNA using Avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI, USA). The cDNA samples were diluted 10‐fold and used as templates.
Inoculation and rice infection assay
Chinese rice cultivar Jijing88 seedlings were grown under standard cultivation conditions. For the cut‐leaf inoculation assay, a conidial suspension (1 × 106 conidia/mL) was sprayed onto 6‐cm leaf fragments cut from 4‐week‐old rice seedlings. Inoculated leaves were placed on plastic plates with wetted filters at 25 °C for 24 h in the dark, and then incubated for a 16‐h photoperiod until most of the enlarged lesions appeared. For inoculation of the intact plant leaves, a conidial suspension (1 × 106 conidia/mL) was sprayed onto the leaves using an air sprayer. Inoculated plants were kept in a humidity chamber at 25 °C and harvested 5–7 days after inoculation. The collected infected leaves were stored at −80 °C for later protein extraction.
Generation of the TAP‐tagged MAP1 vector and transformants
For construction of the TAP‐tagged MAP1 vector, amplification of both MAP1 and TAP was performed using PCR. For MAP1, a DNA fragment that contained the full‐length MAP1 open reading frame sequence and the 2000 bp upstream promoter region was amplified with pM‐forward (F) and pM‐reverse (R) primers derived from M. oryzae genomic DNA. The TAP tag was amplified from a TAP vector with Tag‐F and Tag‐R primers. The MAP1 and TAP PCR products were cut with the restriction enzyme NdeI. After ligation, the MAP1‐fused TAP fragment (6 kb) was amplified with pM‐F and Tag‐R primers. Finally, the EcoRI‐digested fragment was inserted into a pCB1532 vector at the EcoRI restriction site, generating the TAP‐tagged MAP1 vector. The vector containing the TAP‐tagged MAP1 was transformed into protoplasts of the MAP1 deletion mutant (ΔMAP1) (Fig. S1) using polyethyleneglycol (PEG)‐mediated fungal transformation. Transformants grown in sulfonylurea at 100 μg/mL were screened. Transformants were identified by PCR and confirmed by Southern blot analyses. The partial MAP1 fragment amplified with the primer pair HPH‐F and HPH‐R was used as a blotting probe. For further investigation, Western blot analysis was used to examine MAP1 expression. The ability of the transformant strains to infect was compared with that of the wild‐type, and strains with unaltered pathogenicity were used in subsequent experiments.
Protein extraction and in vivo affinity purification of tagged proteins
The leaves severely infected with the TAP‐tagged MAP1 complemented strain were collected as test samples (100 g), and a set of similarly treated leaves infected with the wild‐type strain were collected as controls. Test and control samples were ground in liquid nitrogen, suspended in extraction buffer [50 mm Tris‐HCl, pH 7.4, 250 mm NaCl, 10% glycerol, 0.05% Nonidet P‐40 (NP‐40), 1 mm phenylmethylsulfonyl fluoride (PMSF), 0.2% protease inhibitor, 1 mm benzamidine and 1 mg/mL leupeptin] and centrifuged for 30 min at 15 000 g. The total protein extract was incubated with 1 mL of IgG‐Sepharose 6 Fast Flow beads (GE Healthcare, Little Chalfont, Buckinghamshire, UK) pre‐equilibrated with 10 mL of cold extraction buffer overnight at 4 °C on a rotator (Rigaut et al., 1999). The IgG‐Sepharose beads were loaded onto a 5‐mL Mobicol column and washed with 20 mL of cold IgG wash buffer (10 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 0.1% NP‐40 and 5% ethylene glycol) and 20 mL of cold TEV (Nicotiana tabacum L.) buffer [10 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 0.1% (v/v) NP‐40, 0.5 mm ethylenediaminetetraacetic acid (EDTA), 1 mm PMSF, 1 μm E64 and 5% (v/v) ethylene glycol]. Bound complexes were eluted using AcTEV Protease (twice with 100 units; Invitrogen, Carlsbad, California, USA) for 1 h at 16 °C, followed by two wash steps with 2 mL of calmodulin binding buffer [10 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 0.1% (v/v) NP‐40, 10 mm β‐mercaptoethanol, 1 mm imidazole, 2 mm CaCl2, 1 mm magnesium acetate, complete EDTA‐free protease inhibitor mixture (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) and 5% (v/v) ethylene glycol]. The CaCl2 concentration of the IgG‐eluted fraction was adjusted to 2 mm, and the fraction was incubated for 1 h at 4 °C under gentle rotation with 1 mL of calmodulin–agarose beads (Stratagene, La Jolla, CA, USA) pre‐equilibrated with 20 mL of cold calmodulin binding buffer. The calmodulin–agarose beads were loaded onto another 5‐mL column and washed with 10 mL of cold calmodulin binding buffer. Bound complexes were eluted with 2.5 mL of cold calmodulin elution buffer [10 mm Tris‐HCl, pH 8.0, 150 mm NaCl, 0.1% (v/v) NP‐40, 10 mm β‐mercaptoethanol, 1 mm imidazole, 25 mm ethyleneglycol‐bis(2‐aminoethylether)tetraacetic acid (EGTA) and 5% (v/v) ethylene glycol]. The eluted samples were concentrated using a 10‐kDa‐filtered Microcon tube (Millipore Corporation, Billerica, MA, USA) and subjected to SDS‐PAGE for protein identification.
MS and protein identification
The test and control samples were analysed using SDS‐PAGE. The gel was stained using Coomassie brilliant blue R250. Each detectable band was excised and analysed for protein identification using MS. The gel pieces were washed with Milli‐Q water and lyophilized and then rehydrated in digestion buffer [20 μg/mL of sequencing grade modified trypsin (Promega) with 25 mm H4HCO3] at 37 °C overnight. After a brief centrifugation, the peptides were collected from the supernatant, and were re‐extracted with 5 μL of 0.1% trifluoroacetic acid in 50% acetonitrile to collect the remaining peptides. The peptide extract of each protein band was used for MS analysis.
A MALDI‐TOF/TOF‐MS/MS ABI 4700 proteomics analyser (Applied Biosystems, Foster City, CA, USA) was applied for MS analysis following the manufacturer's instructions. The measured MS data were transferred with GPS Explorer software v3.5 (Applied Biosystems) as inputs to search the National Center for Biotechnology Information non‐redundant (NCBI‐nr) database with the species restricted to M. oryzae using MASCOT search engine version 2.2 (Matrix Science, London, UK). The search parameters were set according to those previously described by Wang et al. (2012).
Yeast two‐hybrid assay
A yeast two‐hybrid system (Clontech) was applied to confirm the interactions of proteins between MAP1 and each of the candidates in vivo. For the construction of the bait vector, the cDNA corresponding to the mature coding region of MAP1 was PCR amplified with forward and reverse primers (MAP1‐F/MAP1‐R) using M. oryzae cDNA as a template. The PCR product, after restriction with NdeI and EcoRI, was then subcloned into the yeast two‐hybrid bait vector pGBKT7 DNA‐BD (630443) to construct pGBKT‐MAP1. For the prey vector of ECH1 (pGADT7‐ECH1), cDNA of the ECH1 gene was amplified with ECH1‐F and ECH1‐R primers. The fragment double digested with XhoI and EcoRI was then ligated to the XhoI and EcoRI restriction sites in the yeast two‐hybrid prey vector pGADT7 AD (630442). For the other prey vectors, a similar construction strategy was used: individual cDNA genes were amplified with corresponding primer pairs and cloned into the pGADT7 AD vector (Procedure S1, see Supporting Information). The primer pairs and corresponding restriction sites used are indicated in Table S1 (see Supporting Information).
The AH109 cells were co‐transformed with the bait and prey vectors according to the manufacturer's instructions (Clontech). The resultant yeast strains were plated on minimal medium agar lacking His, tryptophan and leucine (SD/His–, Trp–, Leu–) in the presence of 20 mm 3‐amino‐1,2,4‐triazole/X‐α‐Gal for 3 days at 30 °C. The interactions between the bait and prey were analysed using colony size and the blue colour as indicators. Yeast strains co‐transformed with pGBKT7‐MAP and the empty vector pGADT7 served as self‐activation controls (CK0), pGBKT7‐53 and pGADT7‐T served as positive controls (CK+), and pGBKT7‐lam and pGADT7‐T served as negative controls (CK–).
Protein expression in E. coli and proteolytic activity analysis
The full‐length cDNA encoding MAP1 cDNA (1119 bp) was cut and recovered from the pGBKT‐MAP1 vector through digestion with the restriction enzymes NdeI and EcoRI, and inserted into the NdeI and EcoRI sites of pET‐28a, resulting in a His‐tagged MAP1 expression vector. For ECH1 expression, full‐length ECH1 cDNA was cut from the pGADT7‐ECH1 vector with XhoI and EcoRI, and ligated to pET‐28a, resulting in the His‐tagged ECH1 expression vector. Using this strategy, the other MAP1‐interacting protein genes were obtained from the corresponding yeast two‐hybrid prey vector pGADT7 and ligated to pET‐28a, resulting in the corresponding His‐tagged expression vectors (Procedure S2, see Supporting Information). Each of the His‐fused protein‐specific polypeptides was expressed in E. coli BL21 with isopropyl β‐d‐1‐thiogalactopyranoside (IPTG, 1 mm) added as an inducer, and then purified following standard methods.
The purified proteins were incubated individually at 25 °C with 2.5 μg of MAP1 in a hydrolysis reaction system of 50 μL (50 mm Tris‐HCl and 10 mm MgCl2 at pH 7.4) supplemented with or without 2.5 mm ATP. The hydrolysis reactions were stopped after a 1‐h incubation by the addition of the SDS‐PAGE loading buffer. Each set of reactions was analysed using SDS‐PAGE followed by Coomassie brilliant blue R‐250 staining. Heat (45 °C)‐pretreated proteins were also detected as described above.
Polyclonal antibody preparation and Western blot analysis
MAP1, C2H2, C3HC, CGT and ECH1 were expressed and purified. Subsequently, each purified protein was injected into rabbits to produce primary polyclonal antibodies.
The total proteins extracted from the wild‐type strain (or TAP‐tagged complemented transformant) and the MAP1‐deleted strain were denatured in SDS‐PAGE loading buffer at 95 °C for 10 min. Western blot analysis was performed using the corresponding antiserum against MAP1, C2H2, C3HC, CGT and ECH1, and visualized with an enhanced horseradish peroxidase‐diaminobenzidine (HRP‐DAB) substrate kit (Tiangen, Beijing China). The Magnaporthe mitochondrial porin (Por1, MGG_00968.6) was used as an internal control as it is not a substrate of Lon (Bayot et al., 2010). Polyclonal antibody against porin was purchased from Qiangyao Peptide Company (Shanghai, China).
Generation of targeted gene‐deleted strains
For biological analyses of proteins interacting with MAP1, a series of the corresponding genes was deleted by replacement with a 1.4‐kb HPH cassette from the pXEH vector. Based on the bioinformatics annotations concerning genes of interacting proteins in the genome of M. oryzae, a series of deletion vectors was constructed. To construct the C2H2 gene replacement vector pC2H2, the 992‐bp upstream and the 970‐bp downstream flanking sequences were amplified using both primer pairs, that is, C2H2‐1F (using XbaI)/C2H2‐1R (using BamHI) and C2H2‐2F (using SacI)/C2H2‐2R (using XhoI), respectively. The double‐restricted flanking fragments were ligated into both sides (XbaI‐BamHI and SacI‐XhoI) of the HPH cassette in the pXEH vector. The primer pair C2H2‐1F and C2H2‐2R was used to confirm the final construct. Linearized pC2H2 vectors were transformed into wild‐type fungal protoplasts. Transformed candidates were selected using hygromycin resistance. The gene deletion transformants were confirmed with reverse transcription (RT)‐PCR of the primer pair MAP1‐1/MAP1‐2 and Southern blot analysis. The partial HPH fragment amplified with the primer pair HPH‐F and HPH‐R was used as a blotting probe. The related transformants were then purified using single conidial isolates.
Supporting information
Fig. S1 Generation of the MAP1 deletion mutant.
Fig. S2 The tandem affinity purification (TAP)‐tagged MAP1 vector and transformants.
Fig. S3 Sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of recombinant MAP1 and its interacting proteins.
Fig. S4 The deletion strategies for MAP1‐interacting protein genes.
Fig. S5 Southern blot analysis for the seven deletion mutants.
Fig. S6 Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis for the seven deletion mutants.
Table S1 Polymerase chain reaction (PCR) primers used in this study.
Table S2 Characterization of the wild‐type and mutant strains.
Procedure S1 Construction of prey vectors.
Procedure S2 Construction of protein expression vector.
Acknowledgements
We thank Dr Hon‐Ming Lam at The Chinese University of Hong Kong for assistance with MALDI‐TOF/TOF‐MS/MS analyses. This work was supported by the National Natural Science Foundation of China (31171794; 31201465) and a Special Fund for Agro‐Scientific Research in the Public Interest (201203014).
References
- Adam, C. , Picard, M. , De'quard‐Chablat, M. , Sellem, C.H. , Denmat, S.H.‐L. and Contamine, V. (2012) Biological roles of the Podospora anserina mitochondrial lon protease and the importance of its N‐domain. PLoS ONE, 7, e38138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz, P.M. , Matheos, D. , Cunningham, K.W. and Estruch, F. (1999) The Saccharomyces cerevisiae RanGTP‐binding protein msn5p is involved in different signal transduction pathways. Genetics, 153, 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D.C. , Zahner, J.E. , Mulholland, J.M. , Pringle, J.R. and Botstein, D. (1997) Aip3p/Bud6p, a yeast actin‐interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell, 8, 729–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila‐Adame, C. and Köller, W. (2002) Disruption of the alternative oxidase gene in Magnaporthe grisea and its impact on host infection. Mol. Plant–Microbe Interact. 15, 493–500. [DOI] [PubMed] [Google Scholar]
- Bayot, A. , Gareil, M. , Rogowska‐Wrzesinska, A. , Roepstorff, P. , Friguet, B. and Bulteau, A.L. (2010) Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP‐dependent Lon‐like protease Pim1. J. Biol. Chem. 285, 11 445–11 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, T. , Lewrenz, I. , Franken, S. , Baitzel, C. and Voos, W. (2011) Mitochondrial enzymes are protected from stress‐induced aggregation by mitochondrial chaperones and the Pim1/LON protease. Mol. Biol. Cell, 22, 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bota, D.A. and Davies, K.J. (2001) Protein degradation in mitochondria: implications for oxidative stress, aging and disease: a novel etiological classification of mitochondrial proteolytic disorders. Mitochondrion, 1, 33–49. [DOI] [PubMed] [Google Scholar]
- Bota, D.A. and Davies, K.J. (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP‐stimulated mechanism. Nat. Cell Biol. 4, 674–680. [DOI] [PubMed] [Google Scholar]
- Braunwarth, A. , Fromont‐Racine, M. , Legrain, P. , Bischoff, F.R. , Gerstberger, T. , Hurt, E. and Kunzler, M. (2003) Identification and characterization of a novel RanGTP‐binding protein in the yeast Saccharomyces cerevisiae . J. Biol. Chem. 278, 15 397–15 405. [DOI] [PubMed] [Google Scholar]
- Bretz, J. , Losada, L. , Lisboa, K. and Hutcheson, S.W. (2002) Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae . Mol. Microbiol. 45, 397–409. [DOI] [PubMed] [Google Scholar]
- Chen, S.H. , Suzuki, C.K. and Wu, S.H. (2008) Thermodynamic characterization of specific interactions between the human Lon protease and G‐quartet DNA. Nucleic Acids Res. 36, 1273–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, J.L. , Katona, L.I. , Kuhlow, C. , Toledo, A. , Okan, N.A. , Tokarz, R. and Benach, J.L. (2009) Evidence that two ATP‐dependent (Lon) proteases in Borrelia burgdorferi serve different functions. PLoS Pathog. 5, e1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis, G.R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825. [DOI] [PubMed] [Google Scholar]
- Ding, S.L. , Liu, W. , Iliuk, A. , Ribot, C. , Vallet, J. , Tao, A. , Wang, Y. , Lebrun, M.H. and Xu, J.R. (2010) The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae . Plant Cell, 22, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, M. , Wang, Z.Y. , Jones, M.A. , Smirnoff, N. and Talbot, N.J. (2007) Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA, 104, 11 772–11 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J. and Wilson, R.A. (2014) Cells in cells: morphogenetic and metabolic strategies conditioning rice infection by the blast fungus Magnaporthe oryzae . Protoplasma, 251, 37–47. [DOI] [PubMed] [Google Scholar]
- Fukuda, R. , Zhang, H. , Kim, J.W. , Shimoda, L. , Dang, C.V. and Semenza, G.L. (2007) HIF‐1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell, 129, 111–122. [DOI] [PubMed] [Google Scholar]
- Glynn, J.M. , Lustig, R.J. , Berlin, A. and Chang, F. (2001) Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11, 836–845. [DOI] [PubMed] [Google Scholar]
- Görlich, D. , Dabrowski, M. , Bischoff, F.R. , Kutay, U. , Bork, P. , Hartmann, E. , Prehn, S. and Izaurralde, E. (1997) A novel class of RanGTP binding proteins. J. Cell Biol. 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond‐Kosack, K.E. and Parker, J.E. (2003) Deciphering plant–pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14, 177–193. [DOI] [PubMed] [Google Scholar]
- Haridas, S. , Wang, Y. , Lim, L. , Massoumi Alamouti, S. , Jackman, S. , Docking, R. , Robertson, G. , Birol, I. , Bohlmann, J. and Breuil, C. (2013) The genome and transcriptome of the pine saprophyte Ophiostoma piceae, and a comparison with the bark beetle‐associated pine pathogen Grosmannia clavigera . BMC Genomics, 4, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, O. , Ichinoda, F. , Tamatani, T. , Yamaguchi, A. , Sato, N. , Ozawa, K. , Kitao, Y. , Miyazaki, M. , Harding, H.P. , Ron, D. , Tohyama, M.M. , Stern, D. and Ogawa, S. (2002) Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J. Cell Biol. 157, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, K. , Czymmek, K.J. , Caplan, J.L. , Sweigard, J.A. and Donofrio, N.M. (2011) HYR1‐mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 7, e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingmer, H. and Brøndsted, L. (2009) Proteases in bacterial pathogenesis. Res. Microbiol. 160, 704–710. [DOI] [PubMed] [Google Scholar]
- Jamai, A. , Salomé, P.A. , Schilling, S.H. , Weber, A.P. and McClung, C.R. (2009) Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxin‐dependent glutamate synthase. Plant Cell, 21, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.L. , Wang, Z.Q. , Qu, H. , Wang, H.X. , Ibrahim, M.M. , Zhang, J. , Huang, Y.H. , Wu, J. , Bai, L.L. , Wang, X.Y. , Meng, J.Y. and Tang, J.W. (2013) Annexin A7 gene is an important factor in the lymphatic metastasis of tumors. Biomed. Pharmacother. 67, 251–259. [DOI] [PubMed] [Google Scholar]
- Kadotani, N. , Nakayashiki, H. , Tosa, Y. and Mayama, S. (2003) RNA silencing in the phytopathogenic fungus Magnaporthe oryzae . Mol. Plant–Microbe Interact. 16, 769–776. [DOI] [PubMed] [Google Scholar]
- Kato, H. and Kozaka, T. (1974) Effect of temperature on lesion enlargement and sporulation of Pricularia oryzae in rice leaves. Phytopathology, 64, 828–830. [Google Scholar]
- Lan, L. , Deng, X. , Xiao, Y. , Zhou, J.M. and Tang, X. (2007) Mutation of Lon protease differentially affects the expression of Pseudomonas syringae type III secretion system genes in rich and minimal media and reduces pathogenicity. Mol. Plant–Microbe Interact. 20, 682–696. [DOI] [PubMed] [Google Scholar]
- Leipelt, M. , Warnecke, D. , Zähringer, U. , Ott, C. , Müller, F. , Hube, B. and Heinz, E. (2001) Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J. Biol. Chem. 276, 33 621–33 629. [DOI] [PubMed] [Google Scholar]
- Li, G. , Zhou, X. and Xu, J.R. (2012) Genetic control of infection‐related development in Magnaporthe oryzae . Curr. Opin. Microbiol. 15, 678–684. [DOI] [PubMed] [Google Scholar]
- Li, J. , Liang, X.L. , Wei, Y. , Liu, J.L. , Lin, F.C. and Zhang, S.H. (2015) An ATP‐dependent protease homolog ensures basic standards of survival and pathogenicity for Magnaporthe oryzae . Eur. J. Plant Pathol. 141, 703–716. doi: 10.1007/s10658-014-0572-9. [DOI] [Google Scholar]
- Losada, L.C. and Hutcheson, S.W. (2005) Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon‐associated degradation. Mol. Microbiol. 55, 941–953. [DOI] [PubMed] [Google Scholar]
- Lu, B. , Yadav, S. , Shah, P.G. , Liu, T. , Tian, B. , Pukszta, S. , Villaluna, N. , Kutejová, E. , Newlon, C.S. , Santos, J.H. and Suzuki, C.K. (2007) Roles for the human ATP‐dependent Lon protease in mitochondrial DNA maintenance. J. Biol. Chem. 282, 17 363–17 374. [DOI] [PubMed] [Google Scholar]
- Matsushima, Y. , Goto, Y. and Kaguni, L.S. (2010) Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc. Natl. Acad. Sci. USA, 107, 18 410–18 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir, S. , Stutzin, A. and Pollard, H.B. (1987) Effect of synexin on aggregation and fusion of chromaffin granule ghosts at pH 6. Biochim. Biophys. Acta, 903, 309–318. [DOI] [PubMed] [Google Scholar]
- Nomura, K.H. , Murata, D. , Hayashi, Y. , Dejima, K. , Mizuguchi, S. , Kage‐Nakadai, E. , Gengyo Ando, K. , Mitani, S. , Hirabayashi, Y. , Ito, M. and Nomura, K. (2011) Ceramide glucosyltransferase of the nematode Caenorhabditis elegans is involved in oocyte formation and in early embryonic cell division. Glycobiology, 21, 834–848. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T. , Brunner, F. , Kemmerling, B. and Piater, L. (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. [DOI] [PubMed] [Google Scholar]
- Osuna‐Canizalez, F.J. , De‐Datta, S.K. and Bonman, J.M. (1991) Nitrogen form and silicon nutrition effects on resistance to blast disease of rice. Plant Soil, 135, 223–231. [Google Scholar]
- Ouyang, T. , Bai, R.Y. , Bassermann, F. , von‐Klitzing, C. , Klumpen, S. , Miething, C. , Morris, S.W. , Peschel, C. and Duyster, J. (2003) Identification and characterization of a nuclear interacting partner of anaplastic lymphoma kinase (NIPA). J. Biol. Chem. 278, 30 028–30 036. [DOI] [PubMed] [Google Scholar]
- Patkar, R.N. , Ramos‐Pamplona, M. , Gupta, A.P. , Fan, Y. and Naqvi, N.I. (2012) Mitochondrial β‐oxidation regulates organellar integrity and is necessary for conidial germination and invasive growth in Magnaporthe oryzae . Mol. Microbiol. 86, 1345–1363. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. (2010) Armed and dangerous. Science, 327, 804–805. [DOI] [PubMed] [Google Scholar]
- Pollard, H.B. , Rojas, E. and Burns, A.L. (1992) Synexin (annexin VII) and membrane fusion during the process of exocytotic secretion. Prog. Brain Res. 92, 247–255. [DOI] [PubMed] [Google Scholar]
- Rigas, S. , Daras, G. , Laxa, M. , Marathias, N. , Fasseas, C. , Sweetlove, L.J. and Hatzopoulos, P. (2009) Role of Lon1 protease in post‐germinative growth and maintenance of mitochondrial function in Arabidopsis thaliana . New Phytol. 181, 588–600. [DOI] [PubMed] [Google Scholar]
- Rigaut, G. , Shevchenko, A. , Rut, B. , Wilm, M. , Mann, M. and Se'raphin, B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rittershaus, P.C. , Kechichian, T.B. , Allegood, J.C. , Merrill, A.H. , Jr Hennig, M. , Luberto, C. and Del Poeta, M. (2006) Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans . J. Clin. Invest. 116, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sando, G.N. , Howard, E.J. and Madison, K.C. (1996) Induction of ceramide glucosyltransferase activity in cultured human keratinocytes. Correlation with culture differentiation. J. Biol. Chem. 271, 22 044–22 051. [DOI] [PubMed] [Google Scholar]
- Su, S. , Stephens, B.B. , Alexandre, G. and Farrand, S.K. (2006) Lon protease of the alpha‐proteobacterium Agrobacterium tumefaciens is required for normal growth, cellular morphology and full virulence. Microbiology, 152, 1197–1207. [DOI] [PubMed] [Google Scholar]
- Sun, C.B. , Suresh, A. , Deng, Y.Z. and Naqvi, N.I. (2006) A multidrug resistance transporter in Magnaporthe is required for host penetration and for survival during oxidative stress. Plant Cell, 18, 3686–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, A. , Suzuki, M. , Matsui, H. , Tomoyasu, T. , Sashinami, H. , Nakane, A. and Yamamoto, T. (2003) Lon, a stress‐induced ATP‐dependent protease, is critically important for systemic Salmonella enterica serovar typhimurium infection of mice. Infect. Immun. 71, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, A. , Tabuchi, F. , Tsuchiya, H. , Isogai, E. and Yamamoto, T. (2008) Negative regulation of quorum‐sensing systems in Pseudomonas aeruginosa by ATP‐dependent Lon protease. J. Bacteriol. 190, 4181–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, K. , Tsuchiya, W. , Noda, N. , Suzuki, R. , Yamazaki, T. and Haas, D. (2014) Lon protease negatively affects GacA protein stability and expression of the Gac/Rsm signal transduction pathway in Pseudomonas protegens . Environ. Microbiol. 16, 2538–2549. [DOI] [PubMed] [Google Scholar]
- Tsilibaris, V. , Maenhaut‐Michel, G. and Van Melderen, L. (2006) Biological roles of the Lon ATP‐dependent protease. Res. Microbiol. 157, 701–713. [DOI] [PubMed] [Google Scholar]
- Van Melderen, L. and Aertsen, A. (2009) Regulation and quality control by Lon‐dependent proteolysis. Res. Microbiol. 160, 645–651. [DOI] [PubMed] [Google Scholar]
- Venkatesh, S. , Lee, J. , Singh, K. , Lee, I. and Suzuki, C.K. (2012) Multitasking in the mitochondrion by the ATP‐dependent Lon protease. Biochim. Biophys. Acta, 1823, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.D. , Wang, X. , Ngai, S.M. and Wong, Y.S. (2012) Proteomics analysis reveals multiple regulatory mechanisms in response to selenium in rice. J. Proteomics, 75, 1849–1866. [DOI] [PubMed] [Google Scholar]
- Yoon, J.H. (2004) Schizosaccharomyces pombe rsm1 genetically interacts with spmex67, which is involved in mRNA export. J. Microbiol. 42, 32–36. [PubMed] [Google Scholar]
- Zhang, H. , Zhao, Q. , Guo, X. , Guo, M. , Qi, Z. , Tang, W. , Dong, Y. , Ye, W. , Zheng, X. , Wang, P. and Zhang, Z. (2014a) Pleiotropic function of the putative zinc‐finger protein MoMsn2 in Magnaporthe oryzae . Mol. Plant–Microbe Interact. 27, 446–460. [DOI] [PubMed] [Google Scholar]
- Zhang, H.L. , Wu, Z.S. , Wang, C.F. , Li, Y. and Xu, J.R. (2014b) Germination and infectivity of microconidia in the rice blast fungus Magnaporthe oryzae . Nat. Commun. 5, 4518. doi: 10.1038/ncomms5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Zhao, J. , Yang, Y. , Chen, C. , Liu, Y. , Jin, X. , Chen, L. , Li, X. , Deng, X.W. , Schumaker, K.S. and Guo, Y. (2012) Ubiquitin‐specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell, 24, 5106–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Generation of the MAP1 deletion mutant.
Fig. S2 The tandem affinity purification (TAP)‐tagged MAP1 vector and transformants.
Fig. S3 Sodium dodecylsulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) of recombinant MAP1 and its interacting proteins.
Fig. S4 The deletion strategies for MAP1‐interacting protein genes.
Fig. S5 Southern blot analysis for the seven deletion mutants.
Fig. S6 Reverse transcription‐polymerase chain reaction (RT‐PCR) analysis for the seven deletion mutants.
Table S1 Polymerase chain reaction (PCR) primers used in this study.
Table S2 Characterization of the wild‐type and mutant strains.
Procedure S1 Construction of prey vectors.
Procedure S2 Construction of protein expression vector.


